- 1Department of Microbiology, University of Manitoba, Winnipeg, MB, Canada
- 2Department of Plant Science, University of Manitoba, Winnipeg, MB, Canada
- 3Department of Biological Science, University of Manitoba, Winnipeg, MB, Canada
In vitro inhibition of the fungal pathogen Sclerotinia sclerotiorum by Pseudomonas chlororaphis PA23 is reliant upon a LysR-type transcriptional regulator (LTTR) called PtrA. In the current study, we show that Sclerotinia stem rot and leaf infection are significantly increased in canola plants inoculated with the ptrA-mutant compared to the wild type, establishing PtrA as an essential regulator of PA23 biocontrol. LTTRs typically regulate targets that are upstream of and divergently transcribed from the LTTR locus. We identified a short chain dehydrogenase (scd) gene immediately upstream of ptrA. Characterization of a scd mutant revealed that it is phenotypically identical to the wild type. Moreover, scd transcript abundance was unchanged in the ptrA mutant. These findings indicate that PtrA regulation does not involve scd, rather this LTTR controls genes located elsewhere on the chromosome. Employing a combination of complementation and transcriptional analysis we investigated whether connections exist between PtrA and other regulators of biocontrol. Besides ptrA, gacS was the only gene able to partially rescue the wild-type phenotype, establishing a connection between PtrA and the sensor kinase GacS. Transcriptomic analysis revealed decreased expression of biosynthetic (phzA, prnA) and regulatory genes (phzI, phzR, rpoS, gacA, rsmX, rsmZ, retS) in the ptrA mutant; conversely, rsmE, and rsmY were markedly upregulated. The transcript abundance of ptrA was nine-fold higher in the mutant background indicating that this LTTR negatively autoregulates itself. In summary, PtrA is an essential regulator of genes required for PA23 biocontrol that is functionally intertwined with GacS.
Introduction
Public concern over the use of chemical pesticides together with the potential for acquiring resistance to these compounds has led to renewed interest in alternative strategies for management of diseases affecting plants. Pseudomonas chlororaphis strain PA23 is a soybean root-tip isolate that demonstrates excellent antifungal (AF) activity against Sclerotinia sclerotiorum (Lib.) de Bary (Savchuk and Fernando, 2004). Strain PA23 produces an arsenal of compounds including the diffusible antibiotics phenazine 1-carboxylic acid (PCA), 2-hydroxyphenazine (2-OH-PHZ), and pyrrolnitrin (PRN) together with degradative enzymes (Poritsanos et al., 2006; Zhang et al., 2006; Selin et al., 2010). PRN is the primary antibiotic responsible for biocontrol; conversely, phenazines (PHZ) are not essential for fungal suppression, but do play a role in biofilm formation (Selin et al., 2010). Despite the fact that many biocontrol agents perform well in the greenhouse, they often exhibit reduced efficacy under field conditions (Cook, 1993; Walsh et al., 2001; Haas and Keel, 2003). Variable expression of genes and gene products required for biocontrol likely contributes to poor performance in the field. It is essential, therefore, that molecular mechanisms underlying biocontrol are well understood so that production of the pathogen-suppressive factors can be optimized in the environment.
In both pathogenic and biocontrol pseudomonads, expression of secondary metabolites is controlled by a multi-tiered network of regulation. Situated at the top of this hierarchy is the GacS/GacA two-component signal transduction system, comprised of the sensor kinase GacS and its cognate response regulator GacA (Heeb and Haas, 2001). For many biocontrol strains, including PA23, a mutation in gacS or gacA leads to a loss of fungal antagonism (Heeb and Haas, 2001; Poritsanos et al., 2006). A second system, called Rsm, functions in concert with Gac and consists of a combination of RsmA-like repressor proteins and untranslated regulatory RNAs. The repressor proteins act at the post-transcriptional level by binding to and blocking the ribosome-binding site (RBS) of target mRNA (Lapouge et al., 2008). The regulatory RNAs antagonize repression by titrating out the RsmA-like proteins, rendering RBSs accessible to the translational machinery (Lapouge et al., 2008). Several additional regulatory components govern expression of PA23 AF metabolites including the PhzR/PhzI quorum-sensing (QS) system (Selin et al., 2012), the stationary phase sigma factor RpoS (Manuel et al., 2012), a regulator of RpoS called PsrA (Selin et al., 2014), and the stringent response (Manuel et al., 2012). Cross-regulation between the regulators themselves adds to the increasingly complex nature of this regulatory hierarchy (Manuel et al., 2012; Selin et al., 2012, 2014).
We have recently identified a novel regulator in PA23 called PtrA (Pseudomonas transcriptional regulator) (Klaponski et al., 2014). The phenotype of a ptrA mutant is very similar to that of a gac-deficient strain exhibiting a complete loss of AF activity (Poritsanos et al., 2006; Selin et al., 2014). PtrA belongs to the LysR-type transcriptional regulator (LTTR) family, which is the most abundant class of transcriptional regulators found among prokaryotes (Schell, 1993). LTTRs act as either activators or repressors and are known to control a diverse range of metabolic functions including cell invasion and virulence, QS, oxidative stress, and amino acid metabolism (Cao et al., 2001; Sperandio et al., 2002; Kim et al., 2004; Heroven and Dersch, 2006; Byrne et al., 2007; Kovaleva and Gelfand, 2007; Hernández-Lucas et al., 2008; Maddocks and Oyston, 2008). Preliminary proteomic and phenotypic analysis of a ptrA mutant revealed 59 differentially expressed proteins together with decreased PHZ and PRN production, consistent with the loss of AF activity (Klaponski et al., 2014).
LTTRs frequently control expression of genes that are upstream of and divergently transcribed from the lttr locus (Schell, 1993; Maddocks and Oyston, 2008). Immediately upstream of ptrA lies a gene encoding a short-chain dehydrogenase, designated scd. At present, it is not known what role scd plays in PA23 biological control. Moreover connections between PtrA and other members of the regulatory network have not been investigated. The aim of the current study was to conduct greenhouse studies to establish whether PtrA is required for PA23-mediated control of Sclerotinia stem rot. We also generated an scd mutant and determined its role in PA23 fungal suppression. Finally, a combination of complementation and transcriptional analysis was used to explore interactions between PtrA and other regulators of PA23 biocontrol.
Materials and Methods
Bacterial Strains and Growth Conditions
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli strains were cultured at 37°C on Lennox Luria Bertani (LB) agar (Difco Laboratories, Detroit, Michigan). P. chlororaphis PA23 and its derivatives were cultured at 28°C on LB agar or M9 minimal media supplemented with 1mM MgSO4 and 0.2% glucose. For AF assays, bacteria were grown on one-fifth potato dextrose agar (PDA; Difco). As required, media were supplemented with the following antibiotics: tetracycline (Tc; 15 μg/mL), gentamicin (Gm; 15 μg/mL), ampicillin (Amp; 100 μg/mL) for E. coli, and rifampicin (Rif; 25 μg/mL), Tc (15 or 100 μg/mL), Gm (20 μg/mL), piperacillin (Pip; 40 or 500 μg/mL) for P. chlororaphis. All antibiotics were obtained from Research Products International Corp. (Mt. Prospect, Illinois).
PCR
Polymerase Chain Reaction (PCR) was performed as follows. Each reaction contained 2.5 μL of both forward and reverse primers (12 μM), 1 μL of template DNA, 10 μL of 10 × Taq Buffer (without Mg added), 1.5 μL of MgSO4, 1 μL of Taq polymerase (Thermo Fisher Scientific, Carlsbad, USA) and nuclease-free water to a final volume of 100 μl. PCR reaction conditions included an initial denaturation at 98°C for 2 min, followed by 30 cycles of 98°C for 30 s, 55°C for 30 s, and 68°C for 1 min/kb, followed by a final extension of 68°C for 5 min.
Nucleic Acid Manipulation
Cloning, purification, electrophoresis, and other manipulations of nucleic acid fragments and constructs were performed using standard techniques (Sambrook et al., 1989).
Generation of a scd Mutant
To generate PA23scd, a 542-bp internal fragment of the scd gene was PCR amplified from PA23 genomic DNA using primers scd-pKNOCK FWD and scd-pKNOCK REV. The amplicon was gel purified and digested with BamHI and XhoI and cloned into the same sites of pKNOCK-Tc. The pKNOCK-scd plasmid was then mobilized into PA23 through triparental mating with the donor strain E. coli DH5αλpir containing pKNOCK-scd and the helper strain DH5α (pRK600). Pseudomonas Isolation Agar (PIA, Difco) supplemented with Tc (150 μg/mL) was used to screen for transconjugants. Insertion of the plasmid into scd was confirmed by PCR using primers tet FWD and new ptrA TL start FWD followed by sequencing of the amplicon.
Plasmid Construction
For complementation analysis, pUCP22-retS and pUCP22-ladS were generated as follows. retS was PCR amplified from PA23 genomic DNA using primers retS-F2 and retS-R2. The 2.9-kb amplicon was digested with BamHI & HindIII before cloning into the same sites of pUCP22. The ladS gene was amplified using primers ladS-F-BamHI and ladS-R-HindIII. The 2.8-kb ladS-containing fragment was subject to digestion with BamHI & HindIII and cloned into pUCP22 digested with the same enzymes. pUCP22-retS and pUCP22-ladS were verified through sequence analysis.
Antifungal Assays
Radial diffusion assays to assess fungal inhibition in vitro were performed according to previously described methods (Poritsanos et al., 2006). Five replicates were analyzed for each strain and assays were repeated three times.
Greenhouse Assays
Strains PA23 (pUCP22), PA23-443 (pUCP22), and PA23-443 (ptrA-pUCP22) were assessed for their ability to control stem rot of canola [Brassica napus (cv. Westar)] under greenhouse conditions. Canola plants were grown in pots (21 × 20 cm) at 24/16°C with a 16-h photoperiod. The plants were sprayed at 30% flowering with bacterial strains (2.0 × 108 CFU mL−1) suspended in sterile distilled water with 0.02% Tween 20 and kept in a growth chamber (24/16°C, 16-h photoperiod). Twenty-four hours after bacterial inoculation, plants were sprayed with ascospores of S. sclerotiorum (8 × 104 spores mL−1) suspended in water containing 0.02% Tween 20. Pathogen control plants were inoculated with ascopores only, while healthy control plants were sprayed with water (0.02% Tween 20). After pathogen inoculation, plants were incubated in a humidity chamber for 72 h, after which they were placed back in the growth chamber. Fourteen days after ascospore inoculation, symptom development on the stem and leaves was scored according to Selin et al. (2010). Ten plants were used for each treatment and the plant studies were repeated two times.
Phenazine Analysis
Production of PCA and 2-OH-PHZ was quantified according to the methods outlined by Chancey et al. (1999). Overnight cultures (5 mL) were grown in M9 minimal media (1 mM MgSO4; 0.2% glucose) and subjected to PHZ extraction. Spectrophotometric quantification was performed at 367 nm and 490 nm for PCA and 2-OH-PHZ, respectively (Maddula et al., 2008). PHZ analysis was performed in triplicate.
Pyrrolnitrin Analysis
Production of the antibiotic PRN was quantified according to the methods outlined by Selin et al. (2010). Briefly, 20 mL cultures of PA23 and its derivatives were grown for 5 days in M9 minimal media (1 mM MgSO4; 0.2% glucose) and PRN was extracted with an equal volume of ethyl acetate. Before extraction, toluene was added to each sample as an internal control. Toluene and PRN UV absorption maxima were recorded at 225 nm with a Varian 335 diode array detector. PRN peaks were detected at 4.7 min. Samples were analyzed in duplicate.
Autoinducer Analysis
The production of AHL was analyzed by spotting 10 μL of an overnight culture onto LB agar plates seeded with C. violaceum CV026. This strain is able to detect exogenous AHLs with carbon chain length structures ranging from C4 to C8, resulting in a purple halo surrounding the colonies. The diameter of the purple zones was measured at 24 h.
Semi-Quantitative Reverse Transcriptase PCR
To monitor expression of genes involved in biocontrol, semi-quantitative reverse transcriptase (RT) PCR was used. PA23 and its derivatives were grown to early stationary phase and total RNA was extracted using a RNeasy Mini Kit (QIAGEN, Valencia, USA). Residual genomic DNA was removed by treatment with TURBO RNAase-free DNAse I (Ambion, Carlsbad, USA) during the RNA isolation procedure. RNA concentrations were measured at 260 and 280 nm and only RNA samples with A260/A280 between 1.8 and 2.0 were used in subsequent steps. cDNA was generated by reverse transcription using the Maxima First Strand cDNA Synthesis Kit (ThermoScientific, Rockford, USA) and random hexamer primers in a 20 μL total reaction volume. The following conditions were employed: initial heating at 25°C for 10 min, 50°C for 15 min for reverse transcription and 85°C for 5 min for enzyme denaturation. Sequences for the genes of interest from PA23 were obtained from GenBank. The primer sequences are listed in Table 1. PCR was performed using a CFX96 Connect™ Real-Time PCR Detection System (Bio-rad, Hercules, USA) and SsoFast™ EvaGreen® Supermix (Bio-rad). The final 10-μL volume mixture in each well contained 0.4 μL of both forward and reverse primers (12 μM), 1 μL of 1:20 diluted cDNA, 5 μL of SsoFast™ EvaGreen® Supermix and 3.4 μL of nuclease-free water. PCR reaction conditions included an initial denaturation at 98°C for 2 min, followed by 39 cycles of 98°C for 5 s, 60°C for 30 s, and 60°C for 5 s. Melt-curve analysis was performed to evaluate the formation of primer dimers and other artifacts to validate results. Each reaction was performed in triplicate and experiments were repeated three times with three biological replicates. Relative gene expression was calculated using the ΔΔCt method as described by Livak and Schmittgen (2001) using rpoB as the reference gene and the CFX Manager™ software (Bio-rad).
Phylogenetic Analysis
Genetic sequences for ptrA homologs were obtained through BLAST (Altschul et al., 1990). The accession number for ptrA (EF054873.1) was used as a query against Pseudomonadales (taxid:72274) sequences in the non-redundant nucleotide collection (nr/nt) employing the blastn algorithm. For amino acid sequences, this query was used against the non-redundant protein database using the blastp algorithm. Forty nucleotide and 41 amino acid sequences were selected with E-values of <2 × 10−100. Both sets of sequences were aligned using Mafft servers (Katoh et al., 2005). Phylogenetic trees were constructed with the MEGA6 program package (Tamura et al., 2013). For nucleotide sequences, the Maximum likelihood method based on the Tamura-Nei model (Tamura and Nei, 1993) was applied, with genetic distances estimated through the Maximum composite likelihood approach. Bootstrap analysis with a thousand substitutions was utilized to test the trees. The nucleotide sequence for a LTTR from Serratia marcescens strain RSC-14 (gi: 926475601) was included as an outgroup for phylogenetic tree construction. For amino acid sequences, the Maximum likelihood method based on the Le-Gascuel 2008 model (Le and Gascuel, 1993) was employed with genetic distances estimated using the JTT model. Bootstrap analysis with a thousand substitutions was used to test the trees. The amino acid sequence for a LTTR from S. marcescens (gi: 759524346) was included as an outgroup for phylogenetic tree construction.
Statistical Analysis
All statistical analysis was performed using unpaired Students's t-test.
Results
PtrA is Essential for PA23 Biocontrol of S. sclerotiorum in the Greenhouse
The wild-type PA23, PA23-443 and the complemented PA23-443 (ptrA-pUCP22) were tested for their ability to protect canola from stem rot disease caused by S. sclerotiorum. Two parameters were evaluated, namely incidence of leaf infection and stem rot disease severity. As illustrated in Figure 1, the ptrA mutant showed a significant reduction in its ability to control fungal infection of leaves and stems and reduce overall disease severity. Compared to the disease control, the ptrA mutant mediated a modest decrease in leaf infection (Figure 1A) and no difference in disease severity (Figure 1B). Addition of ptrA in trans restored the ability of PA23-443 to prevent both leaf infection and stem rot to wild-type levels (Figure 1).
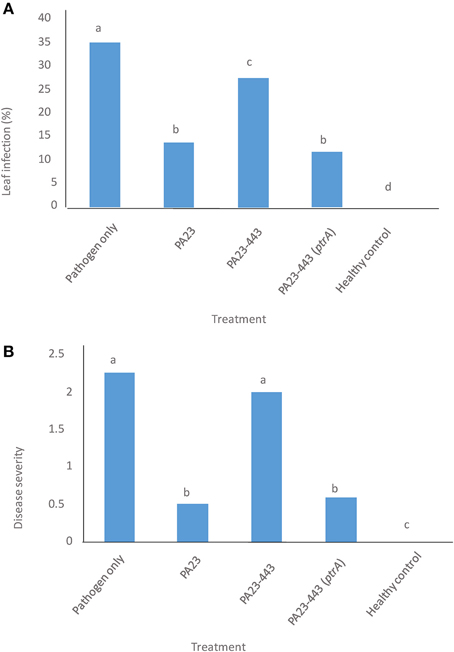
Figure 1. Efficiency of Pseudomonas chlororaphis PA23 (wild type, pUCP22), ptrA-mutant PA23-443 (pUCP22), and ptrA-complemented strain PA23-443 (pUCP22-ptrA) in managing Sclerotinia sclerotiorum ascospore infection on canola plants. (A) Percent incidence of leaf infection. (B) Stem rot disease severity. In all treatments except the healthy control, plants were sprayed with S. sclerotiorum ascospores (8 × 104 spores/ml). The healthy control plants were sprayed with water. Column means labeled with the same letter do not differ significantly by Duncan's Multiple Range Test (DMRT; P > 0.05).
scd, Which Lies Upstream of PtrA, Does Not Appear to Be Involved in PA23 AF Activity
A 115-bp intergenic region separates ptrA and an upstream gene encoding a short-chain dehydrogenase, designated scd. To determine whether this allele is involved in PtrA regulation, an scd insertional mutant was generated. Unlike the ptrA mutant that is devoid of AF activity, the scd mutant showed near wild-type fungal suppression (Figure 2). Due to the plasmid insertion, only the first 100 nt (of 600) of the scd open reading frame remain intact (data not shown). It seems highly unlikely that a functional truncated Scd is being produced, but we cannot rule out this possibility entirely.
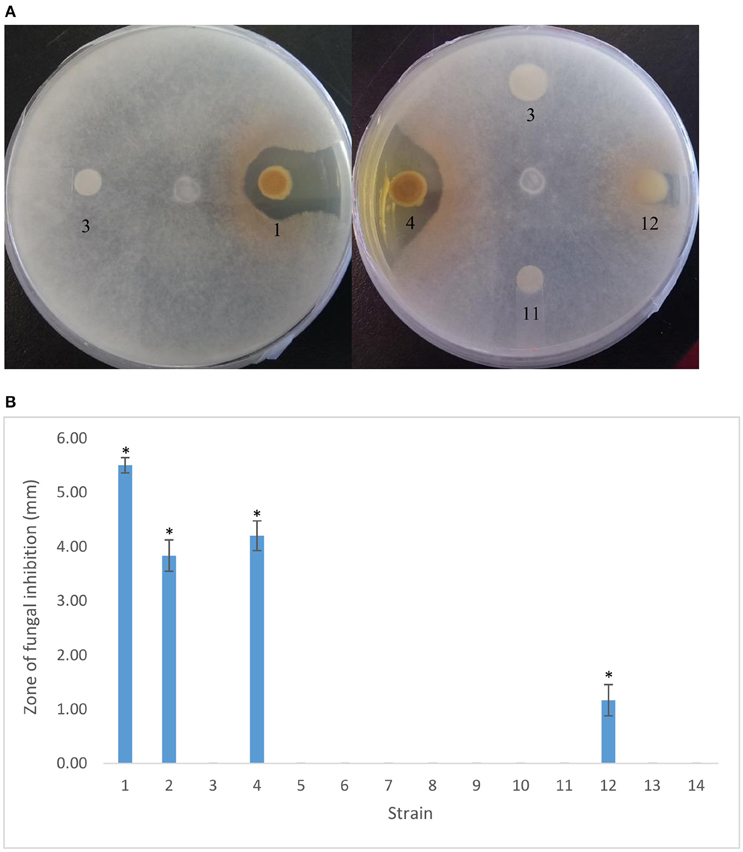
Figure 2. In vitro antifungal activity of PA23 and derivative strains against Sclerotinia sclerotiorum. (A) Radial diffusion assays showing S. sclerotiorum inhibition after 48 h growth on 1/5 PDA at room temperature. (B) Zone of fungal inhibition (mm) surrounding bacterial colonies. Samples are as follows: 1, PA23 (pUCP22); 2, PA23scd (pUCP22); 3, PA23-443 (pUCP22); 4, PA23-443 (pUCP22-ptrA); 5, PA23-443 (pUCP22-rsmA); 6, PA23-443 (pUCP22-rsmE); 7, PA23-443 (pUCP23-rsmZ); 8, PA23-443 (pUCP22-rpoS); 9, PA23-443 (pUCP22-psrA); 10, PA23-443 (pUCP23-phzR); 11, PA23-443 (pUCP22-gacA); 12, PA23-443 (pUCP23-gacS); 13, PA23-443 (pUCP22-retS), PA23-443 (pUCP22-ladS). For strains that differ significantly from PA23-443 (ptrA mutant), columns have been marked with an asterisk (*p < 0.0001).
gacS is Able to Partially Complement the PtrA Mutant
To reveal interactions between PtrA and other members of the regulatory hierarchy overseeing PA23 biocontrol, plasmid-borne copies of regulatory genes constitutively expressed from the lac promoter were transformed into the ptrA mutant. Genes that are able to fully or partially complement the mutant are predicted to lie downstream of PtrA in the regulatory cascade. We began our characterization of the PA23-443 transformants by analyzing AF activity. As expected, providing ptrA in trans restored AF activity close to wild-type levels (Figure 2). The only other gene that resulted in partial complementation of fungal suppression was gacS (Figure 2).
PA23 produces the diffusible antibiotics PHZ and PRN. Antibiotic analysis revealed that strain PA23-443 synthesized markedly lower levels of both compounds, whereas no difference in antibiotic production was observed for the scd mutant (Table 2). Addition of ptrA in trans led to partial and full restoration of PHZ and PRN production, respectively. Consistent with the AF analysis, plasmid-borne gacS resulted in partial complementation of antibiotic synthesis in the ptrA mutant (Table 2). Our protease activity profiles closely mirrored what was observed for the antibiotics. The ptrA mutant was devoid of protease production, whereas the scd mutant showed wild-type activity (Figure 3). Addition of ptrA and gacS in trans lead to full and partial rescue of protease activity in the ptrA mutant background, respectively (Figure 3).
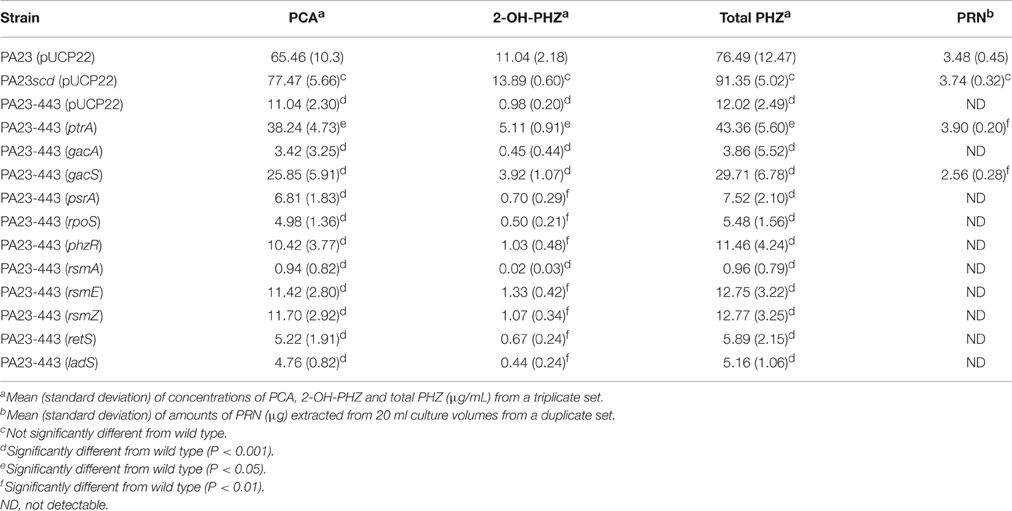
Table 2. Quantification of phenazines and pyrrolnitrin present in cultures of Pseudomonas chlororaphis PA23, PA23scd and PA23-443 harboring empty vector or various overexpression plasmids.
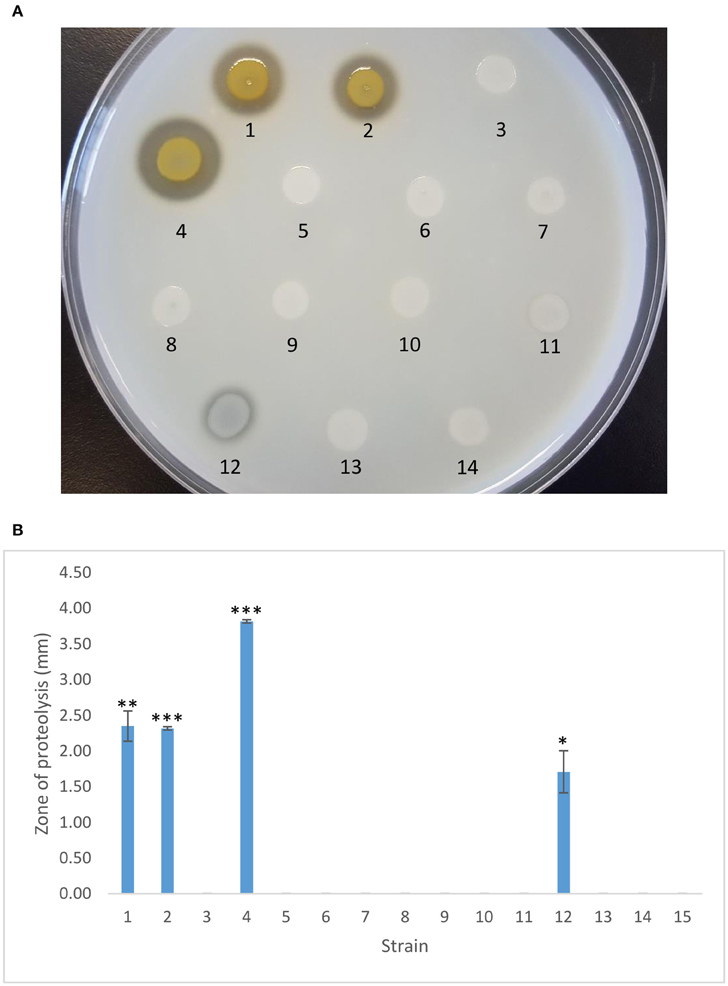
Figure 3. Protease activity exhibited by PA23 and derivative strains. (A) Protease production on 2% skim milk agar. (B) Zone of proteolysis (mm) surrounding bacterial colonies after 48 h incubation at 28°C. Samples are as follows: 1, PA23 (pUCP22); 2, PA23scd (pUCP22); 3, PA23-443 (pUCP22); 4, PA23-443 (pUCP22-ptrA); 5, PA23-443 (pUCP22-rsmA); 6, PA23-443 (pUCP22-rsmE); 7, PA23-443 (pUCP23-rsmZ); 8, PA23-443 (pUCP22-rpoS); 9, PA23-443 (pUCP22-psrA); 10, PA23-443 (pUCP23-phzR); 11, PA23-443 (pUCP22-gacA); 12, PA23-443 (pUCP23-gacS); 13, PA23-443 (pUCP22-retS), PA23-443 (pUCP22-ladS). For strains that differ significantly from PA23-443 (ptrA mutant), columns have been marked with an asterisk (*p < 0.05; **p < 0.01; ***p < 0.0001).
PtrA Regulates AHL Signal Production
The AHL signal generated by the PhzRI QS system activates the CVO26 biosensor resulting in a white to purple color change due to the production of the QS-controlled pigment violacein. While CVO26 cells form a purple halo around colonies of PA23 and the scd mutant, this is not observed for PA23-443 indicating that the latter is AHL deficient. Addition of ptrA, gacA, and gacS in trans rescued AHL production in the ptrA mutant to varying degrees (Figure 4).
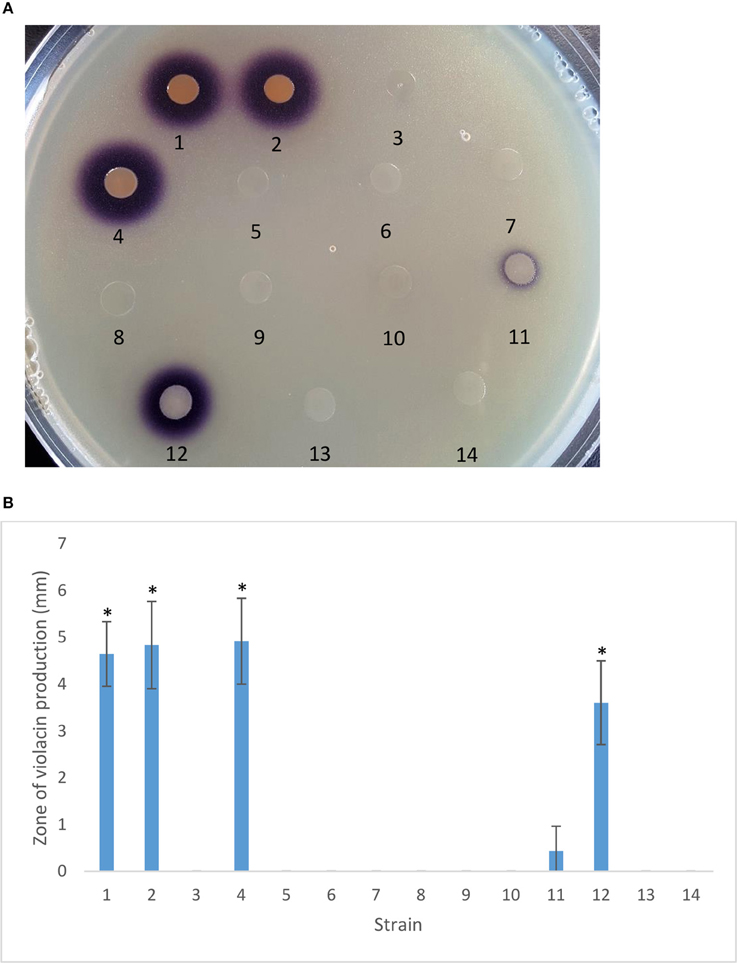
Figure 4. Autoinducer production by PA23 and derivative strains. (A) Autoinducer production by PA23, PA23-443 (ptrA-mutant) and derivative strains determined using Chromobacterium violaceum CVO26-seeded agar. (B) Zone of purple pigmentation (mm) indicative of bacterial autoinducer production after 48 h on CVO26-indicator plates. Samples are as follows: 1, PA23 (pUCP22); 2, PA23scd (pUCP22); 3, PA23-443 (pUCP22); 4, PA23-443 (pUCP22-ptrA); 5, PA23-443 (pUCP22-rsmA); 6, PA23-443 (pUCP22-rsmE); 7, PA23-443 (pUCP23-rsmZ); 8, PA23-443 (pUCP22-rpoS); 9, PA23-443 (pUCP22-psrA); 10, PA23-443 (pUCP23-phzR); 11, PA23-443 (pUCP22-gacA); 12, PA23-443 (pUCP23-gacS); 13, PA23-443 (pUCP22-retS), PA23-443 (pUCP22-ladS). For strains that differ significantly from PA23-443, columns have been marked with an asterisk (*p < 0.0001).
Multiple Genes Show Altered Expression in the PtrA Mutant
To better understand how PtrA functions as a regulator of PA23 biocontrol, expression analysis of biosynthetic and regulatory genes associated with fungal suppression was undertaken (Figure 5). The two biosynthetic genes analyzed, phzA (PHZ) and prnA (PRN), showed a dramatic reduction in expression levels in the ptrA mutant background. In terms of regulatory genes, gacA and gacS exhibited a 50% reduction and no change in expression, respectively. Expression of both phzI, encoding the AHL synthase and phzR, encoding the LysR-type transcriptional regulator PhzR, were significantly downregulated in the ptrA mutant (Figure 5). Interestingly, psrA transcription was modestly increased, while the PsrA-regulated target gene rpoS was down regulated in the mutant background. Analysis of genes encoding Rsm repressor proteins (rsmA, rsmE) and regulatory RNAs (rsmX, rsmY, rsmZ) revealed elevated rsmE and rsmY expression in the ptrA mutant. Conversely rsmA, rsmX and rsmZ were downregulated, indicating positive regulation by PtrA. Next, we examined the orphan sensor kinase-encoding retS and ladS genes. A decrease in retS activity was observed; whereas ladS transcription remained at wild-type levels. The gene showing the most dramatic change in transcript abundance was ptrA. In the PA23-443 background, ptrA expression increased nine fold indicating that this LTTR is subject to negative autoregulation. No change in scd transcription was observed further supporting that PtrA regulation is not mediated through this divergently transcribed gene. Quantitative RT-PCR was used to monitor expression of three genes immediately upstream of scd, encoding a membrane protein (AIC18438.1; primers Up1), a hypothetical protein (AIC18437.1; primers Up2) as well as nhaA (AIC18435.1; primers Up3). No differences in transcript abundance were detected in the ptrA mutant compared to the wild type (data not shown).
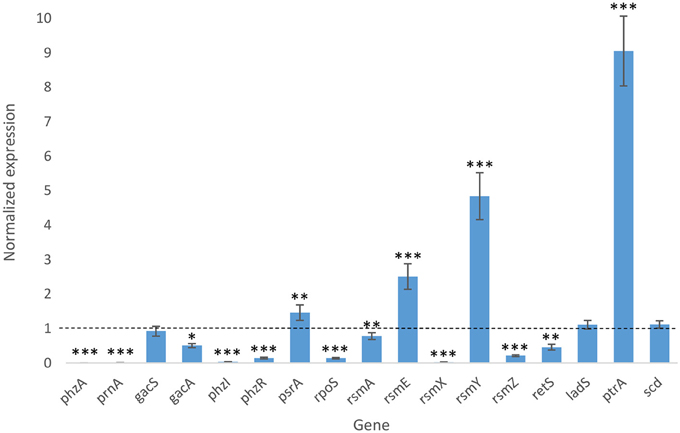
Figure 5. Biocontrol gene expression in PA23-443 compared to wild type. qRT-PCR fold change in gene expression in the ptrA mutant vs. wild type was determined using rpoB as the reference gene. The level of gene expression in the PA23 wild type was normalized to 1.0 (indicated by the dotted line). Gene expression that differs significantly from wild type is indicated with an asterisk (*p < 0.05; **p < 0.01; ***p < 0.0001).
Phylogenetic Analysis of PtrA Homologs
Phylogenetic analysis revealed that PtrA was conserved amongst pseudomonads, including both biocontrol and pathogenic strains. Homologs from well-known biocontrol strains P. chlororaphis O6 and P. protegens Pf-5 were found to cluster with PtrA (Figure 6). A similar pattern was observed when a phylogenetic tree was constructed using genomic sequences (Supplementary Figure 1). As closely related homologs of PtrA are found in several biocontrol strains, they are expected to play a similar role in regulating genes responsible for the production of antifungal compounds. A list of the PtrA homologs used for the phylogenetic analysis can be found in Supplementary Tables 1, 2.
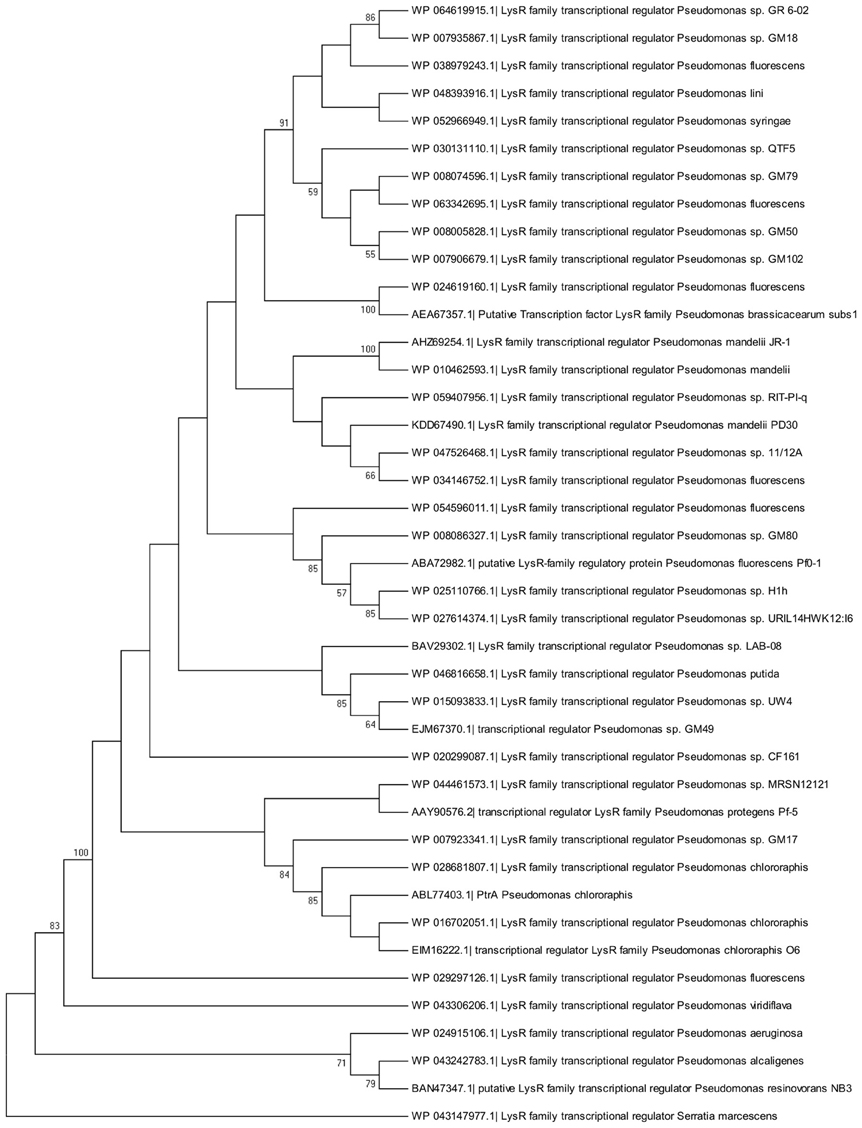
Figure 6. Molecular phylogenetic analysis of PtrA homologs by the Maximum Likelihood method. The evolutionary history was inferred using the Maximum Likelihood method based on the Le Gascuel Method (Le and Gascuel, 1993). This analysis involved 41 amino acid sequences including PtrA. The tree with the highest log likelihood is shown. The percentage of trees in which the associated taxa clustered together is indicated next to the branches.
Discussion
LTTRs, which represent the largest family of prokaryotic transcriptional regulators, frequently regulate divergently transcribed genes; however, targets can be located elsewhere on the chromosome (Schell, 1993; Maddocks and Oyston, 2008). Several pieces of evidence suggest that PtrA fits into this second paradigm, functioning as a global transcriptional regulator. First, 59 differentially regulated proteins distributed across 16 different COG categories were identified in the ptrA mutant (Klaponski et al., 2014) and genes encoding these proteins are scattered about the chromosome (Loewen et al., 2014). Second, insertional inactivation of the divergently transcribed scd gene upstream of ptrA resulted in no observable phenotype. AF activity, antibiotic and AHL production, and protease activity were similar to wild type (Figures 2–4; Table 2). Moreover, expression of scd was unchanged in the ptrA mutant (Figure 5). These findings indicate that scd is not involved in PA23 biocontrol and does not appear to be linked to PtrA. Finally, our qRT-PCR results showed altered expression of several biosynthetic and regulatory genes involved in PA23 biocontrol (Figure 5). For the most part, gene expression profiles corresponded well with the ptrA mutant phenotype. For example, phzA and prnA were both significantly downregulated (Figure 5). Negligible expression of both genes is consistent with the loss of orange pigmentation, fungal suppression (Figure 2) and antibiotic production (Table 2) exhibited by the ptrA mutant. In strain PA23, the Phz QS system regulates expression of antibiotics and degradative enzymes and so it is necessary for biocontrol (Selin et al., 2012). Transcriptional profiling revealed that phzI and phzR were significantly decreased in the ptrA mutant compared to the wild type. Reduced phzI expression coincides with the loss of AHL signal production in this background (Figure 4). While rpoS expression was markedly down in the ptrA-deficient strain, psrA transcription was elevated, albeit modestly (Figure 5). Because RpoS is a negative regulator of PA23 biocontrol (Manuel et al., 2012) the decrease in rpoS transcription was unexpected as the ptrA mutant is no longer capable of fungal suppression. It is important to note, however, that cross regulation occurs between the PhzRI QS system and RpoS (Selin et al., 2012), which may obscure interpretation of findings. We also explored genes belonging to the Gac-Rsm regulatory network. In P. protegens strain CHA0, this regulatory network functions as follows. Upon binding to an unknown signal, the sensor kinase GacS undergoes autophosphorylation and phosphotransfer to the response regulator GacA (Lapouge et al., 2008). Phosphorylated GacA can then activate expression of sRNAs, including RsmXYZ, that titrate out the translational repressors RsmA and RsmE allowing expression of target genes (Lapouge et al., 2008). While all of these components have been identified in strain PA23, the way in which they function has not yet been explored. In the closely related P. chlororaphis 30–84, RsmE was found to repress production of PHZ and AHL signaling molecules; however, RsmA exerted no regulatory effect over these compounds (Wang et al., 2013). Moreover, of the three small RNA molecules, only constitutively expressed rsmZ was able to rescue a gacA mutant for PHZ and AHL production; rsmX exhibited no effect and elevated levels of rsmY were reportedly lethal (Wang et al., 2013). Collectively, these findings suggest that in terms of PHZ and AHL production in P. chlororaphis 30–84, RsmE is the primary repressor protein and RsmZ is the small RNA responsible for lifting repression. If we assume that this circuitry functions in a similar manner in strain PA23, the elevated rsmE expression and reduced rsmZ transcription are consistent with the loss of AF activity exhibited by the ptrA mutant.
Even though gacS is not under PtrA transcriptional control, a regulatory link clearly exists between the two. Our phenotypic assays showed either full or partial complementation of the ptrA mutant by gacS when provided in trans. We hypothesized that PtrA might be controlling expression of regulatory elements that impact signal transduction through the Gac system and that overexpression of gacS is able to overcome this effect. In P. aeruginosa PAO1, the orphan sensor kinases RetS and LadS modulate the Gac-Rsm circuitry (Ventre et al., 2006; Goodman et al., 2009). RetS blocks GacS autophosphorylation and subsequent activation of GacA, while LadS exerts a positive effect on Gac regulation (Ventre et al., 2006; Goodman et al., 2009). To explore whether RetS and LadS are in some way linked to PtrA, we attempted to complement the ptrA mutant by providing retS and ladS in trans. No changes in fungal suppression, protease activity, antibiotic and AHL production were observed in the ptrA mutant harboring plasmid-borne copies of these genes (Figures 2–4; Table 2). We did, however, discover a two-fold decrease in retS transcription in PA23-443 (Figure 5). Because RetS functions to antagonize the Gac system, decreased retS expression is expected to have a positive impact on biocontrol. Taken together, these findings do not support a role for RetS and LadS in the PtrA -GacS interaction.
Phylogenetic analysis illustrates that a wide range of Pseudomonas species harbor a PtrA homolog (Figure 6). For both symbiotic and pathogenic pseudomonads, secreted products play a significant role in the biocontrol and virulence properties of these organisms. In addition, regulatory factors overseeing their expression are in many cases conserved. For example, the Gac two-component system and QS positively regulate exoproducts that play a key role in symbiotic and pathogenic interactions (Heeb and Haas, 2001; Bassler, 2002). It is not surprising, therefore, to find that PtrA is highly conserved amongst Pseudomonas species. To the best of our knowledge, this transcriptional regulator has not been characterized in other pseudomonads.
In summary, we have shown that PtrA is essential for biocontrol of S. sclerotiorum stem rot of canola. PtrA appears to function as a global regulator controlling expression of unlinked genes across the chromosome. Future studies will be directed at analyzing the PtrA transcriptome on a global scale so we can better comprehend how this LTTR is controlling expression of biocontrol factors in PA23. Due to the conserved nature of this regulator, we hypothesize that PtrA governs expression of secondary metabolites in biocontrol strains and pathogenic pseudomonads alike. Elucidating the mechanisms underlying PtrA regulation will have far-reaching implications for our understanding of how these bacteria interact with other members of their environment including prokaryotic and eukaryotic organisms.
Author Contributions
NS, NK, WF, MB, and Td conceived and designed the study. NS and NK drafted the manuscript with input from Td. NS, NK, and RR performed the phenotypic characterization of the ptrA mutant; CS was responsible for the greenhouse analysis. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors gratefully acknowledge financial support for this work through grants awarded to Td from the Natural Sciences and Engineering Research Council (NSERC; RGPIN/249559-2012) Discovery Grants Program and to Td, WF, and MB from the Agri-Food Research and Development Initiative (ARDI; GrwFwd 12-1173).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01512
Supplementary Figure 1. Molecular phylogenetic analysis of Pseudomonas chlororaphis PA23 ptrA by the Maximum Likelihood method. The evolutionary history was inferred using the Maximum Likelihood method based on the Tamura Nei model (Tamura and Nei, 1993). This analysis involved 40 nucleotide sequences including ptrA. The tree with the highest log likelihood is shown. The percentage of trees in which the associated taxa clustered together is indicated next to the branches.
Supplementary Table 1. Protein sequences used in phylogenetic analysis of PtrA and homologs.
Supplementary Table 2. Nucleotide sequences used in the construction of the phylogeny tree in Supplementary Figure 1.
References
Alexeyev, M. F. (1999). The pKNOCK series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of gram-negative bacteria. BioTechniques 26, 824–826, 828.
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Bassler, B. L. (2002). Small talk. Cell-to-cell communication in bacteria. Cell 109, 421–424. doi: 10.1016/S0092-8674(02)00749-3
Byrne, G. A., Russell, D. A., Chen, X., and Meijer, W. G. (2007). Transcriptional regulation of the virR operon of the intracellular pathogen Rhodococcus equi. J. Bacteriol. 189, 5082–5089. doi: 10.1128/JB.00431-07
Cao, H., Krishnan, G., Goumnerov, B., Tsongalis, J., Tompkins, R., and Rahme, L. G. (2001). A quorum sensing-associated virulence gene of Pseudomonas aeruginosa encodes a LysR-type transcriptional regulator with a unique self-regulatory mechanism. Proc. Natl. Acad. Sci. U.S.A. 98, 14613–14618. doi: 10.1073/pnas.251465298
Chancey, S. T., Wood, D. W., and Pierson, L. S. III. (1999). Two-component transcriptional regulation of N-acyl-homoserine lactone production in Pseudomonas aureofaciens. Appl. Environ. Microbiol. 65, 2294–2299.
Cook, R. J. (1993). Making greater use of introduced microorganisms for biological control of plant pathogens. Annu. Rev. Phytopathol. 31, 53–80. doi: 10.1146/annurev.py.31.090193.000413
Finan, T. M., Kunkel, B., De Vos, G. F., and Signer, E. R. (1986). Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J. Bacteriol. 167, 66–72.
Goodman, A. L., Merighi, M., Hyodo, M., Ventre, I., Filloux, A., and Lory, S. (2009). Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev. 23, 249–259. doi: 10.1101/gad.1739009
Haas, D., and Keel, C. (2003). Regulation of antibiotic production in root-colonizing Pseudomonas spp. and relevance for biological control of plant disease. Annu. Rev. Phytopathol. 41, 117–153. doi: 10.1146/annurev.phyto.41.052002.095656
Heeb, S., and Haas, D. (2001). Regulatory roles of the GacS/GacA two-component system in plant-associated and other Gram-negative bacteria. Mol. Plant-Microbe Interact. 14, 1351–1363. doi: 10.1094/MPMI.2001.14.12.1351
Hernández-Lucas, I., Gallego- Hernández, A. L., Encarnación, S., Fernández-Mora, M., Martínez-Batallar, A. G., Salgado, H., et al. (2008). The LysR-type transcriptional regulator LeuO controls expression of several genes in Salmonella enterica serovar Typhi. J. Bacteriol. 190, 1658–1670. doi: 10.1128/JB.01649-07
Heroven, A. K., and Dersch, P. (2006). RovM, a novel LysR-type regulator of the virulence activator gene rovA, controls cell invasion, virulence and motility of Yersinia pseudotuberculosis. Mol. Microbiol. 62, 1469–1483. doi: 10.1111/j.1365-2958.2006.05458.x
House, B. L., Mortimer, M. W., and Kahn, M. L. (2004). New recombination methods for Sinorhizobium meliloti genetics. Appl. Environ. Microbiol. 70, 2806–2815. doi: 10.1128/AEM.70.5.2806-2815.2004
Katoh, K., Kuma, K.-i., Toh, H., and Miyata, T. (2005). MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33, 511–518. doi: 10.1093/nar/gki198
Kim, J., Kim, J. G., Kang, Y., Jang, J. Y., Jog, G. J., Lim, J. Y., et al. (2004). Quorum sensing and the LysR-type transcriptional activator ToxR regulate toxoflavin biosynthesis and transport in Burkholderia glumae. Mol. Microbiol. 54, 921–934. doi: 10.1111/j.1365-2958.2004.04338.x
Klaponski, N., Selin, C., Duke, K., Spicer, V., Fernando, D. W. G., Belmonte, M. F., et al. (2014). The requirement for the LysR-type regulator PtrA for Pseudomonas chlororaphis PA23 biocontrol revealed through proteomic and phenotypic analysis. BMC Microbiol. 14:94. doi: 10.1186/1471-2180-14-94
Kovaleva, G. Y., and Gelfand, M. S. (2007). Transcriptional regulation of the methionine and cysteine transport and metabolism in streptococci. FEMS Microbiol. Lett. 276, 207–215. doi: 10.1111/j.1574-6968.2007.00934.x
Lapouge, K., Schubert, M., Allain, F. H., and Haas, D. (2008). Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Mol. Microbiol. 67, 241–253. doi: 10.1111/j.1365-2958.2007.06042.x
Latifi, A., Winson, M. K., Foglino, M., Bycroft, B. W., Stewart, G. S., Lazdunski, A., et al. (1995). Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PAO1. Mol. Microbiol. 17, 333–343. doi: 10.1111/j.1365-2958.1995.mmi_17020333.x
Le, S. Q., and Gascuel, O. (1993). An improved general amino acid replacement matrix. Mol. Biol. Evol. 25, 1307–1320. doi: 10.1093/molbev/msn067
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Loewen, P. C., Villeneuva, J., Fernando, W. G. D., and de Kievit, T. (2014). Genome sequence of Pseudomonas chlororaphis strain PA23. Genome Announc. 2, e00390–e00314. doi: 10.1128/genomea.00689-14
Maddocks, S. E., and Oyston, P. C. (2008). Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 154, 3609–3623. doi: 10.1099/mic.0.2008/022772-0
Maddula, V. S., Pierson, E. A., and Pierson, L. S. III. (2008). Altering the ratio of phenazines in Pseudomonas chlororaphis (aureofaciens) strain 30-84: effects on biofilm formation and pathogen inhibition. J. Bacteriol. 190, 2759–2766. doi: 10.1128/JB.01587-07
Manuel, J., Selin, C., Fernando, W. G., and de Kievit, T. (2012). Stringent response mutants of Pseudomonas chlororaphis PA23 exhibit enhanced antifungal activity against Sclerotinia sclerotiorum in vitro. Microbiology 158, 207–216. doi: 10.1099/mic.0.053082-0
Poritsanos, N., Selin, C., Fernando, W. G., Nakkeeran, S., and de Kievit, T. R. (2006). A GacS deficiency does not affect Pseudomonas chlororaphis PA23 fitness when growing on canola, in aged batch culture or as a biofilm. Can. J. Microbiol. 52, 1177–1188. doi: 10.1139/w06-079
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. New York, NY: Cold Spring Harbour Press.
Savchuk, S., and Fernando, W. G. D. (2004). Effect of timing of application and population dynamics on the degree of biological control of Sclerotinia sclerotiorum by bacterial antagonists. FEMS Microbiol. Ecol. 49, 379–388. doi: 10.1016/j.femsec.2004.04.014
Schell, M. A. (1993). Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47, 597–626. doi: 10.1146/annurev.mi.47.100193.003121
Selin, C., Fernando, D. W., and de Kievit, T. R. (2012). The PhzI/PhzR quorum-sensing system is required for pyrrolnitrin and phenazine production and exhibits cross regulation with RpoS in Pseudomonas chlororaphis PA23. Microbiology 158, 896–907. doi: 10.1099/mic.0.054254-0
Selin, C., Habibian, R., Poritsanos, N., Athukorala, S. N., Fernando, D., and de Kievit, T. R. (2010). Phenazines are not essential for Pseudomonas chlororaphis PA23 biocontrol of Sclerotinia sclerotiorum, but do play a role in biofilm formation. FEMS Microbiol. Ecol. 71, 73–83. doi: 10.1111/j.1574-6941.2009.00792.x
Selin, C., Manuel, J., Fernando, D. W., and de Kievit, T. (2014). Expression of the Pseudomonas chlororaphis strain PA23 Rsm system is under control of GacA, RpoS, PsrA, quorum sensing and the stringent response. Biol. Control 69, 24–33. doi: 10.1016/j.biocontrol.2013.10.015
Sperandio, V., Li, C. C., and Kaper, J. B. (2002). Quorum-sensing Escherichia coli regulator A: a regulator of the LysR family involved in the regulation of the locus of enterocyte effacement pathogenicity island in enterohemorrhagic E. coli. Infect. Immun. 70, 3085–3093. doi: 10.1128/IAI.70.6.3085-3093.2002
Tamura, K., and Nei, M. (1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10, 512–526.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Ventre, I., Goodman, A. L., Vallet-Gely, I., Vasseur, P., Soscia, C., Molin, S., et al. (2006). Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc. Natl. Acad. Sci. U.S.A. 103, 171–176. doi: 10.1073/pnas.0507407103
Walsh, U. F., Morrissey, J. P., and O'Gara, F. (2001). Pseudomonas for biocontrol of phytopathogens: from functional genomics to commercial exploitation. Curr. Opin. Biotechnol. 12, 289–295. doi: 10.1016/S0958-1669(00)00212-3
Wang, D., Lee, S.-H., Seeve, C., Yu, J. M., and Pierson, L. S. III, Pierson, E. A. (2013). Roles of the Gac-Rsm pathway in the regulation of phenazine biosynthesis in Pseudomonas chlororaphis 30-84. Microbiol. Open 2, 505–524. doi: 10.1002/mbo3.90
West, S. E., Schweizer, H. P., Dall, C., Sample, A. K., and Runyen-Janecky, L. J. (1994). Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148, 81–86. doi: 10.1016/0378-1119(94)90237-2
Keywords: antifungal, pyrrolnitrin, phenazine, degradative enzymes, autoinducer
Citation: Shah N, Klaponski N, Selin C, Rudney R, Fernando WGD, Belmonte MF and de Kievit TR (2016) PtrA Is Functionally Intertwined with GacS in Regulating the Biocontrol Activity of Pseudomonas chlororaphis PA23. Front. Microbiol. 7:1512. doi: 10.3389/fmicb.2016.01512
Received: 02 February 2016; Accepted: 09 September 2016;
Published: 22 September 2016.
Edited by:
Benjamin Gourion, CNRS/INRA, FranceReviewed by:
Muthu Venkateshwaran, University of Wisconsin–Platteville, USAMohd Sayeed Akhtar, Mahatma Jyotiba Phule Rohilkhand University, India
Claire Prigent-Combaret, Centre National de la Recherche Scientifique, France
Copyright © 2016 Shah, Klaponski, Selin, Rudney, Fernando, Belmonte and de Kievit. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Teresa R. de Kievit, dGVyZXNhLmRla2lldml0QGFkLnVtYW5pdG9iYS5jYQ==
†These authors have contributed equally to this work.
 Nidhi Shah
Nidhi Shah Natasha Klaponski1†
Natasha Klaponski1† Carrie Selin
Carrie Selin W. G. Dilantha Fernando
W. G. Dilantha Fernando Mark F. Belmonte
Mark F. Belmonte Teresa R. de Kievit
Teresa R. de Kievit