- Department of Pediatrics, Texas Tech University Health Sciences Center, Amarillo, TX, USA
Child development is a unique and continuous process that is impacted by genetics and environmental factors. Gut microbiome changes with development and depends on the stage of gut maturation, nutrition, and overall health. In spite of emerging data and active study in adults, the gut-renal axis in pediatrics has not been well considered and investigated. This review will focus on the current knowledge of gut microbiota impacts on kidney disease with extrapolation to the pediatric population.
Introduction
Summary of Microbial Activities in Human Metabolism
Knowledge of the microbial communities living in human body is increasing at a fast pace. The Human Microbiome Project demonstrated that humans might be considered superorganisms, composed of human and microbial components, interacting in a symbiotic manner (Turnbaugh et al., 2007). Microbial population has coexisted with humans in a state of mutually beneficial cohabitation and plays an important role in health and disease (Cani and Delzenne, 2009). Inter-organismal crosstalk interruption leads to profound and diverse cellular and metabolic changes observed in gut dysbiosis (Hooper and Gordon, 2001). Gordon et al. showed that the composition of the gut microbiome affects metabolism and energy homeostasis (Ley et al., 2007; Turnbaugh et al., 2007). The gut microbiome is a biotic factor regulating body weight, and thus linked to obesity and other metabolic disorders (Ley et al., 2006) and (Cani and Delzenne, 2009). Maintaining the symbiotic equilibrium between gut microbiome and host in different pathological conditions remains a challenge for the physicians. The understanding of their interaction, manipulation of the structure and functions of human microbiota may allow effective prevention and treatment of many diseases.
Small Intestinal Bacterial Overgrowth may Triggers CKD/ESRD
Dysbiosis can be promoted by any inflammatory reaction, making it difficult to discern cause and consequence of the disease (Sansonetti, 2008; Anders et al., 2013). The gut microbiome could be a trigger for the deregulated immune system in kidney disease (Anders et al., 2013).
Anders et al. (2013) pointed out that the metabolic alterations of uremia favor gut pathogen overgrowth. But this fact has been largely neglected as a trigger for chronic kidney disease (CKD)/end-stage renal disease (ESRD)-related immune derangements (Martin et al., 2009). The concept was that pathobiont overgrowth induces inflammation and loss of barrier function that, results in increased translocation of bacterial components such as LPS and other immune stimulating proteins. This process activates innate immunity characterized by production of pro-inflammatory cytokines and modulates a number of clinically relevant processes in CKD such as the progression of CKD, accelerated atherogenesis, and protein wasting (Anders et al., 2013). Data from a previous studies indicated that toxic products generated by a dysbiotic gut microbiome may contribute to progression of CKD and CKD-related complications (Stecher et al., 2007; Wu et al., 2011; Ramezani and Raj, 2014). As a result, a hypothesis was developed that probiotics and prebiotics reduce progression of CKD and associated uremia by optimizing the gut microbiome (Lin et al., 2011).
With a comprehensive understanding of the structure, density, and function of the gut microbiota, new therapeutic targets could be identified and utilized for a healthier gut. A healthy gut could help improve overall well-being (Vitetta and Gobe, 2013) including health improvement for children with acute and chronic renal conditions.
During development, human microbiome stimulates and synchronizes with the host innate immune functions by the classical complement pathway, the alternate pathway, and the mannose-binding lectin pathway. Kidney disease such as atypical hemolytic uremic syndrome (aHUS) and membranoproliferative glomerulonephritis are closely linked to abnormalities in complement activation due to host genetic anomalies (Sugimoto et al., 2012). Animal experiments have also established that the microbiota modulate the expression of host genes (Bäckhed et al., 2004). Moreover gnotobiotic mouse models revealed that different members of the microbiota induce different types of immune responses (Ivanov et al., 2009). A mixture of Clostridium sp. demonstrated induction of FoxP3+ regulatory T cells and the segmented filamentous bacteria promoted T helper type 17 cell differentiation (Soulage et al., 2013). Animal models have advanced our understanding of the role of microflora in human disease. However, animal conditions can only mimic the features of human disorders (Nagpal et al., 2014). The gut microbiota produces uremic toxins such as advanced glycation end-products, phenols [e.g., p-cresyl sulfate (PCS)], and indoles [e.g., indoxyl sulfate (IS); Bone et al., 1976; Tlaskalová-Hogenová et al., 2011; Hegab et al., 2012]. Animal studies have shown that the biological effect of these molecules include induction of pro-inflammatory responses, leukocyte stimulation, and endothelial dysfunction. These conditions play a substantial role in the development and progression of multiple causes of acute kidney injury (AKI) and CKD (Charney et al., 1993; Motojima et al., 2003; Schepers et al., 2007; Macfarlane and Macfarlane, 2012). IS and PCS are renal and cardiovascular toxins, produced solely by the gut microbiota, which have pro-inflammatory and pro-oxidative properties (Dou et al., 2004). Animal studies have shown that uremic toxins may promote the progression of chronic renal failure by damaging tubular cells (Rossi et al., 2014).
Many reports showed that the composition of gut microbiota is different in CKD patients. Vaziri et al. (2012), using a rat model, showed CKD alters the composition of intestinal microbial flora (Claus et al., 2008). The study revealed substantially less species richness as measured by the number of operational taxonomic units (OTUs) in the nephrectomized rats compared with the controls (Vaziri et al., 2012). The mechanism of bacterial gut alteration was thought to be the result of multiple factors including, but not limited to, metabolic acidosis, volume overload, intestinal wall congestion retention, uremia, frequent use of antibiotics, oral iron, and intestinal ischemia (Kang, 1993; Maslowski and Mackay, 2011; Vaziri et al., 2012). Animal experiments also showed that persistent inflammation contributed to the high rates of CKD development (Vaziri et al., 1985; Stenvinkel et al., 1999).
Some thought that a healthy high fiber diet might promote significant colonization with Shiga toxins-producing Escherichia coli, which can lead to hemolytic-uremic syndrome (HUS; Ivanov et al., 2009). Interesting work by Eaton et al. (2011), showed that the probiotic Lactobacillus reuteri (ATCC PTA 6475) was effective in suppressing disease symptoms of HUS. Their data indicated that L. reuteri partially protected mice from disease manifestations (Eaton et al., 2011). The progression of CKD was hindered by ‘enteric dialysis’ by administering non-pathogenic soil-borne alkalophilic urease-positive bacterium Sporosarcina pasteurii (Sp) in 5/6 nephrectomized rats. That pilot study demonstrated that rats fed with 109 cfu/day of live Sp had reduced blood urea-nitrogen levels and showed significantly prolonged lifespans (Mandal et al., 2013). Furthermore, in germ-free mice that are lacking in immune related to inflammation, the gut microbiome was found to influence kidney homeostasis with elevated levels of key cell volume regulators (betaine, choline, and myo-inositol) in kidneys (Ranganathan et al., 2006).
Obesity and metabolic disorders link tightly to kidney disease. Studies in mice demonstrated that an interrelationship exists between energy balance, diet, and the composition of the gut microbial community. This interrelationship might have significant clinical implications including obesity related complications (Bäckhed et al., 2004, 2007; Ley et al., 2005; Turnbaugh et al., 2006; Clavel and Haller, 2007). In a murine model, gut microbiome product PCS interfered with intracellular insulin signaling pathways and triggered insulin resistance. The treatment of CKD mice with a prebiotic reduced the intestinal production and blood levels of PCS and prevented insulin resistance and lipid abnormalities (Turnbaugh et al., 2008).
To improve animal models to study the human microbiome, a gnotobiotic piglet model was developed (Wang and Donovan, 2015) furthermore human microbiota-associated (HMA) piglets have been established using inocula from infants, children, and adults. The gut microbiota of recipient HMA piglets was more similar to that of the human donor than that of conventionally reared piglets harboring a pig microbiota. Moreover, Bifidobacterium and Bacteroides, two predominant bacterial groups of the infant gut, had been successfully established in the HMA piglets.
What did we Learn About Gut Microbiome and Renal Function From Animal Studies?
Understanding impact of gut microbiome and pathobiome in renal pathology might bring a critical new knowledge in understanding genesis and treatment of kidney disease. The metagenomics approach has the potential to uncover entirely novel genes, gene families, and their encoded proteins, which might be of biotechnological and pharmaceutical relevance (Belizario and Napolitano, 2015). Therefore, animal models strongly proved impact on gut microbiome and renal function, in addition suitable animal models were created to pursue relevant study in pediatric population.
Gut Microbiome and Renal Axis in Adult Population
An adult human’s microbiome is diverse and dependent upon health status, diet, and geographic location. The gut harbors a complex mixture of tens of trillions of microbes, comprised of more than 1,000 diverse species of identified bacteria with over three million genes. In healthy individuals, the phyla Bacteroidetes and Firmicutes contribute >90% of all species, including abundant bacterial genera such as Bacteroides sp., Alistipes sp., Prevotella sp., Porphyromonas sp., Clostridium sp., Dorea sp., Faecalibacterium sp., Eubacterium sp., Ruminococcus sp., and Lactobacillus sp. Other less abundant phyla include the Actinobacteria (that is, Bifidobacterium sp. and Collinsella sp.), Proteobacteria (that is, Enterobacteriaceae, Sutterella sp., and Helicobacter sp.), Verrucomicrobia (that is, Akkermansia sp.), and some others (Qin et al., 2010).
The microbiota colonizing the gut control the normal development and function of the mucosal barriers, support food digestion, and defend the individual from circulation of pathogenic micro-organisms. On the other hand, the break down of proteins and peptides by colonic microorganisms yields a great diversity of end-products, many of which have toxic properties (Mafra et al., 2013).
Adult uremic patients show greatly increased counts of both aerobic and anaerobic organisms in the duodenum and jejunum, portions of the intestines not normally colonized heavily by bacteria in healthy persons (Ramezani and Raj, 2014). With diminished kidney clearance, quantitative, and qualitative alterations in gut microbiota were noted in adult patients with CKD and ESRD (Hida et al., 1996; Simenhoff et al., 1996; Vaziri et al., 2013). Vaziri et al. (2013) showed significant differences in the abundance of 190 microbial OTUs between the patients with ESRD and the normal control individuals. In addition, hemodialysis patients had significantly lower numbers of Bifidobacteria and higher Clostridium perfringens numbers (Rossi et al., 2014).
Intestinal microbial flora in patients with CKD characterized by decreases in both Lactobacillaceae and Prevotellaceae families have been reported (Mafra et al., 2014). As a result, uremic toxins produce bacteria families’ over growth that impacts immune response and inflammatory reactions. Study in 149 CKD patients with a mean estimated Glomerular Filtration Rate (eGFR) of 40 ± 9 mL/min/1.73 m2 showed that serum free and total IS were independently associated with increased levels of serum IL-6, TNF-α, and IFN-γ, whereas serum free and total PCS were independently associated with increased levels of serum IL-6 and pulse wave velocity (Rossi et al., 2014). This study for the first time proved a strong connection between produced and retained uremic toxins and demonstrated strong predictors of cardiovascular mortality in CKD patients. The idea was further supported by a publication of Mafra et al. (2014). They also provided a concise description of the potential role of the CKD-associated changes in the gut microbiome and its potential role in the pathogenesis of inflammation and uremic toxicity (Mafra et al., 2014). PCS also contributed to the development of insulin resistance in patients with CKD (Soulage et al., 2013).
Lipopolysaccharide (LPS)-induced monocyte/macrophage activation could explain systemic inflammation and might be relevant to CKD/ESRD patients’ conditions (Stearns-Kurosawa et al., 2011). Cani et al. (2007) demonstrated in mouse model that the metabolic endotoxemia dysregulates the inflammatory character and activates body weight gain and diabetes. They also observed that lowering plasma LPS concentration could be an effective approach to regulate metabolic diseases such as diabetes (Cani et al., 2007).
We recently reported significant increases in inflammatory markers and markers of endothelial dysfunction in patients with diabetes, CKD, and ESRD on peritoneal dialysis, as well as correlations with levels of LPS and zonulin (a gut permeability marker; Chennasamudram et al., 2013; Nakhla et al., 2014; Singh et al., 2014). Human intestine also acts as an active player by presenting more precursors for fermentation due to disturbances in assimilation caused by uremia, followed by alterations in further processing related to changes in the composition of the fermenting flora (Schepers et al., 2010). Deficiency in vitamin K and calcidiol is a common feature among patients with CKD and ESRD and is likely attributed to gut dysbiosis (LaClair et al., 2005; Pilkey et al., 2007).
The colonic microbiome is an active site of methanol production, which might appear in the exhaled breath of subjects. The study of methanol breath content in ESRD on hemodialysis showed that methanol production could be substantially manipulated by the diet, which changed gut populations per se (Lee et al., 2012).
Strong connection between gut pathobionts and CKD and ESRD in adult population is nowadays undoubtful fact. The next important questions to be answered in nephrology: how gut microbiome diversity impacted by primary renal disease and how it changes with progression of chronic renal impairment.
Pediatric Gut Microbiome
The gastrointestinal tract (GIT) in neonates was thought to be almost sterile at birth and then rapidly colonized by bacteria shortly after delivery until an adult type composition was acquired around the first to the third year of life (Rotimi and Duerden, 1981; Valle et al., 2007; DiGiulio et al., 2008; O’Toole and Claesson, 2010). This view has recently been challenged by reports of detection of diverse microbes in placenta, umbilical cord, amniotic fluid, and meconium (Jiménez et al., 2005, 2008; Dominguez-Bello et al., 2010; Vallés et al., 2012; Gosalbes et al., 2013; Moles et al., 2013; Song et al., 2013; Aagaard et al., 2014). It is now well established that during the first 3 years of life, children experience significant developmental changes that influence their health status as well as their immune system. Using sequential fecal analysis in a large cohort, human microbiome studies across North America, Africa, South America, and Europe, have made it apparent that gut microbiome is highly unstable during the first 3 years of life (De Filippo et al., 2010; Koenig et al., 2011; Yatsunenko et al., 2012). The pattern of GIT microbiome development during child development is summarized in Figure 1.
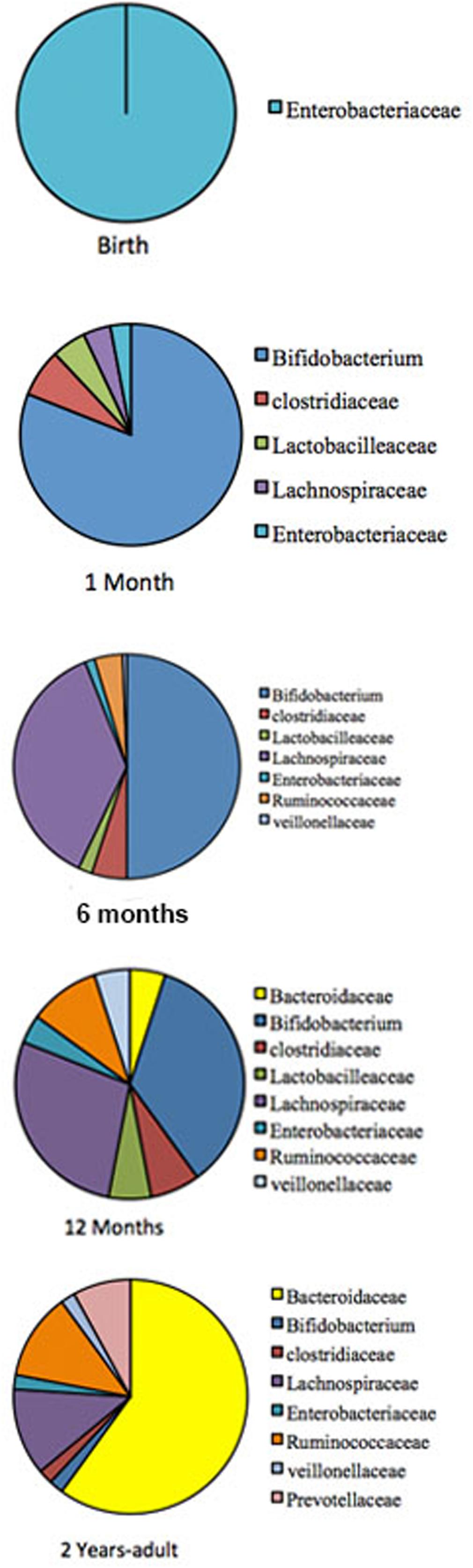
FIGURE 1. The pattern of GIT microbiome development during a child development. Pie charts are showing profusion of the bacterial taxa at different development stage. The gut-microbiome of the new born is primarily colonized by Enterobacteria (Gosalbes et al., 2013). During the first month, Bifidobacterial species pre-dominate in the gut (Koenig et al., 2011), but at around 4–6 months is convoyed by an increase of clostridial species. Later, the microbiome composition consists of mainly Bacteroidaceae, Lachnospiraceae, and Ruminococcaceae, which then remains stable into adulthood (Fanaro et al., 2005; Morelli, 2008; Lozupone et al., 2012).
Gut Microbiome in Children with Kidney Disease
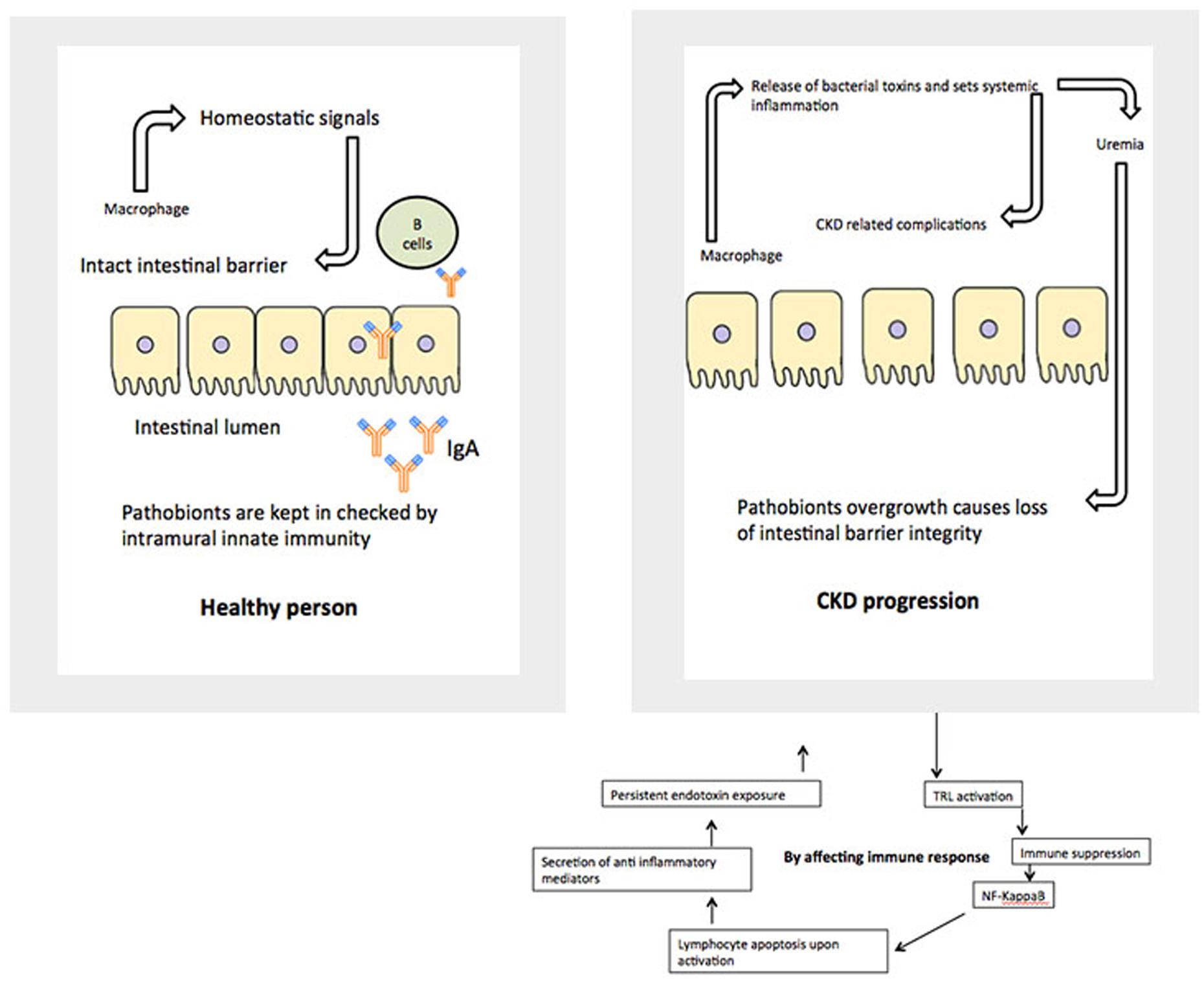
The relationship between alterations in the gut microbiome and its possible involvement in the development of renal disease later in life is an important venue to be investigated. Consideration of GIT barrier structure and it changes in pathological conditions is necessary to understand relationships in the gut-renal axis in pediatrics. The maintenance of a healthy intestinal barrier is extremely important in children. There are multiple layers, which makeup the barrier between gut lumen and the rest of the body. The physical barrier is composed of gut microbiota, mucus, epithelial cells, and the innate and adaptive immune cells forming the gut-associated lymphoid tissue. The internal mucosal layer is dense and does not allow bacteria to penetrate, while the external mucus layer is the habitat of the gut microbiota. Each of those layers is compromised in a uremic condition (Figure 2).
As mentioned above, in a healthy body, normal gut microbiota competes with pathogens for space and energy resources, processes the molecules necessary to maintain mucosal integrity, and modulate the immunological activity of the deep barrier. In CKD and ESRD these functions are greatly compromised, based on animal and adult patient studies. Children with similar renal conditions are usually on a very specific diet, which might include food low in potassium, phosphorus, and salt. They may use phosphate binders and have restrictions on their water consumption. During infancy and early childhood, these restrictions could contribute to the development of an abnormal microbial population and weaken the first line of the gut barrier (Vaziri et al., 2013). This eventually leads to a “leaky gut syndrome.” It also further compromises vitamin absorption necessary for growing bodies, including Vitamin D.
We also presume that many drugs or compounds used in the treatment of CKD and ESRD not only compromise gut microbiome, but may experience abnormal pharmacokinetics in the setting of abnormal intestinal permeability. In addition, the development of new pharmacological approaches to modulate the gut barrier components might contribute to prevention and treatment of extra-intestinal diseases, including kidney pathology.
Does early-life dysbiosis precede and play a role in disease pathogenesis, or simply originate because of the disease process itself? This question has been answered in a few diseases, including inflammatory bowel disease, obesity, and asthma (Arrieta et al., 2014). A paucity of data exists regarding how the gut-renal axis functions in a growing human with renal disease and what the role of the microbiome is in this process. Although, some data gathered from bench research and knowledge obtained from adult populations might be extrapolated to the pediatric population, the unique growing and constantly changing body deserves separate investigation.
One of the challenges in the study of the microbiome in any disease, including kidney disease, in early childhood is the greater variation in strain diversity documented during the first few months and years of life (Viggiano et al., 2015). Gut microbiota evolve and change during that growth. Data showed that individuals harbor their own specific Bifidobacterial microbiota even at the strain level. Thus, 96% of infants possessed their own individual specific Bifidobacterial microbiota (Barrett et al., 2015).
Study of gut microbiome in children with renal disease should focus on four types of dysbiosis: loss of keystone taxa, loss of diversity, shifts in metabolic capacity, and blooms of pathogens. Both genetic and environmental factors should be taken into consideration while studying the development of gut microbiota composition (Spor et al., 2011). For example, one of the relevant papers from China found that gut microbiota was involved in melamine-induced renal injury in infants and children exposed to melamine-tainted milk. Melamine-induced renal toxicity was found to be mediated by the gut microbiota (Zheng et al., 2013).
Preterm children need special consideration relevant to renal pathology. It is known that children born preterm have a high risk of the fast progression of CKD and low nephron numbers (Kandasamy et al., 2012; Brennan and Kandasamy, 2013; Carmody and Charlton, 2013; Mishra et al., 2014). Many infants begin life with a significant number of immature nephrons. They are exposed to a variety of external stressors such as hemodynamic alterations, nephrotoxic medications, infections, and suboptimal nutrition that can delay ongoing kidney development or cause nephron loss. Preterm infants and particularly very low birth weight infants are also at a disadvantage when it comes to the development of a healthy microbiome (Gritz and Bhandari, 2015). The causes of their gut immaturity include preterm rupture of membranes, maternal infection, Cesarean delivery, perinatal and postnatal broad-spectrum antibiotic exposure, exposure to other gut-modifying medications (which might alter gut permeability), periods of fasting, intensive care infection control standards and exposure to resistant microbes and decreased acquaintance of human milk (Scholtens et al., 2012; Berrington et al., 2013; Gupta et al., 2013). Arboleya et al. (2012) compared full-term breastfed vaginally delivered infants with preterm infants with regard to differences in representation of 18 microbial groups within gut flora. They demonstrated that when compared with full-term infants, preterm infants showed increased populations of facultative anaerobes such as Enterococcus, Enterobacter, and Lactobacillus, increased numbers of Staphylococcus, and decreased numbers of anaerobes such as Bifidobacterium, Bacteroides, and Atopobium (Madan et al., 2012; Berrington et al., 2014).
The gut microbiome specifics in relationship to kidney compromised status were not well investigated.
Cross-sectional analysis of pediatric CKD data has revealed valuable information that better defines the prevalence of comorbid conditions and associated risk factors, but not gut microbiome. Hypertension, left ventricular hypertrophy, dyslipidemia, anemia, poor growth, and abnormal neurocognitive development are known comorbidities that accompany CKD (Wong et al., 2012), but pediatric gut microbiome in a uremic milieu is yet to be evaluated.
Conclusion
Extensive microbiome research has been made possible by recent advances in gene sequencing and bioinformatics tools. Further study is required to decipher structures and functions of microbe population in the gut of pediatric AKI, CKD, and ESRD patients. This is a call for concerted efforts of pediatricians, pediatric nephrologists, immunologists, microbiologists, food and nutrition experts, and computational biologists to understand this clinical phenomenon and further applications in developing therapeutic regimens.
Author Contributions
TV and RS contributed to the conception, design of review. TV has drafted the work critically for important intellectual content. TV has provided final approval of the version to be published. Authors have agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Aagaard, K., Ma, J., Antony, K. M., Ganu, R., Petrosino, J., and Versalovic, J. (2014). The placenta harbors a unique microbiome. Sci. Transplant. Med. 6:237ra65. doi: 10.1126/scitranslmed.3008599
Anders, H. J., Andersen, K., and Stecher, B. (2013). The intestinal microbiota, a leaky gut, and abnormal immunity in kidney disease. Kidney Int. 83, 1010–1016. doi: 10.1038/ki.2012.440
Arboleya, S., Binetti, A., Salazar, N., Fernández, N., Solís, G., Hernández-Barranco, A., et al. (2012). Establishment and development of intestinal microbiota in preterm neonates. FEMS Microbiol. Ecol. 79, 763–772. doi: 10.1111/j.1574-6941.2011.01261.x
Arrieta, M. C., Stiemsma, L. T., Amenyogbe, N., Brown, E. M., and Finlay, B. (2014). The intestinal microbiome in early life: health and disease. Front. Immunol. 5:247. doi: 10.3389/fimmu.2014.00427
Bäckhed, F., Ding, H., Wang, T., Hooper, L. V., Koh, G. Y., Nagy, A., et al. (2004). The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. U. S.A. 101, 15718–15723. doi: 10.1073/pnas.0407076101
Bäckhed, F., Manchester, J. K., Semenkovich, C. F., and Gordon, J. I. (2007). Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc. Natl. Acad. Sci. U. S.A. 104, 979–984. doi: 10.1073/pnas.0605374104
Barrett, E., Deshpandey, A. K., Ryan, C. A., Dempsey, E. M., Murphy, B., O’Sullivan, L., et al. (2015). The neonatal gut harbours distinct bifidobacterial strains. Arch. Dis. Child. Fetal Neonatal Ed. 100, F405–F410. doi: 10.1136/archdischild-2014-306110
Belizario, J. E., and Napolitano, M. (2015). Human microbiomes and their roles in dysbiosis, common diseases, and novel therapeutic approaches. Front. Microbiol. 6:1050. doi: 10.3389/fmicb.2015.01050
Berrington, J. E., Stewart, C. J., Cummings, S. P., and Embleton, N. D. (2014). The neonatal bowel microbiome in health and infection. Curr. Opin. Infect. Dis. 27, 236–243. doi: 10.1097/QCO.0000000000000061
Berrington, J. E., Stewart, C. J., Embleton, N. D., and Cummings, S. P. (2013). Gut microbiota in preterm infants: assessment and relevance to health and disease. Arch. Dis. Child. Fetal Neonatal Ed. 98, F286–F290. doi: 10.1136/archdischild-2012-302134
Bone, E., Tamm, A., and Hill, M. (1976). The production of urinary phenols by gut bacteria and their possible role in the causation of large bowel cancer. Am. J. Clin. Nutr. 29, 1448–1454.
Brennan, S., and Kandasamy, Y. (2013). Renal parenchymal thickness as a measure of renal growth in low-birth-weight infants versus normal-birth-weight infants. Ultrasound Med. Biol. 39, 2315–2320. doi: 10.1016/j.ultrasmedbio.2013.07.001
Cani, P. D., Amar, J., Iglesias, M. A., Poggi, M., Knauf, C., and Bastelica, D. (2007). Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 56, 1761–1772. doi: 10.2337/db06-1491
Cani, P. D., and Delzenne, N. M. (2009). The role of the gut microbiota in energy metabolism and metabolic disease. Curr. Pharm. Des. 15, 1546–1558. doi: 10.2174/138161209788168164
Carmody, J. B., and Charlton, J. R. (2013). Short-term gestation, long-term risk: prematurity and chronic kidney disease. Pediatrics 131, 1168–1179. doi: 10.1542/peds.2013-0009
Charney, D. I., Walton, D. F., and Cheung, A. K. (1993). Atherosclerosis in chronic renal failure. Curr. Opin. Nephrol. Hypertens 2, 876–882. doi: 10.1097/00041552-199311000-00004
Chennasamudram, S. P., Noor, T., and Vasylyeva, T. L. (2013). Comparison of sevelamer and calcium carbonate on endothelial function and inflammation in patients on peritoneal dialysis. J. Ren. Care 39, 82–89. doi: 10.1111/j.1755-6686.2013.12009.x
Claus, S. P., Tsang, T. M., Wang, Y., Cloarec, O., Skordi, E., Martin, F. P., et al. (2008). Systemic multicompartmental effects of the gut microbiome on mouse metabolic phenotypes. Mol. Syst. Biol. 4:219. doi: 10.1038/msb.2008.56
Clavel, T., and Haller, D. (2007). Molecular interactions between bacteria, the epithelium, and the mucosal immune system in the intestinal tract: implications for chronic inflammation. Curr. Issues Intest. Microbiol. 8, 25–43.
De Filippo C, Cavalieri, D., Di Paola, M., Ramazzotti, M., Poullet, J. B., Massart, S., et al. (2010). Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. U.S.A. 107, 14691–14696. doi: 10.1073/pnas.1005963107
DiGiulio, D. B., Romero, R., Amogan, H. P., Kusanovic, J. P., Bik, E. M., Gotsch, F., et al. (2008). Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: a molecular and culture-based investigation. PLoS ONE 3:e3056. doi: 10.1371/journal.pone.0003056
Dominguez-Bello, M. G., Costello, E. K., Contreras, M., Magris, M., Hidalgo, G., Fierer, N., et al. (2010). Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. U.S.A. 107, 11971–11975. doi: 10.1073/pnas.1002601107
Dou, L., Bertrand, E., Cerini, C., Faure, V., Sampol, J., Vanholder, R., et al. (2004). The uremic solutes p-cresol and indoxylsulfate inhibit endothelial proliferation and wound repair. Kidney Int. 65, 442–451. doi: 10.1111/j.1523-1755.2004.00399.x
Eaton, K. A., Honkala, A., Auchtung, T. A., and Britton, R. A. (2011). Probiotic Lactobacillus reuteri ameliorates disease due to enterohemorrhagic Escherichia coli in germfree mice. Infect. Immun. 79, 185–191. doi: 10.1128/IAI.00880-10
Fanaro, S., Boehm, G., Garssen, J., Knol, J., Mosca, F., Stahl, B., et al. (2005). Galacto-oligosaccharides and long-chain fructo-oligosaccharides as prebiotics in infant formulas: a review. Acta Paediatr. Suppl. 94, 22–26.
Gosalbes, M. J., Llop, S., Vallès, Y., Moya, A., Ballester, F., and Francino, M. P. (2013). Meconium microbiota types dominated by lactic acid or enteric bacteria are differentially associated with maternal eczema and respiratory problems in infants. Clin. Exp. Allergy 43, 198–211. doi: 10.1111/cea.12063
Gritz, E. C., and Bhandari, V. (2015). The human neonatal gut microbiome: a brief review. Front. Pediatr. 3:17. doi: 10.3389/fped.2015.00060
Gupta, R. W., Tran, L., Norori, J., Ferris, M. J., Eren, A. M., Taylor, C. M., et al. (2013). Histamine-2 receptor blockers alter the fecal microbiota in premature infants. J. Pediatr. Gastroenterol. Nutr. 56, 397–400. doi: 10.1097/MPG.0b013e318282a8c2
Hegab, Z., Gibbons, S., Neyses, L., and Mamas, M. A. (2012). Role of advanced glycation end products in cardiovascular disease. World J. Cardiol. 4, 90–102. doi: 10.4330/wjc.v4.i4.90
Hida, M., Aiba, Y., Sawamura, S., Suzuki, N., Satoh, T., and Koga, Y. (1996). Inhibition of the accumulation of uremic toxins in the blood and their precursors in the feces after oral administration of Lebenin, a lactic acid bacteria preparation, to uremic patients undergoing hemodialysis. Nephron 74, 349–355. doi: 10.1159/000189334
Hooper, L. V., and Gordon, J. I. (2001). Commensal host-bacterial relationships in the gut. Science 292, 1115–1118. doi: 10.1126/science.1058709
Ivanov, I. I., Atarashi, K., Manel, N., Brodie, E. L., Shima, T., Karaoz, U., et al. (2009). Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498. doi: 10.1016/j.cell.2009.09.033
Jiménez, E., Fernández, L., Marín, M. L., Martín, R., Odriozola, J. M., Nueno-Palop, C., et al. (2005). Isolation of commensal bacteria from umbilical cord blood of healthy neonates born by cesarean section. Curr. Microbiol. 51, 270–274. doi: 10.1007/s00284-005-0020-3
Jiménez, E., Marín, M. L., Martín, R., Odriozola, J. M., Olivares, M., Xaus, J., et al. (2008). Is meconium from healthy newborns actually sterile? Res. Microbiol. 159, 187–193. doi: 10.1016/j.resmic.2007.12.007
Kandasamy, Y., Smith, R., and Wright, I. M. R. (2012). Oligonephropathy of prematurity. Am. J. Perinatol. 29, 115–120. doi: 10.1055/s-0031-1295651
Kang, J. Y. (1993). The gastrointestinal tract in uremia. Dig. Dis. Sci. 38, 257–268. doi: 10.1007/BF01307542
Koenig, J. E., Spor, A., Scalfone, N., Fricker, A. D., Stombaugh, J., Knight, R., et al. (2011). Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. U.S.A. 108, 4578–4585. doi: 10.1073/pnas.1000081107
LaClair, R. E., Hellman, R. N., Karp, S. L., Kraus, M., Ofner, S., Li, Q., et al. (2005). Prevalence of calcidiol deficiency in CKD: a cross-sectional study across latitudes in the United States. Am. J. Kidney Dis. 45, 1026–1033. doi: 10.1053/j.ajkd.2005.02.029
Lee, H. J., Pahl, M. V., Vaziri, N. D., and Blake, D. R. (2012). Effect of hemodialysis and diet on the exhaled breath methanol concentration in patients with ESRD. J. Ren. Nutr. 22, 357–364. doi: 10.1053/j.jrn.2011.07.003
Ley, R. E., Bäckhed, F., Turnbaugh, P., Lozupone, C. A., Knight, R. D., and Gordon, J. I. (2005). Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U.S.A. 102, 11070–11075. doi: 10.1073/pnas.0504978102
Ley, R. E., Knight, R., and Gordon, J. I. (2007). The human microbiome: eliminating the biomedical/environmental dichotomy in microbial ecology. Environ. Microbiol. 9, 3–4. doi: 10.1111/j.1462-2920.2006.01222_3.x
Ley, R. E., Turnbaugh, P. J., Klein, S., and Gordon, J. I. (2006). Microbial ecology: human gut microbes associated with obesity. Nature 444, 1022–1023. doi: 10.1038/4441022a
Lin, C. J., Chen, H. H., Pan, C. F., Chuang, C. K., Wang, T. J., Sun, F. J., et al. (2011). p-Cresylsulfate and indoxyl sulfate level at different stages of chronic kidney disease. J. Clin. Labor. Anal. 25, 191–197. doi: 10.1002/jcla.20456
Lozupone, C. A., Stombaugh, J. I., Gordon, J. I., Jansson, J. K., and Knight, R. (2012). Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–30. doi: 10.1038/nature11550
Macfarlane, G. T., and Macfarlane, S. (2012). Bacteria, colonic fermentation, and gastrointestinal health. J. AOAC Int. 95, 50–60. doi: 10.5740/jaoacint.SGE_Macfarlane
Madan, J. C., Farzan, S. F., Hibberd, P. L., and Karagas, M. R. (2012). Normal neonatal microbiome variation in relation to environmental factors, infection and allergy. Curr. Opin. Pediatr. 24, 753–759. doi: 10.1097/MOP.0b013e32835a1ac8
Mafra, D., Barros, A. F., and Fouque, D. (2013). Dietary protein metabolism by gut microbiota and its consequences for chronic kidney disease patients. Future Microbiol. 8, 1317–1323. doi: 10.2217/fmb.13.103
Mafra, D., Lobo, J. C., Barros, A. F., Koppe, L., Vaziri, N. D., and Fouque, D. (2014). Role of altered intestinal microbiota in systemic inflammation and cardiovascular disease in chronic kidney disease. Future Microbiol. 9, 399–410. doi: 10.2217/fmb.13.165
Mandal, A., Das, K., Roy, S., Mondal, KCh, and Nandi, D. K. (2013). In vivo assessment of bacteriotherapy on acetaminophen-induced uremic rats. J. Nephrol. 26, 228–236. doi: 10.5301/jn.5000129
Martin, F. P., Sprenger, N., Yap, I. K., Wang, Y., Bibiloni, R., Rochat, F., et al. (2009). Panorganismal gut microbiome–host metabolic crosstalk. J. Proteome Res. 8, 2090–2105. doi: 10.1021/pr801068x
Maslowski, K. M., and Mackay, C. R. (2011). Diet, gut microbiota and immune responses. Nat. Immunol. 12, 5–9. doi: 10.1038/ni0111-5
Mishra, K., Datta, V., Aarushi, A., Kaur Narula, M., Iyer, R. S., and Nangia, S. (2014). The association between weight for gestational age and kidney volume: a study in newborns in India. Iran. J. Pediatr. 24, 93–99.
Moles, L., Gómez, M., Heilig, H., Bustos, G., Fuentes, S., de Vos, W., et al. (2013). Bacterial diversity in meconium of preterm neonates and evolution of their fecal microbiota during the first month of life. PLoS ONE 8:e66986. doi: 10.1371/journal.pone.0066986
Morelli, L. (2008). Postnatal development of intestinal microflora as influenced by infant nutrition. J. Nutr. 138, 1791S–1795S.
Motojima, M., Hosokawa, A., Yamato, H., Muraki, T., and Yoshioka, T. (2003). Uremic toxins of organic anions up-regulate PAI-1 expression by induction of NF-kappaB and free radical in proximal tubular cells. Kidney Int. 63, 1671–1680. doi: 10.1046/j.1523-1755.2003.00906.x
Nagpal, R., Yadav, H., and Marotta, F. (2014). Gut microbiota: the next-gen frontier in preventive and therapeutic medicine? Front. Med. 1:15. doi: 10.3389/fmed.2014.00015
Nakhla, E., Singh, R., Chennasamudram, S., Vasylyeva, O., Naguib, T., and Vasylyeva, T. L. (2014). Sex disparity in cardiovascular mortality in patient with end-stage renal disease and Type 2 diabetes mellitus. Diabetes Res. Treatm. 1, 111–114.
O’Toole, P. W., and Claesson, M. J. (2010). Gut microbiota: changes throughout the lifespan from infancy to elderly. Int. Dairy J. 20, 281–291. doi: 10.1016/j.idairyj.2009.11.010
Pilkey, R. M., Morton, A. R., Boffa, M. B., Noordhof, C., Day, A. G., Su, Y., et al. (2007). Subclinical vitamin K deficiency in hemodialysis patients. Am. J. Kidney Dis. 49, 432–439. doi: 10.1053/j.ajkd.2006.11.041
Qin, J., Li, R., Raes, J., Arumugam, M., Burgdorf, K. S., Manichanh, C., et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–U70. doi: 10.1038/nature08821
Ramezani, A., and Raj, D. S. (2014). The gut microbiome, kidney disease, and targeted interventions. J. Ame. Soc. Nephrol. 25, 657–670. doi: 10.1681/ASN.2013080905
Ranganathan, N., Patel, B. G., Ranganathan, P., Marczely, J., Dheer, R., Pechenyak, B., et al. (2006). In vitro and in vivo assessment of intraintestinal bacteriotherapy in chronic kidney disease. ASAIO J. 52, 70–79. doi: 10.1097/01.mat.0000191345.45735.00
Rossi, M., Campbell, K. L., Johnson, D. W., Stanton, T., Vesey, D. A., Coombes, J. S., et al. (2014). Protein-bound uremic toxins, inflammation and oxidative stress: a cross-sectional study in stage 3-4 chronic kidney disease. Arch. Med. Res. 45, 309–317. doi: 10.1016/j.arcmed.2014.04.002
Rotimi, V. O., and Duerden, B. I. (1981). The development of the bacterial flora in normal neonates. J. Med. Microbiol. 14, 51–62. doi: 10.1099/00222615-14-1-51
Sansonetti, P. J. (2008). Host-bacteria homeostasis in the healthy and inflamed gut. Curr. Opin. Gastroenterol. 24, 435–439. doi: 10.1097/MOG.0b013e32830007f7
Schepers, E., Glorieux, G., and Vanholder, R. (2010). The gut: the forgotten organ in uremia? Blood Purif. 29, 130–136. doi: 10.1159/000245639
Schepers, E., Meert, N., Glorieux, G., Goeman, J., Van der Eycken, J., and Vanholder, R. (2007). P-cresylsulphate, the main in vivo metabolite of p-cresol, activates leucocyte free radical production. Nephrol. Dial. Transplant. 22, 592–596. doi: 10.1093/ndt/gfl584
Scholtens, P. A., Oozeer, R., Martin, R., Amor, K. B., and Knol, J. (2012). The early settlers: intestinal microbiology in early life. Annu. Rev. Food Sci. Technol. 3, 425–447. doi: 10.1146/annurev-food-022811-101120
Simenhoff, M. L., Dunn, S. R., Zollner, G. P., Fitzpatrick, M. E., Emery, S. M., Sandine, W. E., et al. (1996). Biomodulation of the toxic and nutritional effects of small bowel bacterial overgrowth in end-stage kidney disease using freeze-dried Lactobacillus acidophilus. Miner. Electrolyte Metab. 22, 92–96.
Singh, R., Chennasamudram, S. P., Sheth, S., and Vasylyeva, T. L. (2014). Correlation of fibroblast growth factor 23 with markers of inflammation and endothelial dysfunction in end-stage renal disease and Type 2 diabetes patients on peritoneal dialysis. Diab. Metab. 5, 1–5.
Song, S. J., Dominguez-Bello, M. G., and Knight, R. (2013). How delivery mode and feeding can shape the bacterial community in the infant gut. Can. Med. Assoc. J. 185, 373–374. doi: 10.1503/cmaj.130147
Soulage, C. O., Koppe, L., and Fouque, D. (2013). Protein-bound uremic toxins…new targets to prevent insulin resistance and dysmetabolism in patients with chronic kidney disease. J. Ren. Nutr. 23, 464–466. doi: 10.1053/j.jrn.2013.06.003
Spor, A., Koren, O., and Ley, R. (2011). Unravelling the effects of the environment and host genotype on the gut microbiome. Nat. Rev. Microbiol. 9, 279–290. doi: 10.1038/nrmicro2540
Stearns-Kurosawa, D. J., Osuchowski, M. F., Valentine, C., Kurosawa, S., Remick, D. G., et al. (2011). The pathogenesis of sepsis. Annu. Rev. Pathol. 6, 19–48. doi: 10.1146/annurev-pathol-011110-130327
Stecher, B., Robbiani, R., Walker, A. W., Westendorf, A. M., Barthel, M., Kremer, M., et al. (2007). Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 5:2177–2189. doi: 10.1371/journal.pbio.0050244
Stenvinkel, P., Heimbürger, O., Paultre, F., Diczfalusy, U., Wang, T., Berglund, L., et al. (1999). Strong association between malnutrition, inflammation, and atherosclerosis in chronic renal failure. Kidney Int. 55, 1899–1911. doi: 10.1046/j.1523-1755.1999.00422.x
Sugimoto, K., Fujita, S., Miyazaki, K., Okada, M., and Takemura, T. (2012). C3 glomerulonephritis associated with a missense mutation in the factor H gene. Tohoku J. Exp. Med. 227, 211–215. doi: 10.1620/tjem.227.211
Tlaskalová-Hogenová, H., Stěpánková, R., Kozáková, H., Hudcovic, T., Vannucci, L., Tučková, L., et al. (2011). The role of gut microbiota (commensal bacteria) and the mucosal barrier in the pathogenesis of inflammatory and autoimmune diseases and cancer: contribution of germ-free and gnotobiotic animal models of human diseases. Cell. Mol. Immunol. 8, 110–120. doi: 10.1038/cmi.2010.67
Turnbaugh, P. J., Bäckhed, F., Fulton, L., and Gordon, J. I. (2008). Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 3, 213–223. doi: 10.1016/j.chom.2008.02.015
Turnbaugh, P. J., Ley, R. E., Hamady, M., Fraser-Liggett, C. M., Knight, R., and Gordon, J. I. (2007). The human microbiome project. Nature 449, 804–810. doi: 10.1038/nature06244
Turnbaugh, P. J., Ley, R. E., Mahowald, M. A., Magrini, V., Mardis, E. R., and Gordon, J. I. (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031. doi: 10.1038/nature05414
Valle, J. M, Estepa, R. M., Camacho, R. M., Estrada, R. C., Luna, F. G., and Guitarte, F. B. (2007). Endothelial dysfunction is related to insulin resistance and inflammatory biomarker levels in obese prepubertal children. Eur. J. Endocrinol. 156, 497–502. doi: 10.1530/EJE-06-0662
Vallés, Y., Gosalbes, M. J., de Vries, L. E., Abellán, J. J., and Francino, M. P. (2012). Metagenomics and development of the gut microbiota in infants. Clin. Microbiol. Infect. 18, 21–26. doi: 10.1111/j.1469-0691.2012.03876.x
Vaziri, N. D., Dure-Smith, B., Miller, R., and Mirahmadi, M. K. (1985). Pathology of gastrointestinal tract in chronic hemodialysis patients: an autopsy study of 78 cases. Am. J. Gastroenterol. 80, 608–611.
Vaziri, N. D., Pahl, M. V., Crum, A., and Norris, K. (2012). Effect of uremia on structure and function of immune system. J. Ren. Nutr. 22, 149–156. doi: 10.1053/j.jrn.2011.10.020
Vaziri, N. D., Wong, J., Pahl, M., Piceno, Y. M., Yuan, J., DeSantis, T. Z., et al. (2013). Chronic kidney disease alters intestinal microbial flora. Kidney Int. 83, 308–315. doi: 10.1038/ki.2012.345
Viggiano, D., Ianiro, G., Vanella, G., Bibbò, S., Bruno, G., Simeone, G., et al. (2015). Gut barrier in health and disease: focus on childhood. Eur. Rev. Med. Pharmacol. Sci. 19, 1077–1085.
Vitetta, L., and Gobe, G. (2013). Uremia and chronic kidney disease: the role of the gut microflora and therapies with pro- and prebiotics. Mol. Nutr. Food Res. 57, 824–832. doi: 10.1002/mnfr.201200714
Wang, M., and Donovan, S. M. (2015). Human microbiota-associated swine: current progress and future opportunities. Ilar J. 56, 63–73. doi: 10.1093/ilar/ilv006
Wong, C. J., Moxey-Mims, M., Jerry-Fluker, J., Warady, B. A., and Furth, S. L. (2012). CKiD (CKD in children) prospective cohort study: a review of current findings. Am. J. Kidney Dis. 60, 1002–1011. doi: 10.1053/j.ajkd.2012.07.018
Wu, I. W., Hsu, K. H., Lee, C. C., Sun, C. Y., Hsu, H. J., Tsai, C. J., et al. (2011). p-Cresyl sulphate and indoxyl sulphate predict progression of chronic kidney disease. Nephrol. Dial. Transplant. 26, 938–947. doi: 10.1093/ndt/gfq580
Yatsunenko, T., Rey, F. E., Manary, M. J., Trehan, I., Dominguez-Bello, M. G., Contreras, M., et al. (2012). Human gut microbiome viewed across age and geography. Nature 486, 222–227. doi: 10.1038/nature11053
Keywords: gut microbiome, pediatrics, gut renal axis, inflammatory markers, kidney disease
Citation: Vasylyeva TL and Singh R (2016) Gut Microbiome and Kidney Disease in Pediatrics: Does Connection Exist? Front. Microbiol. 7:235. doi: 10.3389/fmicb.2016.00235
Received: 07 December 2015; Accepted: 15 February 2016;
Published: 03 March 2016.
Edited by:
Joerg Graf, University of Connecticut, USAReviewed by:
David William Waite, Ministry for Primary Industries, New ZealandBenoit Chassaing, Georgia State University, USA
Copyright © 2016 Vasylyeva and Singh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tetyana L. Vasylyeva, dGV0eWFuYS52YXN5bHlldmFAdHR1aHNjLmVkdQ==
 Tetyana L. Vasylyeva
Tetyana L. Vasylyeva Ruchi Singh
Ruchi Singh