- Department of Oral Biology, College of Dentistry, University of Florida, Gainesville, FL, USA
The microbes that inhabit the human oral cavity are subjected to constant fluctuations in their environment. To overcome these challenges and gain a competitive advantage, oral streptococci employ numerous adaptive strategies, many of which appear to be intertwined with the development of genetic competence. Here, we demonstrate that the regulatory circuits that control development of competence in Streptococcus mutans, a primary etiological agent of human dental caries, are integrated with key stress tolerance pathways by the molecular alarmone (p)ppGpp. We first observed that the growth of a strain that does not produce (p)ppGpp (ΔrelAPQ, (p)ppGpp0) is not sensitive to growth inhibition by comX inducing peptide (XIP), unlike the wild-type strain UA159, even though XIP-dependent activation of the alternative sigma factor comX by the ComRS pathway is not impaired in the (p)ppGpp0 strain. Overexpression of a (p)ppGpp synthase gene (relP) in the (p)ppGpp0 mutant restored growth inhibition by XIP. We also demonstrate that exposure to micromolar concentrations of XIP elicited changes in (p)ppGpp accumulation in UA159. Loss of the RelA/SpoT homolog (RSH) enzyme, RelA, lead to higher basal levels of (p)ppGpp accumulation, but to decreased sensitivity to XIP and to decreases in comR promoter activity and ComX protein levels. By introducing single amino acid substitutions into the RelA enzyme, the hydrolase activity of the enzyme was shown to be crucial for full com gene induction and transformation by XIP. Finally, loss of relA resulted in phenotypic changes to ΔrcrR mutants, highlighted by restoration of transformation and ComX protein production in the otherwise non-transformable ΔrcrR-NP mutant. Thus, RelA activity and its influence on (p)ppGpp pools appears to modulate competence signaling and development through RcrRPQ and the peptide effectors encoded within rcrQ. Collectively, this study provides new insights into the molecular mechanisms that integrate intercellular communication with the physiological status of the cells and the regulation of key virulence-related phenotypes in S. mutans.
Introduction
The human oral cavity typically hosts over six billion microbes, with as many as 1,200 distinct taxa being identified in metagenomic studies (Jenkinson, 2011). Oral microbes contend with constant fluctuations in environmental conditions, including pH, oxygen tension, nutrient availability, and carbohydrate source (Lemos and Abranches, 2005). These dynamic perturbations can alter the composition and biochemical activities of the biofilms by disrupting a homeostatic environment and shifting the oral microbiome from a state of health to one of disease. Oral streptococci are particularly abundant members of the oral microbiota and have evolved numerous mechanisms to respond to these environmental challenges in a way that gives them a competitive advantage for persistence in oral biofilms (Lemos and Burne, 2008). Although many oral streptococci have been associated with oral health, Streptococcus mutans often shows the strongest association with human dental caries (Takahashi and Nyvad, 2011).
Under conditions of nutrient limitation, bacteria employ a regulatory mechanism known as the stringent response, which is characterized in part by inhibition of stable RNA, DNA, and cell wall synthesis, an increase in amino acid biosynthesis, and alterations in expression of stress genes (Carneiro et al., 2011). The stringent response is mediated by the nutritional alarmones guanosine 3′-diphosphate 5′-triphosphate and guanosine 3′,5′-bispyrophosphate, which together are referred to as (p)ppGpp (Dalebroux and Swanson, 2012). In the classical stringent response, originally characterized in the Gram-negative bacterium Escherichia coli, (p)ppGpp is synthesized by addition of pyrophosphate from ATP to either GTP or GDP by the RelA synthetase. RelA becomes active in response to deacylated tRNA molecules binding to the ribosomal A-site as amino acids pools are depleted, leading to abortion of translation when the ribosomes stall. In E. coli, the RelA enzyme has only synthetase activity, whereas the SpoT enzyme has weak synthetase activity and efficient (p)ppGpp hydrolase activity. In Gram-positive bacteria, synthetase and hydrolase activities are combined into single, bifunctional enzymes referred to as a Rel/SpoT homolog (RSH) or Rel enzymes (Atkinson et al., 2011). The dual catalytic domains of these bifunctional enzymes are located in the N-terminal domain of the proteins, while the C-terminal domain participates in allosteric regulation. Rel enzymes have the ability to switch between a synthetase-ON/hydrolase-OFF state and a synthetase-OFF/hydrolase-ON state (Mechold et al., 1996, 2002; Hogg et al., 2004). Deletion of the relA gene encoding the Rel enzyme of S. mutans was shown to alter biofilm formation and acid tolerance, and to affect the regulation of sugar catabolism (Lemos et al., 2004; Nascimento et al., 2008). In some bacteria, including S. mutans, there are additional (p)ppGpp synthetases, referred to as small alarmone synthetases (SAS), which lack (p)ppGpp hydrolase domains. The gene encoding the RelP synthetase of S. mutans is located in an operon with a two-component signal transduction system encoded by relRS. RelP is responsible for the majority of (p)ppGpp produced during exponential growth (Lemos et al., 2007). Another synthetase, RelQ, plays an important role in acid and oxidative stress tolerance. The relQ gene is co-transcribed with an NAD kinase (ppnK), a pseudouridine synthase (rluE), and a phosphotransacetylase (pta; Kim et al., 2012). Acetate metabolism and (p)ppGpp accumulation were recently shown to be tightly linked via the Pta-Acetate kinase pathway, and acetyl-phosphate accumulates under conditions of oxidative stress (Kim et al., 2015).
Seaton et al. (2011) characterized the rcrRPQ operon of S. mutans, encoding a MarR-like transcriptional repressor (RcrR) of the rcrR promoter and a pair of ABC exporters (RcrP, RcrQ), that exert significant control over (p)ppGpp production and the development of genetic competence. The gene cluster (SMU.921–923) is situated in close proximity to the relPRS operon (SMU.926–928), and certain mutations in rcrRPQ have negative effects on the expression of relP that lead to a reduction in (p)ppGpp pools during exponential growth (Seaton et al., 2011). Additionally, the expression of the rcrRPQ operon is significantly increased in response to accumulation of (p)ppGpp during a mupirocin-induced stringent response (Nascimento et al., 2008). Thus, regulation of rcrRPQ and (p)ppGpp accumulation are well integrated in S. mutans. In terms of genetic competence, the impact of deletion-replacement mutations of rcrR is strongly dependent on the polarity of the inserted marker, which greatly influences the expression levels of the ABC transporter genes, as well as of two peptide effectors encoded in the 3′ region of rcrQ (Ahn et al., 2014; Seaton et al., 2015). A polar mutation within rcrR (ΔrcrR-P) is constitutively hyper-transformable, but a non-polar mutation (ΔrcrR-NP) is non-transformable. The non-transformable state of the ΔrcrR-NP strain is correlated with overexpression of the rcrPQ gene and rcrQ peptides, and is characterized by a dramatic decrease in the amount of mRNA detectable from the 5′ portion of comX (or σX), which encodes the alternative sigma factor that is the master regulator of late competence gene expression that is absolutely required for cells to progress to competence. Interestingly, the loss of full-length comX mRNA and ComX protein in the ΔrcrR-NP mutant is accompanied by the activation of expression of an open reading frame (ORF) internal to comX that codes for XrpA (ComX regulatory peptide A; Kaspar et al., 2015). If point mutations that block the production of XrpA are present in the ΔrcrR-NP genetic background, expression of a full-length comX transcript, ComX protein levels and transformability are all restored to levels similar to the ΔrcrR-P strain (Kaspar et al., 2015). Although genetic studies clearly show that RcrRPQ, XrpA and the rcrQ-associated peptide effectors work in concert to control competence development, strongly influencing comX expression and ComX protein levels, the mechanisms by which these effectors exert their influence on the competence cascade has not yet been defined.
The early stages of competence development in S. mutans require a complex signaling network that ultimately leads to activation of expression of the gene for ComX. ComX directs transcription of a regulon of late competence genes, many of which are responsible for the uptake of exogenous DNA from the environment and homologous recombination into the bacterial chromosome, when possible (Cvitkovitch, 2001; Merritt et al., 2005b). Regulation of comX expression is modulated directly by the ComRS system (Fontaine et al., 2009; Mashburn-Warren et al., 2010). The comS gene encodes a 17-aa peptide that is exported and processed by an unknown mechanism into the active, 7-aa comX inducing peptide (XIP); XIP is composed of the C-terminal 7 residues of ComS. Externalized XIP is imported by the oligopeptide permease OppA, an ABC transporter, and is bound by the Rgg-like transcriptional activator ComR (Desai et al., 2012; Khan et al., 2012). The ComR–XIP complex activates the promoters for comX and comS, the latter creating a positive feedback loop (Fontaine et al., 2013). Provision of relatively high concentrations of XIP (2–10 μM) to S. mutans can also induce cell death and result in apparent slow growth of the organism (Wenderska et al., 2012). However, the molecular basis for XIP-mediated cell death or growth inhibition is not currently understood. Activation of comX expression by XIP and the ComRS system in a chemically defined medium shows a unimodal response across the entire population (Son et al., 2012), and can be influenced by environmental pH (Guo et al., 2014; Son et al., 2015a). The ComCDE pathway of S. mutans also induces comX expression in a bimodal fashion in complex media that contains peptides, albeit via an indirect mechanism (Hung et al., 2011; Reck et al., 2015; Son et al., 2015b).
In addition to the ComCDE and ComRS pathways, a variety of other gene products in S. mutans can impact transformation efficiency by altering early and/or late competence gene expression, including VicRK, HtrA, CiaRH, HdrM, and others (Ahn et al., 2005; Merritt et al., 2005a; Senadheera et al., 2005, 2012; Abranches et al., 2006; Niu et al., 2008; Banu et al., 2010; Okinaga et al., 2010; Kajfasz et al., 2011; Mair et al., 2012; Kim et al., 2013). Many of these regulatory systems that affect competence have been shown to be critical for adaptation to environmental stress, so there is a considerable body of evidence demonstrating that induction of competence and stress tolerance are tightly intertwined. It was previously reported that there was no reduction in transformation efficiency associated with deletion of relA in S. mutans when cells were made competent in complex medium in the presence or absence of competence stimulating peptide (CSP) (Lemos et al., 2004). However, the recent evidence showing that rcrRPQ gene products affect both (p)ppGpp metabolism and genetic competence, coupled with the discovery of the comRS pathway and its direct and essential role in activation of comX expression call for a reevaluation of the potential linkage of (p)ppGpp metabolism with competence and its relationship to the RcrRPQ system. We report here that gene products involved in (p)ppGpp metabolism can dominantly impact the development of genetic competence and reverse the effect of an rcrR mutation that eliminates transformability, providing critical new insights into the regulation of competence and virulence traits by S. mutans.
Materials and Methods
Bacterial Strains and Growth Conditions
Escherichia coli strain DH10B was grown in Luria broth that was supplemented with 300 μg ml-1 erythromycin and 50 μg ml-1 of kanamycin or spectinomycin, when needed. S. mutans wild-type (WT) strain UA159 and its derivatives (Table 1) were grown in either brain heart infusion (BHI; Difco) or FMC medium (Terleckyj and Shockman, 1975) that was supplemented with 10 μg ml-1 erythromycin and 1 mg ml-1 of kanamycin or spectinomycin, as needed. Synthetic XIP (sXIP, aa sequence = GLDWWSL), corresponding to residues 11–17 of ComS, was synthesized and purified to 96% homogeneity by NeoBioSci (Cambridge, MA, USA). The lyophilized sXIP was reconstituted with 99.7% dimethyl sulfoxide (DMSO) to a final concentration of 2 mM and stored in 40 μl aliquots at -20°C. Unless otherwise noted, cultures were grown overnight in BHI medium with antibiotics, if needed, at 37°C in a 5% CO2 aerobic atmosphere. The cultures were harvested by centrifugation, washed in 1 mL FMC, and resuspended in FMC to remove all traces of BHI before fresh medium was inoculated. For growth rate comparisons, overnight cultures were diluted 1:50 and grown to mid-exponential phase (OD600 = 0.5–0.6). Then, cultures were re-diluted 1:100 into 300 μl of FMC and added to multiwell plates, sXIP was added to the desired final concentrations (0.2, 2, or 20 μM), and cultures were overlaid with 50 μl of sterile mineral oil to reduce the growth inhibitory effects of air on growth of S. mutans. The optical density at 600 nm (OD600) was monitored at 30 min intervals for 24 h using a Bioscreen C lab system (Helsinki, Finland) at 37°C with shaking for 10 s before each reading.
Construction of Strains and DNA Manipulation
Mutant strains of S. mutans were created using a PCR ligation mutagenesis approach (Supplementary Table S1) (Lau et al., 2002). Splice overlap extension was utilized to create single base change mutations within the relA gene using protocols described elsewhere (Ho et al., 1989; Cha et al., 1992; Kaspar et al., 2015). Overexpression of genes was achieved by amplifying the genes of interest from S. mutans UA159 and cloning into the expression vector pIB184 (Biswas et al., 2008; Guo et al., 2014; Kaspar et al., 2015). Transformants were confirmed by PCR and sequencing after selection on BHI agar containing the appropriate antibiotic(s). Plasmid DNA was isolated from E. coli by using QIAGEN (Chatsworth, CA, USA) columns, and restriction and DNA-modifying enzymes were obtained from Invitrogen (Gaithersburg, MD, USA) or New England Biolabs (Beverly, MA, USA). PCRs were carried out with 100 ng of chromosomal DNA using Taq DNA polymerase, and PCR products were purified with the QIAquick kit (QIAGEN). DNA was introduced into S. mutans by natural transformation and into E. coli by the calcium chloride method (Cosloy and Oishi, 1973).
RNA Extraction and Quantitative Real-Time PCR
To measure the expression of genes using quantitative real-time PCR (RT-qPCR), three colonies of S. mutans WT (UA159) and the mutant strains of interest (ΔrelAPQ, ΔrelA) were grown overnight and a 1:50 dilution was added to fresh FMC medium the next day. When cells reached an OD600 of 0.2, cultures were adjusted to a final concentration of 1% DMSO or 2 μM sXIP and incubated at 37°C in a 5% CO2 atmosphere for 1 h before being harvested by centrifugation. Cell lysis was achieved through mechanical disruption (bead beating) and RNA was extracted by acidic phenol phase separation. The RNA was further purified with an RNeasy mini kit (QIAGEN) according to the provided protocol, and treatment with DNase 1 (QIAGEN). Purified RNA (1 μg) was used to generate cDNA from gene-specific primers (Supplementary Table S1) using the Superscript III first-strand synthesis (Invitrogen) reverse transcription protocol. Real-time PCRs were carried out using an iCyclerQ real-time PCR detection system (Bio-Rad) and iQ SYBR green supermix (Bio-Rad) according to the supplier’s protocol. 16S rRNA was used as an internal reference. All assays were performed in triplicate with RNA isolated from three biological replicates.
Transformation Assays
Strains of S. mutans were grown overnight and then diluted 1:50 in 200 μl of FMC medium. When the cultures reached an OD600 = 0.2, either a final concentration of 1% DMSO or 2 μM sXIP was added to the growing culture. After 10 min of incubation, 500 ng of purified plasmid pDL278, which harbors a spectinomycin resistance (SpR) gene, was added. Following 3 h of further incubation, dilutions of the cultures were plated on BHI agar plates with or without 1 mg ml-1 spectinomycin. Colony forming units (CFUs) were counted after 48 h. Transformation efficiency was determined by dividing the number of SpR transformants by the total number of CFU recovered on non-selective medium.
Western Blotting
Overnight cultures of S. mutans were diluted 1:50 into 35 mL of fresh FMC medium and harvested by centrifugation when the cultures reached an OD600 = 0.5. When desired, 1% DMSO or 2 μM sXIP (final concentrations) was added when the cultures reached an OD600 value of 0.2. Cell pellets were washed once with buffer A (0.5 M sucrose; 10 mM Tris–HCl, pH 6.8; 10 mM MgSO4) containing 10 μg ml-1 of phenylmethanesulfonyl fluoride (PMSF; ICN Biomedicals Inc.), collected by centrifugation, and resuspended in 0.5 mL Tris-buffered saline (50 mM Tris–HCl, pH 7.5; 150 mM NaCl). Cells were lysed using a Mini Bead Beater (Biospec Products) in the presence of 1 volume of glass beads (average diameter 0.1 mm) for 30 s intervals, three times, with incubation on ice between homogenizations. Lysates were then centrifuged at 3,000 ×g for 10 min at 4°C. Protein concentrations of the resulting supernates were determined using the bicinchoninic acid assay (BCA; Thermo Scientific) with purified bovine serum albumin as the standard. ComX protein was detected by immunoblotting using a protocol detailed elsewhere (Kaspar et al., 2015). Protein preparations (10 μg) were loaded on a 15% polyacrylamide gel with a 6% stacking gel and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to Immobilon-P polyvinylidene difluoride membranes (Millipore), then treated with primary polyclonal anti-ComX antisera at a 1:1,000 dilution and a secondary peroxidase-labeled, goat anti-rabbit IgG antibody (1:5,000 dilution; Kirkegaard & Perry Laboratories, USA). Detection was performed using a SuperSignal West Pico Chemiluminescent Substrate kit (Thermo Scientific) and visualized with a FluorChem 8900 imaging system (Alpha Innotech, USA).
β-Galactosidase Assays
For measurements of LacZ produced from promoter:reporter gene fusions, strains were grown overnight, washed in FMC, resuspended, and diluted 1:40 into fresh FMC medium. At OD600 = 0.2, sXIP was added to a final concentration of 2 μM, cells were incubated for an additional 90 min, then cells were harvested by centrifugation. β-galactosidase activity was measured by using a modification of the Miller protocol (Zubay et al., 1972). Briefly, cells were washed once with Z buffer [Na-phosphate buffer (pH 7.0), 10 mM KCl, 1 mM MgSO4, 5 mM β-mercaptoethanol), and resuspended in 1.3 ml Z buffer. Part of the sample (500 μl) was vortexed with 25 μl of a toluene–acetone mix (1:9) for 2 min and kept at 37°C, the remaining 0.8 ml of the suspension was used to determine OD600. The LacZ reaction was initiated by the addition of 100 μl of o-nitrophenyl-β-D-galactopyranoside (4 mg ml-1) and was terminated by the addition of 500 μl of 1 M Na2CO3. Samples were centrifuged for 1 min using a tabletop centrifuge and the OD of the supernatant fluid was measured at 420 and 550 nm. Activity was expressed in Miller units (Zubay et al., 1972).
Monitoring (p)ppGpp Accumulation
Visualization of (p)ppGpp accumulation was conducted as described elsewhere (Lemos et al., 2007). Briefly, overnight cultures from strains of interest were diluted 1:50 into a modified FMC medium containing a lower concentration of inorganic phosphate (Lemos et al., 2007). When cells reached an OD600 of 0.2, [32P]-orthophosphate, along with the desired amount of sXIP, was added. Cells were then incubated at 37°C for the specified duration, then harvested by centrifugation. (p)ppGpp was extracted by addition of an equal volume of ice-cold 13 M formic acid, followed by two freeze–thaw cycles in a dry ice/ethanol bath. After extraction, the CPM μl-1 of the supernatant fraction was measured in a scintillation counter and 2.0 × 105 CPM of each sample was spotted onto a polyethyleneimine (PEI) cellulose plate (Selecto Scientific) for thin-layer chromatography (TLC). The TLC plates were chromatographed in one dimension with 1.5 M KH2PO4 (adjusted to pH 3.4 with phosphoric acid), air-dried and exposed to X-ray film at -70°C. The signal density of spots corresponding to ppGpp (GP4) and pppGpp (GP5) was analyzed using ImageJ (v1.47) software1.
Results
A (p)ppGpp0 Mutant of S. mutans Displays Minimal Growth Inhibition by XIP
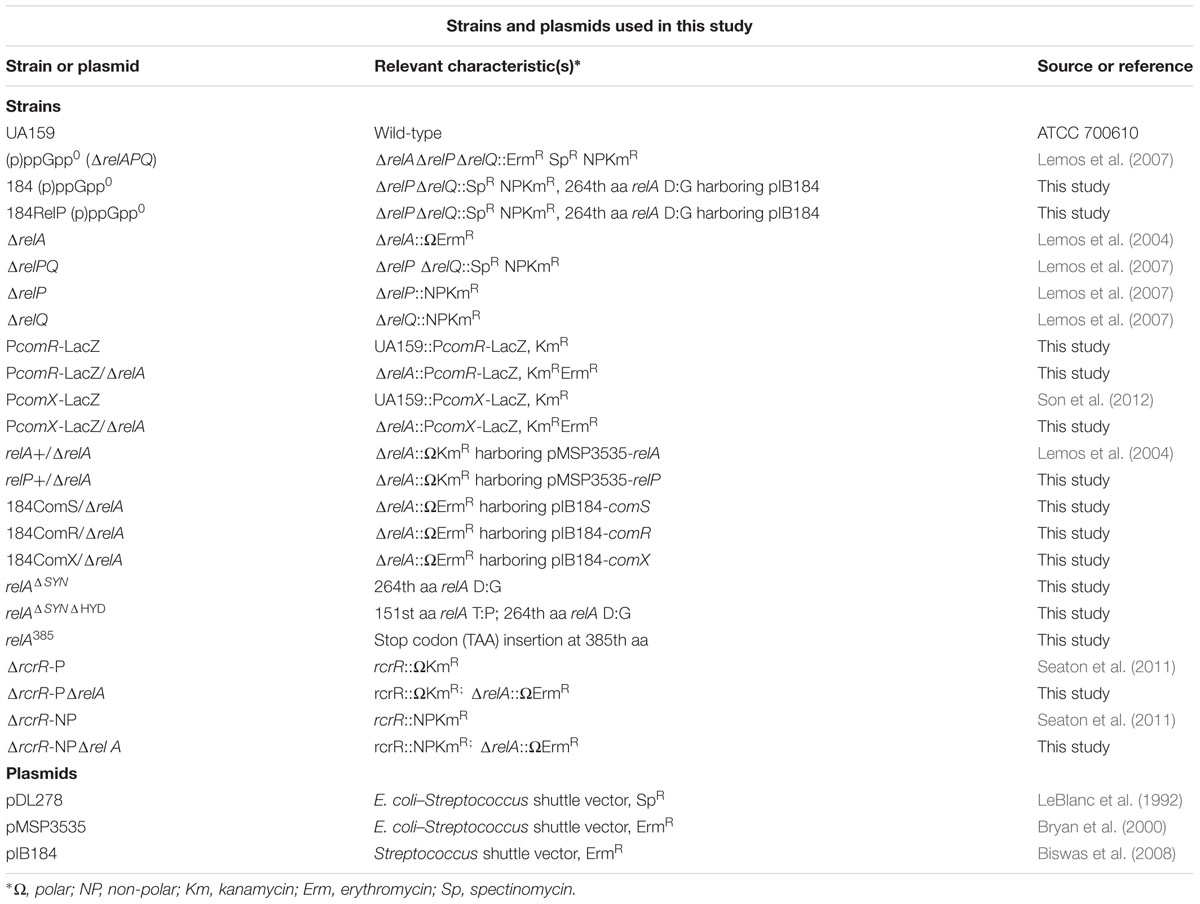
While previous communications indicated that a deletion of the relA gene of S. mutans does not appear to have effects on transformation efficiency (Lemos et al., 2004), those experiments were performed in the complex medium BHI in the presence of CSP. Since that time, it has come to light that the ComRS system is the proximal regulator of alternative sigma factor comX expression, not ComDE, in S. mutans. Importantly, the ComRS system has been shown to function optimally in a chemically defined medium (Son et al., 2012), and requires ComS-derived XIP for ComR-dependent activation of gene expression (Mashburn-Warren et al., 2010). To determine if (p)ppGpp influenced the development of competence by ComRS in a chemically defined medium (FMC), we compared the growth kinetics between the S. mutans WT strain UA159 and a (p)ppGpp0 mutant (ΔrelAPQ) in the presence of various concentrations of sXIP, which was dissolved in DMSO. Cells treated with the same final v/v concentration (1%) of DMSO served as controls. The WT strain UA159 displayed apparent growth inhibition, as assessed by monitoring doubling time and final optical density, as the concentration of sXIP was increased (Wenderska et al., 2012; Guo et al., 2014; Figure 1A). We specify apparent growth inhibition because cell lysis may also influence the total optical density at any given time point. The exponential doubling time of UA159 increased from 66 ± 1 to 204 ± 5 min as the concentration of sXIP was increased from 0.2 to 2 μM. A doubling time of 229 ± 18 min was observed in the presence of 20 μM sXIP. A reduction in the final yield (final OD600 at the 36-h time point) was seen with increasing concentrations of sXIP; 1.08 ± 0.01 with no XIP and 0.45 ± 0.08 at the highest concentration of XIP. Interestingly, the growth inhibitory effects of sXIP were not observed in the (p)ppGpp0 strain (Figure 1B). As sXIP concentrations were increased from 0.2 μM to 2 or 20 μM, there were only minimal increases in the doubling times: 98 ± 2, 111 ± 1, and 109 ± 2 min, respectively. Similarly, there were only marginal reductions in the final yields in the strain lacking (p)ppGpp.
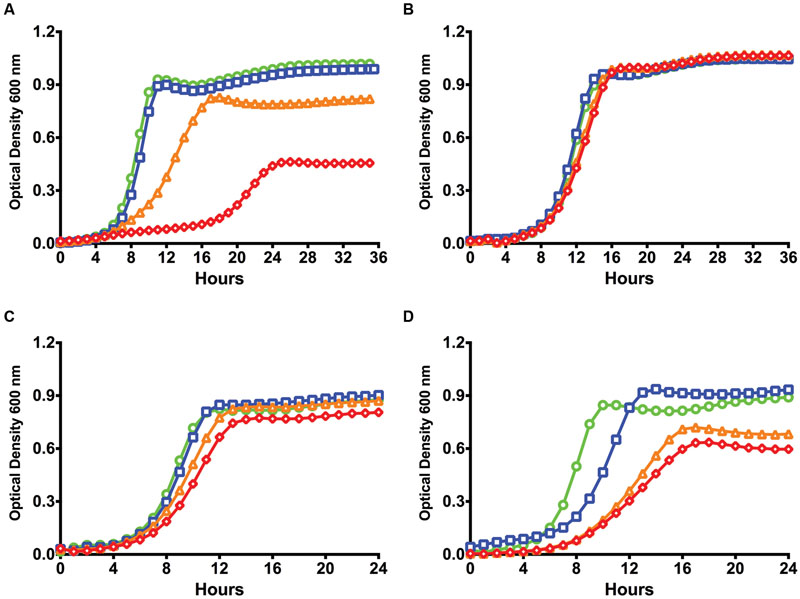
FIGURE 1. Lack of (p)ppGpp abrogates growth inhibition by XIP. Growth curves of (A) S. mutans UA159, (B) a (p)ppGpp0 derivative of UA159 (ΔrelAPQ), (C) pIB184 vector only in the (p)ppGpp0 genetic background, and (D) pIB184RelP in the (p)ppGpp0 genetic background in the presence of different concentrations of sXIP. Bacterial cultures were grown in the chemically defined medium FMC containing 1% DMSO (●; green), or sXIP at final concentrations of 0.2 μM (■; blue), 2 μM (▲; orange), or 20 μM (◆; red). sXIP was dissolved in DMSO, so the final concentration of DMSO in all samples was 1%.
To confirm that changes in growth inhibition were due to a loss of (p)ppGpp, we added back the relP gene, encoding a SAS that lacks hydrolase activity, under the control of the constitutive P23 promoter on a shuttle plasmid (Biswas et al., 2008) and compared the strains growth kinetics to one containing only the empty pIB184 vector. While the (p)ppGpp0 strain containing the empty vector showed limited growth inhibition in the presence of XIP (Figure 1C), addition of relP led to increased doubling times of 208 ± 13 and 273 ± 24 min with addition of 2 or 20 μM sXIP, respectively, compared to 86 ± 3 min with the 1% DMSO control (Figure 1D). Additionally, final yields dropped from 0.88 ± 0.02 at 24 h after growth without sXIP to 0.68 ± 0.03 and 0.60 ± 0.03 with 2 or 20 μM sXIP. These data suggest that modulation of (p)ppGpp levels, as seen with the complementation of the (p)ppGpp0 strain, can lead to phenotypic changes in response to the competence inducing peptide XIP, altering the response level to the signaling peptide.
This lack of growth inhibition phenotype in cells treated with sXIP is commonly associated with strains that have mutations in either comR or comX, indicating a severe reduction in early com gene expression that would lead to the (p)ppGpp0 failing to respond to the sXIP signal (Perry et al., 2009; Wenderska et al., 2012). This same phenotype is also observed in the ΔrcrR-NP strain, which fails to express a full-length comX mRNA (Kaspar et al., 2015). To test for com gene down-regulation, expression was measured by qRT-PCR in the WT and (p)ppGpp0 strains 1 h after addition of 2 μM sXIP to cells at OD600 nm = 0.2. Interestingly the comX gene, encoding the alternative sigma factor required for late competence gene expression (Supplementary Figure S1A), and the late competence gene comYA (Supplementary Figure S1B) were both induced to the same level in the mutant and WT strains treated with XIP. The qRT-PCR results were consistent with the finding that similar levels of ComX protein (Kaspar et al., 2015) were present in lysates of mutant and WT cells that had been treated with XIP (Supplementary Figure S1C). Similar gene expression profiles between the WT and (p)ppGpp0 strains following addition of sXIP were also observed for a direct target of ComE (comD), for the comRS genes, and for rcrR, the first gene in the rcrRPQ operon that has been shown to influence competence, stress tolerance, and (p)ppGpp accumulation (Supplementary Figures S1D–G). Thus, the ComRS signaling pathway was intact and functional in both the WT and (p)ppGpp0 strains, so the specific impact of complete loss of (p)ppGpp on the growth and competence phenotypes apparently does not occur at the level of comRS expression or formation of an active ComR–XIP complex.
(p)ppGpp Levels Are Influenced by XIP Exposure
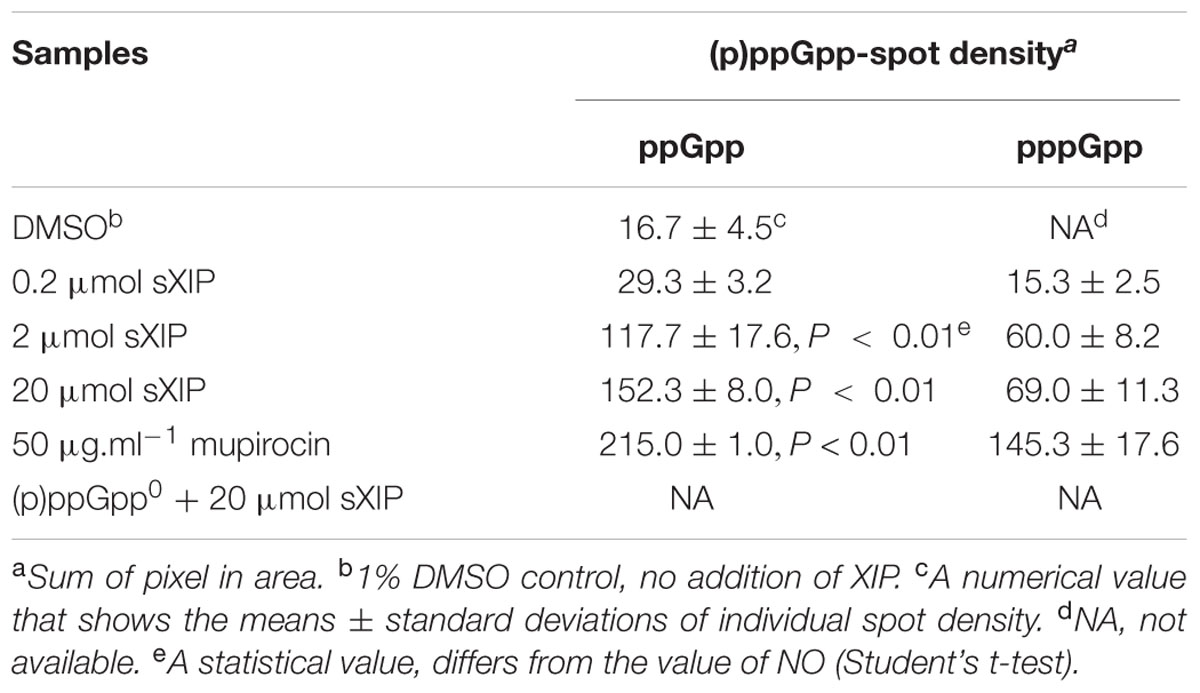
Clearly, (p)ppGpp levels can alter the growth phenotype of cells exposed to sXIP, so we reasoned that addition of sXIP may lead to an increase in internal (p)ppGpp pools. To determine how (p)ppGpp accumulation was affected by the addition of exogenous XIP, the WT strain UA159 was grown in FMC medium and labeled with 32P-orthophosphate in the presence of either 1% DMSO (control) or increasing concentrations of sXIP. More specifically, cultures of S. mutans were grown to OD600 = 0.15 and then 32P-orthophosphate and sXIP were added together. Cultures were incubated for 60 min, cells were harvested, and formic acid extracts of the phosphorylated guanosine nucleotides were prepared and separated by thin layer chromatography (Figure 2A). Addition of mupirocin, which leads to RelA-dependent accumulation of (p)ppGpp, served as a positive control. Only a very modest increase in (p)ppGpp accumulation was evident with addition of 0.2 μM sXIP, compared to the 1% DMSO control. However, there were substantial increases in (p)ppGpp pools in the samples treated with 2 or 20 μM sXIP (Table 2). The latter concentrations of sXIP correspond to those that induce the greatest degree of growth inhibition in UA159, providing further evidence of an essential role for (p)ppGpp in XIP-dependent inhibition of growth. Induction of (p)ppGpp after sXIP exposure is also time-dependent, as cells harvested at 10, 20, 40, and 60 min after addition of 32P-orthophosphate and sXIP showed the greatest accumulation of (p)ppGpp at the 40 and 60 min time points (Supplementary Figure S2). To determine how addition of sXIP to the growth medium might influence expression of individual rel genes, we measured the expression of relP, relQ, and relA mRNA levels in UA159 after addition of 2 μM sXIP (Figure 2B). Expression of the relQ gene was increased 1.9-fold after sXIP addition, compared to the 1% DMSO control, whereas relA gene expression was decreased 2.8-fold with XIP treatment. There were no increases noted in relP mRNA associated with sXIP treatment. However, it was recently reported that there is a ComE binding site in front of the cipI gene (SMU.925, immB; Khan et al., 2016), so we cannot rule out that ComE-dependent activation of cipI following XIP treatment might influence downstream relP (SMU.926) mRNA levels under other conditions that lead to activation of competence. Collectively, though, the results indicate increased production of RelQ and possible allosteric modulation of RelA synthetase/hydrolase activity can impact the quantity of (p)ppGpp in XIP-treated cells.
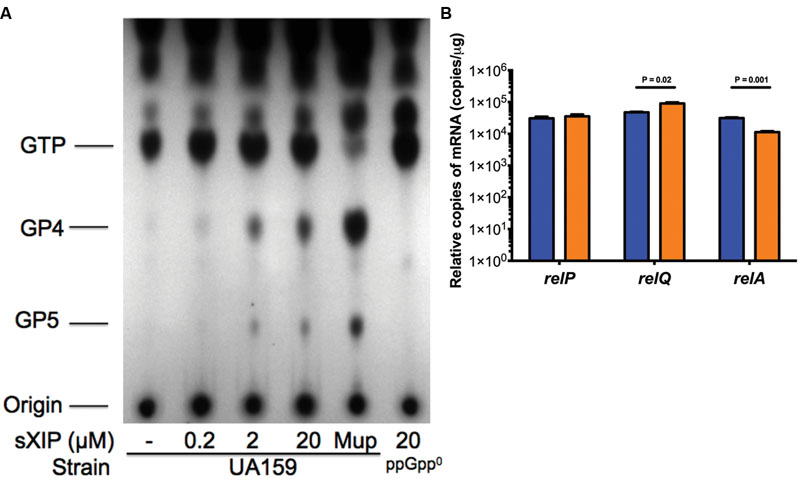
FIGURE 2. Accumulation of (p)ppGpp with addition of sXIP. (A) (p)ppGpp accumulation in S. mutans UA159 with various concentrations of sXIP. Cells were labeled with 32P-orthophosphate in FMC medium when OD600 nm reached 0.15, along with addition of either 1% DMSO, 0.2, 2, or 20 μM sXIP. After an hour, cells were harvested. Nucleotides were extracted by addition of 13 M formic acid, followed by three freeze–thaw cycles. Resulting supernates were spotted onto PEI-cellulose plates for TLC in 1.5 M KH2PO4. Identity of the migrating nucleotides is shown to the left. Mupirocin (Mup; 50 μg ml-1), previously shown to induce a potent stringent response, serves as the positive control for (p)ppGpp accumulation. (B) Measurement of relP, relQ, and relA expression by qRT-PCR of UA159 with addition of either 1% DMSO (blue bars) or 2 μM sXIP (orange bars).
The RelA Enzyme Is Required for Optimal sXIP Signaling
The three (p)ppGpp synthetases in S. mutans appear to contribute to accumulation of alarmone under different circumstances (Lemos et al., 2007), so we explored the behavior of strains carrying individual or selected combinations of RSH and SAS enzymes on competence pathway signaling through ComRS. First, the growth kinetics of strains harboring individual deletion:replacement mutations of relA (Figure 3A), relP, and relQ were evaluated. The ΔrelA strain showed the lowest sensitivity to sXIP, with a doubling time of 99 ± 5 to 101 ± 4 min in the presence of 0.2 and 2 μM sXIP, respectively. A longer doubling time (147 ± 3 min) was observed with 20 μM sXIP. Although doubling times increased with increased concentrations of sXIP, the final yields did not differ (0.70 ± 0.06 to 0.77 ± 0.03 min). In contrast, a strain carrying mutations in both relP and relQ (ΔrelPQ) showed growth inhibitory effects to XIP that were similar to those seen with the WT strain (Figure 3B), as was the case for the strains carrying only relP or relQ mutations (data not shown). The observed doubling times for the ΔrelPQ mutant strain were substantially greater than for the parental strain in the presence of 2 μM (229 ± 39 min) or 20 μM XIP (324 ± 68 min) and final yields were also lower (0.47 ± 0.14, 0.39 ± 0.05). Of note, a lag time of approximately 18 h was seen with 2 μM XIP in the ΔrelPQ strain. When mutants lacking the relA gene were combined with individual relP or relQ mutations (ΔrelAP, ΔrelAQ), the strains grew like the ΔrelA single mutant, providing further evidence that the loss of the Rel (RSH) enzyme alone could alter sensitivity to sXIP.
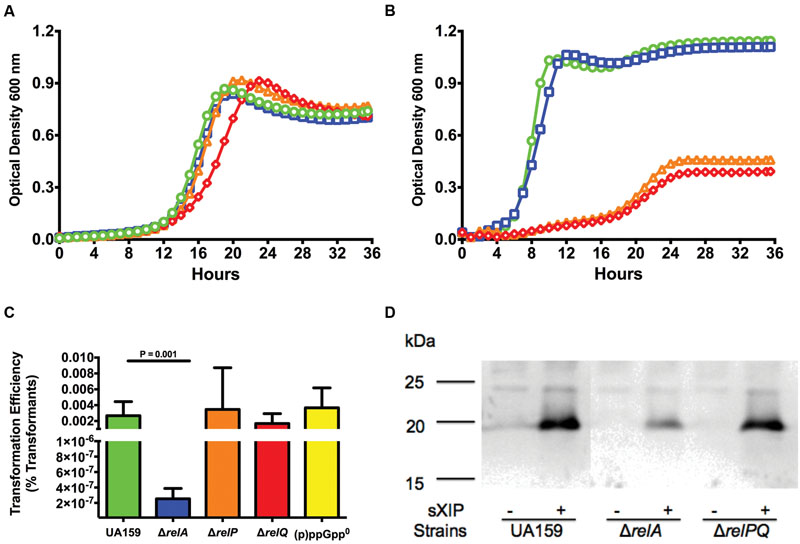
FIGURE 3. Loss of relA results in decreases in sensitivity to XIP and transformation efficiency. Growth curves of (A) ΔrelA and (B) ΔrelPQ strains treated with different concentrations of sXIP. Bacterial cultures were grown in the chemically defined medium FMC containing 1% DMSO (●; green), or 0.2 μM (■; blue), 2 μM (▲; orange), or 20 μM (◆; red) sXIP as in Figure 1. (C) Transformation efficiency of S. mutans UA159 (green bar), ΔrelA (blue bar), ΔrelP (orange bar), ΔrelQ (red bar), and (p)ppGpp0 (ΔrelAPQ; yellow bar) in FMC with addition of 2 μM sXIP. sXIP and plasmid pDL278 (SpR) were added when OD600 reached 0.2. Serial dilutions of the cultures were plated on BHI agar with and without spectinomycin. Transformation efficiency was determined by the CFU of transformants divided by the total CFU of viable bacteria, multiplied by 100 (percent transformants). Data are the averages of three individual experiments with three biological replicates. Statistical analysis was performed by Student’s t-test. (D) Detection of ComX in UA159, ΔrelA, and ΔrelPQ lysates from cells grown in FMC with either 1% DMSO (-) or 2 μM sXIP (+), added when OD600 nm reached 0.2 and followed by 1 h of incubation prior to harvesting and preparation of cell lysates. ComX was detected using a 1:5000 dilution of primary antiserum raised against full-length recombinant ComX from S. mutans. Molecular mass standards (in kDa) are shown to the left. The calculated molecular mass of ComX is 19 kDa.
Fluctuations in sensitivity of strains to sXIP can be a potential indicator for changes in the ability to be transformed by exogenous DNA (Ahn et al., 2014; Kaspar et al., 2015). Transformation efficiency was measured in the ΔrelA, ΔrelP, and ΔrelQ strains grown in FMC and treated with 2 μM XIP to induce competence. The ΔrelA mutant strain showed a five-log reduction in transformation efficiency (2.0 × 10-7), compared to the WT strain (3.8 × 10-2; Figure 3C). In contrast to the ΔrelA mutant, the ΔrelP and ΔrelQ strains showed no reduction in transformation efficiency. To confirm the results of the transformation assay, ComX levels were measured by western blot in the ΔrelA and ΔrelPQ strains. A clear decrease in ComX protein was seen in the ΔrelA strain when treated with sXIP, but ComX protein in the ΔrelPQ and WT strains were similar (Figure 3D). Interestingly, the ΔrelA strain showed a decrease in transformation efficiency of less than one log compared to the WT strain when cells were grown in BHI medium and synthetic competence stimulating peptide (sCSP) was used to induce competence (3.7 × 10-3 to 1.2 × 10-2), comparable to the behavior noted for the ΔrelP strain (3.4 × 10-3; Supplementary Figure S3). It is of interest that the effects of loss of the RSH enzyme were more profound in chemically defined medium. In particular, XIP is known to be ineffective at inducing com gene expression or enhancing transformation in complex medium (BHI), whereas sCSP only induces comX and competence in complex medium. Thus, it is likely that the effects of loss of the RSH enzyme are exerted through the ComRS pathway in a WT genetic background. Importantly, the behavior of the WT strain contrast dramatically with that of the (p)ppGpp0 strain, in which com gene expression was not similarly impacted. This difference in behavior is likely attributable to the fact that the (p)ppGpp0 strain completely lacks (p)ppGpp, whereas the WT and relA mutant have higher basal levels of (p)ppGpp due to a lack of (p)ppGpp hydrolase activity. Thus, com gene expression may only be influenced once (p)ppGpp production reaches a critical threshold and is not impacted when only low levels of (p)ppGpp are present, as is the case for the (p)ppGpp0 strain; highlighting further the critical role of (p)ppGpp in manifestation of these important phenotypes.
comR Expression is Affected by the Loss of RelA
To determine where in the competence signaling cascade the effects of deletion of relA were occurring, the expression of selected com genes was measured by RT-qPCR in cells growing in FMC and treated with 2 μM sXIP. Expression levels of comR were one-log higher in UA159 than in the strain lacking the RSH enzyme (ΔrelA), both in control (1% DMSO) samples and in cells treated with sXIP (Figure 4A). The levels of comR expression were not altered by exposure to XIP in either strain. A different pattern of expression was noted for comX (Figure 4B), comYA (Supplementary Figure S4A) and comS (Supplementary Figure S4B). In particular, expression of comX in the relA-deficient strain was modestly lower than in WT cells in the absence of sXIP treatment and about 1-log lower after treatment with XIP; a similar pattern was noted for comYA. It is significant, though, that induction of comX and comYA still occurs in response to sXIP treatment in the relA-deficient strain, albeit less so than in the WT genetic background. In contrast, the expression of comS, while induced by XIP in both UA159 and the ΔrelA mutant, did not differ between strains (Supplementary Figure S4B). Additionally, expression of comD and cipB were monitored and no differences were detected between UA159 and the ΔrelA strains in cells treated only with DMSO. However, when the ΔrelA strain was treated with sXIP there was no induction of comD and a lower level of induction of cipB, compared to that seen in the WT strain (Supplementary Figures S4C,D). Thus, the data are most consistent with the influences of Rel on competence arising from alterations in the expression of comR, although other inputs outside the ComRS–XIP system may modulate gene expression, sensitivity to XIP, and competence development.
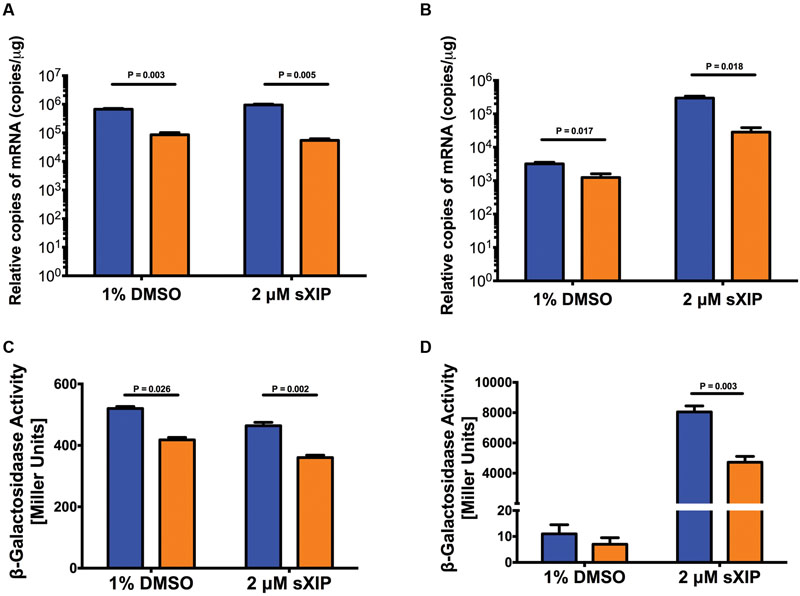
FIGURE 4. Impacts of loss of relA on com gene expression. Expression of comR (A) and comX (B) in S. mutans UA159 (blue bars) and its ΔrelA mutant derivative (orange bars) after addition of 2 μM sXIP. sXIP was added when OD600 nm reached 0.2. After 1 h of incubation, cells were harvested by centrifugation, RNA isolated, and qRT-PCR performed. Gene expression was normalized to 16S rRNA expression. Data represents three biological replicates measured in triplicate. Statistical analysis was performed by Student’s t-test. (C,D) Promoter activity from lacZ gene fusions to PcomR (C) or PcomX (D) in the wild-type (blue bars) and ΔrelA (orange bars) genetic backgrounds determined by β-galactosidase (LacZ) activity in cells grown in FMC and treated with either 1% DMSO or 2 μM sXIP. sXIP was added when OD600 nm reached 0.2. After an hour of incubation, cells were harvested by centrifugation and LacZ assays performed. Data represent three biological replicates assayed in triplicate. Statistical analysis was performed by Student’s t-test.
To examine if the changes in comR mRNA levels were correlated with increased comR promoter activity, PcomR, was fused to a Streptococcus salivarius lacZ gene and integrated in single copy into the chromosome of S. mutans at the mtlA-phn locus (Son et al., 2012; Guo et al., 2014). A significant decrease in LacZ activity was measured in the ΔrelA background, compared to the WT strain (Figure 4C), with or without addition of sXIP. A similar reduction in promoter activity in the relA-deficient strain was also observed using a PcomX-lacZ fusion (Figure 4D). Thus, deletion of relA can influence PcomR activity, with the down-regulation of comR likely accounting for diminished activation of PcomX.
Complementation Analysis of the ΔrelA Mutant
Previously, a complemented ΔrelA strain was constructed by providing an intact copy of relA gene in trans on plasmid pMSP3535, which allows for induction of the gene with subinhibitory concentrations of nisin (Lemos et al., 2004). To confirm that reductions in transformation efficiency and com gene expression were due to the loss of the Rel enzyme, the relevant phenotypes of the complemented strain (ΔrelA/relA+) were compared with those of the relA mutant. The ΔrelA/relA+ strain displayed similar transformation efficiency as the WT strain (1.4 × 10-3 versus 1.5 × 10-3, respectively), whereas a strain in which the relP gene was provided on a plasmid in the relA mutant background (ΔrelA/relP+) displayed transformation efficiencies equivalent to the ΔrelA strain (2.8 × 10-7 to 2.4 × 10-7, respectively; Figure 5A). qRT-PCR of selected com genes in the complemented strains revealed that the ΔrelA/relA+ strain had similar quantities of mRNA as the WT strain for comR (Figure 5B) and comX (Figure 5C), whereas com gene expression in the ΔrelA/relP+ strain remained comparable to that in the ΔrelA mutant. Thus, it was not sufficient to provide a plasmid-borne copy of a gene encoding only (p)ppGpp synthase activity (relP) in the relA mutant to achieve full complementation of RelA deficiency. Rather complementation of the RelA deficit was only achieved with an RSH enzyme containing both (p)ppGpp synthase and hydrolase activities (relA).
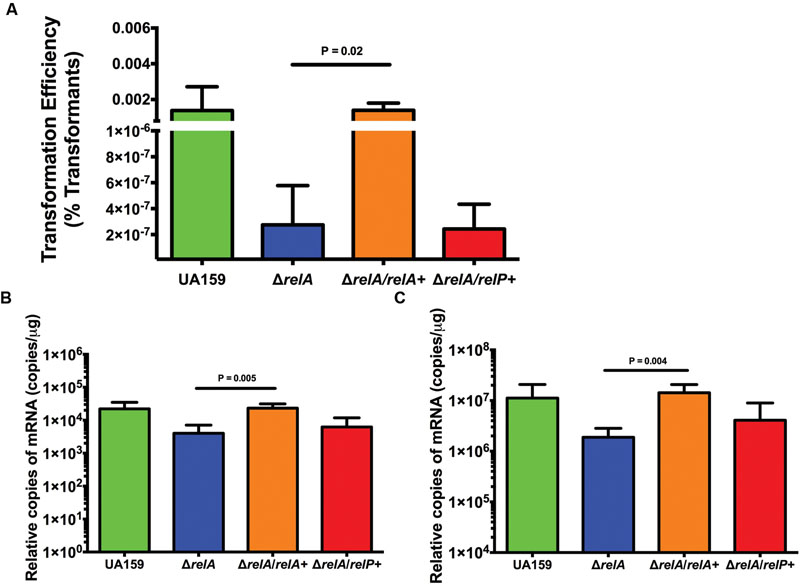
FIGURE 5. Complementation of the ΔrelA mutant with relA restores competence phenotypes. (A) Transformation efficiency between strains UA159 (green bar), ΔrelA (blue bar), relA+/ΔrelA (relA mutant complemented with relA; orange bar), and relP+/ΔrelA (relA mutant complemented with the gene for the small SAS RelP; red bar) in FMC medium with addition of 2 μM sXIP. sXIP and transforming DNA plasmid pDL278 (SpR) were added when OD600 nm reached 0.2. Serial dilutions of cultures were plated on BHI agar. Transformation efficiency was calculated by dividing the number of transformants by the total number of viable bacteria, then multiplying by 100. Data represent the average of three individual experiments conducted in triplicate. Statistical analysis was performed by Student’s t-test. (B,C) Measurement of mRNA by qRT-PCR of comR (B) or comX (C) in UA159 (green bars), ΔrelA (blue bars), relA+/ΔrelA (orange bars), and relP+/ΔrelA (red bars) after addition of 2 μM sXIP. sXIP was added when OD600 nm reached 0.2. After 1 h of incubation, cells were harvested by centrifugation, RNA isolated, and qRT-PCR performed. Gene expression was normalized to 16S rRNA expression. Data are derived from three biological replicates assayed in triplicate. Statistical analysis was performed by Student’s t-test.
While internal accumulation of CipB induced by CSP via the ComDE system is thought to be involved in altruistic cell death and fratricide in a sub-population of cells (Perry et al., 2009), XIP-mediated cell death in chemically defined medium requires the ComRS system (Wenderska et al., 2012). This is apparently attributable to a recently discovered feedback circuit in which ComX binds to the comE promoter and leads to enhanced activity of the ComE regulon (Reck et al., 2015; Son et al., 2015b). Since expression of comR was decreased in the ΔrelA strain, deficiencies in activation of comX by the ComRS system could be responsible for the observed changes in apparent growth inhibition by sXIP and transformation efficiencies. To explore this possible mechanistic linkage, we created strains that overexpressed comS, comR, and comX under the control of the P23 promoter on the shuttle plasmid pIB184 (Biswas et al., 2008) in the ΔrelA genetic background. Growth kinetic studies in response to sXIP were conducted and compared to the ΔrelA strain carrying only the pIB184 vector. Overexpression of comS elicited no changes in growth inhibition by sXIP (Figure 6A). Overexpression of comR in the relA mutant strain (184ComR/ΔrelA) lead to modest growth inhibition in the presence of 2 μM sXIP, with an increase in the doubling time to 109 ± 4 min and reduction in final OD to 0.83 ± 0.05 (Figure 6B). Greater inhibition was observed in the presence of 20 μM sXIP, with an increased lag time (3 h), a doubling time of 194 ± 8 min, and a decrease in final yield to 0.71 ± 0.06. The 184ComX/ΔrelA strain (overexpressing comX) showed the greatest growth inhibition in the presence of sXIP, with lag times of 24 and 29 h in the presence of 2 or 20 μM XIP, respectively (Figure 6C). Growth of the 184ComX/ΔrelA strain in 20 μM sXIP resulted in doubling times of 240 ± 4 min and a final yield of 0.70 ± 0.04 after 48 h, very close to that observed for the WT strain. Thus, the loss of sensitivity of the strain lacking the RelA (RSH) enzyme to sXIP is clearly due to the aberrant decrease in comX mRNA levels and ComX production and can be compensated by overexpression of comR and comX, but not of comS. These results also suggest that the competence deficiencies of ΔrelA stem from a lack of ComR, with enough ComR–XIP complexes present to fully saturate expression from the PcomS promoter, but not the PcomX promoter under similar conditions leading to deviations in transformation efficiency and cell lysis through ComX-dependent mechanisms.
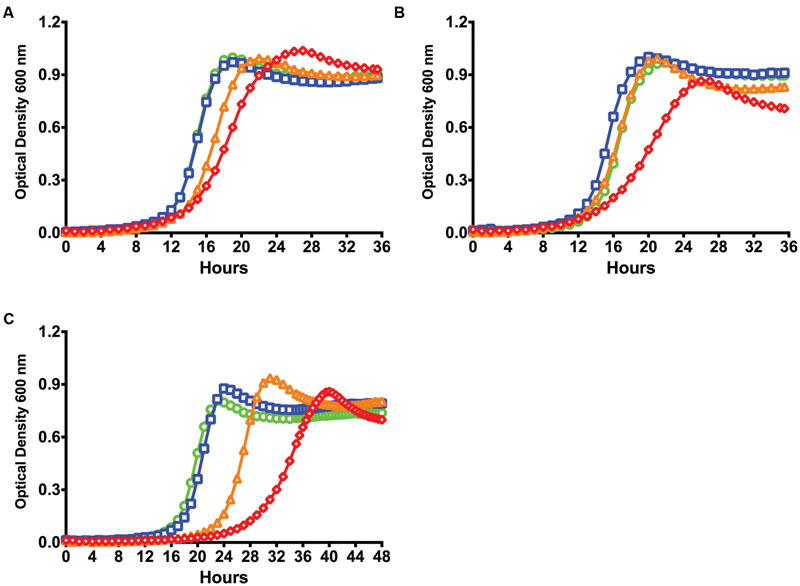
FIGURE 6. Overexpression of comR and comX in a ΔrelA genetic background overexpressing selected genes. Growth kinetics of strains (A) 184ComS/ΔrelA, (B) 184ComR/ΔrelA, and (C) 184ComX/ΔrelA with treatment of differing concentrations of sXIP (see text for more detail). Differing concentrations of sXIP were added to the culture medium: control of 1% DMSO (●; green), or 0.2 μM (■; blue), 2 μM (▲; orange), or 20 μM (◆; red) sXIP.
The Hydrolase Activity of RelA Is Essential for Effects on com Gene Expression
The RelA enzyme can synthesize and degrade (p)ppGpp, with the catalytic domains for these activities located in the N-terminal portion of the enzyme. Synthetase-ON/hydrolase-OFF and synthetase-OFF/hydrolase-ON states are controlled by allosteric regulation mediated by the C-terminal domain (Mechold et al., 1996, 2002). Analysis of the crystal structure of the N-terminal fragment (residues 1–385) of the Rel enzyme of Streptococcus dysgalactiae subsp. equisimilis (RelSeq) revealed amino acid residues critical for the activities of both domains, with mutation of specific amino acids leading to loss of activity in one domain without affecting the function of the other (Hogg et al., 2004). RelA of S. mutans shares 85% identity at the amino acid level with RelSeq, and residues found to play critical roles in the catalytic activity of RelSeq are conserved in S. mutans (data not shown). Amino acid substitution mutations in the RelSeq synthetase and hydrolase domains were recapitulated in the S. mutans RelA enzyme to determine if one or both enzymatic activities were critical for normal com gene expression. Inhibition of the synthetase function was achieved by mutating the aspartate residue at position 264 to glycine (D264G, relAΔSY N; Hogg et al., 2004). When grown with increasing concentrations of sXIP, this strain displayed growth inhibition similar to the WT strain (Figure 7A). Mutation of the catalytic site of the hydrolase domain was achieved by substituting a threonine residue with proline at position 151 (T151P; Hogg et al., 2004). Interestingly, viable colonies containing this single substitution could not be recovered, and DNA sequence analysis of transformants that did contain the desired mutation were found to have secondary mutations in the relA gene; either an additional mutation in the synthetase domain or a frameshift mutation that led to production of a truncated enzyme that was predicted to be non-functional. Thus, we were unable to obtain a relAΔHY D strain. These results suggest that strains of S. mutans in which the loss of function of the RelA (p)ppGpp hydrolase domain has occurred without concurrent loss of the synthetase domain are non-viable. Notably, the relAΔHY D mutation could also not be obtained in a strain lacking relPQ, so synthesis of (p)ppGpp by RelA and the associated over accumulation of alarmone in the absence of the hydrolase domain appears sufficient to account for lack of growth of an otherwise-WT relAΔHY D mutant. We were, however, able to incorporate the T151P mutation into the relAΔSY N strain to create a viable strain expressing a mutant derivative of RelA with no (p)ppGpp synthase or hydrolase activity (relAΔSY NΔHY D). When relAΔSY NΔHY D strain was evaluated for its sensitivity to XIP (Figure 7B), the strain displayed a level of resistance to XIP similar to the ΔrelA mutant, adding further support that loss of the hydrolase domain is responsible for the changes in XIP sensitivity. These observations are consistent with the previous findings (Figure 5) that addition of a synthetase only (relP) back to the ΔrelA strain is unable to complement the reduction in competence activity phenotypes. To further confirm the importance of the hydrolase domain in responses of S. mutans to XIP, we examined a strain (relA385) that includes an insertion of an antibiotic resistance cassette beginning at amino acid 386, resulting in a 385-aa peptide containing only the N-terminal domain of RelA. This strain produces high basal levels of (p)ppGpp (data not shown), with the produced peptide containing no detectable (p)ppGpp degrading activity (Mechold et al., 2002). The relA385 strain showed growth kinetics in the presence of increasing concentrations of sXIP similar to the ΔrelA and relAΔSY NΔHY D strains (Figure 7C). Transformation efficiency (Figure 7D) and ComX levels in each of these strains was consistent with the growth characteristics in the presence of XIP (Figure 7E). Taken together, these results show that the hydrolase domain of RelA plays an essential role in optimizing (p)ppGpp levels to allow for normal responses to XIP and expression of com genes.
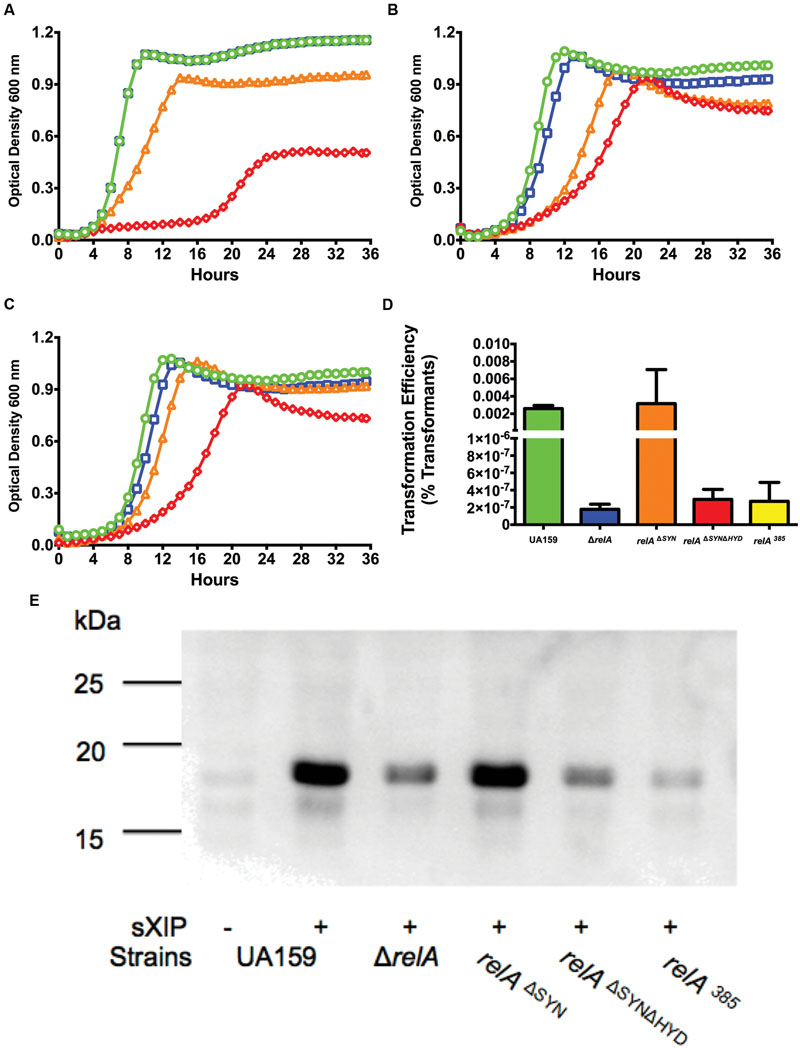
FIGURE 7. RelA hydrolase activity is critical for com gene expression. Growth curves of strains (A) relAΔSY N, (B) relAΔSY NΔHY D, and (C) relA385 treated with different concentrations of sXIP. Bacterial cultures were grown in the chemically defined medium FMC containing 1% DMSO (●; green), or 0.2 μM (■; blue), 2 μM (▲; orange), or 20 μM (◆; red) sXIP. (D) Transformation efficiency of UA159 (green bar), ΔrelA (blue bar), relAΔSY N (orange bar), relAΔSY NΔHY D (red bar), and relA385 (yellow bar) in FMC medium after treatment with 2 μM sXIP. sXIP and purified pDL278 (SpR) were added when OD600 nm reached 0.2. Cultures were diluted and plated, and transformation efficiency was calculated as detailed in Figure 3. Data are the average of three individual experiments conducted in triplicate. (E) Detection of ComX in the relA mutant grown in FMC with either 1% DMSO (-) or 2 μM sXIP (+) added at OD600 nm = 0.2 and then incubated for 1 h. ComX was detected as above, using the same molecular mass standards (in kDa).
Loss of RelA Results in Changes to the Phenotypes of ΔrcrR Strains
Two genes (tpx, cipI) separate the rcrRPQ and relPRS operon, all transcribed in the same direction. The products of rcrRPQ and relP profoundly and concurrently impact stress tolerance, (p)ppGpp production, and genetic competence in S. mutans (Seaton et al., 2011). Moreover, the MarR-type regulator RcrR was found to interact with the relP promoter in vitro (Seaton et al., 2015) and the levels of the ABC transporters RcrP and RcrQ and two small peptides encoded at the 3′ end of rcrQ exert dominant control over competence development (Ahn et al., 2014). Of particular relevance here, a ΔrcrR-P (polar) mutant is hyper-transformable, whereas a ΔrcrR-NP (non-polar) mutant is completely non-transformable, in part due to lack of expression of full-length comX mRNA and production of very low levels of ComX (Kaspar et al., 2015). As the phenotypes displayed by the RcrRPQ operon and (p)ppGpp are intertwined, we reasoned that RcrRPQ and its effectors could be a source for modulation of (p)ppGpp accumulation leading to changes in the transformable state of cells based on environmental signals/conditions. The extremely disparate competence phenotypes displayed by the ΔrcrR polar and non-polar strains provided us the opportunity to explore mechanisms related to RelA activity and its role in com gene expression, which would have been more difficult to analyze in the WT background. Hence, we examined the effects of deleting relA (ΔrelA) in the rcrR mutants. The transformation efficiency of the ΔrcrR-PΔrelA double mutant (Figure 8A) had reduced transformation compared with the ΔrcrR-P mutant (6.8 × 10-4 versus 1.2 × 10-2, respectively). A decrease in ComX protein abundance was also observed in this double mutant (Figure 8B). Interestingly, a complete reversal of the non-transformable phenotype of the ΔrcrR-NP strain was evident when the ΔrelA mutation was present in the same strain. In particular, the ΔrcrR-NP strain alone could not be transformed, but the ΔrcrR-NPΔrelA double mutant displayed transformation efficiency comparable to the hyper-transformable ΔrcrR-P strain (1.3 × 10-2), and ComX levels were also restored to a level similar to those expressed in the ΔrcrR-P strain. Collectively, these data provide further confirmation of the critically important roles of the RelA enzyme and (p)ppGpp in modulation of genetic competence and peptide signaling behaviors in S. mutans.
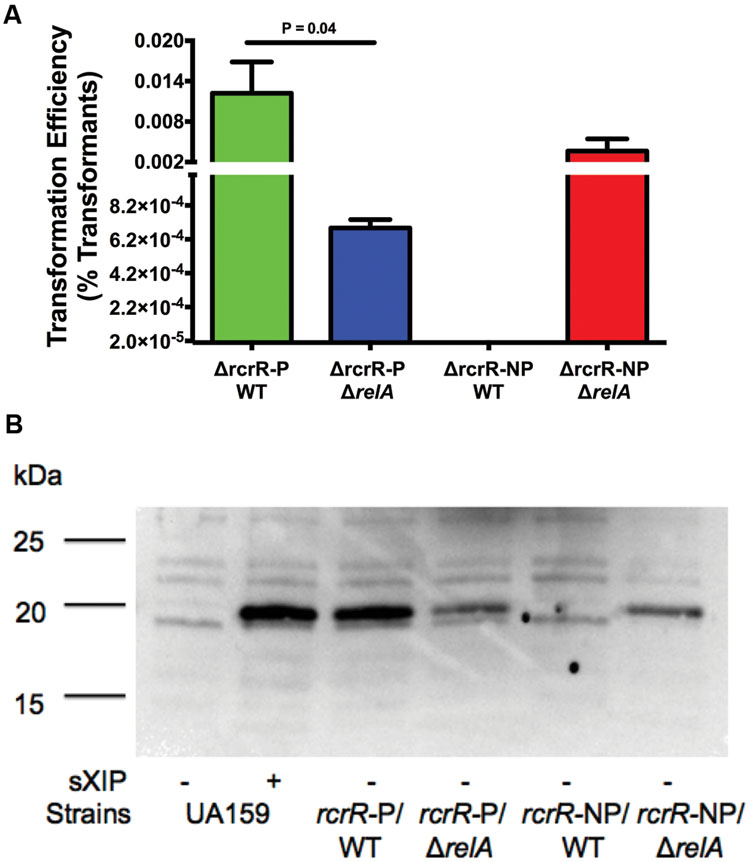
FIGURE 8. Effects of RelA on the phenotypes of ΔrcrR stains. (A) Transformation efficiency of strains ΔrcrR-P (green bar), ΔrcrR-PΔrelA (blue bar), ΔrcrR-NP (no bar because it is completely non-transformable), and ΔrcrR-NPΔrelA (red bar) in FMC medium with addition of 2 μM sXIP. sXIP and transforming DNA plasmid pDL278 (SpR) were added when OD600 nm = 0.2. Cultures were plated and transformation efficiency was determined as above. Data are the average of three individual experiments conducted in triplicate. (B) Detection of ComX in FMC with either 1% DMSO (-) or 2 μM sXIP (+) added at OD600 nm = 0.2 and then incubated for 1 h. ComX was detected as above with the same molecular mass standards (in kDa).
Discussion
Many of the organisms that make up the microbiome of the oral cavity do not survive outside of their host (Lemos et al., 2013). These bacteria have adapted to thrive in specific habitats in the oral cavity and employ substantial phenotypic plasticity to optimize growth in their continually fluctuating environments. To achieve this, the organisms utilize complex regulatory networks to control gene expression in concert with allosteric modulation of enzyme activities to respond to numerous stimuli. From a variety of studies, it appears that S. mutans has evolved in a way that it primarily utilizes a set of global transcriptional regulators, conserved stress-response pathways, monitoring of metabolic status, and carbohydrate transporters as the dominant mechanisms to interpret its environment and make adjustments to its gene expression patterns, physiologic status, and virulence capacities. In this study, we investigated and discovered a novel linkage in S. mutans of gene products that participate in the stringent response and control of growth rate (Lemos et al., 2007) via modulation of (p)ppGpp levels with the regulatory network for genetic competence for natural transformation; the latter benefitting the organism from a nutritional standpoint and by enhancing its genomic diversity.
From the data presented here, we can begin to develop a working model for the connection of (p)ppGpp to the development of genetic competence. In this model, (p)ppGpp concentrations within individual cells within a population serves as a source for “noise” (Lidstrom and Konopka, 2010), creating variance through fluctuations in com gene expression, with comR promoter activity acting as a primary control point (Figure 4C). A similar type of regulatory switch has been documented in E. coli (Maisonneuve et al., 2013), where variations in (p)ppGpp pools within a population lead to a stochastic response in expression of toxin–antitoxin modules, giving rise to a persister population due to growth arrest. A similar phenomenon may govern competence and stress tolerance behaviors in S. mutans, where individual cells must choose to commit to competence development through activation of the comRS pathway, and ultimately activate comX expression, or remain non-competent. A bimodal switch (sub-population response) for comX expression is observed in S. mutans cells growing in a chemically complex medium such as BHI, with proposed autofeedback regulation occurring at the level of ComRS (Son et al., 2012). Through use of promoter fusion strains and quantification of mRNAs and ComX protein, we show here that down-regulation in PcomR activity in a relA mutant can cause a significant drop in comR mRNA, with an associated decrease in ComX levels. These changes should have a direct effect on transformation efficiency of the population, as well as on the proportion of cells that may display growth inhibition or undergo lysis after exposure to peptide signals, consistent with results presented in this communication.
We also propose that an “optimal window” exists for intracellular (p)ppGpp concentrations that allows cells to proceed into the competent state via activation of com gene expression. The existence of such a “window” can be seen in the different behaviors of rel mutant strains, primarily with a relA mutant that accumulates higher basal levels of (p)ppGpp due to loss of hydrolase activity, and a strain lacking the SAS enzymes (ΔrelPQ), which accumulates low basal levels of (p)ppGpp. The relA mutant strain showed a high degree of resistance to the growth inhibition by XIP (Figure 3A), whereas the ΔrelPQ double mutant was very sensitive to 2 μM sXIP (Figure 3B). Thus, the internal concentrations of (p)ppGpp correlate with growth sensitivity to the sXIP peptide, with higher basal levels of (p)ppGpp favoring resistance to the signal and lower levels of (p)ppGpp leading to an increased response. Once outside of the optimal window, cells could potentially change their behavior toward the signal altogether. Such behavior is apparent in the (p)ppGpp0 strain, which is devoid of alarmone and is completely resistant to the apparent growth inhibitory effects of sXIP (Figure 1B). Additionally, after cells reach a high concentration threshold of (p)ppGpp accumulation, com gene expression can be effected. One must also take into account the growth rates of the strains in medium absent XIP to account for their observed behaviors in the presence of sXIP peptide. Specifically, the ΔrelA strain displays a doubling time that is 40 min greater than the WT strain in FMC medium alone, likely due to accumulation of (p)ppGpp in the absence of the hydrolase activity of RelA, whereas relP and/or relQ mutant strains have doubling times comparable to UA159 (Lemos et al., 2007). Thus, the changes in the behavior of the rel mutant strains in response to exogenous XIP is even more profound than if the growth rates of the relA mutant was more similar to those of relP and relQ mutants.
One aspect of our working model (Figure 9) that is not yet well developed is how RcrRPQ and the effector peptides encoded at the end of relQ influence (p)ppGpp pools and, ultimately, competence development. The rcrRPQ operon and (p)ppGpp are tightly connected in their regulation, as previously shown by Seaton et al. (2011). In their study, mutation of the SAS-encoding gene relP resulted in changes in rcrR promoter activity and rcrRPQ expression, which ultimately impacted the expression levels of the RcrPQ ABC-transporters and peptides encoded at the end of rcrQ (Ahn et al., 2014). Importantly, it has been shown that very small fluctuations in the activity of the rcrR promoter or levels of rcrRPQ transcripts can have profound effects on competence-related behaviors (Seaton et al., 2015). While the exact mechanism for how (p)ppGpp and RcrRPQ interact to exert these effects on competence have yet to be disclosed, the significance of this connection is strengthened by data presented here showing that deletion of relA in a ΔrcrR-NP strain completely reverses the non-transformable phenotype (Figure 8). We found previously that the non-transformability of ΔrcrR-NP is accompanied by, and largely attributable to, the loss of a full-length comX mRNA and ComX protein along with activation of expression of an ORF intragenic to comX termed XrpA (Kaspar et al., 2015). When point mutations are introduced to prevent production of XrpA while keeping comX intact in a ΔrcrR-NP background, full-length comX expression is restored, ComX protein reappears and transformability is on par with the rcrR-P hyper-transformable strain (Kaspar et al., 2015). Interestingly, virtually identical changes occur when relA is deleted or RelA enzymatic activity is ablated in the ΔrcrR-NP background. Thus, control of (p)ppGpp levels by the RelA hydrolase play a key role in allowing the cell to determine whether a full-length comX transcript can be produced or whether the transcript will be processed and high levels of the xrpA-specific transcript produced. While the exact mechanism by which (p)ppGpp levels influence this decision remain to be established, the known roles of these alarmones in cellular physiology and gene expression provide evidence that the cells can use (p)ppGpp to integrate decisions to become competent or to select programmed cell death pathways with assessment of the physiologic status of individual cells. In this context, further exploration of how (p)ppGpp levels influence comR transcription, whether (p)ppGpp is an allosteric regulator of RcrR, and if there are interactions between alarmone and the RcrPQ transporters and effector peptides is planned.
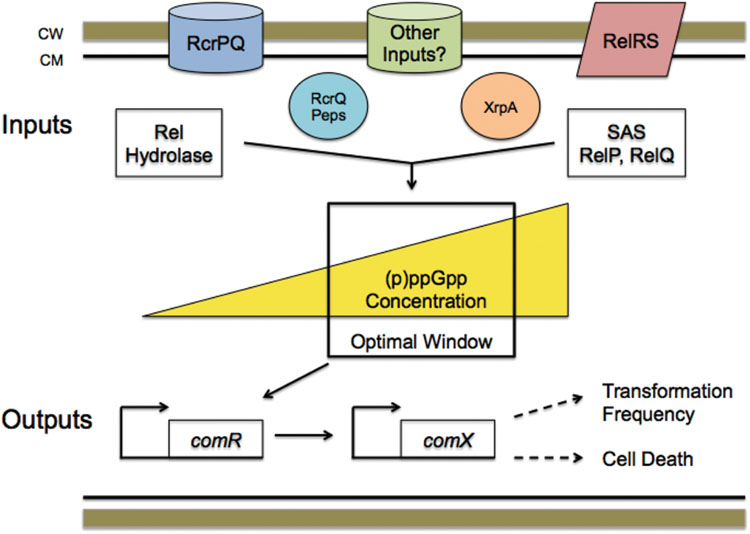
FIGURE 9. Working model integrating (p)ppGpp levels with the development of competence in S. mutans. The model predicts that (p)ppGpp levels play an essential role in determining cell fate after competence gene activation. Numerous inputs that monitor environmental conditions either through cell density or by serving as stress sensors, including RelRS, RcrPQ, and others, e.g., CiaRH and ScnRK, along with the RcrQ peptides (labeled “peps”) and XrpA, all modulate internal (p)ppGpp accumulation via the activity of the RelA hydrolase or the small alarmone synthetases RelP and RelQ. (p)ppGpp fluctuations then serve as a source of “noise” or stochasticity that drives heterogeneity within the population. The internal (p)ppGpp concentration influences PcomR activity, directly or indirectly, with consequential influences on PcomX promoter activity. An “optimal window” in (p)ppGpp concentrations is necessary to achieve “normal” com gene expression responses to peptides. (p)ppGpp levels less than or greater than those in the “optimal window” result in changes in responses to signal inputs that alter com gene expression. Although not shown, ComX stability is also a determining factor for competence and cell death. Thus, shifts in ComX levels and activity, which are controlled transcriptionally and post-transcriptionally, dictate the ratio of cells undergoing transformation or cell death within a population of cells. In this way, (p)ppGpp levels modulated by RelA hydrolase activity allow cells to integrate physiologic status with commitments to signal response, progression to the competent state, or cell death. Other factors, like toxin:antitoxin modules, likely contribute to this decision network in a manner that also depends on (p)ppGpp levels.
We were also able to detect changes in (p)ppGpp accumulation after addition of sXIP to cultures of S. mutans. These changes could be observed 40 min after sXIP addition, and with the greatest effects coming with greater than 1 μM sXIP added to the cultures (Figure 2). It is known that S. mutans displays the greatest growth inhibitory defects in the presence of >1 μM sXIP peptide (Wenderska et al., 2012). One could thus propose that the reduced growth rate exhibited by S. mutans in response to sXIP at these concentrations could be due, at least in part, to the increased accumulation of (p)ppGpp slowing the overall growth rate of the organism.
In Bacillus subtilis, the ComGA protein, an apparent ortholog of ComYA in S. mutans, participates in growth arrest during the K-state by inhibiting replisome assembly in a (p)ppGpp-independent manner. However, ComGA can also interact directly with RelA to increase the pool of (p)ppGpp in cells, inhibiting replication and rRNA synthesis (Haijema et al., 2001; Hahn et al., 2015). The coupling of growth arrest to transformation may allow for cells to overcome the fitness burdens of production of competence proteins and internalization of DNA, while concurrently creating sufficient opportunity for repair of the genome associated with recombination events that occur after DNA uptake (Briley Jr et al., 2011). In this study, we found the increase of (p)ppGpp accumulation upon addition of sXIP is most likely catalyzed by either RelP or RelQ, and not by RelA. When measured by real-time PCR, we were able to detect a 1.89-fold increase in relQ expression, with a concurrent 2.79-fold decrease in relA, whereas relP mRNA levels remained unchanged (Figure 2B). Instead, we measured a significant increase in relQ expression. One possible explanation for the observed changes in relQ expression is that it is co-transcribed with the gene (pta) for phosphotransacetylase, which provides acetyl phosphate that can be used for ATP generation (Kim et al., 2015). It is reasonable to expect that the stress induced by exposure to high levels of XIP could alter metabolism in a way that directs cells to slow growth via RelQ-dependent (p)ppGpp synthesis, while shifting from homofermentative metabolism to acetate production, which yields an additional ATP. And, as proposed above, fluctuations in (p)ppGpp pools as a result of physiologic adaptations when signal molecules are present may help to drive stochastic events that direct segregation of cells into subpopulations that maintain normal growth, undergo autolysis or commit to late competence gene expression (Lemme et al., 2011).
The competence development pathways of S. mutans have been shown to be unusually complex, with additional layers of complexity of the system being revealed as more mutants are evaluated and environmental variables are explored (Ahn et al., 2014; Guo et al., 2014; Kaspar et al., 2015; Son et al., 2015a). Perhaps, though, it is not surprising that the spectrum of variables and inputs governing the competence system, which include RcrRPQ, multiple unique peptides (XrpA, rcrQ-associated peptides) and three (p)ppGpp synthases, is so elaborate in this opportunistic pathogen. Were S. mutans to engage in the activation of the competence cascade in response to peptide signals, with the associated expense of energy to produce and deploy the machinery for DNA uptake and processing, irrespective of physiologic status of the cells, then the fitness of the population could be severely compromised. Instead, a picture is emerging that (p)ppGpp serves a critical function in allowing the cells to distill a diverse set of physiologic parameters (nutrient availability, pH, redox, protein, or envelope damage) when signal peptides reach some critical level to govern genetic responses at the single cell level in a way that is optimal for survival of individual cells and beneficial for the local population of S. mutans. It is also notable that the virulence of S. mutans is so strongly tied to its physiology and metabolism; i.e., damage to the tooth requires acid production through glycolysis and an elaborate regulon that allows for constitutive and adaptive acid tolerance. Perhaps this is why the stress tolerance regulon and genetic competence in this organism are so tightly tied to the organism’s ability to express critical virulence related phenotypes (biofilm formation, acid and oxidative stress tolerance). Thus, dissection of the regulatory network described here, which consists of many effector molecules (e.g., XrpA) that appear to be unique to S. mutans, has tremendous potential for guiding the development of strategies to reduce the incidence of disease caused by this opportunistic pathogen. Future studies will be targeted at how (p)ppGpp, peptide effector molecules, and regulatory proteins interact to collect inputs and govern the behavior of individual cells and populations of S. mutans.
Author Contributions
JK contributed to conception, design, data acquisition and analysis, data interpretation, drafted and critically revised the manuscript; JNK contributed to the design, data acquisition and analysis, data interpretation, and critically revised the manuscript; S-JA contributed to conception, design, data interpretation and critically revised the manuscript; RB contributed to conception, design, data interpretation, drafted and critically revised the manuscript. All authors gave final approval to the manuscript and agree to be accountable for all aspects of the work.
Funding
Research reported in this publication was supported by the National Institute of Dental and Craniofacial Research of the National Institutes of Health under Award Numbers R01 DE13239, T90 DE21990, and F31 DE024416.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Kinda Seaton for technical assistance and advice with experiments reported here. We also thank members of the Burne and Hagen lab, including Robert Shields and Minjun Son, with helpful discussions during the preparation of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01162
Footnotes
References
Abranches, J., Candella, M. M., Wen, Z. T., Baker, H. V., and Burne, R. A. (2006). Different roles of EIIABMan and EIIGlc in regulation of energy metabolism, biofilm development, and competence in Streptococcus mutans. J. Bacteriol. 188, 3748–3756. doi: 10.1128/JB.00169-06
Ahn, S.-J., Kaspar, J., Kim, J. N., Seaton, K., and Burne, R. A. (2014). Discovery of novel peptides regulating competence development in Streptococcus mutans. J. Bacteriol. 196, 3735–3745. doi: 10.1128/JB.01942-14
Ahn, S.-J., Lemos, J. A., and Burne, R. A. (2005). Role of HtrA in growth and competence of Streptococcus mutans UA159. J. Bacteriol. 187, 3028–3038. doi: 10.1128/JB.187.9.3028-3038.2005
Atkinson, G. C., Tenson, T., and Hauryliuk, V. (2011). The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS ONE 6:e23479. doi: 10.1371/journal.pone.0023479
Banu, L. D., Conrads, G., Rehrauer, H., Hussain, H., Allan, E., and van der Ploeg, J. R. (2010). The Streptococcus mutans serine/threonine kinase, PknB, regulates competence development, bacteriocin production, and cell wall metabolism. Infect. Immun. 78, 2209–20. doi: 10.1128/IAI.01167-09
Biswas, I., Jha, J. K., and Fromm, N. (2008). Shuttle expression plasmids for genetic studies in Streptococcus mutans. Microbiology 154, 2275–2282. doi: 10.1099/mic.0.2008/019265-0
Briley, K. Jr., Prepiak, P., Dias, M. J., Hahn, J., and Dubnau, D. (2011). Maf acts downstream of ComGA to arrest cell division in competent cells of B. subtilis. Mol. Microbiol. 81, 23–39. doi: 10.1111/j.1365-2958.2011.07695.x
Bryan, E. M., Bae, T., Kleerebezem, M., and Dunny, G. M. (2000). Improved vectors for nisin-controlled expression in gram-positive bacteria. Plasmid 44, 183–190. doi: 10.1006/plas.2000.1484
Carneiro, S., Lourenço, A., Ferreira, E. C., and Rocha, I. (2011). Stringent response of Escherichia coli: revisiting the bibliome using literature mining. Microb. Inform. Exp. 1:14. doi: 10.1186/2042-5783-1-14
Cha, R. S., Zarbl, H., Keohavong, P., and Thilly, W. G. (1992). Mismatch amplification mutation assay (MAMA): application to the c-H-ras gene. Genome Res. 2, 14–20. doi: 10.1101/gr.2.1.14
Cosloy, S. D., and Oishi, M. (1973). Genetic transformation in Escherichia coli K12. Proc. Natl. Acad. Sci. U.S.A. 70, 84–87. doi: 10.1073/pnas.70.1.84
Cvitkovitch, D. G. (2001). Genetic competence and transformation in oral streptococci. Crit. Rev. Oral Biol. Med. 12, 217–243. doi: 10.1177/10454411010120030201
Dalebroux, Z. D., and Swanson, M. S. (2012). ppGpp: magic beyond RNA polymerase. Nat. Rev. Microbiol. 10, 203–212. doi: 10.1038/nrmicro2720
Desai, K., Mashburn-Warren, L., Federle, M. J., and Morrison, D. A. (2012). Development of competence for genetic transformation of Streptococcus mutans in a chemically defined medium. J. Bacteriol. 194, 3774–3780. doi: 10.1128/JB.00337-12
Fontaine, L., Boutry, C., de Frahan, M. H., Delplace, B., Fremaux, C., Horvath, P., et al. (2009). A novel pheromone quorum-sensing system controls the development of natural competence in Streptococcus thermophilus and Streptococcus salivarius. J. Bacteriol. 192, 1444–1454. doi: 10.1128/JB.01251-09
Fontaine, L., Goffin, P., Dubout, H., Delplace, B., Baulard, A., Lecat-Guillet, N., et al. (2013). Mechanism of competence activation by the ComRS signalling system in streptococci. Mol. Microbiol. 87, 1113–1132. doi: 10.1111/mmi.12157
Guo, Q., Ahn, S.-J., Kaspar, J., Zhou, X., and Burne, R. A. (2014). Growth phase and pH influence peptide signaling for competence development in Streptococcus mutans. J. Bacteriol. 196, 227–236. doi: 10.1128/JB.00995-13
Hahn, J., Tanner, A. W., Carabetta, V. J., Cristea, I. M., and Dubnau, D. (2015). ComGA-RelA interaction and persistence in the Bacillus subtilis K-state. Mol. Microbiol. 97, 454–471. doi: 10.1111/mmi.13040
Haijema, B.-J., Hahn, J., Haynes, J., and Dubnau, D. (2001). A ComGA-dependent checkpoint limits growth during the escape from competence. Mol. Microbiol. 40, 52–64. doi: 10.1046/j.1365-2958.2001.02363.x
Ho, S. N., Hunt, H. D., Horton, R. M., Pullen, J. K., and Pease, L. R. (1989). Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59. doi: 10.1016/0378-1119(89)90358-2
Hogg, T., Mechold, U., Malke, H., Cashel, M., and Hilgenfeld, R. (2004). Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response. Cell 117, 57–68. doi: 10.1016/S0092-8674(04)00260-0
Hung, D. C. I., Downey, J. S., Ayala, E. A., Kreth, J., Mair, R., Senadheera, D. B., et al. (2011). Characterization of DNA binding sites of the ComE response regulator from Streptococcus mutans. J. Bacteriol. 193, 3642–3652. doi: 10.1128/JB.00155-11
Jenkinson, H. F. (2011). Beyond the oral microbiome. Environ. Microbiol. 13, 3077–3087. doi: 10.1111/j.1462-2920.2011.02573.x
Kajfasz, J. K., Abranches, J., and Lemos, J. A. (2011). Transcriptome analysis reveals that ClpXP proteolysis controls key virulence properties of Streptococcus mutans. Microbiology 157, 2880–2890. doi: 10.1099/mic.0.052407-0
Kaspar, J., Ahn, S.-J., Palmer, S. R., Choi, S. C., Stanhope, M. J., and Burne, R. A. (2015). A unique ORF within the comX gene of Streptococcus mutans regulates genetic competence and oxidative stress tolerance. Mol. Microbiol. 96, 463–482. doi: 10.1111/mmi.12948
Khan, R., Rukke, H. V., Høvik, H., Åmdal, H. A., Chen, T., Morrison, D. A., et al. (2016). Comprehensive transcriptome profiles of Streptococcus mutans UA159 map core streptococcal competence genes. mSystems 1:e00038-15. doi: 10.1128/mSystems.00038-15
Khan, R., Rukke, H. V., Ricomini Filho, A. P., Fimland, G., Arntzen, M. Ø., Thiede, B., et al. (2012). Extracellular identification of a processed type II ComR/ComS pheromone of Streptococcus mutans. J. Bacteriol. 194, 3781–3788. doi: 10.1128/JB.00624-12
Kim, J. N., Ahn, S.-J., and Burne, R. A. (2015). Genetics and physiology of acetate metabolism by the Pta-Ack pathway of Streptococcus mutans. Appl. Environ. Microbiol. 81, 5015–5025. doi: 10.1128/AEM.01160-15
Kim, J. N., Ahn, S.-J., Seaton, K., Garrett, S., and Burne, R. A. (2012). Transcriptional organization and physiological contributions of the relQ operon of Streptococcus mutans. J. Bacteriol. 194, 1968–1978. doi: 10.1128/JB.00037-12
Kim, J. N., Stanhope, M. J., and Burne, R. A. (2013). Core-gene-encoded peptide regulating virulence-associated traits in Streptococcus mutans. J. Bacteriol. 195, 2912–2920. doi: 10.1128/JB.00189-13
Lau, P. C., Sung, C. K., Lee, J. H., Morrison, D. A., and Cvitkovitch, D. G. (2002). PCR ligation mutagenesis in transformable streptococci: application and efficiency. J. Microbiol. Methods 49, 193–205. doi: 10.1016/S0167-7012(01)00369-4
LeBlanc, D. J., Lee, L. N., and Abu-Al-Jaibat, A. (1992). Molecular, genetic, and functional analysis of the basic replicon of pVA380-1, a plasmid of oral streptococcal origin. Plasmid 28, 130–145. doi: 10.1016/0147-619X(92)90044-B
Lemme, A., Gröbe, L., Reck, M., Tomasch, J., and Wagner-Döbler, I. (2011). Subpopulation-specific transcriptome analysis of competence-stimulating-peptide-induced Streptococcus mutans. J. Bacteriol. 193, 1863–1877. doi: 10.1128/JB.01363-10
Lemos, J. A., and Abranches, J. (2005). Responses of cariogenic streptococci to environmental stresses. Curr. Issues Mol. Biol. 7, 95–108.
Lemos, J. A., Brown, T. A., and Burne, R. A. (2004). Effects of RelA on key virulence properties of planktonic and biofilm populations of Streptococcus mutans. Infect. Immun. 72, 1431–1440. doi: 10.1128/IAI.72.3.1431-1440.2004
Lemos, J. A., and Burne, R. A. (2008). A model of efficiency: stress tolerance by Streptococcus mutans. Microbiology 154, 3247–3255. doi: 10.1099/mic.0.2008/023770-0
Lemos, J. A., Lin, V. K., Nascimento, M. M., Abranches, J., and Burne, R. A. (2007). Three gene products govern (p)ppGpp production by Streptococcus mutans. Mol. Microbiol. 65, 1568–1581. doi: 10.1111/j.1365-2958.2007.05897.x
Lemos, J. A., Quivey, R. G. Jr., Koo, H., and Abranches, J. (2013). Streptococcus mutans: a new Gram-positive paradigm? Microbiology 159, 436–445. doi: 10.1099/mic.0.066134-0
Lidstrom, M. E., and Konopka, M. C. (2010). The role of physiological heterogeneity in microbial population behavior. Nat. Chem. Biol. 6, 705–712. doi: 10.1038/nchembio.436
Mair, R. W., Senadheera, D. B., and Cvitkovitch, D. G. (2012). CinA is regulated via ComX to modulate genetic transformation and cell viability in Streptococcus mutans. FEMS Microbiol. Lett. 331, 44–52. doi: 10.1111/j.1574-6968.2012.02550.x
Maisonneuve, E., Castro-Camargo, M., and Gerdes, K. (2013). (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell 154, 1140–1150. doi: 10.1016/j.cell.2013.07.048
Mashburn-Warren, L., Morrison, D. A., and Federle, M. J. (2010). A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp. via an Rgg regulator. Mol. Microbiol. 78, 589–606. doi: 10.1111/j.1365-2958.2010.07361.x
Mechold, U., Cashel, M., Steiner, K., Gentry, D., and Malke, H. (1996). Functional analysis of a relA/spoT gene homolog from Streptococcus equisimilis. J. Bacteriol. 178, 1401–1411.
Mechold, U., Murphy, H., Brown, L., and Cashel, M. (2002). Intramolecular regulation of the opposing (p)ppGpp catalytic activities of RelSeq, the Rel/Spo enzyme from Streptococcus equisimilis. J. Bacteriol. 184, 2878–2888. doi: 10.1128/JB.184.11.2878-2888.2002
Merritt, J., Kreth, J., Shi, W., and Qi, F. (2005a). LuxS controls bacteriocin production in Streptococcus mutans through a novel regulatory component. Mol. Microbiol. 57, 960–969. doi: 10.1111/j.1365-2958.2005.04733.x
Merritt, J., Qi, F., and Shi, W. (2005b). A unique nine-gene comY operon in Streptococcus mutans. Microbiology 151, 157–66. doi: 10.1099/mic.0.27554-0
Nascimento, M. M., Lemos, J. A., Abranches, J., Lin, V. K., and Burne, R. A. (2008). Role of RelA of Streptococcus mutans in global control of gene expression. J. Bacteriol. 190, 28–36. doi: 10.1128/JB.01395-07
Niu, G., Okinaga, T., Zhu, L., Banas, J., Qi, F., and Merritt, J. (2008). Characterization of irvR, a novel regulator of the irvA-dependent pathway required for genetic competence and dextran-dependent aggregation in Streptococcus mutans. J. Bacteriol. 190, 7268–7274. doi: 10.1128/JB.00967-08
Okinaga, T., Niu, G., Xie, Z., Qi, F., and Merritt, J. (2010). The hdrRM operon of Streptococcus mutans encodes a novel regulatory system for coordinated competence development and bacteriocin production. J. Bacteriol. 192, 1844–1852. doi: 10.1128/JB.01667-09
Perry, J. A., Jones, M. B., Peterson, S. N., Cvitkovitch, D. G., and Lévesque, C. M. (2009). Peptide alarmone signalling triggers an auto-active bacteriocin necessary for genetic competence. Mol. Microbiol. 72, 905–917. doi: 10.1111/j.1365-2958.2009.06693.x
Reck, M., Tomasch, J., and Wagner-Döbler, I. (2015). The alternative sigma factor SigX controls bacteriocin synthesis and competence, the two quorum sensing regulated traits in Streptococcus mutans. PLoS Genet. 11:e1005353. doi: 10.1371/journal.pgen.1005353
Seaton, K., Ahn, S.-J., and Burne, R. A. (2015). Regulation of competence and gene expression in Streptococcus mutans by the RcrR transcriptional regulator. Mol. Oral Microbiol. 30, 147–159. doi: 10.1111/omi.12079
Seaton, K., Ahn, S.-J., Sagstetter, A. M., and Burne, R. A. (2011). A transcriptional regulator and ABC transporters link stress tolerance, (p)ppGpp, and genetic competence in Streptococcus mutans. J. Bacteriol. 193, 862–874. doi: 10.1128/JB.01257-10
Senadheera, D. B., Cordova, M., Ayala, E. A., Chávez de Paz, L. E., Singh, K., Downey, J. S., et al. (2012). Regulation of bacteriocin production and cell death by the VicRK signaling system in Streptococcus mutans. J. Bacteriol. 194, 1307–1316. doi: 10.1128/JB.06071-11
Senadheera, M. D., Guggenheim, B., Spatafora, G. A., Huang, Y.-C., Choi, J., Hung, D. C., et al. (2005). A VicRK signal transduction system in Streptococcus mutans affects gtfBCD, gbpB, and ftf expression, biofilm formation, and genetic competence development. J. Bacteriol. 187, 4064–4076. doi: 10.1128/JB.187.12.4064-4076.2005
Son, M., Ahn, S.-J., Guo, Q., Burne, R. A., and Hagen, S. J. (2012). Microfluidic study of competence regulation in Streptococcus mutans: environmental inputs modulate bimodal and unimodal expression of comX. Mol. Microbiol. 86, 258–272. doi: 10.1111/j.1365-2958.2012.08187.x
Son, M., Ghoreishi, D., Ahn, S.-J., Burne, R. A., and Hagen, S. J. (2015a). Sharply tuned pH response of genetic competence regulation in Streptococcus mutans: a microfluidic study of the environmental sensitivity of comX. Appl. Environ. Microbiol. 81, 5622–5631. doi: 10.1128/AEM.01421-15
Son, M., Shields, R. C., Ahn, S.-J., Burne, R. A., and Hagen, S. J. (2015b). Bidirectional signaling in the competence regulatory pathway of Streptococcus mutans. FEMS Microbiol. Lett. 362:fnv159. doi: 10.1093/femsle/fnv159
Takahashi, N., and Nyvad, B. (2011). The role of bacteria in the caries process: ecological perspectives. J. Dent. Res. 90, 294–303. doi: 10.1177/0022034510379602
Terleckyj, B., and Shockman, G. D. (1975). Amino acid requirements of Streptococcus mutans and other oral streptococci. Infect. Immun. 11, 656–664.
Wenderska, I. B., Lukenda, N., Cordova, M., Magarvey, N., Cvitkovitch, D. G., and Senadheera, D. B. (2012). A novel function for the competence inducing peptide, XIP, as a cell death effector of Streptococcus mutans. FEMS Microbiol. Lett. 336, 104–112. doi: 10.1111/j.1574-6968.2012.02660.x
Keywords: competence, biofilms, comX, stringent response, (p)ppGpp, dental caries
Citation: Kaspar J, Kim JN, Ahn S-J and Burne RA (2016) An Essential Role for (p)ppGpp in the Integration of Stress Tolerance, Peptide Signaling, and Competence Development in Streptococcus mutans. Front. Microbiol. 7:1162. doi: 10.3389/fmicb.2016.01162
Received: 02 June 2016; Accepted: 12 July 2016;
Published: 28 July 2016.
Edited by:
Ivan Mijakovic, Chalmers University of Technology, SwedenReviewed by:
Maureen Wright, US Department of Agriculture, USAAkos T. Kovacs, University of Jena, Germany
Copyright © 2016 Kaspar, Kim, Ahn and Burne. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Robert A. Burne, cmJ1cm5lQGRlbnRhbC51ZmwuZWR1
 Justin Kaspar
Justin Kaspar Jeong N. Kim
Jeong N. Kim Sang-Joon Ahn
Sang-Joon Ahn Robert A. Burne
Robert A. Burne