- 1Laboratory of Molecular Biology, Department of Biotechnology, Burdwan University, Burdwan, India
- 2Department of Biotechnology, Haldia Institute of Technology, Haldia, India
The gram negative bacterium Stenotrophomonas is rapidly evolving as a nosocomial pathogen in immuno-compromised patients. Treatment of Stenotrophomonas maltophilia infections is problematic because of their increasing resistance to multiple antibiotics. This article aims to review the multi-disciplinary role of Stenotrophomonas in our environment with special focus on their metabolic and genetic potential in relation to bioremediation and phytoremediation. Current and emerging treatments and diagnosis for patients infected with S. maltophilia are discussed besides their capability of production of novel bioactive compounds. The plant growth promoting characteristics of this bacterium has been considered with special reference to secondary metabolite production. Nano-particle synthesis by Stenotrophomonas has also been reviewed in addition to their applications as effective biocontrol agents in plant and animal pathogenesis.
Introduction
Stenotrophomonas maltophilia is an uncommon, aerobic, non-fermentative, gram negative bacterium; motile due to polar flagella, catalase-positive, oxidase-negative slightly smaller (0.7–1.8 × 0.4–0.7 μm; which distinguishes them from most other members of the genus) and have a positive reaction for extracellular DNase. While S. maltophilia is an aerobe, it can still grow using nitrate as a terminal electron acceptor in the absence of oxygen (Crossman et al., 2008). S. maltophilia strains are found to be ubiquitously distributed in the environment with regard to habitat and geography: often associated with roots of many plant species (Ryan et al., 2009).
Growth of S. maltophilia studied in presence of different carbon sources: trichloroethylene (TCE), toluene, phenol, glucose, chloroform, and benzene with 0.1% peptone revealed an interesting growth pattern. Growth in presence of TCE, benzene, and chloroform was almost the same, whereas comparatively less growth was seen in presence of toluene and no growth in phenol even in presence of peptone (Mukherjee and Roy, 2013c). Pompilio et al. (2011), observed the mean growth rate of S. maltophilia obtained from clinical [cystic fibrosis (CF) and non-CF patients] and environmental isolates. CF isolates showed higher mean generation time compared to non-CF ones (3.5 ± 0.5 h vs. 3.1 ± 0.6 h, respectively; p < 0.001). Environmental isolates grown at 37°C exhibited a significantly lower generation time compared to that observed at 25°C (2.5 ± 0.6 h vs. 3.2 ± 0.4 h, respectively; p < 0.05).
Stenotrophomonas maltophilia have specific flagella like structures. The flagella filaments are composed of a 38-kDa subunit, SMFliC, and analysis of its N-terminal amino acid sequence showed considerable sequence identity to the flagellins of Serratia marcescens (78.6%), Escherichia coli, Proteus mirabilis, Shigella sonnei (71.4%), and Pseudomonas aeruginosa (57.2%; de Oliveira-Garcia et al., 2002). de Oliveira-Garcia et al. (2003), were the first to characterize fimbriae in this genus. All so far studied S. maltophilia strains contain multidrug efflux pumps – RND family: SmeABC, SmeDEF, SmeGH, SmeIJK, SmeMN, SmeOP, SmeVWX, and SmeYZ; ABC family: SmrA, MacABCsm; MFS family: EmrCABsm. Evenmore, the intergenic region smet-smeD is considered as a S. maltophilia phylogenetic marker (Alonso and Martínez, 2000, 2001; Alonso et al., 2000; Zhang et al., 2001; Figure 1).
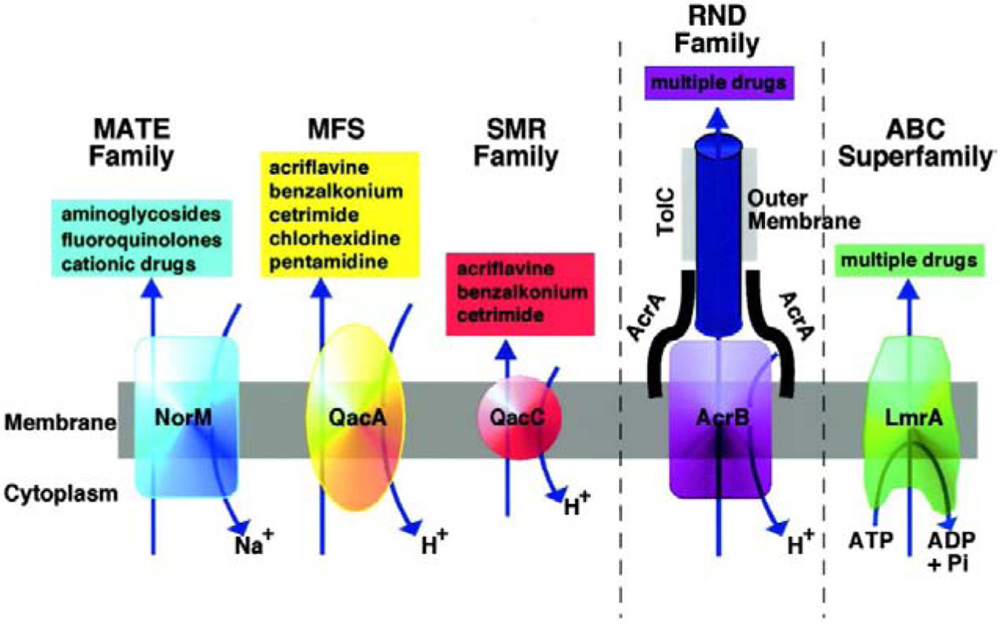
FIGURE 1. Diagrammatic representation of the five families of MDR efflux pumps in bacteria are the resistance nodulation division (RND) family, the major facilitator superfamily (MFS), and the staphylococcal multiresistance (SMR), and multidrug and toxic compound extrusion (MATE) families. A role for ABC (ATP binding cassette) MDR transporters in MDR of clinically relevant bacteria has yet to be established (Piddock, 2006). Reproduced with permission.
Xenobiotic-degrading S. maltophilia have tremendous potential for bioremediation but new modifications are required to make such microorganisms effective and efficient in removing these compounds, which were once thought to be recalcitrant. Metabolic engineering through genomic manipulations might help to improve the efficiency of degradation of toxic compounds by S. maltophilia. However, efficiency of naturally occurring Stenotrophomonas sp. for field bioremediation could be significantly improved by optimizing certain factors such as bioavailability, adsorption and mass transfer. Chemotaxis and microbe–plant interactions could also have an important role in enhancing biodegradation of pollutants.
The great genetic and metabolic diversity within S. maltophilia makes it a “Wonder-bug.” Figure 2 below, describes the multifaceted role of S. maltophilia in our environment:
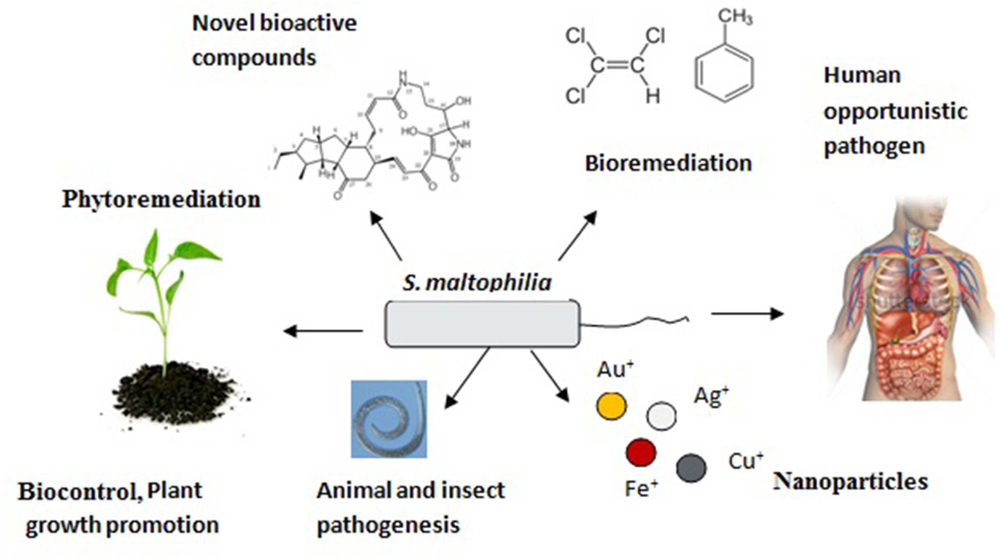
FIGURE 2. Multidimensional role of Stenotrophomonas maltophilia in our environment signifies its substantial genetic, metabolic, and residential diversity.
What are the Rapid S. maltophilia Specific Diagnostic Procedures?
Stenotrophomonas maltophilia may cause nosocomial infections in immune-compromised patients (Elting et al., 1990). Infection is usually facilitated by the presence of prosthetic material (colonizing breathing tubes, endotracheal tubes, or urinary catheters) and removal of the infected prosthesis is sufficient to cure the infection: antibiotics are required if prosthesis cannot be removed. This suggests that its level of invasivity is likely low. A considerable mortality rate (up to 37.5%) can be attributed to S. maltophilia infection (Falagas et al., 2009). The median time to hematological diagnosis and S. maltophilia identification was 2.5 days as reported by Chaplow et al. (2010) in 37 patients with S. maltophilia bacteraemia in a blood and marrow transplant (BMT) and non-BMT hematology conditions, treated with co-trimoxazole or ceftazidime with ciprofloxacin. The median duration of organism-specific treatment was 9 days.
Stenotrophomonas maltophilia infections are laboratory diagnosed using standard techniques as per LiPuma et al. (2007). Gram negative selective medium (GNSA) was developed by Moore et al. (2003) for rapid isolation of gram negative strains like S. maltophilia. Culture based identification is a time taking process and may give false positive results leading to incorrect identification of strains. The availability of the whole DNA sequence of the S. maltophilia strain K279a was utilized to set up fast and accurate PCR-based diagnostic protocols (Emanuela et al., 2008). A novel internally controlled 5-plex real-time PCR nucleic acid diagnostics assay (NAD), was utilized for rapid (<5 h), quantitative detection and identification of S. maltophilia (Minogue et al., 2015). Yang et al. (2014), first described a loop-mediated isothermal amplification (LAMP) method for the rapid detection (<60 min) of metallo-β-lactamase (blaL1) in clinical samples with sensitivity 100-fold greater than that of conventional PCR. Kinoshita et al. (2015) also reported LAMP based rapid detection of S. maltophilia. Another method is target enriched multiplex PCR (TEM-PCR) that was applied for the detection of bloodstream pathogens like S. maltophilia (Stalons et al., 2014). Development of a novel peptide nucleic acid (PNA) probe for S. maltophilia identification by fluorescence in situ hybridization (FISH) was reported (Hansen et al., 2014). PNA FISH is a very fast (less than 90 min) and reliable molecular identification method.
Considering the importance of rapid and accurate diagnosis, PNA probe based identification seems to have some advantages compared to DNA probe based diagnosis methods involving RAPD/NAD/multiplex PCR. PNA probes are small in size with a non-charged polyamide backbone that renders them easy to hybridize and increases the binding strength compared to DNA probes (Pellestor et al., 2008). PNA probes have better sensitivity and specificity, show improved penetration into cells and through biofilm matrices and are not susceptible to bacterial endonucleases which may be present in clinical samples (Bjarnsholt et al., 2009). FISH is comparatively useful for in situ detection of this microorganism directly in clinical samples and mixed bacterial populations without prior cultivation.
The preferred clinical diagnostic protocol of choice depends on several factors. Blood cultures are preferred in most cases though their lengthy incubation time and lack of consistency. For rapid and accurate detection in local hospital laboratory setting, the authors propose PCR to be the tool of choice though FISH in combination with Flow Cytometry is more preferable.
Are New Treatment Strategies Being Developed to Overcome S. maltophilia Infections?
Stenotrophomonas maltophilia are naturally resistant to many broad spectrum antibiotics such as cephalosporins, carbapenems, and aminoglycosides. This means that treatment options are relatively limited According to the World Health Organization (WHO), S. maltophilia is one of the leading drug-resistant pathogens in hospitals worldwide (WHO; Public health importance of antimicrobial resistance1; Brooke, 2014). The treatment of choice for S. maltophilia is trimethoprim-sulfamethoxazole (SXT; Wang et al., 2014). Several combinations of novel agents are currently under investigation, including a β-lactam and dual β-lactamase inhibitor combination (Page et al., 2011) and MD3 (a novel synthetic inhibitor of peptidases) plus colistin (Personne et al., 2014). Most data is collected from case reports; compelling clinical evidence for combination therapies is lacking. Additional published cases and clinical trials are required to formulate a more evidence-based approach for the treatment of patients with S. maltophilia infections.
Quorum sensing (QS) is a bacterial cell–cell communication process that involves the production, detection, and response to extracellular signaling molecules called autoinducers. S. maltophilia has a diffusible signal factor (DSF) that controls cell–cell communication and many functions such as motility, extracellular protease production and microcolonies formation in artificial sputum medium. This DSF signaling also mediates interspecies interactions between S. maltophilia and P. aeruginosa such as susceptibility to polymixin and its influence on biofilm formation. QS-inhibition based drugs need to be developed (Boyen et al., 2009) but DSF-signaling has more potential as drug target for this species. Production and detection of DSF are governed by the rpf cluster, which encodes the synthase RpfF and the sensor RpfC, among other components. Structural analogs of DSF like cis-2-decenoic acid may have a role in control of virulence factor synthesis in different pathogens. Such molecules may represent lead compounds for new drugs. Also, DSF signaling is normally finely balanced during the disease process and such a fine balance can be readily disrupted by either degradation or over-production of the signal (Ryan et al., 2015). Iron, probably through the Fur system, negatively regulates DSF production in S. maltophilia (García et al., 2015). Emodin (an active component of Chinese traditional medicines) was found to inhibit biofilm formation in S. maltophilia and induced proteolysis of the QS signal receptor TraR in E. coli (Ding et al., 2011).
An alternative approach has been to utilize the inherent specificity of immunoglobulins to inhibit the pathogenic functions in S. maltophilia. Another approach has involved screening combinatorial libraries of random peptides. In a study, antibodies to S. maltophilia iron regulated outer membrane proteins (TROMP) were developed that reduced the uptake of iron by blocking the binding of ferric complexes resulting in the inhibition of S. maltophilia proliferation. Growth inhibition studies gave positive results with polyclonal antibodies recovered from rabbits immunized with S. maltophilia membrane associated polypeptides and monoclonal antibodies were also produced using mouse hybridoma technology model (Freeman, 2009).
In another study, in vitro and in vivo activities of epigallocatechin-3-gallate (EGCG), a green tea component, against S. maltophilia isolates from cystic fibrosis patients were analyzed (Vidigal et al., 2014). Essential oils from plants (e.g., orange, bergamot, cinnamon, clove, cypress, eucalyptus, fennel, lavender, lemon, mint, rosemary, sage, and thyme) were investigated and found to demonstrate antibacterial activity against S. maltophilia (Fabio et al., 2007). A surfactant-stabilized oil-in-water nanoemulsion (NB-401) has shown antimicrobial activity against planktonic and biofilm-associated cells of S. maltophilia (LiPuma et al., 2009). This nano-emulsion consists of emulsified cetylpyridinium chloride, poloxamer 407, and ethanol in water with super-refined soybean oil. The interaction of the nano-emulsion with the cell was suggested to result in the fusion of the outer membrane with the nano-emulsion, leading to cell lysis. Bismuth-thiols (BTs) can prevent the formation of microbial biofilms as well as eradicate established biofilms at uncommonly low concentrations. BTs are comprised of a central bismuth atom that is chelated by organic molecules known as thiols. The ability of thiols to chelate bismuth and other metals, has led to long, successful history as antidotes for treatment of heavy metal poisoning. The resulting low toxicity of BTs in mammals and the low cost of production and their stability, make them ideal candidates for development as prescription drugs and as anti-infective medical device coatings (Domenico et al., 2000, 2003; Wu et al., 2002). A device (Podhaler device) that delivers new inhalational tobramycin [tobramycin inhalation powder (TIP)] and attains high drug levels to the lung may be able to exceed current high MICs of tobramycin in S. maltophilia (Ratjen et al., 2015). Waters (2012), suggested a potential role of inhaled aztreonam lysine in the treatment of S. maltophilia pulmonary infection. A Monte Carlo pharmacokinetic/pharmacodynamic simulation was performed that suggested that minocycline may be a proper choice for treatment of HAP caused by S. maltophilia, while tigecycline, moxifloxacin, and levofloxacin may not be optimal as monotherapy (Wei et al., 2015).
The use of phage therapy may be an alternative to the use of antibiotics to treat S. maltophilia infections. A novel giant S. maltophilia phage ΦSMA5 was isolated from sputum samples, pleural effusions, and catheter tips (Chang et al., 2005). This phage was tested against 87 S. maltophilia strains isolated from hospitals and was found to have a narrow host range. A recent review suggested that the use of phages to treat biofilms has potential (Donlan, 2009). To the best of my knowledge, phage therapy is not used in ordinary clinical practice for the treatment of S. maltophilia infections. Together, the observations from the studies described above suggest that it is possible that a cocktail of surfactant, antimicrobial peptides, and phage may provide a suitable alternative to the administration of antibiotics.
Many alternative remedies including biofield energy treatment have recently found their way into the medical mainstream and is widely accepted by most of the healthcare professionals. A change in sensitivity pattern of amikacin from resistant to intermediate along with changes in sensitivity of trimethoprim/sulfamethoxazole and chloramphenicol was observed on biofield treatment (Clarke et al., 2015; Trivedi et al., 2015).
What is the Intra- and Inter-Species Genetic Diversity in S. maltophilia?
With the advance in molecular biology tools and sequencing methods, large repertoires of S. maltophilia strains are easily accessible through NCBI. Molecular dendogram or phylogenetic tree (drawn with Clustalx and MEGA5 softwares), among different Stenotrophomonas groups of bacteria suggests the genetic diversity among the different strains (Figure 3).
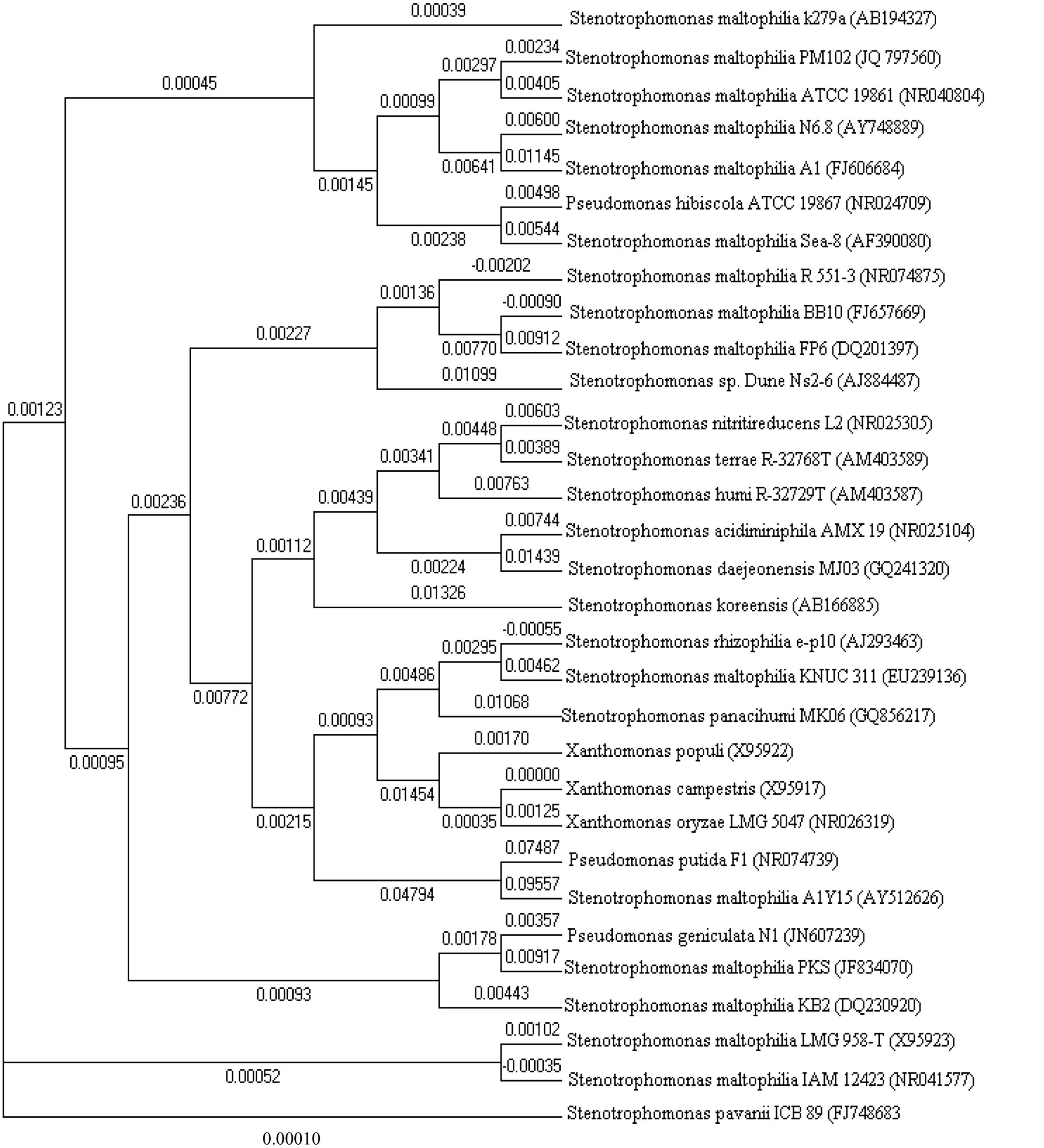
FIGURE 3. Phylogenetic tree constructed with 16S rDNA sequences of various Stenotrophomonas bacteria retrieved from NCBI GenBank.
Stenotrophomonas maltophilia was grouped in the genus Xanthomonas by Swings et al. (1983). However, the proposed reclassification of P. maltophilia as Xanthomonas maltophilia did not meet with universal approval (Palleroni, 1984), and the controversy about the taxonomic status of this bacterium in the genus Xanthomonas remained unresolved (Bradbury, 1984). Several factors requested a reinterpretation of the taxonomic position of X. maltophilia (Van Zyl and Steyn, 1992). When a Xanthomonas-specific 16S rDNA sequence as the primer for PCR was used, a single 480-bp PCR fragment was seen for Xanthomonads; however, X. maltophilia strains produced additional PCR fragments, leading the author to conclude that X. maltophilia does not belong in the genus Xanthomonas (Maes, 1993). Continuing dissatisfaction with the classification of this organism finally gave rise to the proposal in Palleroni and Bradbury (1993) to create the new genus Stenotrophomonas. A proteome driven clustering correctly groups all well-annotated S. maltophilia genomes correctly from the Xanthomonas species.
Neighbor joining tree revealed S. maltophilia clustered homologs were Pseudomonas hibiscola, Stenotrophomonas rhizophilia, Pseudomonas geniculata, and Pseudomonas putida F1. S. pavanii seems to be the most distant in terms of homology. DNA sequence identity values among the S. maltophilia strains ranged from 100 to 98.9%. The dendrogram in this paper reveals 10 different species of the genus Stenotrophomonas and depicts the closest homolog of S. maltophilia PM102 (JQ797560; that was isolated and characterized in our laboratory; being the first report of a Stenotrophomonas species in uptake of trichloroethylene as the sole carbon source) to be S. maltophilia ATCC 19861 (NR040804).
Total genome sequencing of few S. maltophilia strains like AU12-09, k279a, and R551-3 have been undertaken (Figure 4). AU12-09 genome consists of 129,784,052 bp of DNA (GenBank APIT00000000; Zhang et al., 2013). The genome of K279a is 4,851,126 bp and of high G+C content. The sequence reveals an organism with a remarkable capacity for drug and heavy metal resistance: nine resistance-nodulation-division (RND)-type putative antimicrobial efflux systems are present and several mobile DNA segments code for pili/fimbrae involved in adhesion and biofilm formation that contributes to increased antimicrobial drug resistance (Crossman et al., 2008). S. maltophilia R551-3 (Accession no. PRJNA17107) was isolated from the poplar Populus trichocarpa. In the presence of cadmium (Cd), it accumulates cysteine as a reducer in order to undergo chelation, and form CdS, or cadmium sulfide in order to avoid lethal toxicity (Pages et al., 2008). Conchillo-Solé et al. (2015) reported the draft genome sequence of S. maltophilia UV74, isolated from a vascular ulcer. In this isolate, the DSF-mediated QS system is regulated by a new rpf cluster variant (Huedo et al., 2014). Comparative genomic and Transcriptomic approaches have been used to identify significant borders between the MDR S. maltophilia and non-pathogenic plant-associated S. maltophilia R551-3 and S. rhizophila DSM14405. Although, there was significant similarity in host invasive and antibiotic resistance genes, several crucial virulence factor and heat shock protein genes were absent in plant-associated strains (Alavi et al., 2014). Furthermore, an environmental strain of S. maltophilia named BurA1 showed absence of RND pumps (SmeABC) but presence of another MDR RND efflux pump named EbyCAB on a genomic island acquired via Horizontal gene transfer (Youenou et al., 2015).
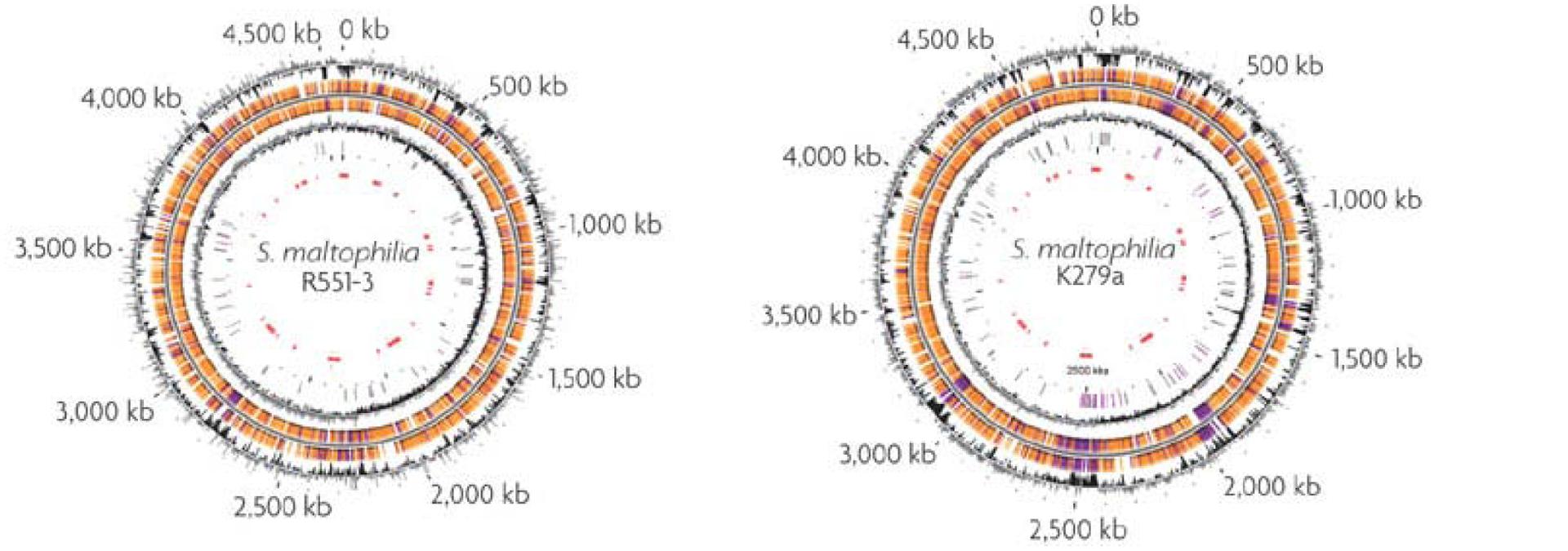
FIGURE 4. Genome maps of the poplar endophyte S. maltophilia R551-3 and of the opportunistic pathogen S. maltophilia k279a. From the outside-in, the circles represent coordinates in kilobase pairs (kbp), % GC content, predicted open reading frames (ORFs) in the clockwise and anticlockwise orientations, GC skew [(G-C and G+C) in a 1000-bp window], transposable elements (pink) and pseudogenes (gray) and the putative S. maltophilia k279a genomic islands (red; Ryan et al., 2009). Reproduced with permission.
What are the Novel Bioactive Compounds and Nanoparticles Synthesized By Stenotrophomonas maltophilia?
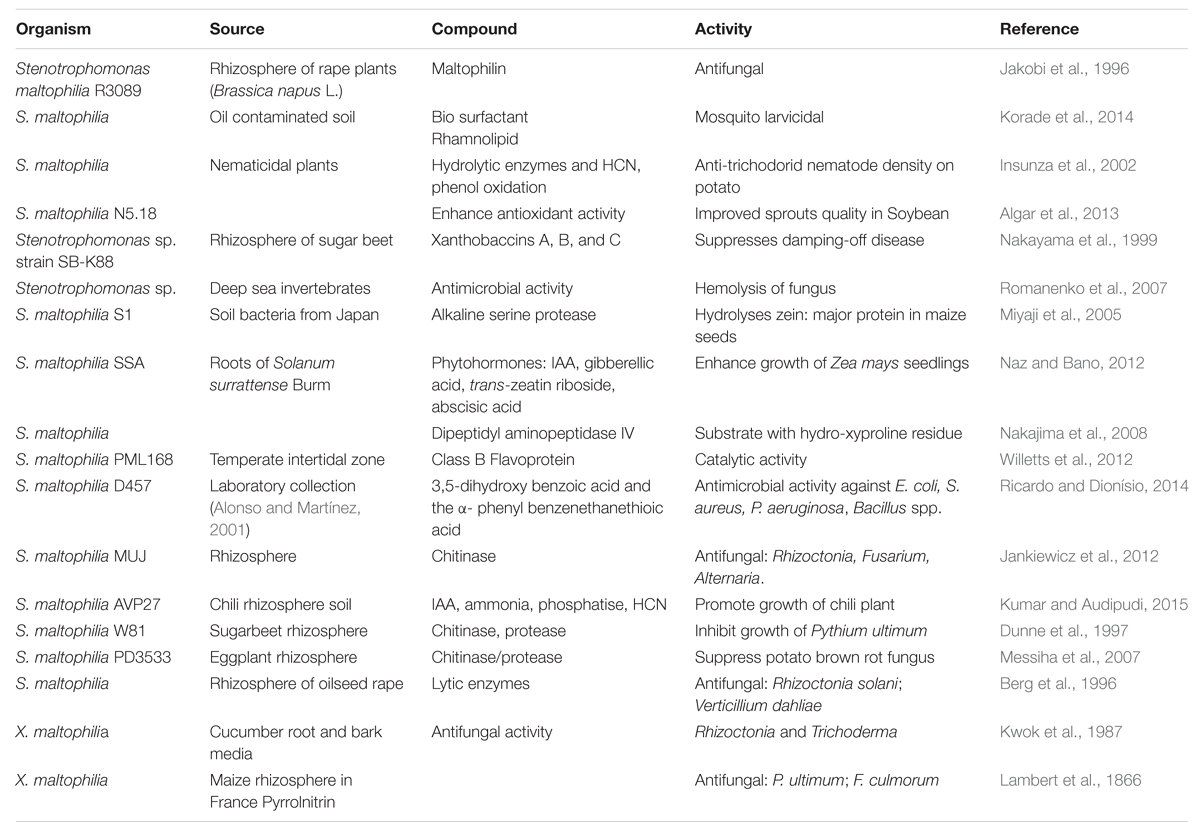
Stenotrophomonas maltophilia have been documented as a potential source for several novel bioactive compounds. These natural chemicals are not only used as bio control agents due to their antifungal, antibacterial and insecticidal properties but also have widespread applications as plant growth promoting substances (PGPR). Rhizobacteria introduced in the rhizosphere of tomato, pepper, melon or bean were found to increase growth of roots/shoots (Elad et al., 1987). Table 1 is listed with the various bioactive compounds from S. maltophilia with their varied applications.
Additionally, S. maltophilia strains have been shown to produce enzymes that play important role in synthesis of compounds with medicinal applications. Production of 2-arylpropanoic acid (NSAID compounds: non-steroidal anti-inflammatory analgesics) was done using lipase obtained from S. maltophilia (Sharma et al., 2001). An hghly thermostable xylanase was reported from Stenotrophomonas. According to recent research from Lucknow, India, a novel psychro-tolerant S. maltophilia (MTCC 7528) with an ability to produce extracellular, cold-active, alkaline and detergent stable protease was isolated from soil of Gangotri Glacier, Western Himalayas, India (Kuddus and Ramteke, 2009).
A novel strain of S. maltophilia was isolated from actual gold enriched soil (Singhbhum gold mines in Jharkhand state of India). After incubation for 8 h in gold chloride (HAuCl4), monodisperse preparation of gold nanoparticles was obtained (Nangia et al., 2009). Another strain of S. maltophilia, isolated from Indian marine origin could synthesize both silver and gold nanoparticles (Malhotra et al., 2013). Role of this bacterium in nanoparticle synthesis (gold and silver being the most important) implicates their importance in biology and medicine (Sperling et al., 2008; Abou El-Nour et al., 2010). In another report, S. maltophilia SELTE02 showed promising transformation of selenite to elemental selenium, accumulating selenium granules in cell cytoplasm or extracellular space (Iravani, 2014). Another S. maltophilia isolated from soil rhizosphere of Astragalus bisulcatus could completely reduce selenite (Vallini et al., 2005). A novel bacterial strain OS4 of S. maltophilia (GenBank: JN247637.1) was isolated. At neutral pH, this Gram negative bacterial strain significantly reduced hexavalent chromium, an important heavy metal contaminant found in the tannery effluents and minings (Oves et al., 2013). Not much is known regarding the mechanism of metal nanoparticle synthesis by bacteria although different hypothesis have been suggested. A promising mechanism for the biosynthesis of gold NPs by S. maltophilia and their stabilization via charge capping was suggested, which involves an NADPH-dependent reductase enzyme that reduces Au3+ to Au0 through electron shuttle enzymatic metal reduction process (Nangia et al., 2009).
Genomic Potential of Stenotrophomonas maltophilia to Locate Enzymes Involved in Bioremediation
The first genomic tool applied when bioremediation is investigated is 16s rDNA sequencing that is applied to identify the organism involved in bioremediation. Isolation of pure cultures and their identification by 16s rDNA analysis is the most important step in the study of biodegradation. A metagenomic approach can also be undertaken in mass identification of environmental samples involved in xenobiotic degradation. The next approach is Genomic analysis to identify the enzymes involved in bioremediation, Genomic analysis showed how various Stenotrophomonas strains are able to make enzymes that play vital role in dechlorination of polychlorinated hydrocarbons or heavy metal uptake by turning the genes on and off as the organism detects something appetizing. Whole genome sequences take this task of enzyme identification a step up further. The draft genome of S. maltophilia strain ZBG7B assembled into 145 contigs with an N50 of 50,104 bp. (Chan et al., 2015) contains a repertoire of biodegradation-related genes: xylosidase, xylanase, xylose isomerise. Stenotrophomonas sp. has been found to play important role in biodegradation of keratin (Yamamura et al., 2002), RDX (Binks et al., 1995), geosmin (Zhou et al., 2011), atrazine (Rousseaux et al., 2001), p-nitrophenol (Liu et al., 2007) and monocyclic hydrocarbons (Urszula et al., 2009), phenanthrene (Gao et al., 2013) and styrene.
Keratin degradation was an outcome of the cooperative action of two types of extracellular proteins: proteolytic (serine protease) and disulfide bond-reducing (disulfide reductase). The best evidence for the pathway of geosmin degradation by bacteria has been provided by Saito et al. (1999), who identified four possible biodegradation products of geosmin (Hoefel et al., 2009). Two of these products were identified as 1, 4a-dimethyl-2, 3, 4, 4a, 5, 6, 7, 8- octahydronaphthalene and enone. Strains belonging to the C. heintzii, A. aminovorans, S. maltophilia, and A. crystallopoietes species are capable of mineralizing or degrading atrazine. Atrazine is catabolized in three enzymatic steps to cyanurate, which can be further metabolized by ring cleavage to carbon dioxide and ammonia. The first enzyme converts atrazine to hydroxyatrazine. Two additional hydrolases continue the process by removing the ethylamine and isopropylamine groups (de Souza et al., 1998; Smith et al., 2005). While some organisms possess all of the required enzymes, other communities degrade atrazine by a community-approach, where different organisms have some of the enzymes, and the intermediates in the pathway are passed between the organisms. These methods undertake a co-metabolic approach of enzyme activity. On the other hand, compounds utilized as sole carbon source involve novel enzymes that are coded by set of genes characteristic of pure cultures. A novel strain PM102 of S. maltophilia was shown to be able to degrade and utilize trichloroethylene as the sole carbon source (Mukherjee and Roy, 2012). A copper enhanced monooxygenase was characterized from the PM102 strain to be involved in the biotransformation of TCE (Mukherjee and Roy, 2013b). Phenanthrene utilization as sole carbon source involved dioxygenation on 1,2-, 3,4-, and 9,10-C, where the 3,4-dioxygenation and subsequent metabolisms were most dominant. The metabolic pathways were further branched by ortho- and meta-cleavage of phenanthrenediols. The KEGG genome map can be retrieved from DBGET integrated genome retrieval system that maps the KEGG pathway off styrene degradation by S. maltophilia K279a.
Stenotrophomonas maltophilia strain (SeITE02) was capable of resistance to high concentrations of selenite [SeO32-, Se (IV)], reducing it to nontoxic elemental selenium under aerobic conditions (Antonioli et al., 2007). Various enzymatic systems, such as nitrite reductase, sulfite reductase, and glutathione (GSH) reductase (GR), have been proposed the reduction of selenite in bacteria, under anaerobic conditions as predicted from the whole genome sequencing of SelTE02. Under aerobic conditions, nitrite reductase was found to play no role but glutathione had some contribution. A study involving bioremediation of copper using copper-resistant bacteria, S. maltophilia PD2 has been reported (Ghosh and Saha, 2013). Metagenomic sequencing has also been used to profile the genes present in microbiomes from Cu-contaminated mining area. A list of genes coding heavy metal uptake efflux pumps could be mapped like P-type ATPases linked to Cu uptake, mdtC in Zn efflux and cusA and ybdB forming cationic efflux systems for Cu and Ag respectively (Van Rossum et al., 2016). Shotgun sequencing of DNA from biofilm samples has facilitated identification of genes that encode for efflux pumps, cell wall components, and metabolic processes for metals tolerance as well. Stenotrophomonas sp. C21 carries plasmids containing the Cu-resistance copA genes. S. maltophilia RSV-2 strain could degrade mixed textile dyes up to 2100 ppm within 67 h and 58% decolorization was obtained through acclimatization study (Rajeswari et al., 2013). Laccase activity of this bacterium has been shown to play a role in dye removal (Galai et al., 2009). copA gene was traced to encode the multicopper oxidase with laccase activity by degenerative PCR; cloned and homologous recombination was used to construct copA mutant strain to confirm laccase activity and dye decolorization of copA in vitro. The biosorption of Pb(II), Zn(II), and Ni(II) from industrial wastewater using S. maltophilia was investigated (Wierzba, 2015). S. maltophilia has been reported to acquire genes involved in heavy metal resistance from Gram-positive bacteria (Alonso et al., 2000).
Stenotrophomonas maltophilia has also been found to play important role in the bioremediation of chlorinated pesticides like Chloropyrifos (Dubey and Fulekar, 2012) and endosulfan (Barragaán-Huerta et al., 2007; Kumar et al., 2007). Chloropyrifos uptake and degradation was mapped to mpd genes of S. maltophilia MHF ENV20. Gene mapping is carried out by PCR amplification using pre-designed primers. In our laboratory, predesigned todc gene primers of Pseudomonas putida were used to map the tce300 and tce350 genes of S. maltophilia PM102 (Mukherjee and Roy, 2013a) and the tce 1 gene of Bacillus cereus 2479 (Mitra and Roy, 2011). The tce300 and tce350 genes were cloned in E. coli and their TCE degradation ability was confirmed by IPTG induction of the recombinant clones.
Soil isolates of Stenotrophomonas degraded dichlorodiphenyl-trichloroethane (DDT) to l, l-dichloro-2, 2-bis (p-chlorophenyl) ethane (DDD; Mwangi et al., 2010). The three main enzyme families implicated in pesticide degradation are esterases, glutathione S-transferases (GSTs) and cytochrome P450 (Ortiz-Hernández et al., 2013). S. maltophilia ZL1 was able to convert steroid hormone: estradiol (E2) to estrone (E1) and finally to amino acid tyrosine (Li et al., 2012). Enzymes involved in protein and lipid biosyntheses were observed to be particularly active. S. maltophilia OG2, isolated from the body microflora of cockroaches, could grow on synthetic pyrethroid insecticides (Chen et al., 2011). Tallur et al. (2008) proposed the pathway of α-cypermethrin biodegradation. It was speculated that the pathway of α-cypermethrin biodegradation in S. maltophilia OG2 is similar. A new feather degrading S. maltophilia DHHJ strain was isolated that produced keratinolytic enzymes (Wu et al., 2012). Thus, S. maltophilia have been shown to possess intrinsic resistance mechanisms against heavy metals and signaling or metabolic pathways as an outcome of their genomic potential toward various environmental pollutants.
Most isolates are considered Biosafety Level 1 (BSL-1) organisms. However, there are notable exceptions, including some commonly studied isolates, which are considered Biosafety Level 2 (BSL-2) pathogens. As a result of the hazard assessment the application of microbes in bioremediation is categorized into one of three risk estimates (Slovic, 1987, 1997): high risk – severe, enduring, or widespread adverse effects are probable; medium risk – adverse effects predicted for probable exposure scenarios may be moderate and self-resolving and low risk – adverse effects predicted for probable exposure scenarios are rare, or mild and self-resolving. Assessment of these risk factors impose an obstacle in registration procedures for release of microbes into the environment for bioremediation purposes in Europe and USA, though in Australia the limit on microbial bioremediation applications is not so tight.
It is much easier and simpler to apply the plant associated naturally occurring S. rhizophilia in bioremediation or rhizoremediation approaches, as pathogenicity to humans for plant-associated S. rhizophila has not been heard of till date. Rather, the association of endophytic bacteria with their plant hosts has been shown to have a growth-promoting effect for many plant species. Endophytic bacteria have several mechanisms by which they can promote plant growth and health on marginal, polluted soils. These include the production of phytohormones or enzymes involved in growth regulator metabolism such as ethylene, 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase, auxins, indoleacetic acid (IAA) or cytokinins. In addition, endophytic bacteria can help their host plants to overcome the phytotoxic effects caused by environmental contamination.
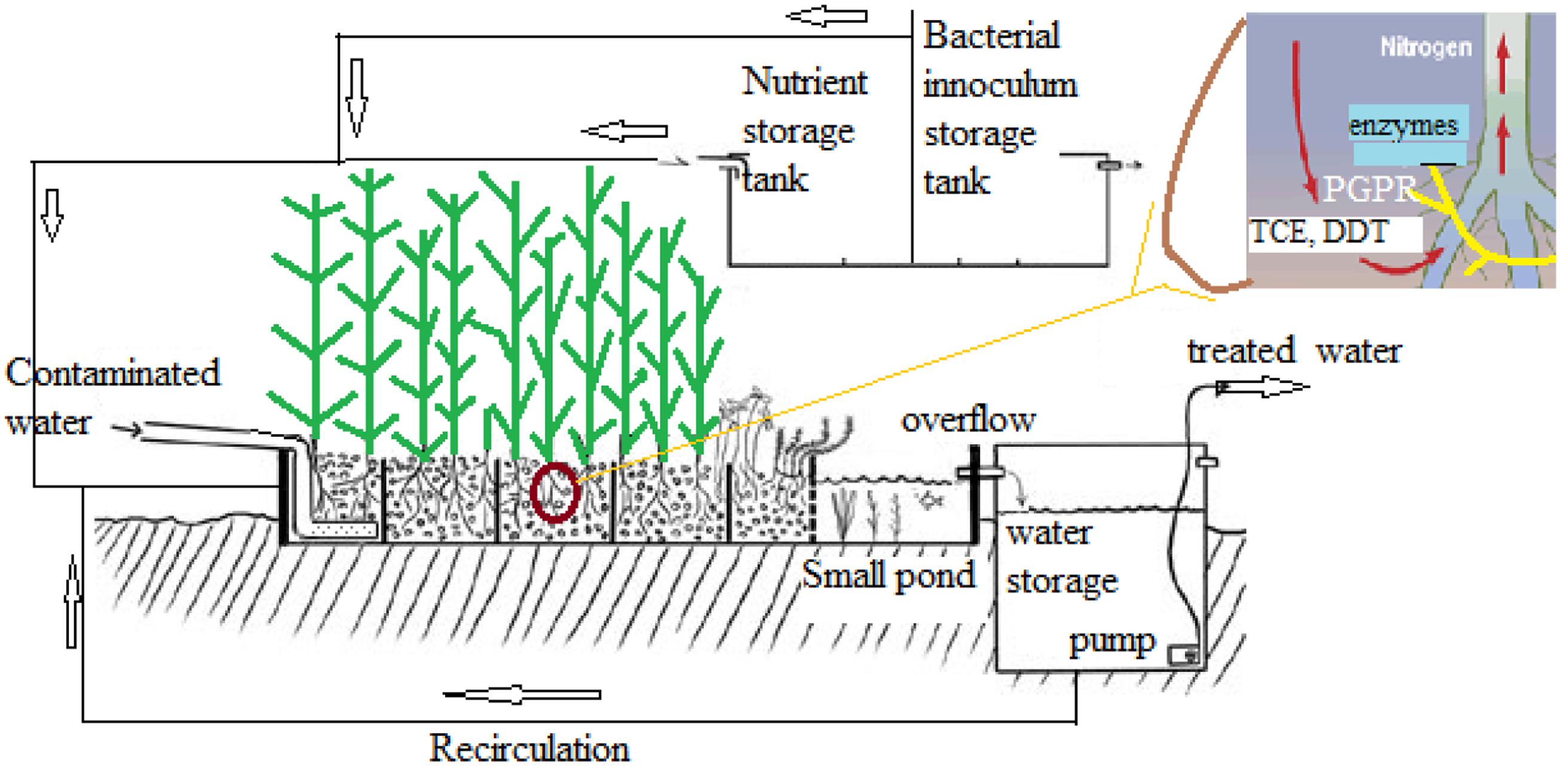
Stenotrophomonas maltophilia R551-3 was isolated from Populus trichocarpa and characterized to improve the growth and phytoremediation potential of poplar on marginal, contaminated soils (Nordberg et al., 2014). Endophytic bacteria equipped with the tom toluene degradation pathway could significantly improve the in planta degradation of BTEX and TCE in poplar, resulting in reduced phytotoxicity and release. Integrated microbial genomes were studied to identify metabolic functions (Markowitz et al., 2006). In a study, an attempt to determine the effect of aromatic compounds of plant origin on nitrophenols degradation by S. maltophilia KB2 strain was made. S. maltophilia KB2 used in this study is known to metabolize broad range of aromatic compounds including phenol, some chloro and methylphenols, benzoic acids, catechols, and others (Gren et al., 2010). A model system for selenium rhizofiltration from the rhizosphere soil of Astragalus bisulcatus, a legume based on plant–rhizobacteria interactions has been proposed (Vallini et al., 2005). Figure 5 represents a model system for the in situ application of plants in the bioremediation of contaminated water and other pollutants like trichloroethylene (TCE).
In another study Pennisetum pedicellatum rhizosphere associated degrading strain MHF ENV20 of S. maltophilia were evaluated for chlorpyrifos remediation (Dubey and Fulekar, 2012). This genus was among several other bacteria isolated as an endophyte in coffee seeds (Vega et al., 2005). Lata et al. (2006) found endophytic bacteria belonging to Stenotrophomonas associated with Echinacea plants. Endophytic bacteria (Stenotrophomonas) associated with sweet potato plants [Ipomoea batatas (L.) Lam] were isolated, identified and tested for their ability to fix nitrogen, produce indole acetic acid (IAA), and exhibit stress tolerance (Khan and Doty, 2009). The multidrug-efflux-pump SmeDEF in S. maltophilia has been shown to play vital role in root colonization in oilseed rape plant (cv. Californium; Kwizda, Austria) rather than its role in resistance toward antibiotic quinolones (Figure 6). Although, naturally occurring plant–microbe interactions are routinely applied in the field of phytoremediation, transgenic plants are also being reported in literature: Brassica juncea for phytoremediation of heavy metals from soil (Dushenkov et al., 1995), Helianthus anus (Dushenkov et al., 1995), Chenopodium amaranticolor (Eapen and D’Souza, 2003) for rhizofiltration of uranium and pumpkin plants for remediation of trichloroethylene. Dicotyledon plant species can be genetically engineered using the Agrobacterium vector system, while most monocotyledon plants can be transformed using particle gun or electroporation techniques.
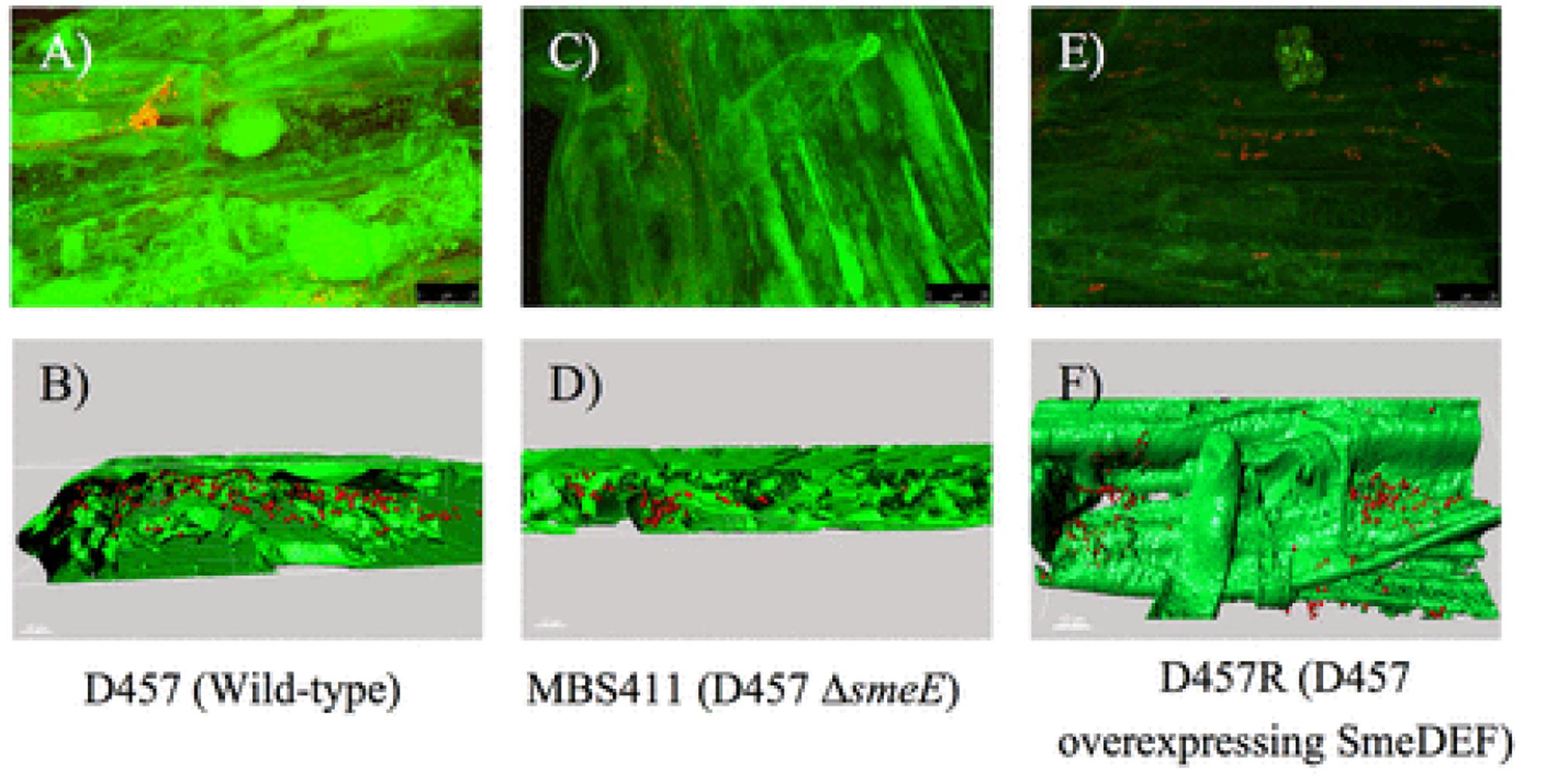
FIGURE 6. Colonization of oilseed rape roots by S. maltophilia visualized by FISH. (A) Wild-type strain D457. (C) sme-E-defective mutant strain MBS411. (E) D457R mutant that overexpresses SmeDEF. 3D analysis of the CLSM stacks of D457 (B), MBS411 (D), D457R, (F) was performed with Imaris 7.0 software (Guillermo et al., 2014). Reproduced with permission.
Conclusion
The advent of new Molecular diagnostic techniques for S. maltophilia, has created the potential to improve clinical outcomes. However, further validation and investigation of clinical correlates (viable bacterial load, antibiotic susceptibility profiles, virulence factor expression and clinical outcomes) is required before routine application. Genome sequencing of various S. maltophilia strains will help to identify the virulence factors, proteins and genes specific for MDR and pave the way for targeted drug delivery in treating S. maltophilia infections. Despite of its initial discovery as a human opportunistic pathogen, the different applications of S. maltophilia has not been left unexplored, although the question of biosafety remains. Non-pathogenic strains can be applied in various environmental issues: being a natural soil bacterium, Stenotrophomonas strains have wide applications in agriculture as potential biocontrol agents in treating fungal infections and plant growth promotion. S. rhizophilia is non-pathogenic and widely associated with plant roots and holds great promise in phytoremediation or rhizoremediation of contaminated ground water. Transgenic plants application to in situ bioremediation is not clear till date as adequate field studies have not been reported. Impact of transgenics to environment like competiveness of transgenic to wild type plants, effect on birds or insects and possibility of gene transfer to other natural plants through pollination are points to be considered. S. maltophilia are ubiquitously distributed in our environment. Pathogenic S. maltophilia might be applied to our environment provided proper measures are taken to eradicate its pathogenicity first. This will involve long term research to identify the genes or proteins of S. maltophilia involved in pathogenicity and MDR. Recombinant S. maltophilia with knockout of such genes might be applied but the problem in changes in present MDR properties through genetic mutation needs to be kept in mind. Lot of work has been done on applications of S. maltophilia in various domains. Still, it is quite impossible to separate these domains and the attitude “favorable” cannot be truly related to pathogenic strains though comparative genomics and transcriptomics helps detect significant borders between pathogenic strains and non-pathogenic plant-associated strains.
Author Contributions
All authors listed, have made substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
Funds were received from The Department of Science and Technology (DST), New Delhi, India as INSPIRE fellowship (Innovation in Science Pursuit for Inspired Research) given to PM to carry out research work on bioremediation of Trichloroethylene by S. maltophilia.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer JM and handling Editor declared a current collaboration and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
Acknowledgments
The authors sincerely acknowledge DST for providing financial support that was received during this research work and review documentation. The authors are also thankful to the Department of Biotechnology, Burdwan University for providing necessary infrastructure and materials that materialized their efforts. The authors sincerely acknowledge the efforts of the reviewers whose valuable comments and critics have greatly helped in correcting and updating the manuscript.
Footnotes
References
Abou El-Nour, K. M. M., Eftaiha, A., Al-Warthan, A., and Ammar, R. A. A. (2010). Synthesis and applications of silver nanoparticles. Arabian J. Chem. 3, 135–140. doi: 10.1016/j.arabjc.2010.04.008
Alavi, P., Starcher, M. R., Thallinger, G. G., Zachow, C., Müller, H., and Berg, G. (2014). Stenotrophomonas comparative genomics reveals genes and functions that differentiate beneficial and pathogenic bacteria. BMC Genomics 15:482. doi: 10.1186/1471-2164-15-482
Algar, E., Ramos-Solano, B., García-Villaraco, A., Sierra, M. D., Gómez, M. S., and Gutiérrez-Mañero, F. J. (2013). Bacterial bioeffectors modify bioactive profile and increase isoflavone content in soybean sprouts (Glycine max var Osumi). Plant Foods Hum. Nutr. 68, 299–305. doi: 10.1007/s11130-013-0373-x
Alonso, A., and Martínez, J. L. (2000). Cloning and characterization of SmeDEF, a novel multidrug efflux pump from Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 44, 3079–3086. doi: 10.1128/AAC.44.11.3079-3086.2000
Alonso, A., and Martínez, J. L. (2001). Expression of multidrug efflux pump SmeDEF by clinical isolates of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 45, 1879–1881. doi: 10.1128/AAC.45.6.1879-1881.2001
Alonso, A., Sanchez, P., and Martínez, J. L. (2000). Stenotrophomonas maltophilia D457R contains a cluster of genes from gram-positive bacteria involved in antibiotic and heavy metal resistance. Antimicrob. Agents Chemother. 44, 1778–1782. doi: 10.1128/AAC.44.7.1778-1782.2000
Antonioli, P., Silvia, L., Irene, C., Giovanni, V., Rinalducci, S., Lello, Z., et al. (2007). Stenotrophomonas maltophilia SeITE02, a new bacterial strain suitable for bioremediation of selenite-contaminated environmental matrices. Appl. Environ. Microbiol. 73, 6854–6863. doi: 10.1128/AEM.00957-07
Barragán-Huerta, B. E., Costa-Perez, C., Peralta-Cruz, J., Barrera-Cortes, J., Esparza-Garcia, F., and Rodriguez-Vazquez, R. (2007). Biodegradation of organochlorine pesticides by bacteria grown in microniches of the porous structure of green bean coffee. Int. Biodeterior. Biodegrad. 59, 239–244. doi: 10.1016/j.ibiod.2006.11.001
Berg, G., Marten, P., and Ballin, G. (1996). Stenotrophomonas maltophilia in the rhizosphere of oilseed rape—occurrence, characterization and interaction with phytopathogenic fungi. Microbiol. Res. 151, 19–27.
Binks, P. R., Nicklin, S., and Bruce, N. C. (1995). Degradation of hexahydro-1, 3, 5-trinitro-1, 3, 5-triazine (RDX) by Stenotrophomonas maltophilia PB1. Appl. Environ. Microbiol. 61, 1318–1322.
Bjarnsholt, T., Jensen, P. O., Fiandaca, M. J., Pedersen, J., Hansen, C. R., Andersen, C. B., et al. (2009). Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatr. Pulmonol. 44, 547–558. doi: 10.1002/ppul.21011
Boyen, F., Eeckhaut, V., Van Immerseel, F., Pasmans, F., Ducatelle, R., and Haesebrouck, F. (2009). Quorum sensing in veterinary pathogens: mechanisms, clinical importance and future perspectives. Vet. Microbiol. 135, 187–195. doi: 10.1016/j.vetmic.2008.12.025
Bradbury, J. F. (1984). “Genus II. Xanthomonas,” in Bergey’s Manual of Systematic Bacteriology, Vol. 1, eds N. R. Krieg and J. G. Holt (Baltimore, MD: The Williams & Wilkins Co), 199–210.
Brooke, J. S. (2014). New strategies against Stenotrophomonas maltophilia: a serious worldwide intrinsically drug-resistant opportunistic pathogen. Expert Rev. Anti Infect. Ther. 12, 1–4. doi: 10.1586/14787210.2014.864553
Chan, K.-G., Chong, T.-M., Adrian, T.-G.-S., Kher, H. L., Hong, K.-W., Grandclément, C., et al. (2015). Whole-genome sequence of Stenotrophomonas maltophilia ZBG7B reveals its biotechnological potential. Genome Announc. 3, e01442-15. doi: 10.1128/genomeA.01442-15
Chang, H.-C., Chen, C. R., Lin, J. W., Shen, G. H., Chang, K. M., Tseng, Y. H., et al. (2005). Isolation and characterization of novel giant Stenotrophomonas maltophilia phage ΦSMA5. Appl. Environ. Microbiol. 71, 1387–1393. doi: 10.1128/AEM.71.3.1387-1393.2005
Chaplow, R., Palmer, B., Heyderman, R., Moppett, J., and Marks, D. I. (2010). Stenotrophomonas maltophilia bacteraemia in 40 haematology patients: risk factors, therapy and outcome. Bone Marrow Transplant. 45, 1109–1110. doi: 10.1038/bmt.2009.274
Chen, S., Yang, L., Hu, M., and Liu, J. (2011). Biodegradation of fenvalerate and 3-phenoxybenzoic acid by a novel Stenotrophomonas sp. strain ZS-S-01 and its use in bioremediation of contaminated soils. Appl. Microbiol. Biotechnol. 90, 755–767. doi: 10.1007/s00253-010-3035-z
Clarke, T. C., Black, L. I., Stussman, B. J., Barnes, P. M., and Nahin, R. L. (2015). Trends in the Use of Complementary Health Approaches Among Adults: Unites States, 2002-2012. National Health Statistics Reports; no 79. Hyattsville, MD: National Center for Health Statistics.
Conchillo-Solé, O., Yero, D., Coves, X., Huedo, P., Martínez-Servat, S., Daura, X., et al. (2015). Draft genome sequence of Stenotrophomonas maltophilia strain UV74 reveals extensive variability within its genomic group. Genome Announc. 3:e00611-15. doi: 10.1128/genomeA.00611-15
Crossman, L. C., Gould, V. C., Dow, J. M., Vernikos, G. S., Okazaki, A., Sebaihia, M., et al. (2008). The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Genome Biol. 9:R74. doi: 10.1186/gb-2008-9-4-r74
de Oliveira-Garcia, D., Dall’Agnol, M., Rosales, M., Azzuz, A. C., Alcántara, N., Martinez, M. B., et al. (2003). Fimbriae and adherence of Stenotrophomonas maltophilia to epithelial cells and to abiotic surfaces. Cell. Microbiol. 5, 625–636. doi: 10.1046/j.1462-5822.2003.00306.x
de Oliveira-Garcia, D., Dall’Agnol, M., Rosales, M., Azzuz, A. C. G. S., Martinez, M. B., and Girón, J. A. (2002). Characterization of flagella produced by clinical strains of Stenotrophomonas maltophilia. Emerg. Infect. Dis. 8, 918–923. doi: 10.3201/eid0809.010535
de Souza, M. L., Seffernick, J., Martinez, B., Sadowsky, M. J., and Wackett, L. P. (1998). The atrazine catabolism genes atzABC are widespread and highly conserved. J. Bacteriol. 180, 1951–1954.
Ding, X., Yin, B., Qian, L., Zeng, Z., Yang, Z., and Li, H. (2011). Screening for novel quorum sensing inhibitors to interfere with the formation of Pseudomonas aeruginosa biofilms. J. Med. Microbiol. 60, 1827–1834. doi: 10.1099/jmm.0.024166-0
Domenico, P., Cunha, B. A., and Salo, R. J. (2000). The potential of bismuth-thiols for treatment and prevention of infection. Infect. Med. 17, 123–127.
Domenico, P., Kazzaz, J. A., and Davis, J. M. (2003). Combating antibiotic resistance with bismuth-thiols. Res. Adv. Antimicrob. Agents Chemother. 3, 79–85.
Donlan, R. M. (2009). Preventing biofilms of clinically relevant organisms using bacteriophage. Trends Microbiol. 17, 66–72. doi: 10.1016/j.tim.2008.11.002
Dubey, K. K., and Fulekar, M. H. (2012). Chlorpyrifos bioremediation in Pennisetum rhizosphere by a novel potential degrader Stenotrophomonas maltophilia MHF ENV20. World J. Microbiol. Biotechnol. 28, 1715–1725. doi: 10.1007/s11274-011-0982-1
Dunne, C., Crowley, J. J., Monne-Loccoz, Y., Dowling, D. N., de Bruijn, F. J., and O’Gara, F. (1997). Biological control of Pythium ultimum by Stenotrophomonas maltophilia W81 is mediated by an extracellular proteolytic activity. Microbiology 143, 1–393.
Dushenkov, V., Kumar, P. B. A. N., Motto, H., and Raskin, I. (1995). Rhizofiltration: the use of plants to remove heavy metals from aqueous streams. Environ. Sci. Technol. 29, 1239–1245. doi: 10.1021/es00005a015
Eapen, S., and D’Souza, S. F. (2003). Prospects of genetic engineering of plants for phytoremediation of toxic metals. Biotech. Adv. 23, 97–114. doi: 10.1016/j.biotechadv.2004.10.001
Elad, Y., Chet, I., and Baker, R. (1987). Increased growth response of plants induced by rhizobacteria antagonistic to soilborne pathogenic fungi. Plant Soil 98, 325–339. doi: 10.1007/BF02378353
Elting, L. S., Khardori, N., Bodey, G. P., and Fainstein, V. (1990). Nosocomial infection caused by Xanthomonas maltophilia: a case-control study of predisposing factors. Infect. Control Hosp. Epidemiol. 11, 134–138. doi: 10.1086/646136
Emanuela, R., Rocco, F., Carlomagno, M. S., Casalino, M., Colonna, B., Zarrilli, R., et al. (2008). PCR-based rapid genotyping of Stenotrophomonas maltophilia isolates. BMC Microbiol. 8:202. doi: 10.1186/1471-2180-8-202
Fabio, A., Cermelli, C., Fabio, G., Nicoletti, P., and Quaglio, P. (2007). Screening of the antibacterial effects of a variety of essential oils on microorganisms responsible for respiratory infections. Phytother. Res. 21, 374–377. doi: 10.1002/ptr.1968
Falagas, M. E., Kastoris, A. C., Vouloumanou, E. K., Rafailidis, P. I., Kapaskelis, A. M., and Dimopoulos, G. (2009). Attributable mortality of Stenotrophomonas maltophilia infections: a systematic review of the literature. Future Microbiol. 4, 1103–1109. doi: 10.2217/fmb.09.84
Freeman, E. S. (2009). Development of an Anti-Stenotrophomonas maltophilia Immunoglobul1n-g (Igg) that Prevents Iron Transport in Gram Negative Bacteria. Ph.D. dissertation, Atlanta University Center, Atlanta, GA, 76.
Galai, S., Limam, F., and Marzouki, M. N. (2009). A new Stenotrophomonas maltophilia strain producing laccase. Use in decolorization of synthetics dyes. Appl. Biochem. Biotechnol. 158, 416–431. doi: 10.1007/s12010-008-8369-y
Gao, S., Seo, J.-S., Wang, J., Keum, Y.-S., Li, J., and Li, Q. X. (2013). Multiple degradation pathways of phenanthrene by Stenotrophomonas maltophilia C6. Int. Biodeterior. Biodegrad. 79, 98–104. doi: 10.1016/j.ibiod.2013.01.012
García, C. A., Alcaraz, E. S., Franco, M. A., and Passerini de Rossi, B. N. (2015). Iron is a signal for Stenotrophomonas maltophilia biofilm formation, oxidative stress response, OMPs expression, and virulence. Front. Microbiol. 6:926. doi: 10.3389/fmicb.2015.00926
Ghosh, A., and Saha, P. D. (2013). Optimization of copper bioremediation by Stenotrophomonas maltophilia PD2. J. Environ. Chem. Eng. 1, 159–163. doi: 10.1016/j.jece.2013.04.012
Gren, I., Danuta, W., Urszula, G., Magdalena, P., and Katarzyna, H.-K. (2010). Enhanced biotransformation of mononitrophenols by Stenotrophomonas maltophilia KB2 in the presence of aromatic compounds of plant origin. World J. Microbiol. Biotechnol. 26, 289–295. doi: 10.1007/s11274-009-0172
Guillermo, G.-L., Alvaro, H., Hernando-Amado, S., Peyman, A., Berg, G., and Martínez, J. L. (2014). A function of SmeDEF, the major quinolone resistance determinant of Stenotrophomonas maltophilia, is the colonization of plant roots. Appl. Environ. Microbiol. 80, 4559–4565.
Hansen, N., Rasmussen, A. K., Fiandaca, M. J., Kragh, K. N., Bjarnsholt, T., Høiby, N., et al. (2014). Rapid identification of Stenotrophomonas maltophilia by peptide nucleic acid fluorescence in situ hybridization. New Microbe New Infect 2, 79–81. doi: 10.1002/nmi2.38
Hoefel, D., Ho, L., Monisa, P. T., Newcombe, G., and Saint, C. P. (2009). Biodegradation of geosmin by a novel Gram-negative bacterium; isolation, phylogenetic characterization and degradation rate determination. Water Res. 43, 2927–2935. doi: 10.1016/j.watres.2009.04.005
Huedo, P., Yero, D., Martínez-Servat, S., Estibariz, I., Planell, R., Martínez, P., et al. (2014). Two different rpfclusters distributed among a population of Stenotrophomonas maltophilia clinical strains display differential diffusible signal factor production and virulence regulation. J. Bacteriol. 196, 2431–2442. doi: 10.1128/JB.01540-14
Insunza, V., Alström, S., and Eriksson, K. B. (2002). Root bacteria from nematicidal plants and their biocontrol potential against trichodorid nematodes in potato. Plant Soil 241, 271–278. doi: 10.1023/A:1016159902759
Iravani, S. (2014). Bacteria in nanoparticle synthesis: current status and future prospects. Int. Sch. Res. Notices 2014, 359316. doi: 10.1155/2014/359316
Jakobi, M., Winkelmann, G., Kaiser, D., Kempler, C., Jung, G., Berg, G., et al. (1996). Maltophilin: a new antifungal compound produced by Stenotrophomonas maltophilia R3089. J. Antibiot. (Tokyo) 49, 1101–1104. doi: 10.7164/antibiotics.49.1101
Jankiewicz, U., Brzezinska, M. S., and Saks, E. (2012). Identification and characterization of a chitinase of Stenotrophomonas maltophilia, a bacterium that is antagonistic towards fungal phytopathogens. J. Biosci. Bioeng. 113, 30–35. doi: 10.1016/j.jbiosc.2011.08.023
Khan, Z., and Doty, S. L. (2009). Characterization of bacterial endophytes of sweet potato plants. Plant Soil 322, 197–207. doi: 10.1007/s11104-009-9908-1
Kinoshita, Y., Niwa, H., and Katayama, Y. (2015). Use of loop-mediated isothermal amplification to detect six groups of pathogens causing secondary lower respiratory bacterial infections in horses. Microbiol. Immunol. 59, 365–370. doi: 10.1111/1348-0421.12257
Korade, D., Puranik, S., and Patil, S. (2014). Larvicidal activity of rhamnolipid biosurfactant produced by Stenotrophomonas maltophilia. Int. J. Sci. Eng. Res. 5, 60–63.
Kuddus, M., and Ramteke, P. W. (2009). Cold-active extracellular alkaline protease from an alkaliphilic Stenotrophomonas maltophilia: production of enzyme and its industrial applications. Can. J. Microbiol. 55, 1294–1301. doi: 10.1139/w09-089
Kumar, K., Devi, S. S., Krishnamurthi, K., Kanade, G. S., and Chakrabarti, T. (2007). Enrichment and isolation of endosulfan degrading and detoxifying bacteria. Chemosphere 68, 317–322. doi: 10.1016/j.chemosphere.2006.12.076
Kumar, N. P., and Audipudi, V. (2015). Exploration of a novel plant growth promoting bacteria Stenotrophomonas maltophilia AVP27 isolated from the chilli rhizosphere. Int. J. Eng. Res. Gen. Sci. 3, 265–276.
Kwok, O. C. H., Fahy, P. C., Hoitink, H. A. J., and Kuter, G. A. (1987). Interactions between bacteria and Trichoderma hamatum in suppression of Rhizoctonia damping-off in bark compost media. Phytopathology 77, 1206–1212. doi: 10.1094/Phyto-77-1206
Lambert, B., Frederik, L., Van Rooyen, L., Gossele, F., Papon, Y., and Swings, J. (1866). Rhizobacteria of maize and their antifungal activities. Appl. Environ. Microbiol. 53, 1866–1871.
Lata, H., Li, X. C., and Silva, B. (2006). Identification of IAA producing endophytic bacteria from micropropagated echinacea plants using 16S rRNA sequencing. Plant Cell Tissue Organ Cult. 85, 353–359. doi: 10.1007/s11240-006-9087-1
Li, Z., Nandakumar, R., Madayiputhiya, N., and Li, X. (2012). Proteomic analysis of 17β-estradiol degradation by Stenotrophomonas maltophilia. Environ. Sci. Technol. 46, 5947–5955. doi: 10.1021/es300273k
LiPuma, J. J., Currie, B. J., Lum, G. D., and Vandamme, P. A. R. (2007). “Burkholderia, Stenotrophomonas, Ralstonia, Cupriavidus, Pandoraea, Brevundimonas, Comamonas, Delftia and Acidovorax,” in Manual of Clinical Microbiology, 9th Edn, eds J. Versalovic, K. C. Carroll, G. Funke, J. H. Jorgensen, M. L. Landry, and D. W. Warnock (Washington, DC: ASM Press), 749–769.
LiPuma, J. J., Rathinavelu, S., Foster, B. K., Keoleian, J. C., Makidon, P. E., Kalikin, L. M., et al. (2009). In vitro activities of a novel nanoemulsion against Burkholderia and other multidrug-resistant cystic fibrosis-associated bacterial species. Antimicrob. Agents Chemother. 53, 249–255. doi: 10.1128/AAC.00691-08
Liu, Z., Yang, C., and Qiao, C. (2007). Biodegradation of p-Ni-trophenol and 4-Chlorophenol by Stenotrophomonas sp. FEMS Microbiol. Ecol. 277, 150–156. doi: 10.1111/j.1574-6968.2007.00940.x
Maes, M. (1993). Fast classification of plant-associated bacteria in the Xanthomonas genus. FEMS Microbiol. Lett. 113, 161–166. doi: 10.1111/j.1574-6968.1993.tb06508.x
Malhotra, A., Kunzes, D., and Roy Choudhury, A. (2013). Biosynthesis of gold and silver nanoparticles using a novel marine strain of Stenotrophomonas. Bioresour. Technol. 142, 727–731. doi: 10.1016/j.biortech.2013.05.109
Markowitz, V. M., Korzeniewski, F., Palaniappan, K., Szeto, E., Werner, G., Padki, A., et al. (2006). The integrated microbial genomes (IMG) system. Nucleic Acids Res 1, 34; D344–348.
Messiha, N. A. S., van Diepeningen, A. D., Farag, N. S., Abdallah, S. A., Janse, J. D., and van Bruggen, A. H. C. (2007). Stenotrophomonas maltophilia: a new potential biocontrol agent of Ralstonia solanacearum, causal agent of potato brown rot. Eur. J. Plant Pathol. 118, 211–225. doi: 10.1007/s10658-007-9136-6
Minogue, E., Tuite, N. L., Smith, C. J., Reddington, K., and Barry, T. (2015). A rapid culture independent methodology to quantitatively detect and identify common human bacterial pathogens associated with contaminated high purity water. BMC Biotechnol. 15:6. doi: 10.1186/s12896-015-0124-1
Mitra, S., and Roy, P. (2011). Molecular phylogeny of a novel trichloroethylene degrading gene of Bacillus cereus 2479. J. Biol. Sci. .11, 58–63. doi: 10.3923/jbs.2011.58.63
Miyaji, T., Otta, Y., Shibata, T., Mitsui, K., Nakagawa, T., Watanabe, T., et al. (2005). Purification and characterization of extracellular alkaline serine protease from Stenotrophomonas maltophilia strain S-1. Lett. Appl. Microbiol. 41, 253–257. doi: 10.1111/j.1472-765X.2005.01750.x
Moore, J. E., Xu, J., Millar, B. C., Courtney, J., and Elborn, J. S. (2003). Development of a gram-negative selective agar (GNSA) for the detection of Gram-negative microflora in sputa in patients with cystic fibrosis. J. Appl. Microbiol. 95, 160–166. doi: 10.1046/j.1365-2672.2003.01956.x
Mukherjee, P., and Roy, P. (2012). Identification and characterisation of a bacterial isolate capable of growth on trichloroethylene as the sole carbon source. Adv. Microbiol. 2, 184–194. doi: 10.4236/aim.2012.23034
Mukherjee, P., and Roy, P. (2013a). Cloning, sequencing and expression of novel trichloroethylene degradation genes from Stenotrophomonas maltophilia PM102: a case of gene duplication. J. Bioremed. Biodegrad. 4, 177.
Mukherjee, P., and Roy, P. (2013b). Copper enhanced mono-oxygenase activity and FT-IR spectroscopic characterisation of biotransformation products in trichloroethylene degrading bacterium: Stenotrophomonas maltophilia PM102. Biomed Res. Int. 2013, 723680. doi: 10.1155/2013/723680
Mukherjee, P., and Roy, P. (2013c). Persistent organic pollutants induced protein expression and immunocrossreactivity by Stenotrophomonas maltophilia PM102: a prospective bioremediating candidate. Biomed Res. Int. 2013, 714232. doi: 10.1155/2013/714232
Mwangi, K., Hamadi, I., Boga Muigai, A. W., Kiiyukia, C., and Tsanuo, M. K. (2010). Degradation of dichlorodiphenyltrichloroethane (DDT) by bacterial isolates from cultivated and uncultivated soil. Afr. J. Microbiol. Res. 4, 185–196.
Nakajima, Y., Ito, K., Toshima, T., Egawa, T., Zheng, H., Oyama, H., et al. (2008). Dipeptidyl aminopeptidase IV from Stenotrophomonas maltophilia exhibits activity against a substrate containing a 4-hydroxyproline residue. J. Bacteriol. 190, 7819–7829. doi: 10.1128/JB.02010-07
Nakayama, T., Homma, Y., Hashidoko, Y., Mizutani, J., and Tahara, S. (1999). Possible role of xanthobaccins produced by Stenotrophomonas sp. strain SB-K88 in suppression of sugar beet damping-off disease. Appl. Environ. Microbiol. 65, 4334–4339.
Nangia, Y., Wangoo, N., Goyal, N., Shekhawat, G., and Suri, R. C. (2009). A novel bacterial isolate Stenotrophomonas maltophilia as living factory for synthesis of gold nanoparticles. Microb. Cell Fact. 8, 39. doi: 10.1186/1475-2859-8-39
Naz, I., and Bano, A. (2012). Assessment Of phytohormones producing capacity of Stenotrophomonas Maltophilia ssa and its interaction with Zea mays L. Pak. J. Bot. 44, 465–469.
Nordberg, H., Cantor, M., Dusheyko, S., Hua, S., Poliakov, A., Shabalov, I., et al. (2014). The genome portal of the department of energy joint genome institute: 2014 updates. Nucleic Acids Res. 42, D26–D31. doi: 10.1093/nar/gkt1069
Ortiz-Hernández, L., Enrique, S.-S., Dantan-Gonzalez, E., and Castrejón-Godínez, M. L. (2013). Pesticide biodegradation: mechanisms, genetics and strategies to enhance the process. biodegradation – life of science. Intech 10, 251–287. doi: 10.5772/56098
Oves, M., Khan, M. S., Zaidi, A., Ahmed, A. S., Ahmed, F., Ahmed, E., et al. (2013). Antibacterial and cytotoxic efficacy of extracellular silver nanoparticles biofabricated from chromium reducing novel OS4 strain of Stenotrophomonas maltophilia. PLoS ONE 8:e59140. doi: 10.1371/journal.pone.0059140
Page, M. G., Dantier, C., Desarbre, E., Gaucher, B., Gebhardt, K., and Schmitt-Hoffmann, A. (2011). In vitro and in vivo properties of BAL30376, a β-lactam and dual beta-lactamase inhibitor combination with enhanced activity against Gram-negative Bacilli that express multiple β-lactamases. Antimicrob. Agents Chemother. 55, 1510–1519. doi: 10.1128/AAC.01370-10
Pages, D., Rose, J., Conrod, S., Cuine, S., Carrier, P., Heulin, T., et al. (2008). Heavy metal tolerance in Stenotrophomonas maltophilia. PLoS ONE 3:e1539. doi: 10.1371/journal.pone.0001539
Palleroni, N. J. (1984). “Genus I. Pseudomonas Migula,” in Bergey’s Manual of Systematic Bacteriology, Vol. 1, eds N. R. Krieg and J. G. Holt (Baltimore, MD: The Williams & Wilkins Co), 141–199.
Palleroni, N., and Bradbury, J. (1993). Stenotrophomonas, a new bacterial genus for Xanthomonas maltophilia (Hugh 1980) Swings et al. 1983. Int. J. Syst. Bacteriol. 43, 606–609. doi: 10.1099/00207713-43-3-606
Pellestor, F., Paulasova, P., and Hamamah, S. (2008). Peptide nucleic acids (PNAs) as diagnostic devices for genetic and cytogenetic analysis. Curr. Pharm. Des. 14, 2439–2444. doi: 10.2174/138161208785777405
Personne, Y., Curtis, M. A., Wareham, D. W., and Waite, R. D. (2014). Activity of the type I signal peptidase inhibitor MD3 against multidrug-resistant Gram-negative bacteria alone and in combination with colistin. J. Antimicrob. Chemother. 69, 3236–3243. doi: 10.1093/jac/dku309
Piddock, L. J. (2006). Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin. Microbiol. Rev. 19, 382–4021. doi: 10.1128/CMR.19.2.382-402
Pompilio, A., Pomponio, S., Crocetta, V., Gherardi, G., Verginelli, F., Fiscarelli, E., et al. (2011). Phenotypic and genotypic characterization of Stenotrophomonas maltophilia isolates from patients with cystic fibrosis: genome diversity, biofilm formation, and virulence. BMC Microbiol. 11:159. doi: 10.1186/1471-2180-11-159
Rajeswari, K., Subashkumar, R., and Vijayaraman, K. (2013). Decolorization and degradation of textile dyes by Stenotrophomonas maltophilia RSV-2. Int. J. Environ. Bioremed. Biodegrad. 1, 60–65. doi: 10.12691/ijebb-1-2-5
Ratjen, A., Yau, Y., Wettlaufer, J., Matukas, L., Zlosnik, J. E., Speert, D. P., et al. (2015). In vitro efficacy of high-dose tobramycin against Burkholderia cepacia complex and Stenotrophomonas maltophilia isolates from cystic fibrosis patients. Antimicrob. Agents Chemother. 59, 711–713. doi: 10.1128/AAC.04123-14
Ricardo, P. M. D., and Dionísio, F. (2014). Screening and Isolation of Compounds with Antimicrobial Activity Produced by Multi-Resistant Bacteria. Master thesis, University of Lisbon, Lisbon.
Romanenko, L. A., Uchino, M., Tanaka, N., Frolova, G. M., Slinkina, N. N., and Mikhailov, V. V. (2007). Occurrence and antagonistic potential of Stenotrophomonas strains isolated from deep-sea invertebrates. Arch. Microbiol. 189, 337–344. doi: 10.1007/s00203-007-0324-8
Rousseaux, S., Hartmann, A., and Soulas, G. (2001). Isolation and characterization of new gram-negative and gram positive atrazine degrading bacteria from different French soils. FEMS Microbiol. Ecol. 36, 211–222. doi: 10.1111/j.1574-6941.2001.tb00842.x
Ryan, R. P., Monchy, S., Cardinale, M., Taghavi, S., Crossman, L., Avison, M. B., et al. (2009). The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat. Rev. Microbiol. 7, 514–525. doi: 10.1038/nrmicro2163
Ryan, R. P., An, S.-Q., Allan, J. H., McCarthy, Y., and Dow, J. M. (2015). The DSF family of cell–cell signals: an expanding class of bacterial virulence regulators. PLoS Pathog. 11:e1004986. doi: 10.1371/journal.ppat.1004986
Saito, A., Tokuyama, T., Tanaka, A., Oritani, T., and Fuchigami, K. (1999). Microbiological degradation of (-)-geosmin. Water Res. 33, 3033–3036. doi: 10.1016/S0043-1354(99)00155-4
Sharma, R., Chisti, Y., and Banerjee, U. C. (2001). Production, purification, characterization, and applications of lipases. Biotechnol. Adv. 19, 627–662. doi: 10.1016/S0734-9750(01)00086-6
Smith, D., Alvey, S., and Crowley, D. E. (2005). Cooperative catabolic pathways within an atrazine-degrading enrichment culture isolated from soil. FEMS Microbiol. Ecol. 1, 265–273. doi: 10.1016/j.femsec.2004.12.011
Sperling, R. A., Rivera Gil, P., Zhang, F., Zanella, M., and Parak, W. J. (2008). Biological applications of gold nanoparticles. Chem. Soc. Rev. 37, 1896–1908. doi: 10.1039/B712170A
Stalons, D., Sesler, C., Green, J., Malone, L., White, H., and Grigorenko, E. (2014). Target Enriched Multiplex PCR (TEM-PCRTM) for Rapid Detection of Bloodstream Infections, 261-267. Available at: www.diatherix.com/assets/pdf/publications/TEM-PCR_Chapter.pdf
Swings, J., De Vos, P., Van den Mooter, M., and De Ley, J. (1983). Transfer of Pseudomonas maltophilia Hugh 1981 to the genus Xanthomonasas Xanthomonas maltophilia (Hugh 1981) comb. nov. Int. J. Syst. Bacteriol. 33, 409–413. doi: 10.1099/00207713-33-2-409
Tallur, P. N., Megadi, V. B., and Ninnekar, H. Z. (2008). Biodegradation of cypermethrin by Micrococcus sp. strain CPN 1. Biodegradation 19, 77–82. doi: 10.1007/s10532-007-9116-8
Trivedi, M. K., Patil, S., Shettigar, H., Gangwar, M., and Jana, S. (2015). An evaluation of biofield treatment on susceptibility pattern of multidrug resistant stenotrophomonas maltophilia : an emerging global opportunistic pathogen. Clin. Microbiol. 4, 211. doi: 10.4172/2327-5073.1000211
Urszula, G., Izabela, G., Danuta, W., and Sylwia, L. (2009). Isolation and characterization of a novel strain of Stenotrophomonas maltophilia possessing various dioxy-genases for monocyclic hydrocarbon degradation. Braz. J. Microbiol. 40, 285–291.
Vallini, G., Di Gregorio, S., and Lampis, S. (2005). Rhizosphere-induced selenium precipitation for possible applications in phytoremediation of se polluted effluents. Z. Naturforsch. C 60, 349–356. doi: 10.1515/znc-2005-3-419
Van Rossum, T., Pylatuk, M. M., Osachoff, H. L., Griffiths, E. J., Lo, R., Quach, M., et al. (2016). Microbiome analysis accross a natural copper gradient at a proposed Northern Canadian Mine. Front. Environ. Sci. 3:84. doi: 10.3389/fenvs.2015.00084
Van Zyl, E., and Steyn, P. L. (1992). Reinterpretation of the taxonomic position of Xanthomonas maltophilia and taxonomic criteria in this genus. Request for an opinion. Int. J. Syst. Bacteriol. 42, 193–198. doi: 10.1099/00207713-42-1-193
Vega, F. E., Ripoll, M. P., Posada, F., and Buyer, J. S. (2005). Endophytic bacteria in Coffea arabica L. J. Basic Microbiol. 45, 371–380. doi: 10.1002/jobm.200410551
Vidigal, P. G., Müsken, M., Becker, K. A., Häussler, S., Wingender, J., Steinmann, E., et al. (2014). Effects of green tea compound epigallocatechin-3-gallate against Stenotrophomonas maltophilia infection and biofilm. PLoS ONE 9:e92876. doi: 10.1371/journal.pone.0092876
Wang, Y. L., Scipione, M. R., Dubrovskaya, Y., and Papadopoulos, J. (2014). Monotherapy with fluoroquinolone or trimethoprim-sulfamethoxazole for treatment of Stenotrophomonas maltophilia infections. Antimicrob. Agents Chemother. 58, 176–182. doi: 10.1128/AAC.01324-13
Waters, V. (2012). New treatments for emerging cystic fibrosis pathogens other than Pseudomonas. Curr. Pharm. Des. 18, 696–725. doi: 10.2174/138161212799315939
Wei, C., Ni, W., Cai, X., and Cui, J. (2015). A Monte Carlo pharmacokinetic/pharmacodynamic simulation to evaluate the efficacy of minocycline, tigecycline, moxifloxacin, and levofloxacin in the treatment of hospital-acquired pneumonia caused by Stenotrophomonas maltophilia. Infect. Dis. (Auckl.) 47, 846–851. doi: 10.3109/23744235.2015.1064542
Wierzba, S. (2015). Biosorption of lead(II), zinc(II) and nickel(II) from industrial wastewater by Stenotrophomonas maltophilia and Bacillus subtilis. Pol. J. Chem. Technol. 17, 79–87. doi: 10.1515/pjct-2015-0012
Willetts, A., Joint, I., Gilbert, J. A., Trimble, W., and Mühling, M. (2012). Isolation and initial characterization of a novel type of Baeyer–Villiger monooxygenase activity from a marine microorganism. Microb. Biotechnol. 5, 549–559. doi: 10.1111/j.1751-7915.2012.00337.x
Wu, C. L., Domenico, P., Hassett, D. J., Beveridge, T. J., Hauser, A. R., and Kazzaz, J. A. (2002). Subinhibitory bismuth-thiols reduce virulence of Pseudomonas aeruginosa. Am. J. Respir. Cell Mol. Biol. 26, 731–738. doi: 10.1165/ajrcmb.26.6.2001-00020oc
Wu, X., Jing, W., Yuan, Z., Zhangjun, C., and Zhou, M. (2012). “Induction and selection of Stenotrophomonas maltophilia DHHJ for feather degradation,” in Proceedings of the International Conference on Biomedical Engineering and Biotechnology ICBEB ’12 (Washington, DC: IEEE Computer Society), 1521–1524.
Yamamura, S., Morita, Y., Hasan, Q., Yokoyama, K., and Tamiya, E. (2002). Keratin degradation: a cooperative action of two enzymes from Stenotrophomonas sp. Biochem. Biophys. Res. Commun. 294, 1138–1143. doi: 10.1016/S0006-291X(02)00580-6
Yang, Z., Liu, W., Cui, Q., Niu, W., Li, H., Zhao, X., et al. (2014). Prevalence and detection of Stenotrophomonas maltophilia carrying metallo-β-lactamase blaL1 in Beijing, China. Front. Microbiol. 5:692. doi: 10.3389/fmicb.2014.200692
Youenou, B., Favre-Bonté, S., Bodilis, J., Brothier, E., Dubost, A., Muller, D., et al. (2015). Comparative genomics of environmental and clinical Stenotrophomonas maltophilia strains with different antibiotic resistance profiles. Genome Biol. Evol. 7, 2484–2505. doi: 10.1093/gbe/evv161
Zhang, L., Li, X. Z., and Poole, K. (2001). Fluoroquinolone susceptibilities of efflux-mediated multidrug-resistant Pseudomonas aeruginosa, Stenotrophomonas maltophilia and Burkholderia cepacia. J. Antimicrob. Chemother. 48, 549–552. doi: 10.1093/jac/48.4.549
Zhang, L., Morrison, M. O., Cuiv, P., Evans, P., and Rickard, C. M. (2013). Genome sequence of Stenotrophomonas maltophilia AU12-09, isolated from an intravascular catheter. Genome Announc. 1, e195–e113. doi: 10.1128/genomeA.00195-13
Keywords: Stenotrophomonas, nosocomial, immunocompromised, multidisciplinary, bioremediation
Citation: Mukherjee P and Roy P (2016) Genomic Potential of Stenotrophomonas maltophilia in Bioremediation with an Assessment of Its Multifaceted Role in Our Environment. Front. Microbiol. 7:967. doi: 10.3389/fmicb.2016.00967
Received: 03 July 2015; Accepted: 03 June 2016;
Published: 22 June 2016.
Edited by:
Giovanni Di Bonaventura, “G. D’Annunzio” University of Chieti-Pescara, ItalyReviewed by:
Paras Jain, Albert Einstein College of Medicine, USAJose L. Martinez, Centro Nacional de Biotecnología, Spain
Copyright © 2016 Mukherjee and Roy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Piyali Mukherjee, cGl5YWxpbWtocmo4QGdtYWlsLmNvbQ==
 Piyali Mukherjee
Piyali Mukherjee Pranab Roy
Pranab Roy