
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 19 August 2016
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 7 - 2016 | https://doi.org/10.3389/fmicb.2016.01268
This article is part of the Research Topic The global challenge posed by the multiresistant international clones of bacterial pathogens View all 23 articles
 Xiaofen Liu1,2
Xiaofen Liu1,2 Huajun Zheng3
Huajun Zheng3 Weipeng Zhang4
Weipeng Zhang4 Zhen Shen1
Zhen Shen1 Miao Zhao1
Miao Zhao1 Yuancheng Chen1,5
Yuancheng Chen1,5 Li Sun1
Li Sun1 Jun Shi2
Jun Shi2 Jing Zhang1,5*
Jing Zhang1,5*Cefoperazone/sulbactam has been shown to be efficacious for the treatment of infections caused by Acinetobacter baumannii; however, the mechanism underlying resistance to this synergistic combination is not well understood. In the present study, two A. baumannii isolates, AB1845 and AB2092, were isolated from a patient with hospital-acquired pneumonia before and after 20 days of cefoperazone/sulbactam therapy (2:1, 3 g every 8 h with a 1-h infusion). The minimum inhibitory concentration (MIC) of cefoperazone/sulbactam for AB1845 and AB2092 was 16/8 and 128/64 mg/L, respectively. Blood samples were collected on day 4 of the treatment to determine the concentration of cefoperazone and sulbactam. The pharmacokinetic/pharmacodynamic (PK/PD) indices (%T>MIC) were calculated to evaluate the dosage regimen and resistance development. The results showed that %T>MIC of cefoperazone and sulbactam was 100% and 34.5% for AB1845, and 0% and 0% for AB2092, respectively. Although there was no available PK/PD target for sulbactam, it was proposed that sulbactam should be administered at higher doses or for prolonged infusion times to achieve better efficacy. To investigate the mechanism of A. baumannii resistance to the cefoperazone/sulbactam combination in vivo, whole-genome sequencing of these two isolates was further performed. The sequencing results showed that 97.6% of the genome sequences were identical and 33 non-synonymous mutations were detected between AB1845 and AB2092. The only difference of these two isolates was showed in sequencing coverage comparison. There was a 6-kb amplified DNA fragment which was three times higher in AB2092, compared with AB1845. The amplified DNA fragment containing the blaOXA-23 gene on transposon Tn2009. Further quantitative real-time PCR results demonstrated that gene expression at the mRNA level of blaOXA-23 was >5 times higher in AB2092 than in AB1845. These results suggested that the blaOXA-23 gene had higher expression level in AB2092 via gene amplification and following transcription. Because gene amplification plays a critical role in antibiotic resistance in many bacteria, it is very likely that the blaOXA-23 amplification results in the development of cefoperazone/sulbactam resistance in vivo.
Acinetobacter baumannii is a non-fermentative, gram-negative opportunistic pathogen that can cause hospital-acquired pneumonia (HAP), blood infections and urinary tract infection, among others (Perez et al., 2007). The rapid spread ofthe multi-drug-resistant A. baumannii has resulted in very limited therapeutic options in the clinic. Sulbactam, a β-lactamase inhibitor, has intrinsic antimicrobial effects against A. baumannii by binding to penicillin-binding protein 2 (Peleg et al., 2008). It is commercially available in a combined formulation of ampicillin and cefoperazone. The clinical efficacy of cefoperazone/sulbactam has been shown in previous work (Xin et al., 2013; Sipahi et al., 2014; Xia et al., 2014). This combination has recently been applied to treat critically ill patients receiving continuous venovenous hemofiltration, and 11 of 14 patients survived (Gao et al., 2016). This combination was also administrated to treat neurosurgical patients in a pilot study, with the results showing that cerebrospinal fluid penetration of cefoperazone/sulbactam could be enhanced after neurosurgical impairment of the blood-brain barrier (Wang et al., 2015). In a retrospective review of the outcomes for patients with cefoperazone/sulbactam treated A. baumannii bacteremia, 77% of the patients (27/35) presented successful clinical efficacy (Choi et al., 2006). Because the combination has been widely used in the clinic, the resistance rate was monitored and observed to increase from 25.0 to 37.7% from 2004 to 2010 in China (Hu et al., 2016). However, the underlying mechanism is not well understood in vitro or in vivo, for sulbactam alone or the combination. Sulbactam alone has been reported to cause resistance in A. baumannii via PBP3 mutation in vitro, despite a lack of information regarding natural pbp3 mutations in clinical isolates (Penwell et al., 2015). The detection and expression of blaTEM-1 has been suggested to relate to the minimum inhibitory concentration (MIC) of sulbactam in A. baumannii (Waltner-Toews et al., 2011). The combination resistance in Klebsiella pneumoniae in an in vitro study was reported owing to two different mechanisms, loss of a 39-kDa outer membrane protein and presence of TEM-2 β-lactamase (Rice et al., 1993).
Whole-genome sequencing is powerful and can reveal a vast amount of DNA information from a global perspective. The published complete genome sequences for many bacteria are beneficial and efficient for strain-to-reference sequencing and bioinformatics analysis (Mardis, 2008). They can be used to identify resistance genes as well as minor changes in the genome due to mutation, gene transfer, gene duplication, and amplification. It has been seen as a basis for whole-genome sequencing revealing the mechanism of bacterial pathogen resistance that develops in patients (Snitkin et al., 2011; Wright et al., 2014; Holt et al., 2015). Wright et al. (2016) sequenced 136 strains of A. baumannii isolated from patients and showed the genome changes occurred mainly due to the single nucleotide variance in protein coding regions and IS element. Genome sequenced A. baumannii showed the colistin resistance was due to the mutations in transcriptional regulatory genes (Cheah et al., 2016). Susceptible S. aureus evolved resistance via a 35-point mutation in 31 loci in a patient with a bloodstream infection receiving vancomycin therapy (Mwangi et al., 2007). Tigecycline resistance to A. baumannii has been reported to be due to the deletion of three contigs in vivo (Hornsey et al., 2011). Additionally, gene amplification was frequently detected due to the antibiotic pressure. The evolution of β-lactamase resistance was studied by exposing Salmonella typhimurium, which contain low level β-lactamase resistance, in an environment of progressively increasing concentrations of cephalosporin. The results showed an amplification of blaTEM-1 gene copy number followed by acquisition of gene mutations (Sun et al., 2009). A genome-wide assay of E. coli strain in 78 different antibiotic environments found that 56 genes were amplified to fit the environment and reproducible increasing in MIC for their corresponding antibiotics was detected (Soo et al., 2011). The amplification of aminoglycoside resistance gene aphA1 in A. baumannii has been considered to be the mechanism underlying the development of tobramycin in vivo (McGann et al., 2014).
In the present study, two A. baumannii isolates, AB1845 and AB2092, were collected before and after cefoperazone/sulbactam therapy from a patient with HAP. Pharmacokinetics/pharmacodynamics (PK/PD) was applied to evaluate the dosage regimen and development of resistance. Moreover, whole-genome sequencing was employed to investigate the mechanism of cefoperazone/sulbactam resistance of the two isolates developed in vivo.
This study was approved by the institutional review board of Huashan Hospital affiliated to Fudan University. Written informed consent was obtained from the patient. A 19-year-old male patient with serious brain trauma was admitted to Huashan Hospital (Shanghai, China). The patient had a high fever with white blood cell counts out of the normal range after being checked into bed for 48 h. Both chest X-Ray radiophotography and computed tomography showed pneumonia. A. baumannii (AB1845) was isolated from the sputum of the patient, revealing bacterial infection. Hence, the patient was treated with meropenem 0.5 g q8h without clinical efficacy. On day 6, the dosage regimen was changed to cefoperazone/sulbactam (2:1, 3 g every 8 h with a 1-h infusion, SULPERAZON®, batch no. 95839116) for 20 days. The treatment was stopped when body temperature and white blood cell counts returned to the normal range; additionally, A. baumannii was not detected in the sputum. However, the patient had a fever, and A. baumannii (AB2092) was again isolated 4 days after cefoperazone/sulbactam treatment. The cefoperazone/sulbactam treatment was finally considered to have failed both clinically and microbiologically efficacy.
Blood samples were collected before the first infusion, immediately after the infusion (0 h), and at 0.5, 1, 2, 4, 6 h afterward on day 4 (i.e., steady state) of the cefoperazone/sulbactam treatment. The drug concentration was determined using the LC-MS/MS method (Zhou et al., 2010), which was validated according to the CFDA guidelines on bioanalytical method validation. The pharmacokinetic parameters of the patients were calculated by Winnonlin (v6.0, phoenix). The patient’s pharmacokinetic profiles of cefoperazone and sulbactam were fit into a well-developed population PK model (Chen et al., 2015). The PK/PD indice of cefoperazone and sulbactam was %T>MIC, namely the percentage of the dosing interval for which the plasma concentration of cefoperazone or sulbactam above the MIC. It was calculated based on the drug pharmacokinetic profile and MICs of the combination (Lips et al., 2014).
Acinetobacter baumannii AB1845 and AB2092 were isolated from the sputum of the patient before and after cefoperazone/sulbactam therapy. Both isolates were stored at -80°C. The MIC was determined in cation-adjusted Mueller-Hinton broth (Becton-Dickinson, Sparks, MD, USA) using a broth microdilution according to Clinical and Laboratory Standards Institute (CLSI) standards. The MICs of the β-lactam/β-lactamase inhibitor combination, including cefoperazone/sulbactam, carbapenems, colistin, and polymyxin B, were determined.
Genomic DNA was extracted using a Genomic DNA Purification Kit (Tiangen, Beijing, China) according to the instruction manual and stored at -80°C before sequencing. A 300-bp paired-end library was constructed for the purified DNA sample following the standard Illumina paired-end protocol. Cluster generation was performed in C-bot, and sequencing was performed on the Illumina Hiseq2500 with 150 cycles. The sequence reads were cleaned using the FASTX toolkit1. Genome assembly was performed using Velvet (Ver 1.0.15) (Zerbino and Birney, 2008). Putative protein-coding sequences were determined by combining the prediction results of the glimmer 3.02 (Delcher et al., 2007) and Z-Curve (Guo and Zhang, 2006) program. The phylogenetic tree was constructed using PHYML (Guindon and Gascuel, 2003) by concatenating orthologs based on the protein sequences in the published genome. Functional annotation of CDS was performed by searching the NCBI non-redundant protein database and the KEGG protein database (Kanehisa et al., 2016). Gene annotation was mapped to the A. baumannii MDR-ZJ06 genome (from the phylogenetic tree) using Bowtie2 with default settings (Langmead and Salzberg, 2012). Single Nucleotide Polymorphisms (SNPs) from alignments were called using Samtools-0.1.16 (Li et al., 2009), and the output was generated in the pileup format. Gene amplification was detected by comparing the coverage of the reads in AB1845 and Ab2092 using the read depth method (Redon et al., 2006).
The mRNA expression level of blaOXA-23 was quantified by real-time PCR. AB1845 and AB2092 were cultured in cation-adjusted Mueller-Hinton broth and collected during the early log phase. Total RNA was extracted using a TaKaRa Mini BEST Universal RNA Extraction Kit (TaKaRa Biotechnology, Dalian, China) according to the manufacturer’s instruction. Quantitative real-time PCR (qPCR) was performed using a two-step process. RNA was first reverse-transcribed to cDNA (TaKaRa Biotechnology, Dalian, China), and real-time PCR was conducted on an ABI Vii7 (Applied Biosystems, Carlsbad, CA, USA) using a TaKaRa SYBR® FAST qPCR Kit with 40 cycles of denaturation for 5 s at 95°C, annealing for 30 s at 50°C, and extension for 20 s at 72°C. The PCR primers for the blaOXA-23 gene were F: CCGAGTCAGATTGTTCAAGGA and R: TGTAGAGGCTGGCACATATTC; and those for 16S rRNA were F: GGCGGCTTATTAAGTCGGATG and R: TTCGTACCTCAGCGTCAGTATT. The melting curve analysis was performed immediately after amplification to verify the specificity of the PCR amplification products.
Fluorescence was measured at the end of the annealing-extension phase of each cycle. A threshold value for the fluorescence of all samples was set manually. The reaction cycle at which the PCR product exceeded this fluorescence threshold was identified as the threshold cycle (CT). The relative quantitation was calculated by the 2-ΔΔCT method (Schmittgen and Livak, 2008). The student t-test was applied to evaluate the significance of the gene expression level of the two isolates.
Minimum inhibitory concentrations were obtained for cefoperazone/sulbactam combinations, carbapenems, colistin and polymyxin B for the pre-therapy isolate AB1845 and the post-therapy isolate AB2092 (Table 2). The MICs of the cefoperazone/sulbactam combination were 16/8 mg/L for AB1845; and the MICs of mono cefoperazone and sulbactam were >128 mg/L and 32 mg/L. The results supported a synergistic effect of the combination (FICI < 0.5, MIC of cefoperazone was set at 128 mg/L for the FICI calculation). However, the MICs of the combination and mono drugs were 128/64, >128, and 128 mg/L for AB2092. The combination did not display a synergistic effect on AB2092.

TABLE 1. The minimum inhibitory concentrations (MICs) of different antibiotics for AB1845 and AB2092.
Interestingly, AB1845 and AB2092 also developed meropenem resistance with MIC increased from 16 to 64 mg/L, although the bacteria only exposed to meropenem for a short term (0.5 g every 8 h for 4 days). The isolates also developed resistance to imipenem and doripenem. Since both cefoperazone and meropenem are β-lactam antibiotics, the increased MIC in both cefoperazone/sulbactam and carbapenems may imply the similar underlying resistance mechanism. The isolates were only sensitive to colistin and polymyxin B, indicating an alternative choice for the AB2092 treatment. The MICs of the antibiotics are shown in Table 1.
Plasma concentrations of cefoperazone and sulbactam at steady state were determined using the validated LC-MS/MS method, and the data were fitted into a developed population PK model (Figure 1). The pharmacokinetic parameters of cefoperazone and sulbactam are summarized in Table 2. The T1/2 of cefoperazone was 4.1 h, which was a bit longer than the reported 1.8 h (Reitberg et al., 1988). This could attribute to liver dysfunction (Boscia et al., 1983) according to the medical records of the patients. The T1/2 of sulbactam was 1.3 h, which was comparable to the published data (Reitberg et al., 1988).
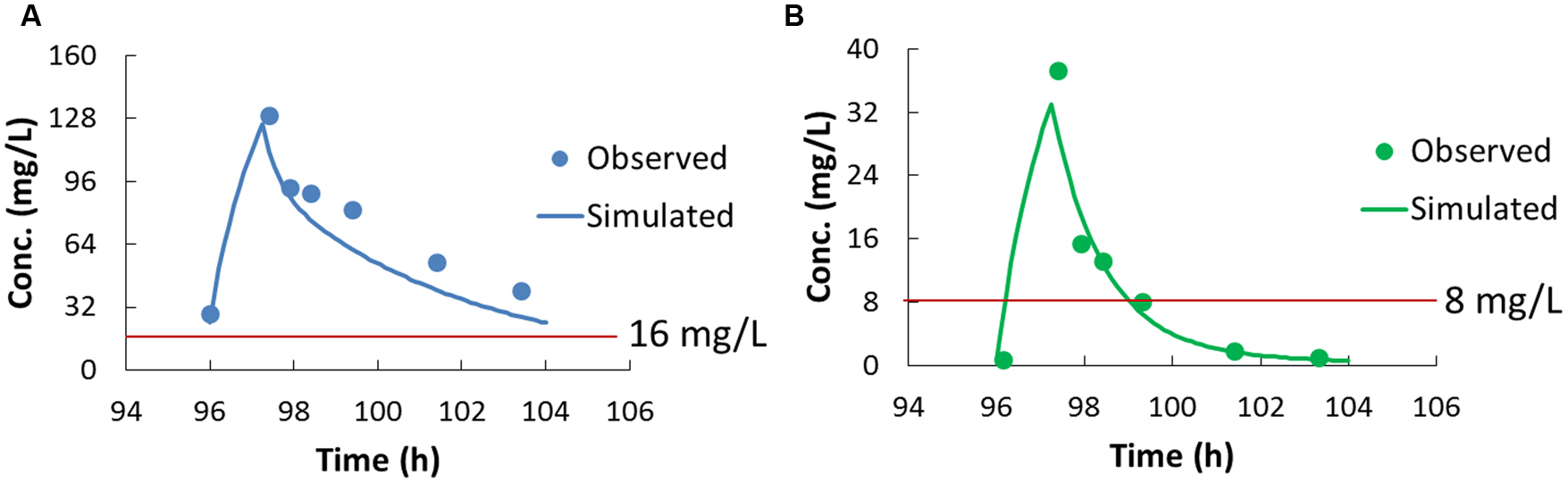
FIGURE 1. The pharmacokinetic profiles of cefoperazone (A) and sulbactam (B). The dots in (A,B) are the concentration determined by LC-MS/MS (before the first infusion on Day 4 and 0.5, 1, 2, 4, 6 h after the infusion), and the curves are the profiles fitted to the population pharmacokinetic model. The red lines were indicated the concentration for the MIC of AB1845.
Pharmacokinetic/pharmacodynamic indices were calculated to evaluate the drug dosage regimen. The %T>MIC was the most predictive PK/PD index for cefoperazone and sulbactam (Cooper et al., 2011; Sy et al., 2015), and the MIC of the combination was used for the %T>MIC calculation. In this study, the %T>MIC was 100% for cefoperazone and 34.5% for sulbactam for AB1845; and the %T>MIC was 0% for both cefoperazone and sulbactam for AB2092.
The two isolates were initially typed by PFGE of ApaI-digested genomic DNA and shown to be the same strain. Whole-genome sequencing produced 12,050,346 and 12,191,576 pairs of 150-bp reads for AB1845 and AB2092, respectively. Assembly of the AB1845 and AB2092 genomes resulted in 132 and 131 contigs larger than 500 bp, comprising 4.0 megabases of sequence and representing a median 930-fold coverage. The AB1845 draft assembly has been deposited in GenBank (accession number LVYA00000000), and raw sequence reads for AB1845 and AB2092 have been submitted to NCBI’s Sequence Read Archive under the study accession number SRP072783.
Automated gene prediction detected 3,811 and 3,810 putative coding sequences (CDSs) for AB1845 and AB2092, respectively. A phylogenetic tree was constructed, and the genome similarity was compared (Figure 2). The 3,718 CDSs were homologous (defined as a BLASTN e-value ≤ 1e-10) to the genome of A. baumannii MDR-ZJ06, which was a multi-drug-resistant isolate belonging to International clone II and wide spreading in China (Zhou et al., 2011). MDR-ZJ06 was used as a reference genome for annotating genes in AB1845 and AB2092 (Figure 3). The results showed that the two isolates possess 97.6% similarity to MDR-ZJ06, with 921 and 915 SNPs, respectively. Certain genes were missing in both AB1845 and AB2092 compared with MDR-ZJ06. For example, 17 genes had a 15-kb fragment that was missing in the resistant island AbaR22, which includes transposition helper proteins, site-specific tyrosine recombinase, transposase protein A and conserved hypothetical proteins, among others.
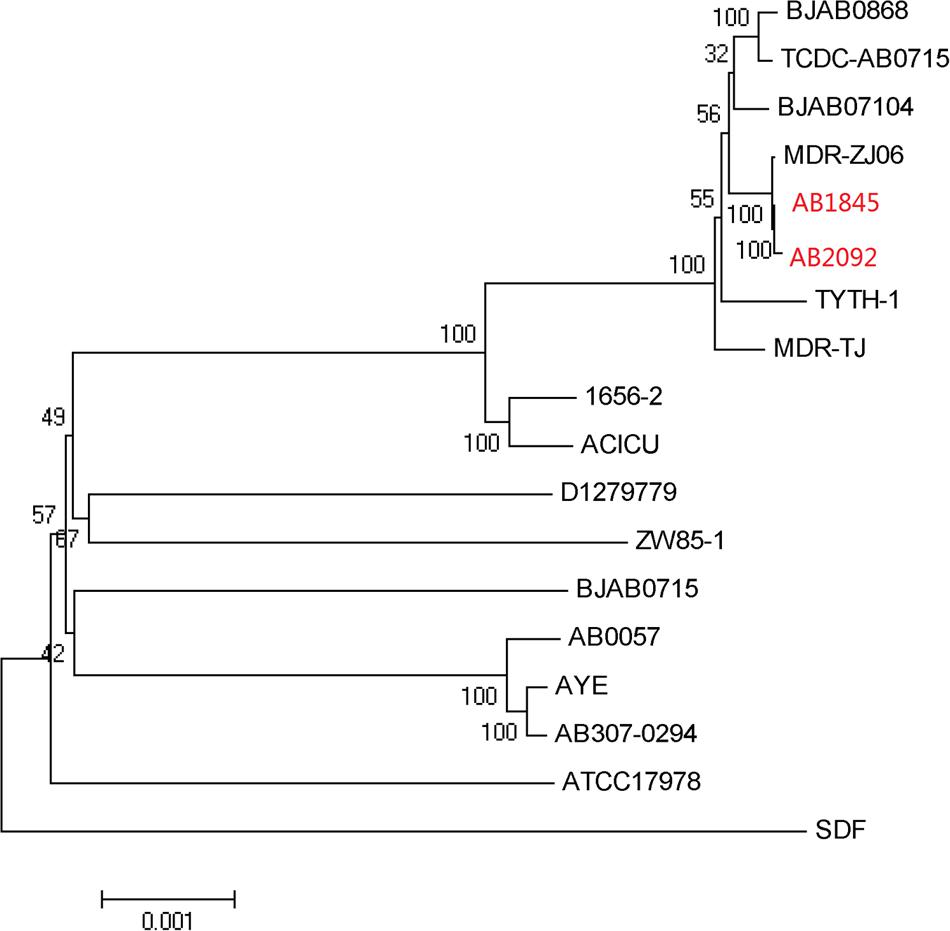
FIGURE 2. Phylogenetic tree of AB1845 and AB2092 with 16 complete genomes of A. baumannii isolates in ftp://ftp.ncbi.nlm.nih.gov/genomes/archive/old_genbank/.
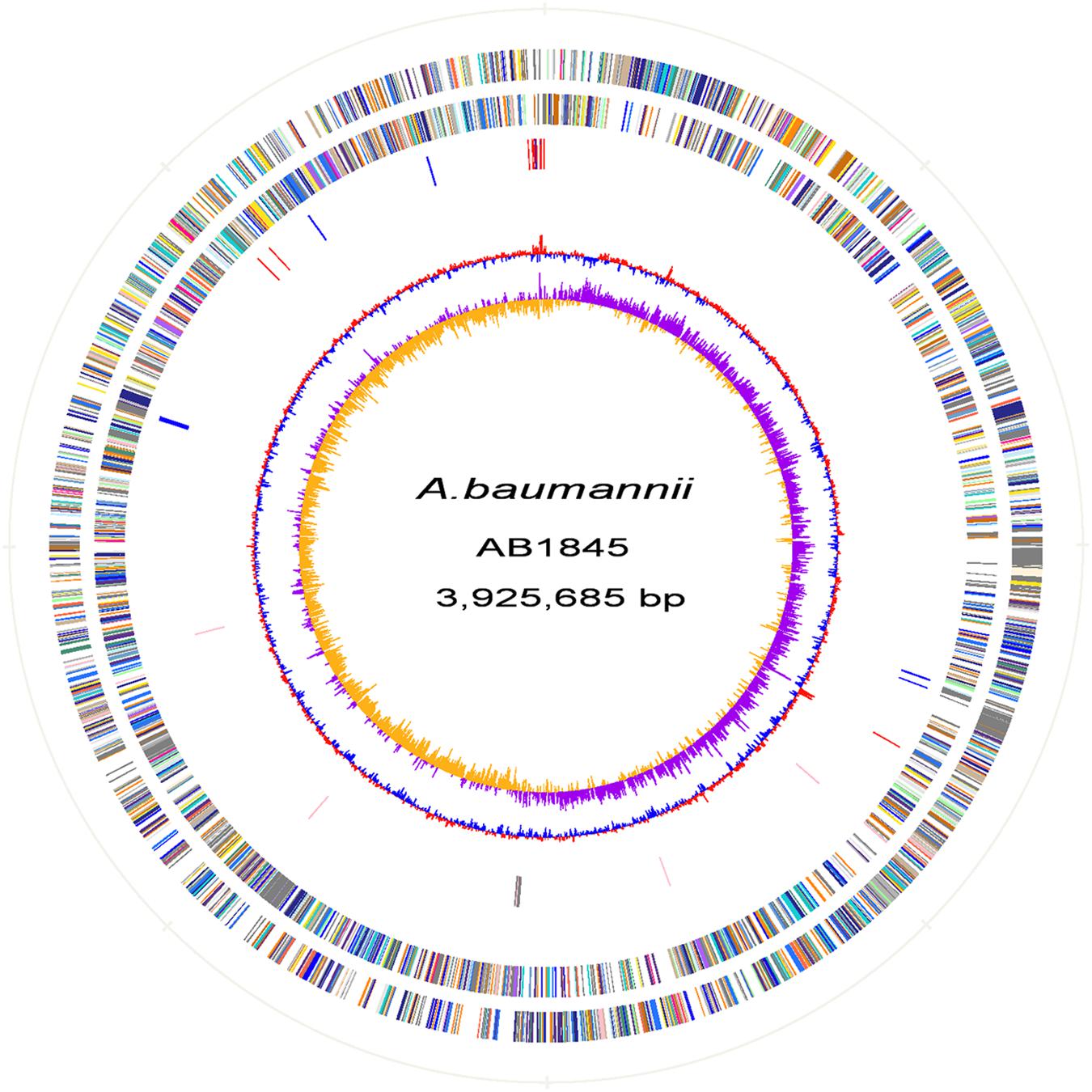
FIGURE 3. Atlas of the A. baumannii AB1845 draft genome. Each concentric circle represents the genomic data for AB1845. The two outer circles illustrate the predicted coding sequences on the plus and minus strands, respectively, colored by functional categories according to the COG classification. The third circle represents the location of the nucleotide substitution between AB1845 and AB2092, with blue representing genes affected by synonymous Single Nucleotide Polymorphisms (SNPs) and red representing SNPs in the intergenic region. The fourth circle displays the loci of the β-lactamase genes (pink) and genes around blaOXA-23 (gray). The fifth circle shows the GC content, and the 6th circle (innermost) represents GC skew (G–C)/(G+C) calculated using a 1-kb window.
The CDS of AB1845 was also mapped to the published plasmid sequence. It showed that certain AB1845 genes mapped to plasmid ABKp1 from the A. baumannii 1656-2 plasmid with 97.6% similarity.
A comparison of the CDS of AB1845 and AB2092 revealed only 33 SNPs. All of these SNPs were synonymous, indicating that SNP was not the main reason for the development of resistance (Supplementary Table S1).
A well-known resistance enzyme for β-lactam antibiotics is β-lactamase. The class A, C, and D classes (blaOXA-23 and blaOXA-51) β-lactamase were identified in both AB1845 and AB2092 (Supplementary Table S2).
Gene amplification was detected by whole-genome sequencing in combination with the read depth method (Cantsilieris et al., 2012). Approximately 99.9% of the genes had a coverage ratio of AB2092 to AB1845 of approximately 1.0, but six genes (out of 3871 genes) had a coverage ratio over 3.0 (Supplementary Table S3). These six genes were AAA ATPase, DEAD/DEAH box helicases, blaOXA-23, and three hypothetical genes. Among these genes, AAA ATPase and DEAD/DEAH box helicases are involved in the function of DNA replication, repair, transcription, and two uncharacterized genes (Johnson and Mckay, 1999; Iyer et al., 2004). The blaOXA-23 was gene encoded class D β-lactamase and conferred resistance to carbapenems and cefoperazone (Pascale and Wright, 2010; Bush, 2013). All six genes were located in a 6-kb sequence fragment in AB1845 designated as Tn2009. Tn2009 was flanked by two ISAba1 elements that contributed to the duplication of Tn2009 in AB2902 (Liu et al., 2015). The amplified genes were transcribed in the same orientation (Figure 4).
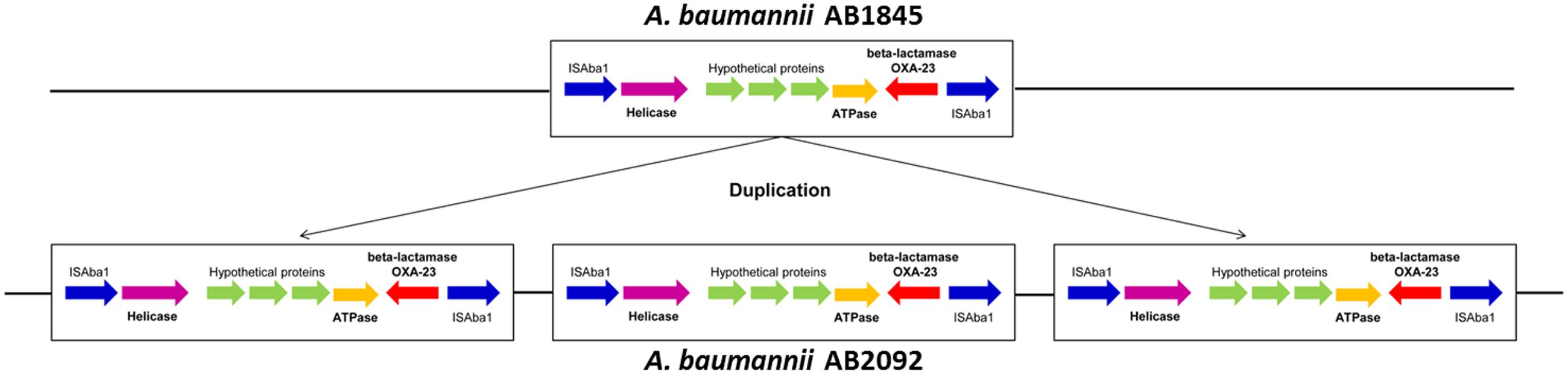
FIGURE 4. The illustration of the 6-kb DNA fragment of Tn2009 in AB1845 and AB2092. The fragment was amplified three times in AB2092.
Quantitative real-time PCR was conducted to measure the mRNA levels of blaOXA-23 in AB1845 and AB2092. Both isolates were collected at the early log phase, and the qPCR results showed that the mRNA level of blaOXA-23 was five times higher in AB2092 than in AB1845 (Figure 5). This finding was consistent with the gene amplification results.
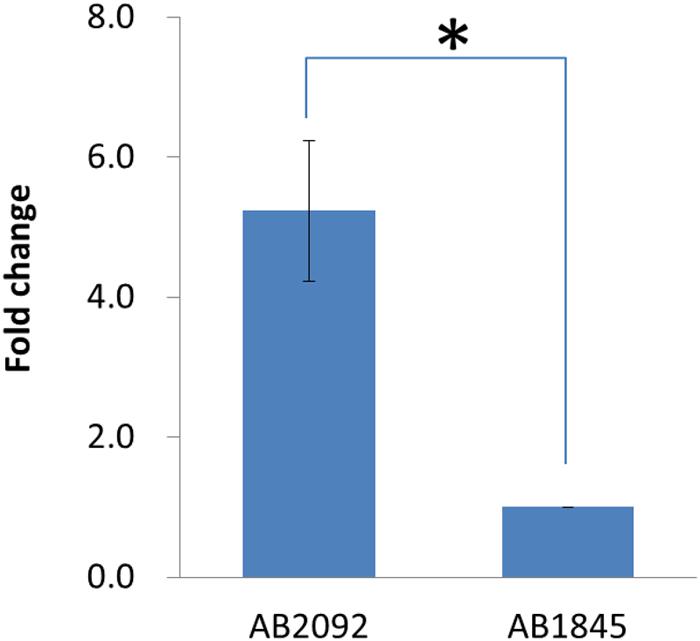
FIGURE 5. Quantitative real-time PCR results for blaOXA-23 in AB1845 and AB2092 (∗student t-test p < 0.05).
Acinetobacter baumannii commonly colonizes in respiratory tract of the hospitalized patients (Peleg et al., 2008). It is difficult to characterize A. baumannii isolated from sputum belongs to the upper airway colonization or the causative pathogen of pneumonia. In this study, the two isolates were considered causative pathogens, as verified by pneumonia testing and symptoms such as cough, fever, and high white blood cell counts. To examine the mechanism of resistance development during cefoperazone/sulbactam treatment, both PK/PD indices and genomic sequencing were employed to understand the problem from a PK/PD and molecular perspective.
Exposure to a sub-optimal antibiotic concentration is the most important factor in the development of bacterial resistance. To optimize the antibiotic dosage regimen and prevent resistance, the PK/PD which bridges drug exposure and effect, has been widely applied in the clinics (Abdulaziz et al., 2015; Asín-Prieto et al., 2015; Monogue et al., 2015). To date, the PK/PD helps adjust dosage based on the evaluation of steady-state drug concentrations (Martinez et al., 2012). In this study, blood samples were collected at steady state, and the MIC of the bacteria was determined. As the most predictive PK/PD index, the %T>MIC of cefoperazone and sulbactam was calculated and compared with published PK/PD targets. The published target %T>MIC of β-lactam antibiotics was 40–70% (Drusano, 2004); however, there was no published target of sulbactam because of a lack of pharmacodynamic data. In our present dosage regimen (3 g q8h, infusion for 1 h), the target was 100% of cefoperazone and 34.5% of sulbactam for AB1845. Cefoperazone clearly reached its target, whereas there were no criteria for sulbactam. In a study of PK/PD simulation for sulbactam in healthy volunteers, in order for A. baumannii with an MIC of 8 mg/L to achieve the target of 40%, the sulbactam had to be administered as a 4-h infusion of 3 g q8h (Jaruratanasirikul et al., 2013). It has even been suggested that the daily dose of sulbactam can be administered up to 6 g for A. baumannii infection (Argyres, 2010) to achieve enhanced efficacy. A comparison of these data revealed that sulbactam should be administered at a much higher dose or prolonged infusion time. A suboptimal dose of sulbactam could lead to the development of resistance during treatment. Additional research to identify the PK/PD target of sulbactam specific to Acinetobacter is needed.
Whole-genome sequencing was further employed to investigate the mechanism of resistance from a molecular perspective. Comparison of the genome between AB1845 and AB2092 revealed no gene deletions or mutations (only 33 synonymous SNPs). However, the reads depth method showed six amplified genes on Tn2009, including blaOXA-23. The blaOXA-23 gene encodes the class D β-lactamase which confers resistance to the last resort carbapenem antibiotics. The OXA-23-producing A. baumannii is widely disseminated in multi-drug-resistant A. baumannii. Although the blaOXA-23 gene has been mainly identified on plasmids, its chromosomal location has also been reported (Zhou et al., 2011). The transportable elements, Tn2006, Tn2007, Tn2008, and Tn2009, play a key role in the transfer of the blaOXA-23 gene to different locations in one bacterium or to different isolates (Corvec et al., 2007; Liu et al., 2015). Tn2008 and Tn2009 have mostly been associated with the transfer of blaOXA-23 in China (Adams-Haduch et al., 2008; Zhou et al., 2011; Liu et al., 2015). IS elements are associated with the genome gain/loss of genes, especially resistant genes. These two transposons share ISAba1 upstream from blaOXA-23, which belongs to the IS4 family, provides promoter for blaOXA-23 (Turton et al., 2006; Mugnier et al., 2009), which could explain the gene duplication or amplification in this study. The mRNA level of blaOXA-23 confirmed the higher expression level of AB2092 and AB1845. There is evidence that gene duplication and amplification in bacteria was directly associated with adaptation to environmental changes, including antibiotic stress (Sandegren and Andersson, 2009). In a study testing the relationships of gene amplification and β-lactams resistance increase showed that 70% colonies of S. Typhimurium with increased resistance to cephalosporin had increased blaTEM gene copy numbers (Sun et al., 2009). Recent study showed the heteroresistance of E. Coli and S. Typhimurium was due to the amplification of pmrD gene which encoded a protein modifying lipid A. Moreover, the heteroresistance phenotype is associated with different copy numbers of the pmrD gene (Hjort et al., 2016). Numerous studies have reported gene duplication and amplification related to bacterial resistance to antibiotics in vitro (Bertini et al., 2007). During the macrolide treatment of a Streptococcus pneumoniae infection, the bacterium developed resistance due to an 18 bp duplication in the rplV gene, leading to the treatment failure. The duplication of rplV gene led to the blocking of macrolide binding site on ribosome (Musher et al., 2002). Another example of gene amplification in bacterial isolates from human patients involves the Streptococcus agalactiae, a leading cause of neonatal infection. The amplification of a four-fold tandemly amplified 13.5-kbp region and a 92 kbp duplication conferred drug resistance to both sulfonamide and trimethoprim (Brochet et al., 2008).
In this study, the isolates developed resistance to cefoperazone/sulbactam as well as carbapenem antibiotics. The whole-genome sequencing showed the amplified blaOXA-23 in the isolates is very likely the contribution to the resistance. It is rational to conferred resistance to carbapenem antibiotics by the amplification of blaOXA-23. However, this is the first time to report the amplification of blaOXA-23 results in cefoperazone/sulbactam combination resistance. Further studies will be focused on collecting more clinical isolates pairs of A. baumannii to support the mechanism proposed.
XL designed the genomic sequencing experiments. XL and HZ performed the data analysis. XL and ZS conducted the qPCR experiments. YC conducted the pharmacokinetic parameter calculation. LS extracted the genomic DNA for sequencing. MZ conducted the MIC test and the PFGE test. JS and JZ conceived of the study and devised the overall study strategy. XL wrote the manuscript with input from all authors.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This research is supported by the National Natural Science Foundation of China (Grant No. 8150130235 and 81373493).
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01268
Abdulaziz, M. H., Lipman, J., Mouton, J. W., Hope, W. W., and Roberts, J. A. (2015). Applying pharmacokinetic/pharmacodynamic principles in critically ill patients: optimizing efficacy and reducing resistance development. Semin. Respir. Crit. Care Med. 36, 136–153. doi: 10.1055/s-0034-1398490
Adams-Haduch, J. M., Paterson, D. L., Sidjabat, H. E., Pasculle, A. W., Potoski, B. A., Muto, C. A., et al. (2008). Genetic basis of multidrug resistance in Acinetobacter baumannii clinical isolates at a tertiary medical center in Pennsylvania. Antimicrob. Agents Chemother. 52, 3837–3843. doi: 10.1128/aac.00570-08
Argyres, M. I. (2010). Hospital-acquired infections due to gram-negative bacteria. New Engl. J. Med. 363, 1483–1483.
Asín-Prieto, E., Rodríguez-Gascón, A., and Isla, A. (2015). Applications of the pharmacokinetic/pharmacodynamic (PK/PD) analysis of antimicrobial agents. J. Infect. Chemother. 21, 319–329. doi: 10.1016/j.jiac.2015.02.001
Bertini, A., Poirel, L., Bernabeu, S., Fortini, D., Villa, L., Nordmann, P., et al. (2007). Multicopy blaOXA-58 gene as a source of high-level resistance to carbapenems in Acinetobacter baumannii. Antimicrob. Agents Chemother. 51, 2324–2328. doi: 10.1128/AAC.01502-06
Boscia, J. A., Korzeniowski, O. M., Snepar, R., Kobasa, W. D., Levison, M. E., and Kaye, D. (1983). Cefoperazone pharmacokinetics in normal subjects and patients with cirrhosis. Antimicrob. Agents Chemother. 23, 385–389. doi: 10.1128/AAC.23.3.385
Brochet, M., Couve, E., Zouine, M., Poyart, C., and Glaser, P. (2008). A naturally occurring gene amplification leading to sulfonamide and trimethoprim resistance in Streptococcus agalactiae. J. Bacteriol. 190, 672–680. doi: 10.1128/jb.01357-07
Bush, K. (2013). The ABCD’s of β-lactamase nomenclature. J. Infect. Chemother. 19, 549–559. doi: 10.1007/s10156-013-0640-7
Cantsilieris, S., Baird, P. N., and White, S. J. (2012). Molecular methods for genotyping complex copy number polymorphisms. Genomics 101, 86–93. doi: 10.1016/j.ygeno.2012.10.004
Cheah, S. E., Johnson, M. D., Zhu, Y., Tsuji, B. T., Forrest, A., Bulitta, J. B., et al. (2016). Polymyxin resistance in Acinetobacter baumannii: genetic mutations and transcriptomic changes in response to clinically relevant dosage regimens. Sci. Rep. 6, 26233. doi: 10.1038/srep26233
Chen, Y.-C., Zhou, Y.-J., Zhang, J., Wu, J.-F., Guo, B.-N., Wu, X.-J., et al. (2015). “Population pharmacokinetics and pharmacodynamics of cefoperazone/sulbactam (2:1) in adult patients with hospital-acquired pneumoniae,” in Proceedings of the Interscience Conference of Antimicrobial Agents and Chemotherapy, San Diego, CA.
Choi, J. Y., Kim, C. O., Park, Y. S., Yoon, H. J., Shin, S. Y., Kim, Y. K., et al. (2006). Comparison of efficacy of cefoperazone/sulbactam and imipenem/cilastatin for treatment of Acinetobacter bacteremia. Yonsei Med. J. 47, 63–69. doi: 10.3349/ymj.2006.47.1.63
Cooper, T. W., Pass, S. E., Brouse, S. D., and Hall, R. G. I. I. (2011). Can pharmacokinetic and pharmacodynamic principles be applied to the treatment of multidrug-resistant Acinetobacter? Ann. Pharmacother. 45, 229–240. doi: 10.1345/aph.1P187
Corvec, S., Poirel, L., Naas, T., Drugeon, H., and Nordmann, P. (2007). Genetics and expression of the carbapenem-hydrolyzing oxacillinase gene blaOXA-23 in Acinetobacter baumannii. Antimicrob. Agents Chemother. 51, 1530–1533. doi: 10.1128/AAC.01132-06
Delcher, A. L., Bratke, K. A., Powers, E. C., and Salzberg, S. L. (2007). Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23, 673–679. doi: 10.1093/bioinformatics/btm009
Drusano, G. L. (2004). Antimicrobial pharmacodynamics: critical interactions of ‘bug and drug.’ Nat. Rev. Microbiol. 2, 289–300. doi: 10.1038/nrmicro862
Gao, C., Tong, J., Yu, K., Sun, Z., An, R., and Du, Z. (2016). Pharmacokinetics of cefoperazone/sulbactam in critically ill patients receiving continuous venovenous hemofiltration. Eur. J. Clin. Pharmacol. 72, 823–830. doi: 10.1007/s00228-016-2045-x
Guindon, S., and Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704. doi: 10.1080/10635150390235520
Guo, F.-B., and Zhang, C.-T. (2006). ZCURVE_V: a new self-training system for recognizing protein-coding genes in viral and phage genomes. BMC Bioinformatics 10:9. doi: 10.1186/1471-2105-7-9
Hjort, K., Nicoloff, H., and Andersson, D. I. (2016). Unstable tandem gene amplification generates heteroresistance (variation in resistance within a population) to colistin in Salmonella enterica. Mol. Microbiol. doi: 10.1111/mmi.13459 [Epub ahead of print].
Holt, K. E., Wertheim, H., Zadoks, R. N., Baker, S., Whitehouse, C. A., Dance, D., et al. (2015). Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc. Natl. Acad. Sci. U.S.A. 112, E3574–E3581. doi: 10.1073/pnas.1501049112
Hornsey, M., Loman, N., Wareham, D. W., Ellington, M. J., Pallen, M. J., Turton, J. F., et al. (2011). Whole-genome comparison of two Acinetobacter baumannii isolates from a single patient, where resistance developed during tigecycline therapy. J. Antimicrob. Chemother. 66, 1499–1503. doi: 10.1093/jac/dkr168
Hu, F. P., Guo, Y., Zhu, D. M., Wang, F., Jiang, X. F., Xu, Y. C., et al. (2016). Resistance trends among clinical isolates in China reported from CHINET surveillance of bacterial resistance, 2005-2014. Clin. Microbiol. Infect. 22(Suppl. 1), S9–S14. doi: 10.1016/j.cmi.2016.01.001
Iyer, L. M., Leipe, D. D., Koonin, E. V., and Aravind, L. (2004). Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 146, 11–31. doi: 10.1016/j.jsb.2003.10.010
Jaruratanasirikul, S., Wongpoowarak, W., Aeinlang, N., and Jullangkoon, M. (2013). Pharmacodynamics modeling to optimize dosage regimens of sulbactam. Antimicrob. Agents Chemother. 57, 3441–3444. doi: 10.1128/AAC.00342-13
Johnson, E. R., and Mckay, D. B. (1999). Crystallographic structure of the amino terminal domain of yeast initiation factor 4A, a representative DEAD-box RNA helicase. RNA 5, 1526–1534. doi: 10.1017/S1355838299991410
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M., and Tanabe, M. (2016). KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462. doi: 10.1093/nar/gkv1070
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Lips, M., Siller, M., Strojil, J., Urbanek, K., Balik, M., and Suchankova, H. (2014). Pharmacokinetics of imipenem in critically ill patients during empirical treatment of nosocomial pneumonia: a comparison of 0.5-h and 3-h infusions. Int. J. Antimicrob. Agents 44, 358–362. doi: 10.1016/j.ijantimicag.2014.05.011
Liu, L. L., Ji, S. J., Ruan, Z., Fu, Y., Fu, Y. Q., Wang, Y. F., et al. (2015). Dissemination of blaOXA-23 in Acinetobacter spp. in China: main roles of conjugative plasmid pAZJ221 and transposon Tn2009. Antimicrob. Agents Chemother. 59, 1998–2005. doi: 10.1128/AAC.04574-14
Mardis, E. R. (2008). The impact of next-generation sequencing technology on genetics. Trends Genet. 24, 133–141. doi: 10.1016/j.tig.2007.12.007
Martinez, M. N., Papich, M. G., and Drusano, G. L. (2012). Dosing regimen matters: the importance of early intervention and rapid attainment of the pharmacokinetic/pharmacodynamic target. Antimicrob. Agents Chemother. 56, 2795–2805. doi: 10.1128/AAC.05360-11
McGann, P., Courvalin, P., Snesrud, E., Clifford, R. J., Yoon, E.-J., Onmus-Leone, F., et al. (2014). Amplification of aminoglycoside resistance gene aphA1 in Acinetobacter baumannii results in tobramycin therapy failure. MBio 5:e00915-14. doi: 10.1128/mBio.00915-14
Monogue, M. L., Kuti, J. L., and Nicolau, D. P. (2015). Optimizing antibiotic dosing strategies for the treatment of gram-negative infections in the era of resistance. Expert Rev. Clin. Pharmacol. 9, 459–476. doi: 10.1586/17512433.2016.1133286
Mugnier, P. D., Poirel, L., and Nordmann, P. (2009). Functional analysis of insertion sequence ISAba1, responsible for genomic plasticity of Acinetobacter baumannii. J. Bacteriol. 191, 2414–2418. doi: 10.1128/JB.01258-08
Musher, D. M., Dowell, M. E., Shortridge, V. D., Flamm, R. K., Jorgensen, J. H., Le Magueres, P., et al. (2002). Emergence of macrolide resistance during treatment of pneumococcal pneumonia. N. Engl. J. Med. 346, 630–631. doi: 10.1056/nejm200202213460820
Mwangi, M. M., Wu, S. W., Zhou, Y., Sieradzki, K., de Lencastre, H., Richardson, P., et al. (2007). Tracking the in vivo evolution of multidrug resistance in Staphylococcus aureus by whole-genome sequencing. Proc. Natl. Acad. Sci. U.S.A. 104, 9451–9456. doi: 10.1073/pnas.0609839104
Pascale, G. D., and Wright, G. D. (2010). Antibiotic resistance by enzyme inactivation: from mechanisms to solutions. Chembiochem 11, 1325–1334. doi: 10.1002/cbic.201000067
Peleg, A. Y., Seifert, H., and Paterson, D. L. (2008). Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21, 538–582. doi: 10.1128/cmr.00058-07
Penwell, W. F., Shapiro, A. B., Giacobbe, R. A., Gu, R. F., Gao, N., Thresher, J., et al. (2015). Molecular mechanisms of sulbactam antibacterial activity and resistance determinants in Acinetobacter baumannii. Antimicrob. Agents Chemother. 59, 1680–1689. doi: 10.1128/aac.04808-14
Perez, F., Hujer, A. M., Hujer, K. M., Decker, B. K., Rather, P. N., and Bonomo, R. A. (2007). Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 51, 3471–3484. doi: 10.1128/aac.01464-06
Redon, R., Ishikawa, S., Fitch, K. R., Feuk, L., Perry, G. H., Andrews, T. D., et al. (2006). Global variation in copy number in the human genome. Nature 444, 444–454. doi: 10.1038/nature05329
Reitberg, D. P., Whall, T. J., Chung, M., Blickens, D., Swarz, H., and Arnold, J. (1988). Multiple-dose pharmacokinetics and toleration of intravenously administered cefoperazone and sulbactam when given as single agents or in combination. Antimicrob. Agents Chemother. 32, 42–46. doi: 10.1128/AAC.32.1.42
Rice, L. B., Carias, L. L., Etter, L., and Shlaes, D. M. (1993). Resistance to cefoperazone-sulbactam in Klebsiella pneumoniae: evidence for enhanced resistance resulting from the coexistence of two different resistance mechanisms. Antimicrob. Agents Chemother. 37, 1061–1064. doi: 10.1128/AAC.37.5.1061
Sandegren, L., and Andersson, D. I. (2009). Bacterial gene amplification: implications for the evolution of antibiotic resistance. Nat. Rev. Microbiol. 7, 578–588. doi: 10.1038/nrmicro2174
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Sipahi, O. R., Arda, B., Nazli-Zeka, A., Pullukcu, H., Tasbakan, M., Yamazhan, T., et al. (2014). Piperacillin/tazobactam vs. cefoperazone/sulbactam in adult low-risk febrile neutropenia cases. Int. J. Clin. Pract. 68, 230–235. doi: 10.1111/ijcp.12279
Snitkin, E. S., Zelazny, A. M., Montero, C. I., Stock, F., Mijares, L., Murray, P. R., et al. (2011). Genome-wide recombination drives diversification of epidemic strains of Acinetobacter baumannii. Proc. Natl. Acad. Sci. U.S.A. 108, 13758–13763. doi: 10.1073/pnas.1104404108
Soo, V. W., Hanson-Manful, P., and Patrick, W. M. (2011). Artificial gene amplification reveals an abundance of promiscuous resistance determinants in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 108, 1484–1489. doi: 10.1073/pnas.1012108108
Sun, S., Berg, O. G., Roth, J. R., and Andersson, D. I. (2009). Contribution of gene amplification to evolution of increased antibiotic resistance in Salmonella typhimurium. Genetics 182, 1183–1195. doi: 10.1534/genetics.109.103028
Sy, S. K., Zhuang, L., and Derendorf, H. (2015). Pharmacokinetics and pharmacodynamics in antibiotic dose optimization. Expert Opin. Drug Metab. Toxicol. 12, 94–114.
Turton, J. F., Ward, M. E., Woodford, N., Kaufmann, M. E., Pike, R., Livermore, D. M., et al. (2006). The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol. Lett. 258, 72–77. doi: 10.1111/j.1574-6968.2006.00195.x
Waltner-Toews, R. I., Paterson, D. L., Qureshi, Z. A., Sidjabat, H. E., Adams-Haduch, J. M., Shutt, K. A., et al. (2011). Clinical characteristics of bloodstream infections due to ampicillin-sulbactam-resistant, non-extended- spectrum-beta-lactamase-producing Escherichia coli and the role of TEM-1 hyperproduction. Antimicrob. Agents Chemother. 55, 495–501. doi: 10.1128/aac.00797-10
Wang, Q., Wu, Y., Chen, B., and Zhou, J. (2015). Drug concentrations in the serum and cerebrospinal fluid of patients treated with cefoperazone/sulbactam after craniotomy. BMC Anesthesiol. 15:33. doi: 10.1186/s12871-015-0012-1
Wright, M. S., Haft, D. H., Harkins, D. M., Perez, F., Hujer, K. M., Bajaksouzian, S., et al. (2014). New insights into dissemination and variation of the health care-associated pathogen Acinetobacter baumannii from genomic analysis. MBio 5:e00963-13. doi: 10.1128/mBio.00963-13
Wright, M. S., Iovleva, A., Jacobs, M. R., Bonomo, R. A., and Adams, M. D. (2016). Genome dynamics of multidrug-resistant Acinetobacter baumannii during infection and treatment. Genome Med. 8, 26. doi: 10.1186/s13073-016-0279-y
Xia, J., Zhang, D., Xu, Y., Gong, M., Zhou, Y., and Fang, X. (2014). A retrospective analysis of carbapenem-resistant Acinetobacter baumannii-mediated nosocomial pneumonia and the in vitro therapeutic benefit of cefoperazone/sulbactam. Int. J. Infect. Dis. 23, 90–93. doi: 10.1016/j.ijid.2014.01.017
Xin, X., Jian, L., Xia, X., Jia, B., Huang, W., Li, C., et al. (2013). A multicentre clinical study on the injection of ceftriaxone/sulbactam compared with cefoperazone/sulbactam in the treatment of respiratory and urinary tract infections. Ann. Clin. Microbiol. Antimicrob. 12, 38. doi: 10.1186/1476-0711-12-38
Zerbino, D. R., and Birney, E. (2008). Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829. doi: 10.1101/gr.074492.107
Zhou, H., Zhang, T., Yu, D., Pi, B., Yang, Q., Zhou, J., et al. (2011). Genomic analysis of the multidrug-resistant Acinetobacter baumannii strain MDR-ZJ06 widely spread in China. Antimicrob. Agents Chemother. 55, 4506–4512.
Zhou, Y., Zhang, J., Guo, B., Yu, J., Shi, Y., Wang, M., et al. (2010). Liquid chromatography/tandem mass spectrometry assay for the simultaneous determination of cefoperazone and sulbactam in plasma and its application to a pharmacokinetic study. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 878, 3119–3124. doi: 10.1016/j.jchromb.2010.09.021
Keywords: cefoperazone/sulbactam, Acinetobacter baumannii, whole-genome sequencing, resistance development, PK/PD indices
Citation: Liu X, Zheng H, Zhang W, Shen Z, Zhao M, Chen Y, Sun L, Shi J and Zhang J (2016) Tracking Cefoperazone/Sulbactam Resistance Development In vivo in A. baumannii Isolated from a Patient with Hospital-Acquired Pneumonia by Whole-Genome Sequencing. Front. Microbiol. 7:1268. doi: 10.3389/fmicb.2016.01268
Received: 17 May 2016; Accepted: 02 August 2016;
Published: 19 August 2016.
Edited by:
Margaret Ip, The Chinese University of Hong Kong, ChinaReviewed by:
Soledad Ramirez, California State University, Fullerton, USACopyright © 2016 Liu, Zheng, Zhang, Shen, Zhao, Chen, Sun, Shi and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing Zhang, emhhbmdqX2Z1ZGFuQGFsaXl1bi5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.