- 1Department of Veterinary Pharmacy, College of Veterinary Medicine, Northeast Agricultural University, Harbin, China
- 2Department of Biotechnology, Heilongjiang Vocational College for Nationalities, Harbin, China
Streptococcus suis (S.suis) is an important zoonotic pathogen that causes severe diseases in humans and pigs. Biofilms of S. suis can induce persistent infections that are difficult to treat. In this study, the effect of tylosin on biofilm formation of S. suis was investigated. 1/2 minimal inhibitory concentration (MIC) and 1/4 MIC of tylosin were shown to inhibit S. suis biofilm formation in vitro. By using the iTRAQ strategy, we compared the protein expression profiles of S. suis grown with sub-MIC tylosin treatment and with no treatment. A total of 1501 proteins were identified by iTRAQ. Ninety-six differentially expressed proteins were identified (Ratio > ±1.5, p < 0.05). Several metabolism proteins (such as phosphoglycerate kinase) and surface proteins (such as ABC transporter proteins) were found to be involved in biofilm formation. Our results indicated that S. suis metabolic regulation, cell surface proteins, and virulence proteins appear to be of importance in biofilm growth with sub-MIC tylosin treatment. Thus, our data revealed the rough regulation of biofilm formation that may provide a foundation for future research into mechanisms and targets.
Introduction
Streptococcus suis (S. suis) is an important zoonotic pathogen that causes a wide range of diseases in pigs, including meningitis, septicaemia, pneumonia, endocarditis, and arthritis (Gottschalk et al., 2010). In addition, this pathogen is an emerging zoonotic agent and an important public health issue in East and Southeast Asia (Sriskandan and Slater, 2006; Gottschalk et al., 2007). During a single outbreak in China in 2005, more than 200 human cases of S. suis were reported, with a death total of 39 (Yang et al., 2006). Current studies show that S. suis can cause persistent infections by forming biofilms in vivo (Wang et al., 2011).
Biofilms are assemblages of microorganisms characterized by cells that are irreversibly attached to a substratum and embedded in a matrix of self-produced extracellular polymeric substances such as exopolysaccharides (EPS), proteins, nucleic acids and other substances; this type of sessile community-based existence is a critical characteristic for bacterial persistence (Davey and O'Toole, 2000). The physical and biological properties of the biofilm, such as slow growth and mechanical barrier, have a substantial role in the development of increased antimicrobial tolerance. Because the bacteria in chronic infections are aggregated and are in close proximity, genes coding for resistance to antimicrobials can be passed horizontally from one bacterium to the another (Bjarnsholt et al., 2013). The bacteria in biofilms could be 1000-times more difficult to kill by antibiotics than the same organism growing planktonically (Gilbert et al., 1997). Therefore, the control of biofilms is understood to be crucial.
Apart from surgical intervention (when applicable), antibiotics are the main option for the treatment of biofilm infections (Bjarnsholt et al., 2013). Previous studies showed that macrolides successfully inhibited Staphylococcus aureus biofilm formation and reduced its virulence when used at sub-inhibitory concentrations (Fujimura et al., 2008). In addition, a sub-minimal inhibitory concentration of erythromycin can inhibit S. suis biofilm formation (Zhao et al., 2015). Tylosin, a macrolide antibiotic produced by Streptomyces fradiae, is widely used as a veterinary medicine. However, there is still not much effective research of sub-MIC tylosin inhibiting biofilm formation of S. suis in vitro.
At present, most of the available information regarding biofilm formation by drug intervention is based on transcriptomic analyses. However, a limitation of transcriptomic analysis for identifying biofilm-regulated gene network raised concerns among investigators. The method of proteomics is thought to be an essential complement to transcriptomic analysis for discovering key regulators of biofilm (Sauer, 2003). Different immunogenic components of planktonically grown S. suis proteins such as secreted or cell wall-associated proteins had been studied by using immunoproteomic assays (Zhang and Lu, 2007a,b; Geng et al., 2008; Zhang et al., 2008). Additionally, our lab found that quorum-sensing played a crucial role leading to biofilm formation through quantitative proteomic analysis of S. suis biofilm inhibited by sub-MIC erythromycin treatment in vitro (Zhao et al., 2015). However, there are no reports regarding the proteomic analysis of sub-MIC tylosin inhibiting biofilm formation of S. suis in vitro.
We identified several proteins in sub-MIC tylosin inhibiting biofilm formation of S. suis by using iTRAQ technology in this study. The findings from the present study may provide a theoretical foundation for therapy of S. suis biofilm infection and provide references for finding new potential therapeutic targets.
Materials and Methods
Growth of S. suis Planktonic Cells
S. suis (ATCC 700794) was grown in Todd-Hewitt yeast Broth (THB; Summus Ltd., Harbin, Heilongjiang, China) for 16–18 h at 37°C with constant shaking for biofilm assays (Wang et al., 2011).
Observation by Scanning Electron Microscopy (SEM)
Mid-exponential growth phase cultures of S. suis ATCC 700794 were adjusted to an optical density of 0.1 at 600 nm (OD600). Then, 2 mL cultures were transferred to the wells of a 6-well microplate containing an 11 × 11 mm sterilized rough glass slide (Mosutech Co., Ltd., Shanghai, China) on the bottom. After culturing for 72 h at 37°C without shaking, the glass slide was removed with tweezers, and the biofilms on the rough glass slide were washed with sterile PBS. The remaining biofilms were fixed with fixative solution [4% (w/v) paraformaldehyde, 2.5% (w/v) glutaraldehyde, 2 mM CaCl2 in 0.2 M cacodylate buffer, pH 7.2] for 6 h and washed three times with 0.1 M PBS 10 min each, then fixed in 2% osmium tetroxide containing 2 mM potassium ferrocyanide and 6% (w/v) sucrose in cacodylate buffer. The samples were dried, gold sputtered with an ion sputtering instrument (current 15 mA, 2 min) and observed using SEM (FEI Quanta, Netherland).
Effect of Tylosin on Biofilm Formation Determined by the TCP Assay
Mid-exponential growth phase cultures of S. suis were adjusted to 0.2 of OD600. Then, 100 μL of cultures were added to each wells of a 96-well microplate with equal volume of tylosin solution with the final concentrations of 1/2 MIC (0.25 μg/mL), 1/4 MIC (0.125 μg/mL), 1/8 MIC (0.0625 μg/mL), and 1/16 MIC (0.03125 μg/mL), respectively. In addition, a negative control (with THB alone) and a positive control (with bacteria alone) were also included. After incubation at 37°C for 72 h without shaking, the medium was removed by aspiration and the wells were washed three times with sterile physiological saline. The remaining attached bacteria were fixed with 200 μL of 99% methanol (Guoyao Ltd., China) per well, and the plates were emptied after 15 min and left to dry. Then, the plates were stained for 5 min with 200 μL of 2% crystal violet (Guoyao Ltd., China) per well. The excess stain was rinsed off by placing the plate under running tap water. After the plates were air dried, the dye bound to the adherent cells was resolubilized with 200 μL of 33% (v/v) glacial acetic acid (Guoyao Ltd., China) per well. The amount of released stain was quantified by measuring the absorbance at 570 nm with a microplate reader (DG5033A, Huadong Ltd., Nanjing, Jiangsu, China). The reported values are the means of three measurements. The experiments were performed in triplicate.
Colony Forming Unit (CFU) Enumeration
Overnight cultures of S. suis were adjusted to an OD 600 of 0.2. Then, the bacteria were inoculated into 96-well microtiter plate wells containing 200 μL of THB alone (untreated wells) or adding the tylosin solution with the final concentrations of 1/4 MIC (0.125 μ g/mL). In addition, a negative control (with THB alone) and a positive control (with bacteria alone) were also included. After incubation at 37°C for 72 h without shaking, the medium was removed by aspiration, and the wells were washed three times with sterile physiological saline. Biofilm cells were removed from wells by sonication for 5 min in 200 μL of THB. The cell suspensions (n = 3) underwent 10-fold dilutions in THB, and 100 μL of each dilution was spot plated onto THB soft-agar plates and incubated at 37°C for 24 h. All the experiments were performed in triplicate.
Preparation of Protein Extracts
For biofilm cultures, S. suis was grown in THB in 100 mm polystyrene petri dishes at 37°C for 24 h. Then, the supernatant was removed and the dishes were washed twice with Tris-HCl buffer (50 mM, pH 7.5). The biofilms were detached by scraping. After being sonicated for 5 min (Bransonic 220; Branson Consolidated Ultrasonic Pvt. Ltd., Australia), the cells were centrifuged at 12,000 × g for 10 min at 4°C. Then, the cell pellets were washed twice with Tris-HCl buffer (Wang et al., 2012).
Protein Digestion and iTRAQ Labeling
Protein digestion was performed according to the reported FASP procedure (Wisniewski et al., 2009). In brief, 200 μg of proteins at two different conditions (1/4 MIC of tylosin treated cells and nontreated cells) were added into 30 μL STD buffer (4% SDS, 100 mM DTT, 150 mM Tris-HCl pH 8.0) and ultrafiltered (Microcon units, 30 kD) with UA buffer (8 M Urea, 150 mM Tris-HCl pH 8.0). To block reduced cysteine residues, 100 μL 0.05 M iodoacetamide was added into UA buffer and incubated for 20 min in the dark. The filters were washed three times with 100 μL UA buffer and twice with 100 μL DS buffer (50 mM triethylammoniumbicarbonate at pH 8.5). Finally, the proteins were digested with 2 μg trypsin (Promega) in 40 μL DS buffer at 37°C for 16–18 h. Then, the resulting peptides were collected as a filtrate. The peptide content was estimated by UV light spectral density at 280 nm using an extinctions coefficient of 1.1 of 0.1% (g/l) solution calculated on the basis of the frequency of tryptophan and tyrosine in vertebrate proteins.
For the iTRAQ labeling, the peptides were labeled with the 8-plex iTRAQ reagent by following the manufacturer's instructions (Applied Biosystems). Each iTRAQ reagent was dissolved in 70 μL of ethanol and added to the respective peptide mixture. The peptides from the S. suis biofilms treated by tylosin were labeled with 115 isobaric reagent, and the peptides from the nontreated S. suis biofilms were labeled with 116 isobaric reagent. Then, the samples were multiplexed and vacuum dried. Three independent biological experiments were performed.
Peptide Fractionation with Strong Cation Exchange (SCX) Chromatography
SCX chromatography using the AKTA Purifier system (GE Healthcare) was used to fractionate the iTRAQ labeled peptides. After being reconstituted and acidified with 2 mL buffer A (10 mM KH2PO4 in 25% of ACN, pH 2.7), the peptides were loaded onto a PolySULFOETHYL 4.6 × 100 mm column (5 μm, 200 Å, PolyLC Inc., Maryland, U.S.A.). Then, the peptides were eluted at 1 ml/min with a gradient of 0–10% buffer B (500 mM KCl, 10 mM KH2PO4 in 25% of ACN, pH 2.7) for 2 min, 10–20% buffer B for 25 min, 20–45% buffer B for 5 min, and 50–100% buffer B for 5 min. The elution was monitored by absorbance at 214 nm, and the fractions were collected after every 1 min. The collected fractions (~30 fractions) were combined into 10 pools and desalted on C18 Cartridges [Empore™ SPE Cartridges C18 (standard density), bed I.D. 7 mm, volume 3 ml, Sigma]. Each pooled fraction was concentrated by vacuum centrifugation and reconstituted in 40 μl of 0.1% (v/v) trifluoroacetic acid and stored at −80°C for LC-MS/MS analysis.
Liquid Chromatography (LC) Electrospray Ionization (ESI) Tandem Ms (MS/MS) Analysis by Q Exactive
Experiments were performed on a Q Exactive mass spectrometer that was coupled to Easy nLC (Thermo Fisher Scientific). A sample (10 μL) of each fraction was injected for the nano LC-MS/MS analysis. The peptide mixture (5 μg) was loaded onto a C18-reversed phase column (Thermo Scientific Easy Column, 10 cm long, 75 μm inner diameter, 3 μm resin) in buffer A (0.1% formic acid) and separated with a linear gradient of buffer B (80% acetonitrile and 0.1% formic acid) at 250 nl/min controlled by IntelliFlow technology for 140 min. MS data were acquired using a data-dependent top10 method dynamically choosing the most abundant precursor ions from the survey scan (300–1800 m/z) for HCD fragmentation. Determination of the target value was based on predictive Automatic Gain Control (pAGC). The dynamic exclusion duration was 60 s. Survey scans were acquired at a resolution of 70,000 at m/z 200, and the resolution for HCD spectra was set to 17,500 at m/z 200. The normalized collision energy was 30 eV, and the underfill ratio, which specifies the minimum percentage of the target value likely to be reached at maximum fill time, was defined as 0.1%. The instrument was run with the peptide recognition mode enabled.
Sequence Database Searching and Data Analysis
The MS/MS spectra were searched using the MASCOT engine (Matrix Science, London, UK; version 2.2) in the Proteome Discoverer 1.3 (Thermo Electron, San Jose, USA.) against the Uniprot S. suis fasta database (38,369 sequences, downloaded March 4th, 2013) and the decoy database. False discovery rates (FDR) were calculated by running all spectra against a decoy database using the MASCOT software. To identify proteins, the following options were used: Peptide mass tolerance = 20 ppm, MS/MS tolerance = 0.1 Da, Enzyme = Trypsin, Missed cleavage = 2, Fixed modification: Carbamidomethyl (C), iTRAQ 8plex (K), iTRAQ 8plex (N-term), Variable modification: Oxidation (M). The quantification was performed based on the peak intensities of the reporter ions in the MS/MS spectra. The ratio of label 115 and 116 represents the expression of proteins with the protein identification confidence of a 1% FDR (Unwin et al., 2010). The proteins were considered over expressed when the iTRAQ ratio was above 1.5 and under expressed when the iTRAQ ratio was lower than 0.67. The Proteome Discoverer tool was used to categorize the proteins detected by Gene Ontology (GO) annotation according to the cellular component, biological process and molecular function.
Results
Effect of Tylosin against Biofilm Formation In vitro by the TCP Assay
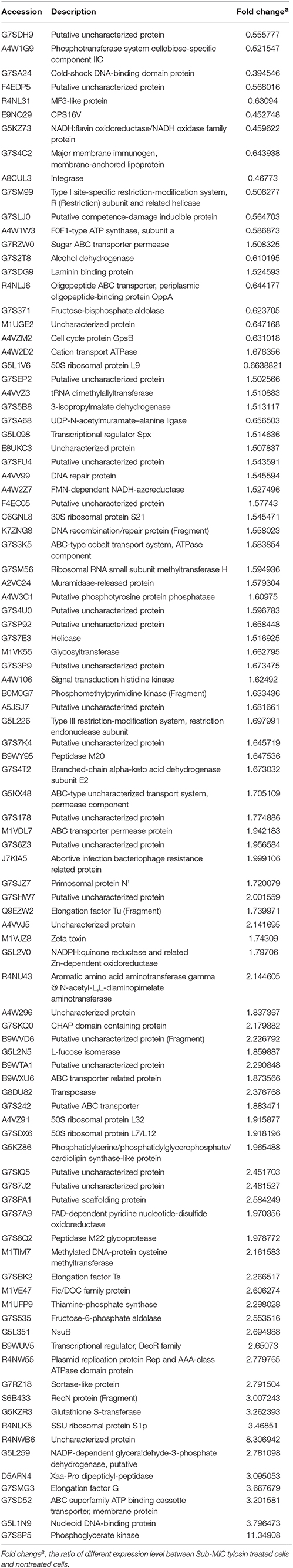
We evaluated the action of tylosin on biofilm growth in vitro. The MIC against S. suis was 0.5 μg·mL−1. Tylosin at 1/2 MIC and 1/4 MIC caused a significantly higher reduction in the biofilm-forming ability of S. suis compared with positive control (p < 0.05). However, there was no pronounced effect for 1/16 MIC and 1/8 MIC of tylosin on biofilm formation of S. suis (p > 0.05; Figure 1).
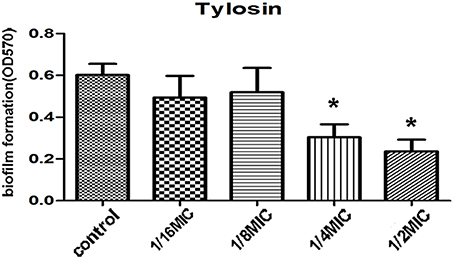
Figure 1. Effect of tylosin at different concentrations on S. suis ATCC 700794 biofilm formation. The data are expressed as the means ± standard deviations. Significant decrease (*p < 0.05) compared with control biofilm formation of S. suis in vitro.
Direct Observation of Biofilm Formation In vitro by Sem
SEM analysis was performed to observe the 1/4 MIC of tylosin treated cells and nontreated cells biofilm formation by S. suis under same growth conditions. As shown in Figure 2A, the surface of the glass slide is almost entirely covered by the aggregates and microcolonies of S. suis when growth was carried out in the culture medium without tylosin. However, when the culture medium was added to 1/4 MIC of tylosin, the biofilms were characterized by the presence of small clusters of cells interspersed amongst individual cells (Figure 2B). This result showed that the biofilm formation of S. suis was inhibited by the tylosin of 1/4 MIC in vitro.
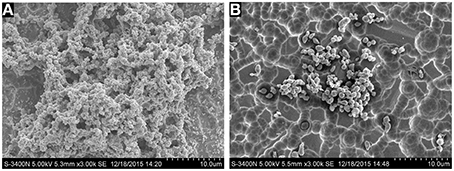
Figure 2. (A) Biofilm formation of S. suis without tylosin. (B) Biofilm formation of S. suis with 1/4 MIC tylosin treatment.
Colony Forming Unit (CFU) Enumeration
To better assess action of tylosin on biofilm quantitatively, the CFUs of S. suis were counted. The viability of S. suis treated with 1/4 MIC of tylosin was different from the viability of untreated S. suis. The number of CFUs/mL in treated biofilms (5.3 log10 CFUs/mL) was significantly fewer than in nontreated biofilms (6.5 log10 CFUs/mL; p < 0.05). The number of CFUs/mL in treated biofilms (5.3 log10 CFUs/mL) was significantly fewer than in nontreated biofilms (6.5 log10 CFUs/mL; p < 0.05) (Figure 3). The findings demonstrated that 1/4 MIC of tylosin remained effective in decreasing the viability of S. suis.
Figure 3. Effect of 1/4 MIC of tylosin on S. suis ATCC 700794 biofilm formation by colony forming unit (CFU) enumeration. The data are expressed as the means ± standard deviations. Significant decrease (*p < 0.05) compared with control biofilm formation of S. suis in vitro.
Sub-MIC Tylosin Inhibits Biofilm Formation and Differentially Expressed Proteins by iTRAQ
A total of 1501 proteins were identified by iTRAQ. Detailed information is shown in the Supplementary File. A ratio of proteins with >1.5 or < 0.67 (p < 0.05) was considered to be differentially expressed. Based on this criterion, 96 differentially expressed proteins were identified in 1/4 MIC tylosin treated cells and nontreated cells. Detailed information could is shown Table 1. These proteins were detected by GO annotation. Thirty-five proteins were deleted from the database. The remaining 61 proteins were classified into biological process, molecular function and cellular component (Figure 4). The results regarding the biological process were as follows: single-organism process (8, 13%), response to stimulus (4, 7%), localization (4, 7%), biological adhesion (1, 2%), cellular component organization or biogenesis (6, 10%), cellular process (25, 41%), biological regulation (3, 5%), metabolic process (30, 49%). The results regarding molecular function were as follows: binding (29, 48%), nucleic acid binding transcription factor activity (1, 2%), transporter activity (3, 5%), structural molecule activity (5, 8%), catalytic activity (25, 41%). The results regarding cellular component were as follows: organelle (6, 10%), virion (1, 2%), cell (13, 21%), extracellular region (1, 2%), membrane (6, 10%), macromolecular complex (8, 13%).
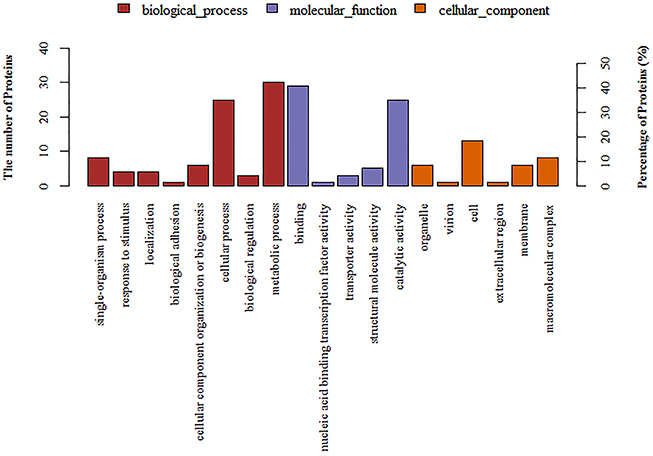
Figure 4. Annotation of differentially regulated protein (ratio >1.5 or < 0.67) functions by Gene Ontology (GO).
Discussion
A TCP assay is based on the ability of bacteria to form biofilms on the bottom of tissue culture plates and is mainly used to identify the formation of bacterial biofilms (Mathur et al., 2006; Okajima et al., 2006; Presterl et al., 2007). Bacteria are grown in cell culture plates, and the quantity of a biofilm is detected by staining the wells with crystal violet according to the correlation between OD values and biofilm formation (Mathur et al., 2006).
To further confirm S. suis biofilm formation, the structure of the S. suis biofilm was identified by scanning electron microscopy (SEM). The biofilm examination relied heavily on SEM. Because of its high magnification, the biofilm microstructure is observed clearly. These findings further confirm that S. suis can form biofilms in vitro. Although these procedures have their advantages, the TCP procedure lacks specificity and sensitivity because crystal violet stains all of the components of the biofilm and could potentially stain non-biofilm material. More specific and sensitive techniques can be performed to assess each of the individual components. This method is good for screening purposes.
Tylosin is a frequently used drug for the treatment of S. suis infection. Upon treatment with this antimicrobial agent, S. suis is inevitably exposed to a sub-inhibitory level of the agent. Therefore, we studied its effect in sub-inhibitory concentrations on S. suis biofilm formation. Tylosin can inhibit S. suis biofilm formation at sub-inhibitory concentrations in a dose-dependent manner. In addition, the inhibition of biofilm formation varied among the antimicrobial agents. Numerous reports have showed biofilm formation in the presence of sub-inhibitory concentrations of antimicrobial agents (Dunne, 1990; Carsenti-Etesse et al., 1993; Rupp and Hamer, 1998; Rachid et al., 2000; Yang et al., 2015). Our results are in agreement with these previous findings. Our results showed a better activity of tylosin against S. suis biofilm formation. However, further studies should be conducted to confirm and clarify the relationship between the relative adherence-inhibiting properties of tylosin and their mechanisms. These results may provide information regarding the clinical use of antimicrobial agents against biofilm-forming bacteria.
Our data identified proteins related to biofilm growth that have previously been uncharacterized. iTRAQ analyses showed that the regulation of metabolism plays a key role during S. suis biofilm growth. First, the iTRAQ quantitative data revealed that carbohydrate metabolism may be particularly important during S. suis biofilm growth. It was reported that fructose bisphosphate aldolase and glyceraldehyde-3-phosphate dehydrogenase were significantly decreased in Streptococcus pneumoniae biofilms (Allan et al., 2014). In our study, there was a >two-fold increase of glyceraldehyde-3-phosphate dehydrogenase in the tylosin inhibiting biofilm formation. In addition, the levels of fructose bisphosphate aldolase (fold change: 0.62) were down-regulated. It was reported that phosphoglycerate kinase was up-regulated in S. suis biofilms compared with planktonic cells by comparative proteomic analysis (Wang et al., 2012). Similarly, biofilms of Pseudomonas aeruginosa (Sauer et al., 2002) and Staphylococcus xylosus (Planchon et al., 2009) exhibit up-regulated phosphoglycerate kinase. There was a >11-fold increase in the level of phosphoglycerate kinase in the tylosin inhibiting biofilm formation. Phosphoglycerate kinase is a glycolytic enzyme that functions in the conversion of glyceraldehyde 3-phosphate into 1, 3-diphosphoglycerate. Glycolysis may play a pivotal role during S. suis biofilm growth. Furthermore, histidine metabolism may play an important role in biofilm formation. Histidine kinase (fold change: 1.62) was up-regulated in the tylosin-inhibited biofilm formation. Compared with other amino acids, L-His had the strongest effect on biofilm induction (Cabral et al., 2011). Histidine metabolism is also found to be involved in biofilm formation and is confirmed by gene disruption (Cabral et al., 2011). Histidine kinase is an important signaling molecule in biofilm formation in gram-positive and negative bacteria (McLoon et al., 2011; Shemesh and Chai, 2013; Yang et al., 2014; Grau et al., 2015). In addition, several ABC transporter system proteins were significantly up-regulated in the tylosin inhibiting biofilm formation. This finding is consistent with the previous results of our laboratory showing that sub-MIC erythromycin inhibits S. suis biofilm formation (Zhao et al., 2015). The carbohydrate substrate selection and fermentation determine ABC transporter proteins were significantly up-regulated in tylosin-treated cells, suggesting that S. suis had the capability of metabolizing a wide range of carbohydrates during biofilm development (Hardy et al., 2001; Marion et al., 2011; Bidossi et al., 2012; Haertel et al., 2012). Moreover, ABC transporters are important because they regulate respiration and biofilm formation, which in turn affect the rate of electricity production and bioremediation (Selvaraj et al., 2014). Furthermore, in the mutants of a pneumococcal biofilm screen, ABC transporters were shown to be defective in colonization (Munoz-Elias et al., 2008).
Cell surface proteins play a crucial role in biofilms. Within the biofilm, bacterial cells are embedded in a self-produced extracellular matrix. This matrix protects bacteria against a number of environmental insults. However, the matrix also confines bacterial access to fresh nutrients. Thus, the verified increase in the expression of transmembrane channels appears to be an essential requirement for the entrance of important nutrient-containing fluid (Costerton et al., 1994). In addition to acting, as channels, porins may act as potential targets for adhesion to other cells and may mediate cell attachment through binding to the proteins released for biofilm formation (Dallo et al., 2010). The outer membrane proteins may mediate cell attachment through binding to the released proteins for biofilm formation. Membrane proteins such as OmpA mediate cell adhesion in Acinetobacter baumannii (Dallo et al., 2010). The key developmental stages of biofilm development have been reported to be adhesion to surfaces, aggregation of micro colonies, and further expansion of the microbial community. The putative involvement of many genes encoding large cell surface proteins is adhesion to the epithelium and biofilm formation (Pridmore et al., 2004; Walter et al., 2005; Frese et al., 2011). Pham (Pham et al., 2010) found increased expression of outer membrane proteins in the Tannerella forsythia biofilm cells by using quantitative non-gel-based proteomic techniques. In our study, the expression of membrane-anchored lipoprotein, phosphoglycerate kinase, sugar ABC transporter permease, ABC superfamily ATP binding cassette transporter membrane protein and L-fucose isomerase changed; these proteins belong to membrane proteins and cell-surface proteins and might be involved in some molecular functions including catalytic activity, motor activity, nucleotide binding, protein binding and transporter activity. For example, phosphoglycerate kinase is a S. suis surface protein that promotes cell adhesion and plays a key role in bacterial infection and invasion (Brassard et al., 2004; Wang and Lu, 2007). We predicted that these membrane proteins might affect the bacterial cell-cell interaction. Thus, membrane proteins might play a significant role in biofilm formation.
S. suis virulence proteins related to infection, persistence and competitive fitness were mostly down regulated when sub-MIC tylosin was used to inhibit biofilm formation. There was a >1.5-fold decrease in the level of NADH oxidase. NADH oxidase regulates competence, virulence, and pneumococcal persistence by its actions as an oxygen sensor, in detoxifying oxygen, and in increasing the efficiency of glucose breakdown (Auzat et al., 1999) and plays an important role in pneumococcal infection in animal models of pneumonia (Yu et al., 2001). NADH oxidase is encoded by nox. The virulence and persistence in mice of a blood isolate was attenuated by a nox insertion mutation (Auzat et al., 1999). Thus, this protein appears to be of major importance in the growth of biofilms.
Conclusions
Our study suggested that sub-MICs of tylosin could inhibit S. suis biofilm formation in vitro. We used a robust and reliable comparative proteomic technique (iTRAQ) to compare the abundances of proteins from 1/4 MIC of tylosin treated cells and nontreated cells. Finally, in sub-MIC tylosin inhibiting cells, we identified 96 differentially expressed proteins when the protein had a fold-change of more than a ratio >1.5 or < 0.67 (p < 0.05). Our proteomic data suggested general changes in metabolism (such as phosphoglycerate kinase) and surface proteins (such as ABC transporter proteins) involved in biofilm formation. Overall, our results indicated that S. suis metabolic regulation, cell-surface proteins, and virulence proteins appear to be of importance in biofilm growth by sub-MIC tylosin treatment. Thus, our data revealed the rough regulation of biofilm formation that might potentially be utilized to manage biofilm infections of S. suis.
Author Contributions
SW the design whole experiment. YL directed the completion of the experiment. YY, YZhao, HZ, JB, JC, YZhou, CW provided help during the experiment.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 31472231) and Guangdong Technology Research Center for Traditional Chinese Veterinary Medicine and Natural Medicine (No.20141020002). We thank Shanghai Applied Protein Technology Co. Ltd. for the help with iTRAQ.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00384
References
Allan, R. N., Skipp, P., Jefferies, J., Clarke, S. C., Faust, S. N., Hall-Stoodley, L., et al. (2014). Pronounced metabolic changes in adaptation to biofilm growth by Streptococcus pneumoniae. PLoS ONE 9:e107015. doi: 10.1371/journal.pone.0107015
Auzat, I., Chapuy-Regaud, S., Le Bras, G., Dos Santos, D., Ogunniyi, A. D., Le Thomas, I., et al. (1999). The NADH oxidase of Streptococcus pneumoniae: its involvement in competence and virulence. Mol. Microbiol. 34, 1018–1028. doi: 10.1046/j.1365-2958.1999.01663.x
Bidossi, A., Mulas, L., Decorosi, F., Colomba, L., Ricci, S., Pozzi, G., et al. (2012). A functional genomics approach to establish the complement of carbohydrate transporters in Streptococcus pneumoniae. PLoS ONE 7:e33320. doi: 10.1371/journal.pone.0033320
Bjarnsholt, T., Ciofu, O., Molin, S., Givskov, M., and Hoiby, N. (2013). Applying insights from biofilm biology to drug development - can a new approach be developed? Nat. Rev. Drug Discov. 12, 791–808. doi: 10.1038/nrd4000
Brassard, J., Gottschalk, M., and Quessy, S. (2004). Cloning and purification of the Streptococcus suis serotype 2 glyceraldehyde-3-phosphate dehydrogenase and its involvement as an adhesin. Vet. Microbiol. 102, 87–94. doi: 10.1016/j.vetmic.2004.05.008
Cabral, M. P., Soares, N. C., Aranda, J., Parreira, J. R., Rumbo, C., Poza, M., et al. (2011). Proteomic and functional analyses reveal a unique lifestyle for Acinetobacter baumannii biofilms and a key role for histidine metabolism. J. Proteome Res. 10, 3399–3417. doi: 10.1021/pr101299j
Carsenti-Etesse, H., Durant, J., Entenza, J., Mondain, V., Pradier, C., Bernard, E., et al. (1993). Effects of subinhibitory concentrations of vancomycin and teicoplanin on adherence of staphylococci to tissue culture plates. Antimicrob. Agents Chemother. 37, 921–923. doi: 10.1128/AAC.37.4.921
Costerton, J. W., Lewandowski, Z., DeBeer, D., Caldwell, D., Korber, D., and James, G. (1994). Biofilms, the customized microniche. J. Bacteriol. 176, 2137–2142.
Dallo, S. F., Denno, J., Hong, S., and Weitao, T. (2010). Adhesion of Acinetobacter baumannii to extracellular proteins detected by a live cell-protein binding assay. Ethn. Dis. 20(1 Suppl. 1):S1-7-11.
Davey, M. E., and O'Toole, G. A. (2000). Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64, 847–867. doi: 10.1128/MMBR.64.4.847-867.2000
Dunne, W. M. Jr. (1990). Effects of subinhibitory concentrations of vancomycin or cefamandole on biofilm production by coagulase-negative staphylococci. Antimicrob. Agents Chemother. 34, 390–393. doi: 10.1128/AAC.34.3.390
Frese, S. A., Benson, A. K., Tannock, G. W., Loach, D. M., Kim, J., Zhang, M., et al. (2011). The evolution of host specialization in the vertebrate gut symbiont Lactobacillus reuteri. PLoS Genet. 7:e1001314. doi: 10.1371/journal.pgen.1001314
Fujimura, S., Sato, T., Mikami, T., Kikuchi, T., Gomi, K., and Watanabe, A. (2008). Combined efficacy of clarithromycin plus cefazolin or vancomycin against Staphylococcus aureus biofilms formed on titanium medical devices. Int. J. Antimicrob. Agents 32, 481–484. doi: 10.1016/j.ijantimicag.2008.06.030
Geng, H., Zhu, L., Yuan, Y., Zhang, W., Li, W., Wang, J., et al. (2008). Identification and characterization of novel immunogenic proteins of Streptococcus suis serotype 2. J. Proteome Res. 7, 4132–4142. doi: 10.1021/pr800196v
Gilbert, P., Das, J., and Foley, I. (1997). Biofilm susceptibility to antimicrobials. Adv. Dent. Res. 11, 160–167. doi: 10.1177/08959374970110010701
Gottschalk, M., Segura, M., and Xu, J. (2007). Streptococcus suis infections in humans: the Chinese experience and the situation in North America. Anim. Health Res. Rev. 8, 29–45. doi: 10.1017/S1466252307001247
Gottschalk, M., Xu, J. G., Calzas, C., and Segura, M. (2010). Streptococcus suis: a new emerging or an old neglected zoonotic pathogen? Future Microbiol. 5, 371–391. doi: 10.2217/fmb.10.2
Grau, R. R., de Ona, P., Kunert, M., Lenini, C., Gallegos-Monterrosa, R., Mhatre, E., et al. (2015). A duo of potassium-responsive histidine kinases govern the multicellular destiny of Bacillus subtilis. mBio 6:e00581–15. doi: 10.1128/mBio.00581-15
Haertel, T., Eylert, E., Schulz, C., Petruschka, L., Gierok, P., Grubmueller, S., et al. (2012). Characterization of central carbon metabolism of Streptococcus pneumoniae by isotopologue profiling. J. Biol. Chem. 287, 4260–4274. doi: 10.1074/jbc.M111.304311
Hardy, G. G., Magee, A. D., Ventura, C. L., Caimano, M. J., and Yother, J. (2001). Essential role for cellular phosphoglucomutase in virulence of type 3 Streptococcus pneumoniae. Infect. Immun. 69, 2309–2317. doi: 10.1128/IAI.69.4.2309-2317.2001
Marion, C., Aten, A. E., Woodiga, S. A., and King, S. J. (2011). Identification of an ATPase, MsmK, which energizes multiple carbohydrate ABC transporters in Streptococcus pneumoniae. Infect. Immun. 79, 4193–4200. doi: 10.1128/IAI.05290-11
Mathur, T., Singhal, S., Khan, S., Upadhyay, D. J., Fatma, T., and Rattan, A. (2006). Detection of biofilm formation among the clinical isolates of Staphylococci: an evaluation of three different screening methods. Indian J. Med. Microbiol. 24, 25–29. doi: 10.4103/0255-0857.19890
McLoon, A. L., Kolodkin-Gal, I., Rubinstein, S. M., Kolter, R., and Losick, R. (2011). Spatial regulation of histidine kinases governing biofilm formation in Bacillus subtilis. J. Bacteriol. 193, 679–685. doi: 10.1128/JB.01186-10
Munoz-Elias, E. J., Marcano, J., and Camilli, A. (2008). Isolation of Streptococcus pneumoniae biofilm mutants and their characterization during nasopharyngeal colonization. Infect. Immun. 76, 5049–5061. doi: 10.1128/IAI.00425-08
Okajima, Y., Kobayakawa, S., Tsuji, A., and Tochikubo, T. (2006). Biofilm formation by Staphylococcus epidermidis on intraocular lens material. Invest. Ophthalmol. Vis. Sci. 47, 2971–2975. doi: 10.1167/iovs.05-1172
Pham, T. K., Roy, S., Noirel, J., Douglas, I., Wright, P. C., and Stafford, G. P. (2010). A quantitative proteomic analysis of biofilm adaptation by the periodontal pathogen Tannerella forsythia. Proteomics 10, 3130–3141. doi: 10.1002/pmic.200900448
Planchon, S., Desvaux, M., Chafsey, I., Chambon, C., Leroy, S., Hebraud, M., et al. (2009). Comparative subproteome analyses of planktonic and sessile Staphylococcus xylosus C2a: new insight in cell physiology of a coagulase-negative staphylococcus in biofilm. J. Proteome Res. 8, 1797–1809. doi: 10.1021/pr8004056
Presterl, E., Suchomel, M., Eder, M., Reichmann, S., Lassnigg, A., Graninger, W., et al. (2007). Effects of alcohols, povidone-iodine and hydrogen peroxide on biofilms of Staphylococcus epidermidis. J. Antimicrob. Chemother. 60, 417–420. doi: 10.1093/jac/dkm221
Pridmore, R. D., Berger, B., Desiere, F., Vilanova, D., Barretto, C., Pittet, A. C., et al. (2004). The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533. Proc. Natl. Acad. Sci. U.S.A. 101, 2512–2517. doi: 10.1073/pnas.0307327101
Rachid, S., Ohlsen, K., Witte, W., Hacker, J., and Ziebuhr, W. (2000). Effect of subinhibitory antibiotic concentrations on polysaccharide intercellular adhesin expression in biofilm-forming Staphylococcus epidermidis. Antimicrob. Agents Chemother. 44, 3357–3363. doi: 10.1128/AAC.44.12.3357-3363.2000
Rupp, M. E., and Hamer, K. E. (1998). Effect of subinhibitory concentrations of vancomycin, cefazolin, ofloxacin, L-ofloxacin and D-ofloxacin on adherence to intravascular catheters and biofilm formation by Staphylococcus epidermidis. J. Antimicrob. Chemother. 41, 155–161. doi: 10.1093/jac/41.2.155
Sauer, K. (2003). The genomics and proteomics of biofilm formation. Genome Biol. 4:219. doi: 10.1186/gb-2003-4-6-219
Sauer, K., Camper, A. K., Ehrlich, G. D., Costerton, J. W., and Davies, D. G. (2002). Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184, 1140–1154. doi: 10.1128/jb.184.4.1140-1154.2002
Selvaraj, A., Sumantran, V., Chowdhary, N., and Kumar, G. R. (2014). Prediction and classification of ABC transporters in Geobacter sulfurreducens PCA using computational approaches. Curr. Bioinform. 9, 166–172. doi: 10.2174/1574893608999140109113236
Shemesh, M., and Chai, Y. (2013). A combination of glycerol and manganese promotes biofilm formation in Bacillus subtilis via histidine kinase KinD signaling. J. Bacteriol. 195, 2747–2754. doi: 10.1128/JB.00028-13
Sriskandan, S., and Slater, J. D. (2006). Invasive disease and toxic shock due to zoonotic Streptococcus suis: an emerging infection in the East? PLoS Med. 3:e187. doi: 10.1371/journal.pmed.0030187
Unwin, R. D., Griffiths, J. R., and Whetton, A. D. (2010). Simultaneous analysis of relative protein expression levels across multiple samples using iTRAQ isobaric tags with 2D nano LC-MS/MS. Nat. Protoc. 5, 1574–1582. doi: 10.1038/nprot.2010.123
Walter, J., Chagnaud, P., Tannock, G. W., Loach, D. M., Dal Bello, F., Jenkinson, H. F., et al. (2005). A high-molecular-mass surface protein (Lsp) and methionine sulfoxide reductase B (MsrB) contribute to the ecological performance of Lactobacillus reuteri in the murine gut. Appl. Environ. Microbiol. 71, 979–986. doi: 10.1128/AEM.71.2.979-986.2005
Wang, K., and Lu, C. (2007). Adhesion activity of glyceraldehyde-3-phosphate dehydrogenase in a Chinese Streptococcus suis type 2 strain. Berl. Munch. Tierarztl. Wochenschr. 120, 207–209. doi: 10.2376/0005-9366-120-207
Wang, Y., Yi, L., Wu, Z., Shao, J., Liu, G., Fan, H., et al. (2012). Comparative proteomic analysis of Streptococcus suis biofilms and planktonic cells that identified biofilm infection-related immunogenic proteins. PLoS ONE 7:e33371. doi: 10.1371/journal.pone.0033371
Wang, Y., Zhang, W., Wu, Z. F., and Lu, C. P. (2011). Reduced virulence is an important characteristic of biofilm infection of Streptococcus suis. FEMS Microbiol. Lett. 316, 36–43. doi: 10.1111/j.1574-6968.2010.02189.x
Wisniewski, J. R., Zougman, A., Nagaraj, N., and Mann, M. (2009). Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362. doi: 10.1038/nmeth.1322
Yang, K., Meng, J., Huang, Y.-C., Ye, L.-H., Li, G.-J., Huang, J., et al. (2014). The role of the QseC quorum-sensing sensor kinase in epinephrine-enhanced motility and biofilm formation by Escherichia coli. Cell Biochem. Biophys. 70, 391–398. doi: 10.1007/s12013-014-9924-5
Yang, W.-Z., Yu, H.-J., Jing, H.-Q., Xu, J.-G., Chen, Z.-H., Zhu, X.-P., et al. (2006). An outbreak of human Streptococcus suis serotype 2 infections presenting with toxic shock syndrome in Sichuan, China. Zhonghua Liu Xing Bing Xue za Zhi 27, 185–191.
Yang, Y. B., Wang, S., Wang, C., Huang, Q. Y., Bai, J. W., Chen, J. Q., et al. (2015). Emodin affects biofilm formation and expression of virulence factors in Streptococcus suis ATCC700794. Arch. Microbiol. 197, 1173–1180. doi: 10.1007/s00203-015-1158-4
Yu, J., Bryant, A. P., Marra, A., Lonetto, M. A., Ingraham, K. A., Chalker, A. F., et al. (2001). Characterization of the Streptococcus pneumoniae NADH oxidase that is required for infection. Microbiology 147(Pt 2), 431–438. doi: 10.1099/00221287-147-2-431
Zhang, A., Xie, C., Chen, H., and Jin, M. (2008). Identification of immunogenic cell wall-associated proteins of Streptococcus suis serotype 2. Proteomics 8, 3506–3515. doi: 10.1002/pmic.200800007
Zhang, W., and Lu, C. P. (2007a). Immunoproteomic assay of membrane-associated proteins of Streptococcus suis type 2 china vaccine strain HA9801. Zoonoses Public Health 54, 253–259. doi: 10.1111/j.1863-2378.2007.01056.x
Zhang, W., and Lu, C. P. (2007b). Immunoproteomics of extracellular proteins of Chinese virulent strains of Streptococcus suis type 2. Proteomics 7, 4468–4476. doi: 10.1002/pmic.200700294
Keywords: proteomics, S. suis, biofilms, tylosin, iTRAQ
Citation: Wang S, Yang Y, Zhao Y, Zhao H, Bai J, Chen J, Zhou Y, Wang C and Li Y (2016) Sub-MIC Tylosin Inhibits Streptococcus suis Biofilm Formation and Results in Differential Protein Expression. Front. Microbiol. 7:384. doi: 10.3389/fmicb.2016.00384
Received: 19 November 2015; Accepted: 11 March 2016;
Published: 30 March 2016.
Edited by:
Etienne Giraud, Institut National de la Recherche Agronomique, FranceReviewed by:
Rui Vitorino, University of Aveiro, PortugalRaymond Allan, University of Southampton, UK
Copyright © 2016 Wang, Yang, Zhao, Zhao, Bai, Chen, Zhou, Wang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanhua Li, bGl5YW5odWExOTcwQDE2My5jb20=
 Shuai Wang1
Shuai Wang1 Yanbei Yang
Yanbei Yang Yulin Zhao
Yulin Zhao Honghai Zhao
Honghai Zhao Jingwen Bai
Jingwen Bai Jianqing Chen
Jianqing Chen Yonghui Zhou
Yonghui Zhou Chang Wang
Chang Wang Yanhua Li
Yanhua Li