- Institute of Medical Microbiology and Hygiene, Technische Universitaet Dresden, Dresden, Germany
In humans of all ages, the cell wall-less and genome-reduced species Mycoplasma pneumoniae can cause infections of the upper and lower respiratory tract. The well-documented occurrence of major peaks in the incidence of community-acquired pneumonia cases reported world-wide, the multifaceted clinical manifestations of infection and the increasing number of resistant strains provide reasons for ongoing interest in the pathogenesis of mycoplasmal disease. The results of recent studies have provided insights into the interaction of the limited virulence factors of the bacterium with its host. In addition, the availability of complete M. pneumoniae genomes from patient isolates and the development of proteomic methods for investigation of mycoplasmas have not only allowed characterization of sequence divergences between strains but have also shown the importance of proteins and protein parts for induction of the immune reaction after infection. This review focuses on selected aspects of the humoral host immune response as a factor that might influence the clinical course of infections, subsequent protection in cases of re-infections and changes of epidemiological pattern of infections. The characterization of antibodies directed to defined antigens and approaches to promote their induction in the respiratory mucosa are also preconditions for the development of a vaccine to protect risk populations from severe disease due to M. pneumoniae.
From an evolutionary point of view, Mycoplasma pneumoniae is a successful minimalized bacterium perfectly adapted to humans as the only known natural host. During the long-term host–pathogen interaction, the genome of these micro-organisms was reduced to only about 816 kb. This includes removal of genes coding for cell wall synthesis as well as a reduction of metabolic capabilities requiring a supply of metabolites from the environment. The adaptation of the bacterium led to distinct tissue tropism using the surface of the respiratory mucosa as the preferred niche, which ensures the survival and parasitic life style of these bacteria. The pathogen possesses a repertoire of factors that cause clinical manifestations after an infection; however, typical virulence determinants of other pathogenic bacteria, such as type III or IV secretion systems, are lacking in M. pneumoniae. The species developed an almost unique adherence apparatus. As the main virulence factor (Atkinson et al., 2008; Atkinson and Waites, 2014), this tip-structured protein complex determines not only the adhesion process to the respiratory epithelium as a first step in host colonization but also supports a gliding mechanism which results in bacterial motility (Miyata, 2010). The close contact of mycoplasmas with epithelial cells reduces the efficiency of mucociliary clearance processes. Damage to epithelial cells of the mucosa causing subsequent symptoms is caused by generation of peroxide and superoxide anion (Hames et al., 2009). In addition, an ADP-ribosylating and vacuolating community-acquired respiratory distress syndrome (CARDS) toxin resembling the pertussis toxin induces ciliastasis and the release of proinflammatory cytokines (Kannan and Baseman, 2006). Further steps in pathogenesis are mainly determined by local inflammation processes and immunomodulation (Meyer Sauteur et al., 2014).
Immunodominant Antigens of M. pneumoniae and Host Response
In addition to the role of virulence factors of M. pneumoniae, the induction of specific antibodies is not only important for serodiagnosis of infections but also for the colonization process. As result of lacking the classical bacterial cell wall, components of the cell membrane are important for interaction of the pathogen with the host. The importance of an intact immune response is demonstrated by the increased occurrence of severe disease, repeated infections or prolonged persistence of M. pneumoniae in patients with deficiencies of humoral immunity (Foy et al., 1973; Taylor-Robinson et al., 1980; Roifman et al., 1986), thus emphasizing the role of specific antibodies for protection. The antigens of M. pneumoniae cells determining the host response include glycolipids as well as proteins (Morrison-Plummer et al., 1986) that induce comparable immune reactions in affected individuals (Jacobs et al., 1986; Vu et al., 1987). In comparison with glycolipids, the more specific proteins were mainly characterized as components of the adhesion apparatus of M. pneumoniae (Razin and Jacobs, 1992). In particular, antibodies to the P1 protein are regularly found in sera of infected patients. The large membrane protein (168 kDa) was characterized as the main adhesin of the bacteria and is also the most antigenic protein, inducing strong and early production of antibodies (Hu et al., 1983). Using different proteomic approaches such as fractionation of whole proteins (Regula et al., 2001), construction of a whole-genome phage display library (Beghetto et al., 2009) or 2D separation of proteins followed by incubation with sera of infected patients (Nuyttens et al., 2010) resulted in the characterizations of further antigens which are membrane-associated and potentially interact with the host immune system. Besides proteins with a confirmed function in adherence, putative lipoproteins, glycolytic enzymes (e.g., pyruvate dehydrogenase subunit B), chaperones (GroEL, DnaK) and proteins of translation/transcription (e.g., elongation factor Tu) were found. Some of these proteins are surface-localized and involved in interactions with components of the human extracellular matrix (Dallo et al., 2002; Gründel et al., 2015). In addition, CARDS toxin as an important virulence factor of M. pneumoniae was characterized as an immune-dominant protein (Kannan and Baseman, 2006). However, the role of antibodies to many of these proteins for the potential to protect the host from re-infections remains to be proved.
With the development of specific tools for investigation of mycoplasmas (Halbedel and Stülke, 2007), such as targeted mutation of TGA triplets coding for tryptophan in M. pneumoniae (Inamine et al., 1990), the recombinant production and analysis of proteins of interest for host–pathogen interaction have accelerated. Regarding naturally infected hosts, Table 1 summarizes defined M. pneumoniae proteins that were found in recent years to elicit a specific and strong immune reaction in humans. These studies confirmed that the immune response is dominated by antibodies against the adhesins and adhesion-related proteins of the bacterium that have limited effect on viability (Krause and Baseman, 1983). It can be suggested that the antibody response results mainly in an influence on the gliding process (Seto et al., 2005) and a decrease of adhesion of bacteria to the target cells of the respiratory mucosa. Studies using quantitative methods to measure the adherence of M. pneumoniae to human cells in vitro showed that specific antisera to total proteins, to adhesins or even to defined regions of adhesins are able to inhibit adhesion to more than 90% in comparison with control sera (Svenstrup et al., 2002; Schurwanz et al., 2009). The importance of the adherence process for further colonization is underlined by the fact that mutants defective in expression of different adhesins and adhesion-related proteins are avirulent (Balish and Krause, 2006).
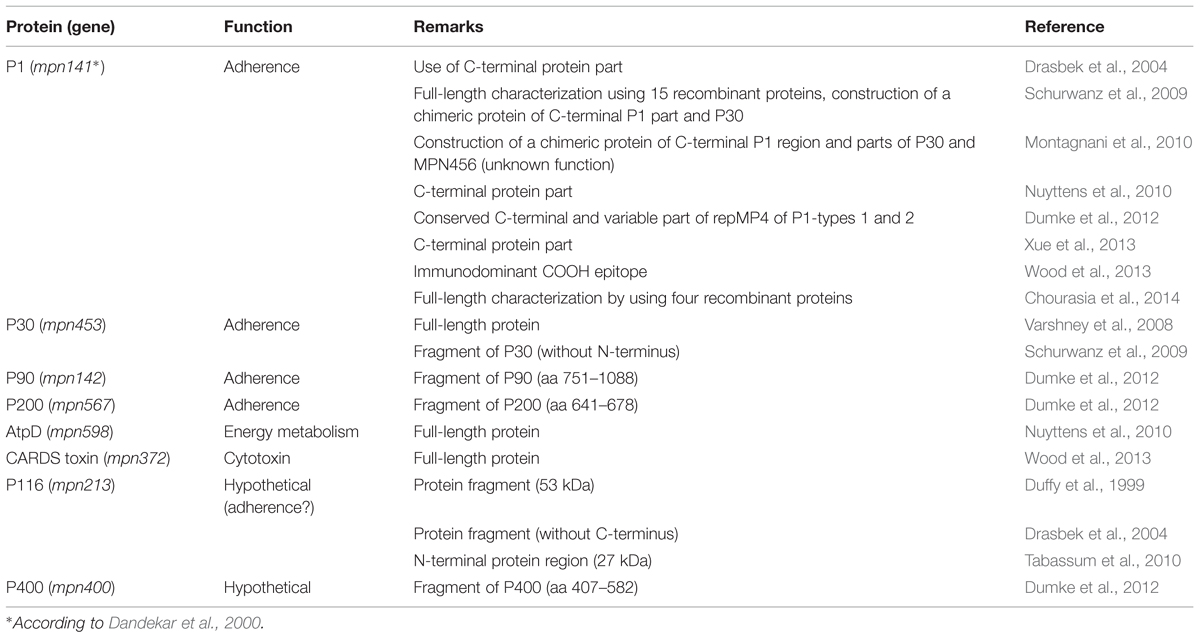
TABLE 1. Recombinant Mycoplasma pneumoniae proteins tested as antigens for detection of specific antibodies in humans.
Beside problems in the sensitivity and specificity of serological assays (Loens et al., 2010; Busson et al., 2013), M. pneumoniae infections are complicated by different host-dependent characteristics, such as variable persistence of antibodies, missing IgM response after re-infection and the infrequent production of IgA antibodies in children (Atkinson et al., 2008). IgM antibodies can be detected 7–10 days after infection and IgG immunoglobulins are measurable approximately 14 days later (Atkinson et al., 2008; Atkinson and Waites, 2014).
Genotype-Specific Immune Response and Influence on the Epidemiology of M. pneumoniae Infections
Genome plasticity and different mechanisms for antigen variation are a typical pattern of different mycoplasma species with pathogenic potential (Citti and Blanchard, 2013). In M. pneumoniae, recent studies comparing whole genomes resulted in a remarkable homology between strains of different origin and isolation period (Lluch-Senar et al., 2015; Xiao et al., 2015). However, isolates of M. pneumoniae or strains in respiratory tract samples from patients show defined sequence variations which can be used for typing by different methods. Multilocus variable number of tandem repeat analysis (Degrange et al., 2009), multilocus sequence typing (Brown et al., 2015) and SNP minisequencing (Touati et al., 2015) have been developed recently using current molecular tools. These approaches investigate variable regions in the genome which are located mainly in intergenic regions and in genes coding for hypothetical proteins or for house-keeping proteins with an intracellular function. Furthermore, the latter two approaches detect small polymorphic sites and even single nucleotide exchanges. Regarding the immune response, an influence of these differences on protective antibodies in infected hosts can hardly be expected. Also, the role in the immune reaction of more different genome regions such as certain genes that contain copies of the repetitive element RepMP1 (Musatovova et al., 2012) is still unclear as the gene products involved and their possible functions have yet to be investigated. Lipoproteins are important antigens in many mycoplasma species showing variations to escape the host immune response (Citti et al., 2010; Szczepanek and Silbart, 2014). In M. pneumoniae, a high proportion (nearly 7%) of genes encoding for putative lipoproteins was confirmed and their expression pattern was investigated (Hallamaa et al., 2006, 2008). So far, strain-specific induction of anti-lipoprotein antibodies during infection has not been demonstrated.
The genome region with the greatest discrimination power in combination with well-characterized gene products is the p1 operon coding for P1 adhesin and the adhesion-related proteins P40 and P90, as well as a phosphoesterase (mpn140). Copies of repetitive elements RepMP2/3 and RepMP4 (p1 gene, mpn141) as well as of RepMP5 (mpn142) can be found, which differ between isolates (Su et al., 1990). Analysis of repetitive elements distributed in variable size and sequence over the genome of M. pneumoniae strains resulted in the characterization of p1 type 1 and p1 type 2 (Spuesens et al., 2009). Despite a high number of RepMP2/3 (n = 10), RepMP4 (8) and RepMP5 (8) copies in the genome of strain M129 (Dandekar et al., 2000), the number of circulating p1 types is relatively low. It can be suggested that the complex functions of the P1 protein limit the possible recombination events that allow effective adhesion, gliding and/or division processes of mycoplasma cells. In addition to the main types 1 and 2, variants differing to a smaller extent from the two p1 types (mainly in the RepMP2/3 copy) were identified after investigation of sequence of p1 genes of clinical strains collected world-wide. According to recent knowledge, these variants cannot be distinguished in the sequence of mpn142, indicating lower variability of proteins P40 and P90. Despite the fact that 76% of the p1 sequence can be assigned to both RepMP copies, the divergence between the amino acid sequences of P1 adhesins of types 1 and 2 strains (1,628 and 1,635 aa) is only about 5%. However, these distinct differences in amino acid sequence may influence the immune response of a colonized host. Indeed, it was shown that the variable parts of the P1 adhesins are antigenic resulting in a type-specific response in immunized animals (Dumke et al., 2008). Thus, recombinantly produced protein parts derived from variable regions of the P1 adhesin can be used for determination of type-specific IgG immunoglobulins (Dumke et al., 2012). Furthermore, guinea pigs infected intranasally with a type 2 strain developed protective immunity to type 2 strains after re-infection with a mixture of types 1 and 2 (Dumke et al., 2004). As a consequence of these findings and because of the dominant role of antibodies to the P1 protein in sera of infected patients, it has been suggested that type-specific immunoglobulins have an influence on subsequent re-infections in affected individuals as well as on the epidemiology of infections in greater populations. The results of different studies confirmed that the epidemic peaks of respiratory infections due to M. pneumoniae occurring at intervals of 3–7 years were correlated with a change of the predominant p1 type (Kenri et al., 2008; Kogoj et al., 2015; Suzuki et al., 2015; Zhao et al., 2015). In other investigated populations, significant differences in the proportion of types 1 and 2 strains during endemic and epidemic periods of infections were not detected (Dumke et al., 2015; Jacobs et al., 2015). Therefore, further long-term epidemiological studies are needed to clarify whether type-specific antibodies induced after an outbreak will protect patients from a re-infection with this genotype of M. pneumoniae. In a study investigating type-specific antibodies in acute-phase sera (IgA and IgG) from pneumonia patients with known p1 type in the respiratory tract, correlation between the occurrence of genotypes and type-specific immune response was lacking (Dumke et al., 2010). Finally, it cannot be excluded that genotype-specific differences in adhesion-related proteins will influence the interaction with target structures for adherence. Interestingly, recent epidemiological reports describe the replacement of p1 type 2 strains by variants of this type in the investigated human population (Jacobs et al., 2015; Suzuki et al., 2015). The reasons for the shift of genotypes remain unclear since it seems unlikely that the small sequence variations in the p1 gene of type 2 and type 2 variant strains will influence the immune response of infected patients. Confirmed type-specific differences in factors that influence the interaction with the host such as biofilm formation (Simmons et al., 2013) or expression of CARDS toxin (Techasaensiri et al., 2010; Lluch-Senar et al., 2015) were demonstrated between types 1 and 2 strains but not between type 2 and type 2 variants. Further studies should be performed to give more insight into the role of the time-variable occurrence of genotype-specific antibodies (host-dependent) and the consequences of sequence differences in the P1 protein for the adherence process (pathogen-dependent) in the epidemiology of infections.
Vaccine Development
Despite the benign course of many infections by M. pneumoniae, the occurrence of severe manifestations and the existence of risk populations with intensive person-to-person contacts (schools, military camps) or with chronic respiratory disease justify efforts to prevent infections. Recently, a further reason resulted from high rates of macrolide-resistant strains circulating mainly in Asia, limiting the antibiotic treatment options especially in pediatric patients. The availability of safe vaccines seems nowadays the most effective measure for control of M. pneumoniae also, and this is supported by the increased knowledge about the virulence factors and by the confirmed genetic homogeneity of this pathogen. Furthermore, the described induction of a strong host immune reaction to defined pathogen factors has encouraged vaccination experiments. However, the results of early studies with volunteers treated with inactivated whole antigen preparations reflect the problems of inducing an immune response that demonstrates effective protection from subsequent infections (summarized in Linchevski et al., 2009). Limited efficacy against pneumonia, adverse reactions and, in some cases, exacerbation of symptoms after infection of immunized individuals have been described and confirmed by recent animal experiments (Sekine et al., 2009; Szczepanek et al., 2012).
Based on the importance of adherence of M. pneumoniae cells for initiating host colonization, recent studies have focused on the induction of adhesion-blocking antibodies. Investigation of components of the adhesion complex resulted in defined protein regions which are antigenic and involved in adherence and which have a conserved sequence (Schurwanz et al., 2009; Nakane et al., 2011). The construction of chimeric antigens composed of shorter protein regions with characterized function in adherence provides an opportunity to target different adhesion-related structures (Schurwanz et al., 2009). Immunization of guinea pigs with a hybrid protein consisting of adherence-related parts of the proteins P1 and P30 led to a significant decrease of specific genome copies in respiratory tract samples from vaccinated and subsequently infected animals (Hausner et al., 2013). Since demonstration of the adherence-blocking properties of sera from immunized animals, the induction of potent stimulation of mucosal immunity can be suggested as a crucial aspect for successful vaccination. This included not only (intranasal) administration of the antigen but also combination with biocompatible adjuvants (Zhu et al., 2012; Hausner et al., 2013).
Carriage of M. pneumoniae
As with other respiratory pathogens such as pneumococci (Donkor, 2013), asymptomatic or convalescent individuals have been confirmed as carriers of M. pneumoniae cells in the upper respiratory tract (Foy, 1993). This is an important fact, not only for the significance of laboratory test results and for epidemiological aspects of transmission but also for evaluation of the role of protective antibodies. Unfortunately, the results of carriage studies are inconsistent. Gnarpe et al. (1992) showed that culturable mycoplasmas occurred in the throats of a relatively high proportion (13.5 and 4.6%) of subjectively healthy adults in two investigation periods. In a recent study, detection of M. pneumoniae infections using ELISA (IgM and IgG) and real-time PCR was similar in groups of asymptomatic children and pediatric patients with respiratory symptoms (Spuesens et al., 2013). In contrast, Jain et al. (2015) reported a low rate (<3%) of PCR-positive results in asymptomatic children in a control group during a period of high incidence of M. pneumoniae. This is in accordance with other studies that demonstrated no detection or only low rates of respiratory tract colonization/infection of asymptomatic patients of different ages (Kumar et al., 2008; Nilsson et al., 2008; Chalker et al., 2011). Not only the occurrence of asymptomatic carriers but also the long-term persistence of M. pneumoniae in the respiratory tract of immunocompetent patients raises doubts regarding the efficiency of the host immune response for clearance of bacteria. Spuesens et al. (2013) reported that 21% of PCR-positive children (with or without symptoms) showed carriage of bacteria for 1–3 months. In a study of 53 patients, the mean carriage time for M. pneumoniae DNA was 7 weeks after disease onset (up to 7 months) after adequate therapy and confirmation of specific antibodies in corresponding sera (Nilsson et al., 2008). Despite the strong immune response after M. pneumoniae infection, the efficiency of specific antibodies to eliminate the bacteria from the upper respiratory tract seems limited in particular individuals. The time-dependent decrease of specific antibodies in combination with the variable, often low production of IgA immunoglobulins in the respiratory tract may contribute to re-infection and long-term carriage.
It should be noted that confirmed persistence and long-term host colonization are typical for bacteria occurring intracellularly. The hypothesis is supported by the aforementioned absence of many metabolic pathways in M. pneumoniae, which requires close association with the host cells for growth and reproduction. However, the intracellular localization of M. pneumoniae has been described in cell culture assays (Meseguer et al., 2003) but so far not in vivo, so the persistence of mycoplasmas in host cells as an aspect of pathogenesis remains hypothetical.
Conclusion
The infection of immunocompetent patients with M. pneumoniae induces a strong antibody response which is mainly directed to surface-located proteins of the adhesion complex of the mycoplasmas. Thus, an influence of specific immunoglobulins that inhibit the adherence of mycoplasmal cells to the respiratory epithelium of the host can be expected. However, the occurrence of long-term carriage of M. pneumoniae, asymptomatic carriers, re-infections as well as a broad spectrum of extra-pulmonary manifestations show that the protective effect of specific serum antibodies is limited as regards the upper respiratory tract. The directed movement of bacteria to the base of the ciliated epithelium of the respiratory mucosa and the specialized adherence to target cells of the host allow the bacteria to reach a protected niche. The role of the local immune response by inhibiting adherence and the immunomodulatory effects of local inflammation on the clinical course have yet to be explained. These factors are also crucial for the development of an effective vaccine. Finally, the confirmed induction of genotype-specific antibodies raises the question of whether this has an influence on re-infections due to different genotypes of M. pneumoniae, which might be important for understanding the special epidemiology of a common human respiratory tract pathogen.
Author Contributions
RD drafted the manuscript. EJ revised and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Atkinson, T. P., Balish, M. F., and Waites, K. B. (2008). Epidemiology, clinical manifestations, pathogenesis and laboratory detection of Mycoplasma pneumoniae infections. FEMS Microbiol. Rev. 32, 956–973. doi: 10.1111/j.1574-6976.2008.00129.x
Atkinson, T. P., and Waites, K. B. (2014). Mycoplasma pneumoniae infections in childhood. Pediatr. Infect. Dis. J. 33, 92–94. doi: 10.1097/INF.0000000000000171
Balish, M. F., and Krause, D. C. (2006). Mycoplasmas: a distinct cytoskeleton for wall-less bacteria. J. Mol. Microbiol. Biotechnol. 11, 244–255. doi: 10.1159/000094058
Beghetto, E., De Paolis, F., Montagnani, F., Cellesi, C., and Gargano, N. (2009). Discovery of new Mycoplasma pneumoniae antigens by use of a whole-genome lambda display library. Microbes Infect. 11, 66–73. doi: 10.1016/j.micinf.2008.10.004
Brown, R. J., Holden, M. T., Spiller, O. B., and Chalker, V. J. (2015). Development of a multilocus sequence typing scheme for molecular typing of Mycoplasma pneumoniae. J. Clin. Microbiol. 53, 3195–3203. doi: 10.1128/JCM.01301-15
Busson, L., van den Wijngaert, S., Dahma, H., Decolvenaer, M., di Cesare, L., Martin, A., et al. (2013). Evaluation of 10 serological assays for diagnosing Mycoplasma pneumoniae infection. Diag. Microbiol. Infect. Dis. 76, 133–137. doi: 10.1016/j.diagmicrobio.2013.02.027
Chalker, V. J., Stocki, T., Mentasti, M., Fleming, D., Sadler, C., Ellis, J., et al. (2011). Mycoplasma pneumoniae infection in primary care investigated by real-time PCR in England and Wales. Eur. J. Clin. Microbiol. Infect. Dis. 30, 915–921. doi: 10.1007/s10096-011-1176-3
Chourasia, B. K., Chaudhry, R., and Malhotra, P. (2014). Delineation of immunodominant and cytadherence segment(s) of Mycoplasma pneumoniae P1 gene. BMC Microbiol. 28:108. doi: 10.1186/1471-2180-14-108
Citti, C., and Blanchard, A. (2013). Mycoplasmas and their host: emerging and re-emerging minimal pathogens. Trends Microbiol. 21, 196–203. doi: 10.1016/j.tim.2013.01.003
Citti, C., Nouvel, L. X., and Baranowski, E. (2010). Phase and antigenic variation in mycoplasmas. Future Microbiol. 5, 1073–1085. doi: 10.2217/fmb.10.71
Dallo, S. F., Kannan, T. R., Blaylock, M. W., and Baseman, J. B. (2002). Elongation factor Tu and E1 β subunit of pyruvate dehydrogenase complex act as fibronectin binding proteins in Mycoplasma pneumoniae. Mol. Microbiol. 46, 1041–1051. doi: 10.1046/j.1365-2958.2002.03207.x
Dandekar, T., Huynen, M., Regula, J. T., Ueberle, B., Zimmermann, C. U., Andrade, M. A., et al. (2000). Re-annotating the Mycoplasma pneumoniae genome sequence: adding value, function and reading frames. Nucleic Acids Res. 28, 3278–3288. doi: 10.1093/nar/28.17.3278
Degrange, S., Cazanave, C., Charron, A., Renaudin, H., Bebear, C., and Bebear, C. M. (2009). Development of multiple-locus variable-number tandem-repeat analysis for molecular typing of Mycoplasma pneumoniae. J. Clin. Microbiol. 47, 914–923. doi: 10.1128/JCM.01935-08
Donkor, E. S. (2013). Understanding the pneumococcus: transmission and evolution. Front. Cell. Infect. Microbiol. 3:7. doi: 10.3389/fcimb.2013.00007
Drasbek, M., Nielsen, P. K., Persson, K., Birkelund, S., and Christiansen, G. (2004). Immune response to Mycoplasma pneumoniae P1 and P116 in patients with atypical pneumonia. BMC Microbiol. 4:7. doi: 10.1186/1471-2180-4-7
Duffy, M. F., Whithear, K. G., Noormohammadi, A. H., Markham, P. F., Catton, M., Leydon, J., et al. (1999). Indirect enzyme-linked immunosorbent assay for detection of immunoglobulin G reactive with a recombinant protein expressed from the gene encoding the 116-kilodalton protein of Mycoplasma pneumoniae. J. Clin. Microbiol. 37, 1024–1029.
Dumke, R., Catrein, I., Herrmann, R., and Jacobs, E. (2004). Preference, adaptation and survival of Mycoplasma pneumoniae subtypes in an animal model. Int. J. Med. Microbiol. 294, 149–155. doi: 10.1016/j.ijmm.2004.06.020
Dumke, R., Schnee, C., Pletz, M. W., Rupp, J., Jacobs, E., Sachse, K., et al. (2015). Mycoplasma pneumoniae and Chlamydia spp. infection in community-acquired pneumonia. Germany 2011–2012. Emerg. Infect. Dis. 21, 426–434. doi: 10.3201/eid2103.140927
Dumke, R., Schurwanz, N., and Jacobs, E. (2008). Characterisation of subtype and variant-specific antigen regions of the P1 adhesin of Mycoplasma pneumoniae. Int. J. Med. Microbiol. 298, 483–491. doi: 10.1016/j.ijmm.2007.06.002
Dumke, R., Strubel, A., Cyncynatus, C., Herrmann, R., Lück, C., and Jacobs, E. (2012). Optimized serodiagnosis of Mycoplasma pneumoniae infections. Diagn. Microbiol. Infect. Dis. 73, 200–203. doi: 10.1016/j.diagmicrobio.2012.02.014
Dumke, R., von Baum, H., Lück, P. C., and Jacobs, E. (2010). Subtypes and variants of Mycoplasma pneumoniae: local and temporal changes in Germany 2003–2006 and absence of a correlation between the genotype in the respiratory tract and the occurrence of genotype-specific antibodies in the sera of infected patients. Epidemiol. Infect. 138, 1829–1837. doi: 10.1017/S0950268810000622
Foy, H. M. (1993). Infections caused by Mycoplasma pneumoniae and possible carrier state in different populations of patients. Clin. Infect. Dis. 17, S37–S46. doi: 10.1093/clinids/17.Supplement_1.S37
Foy, H. M., Ochs, H., Davis, S. D., Kenny, G. E., and Luce, R. R. (1973). Mycoplasma pneumoniae infections in patients with immunodeficiency syndromes: report of four cases. J. Infect. Dis. 127, 388–393. doi: 10.1093/infdis/127.4.388
Gnarpe, J., Lundbäck, A., Sundelöf, B., and Gnarpe, H. (1992). Prevalence of Mycoplasma pneumoniae in subjectively healthy individuals. Scand. J. Infect. Dis. 24, 161–164. doi: 10.3109/00365549209052607
Gründel, A., Friedrich, K., Pfeiffer, M., Jacobs, E., and Dumke, R. (2015). Subunits of the pyruvate dehydrogenase cluster of Mycoplasma pneumoniae are surface-displayed proteins that bind and activate human plasminogen. PLoS ONE 10:e126600. doi: 10.1371/journal.pone.0126600
Halbedel, S., and Stülke, J. (2007). Tools for the genetic analysis of Mycoplasma. Int. J. Med. Microbiol. 297, 37–44. doi: 10.1016/j.ijmm.2006.11.001
Hallamaa, K. M., Browning, G. F., and Tang, S. L. (2006). Lipoprotein multigene families in Mycoplasma pneumoniae. J. Bacteriol. 188, 5393–5399. doi: 10.1128/JB.01819-05
Hallamaa, K. M., Tang, S. L., Ficorilli, N., and Browning, G. F. (2008). Differential expression of lipoprotein genes in Mycoplasma pneumoniae after contact with human lung epithelial cells, and under oxidative and acidic stress. BMC Microbiol. 8:124. doi: 10.1186/1471-2180-8-124
Hames, C., Halbedel, S., Hoppert, M., Frey, J., and Stülke, J. (2009). Glycerol metabolism is important for cytotoxicity of Mycoplasma pneumoniae. J. Bacteriol. 191, 747–753. doi: 10.1128/JB.01103-08
Hausner, M., Schamberger, A., Naumann, W., Jacobs, E., and Dumke, R. (2013). Development of protective anti-Mycoplasma pneumoniae antibodies after immunization of guinea pigs with the combination of a P1-P30 chimeric recombinant protein and chitosan. Microb. Pathog. 64, 23–32. doi: 10.1016/j.micpath.2013.07.004
Hu, P. C., Huang, C. H., Collier, A. M., and Clyde, W. A. Jr. (1983). Demonstration of antibodies to Mycoplasma pneumoniae attachment protein in human sera and respiratory secretions. Infect. Immun. 41, 437–439.
Inamine, J. M., Ho, K. C., Loechel, S., and Hu, P. C. (1990). Evidence that UGA is read as a tryptophan codon rather than as a stop codon by Mycoplasma pneumoniae, Mycoplasma genitalium, and Mycoplasma gallisepticum. J. Bacteriol. 172, 504–506.
Jacobs, E., Bennewitz, A., and Bredt, W. (1986). Reaction pattern of human anti-Mycoplasma pneumoniae antibodies in enzyme-linked immunosorbent assays and immunoblotting. J. Clin. Microbiol. 23, 517–522.
Jacobs, E., Ehrhardt, I., and Dumke, R. (2015). New insights in the outbreak pattern of Mycoplasma pneumoniae. Int. J. Med. Microbiol. 305, 705–708. doi: 10.1016/j.ijmm.2015.08.021
Jain, S., Williams, D. J., Arnold, S. R., Ampofo, K., Bramley, A. M., Reed, C., et al. (2015). Community-acquired pneumonia requiring hospitalization among U.S. children. N. Engl. J. Med. 372, 835–845. doi: 10.1056/NEJMoa1405870
Kannan, T. R., and Baseman, J. B. (2006). ADP-ribosylating and vacuolating cytotoxin of Mycoplasma pneumoniae represents unique virulence determinant among bacterial pathogens. Proc. Natl. Acad. Sci. U.S.A. 103, 6724–6729. doi: 10.1073/pnas.0510644103
Kenri, T., Okazaki, N., Yamazaki, T., Narita, M., Izumikawa, K., Matsuoka, M., et al. (2008). Genotyping analysis of Mycoplasma pneumoniae clinical strains in Japan between 1995 and 2005: type shift phenomenon of M. pneumoniae clinical strains. J. Med. Microbiol. 57, 469–475. doi: 10.1099/jmm.0.47634-0
Kogoj, R., Mrvic, T., Praprotnik, M., and Kese, D. (2015). Prevalence, genotyping and macrolide resistance of Mycoplasma pneumoniae among isolates of patients with respiratory tract infections, Central Slovenia, 2006 to 2014. Euro. Surveill. 20:37. doi: 10.2807/1560-7917.ES.2015.20.37.30018
Krause, D. C., and Baseman, J. B. (1983). Inhibition of Mycoplasma pneumoniae hemadsorption and adherence to respiratory epithelium by antibodies to a membrane protein. Infect. Immun. 39, 1180–1186.
Kumar, S., Wang, L., Fan, J., Kraft, A., Bose, M. E., Tiwari, S., et al. (2008). Detection of 11 common viral and bacterial pathogens causing community-acquired pneumonia or sepsis in asymptomatic patients by using a multiplex reverse transcription-PCR assay with manual (enzyme hybridization) or automated (electronic microarray) detection. J. Clin. Microbiol. 46, 3063–3072. doi: 10.1128/JCM.00625-08
Linchevski, I., Klement, E., and Nir-Paz, R. (2009). Mycoplasma pneumonia vaccine protection efficiency and adverse reactions – systematic review and meta-analysis. Vaccine 27, 2437–2446. doi: 10.1016/j.vaccine.2009.01.135
Lluch-Senar, M., Cozzuto, L., Cano, J., Delgado, J., Llórens-Rico, V., Pereyre, S., et al. (2015). Comparative ”-omics” in Mycoplasma pneumoniae clinical isolates reveals key virulence factors. PLoS ONE 10:e0137354. doi: 10.1371/journal.pone.0137354
Loens, K., Goossens, H., and Ieven, M. (2010). Acute respiratory infection due to Mycoplasma pneumoniae: current status of diagnostic methods. Eur. J. Clin. Microbiol. Infect. Dis. 29, 1055–1069. doi: 10.1007/s10096-010-0975-2
Meseguer, M. A., Alvarez, A., Rejas, M. T., Sánchez, C., Pérez-Díaz, J. C., and Baquero, F. (2003). Mycoplasma pneumoniae: a reduced-genome intracellular bacterial pathogen. Infect. Genet. Evol. 3, 47–55. doi: 10.1016/S1567-1348(02)00151-X
Meyer Sauteur, P. M., van Rossum, A. M., and Vink, C. (2014). Mycoplasma pneumoniae in children: carriage, pathogenesis, and antibiotic resistance. Curr. Opin. Infect. Dis. 27, 220–227. doi: 10.1097/QCO.0000000000000063
Miyata, M. (2010). Unique centipede mechanism of Mycoplasma gliding. Annu. Rev. Microbiol. 64, 519–537. doi: 10.1146/annurev.micro.112408.134116
Montagnani, F., de Paolis, F., Beghetto, E., and Gargano, N. (2010). Use of recombinant chimeric antigens for the serodiagnosis of Mycoplasma pneumoniae infection. Eur. J. Clin. Microbiol. Infect. Dis. 29, 1377–1386. doi: 10.1007/s10096-010-1010-3
Morrison-Plummer, J., Leith, D. K., and Baseman, J. B. (1986). Biological effects of anti-lipid and anti-protein monoclonal antibodies on Mycoplasma pneumoniae. Infect. Immun. 53, 398–403.
Musatovova, O., Kannan, T. R., and Baseman, J. B. (2012). Mycoplasma pneumoniae large DNA repetitive elements RepMP1 show type specific organization among strains. PLoS ONE 7:e47625. doi: 10.1371/journal.pone.0047625
Nakane, D., Adan-Kubo, J., Kenri, T., and Miyata, M. (2011). Isolation and characterization of P1 adhesin, a leg protein of the gliding bacterium Mycoplasma pneumoniae. J. Bacteriol. 193, 715–722. doi: 10.1128/JB.00796-10
Nilsson, A. C., Björkman, P., and Persson, K. (2008). Polymerase chain reaction is superior to serology for the diagnosis of acute Mycoplasma pneumoniae infection and reveals a high rate of persistent infection. BMC Microbiol. 8:93. doi: 10.1186/1471-2180-8-93
Nuyttens, H., Cyncynatus, C., Renaudin, H., Pereyre, S., and Bebear, C. (2010). Identification, expression and serological evaluation of the recombinant ATP synthase beta subunit of Mycoplasma pneumoniae. BMC Microbiol. 10:216. doi: 10.1186/1471-2180-10-216
Razin, S., and Jacobs, E. (1992). Mycoplasma adhesion. J. Gen. Microbiol. 138, 407–422. doi: 10.1099/00221287-138-3-407
Regula, J. T., Boguth, G., Görg, A., Hegermann, J., Mayer, F., Frank, R., et al. (2001). Defining the Mycoplasma “cytoskeleton”: the protein composition of the Triton X-100 insoluble fraction of the bacterium Mycoplasma pneumoniae determined by 2-D gel electrophoresis and mass spectrometry. Microbiology 147, 1045–1057. doi: 10.1099/00221287-147-4-1045
Roifman, C. M., Rao, C. P., Lederman, H. M., Lavi, S., Quinn, P., and Gelfand, E. W. (1986). Increased susceptibility to Mycoplasma infection in patients with hypogammaglobulinemia. Am. J. Med. 80, 590–594. doi: 10.1016/0002-9343(86)90812-0
Schurwanz, N., Jacobs, E., and Dumke, R. (2009). A strategy to create chimeric proteins derived from functional adhesin regions of Mycoplasma pneumoniae for vaccine development. Infect. Immun. 77, 5007–5015. doi: 10.1128/IAI.00268-09
Sekine, H., Taguchi, H., Watanabe, H., Kawai, S., Fujioka, Y., Goto, H., et al. (2009). Immunological analysis and pathological examination of gnotobiotic mice monoassociated with Mycoplasma pneumoniae. J. Med. Microbiol. 58, 697–705. doi: 10.1099/jmm.0.007872-0
Seto, S., Kenri, T., Tomiyama, T., and Miyata, M. (2005). Involvement of P1 adhesin in gliding motility of Mycoplasma pneumoniae as revealed by the inhibitory effects of antibody under optimized gliding conditions. J. Bacteriol. 187, 1875–1877. doi: 10.1128/JB.187.5.1875-1877.2005
Simmons, W. L., Daubenspeck, J. M., Osborne, J. D., Balish, M. F., Waites, K. B., and Dybvig, K. (2013). Type 1 and type 2 strains of Mycoplasma pneumoniae form different biofilms. Microbiology 159, 737–747. doi: 10.1099/mic.0.064782-0
Spuesens, E. B. M., Fraaij, P. L. A., Visser, E. G., Hoogenboezem, T., Hop, W. C. J., van Adrichem, L. N. A., et al. (2013). Carriage of Mycoplasma pneumoniae in the upper respiratory tract of symptomatic and asymptomatic children: an observational study. PLoS Med. 10:e1001444. doi: 10.1371/journal.pmed.1001444
Spuesens, E. B. M., Oduber, M., Hoogenboezem, T., Sluijter, M., Hartwig, N. G., van Rossum, A. M. C., et al. (2009). Sequence variations in RepMP2/3 and RepMP4 elements reveal intragenomic homologous DNA recombination events in Mycoplasma pneumoniae. Microbiology 155, 2182–2196. doi: 10.1099/mic.0.028506-0
Su, C. J., Chavoya, A., Dallo, S. F., and Baseman, J. B. (1990). Sequence divergency of the cytadhesin gene of Mycoplasma pneumoniae. Infect. Immun. 58, 2669–2674.
Suzuki, Y., Seto, J., Itagaki, T., Aoki, T., Abiko, C., and Matsuzaki, Y. (2015). Gene mutations associated with macrolide-resistance and p1 gene typing of Mycoplasma pneumoniae isolated in Yamagata, Japan, between 2004 and 2013. Kansenshogaku Zasshi 89, 16–22.
Svenstrup, H. F., Nielsen, P. K., Drasbek, M., Birkelund, S., and Christiansen, G. (2002). Adhesion and inhibition assay of Mycoplasma genitalium and M. pneumoniae by immunofluorescence microscopy. J. Med. Microbiol. 51, 361–373. doi: 10.1099/0022-1317-51-5-361
Szczepanek, S. M., Majumder, S., Sheppard, E. S., Liao, X., Rood, D., Tulman, E. R., et al. (2012). Vaccination of BALB/c mice with an avirulent Mycoplasma pneumoniae P30 mutant results in disease exacerbation upon challenge with a virulent strain. Infect. Immun. 80, 1007–1014. doi: 10.1128/IAI.06078-11
Szczepanek, S. M., and Silbart, L. K. (2014). “Host immune responses to mycoplasmas,” in Mollicutes Molecular Biology and Pathogenesis, eds G. F. Browning and C. Citti (Norfolk: Caister Academic Press), 273–288.
Tabassum, I., Chaudhry, R., Chourasia, B. K., and Malhotra, P. (2010). Identification of a N-terminal 27 kDa fragment of Mycoplasma pneumoniae P116 protein as specific immunogen in M. pneumoniae infections. BMC Infect. Dis. 13:350. doi: 10.1186/1471-2334-10-350
Taylor-Robinson, D., Webster, A. D., Furr, P. M., and Asherson, G. L. (1980). Prolonged persistence of Mycoplasma pneumoniae in a patient with hypogammaglobulinaemia. J. Infect. 2, 171–175. doi: 10.1016/S0163-4453(80)91284-0
Techasaensiri, C., Tagliabue, C., Cagle, M., Iranpour, P., Katz, K., Kannan, T. R., et al. (2010). Variation in colonization, ADP-ribosylating and vacuolating cytotoxin, and pulmonary disease severity among Mycoplasma pneumoniae strains. Am. J. Respir. Crit. Care Med. 182, 797–804. doi: 10.1164/rccm.201001-0080OC
Touati, A., Blouin, Y., Sirand-Pugnet, P., Renaudin, H., Oishi, T., Vergnaud, G., et al. (2015). Molecular epidemiology of Mycoplasma pneumoniae: genotyping using single nucleotide polymorphisms and SNaPshot technology. J. Clin. Microbiol. 53, 3182–3194. doi: 10.1128/JCM.01156-15
Varshney, A. K., Chaudhry, R., Kabra, S. K., and Malhotra, P. (2008). Cloning, expression, and immunological characterization of the P30 protein of Mycoplasma pneumoniae. Clin. Vaccine Immunol. 15, 215–220. doi: 10.1128/CVI.00283-07
Vu, A. C., Foy, H. M., Cartwright, F. D., and Kenny, G. E. (1987). The principal protein antigens of isolates of Mycoplasma pneumoniae as measured by levels of immunoglobulin G in human serum are stable in strains collected over a 10-year period. Infect. Immun. 55, 1830–1836.
Wood, P. R., Hill, V. L., Burks, M. L., Peters, J. I., Singh, H., Kannan, T. R., et al. (2013). Mycoplasma pneumoniae in children with acute and refractory asthma. Ann. Allergy Asthma Immunol. 110, 328–334. doi: 10.1016/j.anai.2013.01.022
Xiao, L., Ptacek, T., Osborne, J. D., Crabb, D. M., Simmons, W. L., Lefkowitz, E. J., et al. (2015). Comparative genome analysis of Mycoplasma pneumoniae. BMC Genomics 16:610. doi: 10.1186/s12864-015-1801-0
Xue, G., Cao, L., Wang, L., Zhao, H., Feng, Y., Ma, L., et al. (2013). Evaluation of P1 adhesin epitopes for the serodiagnosis of Mycoplasma pneumoniae infections. FEMS Microbiol. Lett. 340, 86–92. doi: 10.1111/1574-6968.12063
Zhao, F., Liu, L., Tao, X., He, L., Meng, F., and Zhang, J. (2015). Culture-independent detection and genotyping of Mycoplasma pneumoniae in clinical specimens from Beijing. China. PLoS ONE 10:e0141702. doi: 10.1371/journal.pone.0141702
Keywords: Mycoplasma pneumoniae, community-acquired pneumonia, immune response, adherence, protective antibodies
Citation: Dumke R and Jacobs E (2016) Antibody Response to Mycoplasma pneumoniae: Protection of Host and Influence on Outbreaks? Front. Microbiol. 7:39. doi: 10.3389/fmicb.2016.00039
Received: 04 December 2015; Accepted: 11 January 2016;
Published: 26 January 2016.
Edited by:
Cécile Bébéar, University of Bordeaux, FranceReviewed by:
Sabine Pereyre, University of Bordeaux, FranceKatherine Loens, Antwerp University Hospital, Belgium
Copyright © 2016 Dumke and Jacobs. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Roger Dumke, cm9nZXIuZHVta2VAdHUtZHJlc2Rlbi5kZQ==
 Roger Dumke
Roger Dumke Enno Jacobs
Enno Jacobs