- 1State Key Laboratory of Bioreactor Engineering and Institute of Applied Chemistry, East China University of Science and Technology, Shanghai, China
- 2School of Biological Sciences, The University of Hong Kong, Hong Kong, China
- 3Shanghai Collaborative Innovation Center for Biomanufacturing Technology, Shanghai, China
In the present study, the microbial community and functional gene composition of a long-term active alkane-degrading methanogenic culture was established after two successive enrichment culture transfers and incubated for a total period of 1750 days. Molecular analysis was conducted after the second transfer (incubated for 750 days) for both the active alkanes-degrading methanogenic enrichment cultures (T2-AE) and the background control (T2-BC). A net increase of methane as the end product was detected in the headspace of the enrichment cultures amended with long-chain n-alkanes and intermediate metabolites, including octadecanoate, hexadecanoate, isocaprylate, butyrate, isobutyrate, propionate, acetate, and formate were measured in the liquid cultures. The composition of microbial community shifted through the successive transfers over time of incubation. Sequences of bacterial and archaeal 16S rRNA gene (16S rDNA) and mcrA functional gene indicated that bacterial sequences affiliated to Thermodesulfovibrio spp. and Anaerolineaceae and archaeal sequences falling within the genus Methanoculleus were the most frequently encountered and thus represented the dominant members performing the anaerobic degradation of long-chain n-alkanes and methanogenesis. In addition, the presence of assA functional genes encoding the alkylsuccinate synthase α subunit indicated that fumarate addition mechanism could be considered as a possible initial activation step of n-alkanes in the present study. The succession pattern of microbial communities indicates that Thermodesulfovibrio spp. could be a generalist participating in the metabolism of intermediates, while Anaerolineaceae plays a key role in the initial activation of long-chain n-alkane biodegradation.
Introduction
Quantitatively, alkanes are one of the most significant component of petroleum hydrocarbons (Head et al., 2010). They were long considered to be recalcitrant to biodegradation in the absence of molecular oxygen, nitrate, and/or sulfate (Heider et al., 1998). In recent years, alkanes biodegradation via methanogenesis has been a topic of increasing interests thanks to the first report of a successful enrichment culture converting long-chain alkanes (specifically hexadecane) to methane (Zengler et al., 1999). Many relevant researches on methanogenic alkanes-degradation studies appeared afterward (Anderson and Lovley, 2000; Jones et al., 2008; Gray et al., 2011; Wang et al., 2011; Li et al., 2012; Mbadinga et al., 2012; Zhou et al., 2012; Aitken et al., 2013; Cheng et al., 2013b; Berdugo-Clavijo and Gieg, 2014; Embree et al., 2014; Sherry et al., 2014; Liang et al., 2015). The initial activation of alkane degradation under anaerobic conditions may involve fumarate addition to the parent alkane, hydroxylation/carboxylation and, in some specific cases, intra-hydroxylation (Callaghan, 2013). Among the above mentioned activation mechanisms, addition to fumarate appear as the most prevalent and almost the best characterized mechanism in anaerobic hydrocarbon degradation (Aitken et al., 2013). The gene assA/masD coding the alkylsuccinate synthetase has eventually been considered as a valuable biomarker for detecting fumarate addition pathway in alkane degradation (Callaghan et al., 2010).
Microbial communities capable of degrading petroleum hydrocarbons under methanogenic conditions are often complex consortia, at least consisting of various fermenting bacteria, syntrophic bacteria and methanogens at least. Generally, the constitution of microbes from different hydrocarbons impacted environments like aquifers, sediments, and soils can be dramatically different. However, among them some microbial taxa appeared with relatively high frequency, a good example of this is that many researches shared the similar microorganisms members of the Syntrophaceae (Smithella/Syntrophus) which was inferred to have the ability of degradation of alkane (Bakermans and Madsen, 2002; Kasai et al., 2005; Allen et al., 2007; Gray et al., 2011; Ramos-Padron et al., 2011; Cheng et al., 2013a; Embree et al., 2014; Tan et al., 2014a,b). A survey that collated published 16S rRNA gene data from 26 culture-independent analyses of methanogenic hydrocarbon impacted environments showed that bacterial sequences affiliated with Firmicutes were detected at the highest frequency followed by γ-proteobacteria, δ-proteobacteria, 𝜀-proteobacteria, β-proteobacteria, Bacteroidetes, Actinobacter, α-proteobacteria, Chloroflexi, Thermotogae, Nitrospira, Spirochaetes, Acidobacter, Planctomycetes and OP11 (Gray et al., 2010). So far, at least 19 anaerobic, alkane-oxidizing microorganisms have been isolated (Webner, 2012; Khelifi et al., 2014; Schouw et al., 2016), while 17 of the isolated strains were affiliated with the phylum of Proteobacteria, other two strains belongs to Firmicutes and Archaeoglobales, respectively. The large proportion of non-cultivable and metabolically inactive organisms interferes with the identification of the active responsible for the degradation. In addition, the various syntrophic associations in methanogenic consortia and the obligate anaerobic conditions make isolation of them in pure cultures difficult and impossible with the current available techniques. Consortium obtained through enrichment culturing with long-term stability for methanogenic alkane degradation can eliminate the inactive members considerably so that the essential ones can be accumulated. In the present research work, a methanogenic alkanes-degrading consortium was established after enrichment culturing and long-term of incubation amended with a mixture of n-alkanes (C15–C20) as the sole sources of carbon and energy. The succession pattern of microbial communities together with the diversity and abundance of potential functional genes were analyzed via PCR based clone libraries construction coupled with quantitative real-time PCR analysis in this study. The degradation intermediates and the final product methane were measured during degradation at the same time.
Materials and Methods
Enrichment Cultures
Inoculum was obtained from an initial methanogenic enrichment culture from Menggulin petroleum reservoir production water (Huabei oilfield, China) amended with n-alkanes as described previously (Li et al., 2012). About 10 ml (20%) of inoculum were transferred into an autoclaved serum bottle (internal volume 120 ml) containing 50 ml of sterilized basal medium prepared by the Hungate technique (Bryant, 1972) and then sealed with a butyl rubber stopper (Bellco Glass, Inc., Vineland, NJ, USA) and aluminum crimp seal. The basal medium composition was described elsewhere (Wang et al., 2012). Active enrichment cultures were amended with the mixture of n-alkanes (C15–C20) as the sole carbon and energy sources (three replicates). Autoclaved controls were prepared in the same way but sterilized three times (three replicates). Background controls were prepared without addition of any organic carbon source (three replicates). The mixture of n-alkanes (C15–C20) contained n-pentadecane (C15; ≥99%), n-hexadecane (C16; ≥99%), n-heptadecane (C17; ≥99%), n-octadecane (C18; ≥99%), n-nonadecane (C19; ≥99%), and n-eicosane (C20; ≥99%) (Sigma–Aldrich, Milwaukee, WI, USA). About 30 μl of the n-alkanes mixture were added to each of the empty serum bottles except the background controls under a stream of N2 gas before sterilization. All of the cultures were incubated at 37°C in the dark.
Chemical Analysis
Gas chromatography (GC) was used to measure the production of methane in the headspace gas of serum bottle during the incubation. Two hundred microliters of the headspace gas taken by gas-tight syringe were injected onto GC by a micro-syringe for analysis. Program setting of the GC analysis was: the initial column temperature at 60°C for 12 min, then increased to 200°C at a rate of 15°C/min, the final temperature at 200°C sustained for 24 min. Temperature of injector and flame ionization detector (FID) was maintained at 200°C. An external standard curve of methane was used for converting peak areas of methane into their respective concentrations (R2 = 0.994, n = 6).
Gas Chromatography-Mass Spectrometer (GC-MS; Agilent Technologies, Inc.) was used for the detection of residual n-alkanes and intermediate metabolites in the aliquot phase through the incubation. For the analysis of intermediate metabolites containing mainly long-chain fatty acids (LCFAs) and volatile fatty acids (VFAs), 5 ml of culture aliquot were taken and the pH was adjusted with ammonia water to >12 and then dried in an oven at 110°C. Esterification was performed by adding 0.5 ml of 10% butanol/sulfate solution at 90°C for 60 min. Extraction of LCFAs and VFAs was conducted with 0.5 ml of n-hexane and 0.5 ml n-dodecane, respectively, and the extracts were injected onto the GC-MS. For LCFAs analysis, program setting was oven temperature maintained at 120°C for 3 min, increased at the rate of 8°C/min to 260°C for 10 min while for VFAs, oven temperature was maintained at 60°C for 1 min and then increased at the rate of 15°C/min to 130°C. Residual n-alkanes in the aliquot samples were extracted with n-hexane as described previously (Wang et al., 2011).
DNA Extraction and PCR Amplification
At the end of incubation period, 10 ml of active culture (three replicates) and the background control (three replicates) were withdrawn from each serum bottle, then centrifuged at 1200 × g for 10 min. The biomass pellet after centrifugation was used for DNA extraction by using AxyPrepTM Bacterial Genomic DNA Maxiprep Kit (Axygen Biosciences, USA) according to the manufacturer’ instructions.
The universal primer sets of 8F/805R (Savage et al., 2010) and 340F/1000R (Gantner et al., 2011) were used for bacterial and archaeal 16S rRNA gene amplification, respectively. For bacterial 16S rRNA gene, PCR amplification reaction was performed according to the followings: 5 min for initial denaturation at 95°C, followed by 38 cycles of 95°C for 30 s, 52°C for 45 s, 72°C for 60 s, and a final elongation step at 72 C for 10 min. For archaeal 16S rRNA gene, PCR amplification conditions were as follows: 5 min for initial denaturation at 95°C, followed by 10 cycles of 95°C for 30 s, 60°C for 30 s (decreased by 0.5°C per cycle to 50°C), 72°C for 60 s. After touchdown, 30 additional cycles at annealing temperature of 50°C were performed, followed by the final elongation step at 72°C for 10 min.
Alkylsuccinate synthetase genes (assA) and methyl coenzyme-M reductase genes (mcrA) as the functional genes in the process of methanogenic degradation of alkanes were also investigated. PCR primer sets of assA2F/assA2R (Callaghan et al., 2010) and ME3MF/ME2R (Hales et al., 1996; Nunoura et al., 2008) were used for the PCR amplification, respectively. PCR thermal cycles for assA were carried out as follows: 5 min for initial denaturation at 95°C, followed by 10 cycles of 95°C for 30 s, 60°C for 30 s (decreased by 0.5°C per cycle to 55°C), 72°C for 60 s. After touchdown, 28 additional cycles at annealing temperature of 55°C were performed, followed by the final elongation step at 72°C for 10 min. PCR cycles for mcrA were as follows: 5 min for initial denaturation at 95°C, followed by 38 cycles of 95°C for 30 s, 52°C for 45 s, 72°C for 60 s, and a final elongation step at 72°C for 10 min.
Construction of 16S rRNA Gene, and assA and mcrA Functional Gene Libraries
After the PCR products were gel purified by using Gel Extraction Kit (Axygen Biosciences, USA), the purified DNA fragments were cloned into E. coli using pMD19®-T simple vector kit (TaKaRa Bio Inc., Japan). The white clones were picked randomly into 1 ml of Luria Broth (LB) medium amended with ampicillin and incubated for 24 h at 37°C. PCR primer set M13-47 (5′-CGCCAGGGTTTTCCCAGTCACGAC-3′) and RV-M (5′-GAGCGGATAACAATTTCACACAGG-3′) was used for positive clone detection. Sequencing of the positive clones was accomplished on an ABI 377 automated sequencer. Chimeric sequences of 16S rRNA gene sequences were excluded by Bellerophon (Huber et al., 2004). Valid sequences with more than 97% similarity were classified through the BLASTclust of MPI bioinformatics toolkit (Biegert et al., 2006) to arrive operational taxonomic unit (OTU). Functional genes were translated through ExPASY translation tool1. Sequences were compared to the GenBank Nucleotide Sequence Database using BLAST (Altschul et al., 1990) to identify the nearest matches in the GenBank database. Phylogenetic and molecular evolutionary analyses were conducted using MEGA6.0 software (Tamura et al., 2013) with neighbor-joining method (Saitou and Nei, 1987) and 1000 bootstrap replicates.
Quantitative PCR
Quantitative PCR of target genes were performed by using SYBR Green I Real-Time PCR (BioRad CFX96 thermocycler, Bio-Rad Laboratories Inc., USA). We amplified 16S rRNA gene of Archaea and Bacteria, each target gene was amplified with specific primers ARC787F/ARC1059R (Yu et al., 2005) and BAC338F/BAC805R (Yu et al., 2005), respectively. These two genes were amplified separately in a SYBR Green I Real-Time PCR reaction with a 10-fold dilution series of plasmids containing target DNA sequences as a calibration standard for establishing the standard curve. Quantitative PCR reaction (20 μl) was composed of SYBR Green Realtime PCR Master Mix-Plus (10 μl; TaKaRa Bio Inc., Japan), Plus Solution 2 μl (TaKaRa Bio Inc., Japan), PCR primers (1 μl of each), 4 μl of sterile water, and 2 μl DNA template (27.4 ng/μl for T2-AE and 15.8 ng/μl for T2-BC). The conditions were as follows: pre-denaturation for 3 min at 95°C, followed by 38 cycles of denaturation at 94°C for 20 s, annealing temperature was 60°C and 57°C, respectively, for 30 s, elongation at 72°C for 60 s.
Nucleotide Sequences Accession Numbers
The sequences generated in this study were deposited in GenBank under accession numbers KP109826-KP109908 and KP341767-KP34176769.
Results
Headspace Methane and Intermediate Metabolites
During the incubation of the second enrichment transfer, the lag phase in the amended enrichment cultures (T2-AE) was only 92 days and 44.76 μmol of methane were generated while in the background control without amendment of any alkanes (T2-BC) approximately 0.28 μmol of methane were detected (Figure 1). Intermediate metabolites including LCFAs (Supplementary Figure S1) and VFAs (Supplementary Figure S2), octadecanoate, hexadecanoate, isocaprylate, butyrate, isobutyrate, propionate, acetate, and formate were detected in the enrichment cultures only. Quantity of residual n-alkanes were detected through the GC-MS with cetyl chloride as the surrogate standard. The net consumption of total n-alkanes was 53.31 μmol by subtracting the residual n-alkanes in the active enrichment cultures from autoclaved controls. Stoichiometry of methane production showed methane accumulated in headspace accounted for 6.23% of theoretically predicted value (Supplementary Table S1).
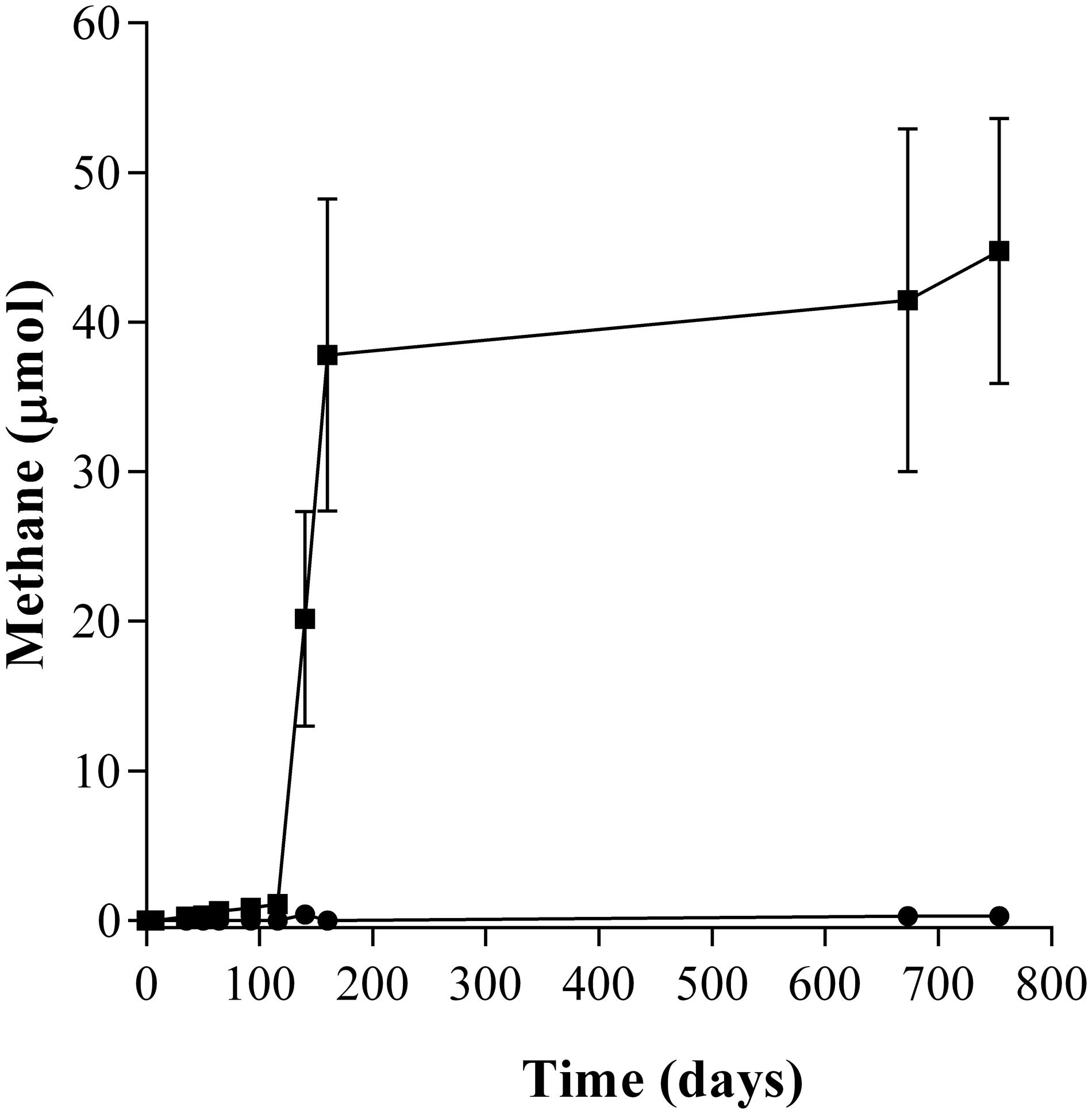
FIGURE 1. Methane production of the n-alkanes degradation consortium under methanogenic conditions. (■) incubated with n-alkanes mixture (C15–C20) as the sole sources of carbon and energy (T2-AE; three replications), and (●) without any n-alkanes and other carbon sources as the background control cultures (T2-BC; three replicates).
Phylogenetic Analysis of Bacteria
A total of 100 positive clones were picked for sequencing bacteria 16S rRNA genes from both the active enrichment (T2-AE) and background control cultures (T2-BC). After removing vector parts and checking chimeras, 81 and 82 valid sequences, we obtained for the active culture and the control, respectively. The valid sequences were classified through BLASTclust of MPI bioinformatics toolkit (Biegert et al., 2006) with 97% similarity and 10 OTUs for T2-AE and seven OTUs for T2-BC were obtained (Figure 2). The OTU of “T2-AE-24 (KP109862)” which contains the most clone sequences belongs to Thermodesulfovibrio within the phylum of Nitrospirae in the active enrichment cultures (T2-AE). Followed by the OTU of “T2-AE-10 (KP109861)” belongs to Anaerolineaceae within the phylum of Chloroflexi which covers 27 clone sequences. Bacteria in the genus of Thermodesulfovibrio having the highest abundance constituted 49% of all of the 81 clones (Figure 2). Family Anaerolineaceae as the dominant bacteria made up 33%. Acetothermia, Aminicenantes, Actinobacteria, and unclassified bacteria composed the remaining 18% (Figure 2). In T2-BC, OTU of “T2-BC-10 (KP109845)” affiliated with Thermodesulfovibrio within the phylum of Nitrospirae representing the most clone sequences of 71 in the whole 82 valid clones (Figure 2). Bacteria in the genus of Thermodesulfovibrio were the most dominant bacteria of 87%, while Anaerolineaceae occupied 1%. The remaining 12% comprised Actinobacteria, Bacteroidetes, Actinobacteria, Synergistia, and Firmicutes (Figure 2).
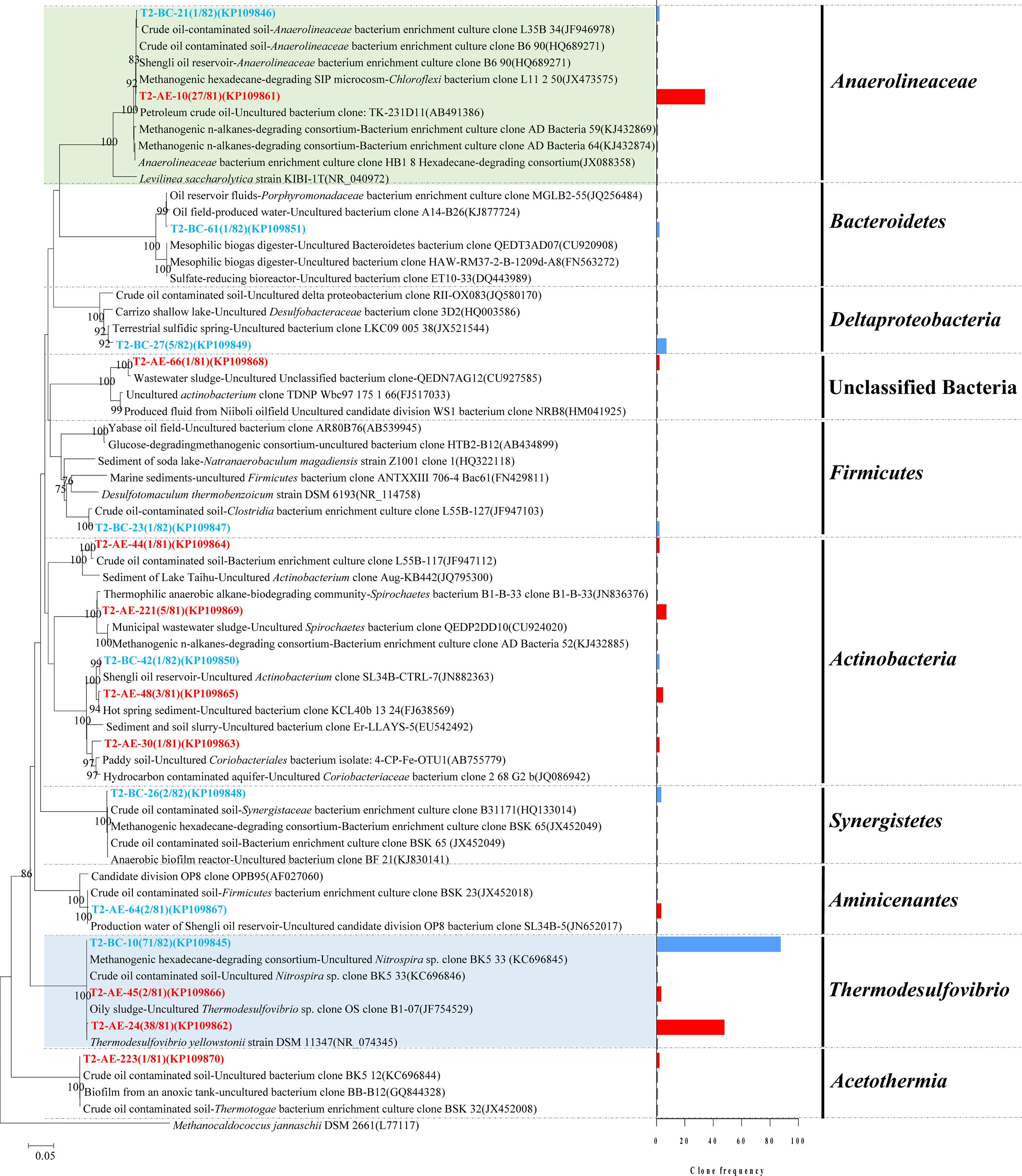
FIGURE 2. Phylogenetic tree of bacterial 16S rRNA gene sequences from methanogenic alkanes-degrading enrichment cultures (T2-AE; in red) and background control cultures (T2-BC; in blue). The tree is rooted with Methanocaldococcus jannaschii DSM 2661 (L77117) as the out group. The OTUs are shown with clone names and accession numbers. One-thousand bootstraps were performed with neighbor-joining method. Bootstrap values below 75% are not shown. Frequency of clones is shown in the bar graph.
Phylogenetic Analysis of Archaea
Positive clones were also picked for sequencing archaea 16S rRNA gene. A total of 61 and 51 valid sequences were obtained for the active enrichment (T2-AE) and background control cultures (T2-BC), respectively, after the removal of vector and chimeras checking. Valid sequences were analysis through BLASTclust of MPI bioinformatics toolkit (Biegert et al., 2006) with 97% similarity to generate the OTUs. There were two OTUs for active enrichment cultures (T2-AE) and four OTUs for background control cultures (T2-BC) (Figure 3A). For the active enrichment cultures (T2-AE), the OTU of “T2-AE-10 (KP109885)” contains the majority 61 clones of the total 62 clones. All of the 62 clone sequences belong to Methanoculleus within the Methanomicrobiales order (Figure 3A). For the background control (T2-BC), the OTU of “T2-BC-13 (KP109880)” had 46 of the total 51 clones. This OTU affiliated with Methanolinea in the order of Methanomicrobiales occupying 90% of all valid clone sequences. Methanobacterium, Methanocella, and Methanomethylovorans constituted the remaining 10% of the clone library (Figure 3A).
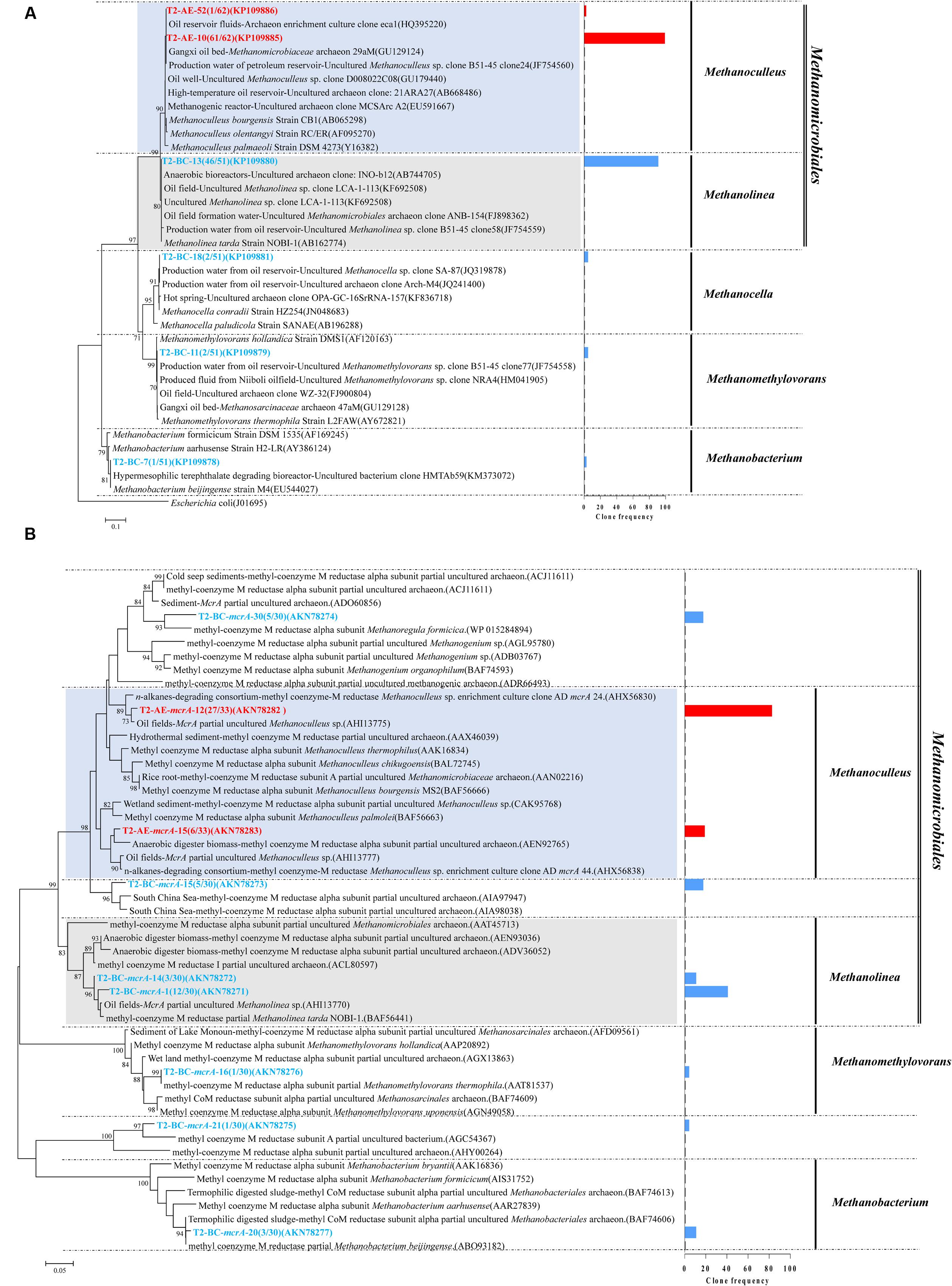
FIGURE 3. Phylogenetic tree of archaeal 16S rRNA gene sequences from methanogenic alkanes-degrading enrichment cultures (T2-AE; in red) and background control cultures (T2-BC; in blue). This tree is rooted with outgroup sequence from Escherichia coli (J01695) as an outgroup. The OTUs are shown with clone names and accession numbers. Topology of the tree was obtained by the neighbor-joining method. One-thousand time bootstraps testing were performed. Bootstrap values below 70% are not shown. Frequency of clones are shown in the bar graph (A); Phylogenetic tree of deduced amino acid sequences of methyl coenzyme-M reductase genes (mcrA) from methanogenic alkanes-degrading enrichment culture (in red) and background control cultures (T2-BC; in blue). Topology of the tree was obtained by the neighbor-joining method. The evolutionary distances were computed using the Poisson correction method. One-thousand time bootstraps testing were performed. Bootstrap values below 70% are not shown (B).
Diversity and Phylogenetic Analysis of assA and mcrA Genes
Functional gene of assA was only detected in the active enrichment cultures (T2-AE) by using the primer set of assA2F/assA2R (Callaghan et al., 2010). Valid sequences were used for phylogenetic analysis and resulted in three OTUs at 97% similarity (Figure 4). Most sequences were in the OTU of “T2-AE-assA-13 (AKK23578)” which contains 32 of the total 39 clone sequences. This OTU has a 100% similarity with the sequence (AIU94859) from production water of an oil reservoir of Jiangsu oil field (Bian et al., 2015). Background control cultures (T2-BC) generated no detectable products by PCR reactions.
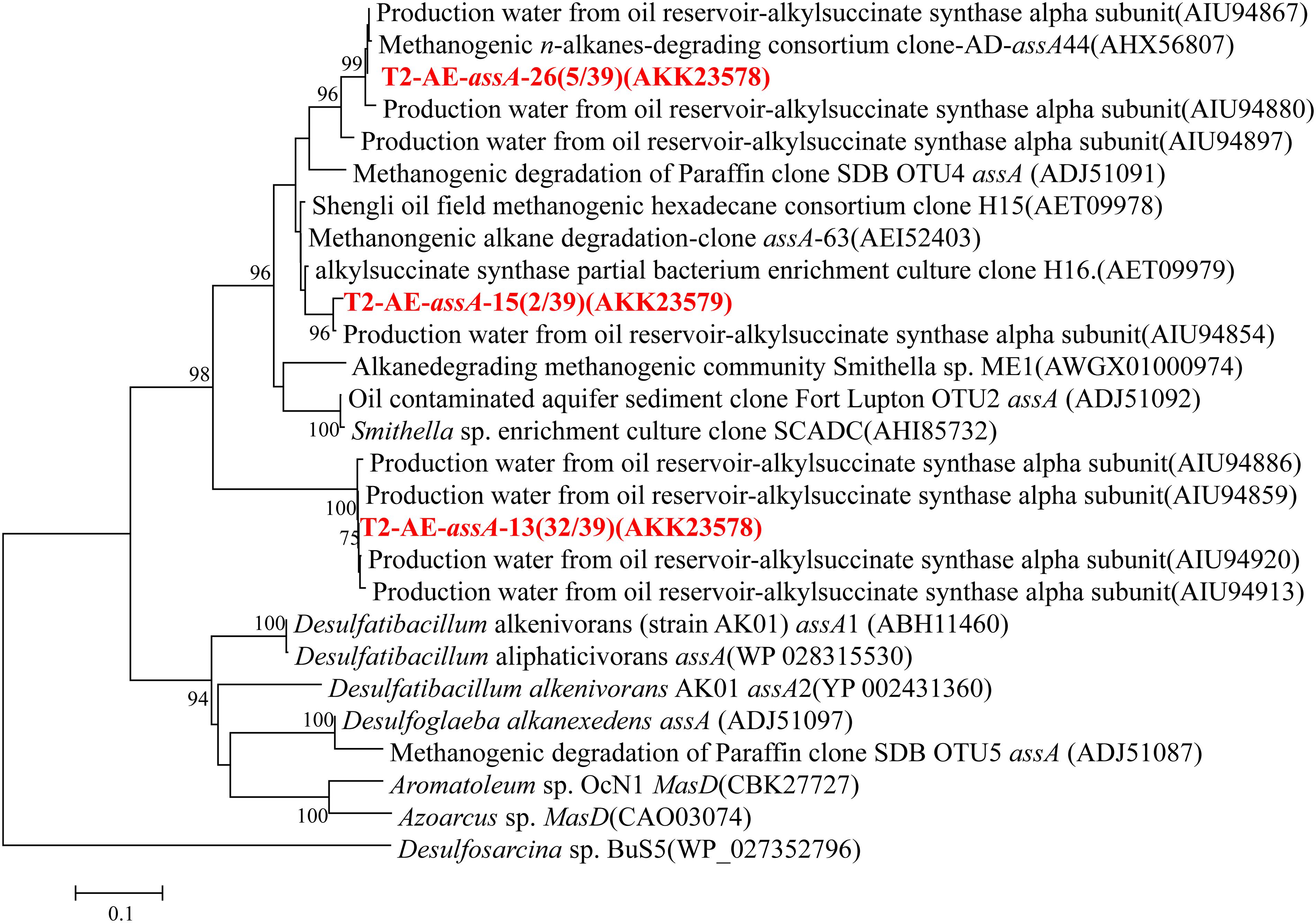
FIGURE 4. Phylogenetic tree of deduced amino acid sequences of alkylsuccinate synthetase genes (assA) genes from methanogenic alkanes-degrading enrichment culture (in red). The topology of the tree was obtained by the maximum likelihood method. Bootstrap values (n = 1000 replicates), values below 75% are not shown.
Both the T2-AE and T2-BC cultures detected the functional gene mcrA by using the prime set of ME3MF/ME2r (Nunoura et al., 2008) and obtained two and seven OTUs, respectively. All of the clone sequences in T2-AE cultures affiliated with Methanoculleus within the order of Methanomicrobiales (Figure 3B). This result was coincident with archaeal 16S rRNA gene clone library. The OTU of “T2-AE-mcrA-12 (AKN78282)” showed 97% similarity with the sequence from the n-alkanes-degrading consortium (AHX56830). Except the OTU of “T2-BC-mcrA-21 (AKN78275),” mcrA functional gene sequences in T2-BC cultures had three clear classifications: Methanomicrobiales, Methanobacteriales, and Methanosarcinales (Figure 3B).
Quantitative PCR
16S rRNA genes of Archaea and Bacteria were amplified for both T2-AE and T2-BC to detect the whole archaea and bacteria 16S rRNA gene copies. Gene abundance (gene copies per milliliter of culture aliquot) of Archaea was 4.17 × 107 (copies/ml) and 1.01 × 106 (copies/ml) in T2-AE and T2-BC, respectively. Gene abundance (gene copies per milliliter of culture aliquot) of Bacteria was 1.48 × 107 (copies/ml) and 1.62 × 106 (copies/ml) for T2-AE and T2-BC, respectively. For Archaea and Bacteria, 16S rRNA gene qPCR reaction, amplification efficiency was 105.6% and 102.3%, R2-values was greater than 0.989 and 0.986, respectively.
Discussion
The entire duration of nearly 5 years of enrichment culturing included the first transfer (about 950 days) and a subsequent second transfer (over 750 days), both of the two transfers and incubations received the mixture of n-alkanes (C15–C20) as the sole carbon and energy sources. During the incubation, the variations of microbial community in different enrichment transfers were investigated. Microbial community of the Menggulin petroleum reservoir production water sample (OS) and the initial transfer of incubation after 400 days (T1-400) were analyzed by our laboratory and reported previously (Li et al., 2012). In the current research, microbial communities in the second transfer from both the active enrichment cultures (T2-AE) and the background control (T2-BC) were investigated after 750 days of incubation.
High Frequency of Thermodesulfovibrio and Anaerolineaceae
Clear variation of bacteria community was observed after the 5 years of methanogenic alkanes degrading enrichment culturing (Figure 5). In the original petroleum reservoir production water sample (OS), the dominant bacteria were Gammaproteobacteria (46.6%) and Betaproteobacteria (32.9%). After nearly 400 days of incubation with addition of a mixture of n-alkanes (T1-400) initially, Actinobacteria (52.2%) and Thermodesulfovibrio (39.1%) were detected as the dominant bacteria. It is clear that the period of Actinobacteria as the dominant bacteria only maintained for a short period of time at the beginning. Then the abundance of Actinobacteria decreased sharply to 6.2% as a minority after another 1350 days of incubation (including a second transfer of incubation for 750 days) in T2-AE, whereas Thermodesulfovibrio remained the most frequently encountered genus and increased gradually to 49.4%. At the same time, Anaerolineaceae rose conspicuously to 33.3% as the dominant bacteria. Similar results were also observed in our earlier n-alkanes-degrading consortium originated from an oily sludge (Liang et al., 2015). Thus, Thermodesulfovibrio and Anaerolineaceae as the most dominant bacteria implies a major role they play in the long-term methanogenic alkanes-degrading consortium.
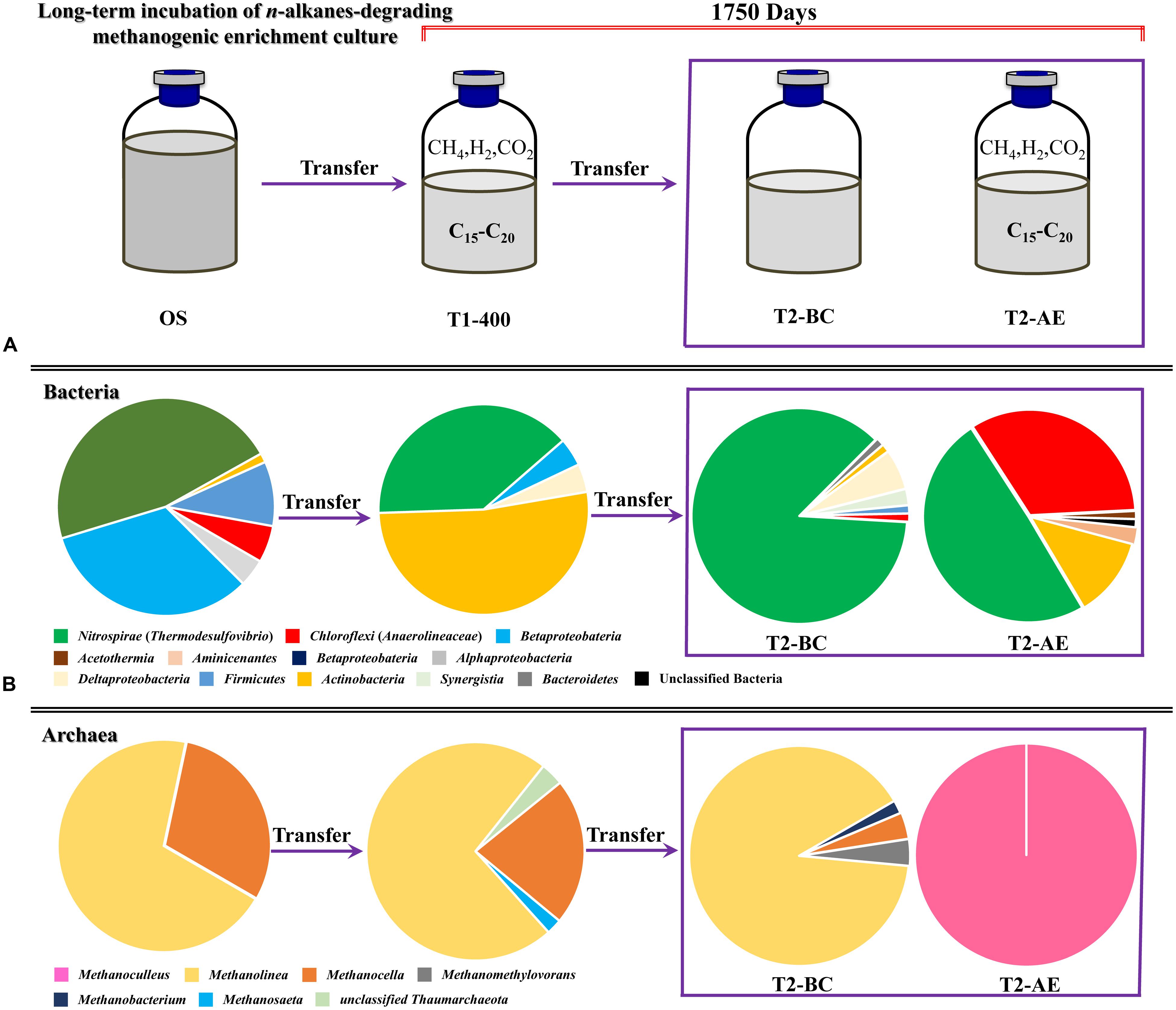
FIGURE 5. The microbial community varied with the time of incubation from the analysis of bacterial and archaeal 16S rRNA gene clone libraries. Relative proportion of bacterial lineages (A) and archaeal lineages (B) during the methanogenic alkanes-degrading enrichment culture. “OS” represents the initial inoculum came from Huabei oilfield petroleum reservoir production water sample in China; “T1-400” represents the initial enrichment culture at 400 days which added with n-alkanes (C15–C20) for details see Li et al. (2012) (initial transferred enrichment culture lasted for 950 days); “T2-AE” and “T2-BC” represent the active enrichment cultures and background control cultures, respectively, in the second transferred enrichment culture amended with n-alkanes as the sole sources of carbon and energy for 750 days.
Thermodesulfovibrio was the only overwhelming majority (86.6%) in the control cultures (T2-BC). Anaerolineaceae occupied only 1.2%, a significant difference from the 33.3% in T2-AE sample. Since both T2-AE and T2-BC were the second enrichment transfers of incubation from the same inoculum initially, such major difference in consortium composition should be attributed to the amendment of organic carbons. When the microorganisms were subject to a starvation condition (T2-BC), abundance of bacteria like Anaerolineaceae and Actinobacteria dropped because of the lack of organic carbons available. Comparing to the background control T2-BC, the dominant Anaerolineaceae in the active enrichment transfer culture T2-AE should be closely related to the alkanes-degrading process. The total bacteria in T2-AE experienced an obvious surge comparing to T2-BC according qPCR analysis showing the alkanes-degrading activity.
Thermodesulfovibrio within the phylum Nitrospirae, is a sulfate-reducing microorganism (SRM) that could use sulfate as the terminal electron acceptor in their energy metabolism (Sherry et al., 2013). OTU of Thermodesulfovibrio in this research shared the highest identity (100%) with Thermodesulfovibrio yellowstonii strain DSM 11347T (NR_074345), which is able to use lactate, pyruvate, and hydrogen as electron donors in the presence of sulfate (Henry et al., 1994). Interestingly, the medium had no sulfate available, and methane was the main end product. Theoretically Thermodesulfovibrio-like species could cooperate with other bacteria or methanogens syntrophically in the alkanes-degrading processes. The presence of this bacterium in similar environments is also reported by others (Sekiguchi et al., 1998; Magot et al., 2000; Pérez-Jiménez et al., 2001; Roest et al., 2005; Li et al., 2007, 2012; Mbadinga et al., 2012; Cheng et al., 2013b; Liu et al., 2014).
Since the general appearance as the majority both in T1-400, T2-AE, and T2-BC, Thermodesulfovibrio as a generalist may participate in the intermediate metabolism, such as the degradation of fatty acids. While Anaerolineaceae as a specialist should be a key player in the degradation process and may likely take part in the initial activation of alkane for degradation since the obvious increase in T2-AE in clear contrasting with OS, T1-400 and T2-BC. This seems logic because many researches have implied the potential of alkane degradation. Chloroflexi was firstly reported to be involved with toluene degradation in a 10-year toluene-degrading methanogenic consortium. Several researches also showed evidence that Anaerolineaceae was one of the frequently encountered microbial species in petroleum hydrocarbon conditions (Winderl et al., 2008; Savage et al., 2010; Gray et al., 2011; Sherry et al., 2013; Sutton et al., 2013; Liang et al., 2015). In an enrichment culture metabolizing low-molecular-weight alkanes (n-propane and n-pentane) under mesophilic sulfate-reducing conditions, Anaerolineaceae as one of the community members showed the potential of hydrocarbon degradation for this lineage organisms (Savage et al., 2010). In a recent study of methanogenic biodegradation of short-chain n-alkanes revealed that members of the Anaerolineaceae may either be directly involved in activation and biodegradation of n-octane and n-decane or act as scavengers of metabolic intermediates (Shahimin et al., 2016). Thus, comparative analysis of 16S rRNA genes sequenced from the cultures with and without alkanes amendment indicated that members of the family Anaerolineaceae, which constitutes more than 33% of the bacterial community 16S rRNA genes in n-alkanes amended culture, may play a key role in the anaerobic metabolism of long-chain alkanes in the long-term incubation.
The obligate anaerobic, non-photosynthetic and multicellular filamentous family Anaerolineaceae in Chloroflexi phylum contains seven isolated strains in the five genera (Sekiguchi et al., 2003; Yamada et al., 2006, 2007; Grégoire et al., 2011). When comparing Anaerolineaceae in T2-AE (KP109861) with the isolated strain, the highest identity is only at 91% with Levilinea saccharolytica strain KIBI-1T (NR_040972), indicating that this Anaerolineaceae in petroleum hydrocarbon environment may be a new division different from the isolated strains. Interestingly, Anaerolineaceae clone (KP109861) in this research has a very high identity (99%) with the Anaerolineaceae clone (KJ432869) in our former n-alkanes-degrading consortium from oily sludge, suggesting that they should be in the same division within the family of Anaerolineaceae.
Methanoculleus in the Methanogenic Process from Long-Term Incubation
Methanogens in the long-term methanogenic alkanes-degrading enrichment culture experienced sharp changes especially in T2-AE sample (Figure 5). Both archaeal 16S rRNA gene clone library and functional gene mcrA clone library showed that the only methanogen in T2-BC sample was Methanoculleus. In contrast, methanogens in T2-BC were mostly Methanolinea (90.2%) and others were Methanobacterium, Methanocella, and Methanomethylovorans which was consistent with the archaeal community in the sample of OS and T1-400 while Methanoculleus was undetected. The phylum Euryarchaeota contains six methanogenic orders: Methanomicrobiales, Methanobacteriales, Methanococcales, Methanosarcinales, Methanopyrales, and Methanocellales (Garrity and Holt, 2001; Ferry, 2010). Both genera of Methanolinea and Methanoculleus are in Methanomicrobiales order. The succession of Methanolinea and Methanoculleus suggests a competitive growth relationship between these two hydrogenotrophic methanogens. Comparing to the isolated methanogens, the OTU of Methanoculleus in T2-AE shows a 99% identity with Methanoculleus receptaculi, while, OTU of Methanolinea in T2-BC has a 99% identity with Methanolinea tarda. M. receptaculi was reported to take acetate as the growth factor with minimal doubling time of 8.2 h (Cheng et al., 2008). M. tarda can use acetate, yeast extract or Coenzyme M stimulates as growth factors with minimal doubling time of 144 h (Imachi et al., 2008). Since acetate and a faster growth rate than Methanolinea was detected in T2-AE, it is natural that Methanoculleus finally turned to be the most dominant methanogens in T2-AE sample.
Possible Metabolic Pathway of Methanogenic Degradation of n-Alkanes
The presence of assA functional gene in the active enrichment cultures (T2-AE) suggests that fumarate addition should be involved in the initial activation of alkanes in this methanogenic alkanes-degrading culture (Figure 6). Moreover, the non-detectable assA functional gene in the background control (T2-BC) confirmed the activity of assA functional gene in T2-AE sample. By contrast, the total amount of Bacteria and Archaea gene copies in T2-AE sample were much higher than those in T2-BC by quantitative PCR analysis. All of these results demonstrate the methanogenic alkanes-degrading activities in T2-AE sample. After initial activation, alkanes converted to LCFAs and VFAs, including octadecanoate, hexadecanoate, isocaprylate, butyrate, isobutyrate, propionate, acetate, and formate. And this intermediate metabolites-degrading phase should be responsible mostly by Chloroflexi (more specifically, Anaerolineaceae) since the community genomic analyses indicated that most members in the phylum of Chloroflexi are anaerobic acetogenic microbes which also have the genomes of complete Wood-Ljungdahl pathway and beta-oxidation of saturated fatty acids (Hug et al., 2013). Growth of some Methanoculleus members requires acetate even though they do not convert it to methane (Barret et al., 2015) and they were reported to produce methane from H2/CO2 and from formate (Oren, 2014). Thus, in methanogenic phase, Methanoculleus as the methanogens generated methane through hydrogenotrophic methanogenesis or methylotrophic methanogenesis (Figure 6).
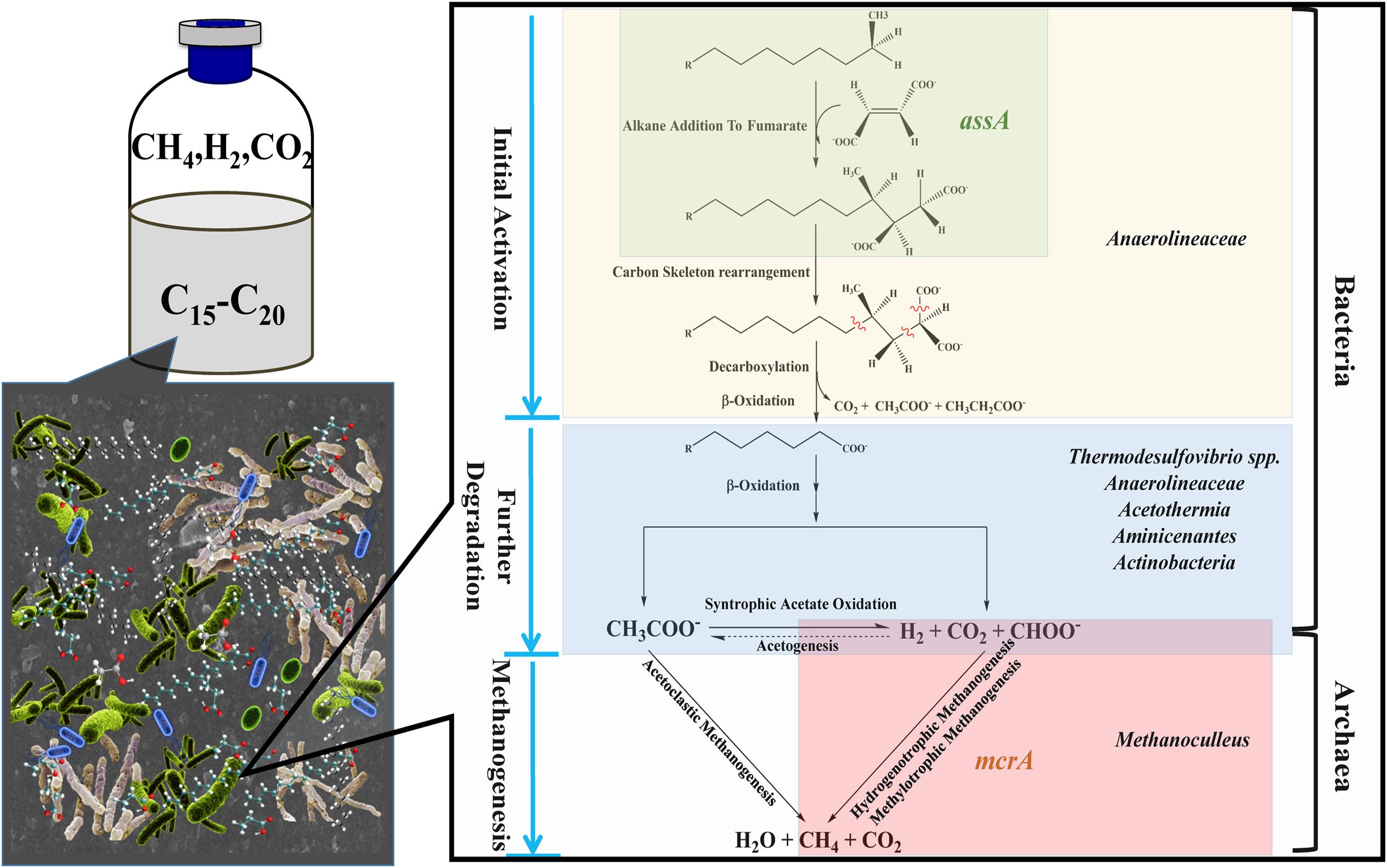
FIGURE 6. Possible metabolic pathway of methanogenic degradation of n-alkanes, based on the pathway proposed by Wilkes et al. (2002) and Callaghan et al. (2012). In the methanogenic n-alkanes-degrading culture, metabolic pathway from n-alkanes to methane can be presumed. Fumarate addition could be involved in the initial activation of alkanes since the detection of assA functional gene. In the phase of methanogenesis, hydrogen, carbon dioxide, and formate were produced through syntrophic acetate oxidation pathway, followed by the production of methane through hydrogenotrophic or methylotrophic methanogenesis. Microorganisms are positioned according their probable role within microbial consortia.
Conclusion
The obvious increase of Thermodesulfovibrio and Anaeroline-aceae in alkanes-dependent methanogenic culture after a long-term of incubation showed that these two bacteria played a crucial role in the degradation process of alkanes. However, whether they act directly on the activation of alkanes is still not clear. It is quite certain that alkanes-degrading bacteria cooperated with methanogens Methanoculleus formed a new methanogenic n-alkanes-degrading microbial community in this long-term incubation consortium. Our knowledge about the key degraders in petroleum hydrocarbon system is still quite limited under anaerobic conditions, further researches should focus on unravel of the specific role of potential alkanes-degrading bacteria.
Author Contributions
L-YW, J-DG, and B-ZM conceived the study. BL and L-YW performed all the experiments and drafted the manuscript. ZZ helped in the experimental part and data analysis. SM was involved in the discussion on the interpretation of the results. LZ helped the analysis of GC-MS data about residual n-alkanes and intermediate metabolites. J-FL and S-ZY were committed to all the experiments. All authors approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
This work was supported by the National Science Foundation of China (No. 41530318, 31200101, 51174092) and the NSFC/RGC Joint Research Fund (No. 41161160560).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01431
Footnotes
References
Aitken, C. M., Jones, D. M., Maguire, M. J., Gray, N. D., Sherry, A., Bowler, B. F. J., et al. (2013). Evidence that crude oil alkane activation proceeds by different mechanisms under sulfate-reducing and methanogenic conditions. Geochim. Cosmochim. Acta 109, 162–174. doi: 10.1016/j.gca.2013.01.031
Allen, J. P., Atekwana, E. A., Atekwana, E. A., Duris, J. W., Werkema, D. D., and Rossbach, S. (2007). The microbial community structure in petroleum-contaminated sediments corresponds to geophysical signatures. Appl. Environ. Microbiol. 73, 2860–2870. doi: 10.1128/aem.01752-06
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Anderson, R. T., and Lovley, D. R. (2000). Biogeochemistry: hexadecane decay by methanogenesis. Nature 404, 722–723. doi: 10.1038/35008145
Bakermans, C., and Madsen, E. L. (2002). Diversity of 16S rDNA and naphthalene dioxygenase genes from coal-tar-waste-contaminated aquifer waters. Microb. Ecol. 44, 95–106. doi: 10.1007/s00248-002-0005-8
Barret, M., Gagnon, N., Morissette, B., Kalmokoff, M. L., Topp, E., Brooks, S. P. J., et al. (2015). Phylogenetic identification of methanogens assimilating acetate-derived carbon in dairy and swine manures. Syst. Appl. Microbiol. 38, 56–66. doi: 10.1016/j.syapm.2014.11.006
Berdugo-Clavijo, C., and Gieg, L. M. (2014). Conversion of crude oil to methane by a microbial consortium enriched from oil reservoir production waters. Front. Microbiol. 5:10. doi: 10.3389/fmicb.2014.00197
Bian, X. Y., Mbadinga, S. M., Liu, Y. F., Yang, S. Z., Liu, J. F., Ye, R. Q., et al. (2015). Insights into the anaerobic biodegradation pathway of n-alkanes in oil reservoirs by detection of signature metabolites. Sci. Rep. 5, 9801. doi: 10.1038/srep09801
Biegert, A., Mayer, C., Remmert, M., Söding, J., and Lupas, A. N. (2006). The MPI Bioinformatics Toolkit for protein sequence analysis. Nucleic Acids Res. 34, W335–W339. doi: 10.1093/nar/gkl217
Bryant, M. P. (1972). Commentary on the Hungate technique for culture of anaerobic bacteria. Am. J. Clin. Nutr. 25, 1324–1328.
Callaghan, A. V. (2013). Enzymes involved in the anaerobic oxidation of n-alkanes: from methane to long-chain paraffins. Front. Microbiol. 4:89. doi: 10.3389/fmicb.2013.00089
Callaghan, A. V., Davidova, I. A., Savage-Ashlock, K., Parisi, V. A., Gieg, L. M., Suflita, J. M., et al. (2010). Diversity of benzyl- and alkylsuccinate synthase genes in hydrocarbon-impacted environments and enrichment cultures. Environ. Sci. Technol. 44, 7287–7294. doi: 10.1021/es1002023
Callaghan, A. V., Morris, B. E. L., Pereira, I. A. C., McInerney, M. J., Austin, R. N., Groves, J. T., et al. (2012). The genome sequence of Desulfatibacillum alkenivorans AK-01: a blueprint for anaerobic alkane oxidation. Environ. Microbiol. 14, 101–113. doi: 10.1111/j.1462-2920.2011.02516.x
Cheng, L., Ding, C., Li, Q., He, Q., Dai, L.-R., and Zhang, H. (2013a). DNA-SIP reveals that Syntrophaceae play an important role in methanogenic hexadecane degradation. PLoS ONE 8:e66784. doi: 10.1371/journal.pone.0066784
Cheng, L., He, Q., Ding, C., Dai, L. R., Li, Q., and Zhang, H. (2013b). Novel bacterial groups dominate in a thermophilic methanogenic hexadecane-degrading consortium. FEMS Microbiol. Ecol. 85, 568–577. doi: 10.1111/1574-6941.12141
Cheng, L., Qiu, T. L., Li, X., Wang, W. D., Deng, Y., Yin, X. B., et al. (2008). Isolation and characterization of Methanoculleus receptaculi sp. nov. from Shengli oil field, China. FEMS Microbiol. Lett. 285, 65–71. doi: 10.1111/j.1574-6968.2008.01212.x
Embree, M., Nagarajan, H., Movahedi, N., Chitsaz, H., and Zengler, K. (2014). Single-cell genome and metatranscriptome sequencing reveal metabolic interactions of an alkane-degrading methanogenic community. ISME J. 8, 757–767. doi: 10.1038/ismej.2013.187
Ferry, J. G. (2010). How to make a living by exhaling methane. Annu. Rev. Microbiol. 64, 453–473. doi: 10.1146/annurev.micro.112408.134051
Gantner, S., Andersson, A. F., Alonso-Sáez, L., and Bertilsson, S. (2011). Novel primers for 16S rRNA-based archaeal community analyses in environmental samples. J. Microbiol. Methods 84, 12–18. doi: 10.1016/j.mimet.2010.10.001
Garrity, G. M., and Holt, J. G. (2001). “The road map to the manual,” in Bergey’s Manual® of Systematic Bacteriology, eds D. Boone, R. Castenholz, and G. Garrity (New York, NY: Springer), 119–166.
Gray, N. D., Sherry, A., Grant, R. J., Rowan, A. K., Hubert, C. R. J., Callbeck, C. M., et al. (2011). The quantitative significance of Syntrophaceae and syntrophic partnerships in methanogenic degradation of crude oil alkanes. Environ. Microbiol. 13, 2957–2975. doi: 10.1111/j.1462-2920.2011.02570.x
Gray, N. D., Sherry, A., Hubert, C., Dolfing, J., and Head, I. M. (2010). “Chapter 5 – Methanogenic degradation of petroleum hydrocarbons in subsurface environments: remediation, heavy oil formation, and energy recovery,” in Advances in Applied Microbiology, eds A. I. Laskin, S. Sariaslani, and M. G. Geoffrey (New York, NY: Academic Press), 137–161.
Grégoire, P., Fardeau, M. L., Joseph, M., Guasco, S., Hamaide, F., Biasutti, S., et al. (2011). Isolation and characterization of Thermanaerothrix daxensis gen. nov., sp. nov., a thermophilic anaerobic bacterium pertaining to the phylum “Chloroflexi”, isolated from a deep hot aquifer in the Aquitaine Basin. Syst. Appl. Microbiol. 34, 494–497. doi: 10.1016/j.syapm.2011.02.004
Hales, B. A., Edwards, C., Ritchie, D. A., Hall, G., Pickup, R. W., and Saunders, J. R. (1996). Isolation and identification of methanogen-specific DNA from blanket bog peat by PCR amplification and sequence analysis. Appl. Environ. Microbiol. 62, 668–675.
Head, I. M., Larter, S. R., Gray, N. D., Sherry, A., Adams, J. J., Aitken, C. M., et al. (2010). “Hydrocarbon degradation in petroleum reservoirs,” in Handbook of Hydrocarbon and Lipid Microbiology, ed. K. Timmis (Berlin: Springer), 3097–3109.
Heider, J., Spormann, A. M., Beller, H. R., and Widdel, F. (1998). Anaerobic bacterial metabolism of hydrocarbons. FEMS Microbiol. Rev. 22, 459–473. doi: 10.1111/j.1574-6976.1998.tb00381.x
Henry, E. A., Devereux, R., Maki, J. S., Gilmour, C. C., Woese, C. R., Mandelco, L., et al. (1994). Characterization of a new thermophilic sulfate-reducing bacterium. Arch. Microbiol. 161, 62–69. doi: 10.1007/BF00248894
Huber, T., Faulkner, G., and Hugenholtz, P. (2004). Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20, 2317–2319. doi: 10.1093/bioinformatics/bth226
Hug, L. A., Castelle, C. J., Wrighton, K. C., Thomas, B. C., Sharon, I., Frischkorn, K. R., et al. (2013). Community genomic analyses constrain the distribution of metabolic traits across the Chloroflexi phylum and indicate roles in sediment carbon cycling. Microbiome 1, 22. doi: 10.1186/2049-2618-1-22
Imachi, H., Sakai, S., Sekiguchi, Y., Hanada, S., Kamagata, Y., Ohashi, A., et al. (2008). Methanolinea tarda gen. nov., sp. nov., a methane-producing archaeon isolated from a methanogenic digester sludge. Int. J. Syst. Evol. Microbiol. 58, 294–301. doi: 10.1099/ijs.0.65394-0
Jones, D. M., Head, I. M., Gray, N. D., Adams, J. J., Rowan, A. K., Aitken, C. M., et al. (2008). Crude-oil biodegradation via methanogenesis in subsurface petroleum reservoirs. Nature 451, 176–180. doi: 10.1038/nature06484
Kasai, Y., Takahata, Y., Hoaki, T., and Watanabe, K. (2005). Physiological and molecular characterization of a microbial community established in unsaturated, petroleum-contaminated soil. Environ. Microbiol. 7, 806–818. doi: 10.1111/j.1462-2920.2005.00754.x
Khelifi, N., Amin Ali, O., Roche, P., Grossi, V., Brochier-Armanet, C., Valette, O., et al. (2014). Anaerobic oxidation of long-chain n-alkanes by the hyperthermophilic sulfate-reducing archaeon, Archaeoglobus fulgidus. ISME J. 8, 2153–2166. doi: 10.1038/ismej.2014.58
Li, H., Yang, S. Z., Mu, B. Z., Rong, Z. F., and Zhang, J. (2007). Molecular phylogenetic diversity of the microbial community associated with a high-temperature petroleum reservoir at an offshore oilfield. FEMS Microbiol. Ecol. 60, 74–84. doi: 10.1111/j.1574-6941.2006.00266.x
Li, W., Wang, L. Y., Duan, R. Y., Liu, J. F., Gu, J. D., and Mu, B. Z. (2012). Microbial community characteristics of petroleum reservoir production water amended with n-alkanes and incubated under nitrate-, sulfate-reducing and methanogenic conditions. Int. Biodeterior. Biodegrad. 69, 87–96. doi: 10.1016/j.ibiod.2012.01.005
Liang, B., Wang, L. Y., Mbadinga, S. M., Liu, J. F., Yang, S. Z., Gu, J. D., et al. (2015). Anaerolineaceae and Methanosaeta turned to be the dominant microorganisms in alkanes-dependent methanogenic culture after long-term of incubation. AMB Express 5, 1–13. doi: 10.1186/s13568-015-0117-4
Liu, C. S., Zhao, D. F., and Zhang, Y. B. (2014). Effects of temperature, acetate and nitrate on methane generation from petroleum hydrocarbons. China Pet. Process. Petrochem. Technol. 4, 24–31. doi: 10.3969/j.issn.1008-6234.2014.04.007
Magot, M., Ollivier, B., and Patel, B. C. (2000). Microbiology of petroleum reservoirs. Antonie Van Leeuwenhoek 77, 103–116. doi: 10.1023/A:1002434330514
Mbadinga, S. M., Li, K. P., Zhou, L., Wang, L. Y., Yang, S. Z., Liu, J. F., et al. (2012). Analysis of alkane-dependent methanogenic community derived from production water of a high-temperature petroleum reservoir. Appl. Microbiol. Biotechnol. 96, 531–542. doi: 10.1007/s00253-011-3828-8
Nunoura, T., Oida, H., Miyazaki, J., Miyashita, A., Imachi, H., and Takai, K. (2008). Quantification of mcrA by fluorescent PCR in methanogenic and methanotrophic microbial communities. FEMS Microbiol. Ecol. 64, 240–247. doi: 10.1111/j.1574-6941.2008.00451.x
Oren, A. (2014). “The family methanomicrobiaceae,” in The Prokaryotes, eds E. Rosenberg, E. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin: Springer), 231–246.
Pérez-Jiménez, J. R., Young, L. Y., and Kerkhof, L. J. (2001). Molecular characterization of sulfate-reducing bacteria in anaerobic hydrocarbon-degrading consortia and pure cultures using the dissimilatory sulfite reductase (dsrAB) genes. FEMS Microbiol. Ecol. 35, 145–150. doi: 10.1111/j.1574-6941.2001.tb00798.x
Ramos-Padron, E., Bordenave, S., Lin, S., Bhaskar, I. M., Dong, X., Sensen, C. W., et al. (2011). Carbon and sulfur cycling by microbial communities in a gypsum-treated oil sands tailings pond. Environ. Sci. Technol. 45, 439–446. doi: 10.1021/es1028487
Roest, K., Altinbas, M., Paulo, P., Heilig, H. G. H. J., Akkermans, A. L., Smidt, H., et al. (2005). Enrichment and detection of microorganisms involved in direct and indirect methanogenesis from methanol in an anaerobic thermophilic bioreactor. Microb. Ecol. 50, 440–446. doi: 10.1007/s00248-005-0237-5
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Savage, K. N., Krumholz, L. R., Gieg, L. M., Parisi, V. A., Suflita, J. M., Allen, J., et al. (2010). Biodegradation of low-molecular-weight alkanes under mesophilic, sulfate-reducing conditions: metabolic intermediates and community patterns. FEMS Microbiol. Ecol. 72, 485–495. doi: 10.1111/j.1574-6941.2010.00866.x
Schouw, A., Leiknes Eide, T., Stokke, R., Birger Pedersen, R., Helene Steen, I., and Bødtker, G. (2016). Abyssivirga alkaniphila gen. nov., sp. nov., an alkane-degrading, anaerobic bacterium from a deep-sea hydrothermal vent system, and emended descriptions of Natranaerovirga pectinivora and Natranaerovirga hydrolytica. Int. J. Syst. Evol. Microbiol. 66, 1724–1734. doi: 10.1099/ijsem.0.000934
Sekiguchi, Y., Kamagata, Y., Syutsubo, K., Ohashi, A., Harada, H., and Nakamura, K. (1998). Phylogenetic diversity of mesophilic and thermophilic granular sludges determined by 16S rRNA gene analysis. Microbiology 144, 2655–2665. doi: 10.1099/00221287-144-9-2655
Sekiguchi, Y., Yamada, T., Hanada, S., Ohashi, A., Harada, H., and Kamagata, Y. (2003). Anaerolinea thermophila gen. nov., sp. nov. and Caldilinea aerophila gen. nov., sp. nov., novel filamentous thermophiles that represent a previously uncultured lineage of the domain Bacteria at the subphylum level. Int. J. Syst. Evol. Microbiol. 53, 1843–1851. doi: 10.1099/ijs.0.02699-0
Shahimin, M. F. M., Foght, J. M., and Siddique, T. (2016). Preferential methanogenic biodegradation of short-chain n-alkanes by microbial communities from two different oil sands tailings ponds. Sci. Total Environ. 553, 250–257. doi: 10.1016/j.scitotenv.2016.02.061
Sherry, A., Grant, R. J., Aitken, C. M., Jones, D. M., Head, I. M., and Gray, N. D. (2014). Volatile hydrocarbons inhibit methanogenic crude oil degradation. Front. Microbiol. 5:9. doi: 10.3389/fmicb.2014.00131
Sherry, A., Gray, N. D., Ditchfield, A. K., Aitken, C. M., Jones, D. M., Röling, W. F. M., et al. (2013). Anaerobic biodegradation of crude oil under sulphate-reducing conditions leads to only modest enrichment of recognized sulphate-reducing taxa. Int. Biodeterior. Biodegrad. 81, 105–113. doi: 10.1016/j.ibiod.2012.04.009
Sutton, N. B., Maphosa, F., Morillo, J. A., Abu Al-Soud, W., Langenhoff, A. A. M., Grotenhuis, T., et al. (2013). Impact of long-term diesel contamination on soil microbial community structure. Appl. Environ. Microbiol. 79, 619–630. doi: 10.1128/aem.02747-12
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tan, B., de Araújo e Silva, R., Rozycki, T., Nesbø, C., and Foght, J. (2014a). Draft genome sequences of three Smithella spp. obtained from a methanogenic alkane-degrading culture and oil field produced water. Genome Announc. 2, e1085–e1014. doi: 10.1128/genomeA.01085-14
Tan, B., Nesbo, C., and Foght, J. (2014b). Re-analysis of omics data indicates Smithella may degrade alkanes by addition to fumarate under methanogenic conditions. ISME J. 8, 2353–2356. doi: 10.1038/ismej.2014.87
Wang, L. Y., Gao, C. X., Mbadinga, S. M., Zhou, L., Liu, J. F., Gu, J. D., et al. (2011). Characterization of an alkane-degrading methanogenic enrichment culture from production water of an oil reservoir after 274 days of incubation. Int. Biodeterior. Biodegrad. 65, 444–450. doi: 10.1016/j.ibiod.2010.12.010
Wang, L. Y., Li, W., Mbadinga, S. M., Liu, J. F., Gu, J. D., and Mu, B. Z. (2012). Methanogenic microbial community composition of oily sludge and its enrichment amended with alkanes incubated for over 500 days. Geomicrobiol. J. 29, 716–726. doi: 10.1080/01490451.2011.619634
Webner, K. (2012). Die Gene der (1-Methylalkyl) succinat-Synthase im Anaeroben n-Alkanabbau des Betaproteobakteriums Stamm HxN1. Ph.D. thesis, University of Bremen, Bremen.
Wilkes, H., Rabus, R., Fischer, T., Armstroff, A., Behrends, A., and Widdel, F. (2002). Anaerobic degradation of n-hexane in a denitrifying bacterium: further degradation of the initial intermediate (1-methylpentyl)succinate via C-skeleton rearrangement. Arch. Microbiol. 177, 235–243. doi: 10.1007/s00203-001-0381-3
Winderl, C., Anneser, B., Griebler, C., Meckenstock, R. U., and Lueders, T. (2008). Depth-resolved quantification of anaerobic toluene degraders and aquifer microbial community patterns in distinct redox zones of a tar oil contaminant plume. Appl. Environ. Microbiol. 74, 792–801. doi: 10.1128/aem.01951-07
Yamada, T., Imachi, H., Ohashi, A., Harada, H., Hanada, S., Kamagata, Y., et al. (2007). Bellilinea caldifistulae gen. nov., sp. nov. and Longilinea arvoryzae gen. nov., sp. nov., strictly anaerobic, filamentous bacteria of the phylum Chloroflexi isolated from methanogenic propionate-degrading consortia. Int. J. Syst. Evol. Microbiol. 57, 2299–2306. doi: 10.1099/ijs.0.65098-0
Yamada, T., Sekiguchi, Y., Hanada, S., Imachi, H., Ohashi, A., Harada, H., et al. (2006). Anaerolinea thermolimosa sp. nov. Levilinea saccharolytica gen. nov., sp. nov. and Leptolinea tardivitalis gen. nov., sp. nov., novel filamentous anaerobes, and description of the new classes Anaerolineae classis nov. and Caldilineae classis nov. in the bacterial phylum Chloroflexi. Int. J. Syst. Evol. Microbiol. 56, 1331–1340. doi: 10.1099/ijs.0.64169-0
Yu, Y., Lee, C., Kim, J., and Hwang, S. (2005). Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol. Bioeng. 89, 670–679. doi: 10.1002/bit.20347
Zengler, K., Richnow, H. H., Rossello-Mora, R., Michaelis, W., and Widdel, F. (1999). Methane formation from long-chain alkanes by anaerobic microorganisms. Nature 401, 266–269. doi: 10.1038/45777
Zhou, L., Li, K. P., Mbadinga, S. M., Yang, S. Z., Gu, J. D., and Mu, B. Z. (2012). Analyses of n-alkanes degrading community dynamics of a high-temperature methanogenic consortium enriched from production water of a petroleum reservoir by a combination of molecular techniques. Ecotoxicology 21, 1680–1691. doi: 10.1007/s10646-012-0949-5
Keywords: alkanes degradation, long-chain alkanes, microbial community, 16S rRNA gene, Thermodesulfovibrio, Anaerolineaceae, Methanoculleus, methanogenesis
Citation: Liang B, Wang L-Y, Zhou Z, Mbadinga SM, Zhou L, Liu J-F, Yang S-Z, Gu J-D and Mu B-Z (2016) High Frequency of Thermodesulfovibrio spp. and Anaerolineaceae in Association with Methanoculleus spp. in a Long-Term Incubation of n-Alkanes-Degrading Methanogenic Enrichment Culture. Front. Microbiol. 7:1431. doi: 10.3389/fmicb.2016.01431
Received: 31 March 2016; Accepted: 29 August 2016;
Published: 16 September 2016.
Edited by:
Gavin Collins, National University of Ireland, Galway, IrelandReviewed by:
Johannes Scholten, Synthetic Genomics Inc., USAPing Li, China University of Geosciences, China
Copyright © 2016 Liang, Wang, Zhou, Mbadinga, Zhou, Liu, Yang, Gu and Mu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bo-Zhong Mu, YnptdUBlY3VzdC5lZHUuY24=
 Bo Liang
Bo Liang Li-Ying Wang
Li-Ying Wang Zhichao Zhou
Zhichao Zhou Serge M. Mbadinga
Serge M. Mbadinga Lei Zhou
Lei Zhou Jin-Feng Liu
Jin-Feng Liu Shi-Zhong Yang
Shi-Zhong Yang Ji-Dong Gu
Ji-Dong Gu Bo-Zhong Mu
Bo-Zhong Mu