
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 31 May 2016
Sec. Food Microbiology
Volume 7 - 2016 | https://doi.org/10.3389/fmicb.2016.00827
This article is part of the Research TopicMicrobiology of Ethnic Fermented Foods and Alcoholic Beverages of the WorldView all 16 articles
Doenjang samples were prepared in triplicate and their microbial abundance, bacterial communities, and metabolites throughout fermentation were analyzed to investigate the functional properties of microorganisms in doenjang. Viable bacterial cells were approximately three orders of magnitude higher than fungal cells, suggesting that bacteria are more responsible for doenjang fermentation. Pyrosequencing and proton nuclear magnetic resonance spectroscopy were applied for the analysis of bacterial communities and metabolites, respectively. Bacterial community analysis based on 16S rRNA gene sequences revealed that doenjang samples included Bacillus, Enterococcus, Lactobacillus, Clostridium, Staphylococcus, Corynebacterium, Oceanobacillus, and Tetragenococcus. These genera were found either in doenjang-meju or solar salts, but not in both, suggesting two separate sources of bacteria. Bacillus and Enterococcus were dominant genera during the fermentation, but their abundances were not associated with metabolite changes, suggesting that they may not be major players in doenjang fermentation. Tetragenococcus was dominant in 108 day-doenjang samples, when lactate, acetate, putrescine, and tyramine increased quickly as glucose and fructose decreased, indicating that Tetragenococcus might be primarily responsible for organic acid and biogenic amine production. Lactobacillus was identified as a dominant group from the 179-day samples, associated with the increase of γ-aminobutyric acid (GABA) and the decrease of galactose, indicating a potential role for this genus as a major GABA producer during fermentation. The results of this study clarified the functional properties of major bacterial communities in the doenjang fermentation process, contributing to the production of safe and high-quality doenjang.
Doenjang is a Korean traditional soybean paste popularly consumed as a condiment for vegetables, fish, and meats or used as a seasoning ingredient in authentic Korean cuisine. The paste has received considerable attention because of numerous reported beneficial human health effects, including antioxidant, fibrinolytic, antimutagenic, and anticancer properties (Kim, 2004; Yun, 2005; Jung et al., 2006; Park et al., 2008; Namgung et al., 2009; Kwon et al., 2010; Tamang et al., 2016a).
Culture-based approaches have been widely applied to bacterial community analysis of doenjang (Yoo et al., 1999; Jeong et al., 2014), but they have produced limited information because culturing is time-consuming and laborious, and because doenjang contains unculturable microbes. Recently, culture-independent methods, such as denaturing gradient gel electrophoresis (DGGE) and pyrosequencing, have been widely used to investigate bacterial communities in doenjang (Cho and Seo, 2007; Kim et al., 2009; Nam et al., 2012). However, previous studies using culture-independent methods have limited their analyses to snapshots of bacterial communities by focusing on short-time frames within the doenjang fermentation process. To the best of our knowledge, thus far, no study has been conducted to investigate microbial community fluctuation over the full doenjang fermentation period. In Korea, traditional doenjang is typically made by further fermentation of the solid parts from a fermented mixture of doenjang-meju (fermented soybean bricks) and brine. The additional fermenting procedure also suggests that the microbial community and indigenous enzymes in doenjang-meju are likely important in determining the microbial community and metabolite change during doenjang fermentation. However, no research exists on how doenjang microbial communities alter when doenjang-meju with known microbial community composition is used.
Traditional doenjang is produced by spontaneous fermentation without the use of starter cultures, leading to the growth of diverse microorganisms. In turn, quality variation of doenjang products tends to result, as well as the occasional production of undesirable metabolites, such as biogenic amines (BAs) or toxins (Cho and Seo, 2007; Shukla et al., 2010; Park et al., 2014). Most previous studies have focused on the analysis of either microbial communities or metabolites in doenjang (Cho and Seo, 2007; Kim et al., 2009; Rhyu and Kim, 2011; Nam et al., 2012), which makes it difficult to investigate microbial functional properties during doenjang fermentation. Instead, examining microbial successions and metabolite changes simultaneously is crucial for a better understanding of microbial community function in doenjang. However, such studies have not yet been performed.
Pyrosequencing based on 16S rRNA gene sequences has been broadly applied to analyze microbial communities in fermented foods, because it yields more detailed data compared with conventional microbiological methods, such as DGGE and culture-based approaches (Nam et al., 2012; Park et al., 2012). Additionally, proton nuclear magnetic resonance (1H NMR) spectroscopy is one of the easiest, yet most comprehensive, and powerful tools to analyze diverse metabolites simultaneously in fermented foods (Jeong et al., 2013; Jung et al., 2013; Lee et al., 2015). In this study, we used pyrosequencing and 1H NMR techniques to investigate microbial succession and metabolite changes, respectively, across the full length of doenjang fermentation. The resultant data will increase our knowledge regarding the functional properties of major microbial communities involved in doenjang fermentation.
Doenjang was prepared in triplicate following a traditional manufacturing method. On January 25, 2013, 90 fermented doenjang-meju bricks from a previous study (Jung et al., 2014) were placed into a large porcelain pot (called jang-dok) filled with 180 L of approximately 20% (w/v) solar salt (salts made by exposing seawater to the sun; Shinan, Korea) solution (Jung et al., 2015). The mixture of doenjang-meju bricks and solar salt solution was stored for 42 days without temperature control in a temporary structure to avoid inclement weather, and then separated into liquid and solid portions. The solid parts (doenjang) were mashed well and equally dispensed into three small porcelain pots, marking the start (0 day) of doenjang fermentation. These pots containing doenjang were stored in the temporary structure without temperature control for 332 days. Doenjang samples were intermittently collected for analysis of viable cell numbers, pH, bacterial communities, and metabolites.
Total viable cells of bacteria and fungi were estimated using a standard counting method as described previously (Jung et al., 2014). Doenjang samples (2 g) were resuspended and serially diluted in PBS buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, and pH 7.2). The diluted supernatants were spread on agar media and incubated at 30°C for 3 days. Respectively, trypticase soy agar (TSA; BD, USA) and potato dextrose agar (PDA; BD, USA), each containing 3% (w/v) NaCl, were used for bacterial and fungal cell counts. Bacterial and fungal cell numbers were counted as colony forming units (CFU) per g-fresh weight of doenjang.
For pH measurements, 10 mL of distilled water was added to 2 g of doenjang samples and vortexed, after which pH values were obtained using a pH meter (Thermo Scientific, USA). For NaCl, concentrations were measured using the Mohr method (AOAC, 2000) and expressed as a percentage (w/w) in the doenjang water phase.
To analyze changes in the bacterial community during doenjang fermentation, 2 g each of doenjang samples were collected from the three porcelain pots and combined. Total genomic DNA was extracted from the combined doenjang samples using the FastDNA Spin kit (MP Biomedical, USA), following manufacturer protocol. The V1–V3 regions of bacterial 16S rRNA genes from total genomic DNA were amplified for barcoded pyrosequencing using Bac9F (5′-adaptor B-AC-GAG TTT GAT CMT GGC TCA G-3′) and Bac541R (5′-adaptor A-X-AC-WTT ACC GCG GCT GCT GG-3′) primers, as described previously (Lee et al., 2012). The “X” denotes 7–10 barcoded sequences for sorting mixed sequencing reads (Supplementary Table S1). The PCR products were purified using a PCR purification kit (Bioneer, Korea), and their concentrations were measured using an ELISA reader equipped with a Take3 multivolume plate (SynergyMx; BioTek). A pooled composite was prepared by mixing equal amounts of the purified PCR products and then sequenced using the 454 GS-FLX Titanium system (Roche, Germany) at Macrogen (Korea).
Pyrosequencing reads were processed and analyzed using RDPipeline tools1 (Cole et al., 2014). The reads were sorted into individual doenjang samples based on their unique barcodes, and then the barcodes were eliminated. Low-quality reads were excluded; these included sequences with more than two ambiguous base calls (“N”), shorter than 300 bp, or average quality scores below 25 (error rate, 0.005). Potential chimeric sequencing reads were also excluded using USEARCH 6.0 available in the RDPipeline (Edgar et al., 2011). The resultant high-quality reads were aligned using the fast, secondary-structure aware INFERNAL aligner (Nawrocki and Eddy, 2007). Their operational taxonomic units (OTUs) and rarefaction curves (Colwell and Coddington, 1994) were calculated at a 97% similarity level using the RDPipeline complete-linkage clustering tool. Shannon–Weaver (Shannon and Weaver, 1963), Chao1 richness (Chao and Bunge, 2002), and evenness indices were also calculated with the RDPipeline. Taxonomic classification of the reads was performed at the phylum and genus levels using the RDP Naïve Bayesian rRNA Classifier 2.5 trained on 16S rRNA training set 9 (Wang et al., 2007) with an 80% confidence threshold.
We used 1H NMR spectroscopy to analyze doenjang metabolites across the entire fermentation period. Metabolites included monosaccharides, organic acids, and nitrogen compounds such as amino acids and BAs. To minimize quantification errors due to large particles, 10 g of doenjang samples were dried in an oven at 80°C for 1 h and ground into a fine power using a pestle and mortar. For sufficient metabolite extraction, 0.2 g of doenjang powder was resuspended in 1.5 mL of 99.9% deuterium oxide (D2O; Sigma–Aldrich, USA) containing 5 mM sodium 2,2-dimethyl-2-silapentane-5-sulfonate (DSS, 97%; Sigma–Aldrich) and incubated on ice with occasional shaking for 1 h. The doenjang powder solutions were centrifuged at 12,000 rpm and 4°C for 10 min, the 600 μL of the supernatant were transferred into NMR tubes. We obtained 1H NMR spectra with a Varian Inova 600-MHz NMR spectrometer (Varian, USA); doenjang metabolites were identified and quantified using the Chenomx NMR Suite program (version 6.1; Chenomx, Canada). Metabolite concentrations were calculated as μmol per g-dry weight doenjang.
The sequence data of the 16S rRNA genes from this study are publicly available in the NCBI Short Read Archive under accession no. SRP072427 (NCBI BioProject PRJNA315598).
The initial pH values of the doenjang samples were approximately 6.4 (Figure 1). During the early fermentation period (0–48 days), pH decreased relatively slowly to approximately 6.0; after 48 days, their drop in pH occurred more quickly. From 108 to 179 days of fermentation, pH remained at around 5.0, after which the samples gradually became more basic again, reaching approximately 6.0 during the late fermentation period (249–332 days of fermentation).
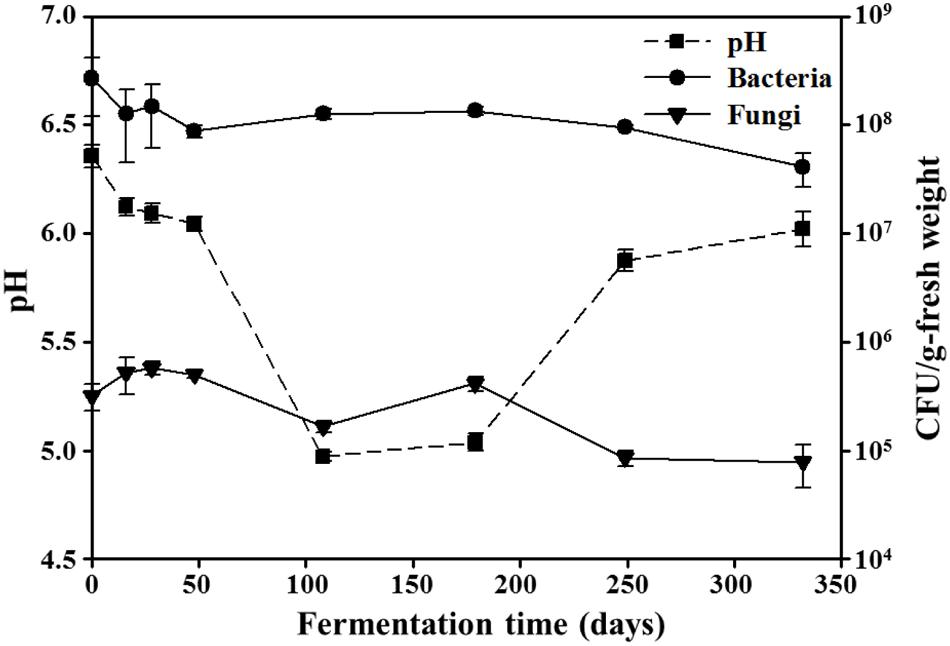
FIGURE 1. Changes in pH, as well as bacterial and fungal abundances, during doenjang fermentation. Data for pH and colony forming units (CFU) are presented as the means ± SD of doenjang samples in triplicate.
Bacterial and fungal viable cells in doenjang were counted on their representative growth agar media, TSA, and PDA, respectively, (Figure 1); bacteria and fungi grown on PDA and TSA, respectively, were excluded from the counting by their colony morphologies. The initial bacterial cells were approximately 2.7 × 108 CFU/g-fresh weight; as fermentation continued, bacterial cell numbers gradually decreased to approximately 4.1 × 107 CFU/g-fresh weight. Similarly, fungal cell counts also experienced a gradual decrease, from an initial number of 3.2 × 105 CFU/g-fresh weight to 7.9 × 104 CFU/g-fresh weight over the course of fermentation. The NaCl concentrations remained relatively constant at approximately 17.5 ± 0.5% (w/w) during the entire fermentation period.
We generated 43,432 sequencing reads from eight doenjang samples using barcoded pyrosequencing. After cleaning, 23,031 high-quality reads, with an average 472-bp length and 2,878 reads per sample, were obtained for the analysis of bacterial diversity and community (Table 1). The rarefaction analysis showed that bacterial diversity fluctuated slightly over the entire doenjang fermentation period (Figure 2), potentially indicating the active occurrence of bacterial succession. Bacterial diversity decreased during the early fermentation period (28 and 48 days) and increase after 48 days until 179 days, only to decrease again during the late fermentation period (249–332 days). All calculated diversity indices (OTU, Shannon–Weaver, Chao1, and evenness) supported the results of the rarefaction curve analysis, although the number of reads obtained affected the bacterial diversity indices (Table 1).
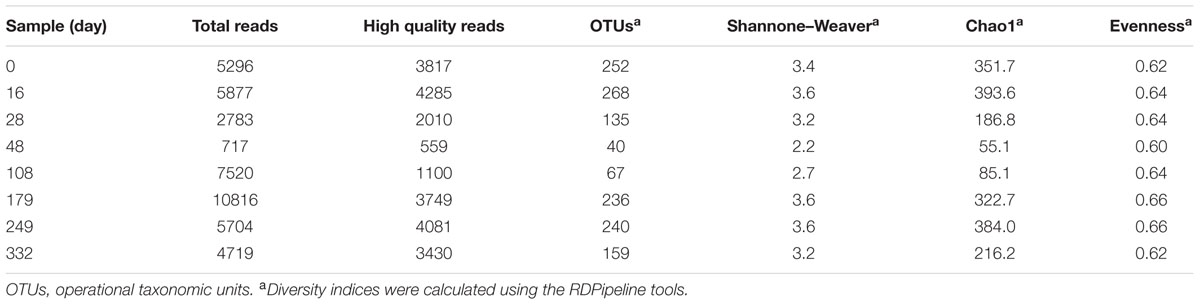
TABLE 1. Bacterial pyrosequencing data sets derived from the doenjang samples and their statistical diversity analysis.
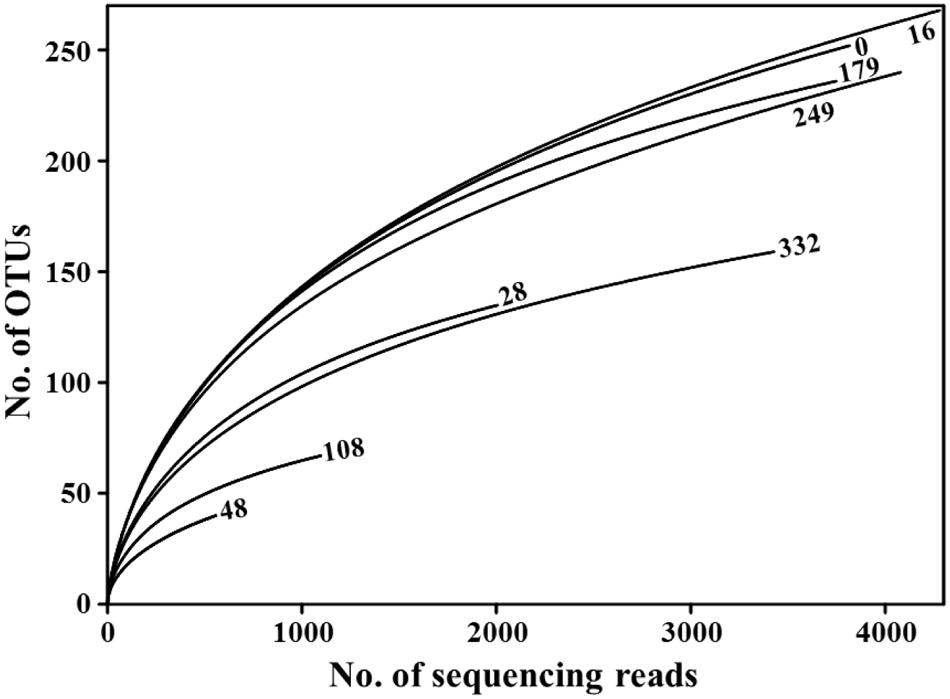
FIGURE 2. Rarefaction curve analysis of 16S rRNA gene sequencing reads showing variation in bacterial diversity throughout the entire doenjang fermentation period. Rarefaction curves were generated with the RDPipeline, using a 97% OTU (operational taxonomic units) cutoff value. Numbers next to the curves indicate doenjang sampling time (days).
Results from phylum- and genus-level classifications of high quality pyrosequencing reads are shown in Figure 3, demonstrating bacterial community fluctuations during doenjang fermentation. The phylum-level analysis revealed that only Firmicutes predominated during the entire fermentation period (Figure 3A). Actinobacteria was also detected as a minor group, with maximum relative abundance reaching approximately 6.5% at 48 days. The genus-level analysis revealed Bacillus to be predominant at a relative abundance of approximately 40%, without an evident fluctuation (Figure 3B). Enterococcus was also identified as a dominant group during early fermentation, but its relative abundance rapidly decreased with the sudden increase of Tetragenococcus at 108 day-doenjang samples. The latter genus was not observed initially but appeared as a dominant group from 108-day samples, and its high relative abundance lasted until the end of fermentation (day 332). Interestingly, Lactobacillus was identified as a dominant group in 179-day samples. Other minor bacterial genera detected in the doenjang samples included Staphylococcus, Clostridium sensu stricto, unclassified Thermoactinomycetaceae 1, and unclassified Bacillales from Firmicutes, as well as Corynebacterium from Actinobacteria; these groups did not exhibit dramatic fluctuations in their relative abundance during fermentation.
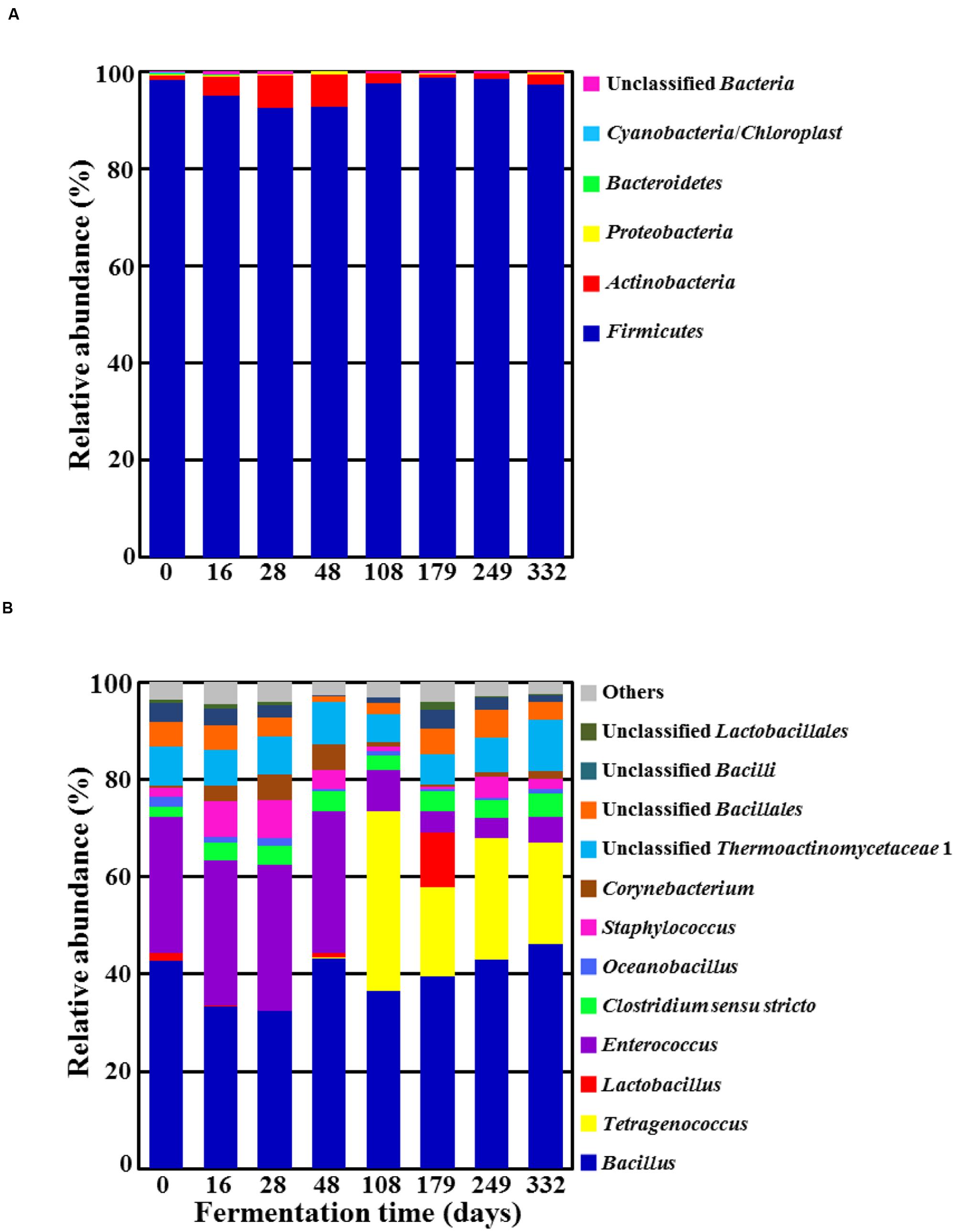
FIGURE 3. Taxonomic classification at the phylum (A) and genus (B) levels showing bacterial community fluctuations during doenjang fermentation. “Others” in (B) refers to genera exhibiting a read percentage<1.0% of the total reads in all doenjang samples.
Results from the 1H NMR analysis of metabolite content throughout doenjang fermentation are presented in Figure 4. Glucose, fructose, galactose, and glycerol were identified as the primary free organic compounds, and their levels increased quickly during the early fermentation period (Figure 4A). However, after approximately 16 days, glucose and fructose concentrations dropped by 108 days; they were almost entirely consumed. In contrast, galactose concentrations continued to increase until 108 days of fermentation, but then began to decrease from 179 days, finally approaching zero at 249 days. Glycerol concentrations reached maximum at 48 days and then experienced a gradual decrease until the end of fermentation.
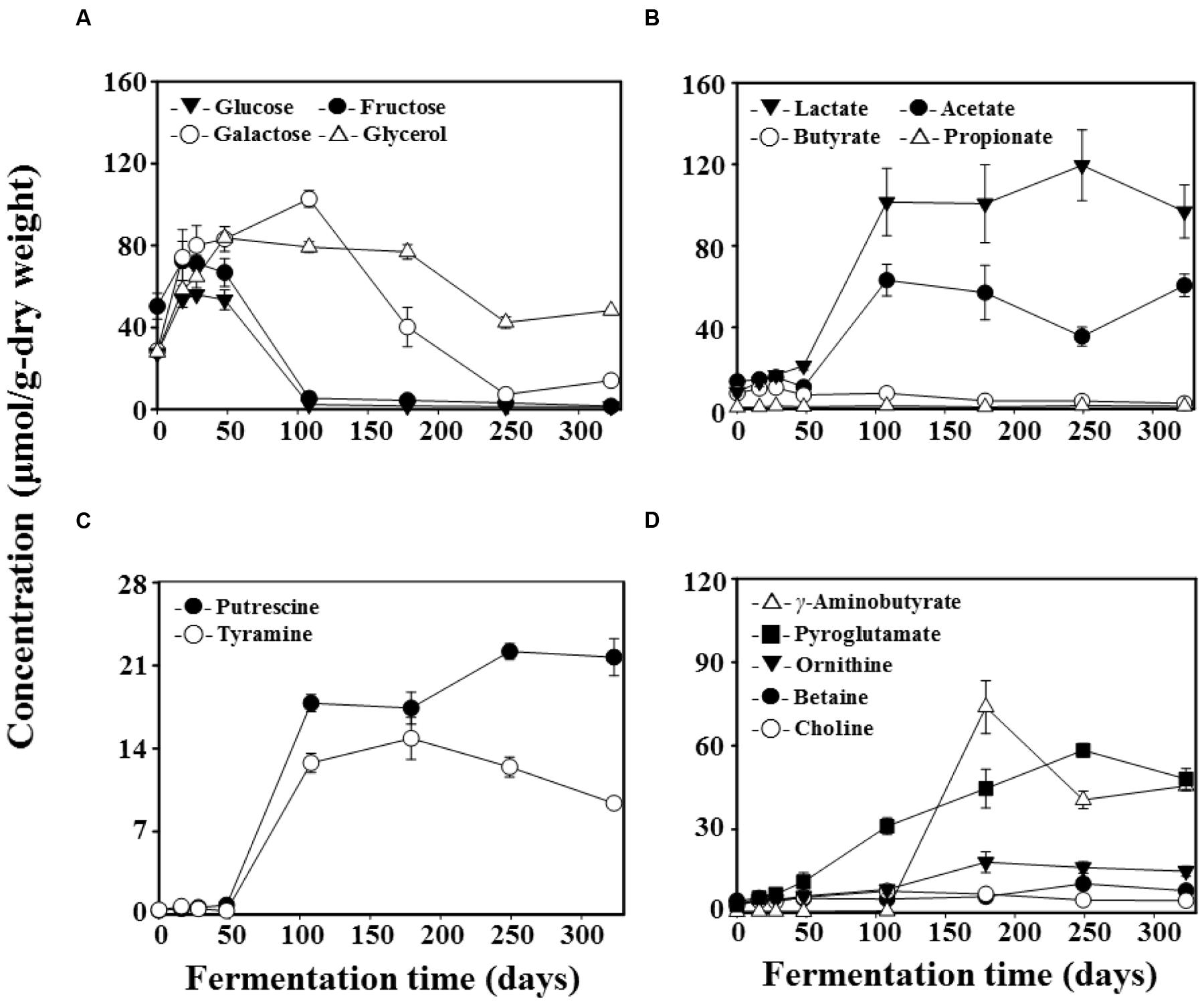
FIGURE 4. Variation in the content of free organic compounds (A), organic acids (B), biogenic amines (C), and nitrogen compounds (D) during doenjang fermentation. Quantifications were performed in the Chenomx NMR suite program (version 6.1, Chenomx, Canada) with sodium 2,2-dimethyl-2-silapentane-5-sulfonate (DSS, 97%) as the internal standard. Data are presented as means ± SD.
Lactate and acetate were identified as the major organic acids during doenjang fermentation (Figure 4B). Both increased rapidly in 108-day samples and then exhibited fairly constant concentrations until the end of fermentation. Minor organic acids found to occur during doenjang fermentation were butyrate and propionate. Next, putrescine and tyramine were identified as the dominant BAs in doenjang; similar to lactate and acetate, these amines also increased rapidly in 108-day samples (Figure 4C). Nitrogen compounds detected in doenjang were ornithine, betaine, choline, γ-aminobutyric acid (GABA), and pyroglutamate (Figure 4D). Pyroglutamate increased continually until 249 days of fermentation. In particular, GABA exhibited a very rapid increase at 179 days.
Finally, the amino acids well known as the main contributors to flavor and taste in doenjang products (Park et al., 2000, 2002; Kim and Lee, 2003) were also major metabolites in our samples (Figure 5). Amino acid concentrations rapidly increased during the early fermentation period. Most (alanine, glutamate, leucine, lysine, and phenylalanine) continued to increase until 108–249 days, and then gradually decreased throughout the remainder of the fermentation.
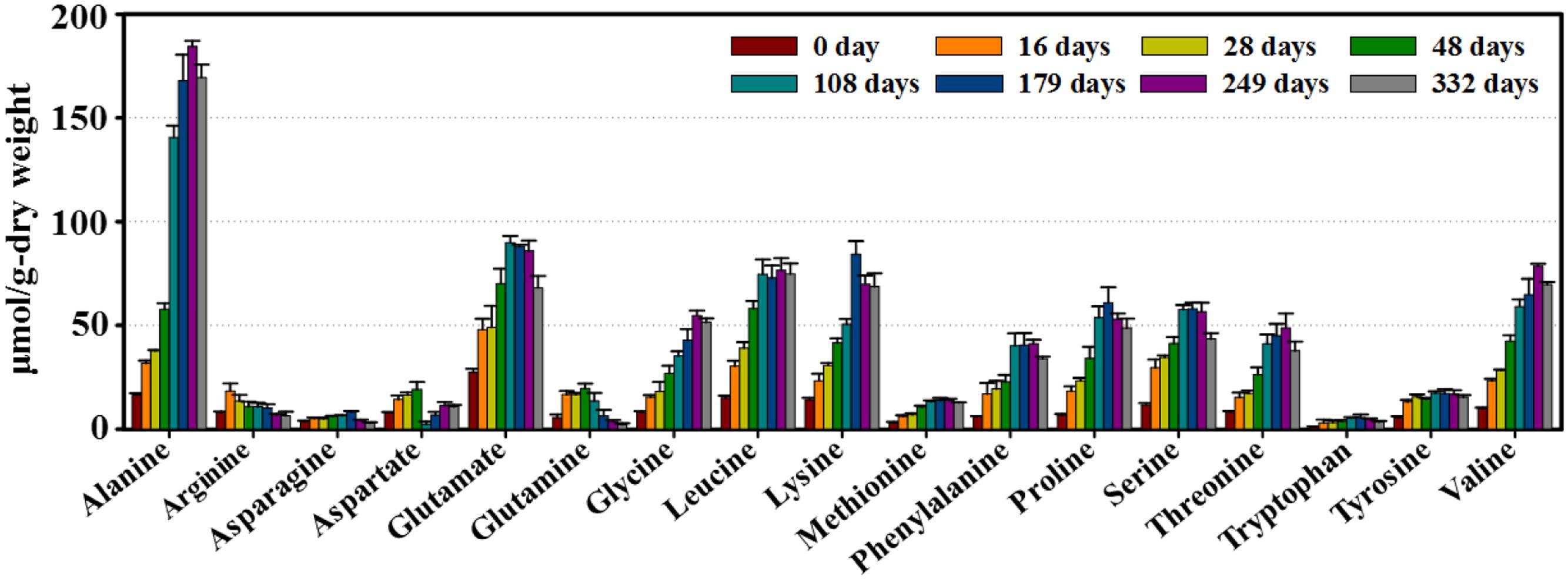
FIGURE 5. Variation in major amino acids identified from doenjang samples during fermentation. Quantifications were performed in the Chenomx NMR Suite (version 6.1, Chenomx, Canada) with sodium 2,2-dimethyl-2-silapentane-5-sulfonate (DSS, 97%) as the internal standard. Data are presented as means ± SD.
Traditional doenjang is produced by a long-term fermentation of solid parts obtained from a doenjang-meju and brine mixture, which is prepared by soaking the fermented doenjang-meju bricks in an 18–20% (w/v) solar salt solution for 40–60 days without the use of starter cultures. Because both bacteria and fungi are involved in doenjang-meju fermentation (Kim et al., 2010b; Lee J.H. et al., 2010; Jung et al., 2014) and have been detected in doenjang samples (Kim et al., 2009; Shukla et al., 2014), researchers have suggested that the two microbial groups may also play important roles in doenjang fermentation. We noted the presence of both bacteria and fungi in our doenjang samples (Figure 1). However, viable bacterial cell counts were approximately three orders of magnitude higher than fungal counts during fermentation. In addition, doenjang fermentation mostly occurs under anaerobic and high moisture conditions. Thus, fungi that prefer dry and aerobic conditions may exhibit lower metabolic activity compared with bacteria during this process. We therefore inferred that the observed fungal growth derived mostly from spores without metabolic activity that had originated in the doenjang-meju. This inference was supported by the lack of hyphae or mycelia in the doenjang samples (data not shown). These results suggest that bacteria are probably more responsible for doenjang fermentation than fungi. Hence, we chose to focus on bacteria in the microbial community analysis.
Although 16S rRNA gene sequencing is the most powerful tool in bacterial taxonomy, it is limitedly used at the species level classification due to the low phylogenetic resolution and poor discriminatory power of 16S rRNA gene sequence at the species level (Fox et al., 1992; Janda and Abbott, 2007). It has been generally accepted that it would be impossible to assign 16S rRNA gene sequences to the level of species because the taxonomies end at the level of genus and DNA relatedness studies are necessary to provide absolute resolution to these taxonomic problems (Schloss and Westcott, 2011). Therefore, in this study we taxonomically classified the 16S rRNA gene sequencing reads at the phylum and genus levels. The genera observed on day 0 of fermentation [Bacillus, Enterococcus, Lactobacillus, Clostridium, Staphylococcus, Corynebacterium, and Oceanobacillus (Figure 3B)] accorded well with genera detected in previous snapshot analyses of doenjang fermentation (Cho and Seo, 2007; Kim et al., 2009; Nam et al., 2012; Jeong et al., 2014). With the exception of Oceanobacillus, all genera had also been identified from doenjang-meju (Jung et al., 2014), despite the preparation step of soaking the bricks in solar salt solution for 42 days. These results suggest that doenjang-meju is a major source of bacteria in doenjang fermentation. In contrast, Oceanobacillus and Tetragenococcus (a dominant group at 108 days of fermentation) likely derived from solar salts because they were not found in doenjang-meju (Jung et al., 2014). Moreover, the two genera are halotolerant or halophilic groups that have been frequently isolated from high-saline environments, including solar salterns (Baati et al., 2010; Lee S.Y. et al., 2010). Thus, solar salts appear to be another important bacterial source in doenjang fermentation.
Bacillus was one of the predominant populations in doenjang during the entire fermentation period (Figure 3B). Due to the well-documented predominance of Bacillus in both culture-dependent and culture-independent studies on doenjang, researchers have generally assumed that this group is primarily responsible for the fermentation process (Yoo et al., 1999; Kim et al., 2010a; Jeon et al., 2016; Tamang et al., 2016b). However, members of Bacillus grow aerobically, and therefore, we consider it unlikely that they can be principal actors in doenjang fermentation, which occurs almost entirely under anaerobic conditions. Furthermore, most Bacillus members do not grow well under conditions more saline than 15% NaCl, although they have been detected from doenjang samples with ∼18% salt concentrations (Jung et al., 2014, 2016). Our data also showed that Bacillus abundance was relatively constant throughout fermentation and was unassociated with fluctuations in metabolite content (Figure 4). Therefore, we infer that Bacillus in doenjang are probably metabolically inactive or exist as spores. Although their relative abundance is high, we infer that this genus is not primarily responsible for doenjang fermentation.
Enterococcus are facultative anaerobic, Gram-positive, cocci-shaped lactic acid bacteria that occurred dominantly alongside Bacillus during the early fermentation period (Figure 3B). This finding corresponds to previous studies that have also reported Enterococcus as a dominant population in doenjang (Cho and Seo, 2007; Kim et al., 2009; Nam et al., 2012). Some members of Enterococcus are pathogenic and have caused infections in humans (Fisher and Phillips, 2009), but other species such as E. faecalis or E. faecium have been frequently isolated from fermented foods and even used as probiotics (M’hir et al., 2012; Todorov and Holzapfel, 2015). Therefore, the dominance of Enterococcus was expected and likely consisted of non-pathogenic species. The high counts of Enterococcus during the early fermentation period was associated with the increase of glucose, fructose, galactose, and glycerol (Figures 3B and 4A), which were probably generated through the hydrolysis of polysaccharides, flavonoid glycosides, and lipids in doenjang-meju. However, most Enterococcus members do not show glyosidic and lipase activities under high saline conditions such as doenjang (Jeong et al., 2014), suggesting that their metabolism was too low during fermentation to hydrolyze the organic carbon compounds. Instead, we speculated that endogenous enzymes derived from doenjang-meju might be responsible for the observed increases in glucose, fructose, galactose, and glycerol. Moreover, despite the dominance of Enterococcus, we only observed slight increases in lactate and acetate during early fermentation, a finding that supports the hypothesis of weak metabolic activity in these lactic acid bacteria (Figures 3B and 4B). However, additional studies will be necessary to investigate how organic carbon compounds increase in doenjang during the early fermentation period.
Bacterial community analysis showed that Staphylococcus increased to approximately 7.6% of the total bacterial abundance at 28 days of fermentation (Figure 3B). Similar to Enterococcus, some members of this genus are pathogenic, but other Staphylococcus species are considered starters for fermentation and accordingly, have been identified from several fermented foods, including doenjang, sausage, and fish sauce (Fontana et al., 2005; Yongsawatdigul et al., 2007; Kim et al., 2009; Guan et al., 2011; Jung et al., 2013; Lee et al., 2015). Therefore, the Staphylococcus species observed in our doenjang samples probably do not have pathogenic properties, but additional research is required to understand their exact role during fermentation, as the function of Staphylococcus was unclear in this study.
Bacterial community analysis demonstrated that Tetragenococcus, a genus of halophilic, Gram-positive lactic acid bacteria, was a dominant group in 108-day samples (Figure 3B). The dominance of Tetragenococcus corresponded well to the rapid increases in lactate and acetate concentrations, as well as the decreases in glucose and fructose, occurring around the same time (Figures 3B and 4A,B). Additionally, the drop in pH at 108 days was in line with heightened lactate and acetate production (Figure 1). Together, these results suggest that Tetragenococcus may be primarily responsible for the production of organic acids during doenjang fermentation. BAs, including putrescine, cadaverine, spermidine, histamine, and tyramine, are low-molecular-weight nitrogenous organic compounds produced via microbial decarboxylation of amino acids and nitrogen compounds during food fermentation (Shalaby, 1996; Park et al., 2000; Moon et al., 2010; Costantini et al., 2013; Jung et al., 2015). These compounds are known to be produced in doenjang (Shukla et al., 2010; Kim and Ji, 2015), but the agents of their production are unknown. The increase of putrescine and tyramine, respectively, generated through ornithine and tyrosine decarboxylation, was also associated with Tetragenococcus dominance (Figures 3B and 4C). These results suggest that Tetragenococcus may be a key agent in biogenic-amine production during doenjang fermentation. Previous studies have also shown that members of Tetragenococcus play important roles in the fermentation of salted goods, including fermented seafood; thus, this genus may be a good bacterial starter for flavor enhancement of fermented foods (Chen et al., 2006; Udomsil et al., 2011; Kim and Park, 2014; Jung et al., 2015, 2016). However, some Tetragenococcus species appear to produce BAs primarily via plasmid-encoded decarboxylation genes (Satomi et al., 2008, 2011, 2014), which increases the difficulty of using Tetragenococcus as starters for salted food fermentation. To address this problem, strains without the ability to produce these compounds have been applied to control biogenic-amine generation during fermentation (Udomsil et al., 2011; Kuda et al., 2012).
Interestingly, Lactobacillus, a group of Gram-positive, facultative anaerobic or microaerophilic, rod-shaped lactic acid bacteria, was identified as a dominant group in 179-day samples (Figure 3B). The non-protein amino acid GABA is a major inhibitory neurotransmitter and is produced through the irreversible α-decarboxylation of L-glutamate by glutamate decarboxylase. Lactobacillus species have been reported as major GABA-producing bacteria during fermentation (Makarova et al., 2006; Cho et al., 2007; Park and Oh, 2007; Siragusa et al., 2007; Komatsuzaki et al., 2008; Jeong et al., 2013). Our metabolite analysis supported these previous findings; galactose decreases and GABA increases were associated with the increase of Lactobacillus (Figures 4A,D), implying that members of Lactobacillus are responsible for GABA production during fermentation. Additionally, the ability of Lactobacillus to metabolize galactose is well-established and members of Lactobacillus have been detected in doenjang (Cho and Seo, 2007; Nam et al., 2012), indicating that lactate production from galactose and GABA synthesis by Lactobacillus are important processes during doenjang fermentation although no report showing their growth in 18% NaCl conditions exists.
To the best of our knowledge, this was the first study to investigate fluctuations in microbial communities and metabolite production simultaneously throughout the entire doenjang fermentation period. Ours was also the first to use doenjang-meju of known bacterial community composition in a study of doenjang fermentation. Here, we suggested that both doenjang-meju and solar salts are important bacterial sources in doenjang fermentation. Furthermore, we proposed that despite their overall abundance, Bacillus may be not as central to doenjang fermentation as previously assumed. Additionally, we showed that solar-salt-derived Tetragenococcus appears to be a primary producer of organic acids and BAs during doenjang fermentation, suggesting that Tetragenococcus strains without this ability are usable as starters, in order to reduce biogenic-amine concentrations. Finally, our results suggested that Lactobacillus is probably one of the major GABA producers during doenjang fermentation. In conclusion, this study contributed to an improved understanding of the biochemical processes underlying doenjang fermentation through exploring the functional properties of major doenjang microbial communities. The data generated should pave the way for additional research employing “omics” technologies, including metagenomics, metatranscriptomics, and metabolomics, which are certain to provide further insights into the production of safe and high-quality doenjang.
CJ conceived the ideas and supervised the work WJ and JJ developed the concepts and performed the experiments WJ and HL analyzed the data and CJ and WJ wrote the manuscript. The manuscript has been reviewed and edited by all authors.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by the Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ00999302), RDA and the Strategic Initiative for Microbiomes in Agriculture and Food, Ministry of Agriculture, Food and Rural Affairs (as part of the multi-ministerial) Genome Technology to Business Translation Program, Republic of Korea.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00827
AOAC (2000). Official Methods of Analysis, 17th Edn. Washington, DC: Association of official analytical chemists.
Baati, H., Amdouni, R., Gharsallah, N., Sghir, A., and Ammar, E. (2010). Isolation and characterization of moderately halophilic bacteria from Tunisian Solar saltern. Curr. Microbiol. 60, 157–161. doi: 10.1007/s00284-009-9516-6
Chao, A., and Bunge, J. (2002). Estimating the number of species in a stochastic abundance model. Biometrics 58, 531–539. doi: 10.1111/j.0006-341X.2002.00531.x
Chen, Y. S., Yanagida, F., and Hsu, J. S. (2006). Isolation and characterization of lactic acid bacteria from dochi (fermented black beans), a traditional fermented food in Taiwan. Lett. Appl. Microbiol. 43, 229–235. doi: 10.1111/j.1472-765X.2006.01922.x
Cho, K. M., and Seo, W. T. (2007). Bacterial diversity in a Korean traditional soybean fermented foods (doenjang and ganjang) by 16S rRNA gene sequence analysis. Food Sci. Biotechnol. 16, 320–324.
Cho, Y. R., Chang, J. Y., and Chang, H. C. (2007). Production of gamma-aminobutyric acid (GABA) by Lactobacillus buchneri isolated from kimchi and its neuroprotective effect on neuronal cells. J. Microbiol. Biotechnol. 17, 104–109.
Cole, J. R., Wang, Q., Fish, J. A., Chai, B., McGarrell, D. M., Sun, Y., et al. (2014). Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 42, D633–D642. doi: 10.1093/nar/gkt1244
Colwell, R. K., and Coddington, J. A. (1994). Estimating terrestrial biodiversity through extrapolation. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 345, 101–118. doi: 10.1098/rstb.1994.0091
Costantini, A., Pietroniro, R., Doria, F., Pessione, E., and Garcia-Moruno, E. (2013). Putrescine production from different amino acid precursors by lactic acid bacteria from wine and cider. Int. J. Food Microbiol. 165, 11–17. doi: 10.1016/j.ijfoodmicro.2013.04.011
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Fisher, K., and Phillips, C. (2009). The ecology, epidemiology and virulence of Enterococcus. Microbiology 155, 1749–1757. doi: 10.1099/mic.0.026385-0
Fontana, C., Sandro Cocconcelli, P., and Vignolo, G. (2005). Monitoring the bacterial population dynamics during fermentation of artisanal Argentinean sausages. Int. J. Food Microbiol. 103, 131–142. doi: 10.1016/j.ijfoodmicro.2004.11.046
Fox, G. E., Wisotzkey, J. D., and Jurtshuk, P. Jr. (1992). How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int. J. Syst. Bacteriol. 42, 166–170. doi: 10.1099/00207713-42-1-166
Guan, L., Cho, K. H., and Lee, J. H. (2011). Analysis of the cultivable bacterial community in jeotgal, a Korean salted and fermented seafood, and identification of its dominant bacteria. Food Microbiol. 28, 101–113. doi: 10.1016/j.fm.2010.09.001
Janda, J. M., and Abbott, S. L. (2007). 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J. Clin. Microbiol. 45, 2761–2764. doi: 10.1128/JCM.01228-07
Jeon, H. H., Jung, J. Y., Chun, B. H., Kim, M. D., Baek, S. Y., Moon, J. Y., et al. (2016). Screening and characterization of potential Bacillus starter cultures for fermenting low salt soybean paste (doenjang). J. Microbiol. Biotechnol. 26, 666–674. doi: 10.4014/jmb.1512.12014
Jeong, D. W., Kim, H. R., Jung, G. S., Han, S. H., Kim, C. T., and Lee, J. H. (2014). Bacterial community migration in the ripening of doenjang, a traditional Korean fermented soybean food. J. Microbiol. Biotechnol. 24, 648–660. doi: 10.4014/jmb.1401.01009
Jeong, S. H., Lee, S. H., Jung, J. Y., Choi, E. J., and Jeon, C. O. (2013). Microbial succession and metabolite changes during long-term storage of kimchi. J. Food Sci. 78, M763–M769. doi: 10.1111/1750-3841.12095
Jung, J. Y., Chun, B. H., and Jeon, C. O. (2015). Chromohalobacter is causing agent for the production of organic acids and putrescine during fermentation of ganjang, a Korean traditional soy sauce. J. Food Sci. 80, M2853–M2859. doi: 10.1111/1750-3841.13114
Jung, J. Y., Lee, H. J., Chun, B. H., and Jeon, C. O. (2016). Effects of temperature on bacterial communities and metabolites during fermentation of myeolchi-Aekjeot, a traditional korean fermented anchovy sauce. PLoS ONE 11:e0151351. doi: 10.1371/journal.pone.0151351
Jung, J. Y., Lee, S. H., and Jeon, C. O. (2014). Microbial community dynamics during fermentation of doenjang-meju, traditional Korean fermented soybean. Int. J. Food Microbiol. 185, 112–120. doi: 10.1016/j.ijfoodmicro.2014.06.003
Jung, J. Y., Lee, S. H., Lee, H. J., and Jeon, C. O. (2013). Microbial succession and metabolite changes during fermentation of saeu-jeot: traditional Korean salted seafood. Food Microbiol. 34, 360–368. doi: 10.1016/j.fm.2013.01.009
Jung, K. O., Park, S. Y., and Park, K. Y. (2006). Longer aging time increases the anticancer and antimetastatic properties of doenjang. Nutrition 22, 539–545. doi: 10.1016/j.nut.2005.11.007
Kim, J. G. (2004). Antigenotoxic effects of water extract from Korean fermented soybean paste (doen-jang). J. Food Prot. 67, 156–161.
Kim, M. S., and Park, E. J. (2014). Bacterial communities of traditional salted and fermented seafoods from Jeju island of Korea using 16S rRNA gene clone library analysis. J. Food Sci. 79, M927–M934. doi: 10.1111/1750-3841.12431
Kim, N. Y., and Ji, G. E. (2015). Characterization of the production of biogenic amines and gamma-aminobutyric acid in the soybean pastes fermented by Aspergillus oryzae and Lactobacillus brevis. J. Microbiol. Biotechnol. 25, 464–468. doi: 10.4014/jmb.1409.09081
Kim, S. H., and Lee, K. A. (2003). Evaluation of taste compounds in water-soluble extract of a doenjang (soybean paste). Food Chem. 83, 339–342. doi: 10.1016/S0308-8146(03)00092-X
Kim, T. W., Kim, Y. H., Kim, S. E., Lee, J. H., Park, C. S., and Kim, H. Y. (2010a). Identification and distribution of Bacillus species in doenjang by whole-cell protein patterns and 16S rRNA gene sequence analysis. J. Microbiol. Biotechnol. 20, 1210–1214. doi: 10.4014/jmb.1002.02008
Kim, T. W., Lee, J. H., Kim, S. E., Park, M. H., Chang, H. C., and Kim, H. Y. (2009). Analysis of microbial communities in doenjang, a Korean fermented soybean paste, using nested PCR-denaturing gradient gel electrophoresis. Int. J. Food Microbiol. 131, 265–271. doi: 10.1016/j.ijfoodmicro.2009.03.001
Kim, T. W., Lee, J. H., Park, M. H., and Kim, H. Y. (2010b). Analysis of bacterial and fungal communities in Japanese- and Chinese-fermented soybean pastes using nested PCR-DGGE. Curr. Microbiol. 60, 315–320. doi: 10.1007/s00284-009-9542-4
Komatsuzaki, N., Nakamura, T., Kimura, T., and Shima, J. (2008). Characterization of glutamate decarboxilase from high gamma-aminobutyric acid (GABA)-producer, Lactobacillus paracasei. Biosci. Biotechnol. Biochem. 72, 278–285. doi: 10.1271/bbb.70163
Kuda, T., Izawa, Y., Ishii, S., Takahashi, H., Torido, Y., and Kimura, B. (2012). Suppressive effect of Tetragenococcus halophilus, isolated from fish-nukazuke, on histamine accumulation in salted and fermented fish. Food Chem. 130, 569–574. doi: 10.1016/j.foodchem.2011.07.074
Kwon, D. Y., Daily, J. W., Kim, H. J., and Park, S. (2010). Antidiabetic effects of fermented soybean products on type 2 diabetes. Nutr. Res. 30, 1–13. doi: 10.1016/j.nutres.2009.11.004
Lee, H. J., Jung, J. Y., Oh, Y. K., Lee, S. S., Madsen, E. L., and Jeon, C. O. (2012). Comparative survey of rumen microbial communities and metabolites across one caprine and three bovine groups, using barcoded pyrosequencing and 1H nuclear magnetic resonance spectroscopy. Appl. Environ. Microbiol. 78, 5983–5993. doi: 10.1128/AEM.00104-12
Lee, J. H., Kim, T. W., Lee, H., Chang, H. C., and Kim, H. Y. (2010). Determination of microbial diversity in meju, fermented cooked soya beans, using nested PCR-denaturing gradient gel electrophoresis. Lett. Appl. Microbiol. 51, 388–394. doi: 10.1111/j.1472-765X.2010.02906.x
Lee, S. H., Jung, J. Y., and Jeon, C. O. (2015). Bacterial community dynamics and metabolite changes in myeolchi-aekjeot, a Korean traditional fermented fish sauce, during fermentation. Int. J. Food Microbiol. 203, 15–22. doi: 10.1016/j.ijfoodmicro.2015.02.031
Lee, S. Y., Oh, T. K., Kim, W., and Yoon, J. H. (2010). Oceanobacillus locisalsi sp. nov., isolated from a marine solar saltern. Int. J. Syst. Evol. Microbiol. 60, 2758–2762. doi: 10.1099/ijs.0.021907-0
Makarova, K. A., Slesarev, Y., Wolf, A., Sorokin, B., Mirkin, E., Koonin, A., et al. (2006). Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. U.S.A. 103, 15611–15616. doi: 10.1073/pnas.0607117103
M’hir, S., Minervini, F., Di Cagno, R., Chammem, N., and Hamdi, M. (2012). Technological, functional and safety aspects of enterococci in fermented vegetable products: a mini-review. Ann. Microbiol. 62, 469–481. doi: 10.1007/s13213-011-0363-x
Moon, J. S., Cho, S. K., Choi, H. Y., Kim, J. E., Kim, S. Y., Cho, K. J., et al. (2010). Isolation and characterization of biogenic amine-producing bacteria in fermented soybean pastes. J. Microbiol. 48, 257–261. doi: 10.1007/s12275-010-0040-y
Nam, Y. D., Lee, S. Y., and Lim, S. I. (2012). Microbial community analysis of Korean soybean pastes by next-generation sequencing. Int. J. Food Microbiol. 155, 36–42. doi: 10.1016/j.ijfoodmicro.2012.01.013
Namgung, H. J., Park, H. J., Cho, I. H., Choi, H. K., Kwon, D. Y., Shim, S. M., et al. (2009). Metabolite profiling of doenjang, fermented soybean paste, during fermentation. J. Sci. Food Agric. 90, 1926–1935. doi: 10.1002/jsfa.4036
Nawrocki, E. P., and Eddy, S. R. (2007). Query-dependent banding (QDB) for faster RNA similarity searches. PLoS Comput. Biol. 3:e56. doi: 10.1371/journal.pcbi.0030056
Park, E. J., Chun, J. S., Cha, C. J., Park, W. S., Jeon, C. O., and Bae, J. W. (2012). Bacterial community analysis during fermentation of ten representative kinds of kimchi with barcoded pyrosequencing. Food Microbiol. 30, 197–204. doi: 10.1016/j.fm.2011.10.011
Park, H. K., Gil, B., and Kim, J. K. (2002). Characteristics of taste components of commercial soybean paste. Food Sci. Biotechnol. 11, 376–379.
Park, H. K., Shukla, S., Lee, J. S., Kim, J. K., and Kim, M. (2014). Reduction of foodborne pathogens and aflatoxins in doenjang samples using defined Meju. J. Food Saf. 34, 161–167. doi: 10.1111/jfs.12109
Park, J. S., Park, H. Y., Kim, D. H., Kim, D. H., and Kim, H. K. (2008). Ortho-dihydroxyisoflavone derivatives from aged doenjang (Korean fermented soypaste) and its radical scavenging activity. Bioorg. Med. Chem. Lett. 18, 5006–5009. doi: 10.1016/j.bmcl.2008.08.016
Park, K. B., and Oh, S. H. (2007). Cloning, sequencing and expression of a novel glutamate decarboxylase gene from a newly isolated lactic acid bacterium, Lactobacillus brevis OPK-3. Bioresour. Technol. 98, 312–319. doi: 10.1016/j.biortech.2006.01.004
Park, S. K., Seo, K. I., Choi, S. H., Moon, J. S., and Lee, Y. H. (2000). Quality assessment of commercial doenjang prepared by traditional method. J. Korean Soc. Food Sci. Nutr. 29, 211–217.
Rhyu, M. R., and Kim, E. Y. (2011). Umami taste characteristics of water extract of doenjang, a Korean soybean paste: low-molecular acidic peptides may be a possible clue to the taste. Food Chem. 127, 1210–1215. doi: 10.1016/j.foodchem.2011.01.128
Satomi, M., Furushita, M., Oikawa, H., Takahashi, M. Y., and Yano, Y. (2008). Analysis of a 30 kbp plasmid encoding histidine decarboxylase gene in Tetragenococcus halophilus isolated from fish sauce. Int. J. Food Microbiol. 126, 202–209. doi: 10.1016/j.ijfoodmicro.2008.05.025
Satomi, M., Furushita, M., Oikawa, H., and Yano, Y. (2011). Diversity of plasmids encoding histidine decarboxylase gene in Tetragenococcus spp. isolated from Japanese fish sauce. Int. J. Food Microbiol. 148, 60–65. doi: 10.1016/j.ijfoodmicro.2011.04.025
Satomi, M., Shozen, K., Furutani, A., Fukui, Y., Kimura, M., Yasuike, M., et al. (2014). Analysis of plasmids encoding the tyrosine decarboxylase gene in Tetragenococcus halophilus isolated from fish sauce. Fish. Sci. 80, 849–858. doi: 10.1007/s12562-014-0756-4
Schloss, P. D., and Westcott, S. L. (2011). Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis. Appl. Environ. Microbiol. 77, 3219–3226. doi: 10.1128/AEM.02810-10
Shalaby, A. R. (1996). Significance of biogenic amines to food safety and human health. Food Res. Int. 29, 675–690. doi: 10.1016/S0963-9969(96)00066-X
Shannon, C. E., and Weaver, W. (1963). The Mathematical Theory of Communication. Urbana, IL: University of Illinois Press.
Shukla, S., Park, H. K., Kim, J. K., and Kim, M. H. (2010). Determination of biogenic amines in Korean traditional fermented soybean paste (doenjang). Food Chem. Toxicol. 48, 1191–1195. doi: 10.1016/j.fct.2010.01.034
Shukla, S., Park, H. K., Lee, J. S., Kim, J. K., and Kim, M. H. (2014). Reduction of biogenic amines and aflatoxins in doenjang samples fermented with various meju as starter cultures. Food Control 42, 181–187. doi: 10.1016/j.foodcont.2014.02.006
Siragusa, S., De Angelis, M., Di Cagno, R., Rizzello, C. G., Coda, R., and Gobbetti, M. (2007). Synthesis of γ-aminobutyric acid by lactic acid bacteria isolated from a variety of Italian cheeses. Appl. Environ. Microbiol. 22, 7283–7290. doi: 10.1128/AEM.01064-07
Tamang, J. P., Shin, D. H., Chae, S., and Jung, S. (2016a). Functional properties of microorganisms in fermented foods. Front. Microbiol. 7:578. doi: 10.3389/fmicb.2016.00578
Tamang, J. P., Watanabe, K., and Holzapfel, W. H. (2016b). Review: diversity of microorganisms in global fermented foods and beverages. Front. Microbiol. 7:377. doi: 10.3389/fmicb.2016.00377
Todorov, S. D., and Holzapfel, W. H. (2015). “Traditional cereal fermented foods as sources of functional (bacteriocinogenic and probiotic) microorganisms,” in Advances in Fermented Foods and Beverages: Improving Quality, Technologies and Health Benefits, ed. W. H. Holzapfel (London: Woodhead), 123–153.
Udomsil, N., Rodtong, S., Choi, Y. J., Hua, Y., and Yongsawatdigul, J. (2011). Use of Tetragenococcus halophilus as a starter culture for flavor improvement in fish sauce fermentation. J. Agric. Food Chem. 59, 8401–8408. doi: 10.1021/jf201953v
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Yongsawatdigul, J., Rodtong, S., and Raksakulthai, N. (2007). Acceleration of Thai fish sauce fermentation using proteinases and bacterial starter cultures. J. Food Sci. 72, M382–M390. doi: 10.1111/j.1750-3841.2007.00532.x
Yoo, S. K., Cho, W. H., Kang, S. M., and Lee, S. H. (1999). Isolation and identification of microorganisms in Korean traditional soybean paste and soybean sauce. Korean J. Appl. Microbiol. Biotechnol. 27, 113–117.
Keywords: doenjang, soybean paste, bacterial community, metabolites, Bacillus, Tetragenococcus, biogenic amine, GABA
Citation: Jung WY, Jung JY, Lee HJ and Jeon CO (2016) Functional Characterization of Bacterial Communities Responsible for Fermentation of Doenjang: A Traditional Korean Fermented Soybean Paste. Front. Microbiol. 7:827. doi: 10.3389/fmicb.2016.00827
Received: 29 March 2016; Accepted: 17 May 2016;
Published: 31 May 2016.
Edited by:
Wilhelm Heinrich Holzapfel, Handong Global University, South KoreaReviewed by:
Jyoti Prakash Tamang, Sikkim University, IndiaCopyright © 2016 Jung, Jung, Lee and Jeon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Che Ok Jeon, Y29qZW9uQGNhdS5hYy5rcg==
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.