- 1State Key Laboratory of Microbial Technology, School of Life Science, Shandong University, Jinan, China
- 2State Key Laboratory of Microbial Technology, School of Life Science, Shandong University-Helmholtz Institute of Biotechnology, Shandong University, Jinan, China
The lack of selective markers has been a key problem preventing multistep genetic engineering in filamentous fungi, particularly for industrial species such as the lignocellulose degrading Penicillium oxalicum JUA10-1(formerly named as Penicillium decumbens). To resolve this problem, we constructed a genetic manipulation system taking advantage of two established genetic systems: the Cre-loxP system and Tet-on system in P. oxalicum JUA10-1. This system is efficient and convenient. The expression of Cre recombinase was activated by doxycycline since it was controlled by Tet-on system. Using this system, two genes, ligD and bglI, were sequentially disrupted by loxP flanked ptrA. The successful application of this procedure will provide a useful tool for genetic engineering in filamentous fungi. This system will also play an important role in improving the productivity of interesting products and minimizing by-product when fermented by filamentous fungi.
Introduction
Filamentous fungi express and secrete valuable primary and secondary metabolites, playing an important role in biotechnology (Gerngross, 2004; Nevalainen and Peterson, 2014; Chávez et al., 2015). Penicillium species are among the best known important strain in industry. Penicillium species have been used widely for producing various products since 1930s, including penicillins (Nijland et al., 2008; Houbraken et al., 2012; Weber et al., 2012) and various cellulolytic enzymes (Adsul et al., 2007; Jung et al., 2015; Liao et al., 2015; Sharma et al., 2015). More than 250 Penicillium species have been identified currently (Houbraken and Samson, 2011) and lots of interesting metabolites are being explored. P. oxalicum was identified with strong cellulolytic ability and has been investigated for more than 30 years in China. Therefore, an efficient and convenient genetic engineering system should be developed in P. oxalicum. Many genetic engineering methods for gene disruption or insertion rely heavily on selection markers for screening target transformants. In filamentous fungi, several selection marker genes (SMGs) such as hygromycin B resistance gene (hph) (Michielse et al., 2005), pyrithiamine resistance gene (ptrA) (Wang F. Z. et al., 2013), and phleomycin resistance gene (Lecordier et al., 2015) are in use currently. However, the markers cannot satisfy the need for complex genetic engineering in one strain. The limitation has hampered the progress of genetic engineering in filamentous fungi.
This limitation has been resolved by taking advantage of the Cre-loxP system used to recycle SMGs in other species. Cre recombinase (Cre) is a member of the bacteriophage λ-integrase family of recombinases which covalently binds to two 34 bp loxP sites and catalyzes recombination between them (Pinkney et al., 2012; Gaj et al., 2014). Cre-loxP system has been used extensively to study gene function and to remove selection markers in many organisms, including mammals such as mouse (Tang et al., 2002; Chen et al., 2009), plants such as tobacco (Luo et al., 2007) and Arabidopsis thaliana (De Buck et al., 2007). Cre-loxP system is also successfully used in fungi such as Trichoderma reesei (Steiger et al., 2011), Saccharomyces cerevisiae (Yamagishi et al., 2008), and Aspergillus species (Forment et al., 2006). In the Cre-loxP system, the constitutive expression of Cre will affect the recycling of SMGs flanked by loxP. Therefore, controlled expression of Cre is necessary. In Saccharomyces spp., genes can replicate and express autonomously in episomal plasmid without integration into genome, and the plasmid is deleted when grown on non-selective medium (Aoki et al., 2010). However, there is no episomal plasmid reported for filamentous fungi. Steiger et al. used xylanase 1 promoter to control expression of Cre in T. reesei (Steiger et al., 2011). However, Ogasawara et al. (2006) reported that xylanase promoters are induced by many kinds of carbohydrates, including D-xylose, Avicel, L-sorbose, sophorose, xylobiose, xylan, cellulose, and sophorose. Derntl et al. reported that the change of xylanase expression has a significant influence on cellulase production (Derntl et al., 2013). Thus, the method with controlling expression of Cre using xylanase promoters is not suitable for Cre-loxP system in cellulase-producing fungi.
The tetracycline (Tet)-inducible system has been used widely to control the expression of transgene efficiently (Barde et al., 2006; Delerue et al., 2014). In the Tet-on system, the reverse tetracycline transactivator (rtTA) interacts with Tet resistance operon (tetO) in the presence of tetracycline or its derivative doxycyline and activates transcription (Stieger et al., 2009). Tet-on system has been used in many species, including mice (Furth et al., 1994; Delerue et al., 2014); Plasmodium falciparum (Dahl et al., 2006) and fungal species such as Candida albicans (Park and Morschhäuser, 2005), Schizosaccharomyces pombe (Faryar and Gatz, 1992), and A. fumigatus (Vogt et al., 2005). Doxycycline has been used to regulate gene expression successfully in C. albicans (Nakayama et al., 2000) and A. niger (Meyer et al., 2011) and has been reported to be the most effective in regulation of tetracycline-regulated promoters in S. Cerevisiae (Garí et al., 1997). P. oxalicum JUA10-1 has been used for the production of lignocellulytic enzymes with good performance at industrial-scale (Wang M. Y. et al., 2013). However, limited genetic modification work was carried out due to the non-recycle use of selection markers.
In this study, we describe a modified Cre-loxP system under the control of Tet-on system for marker gene excision in P. oxalicum JUA10-1. In this system, the expression of Cre is transient only after induction by doxycycline. Since ptrA can be reused, multiple genetic engineering could be carried out to modify P. oxalicum. This system will provide a platform for genetic engineering in filamentous fungi.
Materials and Methods
Strains and Cultivation Conditions
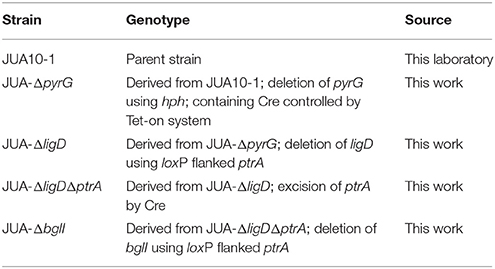
P. oxalicum JUA10-1, which is the catabolite-repression-resistant mutant of P. oxalicum strain 114-2 (CGMCC 5302), is a cellulase hyper-producing strain maintained in our laboratory (Wang M. Y. et al., 2013). All P. oxalicum strains used in this study are listed in Table 1. The modified Mandels' salt solution (MMs) was used as previous described (Liu et al., 2013). The MMs used for cultivation P. oxalicum contained 2% (wt/vol) glucose (GMM), and uridine (10 mM) was added when required. For cellulase protein production of P. oxalicum, the strains were cultivated in liquid MMs added with Avicel (6 g/l), wheat bran (30 g/l), peptone (5 g/l), and tween-80 (3 ml/l).
E.coli strain GB05-dir (Fu et al., 2012) was maintained in Luria-Bertni medium containing appropriate antibiotic (chloramphenicol 15 μg/ml, 100 μg/ml ampicillin) at 37°C.
Plasmids Construction
All primers used in this study are listed in Table 2. PCR amplification was performed using KOD FX DNA polymerase (Toyobo Co., Ltd., Osaka, Japan). The conditions for PCR amplification followed the protocols described by the manufactures.
The Cre expression plasmid pUG6cre (Figure 1A) was constructed by linear-linear homologous recombination mediated by full length RecET in GB05-dir (Fu et al., 2012; Yin et al., 2015). The fragments were obtained as follows. The up- and down-stream of the orotidine 5′-phosphate decarboxylase gene (pyrG, GenBank: GU574814.1) were PCR amplified from P. oxalicum genome using primer pairs G1/G2 and G11/G12, respectively. Hygromycin B resistance gene (hph) with its promoter and terminator was obtained using primer pairs G3/G4 and plasmid pSilent-1 (Nakayashiki, 2005) as template. T. reesei pyruvate kinase gene promoter (Ppki1), which was used to replace PgpdA in Tet-on system, was obtained using primer pairs G5/G6 from T. reesei genome. Tet-on system without PgpdA was amplified by PCR from plasmid pVG2.2 (Ouedraogo et al., 2011) with primer pairs G7/G8, and Cre-encoding sequence was obtained from plasmid pSH65 by PCR with primer pairs G9/G10. The ampicillin resistance gene and replication initiation site was amplified by PCR from pUG6 using primer pairs G13/G14.
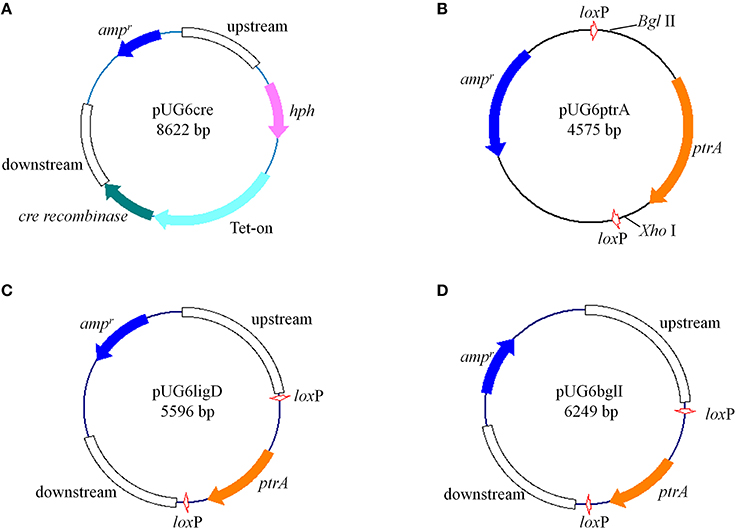
Figure 1. Structures of plasmids constructed in this study (described in “Materials and Methods”) (A) pUG6cre; (B) pUG6ptrA; (C) pUG6ligD; (D) pUG6bglI.
For construction of the loxP-ptrA-loxP cassette, the ptrA gene was inserted into plasmid pUG6 between two loxP sites (Figure 1B). Firstly, the ptrA fragment containing its promoter and terminator was amplified using primers BglIIptrAfw (containing Bgl II recognition sequence) and ptrArevXhoI (containing Xho I recognition sequence). The plasmid pUG6 was then digested with Bgl II-Xho I and the ptrA fragment was ligated into plasmid pUG6 using the corresponding restriction sites, yielding plasmid pUG6ptrA.
The gene encoding P. oxalicum Ligase IV homolog, Ligase D (ligD, GenBank: EPS32530.1) deletion plasmid pUG6ligD (Figure 1C) was constructed based on plasmid pUG6ptrA. The up- and down-stream regions of the ligD were obtained by PCR from P. oxalicum genome with primer pairs L1/L2 and L5/L6, respectively. The ptrA fragment containing two loxP sites were amplified by PCR from plasmid pUG6ptrA with primer pairs L3/L4. The vector fragment containing ampicillin resistance gene and replication initiation site was PCR amplified from pUG6ptrA using primer pairs L7/L8. After purification, the four fragments were used to construct plasmid pUG6ligD in GB05-dir by linear-linear homologous recombination.
The construction of β-glucosidase gene (bglI, GenBank: EU700488.1) deletion plasmid named pUG6bglI (Figure 1D) was similar to the construction of plasmid pUG6ligD. The up- and down-stream regions of the bglI were obtained by PCR from P. oxalicum genome with primer pairs B1/B2 and B5/B6, respectively. The ptrA fragment and the vector fragment were obtained from pUG6ptrA using primer pairs B3/B4 and B7/B8, respectively. The plasmid pUG6bglI was then constructed by linear-linear homologous recombination in GB05-dir.
Transformation of P. oxalicum and Analysis
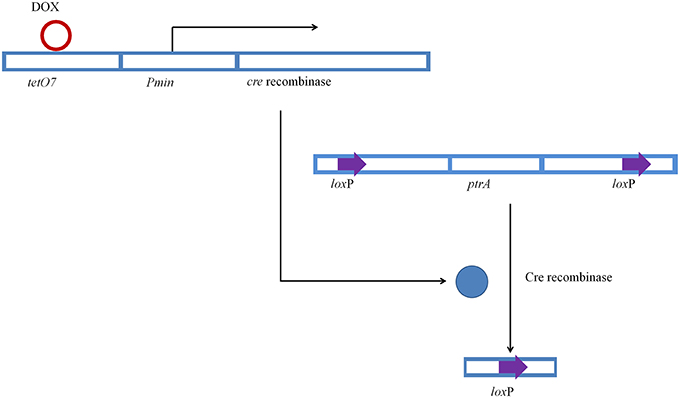
The transformation of P. oxalicum was carried out using the method as previous described (Li et al., 2010). The plasmids, pUG6cre, pUG6ligD, and pUG6bglI, were transformed into P. oxalicum JUA10-1 sequentially. The Cre recombinase under control of Tet-on system and the loxP flanked ptrA were introduced into the P. oxalicum chromosome through homologous recombination method (Figure 2).The plasmid pUG6cre was used to introduce Cre recombinase under control of Tet-on system in site of pyrG through homologous recombination. The plasmid pUG6ligD was used to introduce loxP flanked ptrA in site of ligD through homologous recombination. And the plasmid pUG6bglI was used to introduce loxP flanked ptrA in site of bglI through homologous recombination. After cultivation at 30°C for 5–7 days, the candidate transformants were selected on solid GMM with 10 mM uridine containing 350 μg/ml hygromycin B or 0.3 μg/ml pyrithiamine.
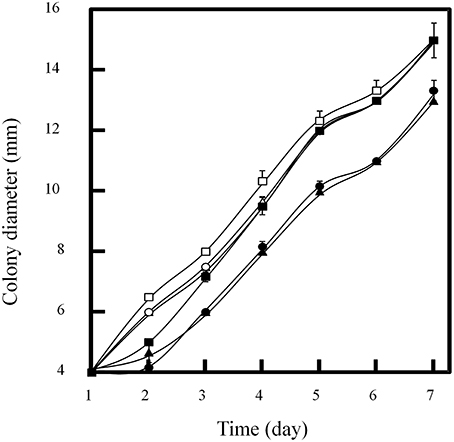
Figure 2. Influence of doxycycline concentrations on colony diameter of P. oxalicum spotted on GMM plate at 30°C. Concentrations of doxycycline (μg/ml): open squar, 0; open triangle, 30; open circle, 50; closed squar, 100; closed triangle, 200; closed circle, 300.
The genomes of candidate transformants were extracted and screened by PCR. To further confirm the validity of the transformants, Southern blotting analysis was performed according to the manufacturer's instruction of a DIG Easy Hyb kit (Roche Diagnostics, Germany). Probe used to analyze deletion of pyrG with primer pairs GS1/GS2 was amplified from P. oxalicum genome. Probe used to analyze disruption of ligD and excision of ptrA was amplified from P. oxalicum genome with primer pairs of LS1/LS2.
Transformants were selected by growth on GMM plate containing 350 μg/ml hygromycin B or 0.3 μg/ml pyrithiamine.
Expression of Cre and Excision of ptrA
Tet-on system induced by the doxycycline was ligated with Cre-encoding sequence for controlled expression of Cre. For expression of Cre, about 100 conidia of the positive transformant containing Cre expression cassette and loxP-ptrA-loxP cassette were inoculated on GMM plate added 50 μg/ml doxycycline (Sigma-Aldrich, St. Louis, MO, USA) for 1 week. The colonies were screened by PCR to test the excision of ptrA.
β-Glucosidase Activity Assay and Protein Quantification
β-glucosidase activity was measured using 5 mM p-nitrophenyl-β- D-glucopyranoside (pNPG) (Sigma-Aldrich, St. Louis, MO, USA) in 5 mM sodium citrate buffer, pH 4.8 as substrate. Fifty microliter substrate and 100 μl sample were incubated at 50°C for 30 min, 150 μl of 10% (wt/vol) Na2CO3 was added to terminate the reaction. Absorbance was read at 420 nm in a microplate using a plate reader (Infinite 200 NanoQuant, Tecan, Zurich, Switzerland).p-nitrophenol was used to prepare a standard curve. One unit (IU) of enzyme activity was defined as the amount of enzyme needed to hydrolyze 1 μmol pNPG in 1 min.
Protein quantification was measured using Bradford with bovine serum albumin (BSA) as standard (Bradford kit, Sangon Biotech, Shanghai, China).
Results
Effects of Doxycycline on the Growth of P. oxalicum
In order to determine the tolerance of P. oxalicum to doxycycline, about 103 conidia were inoculated on GMM plate containing 0–300 μg/ml of doxycycline. The plates were incubated at 30°C for 7 days and the diameters of colonies were measured. There was no significant influence on growth of P. oxalicum when 100 μg/ml of doxycycline was used (Figure 3). However, when the concentration of doxycycline increased to 200 μg/ml, the diameters of colonies decreased from 15 to 13 mm at the 7th day. Therefore, there is no significant influence on P. oxalicum growth when doxycycline under concentration of 100 μg/ml was added.
Introduction of Cre Under Control of Tet-on System in P. oxalicum
The Cre gene expression cassette for deletion pyrG was induced into JUA10-1 with plasmid pUG6cre. The pyrG deletion strain is uridine auxotrophy, thus candidate transformants were grown on GMM plate containing 10 mM uridine. After selection on GMM plate containing 350 μg/ml hygromycin B, the candidate transformants were screened by PCR to detect Cre-encoding sequence with primer pairs G9/G10. We chose one correct strain, called JUA-ΔpyrG, and confirmed the deletion of pyrG in the JUA-ΔpyrG strain using Southern blotting analysis (Figure S1A). The genomes of parent and JUA-ΔpyrG strains were digested by restriction enzyme of Kpn I. Southern blotting analysis showed that the parent chromosome generated a 7.5 kb fragment, while the chromosome of pyrG deletion strain generated a 3.7 kb fragment. The result confirmed that chromosome of JUA-ΔpyrG contains one copy of Cre expression cassette and pyrG was deleted completely.
Deletion of ligD Using loxP Flanked ptrA in P. oxalicum
Furthermore, the ligD gene of JUA-ΔpyrG strain was deleted by loxP flanked ptrA through homologous recombination using plasmid pUG6ligD to improve the homologous recombination frequency with destruction of NHEJ pathway. After selected on GMM plates containing 0.3 μg/ml pyrithiamine and 10 mM uridine, candidate transformants were analyzed by PCR to amplify ptrA fragment with primer pairs L3/L4. We chose one of the correct strains, called JUA-ΔligD, and then confirmed the deletion of ligD by Southern blotting analysis. The genomes of the JUA-ΔpyrG strain and the JUA-ΔligD strain were extracted for Southern blotting analysis. The result of Southern blotting was shown in Figure S1B. After digestion by restriction enzyme of Bgl II, the JUA-ΔpyrG strain generated a 7.1 kb fragment, while the JUA-ΔligD strain generated a 4.6 kb fragment. Our result showed that the JUA-ΔligD strain contained one copy of deletion cassette accompanied by the complete deletion of the ligD allele.
Expression of Cre and Excision of ptrA
Conidia of JUA-ΔligD strain was inoculated on GMM plates containing 50 μg/ml of doxycycline and 10 mM uridine to induce expression of Cre, after which the expressed Cre excised ptrA. We also inoculated JUA-ΔligD strain on GMM plate without doxycycline to test whether the expression of Cre was accurately controlled by Tet-on system. Figure 4 showed the schematic diagram of introduction of Cre and excision of ptrA in JUA-ΔligD strain.
After induction of doxycycline, 20 strains were chosen to extract genomes. PCR with primer pairs L3/L4 was carried out to test the excision of ptrA. The result showed that four of these strains exicised the ptrA. The frequency of excision was about 20%. We chose one of the JUA-ΔligD strains with ptrA excised, and called it JUA-ΔligDΔptrA. Southern blotting analysis was then carried out to further verify deletion of ptrA, as shown in Figure S1C. After digestion of genetic DNA with restriction enzyme of EcoR I, the strain without induction by doxycycline generated an 8.8 kb fragment, while the doxycycline-induced strain generated a 2.8 kb fragment. The result showed that ptrA was completely excised by Cre under the operation of Tet-on system in the presence of doxycycline induction only.
About 100 conidia of JUA-ΔligD and JUA-ΔligDΔptrA strains were inoculated on GMM plates containing 0, 0.3, or 0.6 μg/ml of pyrithiamine and 10 mM uridine. We found that the JUA-ΔligD strains containing ptrA had a normal growth after 5 days cultivation. Meanwhile, the colony numbers of JUA-ΔligDΔptrA on the plates decreased as the concentration of pyrithiamine increased (Figure 5). The result further suggested that the ptrA was excised completely by Cre after induction of doxycycline.
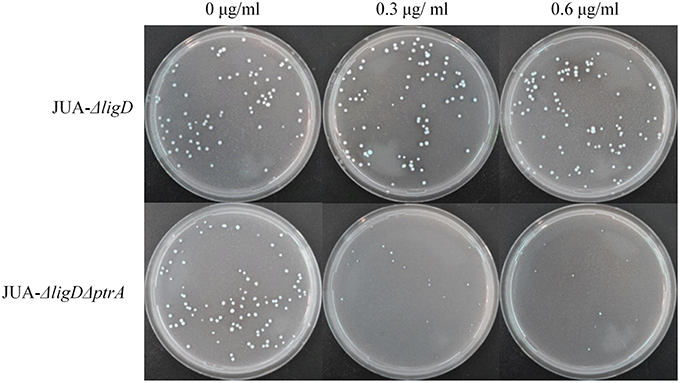
Figure 5. Resistance to pyrithiamine of JUA-ΔligD and JUA-ΔligDΔptrA strains. Concentration of pyrithiamine: 0, 0.3, 0.6 μg/ml.
These results suggested that the expression of Cre is controlled by Tet-on system efficiently.
Reuse of ptrA and Deletion of bglI
After excision of the ptrA, the β-glucosidase (bglI) gene in the JUA-ΔligDΔptrA strain was disrupted by reusing ptrA to testify the recycle use of ptrA because it was easy for measuring the β-glucosidase activity. After transformation with plasmid pUG6bglI, 13 candidate transformants were selected on GMM plates containing 0.3 μg/ml pyrithiamine and 10 mM uridine. PCR analysis was used to test the insertion of ptrA fragment with primer pairs B3/B4 and the result proved that five transformants contained ptrA. Furthermore, another primer pairs B9 (upstream of bglI deletion cassette) and B10 (inside of ptrA) were used to testify the homologous recombination of ptrA using PCR method. The results showed that ptrA was inserted into bglI locus in all five transformants. One correct strain was chosen and called JUA-ΔbglI. Then, we measured the soluble protein concentration and pNPGase activities in JUA-ΔligDΔptrA and JUA-ΔbglI strains at 5, 6, and 7 days. As shown in Figure 6, there was no significant change of soluble protein concentrations between the two strains, while the specific pNPGase activity in JUA-ΔbglI strain was greatly lower than that of JUA-ΔligDΔptrA strain. After cultivation for 7 days, the specific pNPGase activate of JUA-ΔbglI strain was merely 0.034 IU/mg of soluble protein, only 5.8% of that of JUA-ΔligDΔptrA strain (0.58 IU/mg of soluble protein). The result suggested that β-glucosidase encoding gene was disrupted by ptrA successfully.
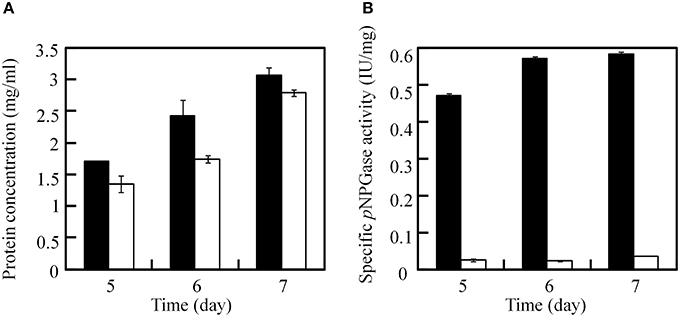
Figure 6. Comparison of soluble protein concentration (A) and specific β-glucosidase activity (B) between JUA-ΔligDΔptrA and JUA-ΔbglI strains. The solid bar represents JUA-ΔligDΔptrA strain, the hollow bar represents JUA-ΔbglI strain.
Discussion
With the broad usage of filamentous fungi, genetic engineering is urgent to modify filamentous fungi, especially for industrial strains. Although many technologies have been employed for genetic engineering for filamentous fungi since the discovery of DNA double helix, only one or two of these technologies are available for a particular species. Recently, the CRSPR-Cas9 system has been used for genetic editing with high efficiency in filamentous fungi. Liu et al. (2015) reported that CRISPR/Cas9 system has been applied for genetic editing in T. reesei and then the promoter of cbh1(Pcbh1) was used to control the expression of Cas9. However, the expression of Cas9 with Pcbh1 has a negative influence on cellulase synthesis. Therefore, it is not suitable for cellulase-producing strains, such as T. reesei and P. oxalicum. Meanwhile, the Tet-on system has no negative effect on cellulase synthesis. Furthermore, the CRSPR/Cas9 system still relies on SMGs for multiple genetic operations, and on the contrary, the Tet-on and Cre-loxP system reduces the limitation of SMGs. In all, the Tet-on and Cre-loxP system still has an advantage in the homologous recombination.
Construction of a system to recycle SMGs is crucial in the extensive engineering of filamentous fungi. The Cre-loxP system has been widely used for filamentous fungi to rescue available SMGs (Mizutani et al., 2012; Zhang et al., 2013; Krappmann, 2014). However, there is no such report of Cre-loxP system used in P. oxalicum. In this paper, we successfully constructed a Cre-loxP system controlled by Tet-on system, and ptrA gene was excised successfully in P. oxalicum for multiple engineering operations.
The accurate control of the expression of Cre is the key in Cre-loxP system to excise the SMGs. During the genetic operation, the constitutive expression of Cre will affect the next gene targeting using loxP flanked marker gene. Therefore, an inducible promoter controlling the expression of Cre should be used. Steiger et al. used xylanase 1 promoter to control expression of Cre in T. reesei (2011). However, the xylanase promoters can be induced by degradation products of lignocelluloses (Ogasawara et al., 2006). Therefore, xylanase promoters are not suitable for cellulase-producing filamentous fungi. Florea et al. used a Cre-expressing plasmid to transiently transfect the target strain (Florea et al., 2009). However, the efficiency was so low that only 0.5–2% of colonies eliminated marker gene with this method. Meanwhile, additional work was needed to transfect the target strain for expression of Cre every time. Zhang et al. constructed a Cre-loxP system to recycle marker gene via anastomosis method in Cryphonectria parasitica (Zhang et al., 2013). In that method, the Cre gene was controlled by a constitutive promoter. One strain was used as the donor to produce Cre. The other strain containing loxP flanked marker gene was used as recipient strain. The SMGs of recipient strain was deleted by Cre from the donor strain with co-cultivation. However, this method was very complicated and time-consuming.
In this study, we used the Tet-on system as the operator to control expression of Cre. The Tet-on system was turned on by induction of doxycycline, and the expression of Cre is activated. After removing doxycycline, the Tet-on system was turned off. This procedure is efficient and convenient. Firstly, the procedure does not need additional transformation for excision of loxP flanked marker gene described as the method by Florea et al. (2009). After cultivation on doxycycline medium, the frequency of excision of ptrA is about 20%, more efficient than the method reported by Florea et al. (2009). And the colonies do not need subculture described as the method by Zhang et al. (2013). Thus, it will be time-saving when extensive genetic engineering work is carried out. Secondly, the expression of Cre is efficiently controlled by the Tet-on system induced by doxycycline. The Tet-on system has been widely used in many species, such as mouse (Milo-Landesman et al., 2001; Giménez et al., 2004), zebrafish (Li et al., 2012), and C. albicans (Oliver et al., 2008). As the inducer of rtTA, the induction of doxycycline has been shown efficiently (Berenjian and Akusjärvi, 2006; Myoung and Ganem, 2011; Chen et al., 2015). Our result showed that Cre was not expressed without induction of doxycycline as shown in Figure S1C. It suggested that the control of Cre-loxP system by Tet-on system was entirely feasible.
In conclusion, we present a Cre-loxP plus Tet-on system for genetic engineering with markerless gene deletion in P. oxalicum. Markerless gene deletion technology is expected to apply for industry strains. The ligD and bglI genes were sequentially knocked out by selection marker ptrA using this system. The ptrA gene was excised by Cre completely and can be reused. Comparing with other Cre-loxP systems in filamentous fungi, this system is efficient and convenient. This modified Cre-loxP system will provide an excellent platform for genetic engineering in filamentous fungi.
Author Contributions
BJ carried out the experiments, analyzed the data, and wrote the draft. XF designed the experiments, analyzed the data, and wrote the manuscript. RZ, DF, FW, KL, YJ, KN, and QY carried out the experiments. MW, HW, and YZ reviewed the paper. All the co-authors have read the manuscript and agreed to publish.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Ms. Shaoli Hou for technical assistance. This work was supported by the National High-Tech R&D Program of China (863 program, No. 2014AA021903), Shandong Scientific and Technology Project (No. 2014CGZH1312), the National Natural Science Foundation of China (No. 31570040), Shandong Province Natural Science Foundation (No. ZR2013CM041), and Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00485
Figure S1. Southern blotting analysis of genes disruption in P. oxalicum. (A) Southern blotting analysis of deletion of pyrG in P. oxalicum. (B) Southern blotting analysis of deletion of ligD. (C) Southern blotting analysis of excision of ptrA.
References
Adsul, M. G., Bastawde, K. B., Varma, A. J., and Gokhale, D. V. (2007). Strain improvement of Penicillium janthinellum NCIM 1171 for increased cellulase production. Bioresour. Technol. 98, 1467–1473. doi: 10.1016/j.biortech.2006.02.036
Aoki, K., Nakajima, R., Furuya, K., and Niki, H. (2010). Novel episomal vectors and a highly efficient transformation procedure for the fission yeast Schizosaccharomyces japonicus. Yeast 27, 1049–1060. doi: 10.1002/syea.1815
Barde, I., Zanta-Boussif, M. A., Paisant, S., Leboeuf, M., Rameau, P., Delenda, C., et al. (2006). Efficient control of gene expression in the hematopoietic system using a single Tet-on inducible lentiviral vector. Mol. Ther. 13, 382–390. doi: 10.1016/j.ymthe.2005.09.012
Berenjian, S., and Akusjärvi, G. (2006). Binary AdEasy vector systems designed for Tet-ON or Tet-OFF regulated control of transgene expression. Virus Res. 115, 16–23. doi: 10.1016/j.virusres.2005.07.001
Chávez, R., Fierro, F., García-Rico, R. O., and Vaca, I. (2015). Filamentous fungi from extreme environments as a promising source of novel bioactive secondary metabolites. Front. Microbiol. 6:903. doi: 10.3389/fmicb.2015.00903
Chen, H., Wang, D., Xia, R., Mao, Q., and Xia, H. (2015). A novel adenoviral vector carrying an all-in-one Tet-On system with an autoregulatory loop for tight, inducible transgene expression. BMC Biotechnol. 15:4. doi: 10.1186/s12896-015-0121-4
Chen, M., Wolfe, A., Wang, X., Chang, C., Yeh, S., and Radovick, S. (2009). Generation and characterization of a complete null estrogen receptor α mouse using Cre/LoxP technology. Mol. Cell Biochem. 321, 145–153. doi: 10.1007/s11010-008-9928-9
Dahl, E. L., Shock, J. L., Shenai, B. R., Gut, J., DeRisi, J. L., and Rosenthal, P. J. (2006). Tetracyclines specifically target the apicoplast of the malaria parasite Plasmodium falciparum. Antimicrob. Agents Chemother. 50, 3124–3131. doi: 10.1128/AAC.00394-06
De Buck, S., Peck, I., De Wilde, C., Marjanac, G., Nolf, J., De Paepe, A., et al. (2007). Generation of single-copy T-DNA transformants in Arabidopsis by the CRE/loxP recombination-mediated resolution system. Plant. Physiol. 145, 1171–1182. doi: 10.1104/pp.107.104067
Delerue, F., White, M., and Ittner, L. M. (2014). Inducible, tightly regulated and non-leaky neuronal gene expression in mice. Transgenic Res. 23, 225–233. doi: 10.1007/s11248-013-9767-7
Derntl, C., Gudynaite-Savitch, L., Calixte, S., White, T., Mach, R. L., and Mach-Aigner, A. R. (2013). Mutation of the Xylanase regulator 1 causes a glucose blind hydrolase expressing phenotype in industrially used Trichoderma strains. Biotechnol. Biofuels 6:62. doi: 10.1186/1754-6834-6-62
Faryar, K., and Gatz, C. (1992). Construction of a tetracycline-inducible promoter in Schizosaccharomyces pombe. Curr. Genet. 21, 345–349. doi: 10.1007/BF00351693
Florea, S., Andreeva, K., Machado, C., Mirabito, P. M., and Schardl, C. L. (2009). Elimination of marker genes from transformed filamentous fungi by unselected transient transfection with a Cre-expressing plasmid. Fungal Genet. Biol. 46, 721–730. doi: 10.1016/j.fgb.2009.06.010
Forment, J. V., Ramón, D., and MacCabe, A. P. (2006). Consecutive gene deletions in Aspergillus nidulans: application of the Cre/loxP system. Curr. Genet. 50, 217–224. doi: 10.1007/s00294-006-0081-2
Fu, J., Bian, X. Y., Hu, S. B., Wang, H. L., Huang, F., Seibert, P. M., et al. (2012). Full-length RecE enhances linear-linear homologous recombination and facilitates direct cloning for bioprospecting. Nat. Biotechnol. 30, 440–446. doi: 10.1038/nbt.2183
Furth, P. A., St Onge, L., Böger, H., Gruss, P., Gossen, M., Kistner, A., et al. (1994). Temporal control of gene expression in transgenic mice by a tetracycline-responsive promoter. Proc. Natl. Acad. Sci. U.S.A. 91, 9302–9306. doi: 10.1073/pnas.91.20.9302
Gaj, T., Sirk, S. J., and Barbas, C. F. (2014). Expanding the scope of site-specific recombinases for genetic and metabolic engineering. Biotechnol. Bioeng. 111, 1–15. doi: 10.1002/bit.25096
Garí, E., Piedrafita, L., Aldea, M., and Herrero, E. (1997). A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast 13, 837–848. doi: 10.1002/(SICI)1097-0061(199707)13:9<837::AID-YEA145>3.0.CO;2-T
Gerngross, T. U. (2004). Advances in the production of human therapeutic proteins in yeasts and filamentous fungi. Nat. Biotechnol. 22, 1409–1414. doi: 10.1038/nbt1028
Giménez, E., Lavado, A., Giraldo, P., Cozar, P., Jeffery, G., and Monliu, L. (2004). A transgenic mouse model with inducible tyrosinase gene expression using the tetracycline (Tet-on) system allows regulated rescue of abnormal chiasmatic projections found in albinism. Pigment Cell Res. 17, 363–370. doi: 10.1111/j.1600-0749.2004.00158.x
Houbraken, J., Frisvad, J. C., Seifert, K. A., Overy, D. P., Tuthill, D. M., Valdez, J. G., et al. (2012). New penicillin-producing Penicillium species and an overview of section Chrysogena. Persoonia 29, 78–100. doi: 10.3767/003158512X660571
Houbraken, J., and Samson, R. A. (2011). Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Stud. Mycol. 70, 1–51. doi: 10.3114/sim.2011.70.01
Jung, Y. R., Park, J. M., Heo, S. Y., Hong, W. K., Lee, S. M., Oh, B. R., et al. (2015). Cellulolytic enzymes produced by a newly isolated soil fungus Penicillium sp. TG2 with potential for use in cellulosic ethanol production. Renew. Energy. 76, 66–71. doi: 10.1016/j.renene.2014.10.064
Krappmann, S. (2014). Genetic surgery in fungi: employing site-specific recombinase for genome manipulation. Appl. Microbial. Biotechnol. 98, 1971–1982. doi: 10.1007/s00253-013-5480-y
Lecordier, L., Uzureau, P., Tebabi, P., Brauner, J., Benghiat, F. S., Vanhollebeke, B., et al. (2015). Adaptation of Trypanosoma rhodesiense to hypohaptoglobinaemic serum requires transcription of the APOL1 resistance gene in a RNA polymerase I locus. Mol. Microbiol. 97, 397–407. doi: 10.1111/mmi.13036
Li, Z., Huang, X. Q., Zhan, H. Q., Zeng, Z. Q., Li, C. X., Spitsbergen, J. M., et al. (2012). Inducible and repressable oncogene-addicted hepatocellular carcinoma in Tet-on xmrk transgenic zebrafish. J. Hepatol. 56, 419–425. doi: 10.1016/j.jhep.2011.07.025
Li, Z. H., Du, C. M., Zhong, Y. H., and Wang, T. H. (2010). Development of a highly efficient gene targeting system allowing rapid genetic manipulations in Penicillium decumbens. Appl. Microbiol. Biotechnol. 87, 1065–1076. doi: 10.1007/s00253-010-2566-7
Liao, H. P., Fan, X. T., Mei, X. L., Wei, Z., Raza, W., Shen, Q. R., et al. (2015). Production and characterization of cellulolytic enzyme from Penicillium oxalicum GZ-2 and its application in lignocellulose saccharification. Biomass Bioenergy 74, 122–134. doi: 10.1016/j.biombioe.2015.01.016
Liu, G. D., Zhang, L., Wei, X. M., Zou, G., Qin, Y. Q., Ma, L., et al. (2013). Genomic and secretomic analyses reveal unique features of the lignocellulolytic enzyme system of Penicillium decumbens. PLoS ONE 8:e55185. doi: 10.1371/journal.pone.0055185
Liu, R., Chen, L., Jiang, Y. P., Zhou, Z. H., and Zou, G. (2015). Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/Cas9 system. Cell Discovery 1:15007. doi: 10.1038/celldisc.2015.7
Luo, K. M., Duan, H., Zhao, D. G., Zheng, X. L., Deng, W., Chen, Y. Q., et al. (2007). ‘GM-gene-deletor’: fused loxP-FRT recognition sequences dramatically improve the efficiency of FLP or Cre on transgene excision from pollen and seed of tobacco plants. Plant Biotechnol. J. 5, 263–374. doi: 10.1111/j.1467-7652.2006.00237.x
Meyer, V., Wanka, F., van Gent, J., Arentshorst, M., van den Hondel, C. A., and Ram, A. F. (2011). Fungal gene expression on demand: an inducible, tunable, and metabolism-independent expression system for Aspergillus niger. Appl. Environ. Microbiol. 77, 2975–2983. doi: 10.1128/AEM.02740-10
Michielse, C. B., Hooykaas, P. J., van den Hondel, C. A., and Ram, A. F. (2005). Agrobacterium-mediated transformation as a tool for functional genomics in fungi. Cur. Genet. 48, 1–17. doi: 10.1007/s00294-005-0578-0
Milo-Landesman, D., Surana, M., Berkovich, I., Compagni, A., Christofori, G., Fleischer, N., et al. (2001). Correction of hyperglycemia in diabetic mice transplanted with reversibly immortalized pancreatic β cells controlled by the tet-on regulatory system. Cell Transplant. 10, 645–650. doi: 10.3727/000000001783986422
Mizutani, O., Masaki, K., Gomi, K., and Iefuji, H. (2012). Modified Cre-loxP recombination in Aspergillus oryzae by direct introduction of Cre recombinase for marker gene rescue. Appl. Environ. Microbiol. 78, 4126–4133. doi: 10.1128/AEM.00080-12
Myoung, J., and Ganem, D. (2011). Generation of a doxycycline-inducible KSHV producer cell line of endothelial origin: maintenance of tight latency with efficient reactivation upon induction. J. Virol. Methods 174, 12–21. doi: 10.1016/j.jviromet.2011.03.012
Nakayama, H., Mio, T., Nagahashi, S., Kokado, M., Arisawa, M., and Aoki, Y. (2000). Tetracycline-regulatable system to tightly control gene expression in the pathogenic fungus Candida albicans. Infect. Immun. 68, 6712–6719. doi: 10.1128/IAI.68.12.6712-6719.2000
Nakayashiki, H. (2005). RNA silencing in fungi: mechanisms and applications. FEBS Lett. 579, 5950–5957. doi: 10.1016/j.febslet.2005.08.016
Nevalainen, H., and Peterson, R. (2014). Making recombinant proteins in filamentous fungi-are we expecting too much? Front. Microbiol. 5:75. doi: 10.3389/fmicb.2014.00075
Nijland, J. G., Kovalchuk, A., van den Berg, M. A., Bovenberg, R. A., and Driessen, A. J. (2008). Expression of the transporter encoded by the cefT gene of Acremonium chrysogenum increases cephalosporin production in Penicillium chrysogenum. Fungal Genet. Biol. 45, 1415–1421. doi: 10.1016/j.fgb.2008.07.008
Ogasawara, W., Shida, Y., Furukawa, T., Shimada, R., Nakagawa, S., Kawamura, M., et al. (2006). Cloning, functional expression and promoter analysis of xylanase III gene from Trichoderma reesei. Appl. Microbiol. Biotechnol. 72, 995–1003. doi: 10.1007/s00253-006-0365-y
Oliver, B. G., Silver, P. M., Marie, C., Hoot, S. J., Leyde, S. E., and White, T. C. (2008). Tetracycline alters drug susceptibility in Candida albicans and other pathogenic fungi. Microbiology 154, 960–970. doi: 10.1099/mic.0.2007/013805-0
Ouedraogo, J. P., Hagen, S., Spielvogel, A., Engelhardt, S., and Meyer, V. (2011). Survival strategies of yeast and filamentous fungi against the antifungal protein AFP. J. Biol. Chem. 286, 13859–13868. doi: 10.1074/jbc.M110.203 588
Park, Y. N., and Morschhäuser, J. (2005). Tetracycline-inducible gene expression and gene deletion in Candida albicans. Eukaryot. Cell 4, 1328–1342. doi: 10.1128/EC.4.8.1328-1342.2005
Pinkney, J. N., Zawadzki, P., Mazuryk, J., Arciszewska, L. K., Sherratt, D. J., and Kapanidis, A. N. (2012). Capturing reaction paths and intermediates in Cre-loxP recombination using single-molecule fluorescence. Proc. Natl. Acad. Sci. U.S.A. 109, 20871–20876. doi: 10.1073/pnas.1211922109
Sharma, B., Agrawal, R., Singhania, R. R., Satlewal, A., Mathur, A., Tuli, D., et al. (2015). Untreated wheat straw: potential source for diverse cellulolytic enzyme secretion by Penicillium janthinellum EMS-UV-8 mutant. Bioresour. Technol. 196, 518–524. doi: 10.1016/j.biortech.2015.08.012
Steiger, M. G., Vitikainen, M., Uskonen, P., Brunner, K., Adam, G., Pakula, T., et al. (2011). Transformation system for Hypocrea jecorina (Trichoderma reesei) that favors homologous integration and employs reusable bidirectionally selection markers. Appl. Environ. Microbiol. 77, 114–121. doi: 10.1128/AEM.02100-10
Stieger, K., Belbellaa, B., Le Guiner, C., Moullier, P., and Rolling, F. (2009). In vivo gene regulation using tetracycline-regulatable systems. Adv. Drug Deliv. Rev. 61, 527–541. doi: 10.1016/j.addr.2008.12.016
Tang, S. H. E., Silva, F. J., Tsark, W. M., and Mann, J. R. (2002). A Cre/loxP-deleter transgenic line in mouse strain 129S1/SvImJ. Genesis 32, 199–202. doi: 10.1002/gene.10030
Vogt, K., Bhabhra, R., Rhodes, J. C., and Askew, D. S. (2005). Doxycycline-regulated gene expression in the opportunistic fungal pathogen Aspergillus fumigates. BMC Microbiol. 5:1. doi: 10.1186/1471-2180-5-1
Wang, F. Z., Liang, Y., Wang, M. Y., Yang, H., Liu, K. M., Zhao, Q. S., and Fang, X. (2013). Functional diversity of the p24γ homologue Erp reveals physiological differences between two filamentous fungi. Fungal Genet. Biol. 61, 15–22. doi: 10.1016/j.fgb.2013.08.017
Wang, M. Y., Mu, Z. M., Wang, J. L., Hou, S. L., Han, L. J., Dong, Y. M., et al. (2013). The identification of and relief from Fe3+ inhibition for both cellulose and cellulase in cellulose saccharification catalyzed by cellulases from Penicillium decumbens. Bioresour. Technol. 133, 507–512. doi: 10.1016/j.biortech.2013.01.172
Weber, S. S., Polli, F., Boer, R., Bovenberg, R. A., and Driessen, A. J. (2012). Increased penicillin production in Penicillium chrysogenum production strains via balanced overexpression of isopenicillin N acyltransferase. Appl. Environ. Microbiol. 78, 7107–7113. doi: 10.1128/AEM.01529-12
Yamagishi, K., Sugiyama, M., Kaneko, Y., Nishizawa, M., and Harashima, S. (2008). Construction and characterization of single-gene chromosomes in Saccharomyces cerevisiae. J. Biosci. Bioeng. 106, 563–567. doi: 10.1263/jbb
Yin, J., Hoffmann, M., Bian, X. Y., Tu, Q., Yan, F., Xia, L. Q., et al. (2015). Direct cloning and heterologous expression of the salinomycin biosynthetic gene cluster from Streptomyces albus DSM41398 in Streptomyces coelicolor A3 (2). Sci. Rep. 5:15081. doi: 10.1038/srep15081
Keywords: selective markers, Penicillium oxalicum, Cre-loxP system, Tet-on system, doxycycline
Citation: Jiang B, Zhang R, Feng D, Wang F, Liu K, Jiang Y, Niu K, Yuan Q, Wang M, Wang H, Zhang Y and Fang X (2016) A Tet-on and Cre-loxP Based Genetic Engineering System for Convenient Recycling of Selection Markers in Penicillium oxalicum. Front. Microbiol. 7:485. doi: 10.3389/fmicb.2016.00485
Received: 27 December 2015; Accepted: 23 March 2016;
Published: 12 April 2016.
Edited by:
Weiwen Zhang, Tianjin University, ChinaReviewed by:
Shuji Tani, Osaka Prefecture University, JapanChun Li, Beijing Institute of Technology, China
Chaoguang Tian, Chinese Academy of Sciences, China
Copyright © 2016 Jiang, Zhang, Feng, Wang, Liu, Jiang, Niu, Yuan, Wang, Wang, Zhang and Fang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xu Fang, ZmFuZ3h1QHNkdS5lZHUuY24=
 Baojie Jiang1
Baojie Jiang1 Quanquan Yuan
Quanquan Yuan Hailong Wang
Hailong Wang Xu Fang
Xu Fang