- 1Laboratory of Molecular Biology and Genetic Engineering, School of Biotechnology, Jawaharlal Nehru University, New Delhi, India
- 2AIRF, Jawaharlal Nehru University, New Delhi, India
Lipid rafts are dynamic, nanoscale assemblies of specific proteins and lipids, distributed heterogeneously on eukaryotic membrane. Flotillin-1, a conserved eukaryotic raft marker protein (RMP) harbor SPFH (Stomatin, Prohibitin, Flotillin, and HflK/C) and oligomerization domains to regulate various cellular processes through its interactions with other signaling or transport proteins. Rafts were thought to be absent in prokaryotes hitherto, but recent report of its presence and significance in physiology of Bacillus subtilis prompted us to investigate the same in pathogenic bacteria (PB) also. In prokaryotes, proteins of SPFH2a subfamily show highest identity to SPFH domain of Flotillin-1. Moreover, bacterial genome organization revealed that Flotillin homolog harboring SPFH2a domain exists in an operon with an upstream gene containing NFeD domain. Here, presence of RMP in PB was initially investigated in silico by analyzing the presence of SPFH2a, oligomerization domains in the concerned gene and NfeD domain in the adjacent upstream gene. After investigating 300 PB, four were found to harbor RMP. Among them, domains of Bas0525 (FlotP) of Bacillus anthracis (BA) showed highest identity with characteristic domains of RMP. Considering the global threat of BA as the bioterror agent, it was selected as a model for further in vitro characterization of rafts in PB. In silico and in vitro analysis showed significant similarity of FlotP with numerous attributes of Flotillin-1. Its punctate distribution on membrane with exclusive localization in detergent resistant membrane fraction; strongly favors presence of raft with RMP FlotP in BA. Furthermore, significant effect of Zaragozic acid (ZA), a raft associated lipid biosynthesis inhibitor, on several patho-physiological attributes of BA such as growth, morphology, membrane rigidity etc., were also observed. Specifically, a considerable decrease in membrane rigidity, strongly recommended presence of an unknown raft associated lipid molecule on membrane of BA. In addition, treatment with ZA decreased secretion of anthrax toxins and FlotP expression, suggesting potential role of raft in pathogenesis and physiology of BA. Thus, the present study not only suggest the existence and role of raft like entity in pathophysiology of BA but also its possible use for the development of novel drugs or vaccines against anthrax.
Introduction
Eukaryotic plasma membrane harbor small (size ~10–100 nm) but complex dynamic assemblies of lipids and proteins constituting membrane microdomains (commonly referred to as lipid rafts), which coordinate membrane signaling and trafficking with raft associated proteins (Bickel et al., 1997; Pike, 2006; Lingwood and Simons, 2010). Moreover, reports on association of lipid raft impairment with various human diseases like Alzheimer, Prion, Parkinson etc., reflect its profound significance in cellular organization as well as physiology (Donovan and Bramkamp, 2009).
Microdomains create heterogeneity on eukaryotic membrane due to high density of specific lipids and proteins, whose stoichiometry in these sub regions make them insoluble in the non-ionic detergents yielding detergent resistant membrane (DRM) and detergent sensitive membrane (DSM) regions (Bickel et al., 1997; Dermine et al., 2001). Flotillin-1, associated with DRMs, has been considered as the bona-fide marker for lipid rafts (Bickel et al., 1997; Dermine et al., 2001; Lingwood and Simons, 2010). Flotillins are conserved, integral membrane proteins with unique SPFH domain (named after Stomatins, Prohibitin, Flotillin, and HflK/C) on its N-terminal followed by Flotillin/oligomerization domain. Flotillin domain has coiled-coil regions which favor its self oligomerization and interaction with other colocalized proteins (Bach and Bramkamp, 2013). It is presumed that interaction between these two domains favors heterogeneous distribution of Flotillins on membrane (Bach and Bramkamp, 2013). In addition, Flotillins have also been shown to be involved in scaffolding of DRMs and recruitment of proteins to lipid rafts to facilitate proper interactions, oligomerization, and functioning such as signal transduction, coupling of membrane-cytoskeleton, and endocytosis (Bickel et al., 1997; Dermine et al., 2001; Krogh et al., 2001; Zhang et al., 2005; López and Kolter, 2010; Zhao et al., 2011; Bach and Bramkamp, 2013). These vital roles of microdomains in membrane organization strongly recommend its need in physiology as well as metabolism.
Till recently, prokaryotes were considered too simple to harbor such complex membrane organization (Bramkamp and Lopez, 2015). However, lipid rafts with homolog of eukaryotic RMP Flotillin-1 have been reported for vital cellular organization and functions such as maintenance of membrane integrity and signaling in non-pathogenic spore forming bacteria, Bacillus halodurans, and B. subtilis (Zhang et al., 2005; Meile et al., 2006; Donovan and Bramkamp, 2009; López and Kolter, 2010). The role of these bacterial rafts in membrane heterogeneity, sporulation, biofilm formation etc., (Donovan and Bramkamp, 2009; Bramkamp and Lopez, 2015) raises the idea that these might be the niche for numerous proteins involved in crucial bacterial physiological processes. Moreover, in bacteria, lipid raft disruption interferes with vital cellular processes without affecting cell viability and thus, is less likely to exhibit spontaneous mutation-acquired resistance against anti-raft compounds like statins that make these drugs less antimicrobial resistant (Donovan and Bramkamp, 2009; Bramkamp and Lopez, 2015). Commercially available anti-raft compounds have already been proved to be non-toxic to humans even at higher concentrations (Bramkamp and Lopez, 2015). In addition, rafts are also known to harbor sterol like molecules. Therefore, inhibitors of enzymes involved in rafts associated lipid biosynthesis have also been shown to affect microdomain organization and thus, physiology of the organism (López and Kolter, 2010). Hence, investigating existence of rafts in pathogenic microbes is likely to reveal hidden insights into bacterial pathophysiology to develop novel drugs or anti-raft compounds that may possess broad spectrum activity, less antimicrobial resistance and minimal host side effects.
Although, variations might exist in microdomains associated proteins or lipids, but features of Flotillin-like proteins remain conserved throughout. Thus, characterization of Flotillin appears to be the most reasonable approach to explore microdomains in bacteria. In this study, we performed in silico analysis to find protein(s) in bacterial pathogens with various primary and secondary structural attributes of Flotillin. The high identity of BAS0525 (FlotP) of Bacillus anthracis to above attributes of Flotillin-1 and importance of anthrax in public health globally prompted us to characterize FlotP (Simons and Sampaio, 2011). Also, it has been seen that biosynthesis of bacterial raft associated lipids involve squalene synthase (López and Kolter, 2010) whose known inhibitor Zaragozic acid (ZA) was found to impede the existence and formation of bacterial raft. Thus, to claim existence of raft, a parallel investigation of raft associated lipid molecule in bacteria would be highly recommended. Therefore, in order to further explore raft, effect of this inhibitor on several pathophysiological attributes of B. anthracis were also studied.
Materials and Methods
In silico Identification of Flotillin Homolog and its Related Gene in Pathogens
For the identification and selection of the most significant Flotillin homolog harboring pathogens from around 300 different pathogenic bacterial strains, following steps were followed. All sequences were retrieved from KEGG database. Initially, orthologs of SPFH2a domain containing homolog of prokaryotic Flotillin were searched by Clustal W using YuaG of B. subtilis as a subject. Since sequences of many of the orthologs showed similarity to B. subtilis YuaG, a criterion of presence of SPFH2a domain was taken. The alignment was used as input for the generation of phylogenetic tree between different species by Clustal Ω. CLUSTAL 2.1 was also employed to determine presence of various sub-domains like oligomerization/flotillin and SPFH2a in selected sequences. Oligomerization/flotillin domain identity was taken as second criteria for screening of suitable pathogens selected by initial analysis. Resulting list of bacteria fulfilling the initial two criteria were compared for the presence of NFeD domain harboring gene adjacent to Flotillin homolog. The values for identity matrix at various stages of analysis were calculated by alignment through Clustal Ω. UPGMA method was employed for phylogenetic tree construction. Bootstrapping of phylogenetic tree was performed by MEGA software 6.06 version with 1000 No. of Bootstrap Replications using Poisson model.
Operon Analysis of flotP and nfeD by RT-PCR
In silico prediction of operon was done using DOOR database (Mao et al., 2009). For validating co-transcription of flotP and nfeD, RNA was first isolated from overnight grown B. anthracis cells by trizol method. DNase treatment was given to remove genomic DNA contamination. cDNA was prepared using 2 μg of DNase treated RNA by High fidelity Reverse Transcriptase kit (Applied Biosystem). Co-transcription was then confirmed by PCR using intergenic primers (Fp: TACTCACTGTGATTTATATC, Rp: TAAAATGAGTAGAATTAAAA) covering a sequence of flotP and nfeD.
Bacterial Strain, Media, and Culture Conditions
Sterne 34F2 (pXO1+ pXO2−) strain of B. anthracis was used for all experiments. Escherichia coli DH5α and BL21(λDE3) were used for cloning and expression of flotP, respectively. For all experiments, E. coli and B. anthracis Sterne strain were grown in Luria Bertini (LB) and Brain Heart Infusion (BHI) medium containing 1% NaHCO3, respectively. Kanamycin (50 μg/ml) and ampicillin (100 μg/ml) were added wherever required. Eighty micrometers of Zaragozic acid (ZA; procured from Sigma) in 95% ethanol was used, for various comparative studies.
Cloning, Expression, Purification of FlotP, and Production of Antibody
Standard procedures were used for cloning. The open reading frames corresponding to BAS 0525 (flotP) was identified and retrieved from NCBI. Full length flotP (1581 bp) from genomic DNA of B. anthracis Sterne was amplified employing forward and reverse primer containing Nco1 and Xho1 restriction sites Amplicon flotP was ligated in pET-28a (Novagen) expression vector digested with same respective enzymes to obtain pET-flot with 6x Histidine tag at N- terminal. E. coli BL21(λDE3) cells were used for protein expression which was achieved by inducing the culture at O.D.600nm ~ 0.6 with the addition of 1 mM IPTG for 6 h at 37°C. Purification of FlotP was done from cytosolic fraction of recombinant E. coli BL21(λDE3) under native condition using Ni2+—NTA affinity chromatography as described previously (Agarwal et al., 2008). The purified protein was then dialyzed against 10 mM Tris (pH 8.0) and 10% glycerol. Protein concentration was determined by using Bradford reagent (Kruger, 2009) taking BSA as a standard. Purified, dialyzed protein was used to immunize Swiss albino mice of 4–6 weeks, procured from the animal house facility of the university, for raising polyclonal sera against specified protein (Sinha and Bhatnagar, 2010). The antibody titers were calculated using standard endpoint ELISA (Frey et al., 1998). Animal experiments were approved by Institutional Animal Ethics Committee of Jawaharlal Nehru University and were done under standard laboratory conditions at animal facility of the university.
Protein Identification by MALDI
Purified rFlotP was run on SDS-PAG followed by coomassie staining. The protein band corresponding to FlotP was excised manually and the sample is processed as described previously (Savary and Vasu, 2012). The MALDI-TOF analysis was executed by using Bruker Daltonic Ultraflex TOF/TOF system. For peptide search, MASCOT search engine and NCBI non-reduntant database were used.
Secondary Structure Prediction and Validation
Secondary structure was predicted by CFSSP server. This helped us to define patterns of various sub-regions of protein. Coiled-coil server was employed to determine the most prominent region of interaction or coiled-coil formation with window size of 14, 21, and 28 amino acids. TopPred software was used to predict transmembrane helix and its orientation on the membrane by calculating hydrophobicity index of the entire sequence. Secondary structure of FlotP was again validated by Circular Dichroism Spectroscopy (CDS) which was done by taking rFlotP at a concentration of 0.2 mg/ml in 10 mM phosphate buffer (pH 7.4). Spectra was obtained by using Jasco Corp., J-710 Spectropolarimeter at 25°C using a 1 mm cell, a wavelength scan from 190 to 240 nm at the rate of 20 nm/min. Minimum 10 scans were taken for each sample and its relevant baseline. The resulting averaged baseline spectrum was then deducted from averaged sample spectrum. The signals thus, obtained were used to calculate the molar ellipticity using the formula: θm = θo × 100/lc, where,
θm, molar ellipticity;
θo, observed ellipticity;
l, path length;
c, molar concentration.
Analysis was done as described previously (Greenfield, 2006; Rahi et al., 2011).
In vivo Expression Study
Quantitative Real Time PCR
To analyze the expression of flotP at transcript level, qRT-PCR was done at different growth stages according to the growth curve of B. anthracis (early exponential, mid exponential, late exponential, and onset of stationary phase). RNA was isolated by trizol method. High capacity cDNA reverse transcription kit (Applied Biosystems) was used for cDNA synthesis taking DNase I treated RNA as a template. PCR reactions were run in ABI PRISM 7500HT sequence detection system from Applied Biosystems. DNA Gyrase gene was taken as internal control for normalization. Result was analyzed by 2−ΔΔCT method. Data was represented as fold change in gene expression profile after normalizing with DNA Gyrase.
For analyzing effect of ZA on flotP expression at transcript level, RNA was isolated from B. anthracis culture grown overnight with or without ZA. cDNA preparation and qRT-PCR analysis was done as described above.
Immunoblotting
B. anthracis cells were allowed to grow till OD reached 0.3, 0.6, 0.9, 1.2, and 1.5 in BHI media. The cells were then pelleted down and subjected to sonication at 30% amplitude for 20 min (750 W Sonic Vibra Cell Sonicator). The lysates were then collected and quantified for their total protein content by Bradford reagent (Sigma). Volume of lysate containing equal protein was then mixed with SDS-loading dye and run on 12% SDS-PAGE followed by immunoblotting using nitrocellulose (NC) membrane. The blot was then developed using protein specific polyclonal antisera as a primary antibody and goat-antimouse IgG-alkaline phosphatase conjugated as secondary antibody.
For analyzing effect of ZA on FlotP expression at protein level, B. anthracis was allowed to grow overnight with or without ZA. Cell lysate preparation and immunoblotting was done as described above.
Localization Studies of FlotP
In Silico Tools
PSORTb and TMHMM were used for predicting localization of FlotP in B. anthracis. TopPred was used for determining hydrophobicity of the protein.
Flow Cytometric Analysis
FACS analysis was done as described previously (Matta et al., 2010) by using FACS Calibur (Beckton Dickinson, Heidelberg, Germany). Anti-rFlotP at a dilution of 1:200 was used. Pre-immune sera (1:200) and only cells were taken as negative control. Anti-rGAP A was used as positive control with the dilution 1:200. IgG-FITC labeled secondary antibody was used with the 1:100 dilution for all samples. Propidium iodide staining was used to differentiate between live and dead bacterial cells. Ten thousand cells were taken to analyze fluorescence and log-side scatter and log-forward scatter dot plot were used to detect the cells. Cell debris and larger cell aggregates were excluded by gating.
Immunolocalization Studies
Confocal fluorescence microscopy
Heterogeneous distribution of FlotP was analyzed by confocal microscopy using anti-rFlotP antibody. For this, B. anthracis cells were allowed to grow to OD600nm ~ 1.2, pelleted down and washed with 1X PBS thrice. Cells were then fixed using 4% paraformaldehyde in 1X PBS for 30 min at room temperature (RT). Washing was then done thrice with 1X PBS. Blocking of cells was done using 2% BSA for 1 h followed by three washes with IX PBS. Cells were incubated with anti-rFlotP antibody at a dilution of 1:200 overnight at 4°C, followed by PBS washing. Cells were then incubated with anti-mouse IgG-FITC labeled antibody (1:100) for 2 h at RT and again followed by three washes with PBS. Finally, the cells were mounted on glass slide and visualized under Olympus FluoView FV1000 Laser Scanning Confocal Microscope.
Immunogold electron microscopy
For imunogold localization studies, B. anthracis cells were harvested from stationary phase culture and fixed with 0.8% glutaraldehyde and 4% paraformaldehyde for 2 h at 4°C followed by washing with 1X PBS. Fixed cells were then blocked with 2% BSA and then imunostained with 1:200 dilution of anti-rFlotP followed by three consecutive PBS washes. The cells were then incubated with gold (10 nm) conjugated anti-mouse secondary antibody at a dilution of 1:100. Three washes with PBS were again given to cells which were then mounted on copper coated grids. Immunostained cells were then visualized using a Jeol 2100F transmission electron microscope (Jeol Analytic Instruments) with the acceleration voltage of 120 KV.
Submembrane Localization in Detergent Resistant (DRM) and Detergent Sensitive (DSM) Fraction
B. anthracis cells were allowed to grow overnight at 37°C in BHI. Cells were collected by centrifugation, washed twice with 1X PBS and finally resuspended in buffer containing 50 mM Tris (pH-8.0) at a cell concentration of nearly 1010 cells/ml. Sonication was then done for 30 s, followed by centrifugation at 4°C. The supernatant containing cytosolic fraction was collected. The pellet was then washed with 50 mM Tris (pH-8.0) followed by resuspension in extraction solution (8 M urea, 4% w/v CHAPS, 40 mM Tris, 2% DTT, and 0.2 w/v Bio-Lyte 3/10) and incubated for 30 min at RT. Supernatant containing the membrane fraction was then collected. The membrane fraction was further fractionated into DRM and DSM using CellLytic MEM protein extraction kit (Sigma) as described previously (López and Kolter, 2010). Quantitation of protein was done by Bradford assay. Equal dilutions of protein from each fraction were then loaded on 12% SDS-PAGE, immunoblotted on NC membrane and detected as described above.
Growth Analysis
Effect of ZA on growth of B. anthracis was assessed by inoculating 1% inoculum from overnight grown culture into fresh BHI containing 1% NaHCO3 with or without ZA. Aliquots were collected from each tube after every hour upto 9 h. Optical density at 600 nm was measured with required dilution periodically.
Morphology Analysis
For microscopic analysis, stationary phase B. anthracis cells grown in BHI containing 1% NaHCO3 with or without ZA were taken, washed with 1X PBS thrice followed by fixation in 1% (v/v) glutaraldehyde for 2 h at 4°C. Washing was done again with 1X PBS. Washed cells were then used for SEM analysis. For analysis, sample was prepared as described previously (Klee et al., 2006). Visualization was done using Zeiss EV040 scanning electron microscope (SEM) at Advance Instrumentation Research Facility (AIRF), JNU. Image J software was used for measuring cell dimensions.
Determination of Change in Membrane Fluidity of B. anthracis
B. anthracis cells were grown in the presence or absence ZA till OD600nm ~ 0.8. Cells were harvested by centrifuging at 10,000 rpm for 10 min followed by washing with 1X PBS twice. The cells were then fixed with 4% paraformaldehyde. Fixed cells were washed again with 1X PBS followed by its dilution in 1X PBS so that the resulting cell count would be equal to 3*108. Fluoresence polarization (Steady state fluorescence) and Time Resolved Florescence Spectrometry (TRFS) were exploited to study effect of inhibitors of sterol biosynthetic pathway on the physical state of the membrane of B. anthracis cells.
Fluorescence Polarization
Fluorescence polarization which gives a measure of membrane fluidity was assessed using fluorescent probe 1, 6-diphenyl- 1, 3, 5-hexatriene (DPH). The method described earlier (Prasad et al., 2006) was followed with little modifications. PBS washed cells were diluted to 1*108 cells/ml and were incubated with 2 μm DPH for 2 h at 30°C. The polarization ratio (p) was then calculated on Perkin-Elmer LS55 spectrofluorimeter (excitation 360 nm, emission 426 nm, and slit size 10 nm for both excitation and emission) as follows (Shinitzky and Barenholz, 1978).
where, IVV = Corrected fluorescence intensity obtained with excitation by vertically polarized light and emission detected by analyzer oriented vertically to the direction of polarized excitation light, IVH = Corrected fluorescence intensity obtained with excitation by vertically polarized light and emission detected by the analyzer oriented horizontal to the direction of polarized excitation light.
Grating Factor G, the correction for optical components of the instrument is calculated as IHV/IHH where subscripts HV and HH indicate the corrected fluorescence intensity values obtained with horizontal-vertical and horizontal-horizontal orientations for the polarizer and analyzer in that order, respectively.
TRFS
TRFS was done using FL920 Edinburgh spectrofluorimeter which was equipped with a xenon arc lamp, a polarizing device and a 375-nm laser diode (NanoLED 375 L, Horiba Ltd.) operated with a pulse frequency of 1 MHz. During the experiment photobleaching was avoided by using extremely low power (6–7 μW).
Decays in nanosecond timescales of DPH were measured in time correlated single photon counting (TCSPC) setup (FL920, Edinburgh Instruments, UK). Samples were excited at 375 nm using picosecond diode laser (pulse width ~100 ps). Fluorescence were dispersed in a monochromator and then collected by a MCP-PMT detector. The time-resolution of TCSPC setup of ~100 ps was determined by measuring the Instrument Response Function (IRF) using LUDOX solution. DPH labeled cells were placed in a quartz cuvette. Fluorescence was emitted at 426 nm and the time 100 ns was split into 4096 channels. Decay was measured at magical angle for 5000 peak counts. The G factor was determined by measuring IHH and IHV between the ranges 400–600 nm at five repeats. The anisotropy decay was determined by convolving the IVH and IVV.
Study of Effect of ZA on Secretion of Toxins (PA, LF, and EF)
The level of primary virulence factors like PA, LF, and EF secreted in the presence of different inhibitors were quantitated using ELISA as described previously (Aggarwal et al., 2015). Briefly, the media supernatants from BHI with 1% NaHCO3 grown B. anthracis cultures containing various inhibitors were first concentrated. Equal volumes of concentrated supernatants were coated in 96-well microtitre plate and kept overnight at 4°C. Plate was then blocked with 2% BSA for 1 h at RT followed by three consecutive washings using 1X PBST containing 0.05% Tween-20. The plate was then incubated with monoclonal goat anti-PA, anti-LF, and anti-EF antibodies in respective wells at a dilution of 1: 15,000, 1: 5000, 1:7000, respectively, and kept at RT for 1 h. The plate was given again three successive washes with 1X PBST followed by incubation with anti-goat-IgG-HRP (1:10,000) dilution at RT for 1 h. Plate was then again washed and colorimetric analysis was done by using TMB substrate and OD was measured at 630 nm.
Statistical Analysis
For analyzing statistical data, Microsoft excel was used. The data was expressed as mean ± S.D. otherwise indicated. The experiments were repeated at least three times and for analyzing the significance (p-value) between different data points, unpaired student t-test was used. p-value < 0.05 was considered as statistically significant.
Results and Discussion
Flotillin Homolog in Pathogenic Bacteria
Flotillin-1 is a member of SPFH superfamily harboring evolutionary conserved SPFH (Stomatin, Prohibitin, Flotillin, and HflK/C protein) domain which shows affinity for membrane microdomains (Tavernarakis et al., 1999; Zhang et al., 2005; Browman et al., 2007). Prokaryotic SPFH super family has been subdivided into 12 subfamilies based on the sequence and operon structure (Hinderhofer et al., 2009) out of which SPFH2a subfamily of proteins show highest similarity to Flotillin-1. A recent study reported presence of Flotillin homolog, YuaG of SPFH2a subfamily as raft marker in a non-pathogenic bacteria B. subtilis (López and Kolter, 2010; Dempwolff et al., 2012; Mielich-Süss et al., 2013). Considering the fact that membrane microdomains harbor variety of signaling proteins, it is of utmost interest to investigate presence of such SPFH2a domain harboring proteins in PB as a key step toward investigation of membrane microdomains in infectious bacteria. This study may unveil some unknown aspects of signaling mechanisms as well as pathophysiology of the pathogens. For identification of the best homolog of raft marker-Flotillin-1, 3 different domains namely SPFH2a, and Flotillin/oligomerization domains of the candidate gene and Nfed domain of upstream gene (belongs to same operon) were taken into consideration. A pictorial representation of these domains taken for the identification of Flotillin-like gene in bacterial pathogens are given in Figure 1A.
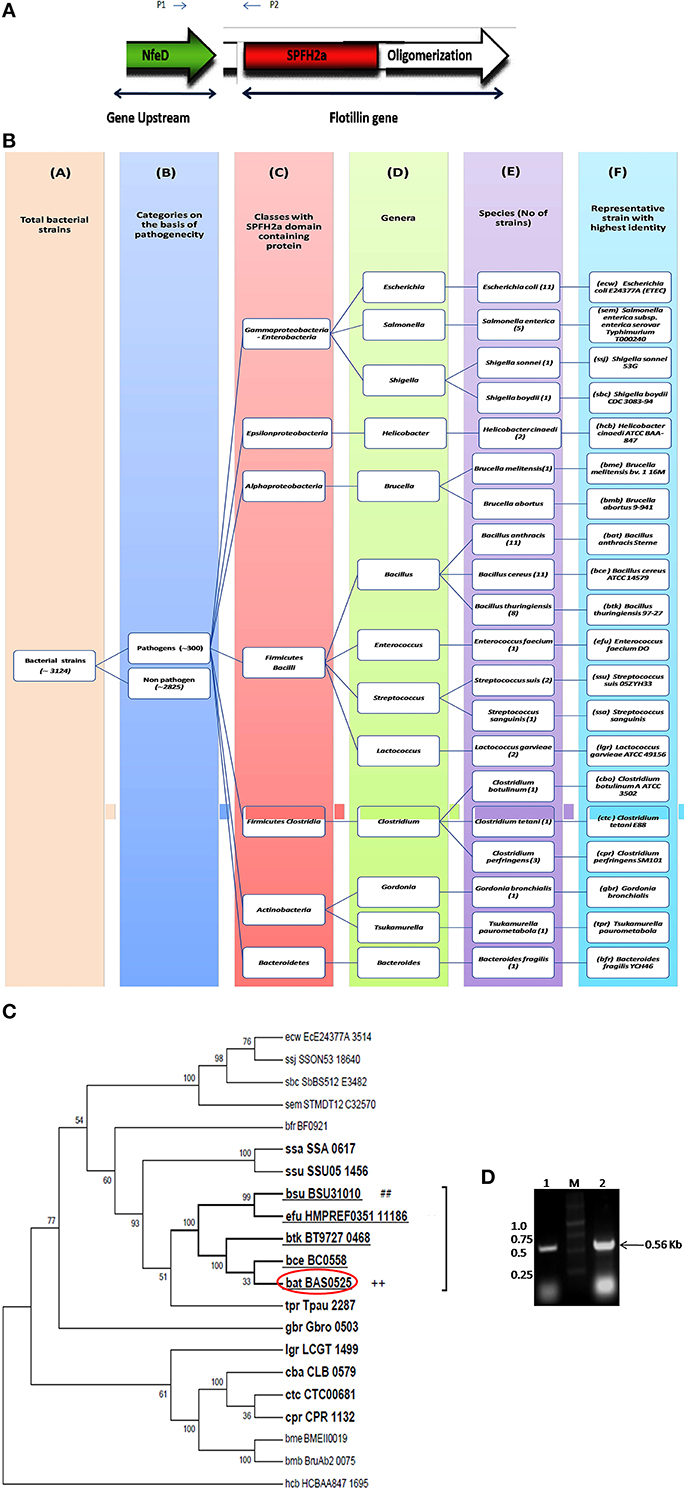
Figure 1. Flotillin-associated domains and identification of flotillin homologs in pathogens. (A) Representation of various domains associated with Raft Marker Protein or Flotillin-1. (B) Schematic representation of 20 shortlisted bacterial pathogens containing proteins of SPFH2a superfamily, each representing individual species. Columns: C, Classes; D, Genera; E, Species; F, Selected strain showing maximum identity with SPFH2a domain. Number in bracket in column E represents total number of strains in that particular species showing identity with SPFH2a domain. (C) Phylogenetic tree showing distance between various PB containing proteins with SPFH2a domain of YuaG of B. subtilis (##), characteristic feature of prokaryotic Flotillin. Among all SPFH2a domain harboring pathogens, the species in bold showed presence of oligomerization domain also. Among them, underlined species showed highest identity with Nfed domain. Encircled species with ++ is finally selected for further charecterization. (D) Co-transcription of Bas0524 (NfeD domain containing gene) and Bas0525, flotP. Amplification with primers P1 and P2 [as represented in (A)] was analyzed when cDNA (Lane 1) and genomic DNA (Lane 2) were taken as template. M represents Marker.
Homology search for SPFH2a domain of YuaG of B. subtilis has been performed in 300 PB strains showed 66 strains containing proteins with SPFH2a domain that belonged to 20 bacterial species. Since, many of these 66 strains belonged to the same species; therefore, the strain from the particular speciesshowing highest similarity to SPFH2a domain was selected as the representative for that species. On this basis, we took a representative pathogen from each of 20 different species for further analysis (Figure 1B). The similarity and distance between all bacterial pathogens harboring SPFH2a domain containing proteins have been represented by the phylogenetic tree in Figure S1. Figures S2A,B, respectively, list 66 bacterial pathogens along with their respective KEGG codes, length of proteins and SPFH2a domain alignment of all pathogens.
Since, Flotillin-1 possesses a characteristic oligomerization/Flotillin domain, our next step was to find this domain in the proteins of shortlisted 20 pathogens. Phylogenetic analysis (Figure 1C) and percent identity matrix (Table 1) further identified only 12 pathogens with more than 25% identity with this domain and interestingly they were all gram positive. Among these, eight were firmicutes from genus Bacillus and showed more than 40% identity.
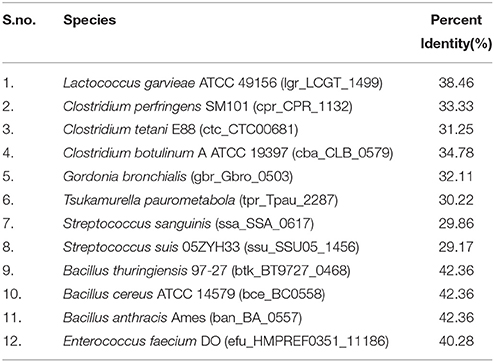
Table 1. List of pathogens showing identity with Flotillin/Oligomerization domain of hsa_10211, Flotillin-1 of Homo sapiens (taken as reference).
In prokaryotes, Flotillin homologs have been shown to be a part of the operon which also contains upstream of Flotillin, a gene with NfeD domain known to help in anchoring of Flotillin within lipid rafts (López and Kolter, 2010; Dempwolff et al., 2012). In non-pathogenic B. subtilis, colocalization of YuaF (NFeD2), and YuaG and their influence on mutual localization appears to favor interplay between Flotillin and NFeD within raft assemblies (Krogh et al., 2001). Therefore, identification of pathogenic bacteria containing Flotillin homolog and NfeD gene in the same operon merits attention. Upon examining the percent identity of NFeD domain in the upstream region of the candidate genes selected for above 12 gram positive bacteria taking B. subtilis YuaF as reference, only four were found to show more than 30% identity (Figure 1C, Table 2). Among the screened organisms, maximum identity (37.36%) was observed for both B. anthracis and B. thuringiensis. Of special mention is the fact that Enterococcus faecium displayed significant identity value of 35.29% even without containing the desired domain (Figure S3). Among the above two pathogens, B. anthracis, a potential biological warfare agent, appears to be an ideal pathogen for such study in order to address global health concerns. Therefore, Flotillin homolog in B. anthracis, Bas0525 (hereafter referred to as FlotP) was further investigated to study rafts in pathogenic bacteria.

Table 2. List of pathogens showing significant identity with NFeD domain harboring gene, bsu_BSU31020 of B. subtilis.
Bas 0525 (flotP) and Bas0524 (nfeD Harboring Gene) Co-Transcribes in the Form of Operon
Studies done so far, our in silico predictions as described above as well as DOOR database for operon analysis suggested the co-transcription of Bas0525 (flotP) and Bas0524 (nfeD harboring gene) in the form of operon at genomic level. Thus, to validate the genomic organization of Bas0525-0524, operon analysis was done by RT-PCR using intergenic primers, represented by P1 and P2 in Figure 1A. On analysis, a band corresponding to the size (~560 bp) of the amplified product was observed with both genomic DNA as well as cDNA (Figure 1D) confirming the genomic organization of the two genes in operon. The result is in consistence with what observed in B. subtilis where co-transcription of Flotillin homolog and NfeD harboring gene was found to have a potential role in proper localization of Flotillin homolog on the bacterial membrane (López and Kolter, 2010; Dempwolff et al., 2012).
MALDI Confirmed Bas0525 Encoded Protein as Flotillin Homolog
FlotP was amplified from B. anthracis genome by gene specific primers yielding a 1581 bp amplicon which was further cloned in pET-28a(+) expression vector adding extra nucleotides corresponding to hexa histidine tag (Figure 2A). The rFlotP was allowed to express in BL21(λDE3) cells in the presence of 1 mM IPTG (Figure 2B). Expressed protein corresponding to ~60 KDa was purified from cytoplasmic fraction by Ni-NTA chromatography (Figure 2C). rFlotP was further confirmed by MALDI analysis (Figure 2D).
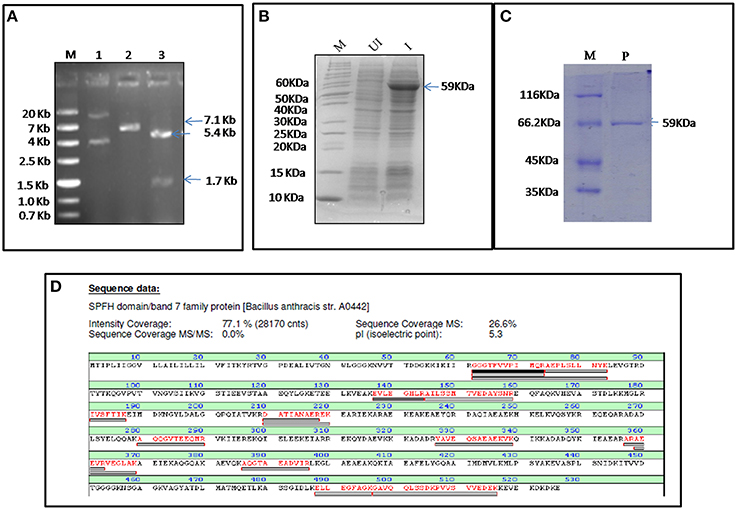
Figure 2. Cloning, expression, and purification of FlotP. (A) Clone confirmation by restriction digestion of pET28a-flotP. Double digestion (DD) confirmed the presence of insert by fall out at 1.7 Kb corresponding to flotp size. Lanes: M- Marker (ZipRuler™; #SM1373), 1- pET28a-flotP, 2- Single digest, 3-double digest. (B) Overexpression of FlotP in BL21 (λDE3) E. coli cells. Lanes: M, Marker (Pageruler unstained; #26614); UI, Uninduced, and I, Induced. (C) Purification by Ni-NTA chromatography. Lanes: M, Marker (Pierce™ Unstained Protein MW Marker; #26610); P, Purified Protein. (D) MALDI analysis showing the sequence of the protein with the peptide coverage marked in red. The peptide hits showed highest similarity to SPFH domain BAND 7 family protein of B. anthracis i.e., FlotP.
flotP of B. anthracis Contains Predominantly α-Helical and Coiled-Coil Structure
Flotillin-1 consists of α-helical region predominantly (Song et al., 2011). To determine secondary structure of B. anthracis FlotP, both in silico and in vitro analysis was performed in this study. CFSSP Server predicted presence of 66.73% α-helix region in the protein (Figure 3A). However, 95% of Flotillin/oligomerization domain comprised of α-helices favoring coiled-coil formation. The presence of coiled-coil regions was confirmed by COILS Server (with window size 14, 21, and 28) which suggest the presence of two coiled-coil regions between amino acid (aa) 211–257 and 290–332 (Figure 3B). Previous studies report that coiled-coil domains are essential for proper localization, oligomerization, and protein-protein interactions in rafts to facilitate functioning of Flotillin-like proteins in bacteria (Krogh et al., 2001; Zhang et al., 2005). Diverse cellular processes associated with Flotillin assemblies, instead of being lipid dependent have been found to depend on the interactions mediated by coiled-coil domain with other raft associated proteins (Krogh et al., 2001). Moreover, cumulative analysis predicted topology of FlotP similar to mitochondrial prohibitin (Sanz et al., 2003) with transmembrane helix (Figures 3C,D) i.e., N-terminal region toward cytoplasm and C-terminal exposed outwards (Figure 3E). The transmembrane region or hydrophobic stretch between aa 4–26 is likely to facilitate the membrane association. C-terminus of FlotP contains a conserved characteristic Flotillin domain with ala-glu-ala-glu (AEAE) repeats (Figure 3E) favoring the formation of coiled-coil like structures similar to eukaryotic Flotillin (Kurrle et al., 2012). CDS of purified protein confirmed α-helical secondary structure of FlotP (Figure 3F).
Figure 3. Prediction and validation of FlotP Topology and Secondary structure. (A) Secondary structure by CFSSP server showing high percentage of α-helical structure (H-Helix; E-Sheet; T-Turns). (B) Prediction of coiled-coil region by Coils server with each size of aa windows. (C) Analysis of hydrophobicity index value by TopPred server predicting the presence of transmembrane region. Y-axis of the graph represents hydrophobicity value predicting the possible location of the particular amino acid stretch of the protein. Value > 1 indicate cytoplasmic fragment; 0–1, transmembrane; < 0, membrane. (D) Prediction of various sub regions by TopPred with their sub-cellular locations. (E) Schematic representation depicting secondary structure and topology of FlotP. Region A in white color represents transmembrane (TM) region (aa 4–26), region B in red color represents SPFH2a domain- characteristic of prokaryotic Flotillin (aa 34–203), region C in yellow color indicates Flotillin or oligomerization domain (aa 203–365) and region D in blue shows domain with unknown function (DUF; aa 365–495). The two shaded boxes (C1- aa 211–257 and C2- aa 290–332) in yellow region C shows two high probability regions of coiled coil or trimer/dimer or interaction. Characteristic 1st, 2nd, and 3rd AEAE repeats are indicated at positions aa 335, 245, and 405. (F) CD spectra of purified rFlotP protein showing absolute probability of α-helix secondary structure.
B. anthracis flotP is Membrane Localized
Lipid raft and associated proteins have been reported to be localized on cell membrane (Lingwood and Simons, 2010). Localization of B. anthracis FlotP was determined through both in silico and in vitro analysis. PSORTb (Gardy et al., 2005) and TMHMM (Krogh et al., 2001) softwares predicted membrane localization for FlotP (Figures S4A,B). In addition, hydropathy index determination by TopPred (Krogh et al., 2001) was in agreement with YuaG (Flotillin homolog) of B. subtilis, confirming the presence of hydrophobic or transmembrane region (Figures 3C,D)
In order to determine the subcellular localization of FlotP in vivo, flow cytometric analysis was done using rFlotP antiserum raised in mice. The data showed positive shift in anti-rFlotP treated bacterial cells in FL1 quadrant which was absent in bacterial cells only (without treating with pre-immune sera) as well as cells treated with pre-immune sera (Figure 4A). Glyceraldehyde 3 phosphate dehydrogenase (GAPDH) is a well-known surface protein of B. anthracis (Matta et al., 2010). Thus, for flow analysis, anti-rGapA (polyclonal sera against one of its isoform) was taken as a positive control which showed a positive shift confirming the surface localization of FlotP (Figure 4A). Figure S5 showed histogram statistics for flow analysis.
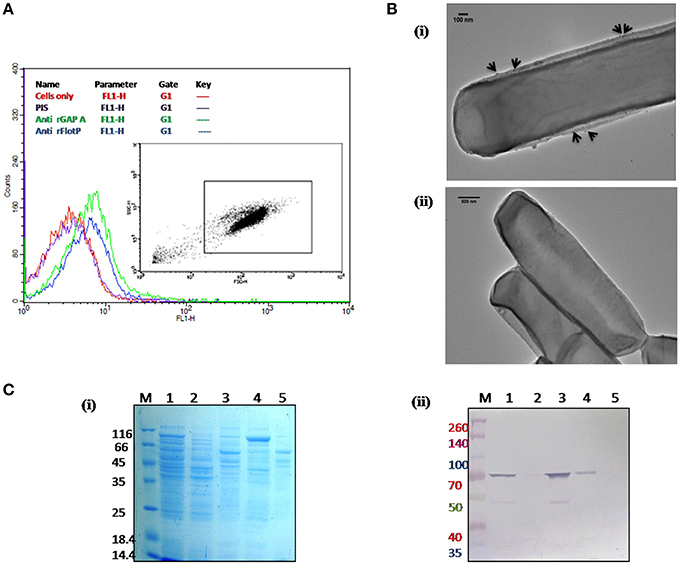
Figure 4. Localization of B. anthracis FlotP. (A) Flow cytometric analysis for B. anthracis cells treated with 1:200 dilution of each anti-rFlotP IgG. anti-rGapA, Pre-immune serum, and only cells were taken as control. After staining with FITC-labeled secondary antibody (1:100), fluorescence was analyzed in FL1 and displayed as a histogram. Inset shows the gated population inside a dot plot between log of both forward scatter (size) and side scatter (granularity/shape). (B) Immunogold localization of FlotP on the membrane of B. anthracis. Whole vegetative cell of B. anthracis was used for TEM analysis. Panel (i) The black arrows indicated the gold particles around the membrane of the cell when anti-rFlotP was used as primary antibody. Images were captured at magnification of 12,000x (ii) No dots were observed when pre-immune sera was used as primary antibody. Images were captured at magnification of 6000x (C) Localization by immunoblotting. Equal amounts of proteins (3 μg) from each fraction were loaded in respective lanes. Panel (i) indicates Coommasie stained SDS-PAGE with all fractions and panel (ii) shows immunoblotting with anti-rFlotP. Lanes: 1, cellular lysate; 2, cytosolic fraction; 3, membrane fraction after ultracentrifugation; 4, DRM; 5, DSM; and M, Marker.
In order to visualize the FlotP localization on the membrane of B. anthracis, immunogold transmission electron microscopy of stationary phase cells was done. Upon visualization, the cells treated with anti-rFlotP and subsequently with Gold conjugated anti-mouse IgG, were found to have gold particles attached with the cell membrane (Figure 4Bi) which were found to be absent in the cells treated with pre-immune sera and then gold conjugated secondary antibody (Figure 4Bii). This further validated our previous observation.
In addition, immunoblotting analysis of various subcellular fractions of B. anthracis was also carried out (Figure 4C) confirming the membrane localization of FlotP.
Presence of Flotillins on the plasma membrane probably allows it to act as an interface between intracellular signaling proteins, signaling receptors, and cytoskeleton to support intra- and inter-cellular communication and activate diverse physiological processes (Donovan and Bramkamp, 2009; Dempwolff et al., 2012).
Heterogenous as Well as Punctuate Distribution of flotP on the Plasma Membrane and its Association with DRM Fraction Favors Microdomain Existence in B. anthracis
Insolubility in the non-ionic detergents due to association with DRMs is a characteristic property of lipid rafts (Brown, 2002; Magee and Parmryd, 2003). Raft based membrane heterogeneity is maintained at nanoscale to form more stable/ordered membrane assemblies which allows partitioning into functionalized sub-compartments (Bickel et al., 1997). The dynamic liquid-liquid immiscibility of DRMs serves as the basis for the concept of sub-compartmentalization in rafts to focus and coordinate membrane bioactivity (Simons and Sampaio, 2011).
DRMs having lipid raft associated proteins can be isolated by floatation on sucrose density gradients owing to their low density and high lipid-to-protein ratio (Brown, 2002; López and Kolter, 2010). For this study, cell membrane fractionations were performed based on the solubility in non-ionic detergents to separate proteins associated with DRM fractions. On analyzing the DRMs and DSMs of B. anthracis on SDS-PAGE, a significant difference in their respective protein profile was observed which clearly implied heterogeneous distribution of proteins with associated macromolecules on B. anthracis membrane (Figure 4Ci). Moreover, FlotP was detected in the DRM fraction using immunoblotting with anti-rFlotP (Figure 4Cii). Figure S6 showed the immunoblotting analysis taking DNA Gyrase and GAPDH as a negative and positive controls, respectively for membrane localization. This result was analogous to the previous reports for Flotillin in eukaryotic lipid rafts, directly favoring our prediction for presence of raft like feature in B. anthracis.
Moreover, in prokaryotes, lipid rafts have been found to be involved in supporting the bacteria to perform or coordinate its vital functions. In B. subtilis a sporulation histidine kinase KinC which is involved in biofilm signaling, shows enhanced activity in the presence of YuaG (Donovan and Bramkamp, 2009). Proteins required for biofilm formation, attachment, signaling and adhesion are found to be present in DRMs in the bacterial pathogen, Staphylococcus aureus (Donovan and Bramkamp, 2009). Similar cellular organization thus might be common for all SPFH2a harboring pathogenic bacteria.
In addition to association with DRM, membrane microdomains provide heterogeneity to the cell membrane (Simons and Vaz, 2004) and associated proteins display punctate distribution. Heterogeneous distribution in specialized membrane microdomains has been observed for many bacterial signaling proteins (Donovan and Bramkamp, 2009). This is supported by the report of punctate and heterogeneous distribution on membrane for Flotillin-like proteins in B. halodurans and B. subtilis (Donovan and Bramkamp, 2009). In order to determine the distribution pattern of FlotP in B. anthracis, we performed indirect immunofluorescence analysis using confocal microscopy. FITC binding was observed as non-uniform green fluorescent patches on the membrane representing the heterogeneous distribution of FlotP (Figure 5C). Anti-GroEL antibody, taken as a negative control showed uniform fluorescence over the entire bacterial surface (Figure 5B) but no such fluorescence was observed with pre-immune sera (Figure 5A). GroEL, a chaperone is known to be distributed uniformly over the cell surface in many bacteria including B. anthracis (Hennequin et al., 2001; Sinha and Bhatnagar, 2010). Thus, the above data confirmed the heterogeneous distribution of FlotP on the cell membrane favoring eukaryotic microdomain-like feature on membrane of prokaryotic pathogen. The role of such proteins in activating signaling cascades and receptor activities might be facilitated by their compartmentalization in restricted membrane domains (Troost et al., 2004).
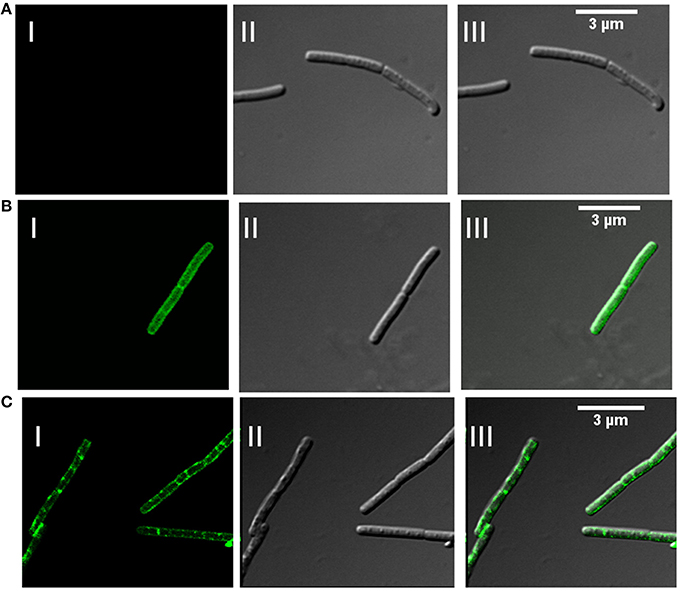
Figure 5. Heterogeneous distribution of FlotP on membrane of B. anthracis by indirect immunofluorescence analysis using Confocal microscopy. (A) B. anthracis cells with pre-immune sera, (B) homogenous distribution of GroEL over the surface of bacteria with anti-rGroEL antibody, and (C) heterogeneous distribution of FlotP with anti-rFlotP antibody. Images were captured at 100X magnification and zoomed twice for final images. Anti-mouse IgG-FITC antibody was used as secondary antibody. In each Figure, panels I, II, and III represents FITC channel, DIC channel, and superimposition of panels I and II, respectively.
FlotP is Constitutively Expressed in B. anthracis at All Stages of Growth
Eukaryotic Flotillin is a highly conserved protein, known to express constitutively in almost all cell types (Babuke and Tikkanen, 2007). Till date this protein has not been reported with any natural mutation (Kokubo et al., 2000). Of note is that changes in mammalian Flotillin-1 expression levels have been found to be associated with Alzheimer's disease (Kokubo et al., 2000) and type 2 diabetes (James et al., 2001).
In this study, growth stage specific expression of FlotP in B. anthracis was analyzed both at protein and transcript levels. For this, different growth stages were selected on the basis of growth curve of B. anthracis viz. early exponential, mid exponential, late exponential, early stationary, and stationary phase. FlotP showed constitutive expression at all stages of growth both at mRNA and protein levels as evident from Figure 6. In parallel, we also analyzed the expression of FlotP at protein level in virulent strain of B. anthracis also Figure S7 which confirm its presence in most pathogenic form of B. anthracis.
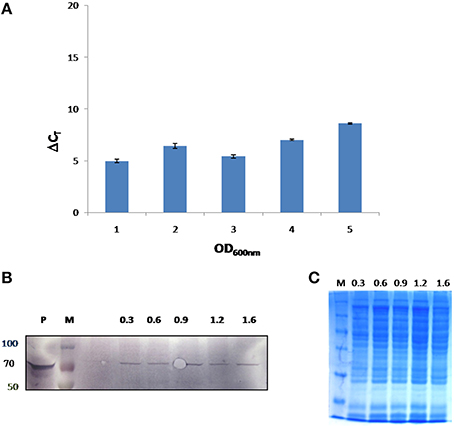
Figure 6. In vivo expression of FlotP during different growth phases of B. anthracis. (A) RT-PCR was done for FlotP transcript using total RNA of various growth stages. Graph represents normalized Ct value with respect to different stages of growth. Data is a representative of three independent experiments showing similar trend. (B) Immunoblotting for in vivo FlotP expression in B. anthracis at different growth stages. Lanes: P- Purified rFlotP, M- marker, and rest indicate OD600 i.e., 0.3, 0.6, 0.9, 1.2, 1.6 for different growth stages taken. (C) SDS page showing loaded protein for immunobloting of each OD600 level.
In eukaryotes, constitutive expression of Flotillin has been implicated in various crucial cellular processes including insulin signaling, membrane trafficking, phagocytosis, endocytosis, epidermal growth factor receptor signaling, T-lymphocyte activation, cell motility, and transformation (Babuke and Tikkanen, 2007). Constitutive expression of FlotP suggests its critical role in various physiological and cellular processes in bacteria such as communication between cell and environment for activation of signaling pathways (Agarwal et al., 2008; Donovan and Bramkamp, 2009). Similar to eukaryotes, Flotillin homologs in non-pathogenic bacteria are active across the membrane to facilitate interaction and oligomerization of proteins (Gardy et al., 2005; Meile et al., 2006). Membrane microdomains in bacteria harboring flotillin homologs known so far have been found to contain Flotillin-1 homologs associated with signaling and transport proteins (Donovan and Bramkamp, 2009). Moreover, studies have also implicated that these proteins are involved in non-redundant and diverse vital functions related to sporulation, cell shape and motility (López and Kolter, 2010). Therefore, the constitutive expression of this protein may be necessitated by pathogen to fulfill their basic structural and functional attributes, making it a significant target for pre or post infection stage in infectious diseases. A graphical representation of the highlights of this part of the study has been depicted in Figure 7.
Figure 7. Graphical representation of the highlights (strategies used at different steps and outcome) of this study. In silico, in vitro, and in vivo approaches were employed for characterization of FlotP, an eukaryotic Flotillin homolog. Numericals represent the different steps of study; bold arrows indicate the domains used for the basis of homology search.
Effect of Raft Associated Lipids Biosynthetic Pathway Inhibitors on B. anthracis
In order to validate our hypothesis of raft existence on B. anthracis membrane, apart from characterization of FlotP, homolog of raft marker protein Flotillin-1, we extended our study toward the search of raft associated lipid candidate. We carried out our study by analyzing the effect of a raft associated lipid biosynthetic pathway inhibitor, Zaragozic acid (ZA). ZA, a compelling inhibitor of squalene synthase, blocks sterol biosynthesis specifically cholesterol synthesis in mammals and ergosterol synthesis in fungi which are the well-known raft associated lipid moieties (Bergstrom et al., 1995; de Souza and Rodrigues, 2009). Although, there is no direct evidence of presence of sterols in bacteria, the foremost report of existence of lipid raft on B. subtilis membrane showed the significant effect of ZA (López and Kolter, 2010) on its physiology. Its effect on lipid raft integrity and thus Flotillin localization suggest the occurrence of similar kind of moieties in bacteria also.
To corroborate the presence of sterol derivative in pathogen we analyzed effect of ZA on several pathophysiological attributes of B. anthracis like growth, morphology, membrane fluidity and toxin secretion.
ZA Slows Down the Growth of B. anthracis
To determine the effect of ZA on growth of bacteria, B. anthracis Sterne was inoculated in BHI media containing 1% NaHCO3 in presence or absence of ZA and the growth was examined periodically. As shown in Figure 8, an insignificant variation was observed for first few hours (till mid log phase). However, a drastic reduction in growth was observed at stationary phase (p < 0.01).
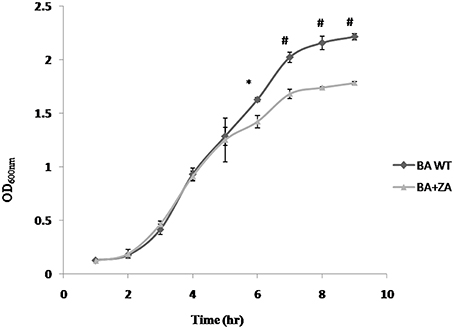
Figure 8. Effect of ZA on growth of B. anthracis. Data is standard mean ± SME of three independent experiments. *indicate p ≤ 0.05 and #indicates p ≤ 0.001.
The observation is completely different from what has been reported in B. subtilis where presence of ZA did not affect the growth (López and Kolter, 2010). However, in yeast (Machida et al., 1999; Hornby et al., 2003) and mammals (Daicho et al., 2007; Joo and Jetten, 2010), ZA has been found to decrease the growth rate by increasing the level of farnesol which affect growth by altering cell cycle signaling mechanism and by apoptosis, respectively. Moreover, in some prokaryotes like S. aureus, farnesol has also been seen to have a negative effect on growth of the cells through some undefined interaction with the cell membrane (Jabra-Rizk et al., 2006) In addition, some studies also suggest that even a little deviation in the membrane integrity of the cell affects rate of replication causing reduction in growth rate (Firshein, 1989; Firshein and Kim, 1997; Nordström, 2003). Thus, we may hypothesize that the above observation may be due to some possible change in the B. anthracis membrane due to the presence of ZA that needs to be further explored.
ZA Increases the Cell Dimension
As per our above observations, any alteration in the membrane integrity affects the growth by affecting rate of replication and hence, it must also affect the cell morphology. Considering this, morphology of ZA treated as well as untreated cells were analyzed by SEM (Figure 9A). Interestingly, cells in presence of ZA were found to increase the mean cell length (Figure 9Bi) as well as width (Figure 9Bii) significantly by ~2 times (p < 0.001) and ~1.2 times (p < 0.01), respectively, as compared to untreated cells. Table 3 indicated the comparison of mean cell dimensions.
Figure 9. Effect of ZA on morphology of B. anthracis. (A) Stationary phase cells from culture grown with (i) or without ZA (ii) were used for SEM analysis. (B) Box-and-whiskers plot to represent the cell dimensions where maximum and minimum size of population, median, and 25th and 75th population percentile is represented by whiskers, center bar, and each box, respectively. (i) cell length, (ii) cell width. **indicates p ≤ 0.01 and #indicates p ≤ 0.001.
Table 3. The cell dimensions (μm) of B. anthracis cells in presence (BA+ZA) or absence (BA WT) of ZA.
Moreover, in yeast also ZA has been found to affect the cell cycle by inhibiting replication (Machida et al., 1999) thus we may hypothesize that the variation observed in cell size might be due to down regulation of replication which might be due to the deviation in organized membrane in presence of ZA. However, detailed study has to be done which is beyond the scope of this study.
ZA Renders Membrane of B. anthracis More Fluid
Fluidity or rigidity are the physical state parameters of membrane (Barrera et al., 2012), which is strongly influenced by raft rigidity that in turn depend on the concentration of sterol derivatives in it (Mattson, 2005). Cells adapt to the changes in environment through modulation of membrane properties and lipid composition resulting in altered membrane fluidity (Barrera et al., 2012). Cell membrane serves as a barrier for entry or exit of biomolecules across the cells and intrinsic (inherent) or acquired cellular changes due to alteration in structure may accompany the cells to modulate the pathogenicity and virulence of the organism (Diakogiannis et al., 2013). In order to determine the effects of endogenous sterol derivatives on membrane fluidity, the biophysical properties of the B. anthracis membrane was accessed in presence of ZA using fluorescence polarization and TRFS.
Steady state fluorescence polarization gives an average measure of the membrane fluidity/rigidity of all the cells. This study revealed a significant change in the fluorescence polarization in B. anthracis in presence of ZA in the media (Figure 10A). The control cells grown without ZA had a higher membrane order and therefore, displayed higher fluorescence polarization values i.e., more rigid membrane than the ZA treated cells.
Figure 10. Effect of ZA on membrane fluidity of B. anthracis. (A) Mean fluorescence polarization “p” values (inversely propotional to membrane fluidity) of B. anthracis grown in absence (control) and presence of ZA. (B) Decay curve for DPH labeled B. anthracis grown with or without ZA. (C) Time dependent Fluorescence decay (with fits) in linear scale with respect to time in nanoseconds of DPH in the ZA treated and untreated cells.
Sterol rich membrane microdomains have been reported to play a crucial role in membrane structure, organization, and function in eukaryotes. As already stated, fluorescence polarization is inversely proportional to membrane fluidity and decrease in fluorescence polarization values are typically due to an increase in rotational mobility of the fluorophore, which is influenced by packing of sterols and fatty acyl chains.
In addition, to further confirm the above observed result, membrane fluidity was also measured by TRFS which measure the time resolved fluorescence decay of fluorescent molecules (DPH) within a specific lipid environment and thus gives a measure of microviscosity of cells. Fluorescence lifetime is a reliable indicator of polarity changes in local environment of the DPH. Life time of DPH has been found to be reduced in presence of water in its immediate environment (Arora et al., 2004).
The data revealed that decay time for DPH in Zaragozic acid treated cells are lower than the decay time in the untreated (control) bacterial cells by 31.6% (Figures 10B,C, Table 4). Water penetration may be more due to lesser rigidifying effect of sterol/other raft associated lipids as revealed by fluorescence polarization. These decay patterns provided the observed changes in the cellular microenvironment of the respective membrane after treatment with various inhibitors
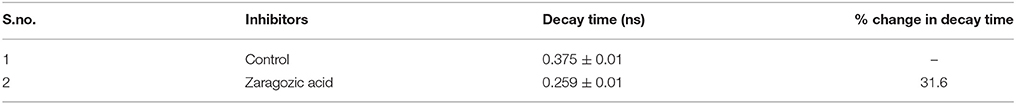
Table 4. Fluorescence lifetime decay of DPH in nanoseconds (ns) at mid log phase untreated cells (control) and B. anthracis cells treated with ZA.
The data obtained from TRFS validated the experiments results of steady state fluorescence polarization. The increase in fluidity for membrane of bacterial cells grown in the presence of zaragozic acid corroborates with the decrease in decay time of DPH in cells grown with these inhibitors. Decay of DPH becomes faster in fluid membrane due to decrease in microviscosity.
These results indicated the possibility of existence of sterol surrogates or lipid candidates in bacterial microdomains similar to eukaryotic sterol asssociated with lipid rafts which has been seen to be affected by fungal sterol biosynthetic pathways inhibitors and thus, treatment with such inhibitors possibly lead to altered composition of such lipid molecules resulting in loose packing of membranes and enhanced membrane fluidity. The observed results may be due to differences in the levels of compaction of cell membrane between untreated and ZA treated bacterial cells clearly suggesting the possibility of existence of sterol surrogates, other lipid candidates and raft associated membrane proteins.
ZA Decreases Toxin Secretion by B. anthracis, thus has Potential to Reduce Virulence
Secretion of virulence factors (PA, Protective antigen; LF, Lethal factor; EF, Edema factor) by B. anthracis in extracellular milieu is a major contributing virulence factor leading to anthrax. Though the mechanism of their secretion is still unknown, a recent report suggested the secretion of these factors in the form of vesicles involving variety of lipids on vesicular membrane (Rivera et al., 2010). Considering the fact that lipid raft like entities possess a large variety of transport or secretory proteins, we were interested in finding whether there is any relation between raft and toxin secretion in B. anthracis. Therefore, we carried out this part of study by determining the effect of ZA on toxin secretion. For this, media supernatants of B. anthracis grown with or without ZA was collected and quantitaion of toxin components was done by ELISA using monoclonal antibodies against PA, LF and EF. In our analysis, ZA was found to decrease significantly the secretion of LF and EF by ~41% (p < 0.05) and ~47% (p < 0.01), respectively whereas, no significant change was observed in PA (p > 0.05; Figure 11).
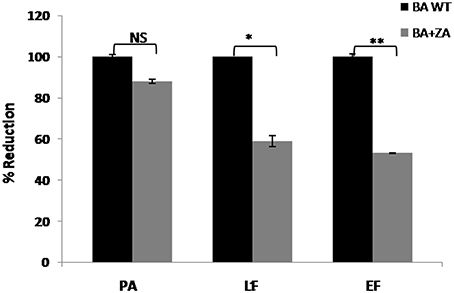
Figure 11. Effect of ZA on toxin production by B. anthracis. Quantitation of major virulence factors (PA, LF, and EF) was done by ELISA. Bars represent the percent reduction of B. anthracis treated with ZA (BA+ZA) with respect to untreated cells (BA WT). NS, non-significant; *p < 0.05 and **p < 0.01.
Although the exact role of ZA in secretion of toxin components is yet to be established but our data suggest the involvement of lipid raft in this crucial physiological process and thus, pathogenesis of B. anthracis. Therefore, we may propose that targeting lipid raft or use of anti-raft compounds may provide alternate treatment strategy against anthrax or other raft harboring pathogens also.
ZA Affects Expression and Localization of FlotP
Our study revealed the membrane localization of FlotP as well as the existence of some unknown sterol derivative on B. anthracis cell surface. To check whether there is any link between FlotP and anonymous lipid moieties, the effect of ZA on FlotP expression was analyzed both at transcript as well as protein level. qRT-PCR analysis showed a significant decrease of approximately Two-fold in flotP transcript level in ZA treated cells (Figure 12A). Interestingly, even at protein level, expression of FlotP was found to be decreased by ~1.8-folds as observed by immunoblotting (Figure 12B). These observations suggest the possibility of some regulatory mechanism to coordinate expression of FlotP according to change in the endogenous availability of sterol derivatives. Thus, we may hypothesize that certain genetic coordination might be existing between raft protein FlotP and sterol kind of lipid moieties in addition to their physical association in the raft of cell.
Figure 12. Effect of ZA on FlotP expression in vivo. (A) Data represents the fold change in flotP expression at mRNA level by qRT–PCR taking dna gyrase as a constitutively expressed gene for normalization of CT values. The experiment was repeated two times biologically with three technical replicates. Data shown here is mean ± SME of one of the two independent biological experiment. **indicates p ≤ 0.01. (B) Expression analysis of FlotP at protein level by immunoblotting. Lanes: 1, Total cell lysate of ZA treated B. anthracis cells; 2, total cell lysate of ZA untreated cells; 3, purified rFlotP. (C) Immunofluorescence analysis of FlotP localization on B. anthracis cell surface using confocal microscopy; BA WT (i), BA+ZA (ii). In each figure, panels I, II, and III represents FITC channel, DIC channel, and superimposition of panels I and II, respectively.
In addition, immunofluorescence analysis revealed disruption in the localization of FlotP on B. anthracis cell surface in the presence of ZA (Figure 12C). The result was in consistence with B. subtilis where continuous treatment of ZA lead to complete abrogation of enzyme involved in the synthesis of anonymous lipid moiety which in turn disturbed the localization of Flotillin homolog.
Thus, these observations indicated the association of FlotP with some unknown sterol derivatives to form raft like entity. This is the preliminary report about the existence of raft like features in B. anthracis. Therefore, the proper mechanism of interaction needs to be explored in order to answer unsolved queries of raft in B. anthracis.
Conclusion
In this study, we have done for the first time, a detailed investigation on the existence of raft like feature in any PB by characterizing it in B. anthracis. Homolog of Flotillin-1, FlotP of B. anthracis, showed similarity in genetic organization, domain architecture as well as distribution pattern with other known RMPs. Constitutive expression of FlotP indicates its possible critical role which might range from basic bacterial metabolism to virulence via regulatory or signaling pathways. Moreover, studies done with anti-raft agent ZA revealed its significant effect on patho-physiology of BA such as growth, morphology, membrane fluidity, toxin secretion, FlotP expression as well as its distribution over cell surface etc. Thus, it may be concluded that lipid rafts have the potential to serve as a new target for controlling bacterial virulence and infections. Lipid raft targeted drugs are likely to have broad spectrum activity, lower microbial resistance and minimal host cell toxicity, thus making them excellent candidates for future use in biomedical applications to address challenges of bacterial infections. Further exploration of the candidate lipid molecule involved in prokaryotic raft formation may prove assistive in identifying other raft harboring pathogens. The specificity of lipid as well as protein of the pathogenic raft would lead to the development of potential anti-infectious agent.
Author Contributions
VS designed, performed and analyzed the experiments; SA performed and analyzed the experiments; DS performed few experiments; VS, SA, TP, RB wrote paper, RB contributed reagents, materials and instruments.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
VS acknowledge the support of CSIR for Senior Research Fellowship. SA and DS thank UGC for Senior Research Fellowships. TP acknowledges support of UPEII scheme of JNU. RB acknowledges fund support from DBT for BSLIII facility and other resources. The authors thank JNU for infrastructural support. Mr. Ashok Kumar Sahu from AIRF- JNU, is duly acknowledged for his support in confocal imaging.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00169
Abbreviations
SPFH, Stomatin, Prohibitin, Flotillin, and HflK/C protein super family; DRM, Detergent Resistant Membrane; DSM, Detergent Sensitive Membrane; CDS, Circular Dichroism Spectroscopy.
References
Agarwal, S., Kulshreshtha, P., Mukku, D. B., and Bhatnagar, R. (2008). α-Enolase binds to human plasminogen on the surface of Bacillus anthracis. Biochim. Biophys. Acta 1784, 986–994. doi: 10.1016/j.bbapap.2008.03.017
Aggarwal, S., Somani, V. K., and Bhatnagar, R. (2015). Phosphate starvation enhances the pathogenesis of Bacillus anthracis. Int. J. Med. Microbiol. 305, 523–531. doi: 10.1016/j.ijmm.2015.06.001
Arora, A., Raghuraman, H., and Chattopadhyay, A. (2004). Influence of cholesterol and ergosterol on membrane dynamics: a fluorescence approach. Biochem. Biophys. Res. Commun. 318, 920–926. doi: 10.1016/j.bbrc.2004.04.118
Babuke, T., and Tikkanen, R. (2007). Dissecting the molecular function of reggie/flotillin proteins. Eur. J. Cell Biol. 86, 525–532. doi: 10.1016/j.ejcb.2007.03.003
Bach, J. N., and Bramkamp, M. (2013). Flotillins functionally organize the bacterial membrane. Mol. Microbiol. 88, 1205–1217. doi: 10.1111/mmi.12252
Barrera, F. N., Fendos, J., and Engelman, D. M. (2012). Membrane physical properties influence transmembrane helix formation. Proc. Natl. Acad. Sci. U.S.A. 109, 14422–14427. doi: 10.1073/pnas.1212665109
Bergstrom, J. D., Dufresne, C., Bills, G. F., Nallin-Omstead, M., and Byrne, K. (1995). Discovery, biosynthesis, and mechanism of action of the zaragozic acids: potent inhibitors of squalene synthase. Annu. Rev. Microbiol. 49, 607–639. doi: 10.1146/annurev.mi.49.100195.003135
Bickel, P. E., Scherer, P. E., Schnitzer, J. E., Oh, P., Lisanti, M. P., and Lodish, H. F. (1997). Flotillin and epidermal surface antigen define a new family of caveolae-associated integral membrane proteins. J. Biol. Chem. 272, 13793–13802. doi: 10.1074/jbc.272.21.13793
Bramkamp, M., and Lopez, D. (2015). Exploring the existence of lipid rafts in bacteria. Microbiol. Mol. Biol. Rev. 79, 81–100. doi: 10.1128/MMBR.00036-14
Browman, D. T., Hoegg, M. B., and Robbins, S. M. (2007). The SPFH domain-containing proteins: more than lipid raft markers. Trends Cell Biol. 17, 394–402. doi: 10.1016/j.tcb.2007.06.005
Brown, D. A. (2002). Isolation and use of rafts. Curr. Protoc. Immunol. 11, 11.10. doi: 10.1002/0471142735.im1110s51
Daicho, K., Maruyama, H., Suzuki, A., Ueno, M., Uritani, M., and Ushimaru, T. (2007). The ergosterol biosynthesis inhibitor zaragozic acid promotes vacuolar degradation of the tryptophan permease Tat2p in yeast. Biochi. Biophys. Acta 1768, 1681–1690. doi: 10.1016/j.bbamem.2007.03.022
Dempwolff, F., Möller, H. M., and Graumann, P. L. (2012). Synthetic motility and cell shape defects associated with deletions of flotillin/reggie paralogs in Bacillus subtilis and interplay of these proteins with NfeD proteins. J. Bacteriol. 194, 4652–4661. doi: 10.1128/JB.00910-12
Dermine, J.-F., Duclos, S., Garin, J., St-Louis, F., Rea, S., Parton, R. G., et al. (2001). Flotillin-1-enriched lipid raft domains accumulate on maturing phagosomes. J. Biol. Chem. 276, 18507–18512. doi: 10.1074/jbc.M101113200
de Souza, W., and Rodrigues, J. C. F. (2009). Sterol biosynthesis pathway as target for anti-trypanosomatid drugs. Interdiscip. Perspect. Infect. Dis. 2009:642502. doi: 10.1155/2009/642502
Diakogiannis, I., Berberi, A., Siapi, E., Arkoudi-Vafea, A., Giannopoulou, L., and Mastronicolis, S. K. (2013). Growth and membrane fluidity of food-borne pathogen Listeria monocytogenes in the presence of weak acid preservatives and hydrochloric acid. Front. Microbiol. 4:152. doi: 10.3389/fmicb.2013.00152
Donovan, C., and Bramkamp, M. (2009). Characterization and subcellular localization of a bacterial flotillin homologue. Microbiology 155, 1786–1799. doi: 10.1099/mic.0.025312-0
Firshein, W. (1989). Role of the DNA/membrane complex in prokaryotic DNA replication. Annu. Rev. Microbiol. 43, 89–120. doi: 10.1146/annurev.mi.43.100189.000513
Firshein, W., and Kim, P. (1997). Plasmid replication and partition in Escherichiacoli: is the cell membrane the key? Mol. Microbiol. 23, 1–10. doi: 10.1046/j.1365-2958.1997.2061569.x
Frey, A., Di Canzio, J., and Zurakowski, D. (1998). A statistically defined endpoint titer determination method for immunoassays. J. Immunol. Methods 221, 35–41. doi: 10.1016/S0022-1759(98)00170-7
Gardy, J. L., Laird, M. R., Chen, F., Rey, S., Walsh, C., Ester, M., et al. (2005). PSORTb v. 2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics 21, 617–623. doi: 10.1093/bioinformatics/bti057
Greenfield, N. J. (2006). Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 1, 2876–2890. doi: 10.1038/nprot.2006.202
Hennequin, C., Porcheray, F., Waligora-Dupriet, A.-J., Collignon, A., Barc, M.-C., Bourlioux, P., et al. (2001). GroEL (Hsp60) of Clostridium difficile is involved in cell adherence. Microbiology 147, 87–96. doi: 10.1099/00221287-147-1-87
Hinderhofer, M., Walker, C. A., Friemel, A., Stuermer, C. A., Möller, H. M., and Reuter, A. (2009). Evolution of prokaryotic SPFH proteins. BMC Evol. Biol. 9:10. doi: 10.1186/1471-2148-9-10
Hornby, J. M., Kebaara, B. W., and Nickerson, K. W. (2003). Farnesol biosynthesis in Candida albicans: cellular response to sterol inhibition by zaragozic acid B. Antimicrob. Agents Chemother. 47, 2366–2369. doi: 10.1128/AAC.47.7.2366-2369.2003
Jabra-Rizk, M., Meiller, T., James, C., and Shirtliff, M. (2006). Effect of farnesol on Staphylococcus aureus biofilm formation and antimicrobial susceptibility. Antimicrob. Agents Chemother. 50, 1463–1469. doi: 10.1128/AAC.50.4.1463-1469.2006
James, D. J., Cairns, F., Salt, I. P., Murphy, G. J., Dominiczak, A. F., Connell, J. M., et al. (2001). Skeletal muscle of stroke-prone spontaneously hypertensive rats exhibits reduced insulin-stimulated glucose transport and elevated levels of caveolin and flotillin. Diabetes 50, 2148–2156. doi: 10.2337/diabetes.50.9.2148
Joo, J. H., and Jetten, A. M. (2010). Molecular mechanisms involved in farnesol-induced apoptosis. Cancer Lett. 287, 123–135. doi: 10.1016/j.canlet.2009.05.015
Klee, S. R., Özel, M., Appel, B., Boesch, C., Ellerbrok, H., Jacob, D., et al. (2006). Characterization of Bacillus anthracis-like bacteria isolated from wild great apes from Cote d'Ivoire and Cameroon. J. Bacteriol. 188, 5333–5344. doi: 10.1128/JB.00303-06
Kokubo, H., Lemere, C. A., and Yamaguchi, H. (2000). Localization of flotillins in human brain and their accumulation with the progression of Alzheimer's disease pathology. Neurosci. Lett. 290, 93–96. doi: 10.1016/S0304-3940(00)01334-3
Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. doi: 10.1006/jmbi.2000.4315
Kruger, N. J. (2009). “The Bradford method for protein quantitation,” in The Protein Protocols Handbook, ed J. M. Walker (Totowa, NJ: Springer), 17–24.
Kurrle, N., John, B., Meister, M., and Tikkanen, R. (2012). Function of Flotillins in Receptor Tyrosine Kinase Signaling and Endocytosis: Role of Tyrosine Phosphorylation and Oligomerization. Rijeka: INTECH Open Access Publisher.
Lingwood, D., and Simons, K. (2010). Lipid rafts as a membrane-organizing principle. Science 327, 46–50. doi: 10.1126/science.1174621
López, D., and Kolter, R. (2010). Functional microdomains in bacterial membranes. Genes Dev. 24, 1893–1902. doi: 10.1101/gad.1945010
Machida, K., Tanaka, T., Yano, Y., Otani, S., and Taniguchi, M. (1999). Farnesol-induced growth inhibition in Saccharomyces cerevisiae by a cell cycle mechanism. Microbiology 145, 293–299. doi: 10.1099/13500872-145-2-293
Magee, A. I., and Parmryd, I. (2003). Detergent-resistant membranes and the protein composition of lipid rafts. Genome Biol. 4, 234–234. doi: 10.1186/gb-2003-4-11-234
Mao, F., Dam, P., Chou, J., Olman, V., and Xu, Y. (2009). DOOR: a database for prokaryotic operons. Nucleic Acids Res. 37(Suppl 1.), D459–D463. doi: 10.1093/nar/gkn757
Matta, S. K., Agarwal, S., and Bhatnagar, R. (2010). Surface localized and extracellular Glyceraldehyde-3-phosphate dehydrogenase of Bacillus anthracis is a plasminogen binding protein. Biochim. Biophys. Acta 1804, 2111–2120. doi: 10.1016/j.bbapap.2010.08.004
Mattson, M. P. (2005). Membrane Microdomain Signaling: Lipid Rafts in Biology and Medicine. Totowa, NJ: Springer Science & Business Media.
Meile, J. C., Wu, L. J., Ehrlich, S. D., Errington, J., and Noirot, P. (2006). Systematic localisation of proteins fused to the green fluorescent protein in Bacillus subtilis: identification of new proteins at the DNA replication factory. Proteomics 6, 2135–2146. doi: 10.1002/pmic.200500512
Mielich-Süss, B., Schneider, J., and Lopez, D. (2013). Overproduction of flotillin influences cell differentiation and shape in Bacillus subtilis. MBio 4, e00719–13. doi: 10.1128/mBio.00719-13
Nordström, K. (2003). The replicon theory 40 years: an EMBO workshop held in Villefranche sur Mer, France, January 18–23, 2003. Plasmid 49, 269–280. doi: 10.1016/s0147-619x(03)00019-2
Pike, L. J. (2006). Rafts defined: a report on the keystone symposium on lipid rafts and cell function. J. Lipid Res. 47, 1597–1598. doi: 10.1194/jlr.E600002-JLR200
Prasad, T., Chandra, A., Mukhopadhyay, C. K., and Prasad, R. (2006). Unexpected link between iron and drug resistance of Candida spp.: iron depletion enhances membrane fluidity and drug diffusion, leading to drug-susceptible cells. Antimicrob. Agents Chemother. 50, 3597–3606. doi: 10.1128/AAC.00653-06
Rahi, A., Rehan, M., Garg, R., Tripathi, D., Lynn, A. M., and Bhatnagar, R. (2011). Enzymatic characterization of Catalase from Bacillus anthracis and prediction of critical residues using information theoretic measure of relative entropy. Biochem. Biophys. Res. Commun. 411, 88–95. doi: 10.1016/j.bbrc.2011.06.099
Rivera, J., Cordero, R. J., Nakouzi, A. S., Frases, S., Nicola, A., and Casadevall, A. (2010). Bacillus anthracis produces membrane-derived vesicles containing biologically active toxins. Proc. Natl. Acad. Sci. U.S.A. 107, 19002–19007. doi: 10.1073/pnas.1008843107
Sanz, M. A., Tsang, W. Y., Willems, E. M., Grivell, L. A., Lemire, B. D., van der Spek, H., et al. (2003). The mitochondrial prohibitin complex is essential for embryonic viability and germline function in Caenorhabditis elegans. J. Biol. Chem. 278, 32091–32099. doi: 10.1074/jbc.M304877200
Savary, B. J., and Vasu, P. (2012). “Routine identity confirmation of recombinant proteins by MALDI-TOF mass spectrometry,” in Recombinant Gene Expression, ed A. Lorence (New York, NY: Springer), 37–50.
Shinitzky, M., and Barenholz, Y. (1978). Fluidity parameters of lipid regions determined by fluorescence polarization. Biochim. Biophys. Acta 515, 367–394. doi: 10.1016/0304-4157(78)90010-2
Simons, K., and Sampaio, J. L. (2011). Membrane organization and lipid rafts. Cold Spring Harb. Perspect. Biol. 3, a004697. doi: 10.1101/cshperspect.a004697
Simons, K., and Vaz, W. L. (2004). Model systems, lipid rafts, and cell membranes 1. Annu. Rev. Biophys. Biomol. Struct. 33, 269–295. doi: 10.1146/annurev.biophys.32.110601.141803
Sinha, K., and Bhatnagar, R. (2010). GroEL provides protection against Bacillus anthracis infection in BALB/c mice. Mol. Immunol. 48, 264–271. doi: 10.1016/j.molimm.2010.08.001
Song, J., Chen, W., Lu, Z., Hu, X., and Ding, Y. (2011). Soluble expression, purification, and characterization of recombinant human flotillin-2 (reggie-1) in Escherichia coli. Mol. Biol. Rep. 38, 2091–2098. doi: 10.1007/s11033-010-0335-4
Tavernarakis, N., Driscoll, M., and Kyrpides, N. C. (1999). The SPFH domain: implicated in regulating targeted protein turnover in stomatins and other membrane-associated proteins. Trends Biochem. Sci. 24, 425–427. doi: 10.1016/S0968-0004(99)01467-X
Troost, J., Lindenmaier, H., Haefeli, W. E., and Weiss, J. (2004). Modulation of cellular cholesterol alters P-glycoprotein activity in multidrug-resistant cells. Mol. Pharmacol. 66, 1332–1339. doi: 10.1124/mol.104.002329
Zhang, H.-M., Li, Z., Tsudome, M., Ito, S., Takami, H., and Horikoshi, K. (2005). An alkali-inducible flotillin-like protein from Bacillus halodurans C-125. Protein J. 24, 125–131. doi: 10.1007/s10930-004-1519-3
Keywords: pathogen, Bacillus anthracis, flotillin, SPFH, DRM, FlotP, microdomain, lipid raft
Citation: Somani VK, Aggarwal S, Singh D, Prasad T and Bhatnagar R (2016) Identification of Novel Raft Marker Protein, FlotP in Bacillus anthracis. Front. Microbiol. 7:169. doi: 10.3389/fmicb.2016.00169
Received: 26 June 2015; Accepted: 01 February 2016;
Published: 17 February 2016.
Edited by:
Bassam Hallis, Public Health England, UKReviewed by:
Paras Jain, Albert Einstein College of Medicine, USAShivani Agarwal, Northwestern University, USA
Rajendra Prasad, Institute of Integrative Sciences and Health and Institute of Biotechnology, India
Copyright © 2016 Somani, Aggarwal, Singh, Prasad and Bhatnagar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rakesh Bhatnagar, cmFrZXNoYmhhdG5hZ2FyQGpudS5hYy5pbg==;
Tulika Prasad, cHJhc2FkdHVsaWthQG1haWwuam51LmFjLmlu;
cHJhc2FkdHVsaWthQGhvdG1haWwuY29t
†These authors have contributed equally to this work.
 Vikas K. Somani1†
Vikas K. Somani1† Somya Aggarwal
Somya Aggarwal Rakesh Bhatnagar
Rakesh Bhatnagar