- Department of Applied BioSciences, Faculty Bioscience Engineering, Ghent University, Ghent, Belgium
Exposure to mycotoxins, secondary metabolites produced by fungi, may infer serious risks for animal and human health and lead to economic losses. Several approaches to reduce these mycotoxins have been investigated such as chemical removal, physical binding, or microbial degradation. This review focuses on the microbial degradation or transformation of mycotoxins, with specific attention to the actual detoxification mechanisms of the mother compound. Furthermore, based on the similarities in chemical structure between groups of mycotoxins and environmentally recalcitrant compounds, known biodegradation pathways and degrading organisms which hold promise for the degradation of mycotoxins are presented.
Introduction
The presence of mycotoxins is inherent to many food and feed products worldwide (Bhat et al., 2010; Marroquín-Cardona et al., 2014). Hallmarks of their presence and their impact on animal and human health are encountered throughout history. Ergotism, also known as “St. Anthony's fire” occurred in several areas in Europe during the tenth century (Schiff, 2006) and was caused by the consumption of rye containing ergot alkaloids, produced by the fungus Claviceps purpurea (Bové, 1970; Beardall and Miller, 1994). In Siberia, a delayed harvest due to the second world war resulted in grains heavily contaminated with trichothecenes produced by Fusarium spp. People later consuming the grain were afflicted with number of nonspecific disorders and mortality mounted up to 10% (Manahan, 2002). In 1962, 100,000 turkeys died in London of Turkey X disease, linked to aflatoxins from Aspergillus flavus (Binder, 2007). These examples mentioned above illustrate the acute impact of high loads of singular mycotoxins on human and animal health. However, longtime exposure to low concentrations of mycotoxins also entail chronic toxicities which often result in non-specific symptoms, difficult to track-and-trace down to mycotoxins. These toxicities include estrogenic gastrointestinal, urogenital, vascular, kidney, and nervous disorders. Some mycotoxins are carcinogenic or immuno-compromising, and as such also promote the development of infectious diseases (Peraica et al., 1999; Hussein and Brasel, 2001; Creppy, 2002; Richard, 2007; Da Rocha et al., 2014).
For many years the research community focused on the occurrence of singular mycotoxins but nowadays scientific interest shifts to studies involving multiple mycotoxins. This new approach is highly relevant as large scale multi-toxin surveys show that a number of mycotoxins tend to co-occur with other sometimes structurally not-related mycotoxins (Gerding et al., 2014; Storm et al., 2014; Vanheule et al., 2014; and many more). In addition, mycotoxins are known to have additive and synergistic effects on human- and animal health (Alassane-Kpembi et al., 2013; Klaric et al., 2013; Clarke et al., 2014).
Research efforts progressively increase to develop mitigation strategies based on risk monitoring, risk characterization, prevention, intervention, and remediation strategies for multiple mycotoxins, which start from critical points along the production chain comprising field, storage, processing, and transportation. However, monitoring and good agricultural, storage, and transportation practices along with an effective Hazard Analysis and Critical Control Point approach do not completely prevent mycotoxin presence in the food or feed chain (Bhat et al., 2010). Decontamination technologies then offer a last resort to salvage contaminated batches along the production chain.
Decontamination strategies to reduce mycotoxins in food- and feed commodities are technologically diverse and based on physical, chemical, or biochemical principles. Some physical processes aim to remove highly contaminated fractions from bulk material (Bullerman and Bianchini, 2007; Cheli et al., 2013; Kaushik, 2015) through sorting (Scudamore et al., 2007), milling (Castells et al., 2007; Khatibi et al., 2014), dehulling (Fandohan et al., 2006; Rios et al., 2009; Matumba et al., 2015), cleaning (van der Westhuizen et al., 2011), heating, irradiation, or combinational approaches (Fandohan et al., 2005; Matumba et al., 2015). Another physical removal strategy is the use of inorganic or organic mycotoxin binders (Ramos et al., 1996; Kolosova and Stroka, 2011). Although these adsorbing binders have some promising features, some may have adverse nutritional effects due to binding of vitamins and minerals (Huwig et al., 2001; Yiannikouris et al., 2006) or reducing the efficacy pharmacokinetics of antibiotics (De Mil et al., 2015).
Chemical remediation strategies involve the conversion of mycotoxins via chemical reactions. Ammoniation (Norred et al., 1991), alkaline hydrolysis, peroxidation, ozonation, and the use of bisulphites are reported to be effective on one or more mycotoxins but a detailed insight into the toxicity of eventual end products or the impact on palatability and nutritive quality is questionable.
Microbial based methods comprise mycotoxin decomposition, transformation, or adsorption. The latter strategy has already been mentioned under physical measures and will not be considered in detail in this review. Focus in this review will be on transformation and biodegradation of the main mycotoxins by microorganisms. Although there are some excellent reviews on biodegradation (Zinedine et al., 2007; Wu et al., 2009; Awad et al., 2010; Jard et al., 2011; Devreese et al., 2013; McCormick, 2013; Hathout and Aly, 2014; Adebo et al., 2015), this review is timely because of two reasons:
Firstly, studies often wrongly identify biodegradation with detoxification, or do not test for toxicity of potential metabolites. Indeed, not all transformation or degradation products are detoxification products. This is nicely illustrated for aflatoxins and zearalenone (ZEN). Aflatoxin M1 (AFM1) is the hydroxylated metabolite of AFB1 and is categorized as possible carcinogenic to humans (Group 2B) by the International Agency for Research on Cancer (IARC; IARC, 2002). Aflatoxicol (or aflatoxin R0), a reduction product of AFB1, has been detected as degradation product by Corynebacterium rubrum, Aspergillus niger, Trichoderma viride, Mucor ambiguous, and Dactylium dendroides (Mann and Rehm, 1976; Wong and Hsieh, 1976). However, Karabulut et al. (2014) concluded that AFB1 and aflatoxicol have similar potency to form an exo-epoxide analog which can bind to DNA. Assessing the ZEN biodegradation capacity of several microorganisms, Hahn et al. (2015) found that many strains were able to convert ZEN to α- and/or β-ZEL, showing similar estrogenic activity compared to ZEN. Aerobic and anaerobic degradation to other uncharacterized metabolites with unidentified toxicity was obtained as well. These results demonstrate the importance of in vitro experiments to critically screen agents claiming mycotoxin detoxification.
Secondly, the available set of mycotoxin degrading microorganisms is limited and their performance is often doubtful when considering multiple mycotoxin degradation. This issue was also nicely illustrated by Hahn et al. (2015). Using an in vitro screening approach, 20 commercially available agents claiming mycotoxin detoxification were tested for their efficacy to inactivate and/or degrade the two structurally not related mycotoxins DON or ZEN. The majority of the agents were not effective or converted the toxins to equally toxic metabolites. Only one of the products efficiently inactivated or degraded the two considered mycotoxins under the tested conditions.
New insights on actual microbial detoxification routes are needed and can be based on known biodegradation metabolisms of non-mycotoxins found in diverse microbial communities, which we chose to identify as “unexplored worlds” to be discovered for the mycotoxin research field. Indeed, many hazardous, undesirable, deleterious, or recalcitrant molecules in other research fields share structural analogies with diverse mycotoxins and are reported to be successfully degraded by microorganisms. These unexplored worlds may serve as resource for cutting edge research in the field of mycotoxin remediation or in the field of metagenomics screening surveys in search for new microbial degraders of mycotoxins.
In this review, We are not only focusing on Fusarium mycotoxins, but also on Aspergillus, Penicillium, and other mycotoxins. This is relevant as independently of the producing genus, mycotoxins often share key-chemical groups responsible for their toxicity and thus biodegrading organisms for one mycotoxin can have their relevance for other mycotoxins produced by distinct fungal genera.
Toxicity and Degradation of Mycotoxins
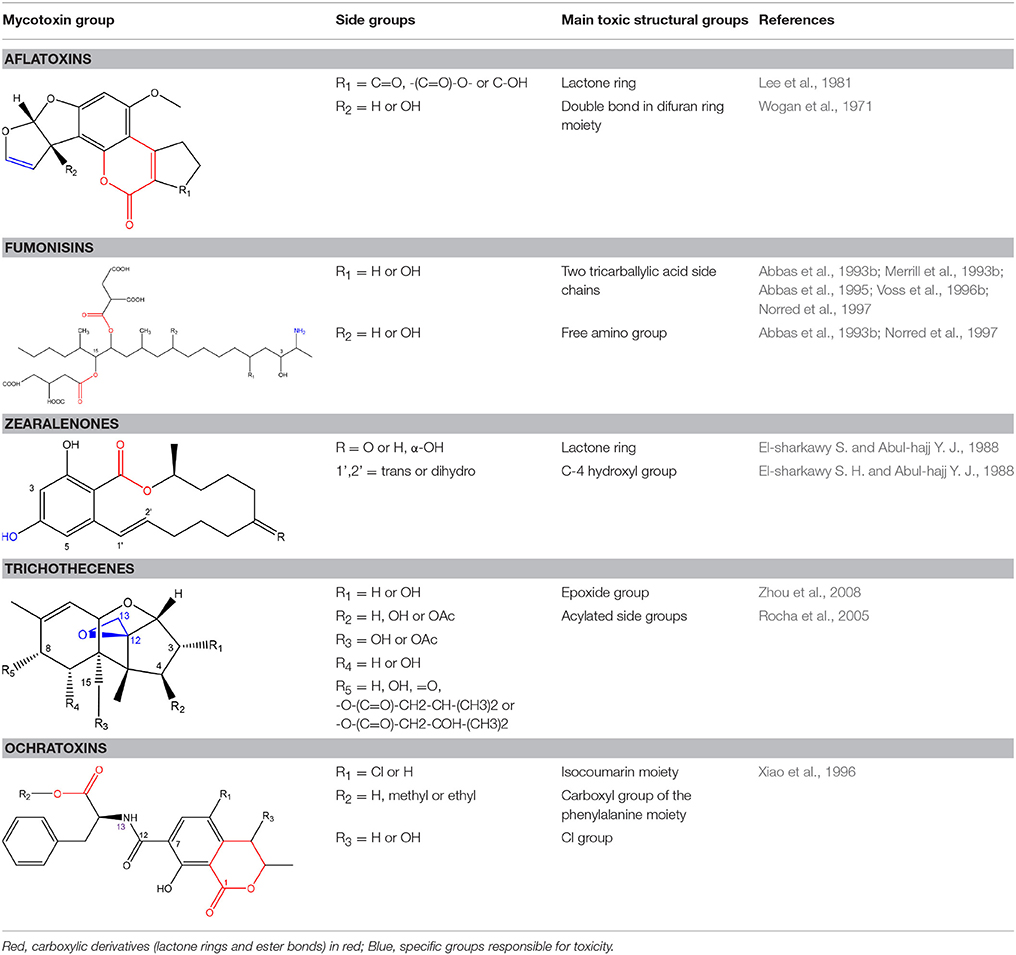
In order to assess detoxification by microorganisms, it is important to pinpoint the actual groups within the chemical structure of each mycotoxin which infer the toxic effects (Table 1). Next to the main toxic structural groups occurring in mycotoxins, structural similarities between mycotoxins are also highlighted; aflatoxins and ochratoxins are both composed of a coumarin moiety, whereas the main structure of aflatoxins, ZEN and ochratoxins is based on a lactone ring (Table 1—Red). Carboxyl derivatives (ester bonds), often playing a role in toxicity, are frequently present, as well in the lactone, as in side groups (Table 1—Red) (observed in fumonisins, ZEN, ochratoxins, and acylated trichothecenes). Each mycotoxin is further characterized concerning specific groups responsible for its toxicity (Table 1—Blue).
From our perspective, there are two ways in which detoxification of the mother compound in a degradation study can be confirmed: (i) the confirmation of reduced toxicity after degradation through one or more actual toxicity assays on particular organisms or cell lines, this is the most convincing proof; (ii) the detection and identification of detoxification products, for which in independent literature has been shown that they confer an lower toxicity to the mother compound. Of course, a combination of both ways provides the most holistic approach. The decreased toxicity of the degradation metabolites listed in Tables 2–7 can therefore be found in Table 1 in Supplementary Material.
Fumonisins
Toxicity
Fumonisins (most importantly FB1, FB2), first described by Gelderblom et al. (1988), are mainly produced by F. verticillioides and F. proliferatum and are structurally similar to sphingolipid long-chain bases such as sphinganine and sphingosine. This feature is tightly related to their toxicity mechanism through the inhibition of the sphingolipid biosynthesis (Merrill et al., 1993a; Soriano et al., 2005) and exposure to fumonisins has been associated a wide variety of diseases in animals as reviewed by Voss et al. (2007), such as liver cancer in rats, equine leukoencephalomalacia, and porcine pulmonary edema.
Specifically, fumonisins are comprised of a 22 carbon aminopentol with two tricarballylate (TCA) side groups, where two structural groups are important in their toxicity mechanism (Table 1). Firstly, the unsubstituted primary amino group at C2 competitively inhibits ceramide synthase, thereby disrupting the de novo biosynthesis of ceramide and sphingolipid metabolism (Voss et al., 2007). This free primary amino group of fumonisin-like compounds is a prerequisite for ceramide synthase inhibition, since N-acetylation of FB1 diminished or removed the toxicity effects in rat liver slices (Norred et al., 1997) and in jimsonweed and several mammalian cell lines (Abbas et al., 1993b). Secondly, the TCA side groups seem to have varying effects on the toxicity. On the one hand, absence of these side groups has been found to reduce both phytotoxicity and mammalian cytotoxicity (Abbas et al., 1995), and the resulting corresponding aminopentol (AP1, AP2) backbones were only 30–40% (Norred et al., 1997) or 10% as potent as the parent toxins (Merrill et al., 1993b). In contrast, removal of the TCA side groups has also been shown to enhance cytotoxicity in certain mammalian cell lines (Abbas et al., 1993b) and AP1 displays renal toxicity comparable to that of FB1 (Voss et al., 1996a).
Degradation: Organisms and Pathways
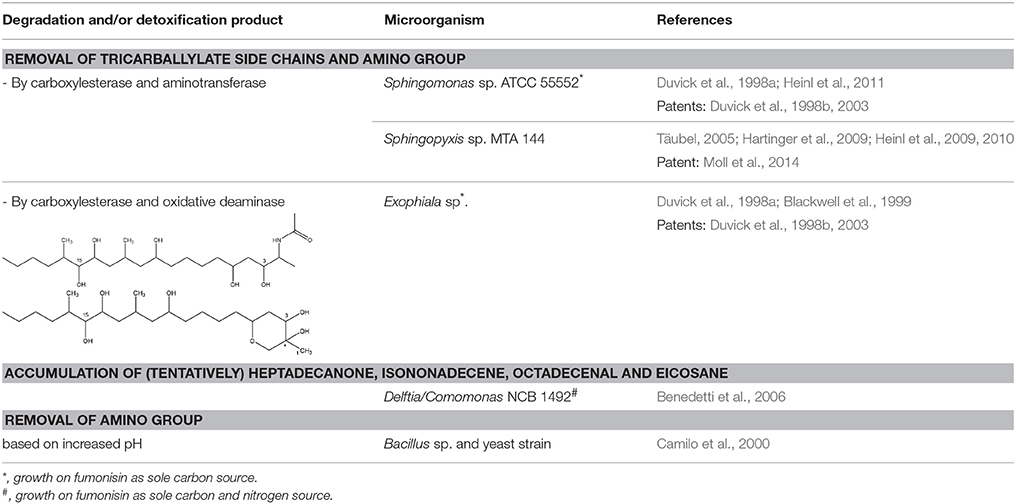
Only a few microorganisms are known to degrade and thereby detoxify fumonisins (Table 2), mostly by removal of the TCA groups as well as the free amino group. Although none of these studies actually determined detoxification of fumonisin B1 by these microorganisms through in vitro assays, based on what is known regarding the role of the TCA groups and the free amino group in inferring the toxicity of FB1 (Table 1, Table 1 in Supplementary Material), we can safely assume detoxification was indeed achieved.
Sphingomonas sp. ATCC 55552 was isolated from field-grown, moldy maize kernels, and stalk tissue (Duvick et al., 1998a) and been shown to degrade fumonisin B1 through the consecutive action of a carboxylesterase (Duvick et al., 2003) and an aminotransferase (Heinl et al., 2011). The same pathway was found in Sphingopyxis sp. MTA 144 isolated from composted earth (Täubel, 2005), in which the gene cluster responsible for fumonisin degradation was identified with fumD, encoding the carboxylesterase and fumI encoding the aminotransferase (Hartinger et al., 2009; Heinl et al., 2009, 2010).
Degradation by Exophiala sp., also isolated from field-grown, moldy maize kernels, and stalk tissue (Duvick et al., 1998a), was shown to be conferred by a carboxylesterase and, in contrast to ATCC 55552 and MTA 144, by an oxidative deaminase. Two degradation products were identified: a new compound, 2-oxo-12,16-dimethyl-3,5,10,14,15-icosanepentol hemiketal, and in smaller amounts the N-acetylated aminopentol backbone (N-acetylAP1).
Strain NCB 1492, isolated from maize field soil and related to the Delftia/Comamonas group, gave rise to four tentative degradation products of fumonisin B1 (C34H59NO15): heptadecanone (C17H34O), isononadecene (C19H38), octadecenal (C18H34O), and eicosane (C20H42) (Benedetti et al., 2006). The first degradative steps are thought to occur extracellularly, with deamination (and possibly esterase) activities followed by a slower degradation of the aliphatic chain.
Insights into the detoxification of fumonisins can also be useful for mycotoxins produced by other fungal genera. In this light, we would like to draw the focus on the Alternaria toxins AAL-TA en -TB, which share with fumonisins a distinct structural similarity and toxicity mechanism (Abbas et al., 1993a; Tsuge et al., 2013). Fumonisins have two TCA side chains esterified to the aminopentol backbone, whereas AALtoxins have only one TCA side group, and are therefore collectively referred to as sphinganine-analog mycotoxins. To the best of our knowledge, there have not been any reports of microbial strains capable of degrading AAL-toxins, but based on their structural similarity it is likely that fumonisin degrading organisms as described above might also be capable of degrading AAL-toxin.
Zearalenone
Toxicity
ZEN is mainly produced by fungi belonging to the genus Fusarium such as F. graminearum and F. culmorum and possesses estrogenic activity in pigs, cattle and sheep (Zinedine et al., 2007). The toxicity of ZEN is mainly conferred by its lactone group and the free C-4 hydroxyl group (Table 1) which is necessary for binding the estrogen receptor (El-sharkawy S. H. and Abul-hajj Y. J., 1988). Many derivatives of ZEN are known and some exhibit a higher estrogenicity than the mother compound (Shier et al., 2001), such as α-zearalenol, α- and β-zearalanol, and zearalanone. Several studies described the microbial transformation of ZEN to such derivatives, but as they do not represent a true detoxification of the compound they are not discussed in this review. Also, cases in which no clear evidence is presented (yet) for true detoxification (e.g., Pseudomonas strains in Tan et al., 2014, 2015) are not discussed in detail.
Degradation: Organisms and Pathways
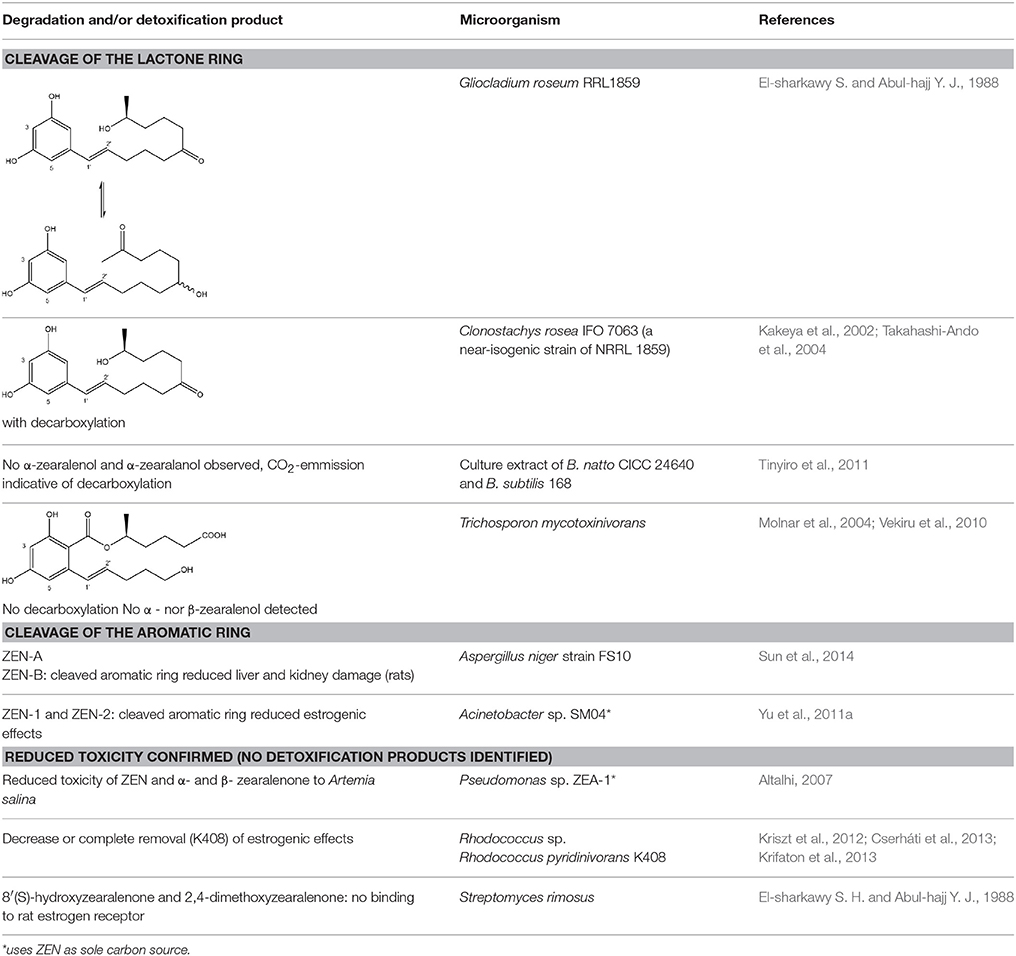
To date, two main detoxification mechanisms are known for ZEN, both cleaving a ring structure (Table 3). The lactone ring can be cleaved by several fungal species through two mechanisms. Degradation by Gliocladium roseum NRRL1859 (El-sharkawy S. and Abul-hajj Y. J., 1988) resulted in a 1:1 mixture of 1-(3,5-dihydroxyphenyl)-10′-hydroxy-1-undecen-6′-one, and 1-(3,5-dihydroxyphenyl)-6′-hydroxy-1-undecen-10′-one. Matthies et al. (2001) showed that production of the ZEN-degrading enzyme in G. roseum DSM 62726 was induced the highest by the derivatives zearalanol and α-zearalanol. Almost similarly, only the first metabolite was observed after degradation by a near isogenic strain of NRRL1859, Clonostachys rosea (synonym: G. roseum, teleomorph: Bionectria ochroleuca) IFO 7063 (Kakeya et al., 2002), resulting in the loss of estrogenic activity in MCF-7 cancer cells (Table 1 in Supplementary Materials), through the activity of a ZEN lactonohydrolase enzyme (zhd101) which catalyzes the hydrolysis of ZEN at the ester bond in the lactone ring, followed by spontaneous decarboxylation (Takahashi-Ando et al., 2004). Based on this knowledge, Popiel et al. (2014) searched a collection of Trichoderma and Clonostachys isolates for functional lactonohydrolase homologs, to find a functional ZEN lactonohydrolase in mycoparasitic Trichoderma aggressivum. A similar pathway might also exist in Bacillus sp., as cell culture extracts of B. natto CICC 24640 and B. subtilis 168 showed complete degradation of ZEN in conjunction with CO2-emmission, indicative of decarboxylation (Tinyiro et al., 2011).
A second cleavage pathway is exhibited by the yeast Trichosporum mycotoxinivorans (Molnar et al., 2004) to ZOM-1 intermediate (cleavage at the C6-ketone group), suggested to take place through a lactone intermediate and subsequent activity by unspecified a/b-hydrolase, but without the decarboxylation as seen in C. rosea. ZOM-1 did not show any estrogenic activity in a yeast bioassay, nor interaction with the human estrogen receptor (Vekiru et al., 2010), nor estrogenic activity with MCF-7 cells (Liu et al., 2001). It is important to notice that T. mycotoxinivorans is well-known in medicine, since it can cause opportunistic infections or induce summer-type hypersensitivity pneumonitis in immune-deficient cystic fibrosis patients (Tintelnot et al., 2011) which can be an impediment for applications.
Detoxification of ZEN contaminated corn steep liquor by A. niger strain FS10 and its culture filtrate, exemplified by less severe liver and kidney damage in rats, was recently reported (Sun et al., 2014). Two intermediate products, ZEN-A and ZEN-B, which inferred reduced liver and kidney damage in rats compared to ZEN, were detected, of which the latter the authors suggested the benzene ring might be cleaved because the UV absorption of ZEN was lost in ZEN-B. Somewhat similarly, two degradation products (ZEN-1 and ZEN-2) were detected after degradation by Acinetobacter sp. SM04 isolated from agricultural soil, for which no equally estrogenic activity could be detected on the basis of the MTT (tetrazolium salt) cell proliferation assay in MCF-7 cell line. Also, UV-Vis spectroscopy indicated cleavage of the benzene ring in these products (Yu et al., 2011a). Interestingly, ZEN and its estrogenic properties were only reduced when degradation tests were performed with extracellular extracts from M1 medium cultures, where sodium acetate is the only extra carbon source, and not from Nutrient Broth cultures, where many different extra carbon sources are present (Yu et al., 2011b). This indicates that the transcription of genes responsible for ZEN degradation may be regulated by catabolite repression.
Pseudomonas sp. ZEA-1, isolated from the rhizosphere of a corn plant, was shown to harbor the responsible degradation genes on a 120 kb plasmid mediating the transformation of ZEN and its derivatives α- and β- ZEN into less toxic products to Artemia salina. The transformation product was not elucidated, other than the specific absorption maximum at 400 nm (Altalhi, 2007). A 5.5 kb fragment containing the gene(s) encoding for ZEN degradation was cloned and actively expressed in Escherichia coli (Altalhi and El-Deeb, 2009).
The complete loss of ZEN estrogenic activity was obtained by several degrading Rhodococcus strains (Kriszt et al., 2012; Cserháti et al., 2013), without the identification of possible metabolites. R. pyridinovorans K408 showed a biodegradation potential of up to 85% and decreased the estrogenicity with 76%. Several strains also simultaneously degraded AFB1, ZEN, and T2-toxin (Cserháti et al., 2013), confirming the status Rhodococcus as a metabolically highly versatile genus with a large potential for degradation of aromatic and other pollutants (Larkin et al., 2005).
Trichothecenes
Toxicity
Trichothecenes are sesquiterpenoids produced by mainly the genera Fusarium, Trichothecium, Myrothecium, Trichoderma, and Stachybotrys fungi (Sudakin, 2003; Kimura et al., 2007; Li et al., 2011). High doses lead to emesis, whereas low doses induce decreased feed consumption and weight gain (Eriksen and Pettersson, 2004). Trichothecenes are characterized by a 12,13-epoxy-trichothec-9-ene nucleus (Hussein and Brasel, 2001). Type A trichothecenes do not contain carbonyl function at C8 (T-2 toxin, HT-2 toxin, T-2 tetraol, T-2 triol, 15-monoacetoxyscirpenol, DAS, neosolaniol, and scirpentriol). Type B trichothecenes have a carbonyl group at C8 [deoxynivalenol (DON), 15-acetyl DON, 3-acetyl DON, nivalenol (NIV), 4-acetyl NIV]. Type C trichothecenes include another epoxide group and type D trichothecenes contains an additional ring system between C4 and C15 position (Zhou et al., 2008; McCormick et al., 2011).
The 12,13-epoxide ring in trichothecenes is essential for their toxicity (Zhou et al., 2008) and has been linked to the cytotoxicity of trichothecenes, namely inhibition of protein, RNA and DNA synthesis (Hussein and Brasel, 2001; Rocha et al., 2005). Trichothecenes bind with the 60S subunit of the ribosome and interfere with the action of peptidyltransferase (Ehrlich and Daigle, 1987). However, the degree of toxicity is dependent on the presence of substituents on C15 and C4 (Cundliffe et al., 1974; Cundliffe and Davies, 1977). The most potent mycotoxin T-2 toxin has acetyl or acyl side groups on C4, C8, and C15 of the basic structure. Loss of a side group from either of these positions resulted in reduced protein synthesis inhibition (T-2 toxin to HT-2 toxin, neosolaniol, or DAS). Further removal of side groups weakens their effect (T-2 triol, T-2 tetraol, 15-monoacetyl DAS, scirpentriol, fusarenon X, and DON) and reduction of hydroxyl groups, forming verrucarol, reduced their effectiveness greatly (Thompson and Wannemacher, 1986; Table 2 in Supplementary Material). De-acylation is clearly a first step toward detoxification, illustrated in Figure 1 in Supplementary Material. This reduced effect of de-acylation of T-2 toxin is also confirmed with human melanoma SK-Mel/27 cell lines (Babich and Borenfreund, 1991) and β-galactosidase activity of Kluyveromyces marxianus (Engler et al., 1999) showing the same tendency (Table 1 in Supplementary Material).
Degradation: Organisms and Pathways
The toxicity of trichothecenes is, next to their epoxide-group, also dependent on their acylated side chains. Therefore, two main groups are distinguished; acylated (e.g., T-2 toxin) and non-acylated trichothecenes (e.g., DON).
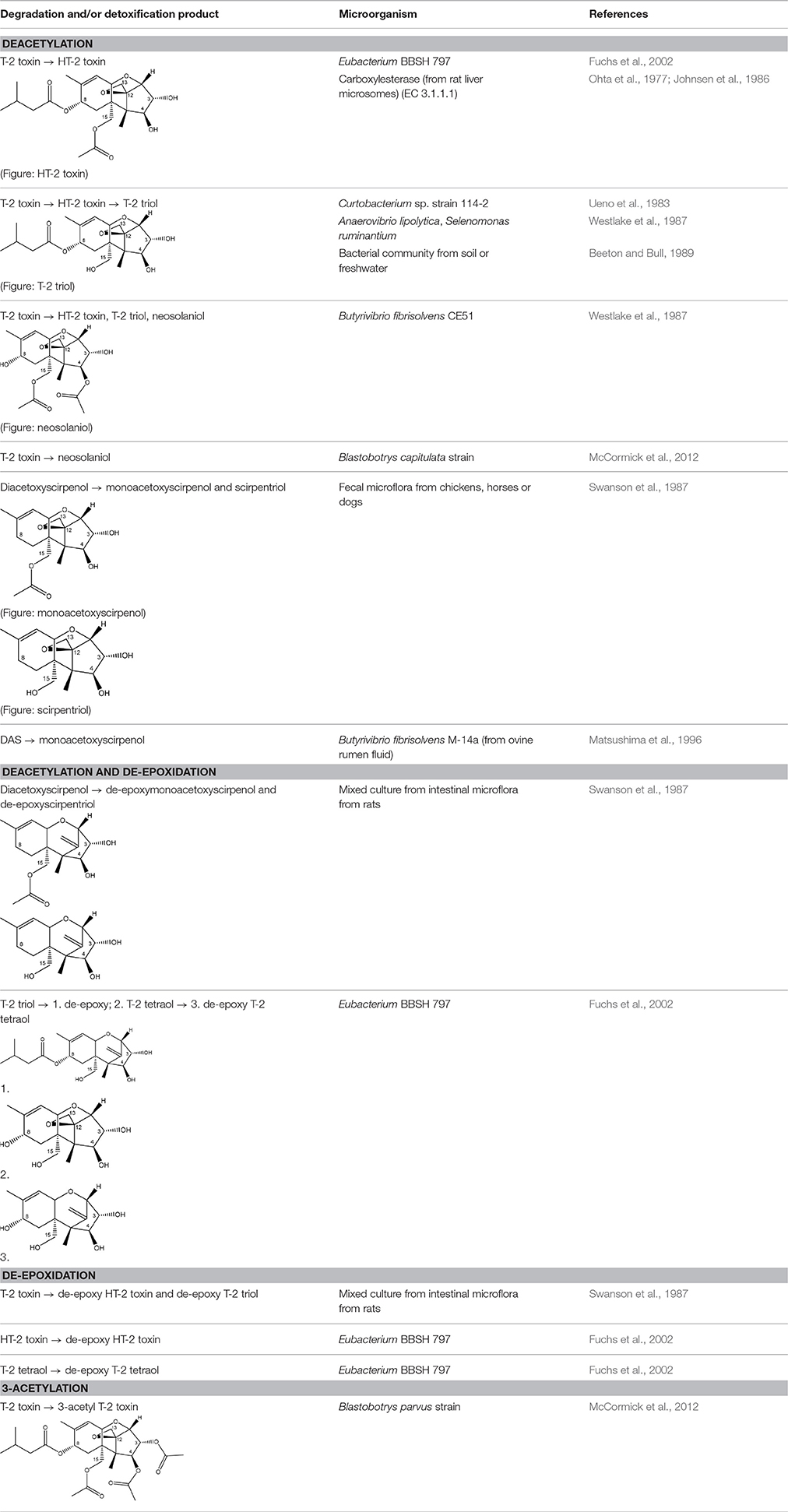
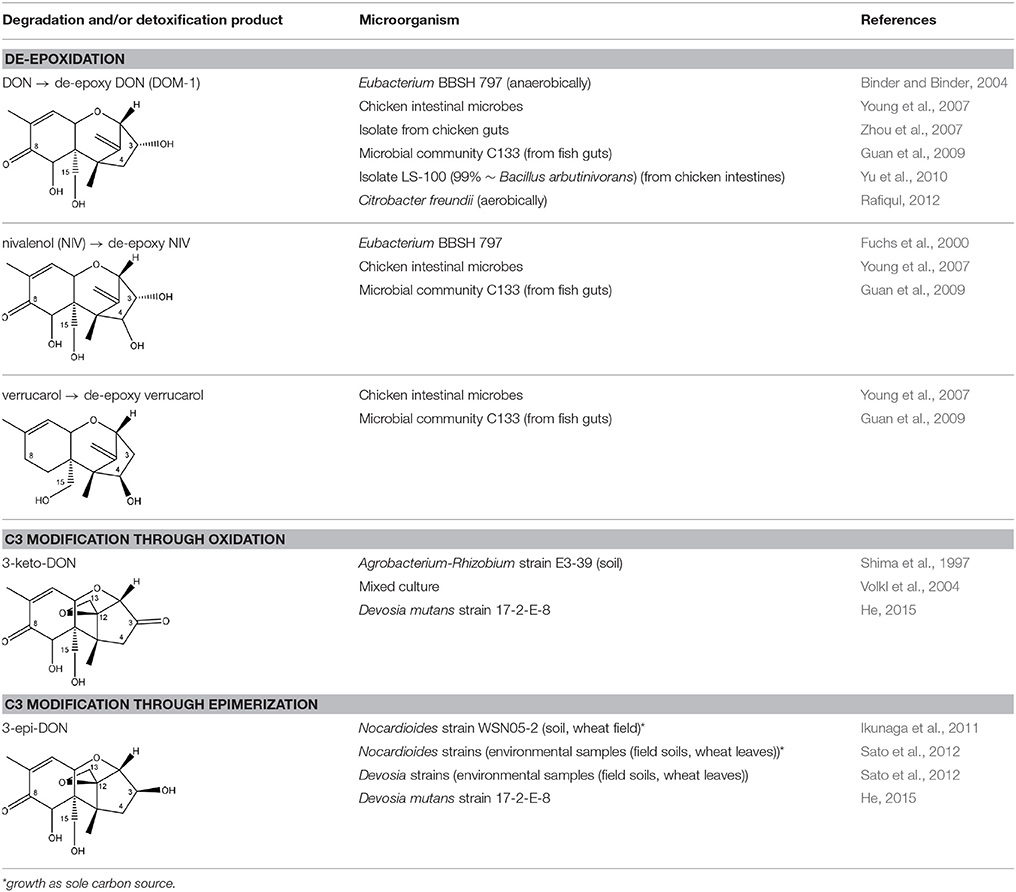
As previously described, de-acylation is the first step in detoxification of acylated trichothecenes. Degradation of T-2 toxin to HT-2 toxin and subsequently to T-2 triol was performed by Curtobacterium sp. strain 114-2 of which the reduced toxicity of T-2 triol was once more confirmed resulting in 23 and 13 times less toxic than, respectively T-2 toxin and HT-2 toxin (Ueno et al., 1983; Table 4). Still, the epoxide group in trichothecenes remains responsible for their toxicity. De-epoxidation is the next step of detoxification trichothecenes. Several studies focuses on the degradation of multiple trichothecenes and the differences between their metabolism by the same organism(s). Young et al. (2007) studied the metabolism of diverse trichothecenes by chicken intestinal microbes. For the non-acylated trichothecenes (4-DON, NIV, and verrucarol) their de-epoxidized metabolites were observed, for DAS, neosolaniol and T-2 toxin only de-acylation was exhibited and for the monoacetyl trichothecenes (3-acetyl DON, 15-acetyl DON, and fusarenon X), de-acylation was the predominant pathway. In another study, pig gastrointestinal microflora transformed 3-acetyl DON into DON and which was further de-epoxidized (Eriksen et al., 2002). Rat intestinal microflora was also able to de-epoxidize T-2 tetraol and scirpentriol, transform T-2 toxin into de-epoxy HT-2 toxin and de-epoxy T-2 triol and DAS into de-epoxymonoacetoxyscirpenol and de-epoxyscirpentriol (Swanson et al., 1987). All above mentioned cases concern degradation by mixed cultures. In contrast, Eubacterium BBSH 797 has the ability to degrade several trichothecenes as pure culture isolated from bovine rumen fluid (Fuchs et al., 2000, 2002; Binder and Binder, 2004) and has been developed into a commercial product (Biomin® BBSH 797) for detoxifying trichothecenes in animal feed (He et al., 2010). It is known for its detoxification capacities of DON into DOM-1 and de-epoxidization of NIV, T-2 tetraol, scirpentriol, and HT-2 toxin. T-2 toxin was de-acylated into HT-2 toxin, whereas degradation of T-2 triol involved the competition of two reactions; (1) de-epoxidation or (2) deacylation into T-2 tetraol and subsequently de-epoxidation into de-epoxy T-2 tetraol (Fuchs et al., 2002). Further, 4-acetyl NIV and 3-acetyl NIV was de-acetylated and/or de-epoxidized (Fuchs et al., 2000).
Degradation of DON occurs through de-epoxidation, oxidation, or isomerization (Table 5). Microbial culture C133 of fish guts transformed DON to DOM-1 (Guan et al., 2009). Eubacterium BBSH 797 is known to degrade DON into DOM-1 anaerobically (Binder and Binder, 2004). Citrobacter freundii could transform DON into DOM-1 aerobically (Rafiqul, 2012). DON can also be oxidized to 3-keto DON which is 10 times less toxic than DON evaluated with a bioassay based on mitogen-induced and mitogen-free proliferations of mouse spleen lymphocytes (Shima et al., 1997). The bacterium strain E3-39 which degraded DON to 3-keto-DON, is belonging to the Agrobacterium-Rhizobium group. A mixed culture from environmental sources could degrade DON into 3-keto-DON, whereas 15-acetyl DON, 3-acetyl DON and fusarenon-X were also transformed (Volkl et al., 2004). Subsequently, He (2015) found the soil bacterium Devosia mutans 17-2-E-8 which transformed DON into 3-epi-DON (major product) and 3-keto-DON (minor product). These metabolites have also been tested on their toxicity with two assays. The IC50 values of 3-epi-DON and 3-keto-DON were 357 and 3 times higher, respectively, than that of DON on the basis of a MTT bioassay using Caco-2 cell line to asses cell viability, and were 1181 and 5 times higher, respectively, than that of DON on the basis of a cell proliferation BrdU bioassay using 3T3 fibroblast cell line to asses DNA synthesis (Table 1 in Supplementary Material). Toxicological effects of 14-day oral exposure of B6C3F1 mouse to DON and 3-epi-DON were also investigated concluding that 3-epi-DON was at least 50 times less toxic than DON (He, 2015). The metabolite 3-epi-DON was also formed by degradation of DON through Nocardioides sp. strain WSN05-2 isolated from a wheat field (Ikunaga et al., 2011). And lastly, nine Nocardioides strains (Gram-positive) and four Devosia strains (Gram-negative) produced 3-epi-DON aerobically. The Gram-positive strains showed DON assimilation, whereas the Gram-negatives did not (Sato et al., 2012).
Further, hydroxylation and glycosylation of trichothecenes are also known for their detoxification capability (He et al., 2010), however these derivatives can be rehydrolyzed or regenerated in the digestive tract of animals and humans losing their detoxification capacity.
Aflatoxins
Toxicity
Aflatoxins are furanocoumarins produced by mainly Aspergillus species (Wu et al., 2009; Samuel et al., 2013). Naturally occurring aflatoxins are categorized by IARC as carcinogenic to humans (Group 1; IARC, 2002). Aflatoxin B1 (AFB1) is activated by cytochrome P450 system to a highly reactive AFB1-8,9-epoxide which can react with DNA (Eaton and Groopman, 1994; Guengerich et al., 1996).
The toxicity of AFB1 is mainly caused by the lactone ring. Cleavage of the lactone ring leads to a non-fluorescent compound with reduced biological activity (Lee et al., 1981). The residual component has a 450 times reduced mutagenicity (measured with the Ames test) and a 18 times reduced toxicity (measured with chicken embryo test; Lee et al., 1981). Also the difuran ring moiety, especially the presence of the double bond in the terminal furan ring, contributes to the toxicity (Wogan et al., 1971) as evidenced by comparing the toxicity of aflatoxins with similar coumarin molecules. Wong and Hsieh (1976) concluded by comparing several aflatoxins and metabolites with the Ames test that the double bond was also involved in both mutagenic and carcinogenic activity of aflatoxins leading that the aflatoxins AFB2 and AFG2 (without a double bond) are much less toxic than AFB1 and AFG1 (with a double bond).
Degradation: Organisms and Pathways
To our knowledge, a first report on the microbial detoxification of AFB1 has been published in 1966, mediated by Flavobacterium aurantiacum (now called Nocardia corynebacterioides; Ciegler et al., 1966; Teniola et al., 2005). Although no detoxification products were analyzed, residual toxicity to ducklings was found to be absent indicating true detoxification (Ciegler et al., 1966). The biosafety of the microorganism was confirmed using an in vivo trial with chickens (Tejada-Castañeda et al., 2008). Since this first report, many studies have focused on the detoxification of AFB1. However, only a few studies detected the degradation products and analyzed their toxicity. Generally, two main detoxification pathways are observed: modification of the difuran ring or modification of the coumarin structure.
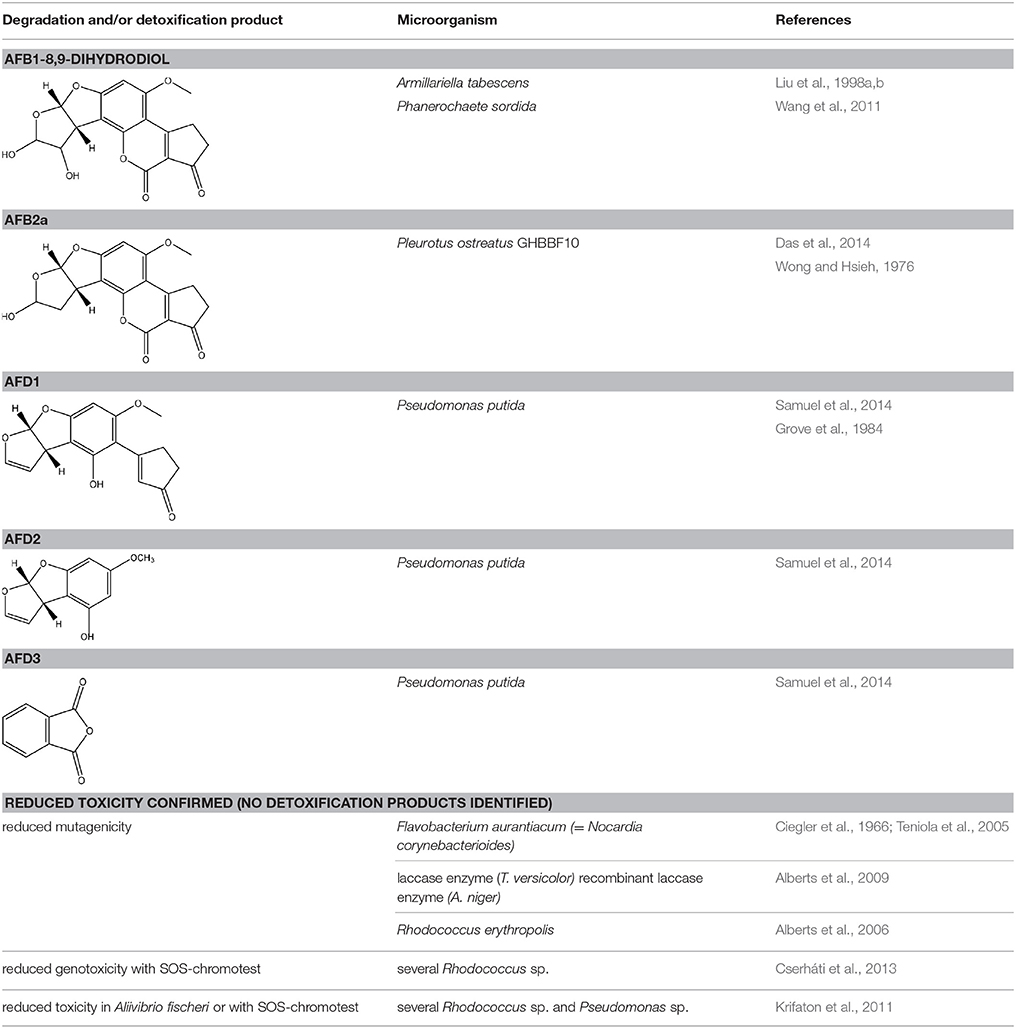
Firstly, modification of the difuran ring moiety was reported in several studies. Degradation of AFB1 into AFB1-8,9-dihydrodiol was performed by manganese peroxidase from the white rot fungi Phanerochaete sordida (Wang et al., 2011) and the “aflatoxin-detoxifizyme (ADTZ)” of fungus Armillariella tabescens (Liu et al., 1998b; Table 6). The authors suggested that AFB1 degradation initially involves formation of AFB1-8,9-epoxide, after which a hydrolysis resulted in a dihydrodiol-derivate. Detoxification was confirmed with a reduced mutagenicity measured by the Ames Salmonella-based test (Liu et al., 1998b, 2001; Wang et al., 2011; Table 1 in Supplementary Material) and reduced toxicity measured with rat liver (Liu et al., 1998b) and chicken embryos (Liu et al., 1998a). Another metabolite was detected with the white rot fungus Pleurotus ostreatus GHBBF10 which degraded 91.76% of AFB1 into a component which could be a hydrolyte of AFB1, namely dihydrohydroxyaflatoxin B1 (AFB2a) (Das et al., 2014; Table 2). AFB2a has also a reduced mutagenicity (Wong and Hsieh, 1976; Table 1 in Supplementary Material).
Secondly, the lactone ring in the coumarin moiety of AFB1 can be changed. A Pseudomonas putida strain has been discovered degrading AFB1 into AFD1 and subsequently into AFD2 (Table 6). The metabolite AFD1 had been previously discovered through ammonization and acidifying AFB1, whereas the difuran ring stays unchanged and the lactone ring is cleaved. AFD1 has a lower mutagenicity and toxicity measured by respectively the Ames Salmonella-based test (Méndez-Albores et al., 2005) and HeLa cells with the MTT [3-(4,5-dimethylthiazole-2-yl)-2,5-diphenyltetrazolium bromide] method (Samuel et al., 2014). The metabolites AFD2 (an aflatoxin metabolite lacking the lactone and cyclopentenone ring) and AFD3 also showed a lower toxicity toward HeLa cells (Table 1 in Supplementary Material; Samuel et al., 2014).
In certain studies, no degradation product was identified, but toxicity tests were performed on the treated AFB1. Similarly to F. aurantiacum as mentioned before, a pure laccase enzyme from Trametes versicolor and a recombinant laccase enzyme produced by A. niger degraded, respectively, 87.34 and 55% of AFB1 with a significant loss of mutagenicity evaluated in the Ames Salmonella-based assay (Alberts et al., 2009). Extracellular enzymes of Rhodococcus eryhtropolis were also able to detoxify AFB1 with a loss of mutagenicity (Alberts et al., 2006).
Ochratoxins
Toxicity
Ochratoxins (OT) are a group of mycotoxins sharing an isocoumarin moiety substituted with a phenylalanine group (OTA, OTB, hydroxyl-OTA), a phenylalanine ester group (OTC, OTA methylester, OTB methyl ester, OTB ethyl ester), or a hydroxyl group (OTα and OTβ). OTA is the most important OT because of its incidence in food- and feed commodities. It is composed of a 7-carboxy-5-chloro-8-hydroxy-3,4-dihydro-3-R-methylisocoumarin (OTα) moiety and the amino acid L- phenylalanine group. Both structures are linked through a carboxy group via an amide bond. OTA is produced by Aspergillus and Penicillium species (Richard, 2007; McCormick, 2013). The mode of action of OTA is broad and therefore the molecule has nephrotoxic, mutagenic, teratogenic, neurotoxic, hepatotoxic, and immunotoxic properties (Pfohl-Leszkowicz and Manderville, 2007). The toxicity of OTA is mainly attributed to its isocoumarin moiety and probably not to the phenylalanine moiety (Table 1; Xiao et al., 1996). The carboxyl group of the phenylalanine moiety and also the Cl group of the other moiety seem to be conducive for the toxicity of OTA.
Degradation: Organisms and Pathways
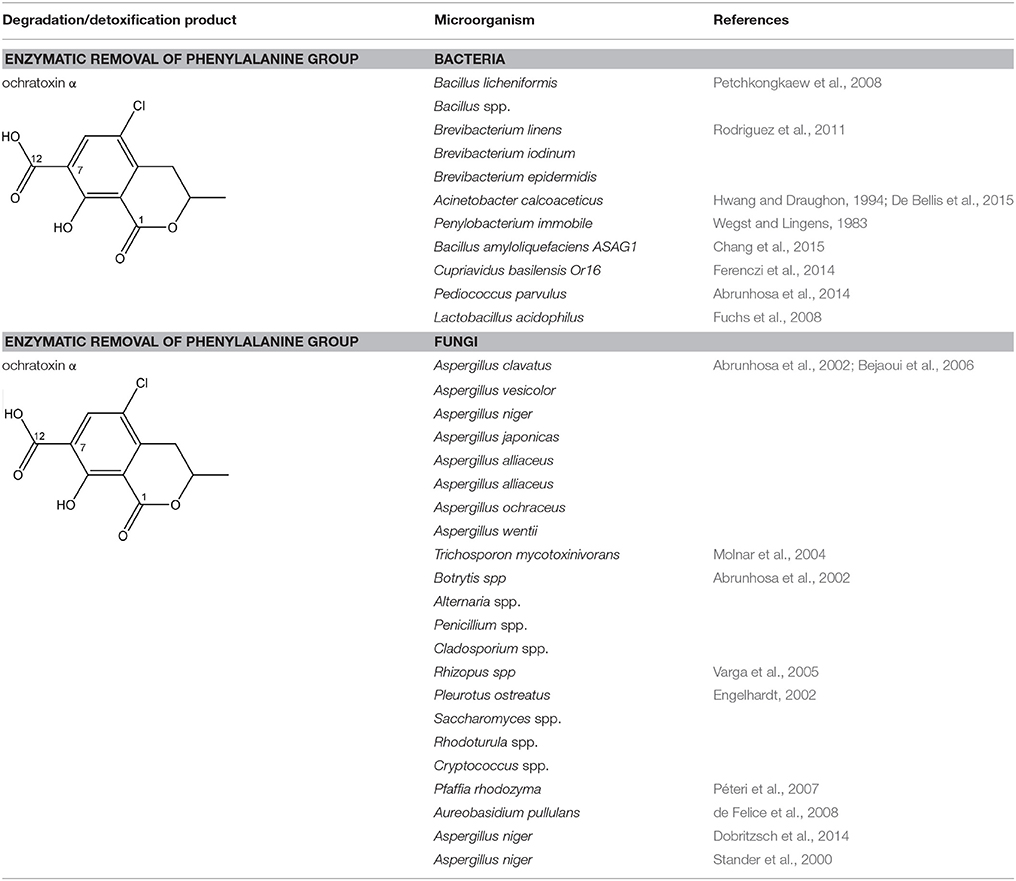
The main detoxification pathway of OTA is the hydrolyzation of the amide bond between the isocoumarin residue and phenylalanine by a carboxypeptidase. Two classes of carboxypeptidases have been associated with degradation of OTA namely Carboxypeptidase A (CPA) (Stander et al., 2001; Chang et al., 2015) and Y (CPY) (Dridi et al., 2015). The main difference between both is the use of a zinc ion within the protein for hydrolysis of the peptide at the C-terminal of the amino acid. Almost all strains that are reported to degrade OTA use this pathway resulting in the formation of L-β- phenylalanine and OTα the former being less toxic than OTA (Table 7; Bruinink and Sidler, 1997). Although this is a very straightforward way of reducing the amount of OTA in food and feed samples, it is important to highlight that the efficient degradation of OTA is depending on the activity of the peptidase enzyme. With this respect, several research groups showed that these carboxypeptidase enzymes tend to have high optimal temperatures (30°C or higher) which might hamper practical applications, observed with Pediococcus parvulus and several yeasts such as Pfaffia rhodozyma (Péteri et al., 2007; Patharajan et al., 2011; Abrunhosa et al., 2014). Other enzymes are also able to carry out this reaction: Deoxygenases, lipases, amidases, and several commercial proteases (Abrunhosa et al., 2006), have also been identified as carrying out this reaction. Although depending on the enzyme, intermediates can be different, the end product is always OTα.
Some interesting strains are highlighted here. Trichosporon mycotoxinivorans was demonstrated to deactivate OTA by conversion into the nontoxic OTα. Even more intriguingly, T. mycotoxinivorans was also able to decarboxylate ZEN (Molnar et al., 2004; Vekiru et al., 2010). After 24 h, ZEN was degraded to carbon dioxide or into metabolites that neither showed fluorescence nor did absorb UV-light. Neither α- nor β-ZEL, other equally estrogenic metabolites of ZEN, could be detected. It is commercially applied as feed additive under the commercial name Biomin® MTV.
Phenylobacterium immobile (Wegst and Lingens, 1983) was also found to convert OTA to OTα through a dioxygenase step on the phenylalanine moiety, a dehydrogenation to catechol, a ring cleavage, and the final formation of OTα via a hydrolase.
Unexplored Worlds that Might Harbor Valuable Mycotoxin Degrading Microorganisms
Targeting Carboxyl Esters
As stated above, fumonisins and acylated trichothecenes share carboxyl-esters which are involved in their toxicity. Detoxification of fumonisins is realized by removal of the tricarballylate side groups via carboxylesterases (EC 3.1.1.1). Similarly, acylated trichothecenes have several side groups where carboxylesterases could attack on the carboxyl group, as observed with carboxylesterases from rat liver microsomes (categorized as EC 3.1.1.1) degrading T-2 toxin into HT-2 toxin (Ohta et al., 1977; Johnsen et al., 1986). Carboxylesterases are multifunctional enzymes that catalyze the hydrolysis of substrates containing ester, amide, and thioester bonds with relatively broad substrate specificity (Bornscheuer, 2002) which is attributed to a large conformable active site that permits entry of numerous structurally diverse substrates. Microbial carboxylesterases have been reported in the degradation of pesticides; some hydrolyze pyrethroids and bind stoichiometrically to carbamates and organophosphates reviewed by Singh (2014). Several organisms have been isolated which degradative capacities of these compounds inferred by the expression of carboxylesterases, for which, bearing in mind their general broad substrate specificity, it might be worthwhile to screen for degradation of fumonisins and acylated trichothecenes. For example, broad-spectrum pyrethroid-hydrolyzing carboxylesterases were identified in the lambda-cyhalothrin degrading Ochrobactrum anthropic YZ-1 strain (Zhai et al., 2012) and Bacillus sp. DG-02, isolated from a pyrethroid-manufacturing wastewater treatment system (Chen et al., 2014). Similarly, an Acinetobacter baumannii strain was shown to degrade a wide range of organophosphorus compounds and evidence for a novel carboxylesterase in this strain was presented. Taking an environmental DNA (eDNA) isolation approach, Rashamuse et al. (2009) screened a microbial community to access novel carboxylesterases from environmental genomes: a carboxylesterase gene with 60% sequence identity to the gene from Ralstonia eutropha was identified, along with subsequent heterologous expression in Escherichia coli in a biologically active form. A similar approach might be taken to discover more mycotoxin-active carboxylases based on sequences of carboxylases present in Sphingomonas sp. ATCC 55552, Exophiala sp., or Sphingopyxis sp. MTA 144.
Targeting a Lactone Ring
The presence of a lactone moiety is shared by OTA, aflatoxins, and ZEN. Lactone chemicals are well-known as auto-regulators in both eukaryotic and prokaryotic cells. A well-known example is acyl homoserine lacton which is a quorum sensing molecule associated with biofilm formation. Because of the detrimental effects of biofilms in many industrial applications, high throughput research initiatives have been undertaken in the past and present in search for enzymes able to degrade these lactone molecules. These, often metagenomics, approaches result in the characterization of new and more efficient lactonase enzymes (Shimizu et al., 2001; Riaz et al., 2008; Schipper et al., 2009). The potential activities of these lactonases with respect to mycotoxins remains elusive but scientific fields studying biofilm issues might offer new microbial consortia ready to be explored for their mycotoxin degrading capacities.
Also targeted analyses can result in the characterization of new and efficient lactonase enzymes. In a screening assay for enzymes able to degrade bio-active lactones, a novel lactonohydrolase, an enzyme that catalyzes the hydrolysis of aldonate lactones to the corresponding aldonic acids, was purified from Fusarium oxysporum AKU 3702. The enzyme irreversibly hydrolyzes a broad spectrum of aromatic lactones, such as dihydrocoumarin and homogentisic-acid lactone (Shimizu et al., 1992; Kobayashi et al., 1998).
New insights for biodegradation of mycotoxins with estrogenic effects such as ZEN might come from studies on the microbial degradation of steroidal estrogens. Several strains have been isolated which are able to degrade the steroidal estrogen estrone (E1), also harboring a lactone ring (Yu et al., 2013), among which Sphingomonas sp. KC8 (Yu et al., 2007), Bacillus subtilis E2Y4 (Jiang et al., 2010), and several Rhodococcus sp. (Yoshimoto et al., 2004), remarkably all isolated from activated sludge. Also, cometabolic degradation of ethinyl estradiol (EE2) was obtained with nitrifying activated sludge (Vader et al., 2000). Therefore, activated sludge might prove to be a rich source of degradation potential for lactone-harboring mycotoxins.
Targeting an Epoxide Moiety
For trichothecenes, the epoxide moiety is an important chemical group associated with toxicity. Microbial transformation of epoxides was studied by Swaving and de Bont (1998) who demonstrated that two types of enzymes were responsible for detoxification of epoxides: glutathione transferases as a class of general detoxifying enzymes and epoxide hydrolases which are specific for detoxification of epoxides. Glutathione transferases (dependent on glutathione as cofactor) are mostly found in aerobic eukaryotes and prokaryotes, such as E. coli and Rhodococcus sp. which degrades a range of epoxides. Epoxide hydrolases are found in many microorganisms, like Flavobacterium, Pseudomonas, Corynebacterium, and Stigmatella species. Other enzymes can also convert an epoxide intermediate via a certain pathway (e.g., alpha-pinene oxide lyase from Nocardia sp. strain P18.3 and Pseudomonas fluorescens NCIMB 11671, styrene oxide isomerase of Pseudomonas species, Xanthobacter 124X or Exophilia jeanselmei, or epoxyalkane-degrading enzyme in Xanthobacter Py2 (Swaving and de Bont, 1998). Broudiscou et al. (2007) proved that mono-and sesquiterpenes were degraded in the presence of mixed rumen microorganisms, corresponding with the isolation origin mostly found for microorganisms degrading trichothecenes. Ptaquiloside, also a sesquiterpene toxin, could be degraded by soil microorganisms (Engel et al., 2007) which can be a new source for biodegradation of trichothecenes.
Targeting Poly-Aromatic Ring Structures
White rot fungi are frequently found for degrading aflatoxins, such as A. tabescens, P. sordida, P. ostreatus, T. versicolor, and Peniophora sp. (Liu et al., 1998b; Motomura et al., 2003; Alberts et al., 2009; Wang et al., 2011; Das et al., 2014; Yehia, 2014). White rot fungi are well-known for their degrading capabilities of their natural substrate lignin and a broad spectrum of structurally diverse toxic environmental pollutants (e.g., munitions waste, pesticides, polychlorinated biphenyls, polycyclic aromatic hydrocarbons, bleach plant effluent, synthetic dyes, synthetic polymers, and wood preservatives; Reddy, 1995; Pointing, 2001). Lignin peroxidases, manganese peroxidases and laccases are the major enzymes involved in lignin degradation based on oxidative mechanisms (Tuor et al., 1995). Laccases and manganese peroxidases of white rot fungi have been reported for degrading aflatoxins which possible can lead to different metabolites (Motomura et al., 2003; Wang et al., 2011). Peniophora sp. SCC0152, P. ostreatus St2-3, and several Trametes sp. strains demonstrated the degradation of Poly R-478 dye and AFB1 (Alberts et al., 2009). Next to white rot fungi, the genus Rhodococcus is also known to have promising degradation capability for xenobiotics (Martínková et al., 2009). Alberts et al. (2006) and Eshelli et al. (2015) suggested that degradation of AFB1 (polyaromatic compound) by a Rhodococcus erythropolis strain could be degraded in a similar way of degrading polyaromatic compounds of which their degradation occurs through a cascade of enzyme reactions (e.g., ring cleavage biphenyl dioxygenases, dihydrodiol dehydrogenases, and hydrolases). Degradation of a wide range of aromatic compounds results in a limited number of central intermediates (catechol, protocatechuate, gentisate) which are further degraded through central pathways for finally entering the citrate cycle (Martínková et al., 2009). In addition, R. erythropolis NI1 strain was found which was capable of degrading AFB1, ZEN, and T-2 toxin at the same time (Cserháti et al., 2013). Hence, various organisms have the potential for degrading multiple mycotoxins or other components, exemplified by. Stenotrophomonas maltophilia, Stenotrophomonas sp. NMO-3, and Pseudomonas aeruginosa which can degrade AFB1 and coumarin (Guan et al., 2008; Liang et al., 2008; Sangare et al., 2015), and Mycobacterium fluoranthenivorans FA4T which can degrade AFB1 and also grow on the polycyclic aromatic hydrocarbon fluoranthene (Hormisch et al., 2004).
Supporting the notion that microorganisms are able to metabolize structurally comparable chemicals from vastly different origins, a mixed enrichment culture capable of removing ZEN as sole carbon source, without the presence of derivatives, was obtained from soil collected at a coal gasification site, which are generally known to be associated with polycyclic aromatic hydrocarbon contamination. Removal of ZEN was enhanced in the presence of phenanthrene through enhanced microbial growth, indicating that organisms capable of using ZEN were also able to metabolize phenanthrene (Megharaj et al., 1997). Building further on this notion, cleaving the aromatic ring of ZEN by A. niger FS10 (Sun et al., 2014) and Acinetobacter sp. SM04 (Yu et al., 2011a), for which no enzymes have been identified, might bear resemblance to the degradation of resorcinol (1,3-dihydroxybenzene) for which degradation is known by P. putida (Chapman Ribbons and Ribbons, 1976) and Azotobacter vinelandii (Groseclose and Ribbons, 1981).
Targeting a Carboxyl/Amide Moiety
Carboxypeptidase A and Y belong to the group of protease enzymes. Great interest in these enzymes comes from the field of wastewater treatments as these enzymes play a vital role in the extracellular catabolism of organic matter in activated sludge. In search of these enzymes, progressively more culture independent screening approaches are being employed as up to 90% of bacteria present in wastewater cannot be cultured and in this way a large reservoir of enzymes is overlooked. In matrices harboring a vast set of microorganisms that cannot be cultured, metagenomics analyses are often the solution to get an in depth insight into the complexity of these enzymes in a certain matrix. Pursuing this approach, a metagenomics analysis of waste water revealed a highly diverse phylogenetic diversity of carboxypeptidase gene sequences including previously undescribed types of carboxypeptidases which might be interesting to be applied for diverse biotechnological applications such as the remediation of OTA contaminated batches (Jin et al., 2014).
Perspectives
Although fumonisins, trichothecenes, ZEN, OTA, and aflatoxins comprise the major mycotoxin groups in food- and feed commodities, there are several other mycotoxins that were not addressed in present review because knowledge on biodegradation and detoxification is scarce. Cyclodepsipeptides such as beauvericin and enniatins are increasingly reported in many countries in several commodities. However, to our knowledge, no reports are available on their biodegradation and detoxification by microorganisms.The same accounts for ergot alkaloids such as lysergic acid, ergine, and ergopeptines. They occur widely but to date, only one paper has recently reported on a R. erythropolis isolate able to degrade these compounds (Thamhesl et al., 2015). Finally, for the Penicllium expansum mycotoxin patulin, recent papers report on biodegradation of this mycotoxin by Pichia caribbica (Cao et al., 2013), Metschnikowia pulcherrima (Reddy et al., 2011), Kodameae ohmeri (Dong et al., 2015), Rhodosporidium spp. (Castoria et al., 2011; Zhu et al., 2015), and Saccharomyces cerevisiae (Moss and Long, 2002). Nevertheless, for these emerging and also for the other mycotoxins, there is still a considerable need for concerted research initiatives to identify new high-performance strains which can be implemented in practice.
Many surveys around the globe illustrate that mycotoxin contaminated batches of food and feed products often contain multiple both structurally related and non-related mycotoxins. An emerging approach to tackle this issue is biodegradation of mycotoxins by microorganisms. In our opinion an ideal biodegrading and detoxification agent should meet following features: (i) a fast and efficient degradation, (ii) of a broad spectrum of toxins, (iii) into non-toxic end products, (iv) by a non-pathogenic strain or consortium (v) under conditions that are relevant for the matrix in which the mycotoxin problem occurs. In order to do so, we urge researchers to look beyond the disappearance of the mother compound to rule out the creation of any lesser evils, and to explore strange new worlds, seek out new organisms and new metabolic pathways.
Author Contributions
KA and LD conceived the idea and scope for the review. IV, KA, and LD all equally contributed to gathering and summarizing the literature, designing the tables and figures, and writing and editing of the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00561
Figure 1. Reduced toxicity of T-2 toxin by subsequent de-acylation.
Table 1. Toxicity data on mycotoxins and their metabolites.
Table 2. Toxicity of trichothecenes (from most potent to almost no effectiveness observed) (Thompson and Wannemacher, 1986).
References
Abbas, H. K., Duke, S. O., and Tanaka, T. (1993a). Phytotoxicity of fumonisins and related-compounds. J. Toxicol.-Toxin Rev. 12, 225–251. doi: 10.3109/15569549309014408
Abbas, H. K., Gelderblom, W. C. A., Cawood, M. E., and Shier, W. T. (1993b). Biological-activities of fumonisins, mycotoxins from fusarium-moniliforme, in jimsonweed (Datura-Stramonium L) and mammalian-cell cultures. Toxicon 31, 345–353. doi: 10.1016/0041-0101(93)90152-9
Abbas, H. K., Tanaka, T., and Shier, W. T. (1995). Biological-activities of synthetic analogs of alternaria-alternata toxin (aal-toxin) and fumonisin in plant and mammalian-cell cultures. Phytochemistry 40, 1681–1689. doi: 10.1016/0031-9422(95)00470-R
Abrunhosa, L., Inês, A., Rodrigues, A. I., Guimaraes, A., Pereira, V. L., Parpot, P., et al. (2014). Biodegradation of ochratoxin A by Pediococcus parvulus isolated from Douro wines. Int. J. Food Microbiol. 188, 45–52. doi: 10.1016/j.ijfoodmicro.2014.07.019
Abrunhosa, L., Serra, R., and Venâncio, A. (2002). Biodegradation of ochratoxin A by fungi isolated from grapes. J. Agric. Food Chem. 50, 7493–7496. doi: 10.1021/jf025747i
Abrunhosa, L., Santos, L., and Venancio, A. (2006). Degradation of ochratoxin A by proteases and by a crude enzyme of Aspergillus niger. Food Biotechnol. 20, 231–242.
Adebo, O. A., Njobeh, P. B., Gbashi, S., Nwinyi, O. C., and Mavumengwana, V. (2015). Review on microbial degradation of aflatoxins. Crit. Rev. Food Sci. Nutr. doi: 10.1080/10408398.2015.1106440. [Epub ahead of print].
Alassane-Kpembi, I., Kolf-Clauw, M., Gauthier, T., Abrami, R., Abiola, F. A., Oswald, I. P., et al. (2013). New insights into mycotoxin mixtures: the toxicity of low doses of Type B trichothecenes on intestinal epithelial cells is synergistic. Toxicol. Appl. Pharmacol. 272, 191–198. doi: 10.1016/j.taap.2013.05.023
Alberts, J. F., Engelbrecht, Y., Steyn, P. S., Holzapfel, W., and van Zyl, W. (2006). Biological degradation of aflatoxin B-1 by Rhodococcus erythropolis cultures. Int. J. Food Microbiol. 109, 121–126. doi: 10.1016/j.ijfoodmicro.2006.01.019
Alberts, J. F., Gelderblom, W. C. A., Botha, A., and van Zyl, W. H. (2009). Degradation of aflatoxin B-1 by fungal laccase enzymes. Int. J. Food Microbiol. 135, 47–52. doi: 10.1016/j.ijfoodmicro.2009.07.022
Altalhi, A. D. (2007). Plasmid-mediated mycotoxin zearalenone in Pseudomonas putida ZEA-1. Am. J. Biotechnol. Biochem. 3, 150–158. doi: 10.3844/ajbbsp.2007.150.158
Altalhi, A. D., and El-Deeb, B. (2009). Localization of zearalenone detoxification gene(s) in pZEA-1 plasmid of Pseudomonas putida ZEA-1 and expressed in Escherichia coli. J. Hazard. Mater. 161, 1166–1172. doi: 10.1016/j.jhazmat.2008.04.068
Awad, W. A., Ghareeb, K., Bohm, J., and Zentek, J. (2010). Decontamination and detoxification strategies for the Fusarium mycotoxin deoxynivalenol in animal feed and the effectiveness of microbial biodegradation. Food Addit. Contam. Part A-Chem. Anal. Control Expo. Risk Assess. 27, 510–520. doi: 10.1080/19440040903571747
Babich, H., and Borenfreund, E. (1991). Cytotoxicity of T-2 toxin and its metabolites determined with the neutral red-cell viability assay. Appl. Environ. Microbiol. 57, 2101–2103.
Beardall, J. M., and Miller, J. D. (1994). “Diseases in humans with mycotoxins as possible causes,” in Mycotoxins in Grain: Compounds Other Than Aflatoxin, eds J. D. Miller and H. L. Trenholm (St. Paul, MN: Eagan Press), 487–539.
Beeton, S., and Bull, A. T. (1989). Biotransformation and detoxification of T-2 toxin by soil and freshwater bacteria. Appl. Environ. Microbiol. 55, 190–197.
Bejaoui, H., Mathieu, F., Taillandier, P., and Lebrihi, A. (2006). Biodegradation of ochratoxin A by Aspergillus section Nigri species isolated from French grapes: a potential means of ochratoxin A decontamination in grape juices and musts. Fems Microbiol. Lett. 255, 203–208. doi: 10.1111/j.1574-6968.2005.00073.x
Benedetti, R., Nazzi, F., Locci, R., and Firrao, G. (2006). Degradation of fumonisin B1 by a bacterial strain isolated from soil. Biodegradation 17, 31–38. doi: 10.1007/s10532-005-2797-y
Bhat, R., Rai, R. V., and Karim, A. A. (2010). Mycotoxins in food and feed: present status and future concerns. Compr. Rev. Food Sci. Food Safety 9, 57–81. doi: 10.1111/j.1541-4337.2009.00094.x
Binder, E. M. (2007). Managing the risk of mycotoxins in modern feed production. Anim. Feed Sci. Technol. 133, 149–166. doi: 10.1016/j.anifeedsci.2006.08.008
Binder, E. M., and Binder, J. (2004). Strain of Eubacterium that Detoxifies Trichothecenes. US Patent 6, 794–175. (Original patent owner: Erber Aktiengesell Schaft). Washington, DC: United States Patent and Trademark Office.
Blackwell, B. A., Gilliam, J. T., Savard, M. E., Miller, J. D., and Duvick, J. P. (1999). Oxidative deamination of hydrolyzed fumonisin B(1) (AP(1)) by cultures of Exophiala spinifera. Nat. Toxins 7, 31–38.
Bornscheuer, U. T. (2002). Microbial carboxyl esterases: classification, properties and application in biocatalysis. FEMS Microbiol. Rev. 26, 73–81. doi: 10.1111/j.1574-6976.2002.tb00599.x
Broudiscou, L. P., Cornu, A., and Rouzeau, A. (2007). in vitro degradation of 10 mono- and sesquiterpenes of plant origin by caprine rumen micro-organisms. J. Sci. Food Agric. 87, 1653–1658. doi: 10.1002/jsfa.2863
Bruinink, A., and Sidler, C. (1997). The neurotoxic effects of ochratoxin-A are reduced by protein binding but are not affected by I-phenylalanine. Toxicol. Appl. Pharmacol. 146, 173–179. doi: 10.1006/taap.1997.8229
Bullerman, L. B., and Bianchini, A. (2007). Stability of mycotoxins during food processing. Int. J. Food Microbiol. 119, 140–146. doi: 10.1016/j.ijfoodmicro.2007.07.035
Camilo, S. B., Ono, C. J., Ueno, Y., and Hirooka, E. Y. (2000). Anti-Fusarium moniliforme activity and fumonisin biodegradation by corn and silage microflora. Braz. Arch. Biol. Technol. 43, 159–164. doi: 10.1590/S1516-89132000000200004
Cao, J., Zhang, H., Yang, Q., and Ren, R. (2013). Efficacy of Pichia caribbica in controlling blue mold rot and patulin degradation in apples. Int. J. Food Microbiol. 162, 167–173. doi: 10.1016/j.ijfoodmicro.2013.01.007
Castells, M., Ramos, A. J., Sanchis, V., and Marín, S. (2007). Distribution of total aflatoxins in milled fractions of hulled rice. J. Agric. Food Chem. 55, 2760–2764. doi: 10.1021/jf063252d
Castoria, R., Mannina, L., Durán-Patrón, R., Maffei, F., Sobolev, A. P., De Felice, D. V., et al. (2011). Conversion of the mycotoxin patulin to the less toxic desoxypatulinic acid by the biocontrol yeast rhodosporidium kratochvilovae strain LS11. J. Agric. Food Chem. 59, 11571–11578. doi: 10.1021/jf203098v
Chang, X., Wu, Z., Wu, S., Dai, Y., and Sun, C. (2015). Degradation of ochratoxin A by Bacillus amyloliquefaciens ASAG1. Food Addit. Contam. Part A-Chem. Anal. Control Expos. Risk Assess. 32, 564–571. doi: 10.1080/19440049.2014.991948
Chapman, P. J., and Ribbons, D. W. (1976). Metabolism of resorcinylic compounds by bacteria - alternative pathways for resorcinol catabolism in Pseudomonas-Putida. J. Bacteriol. 125, 985–998.
Cheli, F., Pinotti, L., Rossi, L., and Dell'orto, V. (2013). Effect of milling procedures on mycotoxin distribution in wheat fractions: a review. LWT-Food Sci. Technol. 54, 307–314. doi: 10.1016/j.lwt.2013.05.040
Chen, S., Chang, C., Deng, Y., An, S., Dong, Y. H., Zhou, J., et al. (2014). Fenpropathrin biodegradation pathway in bacillus sp DG-02 and its potential for bioremediation of pyrethroid-contaminated soils. J. Agric. Food Chem. 62, 2147–2157. doi: 10.1021/jf404908j
Ciegler, A., Lillehoj, E. B., Peterson, R. E., and Hall, H. H. (1966). Microbial detoxification of aflatoxin. Appl. Microbiol. 14, 934–939.
Clarke, R., Connolly, L., Frizzell, C., and Elliott, C. T. (2014). Cytotoxic assessment of the regulated, co-existing mycotoxins aflatoxin B1, fumonisin B1 and ochratoxin, in single, binary and tertiary mixtures. Toxicon 90, 70–81. doi: 10.1016/j.toxicon.2014.07.019
Creppy, E. E. (2002). Update of survey, regulation and toxic effects of mycotoxins in Europe. Toxicol. Lett. 127, 19–28. doi: 10.1016/S0378-4274(01)00479-9
Cserháti, M., Kriszt, B., Krifaton, C., Szoboszlay, S., Hahn, J., Toth, S., et al. (2013). Mycotoxin-degradation profile of Rhodococcus strains. Int. J. Food Microbiol. 166, 176–185. doi: 10.1016/j.ijfoodmicro.2013.06.002
Cundliffe, E., Cannon, M., and Davies, J. (1974). Mechanism of inhibition of eukaryotic protein-synthesis by trichothecene fungal toxins. Proc. Natl. Acad. Sci. U.S.A. 71, 30–34. doi: 10.1073/pnas.71.1.30
Cundliffe, E., and Davies, J. E. (1977). Inhibition of initiation, elongation, and termination of eukaryotic protein-synthesis by trichothecene fungal toxins. Antimicrob. Agents Chemother. 11, 491–499. doi: 10.1128/AAC.11.3.491
Da Rocha, M. E. B., Freire, F. D. O., Maia, F. B. F., Guedes, M. I. F., and Rondina, D. (2014). Mycotoxins and their effects on human and animal health. Food Control 36, 159–165. doi: 10.1016/j.foodcont.2013.08.021
Das, A., Bhattacharya, S., Palaniswamy, M., and Angayarkanni, J. (2014). Biodegradation of aflatoxin B1 in contaminated rice straw by Pleurotus ostreatus MTCC 142 and Pleurotus ostreatus GHBBF10 in the presence of metal salts and surfactants. World J. Microbiol. Biotechnol. 30, 2315–2324. doi: 10.1007/s11274-014-1657-5
De Bellis, P., Tristezza, M., Haidukowski, M., Fanelli, F., Sisto, A., Mulè, G., et al. (2015). Biodegradation of ochratoxin a by bacterial strains isolated from vineyard soils. Toxins 7, 5079–5093. doi: 10.3390/toxins7124864
de Felice, D. V., Solfrizzo, M., De Curtis, F., Lima, G., Visconti, A., and Castoria, R. (2008). Strains of aureobasidium pullulans can lower ochratoxin A contamination in wine grapes. Phytopathology 98, 1261–1270. doi: 10.1094/PHYTO-98-12-1261
De Mil, T., Devreese, M., Broekaert, N., Fraeyman, S., De Backer, P., and Croubels, S. (2015). in vitro adsorption and in vivo pharmacokinetic interaction between doxycycline and frequently used mycotoxin binders in broiler chickens. J. Agric. Food Chem. 63, 4370–4375. doi: 10.1021/acs.jafc.5b00832
Devreese, M., De Backer, P., and Croubels, S. (2013). Different methods to counteract mycotoxin production and its impact on animal health. Vlaams Diergeneeskundig Tijdschrift 82, 181–190.
Dobritzsch, D., Wang, H., Schneider, G., and Yu, S. (2014). Structural and functional characterization of ochratoxinase, a novel mycotoxin-degrading enzyme. Biochem. J. 462, 441–452. doi: 10.1042/BJ20140382
Dong, X., Jiang, W., Li, C., Ma, N., Xu, Y., and Meng, X. (2015). Patulin biodegradation by marine yeast Kodameae ohmeri. Food Addit. Contam. Part A-Chem. Anal. Control Expos. Risk Assess. 32, 352–360. doi: 10.1080/19440049.2015.1007090
Dridi, F., Marrakchi, M., Gargouri, M., Saulnier, J., Jaffrezic-Renault, N., and Lagarde, F. (2015). Comparison of carboxypeptidase Y and thermolysin for ochratoxin A electrochemical biosensing. Anal. Methods 7, 8954–8960. doi: 10.1039/C5AY01905B
Duvick, J., Maddox, J., and Gilliam, J. (2003). Compositions and Methods for Fumonisin Detoxification. US Patent No. 6538177 (Original patent owner: Pioneer Hi-Bred International, Inc.). Charlotte, NC: United States Patent and Trademark Office.
Duvick, J., Rood, T., Maddox, J., and Gilliam, J. (1998a). “Detoxification of mycotoxins in planta as a strategy for improving grain quality and disease resistance: identification of fumonisin-degrading microbes from maize,” in Molecular Genetics of Host-Specific Toxins in Plant Disease, eds K. Kohmoto and O. Yoder (Dordrecht: Kluwer Academic Publishers), 369–381.
Duvick, J., Rood, T., and Wang, N. (1998b). Fumonisin Detoxification Enzymes. US Patent No. 5716820. (Original patent owner: Pioneer Hi-Bred International, Inc.). United States Patent and Trademark Office.
Eaton, D. L., and Groopman, J. D. (1994). The Toxicology of Aflatoxins: Human Health, Veterinary, and Agricultural Significance. San Diego, CA: Academic Press.
Ehrlich, K. C., and Daigle, K. W. (1987). Protein-Synthesis Inhibition by 8-Oxo-12,13-Epoxytrichothecenes. Biochim. Biophys. Acta 923, 206–213. doi: 10.1016/0304-4165(87)90005-5
El-sharkawy, S., and Abul-hajj, Y. J. (1988). Microbial cleavage of zearalenone. Xenobiotica 18, 365–371. doi: 10.3109/00498258809041672
El-sharkawy, S. H., and Abul-hajj, Y. J. (1988). Microbial transformation of zearalenone.2. Reduction, hydroxylation, and methylation products. J. Org. Chem. 53, 515–519. doi: 10.1021/jo00238a008
Engel, P., Brandt, K. K., Rasmussen, L. H., Ovesen, R. G., and Sorensen, J. (2007). Microbial degradation and impact of Bracken toxin ptaquiloside on microbial communities in soil. Chemosphere 67, 202–209. doi: 10.1016/j.chemosphere.2006.08.025
Engelhardt, G. (2002). Degradation of ochratoxin a and b by the white rot fungus Pleurotus ostreatus. Mycol. Res. 18, 37–43. doi: 10.1007/bf02946138
Engler, K. H., Coker, R. D., and Evans, I. H. (1999). A colorimetric technique for detecting trichothecenes and assessing relative potencies. Appl. Environ. Microbiol. 65, 1854–1857.
Eriksen, G. S., and Pettersson, H. (2004). Toxicological evaluation of trichothecenes in animal feed. Anim. Feed Sci. Technol. 114, 205–239. doi: 10.1016/j.anifeedsci.2003.08.008
Eriksen, G. S., Pettersson, H., Johnsen, K., and Lindberg, J. E. (2002). Transformation of trichothecenes in ileal digesta and faeces from pigs. Arch. Tierernahr. 56, 263–274. doi: 10.1080/00039420214343
Eriksen, G. S., Pettersson, H., and Lundh, T. (2004). Comparative cytotoxicity of deoxynivalenol, nivalenol, their acetylated derivatives and de-epoxy metabolites. Food Chem. Toxicol. 42, 619–624. doi: 10.1016/j.fct.2003.11.006
Eshelli, M., Harvey, L., Edrada-Ebel, R., and Mcneil, B. (2015). Metabolomics of the bio-degradation process of aflatoxin B1 by actinomycetes at an initial pH of 6.0. Toxins 7, 439–456. doi: 10.3390/toxins7020439
Fandohan, P., Ahouansou, R., Houssou, P., Hell, K., Marasas, W. F. O., and Wingfield, M. J. (2006). Impact of mechanical shelling and dehulling on Fusarium infection and fumonisin contamination in maize. Food Addit. Contam. 23, 415–421. doi: 10.1080/02652030500442516
Fandohan, P., Zoumenou, D., Hounhouigan, D. J., Marasas, W. F. O., Wingfield, M. J., and Hell, K. (2005). Fate of aflatoxins and fumonisins during the processing of maize into food products in Benin. Int. J. Food Microbiol. 98, 249–259. doi: 10.1016/j.ijfoodmicro.2004.07.007
Ferenczi, S., Cserháti, M., Krifaton, C., Szoboszlay, S., Kukolya, J., Szoke, Z., et al. (2014). A new ochratoxin A biodegradation strategy using Cupriavidus basilensis or16 strain. PLoS ONE 9:e109817. doi: 10.1371/journal.pone.0109817
Fuchs, E., Binder, E., Heidler, D., and Krska, R. (2000). Characterisation of metabolites after the microbial degradation of A- and B-trichothecenes by BBSH 797. Mycotoxin Res. 16, 66–69. doi: 10.1007/BF02942984
Fuchs, E., Binder, E. M., Heidler, D., and Krska, R. (2002). Structural characterization of metabolites after the microbial degradation of type A trichothecenes by the bacterial strain BBSH 797. Food Addit. Contam. 19, 379–386. doi: 10.1080/02652030110091154
Fuchs, S., Sontag, G., Stidl, R., Ehrlich, V., Kundi, M., and Knasmüeller, S. (2008). Detoxification of patulin and ochratoxin A, two abundant mycotoxins, by lactic acid bacteria. Food Chem. Toxicol. 46, 1398–1407. doi: 10.1016/j.fct.2007.10.008
Gelderblom, W. C. A., Jaskiewicz, K., Marasas, W. F. O., Thiel, P. G., Horak, R. M., Vleggaar, R., et al. (1988). Fumonisins - novel mycotoxins with cancer-promoting activity produced by Fusarium-moniliforme. Appl. Environ. Microbiol. 54, 1806–1811.
Gerding, J., Cramer, B., and Humpf, H. U. (2014). Determination of mycotoxin exposure in Germany using an LC-MS/MS multibiomarker approach. Mol. Nutr. Food Res. 58, 2358–2368. doi: 10.1002/mnfr.201400406
Groseclose, E. E., and Ribbons, D. W. (1981). Metabolism of resorcinylic compounds by bacteria - new pathway for resorcinol catabolism in Azotobacter-vinelandii. J. Bacteriol. 146, 460–466.
Grove, M. D., Plattner, R. D., and Peterson, R. E. (1984). Detection of aflatoxin D1 in ammoniated corn by mass-spectrometry mass-spectrometry. Appl. Environ. Microbiol. 48, 887–889.
Guan, S., He, J. W., Young, J. C., Zhu, H. H., Li, X. Z., Ji, C., et al. (2009). Transformation of trichothecene mycotoxins by microorganisms from fish digesta. Aquaculture 290, 290–295. doi: 10.1016/j.aquaculture.2009.02.037
Guan, S., Ji, C., Zhou, T., Li, J. X., Ma, Q., and Niu, T. (2008). Aflatoxin B-1 degradation by Stenotrophomonas maltophilia and other microbes selected using coumarin medium. Int. J. Mol. Sci. 9, 1489–1503. doi: 10.3390/ijms9081489
Guengerich, F. P., Johnson, W. W., Ueng, Y. F., Yamazaki, H., and Shimada, T. (1996). Involvement of cytochrome P450, glutathione S-transferase, and epoxide hydrolase in the metabolism of aflatoxin B-1 and relevance to risk of human liver cancer. Environ. Health Perspect. 104, 557–562.
Hahn, I., Kunz-Vekiru, E., Twaruzek, M., Grajewski, J., Krska, R., and Berthiller, F. (2015). Aerobic and anaerobic in vitro testing of feed additives claiming to detoxify deoxynivalenol and zearalenone. Food Addit. Contam. Part A-Chem. Anal. Control Expos. Risk Assess. 32, 922–933. doi: 10.1080/19440049.2015.1023741
Hartinger, D., Heinl, S., Grabherr, R., Schatzmayr, G., and Moll, W. D. (2009). Heterologous expression of genes from the fumonisin degradation gene cluster of Sphingomonas spp. MTA144 and activity of the catabolic enzymes. New Biotechnol. 25, S132–S133. doi: 10.1016/j.nbt.2009.06.444
Hathout, A. S., and Aly, S. E. (2014). Biological detoxification of mycotoxins: a review. Ann. Microbiol. 64, 905–919. doi: 10.1007/s13213-014-0899-7
He, J. W. (2015). Detoxification of Deoxynivalenol by a Soil Bacterium Devosia mutans 17-2-E-8. Ph.D. thesis, The University of Guelph, Guelph.
He, J. W., Zhou, T., Young, J. C., Boland, G. J., and Scott, P. A. (2010). Chemical and biological transformations for detoxification of trichothecene mycotoxins in human and animal food chains: a review. Trends Food Sci. Technol. 21, 67–76. doi: 10.1016/j.tifs.2009.08.002
Heinl, S., Hartinger, D., Moll, W. D., Schatzmayr, G., and Grabherr, R. (2009). Identification of a fumonisin B1 degrading gene cluster in Sphingomonas spp. MTA144. New Biotechnol. 25, S61–S62. doi: 10.1016/j.nbt.2009.06.290
Heinl, S., Hartinger, D., Thamhesl, M., Schatzmayr, G., Moll, W. D., and Grabherr, R. (2011). An aminotransferase from bacterium ATCC 55552 deaminates hydrolyzed fumonisin B-1. Biodegradation 22, 25–30. doi: 10.1007/s10532-010-9371-y
Heinl, S., Hartinger, D., Thamhesl, M., Vekiru, E., Krska, R., Schatzmayr, G., et al. (2010). Degradation of fumonisin B-1 by the consecutive action of two bacterial enzymes. J. Biotechnol. 145, 120–129. doi: 10.1016/j.jbiotec.2009.11.004
Hormisch, D., Brost, I., Kohring, G. W., Giffhorn, E., Kroppenstedt, R. M., Stackebrandt, E., et al. (2004). Mycobacterium fluoranthenivorans sp nov., a fluoranthene and aflatoxin B-1 degrading bacterium from contaminated soil of a former coal gas plant. Syst. Appl. Microbiol. 27, 653–660. doi: 10.1078/0723202042369866
Hussein, H. S., and Brasel, J. M. (2001). Toxicity, metabolism, and impact of mycotoxins on humans and animals. Toxicology 167, 101–134. doi: 10.1016/S0300-483X(01)00471-1
Huwig, A., Freimund, S., Kappeli, O., and Dutler, H. (2001). Mycotoxin detoxication of animal feed by different adsorbents. Toxicol. Lett. 122, 179–188. doi: 10.1016/S0378-4274(01)00360-5
Hwang, C. A., and Draughon, F. A. (1994). Degradation of ochratoxin-a by Acinetobacter-calcoaceticus. J. Food Prot. 57, 410–414.
IARC (2002). IARCMonographs on the Evaluation of Carcinogenic Risks to Humans: Aflatoxins. Lyon: IARC Press.
Ikunaga, Y., Sato, I., Grond, S., Numaziri, N., Yoshida, S., Yamaya, H., et al. (2011). Nocardioides sp. strain WSN05-2, isolated from a wheat field, degrades deoxynivalenol, producing the novel intermediate 3-epi-deoxynivalenol. Appl. Microbiol. Biotechnol. 89, 419–427. doi: 10.1007/s00253-010-2857-z
Jard, G., Liboz, T., Mathieu, F., Guyonvarc'h, A., and Lebrihi, A. (2011). Review of mycotoxin reduction in food and feed: from prevention in the field to detoxification by adsorption or transformation. Food Addit. Contam. Part A-Chem. Anal. Control Expos. Risk Assess. 28, 1590–1609. doi: 10.1080/19440049.2011.595377
Jiang, L. Y., Yang, J., and Chen, J. M. (2010). Isolation and characteristics of 17 beta-estradiol-degrading Bacillus spp. strains from activated sludge. Biodegradation 21, 729–736. doi: 10.1007/s10532-010-9338-z
Jin, H., Li, B., Peng, X., and Chen, L. M. (2014). Metagenomic analyses reveal phylogenetic diversity of carboxypeptidase gene sequences in activated sludge of a wastewater treatment plant in Shanghai, China. Ann. Microbiol. 64, 689–697. doi: 10.1007/s13213-013-0704-z
Johnsen, H., Odden, E., Lie, O., Johnsen, B. A., and Fonnum, F. (1986). Metabolism of T-2 toxin by rat-liver carboxylesterase. Biochem. Pharmacol. 35, 1469–1473. doi: 10.1016/0006-2952(86)90111-5
Kakeya, H., Takahashi-Ando, N., Kimura, M., Onose, R., Yamaguchi, I., and Osada, H. (2002). Biotransformation of the mycotoxin, zearalenone, to a non-estrogenic compound by a fungal strain of Clonostachys sp. Biosci. Biotechnol. Biochem. 66, 2723–2726. doi: 10.1271/bbb.66.2723
Karabulut, S., Paytakov, G., and Leszczynski, J. (2014). Reduction of aflatoxin B1 to aflatoxicol: a comprehensive DFT study provides clues to its toxicity. J. Sci. Food Agric. 94, 3134–3140. doi: 10.1002/jsfa.6663
Kaushik, G. (2015). Effect of processing on mycotoxin content in grains. Crit. Rev. Food Sci. Nutr. 55, 1672–1683. doi: 10.1080/10408398.2012.701254
Khatibi, P. A., Berger, G., Wilson, J., Brooks, W. S., McMaster, N., Griffey, C. A., et al. (2014). A comparison of two milling strategies to reduce the mycotoxin deoxynivalenol in barley. J. Agric. Food Chem. 62, 4204–4213. doi: 10.1021/jf501208x
Kimura, M., Tokai, T., Takahashi-Ando, N., Ohsato, S., and Fujimura, M. (2007). Molecular and genetic studies of Fusarium trichothecene biosynthesis: pathways, genes, and evolution. Biosci. Biotechnol. Biochem. 71, 2105–2123. doi: 10.1271/bbb.70183
Klaric, M. S., Rašic, D., and Peraica, M. (2013). Deleterious effects of mycotoxin combinations involving ochratoxin A. Toxins 5, 1965–1987. doi: 10.3390/toxins5111965
Kobayashi, M., Shinohara, M., Sakoh, C., Kataoka, M., and Shimizu, S. (1998). Lactone-ring-cleaving enzyme: genetic analysis, novel RNA editing, and evolutionary implications. Proc. Natl. Acad. Sci. U.S.A. 95, 12787–12792. doi: 10.1073/pnas.95.22.12787
Kolosova, A., and Stroka, J. (2011). Substances for reduction of the contamination of feed by mycotoxins: a review. World Mycotoxin J. 4, 225–256. doi: 10.3920/WMJ2011.1288
Krifaton, C., Kriszt, B., Risa, A., Szoboszlay, S., Cserháti, M., Harkai, P., et al. (2013). Application of a yeast estrogen reporter system for screening zearalenone degrading microbes. J. Hazard. Mater. 244, 429–435. doi: 10.1016/j.jhazmat.2012.11.063
Krifaton, C., Kriszt, B., Szoboszlay, S., Cserháti, M., Szucs, A., and Kukolya, J. (2011). Analysis of aflatoxin-B1-degrading microbes by use of a combined toxicity-profiling method. Mutat. Res. 726, 1–7. doi: 10.1016/j.mrgentox.2011.07.011
Kriszt, R., Krifaton, C., Szoboszlay, S., Cserháti, M., Kriszt, B., Kukolya, J., et al. (2012). A new zearalenone biodegradation strategy using non-pathogenic rhodococcus pyridinivorans K408 strain. PLoS ONE 7:e43608. doi: 10.1371/journal.pone.0043608
Larkin, M. J., Kulakov, L. A., and Allen, C. C. R. (2005). Biodegradation and Rhodococcus - masters of catabolic versatility. Curr. Opin. Biotechnol. 16, 282–290. doi: 10.1016/j.copbio.2005.04.007
Lee, L. S., Dunn, J. J., DeLucca, A. J., and Ciegler, A. (1981). Role of lactone ring of aflatoxin-B1 in toxicity and mutagenicity. Experientia 37, 16–17. doi: 10.1007/BF01965543
Li, Y., Wang, Z., Beier, R. C., Shen, J. Z., De Smet, D., De Saeger, S., et al. (2011). T-2 toxin, a trichothecene mycotoxin: review of toxicity, metabolism, and analytical methods. J. Agric. Food Chem. 59, 3441–3453. doi: 10.1021/jf200767q
Liang, Z., Li, J., He, Y., G, S., Wang, N., Ji, C., et al. (2008). AFB1 bio-degradation by a new strain - Stenotrophomonas. sp. Agric. Sci. China 7, 1433–1437. doi: 10.1016/S1671-2927(08)60399-5
Liu, D. L., Ma, L., Gu, L. Q., Liang, R., Yao, D. S., and Chen, W. Q. (1998a). Armillariella tabescen enzymatic detoxification of aflatoxin B-1 - Part III. Immobilized enzymatic detoxification. Enzyme Eng. 864, 592–599. doi: 10.1111/j.1749-6632.1998.tb10387.x
Liu, D. L., Yao, D. S., Liang, R., Ma, L., Cheng, W. Q., and Gu, L. Q. (1998b). Detoxification of aflatoxin B-1 by enzymes isolated from Armillariella tabescens. Food Chem. Toxicol. 36, 563–574. doi: 10.1016/S0278-6915(98)00017-9
Liu, D. L., Yao, D. S., Liang, Y. Q., Zhou, T. H., Song, Y. P., Zhao, L., et al. (2001). Production, purification, and characterization of an intracellular aflatoxin-detoxifizyme from Armillariella tabescens (E-20). Food Chem. Toxicol. 39, 461–466. doi: 10.1016/S0278-6915(00)00161-7
Mann, R., and Rehm, H. J. (1976). Degradation products from aflatoxin-B1 by corynebacterium-rubrum, aspergillus-niger, trichoderma-viride and mucor-ambiguus. (Boca Raton: USA) Eur. J. Appl. Microbiol. 2, 297–306. doi: 10.1007/BF01278613
Marroquín-Cardona, A. G., Johnson, N. M., Phillips, T. D., and Hayes, A. W. (2014). Mycotoxins in a changing global environment - a review. Food Chem. Toxicol. 69, 220–230. doi: 10.1016/j.fct.2014.04.025
Martínková, L., Uhnáková, B., Pátek, M., Nesvera, J., and Kren, V. (2009). Biodegradation potential of the genus Rhodococcus. Environ. Int. 35, 162–177. doi: 10.1016/j.envint.2008.07.018
Matsushima, T., Okamoto, E., Miyagawa, E., Matsui, Y., Shimizu, H., and Asano, K. (1996). Deacetylation of diacetoxyscirpenol to 15-acetoxyscirpenol by rumen bacteria. J. Gen. Appl. Microbiol. 42, 225–234. doi: 10.2323/jgam.42.225
Matthies, I., Woerfel, G., and Karlovsky, P. (2001). Induction of a zearalenone degrading enzyme caused by the substrate and its derivatives. Mycotoxin Res. 17(Suppl. 1), 28–31. doi: 10.1007/BF03036706
Matumba, L., Van Poucke, C., Ediage, E. N., Jacobs, B., and De Saeger, S. (2015). Effectiveness of hand sorting, flotation/washing, dehulling and combinations thereof on the decontamination of mycotoxin-contaminated white maize. Food Addit. Contam. Part A-Chem. Anal. Control Expos. Risk Assess. 32, 960–969. doi: 10.1080/19440049.2015.1029535
McCormick, S. P. (2013). Microbial detoxification of mycotoxins. J. Chem. Ecol. 39, 907–918. doi: 10.1007/s10886-013-0321-0
McCormick, S. P., Price, N. P. J., and Kurtzman, C. P. (2012). Glucosylation and other biotransformations of t-2 toxin by yeasts of the trichomonascus clade. Appl. Environ. Microbiol. 78, 8694–8702. doi: 10.1128/AEM.02391-12
McCormick, S. P., Stanley, A. M., Stover, N. A., and Alexander, N. J. (2011). Trichothecenes: from simple to complex mycotoxins. Toxins 3, 802–814. doi: 10.3390/toxins3070802
Megharaj, M., Garthwaite, I., and Thiele, J. H. (1997). Total biodegradation of the oestrogenic mycotoxin zearalenone by a bacterial culture. Lett. Appl. Microbiol. 24, 329–333. doi: 10.1046/j.1472-765X.1997.00053.x
Méndez-Albores, A., Arámbula-Villa, G., Loarea-Piña, M. G. F., Castaño-Tostado, E., and Moreno-Martínez, E. (2005). Safety and efficacy evaluation of aqueous citric acid to degrade B-aflatoxins in maize. Food Chem. Toxicol. 43, 233–238. doi: 10.1016/j.fct.2004.09.009
Merrill, A. H., Vanechten, G., Wang, E., and Sandhoff, K. (1993a). Fumonisin-B(1) inhibits sphingosine (sphinganine) N-acyltransferase and de-novo sphingolipid biosynthesis in cultured neurons in-situ. J. Biol. Chem. 268, 27299–27306.
Merrill, A. H. Jr., Wang, E., Gilchrist, D. G., and Riley, R. T. (1993b). Fumonisins and other inhibitors of de-novo sphingolipid biosynthesis. Adv. Lipid Res. 26, 215–234.
Moll, W.-D., Hartinger, D., Griebler, K., Binder, E. M., and Schatzmayr, D. (2014). Method for the Production of an Additive for the Enzymatic Decomposition of Mycotoxins, Additive, and Use Thereof. US Patent No. 8703460 (Original patent owner: Erber Aktiengesell Schaft). Washington, DC: United States Patent and Trademark Office.
Molnar, O., Schatzmayr, G., Fuchs, E., and Prillinger, H. (2004). Trichosporon mycotoxinivorans sp nov., a new yeast species useful in biological detoxification of various mycotoxins. Syst. Appl. Microbiol. 27, 661–671. doi: 10.1078/0723202042369947
Moss, M. O., and Long, M. T. (2002). Fate of patulin in the presence of the yeast Saccharomyces cerevisiae. Food Addit. Contam. 19, 387–399. doi: 10.1080/02652030110091163
Motomura, M., Toyomasu, T., Mizuno, K., and Shinozawa, T. (2003). Purification and characterization of an aflatoxin degradation enzyme from Pleurotus ostreatus. Microbiol. Res. 158, 237–242. doi: 10.1078/0944-5013-00199
Norred, W. P., Plattner, R. D., Dombrink-kurtzman, M. A., Meredith, F. I., and Riley, R. T. (1997). Mycotoxin-induced elevation of free sphingoid bases in precision-cut rat liver slices: specificity of the response and structure-activity relationships. Toxicol. Appl. Pharmacol. 147, 63–70. doi: 10.1006/taap.1997.8272
Norred, W. P., Voss, K. A., Bacon, C. W., and Riley, R. T. (1991). Effectiveness of ammonia treatment in detoxification of fumonisin contaminated corn. Food Chem. Toxicol. 29, 815–819. doi: 10.1016/0278-6915(91)90108-J
Ohta, M., Ishii, K., and Ueno, Y. (1977). Metabolism of trichothecene mycotoxins.1. microsomal deacetylation of t-2 toxin in animal-tissues. J. Biochem. 82, 1591–1598.
Patharajan, S., Reddy, K. R. N., Karthikeyan, V., Spadaro, D., Lore, A., Gullino, M. L., et al. (2011). Potential of yeast antagonists on in vitro biodegradation of ochratoxin A. Food Control 22, 290–296. doi: 10.1016/j.foodcont.2010.07.024
Peraica, M., Radic, B., Lucic, A., and Pavlovic, M. (1999). Toxic effects of mycotoxins in humans. Bull. World Health Org. 77, 754–766.
Petchkongkaew, A., Taillandier, P., Gasaluck, P., and Lebrihi, A. (2008). Isolation of Bacillus spp. from Thai fermented soybean (thua-nao): screening for aflatoxin B-1 and ochratoxin A detoxification. J. Appl. Microbiol. 104, 1495–1502. doi: 10.1111/j.1365-2672.2007.03700.x
Péteri, Z., Téren, J., Vágvölgyi, C., and Varga, J. (2007). Ochratoxin degradation and adsorption caused by astaxanthin-producing yeasts. Food Microbiol. 24, 205–210. doi: 10.1016/j.fm.2006.06.003
Pfohl-Leszkowicz, A., and Manderville, R. A. (2007). Ochratoxin A: an overview on toxicity and carcinogenicity in animals and humans. Mol. Nutr. Food Res. 51, 1192–1192. doi: 10.1002/mnfr.200790020
Pointing, S. B. (2001). Feasibility of bioremediation by white-rot fungi. Appl. Microbiol. Biotechnol. 57, 20–33. doi: 10.1007/s002530100745
Popiel, D., Koczyk, G., Dawidziuk, A., Gromadzka, K., Blaszczyk, L., and Chelkowski, J. (2014). Zearalenone lactonohydrolase activity in Hypocreales and its evolutionary relationships within the epoxide hydrolase subset of a/b-hydrolases. BMC Microbiol. 14:82. doi: 10.1186/1471-2180-14-82
Rafiqul, I. (2012). Isolation, Characterization and Genome Sequencing of a Soil-Borne Citrobacter Freundii Strain Capable of Detoxifying Trichothecene Mycotoxins. Ph.D. thesis, The University of Guelph.
Ramos, A. J., Finkgremmels, J., and Hernandez, E. (1996). Prevention of toxic effects of mycotoxins by means of nonnutritive adsorbent compounds. J. Food Prot. 59, 631–641.
Rashamuse, K., Ronneburg, T., Hennessy, F., Visser, D., Van Heerden, E., Piater, L., et al. (2009). Discovery of a novel carboxylesterase through functional screening of a pre-enriched environmental library. J. Appl. Microbiol. 106, 1532–1539. doi: 10.1111/j.1365-2672.2008.04114.x
Reddy, C. A. (1995). The potential for white-rot fungi in the treatment of pollutants. Curr. Opin. Biotechnol. 6, 320–328. doi: 10.1016/0958-1669(95)80054-9
Reddy, K. R. N., Spadaro, D., Gullino, M. L., and Garibaldi, A. (2011). Potential of two Metschnikowia pulcherrima (yeast) strains for in vitro biodegradation of patulin. J. Food Prot. 74, 154–156. doi: 10.4315/0362-028X.JFP-10-331
Riaz, K., Elmerich, C., Moreira, D., Raffoux, A., Dessaux, Y., and Faure, D. (2008). A metagenomic analysis of soil bacteria extends the diversity of quorum-quenching lactonases. Environ. Microbiol. 10, 560–570. doi: 10.1111/j.1462-2920.2007.01475.x
Richard, J. L. (2007). Some major mycotoxins and their mycotoxicoses - an overview. Int. J. Food Microbiol. 119, 3–10. doi: 10.1016/j.ijfoodmicro.2007.07.019
Rios, G., Pinson-Gadais, L., Abecassis, J., Zakhia-Rozis, N., and Lullien-Pellerin, V. (2009). Assessment of dehulling efficiency to reduce deoxynivalenol and Fusarium level in durum wheat grains. J. Cereal Sci. 49, 387–392. doi: 10.1016/j.jcs.2009.01.003
Rocha, O., Ansari, K., and Doohan, F. M. (2005). Effects of trichothecene mycotoxins on eukaryotic cells: a review. Food Addit. Contam. Part A-Chem. Anal. Control Expos. Risk Assess. 22, 369–378. doi: 10.1080/02652030500058403
Rodriguez, H., Reveron, I., Doria, F., Costantini, A., De Las Rivas, B., Munoz, R., et al. (2011). Degradation of ochratoxin a by brevibacterium species. J. Agric. Food Chem. 59, 10755–10760. doi: 10.1021/jf203061p
Samuel, M. S., Aiko, V., Panda, P., and Metha, A. (2013). Aflatoxin B1 occurance, biosynthesis and its degradation. J. Pure Appl. Microbiol. 7, 965–971.
Samuel, M. S., Sivaramakrishna, A., and Mehta, A. (2014). Degradation and detoxification of aflatoxin B1 by Pseudomonas putida. Int. Biodeterior. Biodegradation 86, 202–209. doi: 10.1016/j.ibiod.2013.08.026
Sangare, L., Zhao, Y. J., Folly, Y. M. E., Chang, J. H., Li, J. H., Selvaraj, J. N., et al. (2015). Aflatoxin B-1 degradation by a pseudomonas strain. Toxins 7, 3538–3539. doi: 10.3390/toxins7093538
Sato, I., Ito, M., Ishizaka, M., Ikunaga, Y., Sato, Y., Yoshida, S., et al. (2012). Thirteen novel deoxynivalenol-degrading bacteria are classified within two genera with distinct degradation mechanisms. FEMS Microbiol. Lett. 327, 110–117. doi: 10.1111/j.1574-6968.2011.02461.x
Schatzmayr, G., Heidler, D., Fuchs, E., Mohnl, M., Täubel, M., Loibner, A. P., et al. (2003). Investigation of different yeast strains for the detoxification of ochratoxin A. Mycotoxin Res. 19, 124–128. doi: 10.1007/BF02942950
Schiff, P. L. (2006). Ergot and its alkaloids. Am. J. Pharm. Educ. 70, 98–107. doi: 10.5688/aj700598
Schipper, C., Hornung, C., Bijtenhoorn, P., Quitschau, M., Grond, S., and Streit, W. R. (2009). Metagenome-derived clones encoding two novel lactonase family proteins involved in biofilm inhibition in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 75, 224–233. doi: 10.1128/AEM.01389-08
Scudamore, K. A., Baillie, H., Patel, S., and Edwards, S. G. (2007). Occurrence and fate of Fusarium mycotoxins during commercial processing of oats in the UK. Food Addit. Contam. 24, 1374–1385. doi: 10.1080/02652030701509972
Shier, W. T., Shier, A. C., Xie, W., and Mirocha, C. J. (2001). Structure-activity relationships for human estrogenic activity in zearalenone mycotoxins. Toxicon 39, 1435–1438. doi: 10.1016/S0041-0101(00)00259-2
Shima, J., Takase, S., Takahashi, Y., Iwai, Y., Fujimoto, H., Yamazaki, M., et al. (1997). Novel detoxification of the trichothecene mycotoxin deoxynivalenol by a soil bacterium isolated by enrichment culture. Appl. Environ. Microbiol. 63, 3825–3830.
Shimizu, S., Kataoka, M., Honda, K., and Sakamoto, K. (2001). Lactone-ring-cleaving enzymes of microorganisms: their diversity and applications. J. Biotechnol. 92, 187–194. doi: 10.1016/S0168-1656(01)00359-5
Shimizu, S., Kataoka, M., Shimizu, K., Hirakata, M., Sakamoto, K., and Yamada, H. (1992). Purification and characterization of a novel lactonohydrolase, catalyzing the hydrolysis of aldonate lactones and aromatic lactones, from fusarium-oxysporum. Eur. J. Biochem. 209, 383–390. doi: 10.1111/j.1432-1033.1992.tb17300.x
Singh, B. (2014). Review on microbial carboxylesterase: general properties and role in organophosphate pesticides degradation. Biochem. Mol. Biol. 2, 1–6. doi: 10.12966/bmb.03.01.2014
Soriano, J. M., González, L., and Catala, A. I. (2005). Mechanism of action of sphingolipids and their metabolites in the toxicity of fumonisin B1. Progr. Lipid Res. 44, 345–356. doi: 10.1016/j.plipres.2005.09.001
Stander, M. A., Bornscheuer, U. T., Henke, E., and Steyn, P. S. (2000). Screening of commercial hydrolases for the degradation of ochratoxin A. J. Agric. Food Chem. 48, 5736–5739. doi: 10.1021/jf000413j
Stander, M. A., Steyn, P. S., Van Der Westhuizen, F. H., and Payne, B. E. (2001). A kinetic study into the hydrolysis of the ochratoxins and analogues by carboxypeptidase A. Chem. Res. Toxicol. 14, 302–304. doi: 10.1021/tx000221i
Storm, I. M. L. D., Rasmussen, R. R., and Rasmussen, P. H. (2014). Occurrence of pre- and post-harvest mycotoxins and other secondary metabolites in Danish maize silage. Toxins 6, 2256–2269. doi: 10.3390/toxins6082256
Sudakin, D. L. (2003). Trichothecenes in the environment: relevance to human health. Toxicol. Lett. 143, 97–107. doi: 10.1016/S0378-4274(03)00116-4
Sun, X., He, X., Xue, K., Li, Y., Xu, D., and Qian, H. (2014). Biological detoxification of zearalenone by Aspergillus niger strain FS10. Food Chem. Toxicol. 72, 76–82. doi: 10.1016/j.fct.2014.06.021
Swanson, S. P., Nicoletti, J., Rood, H. D., Buck, W. B., Cote, L. M., and Yoshizawa, T. (1987). Metabolism of 3 trichothecene mycotoxins, t-2 toxin, diacetoxyscirpenol and deoxynivalenol, by bovine rumen microorganisms. J. Chromatogr. 414, 335–342. doi: 10.1016/0378-4347(87)80058-0
Swaving, J., and de Bont, J. A. M. (1998). Microbial transformation of epoxides. Enzyme Microb. Technol. 22, 19–26. doi: 10.1016/S0141-0229(97)00097-5
Takahashi-Ando, N., Ohsato, S., Shibata, T., Hamamoto, H., Yamaguchi, I., and Kimura, M. (2004). Metabolism of zearalenone by genetically modified organisms expressing the detoxification gene from Clonostachys rosea. Appl. Environ. Microbiol. 70, 3239–3245. doi: 10.1128/AEM.70.6.3239-3245.2004
Tan, H., Hu, Y., He, J., Wu, L., Liao, F., Luo, B., et al. (2014). Zearalenone degradation by two Pseudomonas strains from soil. Mycotoxin Res. 30, 191–196. doi: 10.1007/s12550-014-0199-x
Tan, H., Zhang, Z. M., Hu, Y. C., Wu, L., Liao, F., He, J., et al. (2015). Isolation and characterization of Pseudomonas otitidis TH-N1 capable of degrading Zearalenone. Food Control 47, 285–290. doi: 10.1016/j.foodcont.2014.07.013
Täubel, M. (2005). Isolierung und Charakterisierung von Mikroorganismen zur biologischen Inaktivierung von Fumonisinen. Doctoral thesis, University of Natural Resources and Applied Life Sciences, Vienna, Austria.
Tejada-Castañeda, Z. I., Avila-Gonzalez, E., Casaubon-Huguenin, M. T., Cervantes-Olivares, R. A., Vásquez-Peláez, C., Hernandez-Baumgarten, E. M., et al. (2008). Biodetoxification of aflatoxin-contaminated chick feed. Poult. Sci. 87, 1569–1576. doi: 10.3382/ps.2007-00304
Teniola, O. D., Addo, P. A., Brost, I. M., Färber, P., Jany, K. D., Alberts, J. F., et al. (2005). Degradation of aflatoxin B-1 by cell-free extracts of Rhodococcus erythropolis and Mycobacterium fluoranthenivorans sp. nov DSM44556(T). Int. J. Food Microbiol. 105, 111–117. doi: 10.1016/j.ijfoodmicro.2005.05.004
Thamhesl, M., Apfelthaler, E., Schwartz-Zimmermann, H. E., Kunz-Vekiru, E., Krska, R., and Kneifel, W. et al. (2015). Rhodococcus erythropolis MTHt3 biotransforms ergopeptines to lysergic acid. Bmc Microbiol. 15:73. doi: 10.1186/s12866-015-0407-7
Thompson, W. L., and Wannemacher, R. W. (1986). Structure-function-relationships of 12,13-epoxytrichothecene mycotoxins in cell-culture - comparison to whole animal lethality. Toxicon 24, 985–994. doi: 10.1016/0041-0101(86)90004-8
Tintelnot, T., Tegtmeyer, F., and Klotz, M. (2011). Trichosporon mycotoxinovorans -a sword of Damocles for patients with cystic fibrosis. Mycoses 54, 373. doi: 10.1111/j.1439-0507.2011.02066.x
Tinyiro, S. E., Wokadala, C., Xu, D., and Yao, W. (2011). Adsorption and degradation of zearalenone by bacillus strains. Folia Microbiol. 56, 321–327. doi: 10.1007/s12223-011-0047-8
Tsuge, T., Harimoto, Y., Akimitsu, K., Ohtani, K., Kodama, M., Akagi, Y., et al. (2013). Host-selective toxins produced by the plant pathogenic fungus Alternaria alternata. FEMS Microbiol. Rev. 37, 44–66. doi: 10.1111/j.1574-6976.2012.00350.x
Tuor, U., Winterhalter, K., and Fiechter, A. (1995). Enzymes of white-rot fungi involved in lignin degradation and ecological determinants for wood decay. J. Biotechnol. 41, 1–17. doi: 10.1016/0168-1656(95)00042-O
Ueno, Y., Nakayama, K., Ishii, K., Tashiro, F., Minoda, Y., Omori, T., et al. (1983). Metabolism of T-2-Toxin in Curtobacterium Sp Strain-114-2. Appl. Environ. Microbiol. 46, 120–127.
Vader, J. S., van Ginkel, C. G., Sperling, F. M. G. M., de Jong, J., De Boer, W., De Graaf, J. S., et al. (2000). Degradation of ethinyl estradiol by nitrifying activated sludge. Chemosphere 41, 1239–1243. doi: 10.1016/S0045-6535(99)00556-1
van der Westhuizen, L., Shephard, G. S., Rheeder, J. P., Burger, H. M., Gelderblom, W. C. A., Wild, C. P., et al. (2011). Optimising sorting and washing of home-grown maize to reduce fumonisin contamination under laboratory-controlled conditions. Food Control 22, 396–400. doi: 10.1016/j.foodcont.2010.09.009
Vanheule, A., De Boevre, M., Audenaert, K., Beaert, B., De Saeger, S., and Haesaert, G. (2014). Occurence of Fusarium species and their associated mycotoxins in unprocessed cereals, food and feed products in Belgium. Int. J. Food Microbiol. 181, 28–36. doi: 10.1016/j.ijfoodmicro.2014.04.012
Varga, J., Péteri, Z., Tábori, K., Téren, J., and Vagvolgyi, C. (2005). Degradation of ochratoxin A and other mycotoxins by Rhizopus isolates. Int. J. Food Microbiol. 99, 321–328. doi: 10.1016/j.ijfoodmicro.2004.10.034
Vekiru, E., Hametner, C., Mitterbauer, R., Rechthaler, J., Adam, G., Schatzmayr, G., et al. (2010). Cleavage of zearalenone by Trichosporon mycotoxinivorans to a novel nonestrogenic metabolite. Appl. Environ. Microbiol. 76, 2353–2359. doi: 10.1128/AEM.01438-09
Völkl, A., Vogler, B., Schollenberger, M., and Karlovsky, P. (2004). Microbial detoxification of mycotoxin deoxynivalenol. J. Basic Microbiol. 44, 147–156. doi: 10.1002/jobm.200310353
Voss, K. A., Bacon, C. W., Meredith, F. I., and Norred, W. P. (1996a). Comparative subchronic toxicity studies of nixtamalized and water-extracted Fusarium moniliforme culture material. Food Chem. Toxicol. 34, 623–632. doi: 10.1016/0278-6915(96)00024-5
Voss, K. A., Riley, R. T., Bacon, C. W., Chamberlain, W. J., and Norred, W. P. (1996b). Subchronic toxic effects of Fusarium moniliforme and fumonisin B1 in rats and mice. Nat. Toxins 4, 16–23. doi: 10.1002/19960401NT3
Voss, K. A., Smith, G. W., and Haschek, W. M. (2007). Fumonisins: toxicokinetics, mechanism of action and toxicity. Anim. Feed Sci. Technol. 137, 299–325. doi: 10.1016/j.anifeedsci.2007.06.007
Wang, J. Q., Ogata, M., Hirai, H., and Kawagishi, H. (2011). Detoxification of aflatoxin B-1 by manganese peroxidase from the white-rot fungus Phanerochaete sordida YK-624. FEMS Microbiol. Lett. 314, 164–169. doi: 10.1111/j.1574-6968.2010.02158.x
Wegst, W., and Lingens, F. (1983). Bacterial-degradation of ochratoxin-A. FEMS Microbiol. Lett. 17, 341–344. doi: 10.1111/j.1574-6968.1983.tb00433.x
Westlake, K., Mackie, R. I., and Dutton, M. F. (1987). T-2 toxin metabolism by ruminal bacteria and its effect on their growth. Appl. Environ. Microbiol. 53, 587–592.
Wogan, G. N., Newberne, P. M., and Edwards, G. S. (1971). Structure-activity relationships in toxicity and carcinogenicity of aflatoxins and analogs. Cancer Res. 31, 1936–1942.
Wong, J. J., and Hsieh, D. P. H. (1976). Mutagenicity of aflatoxins related to their metabolism and carcinogenic potential. Proc. Natl. Acad. Sci. U.S.A. 73, 2241–2244. doi: 10.1073/pnas.73.7.2241
Wu, Q., Jezkova, A., Yuan, Z., Pavlikova, L., Dohnal, V., and Kuca, K. (2009). Biological degradation of aflatoxins. Drug Metab. Rev. 41, 1–7. doi: 10.1080/03602530802563850
Xiao, H., Madhyastha, S., Marquardt, R. R., Li, S., Vodela, J. K., Frohlich, A. A., et al. (1996). Toxicity of ochratoxin A, its opened lactone form and several of its analogs: structure-activity relationships. Toxicol. Appl. Pharmacol. 137, 182–192. doi: 10.1006/taap.1996.0071
Yehia, R. S. (2014). Aflatoxin detoxification by manganese peroxidase purified from Pleurotus ostreatus. Braz. J. Microbiol. 45, 127–133. doi: 10.1590/S1517-83822014005000026
Yiannikouris, A., Bertin, G., and Jouany, J. P. (2006). Study of the adsorption capacity of Saccharomyces cerevisiae cell wall components toward mycotoxins and the chemical mechanisms invoked. Mycotoxin Factbook 347–361. doi: 10.3920/978-90-8686-587-1
Yoshimoto, T., Nagai, F., Fujimoto, J., Watanabe, K., Mizukoshi, H., Makino, T., et al. (2004). Degradation of estrogens by Rhodococcus zopfii and Rhodococcus equi isolates from activated sludge in wastewater treatment plants. Appl. Environ. Microbiol. 70, 5283–5289. doi: 10.1128/AEM.70.9.5283-5289.2004
Young, J. C., Zhou, T., Yu, H., Zhu, H., and Gong, J. H. (2007). Degradation of trichothecene mycotoxins by chicken intestinal microbes. Food Chem. Toxicol. 45, 136–143. doi: 10.1016/j.fct.2006.07.028
Yu, C. P., Deeb, R. A., and Chu, K. H. (2013). Microbial degradation of steroidal estrogens. Chemosphere 91, 1225–1235. doi: 10.1016/j.chemosphere.2013.01.112
Yu, C. P., Roh, H., and Chu, K. H. (2007). 17 beta-estradiol-degrading bacteria isolated from activated sludge. Environ. Sci. Technol. 41, 486–492. doi: 10.1021/es060923f
Yu, H., Zhou, T., Gong, J. H., Young, C., Su, X. J., Li, X. Z., et al. (2010). Isolation of deoxynivalenol-transforming bacteria from the chicken intestines using the approach of PCR-DGGE guided microbial selection. BMC Microbiol. 10:182. doi: 10.1186/1471-2180-10-182
Yu, Y. S., Qiu, L. P., Wu, H., Tang, Y. Q., Lai, F. R., and Yu, Y. G. (2011a). Oxidation of zearalenone by extracellular enzymes from Acinetobacter sp SM04 into smaller estrogenic products. World J. Microbiol. Biotechnol. 27, 2675–2681. doi: 10.1007/s11274-011-0741-3
Yu, Y. S., Qiu, L., Wu, H., Tang, Y., Yu, Y., Li, X., et al. (2011b). Degradation of zearalenone by the extracellular extracts of Acinetobacter sp SM04 liquid cultures. Biodegradation 22, 613–622. doi: 10.1007/s10532-010-9435-z
Zhai, Y., Li, K., Song, J. L., Shi, Y. H., and Yan, Y. C. (2012). Molecular cloning, purification and biochemical characterization of a novel pyrethroid-hydrolyzing carboxylesterase gene from Ochrobactrum anthropi YZ-1. J. Hazard. Mater. 221, 206–212. doi: 10.1016/j.jhazmat.2012.04.031
Zhou, T., Gong, J., Young, J. C., Yu, H., Li, X. Z., Zhu, H., et al. (2007). “Microorganisms isolated from chicken gut can effectively detoxify DON and other trichothecene mycotoxins,” in 26th Centralia Swine Research Update (Ontario).
Zhou, T., He, J., and Gong, J. (2008). Microbial transformation of trichothecene mycotoxins. World Mycotoxin J. 1, 23–30. doi: 10.3920/WMJ2008.x003
Zhu, R., Feussner, K., Wu, T., Yan, F., Karlovsky, P., and Zheng, X. (2015). Detoxification of mycotoxin patulin by the yeast Rhodosporidium paludigenum. Food Chem. 179, 1–5. doi: 10.1016/j.foodchem.2015.01.066
Keywords: detoxification, microorganisms, mycotoxins, biodegradation, metabolite
Citation: Vanhoutte I, Audenaert K and De Gelder L (2016) Biodegradation of Mycotoxins: Tales from Known and Unexplored Worlds. Front. Microbiol. 7:561. doi: 10.3389/fmicb.2016.00561
Received: 29 January 2016; Accepted: 04 April 2016;
Published: 25 April 2016.
Edited by:
Daniela Gwiazdowska, Poznan University of Economics, PolandReviewed by:
Augusto Schrank, Federal University of Rio Grande do Sul, BrazilOzgur Bayram, Maynooth University, National University of Ireland Maynooth, Ireland
Copyright © 2016 Vanhoutte, Audenaert and De Gelder. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Leen De Gelder, bGVlbi5kZWdlbGRlckB1Z2VudC5iZQ==
 Ilse Vanhoutte
Ilse Vanhoutte Kris Audenaert
Kris Audenaert Leen De Gelder
Leen De Gelder