- 1Division of Medical Microbiology, Department of Pathology, Faculty of Health Sciences, University of Cape Town, Cape Town, South Africa
- 2Centre for Evidence-based Health Care, Faculty of Medicine and Health Sciences, Stellenbosch University, Cape Town, South Africa
- 3Cochrane South Africa, South African Medical Research Council, Cape Town, South Africa
- 4Vaccines for Africa Initiative, Institute of Infectious Disease and Molecular Medicine, University of Cape Town, Cape Town, South Africa
- 5Department of Clinical Epidemiology and Biostatistics, McMaster University, Ontario, Canada
- 6Biostatistics Unit, Father Sean O'SulliVan Research Centre, Ontario, Canada
- 7Division of Immunology, Department of Pathology, Faculty of Health Sciences, University of Cape Town, Cape Town, South Africa
- 8Institute of Infectious Diseases and Molecular Medicine, University of Cape Town, Cape Town, South Africa
- 9International Centre for Genetic Engineering and Biotechnology, University of Cape Town, Cape Town, South Africa
- 10Department of Paediatrics and Child Health, University of Cape Town, Cape Town, South Africa
- 11Red Cross War Memorial Children's Hospital, Cape Town, South Africa
- 12Medical Research Council Unit on Child and Adolescent Health, University of Cape Town, Cape Town, South Africa
- 13National Health Laboratory Service, Groote Schuur Hospital, Cape Town, South Africa
Introduction
Asthma is a complex respiratory condition that involves interplay between genetic predisposition, environmental, and immunological factors (Edwards et al., 2012). It is considered to be one of the most common chronic diseases, affecting ~300 million people (Masoli et al., 2004), and causing an estimated 250,000 deaths annually (Bateman et al., 2008). Furthermore, because of an increased Westernized lifestyle and urbanization in developing countries, it is estimated that by 2025 the global burden of asthma will increase by 100 million people (Masoli et al., 2004).
An increase in the occurrence of allergic diseases, including asthma, was initially attributed to the “hygiene hypothesis,” suggesting that a reduced exposure to microbes during the first years of life plays a role in the development of allergic diseases (Strachan, 1989, 2000). Although this hypothesis is widely accepted, studies showed that reduced microbial exposure cannot fully account for the increased prevalence of asthma, rhinitis, or neurodermitis (Mallol, 2008; Brooks et al., 2013; Kramer et al., 2013). Alternative hypotheses or reformulations of the “hygiene hypothesis” (Hunter, 2012), such as the “microbiota hypothesis” (Wold, 1998), the “old friends hypothesis” (Rook, 2012), the “microbial deprivation hypothesis” (Bloomfield et al., 2006), the “biodiversity hypothesis” (Hanski et al., 2012) and the “disappearing microbiota hypothesis” (Blaser and Falkow, 2009; Taube and Müller, 2012) soon followed; all of which mainly postulate that dysbiosis of the human gastro-intestinal tract (GIT) microbiome may contribute to intra- and extra-intestinal immune-mediated diseases (Penders et al., 2007; Štšepetova et al., 2007; Sekirov et al., 2010; Clemente et al., 2012; Russell and Finlay, 2012). Understanding which bacteria from our GITs contribute to the development or prevention of allergic asthma may result in further research to discern the mechanisms behind bacterial-host interactions and potentially facilitate treatment strategies.
Methods Used to Study the Role of Human Fecal Bacteria in Asthma
Although culture-independent techniques have revolutionized the world of microbiology (Suau et al., 1999; Zoetendal et al., 2006; Rajilić-Stojanović et al., 2007); conventional culture-dependent techniques have been the method widely used to study the role of human fecal bacteria in asthma (Mansson and Colldahl, 1965; Stockert, 2001; Nambu et al., 2008; Vael et al., 2008; Bisgaard et al., 2011). To date, the culture-independent techniques used to characterize fecal bacteria from patients with asthma include quantitative real-time polymerase chain reaction (qPCR) (Van Nimwegen et al., 2011), denaturing gradient gel electrophoresis (DGGE) (Bisgaard et al., 2011; Vael et al., 2011), fluorescent in situ hybridization (FISH) (Salminen et al., 2004), and massively parallel high-throughput sequencing of the 16S ribosomal RNA (rRNA) gene (Arrieta et al., 2015). Despite the advantage of detecting uncultivable bacteria, these culture-independent techniques are not without limitations. Among others, they do not allow for whole community analysis of the microbial population (Sekirov et al., 2010; Fraher et al., 2012; Sankar et al., 2015), which is considered key in determining the patterns of fecal bacteria associated with health and disease states (Schippa and Conte, 2014). For example, qPCR and FISH do not provide identification of novel organisms as they are used to characterize and quantify targeted groups of bacteria (Sekirov et al., 2010; Fraher et al., 2012). DGGE, a band-based method for determining bacterial diversity, does not enable direct identification of bacteria (Sekirov et al., 2010; Fraher et al., 2012). Furthermore, DGGE has low bacterial detection limits and limited phylogenetic resolution (Sekirov et al., 2010). Although massively parallel high-throughput sequencing of the 16S rRNA gene provides an almost comprehensive view of bacterial communities; it does not provide classification at species-level (Gosalbes et al., 2012; Arrieta et al., 2015). The importance of species-level characterization in health and disease states has been demonstrated in murine models of allergic diseases (Karimi et al., 2009; Russell et al., 2012; Kim et al., 2013). An overgrowth of the genus Lactobacillus has been associated with an increased risk of allergic asthma (Russell et al., 2012), while the species L. reuteri and L. rhamnosus provide a protective role in allergic airway disease (Karimi et al., 2009; Kim et al., 2013). In comparison to 16S rRNA gene sequencing techniques; whole genome shotgun (WGS) sequencing offers a higher and more reliable resolution of microbiota profiles at lower taxonomic levels (Morgan and Huttenhower, 2014; Van Dijk et al., 2014; Ranjan et al., 2015). For example, WGS sequencing is able to improve the issue related to the Bifidobacterium amplification bias by certain primer sets (Kurokawa et al., 2007; Sim et al., 2012; Walker et al., 2015). In addition, it allows for determination of the metabolic and functional properties of fecal bacteria which may greatly contribute to our understanding of the role of fecal bacteria in health and disease (Qin et al., 2012; Arrieta et al., 2015; Quince et al., 2015). However, despite the number of advantages that WGS sequencing provides, it has not been incorporated by any of the studies investigating the importance of fecal bacteria in the development of asthma. Furthermore, a causal link between fecal bacterial profiles and asthma in humans has recently been confirmed using murine models (Arrieta et al., 2015). To the best of our knowledge, this is the only report of its kind where the causal role of the fecal bacteria (Lachnospira, Veillonella, Faecalibacterium, and Rothia), which potentially confers protection against the development of asthma in humans, was demonstrated using germ-free mice models (Arrieta et al., 2015).
What Do Studies in Humans Reveal About the Role of Fecal Bacteria in Asthma?
Although prospective longitudinal studies are key in demonstrating the role of fecal bacteria in disease development (Zhao, 2013); we identified only two studies which assessed whether fecal bacterial profiles sampled over time preceded the occurrence of asthma at later stages in life (Bisgaard et al., 2011; Arrieta et al., 2015). Bisgaard et al. (2011) did not report a significant association (Bisgaard et al., 2011). In contrast, Arrieta et al. (2015) found significantly reduced abundances of the bacterial genera Lachnospira, Veillonella, Rothia, and Faecalibacterium in infants at risk for asthma, as evidenced using the Asthma Predictive Index (API) (Arrieta et al., 2015). Moreover, in a prospective birth cohort study conducted in Belgium (using fecal specimens sampled at 3 weeks of age); the detection of Bacteroides (B. fragilis, B. finegoldii, and B. thetaiotaomicron), Ruminococcus (R. productus and R. hansenii), and Clostridium spp. was associated with an increased risk for asthma development (as based on the API) (Vael et al., 2011). At species-level, the prospective birth cohort study by Van Nimwegen et al. (2011) conducted in the Netherlands reported a two-fold increased risk of asthma at 6–7 years in infants colonized with Clostridium difficile at 1 month of life (OR = 2.06; 95% CI 1.16–3.64) (Van Nimwegen et al., 2011).
All prospective birth cohort studies, except for the study by Nambu et al. (2008), made use of the API when assessing asthma as an outcome at < 5 years of age (Vael et al., 2008, 2011; Arrieta et al., 2015). The API, incorporated by three studies cited in this review, is an example of a predictive assessment for asthma development later in life, recommended for young children experiencing recurrent wheeze (Castro-Rodriguez, 2010). Considering that asthma diagnosis in children < 5 years of age is challenging and often based on symptom patterns, clinical assessment of the family history and the presence of atopy (Pedersen, 2007; Sly et al., 2008; Pedersen et al., 2011); the use of predictive assessments, such as the API, is essential. However, despite its success in developed countries, the API should be used with caution in infants from low and middle income countries (LMICs) (Zar and Levin, 2012). This may be explained by the fact that young children from LMICs are more commonly affected by viral lower respiratory tract infections (LRTIs) or pulmonary tuberculosis. Furthermore, it has been suggested that atopy may be less strongly associated with asthma in these settings compared to the more developed countries (Zar and Levin, 2012). This suggests that non-atopic wheeze may be the primary form of asthma in these children, making the API, which relies primarily on the presence of atopy for assessing the risk of asthma, a less reliable predictive assessment tool in LMICs (Zar and Levin, 2012).
Factors Influencing Fecal Bacterial Profiles and Potentially Asthma
Both murine models and human studies have provided evidence that early life changes in the GIT microbiome are most influential in the development of allergic asthma (Russell et al., 2013; Arrieta et al., 2015). Some of the well described factors responsible for these early life changes in fecal bacterial profiles, which have also been associated with childhood asthma, are mode of delivery, feeding practices, and antibiotic use (Kozyrskyj et al., 2011).
Mode of Delivery
A number of childhood studies have reported that infants delivered via cesarean section are at an increased risk for the development of asthma (Thavagnanam et al., 2008). However, these studies do not account for confounding factors that may be associated independently with asthma, as well as changes in fecal bacterial profiles, which will allow for determining the true effect of external factors on fecal microbiota and the resultant health outcome. To date, only a single study using mediation analysis (Van Nimwegen et al., 2011) supported the role of mode and place of delivery (independent variable) in C. difficile colonization (mediator variable), together with its consequent impact on asthma development via modulation of C. difficile profiles (dependent variable).
Feeding Practices
Although, it has been reported that breastfeeding has the potential to protect against allergic airway disease (Dogaru et al., 2014), no studies have used mediation analysis (as performed by Van Nimwegen et al. (2011)) to determine whether bacteria from breast milk protect against asthma via the modulation of infant fecal bacteria.
Antibiotic Use
In humans, a modest increased risk of asthma development, associated with antibiotic use, has been reported (Marra et al., 2009; Risnes et al., 2010; Murk et al., 2011; Penders et al., 2011). To date, only fecal C. difficile colonization has been associated with the occurrence of asthma (Van Nimwegen et al., 2011), which might be explained (among other factors) by a loss of intestinal commensal microbes through the use of antibiotics (Azad and Kozyrskyj, 2012).
Potential Mechanisms Supporting the Role of Gastrointestinal Bacteria in Asthma
The exact mechanisms by which GIT bacteria may influence the development of respiratory diseases are unclear; however recent work has demonstrated that crosstalk between host mucosal immune cells and resident microbes significantly influences the risk for respiratory disease (Forsythe, 2011; Samuelson et al., 2015; Vital et al., 2015). A central player in this regulation of pulmonary immunity by the GIT microbiome are dendritic cells (DCs) (McLoughlin and Mills, 2011) (Figure 1). Intestinal DCs encounter bacterial antigens presented in organized GIT immune tissue (i.e., lamina propria and Peyer's patches) and also directly sample lumen residing bacteria in the GIT by extending their dendrites into the intestinal lumen (Salzman, 2011). This sampling of intestinal bacterial antigens results in DCs co-ordinating B and T cell subset expansion both locally (Peyer's patches and lamina propria) as well as systemically (e.g mesenteric lymph nodes) (Hill and Artis, 2010) (Figure 1). This results in DC-guided local and systemic immune education driven by microbiota associated antigens which has profound effects not just in the intestine but throughout the body (Hill and Artis, 2010; Russell and Finlay, 2012) (Figure 1). An important consequence of this effect of GIT bacteria is manifested in subsequent host T-cell immune responses in the lungs and has been particularly well demonstrated in murine models of asthma (Herbst et al., 2011; Navarro et al., 2011; Konieczna et al., 2012; Oertli et al., 2012). For example, B. fragilis and Clostridium species (cluster IV and XIVa), both intestinally restricted bacteria, can drive induction of T regulatory (Treg) cells and associated elevated secretion of the regulatory cytokine IL-10 in mesenteric lymph nodes to mediate protection against allergic T-helper cell (Th-) 2 airway inflammation (Round et al., 2011) (Figure 1). Other studies have demonstrated that early life depletion of Bacteroidetes species using vancomycin abrogates the ability of mice to launch Treg protection from allergic asthma (Atarashi et al., 2011; Russell et al., 2012). In addition, raised levels of Helicobacter pylori has also been shown to elicit protection against the development of asthma, again, via the induction of Treg cells (Arnold et al., 2011). Interestingly this effect may, in part at least, also be due to de novo production of IL-10 orthologs by H. pylori driving this Treg induction. Moreover, oral administration of probiotics (L. reuteri, L. rhamnosus GG, Bifidobacterium breve, or B. lactis) can impair the onset of ovalbumin induced allergic airway inflammation; again related to the reduced induction of Treg cells (Feleszko et al., 2007; Karimi et al., 2009). Taken together, these and other studies are generating an important profile of the microbial species driving Treg dependent protection against allergic airway inflammation. Other studies have also identified bacteria which may drive the onset of allergic pathology. Segmented filamentous bacteria (SFB), non-cultivable Clostridia-related host-specific species (Gaboriau-Routhiau et al., 2009), and members of the cytophaga-flavobacter-bacteroides (CFB) phylum, for example, have been shown to promote differentiation of pro-inflammatory Th17 cells associated with airway inflammation (Ivanov et al., 2008; Atarashi et al., 2011) (Figure 1).
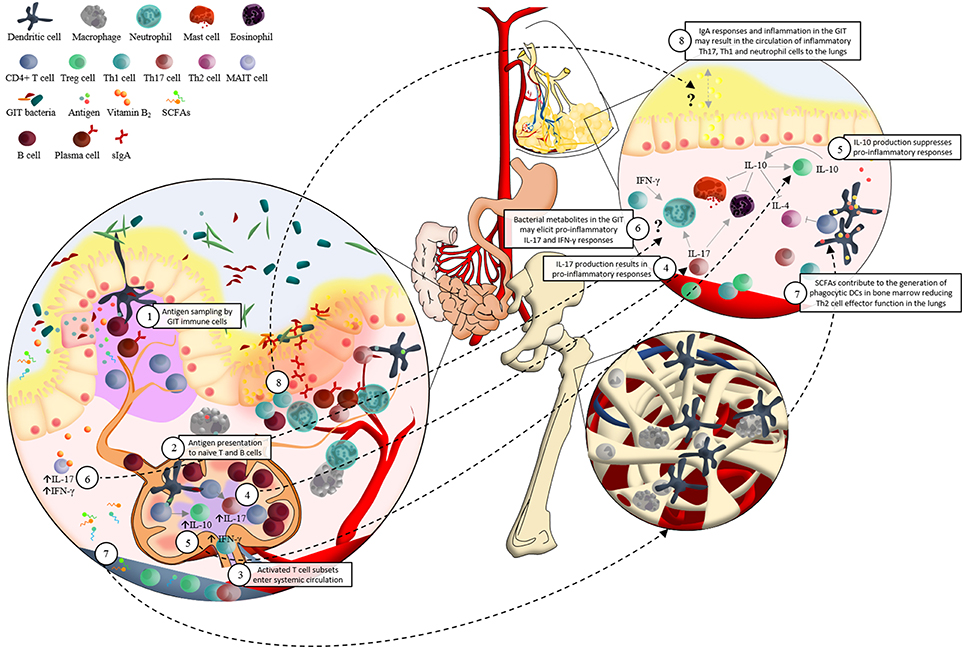
Figure 1. Schematic representation of the potential immunological interaction between the gastrointestinal tract microbiota and the development of asthma. 1. Dendritic cells (DCs) sample antigen in the lamina propria (LP) and Peyer's patches (PP) of the small intestine; and by extending their dendrites into the intestinal lumen. 2. The interactions between DCs and microbial associated molecular patterns (MAMPs) allow DCs to present antigen to naïve lymphocytes in the mesenteric lymph nodes (mLNs). For example, DCs present epitopes together with major histocompatibility complex (MHC) class II and specific immunomodulatory cytokines to naïve CD4+ T cells. This elicits proliferation and activation of various T cell subsets which 3. enter systemic circulation via the efferent lymph, homing to mucosal surfaces inside and outside of the gastrointestinal tract (GIT). 4. TGF-β contributes to the differentiation of Th17 cells, which produce cytokines (such as IL-17) involved in pro-inflammatory responses. 5. IL-12 associated cytokines are responsible for Th1 cell differentiation. This regulates the induction of IL-10, which supresses pro-inflammatory responses. 6. Presentation of vitamin B2, from a wide range of bacteria and fungi via MR1 molecules, to mucosa-associated invariant T (MAIT) cells results in a rapid production of pro-inflammatory Th1/Th17 cytokines. MAIT cells' preferential location in the GIT LP and PP, as well as their pro-inflammatory responses in reaction to bacterial metabolites such as vitamin B2, may support their potential role in asthma pathogenesis via the “gut-lung axis” in a similar manner to what has been proposed for DCs. 7. Circulating short-chain fatty acids (SCFAs) contribute to the protection against allergic airway inflammation via enhanced generation of DC precursors in bone marrow, followed by seeding of the lungs with DCs with high phagocytic capacity and limited ability to promote Th2 cell effector function. 8. Localization of inflammatory GIT bacteria in the GIT mucus layer may induce strong IgA responses and chronic local inflammation. An influx of inflammatory Th17, Th1, and neutrophil cells in the GIT could potentially circulate to the lungs where they may contribute to asthma pathogenesis. This hypothesis may be supported by the strong associations found between irritable bowel disease (IBD) and asthma.
Although the studies described here provide insight into the potential role of GIT bacteria in the development of asthma in murine models; more studies are needed to explore the manner in which whole GIT bacterial communities, from asthmatic and non-asthmatic participants, interact with the innate and adaptive immune cells of the GIT, as well as their subsequent immune effects in the lungs. Besides, studies should also investigate a broader scope of mechanisms to explain the role of GIT bacteria in asthma pathogenesis. For example, a potential mechanism in need of further investigation is the tenable role of IgA-coated inflammatory GIT bacteria in the development of asthmatic responses in the lungs (Figure 1). To date, no clear link between host GIT microbiota-idiopathic intestinal inflammation and allergic lung disease has been demonstrated. However, our recent understanding of the involvement of IgA-coated bacteria in intestinal inflammation (Van der Waaij et al., 2004; Palm et al., 2014), as well as the number of clinical studies denoting an association between inflammatory lung disease and intestinal inflammation (Tulic et al., 2016); provides rationale for investigating the systemic immune effect of IgA-coated GIT bacteria. For example, it is suspected that around 50% of patients suffering from ulcerative colitis and Crohn's disease have subclinical pulmonary abnormalities with low-grade airway inflammation (Kuzela et al., 1999; Mohamed-Hussein et al., 2007). Moreover, a large cohort study, investigating 5260 IBD patients together with 21,040 non-IBD participants, recently provided strong evidence for the association between IBD and an increased risk for asthma (Peng et al., 2015). In support of this, Palm et al. (2014) clearly showed microbial localization of IgA positive bacteria from IBD patients in the normally sterile GIT mucosa of germ-free mice, which was not observed for IgA negative bacteria from IBD patients (Palm et al., 2014). We therefore hypothesize that GIT bacteria characterized by high levels of IgA coating may enter the GIT mucosa (Palm et al., 2014) where they may elicit systemic inflammatory responses at extra-intestinal mucosal sites such as the lungs.
In addition to assessing the immuno-regulatory effect of the composition of GIT bacteria in asthmatic and non-asthmatic participants; studies should also investigate the functional characteristics of GIT bacteria in the occurrence of asthma. In support of this, Trompette et al. (2014) reported the role of circulating short-chain fatty acids (SCFAs) in the protection against allergic airway inflammation (Trompette et al., 2014) (Figure 1). Here, microbiome metabolism in a high fiber diet setting resulted in enhanced SCFA metabolism leading to the generation of myeloid bone marrow precursors that gave rise to populations of pulmonary DCs that protected against Th2 driven allergic airway disease (Trompette et al., 2014). In support of this, Zaiss et al. (2015) demonstrated attenuated allergic airway inflammation via a GPR41 (SCFA receptor) dependent manner, as well as the effect of changes in GIT bacteria on SCFA production (Zaiss et al., 2015). Furthermore, microbial vitamin B2 (riboflavin) metabolites have been shown to activate a subset of innate-like T cells, the mucosa-associated invariant T (MAIT) cells, which are highly abundant in peripheral blood, mucosal tissues, as well as the liver (Treiner et al., 2003; Le Bourhis et al., 2013). Vitamin B2 from a wide range of bacteria and fungi are presented to MAIT cells by MR1 molecules (Kjer-Nielsen et al., 2012; Patel et al., 2013), followed by the rapid production of pro-inflammatory Th1/Th17 cytokines such as interferon-gamma (IFNγ) and IL-17 (Le Bourhis et al., 2011) (Figure 1). MAIT cells' pro-inflammatory responses in reaction to bacterial metabolites, together with their preferential location in the GIT lamina propria and mesenteric lymph nodes (Treiner et al., 2003), may support the “gut-lung axis” theory in a similar manner to what has been proposed for DCs. Therefore, functional properties of GIT bacteria such as dietary fiber metabolism and the production of vitamin B2 may be an important aspect of host microbe crosstalk.
The Potential of Modulating Gastrointestinal Microbiota to Protect Against Asthma
The mechanistic insights into how GIT bacteria may protect or contribute to the development of asthma have provided great potential for the development of intervention studies. For example, the administration of probiotics (beneficial live bacterial species) (De Kivit et al., 2014), prebiotics (non-digestible food ingredients) (Jeurink et al., 2013) or symbiotics (synergistic nutritional supplements combining probiotics and prebiotics) (Van de Pol et al., 2011; Van der Aa et al., 2011) have demonstrated immune-modulatory potential via the restoration of an altered intestinal microbiota. The efficacy of probiotic administration (mainly Lactobacillus or Bifidobacterium spp.) in treatment or prevention of asthma has clearly been demonstrated in animal models (Feleszko et al., 2007; Karimi et al., 2009; MacSharry et al., 2012; Kim et al., 2013); however data in humans are not conclusive (Vliagoftis et al., 2008; Elazab et al., 2013). Nevertheless, probiotic administration needs to be carefully considered as we do not fully understand its effect on GIT bacteria. It may also hold more complex effects for the host (Shenderov, 2013), such as infections (Fijan, 2014) and allergic sensitization (Viljanen et al., 2005; Taylor et al., 2007). Various factors therefore need to be taken into account in the development of probiotics. These include the immunological pathways behind immune responses elicited by live bacteria and bacterial molecules (Caselli et al., 2011); the ongoing research around what a “healthy” GIT profile should look like (Koren et al., 2013; Knights et al., 2014) (prior to considering modulation thereof); what the effect of probiotics are on these “healthy” GIT profiles (Eloe-fadrosh et al., 2015); the inter-individual variability of the human GIT microbiome (De Filippo et al., 2010; Grześkowiak et al., 2012; Yatsunenko et al., 2012; Lin et al., 2013; Ou et al., 2013; Suzuki and Worobey, 2014); the effect of probiotics on host metabolic and signaling pathways (Shenderov, 2013); and whether diversity within specific bacterial taxa is of importance in immunological tolerance (West, 2014). In addition, studies are needed to assess the period, dose and duration of probiotic supplementation. As for probiotics, prebiotic supplementation was not significantly associated with the prevention of asthma in humans (Arslanoglu et al., 2012; Osborn and Sinn, 2013); however, administration of oligosaccharides in mice has been associated with decreased parameters of allergic asthma (Vos et al., 2007).
It is important to also highlight the potential role of vitamin D in modulation of the GIT bacterial community and consequent immune responses such as asthma (Arshi et al., 2014). Vitamin D not only acts on a number of immune cells and processes involved in immune regulation of asthma (Brehm et al., 2009, 2010; Mann et al., 2014), but also has the potential to modulate GIT bacterial profiles and their functions (Mai et al., 2009; Jin et al., 2015). Thus, further exploring the therapeutic potential of vitamin D supplementation, together with pro-, pre- and synbiotic interventions, in modulating the host's GIT microbiota and its subsequent effect on allergic airway diseases such as asthma has merit.
Moreover, understanding the effects of other GIT microbiota, such as fungi and helminths, on the composition of GIT bacteria is also likely to be extremely informative. For example, an overgrowth of commensal fungal Candida species in the GIT, as a result of antibiotic treatment, has been shown to promote M2 macrophage activation in the lungs, as well as increased allergic airway inflammation (Kim et al., 2014). In addition, changes in the GIT bacterial composition of mice following chronic infection with the murine helminth Heligmosomoides polygyrus bakeri have been elegantly shown to protect against house dust mite induced airway inflammation (Zaiss et al., 2015). Importantly this study shows that these changes resulted in elevated SCFA production that actually underlies the protective phenotype (Zaiss et al., 2015). This and work discussed above from the Marsland laboratory provide strong evidence for dietary modulation of the microbiome protecting against allergy.
Conclusion and Perspectives
A systematic search of the literature revealed that studies investigating fecal bacteria from humans and their relationship with asthma have been increasingly published since the beginning of the 21st century. However, reports on the role of fecal bacteria in the development of asthma in humans are limited, and primarily investigate the role of select GIT bacteria in asthma pathogenesis. Large longitudinal prospective cohort studies, with clear definitions of asthmatic outcomes, incorporating high-resolution methods (such as massively parallel 16S rRNA gene sequencing, whole-genome shotgun sequencing or culturomics), are therefore needed to determine the role of fecal bacteria in the development of asthma in both developed and developing countries. Studies also need to assess the impact of covariates (such as mode of delivery and intestinal microbes other than bacteria) on both fecal bacterial profiles and the outcome of interest using rigorous statistical analyses. Furthermore, studies should aim to test the causal link between human fecal bacteria and asthma development using murine models. Finally, the role of GIT bacteria in asthma should be investigated alongside the airway microbiome in order not to mask the importance of the local respiratory microbial-host interactions. In addition, it would be interesting to assess whether GIT bacteria impacts on corticosteroid responsiveness in asthma (Goleva et al., 2013), as well as asthma severity and phenotypes (Zhang et al., 2016).
Literature Search Strategy and Selection Criteria
We systematically searched peer-reviewed articles published on bacteria detected from feces and their association with asthma from six electronic databases (Medline via Pubmed, Scopus via SciVerse, Academic Search Premier, Africa-Wide Information and CINAHL via EBSCOHost and Web of Science via Web of Knowledge), using a combination of keywords [(microbiota* OR metagenome OR microbiome* OR “human microbiota*” OR “human microbiome*” OR “gut microbiota*” OR “gut microbiome*” OR “intestinal flora” OR “digestive flora” OR “gut flora” OR feces OR stool OR faeces OR fecal OR faecal) AND (asthma OR “bronchial asthma” OR “bronchial disease*” OR “respiratory sound*” OR “lung sound*” OR wheez*)]. The last literature search was 19 November 2015. All articles published in English and French were assessed for inclusion in the review. Original research articles investigating bacteria from fecal specimens and their relation to asthma in humans unexposed to antibiotic, pre- or probiotic treatments were included. In addition, we cross-checked the reference lists of all eligible studies included in this review for any additional articles.
Author Contributions
MK, SC, and MN initiated the project. SC extracted the data and reviewed the articles with MK. SC, CW, SM, LT, and MK performed the statistical analysis and interpreted the results. SC, LT, WH, HZ, MN, and MK wrote the manuscript.
Funding
This work was supported by the Bill and Melinda Gates Foundation Global Health Grant (OPP1017641), the National Research Foundation (South Africa), the Carnegie Corporation of New York (United States of America), the US National Institutes of Health (1U01AI110466-01A1), and the Wellcome Trust, United Kingdom (102429/Z/13/Z). The first (SC) and the corresponding author (MK) had full access to the study data. All authors had final responsibility for the decision to submit the article for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
SC is supported by the National Research Foundation and the Drakenstein Child Health Study, University of Cape Town (South Africa), a birth cohort study funded by Bill and Melinda Gates Foundation (OPP1017641). MK was a recipient of Carnegie Corporation of New York (USA) fellowship, and he is currently supported by Wellcome Trust, United Kingdom (102429/Z/13/Z).
References
Arnold, I. C., Dehzad, N., Reuter, S., Martin, H., Becher, B., Taube, C., et al. (2011). Helicobacter pylori infection prevents allergic asthma in mouse models through the induction of regulatory T cells. Clin. Invest J. 121, 3088–3093. doi: 10.1172/JCI45041
Arrieta, M., Stiemsma, L. T., Dimitriu, P. A., Thorson, L., Russell, S., Yurist-doutsch, S., et al. (2015). Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 7, 1–14. doi: 10.1126/scitranslmed.aab2271
Arshi, S., Fallahpour, M., Nabavi, M., Bemanian, M. H., Javad-Mousavi, S. A., Nojomi, M., et al. (2014). The effects of vitamin D supplementation on airway functions in mild to moderate persistent asthma. Ann. Allergy Asthma Immunol. 113, 404–409. doi: 10.1016/j.anai.2014.07.005
Arslanoglu, S., Moro, G. E., Boehm, G., Wienz, F., Stahl, B., and Bertino, E. (2012). Early Neutral Prebiotic Oligosaccharide Supplentation reduces the incidence of some allergic manifestations in the first 5 years of life. J. Biol. Regul. Homeost. Agents 26, 49–59.
Atarashi, K., Tanoue, T., Shima, T., Imaoka, A., Kuwahara, T., Momose, Y., et al. (2011). Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331, 337–341. doi: 10.1126/science.1198469
Azad, M. B., and Kozyrskyj, A. L. (2012). Perinatal programming of asthma: the role of gut microbiota. Clin. Dev. Immunol. 2012:932072. doi: 10.1155/2012/932072
Bateman, E. D., Hurd, S. S., Barnes, P. J., Bousquet, J., Drazen, J. M., FitzGerald, M., et al. (2008). Global strategy for asthma management and prevention: GINA executive summary. Eur. Respir. J. 31, 143–178. doi: 10.1183/09031936.00138707
Bisgaard, H., Li, N., Bonnelykke, K., Chawes, B. L. K., Skov, T., Paludan-Müller, G., et al. (2011). Reduced diversity of the intestinal microbiota during infancy is associated with increased risk of allergic disease at school age. J. Allergy Clin. Immunol. 128, 646–652. doi: 10.1016/j.jaci.2011.04.060
Blaser, M. J., and Falkow, S. (2009). What are the consequences of the disappearing human microbiota? Nat. Rev. Microbiol. 7, 887–894. doi: 10.1038/nrmicro2245
Bloomfield, S., Stanwell-Smith, R., Crevelz, R., and Pickup, J. (2006). Too clean, or not too clean: the Hygiene Hypothesis and home hygiene. Clin. Exp. Allergy 36, 402–425. doi: 10.1111/j.1365-2222.2006.02463.x
Brehm, J. M., Celedón, J. C., Soto-Quiros, M. E., Avila, L., Hunninghake, G. M., and Forno, E. (2009). Serum vitamin D levels and markers of severity of childhood asthma in Costa Rica. Am. J. Respir. Crit. Care Med. 179, 765–771. doi: 10.1164/rccm.200808-1361OC
Brehm, J. M., Schuemann, B., Fuhlbrigge, A. L., Hollis, B. W., Strunk, R. C., Zeiger, R. S., et al. (2010). Serum vitamin D levels and severe asthma exacerbations in the childhood Asthma management program study. J. Allergy Clin. Immunol. 126, 52–58. doi: 10.1016/j.jaci.2010.03.043
Brooks, C., Pearce, N., and Douwes, J. (2013). The hygiene hypothesis in allergy and asthma: an update. Curr. Opin. Allergy Clin. Immunol. 13, 70–77. doi: 10.1097/ACI.0b013e32835ad0d2
Caselli, M., Vaira, G., Calo, G., Papini, F., Holton, J., and Vaira, D. (2011). Structural bacterial molecules as potential candidates for an evolution of the classical concept of probiotics. Adv. Nutr. 2, 372–376. doi: 10.3945/an.111.000604
Castro-Rodriguez, J. A. (2010). The Asthma Predictive Index: a very useful tool for predicting asthma in young children. J. Allergy Clin. Immunol. 126, 212–216. doi: 10.1016/j.jaci.2010.06.032
Clemente, J. C., Ursell, L. K., Parfrey, L. W., and Knight, R. (2012). The impact of the gut microbiota on human health: an integrative view. Cell 148, 1258–1270. doi: 10.1016/j.cell.2012.01.035
De Filippo, C., Cavalieri, D., Di Paola, M., Ramazzotti, M., Poullet, J. B., Massart, S., et al. (2010). Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. U.S.A. 107, 14691–14696. doi: 10.1073/pnas.1005963107
De Kivit, S., Tobin, M. C., Forsyth, C. B., Keshavarzian, A., and Landay, A. L. (2014). Regulation of intestinal immune responses through TLR activation: Implications for pro- and prebiotics. Front. Immunol. 5:60. doi: 10.3389/fimmu.2014.00060
Dogaru, C. M., Nyffenegger, D., Pescatore, A. M., Spycher, B. D., and Kuehni, C. E. (2014). Breastfeeding and childhood asthma: Systematic review and meta-analysis. Am. J. Epidemiol. 179, 1153–1167. doi: 10.1093/aje/kwu072
Edwards, M. R., Bartlett, N. W., Hussell, T., Openshaw, P., and Johnston, S. L. (2012). The microbiology of asthma. Nat. Rev. Microbiol. 10, 459–471. doi: 10.1038/nrmicro2801
Elazab, N., Mendy, A., Gasana, J., Vieira, E. R., Quizon, A., and Forno, E. (2013). Probiotic administration in early life, atopy, and asthma: a meta-analysis of clinical trials. Pediatrics 132, e666–e676. doi: 10.1542/peds.2013-0246
Eloe-fadrosh, E. A., Brady, A., Crabtree, J., Drabek, E. F., Ma, B., Mahurkar, A., et al. (2015). Functional dynamics of the gut microbiome in elderly people during probiotic consumption. MBio 6, 1–12. doi: 10.1128/mBio.00231-15
Feleszko, W., Jaworska, J., Rha, R. D., Steinhausen, S., Avagyan, A., Jaudszus, A., et al. (2007). Probiotic-induced suppression of allergic sensitization and airway inflammation is associated with an increase of T regulatory-dependent mechanisms in a murine model of asthma. Clin. Exp. Allergy 37, 498–505. doi: 10.1111/j.1365-2222.2006.02629.x
Fijan, S. (2014). Microorganisms with claimed probiotic properties: an overview of recent literature. Int. J. Environ. Res. Public Health 11, 4745–4767. doi: 10.3390/ijerph110504745
Fraher, M. H., O'Toole, P. W., and Quigley, E. M. (2012). Techniques used to characterize the gut microbiota: a guide for the clinician. Nat. Rev. Gastroenterol. Hepatol. 9, 312–322. doi: 10.1038/nrgastro.2012.44
Gaboriau-Routhiau, V., Rakotobe, S., Lécuyer, E., Mulder, I., Lan, A., Bridonneau, C., et al. (2009). The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 31, 677–689. doi: 10.1016/j.immuni.2009.08.020
Goleva, E., Jackson, L. P., Harris, J. K., Robertson, C. E., Sutherland, E. R., Hall, C. F., et al. (2013). The effects of airway microbiome on corticosteroid responsiveness in asthma. Am. J. Respir. Crit. Care Med. 188, 1193–1201. doi: 10.1164/rccm.201304-0775OC
Gosalbes, M. J., Llop, S., Vallès, Y., Moya, A., Ballester, F., and Francino, M. P. (2012). Meconium microbiota types dominated by lactic acid or enteric bacteria are differentially associated with maternal eczema and respiratory problems in infants. Clin. Exp. Allergy 43, 198–211. doi: 10.1111/cea.12063
Grześkowiak, Ł., Collado, M. C., Mangani, C., Maleta, K., Laitinen, K., Ashorn, P., et al. (2012). Distinct gut microbiota in southeastern African and northern European infants. J. Paediatr. Gastroenterol. Nutr. 54, 812–816. doi: 10.1097/MPG.0b013e318249039c
Hanski, I., von Hertzen, L., Fyhrquist, N., Koskinen, K., Torppa, K., Laatikainen, T., et al. (2012). Environmental biodiversity, human microbiota, and allergy are interrelated. Proc. Natl. Acad. Sci. U.S.A. 109, 8334–8339. doi: 10.1073/pnas.1205624109
Herbst, T., Sichelstiel, A., Schär, C., Yadava, K., Bürki, K., Cahenzli, J., et al. (2011). Dysregulation of allergic airway inflammation in the absence of microbial colonization. Am. J. Respir. Crit. Care Med. 184, 198–205. doi: 10.1164/rccm.201010-1574OC
Hill, D. A., and Artis, D. (2010). Intestinal bacteria and the regulation of immune cell homeostasis. Annu. Rev. Immunol. 28, 623–667. doi: 10.1146/annurev-immunol-030409-101330
Hunter, P. (2012). The changing hypothesis of the gut. The intestinal microbiome is increasingly seen as vital to human health. EMBO Rep. 13, 498–500. doi: 10.1038/embor.2012.68
Ivanov, I. I., Frutos Rde, L., Manel, N., Yoshinaga, K., Rifkin, D. B., Sartor, R. B., et al. (2008). Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 4, 337–349. doi: 10.1016/j.chom.2008.09.009
Jeurink, P. V., Van Esch, B. C., Rijnierse, A., Garssen, J., and Knippels, L. M. (2013). Mechanisms underlying immune effects of dietary oligosaccharides. Am. J. Clin. Nutr. 98, 572S–577S. doi: 10.3945/ajcn.112.038596
Jin, D., Wu, S., Zhang, Y. G., Lu, R., Xia, Y., Dong, H., et al. (2015). Lack of Vitamin D receptor causes dysbiosis and changes the functions of the murine intestinal microbiome. Clin. Ther. 37, 996–1009. doi: 10.1016/j.clinthera.2015.04.004
Karimi, K., Inman, M. D., Bienenstock, J., and Forsythe, P. (2009). Lactobacillus reuteri–induced regulatory T cells protect against an allergic airway response in mice. Am. J. Respir. Crit. Care Med. 179, 186–193. doi: 10.1164/rccm.200806-951OC
Kim, H. J., Kim, Y. J., Lee, S. -H., Kang, M. J., Yu, H. -S., Jung, Y. H., et al. (2013). Effects of Lactobacillus rhamnosus on asthma with an adoptive transfer of dendritic cells in mice. J. Appl. Microbiol. 115, 872–879. doi: 10.1111/jam.12268
Kim, Y. G., Udayanga, K. G., Totsuka, N., Weinberg, J. B., Núñez, G., and Shibuya, A. (2014). Gut dysbiosis promotes M2 macrophage polarization and allergic airway inflammation via fungi-induced PGE2. Cell Host Microbe 15, 95–102. doi: 10.1016/j.chom.2013.12.010
Kjer-Nielsen, L., Patel, O., Corbett, A. J., Le Nours, J., Meehan, B., Liu, L., et al. (2012). MR1 presents microbial vitamin B metabolites to MAIT cells. Nature 491, 717–723. doi: 10.1038/nature11605
Knights, D., Ward, T. L., Mckinlay, C. E., Miller, H., Gonzalez, A., and Mcdonald, D. (2014). Rethinking “Enterotypes.” Cell Host Microbe 16, 433–437. doi: 10.1016/j.chom.2014.09.013
Konieczna, P., Groeger, D., Ziegler, M., Frei, R., Ferstl, R., Shanahan, F., et al. (2012). Bifidobacterium infantis 35624 administration induces Foxp3 T regulatory cells in human peripheral blood: potential role for myeloid and plasmacytoid dendritic cells. Gut 61, 354–366. doi: 10.1136/gutjnl-2011-300936
Koren, O., Knights, D., Gonzalez, A., Waldron, L., Segata, N., Knight, R., et al. (2013). A guide to enterotypes across the human body: meta-analysis of microbial community structures in human microbiome datasets. PLoS Comput. Biol. 9:e1002863. doi: 10.1371/journal.pcbi.1002863
Kozyrskyj, A. L., Bahreinian, S., and Azad, M. B. (2011). Early life exposures: impact on asthma and allergic disease. Curr. Opin. Allergy Clin. Immunol. 11, 400–406. doi: 10.1097/ACI.0b013e328349b166
Kramer, A., Bekeschus, S., Bröker, B. M., Schleibinger, H., Razavi, B., and Assadian, O. (2013). Maintaining health by balancing microbial exposure and prevention of infection: the hygiene hypothesis versus the hypothesis of early immune challenge. J. Hosp. Infect. 83, S29–S34. doi: 10.1016/S0195-6701(13)60007-9
Kurokawa, K., Itoh, T., Kuwahara, T., Oshima, K., Toh, H., Toyoda, A., et al. (2007). Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 14, 169–181. doi: 10.1093/dnares/dsm018
Kuzela, L., Vavrecka, A., Prikazska, M., Drugda, B., Hronec, J., Senkova, A., et al. (1999). Pulmonary complications in patients with inflammatory bowel disease. Hepatogastroenterology 46, 1714–1719.
Le Bourhis, L., Guerri, L., Dusseaux, M., Martin, E., Soudais, C., and Lantz, O. (2011). Mucosal-associated invariant T cells: unconventional development and function. Trends Immunol. 32, 212–218. doi: 10.1016/j.it.2011.02.005
Le Bourhis, L., Mburu, Y. K., and Lantz, O. (2013). MAIT cells, surveyors of a new class of antigen: development and functions. Curr. Opin. Immunol. 25, 174–180. doi: 10.1016/j.coi.2013.01.005
Lin, A., Bik, E. M., Costello, E. K., Dethlefsen, L., Haque, R., Relman, D. A., et al. (2013). Distinct distal gut microbiome diversity and composition in healthy children from Bangladesh and the United States. PLoS ONE 8:e53838. doi: 10.1371/journal.pone.0053838
MacSharry, J., O'Mahony, C., Shalaby, K. H., Sheil, B., Karmouty-Quintana, H., Shanahan, F., et al. (2012). Immunomodulatory effects of feeding with Bifidobacterium longum on allergen-induced lung inflammation in the mouse. Pulm. Pharmacol. Ther. 25, 325–334. doi: 10.1016/j.pupt.2012.05.011
Mai, V., McCrary, Q. M., Sinha, R., and Glei, M. (2009). Associations between dietary habits and body mass index with gut microbiota composition and fecal water genotoxicity: an observational study in African American and Caucasian American volunteers. Nutr. J. 8:49. doi: 10.1186/1475-2891-8-49
Mallol, J. (2008). Asthma in Latin America: where the asthma causative/protective hypotheses fail. Allergol. Immunopathol. (Madr). 36, 150–153. doi: 10.1016/S0301-0546(08)72540-0
Mann, E. H., Chambers, E. S., Pfeffer, P. E., and Hawrylowicz, C. M. (2014). Immunoregulatory mechanisms of vitamin D relevant to respiratory health and asthma. Ann. N.Y. Acad. Sci. 1317, 57–69. doi: 10.1111/nyas.12410
Mansson, I., and Colldahl, H. (1965). The intestinal flora in patients with bronchial asthma and rheumatoid arthritis. Allergy 20, 94–104. doi: 10.1111/j.1398-9995.1965.tb03360.x
Marra, F., Marra, C. A., Richardson, K., Lynd, L. D., Kozyrskyj, A., Patrick, D. M., et al. (2009). Antibiotic use in children is associated with increased risk of asthma. Pediatrics 123, 1003–1010. doi: 10.1542/peds.2008-1146
Masoli, M., Fabian, D., Holt, S., and Beasley, R. (2004). The global burden of asthma: executive summary of the GINA Dissemination Committee report. Allergy 59, 469–478. doi: 10.1111/j.1398-9995.2004.00526.x
McLoughlin, R. M., and Mills, K. H. (2011). Influence of gastrointestinal commensal bacteria on the immune responses that mediate allergy and asthma. J. Allergy Clin. Immunol. 127, 1097. doi: 10.1016/j.jaci.2011.02.012
Mohamed-Hussein, A. A., Mohamed, N. A., and Ibrahim, M. E. (2007). Changes in pulmonary function in patients with ulcerative colitis. Respir. Med. 101, 977–982. doi: 10.1016/j.rmed.2006.09.005
Morgan, X. C., and Huttenhower, C. (2014). Meta'omic analytic techniques for studying the intestinal microbiome. Gastroenterology 146, 1437–1448. doi: 10.1053/j.gastro.2014.01.049
Murk, W., Risnes, K. R., and Bracken, M. B. (2011). Prenatal or early-life exposure to antibiotics and risk of childhood asthma: a systematic review. Pediatrics 127, 1125–1138. doi: 10.1542/peds.2010-2092
Nambu, M., Shintaku, N., and Ohta, S. (2008). Intestinal microflora at 4 months of age and the development of allergy. Allergol. Int. 53, 121–126. doi: 10.1111/j.1440-1592.2004.00315.x
Navarro, S., Cossalter, G., Chiavaroli, C., Kanda, A., Fleury, S., Lazzari, A., et al. (2011). The oral administration of bacterial extracts prevents asthma via the recruitment of regulatory T cells to the airways. Mucosal Immunol. 4, 53–65. doi: 10.1038/mi.2010.51
Oertli, M., Sundquist, M., Hitzler, I., Engler, D. B., Arnold, I. C., Reuter, S., et al. (2012). DC-derived IL-18 drives Treg differentiation, murine Helicobacter pylori-specific immune tolerance, and asthma protection. J. Clin. Invest. 122, 1082–1096. doi: 10.1172/JCI61029
Osborn, D., and Sinn, J. (2013). Prebiotics in infants for prevention of allergy. Cochrane Database Syst. Rev. 3:CD006474. doi: 10.1002/14651858.CD006474.pub3
Ou, J., Carbonero, F., Zoetendal, E. G., Delany, J. P., Wang, M., Newton, K., et al. (2013). Diet, microbiota, and microbial metabolites in colon cancer risk in rural Africans and African Americans. Am. J. Clin. Nutr. 98, 111–120. doi: 10.3945/ajcn.112.056689
Palm, N. W., De Zoete, M. R., Cullen, T. W., Barry, N. A., Stefanowski, J., Hao, L., et al. (2014). Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 158, 1000–1010. doi: 10.1016/j.cell.2014.08.006
Patel, O., Kjer-Nielsen, L., Le Nours, J., Eckle, S. B. G., Birkinshaw, R., Beddoe, T., et al. (2013). Recognition of vitamin B metabolites by mucosal-associated invariant T cells. Nat. Commun. 4, 2142. doi: 10.1038/ncomms3142
Pedersen, S. (2007). Preschool asthma - not so easy to diagnose. Prim. Care Respir. 16, 4–6. doi: 10.3132/pcrj.2007.00011
Pedersen, S. E., Hurd, S. S., Lemanske, R. F., Becker, A., Zar, H. J., Sly, P. D., et al. (2011). Global strategy for the diagnosis and management of asthma in children 5 years and younger. Pediatr. Pulmonol. 46, 1–17. doi: 10.1002/ppul.21321
Penders, J., Kummeling, I., and Thijs, C. (2011). Infant antibiotic use and wheeze and asthma risk: a systematic review and meta-analysis. Eur. Respir. J. 38, 295–302. doi: 10.1183/09031936.00105010
Penders, J., Stobberingh, E. E., Van den Brandt, P. A., and Thijs, C. (2007). The role of the intestinal microbiota in the development of atopic disorders. Allergy 62, 1223–1236. doi: 10.1111/j.1398-9995.2007.01462.x
Peng, Y. H., Liao, W. C., Su, C. H., Chen, H. J., Hsia, T. C., Chu, C. C., et al. (2015). Association of inflammatory bowel disease with asthma risk: a nationwide cohort study. Allergy Asthma Proc. 36, e92–e98. doi: 10.2500/aap.2015.36.3869
Qin, J., Li, Y., Cai, Z., Li, S. S., Zhu, J., Zhang, F., et al. (2012). A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490, 55–60. doi: 10.1038/nature11450
Quince, C., Ijaz, U. Z., Loman, N., Eren, A. M., Saulnier, D., Russell, J., et al. (2015). Extensive modulation of the fecal metagenome in children with Crohn's Disease during exclusive enteral nutrition. Am. J. Gastroenterol. 1–12. doi: 10.1038/ajg.2015.357
Rajilić-Stojanović, M., Smidt, H., and De Vos, W. M. (2007). Diversity of the human gastrointestinal tract microbiota revisited. Environ. Microbiol. 9, 2125–2136. doi: 10.1111/j.1462-2920.2007.01369.x
Ranjan, R., Rani, A., Metwally, A., McGee, H. S., and Perkens, D. L. (2015). Analysis of the microbiome: advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem. Biophys. Res. Commun. 469, 967–977. doi: 10.1016/j.bbrc.2015.12.083
Risnes, K. R., Belanger, K., Murk, W., and Bracken, M. B. (2010). Antibiotic exposure by 6 months and asthma and allergy at 6 years: Findings in a cohort of 1,401 US children. Am. J. Epidemiol. 173, 310–318. doi: 10.1093/aje/kwq400
Rook, G. (2012). Hygiene hypothesis and autoimmune diseases. Clin. Rev. Allergy Immunol. 42, 5–15. doi: 10.1007/s12016-011-8285-8
Round, J. L., Lee, S. M., Li, J., Tran, G., Jabri, B., Chatila, T. A., et al. (2011). The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 332, 974. doi: 10.1126/science.1206095
Russell, S. L., and Finlay, B. B. (2012). The impact of gut microbes in allergic diseases. Curr. Opin. Gastroenterol. 28, 563–569. doi: 10.1097/MOG.0b013e3283573017
Russell, S. L., Gold, M. J., Hartmann, M., Willing, B. P., Thorson, L., Wlodarska, M., et al. (2012). Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 13, 440–447. doi: 10.1038/embor.2012.32
Russell, S. L., Gold, M. J., Willing, B. P., Thorson, L., McNagny, K. M., and Finlay, B. B. (2013). Perinatal antibiotic treatment affects murine microbiota, immune responses and allergic asthma. Gut Microbes 4, 158–164. doi: 10.4161/gmic.23567
Salminen, S., Gibson, G. R., McCartney, A. L., and Isolauri, E. (2004). Influence of mode of delivery on gut microbiota composition in seven year old children. Gut 53, 1388–1389. doi: 10.1136/gut.2004.041640
Salzman, N. H. (2011). Microbiota-immune system interaction: an uneasy alliance. Curr. Opin. Microbiol. 14, 99–105. doi: 10.1016/j.mib.2010.09.018
Samuelson, D. R., Welsch, D. A., and Shellito, J. E. (2015). Regulation of lung immunity and host defense by the intestinal microbiota. Front. Microbiol. 6:1085. doi: 10.3389/fmicb.2015.01085
Sankar, S. A., Lagier, J. C., Pontarotti, P., Raoult, D., and Fournier, P. E. (2015). The human gut microbiome, a taxonomic conundrum. Syst. Appl. Microbiol. 38, 276–286. doi: 10.1016/j.syapm.2015.03.004
Schippa, S., and Conte, M. (2014). Dysbiotic events in gut microbiota: Impact on human health. Nutrients 6, 5786–5805. doi: 10.3390/nu6125786
Sekirov, I., Russell, S. L., Antunes, L. C. M., and Finlay, B. B. (2010). Gut microbiota in health and disease. Physiol. Rev. 90, 859–904. doi: 10.1152/physrev.00045.2009
Shenderov, B. A. (2013). Metabiotics: novel idea or natural development of probiotic conception. Microb. Ecol. Health Dis. 24, 1–8. doi: 10.3402/mehd.v24i0.20399
Sim, K., Cox, M. J., Wopereis, H., Martin, R., Knol, J., Li, M. S., et al. (2012). Improved detection of bifidobacteria with optimised 16S rRNA-gene based pyrosequencing. PLoS ONE 7:e32543. doi: 10.1371/journal.pone.0032543
Sly, P. D., Boner, A. L., Björksten, B., Bush, A., Custovic, A., Eigenmann, P. A., et al. (2008). Early identification of atopy in the prediction of persistent asthma in children. Lancet 372, 1100–1106. doi: 10.1016/S0140-6736(08)61451-8
Stockert, K. (2001). Physiologische Darmflora bei 6- bis 12-jaehrigen kindern mit asthma bronchiale. Dt. Ztschr. F. Akup. 44, 268–271. doi: 10.1055/s-2001-19471
Strachan, D. P. (1989). Hay fever, hygiene, and household size. BMJ 299, 1259–1260. doi: 10.1136/bmj.299.6710.1259
Strachan, D. P. (2000). Family size, infection and atopy: the first decade of the ‘hygiene hypothesis’. Thorax 55:S2. doi: 10.1136/thorax.55.suppl_1.S2
Štšepetova, J., Sepp, E., Julge, K., Vaughan, E., Mikelsaar, M., and De Vos, W. M. (2007). Molecularly assessed shifts of Bifidobacterium ssp. and less diverse microbial communities are characteristic of 5-year-old allergic children. FEMS Immunol. Med. Microbiol 51, 260–269. doi: 10.1111/j.1574-695X.2007.00306.x
Suau, A., Bonnet, R., Sutren, M., Godon, J. J., Gibson, G. R., Collins, M. D., et al. (1999). Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65, 4799–4807.
Suzuki, T. A., and Worobey, M. (2014). Geographical variation of human gut microbial composition Geographical variation of human gut microbial composition. Biol. Lett. 10, 20131037. doi: 10.1098/rsbl.2013.1037
Taube, C., and Müller, A. (2012). The role of Helicobacter pylori infection in the development of allergic asthma. Expert Rev. Respir. Med. 6, 441–449. doi: 10.1586/ers.12.40
Taylor, A. L., Dunstan, J. A., and Prescott, S. L. (2007). Probiotic supplementation for the first 6 months of life fails to reduce the risk of atopic dermatitis and increases the risk of allergen sensitization in high-risk children: a randomized controlled trial. J. Allergy Clin. Immunol. 119, 184–191. doi: 10.1016/j.jaci.2006.08.036
Thavagnanam, S., Fleming, J., Bromley, A., Shields, M. D., and Cardwell, C. R. (2008). A meta-analysis of the association between Caesarean section and childhood asthma. Clin. Exp. Allergy 38, 629–633. doi: 10.1111/j.1365-2222.2007.02780.x
Treiner, E., Duban, L., Bahram, S., Radosavljevic, M., Wanner, V., Tilloy, F., et al. (2003). Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature 422, 164–169. doi: 10.1038/nature01433
Trompette, A., Gollwitzer, E. S., Yadava, K., Sichelstiel, A. K., Sprenger, N., Ngom-Bru, C., et al. (2014). Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 20, 159–166. doi: 10.1038/nm.3444
Tulic, M. K., Piche, T., and Verhasselt, V. (2016). Lung-gut crosstalk: evidence, mechanisms and implications for the mucosal inflammatory diseases. Clin. Exp. Allergy 46, 519–528. doi: 10.1111/cea.12723
Vael, C., Nelen, V., Verhulst, S. L., Goossens, H., and Desager, K. N. (2008). Early intestinal Bacteroides fragilis colonisation and development of asthma. BMC Pulm. Med. 8:19. doi: 10.1186/1471-2466-8-19
Vael, C., Vanheirstraeten, L., Desager, K. N., and Goossens, H. (2011). Denaturing gradient gel electrophoresis of neonatal intestinal microbiota in relation to the development of asthma. BMC Microbiol. 11:68. doi: 10.1186/1471-2180-11-68
Van de Pol, M. A., Lutter, R., Smids, B. S., Weersink, E. J. M., and Van der Zee, J. S. (2011). Synbiotics reduce allergen-induced T-helper 2 response and improve peak expiratory flow in allergic asthmatics. Allergy 66, 39–47. doi: 10.1111/j.1398-9995.2010.02454.x
Van der Aa, L. B., Van Aalderen, W. M. C., Heymans, H. S. A., Henk Sillevis Smitt, J., Nauta, A. J., Knippels, L. M. J., et al. (2011). Synbiotics prevent asthma-like symptoms in infants with atopic dermatitis. Allergy 66, 170–177. doi: 10.1111/j.1398-9995.2010.02416.x
Van der Waaij, L. A., Kroese, F. G., Visser, A., Nelis, G. F., Westerveld, B. D., Jansen, P. L., et al. (2004). Immunoglobulin coating of faecal bacteria in inflammatory bowel disease. Eur. J. Gastroenterol. Hepatol. 16, 669–674. doi: 10.1097/01.meg.0000108346.41221.19
Van Dijk, E. L., Auger, H., Jaszczyszyn, Y., and Thermes, C. (2014). Ten years of next-generation sequencing technology. Trends Genet. 30, 418–426. doi: 10.1016/j.tig.2014.07.001
Van Nimwegen, F. A., Penders, J., Stobberingh, E. E., Postma, D. S., Koppelman, G. H., Kerkhof, M., et al. (2011). Mode and place of delivery, gastrointestinal microbiota, and their influence on asthma and atopy. J. Allergy Clin. Immunol. 128, 948–955. doi: 10.1016/j.jaci.2011.07.027
Viljanen, M., Pohjavuori, E., Haahtela, T., Korpela, R., Kuitunen, M., Sarnesto, A., et al. (2005). Induction of inflammation as a possible mechanism of probiotic effect in atopic eczema-dermatitis syndrome. J. Allergy Clin. Immunol. 115, 1254–1259. doi: 10.1016/j.jaci.2005.03.047
Vital, M., Harkema, J. R., Rizzo, M., Tiedje, J., and Brandenberger, C. (2015). Alterations of the murine gut microbiome with age and allergic airway disease. J. Immunol. Res. 2015, 1–8. doi: 10.1155/2015/892568
Vliagoftis, H., Kouranos, V. D., Betsi, G. I., and Falagas, M. E. (2008). Probiotics for the treatment of allergic rhinitis and asthma: systematic review of randomized controlled trials. Ann. Allergy Asthma Immunol. 101, 570–579. doi: 10.1016/S1081-1206(10)60219-0
Vos, A. P., Van Esch, B. C., Stahl, B., M'Rabet, L., Folkerts, G., Nijkamp, F. P., et al. (2007). Dietary supplementation with specific oligosaccharide mixtures decreases parameters of allergic asthma in mice. Int. Immunopharmacol. 7, 1582–1587. doi: 10.1016/j.intimp.2007.07.024
Walker, A. W., Martin, J. C., Scott, P., Parkhill, J., Flint, H. J., and Scott, K. P. (2015). 16S rRNA gene-based profiling of the human infant gut microbiota is strongly influenced by sample processing and PCR primer choice. Microbiome 3:26. doi: 10.1186/s40168-015-0087-4
West, C. E. (2014). Gut microbiota and allergic disease: new findings. Curr. Opin. Clin. Nutr. Metab. Care 17, 261–266. doi: 10.1097/MCO.0000000000000044
Wold, A. E. (1998). The hygiene hypothesis revised: is the rising frequency of allergy due to changes in the intestinal flora? Allergy 53, 20–25 doi: 10.1111/j.1398-9995.1998.tb04953.x
Yatsunenko, T., Rey, F. E., Manary, M. J., Trehan, I., Dominguez-Bello, M. G., Contreras, M., et al. (2012). Human gut microbiome viewed across age and geography. Nature 486, 222–227. doi: 10.1038/nature11053
Zaiss, M. M., Rapin, A., Lebon, L., Dubey, L. K., Mosconi, I., Sarter, K., et al. (2015). The intestinal microbiota contributes to the ability of helminths to modulate allergic inflammation. Immunity 43, 998–1010. doi: 10.1016/j.immuni.2015.09.012
Zar, H. J., and Levin, M. E. (2012). Challenges in treating pediatric asthma in developing countries. Pediatr. Drugs 14, 353–359. doi: 10.2165/11597420-000000000-00000
Zhang, Q., Cox, M., Liang, Z., Brinkmann, F., Cardenas, P. A., and Duff, R. (2016). Airway microbiota in severe asthma and relationship to asthma severity and phenotypes. PLoS ONE 11:e0152724. doi: 10.1371/journal.pone.0152724
Zhao, L. (2013). The gut microbiota and obesity: from correlation to causality. Nat. Rev. Microbiol. 11, 639–647. doi: 10.1038/nrmicro3089
Keywords: asthma, fecal bacteria, mechanisms, microbiome, systematic review
Citation: Claassen-Weitz S, Wiysonge CS, Machingaidze S, Thabane L, Horsnell WGC, Zar HJ, Nicol MP and Kaba M (2016) Current Knowledge and Future Research Directions on Fecal Bacterial Patterns and Their Association with Asthma. Front. Microbiol. 7:838. doi: 10.3389/fmicb.2016.00838
Received: 15 January 2016; Accepted: 18 May 2016;
Published: 29 June 2016.
Edited by:
Christine Moissl-Eichinger, Medical University of Graz, AustriaReviewed by:
Geanncarlo Lugo-Villarino, Institut de Pharmacologie et de Biologie Structurale/Centre National de la Recherche Scientifique, FranceJan S. Suchodolski, Texas A&M University, USA
Copyright © 2016 Claassen-Weitz, Wiysonge, Machingaidze, Thabane, Horsnell, Zar, Nicol and Kaba. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mamadou Kaba, bWFtYWRvdS5rYWJhQGhvdG1haWwuY29t
 Shantelle Claassen-Weitz
Shantelle Claassen-Weitz Charles S. Wiysonge
Charles S. Wiysonge Shingai Machingaidze4
Shingai Machingaidze4 William G. C. Horsnell
William G. C. Horsnell Mark P. Nicol
Mark P. Nicol Mamadou Kaba
Mamadou Kaba