
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
METHODS article
Front. Microbiol., 06 May 2016
Sec. Food Microbiology
Volume 7 - 2016 | https://doi.org/10.3389/fmicb.2016.00651
Vibrio alginolyticus is one of the most common pathogenic marine Vibrio species, and has been found to cause serious seafood-poisoning or fatal extra-intestinal infections in humans, such as necrotizing soft-tissue infections, bacteremia, septic shock, and multiple organ failures. Delayed accurate diagnosis and treatment of most Vibrio infections usually result to high mortality rates. The objective of this study was to establish a rapid diagnostic method to detect and identify the presence of V. alginolyticus in different samples, so as to facilitate timely treatment. The widely employed conventional methods for detection of V. alginolyticus include biochemical identification and a variety of PCR methods. The former is of low specificity and time-consuming (2–3 days), while the latter has improved accuracy and processing time. Despite such advancements, these methods are still complicated, time-consuming, expensive, require expertise and advanced laboratory systems, and are not optimal for field use. With the goal of providing a simple and efficient way to detect V. alginolyticus, we established a rapid diagnostic method based on loop-mediated Isothermal amplification (LAMP) technology that is feasible to use in both experimental and field environments. Three primer pairs targeting the toxR gene of V. alginolyticus were designed, and amplification was carried out in an ESE tube scanner and Real-Time PCR device. We successfully identified 93 V. alginolyticus strains from a total of 105 different bacterial isolates and confirmed their identity by 16s rDNA sequencing. We also applied this method on infected mouse blood and contaminated scallop samples, and accurate results were both easily and rapidly (20–60 min) obtained. Therefore, the RT-LAMP assay we developed can be conveniently used to detect the presence of V. alginolyticus in different samples. Furthermore, this method will also fulfill the gap for real-time screening of V. alginolyticus infections especially while on field.
1. A new LAMP method was developed for the detection of Vibrio alginolyticus.
2. It successfully and accurately detected 105 Vibrio strains from different sources.
3. The assay was directly applicable to infected mouse blood and contaminated scallops.
4. The assay was suitable for use both in field and routine laboratories.
Vibrio species are pathogenic bacteria that are implicated in a number of serious infections and illnesses (Vezzulli et al., 2013). One of the most pathogenic species is Vibrio alginolyticus, a gram-negative and curved halophilic bacterium, initially classified as a biological type of Vibrio parahaemolyticus, but was later on listed as an independent species in Buchanan and Gibbons (1974). V. alginolyticus is mainly found in marine estuaries, coastal and aquatic environments (Narracci et al., 2013) with worldwide distribution. It may exist as free-living, a parasite or associated with surfaces of organisms such as marine vertebrates/invertebrates and flora, and even humans (Molitoris et al., 1985; Chen et al., 2011; Schets et al., 2011; Torresi et al., 2011). This species is also highly abundant and usually dominates Vibrio communities (Lai et al., 2004; Schets et al., 2010; Jones et al., 2013).
Vibrio alginolyticus has been found to cause varied infections and inflammation in both humans and animals (Rubin and Tilton, 1975) such as otitis, ocular infections, intracranial infection, peritonitis and osteomyelitis among others (Taylor et al., 1981; Opal and Saxon, 1986; Barbarossa et al., 2002; Feingold and Kumar, 2004; Li et al., 2009). In addition, serious extra-intestinal or seafood borne primary infections, especially in immunocompromised patients, can cause fatal diseases such as necrotizing soft-tissue infections and bacteremia, which are always accompanied by septic shock and multiple organ dysfunction, resulting in high morbidity and mortality (Chien et al., 2002; Hong et al., 2014). The number of cases associated with V. alginolyticus has dramatically increased in the past decades, posing threats to public health and safety, and even food security. In Chinese southeast coastal areas for example, cases of V. alginolyticus-associated wound infections and gastro-intestinal diseases are already considered not uncommon (Ji, 1989; Song et al., 2013). The same is true in the United States and other regions of the world (Weis et al., 2011; Jones et al., 2013). In fact, V. alginolyticus has been observed to cause infections leading to various illnesses in Penaeus vannamei Boone, Epinephelus coioidesi, Crassostrea rhizophorae, Pseudosciaena crocea, and Sparus latus among others (Pereira et al., 2007; Oates et al., 2012).
The severity and life-threatening characteristics of V. alginolyticus related infections necessitate a rapid, low-cost and accurate method of early and specific identification and detection of the pathogen, which would be helpful in saving lives. At present, the widely employed conventional methods include PCR amplification and identification of 16s rRNA gene, and biochemical characterizations (Lai et al., 2014). However, these traditional techniques are still limited with lower accuracy, longer processing time and cost of personnel and reagents.
Loop-mediated isothermal amplification (LAMP) is an innovative isothermal nucleic acid amplification technology that has been applied to gene diagnosis (Notomi et al., 2015). Compared with most known gene diagnostics (i.e., PCR, hybridization), LAMP has low requirement for complex infrastructure with easily readable results without compromising specificity and sensitivity. For example, LAMP has shortened the sample pre-preparation and detection time from 4 to 10 days (collection-extraction-PCR) to only 20–60 min. Because of such apparent advantages, LAMP has been given increased attention by the World Health Organization (WHO), relevant government departments, institutes and researchers all over the world. To date, the method has been successfully used for the rapid diagnosis of microorganisms such as viruses, bacteria and parasites in different samples (Cai et al., 2010; Liew et al., 2014; Ferrara et al., 2015; Hayashida et al., 2015; Liu et al., 2015; Wang et al., 2016).
A crucial parameter in the application of LAMP technology is the design of primers targeting genes of high specificity between species. Among the possible target genes used for the specific Vibrio detection, the concatenated sequences of toxR show a considerable gap between the maximal interspecies similarity and the minimal intraspecies similarity (Pascual et al., 2010). For instance, the similarity between Chromobacterium violaceum, V. parahaemolyticus, and Vibrio vulnificus is only 52–63% (Lin et al., 1993; Lee et al., 2000; Chen et al., 2012). This hypervariable characteristic of toxR gene makes it a good molecular marker for species identification. The toxR gene has been widely used in the identification of Vibrio species, such as V. parahaemolyticus, V. anguillarum, V. vulnificus, Vibrio cholera (Neogi et al., 2010; Crisafi et al., 2011; He et al., 2014). In this study, we aimed to establish a new rapid diagnostic LAMP method to identify the presence of V. alginolyticus by targeting the toxR gene.
A total of 105 marine bacterial strains were used in our study. Ninety-two marine strains were isolated from southeast China coastal waters by Navy General Hospital microbiological experiment. The other six marine bacteria were cultivated from seafood samples and were provided by Beijing Entry-Exit Inspection and Quarantine Bureau. Model and standard strains including V. fluvialis 1A10009, V. parahaemolyticus 1A10122, V. fischeri 1H00021, and V. vulnificus 1H00066 were purchased from MCCC (Marine Culture Collection of China), while V. alginolyticus 17749 was an ATCC standard saved by our laboratory. Escherichia coli MG4 was from Ocean University of China and C. violaceum CV026 was provided by the PLA Academy of Military Sciences.
All the bacteria in this study were pre-identified by physiological and biochemical characterization, and further purified by isolating single colonies. Cultures were routinely maintained and rec-culturing overnight in broth medium (2216E or LB broth) at 30 or 35°C.
Loop-mediated Isothermal amplification primers targeting the toxR gene of V. alginolyticus were designed based on the sequence: GenBank = JF930593.1, using LAMP designer 1.13 (Premier Biosoft, USA). Designed primers and their sequences are summarized in Table 1. The primers were tested in all 105 strains to test for efficiency and specificity.
Two kinds of commercially available DNA extraction kits (BEIJING TIANGEN Biotech, Co., Ltd.) were adopted. TIANamp Bacteria DNA Kit was used for DNA extraction of bacteria grown in the culture medium. Target bacteria were adjusted to 1 × 106 CFU/mL in advance. TIANamp Blood DNA Kit was used for whole blood DNA extraction, and 500 μL blood was needed of each sample. DNA was extracted from both types of samples according to the manufacturer’s protocols. All extracted DNA templates were stored at -20° for later use.
At the meantime, positive and negative controls were set. Plasmid DNA containing the target gene isolated from V. alginolyticus 17749 was used as the positive control for PCR. It was amplified using primers F3 and B3 (Table 1), and then linked to a T vector. Sterile ddH2O was used as the negative control.
Loop-mediated Isothermal amplification assays were carried out using the Bst WarmStartTM DNA polymerase (New England Biolabs; 1 μL) in mixtures added with 22 μL buffer solution containing 0.2 μM F3, 0.2 μM B3, 1.6 μM FIP, 1.6 μM BIP, 0.8 μM FLP, 0.8 μM BIP, 1 M betaine, 8 mM MgSO4, 1.6 mM dNTP, 2.5 μL 10× Thermo pol buffer (New England Biolabs), 5 μM SYTO®-9 (Invitrogen, Life Technology) and ddH2O per reaction. Two microliters of DNA template were used per assay, totaling to 25 μL.
Isothermal amplification was accomplished at 63°C for 30 min while using ESE-Quant tube scanner as one of the two detection platforms, and the whole reaction system was sealed by addition of 200 μL mineral oil (GBCBIO Technologies). The other platform tested measures the amplification as it happens via a Real-Time PCR device (Bio-Rad Laboratories). Reaction started with 45 cycles at 63°C for 15 s (stage 1) and at 63°C for 45 s (stage 2) coupled with detection of fluorescence signals, which were analyzed by Bio-Rad CFX Manager 3.1. Through the two reaction monitoring systems, a result was considered positive when a “S” amplification curve was obtained, while linear or slightly oblique amplification curve showed a negative result.
To validate the results of the LAMP assay, identities of the 105 bacterial strains were confirmed by 16s rDNA identification. Amplification of the target gene was done in 30 μL PCR reaction mixtures following the suggestion of Lane (1991) and Moreno et al. (2002). Primers used for 16s rDNA identification are shown in Table 2.
Quality of the amplified products was checked by DL 2000 Marker (BEIJING TIANGEN Biotech, Co., Ltd.) before being sent to Sino Geno Max (Beijing) for further purification and then sequencing. Raw sequences were manually checked and then aligned using the reference 16s rDNA sequences (accession number: KC768792.1 for V. parahaemolyticus ATCC 17802; JX009143.1 for V. fluvialis NCTC 11327; EU204961.1 for V. furnissii M1; X76333.1 for V. vulnificus ATCC 27562T; DQ068934.1 for V. mimicus UN 11607; JX290082.1 for V. cholera E153) by Sequence Analysis Tool in RDP Release 111.
BALB/c female mice (20–23 g in weight) used in this study were purchased from the Academy of Military Medical Sciences (Beijing, China), and bred under specific-pathogen-free (SPF) conditions (Sakuma et al., 2013). All animal procedures complied with the institutional and national guidelines prescribed by the International Council for Laboratory Animal Science (ICLAS) the ministry of health of the People’s Republic of China. The mouse experiment was approved and supervised by Beijing Institute of Radiation Medicine Experiment Committee (2012-0131). The way of preparation of infected mouse model followed the methods described by Khajanchi et al. (2011), Angkasekwinai et al. (2014), and Liu et al. (2014, 2015).
The mice for virulence testing were divided into six groups (G = 6, n = 4); the bacterial strains injected include V. alginolyticus 17749 (G1), V. fluvialis 1A10009 (G2), V. parahaemolyticus 1A10122 (G3), V. fischeri 1H00021 (G4), V. vulnificus 1H00066 (G5) and non-Vibrio infected mice as the control group (G6). Mice were intraperitoneally injected with the Vibrio species above (1 × 106 CFU/mL) resuspended in 1 × phosphate buffer saline (PBS), while G6 was injected with 1× PBS. After the injection, each BALB/c mouse was fed with sterile water only. Sampling from the infected and control groups were carried out 16 h post-infection by collecting blood. The blood obtained from eyeballs was blended with acid citrate dextrose solution (1.32% M/V sodium citrate, 0.48% M/V citric acid, and 1.47% M/V glucose) as the anticoagulant, and then were prepared for template DNA to run LAMP assay.
To simulate the detection of the pathogens in natural samples (i.e., seafoods), artificially V. alginolyticus-infected scallop samples were developed. Sterile scallop alkaline peptone water culture solutions (samples 4–6) were prepared and then added with V. alginolyticus in gradient. Final bacterial concentrations in solutions were 105 CFU/mL (sample 1), 104 CFU/mL (sample 2), and 103CFU/mL (sample 3). These were then used to assay for the efficiency and sensitivity of the primers by real-time fluorescence LAMP kit method. Positive (bacterial culture) and negative (ddH2O) controls were used.
The limit of detection (LOD) of the LAMP method was tested by serial dilutions, using ESE-Quant tube scanner and qPCR as the detecting device. The 10-fold serial dilution of V. alginolyticus ATCC 17749 with cells ranged from 1 × 100 to 1 × 109 CFU/mL as determined by plate counts. Results showed that no bacteria were detected at 1 × 100 and 1 × 101 CFU/mL DNA templates (Figures 1 and 2), suggesting that the LOD of the LAMP method using ESE-Quant tube scanner and qPCR device in this experiment were both 1 × 102 CFU/mL.
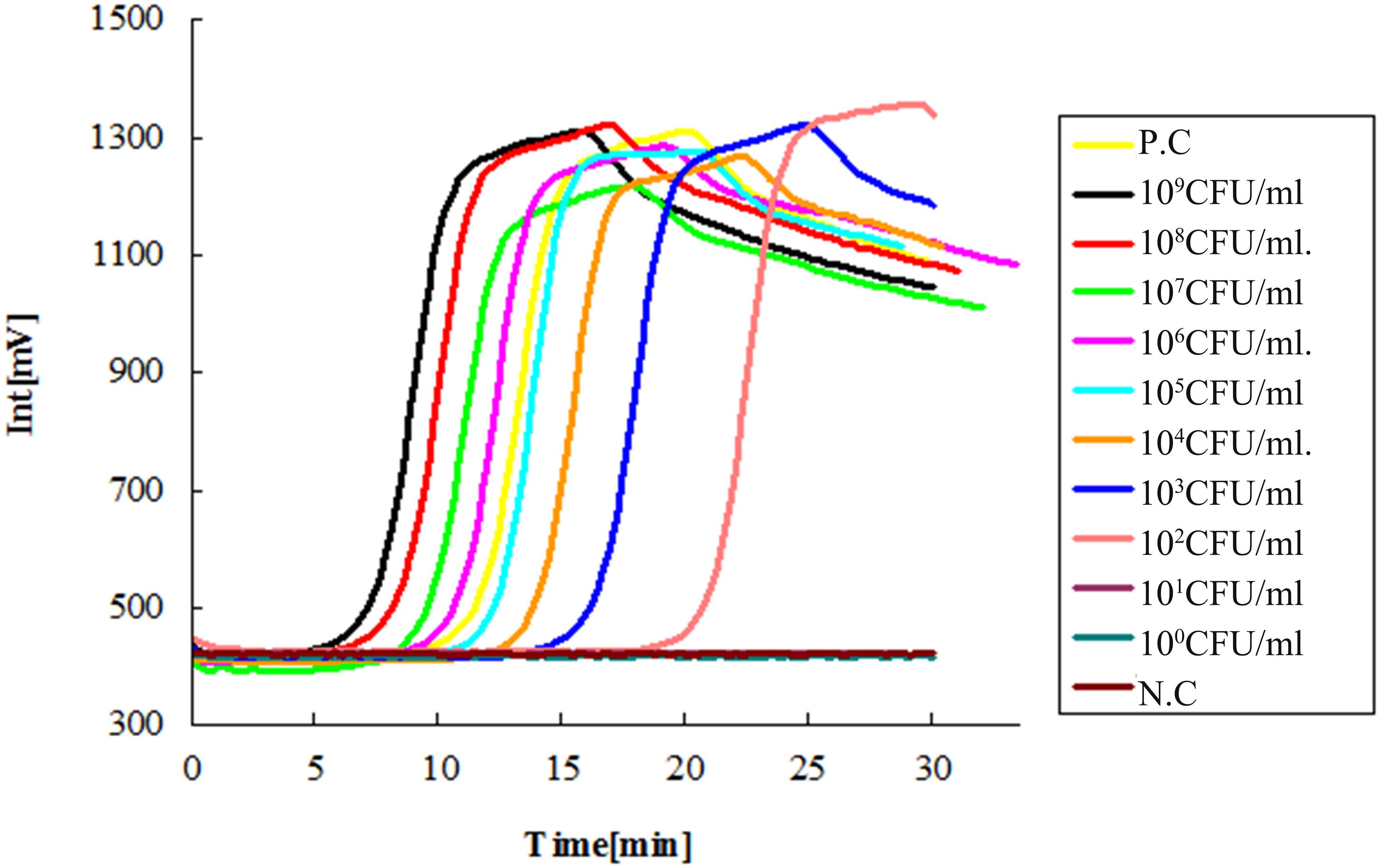
FIGURE 1. Sensitivity of the LAMP assay with ESE-Quant tube scanner. Suspensions of Vibrio alginolyticus ATCC 17749 were 10-fold serially diluted (1 × 100 ∼ 1 × 109 CFU/mL) and detected with the LAMP method using an ESE-Quant tube scanner. No amplification was detected in the 10-fold serial dilution equivalent to 1 × 100 and 1 × 101 CFU/mL DNA templates of V. alginolyticus standard strain ATCC 17749. N.C as negative control, P.C as positive control.
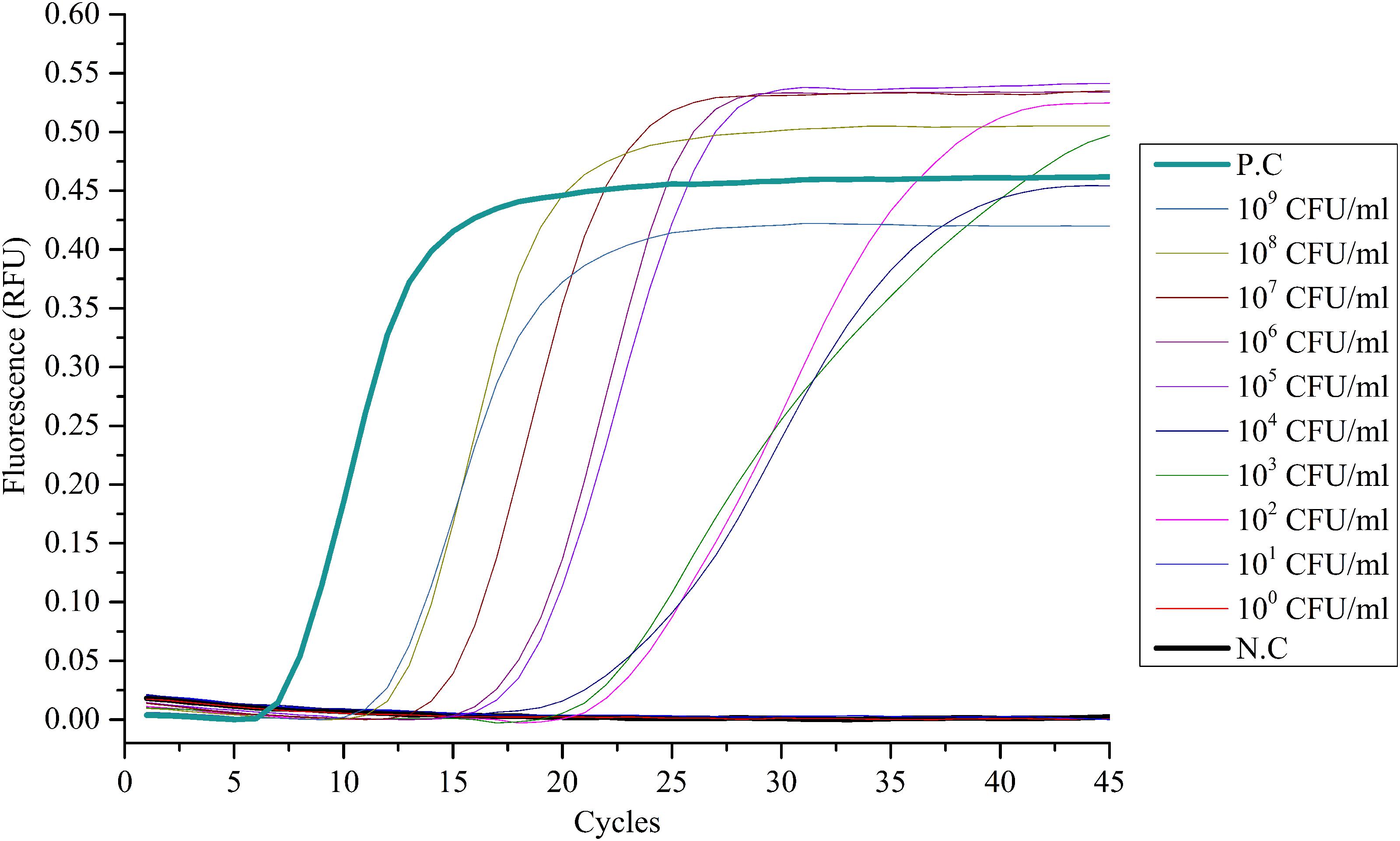
FIGURE 2. Sensitivity of the LAMP assay with qPCR instrument. Suspensions of V. alginolyticus ATCC 17749 were 10-fold serially diluted (1 × 100 ∼ 1 × 109 CFU/mL) and detected with the LAMP method using a qPCR instrument. Only the suspensions of V. alginolyticus ATCC 17749 of 1 × 102 ∼ 1 × 109 CFU/mL could be detected. N.C as negative control, P.C as positive control.
The specificity of the LAMP method using two devices was observed. Altogether, eight different kinds of Vibrio species (12 strains) were used for detection, including two strains of V. alginolyticus, two strains of V. fluvialis, two strains of V. parahaemolyticus, two strains of V. vulnificus, and V. fischeri, V. furnissii, V. mimicus, V. cholera each of one strain. Vibrio strain 030-002 was randomly selected from the 92 strains of V. alginolyticus. Plasmid DNA containing the target gene isolated from V. alginolyticus 17749 was used as the positive control. Sterile ddH2O was used as the negative control. Using the LAMP-ESE-Quant Tube Scanner, aside from the positive control, the only successful amplified results were V. alginolyticus 17749 and the randomly chosen 030-002 strain (Figure 3). Fluorescence voltage increased to 550 mV after 26 min of the reaction which clearly separated the results.
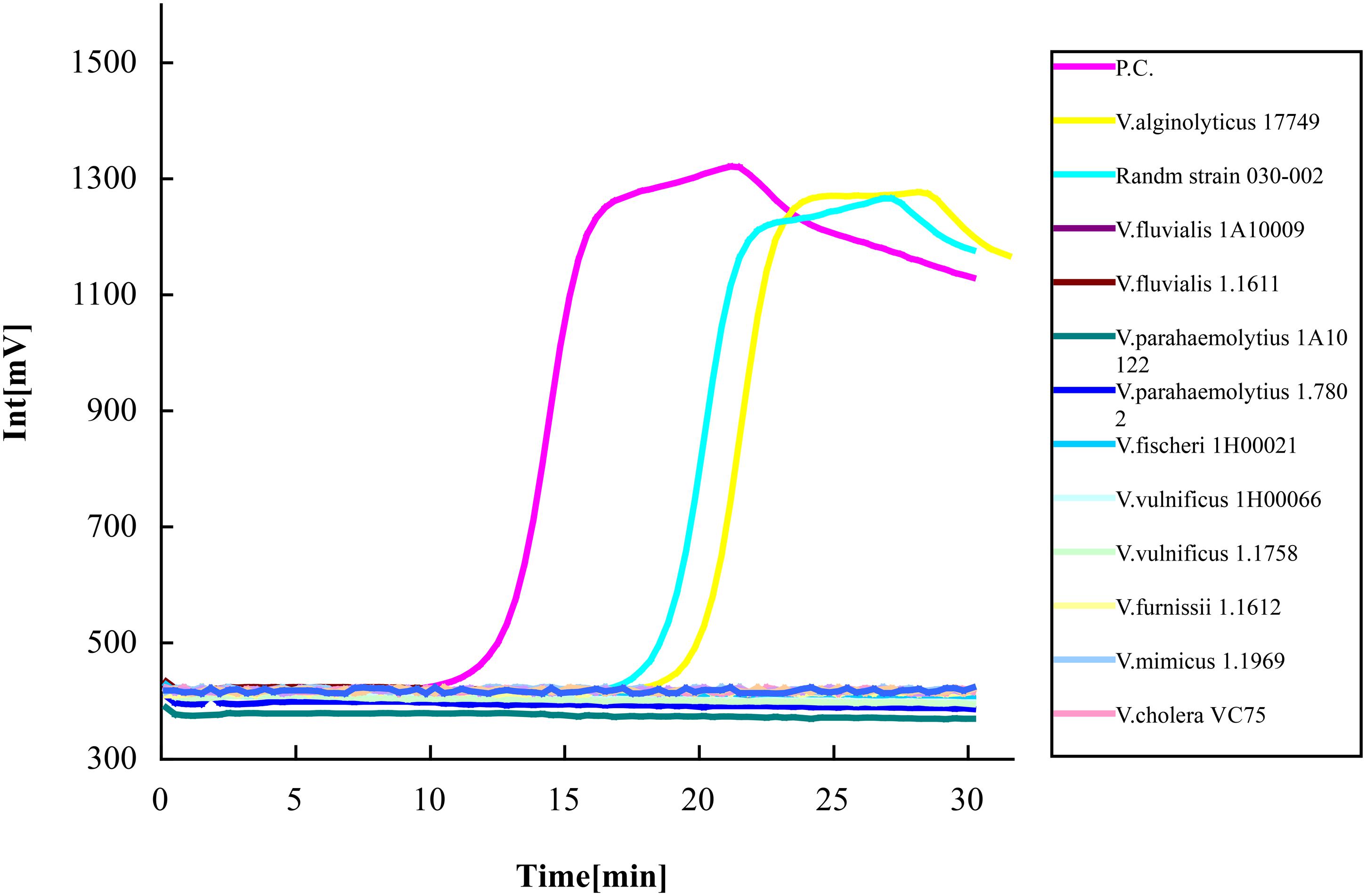
FIGURE 3. Specificity of the LAMP-ESE-Quant Tube Scanner assay. Eight suspensions of different Vibrio species (12 strains) adjusted to 106 CFU/mL were detected with real-time fluorescence loop-mediated isothermal amplification kit using an ESE-Quant tube scanner. Only V. alginolyticus 17749 and strain 030-002 showed amplification curves out of the xx total number of strains tested. N.C as negative control, P.C as positive control.
On the other hand, 14 different bacterial strains and 92 marine-isolated V. alginolyticus strains were detected by the LAMP-qPCR device. Among the 14 strains, an E. coli MG4 strain and a C. violaceum CV026 strain were added in addition to the 12 Vibrio strains used in Figure 3. The positive and negative controls were the same as described above. The results showed only two strains were positively detected, including V. alginolyticus 17749 and the randomly chosen 030-002 strain. The relative fluorescence units (RFU) rose to 0.0096 at the 14th cycle on strain 17749 and 0.0104 at the 10th cycle on the strain 030-002. They later stabilized at 0.10–0.13 from the 20th and 14th cycles, respectively toward the end of the reaction (Figure 4A). Similarly, all of the 92 marine-isolated V. alginolyticus strains tested using LAMP-qPCR device obtained “S” amplified curves in the RT-PCR assay. The intensity of the RFU varied from 0.82 to 0.30 (Figure 4B).
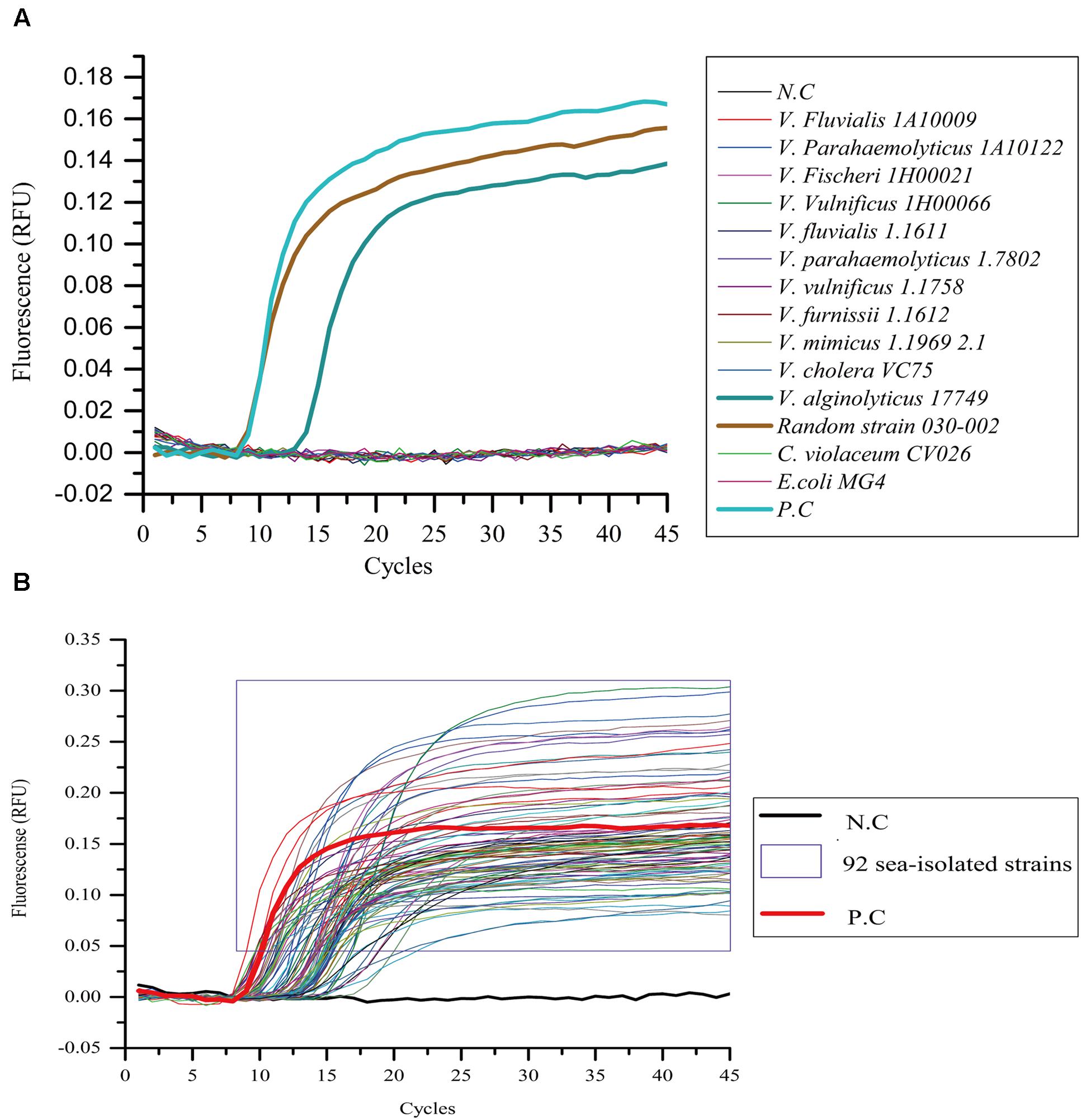
FIGURE 4. Specificity of the LAMP-qPCR device assay. All bacterial suspensions were adjusted to 106 CFU/mL and detected with real-time fluorescence loop-mediated isothermal amplification kit method using a qPCR device. (A) Out of all the strains tested, only V. alginolyticus 17749 and strain 030-002 showed amplification. (B) All the 92 strains showed “S-shaped” amplification curves. N.C as negative control, P.C as positive control.
The 16s rDNA results showed that the sequence homologies of 105 bacterial strains aligned with V. alginolyticus ATCC 17749, V. fluvialis NCTC 11327, V. parahaemolyticus ATCC 17802, V. vulnificus ATCC 27562, V. furnissii M1, V. mimicus UN 11607, and V. cholera E153 were in the range of 99.6–99.9%.
Real-time PCR successfully detected the presence of pathogens in the four replicates of blood infected with V. alginolyticus 17749 (G1), as evident in the obtained “S” curves (Figure 5). Cycle thresholds varied accordingly when the relative fluorescence values were raised to a separable range from the base line. The stable fluorescence values also differed, ranging approximately from 0.11 to 0.19.
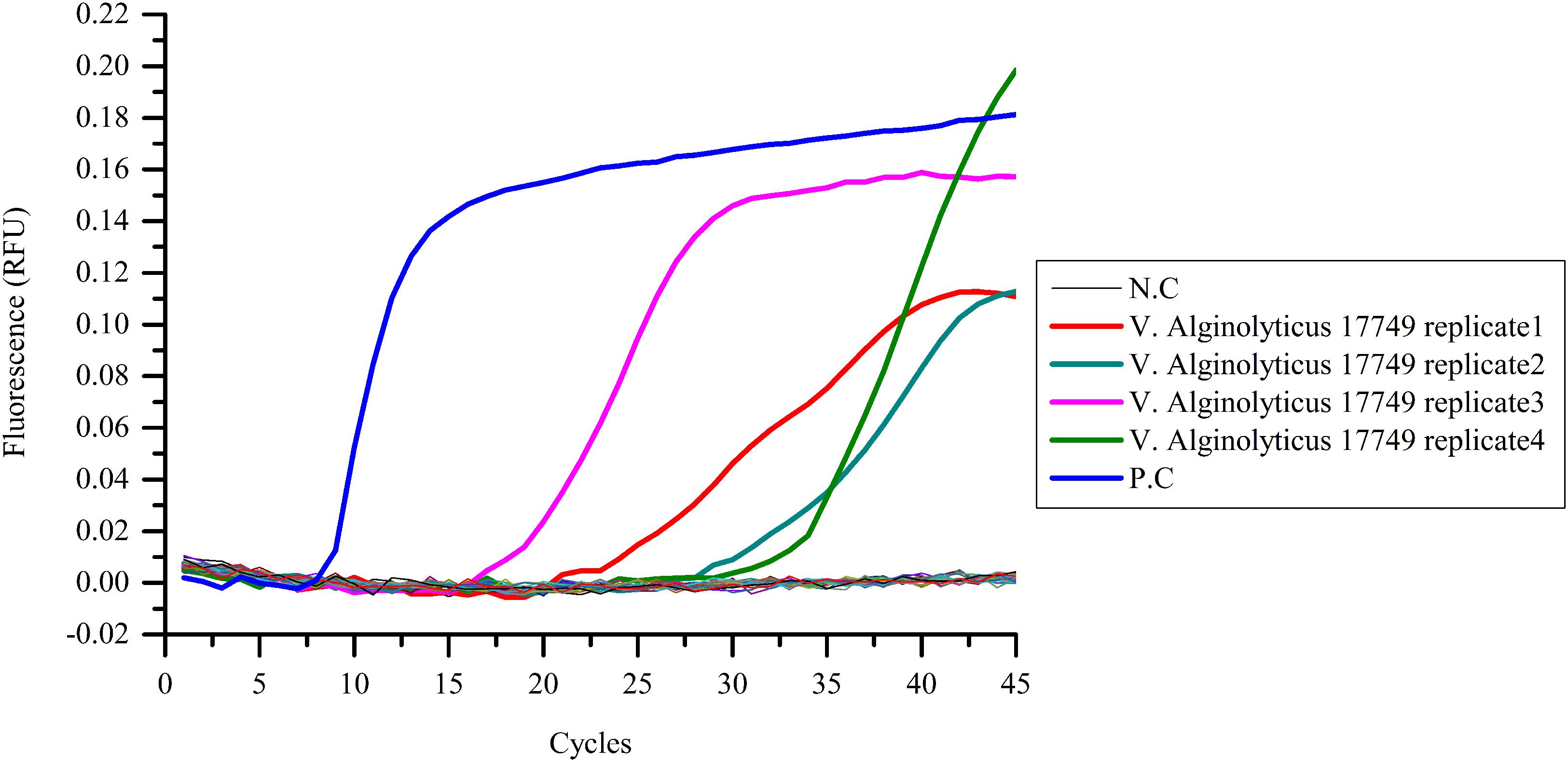
FIGURE 5. Pathogenic bacterial detection in infected blood samples. Blood samples were collected from Vibrio infected and uninfected mice, and template DNA was prepared for the LAMP assay. Among all of the chosen bacteria, amplification curves showed only in four replicates of V. alginolyticus 17749. N.C as negative control, P.C as positive control.
The sterile and V. alginolyticus contaminated scallop alkaline peptone water samples assay results are shown in Table 3. The artificially contaminated samples showed positive amplification and that the results corresponded well to the experimental design, which shows that the detection method is stable and reliable.
Vibrio alginolyticus as one of the most common species of aquatic bacteria and can thrive in oligotrophic conditions. However, once conditions become favorable (e.g., during infection or contamination of host), they exhibit rapid proliferation and growth causing food poisoning, which can easily induce diarrhea upon contact or ingestion (Caccamese and Rastegar, 1999; Chien et al., 2002). Thus, sensitivity and specificity of detection methods or assays are important in devising strategies to identify the presence of bacteria in pure cultures and especially in contaminated samples.
The conventional way of identifying V. alginolyticus is based on 16s rDNA identification accompanied by biochemical characterization. With the increase in the 16s rDNA sequences deposited in reference database, bacterial identification became easier using PCR amplification and sequencing. Phylogenetic and sequence analyses confirmed that 16s rDNA is a reliable indicator of species differentiation with the presence of polymorphic sites within particular organisms (Moreno et al., 2002). Meanwhile, biochemical characterization is based on different bacterial morphology and chemical reaction assays, which also provide a precise detection method. Although widely accepted, these two methods are both logistically limited (e.g., need for sophisticated equipment and facilities), and are very time-consuming (i.e., 3–10 days).
At present, significant improvements in technology for testing and detecting have been performed to help cure, mitigate and prevent the spread of such infections and contaminants. For example, improved PCR-assays using highly specificity primers have been gaining popularity as the accuracy is higher than conventional methods. The advent of molecular approaches such as specific PCR assay (Bej et al., 1999) and DNA probe hybridization (Gooch et al., 2001) paved the way for the development of rapid and easier methods of detection, which increased the efficiency of bacterial identification. Despite such improvements, use of PCR itself is still logistically limited to laboratories and impractical in field conditions and remote areas. Therefore, these molecular identification methods are still time-consuming (2–3 days) and none were appropriate for on field or on the spot monitoring.
The toxR gene was first found in V. cholera and generally exists only in Vibrio species, encoding a transmembrane transcriptional activator protein and mainly regulating the expression of virulence genes (Miller et al., 1987; Chakraborty et al., 2006). The sequence of a membrane tethering region in the toxR gene shows wide differences among Vibrio species, which makes it an excellent molecular target for the identification of Vibrio species (Carlos et al., 2000; Conejero and Hedreyda, 2003). Therefore, the gene polymorphisms in toxR in Vibrio species are greater than that in 16S rDNA.
In this study, based on LAMP technology, a set of six specific primers targeting the toxR gene sequence of V. alginolyticus were designed and tested, which could specifically identify eight independent regions of the targeted gene. Along with Bst WarmStartTM DNA polymerase that had strand substitution activity, strand substitution reaction was successfully carried out along with the synthesis of complementary strand on the target DNA region. Amplification reaction was accomplished at 60–65°C, and the entire detection process was completed within 20–60 min, which was significantly faster compared to previous techniques.
At the same time, using the primers we designed coupled with the RT-PCR and ESE-Quant Tube Scanner, high sensitivity and 100% specificity of the rapid and simple LAMP assay was obtained. The sensitivity of the new method on the two detection platforms (ESE-Quant tube scanner and qPCR device) was highly consistent (Figures 1 and 2). Moreover, compared to conventional 16s rDNA PCR assays, the LAMP assay was more sensitive and the results were more intuitive. In pure cultures, the amplification curves generated via ESE-Quant Tube Scanner clearly showed the separation of V. alginolyticus and non-V. alginolyticus (Figure 3), and the same pattern was observed when qPCR was used (Figure 4). The above results were consistent with results of previous studies (Fall et al., 2008; Han and Ge, 2008; Yamazaki et al., 2008), and also provided data showing the application of this assay not only in field detection but also in routine research laboratories.
Previous investigations found that the LOD for V. parahaemolyticus was 1.1 × 105 CFU/mL (Chen and Ge, 2010), 104 CFU/mL for Bacillus bacilliformis (Angkasekwinai et al., 2014) and 2 CFU/25 g for V. cholerae in food and environmental samples (Garrido-Maestu et al., 2015). Targeting the toxR gene in our study, we showed that the LOD of V. alginolyticus pure culture could be as low as 102 CFU/ml, which was consistent with previous results that detected V. alginolyticus by targeting the gyrB gene using a LAMP method (Cai et al., 2010). This result verifies the hypervariability and heterogeneity of the toxR gene (Chen and Ge, 2010) that can be used to distinguish V. alginolyticus strains. Compared to primers targeting tlh/ldh gene (Yi et al., 2014), our data also showed better accuracy, indicating precision of this rapid and simple LAMP assay.
Previous studies performed in aquatic animal models have tested for the presence of B. bacilliformis in contained conditions. However, the test strain demonstrated limited capacity for growth when artificially fed to L. longipalpis (Angkasekwinai et al., 2014). Meanwhile, Vibrio has strong infectivity and pathogenicity (Oates et al., 2012; Xiong et al., 2014) making it a good model system to study. In fact, the intraperitoneal LD50 in mice reached 1 × 108 to 1 × 109 CFU of V. alginolyticus E0666 (Liu et al., 2014). However, as a relatively newly classified bacterium from V. parahaemolyticus in Buchanan and Gibbons (1974), only few studies testing V. alginolyticus in animal models and much less on mammalian models have been performed.
Our study performed excellently not only in pure bacterial cultures but also in infected mice and artificially contaminated samples. The mice injected with V. alginolyticus and other bacteria immediately exhibited constant chilling and shaking compared with the control group, which were defensive mechanisms against infection. Then, 16 h after injection, physiological states of the infected mice became worse, which varied in between groups. The immune system of the mice played an important role in metabolizing and phagocytosing some of the bacteria, resulting in an expected decrease in the concentration of the pathogens in the blood. In spite of such pathological processes, our LAMP assay still detected the existence of V. alginolyticus in the blood of the mice. To the best of our knowledge, this is the first time that V. alginolyticus has been detected directly from extracted mammalian blood. Using the newly described rapid and simple LAMP assay, we provided a more accurate method for the detection of V. alginolyticus.
Same as the detection in the model animal, V. alginolyticus in artificially contaminated scallop alkaline was also successfully detected. In addition, with the ESE Tube Scanner, our LAMP assay is applicable to rural communities, inaccessible locations and field use. To secure safety and sustainability of marine fisheries, detection and prevention of pathogenic diseases, timely warning and control of food safety problems are of great importance.
Other than high specificity and sensitivity, time-saving and intuitive monitoring of results, the feasibility for the method to be used en masse in the future is highly dependent on availability of the materials such as the fluorochrome SYTO®-9 and ineral oil that could be easily obtained.
The use of two platforms (i.e., Real-Time PCR and ESE-Quant Tube scanner) both obtained 100% inclusion and exclusion. This means that the LAMP assay for V. alginolyticus that we described in this study was very stable and efficient under different environments. The ESE-Quant tube scanner system is the most commonly used LAMP detection platform, which is portable and doesn’t need the experiment operator to have a high level of professional knowledge. On the other hand, as real-time fluorescent quantitative PCR instruments have become common in most laboratories, the subsequent verification experiments were mainly detected on the qPCR platform, so as to provide evidence for its application in laboratories. Therefore our results showed that this method could be carried out both in laboratory and field conditions.
Valued for its sensitivity, specificity, and rapidity in detecting pathogens in pure culture and artificially contaminated samples under different situations, our LAMP assay is a promising tool and which could be used by both professionals and non-professionals worldwide.
All authors were involved in design of the study and interpretation of data. All authors approved the final version. KF and JL helped conceive project, designed and performed experiments and wrote the manuscript. YW conceived project, designed, and interpreted experiments. JL executed experiments and wrote the manuscript. HY conceived and interpreted experiments. LS and LZ helped conceive project and edit the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by National Natural Science Foundation of China (Grant No: 31400107, 81273311, and 31170008).
Angkasekwinai, N., Atkins, E. H., Johnson, R. N., Grieco, J. P., Ching, W. M., and Chao, C. C. (2014). Rapid and sensitive detection of Bartonella bacilliformis in experimentally infected sand flies by loop-mediated isothermal amplification (LAMP) of the Pap31 gene. PLoS Negl. Trop Dis. 8:e3342. doi: 10.1371/journal.pntd.0003342
Barbarossa, V., Kucisec-Tepes, N., Aldova, E., Matek, D., and Stipoljev, F. (2002). Ilizarov technique in the treatment of chronic osteomyelitis caused by Vibrio alginolyticus. Croat. Med. J. 43, 346–349.
Bej, A. K., Patterson, D. P., Brasher, C. W., Vickery, M. C., Jones, D. D., and Kaysner, C. A. (1999). Detection of total and hemolysin-producing Vibrio parahaemolyticus in shellfish using multiplex PCR amplification of tl, tdh and trh. J. Microbiol. Methods 36, 215–225. doi: 10.1016/S0167-7012(99)00037-8
Buchanan, R. E., and Gibbons, N. E. (1974). Bergey’s Manual of Determinative Bacteriology, 8th Edn. Baltimore: Williams & Wilkins Company.
Caccamese, S. M., and Rastegar, D. A. (1999). Chronic diarrhea associated with Vibrio alginolyticus in an immunocompromised patient. Clin. Infect. Dis. 29, 946–947. doi: 10.1086/520473
Cai, S. H., Lu, Y. S., Wu, Z. H., Jian, J. C., Wang, B., and Huang, Y. C. (2010). Loop-mediated isothermal amplification method for rapid detection of Vibrio alginolyticus, the causativeagent of vibriosis in mariculture fish. Lett. Appl. Microbiol. 2010, 480–485. doi: 10.1111/j.1472-765X.2010.02823.x
Carlos, R., Osorio, and Klose, K. E. (2000). A region of the transmembrane regulatory protein toxr that tethers the transcriptional activation domain to the cytoplasmic membrane displays wide divergence among vibrio species. J. Bacteriol. 182, 526–528. doi: 10.1128/JB.182.2.526-528.2000
Chakraborty, R., Sinha, S., Mukhopadhyay, A. K., Asakura, M., Yamasaki, S., Bhattacharya, S. K., et al. (2006). Species-specific identification of Vibrio fluvialis by PCR targeted to the conserved transcriptional activation and variable membrane tether regions of the toxR gene. J. Med. Microbiol. 55(Pt 6), 805–808. doi: 10.1099/jmm.0.46395-0
Chen, C., Wang, Q. B., Liu, Z. H., Zhao, J. J., Jiang, X., Sun, H. Y., et al. (2012). Characterization of role of the toxR gene in the physiology and pathogenicity of Vibrio alginolyticus. Antonie Van Leeuwenhoek 101, 281–288. doi: 10.1007/s10482-011-9632-8
Chen, M. X., Li, H. Y., Li, G., and Zheng, T. L. (2011). Distribution of Vibrio alginolyticus-like species in Shenzhen coastal waters. China. Braz. J. Microbiol. 42, 884–896. doi: 10.1590/S1517-83822011000300007
Chen, S., and Ge, B. (2010). Development of a toxR-based loop-mediated isothermal amplification assay for detecting Vibrio parahaemolyticus. BMC Microbiol. 10:41. doi: 10.1186/1471-2180-10-41
Chien, J. Y., Shih, J. T., Hsueh, P. R., Yang, P. C., and Luh, K. T. (2002). Vibrio alginolyticus as the cause of pleural empyema and bacteremia in an immunocompromised patient. Eur. J. Clin. Microbiol. Infect. Dis. 21, 401–403. doi: 10.1007/s10096-002-0726-0
Conejero, M. J., and Hedreyda, C. T. (2003). Isolation of partial toxR gene of Vibrio harveyi and design of toxR-targeted PCR primers for species detection. J. Appl. Microbiol. 95, 602–611. doi: 10.1046/j.1365-2672.2003.02020.x
Crisafi, F., Denaro, R., Genovese, M., Cappello, S., Mancuso, M., and Genovese, L. (2011). Comparison of 16SrDNA and toxR genes as targets for detection of Vibrio anguillarum in Dicentrarchus labrax kidney and liver. Res. Microbiol. 162, 223–230. doi: 10.1016/j.resmic.2010.11.004
Fall, J., Chakraborty, G., Kono, T., Maeda, M., Itami, T., and Sakai, M. (2008). Establishment of loop-mediated isothermal amplification method (LAMP) for the detection of Vibrio nigripulchritudo in shrimp. FEMS Microbiol. Lett. 288, 171–177. doi: 10.1111/j.1574-6968.2008.01332.x
Feingold, M. H., and Kumar, M. L. (2004). Otitis media associated with Vibrio alginolyticus in a child with pressure-equalizing tubes. Pediatr. Infect. Dis. J. 23, 475–476. doi: 10.1097/01.inf.0000126592.19378.30
Ferrara, M., Perrone, G., Gallo, A., Epifani, F., Visconti, A., and Susca, A. (2015). Development of loop-mediated isothermal amplification (LAMP) assay for the rapid detection of Penicillium nordicum in dry-cured meat products. Int. J. Food Microbiol. 202, 42–47. doi: 10.1016/j.ijfoodmicro.2015.02.021
Garrido-Maestu, A., Chapela, M. J., Vieites, J. M., and Cabado, A. G. (2015). lolB gene, a valid alternative for qPCR detection of Vibrio cholerae in food and environmental samples. Food Microbiol. 46, 535–540. doi: 10.1016/j.fm.2014.09.012
Gooch, J. A., Depaola, A., Kaysner, C. A., and Marshall, D. L. (2001). Evaluation of two direct plating methods using nonradioactive probes for enumeration of Vibrio parahaemolyticus in oysters. Appl. Environ. Microbiol. 67, 721–724. doi: 10.1128/AEM.67.2.721-724.2001
Han, F., and Ge, B. (2008). Evaluation of a loop-mediated isothermal amplification assay for detecting Vibrio vulnificus in raw oysters. Foodborne Pathog. Dis. 5, 311–320. doi: 10.1089/fpd.2008.0084
Hayashida, K., Kajino, K., Hachaambwa, L., Namangala, B., and Sugimoto, C. (2015). Direct blood dry LAMP: a rapid, stable, and easy diagnostic tool for Human African Trypanosomiasis. PLoS Negl. Trop Dis. 9:e0003578. doi: 10.1371/journal.pntd.0003578
He, P., Chen, Z., Luo, J., Wang, H., Yan, Y., Chen, L., et al. (2014). Multiplex real-time PCR assay for detection of pathogenic Vibrio parahaemolyticus strains. Mol. Cell Probes 28, 246–250. doi: 10.1016/j.mcp.2014.06.001
Hong, G. L., Dai, X. Q., Lu, C. J., Liu, J. M., Zhao, G. J., Wu, B., et al. (2014). Temporizing surgical management improves outcome in patients with Vibrio necrotizing fasciitis complicated with septic shock on admission. Burns 40, 446–454. doi: 10.1016/j.burns.2013.08.012
Ji, S. P. (1989). The first isolation of Vibrio alginolyticus from samples which caused food poisoning. Zhonghua Yu Fang Yi Xue Za Zhi 23, 71–73.
Jones, E. H., Feldman, K. A., Palmer, A., Butler, E., Blythe, D., and Mitchell, C. S. (2013). Vibrio infections and surveillance in Maryland, 2002–2008. Public Health Rep. 128, 537–545.
Khajanchi, B. K., Kirtley, M. L., Brackman, S. M., and Chopra, A. K. (2011). Immunomodulatory and protective roles of quorum-sensing signaling molecules N-acyl homoserine lactones during infection of mice with Aeromonas hydrophila. Infect. Immun. 79, 2646–2657. doi: 10.1128/IAI.00096-11
Lai, F. C., Wang, Q., Zhou, Y. P., Mu, C. H., Geng, S. N., Zhang, Y. M., et al. (2004). Investigation of the bacteria in the seawater of Xisha in the South China Sea and their antibiotic sensitivity profile. Di. Yi. Jun. Yi. Da. Xue. Xue. Bao 24, 347–348.
Lai, Y., Tao, J., Sun, C., Shi, C., and Wu, S. (2014). Biological characteristics and histopathological observation of Vibrio alginolyticus from diseased fish. Wei Sheng Wu Xue Bao 54, 1378–1384.
Lane, D. J. (1991). “16S/23S rRNA sequencing,” in Nucleic Acids Techniques in Bacterial Systematics, eds E. Stackebrandt and M. Goodfellow (Chichester: John Wiley & Sons), 115–147.
Lee, S. E., Shin, S. H., Kim, S. Y., Kim, Y. R., Shin, D. H., Chung, S. S., et al. (2000). Vibrio vulnificus has the transmembrane transcription activator ToxRS stimulating the expression of the hemolysin gene vvhA. J. Bacteriol. 182, 3405–3415. doi: 10.1128/JB.182.12.3405-3415.2000
Li, X. C., Xiang, Z. Y., Xu, X. M., Yan, W. H., and Ma, J. M. (2009). Endophthalmitis caused by Vibrio alginolyticus. J. Clin. Microbiol. 47, 3379–3381. doi: 10.1128/JCM.00722-09
Liew, P. S., Teh, C. S., Lau, Y. L., and Thong, K. L. (2014). A real-time loop-mediated isothermal amplification assay for rapid detection of Shigella species. Trop Biomed. 31, 709–720.
Lin, Z., Kumagai, K., Baba, K., Mekalanos, J. J., and Nishibuchi, M. (1993). Vibrio parahaemolyticus has a homolog of the Vibrio cholerae toxRS operon that mediates environmentally induced regulation of the thermostable direct hemolysin gene. J. Bacteriol. 175, 3844–3855.
Liu, D. F., Liu, C. G., Tian, J., Jiang, Y. T., Zhang, X. Z., Chai, H. L., et al. (2015). Establishment of reverse transcription loop-mediated isothermal amplification for rapid detection and differentiation of canine distemper virus infected and vaccinated animals. Infect. Genet. Evol. 32, 102–106. doi: 10.1016/j.meegid.2015.03.002
Liu, X. F., Zhang, H., Liu, X., Gong, Y., Chen, Y., Cao, Y., et al. (2014). Pathogenic analysis of Vibrio alginolyticus infection in a mouse model. Folia Microbiol. (Praha) 59, 167–171. doi: 10.1007/s12223-013-0279-x
Miller, V. L., Taylor, R. K., and Mekalanos, J. J. (1987). Cholera toxin transcriptional activator toxR is a transmembrane DNA binding protein. Cell 48, 271–279. doi: 10.1016/0092-8674(87)90430-2
Molitoris, E., Joseph, S. W., Krichevsky, M. I., Sindhuhardja, W., and Colwell, R. R. (1985). Characterization and distribution of Vibrio alginolyticus and Vibrio parahaemolyticus isolated in Indonesia. Appl. Environ. Microbiol. 50, 1388–1394.
Moreno, C., Romero, J., and Espejo, R. T. (2002). Polymorphism in repeated 16S rRNA genes is a common property of type strains and environmental isolates of the genus Vibrio. Microbiology 148, 1233–1239. doi: 10.1099/00221287-148-4-1233
Narracci, M., Acquaviva, M. I., and Cavallo, R. A. (2013). Mar Piccolo of Taranto: Vibrio biodiversity in ecotoxicology approach. Environ. Sci. Pollut. Res. Int. 21, 2378–2385. doi: 10.1007/s11356-013-2049-3
Neogi, S. B., Chowdhury, N., Asakura, M., Hinenoya, A., Haldar, S., Saidi, S. M., et al. (2010). A highly sensitive and specific multiplex PCR assay for simultaneous detection of Vibrio cholerae, Vibrio parahaemolyticus and Vibrio vulnificus. Lett. Appl. Microbiol. 51, 293–300. doi: 10.1111/j.1472-765X.2010.02895.x
Notomi, T., Mori, Y., Tomita, N., and Kanda, H. (2015). Loop-mediated isothermal amplification (LAMP): principle, features, and future prospects. J. Microbiol. 53, 1–5. doi: 10.1007/s12275-015-4656-9
Oates, S. C., Miller, M. A., Byrne, B. A., Chouicha, N., Hardin, D., Jessup, D., et al. (2012). Epidemiology and potential land-sea transfer of enteric bacteria from terrestrial to marine species in the Monterey Bay Region of California. J. Wildl. Dis. 48, 654–668. doi: 10.7589/0090-3558-48.3.654
Opal, S. M., and Saxon, J. R. (1986). Intracranial infection by Vibrio alginolyticus following injury in salt water. J. Clin. Microbiol. 23, 373–374.
Pascual, J., Macián, M. C., Arahal, D. R., Garay, E., and Pujalte, M. J. (2010). Multilocus sequence analysis of the central clade of the genus Vibrio by using the 16S rRNA, recA, pyrH, rpoD, gyrB, rctB and toxR genes. Int. J. Syst. Evol. Microbiol. 60(Pt 1), 154–165. doi: 10.1099/ijs.0.010702-0
Pereira, C. S., VIana, C. M., and Rodrigues Ddos, P. (2007). Pathogenic Vibrios in oysters (Crassostrea rhizophorae) served at restaurants in Rio de Janeiro: a public health warning. Rev. Soc. Bras. Med. Trop 40, 300–303. doi: 10.1590/S0037-86822007000300010
Rubin, S. J., and Tilton, R. C. (1975). Isolation of Vibrio alginolyticus from wound infections. J. Clin. Microbiol. 2, 556–558.
Sakuma, K., Hayashi, S., Otokuni, K., Matsumoto, I., Matsuoka, H., and Saito, M. (2013). Standard operating procedures for maintaining cleanliness in a novel compact facility for breeding SPF mice. J. Am. Assoc. Lab. Anim. Sci. 52, 717–724.
Schets, F. M., Van Den Berg, H. H., Marchese, A., Garbom, S., and De Roda Husman, A. M. (2011). Potentially human pathogenic vibrios in marine and fresh bathing waters related to environmental conditions and disease outcome. Int. J. Hyg. Environ. Health 214, 399–406. doi: 10.1016/j.ijheh.2011.05.003
Schets, F. M., Van Den Berg, H. H., Rutjes, S. A., and De Roda Husman, A. M. (2010). Pathogenic Vibrio species in dutch shellfish destined for direct human consumption. J. Food Prot. 73, 734–738.
Song, Y., Yu, P., Li, B., Pan, Y., Zhang, X., Cong, J., et al. (2013). The mosaic accessory gene structures of the SXT/R391-like integrative and conjugative elements derived from Vibrio spp. isolated from aquatic products and environment in the Yangtze River Estuary. China. BMC Microbiol. 13:214. doi: 10.1186/1471-2180-13-214
Taylor, R., Mcdonald, M., Russ, G., Carson, M., and Lukaczynski, E. (1981). Vibrio alginolyticus peritonitis associated with ambulatory peritoneal dialysis. Br. Med. J. (Clin. Res. Ed) 283:275. doi: 10.1136/bmj.283.6286.275
Torresi, M., Acciari, V. A., Piano, A., Serratore, P., Prencipe, V., and Migliorati, G. (2011). Detection of Vibrio splendidus and related species in Chamelea gallina sampled in the Adriatic along the Abruzzi coastline. Vet. Ital. 47, 363–370.
Vezzulli, L., Colwell, R. R., and Pruzzo, C. (2013). Ocean warming and spread of pathogenic vibrios in the aquatic environment. Microbiol. Ecol. 65, 817–825. doi: 10.1007/s00248-012-0163-2
Wang, Y., Li, D., Wang, Y., Li, K., and Ye, C. (2016). Rapid and Sensitive Detection of Vibrio parahaemolyticus and Vibrio vulnificus by multiple endonucleaserestriction real-time loop-mediated isothermal amplification technique. Molecules 21:E111. doi: 10.3390/molecules21010111
Weis, K. E., Hammond, R. M., Hutchinson, R., and Blackmore, C. G. (2011). Vibrio illness in Florida, 1998–2007. Epidemiol. Infect. 139, 591–598. doi: 10.1017/S0950268810001354
Xiong, P., Peng, X., Wei, S., Chen, Y., Zhao, H., Tang, S., et al. (2014). Virulence-related genes of Vibrio alginolyticus and its virulence in mice. Wei Sheng Wu Xue Bao 54, 80–88.
Yamazaki, W., Seto, K., Taguchi, M., Ishibashi, M., and Inoue, K. (2008). Sensitive and rapid detection of cholera toxin-producing Vibrio cholerae using a loop-mediated isothermal amplification. BMC Microbiol. 8:94. doi: 10.1186/1471-2180-8-94
Keywords: Vibrio alginolyticus, toxR gene, real-time LAMP, 16s rDNA sequencing, rapid detection
Citation: Fu K, Li J, Wang Y, Liu J, Yan H, Shi L and Zhou L (2016) An Innovative Method for Rapid Identification and Detection of Vibrio alginolyticus in Different Infection Models. Front. Microbiol. 7:651. doi: 10.3389/fmicb.2016.00651
Received: 13 January 2016; Accepted: 18 April 2016;
Published: 06 May 2016.
Edited by:
Abd El-Latif Hesham, Assiut University, EgyptReviewed by:
Maria De Los Angeles Serradell, CCT La Plata-CONICET, Instituto de Ciencias de la Salud-UNAJ, ArgentinaCopyright © 2016 Fu, Li, Wang, Liu, Yan, Shi and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lijun Zhou, aHp6aG91bGpAMTI2LmNvbQ==; Lei Shi, bGVpc2hpQHNjdXQuZWR1LmNu
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.