
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 15 December 2015
Sec. Microbial Physiology and Metabolism
Volume 6 - 2015 | https://doi.org/10.3389/fmicb.2015.01376
This article is part of the Research Topic Secondary Metabolism: An Unlimited Foundation for Synthetic Biology View all 7 articles
The great phylogenetic diversity of microalgae is corresponded by a wide arrange of interesting and useful metabolites. Nonetheless metabolic engineering in microalgae has been limited, since specific transformation tools must be developed for each species for either the nuclear or chloroplast genomes. Microalgae as production platforms for metabolites offer several advantages over plants and other microorganisms, like the ability of GMO containment and reduced costs in culture media, respectively. Currently, microalgae have proved particularly well suited for the commercial production of omega-3 fatty acids and carotenoids. Therefore most metabolic engineering strategies have been developed for these metabolites. Microalgal biofuels have also drawn great attention recently, resulting in efforts for improving the production of hydrogen and photosynthates, particularly triacylglycerides. Metabolic pathways of microalgae have also been manipulated in order to improve photosynthetic growth under specific conditions and for achieving trophic conversion. Although these pathways are not strictly related to secondary metabolites, the synthetic biology approaches could potentially be translated to this field and will also be discussed.
Microalgae, defined as a polyphyletic group of unicellular photosynthetic eukaryotes, are among the most ancient and diverse organisms on the planet. There are at least 40,000–70,000 species belonging to nine different phyla. Additionally, some estimates propose that there could be up to eight times the amount of undiscovered or unclassified species (Norton et al., 1996; Bhattacharya et al., 2004; Guiry, 2012).
Microalgae have evolved to adapt to a wide range of environments and consequently have proven to be a rich source of genetic and chemical diversity (Dufresne et al., 2008; Parker et al., 2008; Tirichine and Bowler, 2011; Blunt et al., 2012; Hildebrand et al., 2013). Figure 1 shows that the polyphyletic nature of microalgae constitutes them as an excellent target for discovering unique groups of protein orthologs (Chen et al., 2006).
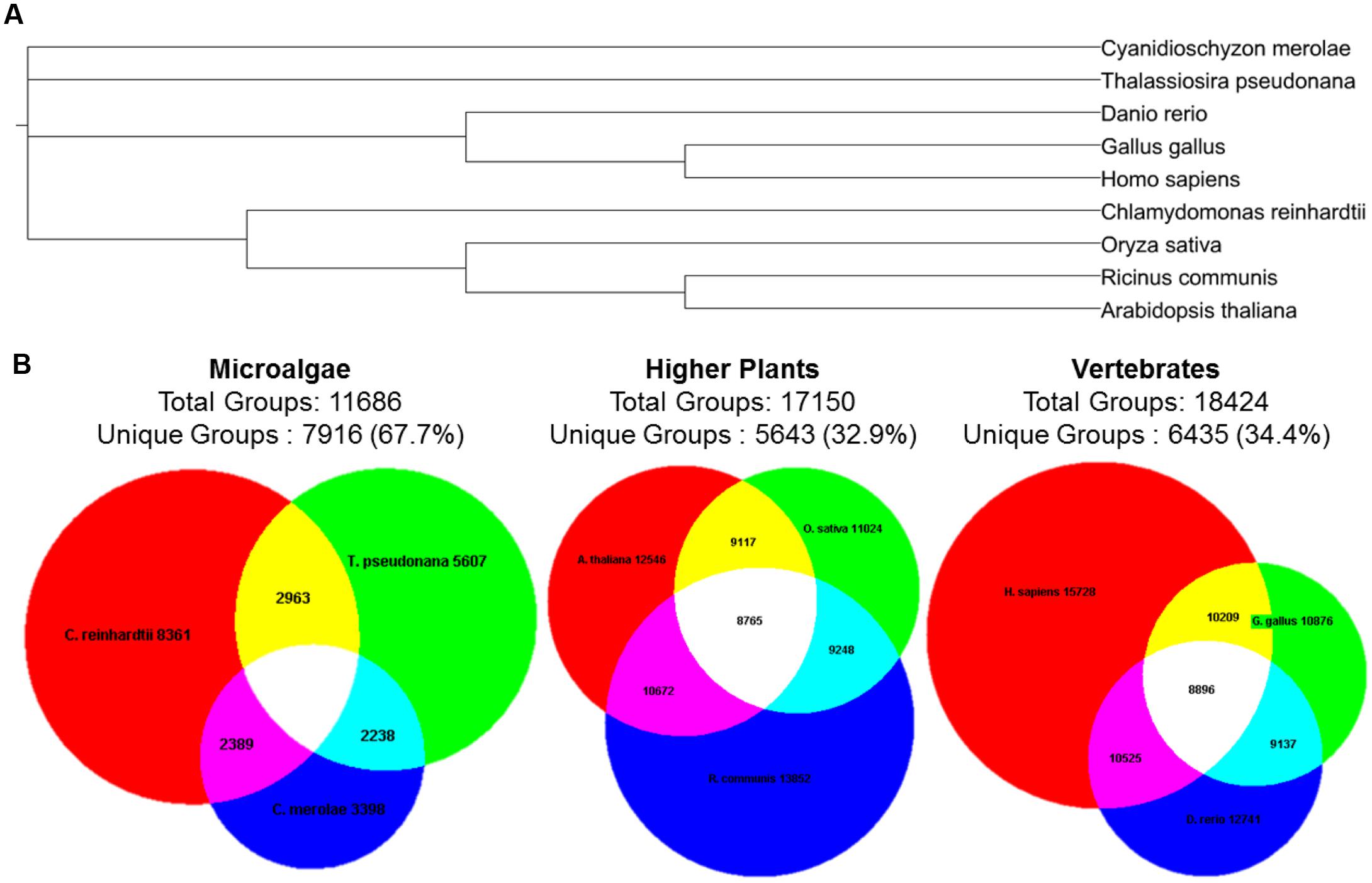
FIGURE 1. Diversity of protein ortholog groups in microalgae, higher plants, and vertebrates. (A) Phylogenetic tree of the selected species according to NCBI taxonomy. The tree was constructed using PhyloT and iTOL (Letunic and Bork, 2011). (B) Unique and shared groups of protein orthologs according to OrthoMCL database (Chen et al., 2006). The Venn diagrams were constructed using Euler3Applet (Chow and Rodgers, 2005). Common names for the species: Chlamydomonas reinhardtii (green alga, red circle); Thalassiosira pseudonana (diatom, green circle) Cyanidioschyzon merolae (red alga, blue circle); Arabidopsis thaliana (thale cress, red circle); Oryza sativa (rice, green circle); Ricinus communis (castor oil plant, blue circle); Homo sapiens (human, red circle); Gallus gallus (chicken, green circle); Danio rerio (zebrafish, blue circle).
Furthermore, algae diversity has been exploited as a unique source of bioactive compounds like carotenoids, fatty acids (FAs), sterols, mycosporine-like amino acids, phycobilins, polyketides, pectins, halogenated compounds, toxins, etc., which have been extensively reviewed previously (Pulz and Gross, 2004; Spolaore et al., 2006; Cardozo et al., 2007; Hallmann, 2007; Milledge, 2011; Borowitzka, 2013; Leu and Boussiba, 2014). Currently, algae are the main sustainable source of commercial carotenoids and omega-3 FAs (Borowitzka, 2013; Leu and Boussiba, 2014). In addition, microalgae have also proven to be cost-effective and safe hosts for expressing a wide array of recombinant proteins, including human and animal therapeutics, and industrial enzymes (Specht et al., 2010; Gong et al., 2011b; Maliga and Bock, 2011; Rosales-Mendoza et al., 2012; Scranton et al., 2015). Furthermore, studies have shown that microalgae have also the potential to be an economically viable source of renewable biofuels (Stephens et al., 2010; Davis et al., 2011; Jones and Mayfield, 2011; Larkum et al., 2011).
While the potential of microalgae as a source of a wide range of products is high, optimization of cultivation and processing technologies will be required before algal-derived biofuels and some of the strain-specific biomolecules can be profitable on a large scale. Techno-economic analysis has identified that three main factors that significantly contribute to the overall cost of production for a microalgal metabolite are product content, growth rate, and cultivation cell density (Davis et al., 2011). These biological outputs are determined by a number of constraints including light intensity, nutrient supply, and the unique metabolism of individual species. Improvements in all these areas will be major drivers in creating the most efficient and economically viable strains of microalgae available for a diverse arrange of biotechnological applications. In this review we will mainly discuss strategies for increasing metabolite product content through genetic engineering. Nonetheless, metabolic engineering of photosynthesis and nutrient assimilation will also be discussed since they directly affect growth rate and cell culture density, thus overall metabolite productivity. Algae biomass can also be converted into biocrude and biogas, through thermochemical conversion and anaerobic digestion, respectively. Since these products do not depend on the accumulation of specific metabolites, engineering of photosynthesis and nutrient assimilation would also be a suitable approach for enhancing productivity of these types of fuels.
Microalgae can be transformed either in the nuclear, chloroplast or mitocondrial genomes (Specht et al., 2010). Most enzymes associated with secondary metabolism are coded in the nuclear genome, but some of them are targeted to the chloroplast for performing their function (Martin, 2010; Terashima et al., 2011; Heydarizadeh et al., 2013). In these cases either the nuclear or plastid genome could be engineered for a desired metabolic trait (Johanningmeier and Fischer, 2010). Electroporation, shaking with glass beads, and particle gun bombardment (biolistic) are the main methods for delivering DNA to microalgae (Coll, 2006), the latter has proven so far the only successful mean for chloroplast transformation (Purton et al., 2013). Nuclear transformation offers the advantages of post-translational modifications of the protein of interest, the possibility of protein targeting to any organelle, simpler transformation protocols and more flexible regulatory sequence recognition, allowing for the use of heterologous promoters and untranslated regions (Leon and Fernandez, 2007; Díaz-Santos et al., 2013; Scaife et al., 2015). It is worth mentioning that the nuclear genomes of some algae also contain efficient microRNA systems for gene silencing, which can also be harnessed for metabolic engineering (Molnar et al., 2009; Cerutti et al., 2011). On the other hand, the main disadvantage of nuclear transformation is the low expression levels of the genes of interest due to silencing and position effects (random integration; Leon and Fernandez, 2007; Specht et al., 2010; Scranton et al., 2015). Recently a system based on the co-translational fusion of an antibiotic-resistance gene with the gene of interest has proven to be effective for alleviating some of these drawbacks for nuclear expression in the model green alga, Chlamydomonas reinhardtii (Rasala et al., 2012). On the other hand, chloroplast gene expression offers higher levels of transgenic protein accumulation (usually ≥1% of total protein) and precise gene targeting given the effective homologous recombination machinery (Johanningmeier and Fischer, 2010; Purton et al., 2013). The latter also enables for efficient gene disruption for metabolic engineering purposes (Fischer et al., 1996). Then again, the disadvantages of plastid-based expression are the lack of post-translational glycosylation, lack of eukaryotic folding machinery, strict codon bias, and the requirement of the sequences for endogenous untranslated regulatory regions and homologous recombination flanking regions (Specht et al., 2010; Gimpel and Mayfield, 2013; Scaife et al., 2015; Scranton et al., 2015).
The great physiologic and genetic diversity of microalgae poses a great challenge for transforming new species given all the variables aforementioned. A proper DNA delivery system and optimized transformation conditions are recognized to be specific for each microalgal specie. Furthermore, the use of endogenous regulatory sequences has always proven to achieve the highest amount of transgenic protein accumulation, highlighting the need for endogenous genomic data (Díaz-Santos et al., 2013; Gimpel and Mayfield, 2013). Additionally, specie-specific codon optimization of the transgene is a critical condition for achieving detectable levels of transgenic protein accumulation (Specht et al., 2010; Gong et al., 2011b). Attempts of circumventing these requirements have resulted in non-reproducible transformation protocols. It is not rare to find past published data of algae transformation that has only been reported once, or by a single laboratory, even though the specie is a highly attractive transformation target for the scientific community (Chow and Tung, 1999; Teng et al., 2002; Coll, 2006; Hallmann, 2007; Chen et al., 2008).
Lipid metabolism in microalgae can be regarded as complex and diverse given the metabolic divergence between the phylogenetic groups. It is clear that for engineering high-yield lipid producing strains, characterization of the specific pathways for any given phylogenetic group of algae will be required (Yu et al., 2011; Hildebrand et al., 2013; Bellou et al., 2014). A simplified scheme for FA, triacylglycerol (TAG), and polyunsaturated fatty acid (PUFA) biosynthesis in green algae is shown in Figure 2. Metabolic engineering strategies for these pathways are summarized in Table 1.
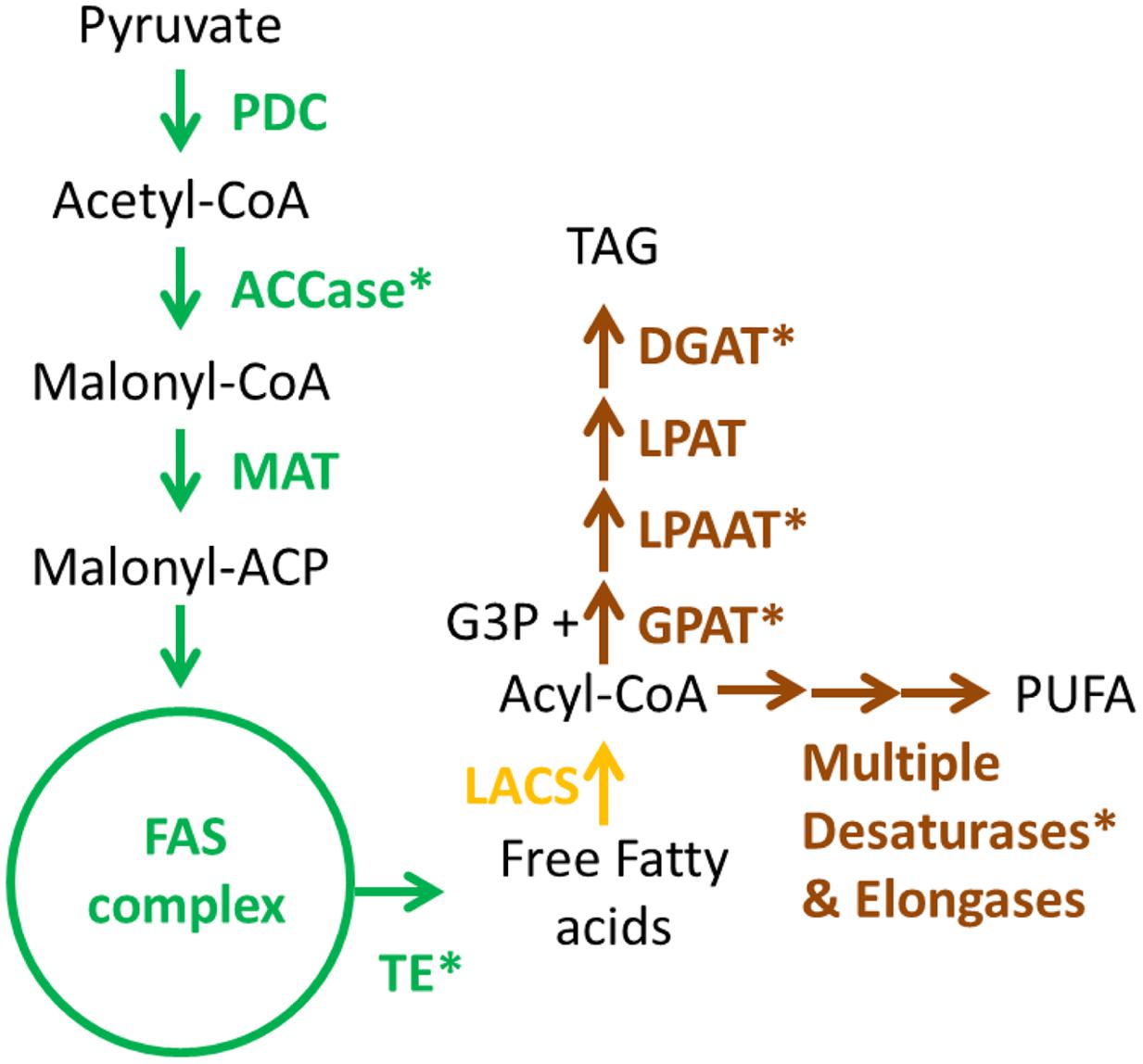
FIGURE 2. Simplified biosynthetic pathways for fatty acids (FAs), triacylglycerols (TAGs) and polyunsaturated FAs in green microalgae. Enzyme names are colored according to their localization. Green: plastid; Yellow: cytoplasm; Brown: endoplasmic reticulum. Asterisks denote enzymes which have been targeted for metabolic engineering, as shown in Table 1. PDC, pyruvate dehydrogenase complex; ACCase, acetyl-CoA carboxylase; MAT, malonyl-CoA/ACP transacylase; ACP, acyl-carrier protein; LACS, long-chain acyl-CoA synthetase; FAS, FA synthase; TE, fatty acyl-ACP thioesterase; GPAT, glycerol-3-phosphate acyltransferase; LPAAT, lyso-phosphatidic acid acyltransferase; LPAT, lyso-phosphatidylcholine acyltransferase; DGAT, diacylglycerol acyltransferase; CoA, coenzyme A; G3P, glycerate-3-phosphate; TAG, triacylglycerol; PUFA, polyunsaturated FA.
The first metabolic engineering report for increasing FA production in algae consisted in the overexpression of the acetyl-CoA carboxylase gene (ACCase) from the diatom Cyclotella cryptica. ACCase codes for the enzyme that carboxylates acetyl-CoA to malonyl-CoA, the first committed step for FA synthesis. Expression vectors and transformation protocols were developed for C. cryptica and the diatom Navicula saprophila. A two–threefold increase in the level of ACCase activity was reported for the transformed diatoms, but no increase in FA accumulation was detected. However, no experimental data was presented for the increase of ACCase activity (Dunahay et al., 1996). Eighteen years later research efforts have been undertaken once again for redirecting carbon and reducing potential toward lipid biosynthesis. Pyruvate can be transformed to acetyl-CoA (substrate for ACCase) through an oxidative decarboxylation reaction catalyzed by the mitochondrial pyruvate dehydrogenase complex (PDC), which is deactivated through phosphorylation by pyruvate carboxylase kinase (PDK). In contrast, plastid PDC is not regulated by a PDK homolog. Using an antisense cDNA construct, PDK expression was knocked down in the diatom Phaeodactylum tricornutum, resulting in up to 82% total neutral lipid increase without changes in the lipid profile (Ma et al., 2014). Malic enzyme (ME) catalyzes the decarboxylation of malate into pyruvate, producing at the same time NADH and carbon dioxide. In addition to pyruvate, NADH is an essential source of reducing power for lipogenesis. Overexpression of the endogenous ME in P. tricornutum resulted in a 2.5-fold increase of total lipid accumulation under nutrient-replete conditions when compared to the control. This great increase didn’t affect growth rate significantly, although cell morphology changes were observed (Xue et al., 2014). Another way of enhancing lipid accumulation is preventing lipid catabolism. After analyzing transcriptomic data from the diatom Thalassiosira pseudonana under silicon-deplete conditions a multifunctional lipase gene was selected as a target for knockdown experiments. RNAi and antisense constructs were transformed; the latter resulted in up to 3.3-fold higher total lipid content than wild-type during the exponential growth phase. Interestingly, the knockdown strains didn’t have a slower growth rate than WT (Trentacoste et al., 2013).
In order to enhance biodiesel cold flow properties, the total amount of short-chain FA (12-14 carbons) has been increased by overexpressing two higher plant FA acyl-carrier protein thioesterases (TE) in P. tricornutum. 75–90% of the short-chain FA were incorporated into TAGs, but there was no significant increase of total lipid accumulation (Radakovits et al., 2011). On the other hand, an endogenous TE was expressed in P. tricornutum (PtTE), resulting in 72% increase of total FA without modification of the FA chain-lengths profile (Gong et al., 2011a). This achievement in total FA increase would have a high impact if it can be reproduced in other species. Furthermore, two TE from higher plants and one endogenous TE gene from C. reinhardtii were overexpressed in this green alga. Only the overexpression of the endogenous gene (CrTE) resulted in higher levels of short FA, while no increase of lipid accumulation was achieved. The latter study also focused in the importance of achieving appropriate protein–protein interactions between the components of the FA synthesis machinery in order to obtain favorable metabolic engineering results (Blatti et al., 2012).
In addition to the modification of FA synthesis, the glycerol acylation steps have also been engineered for improving TAG accumulation. Overexpression of three endogenous Acyl-CoA:diacylglycerol acyltransferases (DGAT) in C. reinhardtii (CrDGAT2a, b, and c) didn’t result in increased TAG accumulation or changes in TAG profiles (La Russa et al., 2012). RNAi silencing of C. reinhardtii CrDGAT2-1 or CrDGAT2-5 resulted in 24 and 37% decrease in lipid accumulation, respectively. Surprisingly, silencing of CrDGAT2-4 caused an increase of up to 34% total lipids. CrDGAT2-2 or CrDGAT2-3 silencing didn’t cause significant changes in lipid accumulation (Deng et al., 2012). In contrast with the results of (La Russa et al., 2012), overexpression of the endogenous DGAT2 in the diatom P. tricornutum resulted in 35% increase of neutral lipid accumulation without significantly affecting growth rate. Furthermore, the valuable omega-3 eicosapentaenoic acid (EPA) accumulated 76% more than in the control (Niu et al., 2013). The simultaneous expression of five TAG biosynthesis-related enzymes derived from the yeasts Saccharomyces cerevisiae and Yarrowia lipolytica (phosphatidic acid phosphatase, lysophosphatidic acid acyltransferase, glycerol-3-phosphate dehydrogenase, glycerol-3-phosphate acyltransferase, and DGAT) in the green microalga Chlorella minutissima resulted in a twofold increase in TAG accumulation. The expression of each enzyme by itself didn’t result in significant changes in lipid accumulation, thus pointing toward the effectiveness of system level approaches (Hsieh et al., 2012).
Transcription factors (TF) can also be manipulated as an alternative to “classic” genetic engineering, which could prove to be more effective for generating global metabolic changes. A patent from Sapphire Energy, Inc. describes that the overexpression of the SN03 TF produces the effect of a “lipid-trigger” in C. reinhardtii, thus resulting in 11% more accumulation of total extractable lipids in nutrient-replete medium. This TF was chosen based on comparative transcriptomic data between nutrient-replete and nitrogen-deplete cells (Yohn et al., 2012). Likewise, a lipogenesis promoting TF from soybean was overexpressed in the green alga C. ellipsoidea, which accumulated up to 52% more total lipids. This TF resulted in the differential expression of 1046 transcripts, including the up-regulation of ACCase (top six up-regulated transcripts were annotated as ACCase; Zhang et al., 2014).
Omega-3 polyunsaturated FAs from microalgae can reach much higher prices than biodiesel. Therefore commercial production of PUFA from genetically engineered algae could be a viable option in a much shorter term, but for a much smaller and more regulated market. Metabolic engineering of PUFA deals mostly with the overexpression of specific FA desaturases and elongases, although TE have also proven useful for this purpose (Niu et al., 2013). In C. reinhardtii, overexpression of the endogenous Δ4 desaturase (CrΔ4FAD) resulted in increased accumulation of its specific product hexadeca-4,7,10,13-tetraenoic acid (16:4). Alternatively, silencing through microRNA resulted in decreased 16:4 accumulation. This shows that classical genetic engineering approaches can result in rational results when modifying the expression of FA desaturases (Zäuner et al., 2012). An endogenous Δ5 desaturase (PtD5b) was overexpressed in P. tricornutum resulting in 58% increased accumulation of EPA. Additional poly and mono unsaturated FA were also significantly augmented. Moreover, this strain accumulated 65% more neutral FA, showing that this strategy could also serve for improving biodiesel yields (Peng et al., 2014).
The complexity of lipid biosynthesis poses a great challenge if the final goal is to enhance accumulation of not only a single type of lipid, but overall lipid accumulation for biofuels. It is then necessary to investigate in further details the regulatory mechanisms in order to deploy system level metabolic engineering of lipid biosynthesis. The studies from (Yohn et al., 2012), and (Zhang et al., 2014) involving TF engineering are great examples in this direction. Additionally there is a lack of efforts toward transforming lipogenic enzymes into the chloroplast genome, given that most of the initial reactions for FA synthesis take place in this organelle (Figure 2).
Green microalgae from the genera Chlamydomona, Scenedesmus, Lobochlamys, and Chlorella can reduce protons to produce hydrogen gas due to their hydrogenase activity, determined to derive predominantly from the [FeFe]-hydrogenase (HYDA1 in C. reinhardtii; Meuser et al., 2009, 2012). Biohydrogen is an attractive fuel alternative because its combustion produces no carbon byproducts and it is a superior fuel for electricity production by fuel cells. Hydrogen production cannot be sustained while photosynthesis is actively occurring because oxygen inactivates hydrogenase. Therefore, a bi-phasic production strategy is necessary, in which algae grow photosynthetically to accumulate biomass, then the cells are exploited for H2 production under anoxic conditions, which can be achieved by inhibition of Photosystem II (PSII) by sulfur deprivation or by using herbicides (oxidation of water by PSII generates oxygen; Melis et al., 2007; Beer et al., 2009). Nonetheless, two novel strains of C. vulgaris have been shown to be able to produce hydrogen under atmospheric oxygen concentrations (Hwang et al., 2014). Increasing hydrogenase activity (or decreasing its oxygen sensitivity), anaerobiosis induction, avoiding competition for electrons from other pathways and increasing the sources of electrons are all steps that could be improved through genetic engineering (Melis et al., 2007; Beer et al., 2009; Esquível et al., 2011; Dubini and Ghirardi, 2014).
Engineering hydrogenase in order to reduce its oxygen sensitivity has yielded only modest improvements, primarily due to the fact that gas channels are formed when the catalytic site is properly folded (Dubini and Ghirardi, 2014). Alternatively, the endogenous hydrogenase has been overexpressed in Chlorella sp. strain DT, resulting in 7–10-fold hydrogen production under semi-aerobic conditions (Chien et al., 2012).
Inactivation of PSII in C. reinhardtii has been achieved by transforming an RNA antisense construct against a sulfate transporter gene (SulP), leading to the accumulation of hydrogen even in the presence of 100 μM sulfate (Chen et al., 2005). Another system for achieving anaerobiosis is based on a copper responsive nuclear transgene that is necessary for the expression of the essential protein D2 of PSII. Upon the addition of copper, the transgene is repressed, along with D2, thus stopping evolution of oxygen through PSII. A brief period of anaerobiosis and hydrogen production is achieved, but it is much shorter than the standard method through sulfur deprivation (Surzycki et al., 2007). In order to reduce PSII efficiency a double mutant of the reaction center protein D1 was generated. The resulting strain could evolve ten times more hydrogen than the control after sulfur deprivation, mainly due to a longer productive period (Torzillo et al., 2009). In Chlorella sp. strain DT expression knock-down of PsbO from PSII (part of the oxygen evolution center) through short interference antisense RNA resulted in up to 10-fold higher hydrogen evolution under semi-aerobic conditions (Lin et al., 2013). The Ligh-Harvesting Complex II (LHCII) captures and channels excitons toward PSII for photochemistry, and thus oxygen evolution. Knock-down of the three major proteins of C. reinhardtii LHCII (LHCMB1, 2, and 3) with three co-transformed RNAi constructs (each one specific for a single gene) resulted in a twofold increase in hydrogen production when compared to the parental strain (Oey et al., 2013).
Oxygen sequestration is another alternative for enhancing hydrogen biosynthesis. Chloroplast transformation of a codon-optimized leghemoglobin protein from soy, which sequesters oxygen in the nitrogen-fixing root nodules of this legume, along with a ferrochelatase from a nitrogen-fixing bacterium (for assembling the heme group) resulted in a fourfold increase of hydrogen production in C. reinhardtii (Wu et al., 2011). Alternatively, an Escherichia coli pyruvate oxidase has been expressed in the nucleus of C. reinhardtii in order to reduce intracellular oxygen concentration. This enzyme decarboxylates pyruvate into acetyl-phosphate while consuming one oxygen molecule. Transgenic algae strains produced up to 2.5-fold more hydrogen than the parental strain under very low light (30 μE m-2 s-1) and sulfur-replete conditions (Xu et al., 2011).
Securing additional reducing power can also result in increased hydrogen yields. The HUP1 hexose transporter from C. kessleri (green alga) was expressed in C. reinhardtii, resulting in a strain that could use glucose as a carbon and electron source (see Trophic Conversion below). This modification also translated into 1.5-fold higher hydrogen production rate (Doebbe et al., 2007). Carbon fixation by Rubisco competes for a significant amount of reducing power. Expression of a mutated small sub-unit of Rubisco (RBCS-Y67A) from the nucleus of an RBCS-deficient C. reinhardtii strain resulted in abolishment of PSII activity along with 10 to 15-fold increase in hydrogen yield in sulfur-deplete medium (Pinto et al., 2013). Ferredoxin-NADP++ reductase (FNR) is notoriously downregulated under sulfur deprivation in C. reinhardtii. RNAi knockdown of FNR resulted in decrease of Rubisco activity (60%) and oxygen evolution (44%), accompanied by increase in starch degradation (140%), under sulfur deprivation. These metabolic changes resulted in 2.5-fold increase in hydrogen production when compared to sulfur-starved WT (Sun et al., 2013).
Despite the limitations in short-term applicability of hydrogen based biofuels when compared to biodesel, algal hydrogen production has received a great amount of scientific attention. The research undergone so far constitutes C. reinhardtii as a model organism for hydrogen biogenesis. Advances in C. reinhardtii engineering could play a key role for developing more sustainable alternatives in the long-term, such as biohydrogen production from wastes degraded by either bacteria or other microalgae species.
Despite the high value of carotenoids and the advantages of microalgal platforms, there have been few reported efforts toward carotenoid production optimization through metabolic engineering in these organisms. Since most of the carotenogenic pathway occurs in the chloroplast of algae, carotenoid metabolic engineering could be achieved by either nuclear or chloroplast transformation (or both; Kempinski et al., 2015). Figure 3 shows the generally accepted pathway for β-carotene and astaxanthin biosynthesis in green microalgae. Table 2 summarizes the metabolic engineering approaches for modifying terpenoid accumulation in these organisms. C. reinhardtii has served as the model organism for studying the effects of genetic engineering in carotenoid accumulation. In the first report of this type, an archeal heat-stable geranylgeranyl-pyrophosphate synthase (involved in the early steps of carotenoid biosynthesis) was expressed in the chloroplast of C. reinhardtii. Unfortunately, there were no measurable effects on the isoprenoid profile of the algae (Fukusaki et al., 2003). Three years later, another group attempted to produce keto-carotenoids (e.g., astaxanthin) in C. reinhardtii by nuclear overexpression of the beta-carotene ketolase genes from H. pluvialis (bkt3) and C. reinhardtii itself (CRBKT). Following several efforts using different expression vectors, no keto-carotenoids could be detected (Wong, 2006). In parallel, (León et al., 2007) followed an analogous approach, but using instead the bkt1 gene from H. pluvialis. In this case a small peak of 4-keto-lutein could be detected, and it was not present in the parental strain. Unfortunately, no peak for astaxanthin was recorded. RNA interference technology has also been used for altering the carotenoid profile of C. reinhardtii. The phytoene desaturase gene (pds, coding for the second step of carotenoid biosynthesis) was targeted, resulting in a 93% reduction of its mRNA. Nonetheless, the carotenoid content didn’t change significantly, pointing toward the existence of additional rate-limiting processes (Vila et al., 2008). Additionally, the phytoene synthase gene (psy), which codes for the committing step enzyme for carotenoid synthesis, has been transformed in the C. reinhardtii nucleus causing an increase in carotenoid accumulation. Transformed strains overexpressing psy from Dunaliella salina and C. zofingiensis stored 2.6 and 2.2-fold more lutein than the wild-type, respectively (Cordero et al., 2011; Couso et al., 2011). Recently, the C. reinhardtii nucleus has been transformed with a point mutant version of its endogenous pds gene. The mutant enzyme had a 27% increase in its desaturase activity in vitro. The algae became resistant to the herbicide norflurazon and accumulated more lutein, beta-carotene, zeaxanthin, and violaxanthin in vivo (Liu et al., 2013).
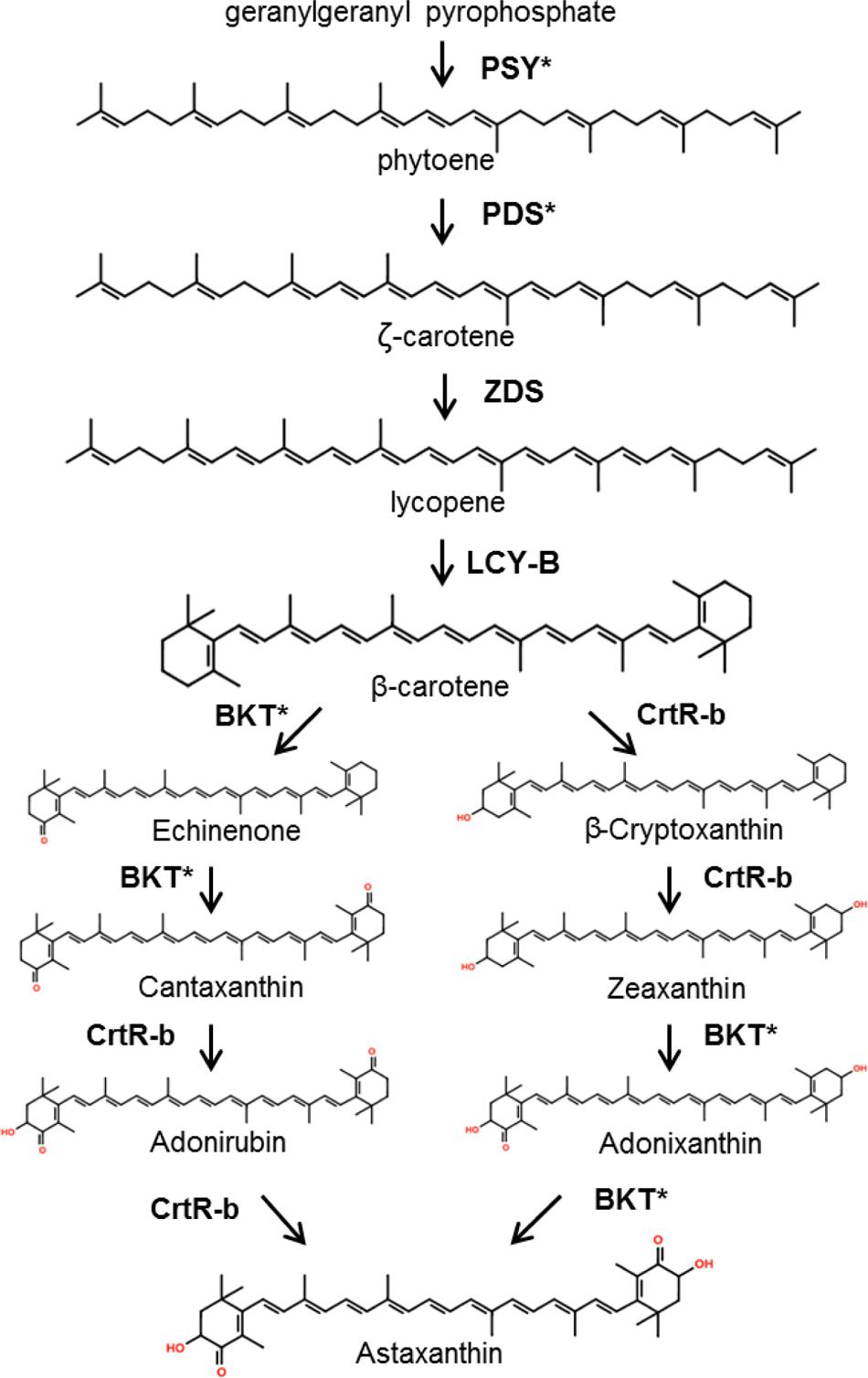
FIGURE 3. Proposed pathway for astaxanthin synthesis in green microalgae. Asterisks denote enzymes which have been targeted for metabolic engineering, as shown in Table 2. PSY, phytoene synthase; PDS, phytoene desaturase; ZDS, ζ-carotene desaturase; LYC-B, lycopene β-cyclase; BKT, β -carotene ketolase; CrtR-b, β-carotene hydroxylase. Modified from (Grünewald et al., 2001; Ye et al., 2008). Synthesis of β -carotene occurs in the chloroplast, while there is evidence that the subsequent steps could also take place in the cytoplasm (Grünewald et al., 2001; Guedes et al., 2011).
Haematococcus pluvialis, D. salina, and Chlorella sp. are highly relevant candidates for carotenoid metabolic engineering given their commercial relevance. In the first attempt of stable nuclear transformation, H. pluvialis was engineered with a mutant pds gene that conferred resistance to norflurazon. Transgenic strains accumulated up to 26% more astaxanthin than the wild-type control after 48 h of induction with high-light (Steinbrenner and Sandmann, 2006). RNAi constructs have been used for reducing the mRNA accumulation of the D. salina pds gene for up to 72%. Intriguingly, the carotenoid content of these strains was not reported (Sun et al., 2008). The most recent metabolic engineering effort involves the development of a nuclear transformation method for C. zofingiensis. A mutant version of the endogenous pds gene was transformed, conferring resistance to norflurazon. The mutant PDS enzyme had 33% higher desaturation activity in vitro. Transformed C. zofingiensis strains accumulated up to 32.1% more total carotenoids and 54.1% more astaxanthin in vivo (Liu et al., 2014).
It is worth mentioning that additional terpenoids from microalgae can also be regarded as highly desirable feedstocks for biofuels and specialty chemicals, which will further broaden the interest for research toward optimizing the production of these compounds (Davies et al., 2014; Heider et al., 2014). Recently, C. reinhardtii has been engineered for accumulating the highly valuable triterpenoid squalene. Wild-type C. reinhardtii contains the necessary genes for squalene synthesis, although it cannot be detected under standard growth conditions. Overexpression of the endogenous squalene synthase didn’t result in squalene accumulation. On the other hand, silencing of squalene epoxidase (56–76% mRNA reduction) resulted in accumulation of up to 1.1 μg/mg dry weight of squalene. Transformation of squalene synthase into these knockdown strains didn’t result in enhanced squalene accumulation (Kajikawa et al., 2015).
Overall, carotenoid metabolic engineering in microalgae has not yielded consistent results. Perhaps the targeted enzymes do not constitute the bottleneck steps, or there are several rate-limiting reactions in the pathway. The future trend in carotenoid engineering should be the simultaneous transformation of three or more enzymes to strengthen the desired metabolic flow. Discovery and engineering of microalgae TFs that regulate terpenoid synthesis should also be considered for this purpose. Use of herbicide resistant phytoene desaturase has proven useful for enhancing carotenoid accumulation (Liu et al., 2014). This system could also be expanded to additional terpenoid biosynthetic enzymes like deoxyxylulose 5-phosphate synthase, deoxyxylulose 5-phosphate reductoisomerase, phytoene synthase, ζ-carotene desaturase, and lycopene β-cyclase, which are sensitive to various bleaching herbicides (Sandmann, 2002; Ferhatoglu and Barrett, 2006).
When microalgae are grown under phototrophic conditions, all newly produced biomass, including lipids, derive from the fixation of CO2 into ribulose-1,5-biphosphate (RuBP) to form 3-phosphoglycerate, catalyzed by the famous enzyme RuBP carboxylase/oxygenase (Rubisco). Additional enzymes are also required to regenerate RuBP in a process named the Calvin–Benson–Bassham cycle (CBB). Significant amounts of ATP and NADPH are also required for this process, which are supplied by the light-driven activity of photosystems I and II (photochemistry; Raines, 2011). Several studies have shown that the activity of Rubisco is the major bottleneck for carbon flux through the CBB when CO2 is not supplied to the media, or under high-light (excess ATP and NADPH) or high-temperature conditions (prevalence of photorespiration, see below), which are all present in commercial scale ponds for algae biomass production in desert areas (Raines, 2011; Ducat and Silver, 2012).
Rubisco is regarded as a “slow and confused” enzyme. Large amounts of this enzyme are required for achieving a sustainable carboxylation rate and it has affinity for oxygen which is used in a futile reaction. The affinity toward oxygen increases at higher temperatures. The consequences of the wasteful oxygenation reaction are partially alleviated by a process named photorespiration (Whitney et al., 2011). C. reinhardtii seems to be a perfect host for engineering Rubisco, since there are Rubisco deficient strains that can complete their life cycle heterotrophically, unlike plants (Whitney et al., 2011). The small subunit of Rubisco genes (rbcS) of Arabidopsis and sunflower have been transformed independently into an rbcS deficient strain of C. reinhardtii (lacking the two nuclear rbcS alleles), while preserving the endogenous large subunit gene in the chloroplast (rbcL). The in vitro CO2/O2 specificity or discrimination factor (Ω) was improved by up to 11% while maintaining the Vmax of carboxylation catalysis (Vc). Nonetheless the cells displayed slower autotrophic growth rates and lacked pyrenoids (carbon-concentration sub-compartments in the algal chloroplast), presumably due to mistargeting of Rubisco caused by the heterologous small subunits (Genkov et al., 2010). Chlamydomonas rbcL has also been subjected to PCR-based gene shuffling with oligonucleotides representing the natural diversity of this gene. Three rounds of gene shuffling and three rounds of strain selection resulted in a Rubisco with increments of 20% Ω and 56% Vc. Some of the enriched mutations were then incorporated into the tobacco rbcL gene and resulted in 14% Ω and 15% Vc increments (Zhu et al., 2010). Another interesting strategy could consist in tuning Rubisco accumulation according to the environmental culture conditions in order to optimize the utilization of energy, carbon and nitrogen. Chlamydomonas strains with different amounts of Rubisco have been engineered by expressing the rbcL mRNA maturation factor MRL1 at different levels in the nuclear genome of an MRL1 deficient strain. Rubisco could be lowered up to 15% that of wild-type while maintaining phototrophic growth. An inducible promoter for MRL1 could be then used to tune Rubisco accumulation according to culture conditions, such as light intensity or CO2 concentration (Johnson, 2011).
The CBB cycle has also been engineered for enhancing carbon fixation. The CBB enzyme sedoheptulose 1,7-bisphosphatase enzyme (part of CBB) from C. reinhardtii has been overexpressed in β–carotene-producing green algae D. bardawil, resulting in a clear increase of oxygen evolution efficiency O2/cell/(μmol photons/m), and in organic carbon content per cell (Fang et al., 2012).
The future trends in Rubisco engineering should consider integrating the advances achieved in both RbcS and RbcL in the same microalgae strain. Future mutational studies should be performed in both genes simultaneously in order to capture co-variations that further enhance functionality. Additionally, there are two wide fields that have not been exploited in eukaryotic microalgae: the engineering of Rubisco activase and the carbon concentrating mechanisms.
Growing microalgae under heterotrophic or photoheterotrophic (mixotrophic) conditions has several advantages over autotrophic growth. Fermentation systems have been widely studied and successfully applied in industry for several years. The culture conditions are highly controlled and reproducible. Moreover, heterotrophic cultures of microalgae achieve higher cell densities, thus resulting in lower harvesting costs. In waste water treatment applications, trophic conversion allows to diversify the nutrients that can be degraded by algae (Chen and Chen, 2006; Chen et al., 2011). In addition, the yield of some metabolites can sometimes increase depending on the rerouting of the metabolic networks involved, as it has been shown for lipid accumulation (Miao and Wu, 2006). However, many microalgae species are strict autotrophs or are highly selective for their organic carbon source (e.g., acetate for C. reinhardtii). Trophic conversion has been achieved, allowing heterotophy in previously obligate phototrophic species as a proof of concept for simple metabolic engineering. V. carteri has been transformed with a hexose transporter from C. kessleri (HUP1, monosaccharide-H+ symporter), resulting in a strain that can survive on glucose under dark conditions (Hallmann and Sumper, 1996). P. tricornutum, Cylindrotheca fusiformis (diatoms) and C. reinhardtii have also been successfully transformed with HUP1, resulting in glucose transport into the cells (Fischer et al., 1999; Zaslavskaia et al., 2001; Doebbe et al., 2007). The human glut1 transporter gene (erythrocyte glucose transporter 1) has also been transformed into P. tricornutum, which could also perform glucose uptake into the cell (Apt et al., 2011). It is worth noting that despite glucose incorporation, the extent of conversion to full heterotrophy is variable between these four algae.
Trophic conversion is a good proof of concept for microalgae metabolic engineering, but despite the advantages of heterotrophic culture, it might not to be optimal for the production of low-value metabolites (e.g., biodiesel). Additional costs for adding a carbon source and the requirement for enclosed bioreactors (given the higher risk of contamination) are major drawbacks compared to phototrophic systems. Furthermore, some metabolites accumulate upon the presence of high-light (e.g., carotenoids), excluding the possibility of saving costs associated with illumination.
Microalgae have evolved large light-harvesting complexes (LHC) for maximizing light absorption in low-light environments, where they naturally occur. Under artificial culture conditions (saturating light) excess energy is dissipated through heat and fluorescence quenching in the LHC. Excess energy that cannot be dissipated usually results in direct photodamage and the production of reactive oxygen species (photoinhibition). The large size of the LHC also limits light penetration into the culture medium, therefore lowering the maximum cell density that can be achieved in large scale facilities (Ort et al., 2011; Wobbe and Remacle, 2015). In the first genetic engineering attempt to overcome this, a single RNAi construct was effectively used for silencing all twenty LHC protein isoforms of C. reinhardtii. These cells have lower mRNA (0.1–26% relative to the control) and protein accumulation for all LHC genes and 68% less chlorophyll than the parental strain, resulting in 290% higher light transmittance in the culture. Furthermore, they present less dissipation energy through fluorescence quenching, which leads to an increase in photosynthetic quantum yield. Under high-light conditions, transformed cells were less susceptible to photoinhibition and grew at a 65% faster rate; however, they did not reach a higher cell density (Mussgnug et al., 2007). Later, the same research group achieved similar results by downregulating LHC expression at the translational level. NAB1 is a translation repressor of the LHCBM family (LHCII; Mussgnug et al., 2005), and its activation is redox dependent (Wobbe et al., 2009). A constitutively activated version of NAB1 (two amino acid mutations) was overexpressed in C. reinhardtii, resulting in a similar phenotype to the one obtained through RNAi. However, the effects were less dramatic, having a chlorophyll/cell reduction of 20% compared to that of 68% using RNAi, and a growth rate increase of 53% compared to the previous 65% (Beckmann et al., 2009). Even so, overexpression of a single repressor would be easier to reproduce in the future than silencing twenty LHC isoforms at the same time. Another group has worked with the TLA1 gene (truncated light-harvesting antenna 1) of C. reinhardtii. Overexpression and silencing (RNAi) of tla1 resulted in 13% increase and 70% reduction of chlorophyll/cell, respectively. This confirms that tla1 is an attractive target for modifying phototrophic growth performance of algae (Mitra et al., 2012). As previously mentioned in the biohydrogen section of this review, the three major proteins of C. reinhardtii LHCII (LHCMB1, 2, and 3) have been knocked-down with three co-transformed RNAi constructs (each one specific for a single gene). This resulted in 50% reduction of chlorophyll/cell, and four times more light penetration at equal cell density. Additionally, the transformed strain grew 85% faster at a 5 mm culture depth under 450 μE m-2 s-1 light intensity (Oey et al., 2013).
One of the main bottlenecks for photosynthetic growth under suboptimal conditions is PSII, the multi-protein complex that performs the light-driven oxidation of water. Degradation of the D1 sub-unit of PSII seems to be predominantly increased when light is in excess or under various abiotic stresses (photoinhibition; Kreslavski et al., 2007; Keren and Krieger-Liszkay, 2011). Rea et al. (2011) have mutated and selected versions of the C. reinhardtii D1 protein that can evolve up to ∼4.5-fold more oxygen in vivo under high-light conditions (50% midday sunlight) compared to the control. This has been achieved by transforming error-prone PCR-amplified D1 coding sequences followed by selection under ionizing radiation. Unfortunately the mutant strains perform slightly worse under laboratory light conditions (10% midday sunlight), suggesting that this strategy may not translate to increased biomass yield for commercial biofuel production. The cyanobacteria Synechoccocus sp. PCC 7942 contains two isoforms of the D1 protein which are differentially expressed under low-light and high-light conditions. These two proteins were expressed independently in C. reinhardtii resulting in reconstitution of the low-light and high-light phenotypes associated with each D1 isoform. Interestingly, C. reinhardtii expressing the cyanobacterial low-light isoform yielded 11% more dry weight biomass than the strains expressing the high-light isoform or the endogenous D1 protein, which is a highly desirable for reducing harvesting costs (Vinyard et al., 2013). Using C. reinhardtii as a transformation host, the same authors determined the precise amino acids that confer the characteristic phenotypes to these two D1 isoforms. The latter will prove very important for designing strategies to optimize D1 and PSII function under specific growth conditions (Vinyard et al., 2014).
In coming years, engineering of the photosynthetic machinery should take into account the adverse environmental conditions that can take place in outdoor ponds, such as high and low irradiance, and very high temperatures, which can all decrease photosynthetic rates (Kreslavski et al., 2007; Keren and Krieger-Liszkay, 2011). Tuning photosystems and light harvesting antennas to perform under specific environmental conditions, or even better, generating regulated systems that can adapt to environmental changes, would be major breakthroughs for large-scale algae cultivation. Furthermore, these advances could potentially be translated to commercial crops. In order to achieve this, researchers will have to consider that the photosynthetic machinery is composed of large complexes of highly interacting proteins. Any major advancement will likely require the engineering of several of these proteins simultaneously, in order to maintain the interactions that have been conserved through the extended evolutionary history of photosynthetic systems.
Highly predictive metabolic models will be required in order to step-up metabolic engineering of microalgae (Veyel et al., 2014). At least eleven genome-wide metabolic network models are available for microalgae, but most of them correspond to C. reinhardtii, even though there are more than 30 sequenced species (Reijnders et al., 2014; Baroukh et al., 2015). Given the great metabolic diversity of microalgae, it is clear that models for at least each phylum will be required in order to give meaningful predictions for the corresponding species within that group (Hildebrand et al., 2013). It is also worth mentioning that metabolic engineering of algae doesn’t have to be circumscribed to metabolites that already exist in these organisms. For example C. reinhardtii has been engineered to produce the five-carbon sugar-alcohol xylitol, an artificial sweetener that doesn’t naturally occur in the alga. Xylose reductase (XR) from the filamentous fungus Neurospora crassa was codon-optimized and expressed in the plastid genome resulting in up to 0.38 g/L xylitol accumulation (Pourmir et al., 2013).
Novel genetic tools will also be a major driving force for metabolic engineering in eukaryotic microalgae (Figure 4). Nuclear genome editing would allow for precise gene deletion and gene integration, therefore enabling to reroute metabolic networks and to obtain predictable expression levels of transgenes (Jinkerson and Jonikas, 2015). Unfortunately, homologous recombination rates in C. reinhardtii are very low, but there are ongoing efforts to circumvent this through zinc-fingers technology (Sizova et al., 2013), and by using the CRISPR/Cas9 system (Jiang et al., 2014). On the other hand, some microalgae appear to have a highly efficient homologous recombination machinery in the nucleus, like for example Nannochloropsis sp. strain W2J3B (Kilian et al., 2011). Targeted induction or constitutive activation of endogenous genes is another valuable tool for modifying metabolic profiles. Activation of endogenous genes through transcription activator-like effectors (TALE) has been achieved in C. reinhardtii for the nuclear genes coding for arylsulfatase (ARS, endogenous colorimetric reporter), and the inorganic carbon membrane transporter HLA3 (Gao et al., 2014, 2015). Chloroplast metabolic engineering could take great advantage of a “shuttle” chloroplast genome that can replicate in C. reinhardtii, S. cerevisiae, and E. coli (O’Neill et al., 2012). Replication in S. cerevisiae allows for extensive DNA manipulations through gene replacement which would be required for simultaneous engineering of multiple enzymes in a metabolic pathway. E. coli replication serves for generating multiple copies of the genome for algae transformation (O’Neill et al., 2012). This “shuttle genome” strategy could also be complemented by a set of synthetic 5′ UTR for C. reinhardtii plastid, which could tune the expression of the multiple components of an engineered pathway (Specht and Mayfield, 2012). Recently, the diatoms P. tricornutum and T. pseudonana have been transformed with DNA episomes that replicate independently in the nucleus. These constructs carry a yeast-derived maintaining region that allows for stable replication even in the absence of antibiotic selection. Furthermore, these vectors can be transformed by trans-kingdom conjugation between E. coli and the diatoms (Karas et al., 2015). These results would allow for gene expression without position effects, and from multiple gene copies. If it is easily reproducible, conjugative transformation would eliminate the requirement for expensive biolistic or electroporation equipment for diatom transformation, thus enabling more laboratories to work in these organisms.
FIGURE 4. Contribution of microalgal genetic tools to enhance the production of secondary metabolites and biomass.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Apt, K. E., Allnutt, F. T., Kyle, D. J., and Lippmeier, J. C. (2011). Trophic Conversion of Obligate Phototrophic Algae Through Metabolic Engineering. Google Patents US Patent number 7939710. Washington, DC: U.S. Patent and Trademark Office.
Baroukh, C., Muñoz-Tamayo, R., Steyer, J.-P., and Bernard, O. (2015). A state of the art of metabolic networks of unicellular microalgae and cyanobacteria for biofuel production. Metab. Eng. 30, 49–60. doi: 10.1016/j.ymben.2015.03.019
Beckmann, J., Lehr, F., Finazzi, G., Hankamer, B., Posten, C., Wobbe, L., et al. (2009). Improvement of light to biomass conversion by de-regulation of light-harvesting protein translation in Chlamydomonas reinhardtii. J. Biotechnol. 142, 70–77. doi: 10.1016/j.jbiotec.2009.02.015
Beer, L. L., Boyd, E. S., Peters, J. W., and Posewitz, M. C. (2009). Engineering algae for biohydrogen and biofuel production. Curr. Opin. Biotechnol. 20, 264–271. doi: 10.1016/j.copbio.2009.06.002
Bellou, S., Baeshen, M. N., Elazzazy, A. M., Aggeli, D., Sayegh, F., and Aggelis, G. (2014). Microalgal lipids biochemistry and biotechnological perspectives. Biotechnol. Adv. 32, 1476–1493. doi: 10.1016/j.biotechadv.2014.10.003
Bhattacharya, D., Yoon, H. S., and Hackett, J. D. (2004). Photosynthetic eukaryotes unite: endosymbiosis connects the dots. Bioessays 26, 50–60. doi: 10.1002/bies.10376
Blatti, J. L., Beld, J., Behnke, C. A., Mendez, M., Mayfield, S. P., and Burkart, M. D. (2012). Manipulating fatty acid biosynthesis in microalgae for biofuel through protein-protein interactions. PLoS ONE 7:e42949. doi: 10.1371/journal.pone.0042949
Blunt, J. W., Copp, B. R., Keyzers, R. A., Munro, M. H. G., and Prinsep, M. R. (2012). Marine natural products. Nat. Prod. Rep. 29, 144–222. doi: 10.1039/c2np00090c
Borowitzka, M. A. (2013). High-value products from microalgae—their development and commercialisation. J. Appl. Phycol. 25, 743–756. doi: 10.1007/s10811-013-9983-9
Cardozo, K. H. M., Guaratini, T., Barros, M. P., Falcão, V. R., Tonon, A. P., Lopes, N. P., et al. (2007). Metabolites from algae with economical impact. Comparative biochemistry and physiology. Toxicol. Pharmacol. 146, 60–78. doi: 10.1016/j.cbpc.2006.05.007
Cerutti, H., Ma, X., Msanne, J., and Repas, T. (2011). RNA-mediated silencing in algae: biological roles and tools for analysis of gene function. Eukaryot. Cell 10, 1164–1172. doi: 10.1128/EC.05106-11
Chen, C. Y., Yeh, K. L., Aisyah, R., Lee, D. J., and Chang, J. S. (2011). Cultivation, photobioreactor design and harvesting of microalgae for biodiesel production: a critical review. Bioresour. Technol. 102, 71–81. doi: 10.1016/j.biortech.2010.06.159
Chen, F., Mackey, A. J., Stoeckert, C. J., and Roos, D. S. (2006). OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 34, D363–D368. doi: 10.1093/nar/gkj123
Chen, G.-Q., and Chen, F. (2006). Growing phototrophic cells without light. Biotechnol. Lett. 28, 607–616. doi: 10.1007/s10529-006-0025-4
Chen, H.-C., Newton, A. J., and Melis, A. (2005). Role of SulP, a nuclear-encoded chloroplast sulfate permease, in sulfate transport and H2evolution in Chlamydomonas reinhardtii. Photosynth. Res. 84, 289–296. doi: 10.1007/s11120-004-7157-y
Chen, H. L., Li, S. S., Huang, R., and Tsai, H. J. (2008). Conditional production of a functional fish growth hormone in the transgenic line of Nannochloropsis oculata (Eustigmatophyceae) 1. J. Phycol. 44, 768–776. doi: 10.1111/j.1529-8817.2008.00508.x
Chien, L.-F., Kuo, T.-T., Liu, B.-H., Lin, H.-D., Feng, T.-Y., and Huang, C.-C. (2012). Solar-to-bioH 2 production enhanced by homologous overexpression of hydrogenase in green alga Chlorella sp. DT. Int. J. Hydrogen Energy 37, 17738–17748. doi: 10.1016/j.ijhydene.2012.09.068
Chow, K.-C., and Tung, W. (1999). Electrotransformation of Chlorella vulgaris. Plant Cell Rep. 18, 778–780. doi: 10.1007/s002990050660
Chow, S., and Rodgers, P. (2005). “Constructing area-proportional Venn and Euler diagrams with three circles,” in Proceedings of the Euler Diagrams 2005, Paris.
Coll, J. (2006). Methodologies for transferring DNA into eukaryotic microalgae: a review. Span. J. Agric. Res. 4, 316–330. doi: 10.5424/sjar/2006044-209
Cordero, B. F., Couso, I., Leon, R., Rodriguez, H., and Vargas, M. A. (2011). Enhancement of carotenoids biosynthesis in Chlamydomonas reinhardtii by nuclear transformation using a phytoene synthase gene isolated from Chlorella zofingiensis. Appl. Microbiol. Biotechnol. 91, 341–351. doi: 10.1007/s00253-011-3262-y
Couso, I., Vila, M., Rodriguez, H., Vargas, M. A., and Leon, R. (2011). Overexpression of an exogenous phytoene synthase gene in the unicellular alga Chlamydomonas reinhardtii leads to an increase in the content of carotenoids. Biotechnol. Prog. 27, 54–60. doi: 10.1002/btpr.527
Davies, F. K., Jinkerson, R. E., and Posewitz, M. C. (2014). Toward a photosynthetic microbial platform for terpenoid engineering. Photosynth. Res. 123, 265–284. doi: 10.1007/s11120-014-9979-6
Davis, R., Aden, A., and Pienkos, P. T. (2011). Techno-economic analysis of autotrophic microalgae for fuel production. Appl. Energy 88, 3524–3531. doi: 10.1016/j.apenergy.2011.04.018
Deng, X.-D., Gu, B., Li, Y.-J., Hu, X.-W., Guo, J.-C., and Fei, X.-W. (2012). The roles of acyl-CoA: diacylglycerol acyltransferase 2 genes in the biosynthesis of triacylglycerols by the green algae Chlamydomonas reinhardtii. Mol. Plant 5, 945–947. doi: 10.1093/mp/sss040
Díaz-Santos, E., Vega, M., Vila, M., Vigara, J., and León, R. (2013). Efficiency of different heterologous promoters in the unicellular microalga Chlamydomonas reinhardtii. Biotechnol. Prog. 29, 319–328. doi: 10.1002/btpr.1690
Doebbe, A., Rupprecht, J., Beckmann, J., Mussgnug, J. H., Hallmann, A., Hankamer, B., et al. (2007). Functional integration of the HUP1 hexose symporter gene into the genome of C-reinhardtii: impacts on biological H-2 production. J. Biotechnol. 131, 27–33. doi: 10.1016/j.jbiotec.2007.05.017
Dubini, A., and Ghirardi, M. L. (2014). Engineering photosynthetic organisms for the production of biohydrogen. Photosynth. Res. 123, 241–253. doi: 10.1007/s11120-014-9991-x
Ducat, D. C., and Silver, P. A. (2012). Improving carbon fixation pathways. Curr. Opin. Chem. Biol. 16, 337–344. doi: 10.1016/j.cbpa.2012.05.002
Dufresne, A., Ostrowski, M., Scanlan, D. J., Garczarek, L., Mazard, S., Palenik, B. P., et al. (2008). Unraveling the genomic mosaic of a ubiquitous genus of marine cyanobacteria. Genome Biol. 9, R90. doi: 10.1186/gb-2008-9-5-r90
Dunahay, T. G., Jarvis, E. E., Dais, S. S., and Roessler, P. G. (1996). Manipulation of microalgal lipid production using genetic engineering. Appl. Biochem. Biotechnol. 57, 223–231. doi: 10.1007/BF02941703
Esquível, M. G., Amaro, H. M., Pinto, T. S., Fevereiro, P. S., and Malcata, F. X. (2011). Efficient H 2 production via Chlamydomonas reinhardtii. Trends Biotechnol. 29, 595–600. doi: 10.1016/j.tibtech.2011.06.008
Fang, L., Lin, H. X., Low, C. S., Wu, M. H., Chow, Y., and Lee, Y. K. (2012). Expression of the Chlamydomonas reinhardtii Sedoheptulose-1, 7-bisphosphatase in Dunaliella bardawil leads to enhanced photosynthesis and increased glycerol production. Plant Biotechnol. J. 10, 1129–1135. doi: 10.1111/pbi.12000
Ferhatoglu, Y., and Barrett, M. (2006). Studies of clomazone mode of action. Pestic. Biochem. Physiol. 85, 7–14. doi: 10.1016/j.pestbp.2005.10.002
Fischer, H., Robl, I., Sumper, M., and Kroger, N. (1999). Targeting and covalent modification of cell wall and membrane proteins heterologously expressed in the diatom Cylindrotheca fusiformis (Bacillariophyceae). J. Phycol. 35, 113–120. doi: 10.1046/j.1529-8817.1999.3510113.x
Fischer, N., Stampacchia, O., Redding, K., and Rochaix, J.-D. (1996). Selectable marker recycling in the chloroplast. Mol. Gen. Genet. 251, 373–380. doi: 10.1007/BF02172529
Fukusaki, E.-I., Nishikawa, T., Kato, K., Shinmyo, A., Hemmi, H., Nishino, T., et al. (2003). Introduction of the archaebacterial geranylgeranyl pyrophosphate synthase gene into Chlamydomonas reinhardtii chloroplast. J. Biosci. Bioeng. 95, 283–287. doi: 10.1016/S1389-1723(03)80030-0
Gao, H., Wang, Y., Fei, X., Wright, D. A., and Spalding, M. H. (2015). Expression activation and functional analysis of HLA3, a putative inorganic carbon transporter in Chlamydomonas reinhardtii. Plant J. 82, 1–11. doi: 10.1111/tpj.12788
Gao, H., Wright, D. A., Li, T., Wang, Y., Horken, K., Weeks, D. P., et al. (2014). TALE activation of endogenous genes in Chlamydomonas reinhardtii. Algal Res. 5, 52–60. doi: 10.1016/j.algal.2014.05.003
Genkov, T., Meyer, M., Griffiths, H., and Spreitzer, R. J. (2010). Functional hybrid rubisco enzymes with plant small subunits and algal large subunits: engineered rbcS cDNA for expression in chlamydomonas. J. Biol. Chem. 285, 19833–19841. doi: 10.1074/jbc.M110.124230
Gimpel, J. A., and Mayfield, S. P. (2013). Analysis of heterologous regulatory and coding regions in algal chloroplasts. Appl. Microbiol. Biotechnol. 97, 4499–4510. doi: 10.1007/s00253-012-4580-4
Gong, Y., Guo, X., Wan, X., Liang, Z., and Jiang, M. (2011a). Characterization of a novel thioesterase (PtTE) from Phaeodactylum tricornutum. J. Basic Microbiol. 51, 666–672. doi: 10.1002/jobm.201000520
Gong, Y., Hu, H., Gao, Y., Xu, X., and Gao, H. (2011b). Microalgae as platforms for production of recombinant proteins and valuable compounds: progress and prospects. J. ind. Microbiol. Biotechnol. 38, 1879–1890. doi: 10.1007/s10295-011-1032-6
Grünewald, K., Hirschberg, J., and Hagen, C. (2001). Ketocarotenoid biosynthesis outside of plastids in the unicellular green alga Haematococcus pluvialis. J. Biol. Chem. 276, 6023–6029. doi: 10.1074/jbc.M006400200
Guedes, A. C., Amaro, H. M., and Malcata, F. X. (2011). Microalgae as sources of carotenoids. Mar. Drugs 9, 625–644. doi: 10.3390/md9040625
Guiry, M. D. (2012). How many species of algae are there? J. Phycol. 48, 1057–1063. doi: 10.1111/j.1529-8817.2012.01222.x
Hallmann, A., and Sumper, M. (1996). The Chlorella hexose/H+ symporter is a useful selectable marker and biochemical reagent when expressed in Volvox. Proc. Natl. Acad. Sci. U.S.A. 93, 669–673. doi: 10.1073/pnas.93.2.669
Heider, S. A., Peters-Wendisch, P., Wendisch, V. F., Beekwilder, J., and Brautaset, T. (2014). Metabolic engineering for the microbial production of carotenoids and related products with a focus on the rare C50 carotenoids. Appl. Microbiol. Biotechnol. 98, 4355–4368. doi: 10.1007/s00253-014-5693-8
Heydarizadeh, P., Poirier, I., Loizeau, D., Ulmann, L., Mimouni, V., Schoefs, B., et al. (2013). Plastids of marine phytoplankton produce bioactive pigments and lipids. Mar. Drugs 11, 3425–3471. doi: 10.3390/md11093425
Hildebrand, M., Abbriano, R. M., Polle, J. E., Traller, J. C., Trentacoste, E. M., Smith, S. R., et al. (2013). Metabolic and cellular organization in evolutionarily diverse microalgae as related to biofuels production. Curr. Opin. Chem. Biol. 17, 506–514. doi: 10.1016/j.cbpa.2013.02.027
Hsieh, H.-J., Su, C.-H., and Chien, L.-J. (2012). Accumulation of lipid production in Chlorella minutissima by triacylglycerol biosynthesis-related genes cloned from Saccharomyces cerevisiae and Yarrowia lipolytica. J. Microbiol. 50, 526–534. doi: 10.1007/s12275-012-2041-5
Hwang, J.-H., Kim, H.-C., Choi, J.-A., Abou-Shanab, R., Dempsey, B. A., Regan, J. M., et al. (2014). Photoautotrophic hydrogen production by eukaryotic microalgae under aerobic conditions. Nat. Commun. 5:3234. doi: 10.1038/ncomms4234
Jiang, W., Brueggeman, A. J., Horken, K. M., Plucinak, T. M., and Weeks, D. P. (2014). Successful transient expression of cas9 and single guide RNA genes in Chlamydomonas reinhardtii. Eukaryot. Cell 13, 1465–1469. doi: 10.1128/EC.00213-14
Jinkerson, R. E., and Jonikas, M. C. (2015). Molecular techniques to interrogate and edit the Chlamydomonas nuclear genome. Plant J. 82, 393–412. doi: 10.1111/tpj.12801
Johanningmeier, U., and Fischer, D. (2010). “Perspective for the use of genetic transformants in order to enhance the synthesis of the desired metabolites: engineering chloroplasts of microalgae for the production of bioactive compounds,” in Bio-Farms for Nutraceuticals, eds M. T. Giardi, G. Rea, and B. Berra (New York, NY: Springer), 144–151.
Johnson, X. (2011). Manipulating RuBisCO accumulation in the green alga, Chlamydomonas reinhardtii. Plant Mol. Biol. 76, 397–405. doi: 10.1007/s11103-011-9783-z
Jones, C. S., and Mayfield, S. P. (2011). Algae biofuels: versatility for the future of bioenergy. Curr. Opin. Biotechnol. 23, 1–6. doi: 10.1016/j.copbio.2011.10.013
Kajikawa, M., Kinohira, S., Ando, A., Shimoyama, M., Kato, M., and Fukuzawa, H. (2015). Accumulation of squalene in a microalga Chlamydomonas reinhardtii by genetic modification of squalene synthase and squalene epoxidase genes. PLoS ONE 10:e0120446. doi: 10.1371/journal.pone.0120446
Karas, B. J., Diner, R. E., Lefebvre, S. C., Mcquaid, J., Phillips, A. P., Noddings, C. M., et al. (2015). Designer diatom episomes delivered by bacterial conjugation. Nat. Commun. 6:6925. doi: 10.1038/ncomms7925.
Kempinski, C., Jiang, Z., Bell, S., and Chappell, J. (2015). “Metabolic engineering of higher plants and algae for isoprenoid production,” in Advances in Biochemical Engineering/Biotechnology, eds J. Schrader and J. Bohlmann (Heidelberg: Springer Berlin), 161–199.
Keren, N., and Krieger-Liszkay, A. (2011). Photoinhibition: molecular mechanisms and physiological significance. Physiol. Plant. 142, 1–5. doi: 10.1111/j.1399-3054.2011.01467.x
Kilian, O., Benemann, C. S., Niyogi, K. K., and Vick, B. (2011). High-efficiency homologous recombination in the oil-producing alga Nannochloropsis sp. Proc. Natl. Acad. Sci. U.S.A. 108, 21265–21269. doi: 10.1073/pnas.1105861108
Kreslavski, V., Carpentier, R., Klimov, V., Murata, N., and Allakhverdiev, S. (2007). Molecular mechanisms of stress resistance of the photosynthetic apparatus. Biochem. (Mosc.) Suppl. Ser. A Membr. Cell Biol. 1, 185–205. doi: 10.1134/S1990747807030014
La Russa, M., Bogen, C., Uhmeyer, A., Doebbe, A., Filippone, E., Kruse, O., et al. (2012). Functional analysis of three type-2 DGAT homologue genes for triacylglycerol production in the green microalga Chlamydomonas reinhardtii. J. Biotechnol. 162, 13–20. doi: 10.1016/j.jbiotec.2012.04.006
Larkum, A. W. D., Ross, I. L., Kruse, O., and Hankamer, B. (2011). Selection, breeding and engineering of microalgae for bioenergy and biofuel production. Trends Biotechnol. 30, 198–205. doi: 10.1016/j.tibtech.2011.11.003
León, R., Couso, I., and Fernández, E. (2007). Metabolic engineering of ketocarotenoids biosynthesis in the unicelullar microalga Chlamydomonas reinhardtii. J. Biotechnol. 130, 143–152. doi: 10.1016/j.jbiotec.2007.03.005
Leon, R., and Fernandez, E. (2007). “Nuclear transformation of eukaryotic microalgae,” in Transgenic Microalgae as Green Cell Factories, eds R. León, A. Galván, and E. Fernández (New York, NY: Springer), 1–11.
Letunic, I., and Bork, P. (2011). Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 39, W475–W478. doi: 10.1093/nar/gkr201
Leu, S., and Boussiba, S. (2014). Advances in the production of high-value products by microalgae. Ind. Biotechnol. 10, 169–183. doi: 10.1089/ind.2013.0039
Lin, H.-D., Liu, B.-H., Kuo, T.-T., Tsai, H.-C., Feng, T.-Y., Huang, C.-C., et al. (2013). Knockdown of PsbO leads to induction of HydA and production of photobiological H 2 in the green alga Chlorella sp. DT. Bioresour. Technol. 143, 154–162. doi: 10.1016/j.biortech.2013.05.101
Liu, J., Gerken, H., Huang, J., and Chen, F. (2013). Engineering of an endogenous phytoene desaturase gene as a dominant selectable marker for Chlamydomonas reinhardtii transformation and enhanced biosynthesis of carotenoids. Process Biochem. 48, 788–795. doi: 10.1016/j.procbio.2013.04.020
Liu, J., Sun, Z., Gerken, H., Huang, J., Jiang, Y., and Chen, F. (2014). Genetic engineering of the green alga Chlorella zofingiensis: a modified norflurazon-resistant phytoene desaturase gene as a dominant selectable marker. Appl. Microbiol. Biotechnol. 98, 5069–5079. doi: 10.1007/s00253-014-5593-y
Ma, Y.-H., Wang, X., Niu, Y.-F., Yang, Z.-K., Zhang, M.-H., Wang, Z.-M., et al. (2014). Antisense knockdown of pyruvate dehydrogenase kinase promotes the neutral lipid accumulation in the diatom Phaeodactylum tricornutum. Microb. Cell Fact. 13, 100. doi: 10.1186/s12934-014-0100-9
Maliga, P., and Bock, R. (2011). Plastid biotechnology: food, fuel, and medicine for the 21st century. Plant Physiol. 155, 1501–1510. doi: 10.1104/pp.110.170969
Martin, W. (2010). Evolutionary origins of metabolic compartmentalization in eukaryotes. Philos. Trans. R. Soc. B Biol. Sci. 365, 847–855. doi: 10.1098/rstb.2009.0252
Melis, A., Seibert, M., and Ghirardi, M. L. (2007). “Hydrogen fuel production by transgenic microalgae,” in Transgenic Microalgae as Green Cell Factories, eds R. León, A. Galván, and E. Fernández (New York, NY: Springer), 110–121.
Meuser, J. E., Ananyev, G., Wittig, L. E., Kosourov, S., Ghirardi, M. L., Seibert, M., et al. (2009). Phenotypic diversity of hydrogen production in chlorophycean algae reflects distinct anaerobic metabolisms. J. Biotechnol. 142, 21–30. doi: 10.1016/j.jbiotec.2009.01.015
Meuser, J. E., D’adamo, S., Jinkerson, R. E., Mus, F., Yang, W., Ghirardi, M. L., et al. (2012). Genetic disruption of both Chlamydomonas reinhardtii [FeFe]-hydrogenases: insight into the role of HYDA2 in H(2) production. Biochem. Biophys. Res. Commun. 417, 704–709. doi: 10.1016/j.bbrc.2011.12.002
Miao, X., and Wu, Q. (2006). Biodiesel production from heterotrophic microalgal oil. Bioresour. Technol. 97, 841–846. doi: 10.1016/j.biortech.2005.04.008
Milledge, J. J. (2011). Commercial application of microalgae other than as biofuels: a brief review. Rev. Environ. Sci. Biotechnol. 10, 31–41. doi: 10.1007/s11157-010-9214-7
Mitra, M., Kirst, H., Dewez, D., and Melis, A. (2012). Modulation of the light-harvesting chlorophyll antenna size in Chlamydomonas reinhardtii by TLA1 gene over-expression and RNA interference. Philos. Trans. R. Soc. B Biol. Sci. 367, 3430–3443. doi: 10.1098/rstb.2012.0229
Molnar, A., Bassett, A., Thuenemann, E., Schwach, F., Karkare, S., Ossowski, S., et al. (2009). Highly specific gene silencing by artificial microRNAs in the unicellular alga Chlamydomonas reinhardtii. Plant J. 58, 165–174. doi: 10.1111/j.1365-313X.2008.03767.x
Mussgnug, J. H., Thomas-Hall, S., Rupprecht, J., Foo, A., Klassen, V., Mcdowall, A., et al. (2007). Engineering photosynthetic light capture: impacts on improved solar energy to biomass conversion. Plant Biotechnol. J. 5, 802–814. doi: 10.1111/j.1467-7652.2007.00285.x
Mussgnug, J. H., Wobbe, L., Elles, I., Claus, C., Hamilton, M., Fink, A., et al. (2005). NAB1 is an RNA binding protein involved in the light-regulated differential expression of the light-harvesting antenna of Chlamydomonas reinhardtii. Plant Cell 17, 3409–3421. doi: 10.1105/tpc.105.035774
Niu, Y.-F., Zhang, M.-H., Li, D.-W., Yang, W.-D., Liu, J.-S., Bai, W.-B., et al. (2013). Improvement of neutral lipid and polyunsaturated fatty acid biosynthesis by overexpressing a type 2 diacylglycerol acyltransferase in marine diatom Phaeodactylum tricornutum. Mar. Drugs 11, 4558–4569. doi: 10.3390/md11114558
Norton, T. A., Melkonian, M., and Andersen, R. A. (1996). Algal biodiversity*. Phycologia 35, 308–326. doi: 10.2216/i0031-8884-35-4-308.1
Oey, M., Ross, I. L., Stephens, E., Steinbeck, J., Wolf, J., Radzun, K. A., et al. (2013). RNAi knock-down of LHCBM1, 2 and 3 increases photosynthetic H2 production efficiency of the green alga Chlamydomonas reinhardtii. PLoS ONE 8:e61375. doi: 10.1371/journal.pone.0061375
O’Neill, B. M., Mikkelson, K. L., Gutierrez, N. M., Cunningham, J. L., Wolff, K. L., Szyjka, S. J., et al. (2012). An exogenous chloroplast genome for complex sequence manipulation in algae. Nucleic Acids Res. 40, 2782–2792. doi: 10.1093/nar/gkr1008
Ort, D. R., Zhu, X., and Melis, A. (2011). Optimizing antenna size to maximize photosynthetic efficiency. Plant Physiol. 155, 79–85. doi: 10.1104/pp.110.165886
Parker, M. S., Mock, T., and Armbrust, E. V. (2008). Genomic insights into marine microalgae. Annu. Rev. Genet. 42, 619–645. doi: 10.1146/annurev.genet.42.110807.091417
Peng, K.-T., Zheng, C.-N., Xue, J., Chen, X.-Y., Yang, W.-D., Liu, J.-S., et al. (2014). Delta 5 fatty acid desaturase upregulates the synthesis of polyunsaturated fatty acids in the marine diatom Phaeodactylum tricornutum. J. Agric. Food Chem. 62, 8773–8776. doi: 10.1021/jf5031086
Pinto, T., Malcata, F., Arrabaça, J., Silva, J., Spreitzer, R., and Esquível, M. (2013). Rubisco mutants of Chlamydomonas reinhardtii enhance photosynthetic hydrogen production. Appl. Microbiol. Biotechnol. 97, 5635–5643. doi: 10.1007/s00253-013-4920-z
Pourmir, A., Noor-Mohammadi, S., and Johannes, T. W. (2013). Production of xylitol by recombinant microalgae. J. Biotechnol. 165, 178–183. doi: 10.1016/j.jbiotec.2013.04.002
Pulz, O., and Gross, W. (2004). Valuable products from biotechnology of microalgae. Appl. Microbiol. Biotechnol. 65, 635–648. doi: 10.1007/s00253-004-1647-x
Purton, S., Szaub, J., Wannathong, T., Young, R., and Economou, C. (2013). Genetic engineering of algal chloroplasts: progress and prospects. Russ. J. Plant Physiol. 60, 491–499. doi: 10.1134/S1021443713040146
Radakovits, R., Eduafo, P. M., and Posewitz, M. C. (2011). Genetic engineering of fatty acid chain length in Phaeodactylum tricornutum. Metab. Eng. 13, 89–95. doi: 10.1016/j.ymben.2010.10.003
Raines, C. A. (2011). Increasing photosynthetic carbon assimilation in C3 plants to improve crop yield: current and future strategies. Plant Physiol. 155, 36–42. doi: 10.1104/pp.110.168559
Rasala, B. A., Lee, P. A., Shen, Z., Briggs, S. P., Mendez, M., and Mayfield, S. P. (2012). Robust expression and secretion of Xylanase1 in Chlamydomonas reinhardtii by fusion to a selection gene and processing with the FMDV 2A peptide. PLoS ONE 7:e43349. doi: 10.1371/journal.pone.0043349
Rea, G., Lambreva, M., Polticelli, F., Bertalan, I., Antonacci, A., Pastorelli, S., et al. (2011). Directed evolution and in silico analysis of reaction centre proteins reveal molecular signatures of photosynthesis adaptation to radiation pressure. PLoS ONE 6:e16216. doi: 10.1371/journal.pone.0016216
Reijnders, M. J., Van Heck, R. G., Lam, C. M., Scaife, M. A., Dos Santos, V. A. M., Smith, A. G., et al. (2014). Green genes: bioinformatics and systems-biology innovations drive algal biotechnology. Trends Biotechnol. 32, 617–626. doi: 10.1016/j.tibtech.2014.10.003
Rosales-Mendoza, S., Paz-Maldonado, L. M. T., and Soria-Guerra, R. E. (2012). Chlamydomonas reinhardtii as a viable platform for the production of recombinant proteins: current status and perspectives. Plant Cell Rep. 31, 479–494. doi: 10.1007/s00299-011-1186-8
Sandmann, G. (2002). “Bleaching herbicides: action mechanism in carotenoid biosynthesis, structural requirements and engineering of resistance,” in Herbicide Classes in Development, eds P. Böger, K. Wakabayashi, and K. Hirai (Heidelberg: Springer), 43–57.
Scaife, M. A., Nguyen, G. T., Rico, J., Lambert, D., Helliwell, K. E., and Smith, A. G. (2015). Establishing Chlamydomonas reinhardtii as an industrial biotechnology host. Plant J. 82, 532–546. doi: 10.1111/tpj.12781
Scranton, M. A., Ostrand, J. T., Fields, F. J., and Mayfield, S. P. (2015). Chlamydomonas as a model for biofuels and bio-products production. Plant J. 82, 523–531. doi: 10.1111/tpj.12780
Sizova, I., Greiner, A., Awasthi, M., Kateriya, S., and Hegemann, P. (2013). Nuclear gene targeting in Chlamydomonas using engineered zinc-finger nucleases. Plant J. 73, 873–882. doi: 10.1111/tpj.12066
Specht, E., Miyake-Stoner, S., and Mayfield, S. (2010). Micro-algae come of age as a platform for recombinant protein production. Biotechnol. Lett. 32, 1373–1383. doi: 10.1007/s10529-010-0326-5
Specht, E. A., and Mayfield, S. P. (2012). Synthetic oligonucleotide libraries reveal novel regulatory elements in Chlamydomonas chloroplast mRNAs. ACS Synth. Biol. 2, 34–46. doi: 10.1021/sb300069k
Spolaore, P., Joannis-Cassan, C., Duran, E., and Isambert, A. (2006). Commercial applications of microalgae. J. Biosci. Bioeng. 101, 87–96. doi: 10.1263/jbb.101.87
Steinbrenner, J., and Sandmann, G. (2006). Transformation of the green alga Haematococcus pluvialis with a phytoene desaturase for accelerated astaxanthin biosynthesis. Appl. Environ. Microbiol. 72, 7477–7484. doi: 10.1128/AEM.01461-06
Stephens, E., Ross, I. L., King, Z., Mussgnug, J. H., Kruse, O., Posten, C., et al. (2010). An economic and technical evaluation of microalgal biofuels. Nat. Biotechnol. 28, 126–128. doi: 10.1038/nbt0210-126
Sun, G., Zhang, X., Sui, Z., and Mao, Y. (2008). Inhibition of pds gene expression via the RNA interference approach in Dunaliella salina (Chlorophyta). Mar. Biotechnol. (N.Y.) 10, 219–226. doi: 10.1007/s10126-007-9056-7
Sun, Y., Chen, M., Yang, H., Zhang, J., Kuang, T., and Huang, F. (2013). Enhanced H 2 photoproduction by down-regulation of ferredoxin-NADP+ reductase (FNR) in the green alga Chlamydomonas reinhardtii. Int. J. Hydrogen Energy 38, 16029–16037. doi: 10.1016/j.ijhydene.2013.10.011
Surzycki, R., Cournac, L., Peltier, G., and Rochaix, J.-D. (2007). Potential for hydrogen production with inducible chloroplast gene expression in Chlamydomonas. Proc. Natl. Acad. Sci. U.S.A. 104, 17548–17553. doi: 10.1073/pnas.0704205104
Teng, C., Qin, S., Liu, J., Yu, D., Liang, C., and Tseng, C. (2002). Transient expression of lacZ in bombarded unicellular green alga Haematococcus pluvialis. J. Appl. Phycol. 14, 497–500. doi: 10.1023/A:1022314815045
Terashima, M., Specht, M., and Hippler, M. (2011). The chloroplast proteome: a survey from the Chlamydomonas reinhardtii perspective with a focus on distinctive features. Curr. Genet. 57, 151–168. doi: 10.1007/s00294-011-0339-1
Tirichine, L., and Bowler, C. (2011). Decoding algal genomes: tracing back the history of photosynthetic life on Earth. Plant J. Cell Mol. Biol. 66, 45–57. doi: 10.1111/j.1365-313X.2011.04540.x
Torzillo, G., Scoma, A., Faraloni, C., Ena, A., and Johanningmeier, U. (2009). Increased hydrogen photoproduction by means of a sulfur-deprived Chlamydomonas reinhardtii D1 protein mutant. Int. J. Hydrogen Energy 34, 4529–4536. doi: 10.1016/j.ijhydene.2008.07.093
Trentacoste, E. M., Shrestha, R. P., Smith, S. R., Glé, C., Hartmann, A. C., Hildebrand, M., et al. (2013). Metabolic engineering of lipid catabolism increases microalgal lipid accumulation without compromising growth. Proc. Natl. Acad. Sci. U.S.A. 110, 19748–19753. doi: 10.1073/pnas.1309299110
Veyel, D., Erban, A., Fehrle, I., Kopka, J., and Schroda, M. (2014). Rationales and approaches for studying metabolism in eukaryotic microalgae. Metabolites 4, 184–217. doi: 10.3390/metabo4020184
Vila, M., Couso, I., and Leon, R. (2008). Carotenoid content in mutants of the chlorophyte Chlamydomonas reinhardtii with low expression levels of phytoene desaturase. Process Biochem. 43, 1147–1152. doi: 10.1016/j.procbio.2008.06.014
Vinyard, D. J., Gimpel, J., Ananyev, G. M., Cornejo, M. A., Golden, S. S., Mayfield, S. P., et al. (2013). Natural variants of photosystem II subunit D1 tune photochemical fitness to solar intensity. J. Biol. Chem. 288, 5451–5462. doi: 10.1074/jbc.M112.394668
Vinyard, D. J., Gimpel, J., Ananyev, G. M., Mayfield, S. P., and Dismukes, G. C. (2014). Engineered photosystem II reaction centers optimize photochemistry versus photoprotection at different solar intensities. J. Am. Chem. Soc. 136, 4048–4055. doi: 10.1021/ja5002967
Whitney, S. M., Houtz, R. L., and Alonso, H. (2011). Advancing our understanding and capacity to engineer nature’s CO2-sequestering enzyme, Rubisco. Plant Physiol. 155, 27–35. doi: 10.1104/pp.110.164814
Wobbe, L., Blifernez, O., Schwarz, C., Mussgnug, J. H., Nickelsen, J., and Kruse, O. (2009). Cysteine modification of a specific repressor protein controls the translational status of nucleus-encoded LHCII mRNAs in Chlamydomonas. Proc. Natl. Acad. Sci. U.S.A. 106, 13290–13295. doi: 10.1073/pnas.0900670106
Wobbe, L., and Remacle, C. (2015). Improving the sunlight-to-biomass conversion efficiency in microalgal biofactories. J. Biotechnol. 201, 28–42. doi: 10.1016/j.jbiotec.2014.08.021
Wong, K. H. (2006). Transgenic chlamydomonas reinhardtii as an Experimental System to Study the Regulation of Carotenoid Biosynthesis in Green Microalgae. Ph.D. thesis, The University of Hong Kong, Hong Kong.
Wu, S., Xu, L., Huang, R., and Wang, Q. (2011). Improved biohydrogen production with an expression of codon-optimized hemH and lba genes in the chloroplast of Chlamydomonas reinhardtii. Bioresour. Technol. 102, 2610–2616. doi: 10.1016/j.biortech.2010.09.123
Xu, F.-Q., Ma, W.-M., and Zhu, X.-G. (2011). Introducing pyruvate oxidase into the chloroplast of Chlamydomonas reinhardtii increases oxygen consumption and promotes hydrogen production. Int. J. Hydrogen Energy 36, 10648–10654. doi: 10.1016/j.ijhydene.2011.05.130
Xue, J., Niu, Y., Huang, T., Yang, W., Liu, J., and Li, H. (2014). Genetic improvement of the microalga Phaeodactylum tricornutum for boosting neutral lipid accumulation. Metab. Eng. 27, 1–9. doi: 10.1016/j.ymben.2014.10.002
Ye, Z.-W., Jiang, J.-G., and Wu, G.-H. (2008). Biosynthesis and regulation of carotenoids in Dunaliella: progresses and prospects. Biotechnol. Adv. 26, 352–360. doi: 10.1016/j.biotechadv.2008.03.004
Yohn, C., Brand, A., Mendez, M., and Behnke, C. A. (2012). Stress-Induced Lipid Trigger. US Patent No 20,120,322,157. Washington, DC: U.S. Patent and Trademark Office.
Yu, W.-L., Ansari, W., Schoepp, N. G., Hannon, M. J., Mayfield, S. P., and Burkart, M. D. (2011). Modifications of the metabolic pathways of lipid and triacylglycerol production in microalgae. Microb. Cell Fact. 10, 91. doi: 10.1186/1475-2859-10-91
Zaslavskaia, L., Lippmeier, J., Shih, C., Ehrhardt, D., Grossman, A., and Apt, K. (2001). Trophic conversion of an obligate photoautotrophic organism through metabolic engineering. Science 292, 2073–2075. doi: 10.1126/science.160015
Zäuner, S., Jochum, W., Bigorowski, T., and Benning, C. (2012). A cytochrome b5-containing plastid-located fatty acid desaturase from Chlamydomonas reinhardtii. Eukaryot. Cell 11, 856–863. doi: 10.1128/EC.00079-12
Zhang, J., Hao, Q., Bai, L., Xu, J., Yin, W., Song, L., et al. (2014). Overexpression of the soybean transcription factor GmDof4 significantly enhances the lipid content of Chlorella ellipsoidea. Biotechnol. Biofuels 7, 128. doi: 10.1186/s13068-014-0128-4
Keywords: microalgae, metabolic engineering, transformation, carotenoids, biodiesel, biohydrogen, PUFA, photosynthesis
Citation: Gimpel JA, Henríquez V and Mayfield SP (2015) In Metabolic Engineering of Eukaryotic Microalgae: Potential and Challenges Come with Great Diversity. Front. Microbiol. 6:1376. doi: 10.3389/fmicb.2015.01376
Received: 30 April 2015; Accepted: 20 November 2015;
Published: 15 December 2015.
Edited by:
Ana Lúcia Leitão, Universidade Nova de Lisboa, PortugalReviewed by:
Zhidan Liu, China Agricultural University, ChinaCopyright © 2015 Gimpel, Henríquez and Mayfield. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stephen P. Mayfield, c21heWZpZWxkQHVjc2QuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.