- 1Laboratory for Molecular Microbiology, Institute of Molecular Genetics and Genetic Engineering, University of Belgrade, Belgrade, Serbia
- 2Bacteriology Group, International Centre for Genetic Engineering and Biotechnology, Area Science Park, Trieste, Italy
- 3Department of Biochemistry, Faculty of Biology, University of Belgrade, Belgrade, Serbia
AggLb is the largest (318.6 kDa) aggregation-promoting protein of Lactobacillus paracasei subsp. paracasei BGNJ1-64 responsible for forming large cell aggregates, which causes auto-aggregation, collagen binding and pathogen exclusion in vitro. It contains an N-terminus leader peptide, followed by six successive collagen binding domains, 20 successive repeats (CnaB-like domains) and an LPXTG sorting signal at the C-terminus for cell wall anchoring. Experimental information about the roles of the domains of AggLb is currently unknown. To define the domain that confers cell aggregation and the key domains for interactions of specific affinity between AggLb and components of the extracellular matrix, we constructed a series of variants of the aggLb gene and expressed them in Lactococcus lactis subsp. lactis BGKP1-20 using a lactococcal promoter. All of the variants contained a leader peptide, an inter collagen binding-CnaB domain region (used to raise an anti-AggLb antibody), an anchor domain and a different number of collagen binding and CnaB-like domains. The role of the collagen binding repeats of the N-terminus in auto-aggregation and binding to collagen and fibronectin was confirmed. Deletion of the collagen binding repeats II, III, and IV resulted in a loss of the strong auto-aggregation, collagen and fibronectin binding abilities whereas the biofilm formation capability was increased. The strong auto-aggregation, collagen and fibronectin binding abilities of AggLb were negatively correlated to biofilm formation.
Introduction
Lactobacillus strains could exhibit probiotic characteristics, which confer a variety of beneficial health effects on the host and they have a number of features that make it particularly suitable for dairy applications (Salminen et al., 1998; Lebeer et al., 2008; Sisto and Lavermicocca, 2012; Giraffa, 2014). Lactobacillus effector molecules that contribute to the health-promoting interactions with the host (intestinal) system are likely located in the bacterial cell envelope (Bron et al., 2004; Kleerebezem et al., 2010; Hymes et al., 2016). It was found that adhesion of lactobacilli to components of the extracellular matrix (ECM) such as mucin, fibronectin, collagen, laminin, or fibrinogen may thus have a direct impact on their probiotic function, e.g., in preventing the adhesion to and the colonization of damaged intestinal tissue sites by invading pathogens (Lorca et al., 2002). It has been reported that damage of the mucosal layer of the ECM can result in its colonization by pathogens, resulting in subsequent infection (Styriak et al., 2003).
The ability of pathogenic bacteria to adhere to distinct components of the ECM, such as collagen and fibronectin, is enabled or facilitated by the expression of ECM-binding proteins, termed adhesins. Adhesins are important virulence factors of pathogens, as they are involved in the initiation of infection (Flock, 1999). Group A streptococci (GAS, Streptococcus pyogenes) have evolved a number of surface-bound and secreted virulence factors, of which the M proteins are probably the best characterized. Binding of GAS to epithelial cells involves an interaction between M protein and fibronectin (Oehmcke et al., 2010). Epithelial cell invasion by Group B Streptococcus (GBS) is associated with expression of alpha C protein (Bolduc and Madoff, 2007). Aggregation protein encoded by asp1 gene of enterococci, characterized as a virulence factor of 142 kDa plays a crucial role in adherence to eukaryotic cells (Galli et al., 1990). In the skin abscess model, a sortase-deficient Staphylococcus aureus strain lacking all of its cell-wall anchored proteins was less virulent than its wild-type strain. Also, strains specifically lacking protein A, fibronectin binding proteins, clumping factor A or surface protein SasF were impaired in their virulence (Josefsson et al., 2008; Kwiecinski et al., 2014). In addition some biofilm factors related to aggregation ability, for example, Bap protein of S. aureus facilitates the persistence in the mammary gland by enhancing adhesion to epithelial cells and prevents cellular internalization through the binding to GP96 host receptor (Taglialegna et al., 2016).
Since systematic analysis of efficacy of probiotic therapy demonstrated that probiotic activities are strain-specific (Hungin et al., 2013; Sanders et al., 2013) the paradigm of probiotic research is rightfully shifting toward understanding the mechanistic action of each specific strain (Johnson and Klaenhammer, 2014). It has been demonstrated that the purified collagen binding protein (Cbp) from L. plantarum 91 possess anti-adhesion activity against the enteric pathogen Escherichia coli 0157:H7 on immobilized collagen (Yadava et al., 2013). Surface fibronectin binding protein from L. casei BL23 participates in cell attachment to immobilized fibronectin (Muñoz-Provencio et al., 2010). Also, binding of immobilized collagen and fibronectin by L. acidophilus CRL 639 depends on cell-surface proteins (Lorca et al., 2002). The S-layer proteins of L. crispatus ZJ001 also inhibited the adhesion of Salmonella typhimurium and E. coli O157:H7 to HeLa cells (Chen et al., 2007). In addition, the S-layer protein associated with moonlighting proteins acted as an adherence factor, which has been evidenced by the high capability of adhesion, auto- and co-aggregation of L. helveticus T159 (Waśko et al., 2014).
The ability of lactobacilli to form multicellular aggregates is an important property for colonization of the oral cavity, human gut or urogenital tract. The underlying mechanisms and the functionality of surface aggregation factors are not fully understood; on the one hand aggregation ability may not be the only components responsible for adhesion, and some of the criteria may be part of a complex mechanism that enables the microorganisms to interact with the host and to exert their beneficial effects (García-Cayuela et al., 2014). On the other hand, important mechanisms involved in this process are thought to include adherence as well as colonization of the GIT (Nazzaro et al., 2012; Skrzypczak et al., 2015). The expression of adhesins on the cell surface could induce cell aggregation visible as auto-aggregation. Aggregation promoting factors of lactobacilli differ in size, from 2 kDa in the strain Lactobacillus gasseri 2459–318.6 kDa in L. paracasei subsp. paracasei BGNJ1-64 (Boris et al., 1997; Miljkovic et al., 2015). Interestingly we have reported a new group of aggregation promoting factors of a high molecular mass, recently discovered in LAB (Kojic et al., 2011; Miljkovic et al., 2015). They differ in size and primary structure; however, they share similar structural organization and functions because they are composed of a large number of collagen-binding and CnaB-like domains (Miljkovic et al., 2015). Currently, no experimental evidence exists concerning the role of these domains in aggregation except for predictions that are based on a S. aureus collagen-binding Cna protein that mediates bacterial adherence to collagen. The major differences between the aggregation factors of the LAB and the Cna protein of S. aureus are that the primary structure of Cna has a non-repetitive collagen binding A region, followed by a repetitive B region (one–four 23 kDa repeating units B1–B4, depending on the strain). It has been suggested that the A region is involved in collagen binding, while the B region acts as a “stalk” that projects the A region from the bacterial surface, facilitating its adherence to collagen (Deivanayagam et al., 2000).
As mentioned above, the AggLb protein is the largest (318.6 kDa) aggregation factor of lactobacilli responsible for auto-aggregation, collagen binding and pathogen exclusion in vitro. AggLb consists of six diverse collagen binding domains (from 13202–15256 Da repeating units) and 20 almost identical CnaB-like domains (a 9916 Da repeating unit). The aim of this study was to investigate the roles of the different domains of the AggLb protein involved in probiotic function; this information might prove useful for its potential application. A series of variants of aggLb gene/protein were constructed, and their capability to induce auto-aggregation, binding to collagen and fibronectin, and biofilm formation was analyzed. It was concluded that AggLb could provide all of these functions: aggregation and binding to collagen and fibronectin as well as biofilm formation. Interestingly, strong auto-aggregation, collagen and fibronectin binding capacities of AggLb are negatively correlated with the ability of biofilm formation.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions
The strains, their derivatives and plasmids used in this study are listed in Table 1. L. paracasei was grown in De Man-Rogosa-Sharpe (MRS; Merck GmbH, Darmstadt, Germany) medium at 30°C. Lactococcus lactis subsp. lactis was grown at 30°C in M17 medium (Merck) supplemented with 0.5% glucose (GM17). Pseudomonas aeruginosa PAO1 and E. coli DH5α and M15 used for cloning and propagation of constructs were routinely grown in Luria-Bertani medium (LB) at 37°C with aeration. To obtain solid medium, agar (15 g/l; Torlak, Belgrade, Serbia) was added. Erythromycin was added to a final concentration of 10 μg/ml and 300 μg/ml for LAB and E. coli, respectively. Ampicillin and kanamycin were added to a final concentration of 100 μg/ml for E. coli. When necessary, 5-bromo-4-chloro-3-indolyl-β-D-galactoside (X-Gal; Fermentas, Vilnius, Lithuania) was added to LB medium plates at a final concentration of 40 μg/ml for blue/white color selection of colonies.
DNA Manipulations
Electrocompetent Lc. lactis subsp. lactis BGKP1-20 cells was prepared as described by Holo and Nes (1989). Transformations were done by electroporation using an Eppendorf Electroporator (Eppendorf, Hamburg, Germany), except E. coli DH5α and M15, which was transformed by heat shock (Hanahan, 1983). Appropriate agar plates with antibiotics were used for the selection of transformants.
Plasmid DNA from E. coli DH5α was isolated by QIAprep Spin Miniprep kit (Qiagen GmBH, Hilden, Germany). Digestion with restriction enzymes was conducted according to the supplier’s instructions (Fermentas). DNA fragments were purified from agarose gels using a QIAquick Gel extraction kit as described by the manufacturer (Qiagen). DNA was ligated with T4 DNA ligase (Agilent technologies, USA) according to the manufacturer’s recommendations.
Specific primers used in this study are listed in section: Construction of the aggLb gene variants. KapaTaq DNA polymerase (Kapa Biosystems, Inc., Boston, MA, USA) was used to amplify DNA fragments by PCR using a GeneAmp PCR system 2700 thermal cycler (Applied Biosystems, Foster City, CA, USA). PCR products were purified with a QiaQuick PCR purification kit (Qiagen) according to the protocol of the supplier and sequenced by the Macrogen Sequencing Service (Macrogen, Netherlands). The DNA Strider program was used for open reading frame (ORF) prediction. Commercial pGEM-T-Easy (Promega, Madison, WI, USA), pCR2.1-TOPO (Thermo Scientific) and pCRII (Thermo Scientific) vectors were used for cloning of PCR products.
Construction of the aggLb Gene Variants
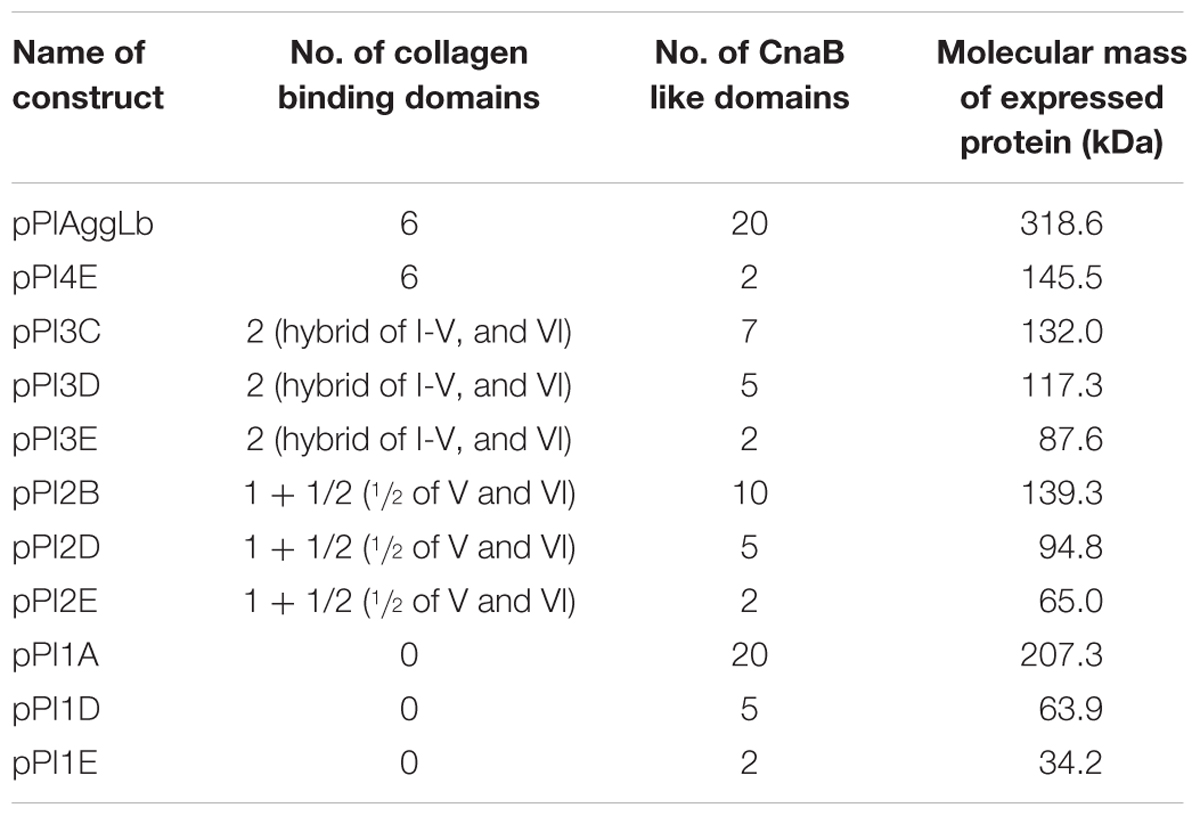
From construct pALb35 (Miljkovic et al., 2015) using XbaI-SalI restriction enzymes we made shorter construct pAggLbXS carrying only aggLb gene, in pAZIL vector (Supplementary Figure 1A). PstI restriction site is located in aggLb gene at position to divide it into two regions: first containing leader peptide sequence and six collagen binding domains and second containing 20 CnaB-like domains and anchor domain (Figure 1). In order to facilitate the construction of a large number of variants, aggLb gene was subcloned from pAggLbXS in two parts into pBScript vector (Agilent technologies): first part as XbaI-PstI (construct pBS-XP) and second as PstI-SalI fragments (construct pBS-PS; Supplementary Figure 1A). Bioinformatic analysis showed that HindIII (in both fragments; Supplementary Figures 1B,D) and SspI (only in XbaI-PstI fragment; Supplementary Figure 1C) restriction enzymes dividing AggLb protein to distinct portions that contain the exact number of codons without free base except one in XbaI-PstI fragment, so that they can be deleted or combined because they provide in frame junction. Constructs pBS-XP [consisting of three HindIII fragments of 820 bp, 821 bp (this two cannot be deleted separately since deletion of each fragment changed frame and introduce frameshift mutation) and 1461 bp] and pBS-PS (consisting of four HindIII fragments of 846 bp, 1266 bp, and two of 1410 bp) were partially digested with HindIII restriction enzyme and ligated. We successfully constructed pBS-XP-1, pBS-XP-4, pBS-PS-A, pBS-PS-B, pBS-PS-C, pBS-PS-D, and pBS-PS-E (for details see Table 1 and Supplementary Figure 1). From construct pBS-XP fragment carrying XbaI/PstI was recloned into pCR2.1-TOPO (since does not contain SspI restriction site; Thermo Scientific, Lithuania) giving construct pCR-XP, which was additionally partially digested with SspI restriction enzyme and ligated (constructs pCR-XP-2 and pCR-XP-3; Supplementary Figure 1). In next step, different constructs containing deletion in first part (pBS-XP-1, pBS-XP-4, pCR-XP-2, and pCR-XP-3) were combined with constructs containing deletion in second part (pBS-PS-A, pBS-PS-B, pBS-PS-C, pBS-PS-D, and pBS-PS-E) in pBScript vector (for details see Table 1 and Figure 1). In order to obtained expression in lactococci, lactococcal promoter PlsbB (Uzelac et al., 2015) was cloned into pAZIL vector together with leader sequence of aggLb gene as SacI-EagI fragment (construct pAZIL-pSE). After that different combinations of variants from pBScript vector were cloned as EagI-SalI fragments into pAZIL-pSE (for details see Table 1 and Figure 1). Lc. lactis subsp. lactis BGKP1-20 was transformed with chosen constructs and expression of different AggLb variants were confirmed by Dot blot analysis using anti-AggLb antibody.
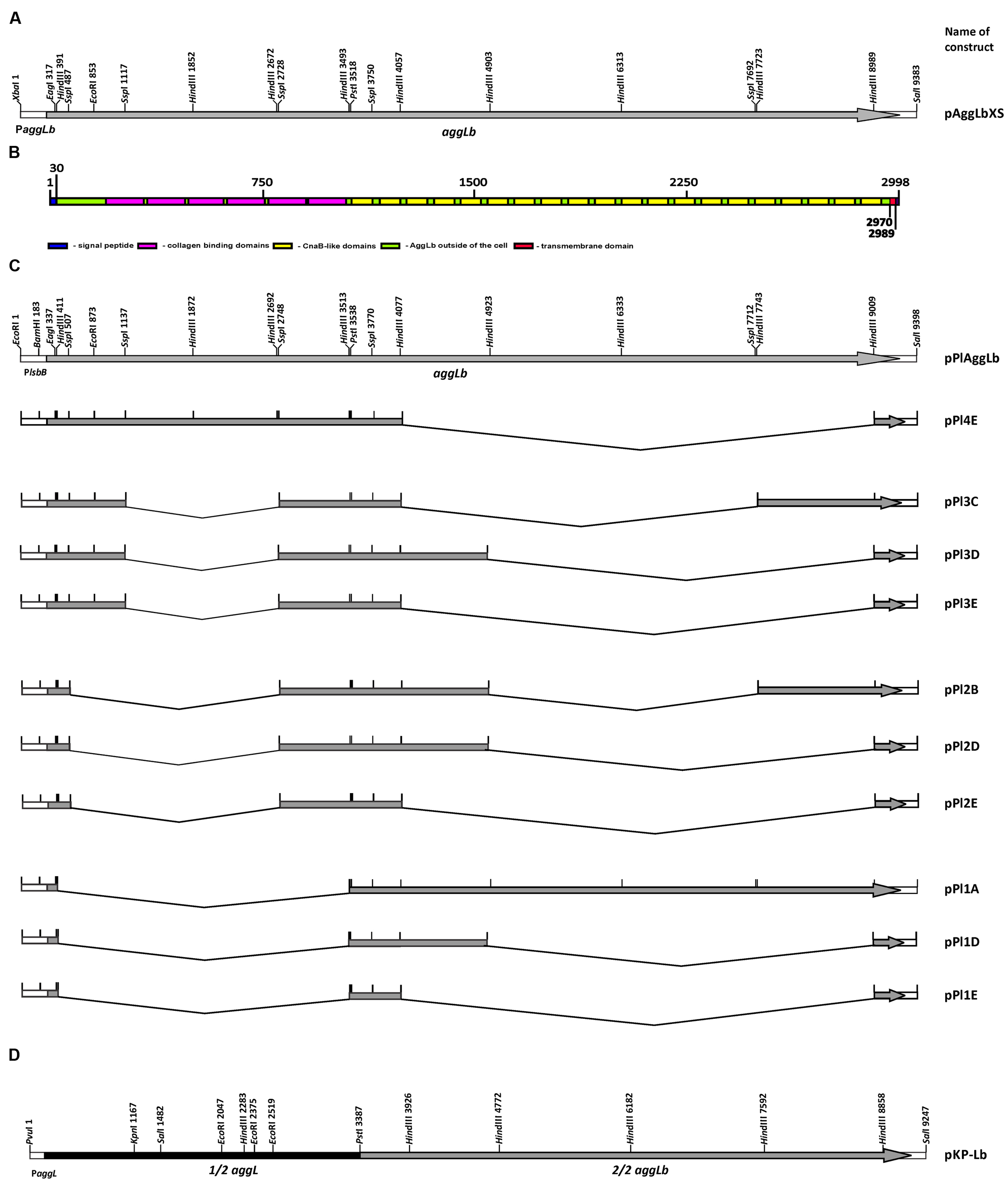
FIGURE 1. Schematic representation of strategy for construction variants of aggLb gene and hybrid construct. (A) aggLb; (B) AggLb domain organization (boxes indicate domains of protein); (C) series of variants expressed using the lactococcal promoter PlsbB; (D) hybrid clone pKP-Lb.
In addition, using template clone KPPvScI (Kojic et al., 2011) and specific set of primers: KPFw (5′GCAAAGCGCCATTCGCC3′), KPPstIRev (5′CGTTCCTTCTGCAGTTCCAC3′), after PCR amplification, we obtained clone pCRII-KPI. BamHI-PstI fragment containing first part of AggL (aggregation factor from Lc. lactis subsp. lactis BGKP1) was recloned from pCRII-KPI into pBS-PS, from which entire hybrid molecule as BamHI/XhoI was transferred to pAZIL vector (digested with BamHI/SalI) and finally obtained clone was named as pKP-Lb (Table 1).
Auto-aggregation Assay
The first step of screening strains was visual auto-aggregation assay. The aggregation phenotype was scored as positive when clearly visible snowflakes-like particles, formed by aggregated cells, gravitated to the bottom of the tube, forming a precipitate and leaving clear supernatant.
The auto-aggregation ability of the selected strains and derivatives was tested according to García-Cayuela et al. (2014) with minor modifications. Briefly, cells of overnight culture were harvested by centrifugation (5000 × g, 10 min, 4°C), washed twice with phosphate-buffered saline – PBS (10 mM Na2HPO4, 1 mM KH2PO4, 140 mM NaCl, 3 mM KCl, pH 7.1) and resuspended in the same buffer. The mixture was vortexed and incubated at 30°C for a period of 5 h. Absorbance (OD600) was measured at different time points. Percentage of auto-aggregation was determined using the equation: [1 - (At/A0) × 100] where At represents the absorbance at different time points (1, 2, 3, 4 and 5 h) and A0 is absorbance at time 0. Auto-aggregation assay was done in three independent experiments. Data are presented as average of absorbance values from three independent experiments per each strain. The significance was determined by Student’s t-test.
Biofilm Formation Assay
The ability of selected strains and derivatives to form biofilm was assayed in microtiter plates as previously described by Peter et al. (2013). P. aeruginosa PAO1 and E. coli DH5α were used as positive and negative control strains, respectively. Additionally, PBS buffer was included to ensure that the influence on biofilm formation by strains (resuspended in the same buffer) not attributed to a non-specific binding effect to crystal violet. The results are presented as average of absorbance values from three independent experiments per each strain. The significance was determined by Student’s t-test.
Collagen and Fibronectin Binding Assays
The wells of Maxisorb plates (Nunc, Roskilde, Denmark) were coated with type I collagen (from rat tail, BD Bioscience, Franklin Lakes, NJ, United States; 100 μg/ml) and human fibronectin (Serva, Heidelberg, Germany; 100 μg/ml) for 16 h at 4°C. The collagen binding ability of the selected strains and derivatives was tested according to Miljkovic et al. (2015), while the ability of tested strains and derivatives to bind to fibronectin was assayed as previously described by Ahmed et al. (2001). After immobilization, wells were washed with PBS and blocked with 2% BSA in PBS. Upon removal of BSA solution and washing wells with PBS, the test cultures (100 μl, 108 CFU/ml) were added and plates were incubated on an orbital platform shaker for 2 h at 37°C. Non-adherent cells were removed by washing the wells three times with 200 μl of PBS. The adhered cells were fixed at 60°C for 20 min and stained with crystal violet (100 μl/well, 0.1% solution) for 45 min. Wells were subsequently washed tree times with PBS to remove the excess stain. The stain bound to the cells was dissolved by 100 μl of citrate buffer (pH 4.3). The absorbance was measured at 570 nm, after 45 min, using the microtiter plate reader. Collagen and fibronectin binding was assayed as described above and the average of absorbance values from three independent experiments per each strain was presented. The significance was determined by Student’s t-test.
Determination of Relationships between Auto-Aggregation, Collagen/Fibronectin Binding, and Biofilm Ability of Transformants Carrying Different Variants of the aggLb Gene
Plots of correlation were produced using Python 2.7.8 and scipy library (version 0.14.0).
Production of Polyclonal Antibody
Since whole AggLb protein was not able to be expressed in E. coli the part of AggLb protein containing the inter region of 190 amino acids between collagen binding and CnaB-like domains (from 1096 aa to 1286 aa) present in all variants was expressed using pQE30 vector with 6 × His tag (Qiagen) for production of anti-AggLb polyclonal antibody. Using clone pALb35 (Miljkovic et al., 2015), HindIII fragment of 560 bp containing PstI restriction site was cloned into pBScript. This fragment was recloned from pBScript vector as BamHI/SalI in frame into expression vector pQE30 with 6 × His tag (pQE30-AggBS). Fusion His-tagged protein was expressed in E. coli M15 cells. His-tag affinity purification of part of AggLb protein was conducted under denaturing conditions: the refolding method using urea to disrupt non-covalent bonds and increase protein solubility was used to solubilise and make the His-tagged AggLb more accessible to the nickel-nitrilotriacetic acid (Ni-NTA) resin. Purification of the fusion protein was applied according to protocol recommended by The QIAexpressionist. The eluted protein was dialyzed by ultrafiltration (Centrifugal Filter Units, Amicon Ultra-15 Centrifugal Filter Devices, 3K, Millipore). Polyclonal antibodies were produced by immunization of mice with the synthetic or purified fusion proteins in animal house of ICGEB, Trieste, Italy.
Dot Blotting
Samples (2 μl of serial dilutions of total proteins dissolved in buffer which contains: 100 mM NaH2PO4, 10 mM Tris-HCl, 8 M urea, pH 8.0) were loaded into a PVDF membrane (Merck Millipore, Darmstadt, Germany) by directly spotted on membrane as described by Niedergang et al. (2000). The same quantity of non-diluted samples was loaded on PAGE-SDS gel stained with Coomassie brilliant blue (Supplementary Figure 2). Membrane was incubated with 10% skim milk diluted in Tris-buffered saline containing 0.1% Tween 20 (TBS-T) over night at 4°C in order to block non-specific reactions. Following blocking, the membrane was incubated 1 h at room temperature with gentle agitation in dilutions of primary antibody (mouse polyclonal antibody anti-AggLb-Ab). Primary antibodies were diluted in 5% skim milk diluted in TBS-T. After washing three times in TBS-T for 15 min, membrane was incubated for 1 h with horseradish peroxidase-labeled anti-mouse IgG (A9044 anti-mouse; Sigma, Germany) at a 1:10000 dilution in 5% skim milk diluted in TBS-T. The blots were washed three times in TBS-T for 15 min. Spots were detected using EMD Millipore ImmobilonTM Western Chemiluminescent HRP Substrate (ECL; Fisher Scientific, USA) following the manufacturer’s instructions.
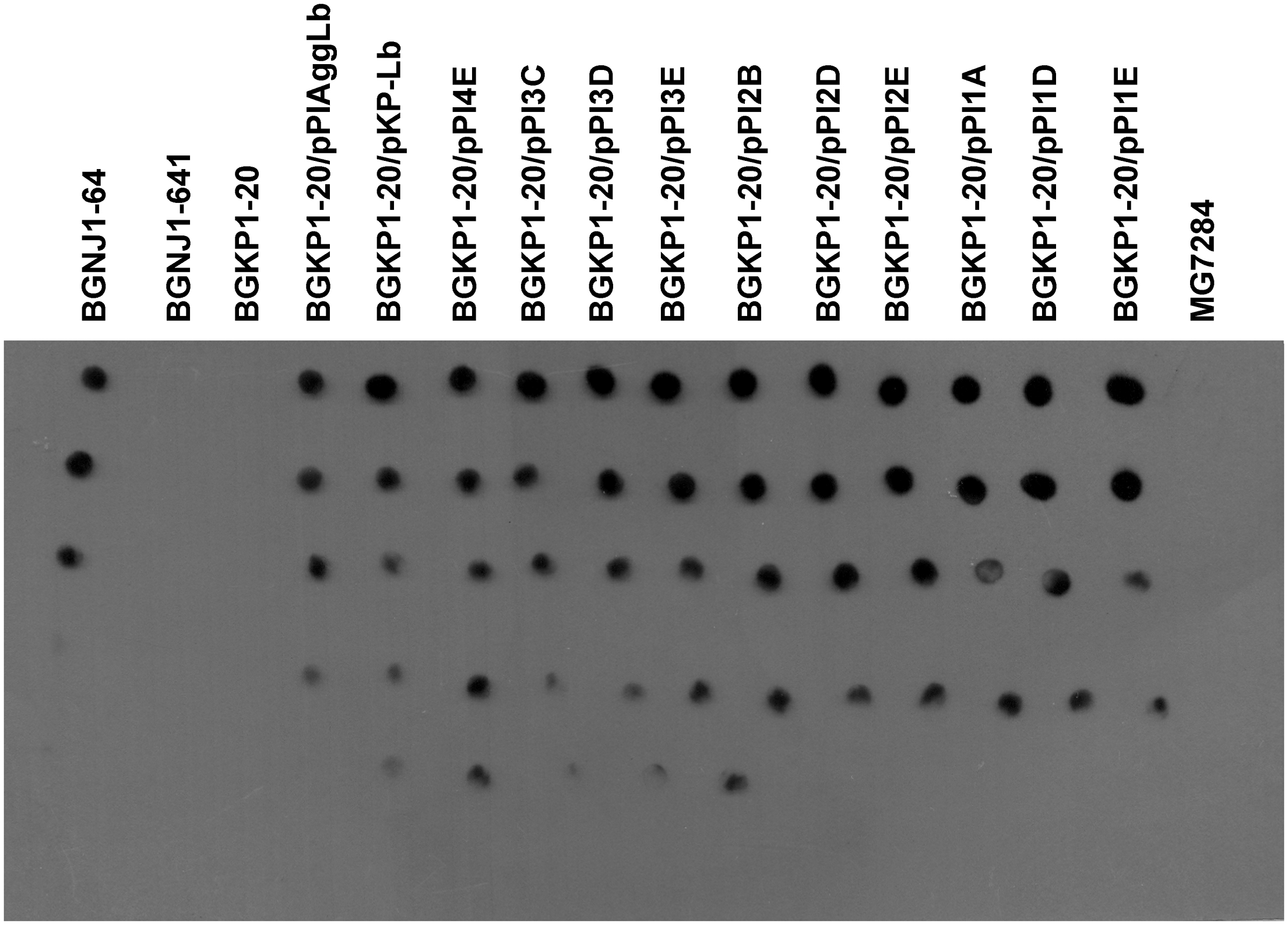
FIGURE 2. Dot blot using anti AggLb antibody. Total proteins of the wild-type strain and of derivatives harboring the different aggLb variants in Lc. lactis subsp. lactis BGKP1-20 strain.
Results
Construction of the AggLb Variants
We performed functional studies of the various domains of the AggLb protein. To produce many different domain variants of the AggLb protein, the aggLb gene was subcloned into two parts SacI-PstI and PstI-SalI fragments, using the pBscript vector. Both cloned fragments first partially digested using the HindIII restriction enzyme, and the first part of the gene was also digested using SspI; importantly, both of these enzymes leave the residual aggLb gene in frame. After obtaining different variants of both fragments they were combined to obtain constructs with different numbers of collagen binding and CnaB-like domains. The construct pPI1E did not contain any collagen binding domains and contained only two CnaB-like domains, whereas pPIAggLb contained the complete aggLb gene. For details of all the constructs see Figure 1 and Table 1. All the different combinations were recloned into the pAZIL vector using the lactococcal promoter PlsbB to provide identical transcription activity of all the constructs (Uzelac et al., 2015). The constructs (Figure 1; Table 1) were transformed into Lc. lactis subsp. lactis BGKP1-20 (the lactococcal derivative BGKP1-20 was used because the original lactobacilli strains had an extremely low efficiency of transformation) and expression was analyzed by Dot blot (Figure 2) using an anti-AggLb antibody raised against the transitional region covering the last part of the first region and the beginning of the second subclone of AggLb because this part is present in all of the constructs. Similar expression was obtained for all of the constructs regardless of the length of the protein (34.2 kDa pPI1E, 63.9 kDa pPI1D, 65.0 kDa pPI2E, 87.6 kDa pPI3E, 94.8 kDa pPI2D, 117.3 kDa pPI3D, 132.0 kDa pPI3C, 139.3 kDa pPI2B, 145.5 kDa pPI4E, 207.3 kDa pPI1A, and 318.6 kDa pPIAggLb). In addition, the hybrid molecule pKP-Lb (314.2 kDa), consisting of the first part of the lactococcal aggL gene from Lc. lactis subsp. lactis BGKP1 (Kojic et al., 2011) as a PvuI-PstI fragment and a second part of the lactobacilli aggLb gene from L. paracasei subsp. paracasei BGNJ1-64 as a PstI-SalI fragment, was constructed (Figure 1D; Table 1). All of the variants constructed were used for functional assays in order to determine the role of various domains of the AggLb aggregation protein. The correct in-frame joining of all the fragments was confirmed by DNA sequencing and expression analysis using a Dot blot (Figure 2; Supplementary Figure 2).
Auto-Aggregation Ability of Transformants Carrying Different Variants of the aggLb Gene
The auto-aggregation ability of the wild-type strain and of the derivatives harboring the different variants of aggLb in the Lc. lactis subsp. lactis BGKP1-20 (see above) was measured for a period of 5 h, and the results are presented in Supplementary Table 1. We concluded that only the constructs carrying all six collagen binding domains and the first two CnaB-like domains were able to strongly auto-aggregate (BGKP1-20/pPI4E; Figure 3; Supplementary Table 1). Alternatively, the absence of the other CnaB-like domains, did not cause a significant effect on auto-aggregation (BGKP1-20/pPI3C, BGKP1-20/pPI3D, BGKP1-20/pPI3E, BGKP1-20/pPI2B, BGKP1-20/pPI2D, BGKP1-20/pPI2E, BGKP1-20/pPI1A, BGKP1-20/pPI1D, and BGKP1-20/pPI1E; Figure 3; Supplementary Table 1). It is also interesting to note that an additive effect dependent on the number of collagen binding domains on auto-aggregation was not linear, indicating that individual collagen binding domains do not have the same contribution. Careful observation revealed that the derivatives BGKP1-20/pPI2E, BGKP1-20/pPI1A, and BGKP1-20/pPI1E formed small aggregates (resembling sand or dust) that did not contribute to the rapid aggregation of the cells. Nevertheless, a negligible level of aggregation that was visible after overnight growth in a test tube was often observed in our collection of LAB. This observation may indicate a relationship between the type and number of collagen binding domains and/or CnaB-like domains within the aggregation factor(s) and the level or types of auto-aggregation. It was, therefore, concluded that the auto-aggregation ability of strains/derivatives was directly dependent on the collagen binding domains, while the 18 C-terminal CnaB-like domains were not required for auto-aggregation. Transformants of Lc. lactis subsp. lactis BGKP1-20 carrying the hybrid construct pKP-Lb composed of the first part of the aggL gene (carrying three collagen binding domains originating from the Lc. lactis subsp. lactis BGKP1) and the second part of the aggLb gene were unable to form big aggregates, which indicated that the resulting hybrid molecule was not functional in strong auto-aggregation, collagen, or fibronectin binding (BGKP1-20/pKP-Lb; Figures 3–5) as wild-type strains (L. paracasei subsp. paracasei BGNJ1-64 and/or Lc. lactis subsp. lactis BGKP1).
FIGURE 3. Comparison of the auto-aggregation ability of the wild-type strain and of derivatives harboring the different aggLb variants in Lc. lactis subsp. lactis BGKP1-20 strain after 5 h incubation at 30°C. Auto-aggregation ability is expressed as percentages. The error bars represent standard deviations of three independent observations.
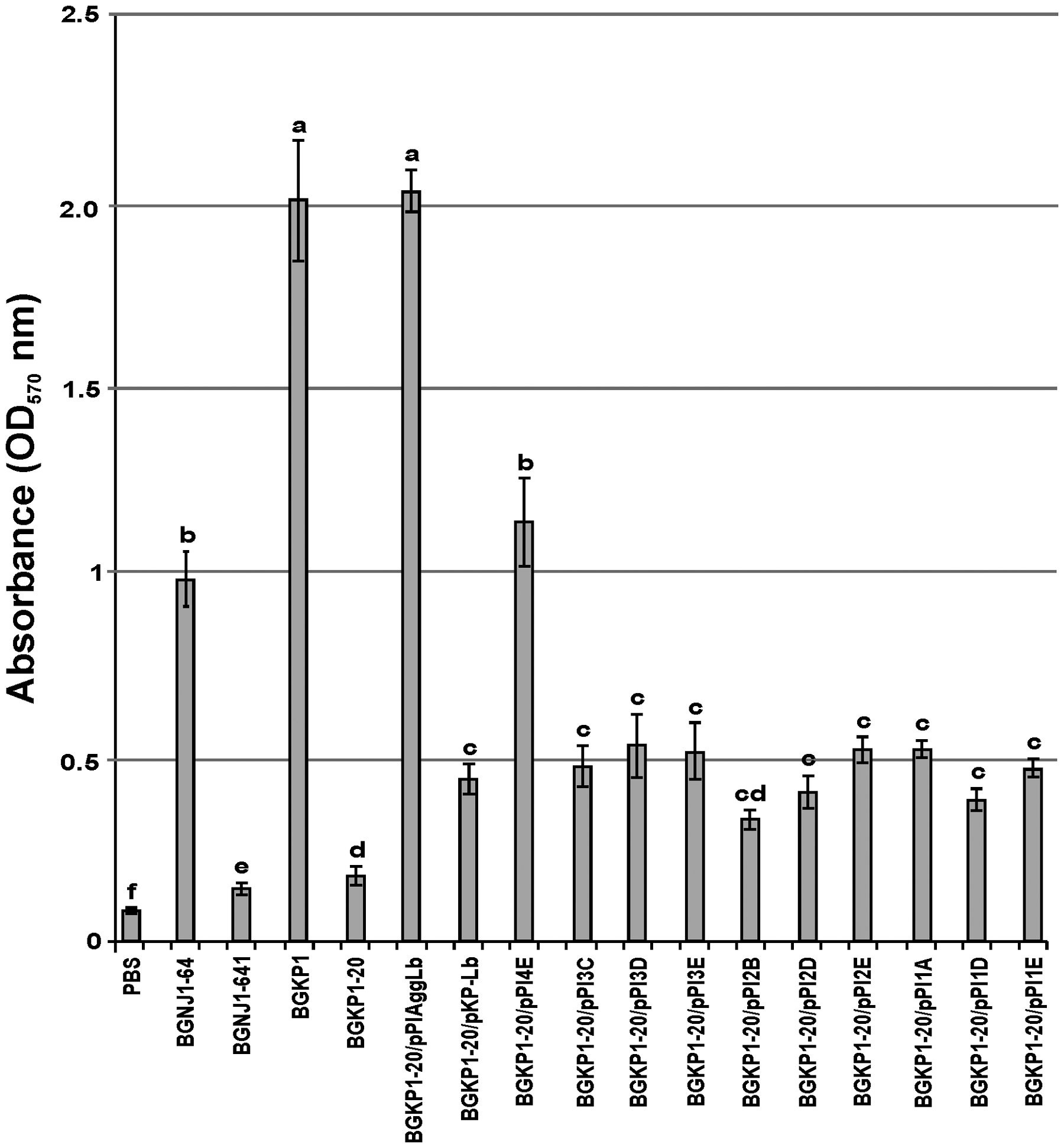
FIGURE 4. Graphical presentation of results obtained in collagen-binding assay of selected strains and derivatives to immobilized collagen in microtiter plates. Results were expressed as average of normalized A570 values. The error bars show the standard deviations. In each column, the values with different superscript letters differ significantly (p < 0.001).
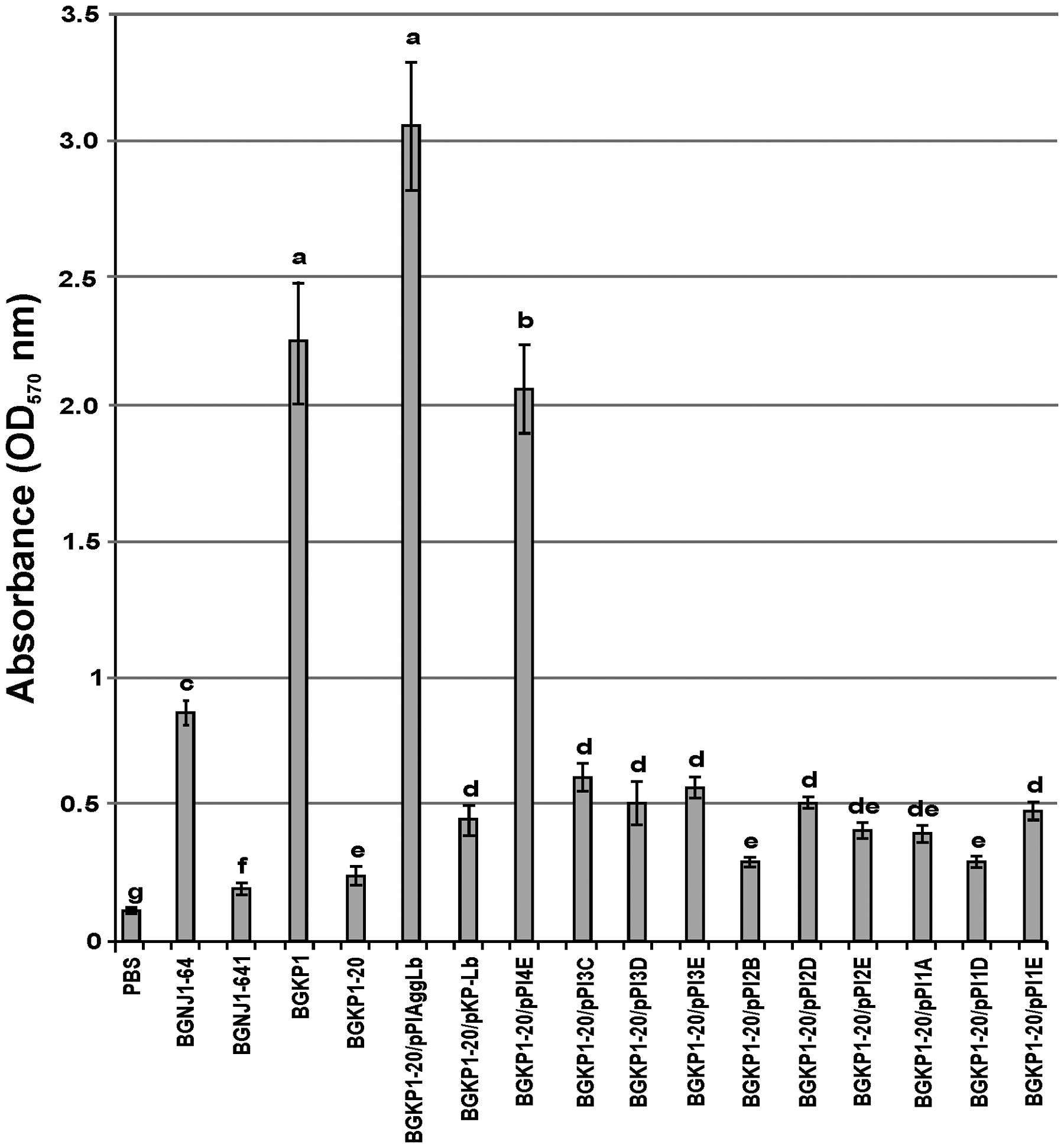
FIGURE 5. Graphical presentation of results obtained in fibronectin-binding assay of selected strains and derivatives to immobilized fibronectin in microtiter plates. Results were expressed as average of normalized A570 values. The error bars show the standard deviations. In each column, the values with different superscript letters differ significantly (p < 0.001).
Collagen and Fibronectin Binding Ability of the Transformants Carrying Different Variants of the aggLb Gene
In our previous studies, we found that isolates carrying the aggL or aggLb genes exhibited a direct correlation between auto-aggregation and their collagen binding ability (Miljkovic et al., 2015). All domain variants of the aggLb gene constructed in this study were tested for the ability to bind to collagen and fibronectin. Transformants carrying the different constructs adhered to immobilized collagen (Figure 4) and fibronectin (Figure 5) to different extents. Significant differences in the adherence to immobilized collagen and fibronectin were apparent between aggregation-positive strains (L. paracasei subsp. paracasei BGNJ1-64 and Lc. lactis subsp. lactis BGKP1) and their aggregation-negative derivatives (L. paracasei subsp. paracasei BGNJ1-641 and Lc. lactis subsp. lactis BGKP1-20) and also between strains carrying the first part of the aggLb gene (consisting of six collagen binding domains and the first two CnaB-like domains; BGKP1-20/pPIAggLb, BGKP1-20/pPI4E) and those variants that had only two or fewer collagen binding domains; these results indicate a role of the collagen binding domains in the interaction with collagen and fibronectin, but the last 18 CnaB-like domains are not indispensable (Figures 4 and 5). As observed in other experiments reported in this study (see above), we noticed that the additive effect dependent on the number of collagen binding domains was much lower than the impact of the specific collagen binding domains (II, III, and IV). The specific binding of AggLb to collagen and fibronectin was dependent on the collagen binding domains in a manner similar to the auto-aggregation ability. It appears that all the three phenotypes (auto-aggregation, collagen and fibronectin binding) are determined by the presence of the same structures of the AggLb protein such as the collagen binding domains.
Biofilm Formation of the Transformants Carrying Different Variants of the aggLb Gene
We determined the role of the AggLb in biofilm formation. Its ability to form biofilms was tested in the wild-type strain, aggregation deficient derivatives and transformants carrying different variants of aggLb using the adherence of the cells to the surfaces of microtiter plates. The strongest biofilm formation was observed for the transformant carrying the construct pPI2D, followed by pPI3C, pPI3D, and finally, pPI3E (Figure 6). A comparative analysis of the variants led to the conclusion that the biofilm formation ability has a negative correlation with auto-aggregation, collagen, and fibronectin binding. It appears that the presence of collagen binding domains determines the formation of certain structures on AggLb that play a role in the interaction with collagen and fibronectin, but simultaneously enable the cells to auto-aggregate (pPI4E). Most likely, the absence of the collagen-binding domain (especially II, III, and IV) allows other structures to come to the fore (i.e., they are unmasked) which promotes biofilm formation. The difference between pPI1D and pPI2D is limited to the presence of a sixth collagen binding domain of AggLb in pPI2D (Figure 1; Table 2); thus, this result indicates that this domain is probably required in combination with the other domain(s) to allow biofilm formation.
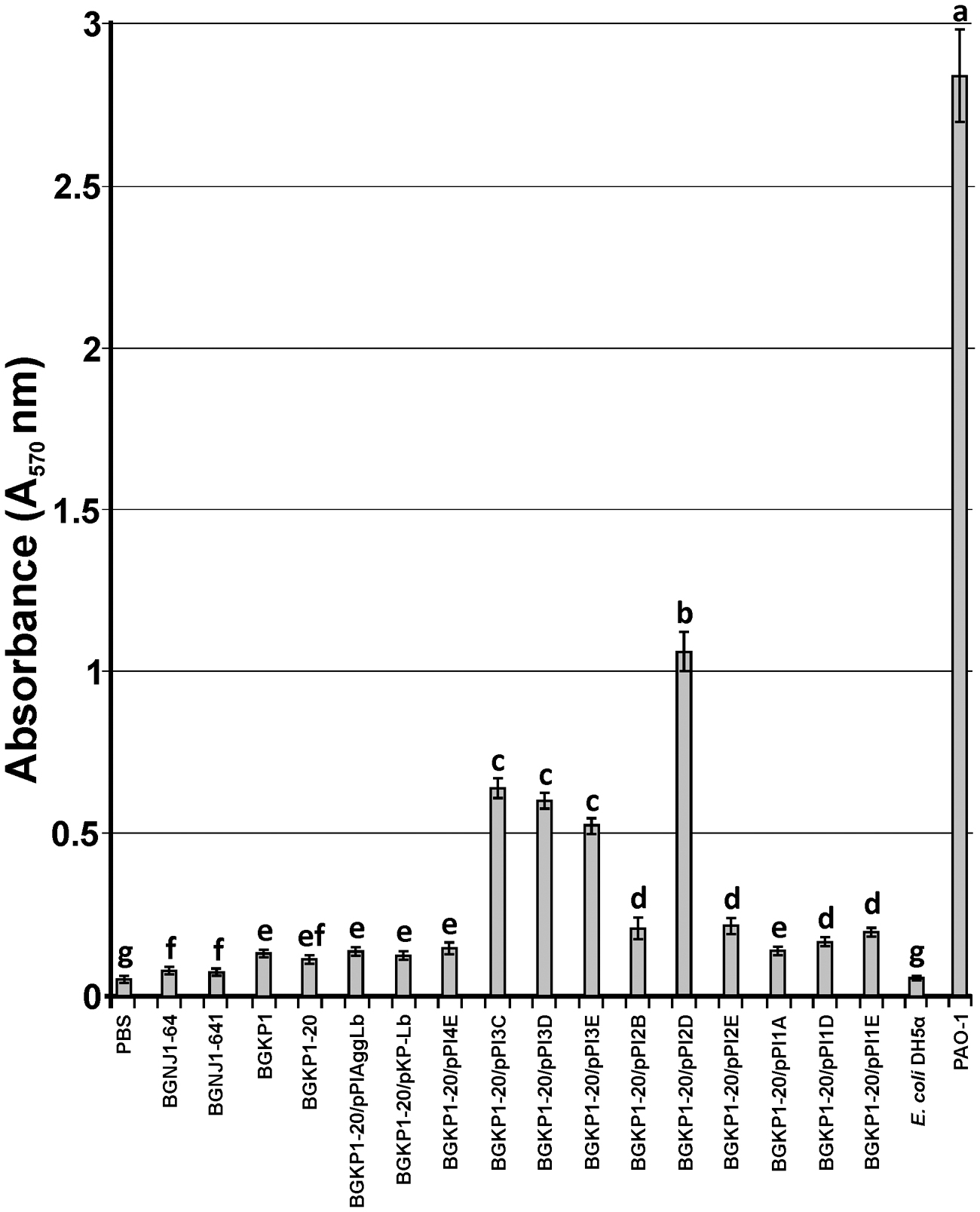
FIGURE 6. Graphical presentation of results obtained in biofilm formation assay of selected strains and derivatives (including control strains) to form biofilms in microtiter plates. Results were expressed as average of normalized A570 values. In each column, the values with different superscript letters differ significantly (p < 0.05).
Relationships between Collagen/Fibronectin Binding and Biofilm Ability of Transformants Carrying Different Variants of the aggLb Gene
We established correlations between auto-aggregation, collagen/fibronectin binding and biofilm formation ability of transformants carrying different variants of the aggLb gene. A comparative analysis of the variants led to the conclusion that the biofilm formation ability has a negative correlation with auto-aggregation – R2 squared 0.312 (Supplementary Figure 3A), binding to collagen – R2 squared 0.260 (Supplementary Figure 3B), binding to fibronectin – R2 squared 0.242 (Supplementary Figure 3C). In addition using Python 2.7.8 and scipy library (version 0.14.0) we proved positive correlation between auto-aggregation and collagen binding – R2 squared 0.652 (Supplementary Figure 3D) and aggregation and fibronectin binding – R2 squared 0.636 (Supplementary Figure 3E).
Discussion
The adhesion of lactic acid bacteria to epithelial and mucosal surfaces is thought to be a rather complex process involving many different factors (Buck et al., 2005). The ability of lactobacilli to aggregate has been linked to their role as probiotic factors (García-Cayuela et al., 2014). The data of the literature suggest that the Apf-like proteins may contribute to the survival of L. acidophilus during its transit through the digestive tract and, potentially, may participate in the interactions with the host intestinal mucosa (Goh and Klaenhammer, 2010). Considering the importance of aggregation phenomena for human health, the experiments described in this study were mainly focused to determine the contribution of the different domains and repeats of the AggLb protein on the modulation of the aggregation phenotype. Additionally, our results have proven the existence of a direct relationship between strong auto-aggregation, collagen or fibronectin binding and biofilm formation.
Biofilms of lactobacilli can be found in many natural environments (Lebeer et al., 2007). Because the gastrointestinal tract is an important target for probiotics, some factors related to this niche have been investigated in the past decade. It was of interest to study the possible relationship between aggregation ability and biofilm formation. It has been reported that the agglutination protein AggA is required for the aggregation and increased biofilm formation of a hyper-aggregating mutant of Shewanella oneidensis MR-1 (De Windt et al., 2006). An insertional mutant of aggA resulted in the loss of aggregation properties and ability to form a biofilm. Additionally, the SasC protein of a pathogenic S. aureus strain was involved in cell aggregation, biofilm formation and colonization during infection. The N-terminal domain of the SasC protein was involved in the production of large cell aggregates, in the attachment to polystyrene, and in increased biofilm formation (Schroeder et al., 2009). Aggregation and biofilm formation are multicellular processes that allow a community to be more resistant to stress conditions. Given that these are similar processes, it is not surprising that the same protein may be involved in both functions. Since biofilm formation is important in food spoilage and pathogenic bacteria because it results in high resistance to different treatments, it is important to identify and characterize the active components that could inhibit bacterial biofilm formation (Söderling et al., 2011; Furukawa, 2015).
The ability to strongly aggregate and adhere to collagen and fibronectin is inversely correlated with the biofilm formation, (if the ability to strongly aggregate and bind collagen and fibronectin is stronger the ability of biofilm formation is less; Figures 3–5; Supplementary Figure 3). Therefore, it seems that the lack of collagen binding domains II, III, and IV in the AggLb protein results in the reduced auto-aggregation, collagen and fibronectin binding and increases the propensity of the cells to form a biofilm. A comparative regression analysis of AggLb variants containing a constant number of CnaB-like domains and a different number of collagen binding domains (pPI4E, pPI3E, pPI2E, and pPI1E; pPI3D, pPI2D, and pPI1D; Figures 4 and 5) showed a correlation of binding to collagen or fibronectin, and an increase in biofilm formation (Supplementary Figure 3).
Our results indicate that the region responsible for the strong auto-aggregation, collagen and fibronectin binding is located on the N-terminus of the AggLb aggregation protein; transformants that carried the construct pPI4E, which contained only the N-terminal part, exhibited a strong aggregation capability, as did as clones that harbored the complete gene. Deletion studies of the AggLb protein showed that all three functions dependent on the collagen binding domains II, III, and IV, and their deletion leads to a complete loss of strong aggregation ability. These three domains are critical for function of AggLb in strong auto-aggregation, binding to collagen and fibronectin, either through direct and specific interaction with proteins of the matrix or by changing the properties of the cell surface. Multiple CnaB-like domains likely function as an antenna which exposes the collagen binding domains to the surface to improve target protein interactions. The CnaB-like domains in AggLb cannot be considered as the domains responsible for the direct interaction with collagen or fibronectin, but they can strengthen the interaction between the collagen binding domains and collagen or fibronectin. Also, we noted that because the first and last CnaB-like domains had sequence heterogeneity compared to the other 18 domains, it is possible they may have a different but not strong effect on AggLb function. We can conclude that the presence of the collagen binding domains predominantly determined the adhesive function of the AggLb protein. In addition, combination of domains from lactobacilli (AggLb) and lactococci (AggL; hybrid molecule – BGKP1–20/pKP-Lb) did not resulted in functional protein in strong auto-aggregation, collagen, or fibronectin binding. The results obtained in this study have demonstrated that a protein may exert different functions depending on physicochemical properties of the bacterial surfaces, and this probably depends on the structure and conformation variants of AggLb. The removal of certain domain(s) not only eliminated certain functions but also resulted in other domain(s) coming to the fore and allowing the protein to assume another function. In our previous publication we have noticed one strain BGGR2-68 that simultaneously exhibits both functions strong auto-aggregation and biofilm formation (Miljkovic et al., 2015). It would be interesting to determine whether these two functions in this strain are associated with one the same protein or independent. This will be the subject of further research.
These results bolster the hypothesis that in the S. aureus collagen-binding Cna protein, the collagen binding A region is responsible and sufficient for collagen binding, while the B region aids as a “stalk” that projects the A region from the bacterial surface to facilitate the bacterial adherence to collagen. Such a B region assembly could result in flexibility, stability, and positioning the ligand-binding A region away from the bacterial cell surface (Deivanayagam et al., 2000). The difference between AggLb and the Cna protein is that the aggregation promoting factor contains repetitive collagen binding domains (six very heterogeneous units with less than 26% identity) that have different contributions to strong auto-aggregation, collagen, and fibronectin binding (II, III, and IV showed the most significant effects), as well as to biofilm formation. It is important to note that even if AggLb is composed of two collagen binding domains, it is not able to provide strong auto-aggregation. In contrast in Cna, this is accomplished with a single domain, indicating that it is important which of the domains is/are present.
Author Contributions
MK conceived, designed, and coordinated this study, interpreted all of results and contributed to the preparation of the figures and wrote this paper. MM designed, performed, analyzed the experiments and wrote this paper. BJ and KN provided experimental assistance and contributed to the preparation of the figures. IB performed one part of experiments of production polyclonal antibody. DF and VV provided technical assistance and contributed to the preparation of this paper. All authors reviewed the results and approved the final version of the manuscript.
Funding
The Ministry of Education and Science of the Republic of Serbia, Republic of Serbia (Grant No. 173019), supported this work.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
The authors thank to the personal of Animal House of ICGEB for excellent technical assistance during immunization of animals and blood sampling.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01422
References
Ahmed, S., Meghji, S., Williams, R. J., Henderson, B., Brock, J. H., and Nair, S. P. (2001). Staphylococcus aureus fibronectin binding proteins are essential for internalization by osteoblasts but do not account for differences in intracellular levels of bacteria. Infect. Immun. 69, 2872–2877. doi: 10.1128/IAI.69.5.2872-2877.2001
Bolduc, G. R., and Madoff, L. C. (2007). The group B streptococcal alpha C protein binds alpha1beta1-integrin through a novel KTD motif that promotes internalization of GBS within human epithelial cells. Microbiology 153, 4039–4049. doi: 10.1099/mic.0.2007/009134-0
Boris, S., Suárez, J. E., and Barbés, C. (1997). Characterization of the aggregation promoting factor from Lactobacillus gasseri, a vaginal isolate. J. Appl. Microbiol. 83, 413–420. doi: 10.1046/j.1365-2672.1997.00250.x
Bron, P. A., Grangette, C., Mercenier, A., de Vos, W. M., and Kleerebezem, M. (2004). Identification of Lactobacillus plantarum genes that are induced in the gastrointestinal tract of mice. J. Bacteriol. 186, 5721–5729. doi: 10.1128/JB.186.17.5721-5729.2004
Buck, B. L., Altermann, E., Svingerud, T., and Klaenhammer, T. R. (2005). Functional analysis of putative adhesion factors in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 71, 8344–8351. doi: 10.1128/AEM.71.12.8344-8351.2005
Chen, X., Xu, J., Shuai, J., Chen, J., Zhang, Z., and Fang, W. (2007). The S-layer proteins of Lactobacillus crispatus strain ZJ001 is responsible for competitive exclusion against Escherichia coli O157:H7 and Salmonella typhimurium. Int. J. Food Microbiol. 115, 307–312. doi: 10.1016/j.ijfoodmicro.2006.11.007
De Windt, W., Gao, H., Kromer, W., van Damme, P., Dick, J., Mast, J., et al. (2006). AggA is required for aggregation and increased biofilm formation of a hyper-aggregating mutant of Shewanella oneidensis MR-1. Microbiology 152, 721–729. doi: 10.1099/mic.0.28204-0
Deivanayagam, C. C., Rich, R. L., Carson, M., Owens, R. T., Danthuluri, S., Bice, T., et al. (2000). Novel fold and assembly of the repetitive B region of the Staphylococcus aureus collagen-binding surface protein. Structure 15, 67–78. doi: 10.1016/S0969-2126(00)00081-2
Flock, J. I. (1999). Extracellular-matrix-binding proteins as targets for the prevention of Staphylococcus aureus infections. Mol. Med. Today 5, 532–537. doi: 10.1016/S1357-4310(99)01597-X
Furukawa, S. (2015). Studies on formation, control and application of biofilm formed by food related microorganisms. Biosci. Biotechnol. Biochem. 79, 1050–1056. doi: 10.1080/09168451.2015.1018126
Galli, D., Lottspeich, F., and Wirth, R. (1990). Sequence analysis of Enterococcus faecalis aggregation substance encoded by the sex pheromone plasmid pAD1. Mol. Microbiol. 4, 895–904. doi: 10.1111/j.1365-2958.1990.tb00662.x
García-Cayuela, T., Korany, A. M., Bustos, I., Gómez de Cadiñanos, L. P., Requena, T., Peláez, C., et al. (2014). Adhesion abilities of dairy Lactobacillus plantarum strains showing an aggregation phenotype. Food Res. Int. 57, 44–50. doi: 10.1016/j.foodres.2014.01.010
Gasson, M. J. (1983). Plasmid complements of Streptococcus lactis NCDO712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154, 1–9.
Giraffa, G. (2014). Lactobacillus helveticus: importance in food and health. Front. Microbiol. 5:338. doi: 10.3389/fmicb.2014.00338
Goh, Y. J., and Klaenhammer, T. R. (2010). Functional roles of aggregation-promoting-like factor in stress tolerance and adherence of Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 76, 5005–5012. doi: 10.1128/AEM.00030-10
Hanahan, D. (1983). Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580. doi: 10.1016/S0022-2836(83)80284-8
Holo, H., and Nes, I. F. (1989). High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 55, 3119–3123.
Hungin, A. P., Mulligan, C., Pot, B., Whorwell, P., Agreus, L., Fracasso, P., et al. (2013). Systematic review: probiotics in the management of lower gastrointestinal symptoms in clinical practice-an evidence-based international guide. Aliment. Pharmacol. Ther. 38, 864–886. doi: 10.1111/apt.12460
Hymes, J. P., Johnson, B. R., Barrangou, R., and Klaenhammer, T. R. (2016). Functional analysis of an S-layer-associated fibronectin-binding protein in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 82, 2676–2685. doi: 10.1128/AEM.00024-16
Johnson, B. R., and Klaenhammer, T. R. (2014). Impact of genomics on the field of probiotic research: historical perspectives to modern paradigms. Antonie Van Leeuwenhoek 106, 141–156. doi: 10.1007/s10482-014-0171-y
Josefsson, E., Higgins, J., Foster, T. J., and Tarkowski, A. (2008). Fibrinogen binding sites P336 and Y338 of clumping factor a are crucial for Staphylococcus aureus virulence. PLoS ONE 3:e2206. doi: 10.1371/journal.pone.0002206
Kleerebezem, M., Hols, P., Bernard, E., Rolain, T., Zhou, M., Siezen, R. J., et al. (2010). The extracellular biology of the lactobacilli. FEMS Microbiol. Rev. 34, 199–230. doi: 10.1111/j.1574-6976.2010.00208.x
Kojic, M., Jovcic, B., Strahinic, I., Begovic, J., Lozo, J., Veljovic, K., et al. (2011). Cloning and expression of novel lactococcal aggregation factor from Lactococcus lactis subsp. lactis BGKP1. BMC Microbiol. 11:265. doi: 10.1186/1471-2180-11-265
Kwiecinski, J., Jin, T., and Josefsson, E. (2014). Surface proteins of Staphylococcus aureus play an important role in experimental skin infection. APMIS 122, 1240–1250. doi: 10.1111/apm.12295
Lebeer, S., Vanderleyden, J., and De Keersmaecker, S. C. J. (2008). Genes and molecules of lactobacilli supporting probiotic action. Microbiol. Mol. Biol. Rev. 72, 728–764. doi: 10.1128/MMBR.00017-08
Lebeer, S., Verhoeven, T. L. A., Velez, M. P., Vanderleyden, J., and De Keersmaecker, S. C. J. (2007). Impact of environmental and genetic factors on biofilm formation by the probiotic strain Lactobacillus rhamnosus GG. Appl. Environ. Microbiol. 73, 6768–6775. doi: 10.1128/AEM.01393-07
Lorca, G., Torino, M. I., Font de Valdez, G., and Ljungh, A. A. (2002). Lactobacilli express cell surface proteins which mediate binding of immobilized collagen and fibronectin. FEMS Microbiol. Lett. 206, 31–37. doi: 10.1111/j.1574-6968.2002.tb10982.x
Miljkovic, M., Strahinic, I., Tolinacki, M., Zivkovic, M., Kojic, S., Golic, N., et al. (2015). AggLb is the largest cell-aggregation factor from Lactobacillus paracasei subsp. paracasei BGNJ1-64, functions in collagen adhesion, and pathogen exclusion in vitro. PLoS ONE. 10:e0126387. doi: 10.1371/journal.pone.0126387
Muñoz-Provencio, D., Pérez-Martínez, G., and Monedero, V. (2010). Characterization of a fibronectin-binding protein from Lactobacillus casei BL23. J. Appl. Microbiol. 108, 1050–1059. doi: 10.1111/j.1365-2672.2009.04508.x
Nazzaro, F., Fratianni, F., Nicolaus, B., Poli, A., and Orlando, P. (2012). The prebiotic source influences the growth, biochemical features and survival under simulated gastrointestinal conditions of the probiotic Lactobacillus acidophilus. Anaerobe 18, 280–285. doi: 10.1016/j.anaerobe.2012.03.002
Niedergang, F., Sirard, J. C., Blanc, C. T., and Kraehenbuhl, J. P. (2000). Entry and survival of Salmonella typhimurium in dendritic cells and presentation of recombinant antigens do not require macrophage-specific virulence factors. Proc. Natl. Acad. Sci. U.S.A. 97, 14650–14655. doi: 10.1073/pnas.97.26.14650
Oehmcke, S., Shannon, O., Mörgelin, M., and Herwald, H. (2010). Streptococcal M proteins and their role as virulence determinants. Clin. Chim. Acta 411, 1172–1180. doi: 10.1016/j.cca.2010.04.032
Peter, A., Zacharia, S., and Mathew, J. (2013). Biofilm formation in enterococci from different source. Int. J. Biopharm 4, 140–144.
Salminen, S., von Wright, A., Morelli, L., Marteau, P., Brassart, D., de Vos, W. M., et al. (1998). Demonstration of safety of probiotics - a review. Int. J. Food Microbiol. 44, 93–106. doi: 10.1016/S0168-1605(98)00128-7
Sanders, M. E., Guarner, F., Guerrant, R., Holt, P. R., Quigley, E. M., Sartor, R. B., et al. (2013). An update on the use and investigation of probiotics in health and disease. Gut 62, 787–796. doi: 10.1136/gutjnl-2012-302504
Schroeder, K., Jularic, M., Horsburgh, S. M., Hirschhausen, N., Neumann, C., Bertling, A., et al. (2009). Molecular characterization of a novel Staphylococcus aureus surface protein (SasC) involved in cell aggregation and biofilm accumulation. PLoS ONE 4:e7567. doi: 10.1371/journal.pone.0007567
Sisto, A., and Lavermicocca, P. (2012). Suitability of a probiotic Lactobacillus paracasei strain as a starter culture in olive fermentation and development of the innovative patented product “probiotic table olives”. Front. Microbiol. 3:174. doi: 10.3389/fmicb.2012.00174
Skrzypczak, K., Gustaw, W., and Waśko, A. (2015). Health-promoting properties exhibited by Lactobacillus helveticus strains. Acta Biochim. Pol. 62, 713–720. doi: 10.18388/abp.2015_1116
Söderling, E. M., Marttinen, A. M., and Haukioja, A. L. (2011). Probiotic lactobacilli interfere with Streptococcus mutans biofilm formation in vitro. Curr. Microbiol. 62, 618–622. doi: 10.1007/s00284-010-9752-9
Styriak, I., Nemcova, R., Chang, Y. H., and Ljungh, A. (2003). Binding of extracellular matrix molecules by probiotic,bacteria. Lett. Appl. Microbiol. 37, 329–333. doi: 10.1046/j.1472-765X.2003.01402.x
Taglialegna, A., Navarro, S., Ventura, S., Garnett, J. A., Matthews, S., Penades, J. R., et al. (2016). Staphylococcal Bap proteins build amyloid scaffold biofilm matrices in response to environmental signals. PLoS Pathog. 12:e1005711. doi: 10.1371/journal.ppat.1005711
Uzelac, G., Miljkovic, M., Lozo, J., Radulovic, Z., Tosic, N., and Kojic, M. (2015). Expression of bacteriocin LsbB is dependent on a transcription terminator. Microbiol. Res. 179, 45–53. doi: 10.1016/j.micres.2015.06.011
Waśko, A., Polak-Berecka, M., Paduch, R., and Jóźwiak, K. (2014). The effect of moonlighting proteins on the adhesion and aggregation ability of Lactobacillus helveticus. Anaerobe 30, 161–168. doi: 10.1016/j.anaerobe.2014.10.002
Yadava, A. K., Tyagia, A., Kaushika, J. K., Saklanib, A. C., Grovera, S., and Batish, V. K. (2013). Role of surface layer collagen binding protein from indigenous Lactobacillus plantarum 91 in adhesion and its anti-adhesion potential against gut pathogen. Microbiol. Res. 168, 639–645. doi: 10.1016/j.micres.2013.05.003
Keywords: AggLb, collagen binding domains, CnaB-like domains, auto-aggregation, biofilm formation
Citation: Miljkovic M, Bertani I, Fira D, Jovcic B, Novovic K, Venturi V and Kojic M (2016) Shortening of the Lactobacillus paracasei subsp. paracasei BGNJ1-64 AggLb Protein Switches Its Activity from Auto-aggregation to Biofilm Formation. Front. Microbiol. 7:1422. doi: 10.3389/fmicb.2016.01422
Received: 29 March 2016; Accepted: 26 August 2016;
Published: 08 September 2016.
Edited by:
Aldo Corsetti, University of Teramo, ItalyReviewed by:
Giuseppe Spano, University of Foggia, ItalyMichael Gänzle, University of Alberta, Canada
Andrea Gomez-Zavaglia, Center for Research and Development in Food Cryotechnology (CIDCA, CONICET), Argentina
Copyright © 2016 Miljkovic, Bertani, Fira, Jovcic, Novovic, Venturi and Kojic. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Milan Kojic, bWtvamljQGltZ2dlLmJnLmFjLnJz
 Marija Miljkovic
Marija Miljkovic Iris Bertani2
Iris Bertani2 Djordje Fira
Djordje Fira Branko Jovcic
Branko Jovcic Katarina Novovic
Katarina Novovic Vittorio Venturi
Vittorio Venturi Milan Kojic
Milan Kojic