- 1Department of Biological Sciences, Auburn University, Auburn, AL, USA
- 2Departamento de Ciencias Médicas Básicas, Facultad de Medicina y Ciencias de la Salud, Institut d’Investigació Sanitària Pere Virgili, Universidad Rovira i Virgili, Reus, Spain
- 3Department of Molecular and Cellular Biology, Kennesaw State University, Kennesaw, GA, USA
The ubiquitous “jack-of-all-trades,” Aeromonas hydrophila, is a freshwater, Gram-negative bacterial pathogen under revision in regard to its phylogenetic and functional affiliation with other aeromonads. While virulence factors are expectedly diverse across A. hydrophila strains and closely related species, our mechanistic knowledge of the vast majority of these factors is based on the molecular characterization of the strains A. hydrophila AH-3 and SSU, which were reclassified as A. piscicola AH-3 in 2009 and A. dhakensis SSU in 2013. Individually, these reclassifications raise important questions involving the applicability of previous research on A. hydrophila virulence mechanisms; however, this issue is exacerbated by a lack of genomic data on other research strains. Collectively, these changes represent a fundamental gap in the literature on A. hydrophila and confirm the necessity of biochemical, molecular, and morphological techniques in the classification of research strains that are used as a foundation for future research. This review revisits what is known about virulence in A. hydrophila and the feasibility of using comparative genomics in light of this phylogenetic revision. Conflicting data between virulence factors, secretion systems, quorum sensing, and their effect on A. hydrophila pathogenicity appears to be an artifact of inappropriate taxonomic comparisons and/or be due to the fact that these properties are strain-specific. This review audits emerging data on dominant virulence factors that are present in both A. dhakensis and A. hydrophila in order to synthesize existing data with the aim of locating where future research is needed.
Introduction
The ubiquitous bacterium Aeromonas hydrophila is a freshwater, facultatively anaerobic, chemoorganoheterotroph (Garrity et al., 2006) and the etiologic agent of disease in amphibians, birds, fishes, mammals, and reptiles, with the most common forms of disease being gastroenteritis, septicemia, and necrotizing fasciitis (Cipriano et al., 1984; Figueras et al., 2007; Monaghan et al., 2008; Janda and Abbott, 2010). Virulence in A. hydrophila is multifactorial, with disease resulting from the production and/or secretion of virulence factors, such as adhesins, cytotoxins, hemolysins, lipases, and proteases as well as the capacity to form biofilms, use specific metabolic pathways, and mediate virulence factor expression through quorum sensing (Allan and Stevenson, 1981; Cahill, 1990; Thornley et al., 1997; Beaz-Hidalgo and Figueras, 2013). The majority of experimental studies on identifying virulence determinants in Aeromonas spp. have been performed in the strain A. hydrophila SSU, which was later recognized to be affiliated with A. dhakensis on the basis of ANI and phylogeny comparisons (Grim et al., 2014). Adding confusion to this complexity, the literature on A. hydrophila is riddled with conflicting reports on the molecular determinants of virulence attributed to this species because of changes in classification and problems stemming from misidentification (Colston et al., 2014; Beaz-Hidalgo et al., 2015). The purpose of this review article is to provide an updated view on what is known about virulence factors in the aftermath of reclassification of A. hydrophila SSU.
Huys et al. (2002) recognized that some diarrheal isolates, while closely related to A. hydrophila, show atypical metabolic activities for urocanic acid (+), L-fucose (-), and L-arabinose (-). On these bases, these strains were classified into a subspecies known as A. hydrophila subsp. dhakensis (Huys et al., 2002). Then, in 2013, A. hydrophila subsp. dhakensis was recognized to be synonymous to A. aquariorum and both were combined under the name A. dhakensis, a species that is functionally divergent from A. hydrophila, based on multilocus phylogenetic analyses and phenotypic characteristics (Beaz-Hidalgo et al., 2013). Studies on the virulence factors expressed by the diarrheal isolate SSU, previously considered to be affiliated to A. hydrophila and now know to be A. dhakensis, are regarded as the seminal literature on molecular pathogenesis of Aeromonas (Grim et al., 2014). Given the turbulent nature of classification within Aeromonas spp., this review aims to clarify which virulence factors have been characterized within current members of A. hydrophila (Supplementary Table S1) by auditing the body of knowledge on the molecular understanding of these genes so that future research can progress from a more solid foundation.
Regulation of Aeromonas Virulence Determinants
Cascades of genetic regulation that lead to situational expression of virulence factors are known to occur in Aeromonas spp., but these interactions remain a relatively uncharted area of research in phylogenetically confirmed A. hydrophila strains. For example, outbreaks of A. hydrophila are generally thought to be linked with changes in host susceptibility caused by environmental changes, such as hypoxic conditions and excessive nitrite levels in farmed fish, as well as increases in temperature, which are linked with the production of virulence factors, such as cyototoxins and hemolysins (Swann and White, 1991; Mateos et al., 1993; Janda and Abbott, 2010). To exploit changes in host susceptibility due to increases in temperature, Aeromonas spp. virulence factors have also evolved temperature-dependent expression (Merino et al., 1992; Gonzalez-Serrano et al., 2002). For example, clinical strains of A. hydrophila can grow at temperatures greater than the isolate’s optimal growth temperature of 28°C (Popoff and Veron, 1976); however, when temperatures increase to 37°C, protease activity decreases and cytotoxin and hemolysin activity increases (Yu et al., 2007). In contrast, environmental isolates are well adapted to low temperatures and can grow uninhibited at temperatures as low as 4°C, a temperature that restricts growth of clinical isolates (Mateos et al., 1993). Some of the better studied regulatory effects are the linkage between quorum sensing and biofilm formation which was shown to not only mediate the expression of virulence factors, but also regulate cell density (Swift et al., 1997; Lynch et al., 2002; Janda and Abbott, 2010). In addition, while polar flagella in A. hydrophila are constitutively expressed, there are well-described regulators that trigger lateral flagella expression such as surface contact and viscosity (Wilhelms et al., 2011, 2013). Another class of regulatory effects includes the upregulation of virulence factors through lysogenic conversion; however, to-date no experimental data has been published on this phenomenon within A. hydrophila. Considering the broad effects that these regulatory factors have on disease, experimental studies that resolve these interactions are fundamental to the advancement of knowledge for the field of A. hydrophila as a whole. A review of known virulence factors and the respective regulatory effects that have been evaluated in Aeromonas spp. and are genetically present within A. hydrophila are presented in Figure 1.
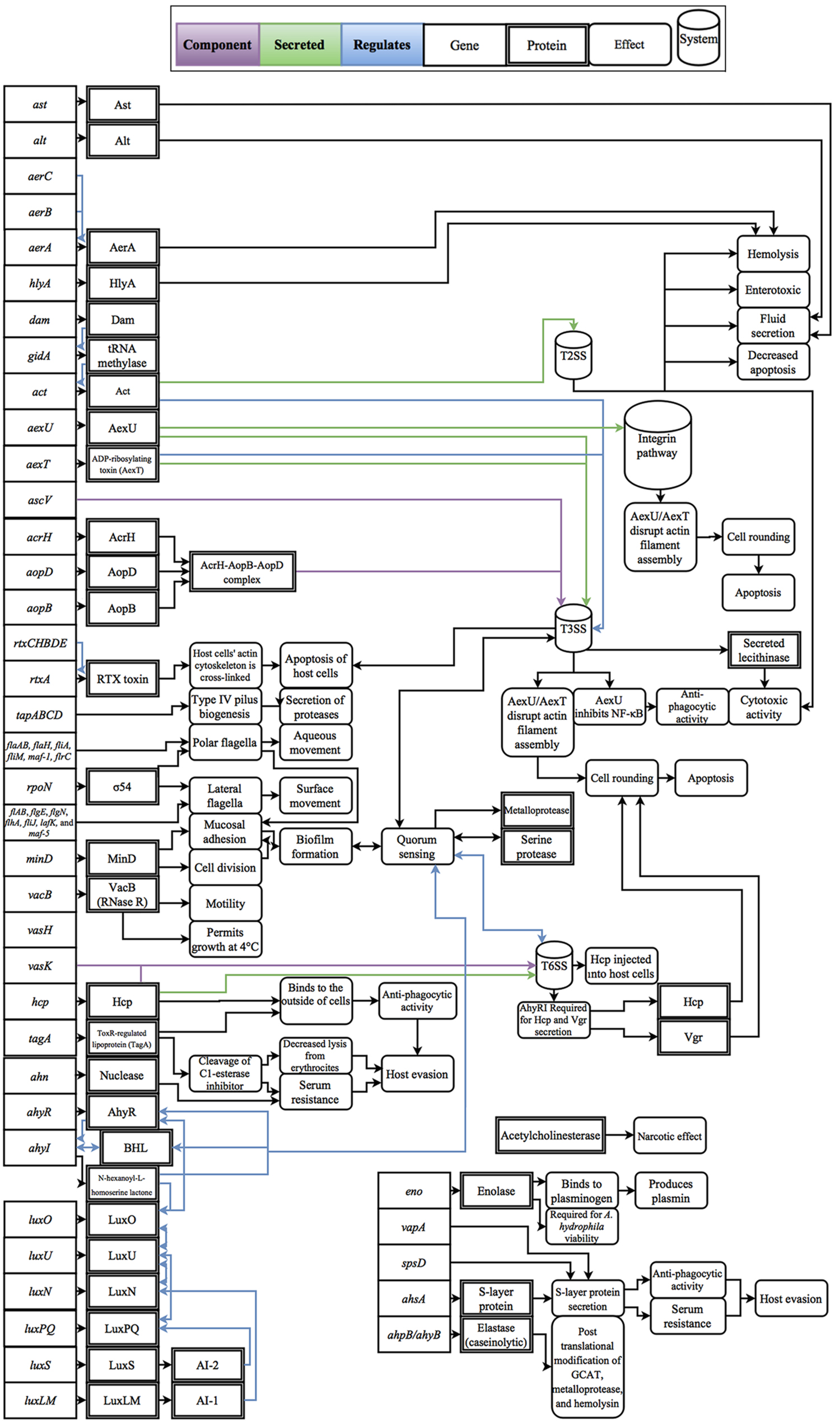
FIGURE 1. Diagram of the gene products, molecular interactions and functions implicated in A. hydrophila virulence. These interactions are based on the collective literature referenced in this manuscript.
Secretion Systems
Type II Secretion System and Effector Proteins
The widely conserved type II secretion system (T2SS) is present in all known members of A. hydrophila and is integral in the extracellular secretion of a wide array of virulence factors including aerolysin, amylases, DNases, and proteases (Sandkvist, 2001; Peabody et al., 2003; Tseng et al., 2009; Pang et al., 2015). In A. dhakensis SSU, the T2SS secretes what is perhaps the most potent virulence factor; the aerolysin-related cytotoxic enterotoxin Act (Galindo et al., 2004). While genes for this and other virulence factors that interact with the T2SS are present in current members of A. hydrophila, the contribution of this system to virulence remains unquantified (Cianciotto, 2005).
Type III Secretion System and Effector Proteins
Found in higher frequency in clinical isolates than in aquatic isolates (Aguilera-Arreola et al., 2005; Pang et al., 2015), the type III secretion system (T3SS) functions as a molecular needle, injecting effector toxins into host cells (Galan and Collmer, 1999; Sha et al., 2002; Sierra et al., 2010). Although no studies have been performed in members of A. hydrophila with publically accessible genomic data, the T3SS has been shown in Aeromonas spp. to be co-regulated by contact with host cells, cytotoxic enterotoxin Act, DNA adenine methyltransferase, flagella, lipopolysaccharides, DNA methylation, temperature, calcium/magnesium levels, and quorum sensing while requiring effectors to have the appropriate secretion signal (Braun et al., 2002; Burr et al., 2003; Ebanks et al., 2006; Erova et al., 2006a,b; Vilches et al., 2009). Because of its strong association with the export of virulence factors by many Aeromonas spp., the experimental manipulation of genes that encode for subunits of this secretion system, which resulted in attenuation of virulence in the reclassified A. piscicola AH-3 (formerly A. hydrophila), may also result in the attenuation of A. hydrophila (Vilches et al., 2004; Sha et al., 2005; Vilches et al., 2008; Beaz-Hidalgo et al., 2009; Vilches et al., 2009; Khajanchi et al., 2010). At the same time, genetic heterogeneity may prevent the translation of this research. For example, calcium chelation promotes T3SS/AexT expression in A. piscicola AH-3 and in A. salmonicida JF2267, but these effects are absent in A. salmonicida A229, A. salmonicida A449 and in A. dhakensis SSU (Burr et al., 2003; Ebanks et al., 2006; Vilches et al., 2008, 2009). On a molecular level, A. salmonicida JF2267 was shown to lose its plasmid, which contains the T3SS genes, at 25°C whereas A. salmonicida A449 conversely increases transcription of T3SS genes between 25 and 28°C (Ebanks et al., 2006). Therefore, while the same system appears, they are different on a procedural level.
Within A. hydrophila, numerous studies linked the T3SS and its effector proteins with virulence. In A. hydrophila AH-1, an isolate of blue gourami (Trichopodus trichopterus) with publically available nucleotide data (whole genome is not available), insertional mutagenesis of aopB (T3SS translocator) and aopD (integral T3SS transmembrane component) causes a reduction in cytotoxicity and an increase in phagocytosis because the T3SS is no longer able to translocate effector proteins (Yu et al., 2004). Similarly, in A. dhakensis SSU, T3SS genes have been linked with virulence that include the T3SS-associated exoenzyme effector (AexU), which increases host evasion, degrades host actin, and is independently lethal (Sierra et al., 2010; Grim et al., 2013). AcrH is a chaperone that complexes with AopB and AopD (Tan et al., 2009); acrH mutants are predicted to have attenuated virulence. Contextually, the aexU and acrH genes are present in a minority of A. hydrophila and no experimental studies have been performed to establish their respective roles in virulence. While no experimental manipulations were performed, a subsequent study compared clinical and environmental isolates of A. hydrophila, showing that T3SS structural genes aopB and ascV are most abundant in A. hydrophila disease isolates (Carvalho-Castro et al., 2010), a link with virulence that is supported by the attenuation of virulence in A. piscicola AH-3 ascV mutants (Vilches et al., 2004). Collectively, these results appear to indicate that the T3SS is a strong contributing factor for virulence of Aeromonas spp. However, genomic analyses of pathogenic A. hydrophila isolates indicate that alternate secretory mechanisms may also be critical for pathogenesis given that hypervirulent isolates of A. hydrophila that infect farmed fish lack T3SS core components (Hossain et al., 2013; Pang et al., 2015).
Previously described in A. salmonicida (Braun et al., 2002), the ADP-ribosylating toxin AexT is present in ~90% of Aeromonas spp. that have a T3SS and when this T3SS effector is abrogated in A. piscicola AH-3, a slight attenuation of virulence has been observed based on virulence assays for cytotoxicity and phagocytosis as well as fish and mice challenges (Vilches et al., 2008). The aexT-like gene aexU shows a stronger contribution to virulence, with aexU mutants having an LD50 of 60% using 2–3 times the dose of wild-type A. dhakensis SSU (Sha et al., 2007; Sierra et al., 2007; Vilches et al., 2008).
The Type VI Secretion System and Effector Proteins
The type VI secretion system (T6SS) functions analogously to a phage tail, allowing injection of virulence factors into host cells via valine glycine repeat G (VrgG) proteins and hemolysin-coregulated protein (Hcp), which functions as an antimicrobial pore-forming protein when secreted or as a structural protein (Bingle et al., 2008). In A. dhakensis SSU, the transcriptional regulator VasH and the helical transmembrane protein VasK are linked with secretion of Hcp, with vasH and vasK mutants resulting in decreased anti-phagocytic activity and attenuated virulence in a septicemic mouse model which serves as a line of evidence that the T6SS is involved in the manifestation of disease (Suarez et al., 2008), but similar to the disparate results of the T3SS, the T6SS is not obligatory for A. hydrophila virulence. For example, some members of the newly described hypervirulent A. hydrophila pathotype of freshwater fishes have a complete T6SS while others retain only 4/13 core components (Pang et al., 2015; Rasmussen-Ivey et al., 2016). With a distribution in 26 out of 37 strains listed as A. hydrophila in GenBank, the T6SS’s role in virulence may be specific to the mode of infection with bacteria that contain a complete T6SS having greater antimicrobial activity, but at the cost of stimulating host defenses. In other bacteria, the T6SS also plays a role in biofilm formation, and evasion of the host immune system, but future research is needed to assess the role(s) of the T6SS within A. hydrophila.
Biofilm Formation
Biofilms provide bacteria with resistance to antimicrobial agents and host defenses (Costerton et al., 1995; Lynch et al., 2002). Aeromonas spp. evolved multiple regulatory mechanisms for biofilm formation that are intimately linked with the production of virulence factors. The quorum sensing response regulator of the reclassified isolate A. piscicola A1 (formerly A. hydrophila), ahyRI, produces LuxRI homologs, N-(butanoyl)-L-homoserine lactones (BHL), and N-hexanoyl-L-homoserine lactones (AHL); autoinducers that regulate cell division (Swift et al., 1997). In A. dhakensis SSU ΔahyRI mutants, T6SS effectors Hcp and Vgr are unable to be secreted which results in decreased biofilm formation (Khajanchi et al., 2009). Interestingly, some strains transcribe ahyRI (e.g., A. hydrophila ATCC 7966), but lack AHL/BHLs, which may indicate an alternate function of ahyRI that has yet to be described (dos Reis Ponce-Rossi et al., 2016). Similarly, the recently characterized autoregulatory two-component signal transduction system QseBC is a widely conserved system within A. hydrophila and was first described in A. dhakensis SSU, as mutants with an inactive response regulator (QseB) have reduced swimming and swarming motility, form thicker biofilms, and secrete fewer virulence factors, which leads to attenuation of virulence. When the gene aha0701h is overexpressed in ΔqseB mutants, biofilm formation decreases, presumably due to dysregulation of genes fleN (regulates flagellar number) and vpsT (transcriptional response regulator; Khajanchi et al., 2012; Kozlova et al., 2012).
Flagella and Pili
A. hydrophila isolates produce lateral flagella for surface movement/swarming and polar flagella for movement in suspension. Polar flagella production has been studied within A. piscicola AH-3, with mutations in flaAB, flaH, fliA, fliM, maf-1, and flrC abolishing production of polar flagella and resulting in decreased adherence and biofilm formation (Canals et al., 2006). Considering that flagellar glycosylation was shown to be linked with the ability to form biofilms as well as adhere to Hep-2 cells, it is important to mention that there are notable differences within Aeromonas species. In addition to having only a single lateral flagellin, polar and lateral flagella are glycosylated in A. piscicola AH-3 whereas A. hydrophila AH-1 has two lateral flagellins and only the polar flagellum is glycosylated. When pseudaminic acid biosynthesis genes pseB and pseI were mutagenized, the result was an inability to produce both polar and lateral flagella in A. piscicola AH-3, but only affected polar flagella production in A. hydrophila AH-1. Therefore, lateral flagella production was unaffected in glycosylation negative A. hydrophila AH-1 mutants (Fulton et al., 2015). Similarly, in the diseased eel isolate A. hydrophila W (no genome submitted), mutations in flgE, flgN, flhA, fliJ, flmB, lafK, and maf-5 result in loss of lateral flagella, which causes decreased motility, biofilm formation, and mucosal adherence (Jiang et al., 2015). While polar and lateral flagella transcriptional hierarchies, regulation, and contribution to virulence are well-described in other species, as the date of this publication, no member of A. hydrophila with a publically accessible genome has undergone genetic manipulations to evaluate the contribution of polar or lateral flagella for virulence.
The A. hydrophila gene cluster tapABCD is responsible for type IV pilus biogenesis and is an integral part of the extracellular secretory pathway. To test for function, the A. hydrophila Ah65 (genome unavailable) gene tapD gene was used to successfully complement a strain of Pseudomonas aeruginosa that lacks PilD (an ortholog of TapD; Pepe et al., 1996). Another type IV pilus is the bundle-forming pilus, which is encoded by bfp and acts an important internal colonization factor for multiple species of Aeromonas (A. hydrophila Ah65 was observed expressing both bfp and tap; Barnett et al., 1997; Kirov et al., 2000). Taken with the observation that TapD is required for secretion of virulence factors, such as aerolysin and proteases, these genes appear to be fundamental for pathogenicity.
Structural Proteins, Phospholipids, and Polysaccharides
Capsules, O-antigens, and S-layer proteins provide mechanisms to evade host defenses. Within Aeromonas spp., capsules also show anti-phagocytic activity, increase resistance to the complement system, and increase adherence (Martinez et al., 1995; Merino et al., 1997). O-antigens are a class of structurally diverse lipopolysaccharides that act as colonization factors. At 20°C O-antigen is produced by A. piscicola AH-3, but not at 37°C, resulting an O-antigen-deficient strains that are unable to colonize hosts and have reduced expression of T3SS components (Merino et al., 1996; Vilches et al., 2009). Across A. hydrophila, eight distinct O-antigen gene clusters are present, with all epidemic strains isolated from channel catfish (Ictalurus punctatus) sharing a homologous O-antigen gene cluster (Hossain, 2012). In A. hydrophila TF7 (genomic data unavailable), the S-layer protein gene (ahsA) encodes an external paracrystalline layer that is lost upon insertional mutagenesis of spsD (S-protein secretion; Thomas and Trust, 1995). Another study of S-layer proteins in five pathogenic human and eel isolates of A. hydrophila (A19, AH290, E37, E40, and TW1; genomic data unavailable) shows that serogroups of A. hydrophila other than O:11 contain S-layer proteins O:14 and O:81 (Esteve et al., 2004).
Hemolysins
Hemolysins are a diverse group of multifunctional enzymes that play a central role in A. hydrophila pathogenesis (Wadstrom et al., 1976; Asao et al., 1984). The extracellular heat-labile hemolysin (AHH1) is the most abundant of several widely distributed hemolysins (AerA, AHH1, AhyA, and Asa1), with the most cytotoxic genotype being a synergistic combination of aerA and ahh1 (Hirono and Aoki, 1991; Wang et al., 2003). In A. media A6 (formerly A. hydrophila) Aerolysin A (aerA) and Hemolysin A (hlyA) comprise another two-component hemolytic system in which virulence is attenuated only when both hlyA and aerA activity is abolished (Wong et al., 1998). In A. dhakensis SSU, the iron dependent, fur and gidA-regulated, enterotoxin Act is the most cytotoxic virulence factor of and a core gene within A. hydrophila, with studies in A. dhakensis SSU demonstrating that Act induces multiple effects including hemolytic, cytotonic, and cytotoxic activities, but unlike other virulence factors exported via the T3SS or T6SS, Act is exported through the T2SS (Chopra et al., 2000; Sha et al., 2001, 2004, 2005; Fadl et al., 2006; Erova et al., 2012).
Collagenase, Serine Protease, Metalloprotease, Enolase, and Lipase
A. hydrophila spp. express diverse degradative enzymes that can contribute to virulence including collagenase, elastase, enolase, lipase, metalloprotease, and serine protease. A. piscicola AH-3 contains a collagenase, which has sequence similarity to the open reading frame AHA_0517 of A. hydrophila ATCC 7966T, and was shown to be cytotoxic to Vero cells, with loss of this enzyme resulting in a 5–15% increase in cell viability; however, this mutation did not result in complete reduction of cytotoxicity (Duarte et al., 2015). The ahpAB genes of A. hydrophila AG2 (genomic data unavailable) produce potent virulence factors: an extracellular protease that is not essential for virulence, but is present in the most virulent pathotypes (AerA+Alt+Ahp+) along with a secreted elastase with caseinolytic and elastolytic activity that correlates with an LD50 100 times more virulent than AhpB- mutants when assayed in rainbow trout (Oncorhynchus mykiss; Rivero et al., 1991; Cascon et al., 2000; Li et al., 2011). Another extracellular protease (epr) was discovered in the soft-shell turtle isolate A. hydrophila AH1 and found to be present in the most common pathotype in diseased fishes (Aer+Alt+Act+EprCAI+Ahp+; Chang et al., 1997; Hu et al., 2012). In the rainbow trout isolate A. hydrophila B32, a novel serine protease (ser) was found that exhibits cytotoxic properties and is thermostable, both of which are characteristics that differentiate this protease from known A. hydrophila α-hemolysins and β-hemolysins (Rodriguez et al., 1992). While four times less active than serine protease, the virulent A. hydrophila EO63 (genomic data unavailable) was shown to produce a thermostable metalloprotease with enzymatic activity on casein and elastin, an optimal pH of 8.0, and an LD50 of 3.5 μg/g (Esteve and Birbeck, 2004). Enolase, a secreted and surface-expressed glycolytic enzyme, was identified as a virulence factor in A. dhakensis SSU, based on binding to human plasminogen which leads to production of plasmin (degrades blood plasma proteins), with previous reports showing that enolase functions as a heat-shock protein and a regulator of transcription by binding host chromatin/cytoskeletal structures as well as being necessary for viability (Sha et al., 2009).
In general, lipases have diverse functions, but are linked with virulence in numerous pathogens (Stehr et al., 2003). An extracellular lipase (EC3.1.1.3) is produced by A. piscicola AH-3 (formerly A. hydrophila); however, the link between virulence and this gene is speculative in A. hydrophila (Anguita et al., 1993). Conversely, the heat-labile lipase Alt and the heat-stable lipase Ast are important cytotonic enterotoxins in the pathogenicity of A. dhakensis SSU, with both being able to cause significant fluid secretion, with only the previously described cytotoxic enterotoxin Act having a greater effect on fluid secretion (Sha et al., 2002; Li et al., 2011). Based on comparative genomics, Alt and Ast are core elements of A. hydrophila; however, no experiments have been performed within existing members of this species to characterize these toxins. Two additional lipases, phospholipase A1 (pla) and phospholipase C (plc), were explored in A. piscicola AH-3, with the finding that pla lacks a significant effect on virulence while plc (lecithinase) was cytotoxic and has LD50 values 10 times more virulent than plc-deficient mutants (Merino et al., 1999).
Other Virulence Factors
The range of virulence factors encoded by A. hydrophila includes adherence proteins, catalysts, nucleases, and toxins that may be expressed differently depending upon the respective environment. The role of the adhesin minD in virulence is its ability to mediate mucosal adherence, increase biofilm formation, and facilitate cell division as well as motility (Huang et al., 2015). The enzyme 5-enolpyruvylshikimate 3-phosphate synthase (encoded by aroA) is required for folate availability and contributes to A. hydrophila AG2 (genomic data unavailable) viability in intraperitoneally injected rainbow trout, with aroA mutants no longer recoverable from fish internal organs because environmentally derived folate is scarce (Hernanz Moral et al., 1998). Another element of host evasion is the nuclease encoded by the ahn gene of A. hydrophila J-1 which shows no significant change in hemolytic activity or growth in vitro; however, when Δahn mutants are introduced into fish and mice models, virulence is attenuated (Ji et al., 2015). Another conserved gene of A. hydrophila that has only been characterized in A. dhakensis SSU is vacB, which encodes RNase R; an exoribonuclease with multiple functions that include permitting growth at 4°C (cold-shock protein) and supporting motility. Isogenic mutants of RNase R show a 70% attenuation in virulence (Erova et al., 2008).
Another virulence factor that is shown to increase host evasion for A. dhakensis SSU is the pore-forming RTX toxin RtxA that requires contact with host cells and is regulated by the rtxACHBDE operon so that production of RtxA coincides with regulation of other cytotoxins, such as aerolysins and hemolysins and acts to covalently cross-link host cytoskeletal-actin, resulting in host cells having a rounded phenotype that leads to apoptosis (McCoy et al., 2010; Suarez et al., 2012). Another conserved virulence factor across all A. hydrophila strains in GenBank is the ToxR-regulated lipoprotein (TagA) of A. dhakensis SSU, which cleaves the complement C1-esterase inhibitor, thereby increasing serum resistance and decreasing erythrocyte lysis (Pillai et al., 2006).
The Role of Horizontal Genetic Transfer in Virulence
The introduction of virulence factors as well as their effects on the alternate regulation within Aeromonas spp. is a recurring theme of crucial importance, yet these elements remain understudied. Plasmids are a confirmed source of multidrug resistance in Aeromonas spp. and have been shown to have the potential to be conjugally transferred between known human pathogens, such as Acinetobacter baumannii AYE and A. hydrophila (Del Castillo et al., 2013). In addition to being used as a “molecular map” to identify ancestral lineages, prophage that contain putative cis-acting elements and trans-acting factors were found to be conserved within hypervirulent strains of A. hydrophila which strongly implies that the differential regulation of virulence factors (and therefore the dramatic increase in virulence) may be caused by the lysogenic conversion of this conserved A. hydrophila lineage by these mobile genetic elements (Hossain et al., 2013). Of note, while A. dhakensis SSU contains the majority of virulence factors that are present within confirmed members of A. hydrophila, all other members of A. dhakensis with fully sequenced genomes appear to lack these genes (Supplementary Table S1). Results of numerous core genome phylogenies and average nucleotide identity analyses support the grouping of A. hydrophila SSU within A. dhakensis (Colston et al., 2014; Beaz-Hidalgo et al., 2015; Rasmussen-Ivey et al., 2016), if virulence factors are introduced or controlled by mobile genetic elements, then taxonomy and functionality demand separate analyses.
Discussion
In Aeromonas spp., as with all pathogens, disease is the result of complex molecular interactions between bacterium, environment, and host; however, the literature on A. hydrophila remains limited by the lack of experimental data on validated members of A. hydrophila. While there numerous virulence factors shared between members of A. hydrophila, A. dhakensis SSU, and A. piscicola AH-3 there are also key examples in the literature that show conflicting data between virulence factors, secretion systems, quorum sensing, and their effect on pathogenicity. This inconsistency is illustrated by the highly virulent catfish isolate A. hydrophila ML09-119 that acts as a primary pathogen when other members of this species act as secondary pathogens (Griffin et al., 2013; Zhang et al., 2013). With that in mind, the thorough research conducted on A. dhakensis SSU by researchers, such as Dr. Ashok Chopra, still holds relevance for A. hydrophila, but future research should be mindful of the phylogenetic reclassification for strains AH-3 and SSU and that there may be significant differences in the molecular determinants of virulence for A. hydrophila.
To compare isolates of A. hydrophila, biochemical, morphological, and molecular techniques are required (Abbott et al., 2003; Martinez-Murcia et al., 2011; Beaz-Hidalgo et al., 2015). As of 2016, few strains exist that have enough supporting data to facilitate comparative studies. There are many sources of uncertainty when comparing A. hydrophila strains, including genetic heterogeneity, the lack of natural models of infection, and reclassification of bacterial strains as new data emerges. Future research should aim to couple typing techniques (e.g., genome sequencing) with experimental data on virulence determinants so that there is a clear phylogenetic context for these studies.
When considering known virulence factors, the definitive biological separation of A. hydrophila, A. piscicola AH-3, and A. dhakensis SSU has yet to be established. In A. hydrophila as in these other species, disease is the result of a molecular symphony, with each virulence factor contributing to a cumulative effect (Figure 1). Research studying novel virulence factors and regulatory effects will help unveil the determinants that allow for infection and what differentiates A. hydrophila from other aeromonads. To better understand A. hydrophila pathogenesis it is imperative that future research develops natural models of infection, assesses the role of mobile genetic elements in virulence, and quantifies the interplay between virulence factors and host response in concert with molecular genetic approaches.
Author Contributions
CR-I, MF, DM, and ML all contributed to the conception, writing and editing of this manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by a grant from the United States Department of Agriculture’s Agriculture and Food Research Initiative (#2013-67015-21313) to MRL and by the Ministerio de Ciencia e Innovación award JPIW2013-095-CO3 to MJF.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01337
TABLE S1 | Gene products implicated in A. hydrophila virulence and the relative coverage and identity of strains listed in GenBank against the strain from which the gene product was characterized.
References
Abbott, S. L., Cheung, W. K., and Janda, J. M. (2003). The genus Aeromonas: biochemical characteristics, atypical reactions, and phenotypic identification schemes. J. Clin. Microbiol. 41, 2348–2357. doi: 10.1128/JCM.41.6.2348-2357.2003
Aguilera-Arreola, M. G., Hernandez-Rodriguez, C., Zuniga, G., Figueras, M. J., and Castro-Escarpulli, G. (2005). Aeromonas hydrophila clinical and environmental ecotypes as revealed by genetic diversity and virulence genes. FEMS Microbiol. Lett. 242, 231–240. doi: 10.1016/j.femsle.2004.11.011
Allan, B. J., and Stevenson, R. M. (1981). Extracellular virulence factors of Aeromonas hydrophila in fish infections. Can. J. Microbiol. 27, 1114–1122. doi: 10.1139/m81-174
Anguita, J., Rodriguez Aparicio, L. B., and Naharro, G. (1993). Purification, gene cloning, amino acid sequence analysis, and expression of an extracellular lipase from an Aeromonas hydrophila human isolate. Appl. Environ. Microbiol. 59, 2411–2417.
Asao, T., Kinoshita, Y., Kozaki, S., Uemura, T., and Sakaguchi, G. (1984). Purification and some properties of Aeromonas hydrophila hemolysin. Infect. Immun. 46, 122–127.
Barnett, T. C., Kirov, S. M., Strom, M. S., and Sanderson, K. (1997). Aeromonas spp. possess at least two distinct type IV pilus families. Microb. Pathog. 23, 241–247. doi: 10.1006/mpat.1997.0152
Beaz-Hidalgo, R., Alperi, A., Figueras, M. J., and Romalde, J. L. (2009). Aeromonas piscicola sp. nov., isolated from diseased fish. Syst. Appl. Microbiol. 32, 471–479. doi: 10.1016/j.syapm.2009.06.004
Beaz-Hidalgo, R., and Figueras, M. J. (2013). Aeromonas spp. whole genomes and virulence factors implicated in fish disease. J. Fish Dis. 36, 371–388. doi: 10.1111/jfd.12025
Beaz-Hidalgo, R., Hossain, M. J., Liles, M. R., and Figueras, M. J. (2015). Strategies to avoid wrongly labelled genomes using as example the detected wrong taxonomic affiliation for Aeromonas genomes in the genbank database. PLoS ONE 10:e0115813. doi: 10.1371/journal.pone.0115813
Beaz-Hidalgo, R., Martinez-Murcia, A., and Figueras, M. J. (2013). Reclassification of Aeromonas hydrophila subsp. dhakensis Huys et al., 2002 and Aeromonas aquariorum Martinez-Murcia et al. 2008 as Aeromonas dhakensis sp. nov. comb nov. and emendation of the species Aeromonas hydrophila. Syst. Appl. Microbiol. 36, 171–176. doi: 10.1016/j.syapm.2012.12.007
Bingle, L. E., Bailey, C. M., and Pallen, M. J. (2008). Type VI secretion: a beginner’s guide. Curr. Opin. Microbiol. 11, 3–8. doi: 10.1016/j.mib.2008.01.006
Braun, M., Stuber, K., Schlatter, Y., Wahli, T., Kuhnert, P., and Frey, J. (2002). Characterization of an ADP-ribosyltransferase toxin (AexT) from Aeromonas salmonicida subsp. salmonicida. J. Bacteriol. 184, 1851–1858. doi: 10.1128/JB.184.7.1851-1858.2002
Burr, S. E., Stuber, K., and Frey, J. (2003). The ADP-ribosylating toxin. AexT, from Aeromonas salmonicida subsp. salmonicida is translocated via a type III secretion pathway. J. Bacteriol. 185, 6583–6591.
Cahill, M. M. (1990). Virulence factors in motile Aeromonas species. J. Appl. Bacteriol. 69, 1–16. doi: 10.1111/j.1365-2672.1990.tb02905.x
Canals, R., Altarriba, M., Vilches, S., Horsburgh, G., Shaw, J. G., Tomas, J. M., et al. (2006). Analysis of the lateral flagellar gene system of Aeromonas hydrophila AH-3. J. Bacteriol. 188, 852–862. doi: 10.1128/JB.188.3.852-862.2006
Carvalho-Castro, G. A., Lopes, C. O., Leal, C. A., Cardoso, P. G., Leite, R. C., and Figueiredo, H. C. (2010). Detection of type III secretion system genes in Aeromonas hydrophila and their relationship with virulence in Nile tilapia. Vet. Microbiol. 144, 371–376. doi: 10.1016/j.vetmic.2010.01.021
Cascon, A., Yugueros, J., Temprano, A., Sanchez, M., Hernanz, C., Luengo, J. M., et al. (2000). A major secreted elastase is essential for pathogenicity of Aeromonas hydrophila. Infect. Immun. 68, 3233–3241. doi: 10.1128/IAI.68.6.3233-3241.2000
Chang, C. Y., Thompson, H., Rodman, N., Bylander, J., and Thomas, J. (1997). Pathogenic analysis of Aeromonas hydrophila septicemia. Ann. Clin. Lab. Sci. 27, 254–259.
Chopra, A. K., Xu, X., Ribardo, D., Gonzalez, M., Kuhl, K., Peterson, J. W., et al. (2000). The cytotoxic enterotoxin of Aeromonas hydrophila induces proinflammatory cytokine production and activates arachidonic acid metabolism in macrophages. Infect. Immun. 68, 2808–2818. doi: 10.1128/IAI.68.5.2808-2818.2000
Cianciotto, N. P. (2005). Type II secretion: a protein secretion system for all seasons. Trends Microbiol. 13, 581–588. doi: 10.1016/j.tim.2005.09.005
Cipriano, R. C., Bullock, G. L., and Pyle, S. W. (1984). Aeromonas hydrophila and Motile Aeromonad Septicemias of Fish. Paper No. 134. Washington, DC: U.S. Fish and Wildlife Service.
Colston, S. M., Fullmer, M. S., Beka, L., Lamy, B., Gogarten, J. P., and Graf, J. (2014). Bioinformatic genome comparisons for taxonomic and phylogenetic assignments using Aeromonas as a test case. MBio 5, e02136. doi: 10.1128/mBio.02136-14
Costerton, J. W., Lewandowski, Z., Caldwell, D. E., Korber, D. R., and Lappin-Scott, H. M. (1995). Microbial biofilms. Annu. Rev. Microbiol. 49, 711–745. doi: 10.1146/annurev.mi.49.100195.003431
Del Castillo, C. S., Hikima, J.-I., Jang, H.-B., Nho, S.-W., Jung, T.-S., Wongtavatchai, J., et al. (2013). Comparative sequence analysis of a multidrug-resistant plasmid from Aeromonas hydrophila. Antimicrob. Agents Chemother. 57, 120–129. doi: 10.1128/AAC.01239-12
dos Reis Ponce-Rossi, A., Pinto, U. M., Ribon, A. D. O. B., Bazzolli, D. M. S., and Vanetti, M. C. D. (2016). Quorum sensing regulated phenotypes in Aeromonas hydrophila ATCC 7966 deficient in AHL production. Ann. Microbiol. 66, 1117–1126. doi: 10.1007/s13213-016-1196-4
Duarte, A. S., Cavaleiro, E., Pereira, C., Merino, S., Esteves, A. C., Duarte, E. P., et al. (2015). Aeromonas piscicola AH-3 expresses an extracellular collagenase with cytotoxic properties. Lett. Appl. Microbiol. 60, 288–297. doi: 10.1111/lam.12373
Ebanks, R. O., Knickle, L. C., Goguen, M., Boyd, J. M., Pinto, D. M., Reith, M., et al. (2006). Expression of and secretion through the Aeromonas salmonicida type III secretion system. Microbiology 152, 1275–1286. doi: 10.1099/mic.0.28485-0
Erova, T. E., Fadl, A. A., Sha, J., Khajanchi, B. K., Pillai, L. L., Kozlova, E. V., et al. (2006a). Mutations within the catalytic motif of DNA adenine methyltransferase (Dam) of Aeromonas hydrophila cause the virulence of the Dam-overproducing strain to revert to that of the wild-type phenotype. Infect. Immun. 74, 5763–5772. doi: 10.1128/IAI.00994-06
Erova, T. E., Kosykh, V. G., Fadl, A. A., Sha, J., Horneman, A. J., and Chopra, A. K. (2008). Cold shock exoribonuclease R (VacB) is involved in Aeromonas hydrophila pathogenesis. J. Bacteriol. 190, 3467–3474. doi: 10.1128/JB.00075-08
Erova, T. E., Kosykh, V. G., Sha, J., and Chopra, A. K. (2012). DNA adenine methyltransferase (Dam) controls the expression of the cytotoxic enterotoxin (act) gene of Aeromonas hydrophila via tRNA modifying enzyme-glucose-inhibited division protein (GidA). Gene 498, 280–287. doi: 10.1016/j.gene.2012.02.024
Erova, T. E., Pillai, L., Fadl, A. A., Sha, J., Wang, S., Galindo, C. L., et al. (2006b). DNA adenine methyltransferase influences the virulence of Aeromonas hydrophila. Infect. Immun. 74, 410–424. doi: 10.1128/IAI.74.1.410-424.2006
Esteve, C., Alcaide, E., Canals, R., Merino, S., Blasco, D., Figueras, M. J., et al. (2004). Pathogenic Aeromonas hydrophila serogroup O:14 and O:81 strains with an S layer. Appl. Environ. Microbiol. 70, 5898–5904. doi: 10.1128/AEM.70.10.5898-5904.2004
Esteve, C., and Birbeck, T. H. (2004). Secretion of haemolysins and proteases by Aeromonas hydrophila EO63: separation and characterization of the serine protease (caseinase) and the metalloprotease (elastase). J. Appl. Microbiol. 96, 994–1001. doi: 10.1111/j.1365-2672.2004.02227.x
Fadl, A. A., Galindo, C. L., Sha, J., Erova, T. E., Houston, C. W., Olano, J. P., et al. (2006). Deletion of the genes encoding the type III secretion system and cytotoxic enterotoxin alters host responses to Aeromonas hydrophila infection. Microb. Pathog. 40, 198–210. doi: 10.1016/j.micpath.2006.01.003
Figueras, M. J., Aldea, M. J., Fernandez, N., Aspiroz, C., Alperi, A., and Guarro, J. (2007). Aeromonas hemolytic uremic syndrome. A case and a review of the literature. Diagn. Microbiol. Infect. Dis. 58, 231–234. doi: 10.1016/j.diagmicrobio.2006.11.023
Fulton, K. M., Mendoza-Barbera, E., Twine, S. M., Tomas, J. M., and Merino, S. (2015). Polar glycosylated and lateral non-glycosylated flagella from Aeromonas hydrophila strain AH-1 (Serotype O11). Int. J. Mol. Sci. 16, 28255–28269. doi: 10.3390/ijms161226097
Galan, J. E., and Collmer, A. (1999). Type III secretion machines: bacterial devices for protein delivery into host cells. Science 284, 1322–1328. doi: 10.1126/science.284.5418.1322
Galindo, C. L., Fadl, A. A., Sha, J., Gutierrez, C. Jr., Popov, V. L., Boldogh, I., et al. (2004). Aeromonas hydrophila cytotoxic enterotoxin activates mitogen-activated protein kinases and induces apoptosis in murine macrophages and human intestinal epithelial cells. J. Biol. Chem. 279, 37597–37612. doi: 10.1074/jbc.M404641200
Garrity, G., Staley, J. T., Boone, D. R., De Vos, P., Goodfellow, M., Rainey, F. A., et al. (2006). Bergey’s Manual® of Systematic Bacteriology: The Proteobacteria, Vol. 2. Berlin: Springer Science & Business Media.
Gonzalez-Serrano, C. J., Santos, J. A., Garcia-Lopez, M. L., and Otero, A. (2002). Virulence markers in Aeromonas hydrophila and Aeromonas veronii biovar sobria isolates from freshwater fish and from a diarrhoea case. J. Appl. Microbiol. 93, 414–419. doi: 10.1046/j.1365-2672.2002.01705.x
Griffin, M. J., Goodwin, A. E., Merry, G. E., Liles, M. R., Williams, M. A., Ware, C., et al. (2013). Rapid quantitative detection of Aeromonas hydrophila strains associated with disease outbreaks in catfish aquaculture. J. Vet. Diagn. Invest. 25, 473–481. doi: 10.1177/1040638713494210
Grim, C. J., Kozlova, E. V., Ponnusamy, D., Fitts, E. C., Sha, J., Kirtley, M. L., et al. (2014). Functional genomic characterization of virulence factors from necrotizing fasciitis-causing strains of Aeromonas hydrophila. Appl. Environ. Microbiol. 80, 4162–4183. doi: 10.1128/AEM.00486-14
Grim, C. J., Kozlova, E. V., Sha, J., Fitts, E. C., Van Lier, C. J., Kirtley, M. L., et al. (2013). Characterization of Aeromonas hydrophila wound pathotypes by comparative genomic and functional analyses of virulence genes. MBio 4, e64–e13. doi: 10.1128/mBio.00064-13
Hernanz Moral, C., Flano Del Castillo, E., Lopez Fierro, P., Villena Cortes, A., Anguita Castillo, J., Cascon Soriano, A., et al. (1998). Molecular characterization of the Aeromonas hydrophila aroA gene and potential use of an auxotrophic aroA mutant as a live attenuated vaccine. Infect. Immun. 66, 1813–1821.
Hirono, I., and Aoki, T. (1991). Nucleotide sequence and expression of an extracellular hemolysin gene of Aeromonas hydrophila. Microb. Pathog. 11, 189–197. doi: 10.1016/0882-4010(91)90049-G
Hossain, M. J. (2012). Molecular Interactions between Phage and the Catfish Pathogen Edwardsiella ictaluri and Comparative Genomics of Epidemic Strains of Aeromonas Hydrophila. Doctoral dissertation, Auburn University, Auburn, AL.
Hossain, M. J., Waldbieser, G. C., Sun, D. W., Capps, N. K., Hemstreet, W. B., Carlisle, K., et al. (2013). Implication of lateral genetic transfer in the emergence of Aeromonas hydrophila isolates of epidemic outbreaks in channel catfish. PLoS ONE 8:e80943. doi: 10.1371/journal.pone.0080943
Hu, M., Wang, N., Pan, Z. H., Lu, C. P., and Liu, Y. J. (2012). Identity and virulence properties of Aeromonas isolates from diseased fish, healthy controls and water environment in China. Lett. Appl. Microbiol. 55, 224–233. doi: 10.1111/j.1472-765X.2012.03281.x
Huang, L., Qin, Y., Yan, Q., Lin, G., Huang, L., Huang, B., et al. (2015). MinD plays an important role in Aeromonas hydrophila adherence to Anguilla japonica mucus. Gene 565, 275–281. doi: 10.1016/j.gene.2015.04.031
Huys, G., Kampfer, P., Albert, M. J., Kuhn, I., Denys, R., and Swings, J. (2002). Aeromonas hydrophila subsp. dhakensis subsp. nov., isolated from children with diarrhoea in Bangladesh, and extended description of Aeromonas hydrophila subsp. hydrophila (Chester 1901) Stanier 1943 (approved lists 1980). Int. J. Syst. Evol. Microbiol. 52, 705–712. doi: 10.1099/ijs.0.01844-0
Janda, J. M., and Abbott, S. L. (2010). The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 23, 35–73. doi: 10.1128/CMR.00039-09
Ji, Y., Li, J., Qin, Z., Li, A., Gu, Z., Liu, X., et al. (2015). Contribution of nuclease to the pathogenesis of Aeromonas hydrophila. Virulence 6, 515–522. doi: 10.1080/21505594.2015.1049806
Jiang, X., Qin, Y. X., Lin, G. F., Huang, L., Huang, B., Huang, W. S., et al. (2015). FlgN plays important roles in the adhesion of Aeromonas hydrophila to host mucus. Genet. Mol. Res. 14, 6376–6386. doi: 10.4238/2015.June.11.13
Khajanchi, B. K., Fadl, A. A., Borchardt, M. A., Berg, R. L., Horneman, A. J., Stemper, M. E., et al. (2010). Distribution of virulence factors and molecular fingerprinting of Aeromonas species isolates from water and clinical samples: suggestive evidence of water-to-human transmission. Appl. Environ. Microbiol. 76, 2313–2325. doi: 10.1128/AEM.02535-09
Khajanchi, B. K., Kozlova, E. V., Sha, J., Popov, V. L., and Chopra, A. K. (2012). The two-component QseBC signalling system regulates in vitro and in vivo virulence of Aeromonas hydrophila. Microbiology 158, 259–271. doi: 10.1099/mic.0.051805-0
Khajanchi, B. K., Sha, J., Kozlova, E. V., Erova, T. E., Suarez, G., Sierra, J. C., et al. (2009). N-acylhomoserine lactones involved in quorum sensing control the type VI secretion system, biofilm formation, protease production, and in vivo virulence in a clinical isolate of Aeromonas hydrophila. Microbiology 155, 3518–3531. doi: 10.1099/mic.0.031575-0
Kirov, S. M., Barnett, T. C., Pepe, C. M., Strom, M. S., and Albert, M. J. (2000). Investigation of the role of type IV Aeromonas pilus (Tap) in the pathogenesis of Aeromonas gastrointestinal infection. Infect. Immun. 68, 4040–4048. doi: 10.1128/IAI.68.7.4040-4048.2000
Kozlova, E. V., Khajanchi, B. K., Popov, V. L., Wen, J., and Chopra, A. K. (2012). Impact of QseBC system in c-di-GMP-dependent quorum sensing regulatory network in a clinical isolate SSU of Aeromonas hydrophila. Microb. Pathog. 53, 115–124. doi: 10.1016/j.micpath.2012.05.008
Li, J., Ni, X. D., Liu, Y. J., and Lu, C. P. (2011). Detection of three virulence genes alt, ahp and aerA in Aeromonas hydrophila and their relationship with actual virulence to zebrafish. J. Appl. Microbiol. 110, 823–830. doi: 10.1111/j.1365-2672.2011.04944.x
Lynch, M. J., Swift, S., Kirke, D. F., Keevil, C. W., Dodd, C. E., and Williams, P. (2002). The regulation of biofilm development by quorum sensing in Aeromonas hydrophila. Environ. Microbiol. 4, 18–28. doi: 10.1046/j.1462-2920.2002.00264.x
Martinez, M. J., Simon-Pujol, D., Congregado, F., Merino, S., Rubires, X., and Tomas, J. M. (1995). The presence of capsular polysaccharide in mesophilic Aeromonas hydrophila serotypes O:11 and O:34. FEMS Microbiol. Lett. 128, 69–73. doi: 10.1016/0378-1097(95)00085-J
Martinez-Murcia, A. J., Monera, A., Saavedra, M. J., Oncina, R., Lopez-Alvarez, M., Lara, E., et al. (2011). Multilocus phylogenetic analysis of the genus Aeromonas. Syst. Appl. Microbiol. 34, 189–199. doi: 10.1016/j.syapm.2010.11.014
Mateos, D., Anguita, J., Naharro, G., and Paniagua, C. (1993). Influence of growth temperature on the production of extracellular virulence factors and pathogenicity of environmental and human strains of Aeromonas hydrophila. J. Appl. Bacteriol. 74, 111–118. doi: 10.1111/j.1365-2672.1993.tb03003.x
McCoy, A. J., Koizumi, Y., Toma, C., Higa, N., Dixit, V., Taniguchi, S., et al. (2010). Cytotoxins of the human pathogen Aeromonas hydrophila trigger, via the NLRP3 inflammasome, caspase-1 activation in macrophages. Eur. J. Immunol. 40, 2797–2803. doi: 10.1002/eji.201040490
Merino, S., Aguilar, A., Nogueras, M. M., Regue, M., Swift, S., and Tomas, J. M. (1999). Cloning, sequencing, and role in virulence of two phospholipases (A1 and C) from mesophilic Aeromonas sp. serogroup O:34. Infect. Immun. 67, 4008–4013.
Merino, S., Aguilar, A., Rubires, X., Abitiu, N., Regue, M., and Tomas, J. M. (1997). The role of the capsular polysaccharide of Aeromonas hydrophila serogroup O:34 in the adherence to and invasion of fish cell lines. Res. Microbiol. 148, 625–631. doi: 10.1016/S0923-2508(97)88086-2
Merino, S., Camprubi, S., and Tomas, J. M. (1992). Effect of growth temperature on outer membrane components and virulence of Aeromonas hydrophila strains of serotype O:34. Infect. Immun. 60, 4343–4349.
Merino, S., Rubires, X., Aguillar, A., Guillot, J. F., and Tomas, J. M. (1996). The role of the O-antigen lipopolysaccharide on the colonization in vivo of the germfree chicken gut by Aeromonas hydrophila serogroup O:34. Microb. Pathog. 20, 325–333. doi: 10.1006/mpat.1996.0031
Monaghan, S. F., Anjaria, D., Mohr, A., and Livingston, D. H. (2008). Necrotizing fasciitis and sepsis caused by Aeromonas hydrophila after crush injury of the lower extremity. Surg. Infect. (Larchmt) 9, 459–467. doi: 10.1089/sur.2007.028
Pang, M. B., Jiang, J. W., Xie, X., Wu, Y. F., Dong, Y. H., Kwok, A. H. Y., et al. (2015). Novel insights into the pathogenicity of epidemic Aeromonas hydrophila ST251 clones from comparative genomics. Sci. Rep. 5, 9833. doi: 10.1038/srep09833
Peabody, C. R., Chung, Y. J., Yen, M. R., Vidal-Ingigliardi, D., Pugsley, A. P., and Saier, M. H. Jr. (2003). Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology 149, 3051–3072. doi: 10.1099/mic.0.26364-0
Pepe, C. M., Eklund, M. W., and Strom, M. S. (1996). Cloning of an Aeromonas hydrophila type IV pilus biogenesis gene cluster: complementation of pilus assembly functions and characterization of a type IV leader peptidase/N-methyltransferase required for extracellular protein secretion. Mol. Microbiol. 19, 857–869. doi: 10.1046/j.1365-2958.1996.431958.x
Pillai, L., Sha, J., Erova, T. E., Fadl, A. A., Khajanchi, B. K., and Chopra, A. K. (2006). Molecular and functional characterization of a ToxR-regulated lipoprotein from a clinical isolate of Aeromonas hydrophila. Infect. Immun. 74, 3742–3755. doi: 10.1128/IAI.00402-06
Popoff, M., and Veron, M. (1976). A taxonomic study of the Aeromonas hydrophila-Aeromonas punctata group. J. Gen. Microbiol. 94, 11–22. doi: 10.1099/00221287-94-1-11
Rasmussen-Ivey, C. R., Hossain, M. J., Odom, S. E., Terhune, J. S., Hemstreet, W. J., Shoemaker, C. A., et al. (2016). Classification of a hypervirulent Aeromonas hydrophila pathotype responsible for epidemic outbreaks in warm-water fishes. Front. Microbiol.
Rivero, O., Anguita, J., Mateos, D., Paniagua, C., and Naharro, G. (1991). Cloning and characterization of an extracellular temperature-labile serine protease gene from Aeromonas hydrophila. FEMS Microbiol. Lett. 65, 1–7. doi: 10.1111/j.1574-6968.1991.tb04702.x
Rodriguez, L. A., Ellis, A. E., and Nieto, T. P. (1992). Purification and characterisation of an extracellular metalloprotease, serine protease and haemolysin of Aeromonas hydrophila strain B32: all are lethal for fish. Microb. Pathog. 13, 17–24. doi: 10.1016/0882-4010(92)90028-M
Sandkvist, M. (2001). Type II secretion and pathogenesis. Infect. Immun. 69, 3523–3535. doi: 10.1128/IAI.69.6.3523-3535.2001
Sha, J., Erova, T. E., Alyea, R. A., Wang, S., Olano, J. P., Pancholi, V., et al. (2009). Surface-expressed enolase contributes to the pathogenesis of clinical isolate SSU of Aeromonas hydrophila. J. Bacteriol. 191, 3095–3107. doi: 10.1128/JB.00005-09
Sha, J., Kozlova, E. V., and Chopra, A. K. (2002). Role of various enterotoxins in Aeromonas hydrophila-induced gastroenteritis: generation of enterotoxin gene-deficient mutants and evaluation of their enterotoxic activity. Infect. Immun. 70, 1924–1935. doi: 10.1128/IAI.70.4.1924-1935.2002
Sha, J., Kozlova, E. V., Fadl, A. A., Olano, J. P., Houston, C. W., Peterson, J. W., et al. (2004). Molecular characterization of a glucose-inhibited division gene, gidA, that regulates cytotoxic enterotoxin of Aeromonas hydrophila. Infect. Immun. 72, 1084–1095. doi: 10.1128/IAI.72.2.1084-1095.2004
Sha, J., Lu, M., and Chopra, A. K. (2001). Regulation of the cytotoxic enterotoxin gene in Aeromonas hydrophila: characterization of an iron uptake regulator. Infect. Immun. 69, 6370–6381. doi: 10.1128/IAI.69.10.6370-6381.2001
Sha, J., Pillai, L., Fadl, A. A., Galindo, C. L., Erova, T. E., and Chopra, A. K. (2005). The type III secretion system and cytotoxic enterotoxin alter the virulence of Aeromonas hydrophila. Infect. Immun. 73, 6446–6457. doi: 10.1128/IAI.73.10.6446-6457.2005
Sha, J., Wang, S. F., Suarez, G., Sierra, J. C., Fadl, A. A., Erova, T. E., et al. (2007). Further characterization of a type III secretion system (T3SS) and of a new effector protein from a clinical isolate of Aeromonas hydrophila–part I. Microb. Pathog. 43, 127–146. doi: 10.1016/j.micpath.2007.05.002
Sierra, J. C., Suarez, G., Sha, J., Baze, W. B., Foltz, S. M., and Chopra, A. K. (2010). Unraveling the mechanism of action of a new type III secretion system effector AexU from Aeromonas hydrophila. Microb. Pathog. 49, 122–134. doi: 10.1016/j.micpath.2010.05.011
Sierra, J. C., Suarez, G., Sha, J., Foltz, S. M., Popov, V. L., Galindo, C. L., et al. (2007). Biological characterization of a new type III secretion system effector from a clinical isolate of Aeromonas hydrophila-part II. Microb. Pathog. 43, 147–160. doi: 10.1016/j.micpath.2007.05.003
Stehr, F., Kretschmar, M., Kroger, C., Hube, B., and Schafer, W. (2003). Microbial lipases as virulence factors. J. Mol. Catal. B Enzym. 22, 347–355. doi: 10.1016/S1381-1177(03)00049-3
Suarez, G., Khajanchi, B. K., Sierra, J. C., Erova, T. E., Sha, J., and Chopra, A. K. (2012). Actin cross-linking domain of Aeromonas hydrophila repeat in toxin A (RtxA) induces host cell rounding and apoptosis. Gene 506, 369–376. doi: 10.1016/j.gene.2012.07.012
Suarez, G., Sierra, J. C., Sha, J., Wang, S., Erova, T. E., Fadl, A. A., et al. (2008). Molecular characterization of a functional type VI secretion system from a clinical isolate of Aeromonas hydrophila. Microb. Pathog. 44, 344–361. doi: 10.1016/j.micpath.2007.10.005
Swann, L., and White, M. (1991). Diagnosis and Treatment of “Aeromonas hydrophila” Infection of Fish. Urbana, IL: Illinois-Indiana Sea Grant Program.
Swift, S., Karlyshev, A. V., Fish, L., Durant, E. L., Winson, M. K., Chhabra, S. R., et al. (1997). Quorum sensing in Aeromonas hydrophila and Aeromonas salmonicida: identification of the LuxRI homologs AhyRI and AsaRI and their cognate N-acylhomoserine lactone signal molecules. J. Bacteriol. 179, 5271–5281.
Tan, Y. W., Yu, H. B., Sivaraman, J., Leung, K. Y., and Mok, Y. K. (2009). Mapping of the chaperone AcrH binding regions of translocators AopB and AopD and characterization of oligomeric and metastable AcrH-AopB-AopD complexes in the type III secretion system of Aeromonas hydrophila. Protein Sci. 18, 1724–1734. doi: 10.1002/pro.187
Thomas, S. R., and Trust, T. J. (1995). Tyrosine phosphorylation of the tetragonal paracrystalline array of Aeromonas hydrophila: molecular cloning and high-level expression of the S-layer protein gene. J. Mol. Biol. 245, 568–581. doi: 10.1006/jmbi.1994.0047
Thornley, J. P., Shaw, J. G., Gryllos, I. A., and Eley, A. (1997). Virulence properties of clinically significant Aeromonas species: Evidence for pathogenicity. Rev. Med. Microbiol. 8, 61–72. doi: 10.1097/00013542-199704000-00002
Tseng, T. T., Tyler, B. M., and Setubal, J. C. (2009). Protein secretion systems in bacterial-host associations, and their description in the Gene Ontology. BMC Microbiol. 9(Suppl. 1):S2. doi: 10.1186/1471-2180-9-S1-S2
Vilches, S., Jimenez, N., Tomas, J. M., and Merino, S. (2009). Aeromonas hydrophila AH-3 type III secretion system expression and regulatory network. Appl. Environ. Microbiol. 75, 6382–6392. doi: 10.1128/AEM.00222-09
Vilches, S., Urgell, C., Merino, S., Chacon, M. R., Soler, L., Castro-Escarpulli, G., et al. (2004). Complete type III secretion system of a mesophilic Aeromonas hydrophila strain. Appl. Environ. Microbiol. 70, 6914–6919. doi: 10.1128/AEM.70.11.6914-6919.2004
Vilches, S., Wilhelms, M., Yu, H. B., Leung, K. Y., Tomas, J. M., and Merino, S. (2008). Aeromonas hydrophila AH-3 AexT is an ADP-ribosylating toxin secreted through the type III secretion system. Microb. Pathog. 44, 1–12. doi: 10.1016/j.micpath.2007.06.004
Wadstrom, T., Ljungh, A., and Wretlind, B. (1976). Enterotoxin, haemolysin and cytotoxic protein in Aeromonas hydrophila from human infections. Acta Pathol. Microbiol. Scand. B 84, 112–114.
Wang, G. H., Clark, C. G., Liu, C. Y., Pucknell, C., Munro, C. K., Kruk, T. M. A. C., et al. (2003). Detection and characterization of the hemolysin genes in Aeromonas hydrophila and Aeromonas sobria by multiplex PCR. J. Clin. Microbiol. 41, 1048–1054. doi: 10.1128/JCM.41.3.1048-1054.2003
Wilhelms, M., Gonzalez, V., Tomas, J. M., and Merino, S. (2013). Aeromonas hydrophila lateral flagellar gene transcriptional hierarchy. J. Bacteriol. 195, 1436–1445. doi: 10.1128/JB.01994-12
Wilhelms, M., Molero, R., Shaw, J. G., Tomas, J. M., and Merino, S. (2011). Transcriptional hierarchy of Aeromonas hydrophila polar-flagellum genes. J. Bacteriol. 193, 5179–5190. doi: 10.1128/JB.05355-11
Wong, C. Y. F., Heuzenroeder, M. W., and Flower, R. L. P. (1998). Inactivation of two haemolytic toxin genes in Aeromonas hydrophila attenuates virulence in a suckling mouse model. Microbiology-Uk 144, 291–298. doi: 10.1099/00221287-144-2-291
Yu, H. B., Kaur, R., Lim, S. M., Wang, X. H., and Leung, K. Y. (2007). Characterization of extracellular proteins produced by Aeromonas hydrophila AH-1. Proteomics 7, 436–449. doi: 10.1002/pmic.200600396
Yu, H. B., Rao, P. S., Lee, H. C., Vilches, S., Merino, S., Tomas, J. M., et al. (2004). A type III secretion system is required for Aeromonas hydrophila AH-1 pathogenesis. Infect. Immun. 72, 1248–1256. doi: 10.1128/IAI.72.3.1248-1256.2004
Keywords: Aeromonas hydrophila, pathogenesis, comparative genomics, bacteria, phylogeny
Citation: Rasmussen-Ivey CR, Figueras MJ, McGarey D and Liles MR (2016) Virulence Factors of Aeromonas hydrophila: In the Wake of Reclassification. Front. Microbiol. 7:1337. doi: 10.3389/fmicb.2016.01337
Received: 30 June 2016; Accepted: 12 August 2016;
Published: 25 August 2016.
Edited by:
Magdalena Popowska, University of Warsaw, PolandReviewed by:
Shivangi Agarwal, Northwestern University, USAAntonio Juárez, University of Barcelona, Spain
Copyright © 2016 Rasmussen-Ivey, Figueras, McGarey and Liles. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mark R. Liles, bGlsZXNtYUBhdWJ1cm4uZWR1
 Cody R. Rasmussen-Ivey1
Cody R. Rasmussen-Ivey1 Maria J. Figueras
Maria J. Figueras Donald McGarey
Donald McGarey Mark R. Liles
Mark R. Liles