- 1Department of Clinical Laboratory, the First Hospital Affiliated to Henan University, Kaifeng, China
- 2State Key Laboratory of Pathogen and Biosecurity, Beijing Institute of Microbiology and Epidemiology, Beijing, China
- 3College of Food Science and Project Engineering, Bohai University, Jinzhou, China
- 4Laboratory Animal Center, Academy of Military Medical Sciences, Beijing, China
- 5Beijing Institute of Genomics (CAS), Beijing, China
The purpose of this study was to characterize mechanisms of plasmid-mediated antimicrobial resistance in Shigella boydii. S. boydii strain 2246 with resistance to ciprofloxacin, ceftriaxone and azithromycin was isolated from a human case of watery diarrhea in a Chinese public hospital. Resistance in strain 2246 to ceftriaxone and azithromycin was attributable to the presence of blaCTX-M-14, and erm(B) and mph(A), respectively, which were co-located on a multidrug-resistant (MDR) plasmid p2246-CTXM. p2246-CTXM represented a novel IncFII-type MDR plasmid with a very complex chimera structure. Its master backbone was genetically closely related to the R100 plasmid, but p2246-CTXM had evolved to integrate additional R100-unrelated backbone regions as well as massive exogenous mobile elements that carried multiple resistance determinants. In p2246-CTXM, erm(B) together with its leading peptide gene erm(C), mph(A) together with its regulatory genes mrx and mphR(A), and blaCTX-M-14 were captured by three different mobile elements Tn6295, the IS26-mph(A)-mrx-mphR(A)-IS6100 unit, and a truncated ISEcp1-blaCTX-M-14-IS903D-iroN transposition unit, respectively, all of which were harbored in a large Tn3-family transposon Tn6285. p2246-CTXM still carried additional resistance determinants mer (mercury resistance), aacA4 (aminoglycoside resistance), cmlA1 (chloramphenicol resistance), and qacED1 (quaternary ammonium compound resistance). This is the first report of identifying a clinical S. boydii strain simultaneously resistant to ciprofloxacin, ceftriaxone, and azithromycin, and determining the complete sequence of a resistance plasmid from S. boydii.
Introduction
Shigellosis remains a common gastrointestinal disease in both developing and industrialized countries. Shigella, the causative agent of shigellosis, can be serologically grouped into four species S. flexneri, S. sonnei, S. boydii, and S. dysenteriae. Being together responsible for about 90% of shigellosis, S. flexneri and S. sonnei are the most prevalent species in developing and developed countries, respectively, but a shift in the dominant species from S. flexneri to S. sonnei has occurred in countries with recent rapid improvement of socioeconomic conditions (Livio et al., 2014; Lima et al., 2015). S. boydii accounts for around 10% of the Shigella isolates found in the samples from the Indian subcontinent that is considered as S. boydii endemic areas, but this pathogen remains very rare (less than 1% of the total Shigella isolates) in other areas (Livio et al., 2014; Lima et al., 2015).
Multidrug-resistant (MDR) isolates of S. flexneri and S. sonnei have been reported worldwide. The previously efficacious old-generation antimicrobials such as ampicillin, chloramphenicol, tetracycline, and sulphonamides have become ineffective in the treatment of shigellosis (Niyogi, 2007; Zhang W. et al., 2011). The situation is getting worse due to the increasing emergence of resistance in Shigella to ciprofloxacin, ceftriaxone, and azithromycin (Klontz and Singh, 2015). The WHO recommends ciprofloxacin as the first choice for the treatment of multidrug-resistant shigellosis, while ceftriaxone and azithromycin can be used as the alternatives for both adults and children and they are preferred among young children because of the concerns regarding the adverse effects of ciprofloxacin in young children (Christopher et al., 2010).
This study describes not only the first reported S. boydii isolate with resistance to ciprofloxacin, ceftriaxone and azithromycin but also the first fully sequenced resistance plasmid from S. boydii. This plasmid, designated p2246-CTXM, is a novel MDR plasmid of the IncFII incompatibility group, and it carries several different resistance determinants especially including blaCTX-M-14 (cephalosporin resistance), and erm(B) and mph(A) (macrolide resistance).
Materials and Methods
Bacterial Identification
The use of human specimens and all related experimental protocols were approved by the Committee on Human Research of indicated institutions and carried out in accordance with the approved guidelines. S. boydii species was identified by slide agglutination using monovalent antisera of Denka Seiken (Tokyo, Japan) and monoclonal antibody reagents of Reagensia AB (Solna, Sweden). The agglutination was sensitively scored according to the following scale: +++, 100% agglutination of the cells; ++, >50% agglutination; +, <50% agglutination; -, no agglutination detected.
Detection of Resistance Genes
The major horizontally acquired quinolone-resistance genes, extended-spectrum β-lactamase (ESBL) genes, and macrolide-resistance genes were screened by PCR (Supplementary Table S1), followed by amplicon sequencing on ABI 3730 Sequencer (LifeTechnologies, Carlsbad, CA, USA).
Plasmid Transfer, Sequencing and Annotation
Plasmid conjugal transfer were performed with Escherichia coli EC600 (LacZ-, NalR, and RifR) being used as recipient for selection of blaCTX-M-positive transconjugants (Chen et al., 2015). Plasmid DNA was isolated from E. coli transconjugant using Qiagen large construct kit (Qiagen, Hilden, Germany), and sequenced by whole-genome shotgun strategy in combination with Illumina HiSeq 2500 (Illumina, San Diego, CA, USA) sequencing technology. The contigs were assembled with Velvet version 1.2, and the gaps were filled through combinatorial PCR and Sanger sequencing on ABI 3730 Sequencer. The genes were predicted with GeneMarkSTM and RAST and further annotated by BLASTP and BLASTN against UniProt and NR databases. Annotation of mobile elements was based on the databases ISfinder, INTEGRALL, Tn Number Registry, and ISCR Elements. Gene organization diagrams were drawn with Inkscape version 0.48.
Phenotypic Analyses
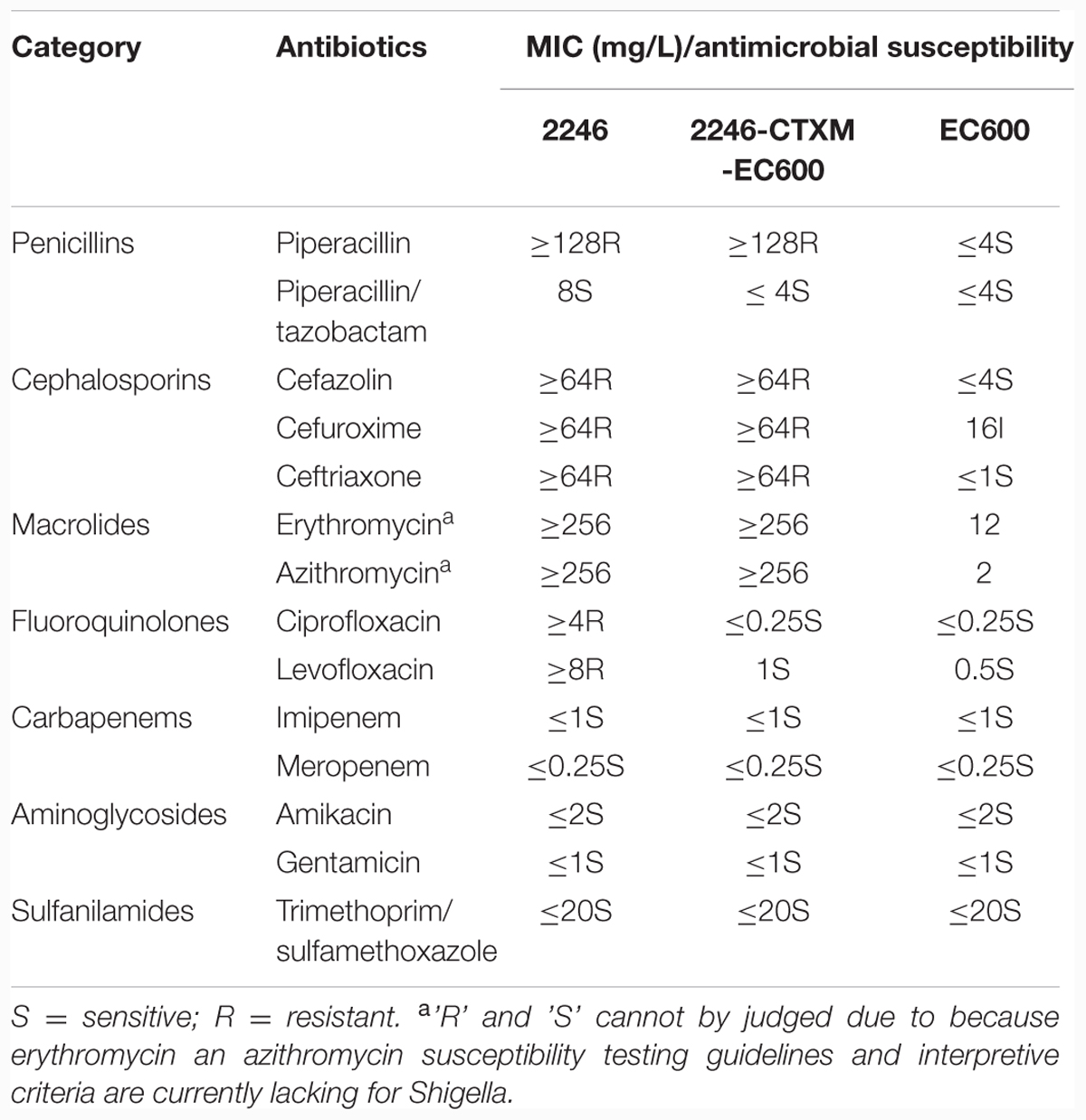
Enzymatic activity of ESBL was determined by the combined disk test as recommended by Clinical and Laboratory Standards Institute (CLSI; CLSI, 2015). Bacterial antimicrobial susceptibility was tested by ETEST (BioMérieux, Hazelwood, MO, USA) for erythromycin and azithromycin and by VITEK 2 (BioMérieux Vitek, Hazelwood, MO, USA) for all the other drugs (Table 1), and interpreted as per CLSI guidelines (CLSI, 2015).
RNA Isolation and Primer Extension Assay
Bacteria were cultured overnight in Mueller-Hinton broth with addition of indicated antibiotics. Total RNAs were extracted from harvested bacterial cells using TRIzol Reagent (Life Technologies). The oligonucleotide primers 5′-GCA CTCTCTTTGTCACCATCTC-3′, 5′-CTACATAACGCATTTGA TAACGC-3′, and 5′-CCATGTCGGGCTGCAAGTGCGTACAG TTGGG-3′ were designed to be complementary to the RNA transcripts of blaCTX-M-14, erm(B)L and mph(A), respectively, and end-labeled with [γ-32P] ATP. About 10 μg of end-labeled primer were annealed with 3 μg of RNA for the primer extension assay with a Primer Extension System (Promega) as described previously (Zhang Y. et al., 2011). The end-labeled primer was used for sequencing the PCR amplicons generated by the primer pairs 5′-TTAAGTATCATTGCAGCAAAG-3′/5′-GCA CTCTCTTTGTCACCATCTC-3′, 5′-AACGCTGAACCCAGCG GTAAATCGT-3′/5′-TACTCCTGAAGTGATTACATCT-3′, and 5′-ATGGCAAACTGAAACGGAT-3′/5′-CCTCTGGTTCGACC TTCG-3′ for blaCTX-M-14, erm(B)L, and mph(A), respectively. DNA sequencing was carried out using the AccuPower & Top DNA Sequencing Kit (Bioneer). The primer extension products and sequencing materials were analyzed on an 8 M urea-6% polyacrylamide gel electrophoresis. Radioactive species were detected by autoradiography.
Nucleotide Sequence Accession Number
The complete sequence of p2246-CTXM was submitted to GenBank under accession number KX646543.
Results
Characterization of S. boydii Strain 2246 and Its Drug Susceptibility
In October 2013, a 51-year-old male with watery diarrhea and liver dysfunction (further diagnosed as liver cirrhosis) was admitted to a public hospital in Beijing, China. Bacterial growth could be observed after culture of fresh stool specimens of the patient on the Salmonella-Shigella agar. The bacterial isolate, designated as 2246, was identified as S. boydii using slide agglutination. Based on the antimicrobial susceptibility test results, the patient received oral administration with piperacillin/tazobactam, and his symptoms of diarrhea progressively disappeared.
As determined by PCR screening for the major plasmid-borne quinolone-resistance, ESBL, and macrolide-resistance genes, strain 2246 harbored blaCTX-M-14, erm(B), and mph(A) rather than any of the other genes tested. These three resistance markers could be co-transferred from strain 2246 into EC600 through conjugation, generating a transconjugant 2246-CTXM-EC600. Strains 2246 and 2246-CTXM-EC600 had the ESBL enzyme activity (Supplementary Figure S1). Both 2246 and 2246-CTXM-EC600 were resistant to piperacillin, cefazolin, cefuroxime, and ceftriaxone, but remained susceptible to piperacillin/tazobactam, imipenem and meropenem, gentamicin and amikacin, and trimethoprim/sulfamethoxazole; 2246 rather than 2246-CTXM-EC600 was resistant to ciprofloxacin and levofloxacin (Table 1). Notably, both 2246 and 2246-CTXM-EC600 possessed greatly elevated minimal inhibitory concentration (MIC) values (≥256 mg/L) against erythromycin and azithromycin (Table 1).
General Features of p2246-CTXM
High-throughput sequencing analysis using plasmid DNA sample isolated from 2246-CTXM-EC600 generated the entire nucleotide sequence of a single plasmid p2246-CTXM (Figure 1) with a mean coverage of 135. The p2246-CTXM genome was manifested as an 111,559 bp circular DNA molecular with a mean G+C content of 51.9% and carried a total of 143 predicted open reading frames (ORFs).
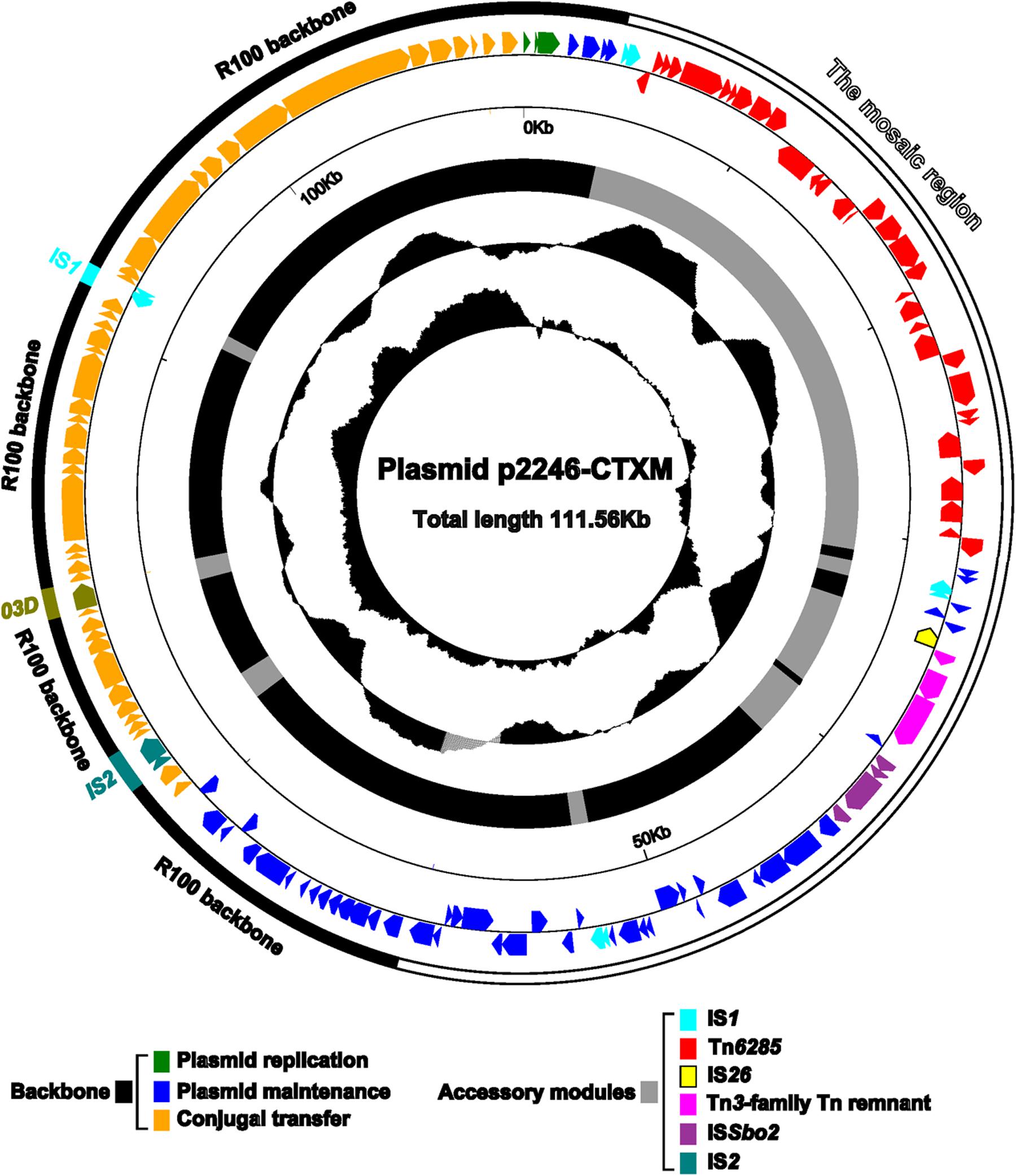
FIGURE 1. Schematic map of p2246-CTXM. Genes are denoted by arrows and colored based on gene function classification. The innermost two circles present the GC-Skew [(G-C)/(G+C)] and the GC contents. Shown also are the backbone and the accessory modules, respectively.
The modular structure of p2246-CTXM could be divided into two major regions, namely, a 55-kb plasmid R100 (accession number AP000342)-derived backbone region with insertion of three different insertion sequences, and a 56.4-kb mosaic region composed of several plasmid backbone sequences and accessory mobile elements of different evolutionary origins. The R100-derived backbone sequences contained DNA regions for plasmid replication (repA2, repA6, and repA1 of the IncFII incompatibility group), maintenance (repA4, tir, pemIK, etc) and conjugal transfer (tra, trb, yfiABC, etc). One copy of each of IS2, IS903D and IS1 was inserted into the R100-derived backbone sequences, disrupting them into four separate portions. The transposition of IS2, IS903D and IS1 leaved three different direct repeats (DRs: target site duplication signals of transposition), i.e., GTTTA, GCATAAATC, and CGCGACGGG, which flanked the corresponding inserted elements, respectively. The insertion of IS2 and IS903D truncated the backbone genes traJ and yfhA, respectively.
Located at different sites of the backbone of R100 were three copies of repeat D, around 438 bp in length, two of which were found in p2246-CTXM. It seemed that the repeat D-mediated homologous recombination led to the deletion of one copy of repeat D together with a 14-kb region containing plasmid maintenance genes ssb, ydeAB, ydgA, yefA, psiBA, sok, mok, hok, and yehA.
The Mosaic Region of p2246-CTXM
The 56.4-kb mosaic region of p2246-CTXM (Figure 2) could be divided into two parts and the first part was composed of an intact IS1 element, and a novel transposon designated Tn6285 bordered by 40 bp imperfect terminal inverted repeats (IRs) at both ends.
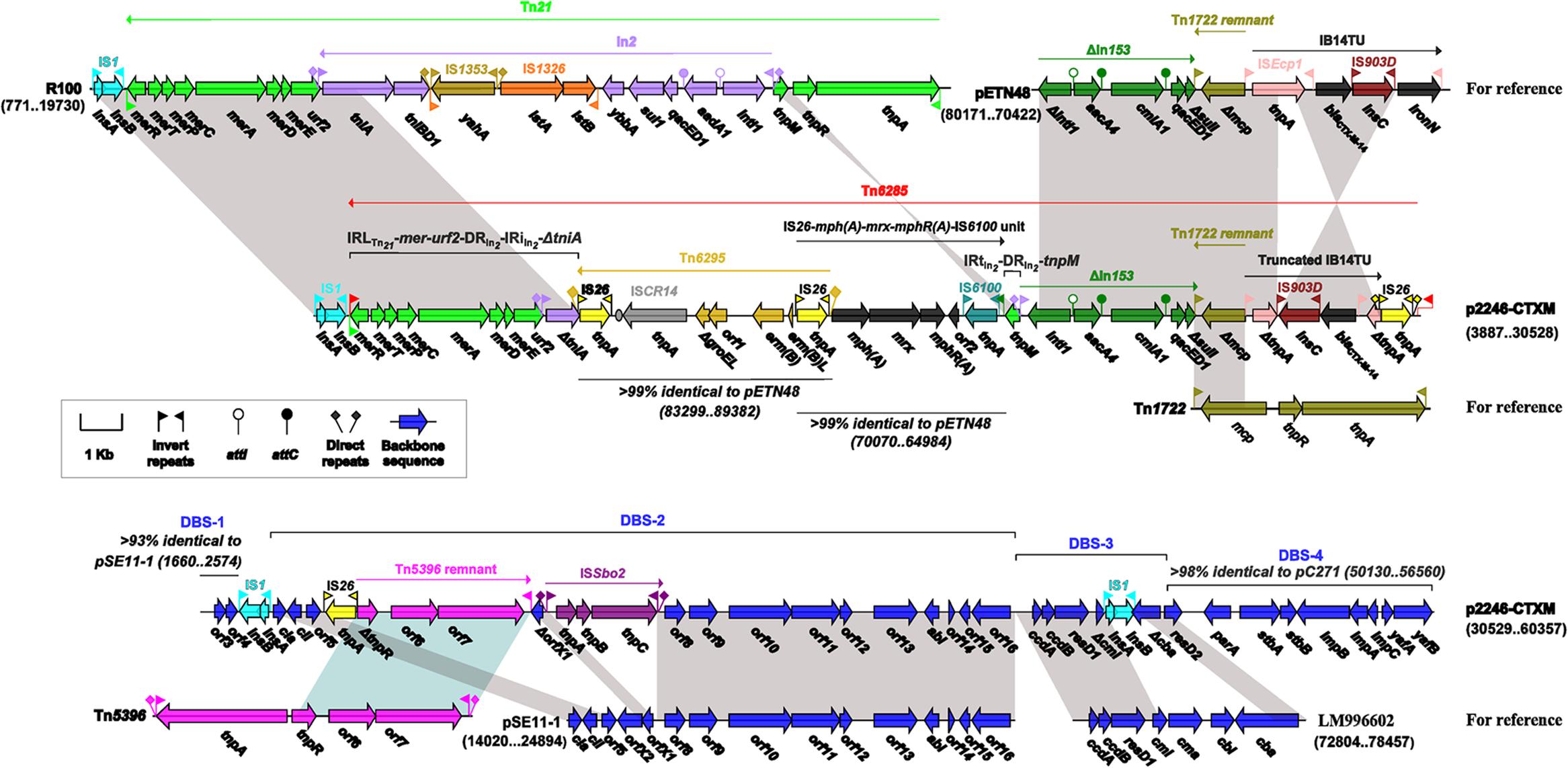
FIGURE 2. The mosaic region of p2246-CTXM with comparison to similar elements. Genes are denoted by arrows and colored based on gene function classification. Shading regions denote shared DNA regions of homology (>95% nucleotide similarity).
Tn21, as observed in plasmid R100, was a Tn3-family unit transposon which was sequentially organized as the core transposition module tnpA (transposase)-tnpR (resolvase), tnpM, the Tn402-related class 1 integron In2 that was delimited by 25 bp IRs [inverted repeat initial (IRi) plus inverted repeat terminal (IRi)] associated 5 bp DRs, urf2, and the mer gene cluster (Liebert et al., 1999).Tn21 was bordered by 38 bp IRs [inverted repeat left (IRL) plus inverted repeat right (IRR)] at both ends. It was though that the insertion of In2 into Tn21 disrupted a presumed ancestral urf2M gene into two separate ones urf2 and tnpM.
A Tn21 remnant, which comprised two separate fractions IRLTn21-mer-urf2-DRIn2- IRiIn2-ΔtniA and IRtIn2-DRIn2-tnpM, was found in Tn6285 (Figure 2). Separation of these two fraction was likely resulted from the insertion of two consecutive resistance elements Tn6295 (name given in this study) and IS26-mph(A)-mrx-mphR(A)-IS6100, which were found at two different locations in pETN48 (accession number FQ482074), a CTX-M-14-encoding plasmid of the FII-FIB replicon type and originating from an E. coli O102-ST405 strain (Billard-Pomares et al., 2011).
Tn6295 (Figure 2) was a composite transposon, which had two flanking copies of IS26 oriented in the same direction and was further delimited by DRs of GGA at both ends; moreover, an ISCR14 element and a macrolide-resistance operon erm(B)L (leading peptide)-erm(B) (methylase) were found in Tn6295.The IS26-mph(A)-mrx- mphR(A)-IS6100 unit was known as a mobile element, which harbored another macrolide-resistance operon mph(A)-mrx-mphR(A) encoding a phosphotransferase, a positive regulator and a negative transcription factor, respectively (Partridge, 2011). Notably, the upstream IS26 copy of Tn6295 and the counterpart of the IS26-mph-IS6100 unit overlapped each other in Tn6285, indicating that the connection of Tn6295 with IS26-mph(A)-mrx-mphR(A)-IS6100 was mediated by homologous recombination in IS26.
Upstream of tnpM in Tn6285 was a 9.5 kb region, which was composed in order of ΔIn153, a Tn1722 remnant, a truncated version of the ISEcp1-blaCTX-M-14-based transposition unit (IB14TU), and IS26 (Figure 2). This region was highly similar to that from nucleotide position 80171–70422 of pETN48. The tnpM gene was flanked by IRtIn2 and IRiIn153, and it appeared that the homologous recombination mediated by these two 25 bp IRs characteristic of class 1 integrons led to the inversion of tnpM in Tn6285 relative to Tn21. The ΔIn153 in Tn6285 contained two resistance gene cassettes aacA4:attCaacA4 and cmlA1:attCcmlA1 and this class 1 integron had undergone the truncation of sulI and the loss of IRt in the 3′-conserved segment. Tn1722 was a Tn3-family cryptic transposon initially identified in E. coli, with a modular structure IRLTn1722-tnpA-tnpR-mcp (methyl-accepting chemotaxis protein)-IRRTn1722 (Allmeier et al., 1992). The Tn1722 remnant in Tn6285 manifested as Δmcp-IRRTn1722.
pETN48 carried a prototype IB14TU, bound by 14 bp terminal IRs, which contained blaCTX-M-14 together with an upstream ISEcp1 and a downstream IS903D followed by ΔiroN. Similar structures of IRLISEcp1-tnpAISEcp1-IRRISEcp1-blaCTX-M-IS903D-ΔiroN- IRRISEcp1 were found in various plasmids of global origin (Zong et al., 2011). Compared with pETN48, Tn6285 harbored a truncated IB14TU element, which had undergone the loss of tnpA574to973 (position 574 to 973 of the total 1263 bp coding region of tnpA in ISEcp1) and ΔiroN-IRRISEcp1, and the inversion of tnpA974to1263-IRRISEcp1-blaCTX-M-14-IS903D (Figure 2).
The second part of the 56.4-kb mosaic region contained four plasmid backbone-derived sequences, designated DBS-1to DBS-4 in the order that they occur (Figure 2). DBS-1 was a 926 bp region with two annotated genes encoding hypothetical proteins, which showed 93% sequence similarity to the IncFII-type plasmid pSE11-1 from E. coli (Oshima et al., 2008). Located between DBS-1 and DBS-2 was an IS1 element. The 18.7-kb DBS-2 contained the colicin Ia activity-immunity system cia–cii, and it was highly similar to a 15-gene region of pSE11-1. Two separate insertions, i.e., IS26 connected with a Tn5396 remnant, and a novel insertion sequence designated ISSbo2, occurred within DBS-2 relative to pSE11-1. Tn5396 was a Tn3-family cryptic transposon initially identified in the small plasmid pEC22 from E. coli, with a modular structure IRLTn5396-tnpA-tnpR-orf6 -orf7-IRRTn5396 (Elhai et al., 1994), and the 3′-end region ΔtnpR-orf6-orf7-IRRTn5396 constituted the Tn5396 remnant observed in p2246-CTXM. The 3.6-kb DBS-3 was highly similar to a 7-gene region of the E. coli genome assembly FHI74 (accession number LM996602), which contained the toxin-antitoxin system ccdBA, the colicin M immunity-activity system cmi–cma and the colicin B system cbi–cba. An IS1 element was inserted into DBS-3, leaving cmi and cba truncated. The 6.4-kb DBS-4 was found in the IncI1-type plasmid pC271 (accession number LN735561) from E. coli and contained several plasmid maintenance genes such as parA, stbAB, and impCAB.
Expression of blaCTX-M-14, erm(B), and mph(A)
Spacer regions between ISEcp1 and blaCTX-M-55 from different ISEcp1-blaCTX-M-55 isoforms displayed three different lengths, namely 45 bp (e.g., blaCTX-M-55p1081-CTXM) (Qu et al., 2014), 48 bp (e.g., blaCTX-M-55p2246-CTXM in this study), and 127 bp (e.g., blaCTX-M-55JQ343851 and blaCTX-M-3AF550415; Figure 3). A total of two promoters P1 and P2 were experimentally identified for blaCTX-M-3AF550415 with a 127 bp spacer and, moreover, the ISEcp1-provided promoter P1 was stronger and more important than the intrinsic P2 promoter in the 127 bp spacer (Ma et al., 2011).
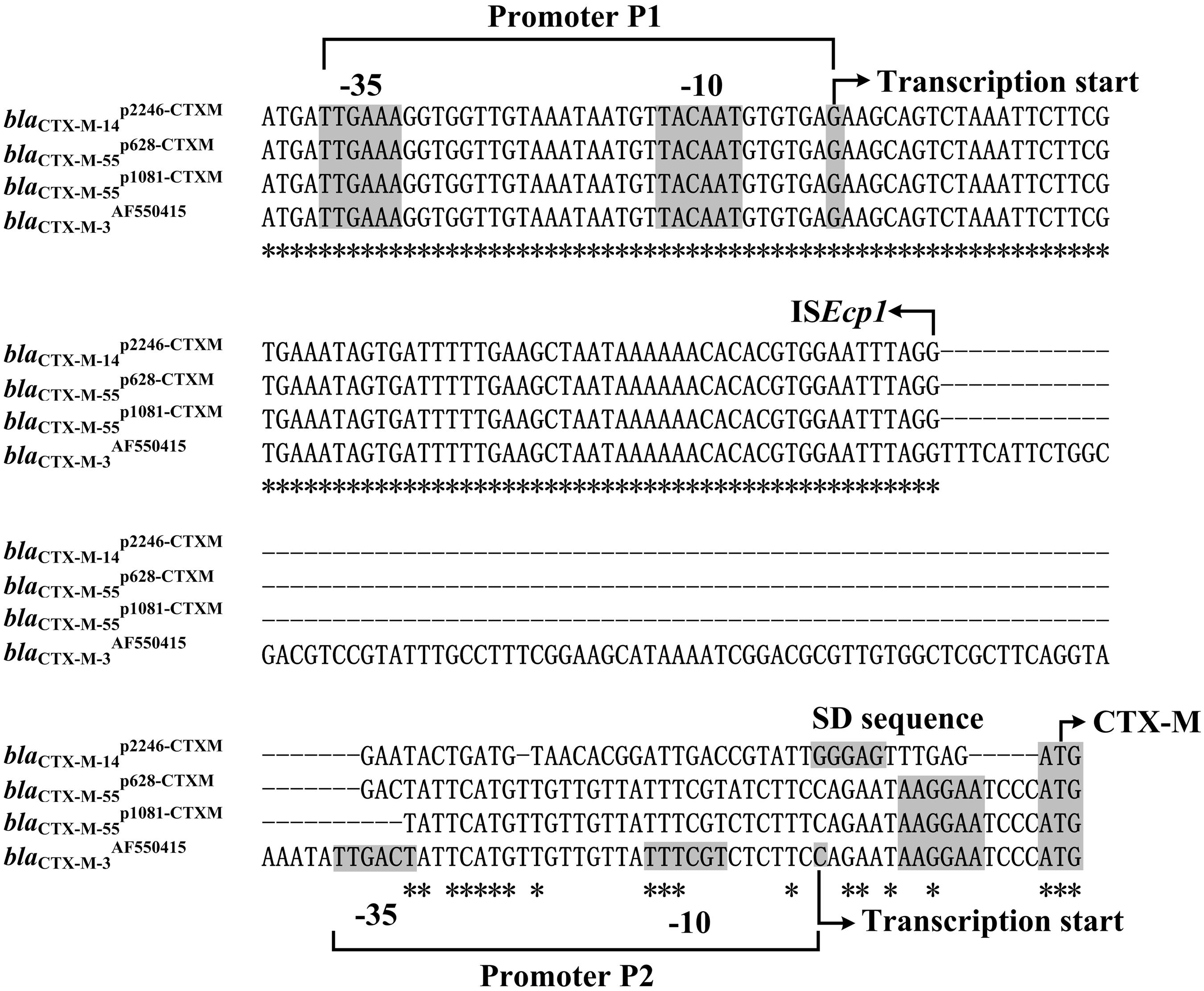
FIGURE 3. Alignment of blaCTX-M upstream sequences. The 150- to 235-bp upstream sequences together with the start codon of the blaCTX-M genes were aligned by MAFFT (http://www.ebi.ac.uk/Tools/msa/mafft/). Shown were core promoter regions, -35 and -10 elements, transcription starts, Shine–Dalgarno (SD) sequences for ribosome recognition, and translation starts.
Compared with the 127 bp spacer, the 45 or 48 bp spacer from blaCTX-M-55p1081-CTXM or blaCTX-M-55p2246-CTXM was a truncated form due to the loss of an 82 or 79 bp region within the spacer, respectively (Figure 3). This deletion impaired the -35 element of P2, most likely making the P2 activity undetectable for blaCTX-M-55p1081-CTXM and blaCTX-M-55p2246-CTXM. In the present study, the primer extension assay detected a single transcription start, i.e., nucleotide G located at 116 bp upstream of blaCTX-M-55 (Figure 4), which corresponded to the P1 promoter shared by blaCTX-M-55p1081-CTXM (Qu et al., 2014) and blaCTX-M-55p2246-CTXM (Figure 3). In addition, the primer extension assay showed that the addition of increasing amounts of ampicillin during cultivation of strains 1081 and 1081-CTXM EC600 had no effect on the activity of blaCTX-M-55p2246-CTXM promoter (Figure 4), denoting the constitutive expression of blaCTX-M-55p2246-CTXM.
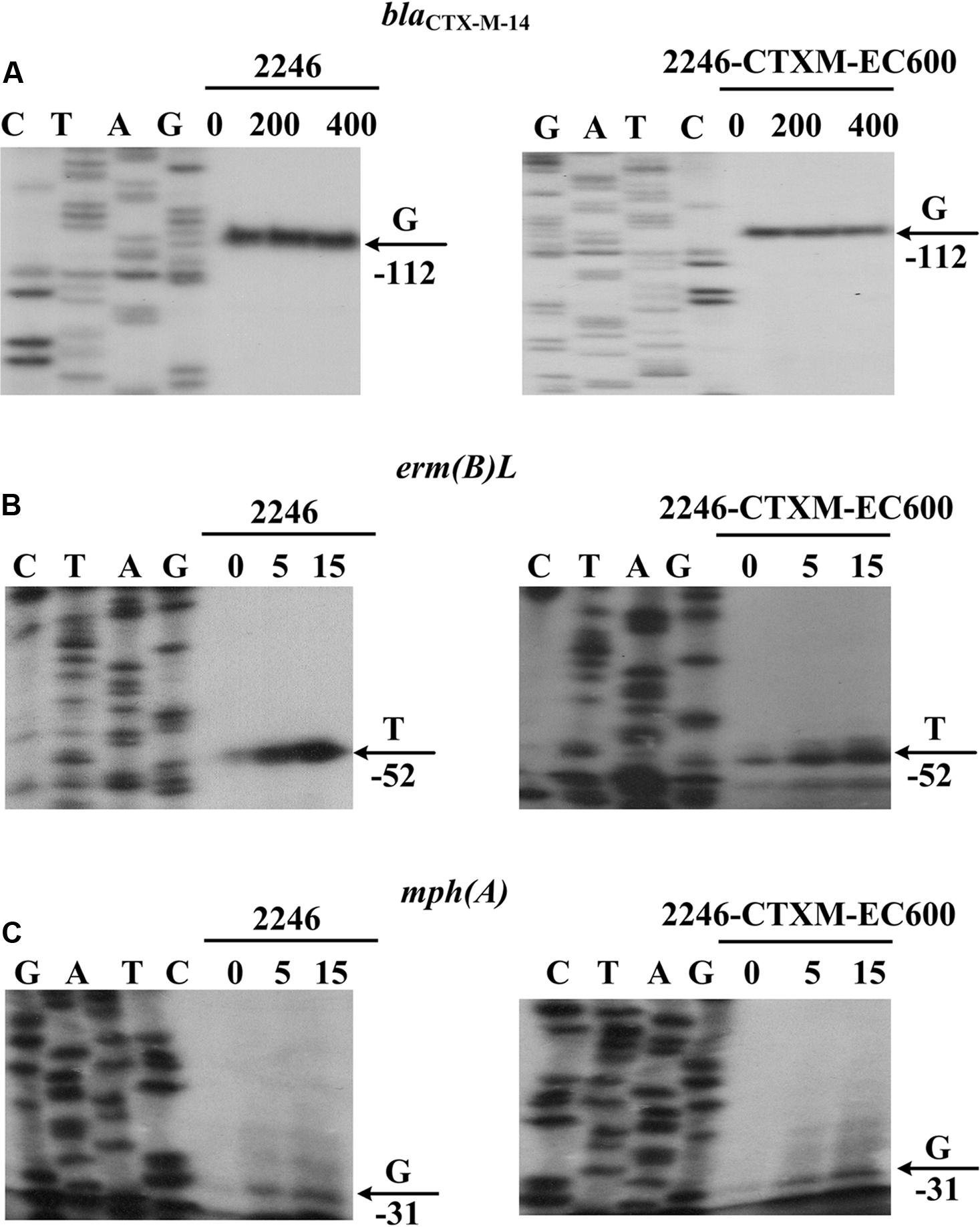
FIGURE 4. Expression of resistance genes. Primer extension assay of the RNA transcript of each of the genes blaCTX-M-14 (A), erm(B)L (B) and mph(A) (C), was performed for the two strains 2246 and 2246-CTXM-EC600 cultured with addition of increasing amounts of indicated antibiotics. Lanes C, T, A and G represent Sanger sequencing reactions. Lanes 0, 200, and 400 stand for 0, 200, and 400 mg/L ampicillin (A), respectively. Lanes 0, 5, and 15 represent 0, 5, and 15 mg/L amikacin (B) or azithromycin (C), respectively. Transcription start of each gene is indicated by arrow with nucleotide, and minus number under arrow indicates nucleotide position upstream of each gene. Representative data from at least two independent biological replicates are shown.
The primer extension assay detected a transcription start for each of erm(B)L and mph(A) from p2246-CTXM, i.e., nucleotide T located at 52 bp upstream of erm(B)L, and G at 31 bp upstream of mph(A); in addition, this assay also showed the inducible expression of erm(B)L and mph(A) upon addition of increasing amounts of amikacin and azithromycin, respectively (Figure 4). The above results were consistent with the expression patterns previously characterized for the two macrolide-resistant operons erm(B)L-erm(B) (Min et al., 2008) and mph(A)-mrx-mphR(A) (Noguchi et al., 2000).
Discussion
The current emergence and spread of resistance in Shigella strains to ciprofloxacin, ceftriaxone and azithromycin hinder the empirical antimicrobial therapy. Antimicrobial resistance patterns and mechanisms are overwhelmingly documented for S. flexneri and S. sonnei, and very little is currently known about S. boydii due to small number of available strains. This work describes a clinical S. boydii strain 2246 simultaneously resistant to ciprofloxacin, ceftriaxone, and azithromycin. As shown by PCR detection, strain 2246 contains none of the known horizontally acquired quinolone-resistance genes (data not shown). It seems that strain 2246 employs chromosomal-borne intrinsic mechanism(s) of quinolone resistance. Resistance in strain 2246 to ceftriaxone and azithromycin is attributable to the presence of blaCTX-M-14, and erm(B) and mph(A), respectively, which are co-located on a MDR plasmid p2246-CTXM.
CTX-M-type ESBLs have the high hydrolytic activity against cefotaxime and they can be divided now into five major phylogenetic groups, i.e, the CTX-M-1 group, the CTX-M-2 group, the CTX-M-8 group, the CTX-M-9 group, and the CTX-M-25 group (D’Andrea et al., 2013). CTX-M enzymes have showed the rapid increasing circulation in Shigella species in China during the past decade, and CTX-M-14 (a member of CTX-M-9 group), and CTX-M-15 and CTX-M-55 (belonging to the CTX-M-1 group) are listed sequentially as the top three common types of ESBL in the cefotaxime-resistant S. flexneri or S. sonnei from China (Zhang W. et al., 2011; Li et al., 2015).
Acquired resistance to macrolides in bacteria mainly result from three distinct mechanisms of resistance (Phuc Nguyen et al., 2009): target ribosome site modification by methylases [erm(A), erm(B), and erm(C)]; macrolide modification by esterases [ere(A) or ere(B)] or by phosphotransferases [mph(A), mph(B), and mph(D)]; and presence of efflux pumps [mef(A) and msr(A)]. Identification of azithromycin-resistant Shigella strains is rare and has been documented for only S. flexneri or S. sonnei from France (Boumghar-Bourtchai et al., 2008), the United States (Howie et al., 2010; Sjolund Karlsson et al., 2013) and Canada (Gaudreau et al., 2014), and all these azithromycin-non-susceptible Shigella isolates harbor the mphA gene. Notably, the p2246-CTXM plasmid coharbors erm(B) and mph(A), which account for two different transferable mechanisms of macrolide resistance.
Up to now, a total of seven virulence or cryptic S. boydii plasmids pBS512_211, pSB4_227, pBS512_2, pBS512_33, pBS512_5, pBS512_7, and pSB13 have their complete nucleotide sequences that ranged from 2 to 211 kb in size. This work presents the first fully sequenced resistance plasmid from S. boydii. p2246-CTXM represents a novel IncFII-type MDR plasmid with a very complex chimera structure. Its master backbone is genetically closely related to the R100 plasmid, but p2246-CTXM has evolved to integrate additional R100-unrelated backbone regions as well as massive exogenous mobile elements that carry multiple resistance determinants.
In p2246-CTXM, erm(B) together with its leading peptide gene erm(C), mph(A) together with its regulatory genes mrx and mphR(A), and blaCTX-M-14 are captured by three different transposable units Tn6295, IS26-mph(A)-mrx-mphR(A)-IS6100, and IB14TU, respectively, all of which are harbored in a large transposon Tn6285. p2246-CTXM still carries several additional resistance determinants mer (mercury resistance), aacA4 (aminoglycoside resistance), cmlA1 (chloramphenicol resistance), and qacED1 (quaternary ammonium compound resistance). The coexistence of multiple resistance genes on the same transmissible plasmid might contribute to bacterial dissemination and persistence in various hosts and environments with different antimicrobial selections. Future surveillance and epidemiological studies are needed to evaluate the prevalence of p2246-CTXM-like plasmids among Shigella isolates in China and other countries.
Author Contributions
DsZ, and YF designed experiments. LW, LL, DL, and ZY performed experiments. DsZ, and LW analyzed data. JF, DfZ, HF, YQ, WC, RY, and JW contributed reagents, materials and analysis tools. DsZ, YF, and LW, wrote this manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work is funded by the National Natural Science Foundation of China (81501779) and the National High-Tech Research and Development Program of China (2014AA021402).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01579
FIGURE S1 | The combined disk test. Experiments are conducted with three plates inoculated with three independent bacterial cultures for each strain, and only a representative figure is shown herein. CTX: 30 μg of cefotaxime; CAZ: 30 μg of ceftazidime; CTX+CA: 30 μg of cefotaxime plus 10 μg of clavulanic acid; CAZ+CA: 30 μg of ceftazidime plus 10 μg of clavulanic acid. Clavulanic acid is a member of the β-lactam family of antibiotics with little or no intrinsic antibacterial activity of its own; instead, it is used to enhance the activity of antibiotics by blocking bacterial β-lactamases as a competitive inhibitor of β-lactamases. A ≥ 5 mm increased in the zone diameter for either cefotaxime (30 μg) or ceftazidime (30 μg) tested in combination with clavulanic acid (10 μg) versus its zone when tested alone indicates the ESBL activity in the indicated strain. The results showed the ESBL activity in strains 2246 and 2246-CTXM-EC600.
References
Allmeier, H., Cresnar, B., Greck, M., and Schmitt, R. (1992). Complete nucleotide sequence of Tn1721: gene organization and a novel gene product with features of a chemotaxis protein. Gene 111, 11–20. doi: 10.1016/0378-1119(92)90597-I
Billard-Pomares, T., Tenaillon, O., Le Nagard, H., Rouy, Z., Cruveiller, S., Medigue, C., et al. (2011). Complete nucleotide sequence of plasmid pTN48, encoding the CTX-M-14 extended-spectrum beta-lactamase from an Escherichia coli O102-ST405 strain. Antimicrob. Agents Chemother. 55, 1270–1273. doi: 10.1128/AAC.01108-10
Boumghar-Bourtchai, L., Mariani-Kurkdjian, P., Bingen, E., Filliol, I., Dhalluin, A., Ifrane, S. A., et al. (2008). Macrolide-resistant Shigella sonnei. Emerg. Infect. Dis. 14, 1297–1299. doi: 10.3201/eid1408.080147
Chen, Z., Li, H., Feng, J., Li, Y., Chen, X., Guo, X., et al. (2015). NDM-1 encoded by a pNDM-BJ01-like plasmid p3SP-NDM in clinical Enterobacter aerogenes. Front. Microbiol. 6:294. doi: 10.3389/fmicb.2015.00294
Christopher, P. R., David, K. V., John, S. M., and Sankarapandian, V. (2010). Antibiotic therapy for Shigella dysentery. Cochrane Database Syst. Rev. 4:CD006784. doi: 10.1002/14651858.CD006784.pub4
CLSI (2015). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Fifth Informational Supplement M100-S25. Wayne, PA: Clinical and Laboratory Standards Institute.
D’Andrea, M. M., Arena, F., Pallecchi, L., and Rossolini, G. M. (2013). CTX-M-type beta-lactamases: a successful story of antibiotic resistance. Int. J. Med. Microbiol. 303, 305–317. doi: 10.1016/j.ijmm.2013.02.008
Elhai, J., Cai, Y., and Wolk, C. P. (1994). Conduction of pEC22, a plasmid coding for MR.EcoT22I, mediated by a resident Tn3-like transposon, Tn5396. J. Bacteriol. 176, 5059–5067.
Gaudreau, C., Barkati, S., Leduc, J. M., Pilon, P. A., Favreau, J., and Bekal, S. (2014). Shigella spp. with reduced azithromycin susceptibility, Quebec, Canada, 2012-2013. Emerg. Infect. Dis. 20, 854–856. doi: 10.3201/eid2005.130966
Howie, R. L., Folster, J. P., Bowen, A., Barzilay, E. J., and Whichard, J. M. (2010). Reduced azithromycin susceptibility in Shigella sonnei, United States. Microb. Drug Resist. 16, 245–248. doi: 10.1089/mdr.2010.0028
Klontz, K. C., and Singh, N. (2015). Treatment of drug-resistant Shigella infections. Expert Rev. Anti Infect. Ther. 13, 69–80. doi: 10.1586/14787210.2015.983902
Li, J., Li, B., Ni, Y., and Sun, J. (2015). Molecular characterization of the extended-spectrum beta-lactamase (ESBL)-producing Shigella spp. in Shanghai. Eur. J. Clin. Microbiol. Infect. Dis. 34, 447–451. doi: 10.1007/s10096-014-2244-2
Liebert, C. A., Hall, R. M., and Summers, A. O. (1999). Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63, 507–522.
Lima, I. F., Havt, A., and Lima, A. A. (2015). Update on molecular epidemiology of Shigella infection. Curr. Opin. Gastroenterol. 31, 30–37. doi: 10.1097/MOG.0000000000000136
Livio, S., Strockbine, N. A., Panchalingam, S., Tennant, S. M., Barry, E. M., Marohn, M. E., et al. (2014). Shigella isolates from the global enteric multicenter study inform vaccine development. Clin. Infect. Dis. 59, 933–941. doi: 10.1093/cid/ciu468
Ma, L., Siu, L. K., and Lu, P. L. (2011). Effect of spacer sequences between bla(CTX-M) and ISEcp1 on bla(CTX-M) expression. J. Med. Microbiol. 60, 1787–1792. doi: 10.1099/jmm.0.033910-0
Min, Y. H., Kwon, A. R., Yoon, E. J., Shim, M. J., and Choi, E. C. (2008). Translational attenuation and mRNA stabilization as mechanisms of erm(B) induction by erythromycin. Antimicrob. Agents Chemother. 52, 1782–1789. doi: 10.1128/AAC.01376-07
Niyogi, S. K. (2007). Increasing antimicrobial resistance–an emerging problem in the treatment of shigellosis. Clin. Microbiol. Infect. 13, 1141–1143. doi: 10.1111/j.1469-0691.2007.01829.x
Noguchi, N., Takada, K., Katayama, J., Emura, A., and Sasatsu, M. (2000). Regulation of transcription of the mph(A) gene for macrolide 2′-phosphotransferase I in Escherichia coli: characterization of the regulatory gene mphR(A). J. Bacteriol. 182, 5052–5058. doi: 10.1128/JB.182.18.5052-5058.2000
Oshima, K., Toh, H., Ogura, Y., Sasamoto, H., Morita, H., Park, S. H., et al. (2008). Complete genome sequence and comparative analysis of the wild-type commensal Escherichia coli strain SE11 isolated from a healthy adult. DNA Res. 15, 375–386. doi: 10.1093/dnares/dsn026
Partridge, S. R. (2011). Analysis of antibiotic resistance regions in Gram-negative bacteria. FEMS Microbiol. Rev. 35, 820–855. doi: 10.1111/j.1574-6976.2011.00277.x
Phuc Nguyen, M. C., Woerther, P. L., Bouvet, M., Andremont, A., Leclercq, R., and Canu, A. (2009). Escherichia coli as reservoir for macrolide resistance genes. Emerg. Infect. Dis. 15, 1648–1650. doi: 10.3201/eid1510.090696
Qu, F., Ying, Z., Zhang, C., Chen, Z., Chen, S., Cui, E., et al. (2014). Plasmid-encoding extended-spectrum beta-lactamase CTX-M-55 in a clinical Shigella sonnei strain, China. Future Microbiol. 9, 1143–1150. doi: 10.2217/fmb.14.53
Sjolund Karlsson, M., Bowen, A., Reporter, R., Folster, J. P., Grass, J. E., Howie, R. L., et al. (2013). Outbreak of infections caused by Shigella sonnei with reduced susceptibility to azithromycin in the United States. Antimicrob. Agents Chemother. 57, 1559–1560. doi: 10.1128/AAC.02360-12
Zhang, W., Luo, Y., Li, J., Lin, L., Ma, Y., Hu, C., et al. (2011). Wide dissemination of multidrug-resistant Shigella isolates in China. J. Antimicrob. Chemother. 66, 2527–2535. doi: 10.1093/jac/dkr341
Zhang, Y., Gao, H., Wang, L., Xiao, X., Tan, Y., Guo, Z., et al. (2011). Molecular characterization of transcriptional regulation of rovA by PhoP and RovA in Yersinia pestis. PLoS ONE 6:e25484. doi: 10.1371/journal.pone.0025484
Keywords: Shigella boydii, p2246-CTXM, blaCTX-M-14, erm(B), mph(A)
Citation: Wang L, Liu L, Liu D, Yin Z, Feng J, Zhang D, Fang H, Qiu Y, Chen W, Yang R, Wang J, Fa Y and Zhou D (2016) The First Report of a Fully Sequenced Resistance Plasmid from Shigella boydii. Front. Microbiol. 7:1579. doi: 10.3389/fmicb.2016.01579
Received: 09 July 2016; Accepted: 21 September 2016;
Published: 06 October 2016.
Edited by:
Peter Mullany, University College London, UKReviewed by:
Juan Wang, University College Dublin, IrelandAshima Kushwaha Bhardwaj, Indian Institute of Advanced Research, India
Copyright © 2016 Wang, Liu, Liu, Yin, Feng, Zhang, Fang, Qiu, Chen, Yang, Wang, Fa and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dongsheng Zhou, ZG9uZ3NoZW5nemhvdTE5NzdAZ21haWwuY29t Yunzhi Fa, eXpjMjAwNEAxMjYuY29t Jinglin Wang, d2psd2psMDgwMUBzaW5hLmNvbQ==
 Li Wang
Li Wang Lei Liu
Lei Liu Dong Liu
Dong Liu Zhe Yin
Zhe Yin Jiao Feng2
Jiao Feng2 Haihong Fang
Haihong Fang Yefeng Qiu
Yefeng Qiu Dongsheng Zhou
Dongsheng Zhou