- 1Department of Immunology and Microbiology, Institutes of Medical Sciences, Shanghai Jiao Tong University School of Medicine, Shanghai, China
- 2Baotou Municipal Center for Disease Control and Prevention, Baotou, China
- 3State Key Laboratory for Infectious Disease Prevention and Control, Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, National Institute for Communicable Disease Control and Prevention, Beijing, China
- 4Department of Population Medicine and Diagnostic Sciences, College of Veterinary Medicine, Cornell University, Ithaca, NY, USA
A newly isolated smooth colony morphology phage-resistant strain 8416 isolated from a 45-year-old cattle farm cleaner with clinical features of brucellosis in China was reported. The most unusual phenotype was its resistance to two Brucella phages Tbilisi and Weybridge, but sensitive to Berkeley 2, a pattern similar to that of Brucella melitensis biovar 1. VITEK 2 biochemical identification system found that both strain 8416 and B. melitensis strains shared positive ILATk, but negative in other B. abortus strains. However, routine biochemical and phenotypic characteristics of strain 8416 were most similar to that of B. abortus biovar 9 except CO2 requirement. In addition, multiple PCR molecular typing assays including AMOS-PCR, B. abortus special PCR (B-ab PCR) and a novel sub-biovar typing PCR, indicated that strain 8416 may belong to either biovar 3b or 9 of B. abortus. Surprisingly, further MLVA typing results showed that strain 8416 was most closely related to B. abortus biovar 3 in the Brucella MLVA database, primarily differing in 4 out of 16 screened loci. Therefore, due to the unusual discrepancy between phenotypic (biochemical reactions and particular phage lysis profile) and molecular typing characteristics, strain 8416 could not be exactly classified to any of the existing B. abortus biovars and might be a new variant of B. abortus biovar 9. The present study also indicates that the present phage typing scheme for Brucella sp. is subject to variation and the routine Brucella biovar typing needs further studies.
Brucellosis is one of the most common zoonotic infectious diseases, causing enormous economic loss in domestic animals and public health problems worldwide (Adone and Pasquali, 2013; Van der Henst et al., 2013). Transmission from animals to human occurs primarily through direct contact with infected animals and ingestion of raw milk or unpasteurized cheese. On the basis of obviously different phenotypic characteristics, host preference, growth and biochemical characteristics including CO2 requirement, substrate utilization and growth on dyes and agglutination with monospecific sera as well as Brucella phage lysis profiles, four main Brucella pathogenic species including Brucella melitensis (sheep and goat), B. suis (pigs), B. abortus (cattle), and B. canis (dogs), a taxonomic scheme can be defined and further divided into multiple biovars. For example, B. abortus is subdivided into eight biovars (biovar 1–7 and 9) (Van der Henst et al., 2013).
Because of unstable phenotypic characteristics among Brucella strains, it is somewhat difficult to define atypical strains into standard biovars. For instance, the susceptibility of smooth B. abortus strains to lysis by most of brucella phages, such as Tbilisi (Tb), Firenze (Fi), Weybridge (Wb), and Berkeley 2 (BK2), is commonly regarded as one of the routine criteria to differentiate this organism from other Brucella species. However, the majority of B. abortus strains resistant to Brucella phage have been currently reported primarily due to variation from smooth to rough form during normal in vitro culture. Since the first smooth phage-resistant strain (SPR) of B. abortus isolated from bovine tissue was reported in 1973 (Corbel and Morris, 1974, 1975), a similar study describing SPR strains has not been reported yet. In this study, we report a newly isolated SPR strain, strain 8416 from a patient with brucellosis in the Inner Mongolia Autonomous Region of China on 2012. Actually, it was the only B. abortus strain among a total of 197 Brucella strains isolated and authenticated by Chinese CDC during this year. The Inner Mongolia Autonomous Region has the highest incidence, responsible for about more than 40% of reported cases in China (Zhang et al., 2010; Chen et al., 2013). Interestingly, the unique phenotypical characteristics of the B. abortus SPR strain 8416, determined by routine biotyping for the identification of Brucella species and biovars, did not completely fit into any of the recognized classification biovars, indicating the potential presence of a new variant of B. abortus biovar 3.
Materials And Methods
Bacterial Isolation and Used Strains
The protocol for this study was approved by ethics committee of local disease control and Prevention Research Center of the Inner Mongolia Autonomous Region and Baotou Municipal Center for Disease Control and Prevention. In June 2012, two workers from a cattle farm in Sichuan province, presenting fever, night sweat and soreness of waist, arthralgia and muscle weakness, were admitted to one local hospital in the Inner Mongolia Autonomous Region. The serum samples from these two patients were strongly positive to Brucella by both Rose-Bengal-plate-agglutination-test (RBPT) and Serum Agglutination Test (SAT) with titers of 1/320 according to standard procedures. Moreover, the two serum samples were also confirmed by positive ELISA results with Brucella IgG (>150 U/ml) and IgM (>60 U/ml) (Brucella IgG and IgM ELISA kits, IBL Germany). At the same time, the blood culture of the two patients were inoculated in a dual-phase coloration blood culture bottle (BioMerieux Inc., Durham, USA) at 37°C for 2–3 weeks at the diagnostic laboratory of Baotou Municipal Center for Disease Control and Prevention, the Inner Mongolia Autonomous Region of China. However, only one blood sample from a 45-year-old male janitor yielded a positive culture result. The isolated strain 8416 displayed smooth, tiny, white, shiny and translucent colonies on solid agar after 3 days of incubation. The strain 8416 was sub-cultured on blood plate with 5% CO2 and displayed typical colonies with small Gram-negative coccobacilli. The strain was sent to department of brucellosis, Chinese Communicable Disease Control and Prevention (Chinese CDC) for further analysis and identification. The reference strains including B. abortus biovar 1 to 7 and 9, strains: 544A (ATCC 23448), 86/8/59 (ATCC 23449), Tulya (ATCC 23450), 292 (ATCC 23451), B3196 (ATCC 23452), 870 (ATCC 23453), 63/75, and C68 (ATCC 23455), B. melitensis biovar 1 to 3, strains: 16M (ATCC 23456), 63/9 (ATCC 23457) and Ether (ATCC 23458)), B. suis biovar 1 to 5, strains: 1330S (ATCC 23444), Thomsen (ATCC 23445), 686 (ATCC 23446), 40 (ATCC 23447), and 513, B. neotomae RM6/66 (ATCC 23365), B. ovis 63/290 (ATCC 25840), and B. canis 5K33 (ATCC 23459) were used as controls for phenotype typing, biochemical and/or molecular analysis.
Analysis of Phenotypic Characteristics
At first, to exclude mixed cultures of different biovars and phage carrier state, the strain used in this study was subjected to a single cloned isolation for successive three times to confirm no variable colonial morphology as described by Jones et al. (1962). The strain was further characterized by using the classical Brucella phenotypic identification procedures, such as CO2 requirement, H2S production, dye sensitivity by basic fuchsin and thionin, agglutination with monospecific antisera, and phage typing as described by Alton GG (Alton et al., 1975). Brucella monospecific antisera to A, M, and R (rough) and Brucella phages Tb, Wb, and Bk2 were used according to standard protocol of the Chinese CDC (Jiang et al., 2013) to characterize this strain. All of phenotypic characterizations in this study were repeated at least three times to make sure the results are repeatable.
Molecular Typing Identification
Brucella strains were inactivated by suspending one loop from a solid bacterial culture in 200 μl DNA storage buffer. Total genomic DNA was extracted using the DNeasy Blood & Tissue Kit (Qiagen China Ltd., Beijing, China) following the manufacture’s instruction. The PCR assay targeting bcsp31, was performed to confirm the Brucella genus as previously described (Bounaadja et al., 2009), and species-level using the routine Abortus-Melitensis-Ovis-Suis PCR (AMOS-PCR) (Bricker and Halling, 1994). Furthermore, B. abortus B-ab PCR and a novel PCR to differentiate B. abortus biovar 3a, 3b, 5, 6, and 9 were performed as previously described (Ocampo-Sosa et al., 2005; Huber et al., 2009).
Multiple Locus Variable Number Tandem Repeat Analysis (MLVA) Genotyping
Multiple locus variable number tandem repeat analysis (MLVA) was performed as previously described by Le Fleche et al. (2006) and by Jiang et al. (2013), respectively. The 16 primer pairs comprised three main groups: panel 1 including bruce06, 08, 11, 12, 42, 43, 45, and 55 for species identification, panel 2A (bruce18, 19, and 21), and panel 2B (bruce04, 07, 09, 16, and 30) for further subspecies differentiation were used.
Biochemical Identification by VITEK 2 System
A total of 47 biochemical reactions of the Brucella strains were analyzed using the standard Gram-negative bacteria identification card on automatic VITEK 2 system according to the manufacturer’s instructions.
Results
Routine Phenotypic Typing Characteristics
According to routine phenotypic analysis, strain 8416 was anti-R negative and H2S positive, agglutination with anti-M serum but not anti-A serum and grew in the presence of thionine and fuchsin dyes (Table 1). Moreover, it was not lysed by Tb and Wb phages both in 1× RTD (Routine Test Dilution) and 104× RTD, but lysed by BK2 phage both in 1× RTD and 102× RTD (Figure 1A). Thus, the particular phenotypic profiles of the strain 8416 were more similar to that of the classic characteristics of B. abortus biovars 9.
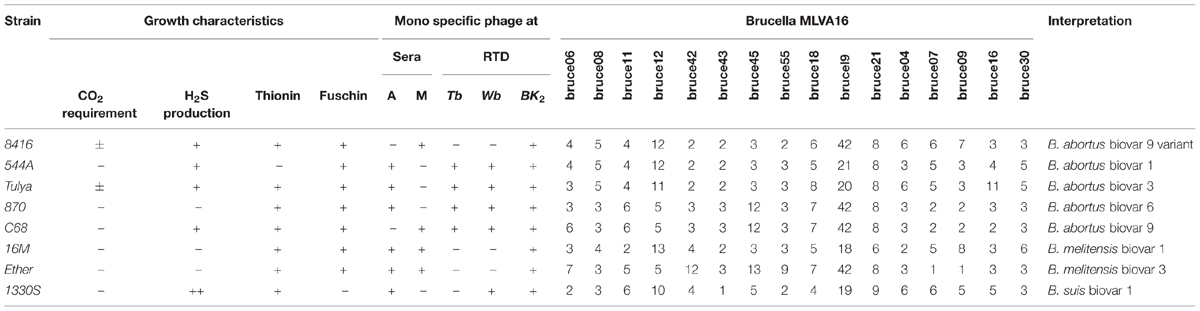
TABLE 1. Comparison of phenotypic characteristics and Brucella phage lysis profiles of Brucella abortus strain 8416 and other Brucella reference strains.
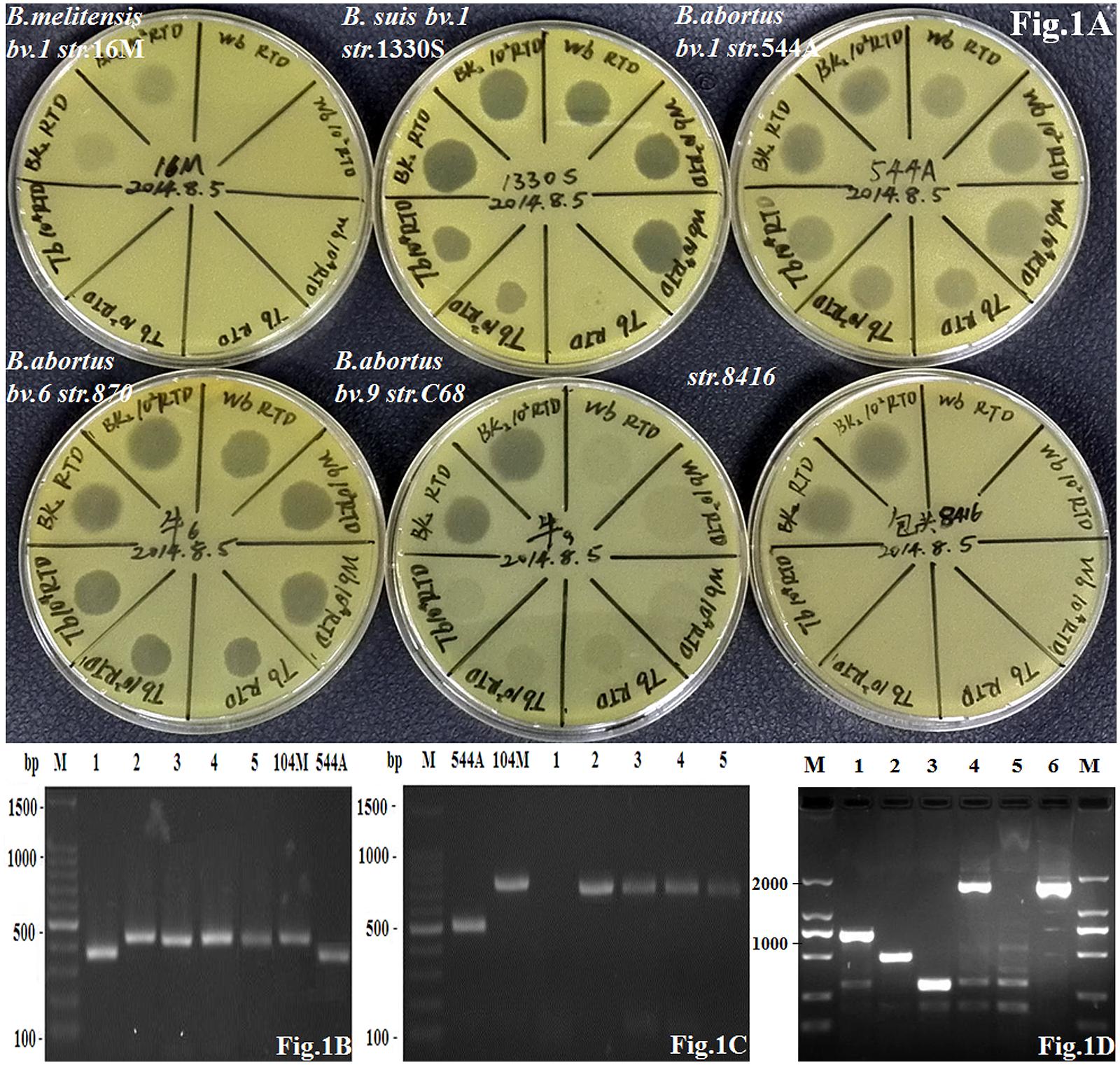
FIGURE 1. (A) The lysis patterns of phage Tb, Wb and Bk2 to Brucella abortus strain 8416, B. abortus biovar 1 strain 544A (A is indicated as B. abortus), B. melitensis biovar 1 strain 16M (M is indicated as B. melitensis), and B. suis biovar 1 strain 1330S (S is indicated as B. suis) as well as B. abortus biovar 6 strain 870 and biovar 9 strain C68; (B) Amplification of DNA fragments from different Brucella strains. Genomic DNA was amplified by the B-ab PCR assay. 1: strain 8416; 2–5: four B. melitensis field strains; 104 M: B. melitensis biovar 1 strain 104M; 544A: B. abortus biovar 1 strain 544A; (C) Amplification of DNA fragments from different Brucella strains. Genomic DNA was amplified by AMOS-PCR assay. 1: strain 8416; 2–5: four B. melitensis field strains; 104 M: B. melitensis biovar 1 strain 104M; 544A: B. abortus biovar 1 strain 544A; (D) Amplification of DNA fragments from different Brucella strains. Genomic DNA was amplified by new PCR assay identifying B. abortus biovar 3b, 5, 6, and 9. 1: B. melitensis biovar 1 strain 16M; 2: B. abortus biovar 1 strain 544A; 3: B. suis biovar 1 strain 1330S; 4: B. abortus biovar 9 strain C68; 5: B. abortus biovar 3a strain Tulya; 6: strain 8416.
Biochemical Identification of Automatic VITEK 2 System
Four biochemical indicators ProA (L-pyrrolydonyl-arylamidase), TyrA (tyrose arylamidase), URE (urease), and GlyA could be used to distinguish Brucella species. All of eight B. abortus reference strains and 21 field strains were positive in ILATk (L-lactate alkalization), but it was negative in strain 8416, three B. melitensis reference strains and 92 field strains (Cui BuYun’s unpublished data). This result indicated that strain 8416 showed special biochemical characteristics distinct from that of B. abortus strains.
Molecular Typing Identification
Strain 8416 was identified as B. abortus by the combination of bcsp31 PCR (223-bp, data not shown) and B-ab PCR (370-bp) (Figure 1B) but not as biovar 1, 2, and 4 of B. abortus according to AMOS-PCR (Figure 1C). The novel PCR assay was used to compare strain 8416 to B. abortus biovar 3b, 5, 6, and 9, and found that the PCR product of 1.7 kb from strain 8416 was similar to B. abortus biovar 3b, 5, 6, and 9, but not to other B. abortus biovars (Figure 1D).
MLVA Genotyping
According to Brucella MLVA typing database (Grissa et al., 2008), 16 loci of MLVA matching results displayed that strain 8416 was closely related to B. abortus biovar 3 (Jiang et al., 2013), but primarily different in four variable loci, bruce04, bruce07, bruce11, and bruce55 (Table 2).
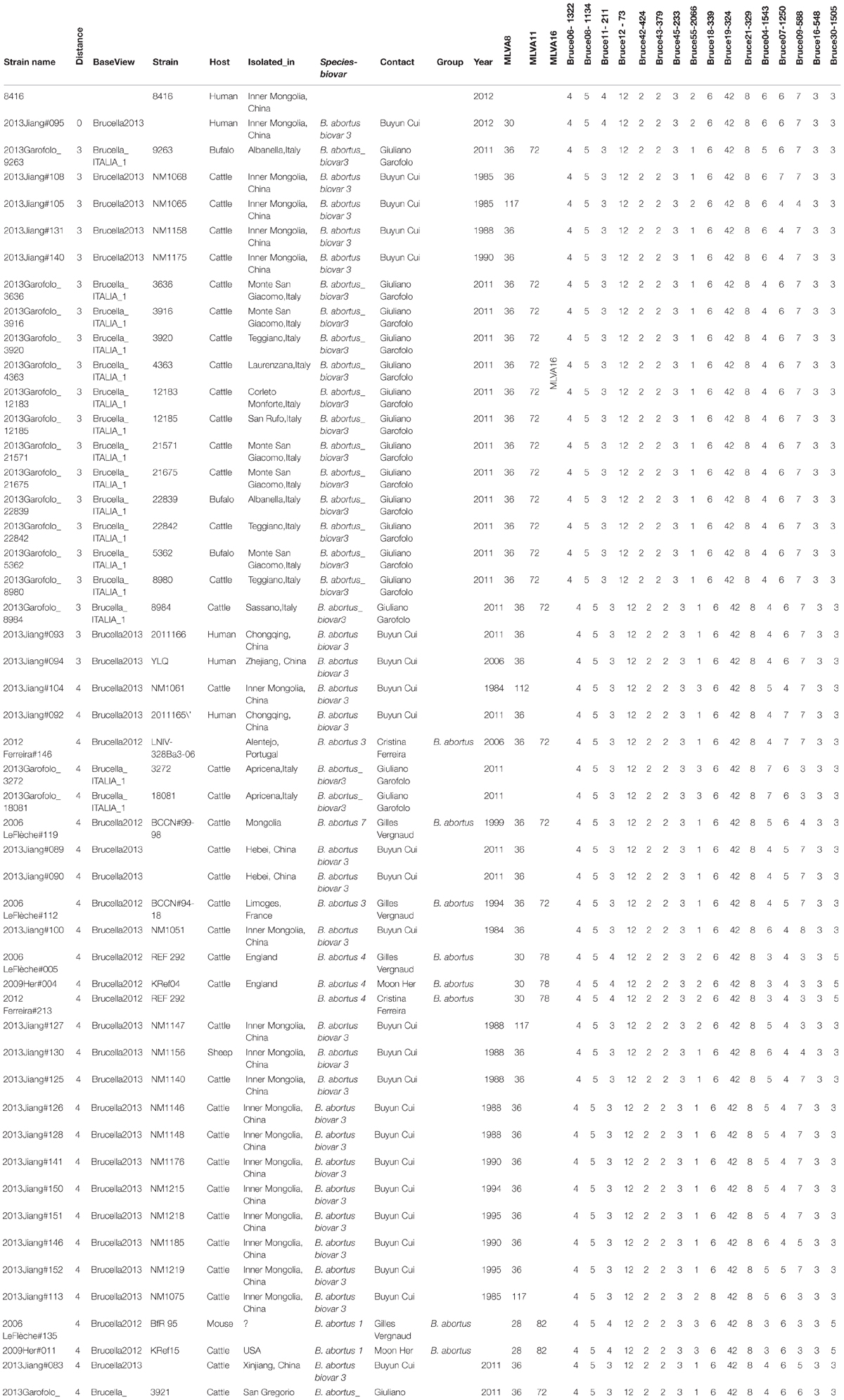
TABLE 2. Comparison of Brucella MLVA typing results of B. abortus strain 8416 and the most closely related B. aborus biovar 3 field strains in the Brucella MLVA database.
Finally, based on these typing results, strain 8416 might be a new variant of B. abortus biovar 9.
Discussion
Until now, the phage resistance mechanism from Brucella SPR strains was poorly understood. In this study, a natural SPR strain of B. abortus isolated from a patient in China was identified. Although SPR strains of B. abortus were rarely isolated from patients, a SPR strain was isolated from a B. abortus phage sensitive parent strain 544 in 1974 and a SPR variant of B. abortus strain 19 was identified in 1976 through the manipulation of laboratory cultures (Corbel and Morris, 1974; Corbel and Thomas, 1976). Compared to the parent strain 544, the SPR strain FS showed no differences in virulence, morphological, cultural, biochemical or metabolic, and serological reactions, but with an altered phage resistance profile (Corbel and Morris, 1974). The potential mechanism of the phage resistance may be due to its failure to penetrate the FS cell wall since the strain FS is more resistant to lysis by phage lysozymes than that of the phage-sensitive parent strain 544 (Corbel and Morris, 1975). Strain 544-FS showed a complete resistance to lysis by many Brucella phages except Bk2 at 1× RTD and 104× RTD. Subsequently, another B. abortus SPR strain with resistance to phage Tb, was isolated from a supramammary lymph node of a cow and it is virulent to guinea-pigs (Harrington et al., 1977). Interestingly, these B. abortus SPR strains mentioned above belonging to B. abortus biovar 1 were identified. However, strain 8416 was significantly different from all of B. abortus biovars by using phenotypic and molecular typing method. However, it shared the same phage lysis profiles to that of B. melitensis biovar 1. In conclusion, strain 8416 is the only SPR strain isolated from the infected human thus far with a similar phage lysis pattern with B. melitensis 16 M. However, despite same resistant to phage Tb, we could not comprehensively compare with phage lysis profiles of the three reported SPR strains due to different Brucella phages tested among them.
Currently, MLVA has been mainly used for tracking the variances of the bacterial genus with a high homology, such as Brucella genus (Haguenoer et al., 2011). The MLVA-16 (panel 1, 2A and 2B) assay was widely used for molecular typing of a larger collection of isolates at both species and biovars level. The panel 1 comprised eight minisatellite markers for species identification (Le Fleche et al., 2006) and the panel 2 markers were found with a higher biovar discriminatory power. Surprisingly, the MLVA-16 typing results showed that strain 8416 was clustered into the Chinese B. abortus biovar 3 strains (Jiang et al., 2013) with four variable loci (bruce04, 07, 11, and 55). Actually, among the four known panel 1 genotypes (28, 30, 112, 116), strain 8416 (genotype 30) was distinct from other 65 Chinese B. abortus biovar 3 strains isolated previously from different geographic origins, suggesting that more B. abortus strains phenotypically identified as biovar 3 are required for the comparison. The MLVA assay confirmed that B. abortus biovar 3 is a heterogeneous group (Le Fleche et al., 2006), and in agreement with the B. abortus biovar 3 divided into two sub-biovar 3a and 3b (Huber et al., 2009).
In this study, an atypical B. abortus strain displaying a phage lysis profile similar to B. melitensis biovars 1 was identified. Most importantly, the lysis pattern by bacteriophages observed in this newly uncovered B. abortus SPR strain. Although phage typing in general can successfully classify Brucella species, our research calls for attention as to conclusions on SPR strains. Further investigation focusing on the strain 8416’s whole genomic variations associated with phage resistance is needed.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by the National Nature Science Foundation (81360444, 81460319) and the Inner Mongolia Autonomous Region of Nature Science Foundation (2013MS1105).
References
Adone, R., and Pasquali, P. (2013). Epidemiosurveillance of brucellosis. Rev. Sci. Tech. 32, 199–205.
Alton, G. G., Jones, L. M., and Pietz, D. E. (1975). Laboratory techniques in brucellosis. Monogr. Ser. No. 55 World Health Organ. 1975, 1–163.
Bounaadja, L., Albert, D., Chenais, B., Henault, S., Zygmunt, M. S., Poliak, S., et al. (2009). Real-time PCR for identification of Brucella spp.: a comparative study of IS711, bcsp31 and per target genes. Vet. Microbiol. 137, 156–164. doi: 10.1016/j.vetmic.2008.12.023
Bricker, B. J., and Halling, S. M. (1994). Differentiation of Brucella abortus bv. 1, 2, and 4, Brucella melitensis, Brucella ovis, and Brucella suis bv. 1 by PCR. J. Clin. Microbiol. 32, 2660–2666.
Chen, Y., Ke, Y., Wang, Y., Yuan, X., Zhou, X., Jiang, H., et al. (2013). Changes of predominant species/biovars and sequence types of Brucella isolates, Inner Mongolia, China. BMC Infect. Dis. 13:514. doi: 10.1186/1471-2334-13-514
Corbel, M. J., and Morris, J. A. (1974). Studies on a smooth phage-resistant variant of Brucella abortus I. Immunological properties. Br. J. Exp. Pathol. 55, 78–87.
Corbel, M. J., and Morris, J. A. (1975). Studies on a smooth phage resistant variant of Brucella abortus II. Mechanism of phage resistance. Br. J. Exp. Pathol. 56, 1–7.
Corbel, M. J., and Thomas, E. L. (1976). The in vivo activity of a smooth phage-resistant variant of Brucella abortus strain 19. Br. Vet. J. 132, 121–123.
Grissa, I., Bouchon, P., Pourcel, C., and Vergnaud, G. (2008). On-line resources for bacterial micro-evolution studies using MLVA or CRISPR typing. Biochimie 90, 660–668. doi: 10.1016/j.biochi.2007.07.014
Haguenoer, E., Baty, G., Pourcel, C., Lartigue, M. F., Domelier, A. S., Rosenau, A., et al. (2011). A multi locus variable number of tandem repeat analysis (MLVA) scheme for Streptococcus agalactiae genotyping. BMC Microbiol. 11:171. doi: 10.1186/1471-2180-11-171
Harrington, R. Jr., Bond, D. R., and Brown, G. M. (1977). Smooth phage-resistant Brucella abortus from bovine tissue. J. Clin. Microbiol. 5, 663–664.
Huber, B., Scholz, H. C., Lucero, N., and Busse, H. J. (2009). Development of a PCR assay for typing and subtyping of Brucella species. Int. J. Med. Microbiol. 299, 563–573. doi: 10.1016/j.ijmm.2009.05.002
Jiang, H., Wang, H., Xu, L., Hu, G., Ma, J., Xiao, P., et al. (2013). MLVA genotyping of Brucella melitensis and Brucella abortus isolates from different animal species and humans and identification of Brucella suis vaccine strain S2 from cattle in China. PLoS ONE 8:e76332. doi: 10.1371/journal.pone.0076332
Jones, L. M., McDuff, C. R., and Wilson, J. B. (1962). Phenotypic alterations in the colonial morphology of Brucella abortus due to a bacteriophage carrier state. J. Bacteriol. 83, 860–866.
Le Fleche, P., Jacques, I., Grayon, M., Al Dahouk, S., Bouchon, P., Denoeud, F., et al. (2006). Evaluation and selection of tandem repeat loci for a Brucella MLVA typing assay. BMC Microbiol. 6:9. doi: 10.1186/1471-2180-6-9
Ocampo-Sosa, A. A., Aguero-Balbin, J., and Garcia-Lobo, J. M. (2005). Development of a new PCR assay to identify Brucella abortus biovars 5, 6 and 9 and the new subgroup 3b of biovar 3. Vet. Microbiol. 110, 41–51. doi: 10.1016/j.vetmic.2005.06.007
Van der Henst, C., De Barsy, M., Zorreguieta, A., Letesson, J. J., and De Bolle, X. (2013). The Brucella pathogens are polarized bacteria. Microbes Infect. 15, 998–1004. doi: 10.1016/j.micinf.2013.10.008
Keywords: B. abortus, smooth phage-resistant (SPR), MLVA typing, unusual biochemical reactions
Citation: Kang Y-X, Li X-M, Piao D-R, Tian G-Z, Jiang H, Jia E-H, Lin L, Cui B-Y, Chang Y-F, Guo X-K and Zhu Y-Z (2015) Typing Discrepancy Between Phenotypic and Molecular Characterization Revealing an Emerging Biovar 9 Variant of Smooth Phage-Resistant B. abortus Strain 8416 in China. Front. Microbiol. 6:1375. doi: 10.3389/fmicb.2015.01375
Received: 30 June 2015; Accepted: 19 November 2015;
Published: 08 December 2015.
Edited by:
Yongqun “Oliver” He, University of Michigan, USAReviewed by:
Jianguo Jian Zhu, Shanghai Jiao Tong University, ChinaMenachem Banai, Kimron Veterinary Institute, Israel
Jiabo Ding, China Institute of Veterinary Drugs Control, China
Copyright © 2015 Kang, Li, Piao, Tian, Jiang, Jia, Lin, Cui, Chang, Guo and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bu-Yun Cui, Y3VpYnV5dW5AaWNkYy5jbg==; Yung-Fu Chang, eWM0MkBjb3JuZWxsLmVkdQ==; Xiao-Kui Guo, bWljcm9iaW9sb2d5QHNqdHUuZWR1LmNu; Yong-Zhang Zhu, eXpoemh1QGhvdG1haWwuY29t
 Yao-Xia Kang1,2
Yao-Xia Kang1,2 Yung-Fu Chang
Yung-Fu Chang Yong-Zhang Zhu
Yong-Zhang Zhu