
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 15 February 2016
Sec. Microbiotechnology
Volume 7 - 2016 | https://doi.org/10.3389/fmicb.2016.00070
This article is part of the Research Topic Biotechnology of microalgae, based on molecular biology and biochemistry of eukaryotic algae and cyanobacteria. View all 17 articles
The current fascination with algal biofuel production stems from a high lipid biosynthetic capacity and little conflict with land plant cultivation. However, the mechanisms which enable algae to accumulate massive oil remain elusive. An enzyme for triacylglycerol (TAG) biosynthesis in Chlamydomonas reinhardtii, CrDGTT2, can produce a large amount of TAG when expressed in yeast or higher plants, suggesting a unique ability of CrDGTT2 to enhance oil production in a heterologous system. Here, we performed metabolic engineering in Saccharomyces cerevisiae by taking advantage of CrDGTT2. We suppressed membrane phospholipid biosynthesis at the log phase by mutating OPI3, enhanced TAG biosynthetic pathway at the stationary phase by overexpressing PAH1 and CrDGTT2, and suppressed TAG hydrolysis on growth resumption from the stationary phase by knocking out DGK1. The resulting engineered yeast cells accumulated about 70-fold of TAG compared with wild type cells. Moreover, TAG production was sustainable. Our results demonstrated the enhanced and sustainable TAG production in the yeast synthetic platform.
Biofuel production is highly demanded as an alternative to the limited fossil fuels. The current fascination with eukaryotic algae as a resource of biofuel production stems from a high lipid biosynthetic capacity and little competition for land required for plant cultivation. However, technical difficulty in gene manipulation of many eukaryotic algae species hampers potential usefulness of these organisms as a synthetic platform of metabolic engineering to produce biofuel. On the other hand, Saccharomyces cerevisiae is an advanced model unicellular eukaryotic microorganism for metabolic engineering as well as basic molecular biological studies on metabolism.
Triacylglycerol (TAG) is a major source of biodiesel (Durrett et al., 2008; Hu et al., 2008), which is synthesized from sn-1,2-diacylglycerol (DAG) by the catalysis of acyltransferase in many organisms such as S. cerevisiae, Chlamydomonas reinhardtii, and Arabidopsis thaliana (Sandager et al., 2002; Zhang et al., 2009; Hung et al., 2013). Because DAG is produced from phosphatidic acid (PA), which is also a substrate for the biosynthesis of major membrane phospholipids, TAG accumulation often competes with membrane lipid biosynthesis (Figure 1). In yeast cells, the primary metabolic flux of glycerolipids differs by the growth status. Biosynthesis of storage lipids such as TAG and membrane lipids such as phospholipids share initial steps of the pathway up to PA production (Henry et al., 2012). In growing cells, newly synthesized glycerolipids are mostly used to produce membrane lipids required for active cell proliferation (Figure 1, green arrow). With entry into the stationary growth phase, the major flux switches to storage lipid biosynthesis. Here, PA is dephosphorylated to DAG by a PA phosphatase encoded by PAH1 (Han et al., 2006), which is subsequently converted to TAG by diacylglycerol acyltransferases (DGATs; Figure 1, brown arrow). Stationary-phase cells can resume their growth once depleted nutrients are added, such as the dilution of cell culture with fresh medium. This growth resumption activates hydrolysis of accumulated TAG to DAG by a set of TAG lipases (Kurat et al., 2009), and DAG is further phosphorylated by DAG kinase 1 (DGK1) to PA (Han et al., 2008a,b) to provide a substrate for membrane lipid biosynthesis (Figure 1, orange arrow). At this phase, DGK1 encodes a key enzyme involved in phospholipid synthesis and recovery after growth resumption (Fakas et al., 2011a).
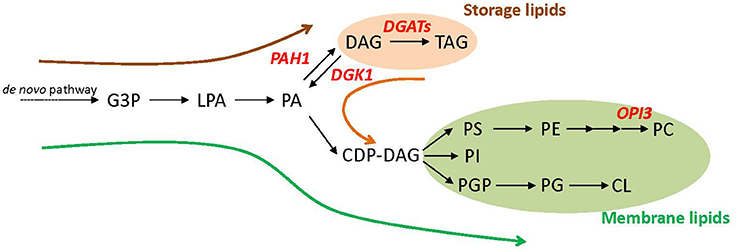
Figure 1. Schematic representation of glycerolipid metabolism in Saccharomyces cerevisiae. Dominant metabolic flux at three different growth phases (log phase, green arrow; stationary phase, brown arrow; growth resumption from the stationary phase, orange arrow) and key metabolic genes engineered in this study (in red letters) are shown. CDP-DAG, cytidine diphosphate-diacylglycerol; CL, cardiolipin; DAG, sn-1,2-diacylglycerol; G3P, glycerol 3-phosphate; LPA lysophosphatidic acid; PA, phosphatidic acid; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PGP, phosphatidylglycerophosphate; PI, phosphatidylinositol; PS, phosphatidylserine; TAG, triacylglycerol.
C. reinhardtii is a model organism for the study of TAG production in the eukaryotic microalgae. DGAT, which represents a major TAG biosynthetic enzyme, catalyzes the transfer of acyl moiety from acyl-CoA to DAG. In C. reinhardtii, DGAT is of 2 types: Type 1 with single isogene (CrDGTT1) and Type 2 with five isogenes (CrDGTT1-5) (Merchant et al., 2012). Interestingly, it was recently reported that heterologous overexpression of CrDGTT2 in S. cerevisiae or A. thaliana increased TAG content by 9- or 25-fold, respectively (Hung et al., 2013; Sanjaya et al., 2013), suggesting highly robust enzyme function of CrDGTT2 in a heterologous system.
In this study, we aimed to perform metabolic engineering that achieves enhanced and sustainable TAG production in the synthetic platform of S. cerevisiae by taking advantage of CrDGTT2 as a robust algal gene resource.
Cells were grown in YPD media (2.0 g of Bacto peptone [Difco 211677], 1.0 g of Bacto yeast extract [Difco 212750], 2.0 g of glucose [Merck 1.08337] in 100 ml of H2O produced by Milli-Q [Millipore]) unless otherwise stated. Cells harboring the URA3 and LEU2 marker plasmids were grown in synthetic complete media lacking uracil and leucine (SC -Ura, -Leu). The OD600 in Figure 3A and Supplementary Figure 1 was measured by diluting fully grown cultures to OD600 of 0.1 and cultured for 60 h at 30°C. In Figures 4A–C, yeast cells were freshly inoculated from glycerol stocks. After reaching the stationary phase, the cell culture was diluted with fresh medium to OD600 of 0.05 and grown for 60 h to the late stationary phase, then time-course observation was started. The dilution was conducted twice at 60 h intervals. Data are mean±SD from three biological replicates.
The yeast expression vector pCH108 was created by replacing the URA3 marker of pCH078 (Hung et al., 2013) with the LEU2 marker derived from pRS315. First, XmaI and BclI restriction sites were created by amplifying the 4895-bp fragment of the pCH078 with the primers CH376 and CH377. Next, a 1574-bp fragment of the LEU2 marker was amplified from pRS315 with the primers CH374 and CH375. The obtained fragments were digested with XmaI and BclI and ligated to construct pCH108. The polymerase chain reaction (PCR) was performed using Phusion polymerase (Finnzymes F-530S, ThermoFisher Scientific) under the following condition; an initial denaturing step of 5 min at 95°C followed by 35 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 3 min and then a final extension at 72°C for 5 min. The plasmids and primers are listed in Table 1 and Supplementary Table 1, respectively.
The 2593-bp open reading frame of ScPAH1 (YMR165C) was amplified with the primers CH367 and CH368, cloned into pENTR/D-TOPO (Invitrogen, Carlsbad, CA), digested with AscI and NotI, and inserted into AscI/NotI sites of pCH108 to construct pCH109. This plasmid contains GPD promoter for constitutive overexpression (Blazeck et al., 2012).
To produce CHY034 (dgk1Δ::KANMX, opi3Δ::HIS), a 932-bp fragment of opi3Δ::HIS3 was amplified with the primers CH430 and CH431 from pRS313, and transformed into dgk1Δ::KANMX. The plasmid pCH109, which overexpresses ScPAH1 (OE-PAH1), was transformed into CHY034 (dgk1Δ::KANMX, opi3Δ::HIS) to produce CHY138 (Δdgk1Δopi3 OE-PAH1). Then, the plasmid pCH091, which overexpresses CrDGTT2 (OE-CrDGTT2), was transformed into CHY138 to produce CHY101, CHY140, CHY141, and CHY142 (Δdgk1Δopi3 OE-PAH1 OE-CrDGTT2). The transformation was performed as described previously by mixing the plasmid with the PLATE reagent, salmon sperm DNA and yeast cells (Gietz and Woods, 2002). The yeast strains are listed in Table 2.
Lipid analysis was performed as follows using yeast cells obtained from 100 ml of culture by centrifugation (3000 × g for 5 min). Cells were lyophilized using freeze drier (Alpha 1-2, Martin Christ Gefriertrocknungsanlagen GmbH, Germany) and dry cell weight was measured with fine weighing scale (BEL engineering, Cat. No. M214Ai, Italy). Prior to the lipid extraction, cells were treated with pre-heated isopropanol containing 0.01% butylated hydroxytoluene at 75°C for 15 min to inactivate phospholipase activity. Total lipids were extracted essentially as described previously using chloroform-methanol solvent system (Folch et al., 1957) with the ratio of solvent (by vol): sample (by weight) as 20:1 for 100 mg of sample. Briefly, cells were suspended in chloroform:methanol (1:2, by vol) to homogenity, followed by addition of chloroform and deionized water (dH2O). Cell suspension was vortexed vigorously and left for 30 min at room temperature. Organic phase was recovered and residual lipids in cell pellet were extracted twice by vigorous vortex with chloroform. The combined organic extracts were vortexed with 1% KCl solution, and separated organic phase was washed once with dH2O. Lipids were dried up under nitrogen stream, dissolved in chloroform:methanol (2:1, by vol) and stored at −30°C till use. Each lipid class was separated by thin layer chromatography (Silica Gel 60 F254 plate, Merck). Separation of TAG was as described previously (Hung et al., 2013) and of phospholipids involved two-dimensional separation with the solvent system of chloroform/methanol/ammonia (65:35:5, by vol) for the first dimension and chloroform/acetone/methanol/acetic acid (50:20:10:10, by vol) for the second dimension based on the previous report (Nelson, 1967). Lipid spots were identified by spraying primuline solution, scraped off and acyl moieties hydrolyzed and methylesterified to fatty acid methyl esters (FAMEs) with HCl-methanol solution by incubating the samples at 85°C for 2 h including pentadecanoic acid (15:0) as internal standard. After the incubation, the FAMEs were extracted with hexane, dried up under the nitrogen gas stream, eluted in 50 μl of hexane, and quantified with gas chromatography (GC-2010; Shimadzu, Kyoto, Japan) with FID detector (FID-2010 Plus; Shimadzu, Kyoto, Japan) equipped with a ULBON HR-SS-10 column (Shinwa Chemical Industries, Japan) (Nakamura et al., 2003). Amount of TAG was shown in % (w/w) of dry cell weight. Composition of fatty acid and phospholipid classes were shown by mol% based on the FAMEs quantified with gas chromatography. Data were averaged by three biological replicates with standard deviations as error bars.
Nile red staining and observation of lipid droplets were as described previously (Hung et al., 2013) using Nile red (Fluka, Cat. No. 72485) and DeltaVision system (Applied Precision) with a 100x objective lens (NA = 1.4) and a CoolSNAP HQ CCD camera (Photomotrics) controlled by softWoRx Suite (Applied Precision). Normarski differential interference contrast (DIC) microscopy image was taken as previously described (Fu et al., 2010).
Yeast cells were grown to the stationary phase in synthetic complete medium lacking uracil and leucine (SC -Ura, -Leu) containing 8% glucose at 30°C for 48 h. Samples were frozen in a high-pressure freezer (Leica EM PACT2) at 2000–2050 bar. Freeze-substitution was performed in anhydrous ethanol (containing 0.2% glutaraldehyde and 0.1% uranyl acetate) with an automatic freeze substitution system (Leica EM AFS2). The samples were first kept at −85°C for 3 days, then switched to −60°C, −20°C, 0°C, and room temperature at 1-day intervals. After 2 times of rinse with ethanol for 12 h, the samples were embedded by infiltrating LR White resin. Ultrathin sections (70–90 nm) were prepared with use of Reichert Ultracut S or Lecia EM UC7 (Leica, Vienna, Austria) and collected on 100-mesh copper grids. The sections were stained with 5% uranyl acetate in 50% methanol for 10 min and 0.4% lead citrate for 4–6 min. Sections were observed under a transmission electron microscope (Philips CM 100) at 80 KV and images were taken with use of a Gatan Orius CCD camera.
To enhance TAG levels by metabolic engineering, we genetically manipulated S. cerevisiae by considering the following 3 points: (1) suppressing metabolic flux to membrane lipid biosynthesis at the log phase, (2) enhancing metabolic flux to TAG biosynthesis at the stationary phase, and (3) suppressing TAG hydrolysis for membrane lipid biosynthesis on growth resumption to maintain accumulated TAG (Figure 2A).
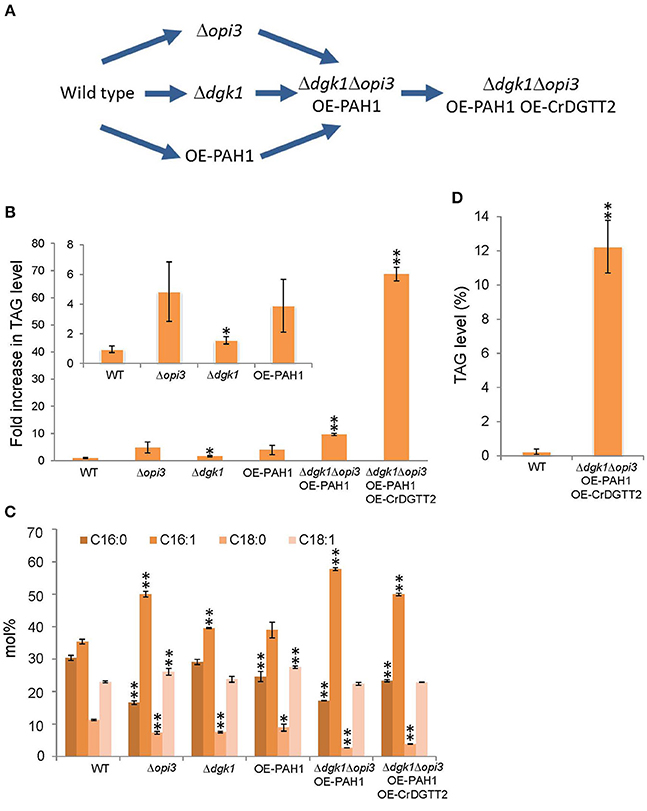
Figure 2. Procedures for metabolic engineering to enhance TAG production in S. cerevisiae. (A) Schematic representation of the procedures. (B) TAG levels of early stationary phase were quantified in the strains of wild type (WT), Δopi3, Δdgk1, OE-PAH1, Δdgk1Δopi3 OE-PAH1, and Δdgk1Δopi3 OE-PAH1 OE-CrDGTT2 shown as fold increase compared to the wild type strain. (C) Fatty acid composition (mol%) of TAG analyzed in (B). (D) TAG production of Δdgk1Δopi3 OE-PAH1 OE-CrDGTT2 strain at stationary phase by dry cell weight basis. Data are mean ± SD from three biological replicates. Asterisks indicate statistical significance by Student's t-test (*P < 0.05; **P < 0.01). WT, wild type; OE, overexpression; 16:0, palmitic acid; 16:1, palmitoleic acid; 18:0, stearic acid; 18:1, oleic acid.
For point 1, to suppress membrane lipid biosynthesis at the log phase, we blocked phosphatidylcholine (PC) biosynthesis by knocking out OPI3, which encodes an enzyme catalyzing the final step of PC biosynthesis (Figure 1). This is because the yeast mutant Δopi3 is viable with abolished PC (McGraw and Henry, 1989), whereas mutation in many other rate-limiting enzymes for phospholipid biosynthesis causes lethal effects on growth (Henry et al., 2012). For example, knocking out CDP-diacylglycerol synthase (CDS), which converts PA into CDP-DAG, causes a lethal effect (Shen et al., 1996). A significant metabolic switch is expected to be in favor of TAG production in Δopi3 because PC is a primary phospholipid of cellular membranes in S. cerevisiae. Indeed, the TAG levels in the stationary phase of Δopi3 cells were 4.8-fold higher than that of the wild type based on a dry cell weight (Figure 2B), which agrees with previous study (Fei et al., 2011). We next analyzed the fatty acid composition of TAG, which determines their quality. The Δopi3 cells showed an increased composition of palmitoleic acid (16:1) as compared with the wild type (Figure 2C). Therefore, partial blockage of primary membrane phospholipid biosynthesis by knocking out OPI3 increases TAG with enriched mono-unsaturated fatty acid, giving better quality for biodiesel use.
For point 2, to enhance metabolic flux to TAG biosynthesis at the stationary phase, we first overexpressed PAH1 (OE-PAH1) to stimulate PA to DAG conversion (Figure 1). The result showed that overexpression of PAH1 in wild-type cells increased TAG levels by 3.9-fold as compared with the wild type (Figure 2B). The fatty acid composition was not altered significantly by overexpressing PAH1 (Figure 2C). Since previous report showed that the Δpah1 mutant has reduced TAG levels (Adeyo et al., 2011; Fakas et al., 2011b), expression level of PAH1 may have dose-dependent effect on TAG accumulation.
For point 3, to suppress TAG hydrolysis on growth resumption, we knocked out DGK1 because previous study demonstrated that DGK1 is required for phospholipid synthesis during growth resumption from stationary phase (Fakas et al., 2011a). In agreement with this observation, TAG content in the Δdgk1 cells was increased 1.6-fold as compared with the wild type (Figure 2B), with no remarkable alteration in fatty acid composition (Figure 2C). Thus, suppressing membrane phospholipid biosynthesis, inducing DAG production from PA, and inhibiting TAG mobilization all increased TAG accumulation to a significant extent.
Next, we combined these three genetic manipulations to construct a strain that overexpresses PAH1 in the Δdgk1Δopi3 (Δdgk1Δopi3 OE-PAH1). The TAG level was increased 9.7-fold in this strain as compared with the wild type (Figures 2A,B). The fatty acid composition of accumulated TAG was similar to that of Δopi3 (Figure 2C), indicating that neither overexpression of PAH1 nor knockout of DGK1 altered the fatty acid composition of TAG. Thus, by altering the expression of 3 genes, OPI3, DGK1, and PAH1, TAG production was increased by 9.7-fold as compared to the wild type in S. cerevisiae.
Here, to further increase TAG contents, we overexpressed CrDGTT2 in the Δdgk1Δopi3 OE-PAH1. The resulting TAG levels in Δdgk1Δopi3 OE-PAH1 OE-CrDGTT2 were further increased more than 7-fold, giving 69-fold increase as compared to the wild type (Figures 2A,B). The fatty acid composition of accumulated TAG was not altered with overexpression of CrDGTT2 in the Δdgk1Δopi3 OE-PAH1 cells (Figure 2C). To confirm the TAG yield of the Δdgk1Δopi3 OE-PAH1 OE-CrDGTT2 cells, we independently established three more strains with the same genotype (CHY140, CHY141, and CHY142) by transforming the plasmid to overexpress CrDGTT2 into the Δdgk1Δopi3 OE-PAH1. When maximal TAG level is produced at late stationary phase (60 h), these cells produced 64.8- to 73.2-fold increase (Supplementary Table 2), indicating reproducibly high yield of TAG in this engineered strain. This TAG amount was estimated to be 12.2% of total dry cell biomass (Figure 2D). This corresponds to 166 mg of TAG per liter of liquid broth culture, which produces about 1.35 g/L of dry cell weight. Thus, our metabolic engineering greatly increased TAG contents using a robust algal gene CrDGTT2.
We found that the Δdgk1Δopi3 OE-PAH1 OE-CrDGTT2 cells showed reduced growth, because culture of the Δdgk1Δopi3 OE-PAH1 OE-CrDGTT2 reached a plateau at OD600 of 4 while that of the wild type reached a plateau at OD600 of 10 in synthetic complete media lacking Ura and Leu (Figure 3A; see Supplementary Figure 1 for the growth curve of the strains). This growth retardation may be due to suppressed membrane lipid biosynthesis rather than massive accumulation of TAG, because the value of OD600 was comparable to that of the Δdgk1Δopi3 cells (Figure 3A). Indeed, composition of membrane phospholipids was greatly altered as compared to the wild type (Figure 3B; Supplementary Table 3). We also measured level of free fatty acid; wild type cells had 5.8 ± 0.7 μg/mg dry cell weight whereas the Δdgk1Δopi3 OE-PAH1 OE-CrDGTT2 cells had 38.2 ± 9.6 μg/mg dry cell weight. Nile red staining for lipid droplets was greater in the Δdgk1Δopi3 OE-PAH1 OE-CrDGTT2 cells than wild type cells (Figures 3C,E), which supports massive production of TAG (Figure 2; Supplementary Table 2). The cell size of Δdgk1Δopi3 OE-PAH1 OE-CrDGTT2 with TAG accumulation at the stationary phase was larger than wild-type cells (Figures 3D,F). To test whether the Δdgk1Δopi3 OE-PAH1 OE-CrDGTT2 cells are still capable of accumulating TAG, we shifted glucose concentration of culture media from 2 to 8% at the end of log phase to trigger extra carbon uptake and fixation to carbon-containing metabolite reserves, including TAG (Kamisaka et al., 2013). As compared with the normal 2% glucose condition, this “sugar boost” further increased TAG level by 3.2-fold (Figure 3G; Supplementary Table 4). These data suggest that the Δdgk1Δopi3 OE-PAH1 OE-CrDGTT2 cells have capacity to accumulate extra TAG in response to external glucose supply. We observed the ultrastructure of the Δdgk1Δopi3 OE-PAH1 OE-CrDGTT2 cells accumulating TAG under the 8% glucose condition. Compared to the wild type (Figure 3H), the Δdgk1Δopi3 OE-PAH1 OE-CrDGTT2 produced super-sized lipid droplets that occupied the primary space in the intracellular compartment (Figure 3I).
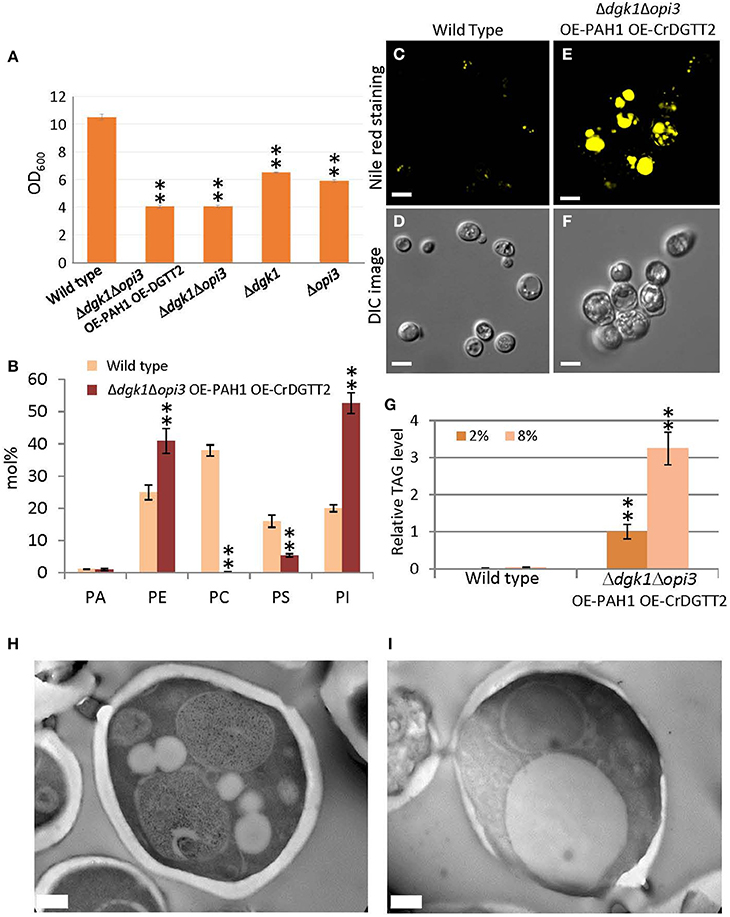
Figure 3. Phenotypes of the Δdgk1Δopi3 OE-PAH1 OE-CrDGTT2 strain. (A) The OD600 value of culture at stationary phase of wild type, Δdgk1Δopi3 OE-PAH1 OE-CrDGTT2, Δdgk1Δopi3, Δdgk1, and Δopi3. Fully grown yeast cell cultures were diluted to OD600 of 0.1 in 10 ml synthetic complete media at 30°C, and OD600 was measured after 72 h of incubation, at which culture reached a plateau. Data are mean ± SD from three biological replicates. (B) Membrane lipid composition (mol%) of the Δdgk1Δopi3 OE-PAH1 OE-CrDGTT2 strain. Wild type, yellow bars; the Δdgk1Δopi3 OE-PAH1 OE-CrDGTT2 strain, brown bars. See Supplementary Table 3 for numerical values. (C–F) The Δdgk1Δopi3 OE-PAH1 OE-CrDGTT2 strain at the beginning of stationary phase by Nile red staining (E) and Normarski differential interference contrast (DIC) microscopy image (F) compared with the wild type (C,D). (G) Fold increase of TAG levels in response to glucose boost from 2 to 8% at the beginning of stationary phase. TAG level of Δdgk1Δopi3 OE-PAH1 OE-CrDGTT2 strain under 2% glucose condition was set as 1. See Supplementary Table 4 for numerical values. (H,I) Transmission electron microscope images of the ultrastructure of wild type (H) and the Δdgk1Δopi3 OE-PAH1 OE-CrDGTT2 (I) cells in response to glucose boost from 2 to 8% at the end of log phase. Data in (A,B) and (G) are mean ± SD from three biological replicates. Asterisks indicate statistical significance by Student's t-test (**P < 0.01). See Figure 1 for abbreviations. Bars in (C–F) are 5 μm and (H–I) are 0.5 μm.
A major challenge in TAG metabolic engineering has been how to maintain accumulated TAG with continuous cell growth. This is because TAG accumulation occurs during the stationary phase in wild-type cells, when cell numbers are no longer increased. However, on growth resumption from the stationary phase, accumulated TAG is mostly hydrolyzed to DAG (TAG mobilization) as a substrate to produce phospholipids which are required for rapid growth of cellular membranes (Athenstaedt and Daum, 2005; Kurat et al., 2009).
To demonstrate whether sustainable TAG production is possible with the Δdgk1Δopi3 OE-PAH1 OE-CrDGTT2 cells, quantity (Figure 4B) and quality (Figure 4C) of TAG were profiled along with the cell growth (Figure 4A) through three consecutive repeats of growth resumption. Upon growth resumption from the stationary phase, the growth rate was slower in the Δdgk1Δopi3 OE-PAH1 OE-CrDGTT2 than the wild type (Figure 4A); the Δdgk1Δopi3 OE-PAH1 OE-CrDGTT2 cells reached stationary phase at 48 h after growth resumption with OD600 of 4, whereas wild type cells reached stationary phase at 36 h with OD600 of 10. At 60 h, these cultures were diluted again to resume the growth, which reproduced similar growth profile through the following two repeats of growth resumption. TAG level of the Δdgk1Δopi3 OE-PAH1 OE-CrDGTT2 cells in full growth at 60 h was 13.2 to 14.5% of dry cell weight (Figure 4B; Supplementary Table 5). Twenty-four hours after growth resumption, TAG level was reduced transiently but was recovered to the initial levels by 60 h. Repeated dilution reproduced the same extent of TAG levels after 60 h for two more times (Figure 4B; Supplementary Table 5). Moreover, fatty acid composition of TAG was maintained throughout the different time points (Figure 4C, Supplementary Table 6), indicating stable quality of TAG. Thus, the Δdgk1Δopi3 OE-PAH1 OE-CrDGTT2 cells could grow with reduced degradation of accumulated TAG, which favored sustainable TAG production.
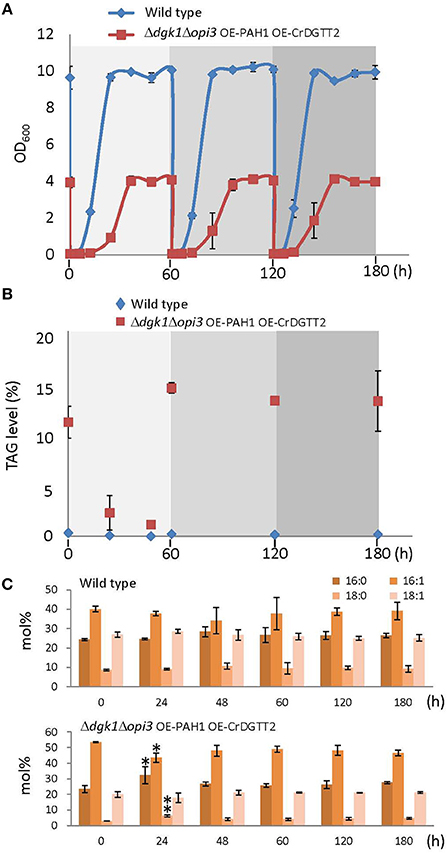
Figure 4. Sustainable TAG production in the Δdgk1Δopi3 OE-PAH1 OE-CrDGTT2 strain. (A) Growth profile of the Δdgk1Δopi3 OE-PAH1 OE-CrDGTT2 strain with repeated resumption of growth. One hundred milliliter of cells at full growth was diluted to OD600 of 0.05 at 0, 60, and 120 h. (B,C) TAG levels (B) and fatty acid composition (C) of wild type and the Δdgk1Δopi3 OE-PAH1 OE-CrDGTT2 strain with repeated resumption of growth. Data in (A–C) are mean ± SD from three biological replicates. Asterisks indicate statistical significance by Student's t-test (*P < 0.05; **P < 0.01). See Supplementary Tables 5, 6 for numerical values.
Our metabolic engineering to overexpress an algal gene in genetically manipulated yeast S. cerevisiae resulted in about 70-fold increase in TAG level, which is estimated to be 12.2% of dry cell weight (Figure 2D). This yield is superior to recently achieved TAG engineering in S. cerevisiae. For example, overexpression of a glycerol kinase and DAG acyltransferases achieved 8.2% of TAG with 2% glycerol as carbon source (Yu et al., 2013). In addition, overexpression of fatty acid biosynthesis genes produced 17% of total lipid fraction including TAG and other lipid classes (Runguphan and Keasling, 2014); because S. cerevisiae produces similar amounts of TAG and sterylesters as major lipid content (Beopoulos et al., 2009), our metabolic engineering may produce higher TAG contents. Moreover, our engineered S. cerevisiae strain diluted from full growth resumed growth with TAG accumulated (Figure 4). This sustainable TAG production may be an intriguing feature of this engineered strain from the viewpoint of metabolic engineering, although it may have little advantage in real industrial application since dilution requires cost of new medium. Oily yeasts such as Lipomyces starkeyi, Rhodosporidium toruloides, Rhodotorula glutinis, and Yarrowia lipolytica can produce up to 20% lipids per dry biomass (Runguphan and Keasling, 2014). For oleaginous algae, lipid levels commonly range from 20 to 50% per dry biomass, exceeding the usual yield of oleaginous yeasts (Beopoulos et al., 2009). Although levels of TAG accumulation in the Δdgk1Δopi3 OE-PAH1 OE-CrDGTT2 cells were not superior to these oleaginous microorganisms, several advantages of using S. cerevisiae over other yeasts or microalgae have been pointed, such as genetic tractability, commercially available whole-genome deletion strain collection or a proven track record in different industrial applications (Tang et al., 2013). Our strategy of metabolic engineering could be applicable to these oleaginous microorganisms to further improve their lipid yields. Molecular engineering of eukaryotic algae still awaits technical developments despite that engineered algae may contribute to the carbon neutrality owing to photosynthesis. Therefore, our TAG engineering using CrDGTT2 provides a new strategy to enhance oil production in S. cerevisiae. Since algal species are highly diverse, a robust enzyme similar to CrDGTT2 may be found in other oleaginous algal species. Since CrDGTT2 showed much higher expression level than any other DGTT isoforms in S. cerevisiae even though they were all expressed by the same promoter system, it is possible that the robustness of CrDGTT2 may be due to higher stability of mRNA in a heterologous system (Hung et al., 2013). Using algal gene resources to engineer established model microorganism can contribute to develop an innovative approach in the field of synthetic biology and has potential to produce TAG or other value-added oils for industrial demand.
In conclusion, we employed an algal gene resource, CrDGTT2, to reconstitute enhanced and sustainable TAG production in a yeast S. cerevisiae by metabolic engineering.
YN conceived research; KK and YN designed experiments; CH performed experiments and analyzed data; KK provided technical assistance to CH, KK, and YN wrote the manuscript; all author commented on the manuscript and approved the contents.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Wann-Nen Jane and Mei-Jane Fang (Institute of Plant and Microbial Biology, Academia Sinica) for technical support with microscopy. This research was supported by PRESTO, Japan Science and Technology Agency, Ministry of Science and Technology Taiwan (Grant ID: 103-2311-B-001-029-MY3), and a core operation budget from the Institute of Plant and Microbial Biology, Academia Sinica.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00070
CDP-DAG, cytidine diphosphate diacylglycerol; CL, cardiolipin; DAG, sn-1,2-diacylglycerol; G3P, glycerol 3-phosphate; LPA, lysophosphatidic acid; PA, phosphatidic acid; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PGP, phosphatidylglycerophosphate; PI, phosphatidylinositol; PS, phosphatidylserine; TAG, triacylglycerol.
Adeyo, O., Horn, P. J., Lee, S., Binns, D. D., Chandrahas, A., Chapman, K. D., et al. (2011). The yeast lipin orthologue Pah1p is important for biogenesis of lipid droplets. J. Cell Biol. 192, 1043–1055. doi: 10.1083/jcb.201010111
Athenstaedt, K., and Daum, G. (2005). Tgl4p and Tgl5p, two triacylglycerol lipases of the yeast Saccharomyces cerevisiae are localized to lipid particles. J. Biol. Chem. 280, 37301–37309. doi: 10.1074/jbc.M507261200
Beopoulos, A., Cescut, J., Haddouche, R., Uribelarrea, J. L., Molina-Jouve, C., and Nicaud, J. M. (2009). Yarrowia lipolytica as a model for bio-oil production. Prog. Lipid Res. 48, 375–387. doi: 10.1016/j.plipres.2009.08.005
Blazeck, J., Garg, R., Reed, B., and Alper, H. S. (2012). Controlling promoter strength and regulation in Saccharomyces cerevisiae using synthetic hybrid promoters. Biotechnol. Bioeng. 109, 2884–2895. doi: 10.1002/bit.24552
Durrett, T. P., Benning, C., and Ohlrogge, J. (2008). Plant triacylglycerols as feedstocks for the production of biofuels. Plant J. 54, 593–607. doi: 10.1111/j.1365-313X.2008.03442.x
Fakas, S., Konstantinou, C., and Carman, G. M. (2011a). DGK1-encoded diacylglycerol kinase activity is required for phospholipid synthesis during growth resumption from stationary phase in Saccharomyces cerevisiae. J. Biol. Chem. 286, 1464–1474. doi: 10.1074/jbc.M110.194308
Fakas, S., Qiu, Y., Dixon, J. L., Han, G. S., Ruggles, K. V., Garbarino, J., et al. (2011b). Phosphatidate phosphatase activity plays key role in protection against fatty acid-induced toxicity in yeast. J. Biol. Chem. 286, 29074–29085. doi: 10.1074/jbc.M111.258798
Fei, W., Shui, G., Zhang, Y., Krahmer, N., Ferguson, C., Kapterian, T. S., et al. (2011). A role for phosphatidic acid in the formation of “supersized” lipid droplets. PLoS Genet. 7:e1002201. doi: 10.1371/journal.pgen.1002201
Folch, J., Lees, M., and Sloane Stanley, G. H. (1957). A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226, 497–509.
Fu, D., Oh, S., Choi, W., Yamauchi, T., Dorn, A., Yaqoob, Z., et al. (2010). Quantitative DIC microscopy using an off-axis self-interference approach. Opt. Lett. 35, 2370–2372. doi: 10.1364/OL.35.002370
Gietz, R. D., and Woods, R. A. (2002). Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Meth. Enzymol. 350, 87–96. doi: 10.1016/s0076-6879(02)50957-5
Han, G. S., O'Hara, L., Carman, G. M., and Siniossoglou, S. (2008a). An unconventional diacylglycerol kinase that regulates phospholipid synthesis and nuclear membrane growth. J. Biol. Chem. 283, 20433–20442. doi: 10.1074/jbc.M802903200
Han, G. S., O'Hara, L., Siniossoglou, S., and Carman, G. M. (2008b). Characterization of the yeast DGK1-encoded CTP-dependent diacylglycerol kinase. J. Biol. Chem. 283, 20443–20453. doi: 10.1074/jbc.M802866200
Han, G. S., Wu, W. I., and Carman, G. M. (2006). The Saccharomyces cerevisiae Lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J. Biol. Chem. 281, 9210–9218. doi: 10.1074/jbc.M600425200
Henry, S. A., Kohlwein, S. D., and Carman, G. M. (2012). Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics 190, 317–349. doi: 10.1534/genetics.111.130286
Hu, Q., Sommerfeld, M., Jarvis, E., Ghirardi, M., Posewitz, M., Seibert, M., et al. (2008). Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. Plant J. 54, 621–639. doi: 10.1111/j.1365-313X.2008.03492.x
Hung, C. H., Ho, M. Y., Kanehara, K., and Nakamura, Y. (2013). Functional study of diacylglycerol acyltransferase type 2 family in Chlamydomonas reinhardtii. FEBS Lett. 587, 2364–2370. doi: 10.1016/j.febslet.2013.06.002
Kamisaka, Y., Kimura, K., Uemura, H., and Yamaoka, M. (2013). Overexpression of the active diacylglycerol acyltransferase variant transforms Saccharomyces cerevisiae into an oleaginous yeast. Appl. Microbiol. Biotechnol. 97, 7345–7355. doi: 10.1007/s00253-013-4915-9
Kurat, C. F., Wolinski, H., Petschnigg, J., Kaluarachchi, S., Andrews, B., Natter, K., et al. (2009). Cdk1/Cdc28-dependent activation of the major triacylglycerol lipase Tgl4 in yeast links lipolysis to cell-cycle progression. Mol. Cell 33, 53–63. doi: 10.1016/j.molcel.2008.12.019
McGraw, P., and Henry, S. A. (1989). Mutations in the Saccharomyces cerevisiae opi3 gene: effects on phospholipid methylation, growth and cross-pathway regulation of inositol synthesis. Genetics 122, 317–330.
Merchant, S. S., Kropat, J., Liu, B., Shaw, J., and Warakanont, J. (2012). TAG, you're it! Chlamydomonas as a reference organism for understanding algal triacylglycerol accumulation. Curr. Opin. Biotechnol. 23, 352–363. doi: 10.1016/j.copbio.2011.12.001
Nakamura, Y., Arimitsu, H., Yamaryo, Y., Awai, K., Masuda, T., Shimada, H., et al. (2003). Digalactosyldiacylglycerol is a major glycolipid in floral organs of Petunia hybrida. Lipids 38, 1107–1112. doi: 10.1007/s11745-006-1166-x
Nelson, G. J. (1967). The phospholipid composition of plasma in various mammalian species. Lipids 2, 323–328. doi: 10.1007/BF02532119
Runguphan, W., and Keasling, J. D. (2014). Metabolic engineering of Saccharomyces cerevisiae for production of fatty acid-derived biofuels and chemicals. Metab. Eng. 21, 103–113. doi: 10.1016/j.ymben.2013.07.003
Sandager, L., Gustavsson, M. H., Ståhl, U., Dahlqvist, A., Wiberg, E., Banas, A., et al. (2002). Storage lipid synthesis is non-essential in yeast. J. Biol. Chem. 277, 6478–6482. doi: 10.1074/jbc.M109109200
Sanjaya, M. R., Durrett, T. P., Kosma, D. K., Lydic, T. A., Muthan, B., Koo, A. J., et al. (2013). Altered lipid composition and enhanced nutritional value of Arabidopsis leaves following introduction of an algal diacylglycerol acyltransferase 2. Plant Cell 25, 677–693. doi: 10.1105/tpc.112.104752
Shen, H., Heacock, P. N., Clancey, C. J., and Dowhan, W. (1996). The CDS1 gene encoding CDP-diacylglycerol synthase in Saccharomyces cerevisiae is essential for cell growth. J. Biol. Chem. 271, 789–795. doi: 10.1074/jbc.271.2.789
Tang, X., Feng, H., and Chen, W. N. (2013). Metabolic engineering for enhanced fatty acids synthesis in Saccharomyces cerevisiae. Metab. Eng. 16, 95–102. doi: 10.1016/j.ymben.2013.01.003
Yu, K. O., Jung, J., Ramzi, A. B., Choe, S. H., Kim, S. W., Park, C., et al. (2013). Development of a Saccharomyces cerevisiae strain for increasing the accumulation of triacylglycerol as a microbial oil feedstock for biodiesel production using glycerol as a substrate. Biotechnol. Bioeng. 110, 343–347. doi: 10.1002/bit.24623
Keywords: triacylglycerol, Chlamydomonas reinhardtii, Saccharomyces cerevisiae, diacylglycerol acyltransferase type 2, metabolic engineering
Citation: Hung C-H, Kanehara K and Nakamura Y (2016) In vivo Reconstitution of Algal Triacylglycerol Production in Saccharomyces cerevisiae. Front. Microbiol. 7:70. doi: 10.3389/fmicb.2016.00070
Received: 05 November 2015; Accepted: 15 January 2016;
Published: 15 February 2016.
Edited by:
Pankaj Kumar Arora, Yeungnam University, South KoreaReviewed by:
Shawn Chen, Ohio University, USACopyright © 2016 Hung, Kanehara and Nakamura. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuki Nakamura, bmFrYW11cmFAZ2F0ZS5zaW5pY2EuZWR1LnR3
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.