
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 13 October 2016
Sec. Microbial Symbioses
Volume 7 - 2016 | https://doi.org/10.3389/fmicb.2016.01569
This article is part of the Research Topic Experimental models in animal-associated microbiota View all 36 articles
Digestive-tract microbiota exert tremendous influence over host health. Host-symbiont model systems are studied to investigate how symbioses are initiated and maintained, as well as to identify host processes affected by resident microbiota. The medicinal leech, Hirudo verbana, is an excellent model to address such questions owing to a microbiome that is consistently dominated by two species, Aeromonas veronii and Mucinivorans hirudinis, both of which are cultivable and have sequenced genomes. This review outlines current knowledge about the dynamics of the H. verbana microbiome. We discuss in depth the factors required for A. veronii colonization and proliferation in the leech crop and summarize the current understanding of interactions between A. veronii and its annelid host. Lastly, we discuss leech usage in modern medicine and highlight how leech-therapy associated infections, often attributable to Aeromonas spp., are of growing clinical concern due in part to an increased prevalence of fluoroquinolone resistant strains.
The human digestive-tract microbiota is implicated in affecting circadian rhythms, cancer, obesity, pharmacokinetics, and even mental health (Spanogiannopoulos et al., 2016). Investigating naturally occurring, simple, tractable model symbioses allows the identification of molecular mechanisms that through comparative studies can be generalized (Ruby, 2008) and applied to humans. One such model is the medicinal leech. The leech has a number of aspects making it suitable for molecular studies, including the presence of a simple microbial community whose dominant members can be cultured (Graf et al., 2006; Nelson and Graf, 2012).
Leeches are fascinating animals whose ability to consume blood from vertebrate hosts has been used to treat a wide range of diseases for millennia (Graf, 2000; Müller, 2000). Since the 1980’s, medicinal leech use in Western Europe and the United States has made a resurgence, especially in the treatment of vascular congestion after reconstructive surgery (de Chalain, 1996; Whitaker et al., 2004a, 2011). However, clinical use of leeches in many cases leads to wound infections that are presumably caused by digestive-tract symbiont(s) (Whitlock et al., 1983; Lineaweaver et al., 1992; Bauters et al., 2007; Whitaker et al., 2011). Historically, Pseudomonas hirudinis (now reclassified as Aeromonas hydrophila) was reported to be the only culturable bacterium from the crop (Büsing, 1951). More recent studies reveal a moderately complex community dominated by Aeromonas veronii and Mucinivorans hirudinis (Graf, 1999; Worthen et al., 2006; Maltz et al., 2014).
Studies of microbe–host interactions are aided by an ability to culture the symbionts, manipulate associations, perform genetics on the partners, and have access to the partners’ genome sequences (Ruby, 2008; Graf, 2016). H. verbana and its symbionts meet many of these criteria (Nelson and Graf, 2012): dominant symbionts are culturable (Graf, 1999; Bomar et al., 2011); genetic tools are available for A. veronii (Rio et al., 2007; Silver et al., 2007b; Maltz et al., 2015); the microbe–host association can be manipulated through antibiotic treatment and feeding of microbial species of interest (Graf, 1999; Mumcuoglu et al., 2010); genomes, metagenomes, and metatranscriptomes for the symbionts are available (Bomar et al., 2011, 2013; Bomar and Graf, 2012; Maltz et al., 2014; Nelson et al., 2015a); and an EST library for the host is also available (Macagno et al., 2010). The successful application of these tools has made the leech an amenable and powerful model for studying digestive-tract symbioses. In this review we outline current knowledge regarding microbial symbioses within the leech digestive tract, summarize known colonization factors of the dominant symbiont, A. veronii, and discuss current practices and precautions associated with medicinal leech treatment.
The most commonly available medicinal leech in the United States is H. verbana, although it is often mislabeled as H. medicinalis by medical suppliers (Siddall et al., 2007a). This confusion stems from a recent clarification of Hirudo taxonomy and the challenge of differentiating species solely based on pigmentation patterns. Hirudo species are native to Africa, Asia and Europe: H. orientalis (Transcaucasia and Iran), H. nipponia (East Asia), H. troctina (North Africa), H. verbana (Southeastern Europe and Turkey), and H. medicinalis (continental Europe and Britain) (Sawyer, 1986; Siddall et al., 2007a; Trontelj and Utevsky, 2012). In order to accurately identify a given species, DNA barcoding using the cytochrome C oxidase subunit 1 gene is recommended (Siddall et al., 2007a). Although leech species differ in salivary protein (Baskova et al., 2008; Siddall et al., 2011) and gut microbiota composition (Graf, 1999; Siddall et al., 2007b; Laufer et al., 2008; Whitaker et al., 2014), it remains unknown whether or not the efficacy of leech therapy is dependent on the leech species used.
The leech digestive tract is comprised of three major regions, the pharynx, crop, and intestinum, with each region performing distinct functions (Figure 1) (Sawyer, 1986). The pharynx is a muscular region located immediately downstream of the jaws and adjacent to the salivary glands. The largest compartment of the digestive tract is the crop, where ingested blood meals are stored and from which water and osmolytes are removed (Wenning et al., 1980). The removal of water concentrates the blood meal and forms a highly viscous intraluminal fluid (ILF). Pairs of bladders flank each cecum in the crop, facilitate the removal of water, and are themselves colonized by a distinct microbial community (Wenning and Cahill, 1989; Kikuchi et al., 2009). Digestion occurs over several weeks and is thought to occur mostly in the intestinum. The leech’s anatomy allows it to ingest a sizeable blood meal upon encountering its prey, accommodating up to five times its body weight of blood in a single meal (Wenning et al., 1980). Ingested erythrocytes are stored in the crop, remaining visually intact over prolonged time periods despite the presence of bacteria capable of β-hemolysis (Figure 1). Due to effective storage and slow digestion, the leech can go for 6 months between feedings (Sawyer, 1986).
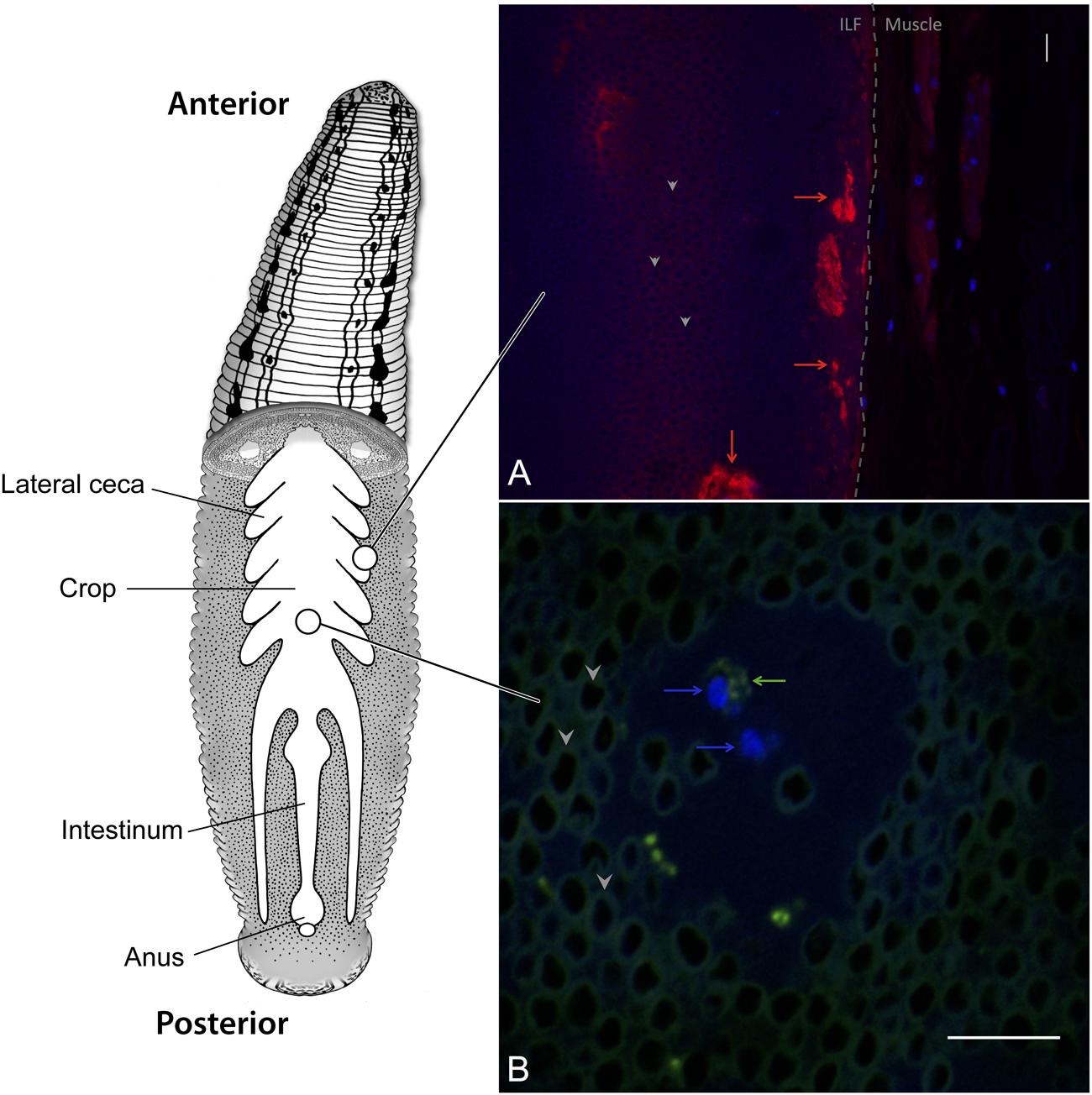
FIGURE 1. Hirudo verbana Digestive Tract. Schematic of the leech digestive tract (modified from Nelson and Graf, 2012 and Maltz et al., 2014). The ingested blood meal is stored in the crop where it forms a highly viscous intraluminal fluid (ILF) consisting of densely packed erythrocytes (dark circles surrounded by autofluorescence, examples indicated with arrow heads in insets). Fluorescence in situ hybridization micrographs of the leech crop describe (A) thick layers of mucus (red arrows) near the crop epithelium (dashed line) that develop after feeding and (B) circulating hemocytes (blue arrows) within the ILF that contain bacterial cells (green arrows). DAPI (blue), sWGA (red), and EUB338 (green). Scale bars = 10 μm.
To date, the composition of the gut microbiota from H. verbana, H. medicinalis, and H. orientalis have been studied. In each host species, the microbial community is dominated by Aeromonas and Bacteroidetes spp. (Worthen et al., 2006; Siddall et al., 2007b, 2011; Laufer et al., 2008; Whitaker et al., 2014). In H. verbana, the predominant Bacteroidetes species was initially termed Rikenella-like and was recently renamed as Mucinivorans hirudinis, a member of the Rickenellaceae (Worthen et al., 2006; Nelson et al., 2015b). Phylogenetic analysis of 16S rRNA gene sequences of Bacteroidetes isolates from hirudiniform leeches suggests close evolutionary relationships between the leech species and their Bacteroidetes symbionts (Siddall et al., 2011).
M. hirudinis is a member of the family Rikenellaceae along with Alistipes, Anaerocella, and Rikenella (Graf, 2014). These bacteria are capable of anaerobic metabolism and utilize carbohydrates as carbon and energy sources. M. hirudinis can ferment glucose, lactose, mannose, and melibiose and the metabolic endproducts include alcohols, acetic acid, proprionic acid, and succinic acid (Nelson et al., 2015b). In addition, M. hirudinis can metabolize mucus (Bomar et al., 2011), which likely provides it with an advantage in colonizing digestive tracts.
In contrast to highly specialized symbiotic bacteria that must live in close association with their host, Aeromonas spp. can succeed in a wide range of habitats (Martin-Carnahan and Joseph, 2005). A. veronii is capable of proliferating as a symbiont in digestive tracts of leeches and zebrafish (Bates et al., 2007; Roeselers et al., 2011; Nelson and Graf, 2012), subsisting as free-living cells within aquatic environments, and causing diseases in fish and mammals (Janda and Abbott, 2010; Beaz-Hidalgo and Figueras, 2013; Hossain et al., 2014). This ability to associate with different hosts provides an excellent opportunity to compare mechanistic aspects of bacterial virulence and mutualistic associations (Hentschel et al., 2000; Silver et al., 2007a).
The origin of bacterial symbionts in the leech crop has been evaluated using diagnostic PCR-based to determine whether symbionts are transmitted vertically from parent leech to offspring. The results suggested that Aeromonas is already associated with leech embryos inside the cocoon and that Mucinivorans reached detectable levels after hatching (Rio et al., 2009). Two additional studies using GFP-labeled A. veronii indicated that bacteria present on the mucosal castings could enter the digestive tract of antibiotic-cured adult leeches (Ott et al., 2014, 2016). These findings suggest that multiple modes of transmission of Aeromonas to juveniles may exist.
Because H. verbana has been studied in the greatest detail, this review will focus specifically on the symbionts of this host. In addition to the two dominant symbionts, A. veronii and M. hirudinis, other reported genera include: Morganella, Clostridium, Erysipelothrix, Desulfovibrio, and Fusobacterium (Worthen et al., 2006; Maltz et al., 2014). Of these bacteria, Aeromonas, Mucinivorans, Morganella, Clostridium, and Desulfovibrio were also found in H. orientalis (Whitaker et al., 2014). The prevalence of these bacteria in hirudiniform leeches immediately after being captured in the wild needs to be evaluated to gather a better understanding of naturally occurring microbial diversity.
Bacteria capable of occupying multiple habitats must generally be able to acquire and metabolize diverse nutrients. One of the most basic colonization barriers to symbionts and non-symbionts alike is the need to compete for and utilize available food sources (Graf, 2016). One approach to identify these resources is to screen for mutants with a reduced ability to colonize. Using signature-tagged mutagenesis (STM), one can screen multiple mutants in one animal. A mixture of mutants is introduced and they compete for nutrients and other resources inside the host (Hensel et al., 1995). For verifying the colonization capability of mutants, an individual mutant and a competitor strain are fed to the leech and forced to compete against each other and the native microbiota. The output and input ratios are used to calculate a competitive index (CI) where a ratio of less than one indicates a colonization defect of the mutant. The metabolic capacity of the organism and the resulting ability to outcompete other organisms inside the host niche are an important determinant of successful host colonization.
The digestive tracts of many animals are lined with mucus, which protects the underlying epithelium and serves as a nutrient source for some digestive-tract symbionts, (e.g., Bacteroides thetaiotaomicron and Akkermansia muciniphila) (Tailford et al., 2015). The epithelium of the leech crop is also covered with mucus, which increases in thickness after feeding (Bomar et al., 2011) (Figure 1A). Fluorescence in situ hybridization (FISH) imaging shows that M. hirudinis associates with mucus lining the leech crop epithelial wall and the abundance of surface-associated cells increases after the leech consumes a blood meal (Kikuchi and Graf, 2007). A metatranscriptomic analysis of the H. verbana ILF revealed that M. hirudinis expresses genes involved in mucin and glycan utilization at a level exceeding that of ribosomal protein coding genes. This information was exploited to design media containing mucin as the sole carbon source in order to propagate this bacterium. This optimized media was successfully used to culture the bacterium, which had previously proven recalcitrant to cultivation (Bomar et al., 2011). It is hypothesized that acetate, a fermentation product released by M. hirudinis is used as an energy source by A. veronii (Bomar et al., 2011). It is unknown whether increased mucus expression is driven by the utilization of mucin by M. hirudinis, as was shown for B. thetaiotaomicron inducing the biosynthesis of fucose in the mouse gut (Hooper et al., 1999), or if the leech simply produces mucus after feeding to protect its epithelium.
Blood is a rich nutrient source high in proteins, particularly albumin. Two lines of evidence suggest that A. veronii catabolizes the highly concentrated proteins in the leech crop. Firstly, a STM screen identified a mutant with a significantly lower CI in the crop (Table 1). This mutant had a disruption of tdcC, a conserved, anaerobically induced, threonine/serine transporter (Silver et al., 2007b), suggesting an increased competition for these amino acids in the leech crop. The second line of evidence for proteins being utilized as a nutrient in the leech crop is based on an analysis of metatranscriptome data. In this study high expression levels of genes associated with arginine catabolism, arcABCD, were detected during colonization (Bomar et al., 2011). These genes encode proteins whose products have roles in catabolism of arginine, a poor energy source, via the arginine deiminase pathway (Bomar et al., 2011). Collectively these data suggest that the proteins present in blood are an important nutrient for the digestive–tract symbionts.
A major source of nutrients in the blood meal is erythrocytes. Interestingly the erythrocytes in the leech crop are maintained intact for months after feeding despite the presence of bacteria capable of lysing blood cells (Sawyer, 1986; Lent et al., 1988; Maltz and Graf, 2011). Insight into maintenance of erythrocyte integrity was provided by a transposon mutant screen, which generated a single Aeromonas mutant unable to perform β-hemolysis (Maltz and Graf, 2011). The mutation mapped to exeM, a type 2 secretion system (T2SS) component, and resulted in a significantly reduced ability to colonize the leech crop (Table 1). The colonization defect was alleviated by feeding leeches partially lysed blood. A likely explanation of this phenotypic complementation is that the loss of β-hemolysis was responsible for the colonization defect due to the lack of liberated protein, lipids, and heme from the lysed erythrocytes.
The lysis of ingested erythrocytes likely provides an ample source of lipids. A. veronii colonizing the leech crop show elevated expression of malate synthase and isocitrate lyase. These enzymes are critical in the glyoxylate shunt, responsible for lipid metabolism (Bomar et al., 2011), and their expression suggests that in the leech crop A. veronii utilizes short-chained fatty acids (SCFA), such as acetate, and/or β-oxidation of fatty acids (Bomar et al., 2011). These nutrients are likely by-products of glycan fermentation by M. hirudinis or obtained from erythrocyte membranes, respectively.
Although blood is a high-energy nutrient source it is notably deficient in B-vitamins (Lehane, 1991). Biosynthesis of these vitamins by symbiotic bacteria is thought to supplement the dietary requirements of exclusively sanguivorous organisms. In order to foster this association, some leeches possess a specialized organ, a mycetome, which houses highly adapted intracellular bacteria (Kikuchi and Fukatsu, 2002; Siddall et al., 2004). An example of such a highly adapted symbiont is Providencia siddallii, an endosymbiont of Haementeria officinalis, with a reduced genome lacking canonical synthesis pathways for all essential amino acids while maintaining those which produce most cofactors and B-vitamins (Manzano-Marin et al., 2015). The capacity for biosynthesis of B-vitamins is observed in the endosymbionts of many obligate blood-feeders (Manzano-Marin et al., 2015) and is believed to be possessed by the digestive-tract symbionts in H. verbana, which lacks a mycetome (Maltz et al., 2014; Nelson et al., 2015a). For instance, the genome of M. hirudinis suggests a capability of producing cobinamide, a precursor of the vitamin B12 coenzyme (Nelson et al., 2015a). Without its microbial symbionts, subsisting exclusively on a blood diet would probably be impossible for H. verbana.
As blood contains very high levels of heme, organisms that feed on blood must have mechanisms to counter heme toxicity (Graça-Souza et al., 2006). One aspect of this damage is Fenton reaction-mediated oxidative stress due to the release of iron from the heme moieties. In some hemipterans, haemoxisomes in epithelial cells lining the gut protect the animal by sequestering heme (Silva et al., 2006). In the North American leech, Macrobdella decora, this is accomplished with His-rich proteins (Min et al., 2010), though whether H. verbana possesses homologs has yet to be ascertained. Sequestration of iron by transferrin in plasma and by hemoglobin in erythrocytes not only prevents oxidative damage to the host, but also restricts an essential nutrient for bacterial growth (Schaible and Kaufmann, 2004). Bacteria can acquire protein-bound iron either by producing high affinity siderophores or proteins that bind iron-containing host proteins and mediate their uptake (Byers et al., 1991).
The ability to lyse erythrocytes in order to acquire iron from the released heme is critical for allowing rapid proliferation of Aeromonas during leech colonization (Maltz and Graf, 2011; Maltz et al., 2015). Disruption of hgpB, an outer membrane heme receptor, or an associated transcriptional activator, hgpR, prevents Aeromonas from obtaining heme-associated iron and colonizing the leech digestive tract (Maltz et al., 2015). In contrast, disrupting viuB (vibrobactin utilization protein), which mediates transport of siderophore acquired iron, did not affect colonization of the crop (Maltz et al., 2015). These data suggest that attaining iron from heme is crucial for the ability of A. veronii to colonize the leech. Interestingly, heme utilization genes are widely distributed among Aeromonas species and isolates obtained from different sources, suggesting that they may play an additional role outside of symbiosis (Maltz et al., 2015).
In addition to carbon and energy sources, sufficient amounts of minor nutrients and metals can also be important for rapid growth inside the host. The STM screen identified two additional strains with nutrient-acquistion related genes that were disrupted (Silver et al., 2007b). JG537 is mutated in a pstC homolog, encoding a phosphate specific ABC transporter permease (Table 1). While the observed competition defect may be attributable to phosphate starvation, an alternative explanation of decreased membrane stability cannot be ruled out since both phenotypes are linked to disruption of this apparatus (Rao and Torriani, 1990; Daigle et al., 1995; Aguena et al., 2002; Lamarche et al., 2005). The other mutant, JG698, has a transposon within a gene encoding a ZupT ZIP protein family permease (Table 1). ZupT from Escherichia coli has been shown to have broad specificity for diverse cations, with an overall preference for zinc (Grass et al., 2002, 2005; Taudte and Grass, 2010). The reduced CI in both the leech and blood control of this mutant indicate that this transporter has a more general growth defect rather than being leech-specific (Silver et al., 2007b). However, if the leech locally restricts cation availability in the crop in response to colonization, the ZupT homolog may have a role in overcoming that response.
The growth of symbionts in vivo is affected by the ability to utilize nutrients, evade the host immune response, and compete with resident microbes. In the leech, the infrequent consumption of blood meals leads to particular bacterial growth dynamics inside the crop. After feeding, A. veronii and M. hirudinis rapidly proliferate for ∼3 days before entering a quiescent state marked by an increase in expression of stress-response related genes and a gradual decrease in population size of A. veronii while M. hirudinis continues to increase more gradually, peaking at ∼7 days (Kikuchi and Graf, 2007; Bomar and Graf, 2012). After the initial rapid proliferation, the populations of both symbionts gradually decrease in abundance and return to levels found in the starved state. For A. veronii this decline occurs within 14 days while the abundance of M. hirudinis drops at a slower rate (Kikuchi and Graf, 2007).
A major hallmark of the A. veronii transition into a quiescent state is the upregulation of the ncRNAs CsrB and CsrC (carbon starvation response). Both ncRNAs negatively regulate the mRNA binding translational regulator CsrA. CsrA/B/C homologs are found in diverse bacteria and control numerous processes such as metabolism, biofilm formation, and virulence factor production (Vakulskas et al., 2015). CsrA binds and regulates translation of target mRNA transcripts, but is antagonized by CsrB/C, which contain high affinity CsrA binding sites. A comparative in vitro and in vivo RNAseq transcriptome analysis revealed dramatically higher levels of CsrB and CsrC (>50 fold) inside the leech crop than when cultured to stationary phase in a rich medium (Bomar and Graf, 2012). This observation illustrates that the global control over mRNA and protein production is a vital aspect of the processes by which Aeromonas adapts to growth inside the leech crop.
In persistent symbiotic relationships between bacteria and animals, the symbionts are in a stalemate or a détente (McFall-Ngai, 2000). The host shapes the bacterial population by providing nutrients to allow proliferation while affecting immune responses to limit population size and restrict areas of colonization (Silver and Graf, 2011; Nyholm and Graf, 2012; Graf, 2016). Bacteria modify the host’s response by inducing or limiting the expression of proteins, surface structures and signaling molecules. In leeches, different components of the innate immune system and ingested blood meal act in concert to limit microbe colonization and expansion (Indergand and Graf, 2000; Silver and Graf, 2011; Tasiemski et al., 2015).
Many innate immune system components likely to play a role in dominant symbiont selection have been identified in Hirudo spp. (Nyholm and Graf, 2012). In the crop, bacteria are phagocytosed by circulating hemocytes (Figure 1B). A. veronii expresses a type 3 secretion system (T3SS) that is critical for avoiding this phagocytosis (Silver et al., 2007a). In addition, a number of antimicrobial peptides have been identified in H. medicinalis and related hirudiniform leeches (Tasiemski et al., 2004, 2015; Schikorski et al., 2008) including salivary lectins (Min et al., 2010), theromyzin (TMZ), theromacin (TMC), allograft inflammatory factor-1 (AIF-1), neuromacin (NMC), lumbricin (LUMB). TMC and NMC are active against both Gram-positive and -negative bacteria through pore-forming and aggregate-forming mechanisms respectively (Jung et al., 2012). TMZ is active against Gram-positive bacteria while LUMB is active against a broad range of microorganisms (Cho et al., 1998). NMC, TMZ, and LUMB are all repressed in the leech crop when A. veronii is present (Tasiemski et al., 2015), suggesting that their expression may be partially regulated by this bacterium.
Putative lipopolysaccharide binding/bactericidal perme-ability increasing proteins (LBP/BPI) have been identified that may be involved in signaling or bacteriolysis. Four putative toll-like receptors (TLRs) have also been identified in the host transcriptome and may be important in recognizing microbe-associated molecular patterns (Schikorski et al., 2008; Macagno et al., 2010; Hibsh et al., 2015). A number of anti-human CD antibodies indicative of macrophages and natural killer cells also cross-react with leech hemocytes, suggesting the ability of self/non-self recognition (de Eguileor et al., 2000a,b).
In addition to leech-produced compounds, factors present in the ingested blood meal and those produced by symbionts help to shape the microbiome. The complement from ingested blood is active inside the leech gut and restricts the proliferation of certain non-symbiotic species and serum-sensitive Aeromonas mutants (Indergand and Graf, 2000; Braschler et al., 2003). A recent study by Tasiemski et al. (2015) suggested that antimicrobial peptides produced by A. veronii also contribute to restricting the species diversity of the leech microbiome. This observation confirms early findings by Büsing et al. (1953) who suggested that the leech digestive-tract symbionts prevent other bacteria from colonizing the leech digestive tract.
The combination of host-derived innate immune response, blood-meal-derived innate immune components, and symbiont-produced compounds suggests that a complicated network of factors controls the composition and density of symbionts. While the involvement of most factors remains to be verified, these data suggest that even in the absence of the canonical adaptive immune system there are many layers of antimicrobial compounds that a successful symbiont must overcome.
Bacteria continually monitor and respond to environmental cues in order to adapt to changing conditions. The ability of A. veronii isolates to beneficially associate with leeches and zebrafish (Graf, 1999; Roeselers et al., 2011), persist as free-living bacteria in freshwater aquatic environments, and exhibit virulence toward various vertebrates (Janda and Abbott, 2010) suggests the need for regulatory systems which detect different environments and regulate gene expression accordingly. Three predicted A. veronii Hm21 regulatory elements are implicated in colonization of the leech crop through mini-Tn5 STM (Table 1) (Silver et al., 2007b).
An encoded RNase II disrupted in JG574 is a member of the RNase II/RNB-family of 3′-5′ exoribonucleases, which function in mRNA turnover (Table 1). Several enzymes from this group, particularly RNase R homologs, are implicated as virulence factors in various pathogens (Matos et al., 2014). Interestingly, an RNAse II from the nematode symbiont Photorhabdus temperata is needed for full insect virulence but not symbiosis (Hurst et al., 2015). While the specific role for RNase II in A. veronii symbiosis is unknown, it may act through a mechanistic pathway similar to that occurring in other bacterial pathogens.
Another factor with a somewhat ambiguous function is a YchF-GTPase homolog disrupted in JG697, a member of a group of highly conserved GTPases having unique substrate specificity for ATP over GTP (Table 1). YchF associates with ribosomes in E. coli, though the importance of this interaction is unclear (Verstraeten et al., 2011). Interestingly, the ATPase activity of this enzyme was recently shown to be redox regulated, and a wider role for YchF in inhibition of the oxidative stress response has been proposed (Wenk et al., 2012; Hannemann et al., 2016). Although the function and impact of the A. veronii Hm21 YchF homolog on leech colonization remains unexplored, since the mutant displayed similarly reduced CI values in both the leech and blood a general defect may be present (Silver et al., 2007b).
The ability of A. veronii to proliferate as a symbiont in digestive tracts of leeches and zebrafish (Bates et al., 2007; Roeselers et al., 2011; Nelson and Graf, 2012) and cause diseases in fish and mammals (Janda and Abbott, 2010; Beaz-Hidalgo and Figueras, 2013; Hossain et al., 2014) provides an opportunity to identify colonization factors for beneficial associations as well as pathogenic ones (Hentschel et al., 2000; Silver et al., 2007a). A. veronii Hm21, a strain isolated from H. verbana, displays virulence in multiple model systems including Galleria mellonella (wax-worms), intraperitoneal mouse injections, and an in vitro mammalian cell cytotoxicity model (Silver et al., 2007a, 2011). Unlike other pathogens that are highly host-specific, there is strong phylogenetic evidence for host-switching and virulence factor horizontal transmission between Aeromonas species (Silver et al., 2011; Martino et al., 2013). This high frequency of horizontal gene transfer makes ascribing clear ‘species-edge’ delineations very difficult when using a single or few housekeeping genes (Martino et al., 2013; Colston et al., 2014; Beaz-Hidalgo et al., 2015).
Several secretion systems have been identified as being necessary for virulence and symbiosis, including the T2SS and T3SS. The importance of the T2SS in colonization with regards to erythrocyte lysis was discussed earlier. Another STM mutant (JG752) was disrupted in a T3SS structural component and was unable to colonize the leech (Table 1). Unlike wild-type cells, JG752 was phagocytosed by leech hemocytes, indicating a specific role for the T3SS in Aeromonas evasion of the host immune system (Silver et al., 2007a). JG752 also exhibited decreased lysis of murine macrophage cells and decreased virulence in mice relative to the wild-type strain (Silver et al., 2007a), illustrating the dual importance of this factor in both symbiosis and pathogenesis. Identification and characterization of the complement of T3SS effectors will be crucial for understanding how A. veronii specifically utilizes this colonization/virulence factor to either promote persistence in the leech or opportunistic virulence in other organisms.
An additional secretion system, the T6SS, is an important factor used by bacteria to attack both prokaryotic and eukaryotic cells. New and diverse effector/immunity proteins for the T6SS are routinely being discovered (Cianfanelli et al., 2016). A re-evaluation of the sequence surrounding the disrupted locus of JG573 identified a rearrangement hotspot protein (Rhs) encoding gene located within a cluster of T6SS genes which include an encoded hemolysin co-regulated protein (Hcp), a valine-glycine repeat protein G (VgrG), and a PAAR-repeat protein (Table 1). These are all integral components of the T6SS delivery apparatus and can associate with T6SS effectors. Rhs proteins typically contain multiple domains and may possess a range of effector domains, which disrupt target cell processes (Koskiniemi et al., 2013). In A. hydrophila, the T6SS has been shown to be important in virulence (Suarez et al., 2010). The severe competition defect (>100-fold) displayed by the JG573 mutant indicates that the T6SS is an important Aeromonas colonization factor in the leech, though whether it is required for interaction with other bacterial cells or those of the host immune system remains to be seen.
Leech use as a medical practice dates back to ancient Egypt (Whitaker et al., 2004b). Since then, blood-letting has transformed from a religious experience to rid the body of disease and ‘ill humors’ to the contemporary practice of hirudotherapy. H. medicinalis leeches gained FDA-approval in 2004 and today leeches are widely used in US hospitals for treatment of compromised vasculature. Leeches are especially used after free-tissue transfer such as replanted digits, ears, facial and breast tissue (Whitaker et al., 2004a,b, 2012; Nelson and Graf, 2012). Leech therapy provides great therapeutic benefits during post-operative remediation, with studies demonstrating an associated decrease in the rate of graft failures and risk of amputation (Whitaker et al., 2011, 2012). Leeches are applied to the venous-congested sites and bite the tissue to withdraw obstructive blood while simultaneously secreting an anticoagulating agent and vasodilators to further reduce circulatory obstruction and facilitate blood flow through the area (Michalsen et al., 2008; Whitaker et al., 2011, 2012). One survey analyzed 277 case reports to quantify the efficacy of leech therapy and found that 78% of cases resulted in success, where transferred tissue was salvaged and no complications occurred (Whitaker et al., 2012). However, the advantages of leech therapy are confounded by more recent and widely reported occurrences of leech-borne infections at the bite wound, which may cause septicemia in the patient when left untreated.
Complications of leech therapy occur in part due to bacterial infections, which are thought to originate from the microbial community of the H. verbana crop. The incidence of infections in the literature ranges from 2 to 36% of cases (Whitaker et al., 2011). Prophylactic antibiotics reduce incidence of infections to the lower end of this range, though in some clinical settings no prophylactics are used at all (Whitaker et al., 2011). The occurrence of infections dramatically reduces the ability to salvage new tissue and thus jeopardizes the successful outcome of the surgery (Whitaker et al., 2011). Even with prophylactic antibiotic use, recent case reports describe severe infection of the tissue graft, which in some cases resulted in amputation of the limb or digit (Table 2).
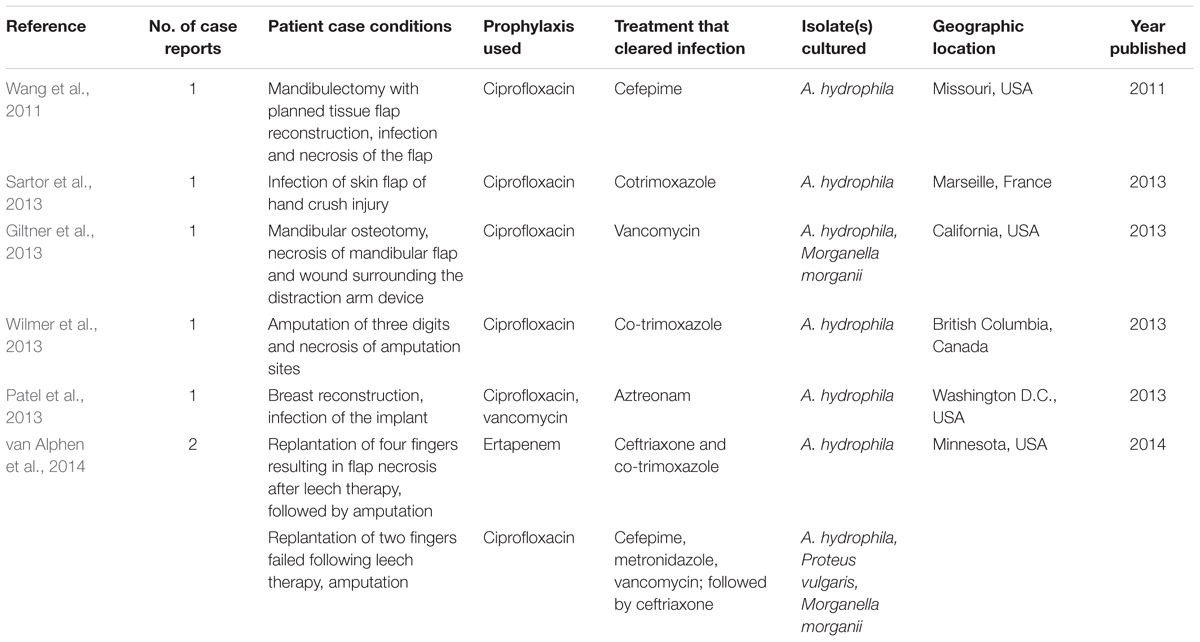
TABLE 2. Recently published case reports of CiprofloxacinR Aeromonas spp. cultured in association with medicinal leech therapy.
The most commonly isolated bacteria from infected leech bite wounds belong to the Aeromonas genus, including the fish and human pathogen A. hydrophila (Whitaker et al., 2012). A. hydrophila was reported in 88% of case reports involving infections, followed by A. veronii and A. sobria (Whitaker et al., 2012). These numbers may be affected by species-misidentifications resulting from inadequate characterization methods of the strains within this genus (Silver et al., 2011; Colston et al., 2014). Surprisingly, a recent Aeromonas infection following a pharyngectomy was reported to cause pneumonia in addition to tissue flap infection (Van Derick and Dasgupta, 2016). The risks associated with leech-borne Aeromonas infections have led many hospitals to adopt the use of ciprofloxacin (Cp), for prophylactic treatment as a standard practice before leech application. Cp is a widely used broad-spectrum fluoroquinolone and has been shown to inhibit Aeromonas, making it very useful in leech therapy prophylaxis (Whitaker et al., 2011). Until the 2000s, Aeromonas resistance to Cp was largely unreported, and to our knowledge, no CpR (Cp resistance) cases associated with leech therapy were published until 2011.
From 2011 to 2016, infections by CpR A. hydrophila were reported in eight patients following leech therapy where Cp was used as prophylaxis in the United States, Canada, and France (Wang et al., 2011; Giltner et al., 2013; Patel et al., 2013; Sartor et al., 2013; Wilmer et al., 2013; van Alphen et al., 2014), contributing to concerns of a rise in antibiotic resistant-Aeromonas infections (Table 2). These infections occurred at a range of body sites and to date have been successfully controlled by administering either individual antibiotics or combinations (see Table 2 for details). However, serious consequences of CpR Aeromonas infections following leech therapy can occur, including complete graft necrosis and amputation. For example, in 2013 a patient receiving leech therapy after mandibular surgery acquired an infection that resulted in tissue necrosis and required immediate treatment with more effective antibiotics (Van Derick and Dasgupta, 2016). In light of these nosocomial infections caused by Aeromonas strains, some research has been done to determine the genetic factors underlying an increase in CpR. Several studies suggested the importance of point mutations in gyrA and parC as well as the acquisition of plasmid encoded resistance genes such as qnrS in CpR (Giraud et al., 2004; Arias et al., 2010). Sartor et al. (2013) hypothesize a rise in resistance could originate from exposure to fluoroquinolones present in the blood of poultry used to feed leeches at the raising facility. However, further work needs to be performed to directly link clinical isolates to leeches and the genetic basis of the CpR in these strains remains to be determined.
A microbiome that is consistently dominated by two species, access to the genome sequences, culturability of dominant symbionts, and an ability to genetically manipulate Aeromonas are reasons for which the medicinal leech is an excellent model for studying the microbe–host interactions in digestive-tract symbioses. Owing in part to ease of culturing and genetic manipulation of Aeromonas, the nutrition, colonization, and persistence factor requirements of this symbiont are much better understood than others, such as M. hirudinis.
Global interrogative methods such as metagenomic and metatranscriptomic analyses have proven invaluable in identifying host and symbiont responses relating to altered gut microbiome composition and physiology. Future elucidation of more complex interactions and interrelations amongst symbionts and the leech host, such as nutrient metabolic cascades and specific immune responses, will require increased application of biochemical, molecular and genetic tools. We now have a substantial understanding and appreciation of the diversity of the leech microbiome. Future research should aim to identify parameters that contribute to the establishment of the leech gut microbiome.
Lastly, despite the proven medical benefits of leech therapy, recognition of the leech as a vector for wound infections following reconstructive surgery has led to a greater appreciation for a need to proactively minimize this undesired outcome. To this end, prophylactic administration of ciprofloxacin is common practice in leech therapy. However, since we now know medicinal leech associated Aeromonad fluoroquinolone resistance is on the rise, medical practices will need to be modified to prevent avoidable infections.
All authors listed have made substantial, direct and intellectual contribution to the work, and approved it for publication.
This work was supported by NIH RO1 GM095390 to JG and NSF 1447711 to Sanguthevar Rajasekaran.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Aguena, M., Yagil, E., and Spira, B. (2002). Transcriptional analysis of the pst operon of Escherichia coli. Mol. Genet. Genomics 268, 518–524. doi: 10.1007/s00438-002-0764-4
Arias, A., Seral, C., Gude, M. J., and Castillo, F. J. (2010). Molecular mechanisms of quinolone resistance in clinical isolates of Aeromonas caviae and Aeromonas veronii bv. sobria. Int. Microbiol. 13, 135–141.
Baskova, I. P., Kostrjukova, E. S., Vlasova, M. A., Kharitonova, O. V., Levitskiy, S. A., Zavalova, L. L., et al. (2008). Proteins and peptides of the salivary gland secretion of medicinal leeches Hirudo verbana. H. medicinalis, and H. orientalis. Biochemistry (Mosc.) 73, 315–320. doi: 10.1134/S0006297908030127
Bates, J. M., Akerlund, J., Mittge, E., and Guillemin, K. (2007). Intestinal alkaline phosphatase detoxifies lipopolysaccharide and prevents inflammation in zebrafish in response to the gut microbiota. Cell Host Microbe 2, 371–382. doi: 10.1016/j.chom.2007.10.010
Bauters, T. G., Buyle, F. M., Verschraegen, G., Vermis, K., Vogelaers, D., Claeys, G., et al. (2007). Infection risk related to the use of medicinal leeches. Pharm. World Sci. 29, 122–125. doi: 10.1007/s11096-007-9105-3
Beaz-Hidalgo, R., and Figueras, M. J. (2013). Aeromonas spp. whole genomes and virulence factors implicated in fish disease. J. Fish Dis. 36, 371–388. doi: 10.1111/jfd.12025
Beaz-Hidalgo, R., Hossain, M. J., Liles, M. R., and Figueras, M. J. (2015). Strategies to avoid wrongly labelled genomes using as example the detected wrong taxonomic affiliation for Aeromonas genomes in the GenBank database. PLoS ONE 10:e0115813. doi: 10.1371/journal.pone.0115813
Bomar, L., and Graf, J. (2012). Investigation into the physiologies of Aeromonas veronii in vitro and inside the digestive tract of the medicinal leech using RNA-seq. Biol. Bull. 223, 155–166.
Bomar, L., Maltz, M., Colston, S., and Graf, J. (2011). Directed culturing of microorganisms using metatranscriptomics. MBio 2, e12-11. doi: 10.1128/mBio.00012-11
Bomar, L., Stephens, W. Z., Nelson, M. C., Velle, K., Guillemin, K., and Graf, J. (2013). Draft genome sequence of Aeromonas veronii Hm21, a symbiotic isolate from the medicinal leech digestive tract. Genome Announc. 1, e00800-13. doi: 10.1128/genomeA.00800-13
Braschler, T. R., Merino, S., Tomas, J. M., and Graf, J. (2003). Complement resistance is essential for colonization of the digestive tract of Hirudo medicinalis by Aeromonas strains. Appl. Environ. Microbiol. 69, 4268–4271. doi: 10.1128/AEM.69.7.4268-4271.2003
Büsing, K.-H. (1951). Pseudomonas hirudinis, ein bakterieller Darmsymbiont des Blutegels (Hirudo officinalis). Zentralbl. Bakteriol. 157, 478–485.
Büsing, K.-H., Döll, W., and Freytag, K. (1953). Die Bakterienflora der medizinischen Blutegel. Arch. Mikrobiol. 19, 52–86. doi: 10.1007/BF00412315
Byers, B. R., Massad, G., Barghouthi, S., and Arceneaux, J. E. (1991). Iron acquisition and virulence in the motile aeromonads: siderophore-dependent and -independent systems. Experientia 47, 416–418.
Cho, J. H., Park, C. B., Yoon, Y. G., and Kim, S. C. (1998). Lumbricin I, a novel proline-rich antimicrobial peptide from the earthworm: purification, cDNA cloning and molecular characterization. Biochim. Biophys. Acta 1408, 67–76. doi: 10.1016/S0925-4439(98)00058-1
Cianfanelli, F. R., Monlezun, L., and Coulthurst, S. J. (2016). Aim, load, fire: the type VI secretion system, a bacterial nanoweapon. Trends Microbiol. 24, 51–62. doi: 10.1016/j.tim.2015.10.005
Colston, S. M., Fullmer, M. S., Beka, L., Lamy, B., Gogarten, J. P., and Graf, J. (2014). Bioinformatic genome comparisons for taxonomic and phylogenetic assignments using Aeromonas as a test case. MBio 5:e02136-14. doi: 10.1128/mBio.02136-14
Daigle, F., Fairbrother, J. M., and Harel, J. (1995). Identification of a mutation in the pst-phoU operon that reduces pathogenicity of an Escherichia coli strain causing septicemia in pigs. Infect. Immun. 63, 4924–4927.
de Chalain, T. M. (1996). Exploring the use of the medicinal leech: a clinical risk-benefit analysis. J. Reconstr. Microsurg. 12, 165–172. doi: 10.1055/s-2007-1006471
de Eguileor, M., Grimaldi, A., Tettamanti, G., Valvassori, R., Cooper, E. L., and Lanzavecchia, G. (2000a). Different types of response to foreign antigens by leech leukocytes. Tissue Cell 32, 40–48. doi: 10.1054/tice.1999.0085
de Eguileor, M., Grimaldi, A., Tettamanti, G., Valvassori, R., Cooper, E. L., and Lanzavecchia, G. (2000b). Lipopolysaccharide-dependent induction of leech leukocytes that cross-react with vertebrate cellular differentiation markers. Tissue Cell 32, 437–445. doi: 10.1054/tice.2000.0132
Giltner, C. L., Bobenchik, A. M., Uslan, D. Z., Deville, J. G., and Humphries, R. M. (2013). Ciprofloxacin-resistant Aeromonas hydrophila cellulitis following leech therapy. J. Clin. Microbiol. 51, 1324–1326. doi: 10.1128/JCM.03217-12
Giraud, E., Blanc, G., Bouju-Albert, A., Weill, F. X., and Donnay-Moreno, C. (2004). Mechanisms of quinolone resistance and clonal relationship among Aeromonas salmonicida strains isolated from reared fish with furunculosis. J. Med. Microbiol. 53, 895–901. doi: 10.1099/jmm.0.45579-0
Graça-Souza, A. V., Maya-Monteiro, C., Paiva-Silva, G. O., Braz, G. R. C., Paes, M. C., Sorgine, M. H. F., et al. (2006). Adaptations against heme toxicity in blood-feeding arthropods. Insect Biochem. Mol. Biol. 36, 322–335. doi: 10.1016/j.ibmb.2006.01.009
Graf, J. (1999). Symbiosis of Aeromonas veronii biovar sobria and Hirudo medicinalis, the medicinal leech: a novel model for digestive tract associations. Infect. Immun. 67, 1–7.
Graf, J. (2000). The symbiosis of Aeromonas and Hirudo medicinalis, the medicinal leech. ASM News 66, 147–153.
Graf, J. (2014). “Rikenellaceae,” in The Prokaryotes, 4th Edn, eds E. Rosenberg, E. F. Delong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin: Springer), 927.
Graf, J. (2016). Lessons from digestive-tract symbiosis between bacteria and invertebrates. Annu. Rev. Microbiol. 70, 375–393. doi: 10.1146/annurev-micro-091014-104258
Graf, J., Kikuchi, Y., and Rio, R. V. M. (2006). Leeches and their microbiota: naturally simple symbiosis models. Trends Microbiol. 14, 365–371. doi: 10.1016/j.tim.2006.06.009
Grass, G., Franke, S., Taudte, N., Nies, D. H., Kucharski, L. M., Maguire, M. E., et al. (2005). The metal permease ZupT from Escherichia coli is a transporter with a broad substrate spectrum. J. Bacteriol. 187, 1604–1611. doi: 10.1128/JB.187.5.1604-1611.2005
Grass, G., Wong, M. D., Rosen, B. P., Smith, R. L., and Rensing, C. (2002). ZupT is a Zn(II) uptake system in Escherichia coli. J. Bacteriol. 184, 864–866. doi: 10.1128/JB.184.3.864-866.2002
Hannemann, L., Suppanz, I., Ba, Q., Macinnes, K., Drepper, F., Warscheid, B., et al. (2016). Redox activation of the universally conserved ATPase YchF by Thioredoxin 1. Antioxid. Redox. Signal. 24, 141–156. doi: 10.1089/ars.2015.6272
Hensel, M., Shea, J. E., Gleeson, C., Jones, M. D., Dalton, E., and Holden, D. W. (1995). Simultaneous identification of bacterial virulence genes by negative selection. Science 269, 400–403. doi: 10.1126/science.7618105
Hentschel, U., Steinert, M., and Hacker, J. (2000). Common molecular mechanisms of symbiosis and pathogenesis. Trends Microbiol. 8, 226–231. doi: 10.1016/S0966-842X(00)01758-3
Hibsh, D., Schori, H., Efroni, S., and Shefi, O. (2015). De novo transcriptome assembly databases for the central nervous system of the medicinal leech. Sci. Data 2:150015. doi: 10.1038/sdata.2015.15
Hooper, L. V., Xu, J., Falk, P. G., Midtvedt, T., and Gordon, J. I. (1999). A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc. Natl. Acad. Sci. U.S.A. 96, 9833–9838. doi: 10.1073/pnas.96.17.9833
Hossain, M. J., Sun, D., Mcgarey, D. J., Wrenn, S., Alexander, L. M., Martino, M. E., et al. (2014). An Asian origin of virulent Aeromonas hydrophila responsible for disease epidemics in United States-farmed catfish. MBio 5, e848-14. doi: 10.1128/mBio.00848-14
Hurst, S. T., Rowedder, H., Michaels, B., Bullock, H., Jackobeck, R., Abebe-Akele, F., et al. (2015). Elucidation of the Photorhabdus temperata genome and generation of a transposon mutant library to identify motility mutants altered in pathogenesis. J. Bacteriol. 197, 2201–2216. doi: 10.1128/JB.00197-15
Indergand, S., and Graf, J. (2000). Ingested blood contributes to the specificity of the symbiosis of Aeromonas veronii biovar sobria and Hirudo medicinalis, the medicinal leech. Appl. Environ. Microbiol. 66, 4735–4741. doi: 10.1128/AEM.66.11.4735-4741.2000
Janda, J. M., and Abbott, S. L. (2010). The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 23, 35–73. doi: 10.1128/CMR.00039-09
Jung, S., Sonnichsen, F. D., Hung, C. W., Tholey, A., Boidin-Wichlacz, C., Haeusgen, W., et al. (2012). Macin family of antimicrobial proteins combines antimicrobial and nerve repair activities. J. Biol. Chem. 287, 14246–14258. doi: 10.1074/jbc.M111.336495
Kikuchi, Y., Bomar, L., and Graf, J. (2009). Stratified bacterial community in the bladder of the medicinal leech, irudo verbana. Environ. Microbiol. 11, 2758–2770. doi: 10.1111/j.1462-2920.2009.02004.x
Kikuchi, Y., and Fukatsu, T. (2002). Endosymbiotic bacteria in the esophageal organ of glossiphoniid leeches. Appl. Environ. Microbiol. 68, 4637–4641. doi: 10.1128/AEM.68.9.4637-4641.2002
Kikuchi, Y., and Graf, J. (2007). Spatial and temporal population dynamics of a naturally occurring two-species microbial community inside the digestive tract of the medicinal leech. Appl. Environ. Microbiol. 73, 1984–1991. doi: 10.1128/AEM.01833-06
Koskiniemi, S., Lamoureux, J. G., Nikolakakis, K. C., T’kint De Roodenbeke, C., Kaplan, M. D., Low, D. A., et al. (2013). Rhs proteins from diverse bacteria mediate intercellular competition. Proc. Natl. Acad. Sci. U.S.A. 110, 7032–7037. doi: 10.1073/pnas.1300627110
Lamarche, M. G., Dozois, C. M., Daigle, F., Caza, M., Curtiss, R. III, Dubreuil, J. D., et al. (2005). Inactivation of the pst system reduces the virulence of an avian pathogenic Escherichia coli O78 strain. Infect. Immun. 73, 4138–4145. doi: 10.1128/IAI.73.7.4138-4145.2005
Laufer, A. S., Siddall, M. E., and Graf, J. (2008). Characterization of the digestive-tract microbiota of Hirudo orientalis, a European medicinal leech. Appl. Environ. Microbiol. 74, 6151–6154. doi: 10.1128/AEM.00795-08
Lehane, M. J. (1991). “Managing the blood meal,” in Biology of Blood-Sucking Insects. (Netherlands: Springer), 79–110. Available at: http://www.springer.com/us/book/9789401179553
Lent, C. M., Fliegner, K. H., Freedman, E., and Dickinson, M. H. (1988). Ingestive behaviour and physiology of the medicinal leech. J. Exp. Biol. 137, 513–527.
Lineaweaver, W. C., Hill, M. K., Buncke, G. M., Follansbee, S., Buncke, H. J., Wong, R. K., et al. (1992). Aeromonas hydrophila infections following use of medicinal leeches in replantation and flap surgery. Ann. Plast. Surg. 29, 238–244. doi: 10.1097/00000637-199209000-00008
Macagno, E. R., Gaasterland, T., Edsall, L., Bafna, V., Soares, M. B., Scheetz, T., et al. (2010). Construction of a medicinal leech transcriptome database and its application to the identification of leech homologs of neural and innate immune genes. BMC Genomics 11:407. doi: 10.1186/1471-2164-11-407
Maltz, M., and Graf, J. (2011). The type II secretion system is essential for erythrocyte lysis and gut colonization by the leech digestive tract symbiont Aeromonas veronii. Appl. Environ. Microbiol. 77, 597–603. doi: 10.1128/AEM.01621-10
Maltz, M., Levarge, B., and Graf, J. (2015). Identification of iron and heme utilization genes in Aeromonas and their role in the colonization of the leech digestive tract. Front. Microbiol. 6:763. doi: 10.3389/fmicb.2015.00763
Maltz, M. A., Bomar, L., Lapierre, P., Morrison, H. G., Mcclure, E. A., Sogin, M. L., et al. (2014). Metagenomic analysis of the medicinal leech gut microbiota. Front. Microbiol. 5:151. doi: 10.3389/fmicb.2014.00151
Manzano-Marin, A., Oceguera-Figueroa, A., Latorre, A., Jimenez-Garcia, L. F., and Moya, A. (2015). Solving a bloody mess: B-vitamin independent metabolic convergence among gammaproteobacterial obligate endosymbionts from blood-feeding arthropods and the leech Haementeria officinalis. Genome Biol. Evol. 7, 2871–2884. doi: 10.1093/gbe/evv188
Martin-Carnahan, A., and Joseph, S. W. (eds). (2005). Genus I. Aeromonas Stanier 1943, 213AL. New York, NY: Springer.
Martino, M. E., Fasolato, L., Montemurro, F., Novelli, E., and Cardazzo, B. (2013). Aeromonas spp.: ubiquitous or specialized bugs? Environ. Microbiol. 16, 1005–1018.
Matos, R. G., Barria, C., Moreira, R. N., Barahona, S., Domingues, S., and Arraiano, C. M. (2014). The importance of proteins of the RNase II/RNB-family in pathogenic bacteria. Front. Cell Infect. Microbiol. 4:68. doi: 10.3389/fcimb.2014.00068
McFall-Ngai, M. J. (2000). Negotiations between animals and bacteria: the ‘diplomacy’ of the squid-vibrio symbiosis. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 126, 471–480. doi: 10.1016/S1095-6433(00)00233-6
Michalsen, A., Lüdtke, R., Cesur, O., Afra, D., Musial, F., Baecker, M., et al. (2008). Effectiveness of leech therapy in women with symptomatic arthrosis of the first carpometacarpal joint: a randomized controlled trial. Pain 137, 452–459. doi: 10.1016/j.pain.2008.03.012
Min, G. S., Sarkar, I. N., and Siddall, M. E. (2010). Salivary transcriptome of the North American medicinal leech, Macrobdella decora. J. Parasitol. 96, 1211–1221. doi: 10.1645/GE-2496.1
Mumcuoglu, K. Y., Huberman, L., Cohen, R., Temper, V., Adler, A., Galun, R., et al. (2010). Elimination of symbiotic Aeromonas spp. from the intestinal tract of the medicinal leech, Hirudo medicinalis, using ciprofloxacin feeding. Clin. Microbiol. Infect. 16, 563–567. doi: 10.1111/j.1469-0691.2009.02868.x
Nelson, M. C., Bomar, L., and Graf, J. (2015a). Complete genome sequence of the novel leech symbiont Mucinivorans hirudinis M3T. Genome Announc. 3, e1530–e1514. doi: 10.1128/genomeA.01530-14
Nelson, M. C., Bomar, L., Maltz, M., and Graf, J. (2015b). Mucinivorans hirudinis gen. nov., sp. nov., an anaerobic, mucin-degrading bacterium isolated from the digestive tract of the medicinal leech, Hirudo verbana. Int. J. Syst. Evol. Microbiol. 65, 990–995. doi: 10.1099/ijs.0.000052
Nelson, M. C., and Graf, J. (2012). Bacterial symbioses of the medicinal leech Hirudo verbana. Gut Microbes 3, 322–331. doi: 10.4161/gmic.20227
Nyholm, S. V., and Graf, J. (2012). Knowing your friends: invertebrate innate immunity fosters beneficial bacterial symbioses. Nat. Rev. Microbiol. 10, 815–827. doi: 10.1038/nrmicro2894
Ott, B. M., Cruciger, M., Dacks, A. M., and Rio, R. V. (2014). Hitchhiking of host biology by beneficial symbionts enhances transmission. Sci. Rep. 4:5825. doi: 10.1038/srep05825
Ott, B. M., Dacks, A. M., Ryan, K. J., and Rio, R. V. (2016). A tale of transmission: Aeromonas veronii activity within leech-exuded mucus. Appl. Environ. Microbiol. 82, 2644–2655. doi: 10.1128/AEM.00185-16
Patel, K. M., Svestka, M., Sinkin, J., and Ruff, P. T. (2013). Ciprofloxacin-resistant Aeromonas hydrophila infection following leech therapy: a case report and review of the literature. J. Plast. Reconstr. Aesthet. Surg. 66, e20-2. doi: 10.1016/j.bjps.2012.10.002
Rao, N. N., and Torriani, A. (1990). Molecular aspects of phosphate transport in Escherichia coli. Mol. Microbiol. 4, 1083–1090. doi: 10.1111/j.1365-2958.1990.tb00682.x
Rio, R. V., Maltz, M., Mccormick, B., Reiss, A., and Graf, J. (2009). Symbiont succession during embryonic development of the European medicinal leech, Hirudo verbana. Appl. Environ. Microbiol. 75, 6890–6895. doi: 10.1128/AEM.01129-09
Rio, R. V. M., Anderegg, M., and Graf, J. (2007). Characterization of a catalase gene from Aeromonas veronii, the digestive-tract symbiont of the medicinal leech. Microbiology 153, 1897–1906. doi: 10.1099/mic.0.2006/003020-0
Roeselers, G., Mittge, E. K., Stephens, W. Z., Parichy, D. M., Cavanaugh, C. M., Guillemin, K., et al. (2011). Evidence for a core gut microbiota in the zebrafish. ISME J. 5, 1595–1608. doi: 10.1038/ismej.2011.38
Ruby, E. G. (2008). Symbiotic conversations are revealed under genetic interrogation. Nat. Rev. Microbiol. 6, 752–762. doi: 10.1038/nrmicro1958
Sartor, C., Bornet, C., Guinard, D., and Fournier, P. E. (2013). Transmission of Aeromonas hydrophila by leeches. Lancet 381:1686. doi: 10.1016/S0140-6736(13)60316-5
Schaible, U. E., and Kaufmann, S. H. (2004). Iron and microbial infection. Nat. Rev. Microbiol. 2, 946–953. doi: 10.1038/nrmicro1046
Schikorski, D., Cuvillier-Hot, V., Leippe, M., Boidin-Wichlacz, C., Slomianny, C., Macagno, E., et al. (2008). Microbial challenge promotes the regenerative process of the injured central nervous system of the medicinal leech by inducing the synthesis of antimicrobial peptides in neurons and microglia. J. Immunol. 181, 1083–1095. doi: 10.4049/jimmunol.181.2.1083
Siddall, M. E., Min, G. S., Fontanella, F. M., Phillips, A. J., and Watson, S. C. (2011). Bacterial symbiont and salivary peptide evolution in the context of leech phylogeny. Parasitology 138, 1815–1827. doi: 10.1017/S0031182011000539
Siddall, M. E., Perkins, S. L., and Desser, S. S. (2004). Leech mycetome endosymbionts are a new lineage of alphaproteobacteria related to the Rhizobiaceae. Mol. Phylogenet. Evol. 30, 178–186. doi: 10.1016/S1055-7903(03)00184-2
Siddall, M. E., Trontelj, P., Utevsky, S. Y., Nkamany, M., and Macdonald, K. S. (2007a). Diverse molecular data demonstrate that commercially available medicinal leeches are not Hirudo medicinalis. Proc. Biol. Sci. 274, 1481–1487. doi: 10.1098/rspb.2007.0248
Siddall, M. E., Worthen, P. L., Johnson, M., and Graf, J. (2007b). Novel role for Aeromonas jandaei as a digestive tract symbiont of the North American medicinal leech. Appl. Environ. Microbiol. 73, 655–658. doi: 10.1128/AEM.01282-06
Silva, J. R., Gomes-Silva, L., Lins, U. C., Nogueira, N. F., and Dansa-Petretski, M. (2006). The haemoxisome: a haem-iron containing structure in the Rhodnius prolixus midgut cells. J. Insect Physiol. 52, 542–550. doi: 10.1016/j.jinsphys.2006.01.004
Silver, A. C., and Graf, J. (2011). Innate and procured immunity inside the digestive tract of the medicinal leech. Inv. Surviv. J. 8, 173–178.
Silver, A. C., Kikuchi, Y., Fadl, A. A., Sha, J., Chopra, A. K., and Graf, J. (2007a). Interaction between innate immune cells and a bacterial type III secretion system in mutualistic and pathogenic associations. Proc. Natl. Acad. Sci. U.S.A. 104, 9481–9486. doi: 10.1073/pnas.0700286104
Silver, A. C., Rabinowitz, N. M., Kuffer, S., and Graf, J. (2007b). Identification of Aeromonas veronii genes required for colonization of the medicinal leech, Hirudo verbana. J. Bacteriol. 189, 6763–6772. doi: 10.1128/JB.00685-07
Silver, A. C., Williams, D., Faucher, J., Horneman, A. J., Gogarten, J. P., and Graf, J. (2011). Complex evolutionary history of the Aeromonas veronii group revealed by host interaction and DNA sequence data. PLoS ONE 6:e16751. doi: 10.1371/journal.pone.0016751
Spanogiannopoulos, P., Bess, E. N., Carmody, R. N., and Turnbaugh, P. J. (2016). The microbial pharmacists within us: a metagenomic view of xenobiotic metabolism. Nat. Rev. Microbiol. 14, 273–287. doi: 10.1038/nrmicro.2016.17
Suarez, G., Sierra, J. C., Erova, T. E., Sha, J., Horneman, A. J., and Chopra, A. K. (2010). A type VI secretion system effector protein, VgrG1, from Aeromonas hydrophila that induces host cell toxicity by ADP ribosylation of actin. J. Bacteriol. 192, 155–168. doi: 10.1128/JB.01260-09
Tailford, L. E., Crost, E. H., Kavanaugh, D., and Juge, N. (2015). Mucin glycan foraging in the human gut microbiome. Front. Genet. 6:81. doi: 10.3389/fgene.2015.00081
Tasiemski, A., Massol, F., Cuvillier-Hot, V., Boidin-Wichlacz, C., Roger, E., Rodet, F., et al. (2015). Reciprocal immune benefit based on complementary production of antibiotics by the leech Hirudo verbana and its gut symbiont Aeromonas veronii. Sci. Rep. 5:17498. doi: 10.1038/srep17498
Tasiemski, A., Vandenbulcke, F., Mitta, G., Lemoine, J., Lefebvre, C., Sautière, P.-E., et al. (2004). Molecular characterization of two novel antibacterial peptides inducible upon bacterial challenge in an annelid, the leech Theromyzon tessulatum. J. Biol. Chem. 279, 30973–30982. doi: 10.1074/jbc.M312156200
Taudte, N., and Grass, G. (2010). Point mutations change specificity and kinetics of metal uptake by ZupT from Escherichia coli. Biometals 23, 643–656. doi: 10.1007/s10534-010-9319-z
Trontelj, P., and Utevsky, S. Y. (2012). Phylogeny and phylogeography of medicinal leeches (genus Hirudo): fast dispersal and shallow genetic structure. Mol. Phylogenet. Evol. 63, 475–485. doi: 10.1016/j.ympev.2012.01.022
Vakulskas, C. A., Potts, A. H., Babitzke, P., Ahmer, B. M., and Romeo, T. (2015). Regulation of bacterial virulence by Csr (Rsm) systems. Microbiol. Mol. Biol. Rev. 79, 193–224. doi: 10.1128/MMBR.00052-14
van Alphen, N. A., Gonzalez, A., Mckenna, M. C., Mckenna, T. K., Carlsen, B. T., and Moran, S. L. (2014). Ciprofloxacin-resistant Aeromonas infection following leech therapy for digit replantation: report of 2 cases. J. Hand Surg. Am. 39, 499–502. doi: 10.1016/j.jhsa.2013.11.041
Verstraeten, N., Fauvart, M., Versees, W., and Michiels, J. (2011). The universally conserved prokaryotic GTPases. Microbiol. Mol. Biol. Rev. 75, 507–542. doi: 10.1128/MMBR.00009-11
Wang, E. W., Warren, D. K., Ferris, V. M., Casabar, E., and Nussenbaum, B. (2011). Leech-transmitted ciprofloxacin-resistant Aeromonas hydrophila. Arch. Otolaryngol. Head Neck Surg. 137, 190–193. doi: 10.1001/archoto.2010.257
Wenk, M., Ba, Q., Erichsen, V., Macinnes, K., Wiese, H., Warscheid, B., et al. (2012). A universally conserved ATPase regulates the oxidative stress response in Escherichia coli. J. Biol. Chem. 287, 43585–43598. doi: 10.1074/jbc.M112.413070
Wenning, A., and Cahill, M. A. (1989). Structural and ultrastructural features of the inlet and outlet regions of the urinary bladders of the leech, Hirudo medicinalis L. J. Morphol. 201, 285–291. doi: 10.1002/jmor.1052010307
Wenning, A., Zerbst-Boroffka, I., and Bazin, B. (1980). Water and salt excretion in the leech. J. Comp. Physiol. B 139, 97–102. doi: 10.1007/BF00691022
Whitaker, I. S., Izadi, D., Oliver, D. W., Monteath, G., and Butler, P. E. (2004a). Hirudo medicinalis and the plastic surgeon. Br. J. Plast. Surg. 57, 348–353. doi: 10.1016/j.bjps.2003.12.016
Whitaker, I. S., Josty, I. C., Hawkins, S., Azzopardi, E., Naderi, N., Graf, J., et al. (2011). Medicinal leeches and the microsurgeon: a four-year study, clinical series and risk benefit review. Microsurgery 31, 281–287. doi: 10.1002/micr.20860
Whitaker, I. S., Maltz, M., Siddall, M. E., and Graf, J. (2014). Characterization of the digestive tract microbiota of Hirudo orientalis (medicinal leech) and antibiotic resistance profile. Plast. Reconstr. Surg. 133, 408e–418e. doi: 10.1097/01.prs.0000438461.06217.bb
Whitaker, I. S., Oboumarzouk, O., Rozen, W. M., Naderi, N., Balasubramanian, S. P., Azzopardi, E. A., et al. (2012). The efficacy of medicinal leeches in plastic and reconstructive surgery: a systematic review of 277 reported clinical cases. Microsurgery 32, 240–250. doi: 10.1002/micr.20971
Whitaker, I. S., Rao, J., Izadi, D., and Butler, P. E. (2004b). Historical article: Hirudo medicinalis: ancient origins of, and trends in the use of medicinal leeches throughout history. Br. J. Oral Maxillofac. Surg. 42, 133–137. doi: 10.1016/S0266-4356(03)00242-0
Whitlock, M. R., Pm, O. H., Sanders, R., and Morrow, N. C. (1983). The medicinal leech and its use in plastic surgery: a possible cause for infection. Br. J. Plast. Surg. 36, 240–244. doi: 10.1016/0007-1226(83)90100-5
Wilmer, A., Slater, K., Yip, J., Carr, N., and Grant, J. (2013). The role of leech water sampling in choice of prophylactic antibiotics in medical leech therapy. Microsurgery 33, 301–304. doi: 10.1002/micr.22087
Keywords: Aeromonas, Hirudo, digestive-tract symbiosis, bacteroidetes, leech therapy, mucinivorans, beneficial bacteria
Citation: Marden JN, McClure EA, Beka L and Graf J (2016) Host Matters: Medicinal Leech Digestive-Tract Symbionts and Their Pathogenic Potential. Front. Microbiol. 7:1569. doi: 10.3389/fmicb.2016.01569
Received: 30 June 2016; Accepted: 20 September 2016;
Published: 13 October 2016.
Edited by:
Thomas Carl Bosch, University of Kiel, GermanyReviewed by:
Mark J. Mandel, Northwestern University, USACopyright © 2016 Marden, McClure, Beka and Graf. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joerg Graf, am9lcmcuZ3JhZkB1Y29ubi5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.