- 1Medical Mycology Research Center, Chiba University, Chiba, Japan
- 2Faculdade de Ciências Farmacêuticas de Ribeirão Preto, Universidade de São Paulo, Ribeirão Preto, Brazil
Invasive aspergillosis is a life-threatening mycosis caused by the pathogenic fungus Aspergillus. The predominant causal species is Aspergillus fumigatus, and azole drugs are the treatment of choice. Azole drugs approved for clinical use include itraconazole, voriconazole, posaconazole, and the recently added isavuconazole. However, epidemiological research has indicated that the prevalence of azole-resistant A. fumigatus isolates has increased significantly over the last decade. What is worse is that azole-resistant strains are likely to have emerged not only in response to long-term drug treatment but also because of exposure to azole fungicides in the environment. Resistance mechanisms include amino acid substitutions in the target Cyp51A protein, tandem repeat sequence insertions at the cyp51A promoter, and overexpression of the ABC transporter Cdr1B. Environmental azole-resistant strains harboring the association of a tandem repeat sequence and punctual mutation of the Cyp51A gene (TR34/L98H and TR46/Y121F/T289A) have become widely disseminated across the world within a short time period. The epidemiological data also suggests that the number of Aspergillus spp. other than A. fumigatus isolated has risen. Some non-fumigatus species intrinsically show low susceptibility to azole drugs, imposing the need for accurate identification, and drug susceptibility testing in most clinical cases. Currently, our knowledge of azole resistance mechanisms in non-fumigatus Aspergillus species such as A. flavus, A. niger, A. tubingensis, A. terreus, A. fischeri, A. lentulus, A. udagawae, and A. calidoustus is limited. In this review, we present recent advances in our understanding of azole resistance mechanisms particularly in A. fumigatus. We then provide an overview of the genome sequences of non-fumigatus species, focusing on the proteins related to azole resistance mechanisms.
Introduction
The incidence of fungal infection has increased over the past three decades (Denning, 1998; Dasbach et al., 2000; Kousha et al., 2011; Suzuki et al., 2013; Bitar et al., 2014). This is largely due to the increased number of patients at risk who have received hematopoietic stem cell or solid organ transplantation and immunosuppressive therapy. Although new antifungals have been developed, fungal infections remain a threat to human health. Among filamentous fungal infections, those caused by Aspergillus species are the most common. The mortality and morbidity associated with such infections are relatively high, and the predominant causal agent is Aspergillus fumigatus (Steinbach et al., 2012). Accordingly, this fungus is regarded as a model pathogen to study many aspects of aspergillosis, such as fungal virulence factors, immune responses, pathology, and drug efficacy. The genome sequence of A. fumigatus was determined in 2005 (Nierman et al., 2005), which accelerated our understanding of the molecular mechanisms underlying its pathogenicity (Kwon-Chung and Sugui, 2013; Perez-Nadales et al., 2014; Brown and Goldman, 2016). However, there is still much to be elucidated and the issue of drug resistance has only emerged during the past decade.
The antifungal drugs currently available for the treatment of aspergillosis fall into four categories: pyrimidine, echinocandin, polyene, and azole drugs. Among these, azoles are the first choice drugs in the management and prophylaxis of aspergillosis. Since the first report of an azole-resistant A. fumigatus strain in 1997 (Denning et al., 1997), resistant isolates have been detected with increasing incidence worldwide (Chen et al., 2005; Verweij et al., 2007; Howard et al., 2009; van der Linden et al., 2015). It is now widely accepted that azole resistance can develop upon prolonged exposure to azoles at a sub-lethal concentration during the therapy of patients with aspergillosis (particularly with chronic aspergillosis), which is supported by the findings of several studies (Howard et al., 2009; Tashiro et al., 2012; Hagiwara et al., 2014). In addition, environmentally-derived resistance mutations have emerged as a major cause of resistance among strains over the last decade (Snelders et al., 2008; Chowdhary et al., 2013). These mutations involve a combination of tandem repeats (TR) in the cyp51A promoter region and amino acid substitution(s) (TR34/L98H and TR46/Y121F/T289A). The strains harboring such resistance mutations were prevalent among both clinical and environmental settings. As azole resistance is correlated with aspergillosis treatment failure (Howard et al., 2009), a number of studies have focused on the epidemiology, molecular mechanisms, and diagnostic methods relating to this type of resistance. Another emerging issue in the pathology of aspergillosis is that non-fumigatus species are being increasingly identified as causal agents of invasive aspergillosis; these include A. flavus, A. niger, A. terreus, and A. calidoustus as well as other Aspergillus species that belong to Aspergillus section Fumigati (Baddley et al., 2009; Balajee et al., 2009; Krishnan et al., 2009; Tashiro et al., 2011). These species show variable drug susceptibility profiles, imposing the need for clinicians to perform accurate identification and drug susceptibility testing of isolates. Conventional culture methods do not provide an adequate level of specificity and/or sensitivity for accurate diagnosis, making species identification and determination of azole-resistance mutations in the cultured isolates a major challenge. The lack of accurate diagnostic techniques also affects management of patients with aspergillosis caused by azole-resistant Aspergillus (Verweij et al., 2015).
Recent advances in DNA sequencing technology have yielded several genomes of Aspergillus pathogens and the subsequent detection of sequence variations associated with azole resistance mechanisms. Taken together with epidemiological data, genomic approaches are increasing our understanding of emerging issues in Aspergillus pathogenesis. This review aims to summarize the recent findings on azole resistance in A. fumigatus, as well as in other related Aspergillus pathogens.
How Widely has Azole-Resistant Aspergillus Fungi Spread?
Epidemiology of Clinically Isolated Aspergillus Fungi
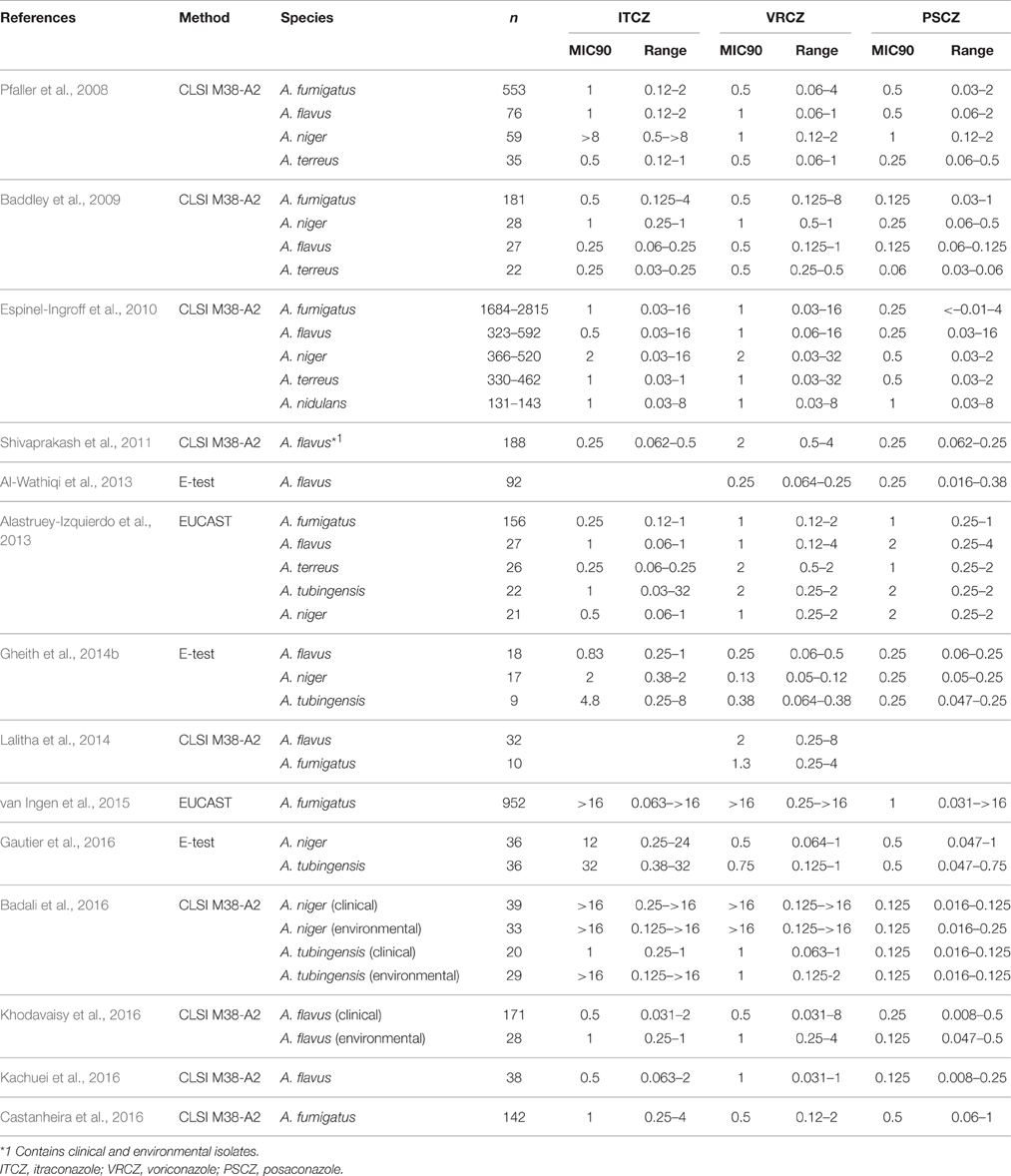
The genus Aspergillus comprises 344 species (Samson et al., 2014), and several species have been reported to be pathogenic in humans and animals. It is clear from epidemiological data that A. fumigatus is the predominant etiological agent isolated from immune-compromised patients. The second leading cause of aspergillosis is reportedly either A. flavus or A. niger. Due to the different sources of isolation (country, region, hospital, or ward), and the different clinical manifestations, comparing the frequencies of isolation among different studies is difficult. However, to gain a generalized overview or a trend of the incidence rates, epidemiological data collected from multicenter studies are valuable. Recently, epidemiology data for invasive aspergillosis (IA) was reported, which included a total of 563 patients from 30 intensive care units (ICUs) in eight countries (Taccone et al., 2015). In this study, A. fumigatus was the most commonly isolated fungus (n = 512, 92%), followed by A. flavus (n = 19, 3%) and A. niger (n = 7, 1%). In a 1-year (April 2011 to April 2012) prospective multicenter (18 Belgian hospitals) cohort study, 192 isolates of the A. fumigatus complex (87.3%), 13 of the A. flavus complex (5.9%), and 10 of the A. niger complex (4.5%) were isolated (Vermeulen et al., 2015). In another study of 29 Spanish hospitals (two periods; October 2010 and May 2011), among 278 Aspergillus isolates, 156 were identified as A. fumigatus (56.1%), 27 were A. flavus (9.7%), 26 were A. terreus (9.4%), 22 were A. tubingensis (7.9%), and 21 were A. niger (7.6%; Alastruey-Izquierdo et al., 2013). Prospective cohort studies of transplant-associated fungal infections in the United States carried out by the Transplant-Associated Infection Surveillance Network (TRANSNET) have also revealed that A. fumigatus is the leading cause of aspergillosis, followed by A. flavus, A. niger, and A. terreus (Kontoyiannis et al., 2010; Pappas et al., 2010). Analyzing these data collectively, it is clear that A. fumigatus is the predominant causative agent of aspergillosis, but several non-fumigatus species were also isolated from patients.
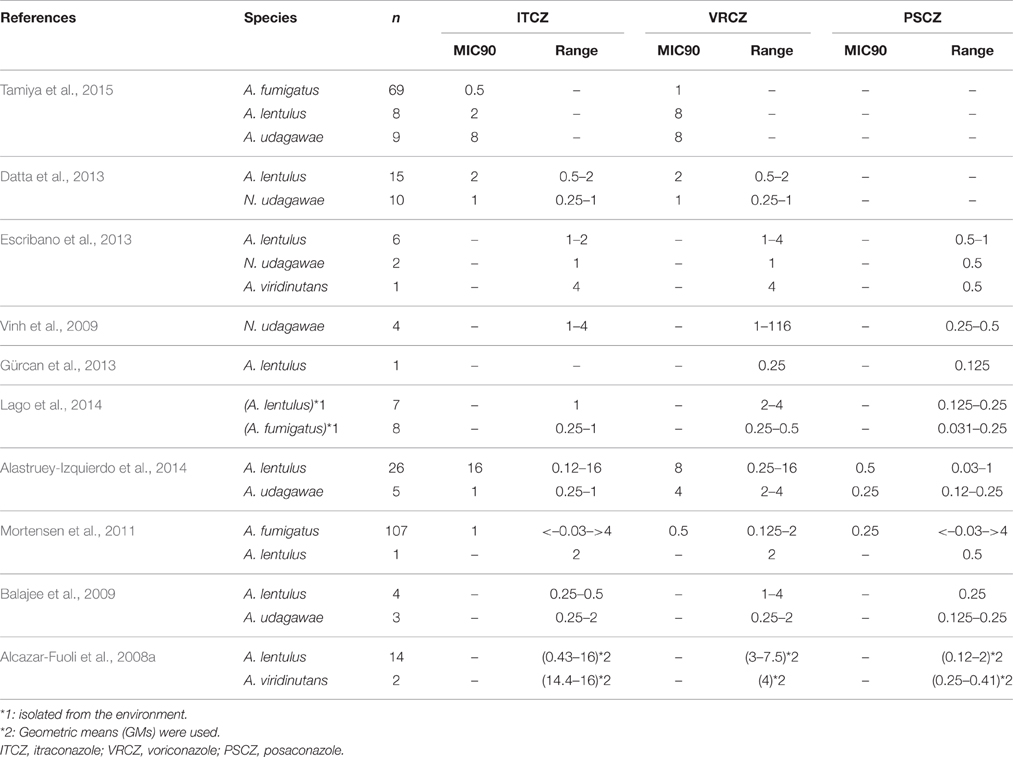
Recently, cryptic species of Aspergilli have received much attention owing to advances in the molecular tools for identification. Species that cannot be morphologically distinguished from the leading pathogen of their section are defined as “cryptic” species. Aspergillus section Nigri, whose members are known as the black aspergilli, is represented by A. niger, and includes more than 15 species, including A. tubingensis (Abarca et al., 2004). Within the section Nigri, A. niger, and A. tubingensis are the most common etiological agents of otomycosis, onchomycosis, pulmonary aspergillosis, and aspergilloma (Pappas et al., 2010; Alastruey-Izquierdo et al., 2014; Gheith et al., 2014a; Gautier et al., 2016). Aspergillus section Fumigati, comprising more than 60 species, also has important clinical implications, with 15 of its species having been reported to be isolated in clinical specimens (Alcazar-Fuoli et al., 2008a; Alastruey-Izquierdo et al., 2014; Lamoth, 2016). Misidentification within the section Fumigati has been increasingly reported, with A. lentulus, A. viridinutans, A. fumigatiaffinis, A. fumisynnematus, A. pseudofischeri, A. hiratsukae, and A. udagawae frequently being reported as A. fumigatus (Balajee et al., 2005a,b, 2006; Howard, 2014). Indeed, recent reports demonstrated that the cryptic species including A. lentulus and A. udagawae accounted for 3–6% of the collection of Aspergillus section Fumigati isolates (Balajee et al., 2009; Alastruey-Izquierdo et al., 2013; Escribano et al., 2013). These cryptic species in section Nigri and section Fumigati sometimes show different drug susceptibility profiles and different levels of pathogenicity from those of A. fumigatus (Vinh et al., 2009; Coelho et al., 2011; Alastruey-Izquierdo et al., 2014).
Regarding azole resistance, Aspergillus calidoustus, belonging to section Usti, should be noted, as it shows intrinsic pan-azole resistance. Prior to A. calidoustus being identified by specific sequencing as the only pathogenic species in this section, it was often reported as Aspergillus ustus (Varga et al., 2008). The incidence of infection caused by A. calidoustus was increased in transplant patients under azole prophylaxis (Egli et al., 2012). The TRANSNET study showed that A. calidoustus accounted for 2.7% (6/218) of Aspergillus species isolates (Balajee et al., 2009), while a population-based survey performed in Spain found 1.4% of the 278 Aspergillus species isolates were A. calidoustus (Alastruey-Izquierdo et al., 2013). According to these studies, some of these intrinsically azole-resistant A. calidoustus strains emerged in the setting of invasive aspergillosis. The growing recognition of azole-resistant cryptic species highlights the clinical need for full and accurate identification and susceptibility testing. In parallel, more focused studies are required to develop a better understanding of these species in the future.
Prevalence of Azole-Resistant Aspergillus Species
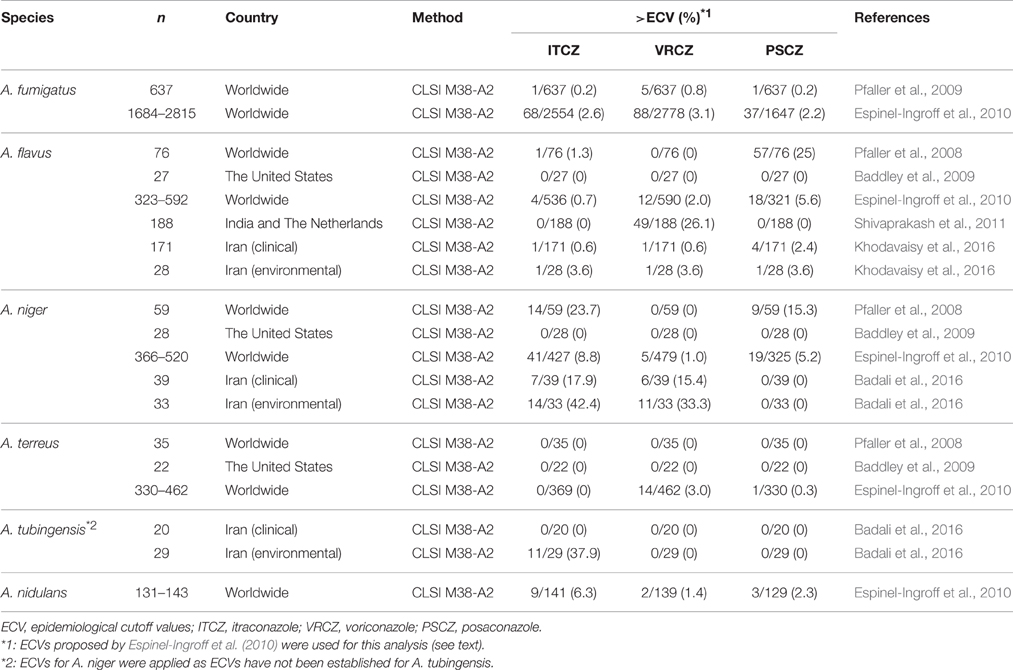
The predominant pathogen among Aspergillus species, A. fumigatus, is intrinsically susceptible to medical azoles. The epidemiological cutoff values (ECVs) for three triazoles, determined by Clinical and Laboratory Standards Institute broth microdilution (CLSI BMD) methods, have been proposed by Pfaller et al. (2009): itraconazole (1 mg/L), voriconazole (1 mg/L), and posaconazole (0.25 mg/L). They showed that 0.2, 0.2, and 0.8% of isolates in a large collection of Aspergillus species (n = 637) had itraconazole, voriconazole, and posaconazole minimum inhibitory concentrations (MICs) above the ECVs. In a subsequent study (n = 1647–2778) by Espinel-Ingroff et al. (2010), the ECVs for A. fumigatus were determined by CLSI method as itraconazole (1 mg/L), voriconazole (1 mg/L), and posaconazole (0.5 mg/L). By these criteria, the rates of isolates with itraconazole, voriconazole, and posaconazole MICs outside of the ECVs were 2.6, 3.1, and 2.2%, respectively. The sample size and regional differences in the collections might affect the prevalence of azole-resistant isolates. This study also proposed the ECVs (itraconazole, voriconazole, and posaconazole) for other Aspergillus species including A. flavus (1, 1, and 0.25 mg/L), A. terreus (1, 1, and 0.5 mg/L), A. niger (2, 2, and 0.5 mg/L), and A. nidulans (1, 2, and 1 mg/L; Espinel-Ingroff et al., 2010). These ECV data could help to characterize Aspergillus isolates and to monitor the emergence of azole-resistant strains by in vitro antifungal susceptibility testing with CLSI BMD method.
Over the past decade, ongoing azole resistance surveillance reports have been published by several research groups from different countries. A summary of recent reports on the surveillance of major Aspergillus species is shown in Table 1. The prevalence of azole-resistant A. fumigatus and A. terreus strains appears to be largely consistent between the studies. Based on the ECVs determined in the study by Espinel-Ingroff et al. (2010), the prevalence rates for azole-resistant isolates determined using the CLSI method in these studies are listed in Table 2. In these studies, 0.2–2.6, 0.8–3.1, and 0.2–2.2% of A. fumigatus isolates were resistant to itraconazole, voriconazole, and posaconazole, respectively, while 0, 0–3.0, and 0–0.3% of A. terreus isolates were resistant to itraconazole, voriconazole, and posaconazole, respectively (Table 2). It is notable that A. flavus, A. niger, and A. tubingensis showed variations in the prevalence of azole resistance between the studies. In particular, high rates of resistance to itraconazole were demonstrated amongst A. niger strains, suggesting intrinsic resistance to itraconazole in this species. However, this remains controversial as the effects of regional and individual laboratory conditional differences between the studies cannot be ruled out. Thus, further studies are necessary to draw definite conclusions.
In addition to the non-fumigatus species, the MICs for azoles were determined in some species of Aspergillus section Fumigati (Table 3). Isolates of A. lentulus, A. udagawae, and A. viridinutans with high MICs (>1 mg/L) for itraconazole and/or voriconazole were detected in most of these studies. Although these data were suggestive of intrinsic resistance to the azoles in these species, it should not be overlooked that the susceptibilities were variable among isolates. It is particularly noteworthy that posaconazole appears to retain remarkable antifungal activity against these cryptic species, and that novel azole drug isavuconazole also shows significant activity in these species (Datta et al., 2013).
Many surveys focused particularly on A. fumigatus have been conducted, as azole resistance mechanisms have been intensively investigated in this fungus. An overview of the prevalence of azole-resistant strains was presented in two excellent reviews both published in 2013 (Lelièvre et al., 2013; Vermeulen et al., 2013). The literature cited in the review by Vermeulen et al. (2013) revealed that the overall azole resistance rate of A. fumigatus ranged from 0.6 to 27.8% across the studies. Again the prevalence rates varied between studies (countries), likely due to differences in region and disease manifestation in patients. The emergence and spread of “environmental resistance mechanisms” during the past decade might also have affected the prevalence data. Accordingly, a research group from the Netherlands most recently published a survey of azole-resistant A. fumigatus, in which 364 of 952 clinical strains (38.2%) isolated by, or referred to, the laboratory from 2010 to 2013 were resistant to azoles (van Ingen et al., 2015). Among them, 225 (23.6%) and 98 (10.3%) strains possessed the environmental resistance mechanisms TR34/L98H and TR46/Y121F/T289A, respectively. Another group from the Netherlands reported that 21 azole-resistant strains (20.0%) out of 105 isolates included 13 strains harboring the TR34/L98H allele (12.4%) and three strains harboring the TR46/Y121F/T289A allele (2.9%; Fuhren et al., 2015). The isolation rate of environmentally-derived azole-resistant A. fumigatus strains seems to be constantly increasing, threatening the effectiveness of current frontline antifungal therapy against aspergillosis.
A multicenter epidemiological study carried out by the Surveillance Collaboration on Aspergillus Resistance in Europe (SCARE) network provided a more general understanding of the prevalence of azole-resistant Aspergillus strains (van der Linden et al., 2015). In total, 22 centers from 19 countries participated the study, and 3788 Aspergillus isolates were collected between January 2009 and January 2011. Of these, 2941 isolates (77.6%) belonged to the A. fumigatus complex, 60 of which showed azole resistance. Forty-seven of these azole-resistant isolates were A. fumigatus sensu stricto, and strains with environmental resistance mechanisms TR34/L98H or TR46/Y121F/T289A accounted for 55.3% (n = 26) of isolates. These strains were recovered from six countries: Austria, Belgium, Denmark, France, Italy, and The Netherlands. Recently, strains containing the TR34/L98H and TR46/Y121F/T289A mutations have also been identified in the United States, Columbia, Taiwan, and Japan (Wu et al., 2015; Hagiwara et al., 2016; Le Pape et al., 2016; Wiederhold et al., 2016). These reports highlight a global risk for resistant A. fumigatus strains.
Azole Resistance Mechanisms in A. fumigatus
The basic resistance mechanisms of microbial cells to growth-inhibiting drugs are depicted in Figure 1. The mechanisms are categorized as: (1) reduced interaction affinity of the target protein to the drugs, (2) overexpression of the target protein in the cells, (3) decreased drug concentration by boosted efflux system, (4) intra- or extra-cellular degradation of the drugs, and (5) alternative pathways bypassing the drug effects. The following section summarizes recent advances in our understanding of each of these resistance mechanisms, particularly in regard to A. fumigatus. Furthermore, we review several attempts to reproduce the resistance mutations under laboratory conditions, and new approaches to identifying the responsible mutations by genome comparison.
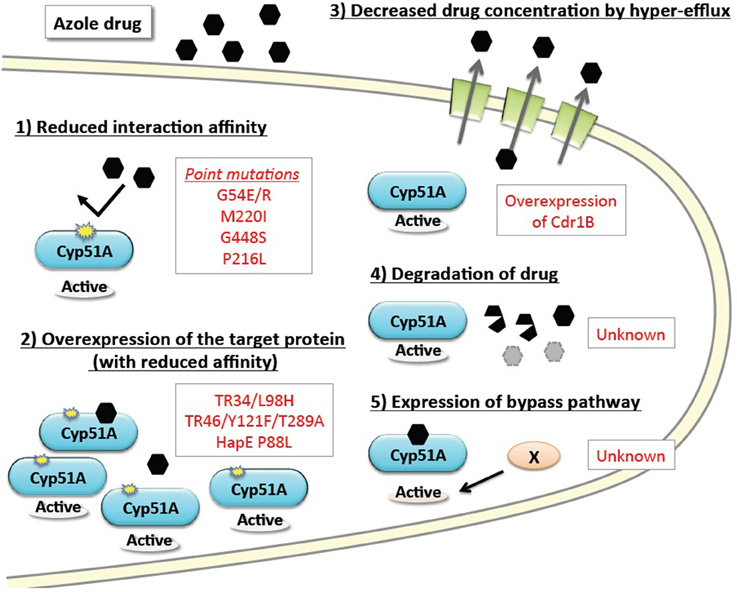
Figure 1. Basic drug resistance mechanisms and the corresponding azole resistance mechanisms in A. fumigatus. Drug resistance mechanisms are categorized into five basic types. The underlying molecular mechanisms identified in A. fumigatus are shown in red text.
CYP51 Proteins
The main mechanisms of azole resistance were elucidated based on the identification of numerous mutations in resistant A. fumigatus isolates. The most frequent mutations detected were related to the target protein Cyp51A: 14α-demethylase. This enzyme is involved in ergosterol biosynthesis and sterol metabolism, and plays an important role in Aspergillus. A. fumigatus has a paralogous protein Cyp51B. Both proteins are capable of complementing the lethality of a Saccharomyces cerevisiae erg11/cyp51 (sterol 14α-demethylase) mutant (Martel et al., 2010), indicating the functionality of the Cyp51 proteins. Neither Cyp51A nor Cyp51B is individually essential for A. fumigatus growth (Garcia-Effron et al., 2005; Mellado et al., 2005), but the attempted inactivation of both genes was lethal (Hu et al., 2007). Azoles interact with and inhibit the Cyp51 proteins (Warrilow et al., 2015), which in turn reduce the ergosterol content and impair sterol metabolism in the cells (Alcazar-Fuoli et al., 2008b), resulting in a fungistatic (under low azole concentrations), or a fungicidal (under high azole concentrations) effect. Single amino acid substitutions at G54, P216, F219, M220, and G448 in the Cyp51A protein have been well-described to confer azole resistance (Mann et al., 2003; Mellado et al., 2004; Camps S. M. et al., 2012; Krishnan-Natesan et al., 2012; Figure 2A). Besides these “hotspots,” amino acid changes at other positions (Y121, G138, and Y431) were also found in the Cyp51A protein of resistant strains (Albarrag et al., 2011; Lescar et al., 2014). These resistance mutations tend to arise during prolonged treatment of chronic aspergillosis with azole drugs. This was well-exemplified by several cases, in which azole-susceptible and azole-resistant isolates were serially isolated from one individual patient (Howard et al., 2009; Hagiwara et al., 2014). It is worth noting that such resistance mutations have never been reported to date in the Cyp51B protein. Instead, high-level induced expression and constitutive overexpression of the cyp51B gene were observed in azole-resistant clinical isolates (Buied et al., 2013). Although these findings await further genetic verification, they raise the possibility of the involvement of Cyp51B in azole resistance as a non-Cyp51A-mediated resistance mechanism in clinical settings.
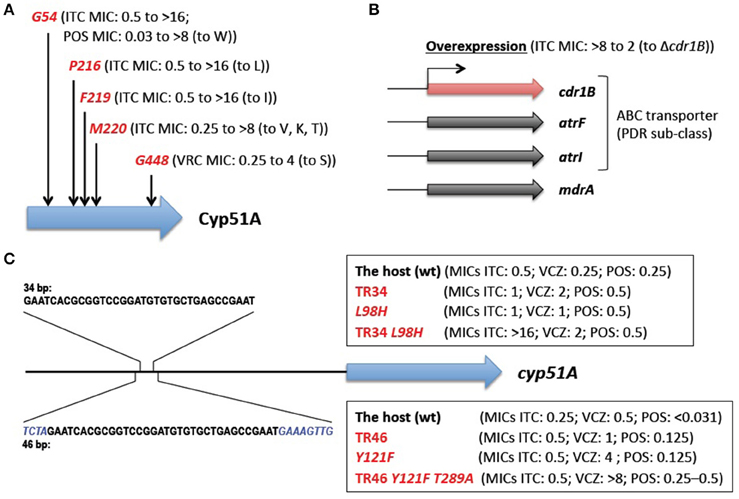
Figure 2. Known azole resistance mechanisms in A. fumigatus. (A) Amino acid substitutions responsible for azole resistance. G54, P216, F219, M220, and G448 show the position at which amino acid changes resulted in azole resistance. The MICs of the strains harboring the change (to the indicated amino acid) are shown in parentheses (Mann et al., 2003; Mellado et al., 2004; Camps S. M. et al., 2012; Krishnan-Natesan et al., 2012). (B) The efflux transporters related to azole resistance. The MIC of the strain with overexpression of the cdr1B gene is shown in parentheses (Fraczek et al., 2013). (C) The tandem repeat sequences in a cyp51A promoter. The none-overlapping bases are shown in italics. The MICs of the strains harboring TR34 or TR46 and/or amino acid change(s) are shown in parentheses (Snelders et al., 2011, 2015). ITCZ, itraconazole; PSCZ, posaconazole; VRCZ, voriconazole.
Efflux Transporters
The overexpression of efflux transporters has been well-documented in yeasts. A. fumigatus contains at least 49 genes encoding the ATP-binding cassette (ABC) transporter (Lelièvre et al., 2013), among which 12 genes show high homology (>30% identities and >50% positive results for >80% of the query protein sequences) with the S. cerevisiae PDR5 and PDR15 proteins that are involved in azole resistance (Paul and Moye-Rowley, 2014). These 12 transporters are included in a PDR (also called an ABC-G) sub-class of ABC transporters. Among them, Cdr1B was identified to be overexpressed in azole-resistant strains, and deleting the cdr1B gene in one such strain resulted in increased susceptibility to itraconazole (Fraczek et al., 2013; Figure 2B). The other strains with the cdr1B gene deleted showed azole hyper-sensitivity (Paul et al., 2013), which indicated that Cdr1B is responsible for azole resistance in A. fumigatus. Recently, Dr. Sanglard and his colleagues demonstrated that deletion mutants of two distinct ABC transporters (AtrF, AtrI) and a major facilitator superfamily transporter (MdrA) also showed sensitivity to azoles (Meneau et al., 2016). The expression levels of the cdr1B gene (also called abcB) were slightly induced upon voriconazole treatment (Paul et al., 2013), whereas some of the other transporter genes (abcB/Afu1g10390, abcE, mfsA, mfsB, and mfsC) were shown to be upregulated in response to voriconazole (da Silva Ferreira et al., 2006). It therefore appears that other efflux transporters may potentially be involved in clinical azole resistance.
Environmentally-Derived Resistance Mechanisms
During this decade, A. fumigatus azole-resistant strains with a combination of a TR in the promoter region of cyp51A and amino acid mutation(s) (TR34/L98H and TR46/Y121F/T289A) have been increasingly reported. These strains have been isolated from patients regardless of their azole treatment history, as well as from the environment. The strains harboring such mutations were isolated in many countries from several continents (Mellado et al., 2007; Snelders et al., 2008; Vermeulen et al., 2012; Chowdhary et al., 2014; Wu et al., 2015; Hagiwara et al., 2016; Wiederhold et al., 2016). The high incidence of resistant strains reported in the Netherlands is particularly alarming (Fuhren et al., 2015; van Ingen et al., 2015). It is now widely accepted that such resistance mechanisms were derived from exposure to azole fungicides in the environment (Snelders et al., 2008).
The molecular mechanisms underlying environmentally-derived mutations were intensively studied by Dr. Melchers's group (Figure 2C). A. fumigatus recombinants with different cyp51A amino acid substitutions and/or a promoter insertion were constructed (Snelders et al., 2011, 2015). The recombinants harboring TR34 or TR46 showed increased expression of the cyp51A gene compared with the parental strains, however, the TR by itself only had a moderate effect on azole susceptibility (itraconazole (ITCZ) MIC: 0.25–0.5 to 0.5–1 mg/L; voriconazole (VRCZ) MIC: 0.25–0.5 to 1–2 mg/L). Site-directed mutagenesis of L98H or Y121F alone also showed a moderate increase in the MICs (ITCZ MIC: 0.25–0.5 to 0.5–1 mg/L; VRCZ MIC: 0.25–0.5 to 1–4 mg/L). Azole resistance levels comparable to those of the clinical isolates (TR34/L98H or TR46/Y121F/T289A) were achieved by a combination of the TR and an amino acid substitution; the recombinant with TR34/L98H showed MICs >16 mg/L for ITCZ and 2 mg/L for VRCZ, and the recombinant with TR46/Y121F showed MICs >8 mg/L for ITCZ and >8 mg/L for VRCZ. Interestingly, the results suggested that T289A was dispensable for full resistance to azoles. Paul et al. (2012) also evaluated the effect of the TR34 element on cyp51A gene expression using a promoter-luciferase reporter system. The lack of the 34-bp sequence led to a 90% reduction in the expression level compared with the wild-type promoter, indicating that the 34-bp element played a critical role in maintaining wild-type expression of the cyp51A gene. Although repeats of the 34-bp element resulted in a modest increase in expression, the sequence may function as an enhancer element for cyp51A gene expression.
Several reports of similar Cyp51 overexpression were also found in plant fungal pathogens (Becher and Wirsel, 2012). The high expression was correlated with the presence of insertions in the promoter region of the cyp51 gene. A five-time TR of a unique 126-bp sequence was found in the cyp51A promoter region in Penicillium digitatum (Hamamoto et al., 2000). Furthermore, 553-, 120-, and 65-bp insertions were found in the cyp51 promoter region in Venturia inaequalis, Mycosphaerella graminicola, and Monilinia fructicola, respectively (Schnabel and Jones, 2000; Luo and Schnabel, 2008; Cools et al., 2012). Changes in the promoter region of the cyp51 gene by the presence of insertions or TRs may frequently occur under certain conditions such as long-term exposure to azole fungicides in environmental niches.
Lessons from In vitro Evolution and Perspectives
To broaden our knowledge of azole resistance mutations, attempts to evolve drug-resistant A. fumigatus strains in vitro by transferring strains onto plates containing sub-lethal concentration of azoles have been undertaken by several groups. da Silva Ferreira et al. (2004) generated 10 itraconazole-resistant strains (nine showed >16 mg/L of ITCZ MIC and one showed 1 mg/L) by in vitro evolution procedure, three of these possessed a G54R point mutation in the Cyp51A protein, and two possessed a M220I point mutation in this protein. Novel mutations (positions N22, T440, and Y491) were also found in the Cyp51A protein of itraconazole-resistant strains, among which the N22D substitution was shown to confer itraconazole resistance to wild-type A. fumgiatus by genetic transformation. They also demonstrated that these resistant mutants tended to show increased expression levels of efflux transporter genes. Upon voriconazole exposure in another study, six resistant strains were obtained from three different parental strains under laboratory conditions (Krishnan-Natesan et al., 2012). All of these strains harbored a G448S mutation in the Cyp51A protein and showed MICs of 2–8 mg/L for VRCZ, whereas the ITCZ MIC ranged from 0.25 to 2 mg/L and the PSCZ MIC from 0.0625 to 0.15 mg/L. This suggested that G448S is the predominant mutation conferring resistance to voriconazole. Snelders et al. (2012) exposed distinct A. fumigatus strains (wild-type, the strain with TR34, and the strain with L98H) to itraconazole, azole fungicides (bromuconazole, difenoconazole, epoxiconazole, propiconazole, or tebuconazole), or a mixture of the fungicides. In the case of itraconazole exposure, G138C and P216L were identified in the wild-type strain that had evolved by more than three transfers. However, none of the strains gained “environmentally-derived mutations” such as TR34/L98H and TR46/Y121F/T289A. The authors hypothesized that the development of TR34/L98H and TR46/Y121F/T289A mutations might be extremely infrequent in the environment. Recent microsatellite genotyping studies showed that less genetic variation was found among strains harboring TR34/L98H or TR46/Y121F/T289A mutations compared with the set of wild-type isolates (Snelders et al., 2009; Hagiwara et al., 2016). From these studies, it is tempting to speculate that the TR34/L98H and TR46/Y121F/T289A-containing isolates did not arise in distinct strains in various regions, but might have originated from common ancestors. If this is the case, the rapid spread of these resistant strains across the globe in a short period of time is quite surprising and worrisome. At this moment, however, the possibility that there is a preferential genetic background in which “environmental mutations” occur more frequently cannot be ruled out. More extensive studies, potentially including population genetic analyses, might provide insight into this important issue.
Exploring Resistance Mutations by Whole-Genome Comparisons
As stated earlier, azole resistance mutations tend to occur during long-term azole treatment (Tashiro et al., 2012). In some cases, multiple strains are serially isolated from one patient at different time points. Resistant isolates theoretically possess the mutation attributed to resistance, which does not exist in the corresponding sensitive isolates. Inspection of the cyp51A gene sequence between these isolates has uncovered several mutations in the Cyp51A protein. Furthermore, to identify novel non-cyp51A resistance mutations, whole-genome comparisons were conducted between azole-sensitive and azole-resistant cognate isolates. Camps S. M. T. et al. (2012) identified a new mutation in the hapE gene by whole-genome comparison and verified its involvement by sexual crossing. HapE encodes a subunit of the CCAAT-binding transcription factor complex, which plays a regulatory role in a wide array of fungal phenotypes. The cyp51A expression level in strains with the hapE mutation was higher than in the corresponding wild-type strain, which suggested that HapE plays a role in regulating cyp51A gene expression. As a presumed CCAAT-box is present in the promoter region of cyp51A, further analysis may uncover the mechanism of regulation of the cyp51A gene. In addition to this novel resistance mutation, they also detected at least 22 mutations in a pair of strains that were recovered 17 weeks apart. Among them, five of these mutations appeared to be in non-coding regions and 11 were synonymous mutations. This indicated that dynamic alterations, likely irrespective of azole resistance, occur in the A. fumigatus genome within its host during infection and treatment. In another study, a large genomic deletion in a region containing 11 genes was identified by whole-genome comparison (Hagiwara et al., 2014). Accordingly, whole-genome comparison analysis can provide interesting insight into genetic variation provoked during infection, and is therefore a powerful tool for further understanding genome-scale azole resistance mechanisms.
Frontiers of Azole Resistance in Non-fumigatus Aspergillus Species
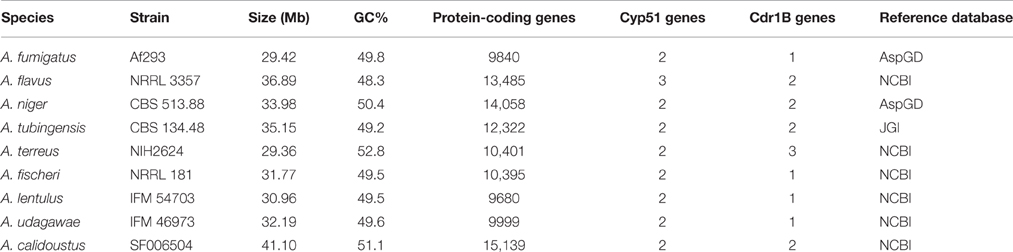
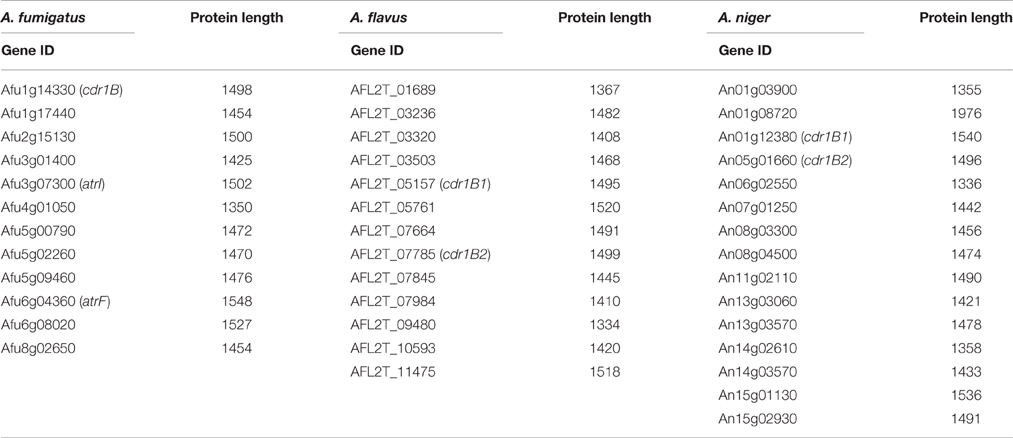
As is the case with A. fumigatus, acquired antifungal resistance is potentially able to arise in non-fumigatus species, whether in the environment or within a host. However, reports identifying antifungal resistance in such species are so far limited. Genome data for several Aspergillus species are available in databases, and the genomes of the cryptic species A. lentulus and A. udagawae as well as A. calidoustus were recently added (Kusuya et al., 2015, 2016; Horn et al., 2016). Thus, there is an opportunity to study pathogenic Aspergillus genomes with special reference to azole resistance. The characteristics of pathogenic Aspergillus genomes are shown in Table 4.
The sequences of the Cyp51 and Cdr1B proteins of A. flavus, A. niger, A. tubingensis, A. terreus, A. fischeri, A. lentulus, A. udagawae, A. calidoustus as well as A. fumigatus, were retrieved from the NCBI (http://www.ncbi.nlm.nih.gov/) and AspGD (http://www.aspgd.org/) databases and compared. Phylogenetic trees constructed based on the Cyp51 and Cdr1B protein sequences are depicted in Figures 3A,B, respectively. All of the species listed above, except for A. flavus, possess two Cyp51 proteins, Cyp51A and Cyp51B, which form distinct sub-groups in the phylogenetic tree. The third Cyp51 protein, Cyp51C, is found only in A. flavus, and is relatively similar to Cyp51A (Figure 3A). The Y319H mutation in A. flavus Cyp51C was found specifically in an azole-resistant clinical isolate, suggesting a role in the resistance mechanism (Paul et al., 2015). Through an in vitro evolution experiment, several mutations in Cyp51A (K197N, D282E, M288L, Y132N, and T469S) and Cyp51B (H399P, D411N, T454P, and T486P) were identified in multi-azole-resistant A. flavus, which warranted further study (Krishnan-Natesan et al., 2008). It is interesting to note that intrinsically azole-resistant species A. calidoustus has two Cyp51 proteins. One of these, a Cyp51A-like protein, shows relatively distant homology to A. fumigatus Cyp51A, whereas the other falls into the Cyp51B group (Figure 3A). When the A. calidoustus Cyp51A-like protein was aligned with Cyp51A proteins from other Aspergillus species, the methionine at position 220 in A. fumigatus Cyp51A was replaced with a valine in A. calidoustus Cyp51A (Figure 3C). As M220 in the A. fumigatus Cyp51A protein is involved in azole resistance, this replacement may cause the observed insensitivity to azole drugs in this fungus. It is yet to be confirmed whether the varied azole susceptibility among species is indeed derived from the sequence difference in the Cyp51A protein. Regarding the protein sequences of Cyp51A and Cyp51B in A. lentulus, differences of 16 and 12 amino acids, respectively, were detected compared with the corresponding sequences in A. fumigatus. Mellado et al. showed that heterologous expression of the A. lentulus cyp51A gene in the A. fumigatus cyp51A deletion mutant resulted in an A. lentulus level azole-resistant phenotype (Mellado et al., 2011). This elegantly demonstrated that Cyp51A is responsible for the differences in azole resistance between A. fumgiatus and A. lentulus.
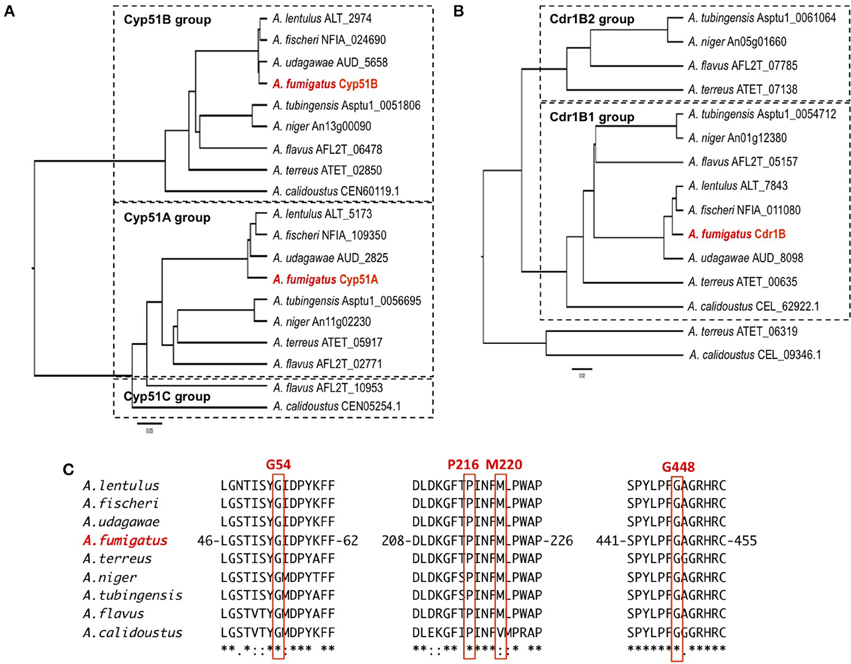
Figure 3. The molecular genetic phylogenetic trees of the Cyp51A and Cdr1B proteins of pathogenic Aspergillus fungi. The sequences were retrieved from the AspGD and NCBI databases according to sequence similarity. The protein sequences of Cyp51A (A) and Cdr1B (B) were aligned using the ClustalW software, and the phylogenetic trees were constructed by the UPGMA method. The trees were drawn using FigTree v1.4.2 software. The IDs shown behind a species name are associated with the database from which the sequences were retrieved. (C) Amino acid sequence alignment of Cyp51A. The sequence surrounding the azole resistance-related amino acids (G54, P216, M220, and G448) is depicted. The numbers indicate amino acid position in the A. fumigatus Cyp51A protein.
As described above, A. fumigatus has 12 ABC transporters in a PDR sub-group that includes the Cdr1B protein. Likewise, according to sequence similarity, A. flavus and A. niger have 13 and 15 PDR-type ABC transporters, respectively (Table 5). The Cdr1B ortholog is duplicated in A. flavus and A. niger. From the genome sequence data, one Cdr1B protein is present in A. fumigatus, A. fischeri, A. lentulus, and A. udagawae, whereas A. flavus, A. niger, A. tubingensis, and A. calidoustus possess two Cdr1B proteins, and A. terreus has three such proteins (Figure 3B). The distribution of Cdr1B proteins therefore varies among Aspergillus species/complexes.
Conclusion and Perspectives
Recent genetic and genomic studies have provided important insights into azole resistance mechanisms in Aspergillus. In particular, whole-genome comparison has proven to be a powerful tool for discovering novel mutations responsible for drug resistance. The analysis of genome sequences has also advanced our understanding of the important diversity of Aspergilli. However, there remains much to be investigated in A. flavus and A. niger, the second most frequent causative agents of aspergillosis. A comprehensive investigation of the genes related to azole resistance among a large population of clinical isolates would broaden our knowledge. In addition, the variable drug susceptibility within the cryptic species may form an important focus of future studies. A more complete understanding of the mechanisms underlying azole resistance will aid the development of new therapeutic drugs against azole-resistant Aspergillus fungi/strains.
Author Contributions
DH, AW, KK, GG: Designed the study. DH: Wrote the manuscript.
Funding
This work was also supported in part by the Japanese Ministry of Education, Culture, Sports, Science, and Technology (MEXT) Special Budget for Research Project: The Project on Controlling Aspergillosis and the Related Emerging Mycoses. GG thanks for Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP) and Conselho Nacional de Desenvolvimento Cientifico e Tecnologico (CNPq), both from Brazil, for providing resources for the research in his laboratory.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abarca, M. L., Accensi, F., Cano, J., and Cabañes, F. J. (2004). Taxonomy and significance of black aspergilli. Antonie Van Leeuwenhoek. 86, 33–49. doi: 10.1023/B:ANTO.0000024907.85688.05
Alastruey-Izquierdo, A., Alcazar-Fuoli, L., and Cuenca-Estrella, M. (2014). Antifungal susceptibility profile of cryptic species of Aspergillus. Mycopathologia 178, 427–433. doi: 10.1007/s11046-014-9775-z
Alastruey-Izquierdo, A., Mellado, E., Peláez, T., Pemán, J., Zapico, S., Alvarez, M., et al. (2013). Population-based survey of filamentous fungi and antifungal resistance in Spain (FILPOP Study). Antimicrob. Agents Chemother. 57, 3380–3387. doi: 10.1128/AAC.00383-13
Albarrag, A. M., Anderson, M. J., Howard, S. J., Robson, G. D., Warn, P. A., Sanglard, D., et al. (2011). Interrogation of related clinical pan-azole-resistant Aspergillus fumigatus strains: G138C, Y431C, and G434C single nucleotide polymorphisms in cyp51A, upregulation of cyp51A, and integration and activation of transposon Aft1 in the cyp51A promoter. Antimicrob. Agents Chemother. 55, 5113–5121. doi: 10.1128/AAC.00517-11
Alcazar-Fuoli, L., Mellado, E., Alastruey-Izquierdo, A., Cuenca-Estrella, M., and Rodriguez-Tudela, J. (2008a). Aspergillus section Fumigati: antifungal susceptibility patterns and sequence based identification. Antimicrob. Agents Chemother. 52, 1244–1251. doi: 10.1128/AAC.00942-07
Alcazar-Fuoli, L., Mellado, E., Garcia-Effron, G., Lopez, J. F., Grimalt, J. O., Cuenca-Estrella, J. M., et al. (2008b). Ergosterol biosynthesis pathway in Aspergillus fumigatus. Steroids 73, 339–347. doi: 10.1016/j.steroids.2007.11.005
Al-Wathiqi, F., Ahmad, S., and Khan, Z. (2013). Molecular identification and antifungal susceptibility profile of Aspergillus flavus isolates recovered from clinical specimens in Kuwait. BMC Infect. Dis. 13:126. doi: 10.1186/1471-2334-13-126
Badali, H., Fakhim, H., Zarei, F., Nabili, M., Vaezi, A., Poorzad, N., et al. (2016). In vitro activities of five antifungal drugs against opportunistic agents of Aspergillus nigri complex. Mycopathologia 181, 235–240. doi: 10.1007/s11046-015-9968-0
Baddley, J. W., Marr, K. A., Andes, D. R., Walch, T. J., Kauffman, C. A., Kontoyiannis, D. P., et al. (2009). Patterns of susceptibility of Aspergillus isolates recovered from patients enrolled in the transplant-associated infection surveillance network. J. Clin. Microbiol. 47, 3271–3275. doi: 10.1128/JCM.00854-09
Balajee, S. A., Gribskov, J., Brandt, M., Ito, J., Fothergill, A., and Marr, K. A. (2005a). Mistaken identity: Neosartorya pseudofischeri and its anamorph masquerading as Aspergillus fumigatus. J. Clin. Microbiol. 43, 5996–5999. doi: 10.1128/JCM.43.12.5996-5999.2005
Balajee, S. A., Gribskov, J., Hanley, E., Nickle, D., and Marr, K. A. (2005b). Aspergillus lentulus sp. nov., a new sibling species of A. fumigatus. Eukaryot. Cell 4, 625–632. doi: 10.1128/EC.4.3.625-632.2005
Balajee, S. A., Kano, R., Baddley, J. W., Moser, S. A., Marr, K. A., Alexander, B. D., et al. (2009). Molecular identification of Aspergillus species collected for the transplant-associated infection surveillance network. J. Clin. Microbiol. 47, 3138–3141. doi: 10.1128/JCM.01070-09
Balajee, S. A., Nickle, D., Varga, J., and Marr, K. A. (2006). Molecular studies reveal frequent misidentification of Aspergillus fumigatus by morphotyping. Eukaryot. Cell 5, 1705–1712. doi: 10.1128/EC.00162-06
Becher, R., and Wirsel, S. G. R. (2012). Fungal cytochrome P450 Sterol 14α-Demethylase (CYP51) and azole resistance in plant and human pathogens. Appl. Microbiol. Biotechnol. 95, 825–840. doi: 10.1007/s00253-012-4195-9
Bitar, D., Lortholary, O., Le Strat, Y., Nicolau, J., Coignard, B., and Tattevin, P. (2014). Population-based analysis of invasive fungal infections, France, 2001-2010. Emerging Infect. Dis. 20, 1149–1155. doi: 10.3201/eid2007.140087
Brown, N. A., and Goldman, G. H. (2016). The contribution of Aspergillus fumigatus stress responses to virulence and antifungal resistance. J. Microbiol. (Seoul, Korea) 54, 243–253. doi: 10.1007/s12275-016-5510-4
Buied, A., Moore, C. B., Denning, D. W., and Bowyer, P. (2013). High-level expression of cyp51B in azole-resistant clinical Aspergillus fumigatus isolates. J. Antimicrob. Chemother. 68, 512–514. doi: 10.1093/jac/dks451
Camps, S. M. T., Dutilh, B. E., Arendrup, M. C., Rijs, A. J. M. M., Snelders, E., Huynen, M. A., et al. (2012). Discovery of a hapE mutation that causes azole resistance in Aspergillus fumigatus through whole genome sequencing and sexual crossing. PLoS ONE 7:e50034. doi: 10.1371/journal.pone.0050034
Camps, S. M., van der Linden, J. W., Li, Y., Kuijper, E. J., van Dissel, J. T., Verweij, P. E., et al. (2012). Rapid induction of multiple resistance mechanisms in Aspergillus fumigatus during azole therapy: a case study and review of the literature. Antimicrob. Agents Chemother. 56, 10–16. doi: 10.1128/AAC.05088-11
Castanheira, M., Messer, S. A., Rhomberg, P. R., and Pfaller, M. A. (2016). Antifungal susceptibility patterns of a global collection of fungal isolates: results of the SENTRY Antifungal Surveillance Program (2013). Diagn. Microbiol. Infect. Dis. 85, 200–204. doi: 10.1016/j.diagmicrobio.2016.02.009
Chen, J., Li, H., Li, R., Bu, D., and Wan, Z. (2005). Mutations in the cyp51A gene and susceptibility to itraconazole in Aspergillus fumigatus serially isolated from a patient with lung aspergilloma. J. Antimicrob. Chemother. 55, 31–37. doi: 10.1093/jac/dkh507
Chowdhary, A., Kathuria, S., Xu, J., and Meis, J. F. (2013). Emergence of azole-resistant Aspergillus fumigatus strains due to agricultural azole use creates an increasing threat to human health. PLoS Pathog. 9:e1003633. doi: 10.1371/journal.ppat.1003633
Chowdhary, A., Sharma, C., Kathuria, S., Hagen, F., and Meis, J. F. (2014). Azole-resistant Aspergillus fumigatus with the environmental TR46/Y121F/T289A mutation in India. J. Antimicrob. Chemother. 69, 555–557. doi: 10.1093/jac/dkt397
Coelho, D., Silva, S., Vale-Silva, L., Gomes, H., Pinto, E., Sarmento, A., et al. (2011). Aspergillus viridinutans: an agent of adult chronic invasive aspergillosis. Med. Mycol. 49, 755–759. doi: 10.3109/13693786.2011.556672
Cools, H. J., Bayon, C., Atkins, S., Lucas, J. A., and Fraaije, B. A. (2012). Overexpression of the sterol 14α-Demethylase gene (MgCYP51) in Mycosphaerella graminicola isolates confers a novel azole fungicide sensitivity phenotype. Pest Manag. Sci. 68, 1034–1040. doi: 10.1002/ps.3263
Dasbach, E. J., Davies, G. M., and Teutsch, S. M. (2000). Burden of aspergillosis-related hospitalizations in the United States. Clin. Infect. Dis. 31, 1524–1528. doi: 10.1086/317487
da Silva Ferreira, M. E., Capellaro, J. L., dos Reis Marques, E., Malavazi, I., Perlin, D., Park, S., et al. (2004). In vitro evolution of itraconazole resistance in Aspergillus fumigatus involves multiple mechanisms of resistance. Antimicrob. Agents Chemother. 48, 4405–4413. doi: 10.1128/AAC.48.11.4405-4413.2004
da Silva Ferreira, M. E., Malavazi, I., Savoldi, M., Brakhage, A. A., Goldman, M. H. S., Kim, H. S., et al. (2006). Transcriptome analysis of Aspergillus fumigatus exposed to voriconazole. Curr. Genet. 50, 32–44. doi: 10.1007/s00294-006-0073-2
Datta, K., Rhee, P., Byrnes, E., Garcia-Effron, G., Perlin, D. S., Staab, J. F., et al. (2013). Isavuconazole activity against Aspergillus lentulus, Neosaartorya udagawae, and Cryptococcus gattii, emerging fungal pathogens with reduced azole susceptibility. J. Clin. Microbiol. 51, 3090–3093. doi: 10.1128/JCM.01190-13
Denning, D. W., Venkateswarlu, K., Oakley, K. L., Anderson, M. J., Manning, N. J., Stevens, D. A., et al. (1997). Itraconazole resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 41, 1364–1368.
Egli, A., Fuller, J., Humar, A., Lien, D., Weinkauf, J., Nador, R., et al. (2012). Emergence of Aspergillus calidoustus infection in the era of posttransplantation azole prophylaxis. Transplantation 94, 403–410. doi: 10.1097/TP.0b013e31825992f0
Escribano, P., Pelaez, T., Munoz, P., Bouza, E., and Guinea, J. (2013). Is azole resistance in Aspergillus fumigatus a probrem in Spain? Antimicrobiol. Agents Chemother. 57, 2815–2820. doi: 10.1128/AAC.02487-12
Espinel-Ingroff, A., Diekema, D. J., Fothergill, A., Johnson, E., Pelaez, T., Pfaller, M. A., et al. (2010). Wild-type MIC distributions and epidemiological cutoff values for the triazoles and six Aspergillus spp. for the CLSI broth microdilution method (M38-A2 document). J. Clin. Microbiol. 48, 3251–3257. doi: 10.1128/JCM.00536-10
Fraczek, M. G., Bromley, M., Buied, A., Moore, C. B., Rajendran, R., Rautemaa, R., et al. (2013). The cdr1B efflux transporter is associated with Non-cyp51a-mediated itraconazole resistance in Aspergillus fumigatus. J. Antimicrob. Chemother. 68, 1486–1496. doi: 10.1093/jac/dkt075
Fuhren, J., Voskuil, W. S., Boel, C. H. E., Haas, P. J. A., Hagen, F., Meis, J. F., et al. (2015). High prevalence of azole resistance in Aspergillus fumigatus isolates from high-risk patients. J. Antimicrob. Chemother. 70, 2894–2898. doi: 10.1093/jac/dkv177
Garcia-Effron, G., Mellado, E., Gomez-Lopez, A., Alcazar-Fuoli, L., Cuenca-Estrella, M., and Rodriguez-Tudela, J. L. (2005). Differences in interactions between azole drugs related to modifications in the 14-Alpha sterol demethylase gene (cyp51A) of Aspergillus fumigatus. Antimicrob. Agents Chemother. 49, 2119–2121. doi: 10.1128/AAC.49.5.2119-2121.2005
Gautier, M., Normand, A., L'Ollivier, C., Cassagne, C., Reynaud-Gaubert, M., Dubus, J. C., et al. (2016). Aspergillus tubingensis: a major filamentous fungus found in the airways of patients with lung disease. Med. Mycol. 54, 459–470. doi: 10.1093/mmy/myv118
Gheith, S., Saghrouni, F., Bannour, W., Ben Youssef, Y., Khelif, A., Normand, A., et al. (2014a). Characteristics of invasive aspergillosis in neutropenic haematology patients (Sousse, Tunisia). Mycopathologia 177, 281–289. doi: 10.1007/s11046-014-9742-8
Gheith, S., Saghrouni, F., Bannour, W., Ben Youssef, Y., Khelif, A., Normand, A., et al. (2014b). In vitro susceptibility to amphotericin B, itraconazole, voriconazole, posaconazole and caspofungin of Aspergillus spp. Isolated from patients with haematological malignancies in Tunisia. Springerplus 3:19. doi: 10.1186/2193-1801-3-19
Gürcan, S., Tikveşli, M., Ustündağ, S., and Ener, B. (2013). A case report on Aspergillus lentulus pneumonia. Balkan Med. J. 30, 429–431. doi: 10.5152/balkanmedj.2013.8572
Hagiwara, D., Takahashi, H., Fujimoto, M., Sugahara, M., Misawa, Y., Gonoi, T., et al. (2016). Multi-azole resistant Aspergillus fumigatus harboring Cyp51A TR46/Y121F/T289A isolated in Japan. J. Infect. Chemother. 22, 577–579. doi: 10.1016/j.jiac.2016.01.015
Hagiwara, D., Takahashi, H., Watanabe, A., Takahashi-Nakaguchi, A., Kawamoto, S., Kamei, K., et al. (2014). Whole-genome comparison of Aspergillus fumigatus strains serially isolated from patients with aspergillosis. J. Clin. Microbiol. 52, 4202–4209. doi: 10.1128/JCM.01105-14
Hamamoto, H., Hasegawa, K., Nakaune, R., Lee, Y. J., Makizumi, Y., Akutsu, K., et al. (2000). Tandem repeat of a transcriptional enhancer upstream of the sterol 14α-demethylase gene (CYP51) in Penicillium digitatum. Appl. Environ. Microbiol. 66, 3421–3426. doi: 10.1128/AEM.66.8.3421-3426.2000
Horn, F., Linde, J., Mattern, D. J., Walther, G., Guthke, R., Scherlach, K., et al. (2016). Draft genome sequences of fungus Aspergillus calidoustus. Genome Announc. 4, e00102–e00116. doi: 10.1128/genomeA.00102-16
Howard, S. J. (2014). Multi-resistant aspergillosis due to cryptic species. Mycopathologia 178, 435–439. doi: 10.1007/s11046-014-9774-0
Howard, S. J., Cerar, D., Anderson, M. J., Albarrag, A., Fisher, M. C., Pasqualotto, A. C., et al. (2009). Frequency and evolution of azole resistance in Aspergillus fumigatus associated with treatment failure. Emer. Infect. Dis. 15, 1068–1076. doi: 10.3201/eid1507.090043
Hu, W., Sillaots, S., Lemieux, S., Davison, J., Kauffman, S., Breton, A., et al. (2007). Essential gene identification and drug target prioritization in Aspergillus fumigatus. PLoS Pathog. 3:e24. doi: 10.1371/journal.ppat.0030024
Kachuei, R., Khodavaisy, S., Rezaie, S., and Sharifynia, S. (2016). In vitro antifungal susceptibility of clinical species belonging to Aspergillus genus and Rhizopus oryzae. J. Mycol. Med. 26, 17–21. doi: 10.1016/j.mycmed.2015.12.002
Khodavaisy, S., Badali, H., Hashemi, S. J., Aala, F., Nazeri, M., Nouripour-Sisakht, S., et al. (2016). In vitro activities of five antifungal agents against 199 clinical and environmental isolates of Aspergillus flavus, an opportunistic fungal pathogen. J. Mycol. Med. 26, 116–121. doi: 10.1016/j.mycmed.2016.01.002
Kontoyiannis, D. P., Marr, K. A., Park, B. J., Alexander, B. D., Anaissie, E. J., Walsh, T. J., et al. (2010). Prospective surveillance for incasive fungal infections in hematopoietic stem cell transplantat recipients, 2001–2006: overview of the Transplant-Associated Infection Surveillance Network (TRANSNET) database. Clin. Infect. Dis. 50, 1091–1100. doi: 10.1086/651263
Kousha, M., Tadi, R., and Soubani, A. O. (2011). Pulmonary aspergillosis: a clinical review. Eur. Respir. Rev. 20, 156–174. doi: 10.1183/09059180.00001011
Krishnan, S., Manavathu, E. K., and Chandrasekar, P. H. (2009). Aspergillus flavus: an emerging non-fumigatus Aspergillus species of significance. Mycoses 52, 206–222. doi: 10.1111/j.1439-0507.2008.01642.x
Krishnan-Natesan, S., Chandrasekar, P. H., Alangaden, G. J., and Manavathu, E. K. (2008). Molecular characterisation of cyp51A and cyp51B genes coding for P450 14α-lanosterol demethylases A (CYP51Ap) and B (CYP51Bp) from voriconazole-resistant laboratory isolates of Aspergillus flavus. Int. J. Antimicrob. Agents 32, 519–524. doi: 10.1016/j.ijantimicag.2008.06.018
Krishnan-Natesan, S., Wu, W., Cutright, J. L., and Chandrasekar, P. H. (2012). In vitro-in vivo correlation of voriconazole resistance due to G448S mutation (cyp51A Gene) in Aspergillus fumigatus. Diagn. Microbiol. Infect. Dis. 74, 272–277. doi: 10.1016/j.diagmicrobio.2012.06.030
Kusuya, Y., Sakai, K., Kamei, K., Takahashi, H., and Yaguchi, T. (2016). Draft genome sequence of the pathogenic filamentous fungus Aspergillus lentulus IFM 54703T. Genome Announc. 4, e01568–e01515. doi: 10.1128/genomeA.01568-15
Kusuya, Y., Takahashi-Nakaguchi, A., Takahashi, H., and Yaguchi, T. (2015). Draft genome sequence of the pathogenic filamentous fungus Aspergillus udagawae strain IFM 46973T. Genome Announc. 3, e00834–e00815. doi: 10.1128/genomeA.00834-15
Kwon-Chung, K. J., and Sugui, J. A. (2013). Aspergillus fumigatus-what makes the species a ubiquitous human fungal pathogen? PLoS Pathog. 9:e1003743. doi: 10.1371/journal.ppat.1003743
Lago, M., Aguiar, A., Natário, A., Fernandes, C., Faria, M., and Pinto, E. (2014). Does fungicide application in vineyards induce resistance to medical azoles in Aspergillus species? Environ. Monit. Assess. 186, 5581–5593. doi: 10.1007/s10661-014-3804-8
Lalitha, P., Sun, C. Q., Prajna, N. V., Karpagam, R., Geetha, M., O'Brien, K. S., et al. (2014). In vitro susceptibility of filamentous fungal isolates from a corneal ulcer clinical trial. Am. J. Ophthalmol. 157, 318–326. doi: 10.1016/j.ajo.2013.10.004
Lamoth, F. (2016). Aspergillus fumigatus-related species in clinical practice. Front. Microbiol. 7:683. doi: 10.3389/fmicb.2016.00683
Lelièvre, L., Groh, M., Angebault, C., Maherault, A. C., Didier, E., and Bougnoux, M. E. (2013). Azole resistant Aspergillus fumigatus: an emerging problem. Med. Mal. Infect. 43, 139–145. doi: 10.1016/j.medmal.2013.02.010
Le Pape, P., Lavergne, R. A., Morio, F., and Alvarez-Moreno, C. (2016). Multiple fungicide-driven alterations in azole-resistant Aspergillus fumigatus, Colombia, 2015. Emerg. Infect. Dis. 22, 156–157. doi: 10.3201/eid2201.150978
Lescar, J., Meyer, I., Akshita, K., Srinivasaraghavan, K., Verma, C., Palous, M., et al. (2014). Aspergillus fumigatus harboring the sole Y121F mutation shows decreased susceptibility to voriconazole but maintained susceptibility to itraconazole and posaconazole. J. Antimicrob. Chemother. 69, 3244–3247. doi: 10.1093/jac/dku316
Luo, C., and Schnabel, G. (2008). The cytochrome P450 lanosterol 14α-demethylase gene is a demethylation inhibitor fungicide resistance determinant in monilinia fructicola field isolates from Georgia. Appl. Environ. Microbiol. 74, 359–366. doi: 10.1128/AEM.02159-07
Mann, P. A., Parmegiani, R. M., Wei, S. Q., Mendrick, C. A., Li, X., Loebenberg, D., et al. (2003). Mutations in Aspergillus fumigatus resulting in reduced susceptibility to posaconazole appear to be restricted to a single amino acid in the cytochrome P450 14α-demethylase. Antimicrob. Agents Chemother. 47, 577–581. doi: 10.1128/AAC.47.2.577-581.2003
Martel, C. M., Parker, J. E., Warrilow, A. G. S., Rolley, N. J., Kelly, S. L., and Kelly, D. E. (2010). Complementation of a Saccharomyces cerevisiae ERG11/CYP51 (sterol 14α-demethylase) doxycycline-regulated mutant and screening of the azole sensitivity of Aspergillus fumigatus isoenzymes CYP51A and CYP51B. Antimicrob. Agents Chemother. 54, 4920–4923. doi: 10.1128/AAC.00349-10
Mellado, E., Alcazar-Fuoli, L., Cuenca-Estrella, M., and Rodriguez-Tudela, J. L. (2011). Role of Aspergillus lentulus 14-α sterol demethylase (Cyp51A) in azole drug susceptibility. Antimicrob. Agents Chemother. 55, 5459–5468. doi: 10.1128/AAC.05178-11
Mellado, E., Garcia-Effron, G., Alcazar-Fuoli, L., Cuenca-Estrella, M., and Rodriguez-Tudela, J. L. (2004). Substitutions at methionine 220 in the 14α-sterol demethylase (cyp51A) of Aspergillus fumigatus are responsible for resistance in vitro to azole antifungal drugs. Antimicrob. Agents Chemother. 48, 2747–2750. doi: 10.1128/AAC.48.7.2747-2750.2004
Mellado, E., Garcia-Effron, G., Alcázar-Fuoli, L., Melchers, W. J. G., Verweij, P. E., Cuenca-Estrella, M., et al. (2007). A new Aspergillus fumigatus resistance mechanism conferring in vitro cross-resistance to azole antifungals involves a combination of cyp51A alterations. Antimicrob. Agents Chemother. 51, 1897–1904. doi: 10.1128/AAC.01092-06
Mellado, E., Garcia-Effron, G., Buitrago, M. J., Alcazar-Fuoli, L., Cuenca-Estrella, M., and Rodriguez-Tudela, J. L. (2005). Targeted gene disruption of the 14-α sterol demethylase (cyp51A) in Aspergillus fumigatus and its role in azole drug susceptibility. Antimicrob. Agents Chemother. 49, 2536–2538. doi: 10.1128/AAC.49.6.2536-2538.2005
Meneau, I., Coste, A. T., and Sanglard, D. (2016). Identification of Aspergillus fumigatus multidrug transporter genes and their potential involvement in antifungal resistance. Med. Mycol. 54, 616–627. doi: 10.1093/mmy/myw005
Mortensen, K. L., Johansen, H. K., Fuursted, K., Knudsen, J. D., Gahrn-Hansen, B., and Jensen, R. H. (2011). A prospective survey of Aspergillus spp. in respiratory tract samples: prevalence, clinical impact and antifungal susceptibility. Eur. J. Clin. Microbiol. Infect. Dis. 30, 1355–1363. doi: 10.1007/s10096-011-1229-7
Nierman, W. C., Pain, A., Anderson, M. J., Wortman, J. R., Kim, H. S., Arroyo, J., et al. (2005). Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438, 1151–1156. doi: 10.1038/nature04332
Pappas, P. G., Alexander, B. D., Andes, D. R., Hadley, S., Kauffman, C. A., Freifeld, A., et al. (2010). Invasive fungal infections among organ transplant recipients: results of the Transplant-Associated Infection Surveillance Network (TRANSNET). Clin. Infect. Dis. 50, 1101–1111. doi: 10.1086/651262
Paul, R. A., Rudramurthy, S. M., Meis, J. F., Mouton, J. W., and Chakrabarti, A. (2015). A Novel Y319H substitution in CYP51C associated with azole resistance in Aspergillus flavus. Antimicrob. Agents Chemother. 59, 6615–6619. doi: 10.1128/AAC.00637-15
Paul, S., Diekema, D., and Moye-Rowley, W. S. (2013). Contributions of Aspergillus fumigatus ATP-binding cassette transporter proteins to drug resistance and virulence. Eukaryotic Cell 12, 1619–1628. doi: 10.1128/EC.00171-13
Paul, S., Klutts, J. S., and Moye-Rowleya, W. S. (2012). Analysis of promoter function in Aspergillus fumigatus. Eukaryot. Cell 11, 1167–1177. doi: 10.1128/EC.00174-12
Paul, S., and Moye-Rowley, W. S. (2014). Multidrug resistance in fungi: regulation of transporter-encoding gene expression. Front. Physiol. 5:143 doi: 10.3389/fphys.2014.00143
Perez-Nadales, E., Nogueira, M. F. A., Baldin, C., Castanheira, S., Ghalid, M. L., Grund, E., et al. (2014). Fungal model systems and the elucidation of pathogenicity determinants. Fungal Genet. Biol. 70, 42–67. doi: 10.1016/j.fgb.2014.06.011
Pfaller, M. A., Diekema, D. J., Ghannoum, M. A., Rex, J. H., Alexander, B. D., Andes, D., et al. (2009). Wild-type MIC distribution and epidemiological cutoff values for Aspergillus fumigatus and three triazoles as determined by the Clinical and Laboratory Standards Institute broth microdilution methods. J. Clin. Microbiol. 47, 3142–3146. doi: 10.1128/JCM.00940-09
Pfaller, M. A., Messer, S. A., Boyken, L., Rice, C., Tendolkar, S., Hollis, R. J., et al. (2008). In vitro survey of triazole cross-resistance among more than 700 clinical isolates of Aspergillus species. J. Clin. Microbiol. 46, 2568–2572. doi: 10.1128/JCM.00535-08
Samson, R. A., Visagie, C. M., Houbraken, J., Hong, S.-B., Hubka, V., Klaassen, C. H. W., et al. (2014). Phylogeny, identification and nomenclature of the genus Aspergillus. Stud. Mycol. 78, 141–173. doi: 10.1016/j.simyco.2014.07.004
Schnabel, G., and Jones, A. L. (2000). The 14α − demethylase (CYP51A1) gene is overexpressed in Venturia inaequalis strains resistant to myclobutanil. Phytopathology 91, 102–110. doi: 10.1094/PHYTO.2001.91.1.102
Shivaprakash, M. R., Geertsen, E., Chakrabarti, A., Mouton, J. W., and Meis, J. F. (2011). In vitro susceptibility of 188 clinical and environmental isolates of Aspergillus flavus for the new triazole isavuconazole and seven other antifungal drugs. Mycoses 54, e583–e589. doi: 10.1111/j.1439-0507.2010.01996.x
Snelders, E., Camps, S. M. T., Karawajczyk, A., Rijs, A. J. M. M., Zoll, J., Verweij, P. E., et al. (2015). Genotype-phenotype complexity of the TR46/Y121F/T289A cyp51A azole resistance mechanism in Aspergillus fumigatus. Fungal Genet. Biol. 82, 129–135. doi: 10.1016/j.fgb.2015.06.001
Snelders, E., Camps, S. M. T., Karawajczyk, A., Schaftenaar, G., Kema, G. H. J., van der Lee, H. A., et al. (2012). Triazole fungicides can induce cross-resistance to medical triazoles in Aspergillus fumigatus. PLoS ONE 7:e31801. doi: 10.1371/journal.pone.0031801
Snelders, E., Huis In't Veld, R. A. G., Rijs, A. J. M. M., Kema, G. H. J., Melchers, W. J. G., and Verweij, P. E. (2009). Possible environmental origin of resistance of Aspergillus fumigatus to medical triazoles. Appl. Environ. Microbiol. 75, 4053–4057. doi: 10.1128/AEM.00231-09
Snelders, E., Karawajczyk, A., Verhoeven, R. J. A., Venselaar, H., Schaftenaar, G., Verweij, P. E., et al. (2011). The structure-function relationship of the Aspergillus fumigatus cyp51A L98H conversion by site-directed mutagenesis: the mechanism of L98H azole resistance. Fungal Genet. Biol. 48, 1062–1070. doi: 10.1016/j.fgb.2011.08.002
Snelders, E., Van Der Lee, H. A. L., Kuijpers, J., Rijs, A. J. M. M., Varga, J., Samson, R. A., et al. (2008). Emergence of azole resistance in Aspergillus fumigatus and spread of a single resistance mechanism. PLoS Med. 5:e219. doi: 10.1371/journal.pmed.0050219
Steinbach, W. J., Marr, K. A., Anaissie, E. J., Azie, N., Quan, Q., Meier-Kriesche, H. U., et al. (2012). Clinical epidemiology of 960 patients with invasive aspergillosis from the PATH alliance registry. J. Infect. 65, 453–464. doi: 10.1016/j.jinf.2012.08.003
Suzuki, Y., Kume, H., Togano, T., Kanoh, Y., and Ohto, H. (2013). Epidemiology of visceral mycoses in autopsy cases in Japan: the data from 1989 to 2009 in the annual of pathological autopsy cases in Japan. Med. Mycol. 51, 522–526. doi: 10.3109/13693786.2012.755574
Taccone, F. S., Van den Abeele, A. M., Bulpa, P., Misset, B., Meersseman, W., Cardoso, T., et al. (2015). Epidemiology of invasive aspergillosis in critically ill patients: clinical presentation, underlying conditions, and outcomes. Crit. Care 19:7. doi: 10.1186/s13054-014-0722-7
Tamiya, H., Ochiai, E., Kikuchi, K., Yahiro, M., Toyotome, T., Watanabe, A., et al. (2015). Secondary metabolite profiles and antifungal drug susceptibility of Aspergillus fumigatus and closely related species, Aspergillus lentulus, Aspergillus udagawae, and Aspergillus viridinutans. J. Infect. Chemother. 21, 385–391. doi: 10.1016/j.jiac.2015.01.005
Tashiro, M., Izumikawa, K., Hirano, K., Ide, S., Mihara, T., Hosogaya, N., et al. (2012). Correlation between triazole treatment history and susceptibility in clinically isolated Aspergillus fumigatus. Antimicrob. Agents Chemother. 56, 4870–4875. doi: 10.1128/AAC.00514-12
Tashiro, T., Izumikawa, K., Tashiro, M., Takazono, T., Morinaga, Y., Yamamoto, K., et al. (2011). Diagnostic significance of Aspergillus species isolated from respiratory samples in an adult pneumology ward. Med. Mycol. 49, 581–587. doi: 10.3109/13693786.2010.548084
van der Linden, J. W. M., Arendrup, M. C., Warris, A., Lagrou, K., Pelloux, H., Hauser, P. M., et al. (2015). Prospective multicenter international surveillance of azole resistance in Aspergillus fumigatus. Emer. Infect. Dis. 21, 1041–1044. doi: 10.3201/eid2106.140717
van Ingen, J., van der Lee, H. A., Rijs, T. A. J., Zoll, J., Leenstra, T., Melchers, W. J. G., et al. (2015). Azole, polyene and echinocandin MIC distributions for wild-type, TR34/L98H and TR46/Y121F/T289A Aspergillus fumigatus isolates in the Netherlands. J. Antimicrob. Chemother. 70, 178–181. doi: 10.1093/jac/dku364
Varga, J., Houbraken, J., van der Lee, H. A., Verweij, P. E., and Samson, R. A. (2008). Aspergillus calidoustus sp. nov., causative agent of human infections previously assigned to Aspergillus ustus. Eukaryot. Cell 7, 630–638. doi: 10.1128/EC.00425-07
Vermeulen, E., Lagrou, K., and Verweij, P. E. (2013). Azole resistance in Aspergillus fumigatus: a growing public health concern. Curr. Opin. Infect. Dis. 26, 493–500. doi: 10.1097/QCO.0000000000000005
Vermeulen, E., Maertens, J., De Bel, A., Nulens, E., Boelens, J., Surmont, I., et al. (2015). Nationwide surveillance of azole resistance in Aspergillus diseases. Antimicrob. Agents Chemother. 59, 4569–4576. doi: 10.1128/AAC.00233-15
Vermeulen, E., Maertens, J., Schoemans, H., and Lagrou, K. (2012). Azole-resistant Aspergillus fumigatus due to TR46/Y121F/T289A mutation emerging in Belgium, July 2012. Euro Surveill. 17, pii 20326. Available online at: http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20326
Verweij, P. E., Ananda-Rajah, M., Andes, D., Arendrup, M. C., Brüggemann, R. J., Chowdhary, A., et al. (2015). International expert opinion on the management of infection caused by azole-resistant Aspergillus fumigatus. Drug. Resist. Updat. 21–22, 30–40. doi: 10.1016/j.drup.2015.08.001
Verweij, P. E., Mellado, E., and Melchers, W. J. G. (2007). Multiple-triazole-resistant aspergillosis. N. Engl. J. Med. 356, 1481–1483. doi: 10.1056/NEJMc061720
Vinh, D. C., Shea, Y. R., Sugui, J. A., Parrilla-Castellar, E. R., Freeman, A. F., Campbell, J. W., et al. (2009). Invasive aspergillosis due to Neosartorya udagawae. Clin. Infect. Dis. 49, 102–111. doi: 10.1086/599345
Warrilow, A. G. S., Parker, J. E., Price, C. L., Nes, W. D., Kelly, S. L., and Kelly, D. E. (2015). In vitro biochemical study of CYP51-mediated azole resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 59, 7771–7778. doi: 10.1128/AAC.01806-15
Wiederhold, N. P., Gil, V. G., Gutierrez, F., Lindner, J. R., Albataineh, M. T., McCarthy, D. I., et al. (2016). First detection of TR34 L98H and TR46 Y121F T289A Cyp51 mutations in Aspergillus fumigatus isolates in the United States. J. Clin. Microbiol. 54, 168–171. doi: 10.1128/JCM.02478-15
Keywords: Aspergillus fumigatus, azole resistance, Cyp51A, Cdr1B, tandem repeats, A. flavus, A. niger, Aspergillus section Fumigati
Citation: Hagiwara D, Watanabe A, Kamei K and Goldman GH (2016) Epidemiological and Genomic Landscape of Azole Resistance Mechanisms in Aspergillus Fungi. Front. Microbiol. 7:1382. doi: 10.3389/fmicb.2016.01382
Received: 17 June 2016; Accepted: 22 August 2016;
Published: 21 September 2016.
Edited by:
Dominique Sanglard, University of Lausanne, SwitzerlandReviewed by:
Siddharth Kaushal Tripathi, University of Mississippi, USAFrederic Lamoth, Lausanne University Hospital, Switzerland
Copyright © 2016 Hagiwara, Watanabe, Kamei and Goldman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daisuke Hagiwara, ZGhhZ2lAY2hpYmEtdS5qcA==
 Daisuke Hagiwara
Daisuke Hagiwara Akira Watanabe
Akira Watanabe Katsuhiko Kamei1
Katsuhiko Kamei1 Gustavo H. Goldman
Gustavo H. Goldman