
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol., 03 August 2016
Sec. Microbial Symbioses
Volume 7 - 2016 | https://doi.org/10.3389/fmicb.2016.01193
This article is part of the Research TopicBifidobacteria and their role in the human gut microbiotaView all 23 articles
Bifidobacteria are commensal microoganisms found in the gastrointestinal tract. Several strains have been attributed beneficial traits at local and systemic levels, through pathogen exclusion or immune modulation, among other benefits. This has promoted a growing industrial and scientific interest in bifidobacteria as probiotic supplements. However, the molecular mechanisms mediating this cross-talk with the human host remain unknown. High-throughput technologies, from functional genomics to transcriptomics, proteomics, and interactomics coupled to the development of both in vitro and in vivo models to study the dynamics of the intestinal microbiota and their effects on host cells, have eased the identification of key molecules in these interactions. Numerous secreted or surface-associated proteins or peptides have been identified as potential mediators of bifidobacteria-host interactions and molecular cross-talk, directly participating in sensing environmental factors, promoting intestinal colonization, or mediating a dialogue with mucosa-associated immune cells. On the other hand, bifidobacteria induce the production of proteins in the intestine, by epithelial or immune cells, and other gut bacteria, which are key elements in orchestrating interactions among bifidobacteria, gut microbiota, and host cells. This review aims to give a comprehensive overview on proteinaceous molecules described and characterized to date, as mediators of the dynamic interplay between bifidobacteria and the human host, providing a framework to identify knowledge gaps and future research needs.
The human gut is inhabited by a trillion of microorganisms which constitute the gut microbiota. These microorganisms are in close contact with the intestinal mucosa, which represents the largest extension of the human body exposed to external stimuli. A complex molecular interplay is established among microbiota, dietary components and host cells, which regulates immune and metabolic functions in the host (Furusawa et al., 2015). Dysbiosis, defined as changes in the gut microbiota structure associated to healthy individuals, disrupts the microbiome-host cross-talk homeostasis and correlates with metabolic and inflammatory disorders (Evans et al., 2013; Patel et al., 2013; Levy et al., 2015).
Pro- and pre-biotics can improve host health through microbiota modulation and immune system boosting (Picard et al., 2005). Some strains of bifidobacteria, which are among the first colonizers of the human intestine and one of the dominant groups in the breast-fed infant microbiota (Garrido et al., 2012), have been attributed several health benefits, encouraging interest in their use as probiotics. Pathogen inhibition and diarrhea amelioration are their best established outcomes and have been related to the production of organic acids (Fukuda et al., 2011), antibacterial peptides (Moroni et al., 2006), quorum-sensing inhibitors (Cotar et al., 2010), pathogen displacement (Ruas-Madiedo et al., 2006), and virulence attenuation (Tanner et al., 2016). Bifidobacteria also regulate host functions and ferment complex polysaccharides from our diet (Ménard et al., 2005; Heuvelin et al., 2009, 2010; Bermudez-Brito et al., 2013; Furusawa et al., 2015), although there is still limited knowledge on the molecular mechanisms triggering these effects.
Delineating the specific molecules mediating Bifidobacterium cross-talk with the host, will help to understand their beneficial effects and establish microbiome-targeted therapies for human diseases. This review gives an overview on molecules behind the bifidobacterial-host cross-talk, providing a framework to design safe and efficacious probiotic-derived supplements (Licciardi et al., 2010; Shenderov, 2013).
Bacterial adhesion to the intestinal surface is mediated by non-specific, hydrophobic or electrostatic interactions, and specific mechanisms involving macromolecular interactions between bacterial and host receptors. Functionally characterized Bifidobacterium adhesins, surface-associated proteins that facilitate bifidobacteria attachment to intestinal cells and/or the extracellular matrixes surrounding them, are reviewed below and summarized in Figure 1.
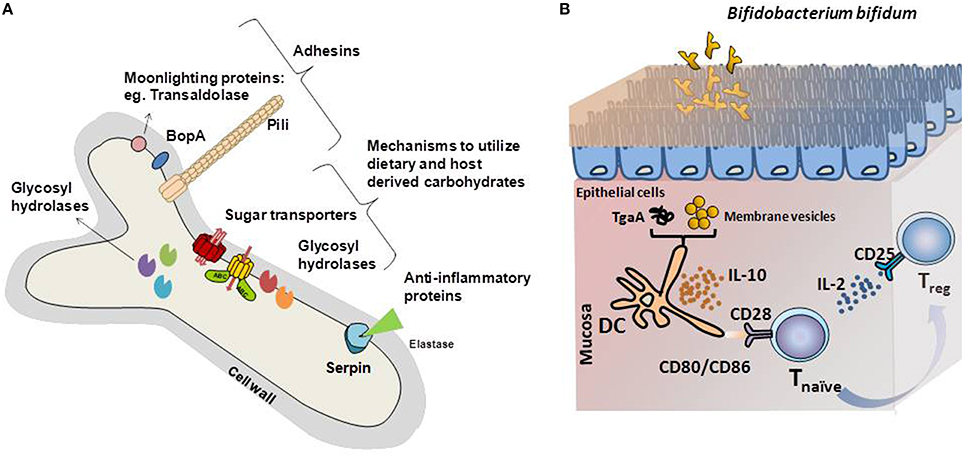
Figure 1. (A) Schematic representation of the bifidobacterial proteins identified as key mediators of the cross-talk mechanisms with the intestinal environment. Adhesin-like factors, proteins with immunomodulatory capabilities and glycosyl hydrolases specific for carbon sources encountered in the gastrointestinal tract are represented. (B) Graphical illustration of the immunomodulatory mechanisms driven by model Bifidobacterium bifidum strains. Different B. bifidum fractions and molecules induces Treg response, key in maintaining the balance of effector T-cell responses. Membrane vesicles or the extracellular protein TgaA affects dendritic cells, which induces Treg differentiation after interaction with naïve T-cells. In this process, increased IL-10 secretion, recognition of CD80, and CD86 by CD28 in naïve T-cells and release of IL-2 are key for Treg response development.
Involvement of pili in bifidobacterial adhesion has been studied in Bifidobacterium bifidum and Bifidobacterium breve (O'Connell Motherway et al., 2011b; Turroni et al., 2013). The B. bifidum PRL2010 genome harbors three pilus clusters. Of these, pil2 and pil3, encode putative sortase-dependent pili that are expressed under both in vitro and in vivo conditions. Heterologous expression of pil3 in Lactococcus lactis significantly increased this bacterium adhesion to the human epithelial cell line Caco-2 (Turroni et al., 2013). The type IVb tight adherence (Tad) pilus-encoding gene cluster from B. breve UCC2003, was found to be essential for the colonization of, and persistence in, the murine gut. Tad inactivation impaired the strain ability to stably colonize the murine intestine, as reflected by reduced shedding level and bifidobacterial numbers in the gut (O'Connell Motherway et al., 2011b). Another surface-anchored protein potentially involved in intestinal adhesion of bifidobacteria is BopA. This purified lipoprotein competes with B. bifidum MIMBb75 adhesion to Caco-2 cells, and probably facilitates B. bifidum adhesion (Guglielmetti et al., 2008). Indeed, Bifidobacterium strains overexpressing bopA adhere better (Gleinser et al., 2012). However, B. bifidum treatment with anti-BopA antibodies does not reduce the attachment to intestinal cells (Kainulainen et al., 2013), thus the specific adhesion mechanism mediated through BopA must be further elucidated.
Remarkably, bifidobacteria can use some moonlighting proteins, those with multiple functions (Jeffery, 2003), as adhesin-like factors. Surface-exposed glycolytic enzymes, including transaldolase from B. bifidum and enolase from Bifidobacterium animalis, are adhere to mucin and plasminogen, respectively (Candela et al., 2009; González-Rodríguez et al., 2012). Other surface-exposed moonlighting proteins, including the chaperone DnaK from B. animalis and the elongation factor Tu from Bifidobacterium longum, showed high affinity for human plasminogen in vitro and have been proposed as mediators of intestinal attachment (Candela et al., 2010; Wei et al., 2014).
It is worth highlighting that most of the adhesins herein described have been identified on in vitro assays and their relevance for intestinal colonization has not been tested in vivo. Since laboratory models do not accurately mimic all the factors that can affect bacterial attachment to the intestinal mucosa (e.g., due to the absence of a mucus layer and resident microbiota), functional confirmation of the adhesion capacity in vivo is still required (Ouwehand and Salminen, 2003).
A single layer of epithelial cells provides a selective barrier separating intestinal lumen from subjacent tissues. Tight-junctions (TJ) are multi-protein complexes that control molecule translocation across this barrier (Lee, 2015), and their disruption leads to uncontrolled trafficking of noxious molecules triggering inflammation (Bergmann et al., 2013).
Specific Bifidobacterium strains promote TJ enhancing epithelial barrier integrity (Ohland and Macnaughton, 2010; Mokkala et al., 2016). In animal models, B. bifidum and B. longum strains preserved TJ localization, attenuating intestinal permeability, and decreasing necrotizing enterocolitis incidence (Khailova et al., 2009; Bergmann et al., 2013; Srutkova et al., 2015). Preliminary work suggested that B. bifidum metabolites, like acetate, induced TJ expression in intestinal cells (Hsieh et al., 2015). Soluble factors present in B. longum lysates or secreted by B. infantis or B. breve strains, also mediate epithelial barrier maintenance (Ménard et al., 2005; Ewaschuk et al., 2008; Sultana et al., 2013). Further research to identify the specific molecules mediating this TJ promotion is needed.
The spatial distribution of bacteria throughout the gastrointestinal tract, is partly controlled by nutrients availability for resident microbiota (Donaldson et al., 2016). Indeed, our wellbeing relates with the nutrient harvesting capability of our gut microbes. These microorganisms, overall, can utilize dietary and host carbohydrates, and glycans produced by other gut bacteria. Indeed, 89 “carbohydrate active enzymes” (CAZyme) have recently been identified in 85% of the microbiomes obtained from 488 individuals (Bhattacharya et al., 2015), suggesting that gut bacteria are highly specialized in using available glycans as their main sustenance. Bifidobacterial genomes are abundant in saccharolytic features whose expression is tightly regulated by available carbohydrates (Khoroshkin et al., 2016), supporting that host glycans were a potent evolutionary force driving their successful gut colonization (Sánchez et al., 2013; Milani et al., 2016).
Numerous studies demonstrated bifidobacteria's capability to use dietary non-digestible oligosaccharides, which is on the basis of the prebiotic concept (Rastall and Gibson, 2015). Glycosyl hydrolases (GH, or glycosidades), many of which are extracellular, have high specificity for the oligosaccharides constituents and cleave the glycosidic bonds (Table 1). Special attention has been paid to the GH acting on human milk oligosaccharides (HMO) since these serve as substrates for bifidobacteria, which are the initial gut colonizers in breast-fed infants. HMO are structurally diverse and composed of several monosaccharides (glucose, galactose, N-acetylglucosamine, fucose, or sialic acid). They mainly consist of a lactose core linked to units (n = 0–15) of lacto-N-biose (type I) or to N-acetyl-lactosamine (type II; Smilowitz et al., 2014). Bifidobacteria secrete GH that cleave specific linkages within the HMO molecules and the best characterized are those synthesized by B. bifidum which, together with B. longum subsp. infantis, are two abundant species in breast-fed neonates (Table 1). These species employ different strategies for HMO utilization. Whereas B. bifidum has an array of membrane-associated GH, B. longum subsp. infantis is specialized in the import and intracellular breakdown of HMO (Garrido et al., 2013; Jae-Han et al., 2013). Moreover B. longum strains have similar HMO-utilization patterns, whilst B. bifidum strains are more diverse with some unable to use fucosylated or sialylated HMO (Garrido et al., 2015). Similarly, the B. breve HMO utilization profile is strain dependent and, contrary to B. bifidum, some strains consume fucosylated or sialilated HMOs. B. breve's capability to use these HMOs explains its abundance in breast-fed babies (Ruiz-Moyano et al., 2013).

Table 1. Gycosyl hydrolases and sugar transporters characterized and/or described in Bifidobacterium genus.
Some bifidobacteria can also utilize mucins from the mucus layer coating the intestine. Mucin composition and structure resemble that of HMO; consisting of a core of different O-glycans, built on α- and β-linked N-acetyl-galactosamine, galactose, and N-acetyl-glucosamine residues, which can incorporate fucose and sialic acid residues (Tailford et al., 2015). B. longum and B. breve strains' capability to effectively use mucin carbohydrates, has been confirmed in vitro (Ruas-Madiedo et al., 2008). However, GH able to degrade mucins have only been described in B. bifidum (Table 1). Indeed, in a comparative genomic study 60% of the GH-encoding genes from B. bifidum were predicted to breakdown mucin-like glycans and most of them were exclusively present in this bifidobacterial species (Turroni et al., 2014). Remarkably, other species could use the mono- and oligosaccharides released by B. bifidum GH thus evidencing the existence of cross-feeding mechanisms, as it has been demonstrated in B. breve and B. bifidum co-cultures (Egan et al., 2014).
Bifidobacterium capacity to metabolize specific dietary and host-derived carbohydrates is also dependent on the presence of specific sugar transport systems. These are crucial for their competitive establishment in the gut, thus representing one of the molecular mechanisms by which bifidobacteria interact with the host. Import sugar mechanisms in bifidobacteria are herein described (Bottacini et al., 2014).
First, ATP-binding cassette (ABC) systems are active transporters which couple ATP hydrolysis to translocation uptake across the cell membrane. They are the most frequent sugar transporters in bifidobacteria and have been described for mono- and oligosaccharides in different species (Nishimoto and Kitaoka, 2007; Wada et al., 2008; Wei et al., 2012), although only a few of them have been functionally characterized at protein level (Suzuki et al., 2008; Ejby et al., 2013).
Secondly, some secondary transporters, predicted to consist of single integral membrane-associated proteins, have been characterized at protein level in bifidobacteria. These include permease systems for the uptake of lactose, glucose, and sucrose (Parche et al., 2006). Secondary transporters encoding genes have been identified in different bifidobacterial species, although most of them have not been characterized at protein level (Turroni et al., 2012).
Proton symporters of the glycoside-pentoside-hexuronide (GPH) cation symporter family for melibiose and pentosides were also described in Bifidobacterium (Lee and O'sullivan, 2010; Turroni et al., 2012), although they remain to be characterized.
Finally, phosphoenolpyruvate-phosphotransferase (PEP-PTS) systems were first characterized in the 90's in B. breve and B. bifidum at protein level (Lee and O'sullivan, 2010). Later, genome sequence availability revealed their wide spread distribution in bifidobacteria. In particular, B. breve UCC2003 genome contains four PEP-PTS systems, one of which has been characterized as a fructose-specific transporter (Mazé et al., 2007). Also, in silico analysis revealed a putative glucose-specific PEP-PTS uptake system in B. longum (Lorca et al., 2007). However, genome analysis of different B. longum strains showed that glucose-specific PTS transporters are minor in comparison with ABC transporters (Pokusaeva et al., 2011), thus glucose may be transported preferentially by secondary permeases (Parche et al., 2006).
Comparative genomic analysis revealed that sugar PEP-PTS systems are present in all bifidobacterial genomes, except for B. animalis subsp. lactis (Lee and O'sullivan, 2010) which is hypothesized to have lost most of their carbohydrate transporters due to extended cultivation under industrial conditions. In fact, the capability to utilize variable carbon sources is considered an adaptation to the gut environment. For instance, the dominant Bifidobacterium species in infant fecal samples (B. longum and B. bifidum) is consistent with their inherent ability to use host-derived oligosaccharides such as mucin and HMO (Bottacini et al., 2014), and their possession of a wide range of host-derived carbohydrate transporters, such as those involved in N-biose import (Suzuki et al., 2008).
In the absence of disease, the ensemble of molecular interactions taking place in the human gut results in the intestinal homeostasis. Specialized epithelial cells denominated M-cells and antigen presenting cells (APCs) from the gut-associated lymphoid tissue (GALT) continuously sample the intestinal content. Interaction of APCs with the rest of GALT effectors, mainly T and B cells, leads to immunotolerance against commensal microbes and dietary components, whilst the capacity of mounting an acute, quick, and powerful response against enteropathogens is developed.
Differentiation of commensal and pathogenic bacteria is based on the presence of pattern recognition receptors (PRR) on the APC and epithelial cell surfaces. Among them, Toll-like receptors, NOD-like receptors, C-type lectin receptors, and RIG-I-like receptors are in charge of recognizing specific microbial-associated molecular patterns (MAMPs), such as flagellin, teichoic acids, or lipopolysaccharide among others. The type and intensity of the downstream and intracellular signaling cascades deployed after MAMPs-PRR interaction is essential for the APCs interaction with T-cells, which will finally determine the nature of the T-cell response. Roughly, T-cell responses are divided into effector (Th) and regulatory (Treg), its balance being key in the intestinal homeostasis maintenance (Maloy and Powrie, 2011). It is generally accepted that commensal microbiota, by inducing Treg response, modulates the Th1/Th2 balance favoring immune tolerance against the gut microbiota (Ventura et al., 2012). Indeed, the classical MAMP triggering Treg response is the exopolysaccharide A of the commensal bacterium Bacteroides fragilis, molecule also involved in the GALT maturation (Mazmanian et al., 2008).
Bifidobacteria may drive species-specific T-cell responses, as it was revealed by a series of experiments in which the cytokine secretion profiles of monocyte-derived dendritic cells (MoDCs) and full fractions of peripheral blood mononuclear cells (PBMCs) were determined (López et al., 2010). Relative levels of key cytokines (IL-10, IL-17, TNFα among others) suggested a specific immunomodulation mechanism for each species, as reported recently for probiotics (Hill et al., 2014). Challenging immature MoDCs with different strains, followed by co-culture with allogeneic naïve CD4+ cells and cytokine determination, further confirmed this effect (López et al., 2011).
Remarkably, B. bifidum LMG13195 appeared to induce a Treg response in vitro (Figure 1; López et al., 2011). Dendritic cells challenged with membrane vesicles from this strain induced naïve CD4+ cells polarization into Treg, as deduced from the increases in the expression of foxP3 regulation factor and the CD25 marker (López et al., 2012). Most likely, surface-associated proteins play a role in this process. Several proteins have been identified in the bifidobacterial membrane, among which moonlighting proteins such as fructose-6-phosphate phosphoketolase or enolase, might be behind the immunomodulatory effects of the membrane vesicles (Sánchez et al., 2004). However, the particular proteins involved in this T-cell polarization have not been identified.
Other immunogenic extracellular proteins are pili, proteinaceous structures that self-assemble into filaments on the bacterial surface (Ventura et al., 2012). Specifically, one sortase-dependent pili from B. bifidum PRL2010 induced TNFα production during transient colonization of the murine mucosa, which acted as a macrophage-activating factor during Th1 (Turroni et al., 2013). Another surface-protein able to influence T-cell responses is TgaA from B. bifidum, a peptidoglycan-derived enzyme able to induce DC activation and IL-2 production (Guglielmetti et al., 2014). IL-2 is one of the main cytokines supporting Treg proliferation, which are characterized by the presence of CD25, the T-cell receptor for that interleukin (Zelante et al., 2012). Despite the evidence supporting an immunomodulation role of bifidobacteria, only a few surface-associated proteins have been identified as possible mediators of this effect on a limited number of in vitro experiments. Identifying the molecules behind this effect and confirming their efficacy in clinical trials, might provide keys to ameliorate diseases characterized by exacerbated immune responses.
Immunoglobulin A (IgA) is the most abundant antibody in human mucosa and modulates immune responses against commensal bacteria, preventing direct contact with immune cells (Peterson et al., 2007; Brandtzaeg, 2013). Globally, 40% of gut bacteria are IgA-coated although these values are species- and strain-dependent (Talja et al., 2014). In healthy individuals, IgA coating of bifidobacteria is higher than that of other commensals (van der Waaij et al., 2004; De Palma et al., 2010), explaining the immune tolerance to high densities of bifidobacteria. In fact, 44 proteins from B. longum and 24 from B. adolescentis were recognized by IgA (Talja et al., 2014). IgA-coated bifidobacteria also enhanced probiotic attachment to Caco-2 cells and increased production of mucosal defense molecules (Mathias et al., 2010).
Levels of IgA-coated gut commensals are altered in dysbiosis states such as those described in coeliac disease (De Palma et al., 2010), inflammatory bowel disease (van der Waaij et al., 2004), or autoimmunity disorders (Talja et al., 2014). Coeliac children showed reduced levels of bifidobacteria and IgA-coated bacteria (De Palma et al., 2010). Conversely, IgA from children developing islet autoimmunity, bound to more B. adolescentis antigens than those from healthy controls (Talja et al., 2014). Morevoer, bifidobacterial supplements modulate IgA production (Holscher et al., 2012; Kandasamy et al., 2014). A probiotic mixture containing bifidobacteria increased IgA and reduced diarrhea following rotavirus vaccination in a gnotobiotic pig model (Kandasamy et al., 2014). B. animalis Bb12 supplementation to formula-fed infants increased IgA in feces and, in those delivered by C-section, enhanced immune responses as reflected by higher anti-rotavirus and anti-poliovirus IgA production following vaccination (Holscher et al., 2012). The bifidogenic effect of galactooligosaccharides also correlated to increased IgA production (Vulevic et al., 2013; Paineau et al., 2014). Further, research to specifically delineate the bifidobacterial molecules mediating IgA induction and interaction is necessary.
A few extracellular proteins, with important physiological roles not discussed in the previous sections deserve further attention. Some Bifidobacterium strains produce surface-exposed Serine Protease Inhibitors of proteinaceous nature (serpins), which participate in a variety of physiological processes. The serpin produced by B. longum NCC2705 inhibits elastase-like proteases, including neutrophil or pancreatic elastases, thus suggesting a role in protecting bifidobacteria against exogenous proteases and potential anti-inflammatory activity (Ivanov et al., 2006). Remarkably, serpins are widely distributed in bifidobacteria and several species harbor serpin-encoding genes in their genomes (Turroni et al., 2010).
Some Bifidobacterium proteins have been identified as mediators of the cross-talk bifidobacteria-host, providing bases to understand their beneficial traits and opening new avenues to conceive bifidobacterial-based therapeutic strategies. However, in most cases, the molecular mechanisms triggered remain unknown what has limited their translation into improved functional supplements. Identifying targets for intervention at intestinal level and developing appropriate models to search for bifidobacterial mediators is required to delineate strategies that fine-tune disease-associated alterations in the microbiota-host interplay.
LR, SD, PM, AM, and BS contributed to the design and organization of the manuscript, drafted, reviewed, and accepted the final version of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This research was funded by Grant AGL2013-44039-R from the Spanish “Plan Estatal de I+D+I.” LR has received funding from the People Programme (Marie Curie Actions) of the European Union's Seventh Framework Programme FP7/2007-2013/ under REA grant agreement n°624773. BS was recipient of a Ramón y Cajal postdoctoral contract from the Spanish Ministry of Economy and Competitiveness. SD is hired on a contract supported by BIO2014-55019-JIN project from the Spanish “Plan Estatal de I+D+I.”
Alegría, A., Delgado, S., Guadamuro, L., Flórez, A. B., Felis, G. E., Torriani, S., et al. (2014). The genome of Bifidobacterium pseudocatenulatum IPLA 36007, a human intestinal strain with isoflavone-activation activity. Gut Pathog. 6:31. doi: 10.1186/1757-4749-6-31
Andersen, J. M., Barrangou, R., Abou Hachem, M., Lahtinen, S. J., Goh, Y. J., Svensson, B., et al. (2013). Transcriptional analysis of oligosaccharide utilization by Bifidobacterium lactis Bl-04. BMC Genomics 14:12. doi: 10.1186/1471-2164-14-312
Ashida, H., Miyake, A., Kiyohara, M., Wada, J., Yoshida, E., Kumagai, H., et al. (2009). Two distinct α-L-fucosidases from Bifidobacterium bifidum are essential for the utilization of fucosylated milk oligosaccharides and glycoconjugates. Glycobiology 9, 1010–1017. doi: 10.1093/glycob/cwp082
Bang, S.-H., Hyun, Y. J., Shim, J., Hong, S.-W., and Kim, D. H. (2015). Metabolism of rutin and poncirin by human intestinal microbiota and cloning of their metabolizing α-L-rhamnosidase from Bifidobacterium dentium. J. Microbiol. Biotechnol. 25, 18–25. doi: 10.4014/jmb.1404.04060
Bergmann, K. R., Liu, S. X., Tian, R., Kushnir, A., Turner, J. R., Li, H. L., et al. (2013). Bifidobacteria stabilize claudins at tight junctions and prevent intestinal barrier dysfunction in mouse necrotizing enterocolitis. Am. J. Pathol. 182, 1595–1606. doi: 10.1016/j.ajpath.2013.01.013
Bermudez-Brito, M., Muñoz-Quezada, S., Gomez-Llorente, C., Matencio, E., Bernal, M. J., Romero, F., et al. (2013). Cell-free culture supernatant of Bifidobacterium breve CNCM I-4035 decreases pro-inflammatory cytokines in human dendritic cells challenged with Salmonella typhi through TLR activation. PLoS ONE 8:e59370. doi: 10.1371/journal.pone.0059370
Bhattacharya, T., Ghosh, T. S., and Mande, S. S. (2015). Global profiling of carbohydrate active enzymes in human gut microbiome. PLoS ONE 10:e0142038. doi: 10.1371/journal.pone.0142038
Bottacini, F., Ventura, M., van Sinderen, D., and O'Connell Motherway, M. (2014). Diversity, ecology and intestinal function of bifidobacteria. Microb. Cell Fact. 13(Suppl. 1):S4. doi: 10.1186/1475-2859-13-S1-S4
Brandtzaeg, P. (2013). Secretory IgA: designed for anti-microbial defense. Front. Immunol. 4:222. doi: 10.3389/fimmu.2013.00222
Briczinski, E. P., Phillips, A. T., and Roberts, R. F. (2008). Transport of glucose by Bifidobacterium animalis subsp. lactis occurs via facilitated diffusion. Appl. Environ. Microbiol. 74, 6941–6948. doi: 10.1128/AEM.01280-08
Bujacz, A., Jedrzejczak-Krzepkowska, M., Bielecki, S., Redzynia, I., and Bujacz, G. (2011). Crystal structures of the apo form of β-fructofuranosidase from Bifidobacterium longum and its complex with fructose. FEBS J. 278, 1728–1744. doi: 10.1111/j.1742-4658.2011.08098
Candela, M., Biagi, E., Centanni, M., Turroni, S., Vici, M., Musiani, F., et al. (2009). Bifidobacterial enolase, a cell surface receptor for human plasminogen involved in the interaction with the host. Microbiology 155, 3294–3303. doi: 10.1099/mic.0.028795-0
Candela, M., Centanni, M., Fiori, J., Biagi, E., Turroni, S., Orrico, C., et al. (2010). DnaK from Bifidobacterium animalis subsp. lactis is a surface-exposed human plasminogen receptor upregulated in response to bile salts. Microbiology 156, 1609–1618. doi: 10.1099/mic.0.038307-0
Cotar, A. I., Chifiriuc, M. C., Dinu, S., Pelinescu, D., Banu, O., and Lazăr, V. (2010). Quantitative real-time PCR study of the influence of probiotic culture soluble fraction on the expression of Pseudomonas aeruginosa quorum sensing genes. Roum. Arch. Microbiol. Immunol. 69, 213–223.
De Palma, G., Nadal, I., Medina, M., Donat, E., Ribes-Koninckx, C., Calabuig, M., et al. (2010). Intestinal dysbiosis and reduced immunoglobulin-coated bacteria associated with coeliac disease in children. BMC Microbiol. 10:63. doi: 10.1186/1471-2180-10-63
Donaldson, G. P., Lee, S. M., and Mazmanian, S. K. (2016). Gut biogeography of the bacterial microbiota. Nature Rev. Microbiol. 14, 20–32. doi: 10.1038/nrmicro3552
Duranti, S., Turroni, F., Lugli, G. A., Milani, C., Viappiani, A., Mangifesta, M., et al. (2014). Genomic characterization and transcriptional studies of the starch-utilizing strain Bifidobacterium adolescentis 22L. Appl. Environ. Microbiol. 80, 6080–6090. doi: 10.1128/AEM.01993-14
Egan, M., O'Connell-Motherway, M., Kilcoyne, M., Kane, M., Joshi, L., Ventura, M., et al. (2014). Cross-feeding by Bifidobacterium breve UCC2003 during co-cultivation with Bifidobacterium bifidum PRL2010 in a mucin-based medium. BMC Microbiol. 14:282. doi: 10.1186/s12866-014-0282-7
Ejby, M., Fredslund, F., Vujicic-Zagar, A., Svensson, B., Slotboom, D. J., and Abou Hachem, M. (2013). Structural basis for arabinoxylo-oligosaccharide capture by the probiotic Bifidobacterium animalis subsp. lactis Bl-04. Mol. Microbiol. 90, 1100–1112. doi: 10.1111/mmi.12419
Evans, J. M., Morris, L. S., and Marchesi, J. R. (2013). The gut microbiome: the role of a virtual organ in the endocrinology of the host. J. Endocrinol. 218, R37–R47. doi: 10.1530/JOE-13-0131
Ewaschuk, J. B., Diaz, H., Meddings, L., Diederichs, B., Dmytrash, A., Backer, J., et al. (2008). Secreted bioactive factors from Bifidobacterium infantis enhance epithelial cell barrier function. Am. J. Physiol. Gastrointest. Liver Physiol. 295, G1025–G1034. doi: 10.1152/ajpgi.90227.2008
Fujita, K., Oura, F., Nagamine, N., Katayama, T., Hiratake, J., Sakata, K., et al. (2005). Identification and molecular cloning of a novel glycoside hydrolase family of core 1 type O-glycan-specific endo-alpha-N-acetylgalactosaminidase from Bifidobacterium longum. J. Biol. Chem. 280, 37415–37422. doi: 10.1074/jbc.M506874200
Fujita, K., Sakaguchi, T., Sakamoto, A., Shimokawa, M., and Kithara, K. (2014). Bifidobacterium longum subsp. longum exo-β-1,3-Galactanase, an enzyme for the degradation of type II arabinogalactan. Appl. Environ. Microbiol. 80, 4577–4584. doi: 10.1128/AEM.00802-14
Fujita, K., Sakamoto, S., Ono, Y., Wakao, M., Suda, Y., Kitahara, K., et al. (2011). Molecular cloning and characterization of a β-L-arabinobiosidase in Bifidobacterium longum that belongs to a novel glycoside hydrolase family. J. Biol. Chem. 286, 5143–5150. doi: 10.1074/jbc.M110.190512
Fukuda, S., Toh, H., Hase, K., Oshima, K., Nakanishi, Y., Yoshimura, K., et al. (2011). Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 469, 543–547. doi: 10.1038/nature09646
Furusawa, Y., Obata, Y., and Hase, K. (2015). Commensal microbiota regulates T cell fate decision in the gut. Semin. Immunopathol. 37, 17–25. doi: 10.1007/s00281-014-0455-3
Garrido, D., Barile, D., and Mills, D. A. (2012). A molecular basis for bifidobacterial enrichment in the infant gastrointestinal tract. Adv. Nutr. 3, 415S–421S. doi: 10.3945/an.111.001586
Garrido, D., Dallas, D. C., and Mills, D. A. (2013). Consumption of human milk glycoconjugates by infant-associated bifidobacteria: mechanisms and implications. Microbiology 159, 649–664. doi: 10.1099/mic.0.064113-0
Garrido, D., Kim, J. H., German, J. B., Raybould, H. E., and Mills, D. A. (2011). Oligosaccharide binding proteins from Bifidobacterium longum subsp. infantis reveal a preference for host glycans. PLoS ONE 6:e17315. doi: 10.1371/journal.pone.0017315
Garrido, D., Ruiz-Moyano, S., Lemay, D. G., Sela, D. A., German, J. B., and Mills, D. A. (2015). Comparative transcriptomics reveals key differences in the response to milk oligosaccharides of infant gut-associated bifidobacteria. Sci. Rep. 5:13517. doi: 10.1038/srep13517
Gilad, O., Jacobsen, S., Stuer-Lauridsen, B., Pedersen, M. B., and Svensson, B. (2010). Combined transcriptome and proteome analysis of Bifidobacterium animalis subsp. lactis BB-12 grown on xylo-oligosaccharides and a model of their utilization. Appl. Environ. Microbiol. 76, 7285–7291. doi: 10.1128/AEM.00738-10
Gleinser, M., Grimm, V., Zhurina, D., Yuan, J., and Riedel, C. U. (2012). Improved adhesive properties of recombinant bifidobacteria expressing the Bifidobacterium bifidum-specific lipoprotein BopA. Microb. Cell Fact. 11:80. doi: 10.1186/1475-2859-11-80
González-Rodríguez, I., Sánchez, B., Ruiz, L., Turroni, F., Ventura, M., Ruas-Madiedo, P., et al. (2012). Role of extracellular transaldolase from Bifidobacterium bifidum in mucin adhesion and aggregation. Appl. Environ. Microbiol. 78, 3992–3998. doi: 10.1128/AEM.08024-11
Guglielmetti, S., Tamagnini, I., Mora, D., Minuzzo, M., Scarafoni, A., Arioli, S., et al. (2008). Implication of an outer surface lipoprotein in adhesion of Bifidobacterium bifidum to Caco-2 cells. Appl. Environ. Microbiol. 74, 4695–4702. doi: 10.1128/AEM.00124-08
Guglielmetti, S., Zanoni, I., Balzaretti, S., Miriani, M., Taverniti, V., De Noni, I., et al. (2014). Murein lytic enzyme TgaA of Bifidobacterium bifidum MIMBb75 modulates dendritic cell maturation through its cysteine- and histidine-dependent amidohydrolase/peptidase (CHAP) amidase domain. Appl. Environ. Microbiol. 80, 5170–5177. doi: 10.1128/AEM.00761-14
Heuvelin, E., Lebreton, C., Bichara, M., Cerf-Bensussan, N., and Heyman, M. A. (2010). Bifidobacterium probiotic strain and its soluble factors alleviate chloride secretion by human intestinal epithelial cells. J. Nutr. 140, 7–11. doi: 10.3945/jn.109.114553
Heuvelin, E., Lebreton, C., Grangette, C., Pot, B., Cerf-Bensussan, N., and Heyman, M. (2009). Mechanisms involved in alleviation of intestinal inflammation by Bifidobacterium breve soluble factors. PLoS ONE 4:e5184. doi: 10.1371/journal.pone.0005184
Hill, C., Guarner, F., Reid, G., Gibson, G. R., Merenstein, D. J., Pot, B., et al. (2014). Expert consensus document: the international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 11, 9. doi: 10.1038/nrgastro.2014.66
Hinz, S. W. A., Pastink, M. I., van den Broek, L. A. M., Vincken, J.-P., and Voragen, A. G. J. (2005). Bifidobacterium longum endogalactanase liberates galactotriose from type I galactans. Appl. Environ. Microbiol. 71, 5501–5510. doi: 10.1128/AEM.71.9.5501-5510.2005
Holscher, H. D., Czerkies, L. A., Cekola, P., Litov, R., Benbow, M., Santema, S., et al. (2012). Bifidobacterium lactis Bb12 enhances intestinal antibody response in formula-fed infants: a randomized, double-blind, controlled trial. J. Parenter. Enteral Nutr. 36(1 Suppl.), 106S–117S. doi: 10.1177/0148607111430817
Hsieh, C. Y., Osaka, T., Moriyama, E., Date, Y., Kikuchi, J., and Tsuneda, S. (2015). Strengthening of the intestinal epithelial tight junction by Bifidobacterium bifidum. Physiol. Rep. 3:e12327. doi: 10.14814/phy2.12327
Hyun, Y. J., Kim, B., and Kim, D. H. (2012). Cloning and characterization of ginsenoside Ra1-hydrolyzing beta-D-xylosidase from Bifidobacterium breve K-110. J. Microbiol. Biotechnol. 22, 535–540.
Ito, T., Katayama, T., Hattie, M., Sakurama, H., Wada, J., Suzuki, R., et al. (2013). Crystal structures of a glycoside hydrolase family 20 lacto-N-biosidase from Bifidobacterium bifidum. J. Biol. Chem. 288, 11795–11706. doi: 10.1074/jbc.M112.420109
Ivanov, D., Emonet, C., Foata, F., Affolter, M., Delley, M., Fisseha, M., et al. (2006). A serpin from the gut bacterium Bifidobacterium longum inhibits eukaryotic elastase-like serine proteases. J. Biol. Chem. 281, 17246–17252. doi: 10.1074/jbc.M601678200
Jae-Han, K., An, H. J., Garrido, D., German, J. B., Lebrilla, C. B., and Mills, D. A. (2013). Proteomic analysis of Bifidobacterium longum subsp. infantis reveals the metabolic insight on consumption of prebiotics and host glycans. PLoS ONE 8:e57535. doi: 10.1371/journal.pone.0057535
Jeffery, C. J. (2003). Moonlighting proteins: old proteins learning new tricks. Trends Genet. 19, 415–417. doi: 10.1016/S0168-9525(03)00167-7
Kainulainen, V., Reunanen, J., Hiippala, K., Guglielmetti, S., Vesterlund, S., Palva, A., et al. (2013). BopA does not have a major role in the adhesion of Bifidobacterium bifidum to intestinal epithelial cells, extracellular matrix proteins, and mucus. Appl. Environ. Microbiol. 79, 6989–6997. doi: 10.1128/AEM.01993-13
Kandasamy, S., Chattha, K. S., Vlasova, A. N., Rajashekara, G., and Saif, L. J. (2014). Lactobacilli and Bifidobacteria enhance mucosal B cell responses and differentially modulate systemic antibody responses to an oral human rotavirus vaccine in a neonatal gnotobiotic pig disease model. Gut Microbes 5, 639–651. doi: 10.4161/19490976.2014.969972.
Katayama, T., Sakuma, A., Kimura, T., Makimura, Y., Hiratake, J., Sakata, K., et al. (2004). Molecular cloning and characterization of Bifidobacterium bifidum 1,2-α-L-Fucosidase (AfcA), a novel inverting glycosidase (Glycoside Hydrolase family 95). J. Bacteriol. 186, 4885–4893. doi: 10.1128/JB.186.15.4885-4893.2004
Kelly, E. D., Bottacini, F., O'Callaghan, J., O'Connell Motherway, M., O'Connell, K. J., Stanton, K., et al. (2016). Glycoside hydrolase family 13 α-glucosidases encoded by Bifidobacterium breve UCC2003. A comparative analysis of function, structure and phylogeny. Int. J. Food Microbiol. 224, 55–65. doi: 10.1016/j.ijfoodmicro.2016.02.014
Khailova, L., Dvorak, K., Arganbright, K. M., Halpern, M. D., Kinouchi, T., Yajima, M., et al. (2009). Bifidobacterium bifidum improves intestinal integrity in a rat model of necrotizing enterocolitis. Am. J. Physiol. Gastrointest. Liver Physiol. 297, G940–G949. doi: 10.1152/ajpgi.00141.2009
Khoroshkin, M. S., Leyn, S. A., Van Sinderen, D., and Rodionov, D. A. (2016). Transcriptional regulation of carbohydrate utilization pathways in the Bifidobacterium genus. Front. Microbiol. 7:120. doi: 10.3389/fmicb.2016.00120
Kim, J. Y., Wang, Y., Park, S. J., Ji, G. E., and Park, M. S. (2012). Cloning and expression of β-glucosidases from Bifidobacterium lactis AD011. Food Sci. Biotechnol. 21, 731–738. doi: 10.1007/s10068-012-0095-0
Kiyohara, M., Nakatomi, T., Kurihara, S., Fushinobu, S., Suzuki, H., Tanaka, T., et al. (2012). α-N-acetylgalactosaminidase from infant-associated bifidobacteria belonging to novel glycoside hydrolase family 129 is implicated in alternative mucin degradation pathway. J. Biol. Chem. 287, 693–700. doi: 10.1074/jbc.M111.277384
Kiyohara, M., Tanigawa, K., Chaiwangsri, T., Katayama, T., Ashida, H., and Yamamoto, K. (2011). An exo-alpha-sialidase from bifidobacteria involved in the degradation of sialyloligosaccharides in human milk and intestinal glycoconjugates. Glycobiology. 21, 437–447. doi: 10.1093/glycob/cwq175
Lee, J. H., and O'sullivan, D. J. (2010). Genomic insights into bifidobacteria. Microbiol. Mol. Biol. Rev. 74, 378–416. doi: 10.1128/MMBR.00004-10
Lee, S. H. (2015). Intestinal permeability regulation by tight junction: implication on inflammatory bowel diseases. Intest. Res. 13, 1–8. doi: 10.5217/ir.2015.13.1.11
Levy, M., Thaiss, C. A., and Elinav, E. (2015). Metagenomic cross-talk: the regulatory interplay between immunogenomics and the microbiome. Genome Med. 7, 120. doi: 10.1186/s13073-015-0249-9
Licciardi, P. V., Wong, S. S., Tang, M. L., and Karagiannis, T. C. (2010). Epigenome targeting by probiotic metabolites. Gut Pathog. 2:24. doi: 10.1186/1757-4749-2-24
Liu, D., Wang, S., Xu, B., Guo, Y., Zhao, J., Liu, W., et al. (2011). Proteomics analysis of Bifidobacterium longum NCC2705 growing in glucose, fructose, mannose, xylose, ribose, and galactose. Proteomics 11, 2628–2638. doi: 10.1002/pmic.201100035
Liu, S., Ren, F., Zhao, L., Jiang, L., Hao, Y., Jin, J., et al. (2015). Starch and starch hydrolysates are favorable carbon sources for bifidobacteria in the human gut. BMC Microbiol. 15:54. doi: 10.1186/s12866-015-0362-3
López, P., González-Rodríguez, I., Gueimonde, M., Margolles, A., and Suárez, A. (2011). Immune response to Bifidobacterium bifidum strains support Treg/Th17 plasticity. PLoS ONE 6:e24776. doi: 10.1371/journal.pone.0024776
López, P., González-Rodríguez, I., Sánchez, B., Gueimonde, M., Margolles, A., and Suárez, A. (2012). Treg-inducing membrane vesicles from Bifidobacterium bifidum LMG13195 as potential adjuvants in immunotherapy. Vaccine 30, 825–829. doi: 10.1016/j.vaccine.2011.11.115
López, P., Gueimonde, M., Margolles, A., and Suárez, A. (2010). Distinct Bifidobacterium strains drive different immune responses in vitro. Int. J. Food Microbiol. 138, 157–165. doi: 10.1016/j.ijfoodmicro.2009.12.023
Lorca, G. L., Barabote, R. D., Zlotopolski, V., Tran, C., Winnen, B., Hvorup, R. N., et al. (2007). Transport capabilities of eleven gram-positive bacteria: comparative genomic analyses. Biochim. Biophys. Acta 1768, 1342–1366. doi: 10.1016/j.bbamem.2007.02.007
Maloy, K. J., and Powrie, F. (2011). Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature 474, 298–306. doi: 10.1038/nature10208
Mathias, A., Duc, M., Favre, L., Benyacoub, J., Blum, S., and Corthésy, B. (2010). Potentiation of polarized intestinal Caco-2 cell responsiveness to probiotics complexed with secretory IgA. J. Biol. Chem. 285, 33906–33913. doi: 10.1074/jbc.M110.135111
Mazé, A., O'Connell-Motherway, M., Fitzgerald, G. F., Deutscher, J., and van Sinderen, D. (2007). Identification and characterization of a fructose phosphotransferase system in Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 73, 545–553. doi: 10.1128/AEM.01496-06
Mazmanian, S. K., Round, J. L., and Kasper, D. L. (2008). A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453, 620–625. doi: 10.1038/nature07008
Ménard, S., Laharie, D., Asensio, C., Vidal-Martinez, T., Candalh, C., Rullier, A., et al. (2005). Bifidobacterium breve and Streptococcus thermophilus secretion products enhance T helper 1 immune response and intestinal barrier in mice. Exp. Biol. Med. 230, 749–756.
Michlmayr, H., Varga, E., Malachova, A., Nguyen, N. T., Lorenz, C., Haltrich, D., et al. (2015). A versatile family 3 Glycoside Hydrolase from Bifidobacterium adolescentis hydrolyzes β-glucosides of the Fusarium mycotoxins deoxynivalenol, nivalenol, and HT-2 toxin in cereal matrices. Appl. Environ. Microbiol. 81, 4885–4893. doi: 10.1128/AEM.01061-15
Milani, C., Turroni, F., Duranti, S., Lugli, G. A., Mancabelli, L., Ferrario, C., et al. (2016). Genomics of the genus Bifidobacterium reveals species-specific adaptation to the glycan-rich gut environment. Appl. Environ. Microbiol. 82, 980–991. doi: 10.1128/AEM.03500-15
Miwa, M., Horimoto, T., Kiyohara, M., Katayama, T., Kitaoka, M., Ashida, H., et al. (2010). Cooperation of β-galactosidase and β-N-acetylhexosaminidase from bifidobacteria in assimilation of human milk oligosaccharides with type 2 structure. Glycobiology 20, 1402–1409. doi: 10.1093/glycob/cwq101
Mokkala, K., Laitinen, K., and Röytiö, H. (2016). Bifidobacterium lactis 420 and fish oil enhance intestinal epithelial integrity in Caco-2- cells. Nutr. Res. 36, 246–252. doi: 10.1016/j.nutres.2015.11.014
Moroni, O., Kheadr, E., Boutin, Y., Lacroix, C., and Fliss, I. (2006). Inactivation of adhesion and invasion of food-borne Listeria monocytogenes by bacteriocin-producing Bifidobacterium strains of human origin. Appl. Environ. Microbiol. 72, 6894–6901. doi: 10.1128/AEM.00928-06
Morrill, J., Kulcinskaja, E., Sulewska, A. M., Lahtinen, S., Stalbrand, H., Svensson, B., et al. (2015). The GH5 1,4-β-mannanase from Bifidobacterium animalis subsp. lactis Bl-04 possesses a low-affinity mannan-binding module and highlights the diversity of mannanolytic enzymes. BMC Biochem. 16:26. doi: 10.1186/s12858-015-0055-4
Nishimoto, M., and Kitaoka, M. (2007). Identification of N-acetylhexosamine 1-kinase in the complete lacto-N-biose I/galacto-N-biose metabolic pathway in Bifidobacterium. Appl. Environ. Microbiol. 73, 6444–6449. doi: 10.1128/AEM.01425-07
O'Connell Motherway, M., Fitzgerald, G. F., and van Sinderen, D. (2011a). Metabolism of a plant derived galactose-containing polysaccharide by Bifidobacterium breve UCC2003. Microb. Biotechnol. 4, 403–416. doi: 10.1111/j.1751-7915.2010.00218
O'Connell Motherway, M., Zomer, A., Leahy, S. C., Reunanen, J., Bottacini, F., Claesson, M. J., et al. (2011b). Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc. Natl. Acad. Sci. U.S.A. 108, 11217–11222. doi: 10.1073/pnas.1105380108
Ohland, C. L., and Macnaughton, W. K. (2010). Probiotic bacteria and intestinal epithelial barrier function. Am. J. Physiol. Gastrointest. Liver Physiol. 298, G807–G819. doi: 10.1152/ajpgi.00243.2009
Ouwehand, A. C., and Salminen, S. (2003). In vitro adhesion assay for probiotics and their in vivo relevance: a review. Microb. Ecol. Health Dis. 15, 175–184. doi: 10.1080/08910600310019886
Paineau, D., Respondek, F., Menet, V., Sauvage, R., Bornet, F., and Wagner, A. (2014). Effects of short-chain fructooligosaccharides on faecal bifidobacteria and specific immune response in formula-fed term infants: a randomized, double-blind, placebo-controlled trial. J. Nutr. Sci. Vitaminol. 60, 167–175. doi: 10.3177/jnsv.60.167
Parche, S., Amon, J., Jankovic, I., Rezzonico, E., Beleut, M., Schendel, I., et al. (2007). Sugar transport systems of Bifidobacterium longum NCC2705. J. Mol. Microbiol. Biotechnol. 12, 9–19. doi: 10.1159/000096455
Parche, S., Beleut, M., Rezzonico, E., Jacobs, D., Arigoni, F., Titgemeyer, F., et al. (2006). Lactose-over-glucose preference in Bifidobacterium longum NCC2705: glcP, encoding a glucose transporter, is subject to lactose repression. J. Bacteriol. 188, 1260–1265. doi: 10.1128/JB.188.4.1260-1265.2006
Patel, P. H., Maldera, J. A., and Edgar, B. A. (2013). Stimulating cROSstalk between commensal bacteria and intestinal stem cells. EMBO J. 32, 3009–3010. doi: 10.1038/emboj.2013.244
Peterson, D. A., McNulty, N. P., Guruge, J. L., and Gordon, J. I. (2007). IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe 2, 328–339. doi: 10.1016/j.chom.2007.09.013
Picard, C., Fioramonti, J., Francois, A., Robinson, T., Neant, F., and Matuchansky, C. (2005). Review article: bifidobacteria as probiotic agents — physiological effects and clinical benefits. Aliment. Pharmacol. Ther. 22, 495–512. doi: 10.1111/j.1365-2036.2005.02615.x
Pokusaeva, K., Fitzgerald, G. F., and van Sinderen, D. (2011). Carbohydrate metabolism in Bifidobacteria. Genes Nutr. 6, 285–306. doi: 10.1007/s12263-010-0206-6
Pokusaeva, K., Neves, A. R., Zomer, A., O'Connell-Motherway, M., MacSharry, J., Curley, P., et al. (2010). Ribose utilization by the human commensal Bifidobacterium breve UCC2003. Microb. Biotechnol. 3, 311–323. doi: 10.1111/j.1751-7915.2009.00152
Pokusaeva, K., O'Connell-Motherway, M., Zomer, A., Fitzgerald, G. F., and van Sinderen, D. (2009). Characterization of two Novel α-glucosidases from Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 75, 1135–1143. doi: 10.1128/AEM.02391-08
Rastall, R. A., and Gibson, G. R. (2015). Recent developments in prebiotics to selectively impact beneficial microbes and promote intestinal health. Curr. Opin. Biotechnol. 32, 42–49. doi: 10.1016/j.copbio.2014.11.002
Ruas-Madiedo, P., Gueimonde, M., Fernández-García, M., de los Reyes-Gavilán, C. G., and Margolles, A. (2008). Mucin degradation by Bifidobacterium strains isolated from the human intestinal microbiota. Appl. Environ. Microbiol. 74, 1936–1940. doi: 10.1128/AEM.02509-07
Ruas-Madiedo, P., Gueimonde, M., Margolles, A., de los Reyes-Gavilán, C. G., and Salminen, S. (2006). Exopolysaccharides produced by probiotic strains modify the adhesion of probiotics and enteropathogens to human intestinal mucus. J. Food Prot. 69, 2011–2015.
Ruiz-Moyano, S., Totten, S. M., Garrido, D. A., Smilowitz, J. T., German, J. B., Lebrilla, C. B., et al. (2013). Variation in consumption of human milk oligosaccharides by infant gut-associated strains of Bifidobacterium breve. Appl. Environ. Microbiol. 79, 6040–6049. doi: 10.1128/AEM.01843-13
Sánchez, B., Noriega, L., Ruas-Madiedo, P., de los Reyes-Gavilán, C. G., and Margolles, A. (2004). Acquired resistance to bile increases fructose-6-phosphate phosphoketolase activity in Bifidobacterium. FEMS Microbiol. Lett. 235, 35–41. doi: 10.1016/j.femsle.2004.04.009
Sánchez, B., Ruiz, L., Gueimonde, M., Ruas-Madiedo, P., and Margolles, A. (2013). Adaptation of bifidobacteria to the gastrointestinal tract and functional consequences. Pharmacol. Res. 69, 127–136. doi: 10.1016/j.phrs.2012.11.004
Sela, D. A., Garrido, D., Lerno, L., Wu, S., Tan, K., Eom, H.-J., et al. (2011). Bifidobacterium longum subsp. infantis ATCC 15697 α-fucosidases are active on fucosylated human milk oligosaccharides. Appl. Environ. Microbiol. 78, 795–803. doi: 10.1128/AEM.06762-11
Shenderov, B. A. (2013). Metabiotics: novel idea or natural development of probiotic conception. Microb. Ecol. Health Dis. 24:8. doi: 10.3402/mehd.v24i0.20399
Shigehisa, A., Sotoya, H., Sato, T., Hara, T., Matsumoto, H., and Matsuki, T. (2015). Characterization of a bifidobacterial system that utilizes galacto-oligosaccharides. Microbiology 161(Pt 7), 1463–1470. doi: 10.1099/mic.0.000100
Shimada, Y., Watanabe, Y., Wakinaka, T., Funeno, Y., Kubota, M., Chaiwangsri, T., et al. (2015). α-N-Acetylglucosaminidase from Bifidobacterium bifidum specifically hydrolyzes α-linked N-acetylglucosamine at nonreducing terminus of O-glycan on gastric mucin. Appl. Microbiol. Biotechnol. 99, 3941–3948. doi: 10.1007/s00253-014-6201
Smilowitz, J. T., Lebrilla, C. B., Mills, D. A., German, J. B., and Freeman, S. L. (2014). Breast milk oligosaccharides: structure-function relationships in the neonate. Annu. Rev. Nutr. 34, 143–169. doi: 10.1146/annurev-nutr-071813-105721
Srutkova, D., Schwarzer, M., Hudcovic, T., Zakostelska, Z., Drab, V., Spanova, A., et al. (2015). Bifidobacterium longum CCM 7952 promotes epithelial barrier function and prevents acute DSS-induced colitis in strictly strain-specific manner. PLoS ONE 10:e0134050. doi: 10.1371/journal.pone.0134050
Sultana, R., McBain, A. J., and O'Neill, C. A. (2013). Strain-dependent augmentation of tight-junction barrier function in human primary epidermal keratinocytes by Lactobacillus and Bifidobacterium lysates. Appl. Environ. Microbiol. 79, 4887–4894. doi: 10.1128/AEM.00982-13
Suzuki, H., Murakami, A., and Yoshida, K. (2013). Motif-guided identification of a glycoside hydrolase family 1 α-L-arabinofuranosidase in Bifidobacterium adolescentis. Biosci. Biotechnol. Biochem. 77, 1709–1714. doi: 10.1271/bbb.130279
Suzuki, R., Wada, J., Katayama, T., Fushinobu, S., Wakagi, T., Shoun, H., et al. (2008). Structural and thermodynamic analyses of solute-binding Protein from Bifidobacterium longum specific for core 1 disaccharide and lacto-N-biose I. J. Biol. Chem. 283, 13165–13173. doi: 10.1074/jbc.M709777200
Tailford, L. E., Crost, E. H., Kavanaugh, D., and Juge, N. (2015). Mucin glycan foraging in the human gut microbiome. Front. Genet. 6:81. doi: 10.3389/fgene.2015.00081
Talja, I., Kubo, A. L., Veijola, R., Knip, M., Simell, O., Ilonen, J., et al. (2014). Antibodies to Lactobacilli and Bifidobacteria in young children with different propensity to develop islet autoimmunity. J. Immunol. Res. 2014:325938. doi: 10.1155/2014/325938
Tanner, S. A., Chassard, C., Rigozzi, E., Lacroix, C., and Stevens, M. J. (2016). Bifidobacterium thermophilum RBL67 impacts on growth and virulence gene expression of Salmonella enterica subsp. enterica serovar Typhimurium. BMC Microbiol. 16:46. doi: 10.1186/s12866-016-0659-x
Turroni, F., Duranti, S., Bottacini, F., Guglielmetti, S., van Sinderen, D., and Ventura, M. (2014). Bifidobacterium bifidum as an example of a specialized human gut commensal. Front. Microbiol. 5:437. doi: 10.3389/fmicb.2014.00437
Turroni, F., Foroni, E., O'Connell-Motherway, M., Bottacini, F., Giubellini, V., and Zomer, A. (2010). Characterization of the serpin-encoding gene of Bifidobacterium breve 210B. Appl. Environ. Microbiol. 76, 3206–3219. doi: 10.1128/AEM.02938-09
Turroni, F., Serafini, F., Foroni, E., Duranti, S., O'Connell Motherway, M., and Taverniti, V. (2013). Role of sortase-dependent pili of Bifidobacterium bifidum PRL2010 in modulating bacterium-host interactions. Proc. Natl. Acad. Sci. U.S.A. 110, 11151–11156. doi: 10.1073/pnas.1303897110
Turroni, F., Strati, F., Foroni, E., Serafini, F., Duranti, S., van Sinderen, D., et al. (2012). Analysis of predicted carbohydrate transport systems encoded by Bifidobacterium bifidum PRL2010. Appl. Environ. Microbiol. 78, 5002–5012. doi: 10.1128/AEM.00629-12
van den Broek, L. A. M., Lloyd, R. M., Beldman, G., Verdoes, J. C., McCleary, B. V., and Voragen, A. G. J. (2005). Cloning and characterization of arabinoxylan arabinofuranohydrolase-D3 (AXHd3) from Bifidobacterium adolescentis DSM20083. Appl. Microbiol. Biotechnol. 67, 641–647. doi: 10.1007/s00253-004-1850-9
van der Waaij, L. A., Kroese, F. G., Visser, A., Nelis, G. F., Westerveld, B. D., Jansen, P. L., et al. (2004). Immunoglobulin coating of faecal bacteria in inflammatory bowel disease. Eur. J. Gastroenterol. Hepatol. 16, 669–674. doi: 10.1097/01.meg.0000108346.41221.19
Ventura, M., Turroni, F., Motherway, M. O. C., MacSharry, J., and van Sinderen, D. (2012). Host-microbe interactions that facilitate gut colonization by commensal bifidobacteria. Trends Microbiol. 20, 467–476. doi: 10.1016/j.tim.2012.07.002
Viborg, A. H., Sørensen, K. I., Gilad, O., Steen-Jensen, D. B., Dilokpimol, A., Jacobsen, S., et al. (2013). Biochemical and kinetic characterisation of a novel xylooligosaccharide-upregulated GH43 β-Dxylosidase/ α-L-arabinofuranosidase (BXA43) from the probiotic Bifidobacterium animalis subsp. lactis BB-12. AMB Express 3:56. doi: 10.1186/2191-0855-3-56
Vulevic, J., Juric, A., Tzortzis, G., and Gibson, G. R. (2013). A mixture of trans-galactooligosaccharides reduces markers of metabolic syndrome and modulates the fecal microbiota and immune function of overweight adults. J. Nutr. 143, 324–331. doi: 10.3945/jn.112.166132
Wada, J., Ando, T., Kiyohara, M., Ashida, H., Kitaoka, M., Yamaguchi, M., et al. (2008). Bifidobacterium bifidum lacto-N-biosidase, a critical enzyme for the degradation of human milk oligosaccharides with a Type 1 structure. Appl. Environ. Microbiol. 74, 3996–3904. doi: 10.1128/AEM.00149-08
Wada, J., Suzuki, R., Fushinobu, S., Kitaoka, M., Wakagi, T., Shoun, H., et al. (2007). Purification, crystallization and preliminary X-ray analysis of the galacto-N-biose/lacto-N-biose I- binding protein (GL-BP) of the ABC transporter from Bifidobacterium longum JCM1217. Acta Crystallogr. Sect. F. Struct. Biol. Cryst. Commun. 63(Pt 9), 751–753. doi: 10.1107/S1744309107036263
Wei, X., Guo, Y., Shao, C., Sun, Z., Zhurina, D., Liu, D., et al. (2012). Fructose uptake in Bifidobacterium longum NCC2705 is mediated by an ATP-binding cassette transporter. J. Biol. Chem. 287, 357–367. doi: 10.1074/jbc.M111.266213
Wei, X., Yan, X., Chen, X., Yang, Z., Li, H., Zou, D., et al. (2014). Proteomic analysis of the interaction of Bifidobacterium longum NCC2705 with the intestine cells Caco-2 and identification of plasminogen receptors. J. Proteome Res. 7, 375–385. doi: 10.1016/j.jprot.2014.04.038
Yoshida, E., Sakurama, H., Kiyohara, M., Nakajima, M., Kitaoka, M., Ashida, H., et al. (2012). Bifidobacterium longum subsp. infantis uses two different β-galactosidases for selectively degrading type-1 and type-2 human milk oligosaccharides. Glycobiol. 22, 361–368. doi: 10.1093/glycob/cwr116
Zelante, T., Fric, J., Wong, A. Y. W., and Ricciardi-Castagnoli, P. (2012). Interleukin-2 production by dendritic cells and its immuno-regulatory functions. Front. Immunol. 3:161. doi: 10.3389/fimmu.2012.00161
Keywords: Bifidobacterium, host interaction, proteome, immunomodulation, adhesin
Citation: Ruiz L, Delgado S, Ruas-Madiedo P, Margolles A and Sánchez B (2016) Proteinaceous Molecules Mediating Bifidobacterium-Host Interactions. Front. Microbiol. 7:1193. doi: 10.3389/fmicb.2016.01193
Received: 26 April 2016; Accepted: 19 July 2016;
Published: 03 August 2016.
Edited by:
Marco Ventura, University of Parma, ItalyReviewed by:
Ines Martinez, University of Nebraska—Lincoln, USACopyright © 2016 Ruiz, Delgado, Ruas-Madiedo, Margolles and Sánchez. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lorena Ruiz, bG9ydWl6QHVjbS5lcw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.