- 1Department of Microbiology, The Forsyth Institute, Cambridge, MA, USA
- 2Department of Oral Medicine, Infection, and Immunity, Harvard School of Dental Medicine, Boston, MA, USA
- 3Department of Applied Oral Sciences, The Forsyth Institute, Cambridge, MA, USA
- 4Department of Surgery, Texas Tech University Health Sciences Center, Lubbock, TX, USA
- 5Division of Infectious Diseases, Boston Children’s Hospital, Harvard Medical School, Boston, MA, USA
Staphylococcus aureus–human interactions result in a continuum of outcomes from commensalism to pathogenesis. S. aureus is a clinically important pathogen that asymptomatically colonizes ~25% of humans as a member of the nostril and skin microbiota, where it resides with other bacteria including commensal Corynebacterium species. Commensal Corynebacterium spp. are also positively correlated with S. aureus in chronic polymicrobial diabetic foot infections, distinct from acute monomicrobial S. aureus infections. Recent work by our lab and others indicates that microbe–microbe interactions between S. aureus and human skin/nasal commensals, including Corynebacterium species, affect S. aureus behavior and fitness. Thus, we hypothesized that S. aureus interactions with Corynebacterium spp. diminish S. aureus virulence. We tested this by assaying for changes in S. aureus gene expression during in vitro mono- versus coculture with Corynebacterium striatum, a common skin and nasal commensal. We observed a broad shift in S. aureus gene transcription during in vitro growth with C. striatum, including increased transcription of genes known to exhibit increased expression during human nasal colonization and decreased transcription of virulence genes. S. aureus uses several regulatory pathways to transition between commensal and pathogenic states. One of these, the quorum signal accessory gene regulator (agr) system, was strongly inhibited in response to Corynebacterium spp. Phenotypically, S. aureus exposed to C. striatum exhibited increased adhesion to epithelial cells, reflecting a commensal state, and decreased hemolysin activity, reflecting an attenuation of virulence. Consistent with this, S. aureus displayed diminished fitness in experimental in vivo coinfection with C. striatum when compared to monoinfection. These data support a model in which S. aureus shifts from virulence toward a commensal state when exposed to commensal Corynebacterium species.
Introduction
The bacterium Staphylococcus aureus is a common member of the human microbiota on the skin of the nasal vestibules (nostrils), where it colonizes more than a quarter of the U.S. population (Gorwitz et al., 2008), as well as on other skin surfaces. S. aureus is also a common human pathogen that causes a range of diseases from mild skin infections to lethal bacteremias (Lowy, 1998). S. aureus nostril colonization correlates with an increased risk of S. aureus infection (Wertheim et al., 2005) and approximately 80% of bloodstream infection isolates match nostril strains (Wertheim et al., 2004). In the past decade, methicillin-resistant S. aureus (MRSA) has emerged as an important public health issue; from 2005 to 2013, MRSA was responsible for nearly 10,000 deaths annually in the United States (CDC, 2005–2013). The possibility that S. aureus might acquire or evolve resistance to antibiotics beyond β-lactams, such as methicillin, is a grave concern in medicine and public health. This underlies the urgent need for research on novel antimicrobial (Conlon et al., 2013) and antivirulence therapies (Murray et al., 2014; Nielsen et al., 2014; Sully et al., 2014). Specific mechanisms of virulence in S. aureus have been studied for decades and are well characterized. Yet factors that influence the maintenance of harmless colonization (commensalism) and the transition from commensalism to virulence are still being defined.
Staphylococcus aureus possesses a broad array of colonization and virulence factors that interact with the human host; these include cytolysins, macromolecule degrading enzymes and immune evasion machinery (Lowy, 1998; Otto, 2010). S. aureus virulence is heavily affected by expression of the quorum sensing-controlled accessory gene regulator (agr) genetic locus, which has been studied extensively (for reviews Novick and Geisinger, 2008; Thoendel et al., 2011). The agr locus is divided into two divergent transcripts, RNAII and RNAIII, which comprise the agrBDCA operon and RNAIII regulatory RNA, respectively. The genes of the agrBDCA operon encode AgrB, which processes and exports an autoinducing peptide signal (AIP) derived from AgrD; and the AgrC sensor kinase with its cognate response regulator AgrA, which, when activated at high cell density, induces RNAII and RNAIII expression. Increased RNAIII transcription ultimately leads to the repression of adhesins and other surface proteins and the induction of capsule synthesis, toxins, proteases and other extracellular virulence factor production. Thus, agr activation is postulated to play a key role in S. aureus’ transition from an adherent commensal lifestyle to an invasive pathogenic lifestyle (Novick and Geisinger, 2008; Thoendel et al., 2011).
As a member of the healthy skin microbiota, S. aureus interacts with a diverse array of other bacterial constituents; e.g., S. aureus primarily colonizes the nostrils (a.k.a. anterior nares) where it is detected in conjunction with members of the genera Corynebacterium and Propionibacterium (Uehara et al., 2000; Lina et al., 2003; Frank et al., 2010; Wos-Oxley et al., 2010; Oh et al., 2012; Yan et al., 2013). S. aureus also overlaps with other bacteria in various infection environments. For example, in chronic, polymicrobial diabetic foot infections (DFI) S. aureus is detected alongside numerous other bacterial species (Citron et al., 2007; Gardner et al., 2013); in particular, there is a positive correlation between S. aureus and Corynebacterium spp. in DFIs (Gardner et al., 2013). Recent work by us and others has begun to characterize specific microbe–microbe interactions of S. aureus with either Propionibacterium spp. (Wang et al., 2014; Wollenberg et al., 2014) or Corynebacterium spp. (Yan et al., 2013). We, and others, hypothesize that commensal bacteria play a role in maintaining health either by influencing S. aureus gene expression toward a commensal lifestyle or by limiting the expansion of S. aureus, both of which would limit the risk of acute infection.
In this study, we tested the hypothesis that S. aureus interactions with Corynebacterium spp. limit S. aureus virulence. Using a reductionist approach to mechanistically characterize interactions, we focused on S. aureus and Corynebacterium striatum, a skin commensal also commonly reported in DFIs (Citron et al., 2007). To assess how S. aureus responds to growth with C. striatum, we examined the S. aureus transcriptomes in mono- versus coculture in vitro with C. striatum. Growth with C. striatum resulted in global changes in S. aureus transcript abundance, including decreased expression of many genes induced by the agr QS system and increased expression of genes upregulated during in vivo nasal colonization (Burian et al., 2010a,b; Krismer et al., 2014). We found that exposure to cell-free conditioned medium (CFCM) from C. striatum and other Corynebacterium spp. was sufficient to decrease expression of an agr-induced promoter in S. aureus. When exposed to C. striatum CFCM, S. aureus displayed increased epithelial cell-adhesion activity and decreased hemolysin activity. In a murine subcutaneous abscess model, we observed that, compared to monoinfection, S. aureus abundance in vivo was diminished during coinfection with C. striatum, i.e., S. aureus was a less successful pathogen. These data demonstrate diminished virulence of S. aureus in response to commensal Corynebacterium spp., which is consistent with a shift to a commensal lifestyle when in the presence of a healthy microbiota rich with Corynebacterium spp. Overall, these results suggest a beneficial role for non-pathogenic members of the Corynebacterium genus in limiting S. aureus virulence.
Materials and Methods
Strains and Media
Strains used in this study are described in Supplementary Table S2. Unless indicated, Staphylococcus spp. and Corynebacterium spp. were routinely cultured using Brain Heart Infusion (BHI) broth or solid agar medium and Escherichia coli was grown on Lysogeny Broth (LB) medium. For indicated experiments, a chemically defined medium (CDM) (Brown and Whiteley, 2007) was used with the addition of 25 mM 2-(N-morpholino)ethanesulfonic acid (MES) buffer at pH 6 or pH 7.5. All cultures were grown at 37°C in standard atmosphere; liquid cultures were shaken at 250 RPM. Antibiotics were used at the following concentrations: ampicillin 100 μg/ml, erythromycin 10 μg/ml, tetracycline 3 μg/ml, fosfomycin 20 μg/ml and chloramphenicol 20 μg/ml for E. coli and 10 μg/ml for S. aureus. We selectively enumerated S. aureus on Mannitol Salts Agar (MSA) and Corynebacterium spp. on BHI agar with 20 μg/ml fosfomycin using colony-forming unit (CFU) measurement via track dilution as described previously (Jett et al., 1997).
DNA and Plasmid Manipulations
DNA and plasmid isolation were performed using standard methods (Ausubel et al., 2002). Restriction endonucleases and DNA modification enzymes were purchased from New England Biolabs. Chromosomal DNA from all bacteria was isolated using DNeasy tissue kits (Qiagen); plasmid isolations were performed using QIAprep spin miniprep kits (Qiagen). For both kits, S. aureus cells were lysed by adding 25 μg of lysostaphin (Sigma–Aldrich) to the kit lysis buffer and incubating at 37°C for 30 m prior to following the manufacturer’s protocol. DNA fragments were purified using QIAquick mini-elute PCR purification kits, and PCR was performed using GoTaq Green (Promega). Primers used in this study are indicated in Supplementary Table S3. DNA sequencing was performed by automated sequencing technology through Macrogen Co. (Macrogen USA).
Markerless Gene Deletion Construction in S. aureus
Markerless gene deletions were generated using a previously established protocol with derivatives of the temperature sensitive S. aureus shuttle vector pKFT (Kato and Sugai, 2011). Surface protein A (spa) deletion was accomplished using pLF048, a pKFT derivative (Foulston et al., 2014). pKFT-derivative vectors were electroporated into S. aureus RN4220 and grown at room temperature for 2 h before plating on BHI tetracycline 3 μg/ml agar medium and incubated at 30°C for ~36 h. Replicating plasmids were purified from S. aureus RN4220 prior to electroporation into S. aureus JE2 and treated as previously described (Kato and Sugai, 2011). Markerless tetS mutants were confirmed by PCR using primers external to flanking regions of the pKFT derivatives (oKL377 and oKL378 for the spa deletion) followed by amplicon sequencing to verify loss of the targeted gene and religation of the flanking regions.
Construction of the spa-Expression Vector
To express spa in trans in the Δspa mutant, we cloned the spa gene with its native promoter region into pEPSA5 (Forsyth et al., 2002). Using primers oKL554 and oKL555, we amplified the Pspa-spa-containing fragment, then digested it and pEPSA5 with SalI and BamHI (NEB). We electroporated pEPSA5 and pEPSA5-spa into S. aureus RN4220 then repurified and transformed each respectively into KPL2389 to generate the Δspa mutant with the empty vector control (KPL2496) and spa-complemented strain (KPL 2497).
Mono- and Coculture Assays
Colony cultures (Zheng and Stewart, 2002) were prepared similar to previously described on agarose medium (Ramsey and Whiteley, 2009). Briefly, CDM 1% agarose solid medium was prepared using 25 mM MES buffered CDM at pH 6.0. Sterile 25 mm 0.2 μm polycarbonate membranes (Millipore #GTTP02500) were placed on solid agarose medium and inoculated with 10 μl of liquid CDM containing either 5 × 106 C. striatum or 1 × 104 S. aureus separately in monoculture or combined in coculture. Inoculated spots were allowed to briefly air dry then transferred to 37°C for 18 h. Next, membranes were aseptically transferred to fresh medium and incubated at 37°C for another 4 h. To harvest cells, membranes were either immediately transferred to RNALater (Ambion) for RNAseq experiments (described below); transferred to 1 ml sterile Phosphate Buffered Saline (PBS), vortexed for 30 s and serially diluted for CFU enumeration; or transferred to 1 ml sterile PBS, vortexed for 30 s, pelleted by centrifugation at 10,000 g for 5 m then resuspended in 300 μl of lysis buffer for β-galactosidase assays (described below).
RNA Isolation
Bacterial cells resuspended in 1 ml of RNALater (Ambion) (see above) were pelleted by centrifugation (10,000 g at 4°C for 10 m). Cell pellets were stored at -20°C. For lysis, cells were resuspended in 100 μL TE buffer with 10 mg/ml lysozyme and 25 μg/ml lysostaphin and incubated 20 m at 37°C. After adding 900 μl of RNA Bee (Amsbio), the mixture was transferred to 2 ml Lysing Matrix B bead tubes (MP Biomedicals) and bead beat in a FastPrep-24 (MP Biomedicals) 6x at the highest setting for 30 s each, with 30 s of rest on ice between each cycle. Next, 0.2 ml of chloroform was added to each tube and vortexed for 30 s then stored on ice for 5 m prior to centrifugation (12,000 g for 15 m at 4°C). The (upper) aqueous phase was removed and mixed with 0.5 ml of isopropanol in a microcentrifuge tube and incubated 10 m at room temperature (RT). After centrifugation at 12,000 g for 5 m at RT, the supernatant was carefully removed. One ml of ice-cold 75% ethanol was added to each tube, vortexed briefly and then centrifuged for 5 m at 7,500 g. This 75% ethanol wash was repeated, then RNA pellets were air dried at RT for 5–10 m. RNA was resuspended in 30 μl of nuclease-free ddH2O. DNA contamination was removed by digestion with 10 U of RQ1 DNAse (Promega) in a 100 μl reaction for 30 m at 37°C. RNA was then re-purified using RNABee as in the steps above. Final RNA samples were resuspended in 30 μl of nuclease-free H2O. RNA integrity was verified by visualization on a SYBR-Safe (Invitrogen) stained agarose gel and RNA quantity was determined by spectrophotometric analysis.
RNAseq Library Preparation
RNAseq libraries were prepared using methods adapted from those described previously (Jorth et al., 2014). Ribosomal RNA was removed with the RiboZero Bacterial kit (Epicentre) per the manufacturer’s instructions. RNA was then fragmented with the NEBNext Magnesium RNA Fragmentation Module (NEB) and ethanol Na-acetate precipitated with the addition of 2 μl of linear acrylamide (NEB). Precipitated RNA was resuspended in 14 μl of nuclease-free H2O. Sequencing libraries of fragmented RNA were assembled using the NEBNext sRNA library prep kit (NEB) per the manufacturer’s instructions. Assembled libraries were run on 6% polyacrylamide TBE gels and bands between 130 and 300 bp were gel purified and extracted as per the NEBNext sRNA library kit instructions. Libraries were assayed for concentration and integrity on a Bioanalyzer 2100 (Agilent). RNAseq libraries were submitted to Tufts University Core Facility Genomics for sequencing using single-end 50 bp reads on an Illumina HiSeq 2500 system.
RNAseq Results Validation
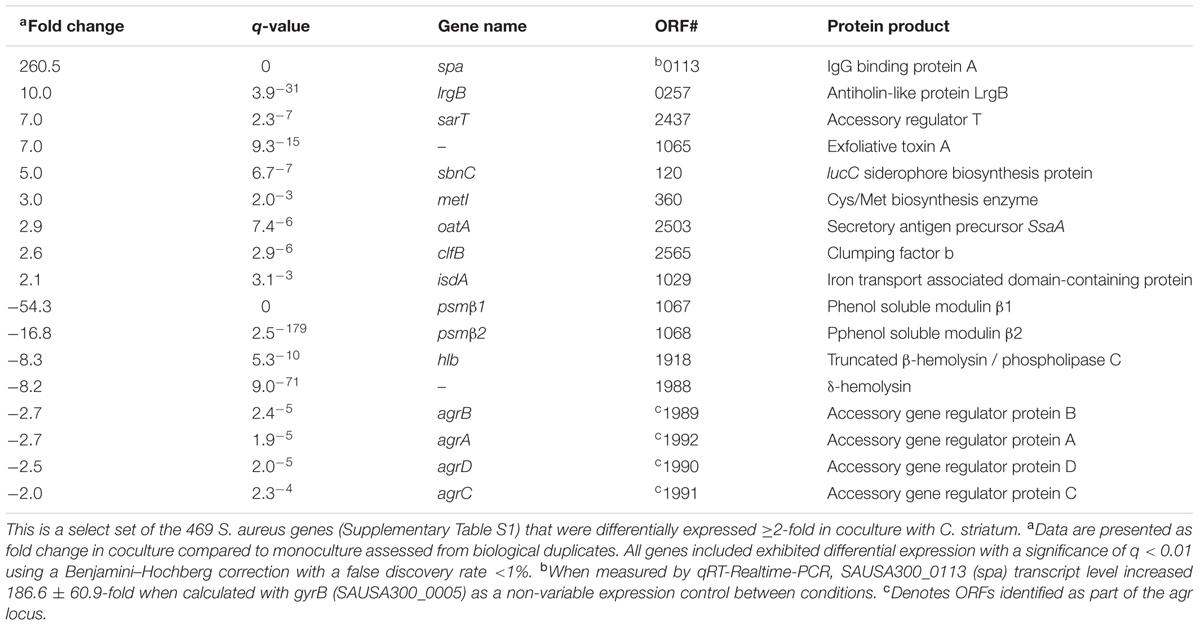
Separate quantification of gene expression from S. aureus JE2 was determined by quantitative Reverse Transcriptase PCR (qRT-PCR) from the original non-rRNA-depleted RNA samples and additional experimental samples generated under identical conditions. RNA preparation with DNA removal was performed as above and 0.5 ng of total RNA was used for qRT-PCR with a qScript One-Step SYBR Green qRT-PCR kit (Quanta Biosciences) to determine overall transcript abundance in each experimental condition. We used primers oKL311 and oKL312 to measure spa transcript abundance and oKL339 and oKL340 to measure gyrB abundance as an internal control. Controls lacking reverse transcriptase were also performed to verify lack of contaminating DNA. Comparisons of relative abundance of spa expression normalized to gyrB expression per sample were used to calculate fold change of spa expression and compared to RNAseq data under the same conditions (Table 1).
RNAseq Data Analysis
The S. aureus USA300 FPR3757 genome sequence (Diep et al., 2006) (NCBI accession #NC_007793) was used for RNAseq read alignment. FASTQ files were aligned to the S. aureus JE2 (NC_007793) genome using Rockhopper (McClure et al., 2013) with default settings.
Nucleotide Sequence Accession Numbers
RNAseq sequencing data are available at NCBI in the sequence read archive under BioProject accession number PRJNA335565.
AIP-1 Preparation
AIP-1 production was performed similarly to methods described previously (Thoendel and Horswill, 2009). Briefly, E. coli strain AH594 carrying an arabinose-inducible AIP-1 synthesizing gene locus (PBAD-agrBD) was grown overnight at 37°C shaking at 200 RPM in 5 ml of LB with 100 μg/ml ampicillin. A 1:50 dilution of overnight culture was added to 50 ml LB without ampicillin in a 250 ml flask and was shaken at 200 RPM incubated at 37°C for 90 min prior to addition of arabinose to a final concentration of 0.02%. The induced culture was further incubated in the same conditions for 4 h. Next the cell culture was transferred to a sterile 50 ml conical centrifuge tube and cell supernatants were removed after centrifugation at 7,000 g for 10 m at 4°C. Cell supernatants were next passed through a 0.2 μm filter (Millipore, #SCGP00525) to remove remaining cell debris and then passed through a 3 kDa-cutoff size exclusion filter (Millipore #UFC900324). Filtrates were prepared for storage by addition of sterile glycerol and dithiothreitol to concentrations of 10% and 1 mM respectively. Aliquots of 1.5 ml each were stored at -80°C and exhibited full activity for >30 days.
Corynebacterium spp. CFCM Preparation
Cell-free conditioned medium was generated from each indicated Corynebacterium spp. after inoculation into BHI cultures and growth at 37°C shaking at 200 RPM for 48 h. Conditioned medium was removed after centrifugation of 48-h cultures for 10 m at 7,000 g and then passed through a 0.2 μm filter (Millipore, #SCGP00525) yielding CFCM, which was either used directly for experiments, frozen at -80°C prior to use or further passed through a 3 kDa molecular weight size exclusion filter (Millipore #UFC900324) then frozen at -80°C. For some samples, CFCM was heat treated at 95°C for 15 m. For other samples, CFCM was Proteinase K (Promega #V3021) treated by addition of 100 μg in 1 ml of CFCM prior to incubation at 55°C for 1 h followed by a 95°C heat inactivation for 10 m prior to testing for the effect on AIP-1.
S. aureus AIP-1 Luminescence Detection Assays
AIP-1-dependent luminescence assays were performed similarly to those described previously (Jensen et al., 2008; Thoendel and Horswill, 2009). Briefly, 100 μl of an overnight culture of the AIP-1-responsive agrP3-lux luminescence reporter strain ROJ143 grown in 3 ml BHI with 10 μg/ml chloramphenicol was inoculated into 5 ml BHI with 10 μg/ml chloramphenicol and incubated at 37°C, shaking at 200 RPM for 1.5 h. Optical density at 600 nm (OD600) was measured and cells were then diluted in BHI to an OD600 of 0.1 and 100 μl were added to each well of a 96-well plate to be combined with mixes of CFCM for testing. Meanwhile, 150 μL of E. coli AIP-1-containing CFCM was incubated in a 1.5 ml microcentrifuge tube for 2 h at 37°C with an equal volume of one of the following: BHI or Corynebacterium spp. CFCM. Next, 25 μl of each incubated solution was added to four separate wells containing 100 μl of the ROJ143 reporter strain and incubated for 3 h at 37°C prior to reading luminescence on a BioTek Synergy HT illuminometer with a 0.5 s integration time. Luminescence was quantified as the average of four replicate wells each from 3 independent experiments.
S. aureus Human Epithelial Cell Attachment Assay
We adapted this assay from a previously published protocol (Weidenmaier et al., 2004) and A549 cell cultures were performed identically with the exception that F-12K medium and 10% Fetal Bovine Serum (Life Technologies) was used. A549 cells were grown to 90–95% confluency (5 × 104/well) in 24-well plates and washed 3x in serum-free F-12K medium before addition of S. aureus. We added a 1:10 dilution of E. coli CFCM containing AIP-1 into 1.5 ml cultures containing 1:100 inoculums from overnight cultures of either wild-type or agrA-deficient S. aureus JE2. A separate set of tubes was prepared identically plus the addition of a 1:2 dilution of 3 kDa-filtered C. striatum CFCM. These cultures were incubated at 37°C shaking at 200 RPM for 4 h. Late exponential cells were washed 3x in BHI and then diluted in serum-free 37°C F-12K medium and added to each well in 500 μl volumes at an MOI of 25. S. aureus cells from each growth condition and A549 cells were coincubated for 1h at 37°C and 5% CO2. Aliquots of planktonic cells were removed after incubation for CFU enumeration and A549 cell cultures were then washed 3x with sterile PBS. Five hundred microliter of PBS with 0.04% Triton-X100 was added to each well and incubated for 5 m at ambient temperature before epithelial cells were disrupted by vigorous micropipette aspiration. S. aureus-containing suspensions were transferred to microcentrifuge tubes and vortexed for 30 s each before being serially diluted for CFU enumeration. CFU values for attached cells were divided by their respective planktonic CFU values to determine % attachment for each condition. Percent attachment was divided by the % attachment for the WT S. aureus condition to determine fold change differences in attachment for each condition. Attachment was measured from technical triplicates for each condition across three biological replicates from separate A549 cell cultures.
Immunofluorescence IgG-Capture Assay, Microscopy and Image Analysis
Cultures were grown as described in ‘mono- and coculture assays.’ Polycarbonate membranes were transferred to 1.5 ml microcentrifuge tubes containing 1 ml of ice-cold 95% methanol and stored at -20°C. Cells were removed from membranes by gentle agitation and membranes were discarded. We added 5 μl of cell suspension to poly-lysine coated slides (Corning). These were air dried for ~5 m. To block, 100 μl of PBS with 1% BSA was added to each sample and incubated for 1 h at RT. Blocking solution was removed by pipette after 1 h and an equal volume was added then removed immediately after. Fifty microliter of rabbit α-goat FITC-conjugated IgG antibody (Life Technologies) diluted 1:500 in PBS with 1% BSA was added to the sample and incubated in the dark at RT for 1 h. Antibody solution was removed by pipette and the sample was rinsed 3x with 200 μl PBS with 0.1% Triton X100 which was gently added and removed by pipette. The sample was rinsed 1x with 100 μl PBS and excess liquid was removed by pipette without allowing the sample to air dry. Seven microliter of DAPI mounting medium (Southern Biotech) was quickly added and the sample was covered with a 25 × 25 mm #1 coverslip and incubated in the dark for >10 m at RT.
Each sample was imaged at 1000x magnification on a Zeiss Axio Observer using manufacturers filter settings for DAPI and FITC acquisition and identical exposure times for all samples based on autofluorescence of a non-FITC stained control sample. Three randomly selected fields of view were taken of each sample from three biological replicates for each growth condition. Images were analyzed using ImageJ software (NIH). Due to pronounced differences in DAPI-staining intensity between species, S. aureus was easily quantified from coculture samples by thresholding images in the DAPI channel. Cells positively stained for FITC were also identified by thresholding based on a non-FITC stained control. Using identical thresholding settings, total S. aureus (DAPI) versus SpA-expressing S. aureus (FITC) were counted in all fields of view using the ‘Analyze Particles’ function.
S. aureus Rabbit Erythrocyte Lysis Assay
This assay was adapted from a previously published protocol (Pang et al., 2010). We added a 1:10 dilution of E. coli CFCM containing AIP-1 into 1.5 ml cultures containing 1:100 inoculums of either wild-type or agrA-deficient S. aureus JE2. A separate set of tubes was prepared identically plus the addition of a 1:2 dilution of 3 kDa-filtered C. striatum CFCM. These cultures were incubated at 37°C shaking at 200 RPM for 4 h. After incubation, CFCM was generated from each S. aureus culture by passage through a 0.2 μm filter (Millipore, #SCGP00525). Rabbit blood (Hemostat Laboratories, USA) was diluted v/v to 3% in sterile PBS then 70 μl was combined with 30 μl of each S. aureus CFCM in eight replicates for each condition in a 96-well plate. Blood was also combined at the same ratio with BHI as a negative control and BHI was used as a blank for OD630 measurements. Plates were incubated at 37°C for 40 m then OD630 was read to determine rabbit erythrocyte lysis.
In vivo Murine Abscess Growth
Murine abscesses were generated essentially as described previously (Mastropaolo et al., 2005). Briefly, 6–8 week-old, female, Swiss Webster mice were anesthetized with an intraperitoneal injection of Nembutal (50 mg/kg). The hair on the left inner thigh of each mouse was shaved and the skin was disinfected with 70% alcohol. Mice were injected subcutaneously in the inner thigh with ~106 CFU either S. aureus (wt or Δspa) or ~106 C. striatum, or a combination of both. At 4 days post-infection, mice were euthanized and intact abscesses were harvested, weighed and placed into 2 ml of sterile PBS. Tissues were homogenized, serially diluted and plated on BHI agar with 25 μg/ml fosfomycin for C. striatum enumeration or MSA for S. aureus enumeration, to determine bacterial CFU/abscess. Experimental protocols involving mice were examined and approved by the Texas Tech University HSC Institutional Animal Care and Use Committee.
S. aureus AIP1-4 GFP Fluorescence Assays
Assays to detect AIP induction were carried out similarly to those described previously, (Kavanaugh et al., 2007) using the agrP3-gfp AIP inducible reporter vector pDB59 (Yarwood et al., 2004). Briefly, S. aureus reporter strains AH1677, AH430, AH1747 and AH1871 (representing agr types I–IV respectively) were grown overnight in 5 ml BHI cultures at 37°C shaking at 200 RPM. From each culture, 3 ml were passed through a 0.2 μm membrane to generate S. aureus CFCM containing AIP produced by post-exponential phase cultures. We then generated 1.5 ml BHI cultures, which were separately inoculated with a 1:100 dilution of each overnight culture and contained a 1:10 dilution of the cognate AIP-containing overnight CFCM to induce agr-dependent expression of the GFP reporter as positive controls. A separate set of tubes was prepared similarly but with the addition of a 1:10 dilution of S. aureus CFCM containing an AIP type known to inhibit the agr type of the respective reporter strain; these served as negative controls for agr induction. E. coli AIP-1-containing CFCM was used to inhibit agr types II and III while S. aureus CFCM containing AIP-2 was used to inhibit agr types I and IV. Finally, another set of tubes was prepared identically to the positive controls except that these contained a 1:2 dilution of C. striatum CFCM passed through a 3 kDa-cutoff size exclusion filter. These cultures were incubated identically to the overnight cultures for either 4 or 6 h and examined for GFP fluorescence. To quantify GFP fluorescence, 25 μl of culture was combined with 75 μl of sterile BHI in a single well of a 96-well plate with four replicates per culture. GFP activity was determined by first measuring OD at 630 nm (OD630) then measuring GFP emission through 485 ± 20 nm excitation and 528 ± 20 nm emission filters on a BioTek Synergy HT plate reader. Fluorescence measurements were divided by OD630 to normalize for changes in culture density. Strains of agr-I, III, and IV showed maximal induction 6 h after supernatant additions whereas agr-II was induced at 4 h.
Ethics Statement
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Institutional Animal Care and Use Committee of Texas Tech University Health Sciences Center (Protocol Number: 09039).
Results
Cocultivation of S. aureus with C. striatum Affects the Expression of Genes Involved in Virulence and Colonization
We hypothesized that S. aureus gene expression would change in response to C. striatum. To test this hypothesis, we assessed gene expression using RNA sequencing (RNAseq) after growing S. aureus JE2 (Fey et al., 2013) (a USA300 LAC derivative) in either mono- or coculture with C. striatum ATCC 6940 on solid (1% agarose) CDM (Brown and Whiteley, 2007) at pH 6 without glucose at 37°C. This solid-phase, low pH medium partially approximates human skin-surface conditions. There were no significant changes in growth yield, as determined by CFU (colony forming unit) measurement, between mono- and coculture conditions for either species. Table 1 contains select S. aureus genes that were differentially expressed in coculture with C. striatum as compared to monoculture. Overall, 469 genes were differentially expressed ≥2-fold, with statistical significance (Supplementary Table S1).
A striking result from the S. aureus RNAseq data was that roughly half of the differentially expressed genes in cocultivation with C. striatum are also members of the S. aureus agr regulon (Cassat et al., 2006; Queck et al., 2008). The gene whose transcript was most decreased, psmβ1, encodes a phenol-soluble modulin toxin, which is positively regulated by the agr regulon, and psmβ1 downregulation in coculture with C. striatum was validated using a lacZ reporter construct based on its own promoter (Supplementary Figure S1). Further, expression of the entire S. aureus agr operon was decreased in coculture with C. striatum (Table 1). Similarly, the gene whose transcript was most increased in coculture (up 260-fold) is indirectly influenced by the S. aureus agr QS system (Schmidt et al., 2003; Novick and Geisinger, 2008). This is the staphylococcal protein A (spa) gene, which encodes surface protein A (SpA). We validated this increase in spa transcription by qRT-PCR (Table 1). SpA is characterized as an immunoprotective protein that inhibits opsonization and phagocytosis (Forsgren and Sjoquist, 1966; Peterson et al., 1977; Spika et al., 1981) and recent research indicates a role for SpA during nasal colonization. Transcription of spa is elevated in both humans and rodents during nasal colonization compared to in vitro culture (Burian et al., 2010a,b; Krismer et al., 2014). In addition, during experimental in vivo human nasal colonization, spa-deficient mutants are cleared more rapidly than wild-type S. aureus in hosts with a robust nasal immune response and SpA protein levels positively correlate with duration of colonization (Cole et al., 2016). Interestingly, in addition to spa, C. striatum also induced several other S. aureus genes whose expression is increased during in vivo nasal colonization of humans (and of cotton rats), including metI, sbnC, clfB, isdA, and oatA (Table 1) (Burian et al., 2010a,b; Krismer et al., 2014). Overall, we observed altered levels of S. aureus transcripts regulated by agr QS during cocultivation with C. striatum with increased expression of genes known to be upregulated during in vivo nasal colonization and decreased expression of genes known to be upregulated during invasive infection. Based on these data, we hypothesized that, in response to the commensal C. striatum, S. aureus shifts to a commensal state with a decrease in activities positively regulated by the agr QS system and an increase in activities negatively regulated by the agr QS, i.e., diminished expression of virulence factors needed for successful invasive infection and increased expression of surface-associated adhesion factors important for host colonization.
Exposure to C. striatum Cell-Free Conditioned Medium Is Sufficient to Alter S. aureus agr-Dependent Gene Expression
As the first step toward testing the hypotheses above, we asked whether the decrease in expression of genes positively regulated by agr QS in coculture with C. striatum requires cell-cell contact or whether exposure to C. striatum CFCM is sufficient. To address this question, we used a luminescent agrP3 promoter reporter assay (Jensen et al., 2008), which produces light only when agr QS is activated, and measured the effect of C. striatum CFCM on agr QS (Figure 1). This also allowed us to confirm that the S. aureus response is mediated via agr QS. In this assay, S. aureus autoinducing peptide type 1 (AIP-1) that has been heterologously produced in E. coli (Thoendel and Horswill, 2009) induces an AIP-1-responsive agrP3-luxCDABE transcriptional fusion in a S. aureus mutant unable to produce AIP-1 (Jensen et al., 2008). If C. striatum CFCM is sufficient to decrease agr QS then addition of CFCM will decrease luminescence. C. striatum CFCM samples were adjusted to neutral pH to avoid inhibition of agr activity by pH extremes (Regassa et al., 1992). CFCM from C. striatum stationary phase cultures (>36 h old) was sufficient to inhibit S. aureus agr QS (Figure 1, leftmost light gray bar), indicating that cell–cell contact is not required for the observed interaction between S. aureus and C. striatum. In contrast, neither CFCM from the parent E. coli strain used to produce AIP-1 nor a S. aureus mutant with a transposon insertion in agrB (AgrB-), which is unable to produce AIP, resulted in inhibition of agrP3-lux activity to the extent of C. striatum CFCM (Figure 1, white bars). Further fractionation of C. striatum CFCM revealed that the inhibitory activity passed through a 3 kDa-molecular weight filter and also remained functional after heat treatment at 95°C for 15 min and was resistant to Proteinase K digestion (Figure 1, medium gray bars). Maximal agr QS inhibitory activity required preincubation of AIP-1 with C. striatum CFCM for at least 2 h prior to addition to the reporter strain (Supplementary Figure S2). This eliminates the possibility that inhibition is due to production of a competitive inhibitor of the AgrC sensor kinase by C. striatum, because an inhibitor that binds directly to AgrC would not require preincubation with AIP-1 for activity. These data established that S. aureus decreases activation of the agr QS system in response to C. striatum CFCM likely due to the presence of a small molecule that acts on AIP-1 itself. In subsequent experiments, we assayed for changes in S. aureus’ activity (behavior) after exposure to C. striatum by using CFCM, rather than cocultivation.
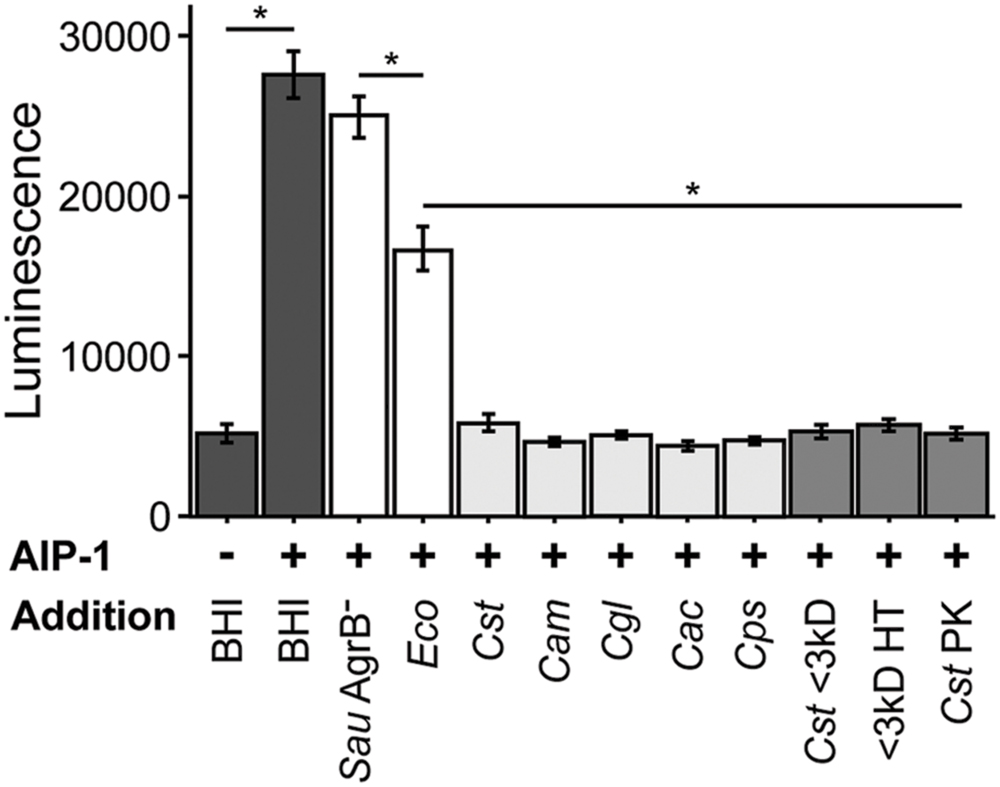
FIGURE 1. Staphylococcus aureus agr quorum sensing decreases in response to Corynebacterium spp. Using a luminescent agr-I activity assay, we tested the ability of CFCM from the indicated species to inhibit activation of agrP3 after exposure to exogenous AIP-1. Dark gray bars indicate AIP-1 negative and positive controls. White bars indicate CFCM controls from S. aureus JE2 containing a Tn insertion in agrB and the plasmid-free derivative of the E. coli strain used to exogenously produce AIP-1. Light gray bars indicate CFCM from 48 h cultures of C. striatum, C. amycolatum, C. glutamicum, C. accolens, and C. pseudodiphtheriticum, respectively. Medium gray bars indicate C. striatum (Cst) CFCM treatments, ‘<3 kD’ indicates sub 3 kilodalton fractions, ‘HT’ is CFCM heat treated at 95°C for 15 m, ‘PK’ indicates Proteinase K digested CFCM. For each bar, n = 3 and error bars represent SEM. For each result between the indicated samples (horizontal line), *p < 0.05 by two-tailed Student’s t-test with Bonferroni correction for multiple testing.
S. aureus Exhibits Decreased Activation of agr QS in Response to Multiple Commensal Species of Corynebacterium
In addition to C. striatum, S. aureus encounters a number of other commensal species of Corynebacterium in the nasal passages, on skin and in DFIs, e.g., (Citron et al., 2007; Frank et al., 2010; Wos-Oxley et al., 2010; Oh et al., 2012; Kaspar et al., 2016). To determine whether S. aureus responds similarly to other commensal Corynebacterium spp., we selected C. amycolatum, C. accolens and C. pseudodiphtheriticum, all three of which are common members of nasal microbiota (Yan et al., 2013; Kaspar et al., 2016), as is S. aureus. Also, like C. striatum, C. amycolatum is associated with DFIs (Citron et al., 2007). In addition, we tested a strain of C. glutamicum, which was originally isolated from soil (Abe et al., 1967) and is not associated with human colonization. When exposed to CFCM from these Corynebacterium spp., S. aureus again displayed reduced agrP3-lux reporter activity (Figure 1, light gray bars). These results are consistent with a conserved genus-level interaction between Corynebacterium spp. and S. aureus that is predicted to result in a shift in S. aureus behavior toward colonization and away from virulence. We selected C. striatum for all subsequent experiments to characterize the S. aureus phenotypic response to commensal Corynebacterium spp.
S. aureus Adhesion to Epithelial Cells Increases When Exposed to C. striatum
Many of the cell surface proteins that are expressed when the S. aureus agr QS is not activated are involved in adhesion to host cells (Burian et al., 2010b; Foster et al., 2014), and are negatively regulated by activation of agr QS. Therefore, we hypothesized that when grown to high cell density in the presence of C. striatum CFCM, S. aureus would exhibit increased adhesion to host epithelial cells, a phenotype that can serve as a proxy for colonization or commensal behavior (Foster et al., 2014). To test this, we measured S. aureus attachment to A549 human airway epithelial cells, similarly to previously described (Weidenmaier et al., 2004). We cultured wild-type S. aureus in the presence of exogenous AIP-1 plus or minus C. striatum CFCM. As a positive control, we performed the identical experiment with an isogenic S. aureus agrA-deficient mutant in the presence of AIP-1. S. aureus cells in late exponential phase from each condition were normalized to identical optical densities and then added to epithelial cell monolayers at a multiplicity of infection (MOI) of 25. Next, we enumerated total planktonic and epithelial-cell-attached S. aureus by CFU measurement. We compared the proportion of planktonic versus epithelial cell-attached CFUs in each condition and expressed the results as fold change of attached cells from each condition compared to the S. aureus WT. After growth with C. striatum CFCM, the wild-type S. aureus (WT; dark gray bars in Figure 2) exhibited increased adhesion to the respiratory epithelial cells; this increase was comparable to that of an isogenic agrA-deficient mutant (AgrA-, white bar in Figure 2). The increase in S. aureus adhesion to epithelial cells when exposed to C. striatum is consistent with our hypothesis that S. aureus shifts toward a commensal state in the presence of C. striatum.
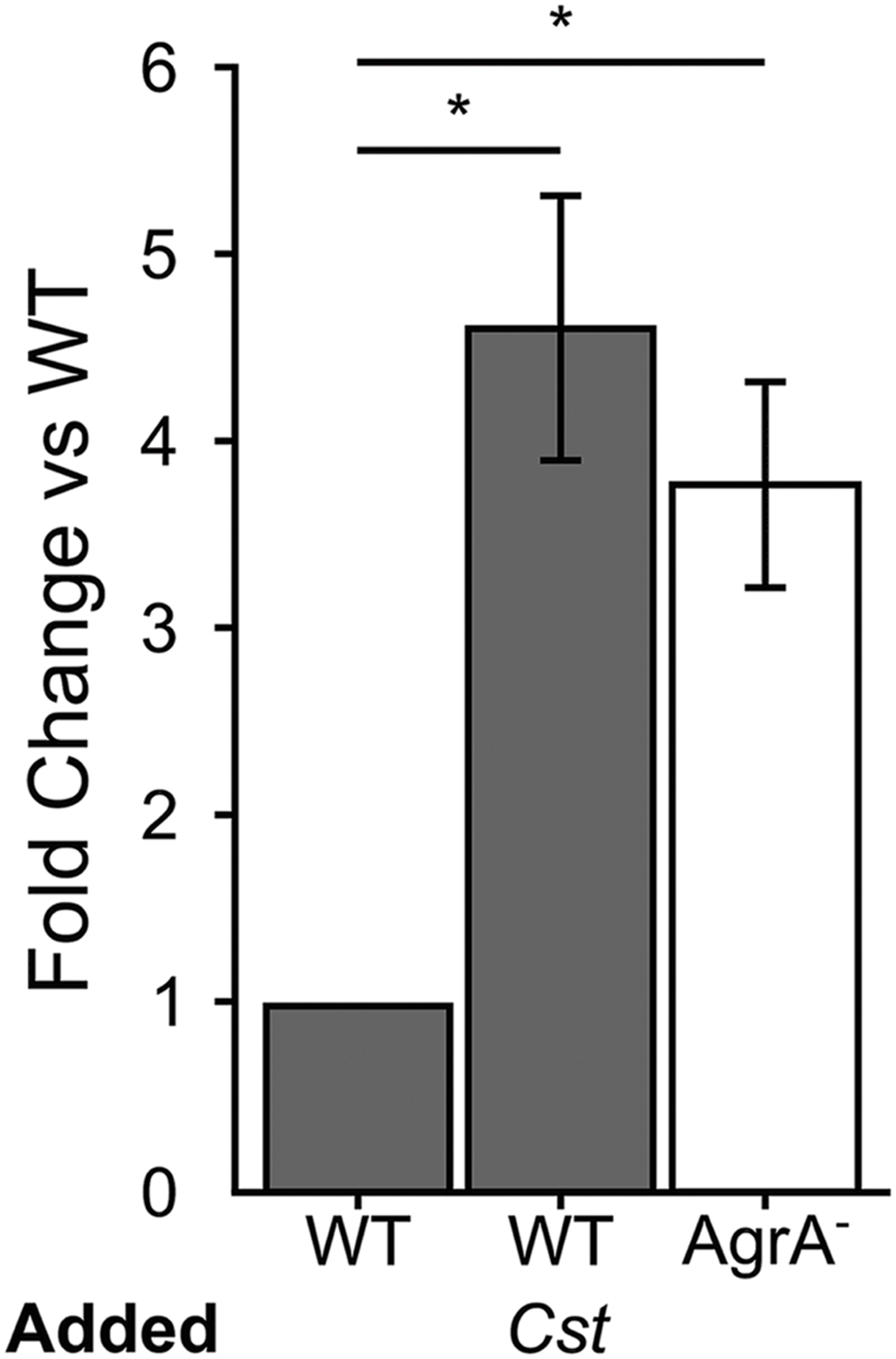
FIGURE 2. Staphylococcus aureus attachment to human epithelial cells increases in response to C. striatum CFCM. When S. aureus WT (gray bars) and AgrA- (white bar) cells were exposed to AIP-1 plus or minus C. striatum CFCM (Cst), we observed 4.5- and 3.6-fold increases in attachment respectively. These increases were statistically significant (*) when compared to the WT alone. S. aureus attachment to human A549 epithelial cells was quantified after 1 h of exposure. Attachment was measured as the percentage of attached cells divided by the total number of S. aureus planktonic cells added, as determined by CFU enumeration. Fold change was determined by dividing the percent attached for each condition by that of the % attached for WT exposed only to AIP. Error bars were omitted from the WT normalized data for clarity. Data were analyzed by two-tailed Student’s t-test with Bonferroni correction for multiple testing (*p < 0.03). Error bars represent SEM.
S. aureus Increases SpA on Its Surface in Response to C. striatum
The S. aureus spa gene transcript was the most highly increased in coculture with C. striatum compared to monoculture. Traditionally, SpA has been studied in the context of invasive infection; however, using qRT-PCR, Burian et al. (2010b) report that transcript levels of spa are increased in vivo during nasal colonization compared to in vitro culture indicating SpA might play a role in commensal interactions with the host. More recently, Cole et al. (2016) found that increased levels of SpA correlate with longer duration of S. aureus colonization during experimental human nasal inoculation and that spa-deficient mutants are less effective than the wild-type at colonizing some humans. Based on these data, which indicate a likely role for SpA in colonization, we proceeded to explore the effect of the observed increase in spa transcription in response C. striatum. Regulation of spa transcript levels is well studied and complex, depending on several elements (Schmidt et al., 2003; Gustafsson et al., 2009). Like many adhesion factors, SpA is a cell-surface protein that is negatively regulated through agr QS, albeit indirectly. Therefore, we first verified that the increase in spa transcription during cocultivation with C. striatum was indeed dependent on agrA (Supplementary Figure S3). We hypothesized that the increase in spa transcript level when grown with C. striatum would translate to an increase in S. aureus production of functional SpA. To test this hypothesis, we investigated SpA abundance and activity in S. aureus using immunoblot and immunofluorescence assays. First, we stained for SpA via immunoblot with an α-SpA antibody using cell lysates from cultures incubated in the presence of AIP-1 alone or with exposure to C. striatum CFCM. SpA abundance qualitatively increased upon treatment with C. striatum CFCM and SpA was absent in a Δspa mutant (Supplementary Figure S4). We then assayed for SpA activity with a standard IgG binding assay. Using solid-phase culture conditions identical to those in our RNAseq experiments, we quantified the proportion of S. aureus cells exhibiting SpA IgG binding activity in mono- versus coculture by measuring SpA capture of FITC-conjugated IgG antibodies. We found that 60% of wt S. aureus cells had detectable IgG capture in coculture compared to only 10% in monoculture (Figure 3A, dark bars). IgG capture was visible as a ring around cells in both mono- and coculture with C. striatum, consistent with cell-surface expression of SpA (Figures 3B,C). No FITC fluorescence was detected in a Δspa mutant in mono- or coculture (Figure 3A). Fluorescence signal was restored when spa was complemented in trans. The greater proportion of cells with detected surface IgG capture in the Δspa mutant containing a multicopy plasmid with spa under its native promoter suggests that the construct results in expression of spa above wt levels (Figure 3A, white bars). Together these data demonstrate that the C. striatum-dependent increase in S. aureus spa transcript levels (Table 1) results in a phenotypic change with an increased number of cells displaying active SpA on their surface. Active SpA can diminish opsonization and phagocytosis via binding of the Fc portion of IgG, inhibiting serum complement pathway activation (Forsgren and Sjoquist, 1966; Peterson et al., 1977; Spika et al., 1981). Therefore, we also confirmed that the increased level of SpA activity in coculture corresponded to decreased S. aureus phagocytosis (Supplementary Figure S5). Together with the evidence for increased epithelial cell adhesion after exposure to C. striatum, these results, along with reports that spa expression and SpA abundance increases during in vivo nasal colonization (Burian et al., 2010b; Cole et al., 2016), support our hypothesis that S. aureus shifts toward a colonization (commensal) state in the presence of Corynebacterium spp.
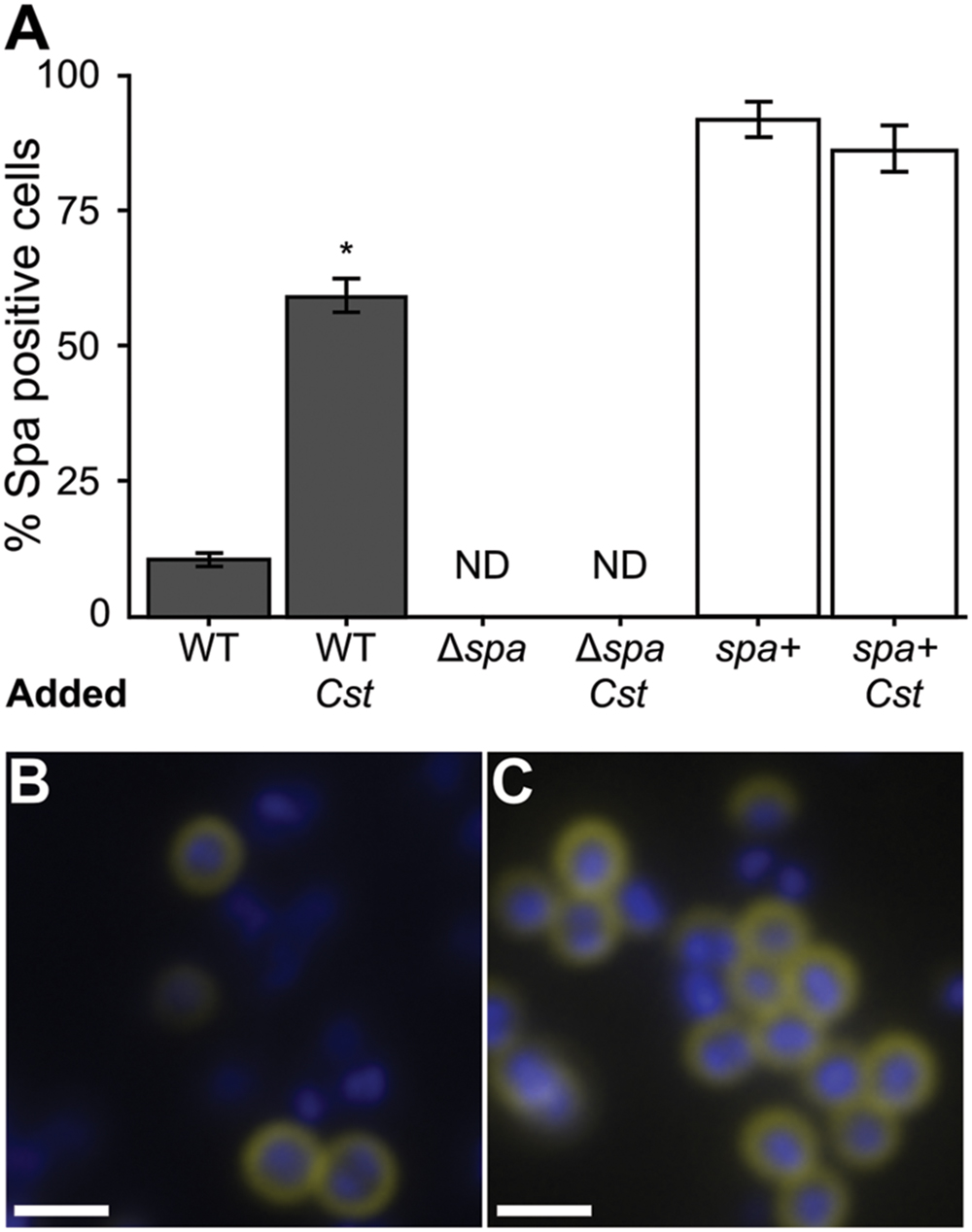
FIGURE 3. The proportion of S. aureus cells with active surface-associated SpA increases in coculture with C. striatum. S. aureus strains were grown in mono- versus coculture with C. striatum under conditions identical to those used in Table 1. SpA activity was detected by observing SpA-mediated capture of FITC-conjugated goat IgG (yellow in B,C). Cells were counterstained with DAPI (blue in B,C). Cell intensity was quantified for all S. aureus strains in three fields of view at 1000x magnification each from 3 biological replicates under identical exposure settings. (A) ImageJ software was used to count total cells versus FITC-positive cells. 10% of wild-type S. aureus (WT) were positive for SpA activity in monoculture compared to 60% in coculture with C. striatum (Cst). The spa-deletion mutant carrying the empty expression vector (Δspa) had no detectable FITC staining (ND). The spa-deletion mutant carrying an expression vector expressing spa from its native promoter (spa+) had FITC-positive cells in both conditions consistent with this construct producing SpA above wt levels. Error bars represent SEM. Data were analyzed by two-tailed Student’s t-test with Bonferroni correction for multiple testing (*p < 0.003). Representative micrographs of wild-type S. aureus cells (blue) in mono- (B) versus coculture (C) stained for SpA (yellow) at the cell surface. Scale bars represent 1.5 μm.
S. aureus agr-Dependent Hemolytic Activity Decreases in Response to C. striatum
The other side of agr QS-regulated activities in S. aureus is an increase in the production of secreted virulence factors at high cell density when agr QS is activated (Novick and Geisinger, 2008; Thoendel et al., 2011). Hemolysis has traditionally served as an approximation of S. aureus virulence factor production and is typically diminished in agr mutants (Blevins et al., 2002). Therefore, we assayed for the rabbit erythrocyte hemolytic activity of CFCM from S. aureus grown with or without C. striatum CFCM. Based on our RNAseq data, we predicted that S. aureus exposed to C. striatum would exhibit decreased hemolysin activity due to repression of genes encoding β and δ-hemolysins (Table 1), which are both positively regulated by agr QS. Using a published method (Pang et al., 2010), we quantified hemolysis as the loss of optical density at 630 nm of rabbit erythrocytes when mixed with CFCM harvested from each S. aureus strain induced with AIP-1, in late-exponential phase, in the presence or absence of C. striatum CFCM (Cst). An agrA-deficient mutant served as a non-hemolytic control (Figure 4, light gray bars). In this assay, wild-type S. aureus production of hemolytic activity was strongly diminished by exposure to C. striatum CFCM (Figure 4, dark gray bars). These results further support our hypothesis that S. aureus shifts away from virulence in the presence of C. striatum.
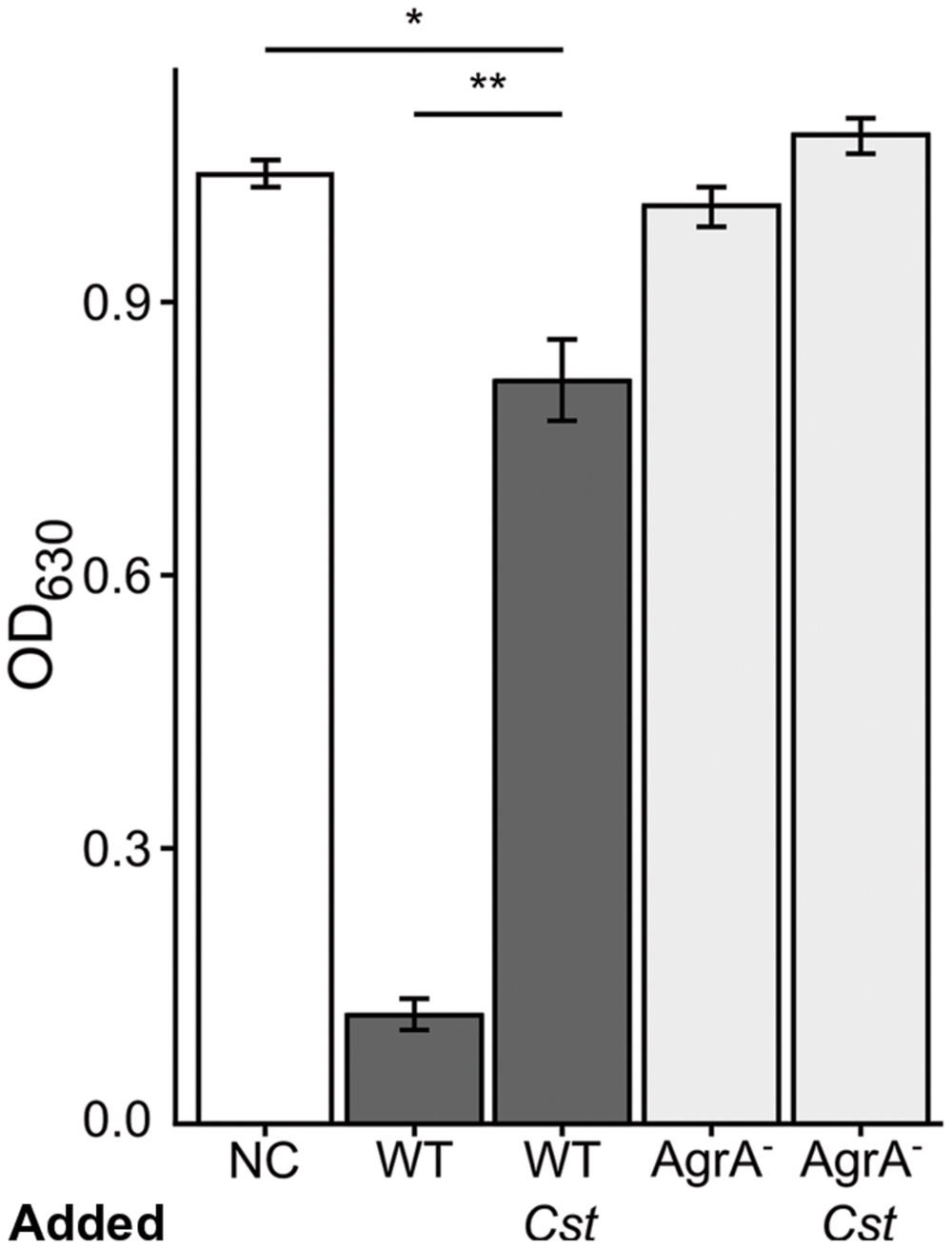
FIGURE 4. Staphylococcus aureus exhibits decreased hemolytic activity when grown with C. striatum CFCM. Hemolysis of rabbit erythrocytes was quantified 30 m after exposure to BHI, as the negative control (NC), or CFCM from S. aureus strains grown in the presence of AIP-1 alone or plus the addition of C. striatum CFCM (Cst) for the wild-type (WT) and agrA::Tn mutant (AgrA-), n = 3 each. Decreased OD630 is indicative of S. aureus hemolytic activity. Growing S. aureus in the presence of C. striatum CFCM significantly diminished the hemolytic activity produced by the WT. In contrast, the agrA::Tn mutant (AgrA-) was incapable of significant hemolysis in either condition. Data were analyzed by two-tailed Student’s t-test with Bonferroni correction for multiple testing (*p < 0.005, **p < 0.00005). Error bars represent SEM.
S. aureus Numbers Decrease during Coinfection with C. striatum in a Murine Abscess Model
We have shown that S. aureus agr QS-dependent gene expression is altered in response to commensal Corynebacterium spp. in a manner that shifts its phenotypic behavior toward colonization and away from virulence. Therefore, we hypothesized that in vivo coinfection with C. striatum would attenuate S. aureus’ success when compared to monoinfection. To test this, we compared colony-forming units (CFUs) from mono- and coinfection of S. aureus and C. striatum in a previously established murine subcutaneous abscess model (Mastropaolo et al., 2005; Ramsey et al., 2011). We measured abscess weight and bacterial CFUs 4 days after subcutaneous injection of single- versus dual-species infections. S. aureus CFUs decreased 6.2-fold in coinfection with C. striatum (Figure 5A), whereas C. striatum CFUs increased 20.7-fold in coinfection with S. aureus (Figure 5B). Thus, coinfection with S. aureus and C. striatum resulted in a disadvantage for S. aureus while providing an advantage to C. striatum compared to monoinfection, implying that S. aureus responds to the presence of C. striatum with changes in gene/protein expression that decrease its fitness for invasive infection while C. striatum capitalizes on the presence of S. aureus in this infection model.
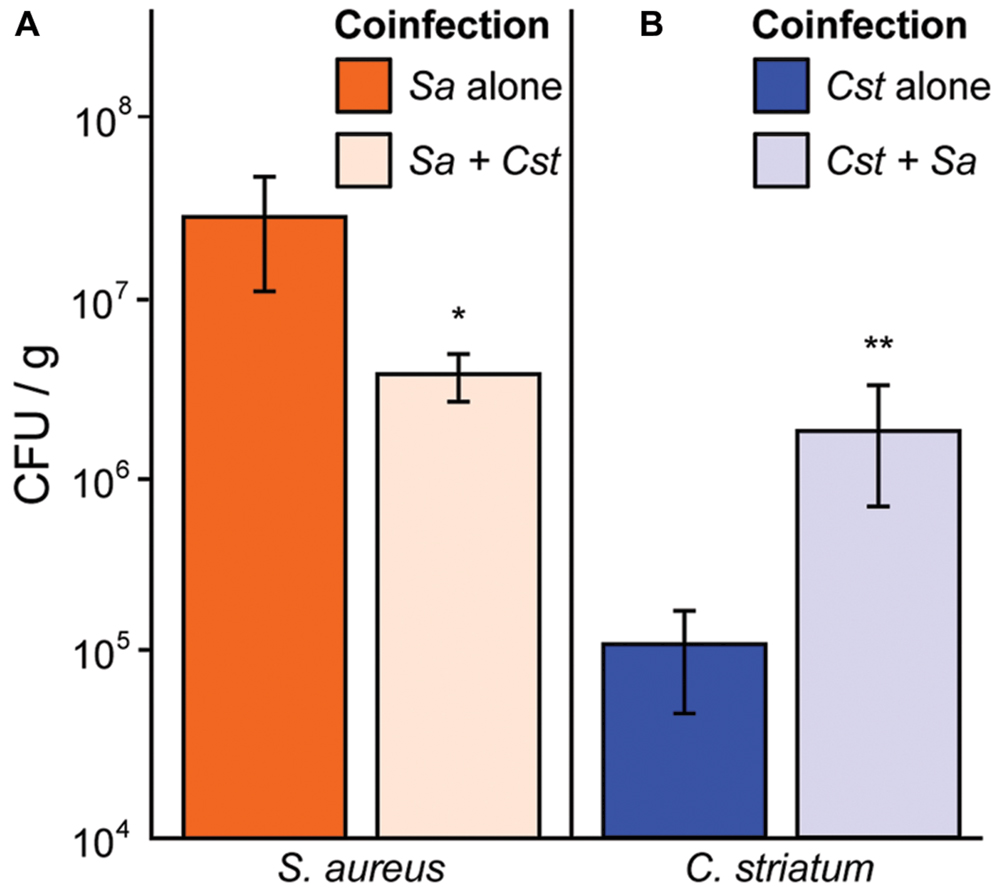
FIGURE 5. Staphylococcus aureus abundance decreases in vivo when coinfected with C. striatum in a murine abscess model. (A) In a murine abscess infection model 4 days post-infection, wild-type S. aureus showed reduced numbers (CFU/g) during coinfection with C. striatum (light orange bar; Sa + Cst) compared to monoinfection (orange bar; Sa alone). (B) In the same model, C. striatum numbers increased significantly when coinfected with S. aureus (light blue bar; Cst + Sa) when compared to monoinfection (blue bar; Cst alone). For each bar, n = 9. Data were analyzed using the Mann Whitney U-test (*p < 0.03, **p < 0.02). Error bars represent SEM.
S. aureus AIP Types 1, 2, and 3 Respond Similarly to C. striatum
Staphylococcus aureus strain JE-2, which was used for all of the above experiments, is an AIP type I strain. S. aureus has four characterized agr types, (agr I–IV) that each make a specific AIP molecule (AIP 1–4) along with its cognate AgrC (reviewed in Novick and Geisinger, 2008; Thoendel et al., 2011). Each AIP type acts as a potent quorum signal inhibitor for other agr type strains, with the exception of AIP-1 and AIP-4 whose structures differ by only a single peptide (D-Y substitution) and which do not inhibit each other (Novick and Geisinger, 2008; Thoendel et al., 2011). We hypothesized that multiple agr types would respond similarly to C. striatum and tested this using a published assay (Kavanaugh et al., 2007) with an AIP-inducible agrP3-gfp transcriptional fusion to monitor agr QS-dependent activation in strains representing all four agr types. In liquid culture, the agr QS system is activated in stationary phase when AIP levels are highest; therefore, as a positive control, we added stationary-phase CFCM from each cognate AIP-type strain as an inducer to mid-exponential-phase cells of the same strain and measured GFP fluorescence (Figure 6, dark gray bars). Strains incubated with a combination of the cognate S. aureus AIP CFCM and an inhibitory S. aureus AIP-type CFCM served as negative controls (Figure 6, white bars). Reporter cultures were incubated with a mix of cognate inducing S. aureus AIP CFCM and C. striatum CFCM to test for inhibition. In response to C. striatum CFCM, the agrP3 activity of agr types I, II, and III decreased, but type IV’s did not (Figure 6, light gray bars). These results indicate that in response to C. striatum, and likely Corynebacterium spp. in general, agr QS decreases in multiple S. aureus agr lineages.
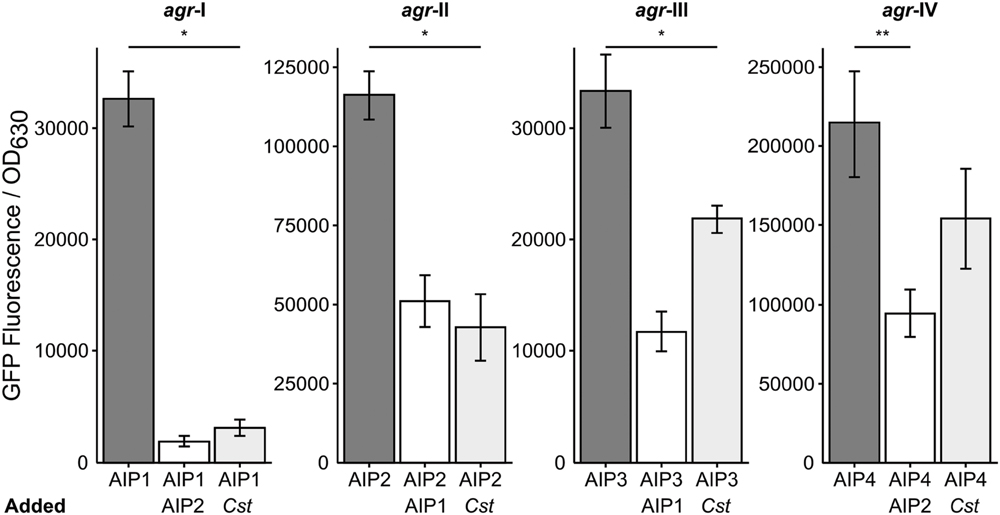
FIGURE 6. Corynebacterium striatum CFCM inhibits signaling of S. aureus agr types I, II, and III. For each respective agr type, post-exponential phase S. aureus CFCM from that same type was used to induce an agrP3:gfp reporter. CFCM containing AIP-1 was used to inhibit S. aureus agr types II and III strains and CFCM containing AIP-2 was used to inhibit S. aureus agr types I and IV strains (white bars). Cultures grown in the presence of inducing AIP-containing S. aureus CFCM and C. striatum CFCM showed pronounced inhibition of agr types I, II, and III and no significant inhibition of agr type IV (light gray bars). GFP was measured as fluorescence/OD630 (n = 3). Error bars represent SEM. Data were analyzed by two-tailed Student’s t-test with Bonferroni correction for multiple testing (*p < 0.005 and **p < 0.01).
Discussion
Staphylococcus aureus is a common cause of infection in humans, and nasal colonization correlates with an increased risk of infection (Wertheim et al., 2005). S. aureus infections range from chronic and indolent, e.g., polymicrobial DFIs, to acute and aggressive, e.g., monomicrobial bacteremia. However, S. aureus is typically an asymptomatic colonizer of humans (Kuehnert et al., 2006; Gorwitz et al., 2008), coexisting in the nasal and skin microbiota with other bacteria, including members of the genus Corynebacterium, e.g., (Uehara et al., 2000; Lina et al., 2003; Wos-Oxley et al., 2010; Oh et al., 2012; Yan et al., 2013). Therefore, from both a clinical and public health perspective, it is important to identify factors that influence whether S. aureus behaves as a commensal or a pathogen. This is highlighted by the fact that S. aureus has eluded repeated attempts at vaccine development (Proctor, 2012; Jansen et al., 2013), accentuating the need for alternative approaches to prevent S. aureus infections. Corynebacterium spp. commonly coexist with S. aureus on epithelial surfaces of the nasal passages and skin, as well as in DFIs (Uehara et al., 2000; Lina et al., 2003; Citron et al., 2007; Frank et al., 2010; Wos-Oxley et al., 2010; Oh et al., 2012; Gardner et al., 2013; Yan et al., 2013); yet, little is known about how S. aureus interacts with Corynebacterium spp. Here, we have demonstrated that S. aureus responds to commensal Corynebacterium spp. with altered expression of genes involved in colonization and virulence (Table 1, Supplementary Table S1), and does so in a manner that is similar to the transcriptomes of agr QS loss-of-function mutants (Dunman et al., 2001; Cassat et al., 2006; Queck et al., 2008). This transcriptional response translates to an increase in cell-surface activities associated with colonization, e.g., epithelial-cell adhesion and SpA activity (Figures 2 and 3), and a decrease in production of secreted virulence factors, e.g., hemolysin (Figure 4). During in vivo infection, these result in decreased success during coinfection with C. striatum. In total, the data presented here indicate that S. aureus responds to commensal Corynebacterium with a shift to commensalism. This opens up the possibility that commensal Corynebacterium spp. are an unexplored source for new antivirulence therapies that limit activation of S. aureus agr QS as a means to control and/or prevent S. aureus infection, a goal that has been actively explored through other approaches (Murray et al., 2014; Nielsen et al., 2014; Sully et al., 2014).
Our in vitro coculture RNAseq data showed striking similarities to S. aureus gene expression during in vivo nasal colonization of humans, and rodent models of S. aureus nasal colonization (Kiser et al., 1999; Burian et al., 2010a,b; Krismer et al., 2014) (Figure 7). For example, Krismer et al. (2014) suggest that methionine competition and synthesis, along with oligopeptide transport and iron acquisition, might be critical for S. aureus colonization, based on finding that metI, which encodes a methionine biosynthesis gene; oppB, which encodes an oligopeptide transporter; and sbnC, which encodes an iron transport protein, are induced during human nostril colonization. Indeed, a metI-deficient mutant has strongly reduced colonization capacity in the cotton rat model of S. aureus nasal colonization (Krismer et al., 2014). Our transcriptome data show that the levels of metI, and the methionine synthesis operon it is part of (SAUSA300_0357-360; Table 1/Supplementary Table S1), are increased in the presence of C. striatum suggesting that S. aureus and Corynebacterium may compete for methionine in vitro in methionine-replete medium, as well as in vivo. We did not detect significant differential expression of oppB; however; we did observe upregulation of six other opp genes (Supplementary Table S1), two of which are adjacent to oppB, suggesting S. aureus may respond similarly during coculture with C. striatum and in vivo nasal colonization. We also found that sbnC and its operon (SAUSA300_0118-122; Table 1/Supplementary Table S1) were upregulated in coculture with C. striatum along with iron-regulated surface determinant adhesin-encoding isdA (Table 1/Supplementary Table S1) indicating another possible competition for iron.
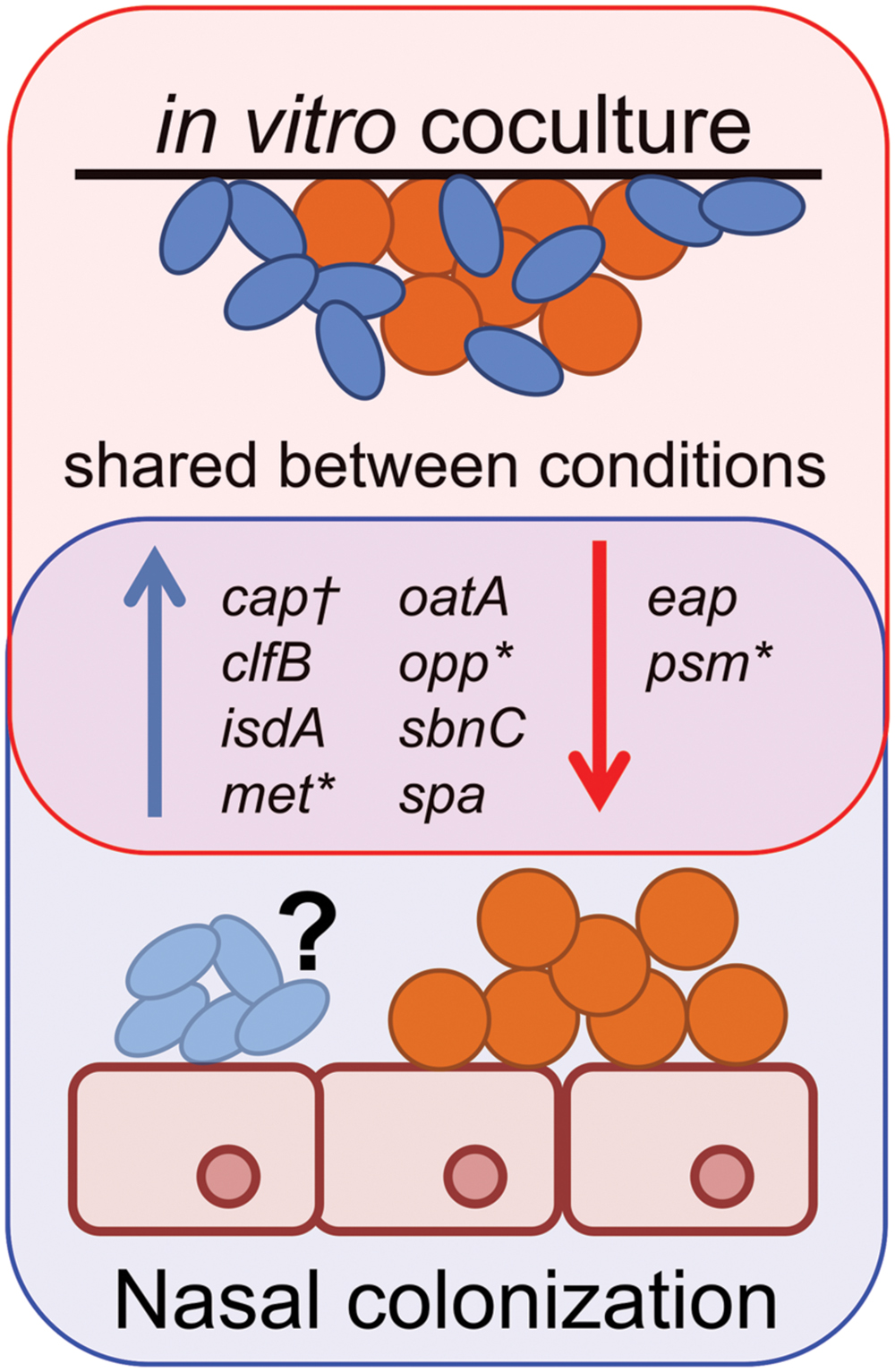
FIGURE 7. Staphylococcus aureus gene expression during in vivo colonization is similar to in vitro coculture with C. striatum. Gene expression data from S. aureus (orange spheres) in vivo colonization of humans (Burian et al., 2010b; Krismer et al., 2014), cotton rats (Burian et al., 2010a) or in vitro coculture with C. striatum (blue ovals) (Table 1/Supplementary Table S1) is depicted as upregulated (blue arrows) or downregulated (red arrows) in comparison to in vitro monoculture. Genes quantified by qRT-PCR in vivo share similar patterns of expression with those detected with RNAseq during in vitro coculture with C. striatum (“shared between conditions”). *In several cases, individual genes identified by qRT-PCR were part of operons whose members were also differentially regulated in our RNASeq results (Supplementary Table S1). †Type 5 capsule (CP5) production was quantified by ELISA in a mouse nasal colonization model (Kiser et al., 1999) and was overrepresented in vivo versus in vitro. We observed upregulation of several CP5 synthesis genes in coculture with C. striatum (Supplementary Table S1). It is unknown (“?”) whether or not Corynebacterium spp. were present in the referenced in vivo experiments. These data demonstrate the similarities in S. aureus gene expression during host commensal in vivo colonization and in vitro growth with Corynebacterium spp.
In another study, Burian et al. (2010b) selected target genes to reflect functions that distinguish colonization from invasive infections, e.g., adhesins versus secreted toxins (Burian et al., 2010b), and then used qRT-PCR to examine transcription of these 30 S. aureus genes during human nostril colonization. They report increased transcript levels of spa, the clumping factor B adhesin-encoding clfB [which is reported to be a major determinant in S. aureus human nasal colonization (Wertheim et al., 2008)], isdA and the secretory antigen oatA in human nostrils compared to in vitro culture, along with decreased expression of the psmβ1-2 genes (Burian et al., 2010b). Again, these changes in S. aureus transcription are similar to our in vitro coculture data (Table 1; Supplementary Table S1, Figure 1; Supplementary Figure S1). However, unlike their in vivo colonization data, we did not observe differential expression of sceD, atlA, sak, hla and wall teichoic acid related genes (tagO and tarK) (Supplementary Table S1), which suggests that the increase in these might be a response to the host environment or to other commensal bacteria.
Staphylococcus aureus nasal colonization has also been studied in rodent models. (Of note, at present, there are no established models for Corynebacterium spp. nasal colonization.) During stable late-term S. aureus nasal colonization of the cotton rat, another study found increased expression of clfB and isdA (Burian et al., 2010a), both of which were upregulated in coculture with C. striatum in vitro (Table 1/Supplementary Table S1). Additionally, Kiser et al. (1999) observed that S. aureus type-5 capsule (CP5) is produced in greater abundance in a mouse nasal colonization model than in vitro and that CP5 is necessary for abundant nasal colonization. Consistent with this, we detected five S. aureus CP5 biosynthesis genes upregulated in coculture with C. striatum (Supplementary Table S1).
There are two caveats in comparing our in vitro transcription data to data from in vivo nasal colonization. First, we do not know the composition of the nasal microbiota in the human or rodent colonization studies. Second, our conditions do not perfectly mimic the host environment. However, our data demonstrate that many of the key genes expressed during S. aureus nasal colonization of humans and rodents are also expressed in the presence of C. striatum in the complete absence of a mammalian host. This suggests the S. aureus response to Corynebacterium spp. in the microbiota contributes to reported in vivo expression changes. This type of observation is not unprecedented; Ramsey and Whiteley (2009) have previously shown in an unrelated study that a commensal bacterium alone induces an immunoprotective response in an opportunistic pathogen in vitro. In fact, it is possible that many of the transcriptional responses observed in the microbiota may be driven by microbe–microbe interactions and not simply by host–microbe interactions.
Based on results presented here, we propose that the S. aureus response to commensal Corynebacterium in polymicrobial infections dampens its virulence with implications for its behavior during both colonization and polymicrobial infection. S. aureus is capable of causing acute, destructive monomicrobial infections through the production of secreted virulence factors regulated by agr QS and other pathways (Lowy, 1998; Otto, 2010). However, in some chronic infections, S. aureus displays a loss of agr function, e.g., persistent bacteremia (Fowler et al., 2004) and infections of the cystic fibrosis lung (Goerke et al., 2000). Another type of chronic infection is the DFI, where S. aureus is a prominent member and positively correlates with the presence of Corynebacterium spp. (Gardner et al., 2013). We observed that S. aureus agr QS is mitigated in response to multiple Corynebacterium spp. across several agr classes (Figures 1 and 6) and that this results in a lack of hemolytic activity, which represents a lack of production of secreted virulence factors (Figure 4) and decreased success during in vivo coinfection in a mouse subcutaneous abscess model (Figure 5). Thus, in response to phylogenetically diverse Corynebacterium spp., S. aureus shifts toward a commensal (i.e., less virulent) state reminiscent of agr-defective mutants. We speculate that this contributes to a shift from acute to chronic S. aureus infection in polymicrobial settings and, along with an increase in the abundance of C. striatum in coinfection (Figure 5), may partly explain the positive correlation between S. aureus and Corynebacterium spp. in chronic DFIs.
Although the mechanism remains to be determined, S. aureus’ response to C. striatum is reminiscent of how Lactobacillus reuteri-produced cyclic dipeptides inhibit S. aureus agr QS and diminish TSS-1 production (Li et al., 2011). A number of distinct mechanisms could result in a similar diminution of agr QS and we are actively pursuing the identity and mechanism of the activity in C. striatum CFCM that triggers this response.
Overall, our results point to the potential to develop antivirulence therapies against S. aureus from Corynebacterium-produced products and also suggest a potential reason for the high frequency of commensal behavior by S. aureus during human nasal colonization. Research on nostril microbiota composition and observed correlations, both positive and negative, between the presence/relative abundance of S. aureus and commensal Corynebacterium spp., e.g., (Uehara et al., 2000; Wos-Oxley et al., 2010; Yan et al., 2013; Kaspar et al., 2016), have sparked renewed interest in the potential use of commensal Corynebacterium spp. as probiotics to eradicate S. aureus nostril colonization, and there is precedent for this in a small cohort of adults (Uehara et al., 2000). Our findings suggest the possibility of an alternative or additional role of probiotic Corynebacterium spp. in limiting S. aureus virulence, e.g., in persistent carriers. In addition, our results provide an additional impetus for the development of an animal model of Corynebacterium spp. nasal colonization. Future efforts to fully characterize and manage Corynebacterium–S. aureus interactions have the potential to either maintain healthy microbiota composition or attenuate local S. aureus infections and may lead to new minimally invasive therapeutic adjuncts and/or alternatives to antibiotic treatment.
Author Contributions
Conceptualization, MR and KL; Methodology, MR, KL, KR, and MF; Investigation, MR, MF, and RG; Writing – Original Draft, MR and KL; Writing – Review and Editing, MR., KL, KR; Funding Acquisition, KL and KR; Supervision, KL and KR. All authors agree to be accountable for the content of the work.
Funding
This work was supported by the National Institutes of Health NIAID grants F32 AI102498 (MR), R00 DE023584 (MF), R15 AI105763 (KR) and R01 AI101018 (KL). The funders had no role in study design, data collection and interpretation or the decision to submit the work for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Lucy Foulston and Alex Horswill for numerous S. aureus strains and suggestions, and Susan R. Rittling for help with the attachment assay. We thank Michael R. Wessels, Silvio D. Brugger, Kathryn Ramsey, Gleb Pishchany, Megan A. Lambert, Jennifer Spagnolo, Isabel F. Escapa and Lindsey Bomar for valuable advice and manuscript edits, as well as others members of the Lemon Lab for suggestions.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01230
References
Abe, S., Takayama, K.-I., and Kinoshita, S. (1967). Taxonomical studies on glutamic acid-producing bacteria. J. Gen. Appl. Microbiol. 13, 279–301. doi: 10.2323/jgam.13.279
Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J., Smith, J. A., et al. (2002). Short Protocols in Molecular Biology: A Compendium of Methods from Current Protocols in Molecular Biology. New York, NY: Wiley.
Blevins, J. S., Beenken, K. E., Elasri, M. O., Hurlburt, B. K., and Smeltzer, M. S. (2002). Strain-dependent differences in the regulatory roles of sarA and agr in Staphylococcus aureus. Infect. Immun. 70, 470–480. doi: 10.1128/IAI.70.2.470-480.2002
Brown, S. A., and Whiteley, M. (2007). A novel exclusion mechanism for carbon resource partitioning in Aggregatibacter actinomycetemcomitans. J. Bacteriol. 189, 6407–6414. doi: 10.1128/JB.00554-07
Burian, M., Rautenberg, M., Kohler, T., Fritz, M., Krismer, B., Unger, C., et al. (2010a). Temporal expression of adhesion factors and activity of global regulators during establishment of Staphylococcus aureus nasal colonization. J. Infect. Dis. 201, 1414–1421. doi: 10.1086/651619
Burian, M., Wolz, C., and Goerke, C. (2010b). Regulatory adaptation of Staphylococcus aureus during nasal colonization of humans. PLoS ONE 5:e10040. doi: 10.1371/journal.pone.0010040
Cassat, J., Dunman, P. M., Murphy, E., Projan, S. J., Beenken, K. E., Palm, K. J., et al. (2006). Transcriptional profiling of a Staphylococcus aureus clinical isolate and its isogenic agr and sarA mutants reveals global differences in comparison to the laboratory strain RN6390. Microbiology 152, 3075–3090. doi: 10.1099/mic.0.29033-0
CDC (2005–2013). Active Bacterial Core Surveillance Report, Emerging Infectious Program Network, Methicillin-Resistant Staphylococcus aureus. Atlanta, GA: Centers for Disease Control and Prevention.
Citron, D. M., Goldstein, E. J., Merriam, C. V., Lipsky, B. A., and Abramson, M. A. (2007). Bacteriology of moderate-to-severe diabetic foot infections and in vitro activity of antimicrobial agents. J. Clin. Microbiol. 45, 2819–2828. doi: 10.1128/JCM.00551-07
Cole, A. L., Muthukrishnan, G., Chong, C., Beavis, A., Eade, C. R., Wood, M. P., et al. (2016). Host innate inflammatory factors and staphylococcal protein A influence the duration of human Staphylococcus aureus nasal carriage. Mucosal Immunol. doi: 10.1038/mi.2016.2 [Epub ahead of print].
Conlon, B. P., Nakayasu, E. S., Fleck, L. E., Lafleur, M. D., Isabella, V. M., Coleman, K., et al. (2013). Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature 503, 365–370. doi: 10.1038/nature12790
Diep, B. A., Gill, S. R., Chang, R. F., Phan, T. H., Chen, J. H., Davidson, M. G., et al. (2006). Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367, 731–739. doi: 10.1016/S0140-6736(06)68231-7
Dunman, P. M., Murphy, E., Haney, S., Palacios, D., Tucker-Kellogg, G., Wu, S., et al. (2001). Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 183, 7341–7353. doi: 10.1128/JB.183.24.7341-7353.2001
Fey, P. D., Endres, J. L., Yajjala, V. K., Widhelm, T. J., Boissy, R. J., Bose, J. L., et al. (2013). A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. MBio 4:e00537-00512. doi: 10.1128/mBio.00537-12
Forsgren, A., and Sjoquist, J. (1966). “Protein A” from S. aureus. I. Pseudo-immune reaction with human gamma-globulin. J. Immunol. 97, 822–827.
Forsyth, R. A., Haselbeck, R. J., Ohlsen, K. L., Yamamoto, R. T., Xu, H., Trawick, J. D., et al. (2002). A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol. Microbiol. 43, 1387–1400. doi: 10.1046/j.1365-2958.2002.02832.x
Foster, T. J., Geoghegan, J. A., Ganesh, V. K., and Hook, M. (2014). Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 12, 49–62. doi: 10.1038/nrmicro3161
Foulston, L., Elsholz, A. K., Defrancesco, A. S., and Losick, R. (2014). The extracellular matrix of Staphylococcus aureus biofilms comprises cytoplasmic proteins that associate with the cell surface in response to decreasing pH. MBio 5:e01667-14. doi: 10.1128/mBio.01667-14
Fowler, V. G. Jr., Sakoulas, G., Mcintyre, L. M., Meka, V. G., Arbeit, R. D., Cabell, C. H., et al. (2004). Persistent bacteremia due to methicillin-resistant Staphylococcus aureus infection is associated with agr dysfunction and low-level in vitro resistance to thrombin-induced platelet microbicidal protein. J. Infect. Dis. 190, 1140–1149. doi: 10.1086/423145
Frank, D. N., Feazel, L. M., Bessesen, M. T., Price, C. S., Janoff, E. N., and Pace, N. R. (2010). The human nasal microbiota and Staphylococcus aureus carriage. PLoS ONE 5:e10598. doi: 10.1371/journal.pone.0010598
Gardner, S. E., Hillis, S. L., Heilmann, K., Segre, J. A., and Grice, E. A. (2013). The neuropathic diabetic foot ulcer microbiome is associated with clinical factors. Diabetes 62, 923–930. doi: 10.2337/db12-0771
Goerke, C., Campana, S., Bayer, M. G., Doring, G., Botzenhart, K., and Wolz, C. (2000). Direct quantitative transcript analysis of the agr regulon of Staphylococcus aureus during human infection in comparison to the expression profile in vitro. Infect. Immun. 68, 1304–1311. doi: 10.1128/IAI.68.3.1304-1311.2000
Gorwitz, R. J., Kruszon-Moran, D., Mcallister, S. K., Mcquillan, G., Mcdougal, L. K., Fosheim, G. E., et al. (2008). Changes in the prevalence of nasal colonization with Staphylococcus aureus in the United States, 2001-2004. J. Infect. Dis. 197, 1226–1234. doi: 10.1086/533494
Gustafsson, E., Karlsson, S., Oscarsson, J., Sogard, P., Nilsson, P., and Arvidson, S. (2009). Mathematical modelling of the regulation of spa (protein A) transcription in Staphylococcus aureus. Int. J. Med. Microbiol. 299, 65–74. doi: 10.1016/j.ijmm.2008.05.011
Jansen, K. U., Girgenti, D. Q., Scully, I. L., and Anderson, A. S. (2013). Vaccine review: “Staphyloccocus aureus vaccines: problems and prospects”. Vaccine 31, 2723–2730. doi: 10.1016/j.vaccine.2013.04.002
Jensen, R. O., Winzer, K., Clarke, S. R., Chan, W. C., and Williams, P. (2008). Differential recognition of Staphylococcus aureus quorum-sensing signals depends on both extracellular loops 1 and 2 of the transmembrane sensor AgrC. J. Mol. Biol. 381, 300–309. doi: 10.1016/j.jmb.2008.06.018
Jett, B. D., Hatter, K. L., Huycke, M. M., and Gilmore, M. S. (1997). Simplified agar plate method for quantifying viable bacteria. Biotechniques 23, 648–650.
Jorth, P., Turner, K. H., Gumus, P., Nizam, N., Buduneli, N., and Whiteley, M. (2014). Metatranscriptomics of the human oral microbiome during health and disease. MBio 5:e01012-01014. doi: 10.1128/mBio.01012-14
Kaspar, U., Kriegeskorte, A., Schubert, T., Peters, G., Rudack, C., Pieper, D. H., et al. (2016). The culturome of the human nose habitats reveals individual bacterial fingerprint patterns. Environ Microbiol. 18, 2130–2142. doi: 10.1111/1462-2920.12891
Kato, F., and Sugai, M. (2011). A simple method of markerless gene deletion in Staphylococcus aureus. J. Microbiol. Methods 87, 76–81. doi: 10.1016/j.mimet.2011.07.010
Kavanaugh, J. S., Thoendel, M., and Horswill, A. R. (2007). A role for type I signal peptidase in Staphylococcus aureus quorum sensing. Mol. Microbiol. 65, 780–798. doi: 10.1111/j.1365-2958.2007.05830.x
Kiser, K. B., Cantey-Kiser, J. M., and Lee, J. C. (1999). Development and characterization of a Staphylococcus aureus nasal colonization model in mice. Infect. Immun. 67, 5001–5006.
Krismer, B., Liebeke, M., Janek, D., Nega, M., Rautenberg, M., Hornig, G., et al. (2014). Nutrient limitation governs Staphylococcus aureus metabolism and niche adaptation in the human nose. PLoS Pathog. 10:e1003862. doi: 10.1371/journal.ppat.1003862
Kuehnert, M. J., Kruszon-Moran, D., Hill, H. A., Mcquillan, G., Mcallister, S. K., Fosheim, G., et al. (2006). Prevalence of Staphylococcus aureus nasal colonization in the United States, 2001-2002. J. Infect. Dis. 193, 172–179. doi: 10.1086/499632
Li, J., Wang, W., Xu, S. X., Magarvey, N. A., and Mccormick, J. K. (2011). Lactobacillus reuteri-produced cyclic dipeptides quench agr-mediated expression of toxic shock syndrome toxin-1 in staphylococci. Proc. Natl. Acad. Sci. U.S.A. 108, 3360–3365. doi: 10.1073/pnas.1017431108
Lina, G., Boutite, F., Tristan, A., Bes, M., Etienne, J., and Vandenesch, F. (2003). Bacterial competition for human nasal cavity colonization: role of Staphylococcal agr alleles. Appl. Environ. Microbiol. 69, 18–23. doi: 10.1128/AEM.69.1.18-23.2003
Lowy, F. D. (1998). Staphylococcus aureus Infections. N. Engl. J. Med. 339, 520–532. doi: 10.1056/NEJM199808203390806
Mastropaolo, M. D., Evans, N. P., Byrnes, M. K., Stevens, A. M., Robertson, J. L., and Melville, S. B. (2005). Synergy in polymicrobial infections in a mouse model of type 2 diabetes. Infect. Immun. 73, 6055–6063. doi: 10.1128/IAI.73.9.6055-6063.2005
McClure, R., Balasubramanian, D., Sun, Y., Bobrovskyy, M., Sumby, P., Genco, C. A., et al. (2013). Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res. 41:e140. doi: 10.1093/nar/gkt444
Murray, E. J., Crowley, R. C., Truman, A., Clarke, S. R., Cottam, J. A., Jadhav, G. P., et al. (2014). Targeting Staphylococcus aureus quorum sensing with nonpeptidic small molecule inhibitors. J. Med. Chem. 57, 2813–2819. doi: 10.1021/jm500215s
Nielsen, A., Mansson, M., Bojer, M. S., Gram, L., Larsen, T. O., Novick, R. P., et al. (2014). Solonamide B inhibits quorum sensing and reduces Staphylococcus aureus mediated killing of human neutrophils. PLoS ONE 9:e84992. doi: 10.1371/journal.pone.0084992
Novick, R. P., and Geisinger, E. (2008). Quorum sensing in staphylococci. Annu. Rev. Genet. 42, 541–564. doi: 10.1146/annurev.genet.42.110807.091640
Oh, J., Conlan, S., Polley, E. C., Segre, J. A., and Kong, H. H. (2012). Shifts in human skin and nares microbiota of healthy children and adults. Genome Med. 4:77. doi: 10.1186/gm378
Otto, M. (2010). Basis of virulence in community-associated methicillin-resistant Staphylococcus aureus. Annu. Rev. Microbiol. 64, 143–162. doi: 10.1146/annurev.micro.112408.134309
Pang, Y. Y., Schwartz, J., Thoendel, M., Ackermann, L. W., Horswill, A. R., and Nauseef, W. M. (2010). agr-Dependent interactions of Staphylococcus aureus USA300 with human polymorphonuclear neutrophils. J Innate Immun. 2, 546–559. doi: 10.1159/000319855
Peterson, P. K., Verhoef, J., Sabath, L. D., and Quie, P. G. (1977). Effect of protein A on staphylococcal opsonization. Infect. Immun. 15, 760–764.
Proctor, R. A. (2012). Challenges for a universal Staphylococcus aureus vaccine. Clin. Infect. Dis. 54, 1179–1186. doi: 10.1093/cid/cis033
Queck, S. Y., Jameson-Lee, M., Villaruz, A. E., Bach, T. H., Khan, B. A., Sturdevant, D. E., et al. (2008). RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol. Cell 32, 150–158. doi: 10.1016/j.molcel.2008.08.005
Ramsey, M. M., Rumbaugh, K. P., and Whiteley, M. (2011). Metabolite cross-feeding enhances virulence in a model polymicrobial infection. PLoS Pathog. 7:e1002012. doi: 10.1371/journal.ppat.1002012
Ramsey, M. M., and Whiteley, M. (2009). Polymicrobial interactions stimulate resistance to host innate immunity through metabolite perception. Proc. Natl. Acad. Sci. U.S.A. 106, 1578–1583. doi: 10.1073/pnas.0809533106
Regassa, L. B., Novick, R. P., and Betley, M. J. (1992). Glucose and nonmaintained pH decrease expression of the accessory gene regulator (agr) in Staphylococcus aureus. Infect. Immun. 60, 3381–3388.
Schmidt, K. A., Manna, A. C., and Cheung, A. L. (2003). SarT influences sarS expression in Staphylococcus aureus. Infect. Immun. 71, 5139–5148. doi: 10.1128/IAI.71.9.5139-5148.2003
Spika, J. S., Verbrugh, H. A., and Verhoef, J. (1981). Protein A effect on alternative pathway complement activation and opsonization of Staphylococcus aureus. Infect. Immun. 34, 455–460.
Sully, E. K., Malachowa, N., Elmore, B. O., Alexander, S. M., Femling, J. K., Gray, B. M., et al. (2014). Selective chemical inhibition of agr quorum sensing in Staphylococcus aureus promotes host defense with minimal impact on resistance. PLoS Pathog. 10:e1004174. doi: 10.1371/journal.ppat.1004174
Thoendel, M., and Horswill, A. R. (2009). Identification of Staphylococcus aureus AgrD residues required for autoinducing peptide biosynthesis. J. Biol. Chem. 284, 21828–21838. doi: 10.1074/jbc.M109.031757
Thoendel, M., Kavanaugh, J. S., Flack, C. E., and Horswill, A. R. (2011). Peptide signaling in the staphylococci. Chem. Rev. 111, 117–151. doi: 10.1021/cr100370n
Uehara, Y., Nakama, H., Agematsu, K., Uchida, M., Kawakami, Y., Abdul Fattah, A. S., et al. (2000). Bacterial interference among nasal inhabitants: eradication of Staphylococcus aureus from nasal cavities by artificial implantation of Corynebacterium sp. J. Hosp. Infect. 44, 127–133. doi: 10.1053/jhin.1999.0680
Wang, Y., Dai, A., Huang, S., Kuo, S., Shu, M., Tapia, C. P., et al. (2014). Propionic acid and its esterified derivative suppress the growth of methicillin-resistant Staphylococcus aureus USA300. Benef. Microbes 5, 161–168. doi: 10.3920/BM2013.0031
Weidenmaier, C., Kokai-Kun, J. F., Kristian, S. A., Chanturiya, T., Kalbacher, H., Gross, M., et al. (2004). Role of teichoic acids in Staphylococcus aureus nasal colonization, a major risk factor in nosocomial infections. Nat. Med. 10, 243–245. doi: 10.1038/nm991
Wertheim, H. F., Melles, D. C., Vos, M. C., Van Leeuwen, W., Van Belkum, A., Verbrugh, H. A., et al. (2005). The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis. 5, 751–762. doi: 10.1016/S1473-3099(05)70295-4
Wertheim, H. F., Vos, M. C., Ott, A., Van Belkum, A., Voss, A., Kluytmans, J. A., et al. (2004). Risk and outcome of nosocomial Staphylococcus aureus bacteraemia in nasal carriers versus non-carriers. Lancet 364, 703–705. doi: 10.1016/S0140-6736(04)16897-9
Wertheim, H. F., Walsh, E., Choudhurry, R., Melles, D. C., Boelens, H. A., Miajlovic, H., et al. (2008). Key role for clumping factor B in Staphylococcus aureus nasal colonization of humans. PLoS Med. 5:e17. doi: 10.1371/journal.pmed.0050017
Wollenberg, M. S., Claesen, J., Escapa, I. F., Aldridge, K. L., Fischbach, M. A., and Lemon, K. P. (2014). Propionibacterium-produced coproporphyrin III induces Staphylococcus aureus aggregation and biofilm formation. MBio 5:e01286-01214. doi: 10.1128/mBio.01286-14
Wos-Oxley, M. L., Plumeier, I., Von Eiff, C., Taudien, S., Platzer, M., Vilchez-Vargas, R., et al. (2010). A poke into the diversity and associations within human anterior nare microbial communities. ISME J. 4, 839–851. doi: 10.1038/ismej.2010.15
Yan, M., Pamp, S. J., Fukuyama, J., Hwang, P. H., Cho, D. Y., Holmes, S., et al. (2013). Nasal microenvironments and interspecific interactions influence nasal microbiota complexity and S. aureus carriage. Cell Host Microbe 14, 631–640. doi: 10.1016/j.chom.2013.11.005
Yarwood, J. M., Bartels, D. J., Volper, E. M., and Greenberg, E. P. (2004). Quorum sensing in Staphylococcus aureus biofilms. J. Bacteriol. 186, 1838–1850. doi: 10.1128/JB.186.6.1838-1850.2004
Keywords: quorum sensing (QS), Corynebacterium, Staphylococcus aureus, agr system, microbiome, commensal bacteria
Citation: Ramsey MM, Freire MO, Gabrilska RA, Rumbaugh KP and Lemon KP (2016) Staphylococcus aureus Shifts toward Commensalism in Response to Corynebacterium Species. Front. Microbiol. 7:1230. doi: 10.3389/fmicb.2016.01230
Received: 01 June 2016; Accepted: 25 July 2016;
Published: 17 August 2016.
Edited by:
Joerg Graf, University of Connecticut, USAReviewed by:
Nicola Segata, University of Trento, ItalyIlana Kolodkin-Gal, Weizmann Institute of Science, Israel
Copyright © 2016 Ramsey, Freire, Gabrilska, Rumbaugh and Lemon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Katherine P. Lemon, a2xlbW9uQGZvcnN5dGgub3Jn
 Matthew M. Ramsey
Matthew M. Ramsey Marcelo O. Freire
Marcelo O. Freire Rebecca A. Gabrilska
Rebecca A. Gabrilska Kendra P. Rumbaugh
Kendra P. Rumbaugh Katherine P. Lemon
Katherine P. Lemon