
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 12 January 2016
Sec. Infectious Agents and Disease
Volume 6 - 2015 | https://doi.org/10.3389/fmicb.2015.01548
 DoKyung Lee1,2†
DoKyung Lee1,2† Eun Jin Kim1,2†
Eun Jin Kim1,2† Paul E. Kilgore3
Paul E. Kilgore3 Hideyuki Takahashi4
Hideyuki Takahashi4 Makoto Ohnishi4
Makoto Ohnishi4 Jun Tomono5
Jun Tomono5 Shigehiko Miyamoto5
Shigehiko Miyamoto5 Daisuke Omagari6,7
Daisuke Omagari6,7 Dong Wook Kim1,2*
Dong Wook Kim1,2* Mitsuko Seki6,7*
Mitsuko Seki6,7*We have developed a novel Neisseria meningitidis serogroup-specific loop-mediated isothermal amplification (LAMP) assay for six of the most common meningococcal serogroups (A, B, C, W, X, and Y). The assay was evaluated using a set of 31 meningococcal LAMP assay positive cerebrospinal fluid (CSF) specimens from 1574 children with suspected meningitis identified in prospective surveillance between 1998 and 2002 in Vietnam, China, and Korea. Primer specificity was validated using 15 N. meningitidis strains (including serogroups A, B, C, E, W, X, Y, and Z) and 19 non-N. meningitidis species. The N. meningitidis serogroup LAMP detected down to ten copies and 100 colony-forming units per reaction. Twenty-nine CSF had N. meningitidis serogroup identified by LAMP compared with two CSF in which N. meningitidis serogroup was identified by culture and multi-locus sequence typing. This is the first report of a serogroup-specific identification assay for N. meningitidis using the LAMP method. Our results suggest that this assay will be a rapid, sensitive, and uniquely serogroup-specific assay with potential for application in clinical laboratories and public health surveillance systems.
Neisseria meningitidis is a gram-negative β-proteobacterium and member of the bacterial family Neisseriaceae (Stephens et al., 2007). The meningococcal polysaccharide capsule and outer membrane proteins have been identified as N. meningitidis virulence factors (Stephens, 2009). N. meningitidis is principally subdivided into 12 serogroups based on the capsular polysaccharide type (A, B, C, E, H, I, K, L, W, X, Y, and Z; Harrison et al., 2013). Globally, the majority of invasive meningococcal disease has been attributable to six serogroups designated A, B, C, W, X, and Y (Stephens et al., 2007; Stephens, 2009; Verma and Khanna, 2012).
The geographic distribution and epidemic potential of N. meningitidis may vary by serogroup. In Europe and other industrialized countries, serogroups B and C are major causes of invasive meningococcal disease (Harrison et al., 2009). The majority of meningococcal disease in European countries, which ranges in incidence from 0.2 to 14 cases per 100,000 (Harrison et al., 2009), is caused by serogroup B strains, particularly in countries that have introduced meningococcal serogroup C conjugate vaccines. More recent reports have noted increases in serogroup W and Y meningococcal disease (Campbell et al., 2009; Ladhani et al., 2012). In the Americas, the majority of meningococcal disease is caused by serogroups B and C (Stephens et al., 2007). The emergence of serogroup W and Y has been noted in some countries where surveillance is present (Chiavetta et al., 2007). In Africa, N. meningitidis is the leading cause of severe, life-threatening meningitis and has been responsible for thousands of cases and scores of deaths across sub-Saharan meningitis belt countries (Alonso et al., 2006). Seasonal large-scale epidemics caused by N. meningitidis serogroup A have been frequent in Sub-Saharan Africa ranging from Senegal to Ethiopia (WHO, 2011). The introduction of meningococcal serogroup A conjugate vaccines in countries of Sub-Saharan Africa has led to significant reductions in serogroup A epidemics (Kristiansen et al., 2014). Serogroup C, serogroup X and, most recently, serogroup W disease have also been reported (Harrison et al., 2009). The emergence of serogroup W meningococcal disease in Africa was associated with a large epidemic among Hajj pilgrims in 2000 and a large-scale meningitis outbreak in Burkina Faso during 2002 (Alonso et al., 2006). In Asia, serogroup B has been identified as the leading cause of meningococcal disease in Thailand and Taiwan (Harrison et al., 2009). N. meningitidis serogroups A, C, X, and Y have been identified in these and other Asian countries (Stephens et al., 2007; Harrison et al., 2009; Kim et al., 2012b).
Traditionally, invasive disease due to N. meningitidis has been diagnosed using specialized bacterial culture media and reagents (e.g., anti-sera) used for identifying N. meningitidis serogroups (Corless et al., 2001). While traditional gram staining and observation using light microscopy are used for N. meningitis detection, these techniques do not permit serogroup identification (Chanteau et al., 2006). The diagnosis of N. meningitidis invasive disease by culture testing requires a well-equipped laboratory with appropriate biosafety equipment and procedures (Borrow et al., 2014). In some settings, rapid immunochromatographic antigen detection testing has been applied for detection of N. meningitidis but these assays are limited by relatively low diagnostic sensitivity (Chanteau et al., 2006). Traditional diagnostic assays also have limited utility in many populations and health systems where inappropriate use of antibiotics continues despite global increases in antibiotic resistance (Levy and Marshall, 2004). In such settings, detection of N. meningitidis may be problematic due to inhibition of bacterial growth due to antibiotics present in human clinical samples (Laxminarayan et al., 2013). In such cases, isolation of bacteria from patients’ cerebrospinal fluid (CSF) is time-consuming and may yield little or no growth.
Given the geographic and temporal variations in meningococcal strains causing both epidemic and sporadic disease, the serogroup identification of N. meningitidis to identify circulating serogroups is important as a part of disease control and surveillance programs. Efforts to improve diagnostics for N. meningitidis have included development of a rapid immunochromatographic assay that uses monoclonal antibodies against polysaccharides for identification of serogroups A, W, C, and Y. Use of this assay has been most prominent in endemic regions of Africa (Chanteau et al., 2006). A number of PCR-based methods have been developed to identify major N. meningitidis serogroups by targeting the polysialyltransferase siaD gene or other genes involved in serogroup-specific capsule synthesis (Taha et al., 2005; Fraisier et al., 2009). In addition, real-time and multiplex PCR assays for serogroup identification of N. meningitidis have been developed (Fraisier et al., 2009; Zhu et al., 2012; Doyle and Jennison, 2013). However, equipment required for conventional and real-time PCR assays is relatively expensive, and PCR methods are complex to perform in resource-limited laboratory settings found in many developing countries where N. meningitidis infections have been reported.
The loop-mediated isothermal amplification (LAMP) method (Notomi et al., 2000, 2015) offers an attractive alternative to bacterial cultivation and PCR. The LAMP assay uses four different primers to identify six distinct regions on the target gene, resulting in a greater specificity than conventional PCR. Additional primers (i.e., the loop primers designated LF and LB) designed to anneal the loop structure in LAMP can be used to increase sensitivity and specificity of the LAMP reaction (Tomita et al., 2008). During the LAMP reaction, a small amount of DNA can be detected due to the high amplification capacity of the assay. In addition, the LAMP assay does not require the use of complicated procedures, equipment, or machines. For these reasons, the LAMP assay is considered more rapid, efficient, simpler, and more economical than other diagnostic methods.
Recently, we developed a LAMP-based assay (Lee et al., 2015) that demonstrated high sensitivity and specificity for N. meningitidis detection in CSF. In the present study, we report the development of a LAMP-based N. meningitidis serogroup identification assay and the evaluation of this assay’s ability to detect specific N. meningitidis serogroups in CSF. This is the first report of N. meningitidis serogroup-specific identification using the LAMP method.
This study used 35 standard reference strains, including 15 N. meningitidis; serogroups A (HY0001 and NIID1), B (HY0002, H44/76, and NIID2), C (HY0003 and NIID3), E (NIID8), W (HY0006 and NIID93), X (HY0004 and NIID4), Y (HY0005 and NIID5), and Z (NIID6): nine non-meningococcal Neisseria species; N. gonorrhoeae NIID9, N. flavescens NIID10, N. denitrificans NIID11, N. elongata NIID12, N. canis NIID13, N. cinerea NIID14, N. lactamica NIID85, N. mucosa NIID16, and N. sicca NIID17: and 10 other bacterial strains; Streptococcus pneumoniae ATCC 49619, Staphylococcus aureus ATCC 29212, Klebsiella pneumoniae ATCC 700603, K. oxytoca ATCC 700324, Pseudomonas aeruginosa ATCC 27853, Escherichia coli ATCC 25922, Enterococcus faecalis ATCC 700324, Mycobacterium tuberculosis ATCC 27294, and Haemophilus influenzae ATCC 9007 and IID984 (Lee et al., 2015; Supplemental Materials, Table 1).
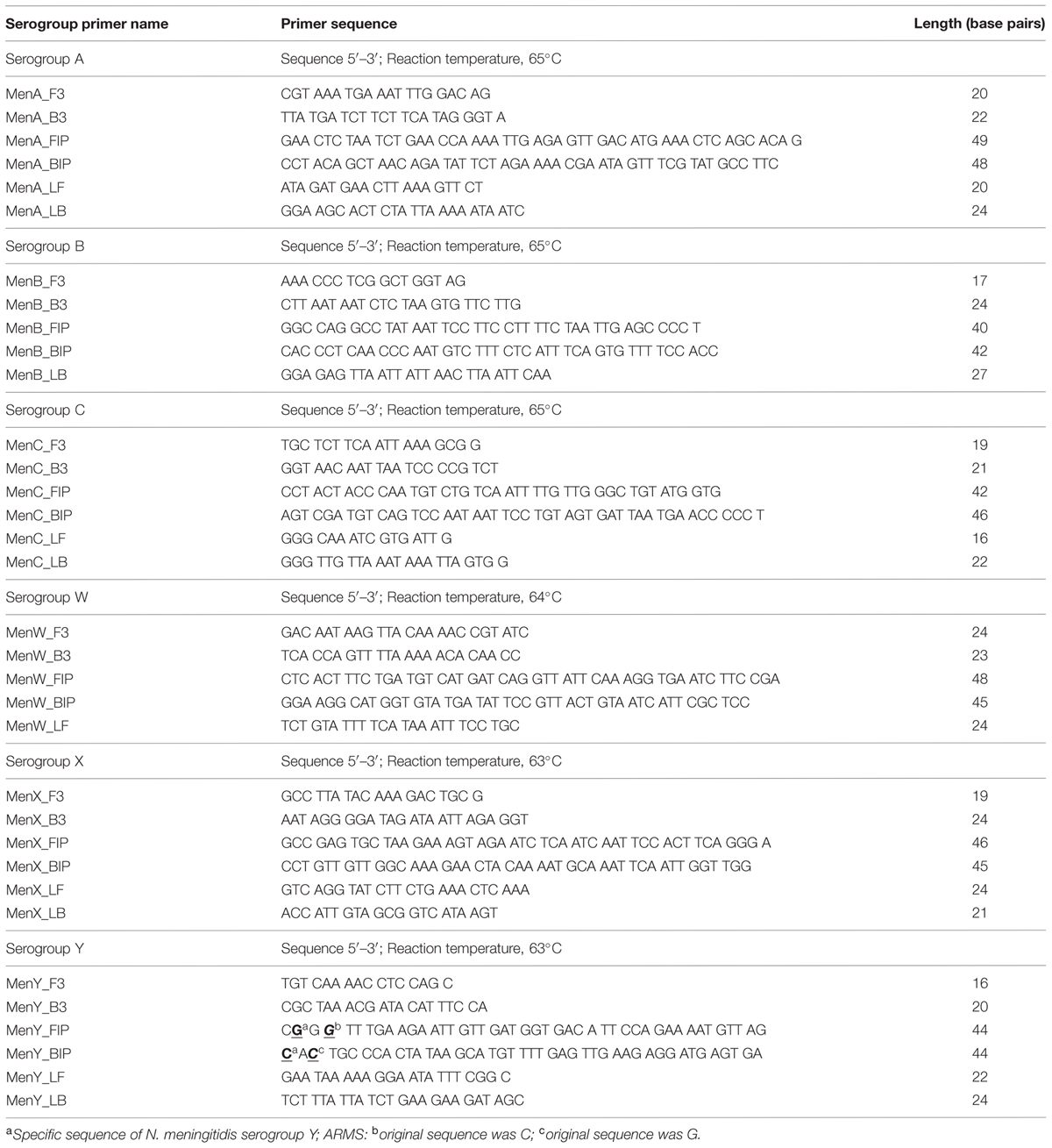
TABLE 1. Loop-mediated isothermal amplification (LAMP) primer sets for serogroup identification of N. meningitidis.
For the detection limit study; we used six N. meningitidis strains; serogroups A (HY0001), B (HY0002), C (HY0003), W (HY0006), X (HY0004), and Y (HY0005). Bacterial colony forming unit (CFU) was determined as follows: bacteria were inoculated on chocolate agar culture plate (Becton, Dickinson & Co., Franklin Lakes, NJ, USA) and further incubated in 5% CO2 at 37°C for 24 h. The bacteria were collected in 1 ml PBS and CFUs were determined by serial dilutions of culture suspensions inoculated on chocolate agar plate.
Genomic DNA was extracted from the 35 standard reference strains by the phenol-chloroform method (Lee et al., 2015). For detection limit analysis, genomic DNAs from N. meningitidis serogroups A (HY0001), B (HY0002), C (HY0003), W (HY0006), X (HY0004), and Y (HY0005) were used, and the concentration was determined using a NanoDrop 1000 (Thermo Fisher Scientific, Inc., Waltham, MA, USA). Approximately 2.5 fg/2 μl of a DNA template was taken as one copy of the N. meningitidis genome per reaction, based on the genome of N. meningitidis serogroup B strain MC58 [accession number AE002098; 2272351 bp (Tettelin et al., 2000)]. To ascertain the detection limit of the LAMP assay, serial 10-fold dilutions of the genomic DNA were tested in each LAMP reaction, and the results were compared with those of each culture test as described previously (Kim et al., 2012b).
For the detection limit study, triplicate LAMP testing was performed using 10-fold dilutions of genomic DNA. Two technicians tested the same samples independently to confirm the reproducibility of LAMP results as described previously (Kim et al., 2012a; Lee et al., 2015).
Using PrimerExplorer V4 software1, we designed six serogroup identification LAMP primer sets (Table 1) targeting the genome sequence of each serogroup-specific gene identified (Taha et al., 2005; Fraisier et al., 2009): sacB (GenBank accession number FR774048) for serogroup A, siaD (GenBank accession number CP002424) for serogroup B, siaD (GenBank accession number AM421808) for serogroup C, synG (GenBank accession number AY234197) for serogroup W, ctrA (GenBank accession number AY289931) for serogroup X, and synF (GenBank accession number AY234201)for serogroup Y.
Due to a high level of sequence identity between synF and synG genes (Zhu et al., 2012; Doyle and Jennison, 2013), we used the LAMP method along with an amplification refractory mutation system (ARMS; Newton et al., 1989; Ikeda et al., 2007) to detect the specific sequences of synF (Table 1). The ARMS uses uniquely designed primers that enable detection of mutations (Newton et al., 1989). As a target of the present investigation, we chose two specific sequences of the serogroup Y synF gene. Based on the principle of ARMS, one of the two specific sequences was designed in the 5′ end of the BIP primer, and the other was designed in the second sequence of the 5′ end of the FIP primer (Table 1). We then added one mutation in the third sequence of the 5′ end of the BIP primer (from G to C), and an additional mutation in the fourth sequence of the 5′ end of the FIP primer (from C to G).
The LAMP reaction was performed with 25 μl of a mixture containing 1.6 μM each of primers FIP and BIP, 0.2 μM of primers F3 and B3, 0.4 μM of primers LF and LB, 8 U of the Bst DNA polymerase large fragment (New England Biolabs, Ipswich, MA, USA), 1.4 mM each of the four deoxynucleoside triphosphates, 0.8 M betaine, 20 mM Tris-HCl (pH 8.8), 10 mM KCl, 10 mM (NH4)2SO4, 8 mM MgSO4, 0.1% Tween 20, and 2 μl of template. The high-performance liquid chromatography-purified primers were dissolved in Tris-EDTA buffer. The mixture was incubated at 63–65°C (Table 1) for 60 min and then heated at 80°C for 2 min to terminate the reaction. For the detection limit study, PCR was performed as described previously (Fraisier et al., 2009).
Neisseria meningitidis serogroup detection was observed by the visual inspection based on its generation of turbidity proportional to the amount of amplified DNA (Mori et al., 2004; Lee et al., 2015). A Loopamp real-time turbidimeter (LA-200 real-time turbidimeter; Eiken Chemical, Co., Ltd., Tokyo, Japan) was used to monitor the turbidity in the reaction tube in real-time by reading the OD650 every 6 s. According to the manufacturer’s protocol (Mori et al., 2004), we used the application software for the turbidimeter to obtain the amplification time required to exceed a turbidity level of 0.1 (Tt). For the detection limit and specificity study, we also used electrophoretic analysis and a colorimetric visual inspection dye, Leuco Crystal Violet (LCV; D-Quick; Kaneka, Co., Ltd., Osaka, Japan; Miyamoto et al., 2015). LCV was dried down in the caps of the reaction tubes. After reactions were completed, the LAMP amplicons were mixed by inverting the tubes, and changes in color were observed to obtain test results (negative, remained colorless; positive, color change to blue: Figure 1).
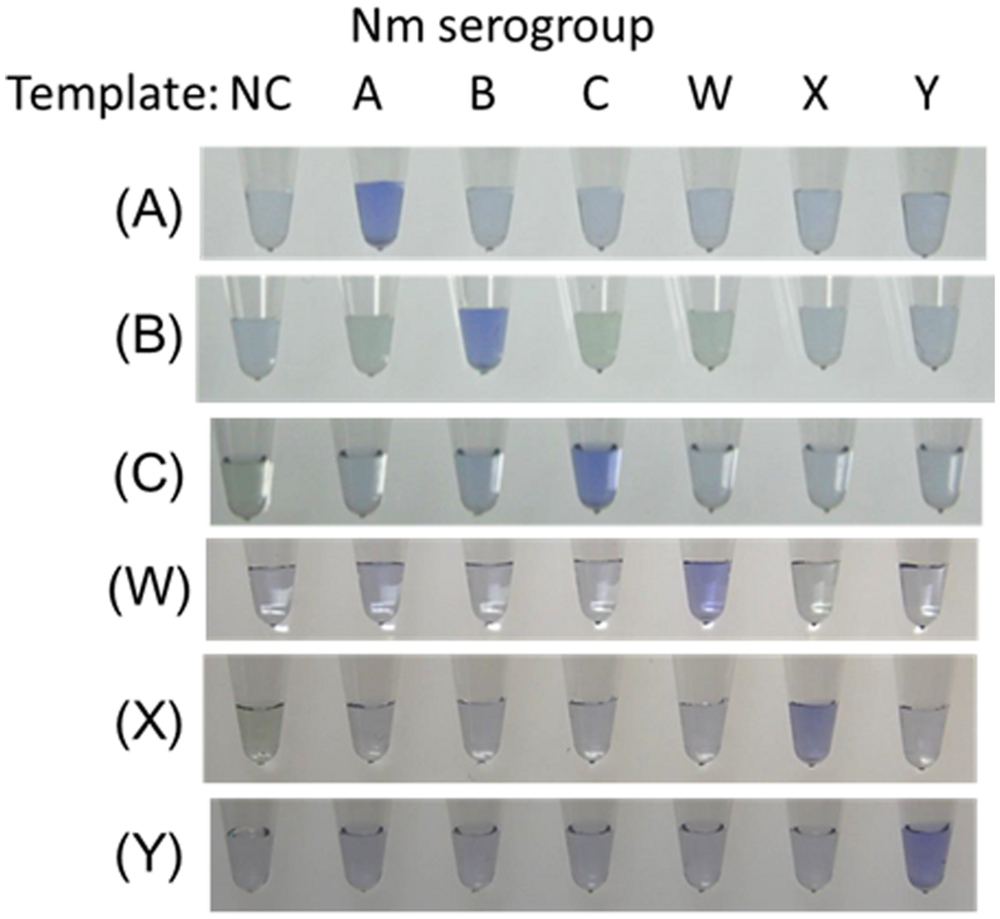
FIGURE 1. Visual inspection of dye-mediated monitoring of the N. meningitidis serogroup-specific loop-mediated isothermal amplification (LAMP) assay. The original colorless appearance of the visual inspection dye (Kaneka, Co., Ltd., Osaka, Japan) changed to blue if the reaction was positive; if reaction was negative, the dye remained colorless. (A), Results of N. meningitidis serogroup A-LAMP assay. (B), Results of N. meningitidis serogroup B-LAMP assay. (C), Results of N. meningitidis serogroup C-LAMP assay. (W), Results of N. meningitidis serogroup W-LAMP assay. (X), Results of N. meningitidis serogroup X-LAMP assay. (Y), Results of N. meningitidis serogroup Y-LAMP assay. NC, negative control.
To verify the structure of LAMP amplified products, the amplified LAMP products were sequenced using a BigDye Terminator v. 3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA, USA) and an ABI PRISM 377 DNA sequencer (Applied Biosystems) according to the manufacturer’s instructions (Kim et al., 2012a; Lee et al., 2015). The target region was between F2 and B2, and the primer sequences were from the F2 and B2 regions.
Children with suspected meningitis who were less than 5 years of age were prospectively enrolled at the participating hospitals between 1998 and 2002 (Kim et al., 2012b). CSF was streaked on commercial blood agar culture medium (Becton, Dickinson & Co., Franklin Lakes, NJ, USA), incubated in 5% CO2 at 37°C for 3 days, and checked daily for bacterial growth (Kim et al., 2012b). Isolates were identified using standard microbiological criteria (Isenberg, 1998). The established multi-locus sequence typing (MLST) scheme2 for Neisseria sp. was used for further genotype identification.
As described previously (Kim et al., 2012a; Lee et al., 2015), to extract bacterial DNA, CSF specimens were heated at 95°C for 3 min and centrifuged at 13,000 × g for 5 min. The samples were stored at -80°C. Two microliters of the supernatant were used for LAMP assays. In the previous study (Lee et al., 2015), we had tested 1,574 randomly selected CSF specimens in Korea (n = 470), China (n = 536), and Vietnam (n = 568) using the Nm-LAMP assay targeting specific sequences of N. meningitidis ctrA gene. In this study, we evaluated the serogroup identification LAMP assay using the set of 31 Nm-LAMP positive CSF specimens.
We utilized CSF specimens preserved from our previous surveillance study (Kennedy et al., 2007). All CSF specimens utilized in this study were de-identified prior to laboratory processing and analysis. Ethical approvals for patient specimen collection during surveillance were obtained from the following ethics review committees: the Institutional Review Board of International Vaccine Institute, Seoul, Korea; the Institutional Review Board at National Institute of Hygiene and Epidemiology, Hanoi, Vietnam; and the Guangxi Zhuang Autonomous Region Center for Disease Control, Nanning, China. Each institution participated in prospective, population-based surveillance for childhood meningitis from 1999 to 2002 (Kim et al., 2004; Anh et al., 2006). During those surveillance studies, written consent was not obtained as the CSF collection was considered routine standard care for hospitalized children with suspected bacterial meningitis. For this reason, verbal consent of the parent or legal guardian present with the child during the period of hospitalization was recorded in the patient’s medical chart at the time of the clinical lumbar puncture procedure. This consent procedure was approved by these local scientific ethics review committees of the participating institutions.
To evaluate the species specificity of the N. meningitidis LAMP primer set, we tested 35 reference strains, as described above. When approximately 1-ng whole-cell DNA (106 copies of genomic DNA of each strain) was used in the LAMP reaction, amplification products from each of the related serogroup strains were observed within 30 min, whereas none of the other strains resulted in amplification products even after 60 min of incubation (Figure 1). We confirmed that the amplification products corresponded to the selected sequence target by sequencing using primers F2 and B2. The LAMP reaction is highly specific for each serogroup.
The detection limits for the LAMP assays were 10 or 100 genome copies and 100 to 1,000 CFUs (25 μL of reaction mix, triplicate trials) as described in Table 2. The detection limits of the PCR assays were 103 to 104 genome copies per reaction (Table 2). Throughout the study, the evaluation of the LAMP reactions showed complete agreement between a white precipitate recorded by visual inspection, the real-time turbidimeter, electrophoretic analysis, and LCV. Using LCV, the colorless dye changed to blue color indicating a positive reaction. These results were evaluable under natural light without the need for UV light (Figure 1).
A group of 31 known Nm-LAMP-positive specimens (Lee et al., 2015) were also analyzed by N. meningitidis serogroup-specific LAMP tests. The LAMP assay identified N. meningitidis serogroup for 29 of the 31 N. meningitidis-positive CSF specimens (Table 3). Using a novel LAMP assay based on the ARMS principle, we succeeded in differentiating N. meningitidis serogroup Y from N. meningitidis serogroup W (Figure 1). Five serogroup A (5/31, 16.1%), five serogroup B (5/31, 16.1%), three serogroup C (3/31, 9.7%), five serogroup X (5/31, 16.1%), five serogroup Y (5/31, 16.1%), and six serogroup W (6/31, 19.4%) specimens were identified. Two samples were non-typable for N. meningitidis (2/31, 6.5%). Each serogroup-specific LAMP-amplified product was confirmed by sequencing, and the sequence results were identical to the reference sequences.
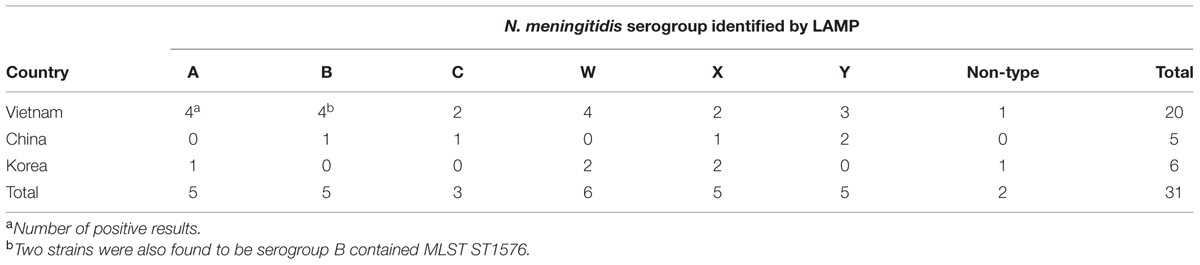
TABLE 3. Results of the serogroup-specific LAMP assay of 31 CSF samples of N. meningitidis ctrA positive.
In the previous study (Kennedy et al., 2007), three N. meningitidis culture-positive (3/31, 9.7%) were confirmed and two of the three specimens were serogroup B positive using MLST (Table 3). The MLST scheme and database of Neisseria sp. are well-established and publically available at http://pubmlst.org/neisseria/. The MLST scheme and methods previously described were applied to the N. meningitidis isolates. Seven MLST loci were amplified by using PCR primers and the DNA sequences of each locus were compared to the same locus from the database. Two serogroup B N. meningitidis isolates contained an allele type 140, 5, 9, 173, 175, 34, 165 (in the order abcZ, adk, aroE, fumC, gdh, pdhC, and pgm), which has been designated as sequence type 1576 (ST1576).
Compared to the culture method, the sensitivity and specificity of the N. meningitidis serogroup B LAMP assay were 100% (2/2) and 89.7% (26/29).
The novel N. meningitidis serogroup-specific LAMP assay reported here demonstrated high detection rates as well as high test specificity and sensitivity. These higher detection rates found with the LAMP assay compared with conventional PCR is consistent with previous studies of LAMP assays (Kim et al., 2011, 2012a). The robust performance of the LAMP assay is made possible by the use of four different primers to identify six distinct regions on the target gene. The design of our primers for the assay described here are unique and enable clear differentiation between six major serogroups of N. meningitidis responsible for invasive meningococcal disease (including meningitis) found around the world.
We successfully differentiated N. meningitidis serogroup Y from N. meningitidis serogroup W using the ARMS principle (Newton et al., 1989). The first application of ARMS in LAMP primer design to detect a point mutation of target sequences was reported by Ikeda et al. (2007). The investigator added one mutation at the 5′ end of BIP primer that permitted successful detection of the point mutation. In our experiments, optimizing the design of the LAMP primer set for serogroup Y proved to be challenging because key target gene sequences (synF from serogroup Y and synG from serogroup W) share a high degree of similarity that has been also noted by other investigators (Zhu et al., 2012; Doyle and Jennison, 2013). To successfully differentiate and accurately identify serogroup Y and W strains, we created mutations at the 5′ end of both the FIP and BIP primers.
The detection limits for the serogroup-specific LAMP assays were 10 or 100 genome copies and 100 to 1,000 CFUs, and the conventional PCR assays had much lower sensitivity (103 to 104 genome copies) than LAMP. The evaluation of the LAMP reactions showed complete agreement between a white precipitate recorded by visual inspection, the real-time turbidimeter, electrophoretic analysis, and LCV. The results of LCV were evaluable under natural light without the need for UV light and the color change remained visible for at least 1 week (Lee et al., 2015). The experience using LCV colorimetric visual inspection dye reported here can inform and facilitate future development and application of the LAMP assay in resource-limited settings.
We used preserved de-identified CSF specimens that were collected between 1998 and 2002 in our previous study of bacterial meningitis (Kim et al., 2012b). In the previous study (Lee et al., 2015), using 1574 randomly selected CSF specimens, we detected 31 Nm-LAMP assay positive CSF specimens including three N. meningitidis culture-positive (3/31, 9.7%). Two of the three specimens were confirmed as serogroup B positive using MLST.
Using the meningococcus serogroup-specific LAMP assay, 29 of the 31 known Nm-LAMP-positive specimens (Lee et al., 2015) were one of six serogroup-specific positive CSF specimens. Five serogroup A (5/31, 16.1%), five serogroup B (5/31, 16.1%), three serogroup C (3/31, 9.7%), five serogroup X (5/31, 16.1%), five serogroup Y (5/31, 16.1%), and six serogroup W (6/31, 19.4%) specimens were identified and two samples were non-typable for N. meningitidis (2/31, 6.5%). In these results, the observation of two non-typable specimens may be due to potential discrepancies in the sensitivity of the Nm-LAMP (10 copies; Lee et al., 2015) compared with the N. meningitidis serogroup-specific LAMP (10 or 100 copies) assays. Other possible explanations for the observation of non-typable strains include the degradation of DNA material in stored specimens as well as the presence of N. meningitidis serogroups not covered by designed LAMP assays.
In settings of many developing countries where empiric use of broad-spectrum antibiotics is common, conventional bacterial culture methods have low yield and are likely to underestimate the true burden of N. meningitidis serogroups. During the original prospective surveillance studies, field surveys showed that over-the-counter use of antibiotics without a prescription was common in each country (Kim et al., 2012b). Thus, to better understand the epidemiology bacterial meningitis pathogens such as N. meningitidis, the meningococcal serogroup-specific LAMP offers an accurate alternative diagnostic test. Our results suggest that the meningococcal serogroup LAMP may provide a useful tool in areas where public health agencies wish to measure the burden of N. meningitidis following introduction of meningococcal vaccines.
Based on our experience in this study, the N. meningitidis serogroup-specific LAMP assay was easy to set-up and required no special equipment. In addition, this LAMP assay provides high-quality test results within 2 h that make it feasible and cost-effective for use. Given the simple format for the LAMP assay, this test also has potential for performance in district-level health facilities in developing countries. To confirm the LAMP assay performance characteristics compared with bacterial culture, antigen detection and PCR, further evaluation of the N. meningitidis serogroup identification LAMP in prospective studies are now planned.
PK, HT, MO, DK, MS, contributed the conception of this study; SM, JT, DK, MS designed the experiments; DL, EK, SM, DK, MS, DO performed the experiments; PK, HT, MO acquired samples; DL, EK, JT, SM, DO, PK, DK, MS analyzed data; DL, EK, JT, SM, DO, PK, DK, MS interpreted data; DL, EK, PK, HT, DK, MS drafted the manuscript; and DO, MO, SM, JT approved the manuscript.
This work was supported by Japan Society for the Promotion of Science (JSPS) KAKENHI Grant Number 10201004, the grant 2015R1A2A2A01007297 from National Research Foundation (NRF) of Korea, and the JSPS and NRF under the Japan – Korea Basic Scientific Cooperation Program (NRF-2014K2A2A4001480).
Mitsuko Seki has received research grant funding from Kaneka, Co., Ltd. Jun Tomono and Shigehiko Miyamoto are employees of Kaneka, Co., Ltd. Paul E. Kilgore serves on meningococcal vaccine speakers bureau for Pfizer, Inc. The following authors have no conflict of interests or financial disclosures to declare: DoKyung Lee, Eun Jin Kim, Hideyuki Takahashi, Makoto Ohnishi, and DongWook Kim. These statements do not alter our adherence to the journal policies on sharing data and materials.
We are grateful to Dr. Soon Ae Kim for her helpful advice. We are especially grateful to Dr. Dang Duc Anh, National Institute of Hygiene and Epidemiology, Hanoi, Vietnam; Dr. Bai Qing Dong, Guangxi Zhuang Autonomous Region Health Bureau, Nanning, Guangxi, China; and Prof. Jung Soo Kim, Chonbuk National University School of Medicine, Jeonju, Korea for their support in this study.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.01548
Nm-LAMP, meningococcal LAMP.
Alonso, J. M., Bertherat, E., Perea, W., Borrow, R., Chanteau, S., Cohet, C., et al. (2006). From genomics to surveillance, prevention and control: new challenges for the African meningitis belt. Vaccine 24, 4279–4284. doi: 10.1016/j.vaccine.2006.03.037
Anh, D. D., Kilgore, P. E., Kennedy, W. A., Nyambat, B., Long, H. T., Jodar, L., et al. (2006). Haemophilus influenzae type B meningitis among children in Hanoi, Vietnam: epidemiologic patterns and estimates of H. Influenzae type B disease burden. Am. J. Trop. Med. Hyg. 74, 509–515.
Borrow, R., Findlow, J., Gray, S., Taylor, S., and Kaczmarski, E. (2014). Safe laboratory handling of Neisseria meningitidis. J. Infect. 68, 305–312. doi: 10.1016/j.jinf.2014.01.003
Campbell, H., Borrow, R., Salisbury, D., and Miller, E. (2009). Meningococcal C conjugate vaccine: the experience in England and Wales. Vaccine 27(Suppl. 2), B20–B29. doi: 10.1016/j.vaccine.2009.04.067
Chanteau, S., Dartevelle, S., Mahamane, A. E., Djibo, S., Boisier, P., and Nato, F. (2006). New rapid diagnostic tests for Neisseria meningitidis serogroups A, W135, C, and Y. PLoS Med. 3:e337. doi: 10.1371/journal.pmed.0030337
Chiavetta, L., Chavez, E., Ruzic, A., Mollerach, M., and Regueira, M. (2007). [Surveillance of Neisseria meningitidis in Argentina, 1993-2005: distribution of serogroups, serotypes and serosubtypes isolated from invasive disease]. Rev. Argent. Microbiol. 39, 21–27.
Corless, C. E., Guiver, M., Borrow, R., Edwards-Jones, V., Fox, A. J., and Kaczmarski, E. B. (2001). Simultaneous detection of Neisseria meningitidis, Haemophilus influenzae, and Streptococcus pneumoniae in suspected cases of meningitis and septicemia using real-time PCR. J. Clin. Microbiol. 39, 1553–1558. doi: 10.1128/JCM.39.4.1553-1558.2001
Doyle, C. J., and Jennison, A. V. (2013). Novel real-time polymerase chain reactions for serogroup specific gene detection of Neisseria meningitidis serogroups B, C, W-135 and Y. J. Microbiol. Methods 94, 83–85. doi: 10.1016/j.mimet.2013.04.011
Fraisier, C., Stor, R., Tenebray, B., Sanson, Y., and Nicolas, P. (2009). Use of a new single multiplex PCR-based assay for direct simultaneous characterization of six Neisseria meningitidis serogroups. J. Clin. Microbiol. 47, 2662–2666. doi: 10.1128/JCM.02415-08
Harrison, L. H., Trotter, C. L., and Ramsay, M. E. (2009). Global epidemiology of meningococcal disease. Vaccine 27(Suppl. 2), B51–B63. doi: 10.1016/j.vaccine.2009.04.063
Harrison, O. B., Claus, H., Jiang, Y., Bennett, J. S., Bratcher, H. B., Jolley, K. A., et al. (2013). Description and nomenclature of Neisseria meningitidis capsule locus. Emerg. Infect. Dis. 19, 566–573. doi: 10.3201/eid1904.111799
Ikeda, S., Takabe, K., Inagaki, M., Funakoshi, N., and Suzuki, K. (2007). Detection of gene point mutation in paraffin sections using in situ loop-mediated isothermal amplification. Pathol. Int. 57, 594–599. doi: 10.1111/j.1440-1827.2007.02144.x
Kennedy, W. A., Chang, S. J., Purdy, K., Le, T., Kilgore, P. E., Kim, J. S., et al. (2007). Incidence of bacterial meningitis in Asia using enhanced CSF testing: polymerase chain reaction, latex agglutination and culture. Epidemiol. Infect. 135, 1217–1226. doi: 10.1017/S0950268806007734
Kim, D. W., Kilgore, P. E., Kim, E. J., Kim, S. A., Anh, D. D., Dong, B. Q., et al. (2012a). The enhanced pneumococcal LAMP assay: a clinical tool for the diagnosis of meningitis due to Streptococcus pneumoniae. PLoS ONE 7:e42954. doi: 10.1371/journal.pone.0042954
Kim, S. A., Kim, D. W., Dong, B. Q., Kim, J. S., Anh, D. D., and Kilgore, P. E. (2012b). An expanded age range for meningococcal meningitis: molecular diagnostic evidence from population-based surveillance in Asia. BMC Infect. Dis. 12:310. doi: 10.1186/1471-2334-12-310
Kim, D. W., Kilgore, P. E., Kim, E. J., Kim, S. A., Anh, D. D., and Seki, M. (2011). Loop-mediated isothermal amplification assay to detect Haemophilus influenzae type b in cerebrospinal fluid. J. Clin. Microbiol. 49, 3621–3626. doi: 10.1128/JCM.00515-11
Kim, J. S., Jang, Y. T., Kim, J. D., Park, T. H., Park, J. M., Kilgore, P. E., et al. (2004). Incidence of Haemophilus influenzae type b and other invasive diseases in South Korean children. Vaccine 22, 3952–3962. doi: 10.1016/j.vaccine.2004.04.003
Kristiansen, P. A., Ba, A. K., Ouedraogo, A. S., Sanou, I., Ouedraogo, R., Sangare, L., et al. (2014). Persistent low carriage of serogroup A Neisseria meningitidis two years after mass vaccination with the meningococcal conjugate vaccine, MenAfriVac. BMC Infect. Dis. 14:663. doi: 10.1186/s12879-014-0663-4
Ladhani, S. N., Flood, J. S., Ramsay, M. E., Campbell, H., Gray, S. J., Kaczmarski, E. B., et al. (2012). Invasive meningococcal disease in England and Wales: implications for the introduction of new vaccines. Vaccine 30, 3710–3716. doi: 10.1016/j.vaccine.2012.03.011
Laxminarayan, R., Duse, A., Wattal, C., Zaidi, A. K., Wertheim, H. F., Sumpradit, N., et al. (2013). Antibiotic resistance-the need for global solutions. Lancet Infect. Dis. 13, 1057–1098. doi: 10.1016/S1473-3099(13)70318-9
Lee, D. K., Kim, E. J., Kilgore, P. E., Kim, S. A., Takahashi, H., Ohnishi, M., et al. (2015). Clinical evaluation of a Loop-Mediated Isothermal Amplification (LAMP) assay for rapid detection of Neisseria meningitidis in cerebrospinal fluid. PLoS ONE 10:e0122922. doi: 10.1371/journal.pone.0122922
Levy, S. B., and Marshall, B. (2004). Antibacterial resistance worldwide: causes, challenges and responses. Nat. Med. 10, S122–S129. doi: 10.1038/nm1145
Miyamoto, S., Sano, S., Takahashi, K., and Jikihara, T. (2015). Method for colorimetric detection of double-stranded nucleic acid using leuco triphenylmethane dyes. Anal. Biochem. 473, 28–33. doi: 10.1016/j.ab.2014.12.016
Mori, Y., Kitao, M., Tomita, N., and Notomi, T. (2004). Real-time turbidimetry of LAMP reaction for quantifying template DNA. J. Biochem. Biophys. Methods 59, 145–157. doi: 10.1016/j.jbbm.2003.12.005S0165022X03002173
Newton, C. R., Graham, A., Heptinstall, L. E., Powell, S. J., Summers, C., Kalsheker, N., et al. (1989). Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res. 17, 2503–2516. doi: 10.1093/nar/17.7.2503
Notomi, T., Mori, Y., Tomita, N., and Kanda, H. (2015). Loop-mediated isothermal amplification (LAMP): principle, features, and future prospects. J. Microbiol. 53, 1–5. doi: 10.1007/s12275-015-4656-9
Notomi, T., Okayama, H., Masubuchi, H., Yonekawa, T., Watanabe, K., Amino, N., et al. (2000). Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28, E63. doi: 10.1093/nar/28.12.e63
Stephens, D. S. (2009). Biology and pathogenesis of the evolutionarily successful, obligate human bacterium Neisseria meningitidis. Vaccine 27(Suppl. 2), B71–B77. doi: 10.1016/j.vaccine.2009.04.070
Stephens, D. S., Greenwood, B., and Brandtzaeg, P. (2007). Epidemic meningitis, meningococcaemia, and Neisseria meningitidis. Lancet 369, 2196–2210. doi: 10.1016/S0140-6736(07)61016-2
Taha, M. K., Alonso, J. M., Cafferkey, M., Caugant, D. A., Clarke, S. C., Diggle, M. A., et al. (2005). Interlaboratory comparison of PCR-based identification and genogrouping of Neisseria meningitidis. J. Clin. Microbiol. 43, 144–149. doi: 10.1128/JCM.43.1.144-149.2005
Tettelin, H., Saunders, N. J., Heidelberg, J., Jeffries, A. C., Nelson, K. E., Eisen, J. A., et al. (2000). Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287, 1809–1815. doi: 10.1126/science.287.5459.1809
Tomita, N., Mori, Y., Kanda, H., and Notomi, T. (2008). Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat. Protoc. 3, 877–882. doi: 10.1038/nprot.2008.57
Verma, R., and Khanna, P. (2012). Meningococcal vaccine: a new vaccine to combat meningococcal disease in India. Hum. Vaccin. Immunother. 8, 1904–1906. doi: 10.4161/hv.21666
WHO (2011). Meningococcal vaccines: WHO position paper, November 2011. Wkly Epidemiol. Rec. 86, 521–539.
Keywords: Neisseria meningitidis, serogroup identification, loop-mediated isothermal amplification, meningitis, cerebrospinal fluid, children
Citation: Lee D, Kim EJ, Kilgore PE, Takahashi H, Ohnishi M, Tomono J, Miyamoto S, Omagari D, Kim DW and Seki M (2016) A Novel Loop-Mediated Isothermal Amplification Assay for Serogroup Identification of Neisseria meningitidis in Cerebrospinal Fluid. Front. Microbiol. 6:1548. doi: 10.3389/fmicb.2015.01548
Received: 07 August 2015; Accepted: 21 December 2015;
Published: 12 January 2016.
Edited by:
Awdhesh Kalia, University of Texas MD Anderson Cancer Center, USAReviewed by:
Paras Jain, Albert Einstein College of Medicine, New York, USACopyright © 2016 Lee, Kim, Kilgore, Takahashi, Ohnishi, Tomono, Miyamoto, Omagari, Kim and Seki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dong Wook Kim, ZG9uZ3dvb2tAaGFueWFuZy5hYy5rcg==; Mitsuko Seki, c2VraS5taXRzdWtvQG5paG9uLXUuYWMuanA=
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.