- 1Department of Microbial Ecology, Netherlands Institute of Ecology (NIOO-KNAW), Wageningen, Netherlands
- 2Laboratory of Phytopathology, Wageningen University, Wageningen, Netherlands
- 3Plant Research International, Business Unit Bioscience, Wageningen University and Research Centre, Wageningen, Netherlands
- 4Centre for Biosystems Genomics, Wageningen, Netherlands
- 5Molecular Biotechnology, Institute of Biology, Leiden University, Leiden, Netherlands
In disease-suppressive soils, plants are protected from infections by specific root pathogens due to the antagonistic activities of soil and rhizosphere microorganisms. For most disease-suppressive soils, however, the microorganisms and mechanisms involved in pathogen control are largely unknown. Our recent studies identified Actinobacteria as the most dynamic phylum in a soil suppressive to the fungal root pathogen Rhizoctonia solani. Here we isolated and characterized 300 isolates of rhizospheric Actinobacteria from the Rhizoctonia-suppressive soil. Streptomyces species were the most abundant, representing approximately 70% of the isolates. Streptomyces are renowned for the production of an exceptionally large number of secondary metabolites, including volatile organic compounds (VOCs). VOC profiling of 12 representative Streptomyces isolates by SPME-GC-MS allowed a more refined phylogenetic delineation of the Streptomyces isolates than the sequencing of 16S rRNA and the house-keeping genes atpD and recA only. VOCs of several Streptomyces isolates inhibited hyphal growth of R. solani and significantly enhanced plant shoot and root biomass. Coupling of Streptomyces VOC profiles with their effects on fungal growth, pointed to VOCs potentially involved in antifungal activity. Subsequent assays with five synthetic analogs of the identified VOCs showed that methyl 2-methylpentanoate, 1,3,5-trichloro-2-methoxy benzene and the VOCs mixture have antifungal activity. In conclusion, our results point to a potential role of VOC-producing Streptomyces in disease suppressive soils and show that VOC profiling of rhizospheric Streptomyces can be used as a complementary identification tool to construct strain-specific metabolic signatures.
Introduction
Disease-suppressive soils are soils in which plants are effectively protected from infections by specific root pathogens due to antagonistic activities of soil and rhizosphere (micro)organisms (Hornby, 1983; Weller et al., 2002). This phenomenon has been described worldwide, but the responsible (micro)organisms and underlying mechanisms are largely unknown for most suppressive-soils (Weller et al., 2002; Mendes et al., 2011; Chapelle et al., 2015). In recent studies, we identified the microbiome of a soil suppressive to Rhizoctonia solani, an economically important soil-borne fungal pathogen of many crops including sugar beet, potato, and rice (Mendes et al., 2011; Chapelle et al., 2015). PhyloChip-based metagenomics detected more than 33000 bacterial and archaeal taxa in the rhizosphere of sugar beet seedlings grown in the Rhizoctonia-suppressive soil and revealed bacterial groups consistently associated with the disease suppressive state. Among the top 10% of most dynamic taxa (i.e., taxa relatively more abundant in suppressive than in non-suppressive soil), Actinobacteria were the most dynamic phylum found in the rhizosphere of sugar beet seedlings growing in the suppressive soil.
Actinobacteria are ubiquitously found in nature and the phylum comprises more than 500 formally described species (Goodfellow, 2012; Labeda et al., 2012). Many Actinobacteria are multicellular bacteria with a complex life cycle and are renowned for the production of an exceptionally large number of bioactive metabolites (Claessen et al., 2014). Members of the genus Streptomyces produce over 10000 secondary metabolites, including volatile organic compounds (VOCs) (Bérdy, 2005; Hopwood, 2007; van Wezel et al., 2009). Approximately 1000 microbial VOCs have been identified to date (Piechulla and Degenhardt, 2014). Although the production of VOCs by microorganisms is known for many years (Zoller and Clark, 1921; Stotzky and Schenck, 1976), it is only since the last decade that an increasing number of studies have reported on the diversity and potential functions of these compounds. The blend of VOCs released by microorganisms is diverse and complex. Microbial VOCs belong to different classes of compounds such as alkenes, alcohols, ketones, terpenes, benzenoids, aldehydes, pyrazines, acids, esters, and sulfur-containing compounds (Effmert et al., 2012). The same VOCs can be found for different, often unrelated, microorganisms but some VOCs are unique to specific microorganisms (Schulz and Dickschat, 2007; Garbeva et al., 2014). Microbial VOCs display versatile functions: they inhibit bacterial and fungal growth, promote or inhibit plant growth, trigger plant resistance and attract other micro- and macro-organisms (Ryu et al., 2003, 2004; Vespermann et al., 2007; Kai et al., 2009; Verhulst et al., 2009; Bailly and Weisskopf, 2012; Hagai et al., 2014; Schmidt et al., 2015). Furthermore, VOCs have been proposed to function as signaling molecules in inter- and intra-specific interactions and in cell-to-cell communication. To date, however, the natural functions of microbial VOCs and their modes of action remain largely unknown (Kai et al., 2009; Kim et al., 2012; Schmidt et al., 2015).
Here we studied the diversity and functions of VOCs produced by different Streptomyces from the rhizosphere of sugar beet seedlings grown in a Rhizoctonia-suppressive soil. We first isolated and characterized 300 Actinobacteria. As Streptomyces represented almost 70% of all isolates, subsequent VOC analyses, phylogeny, antifungal activity and plant growth assays were conducted with this group of Actinobacteria. By coupling SPME-GC-MS and hierarchical clustering of VOC profiles, we identified VOCs potentially involved in antifungal activity.
Materials and Methods
Selective Isolation of Actinobacteria
Actinobacteria were isolated from the rhizosphere (roots with adhering soil) of sugar beet plants grown in a soil suppressive to R. solani. The soil was previously collected in 2003 and 2004 from an agricultural sugar beet field close to the town of Hoeven, the Netherlands (51°35′10″N 4°34″44′E). For the collection of Actinobacteria from the rhizosphere, sugar beet seeds (cultivar Alligator) were sown in square PVC pots containing 250 g of field soil with an initial moisture content of 10% (v/w). Plants were grown in a growth chamber (24°C/24°C day/night temperatures; 180 μmol light m−2 s−1 at plant level during 16 h/d; 70% relative humidity) and watered weekly with standard Hoagland solution (macronutrients only). After 3 weeks of plant growth, 1 g of sugar beet roots with adhering soil was suspended in 5 mL of potassium-phosphate buffer (pH 7.0). Samples were vortexed and sonicated for 1 min. To enrich for different genera of Actinobacteria, a number of treatments were applied to the soil suspension (Supplementary Table S1). Single colonies were picked based on the morphology and purified on fresh agar plates. Isolates were stored in glycerol (20%, v/v) at −20 and −80°C.
Characterization of Actinobacteria
All 300 Actinobacterial isolates were characterized by 16S rRNA gene sequencing. PCR amplifications were conducted using primers 8F (5′- AGAGTTTGATC CTGGCTCAG - 3′) and 1392R (5′- ACGGGCGGT GTGTACA - 3′) or 27F (5′- GAGTTTGATCCTG GCTCAG - 3′) and 1492R (5′- ACCTTGTTACGACGACTT - 3′) (Lane, 1991; Deangelis et al., 2009). For obtaining DNA, bacterial cells were disrupted by heating at 95°C for 10 min. For spore forming isolates, cells were disrupted in the microwave at 650 W for 30 s in TE buffer. Suspensions were centrifuged at 13000 rpm for 10 min. After centrifugation, 2 μl of the supernatants were used for the PCR reactions. PCR products were purified and sequenced at Macrogen Inc. Isolates were characterized based on sequence identity with 16S rRNA gene sequences in the Greengenes database (McDonald et al., 2012) (http://greengenes.lbl.gov/).
Coupling Streptomyces Isolates to OTUs Detected by PhyloChip
16S rRNA gene sequences of 173 Streptomyces isolates were compared with the 16S rRNA gene sequences of Streptomyces OTUs previously identified by PhyloChip-based metagenomic analysis as the top 10% of most abundant taxa associated with disease suppressiveness (Mendes et al., 2011). Phylogenetic analysis was performed with Muscle in MEGA6 (Tamura et al., 2013) and iTOL (Letunic and Bork, 2011) (http://itol.embl.de/). A Neighbor-joining consensus tree (Saitou and Nei, 1987) with 1000 bootstrap replicates (Felsenstein, 1985) was constructed using Tamura-Nei model (Tamura and Nei, 1993) with gamma distribution. A total of 11 isolates, which were closely related to the isolates detected by PhyloChip, was selected to study the composition of emitted VOCs and their in vitro effects on fungal and plant growth. Streptomyces lividans 1326 (Cruz-Morales et al., 2013) was used as a reference strain.
Characterization of Selected Streptomyces Isolates
The 11 Streptomyces isolates were characterized based on colony morphology and by sequence analysis of the house-keeping genes recA (recombinase A) and atpD (ATP synthase subunit B). These genes were amplified and sequenced as previously described (Guo et al., 2008). Partial sequences of recA (500 bp), atpD (423 bp), and 16S rRNA (516 bp) genes of Streptomyces were concatenated to yield an alignment of 1439 sites. A concatenated phylogenetic tree supplemented with sequences of Streptomyces strains with a sequenced genome (NCBI database) was constructed using UPGMA with the Tamura-3 parameter calculation model with gamma distribution and 1.000 bootstrap replicates. All sequences were deposited to GenBak and have been assigned to accession numbers: KT60032-KT600042 (16S rRNA gene), KT600043-KT600053 (recA gene), and KT600054-KT600064 (atpD gene).
Collection and Analysis of Streptomyces VOCs
For trapping the VOCs, the Streptomyces isolates were inoculated individually in 10 ml sterile glass vials containing 2.5 ml of GA medium (Zhang, 1990) with three replicates each. Vials containing medium only served as controls. All vials were closed and incubated at 30°C. After 7 days, VOCs from the headspace of each vial were collected by solid phase microextraction (SPME) with a 65-mm polydimethylsiloxane-divinylbenzene fiber (Supelco, Bellefonte, USA).
Streptomyces VOCs were analyzed by GC-MS (Agilent GC7890A with a quadrupole MSD Agilent 5978C). VOCs were thermally desorbed at 250°C by inserting the fiber for 2 min into the hot GC injection port. The compounds released were transferred onto the analytical column (HP-5MS, 30 m × 0.25 mm ID, 0.25 μm—film thickness) in splitless mode. The temperature program of the GC oven started at 45°C (2-min hold) and rose with 10°C min−1 to 280°C (3-min hold). Mass scanning was done from 33 to 300 m/z with a scan time of 2.8 scans s−1. GC-MS raw data were processed by an untargeted metabolomics approach. MetAlign software (Lommen and Kools, 2012) was used to extract and align the mass signals (s/n = 3). MSClust was used to remove signal redundancy per metabolite and to reconstruct compound mass spectra as previously described (Tikunov et al., 2012). VOCs were tentatively annotated by comparing their mass spectra with those of commercial (NIST08) and in-house mass spectral libraries. Linear retention indices (RI) of VOCs were calculated as previously described (Strehmel et al., 2008) and compared with those in the literature. VOCs selected for in vitro antifungal assays [methyl butanoate (≥98%), methyl 2-methylpentanoate (≥98%), methyl 3-methylpentanoate (≥97%), 1,3,5-trichloro-2-methoxy benzene (99%) and 3-octanone (≥98%)] were confirmed with authentic reference standards obtained at Sigma-Aldrich. Processed VOC data were log transformed and auto-scaled using the average as an offset and the standard deviation as scale [raw value-average (offset)/SD (scale)]. Log transformed data were then subjected to multivariate statistical analysis. One-way ANOVA was performed with GeneMaths XT Version 2.11 (Applied Maths, Belgium) to identify VOCs significantly different from the control (medium only) [p < 0.05; with false discovery rate (FDR) correction]. After that, hierarchical cluster analysis (HCA) using Pearson's correlation coefficient with UPGMA algorithm was performed.
VOC-mediated Antifungal Activity
The effect of Streptomyces VOCs on the growth of the fungus R. solani was investigated using the bottoms of two 90-mm-diameter Petri dishes allowing physical separation between the bacteria and the fungus. One bottom contained a Streptomyces isolate on GA medium, previously incubated at 30°C for 4 days. The other bottom contained a plug of R. solani mycelium on 1/10th Tryptone Soy Agar (TSA, Oxoid). Both Petri dishes were sealed facing each other and incubated at 25°C with the Petri dish containing the Streptomyces on the bottom to avoid spores transferring to the plate with the fungus. As a control, the Petri dish containing R. solani was exposed to a Petri dish containing GA medium only. Fungal growth inhibition was calculated by measuring the radial growth of the fungal hyphae after 1, 2, and 3 days of incubation. Percentage of inhibition was calculated as [(diameter of fungus in control − diameter of fungus exposed to VOCs)*100/diameter of fungus in control] for each of the 3 replicates. Student's t-Test was performed to determine statistically significant differences compared to the control (p < 0.05, n = 3).
Antifungal Activity of Synthetic VOCs
Methyl butanoate (≥98%), methyl 2-methylpentanoate (≥98%), methyl 3-methylpentanoate (≥97%), 1,3,5-trichloro-2-methoxy benzene (99%), and 3-octanone (≥98%) were obtained at Sigma-Aldrich. All VOCs were dissolved in methanol with final concentrations ranging from 1 M to 1 nM (10-fold dilutions). Assays were performed using a standard 90 mm-diameter Petri dish with the fungal plug on 1/10th TSA medium on top and with a sterile paper filter (1.5 × 1.5 cm) on the bottom. Twenty microliters of each VOC dissolved in methanol were applied on the paper filter, plates were immediately sealed and incubated at 25°C. Radial hyphal growth of the fungus was measured after 1 and 2 days of exposure to single or a mixture of the 5 VOCs and compared to control (empty top of a Petri dish). To check whether the solvent itself had any effect on growth of the fungus, R. solani was also exposed to methanol alone. Student's t-Test was performed to determine statistically significant differences compared to the control (p < 0.05, n = 3 − 5).
VOC-mediated Plant Growth Promotion
To determine whether Streptomyces VOCs had an effect on plant growth, Arabidopsis thaliana seedlings were exposed to the VOCs emitted by the different isolates. A. thaliana seeds (wild-type Col-0) were surface sterilized as previously described (van de Mortel et al., 2012) and sown on 90-mm-diameter Petri dishes containing 50 ml of 0.5X Murashige and Skoog medium (Murashige and Skoog, 1962) supplemented with 0.5% (w/v) sucrose. The 90-mm-diameter Petri dishes were placed inside a 145-mm-diameter Petri dish, sealed and incubated in a climate chamber (21°C/21°C day/night temperatures; 180 μmol light m−2 s−1 at plant level during 16 h/d; 70% relative humidity). After 7 days, 35-mm-diameter Petri dishes containing Streptomyces isolates growing on GA medium (previously incubated at 30°C for 1 week) were added to the 145-mm Petri dishes with the A. thaliana seedlings. Plates were sealed and kept at 21°C. After 14 days, plant fresh weight was determined. In addition, plant dry weight was measured after drying shoots and roots overnight in an incubator at 65°C. Student's t-Test was performed to determine statistically significant differences compared to the control treatment (plants exposed to medium only).
Results
Diversity of Actinobacteria Isolated from Suppressive Soil
Using PhyloChip-based metagenomic analyses, we previously described the diversity of the bacterial community associated with the rhizosphere of sugarbeet plants grown in a Rhizoctonia-suppressive soil (Mendes et al., 2011). Actinobacteria were prominently more represented in the suppressive soil than in the non-suppressive (conducive) soil. Bacterial diversity detected by the PhyloChip used in the aforementioned study is displayed in Figure 1A. To select as many Actinobacterial isolates as possible, several pre-treatments of the rhizospheric soil and different selective media were used for their isolation (Supplementary Table S1). A total of 300 Actinobacterial isolates were obtained and characterized by 16S rRNA gene sequencing. Based on the sequence similarities (95–100%) to the 16S rRNA gene sequences available in the Greengenes database (used as reference in the PhyloChip analyses), 18 different genera of Actinobacteria were identified. These were Streptomyces, Microbacterium, Rhodococcus, Micromonospora, Microbispora, Kribbella, Pseudonocardia, Cellulomonas, Mycobacterium, Actinoplanes, Arthrobacter, Actinomadura, Amycolaptosis, Nocardioides, Nonomureae, Streptosporangium, Micrococcus, and Rothia (Figure 1B). The genus Streptomyces was the most abundant, representing 69% of all isolates and at least 25 different species based on 16S rRNA gene sequences (Figure 1C).
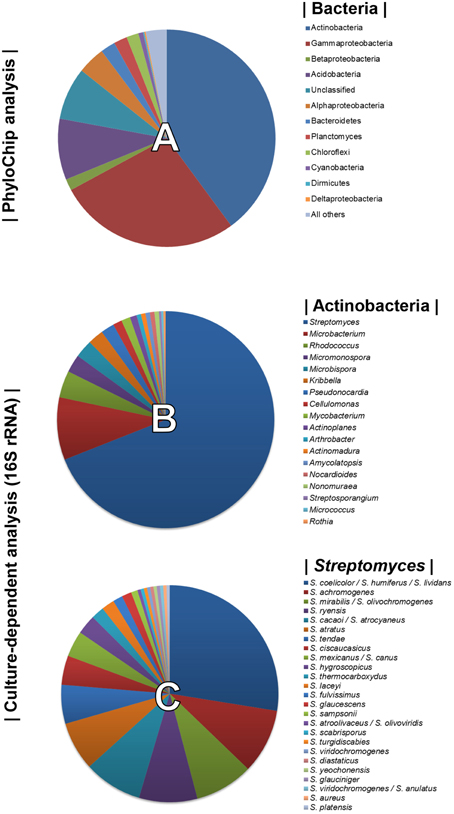
Figure 1. Top 10% most dynamic bacterial (and archaeal) phyla detected by PhyloChip analysis of the rhizosphere microbiome of sugar beet seedlings grown in Rhizoctonia-suppressive soil (pie chart A, adapted from Mendes et al., 2011). Diversity of Actinobacteria (pie chart B) and of Streptomyces species (pie chart C) isolated from the rhizosphere of sugar beet seedlings grown in Rhizoctonia-suppressive soil (this study).
Phylogenetic Analysis of Streptomyces Isolates
To select Streptomyces isolates for VOC and functional analyses, 16S rRNA gene sequences of the Streptomyces isolates (n = 173) obtained in this study were compared with those of the representative Streptomyces OTUs (n = 430) originally detected by PhyloChip (Mendes et al., 2011). A phylogenetic tree was constructed using these sequences and the sequences of different Streptomyces type strains (Figure 2). This comparison led to the selection of 11 isolates (Figure 3). We then constructed phylogenetic trees with these 11 isolates, their closest type strains, other Streptomyces species with sequenced genomes and the reference strain Streptomyces lividans 1326 (Supplementary Figure S1A). Additionally, we sequenced the house-keeping genes atpD and recA (Supplementary Figure S1B). Concatenation of atpD, recA, and 16S sequences allowed a better resolution of the different Streptomyces isolates than based on 16S sequences only. However, closely related but phenotypically different isolates, like Streptomyces strains W75.5 and W126 (Figure 3), could not be distinguished based on these three molecular markers.
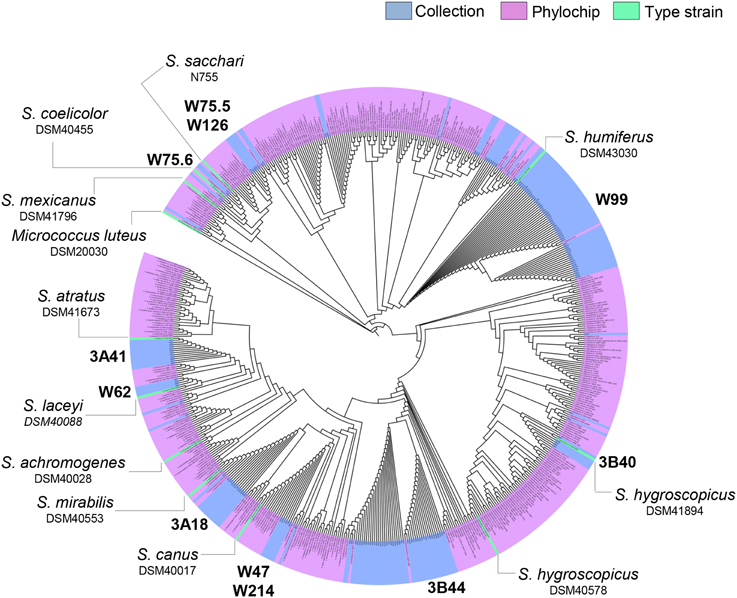
Figure 2. Neighbor-joining phylogenetic tree based on 16S rRNA gene sequences of the Streptomyces collection obtained in this study (in blue), Streptomyces detected by Phylochip analysis (in pink), and Streptomyces type strains (in green). Streptomyces isolates selected for VOC analysis are indicated in bold.
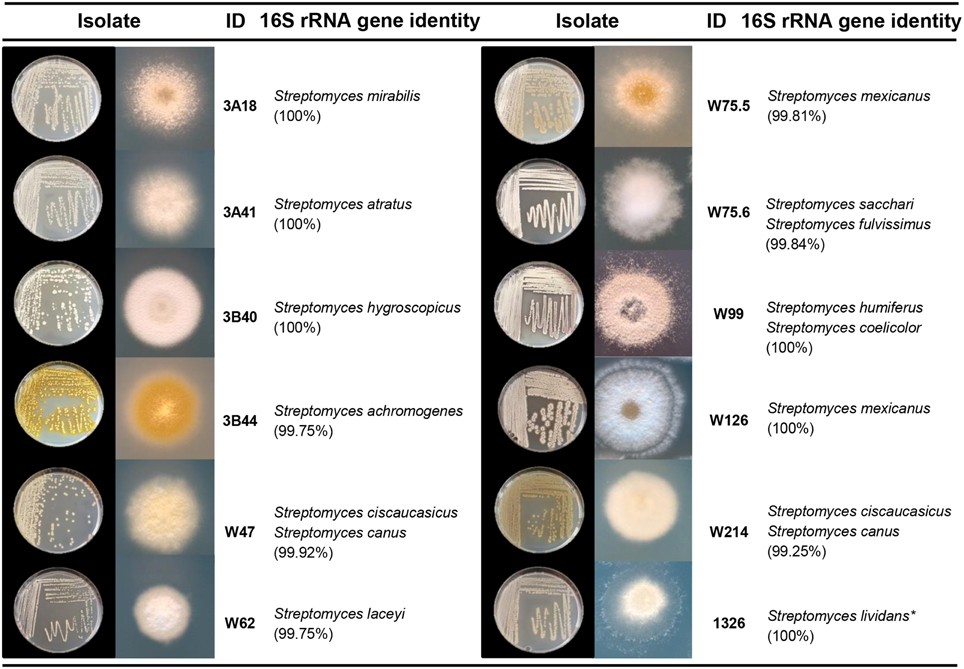
Figure 3. Characterization of Streptomyces isolates used in this study. Species names are based on 16S rRNA gene sequence comparison using the Greengenes database. Pictures depict 4–7 day-old isolates grown on GA medium. *S. lividans 1326 refers to John Innes Center collection number and corresponds to S. lividans 66 (Hopwood et al., 1983).
VOC Profiling of Streptomyces Isolates
For the 12 Streptomyces isolates (11 rhizosphere isolates and reference strain S. lividans 1326) grown on GA medium and the medium alone (control), a total of 536 VOCs were detected in the headspace. Out of these, 381 VOCs that were significantly different (ANOVA, p < 0.05) and detected at intensities at least twice as high as in the control were considered for further analyses. The diversity of VOCs produced by the different Streptomyces isolates is shown in Supplementary Table S2 and highlighted in the heat-map (Figure 4). The VOCs detected belong to diverse classes of compounds such as alcohols, aldehydes, carboxylic acids, esters, ketones, sulfur compounds, and several terpenes (Supplementary Table S2). Most VOCs were found to be specific for some Streptomyces isolates and 45 VOCs were found to be commonly produced by all isolates tested. Geosmin (trans-1,10-dimethyl-trans-9-decalol, RI 1423; Supplementary Table S2) was one of these common VOCs. HCA of the VOC profiles resulted in a similar clustering of the 12 Streptomyces isolates as the clustering based on the different molecular markers (Figure 5). In contrast to the molecular markers, however, VOC profiling allowed differentiation between closely related Streptomyces isolates such as Streptomyces strains W75.5 and W126 as well as Streptomyces strains W47 and W214.
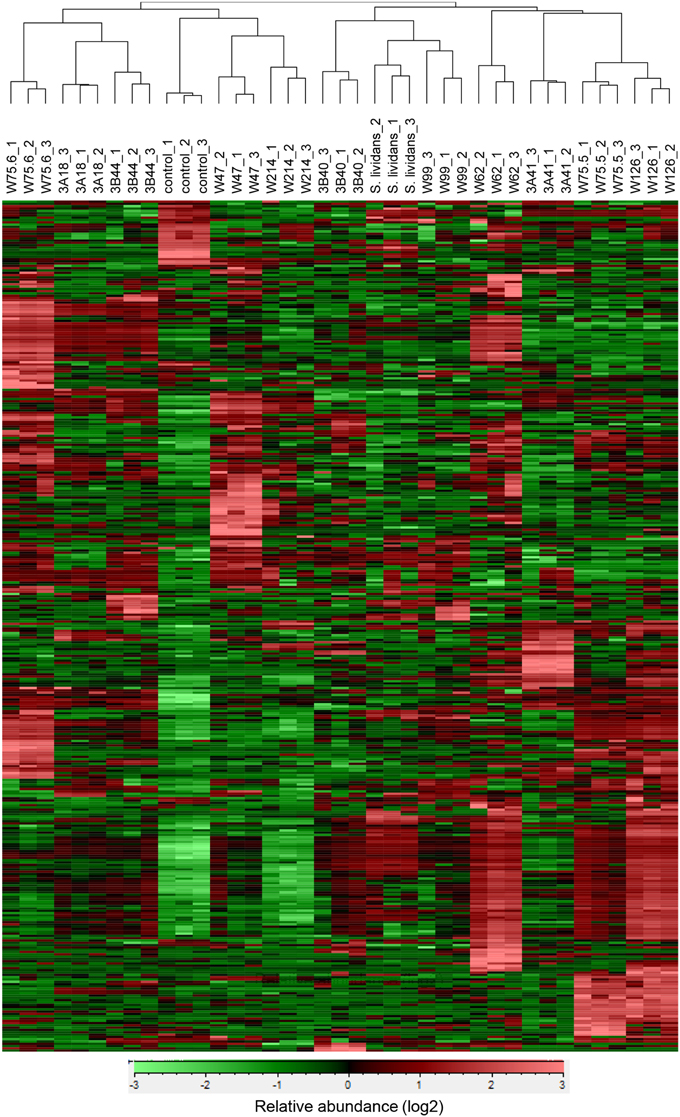
Figure 4. Hierarchical cluster and heat-map analyses of VOC profiles of the selected Streptomyces isolates. Columns represent three replicate VOC measurements of each of the 12 isolates and the medium alone (control). Rows represent the different VOCs (green, low abundance; red, high abundance), several of which were putatively annotated (see Supplementary Table S2).
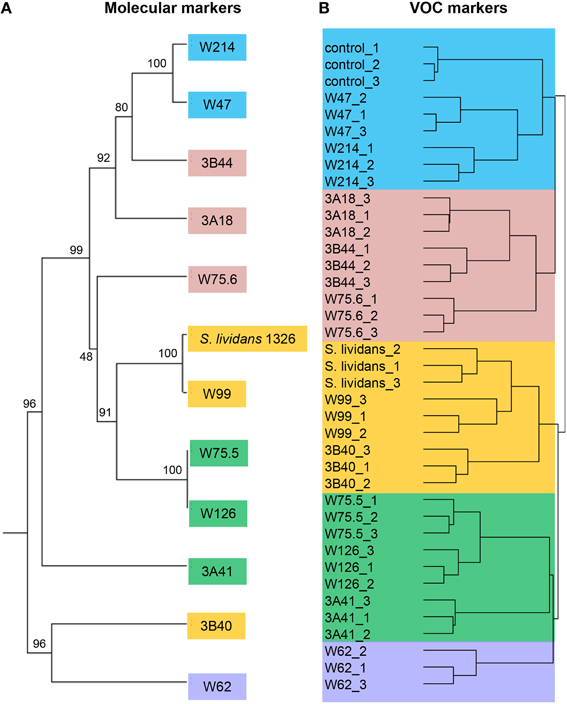
Figure 5. (A) Phylogenetic tree of concatenated partial sequences of 16S rRNA, atpD and recA genes of 11 Streptomyces isolates from the Rhizoctonia-suppressive soil and the reference strain S. lividans 1326. The tree was constructed using UPGMA method and Tamura-3 parameter calculation model with gamma distribution and 1000 bootstrap replicates. (B) Hierarchical cluster analysis (HCA) of Streptomyces VOCs with UPGMA method and Pearson's correlation coefficient. Different colors indicate different clusters of isolates based on VOC profiles.
Effect of Streptomyces VOCs on Fungal and Plant Growth
To test the antifungal activity of VOCs produced by the Streptomyces isolates from disease suppressive soil, hyphal growth of R. solani was measured during exposure to VOCs from each of the isolates. In the control, fungal hyphae reached the edge of the agar plates after 2 days of incubation. All Streptomyces strains were able to significantly retard the growth of R. solani. Streptomyces strains W47 and W214 were the most inhibitory. When exposed for 2 days to the VOCs produced by these isolates, radial hyphal growth was reduced by 57 and 41%, respectively (Figure 6A).
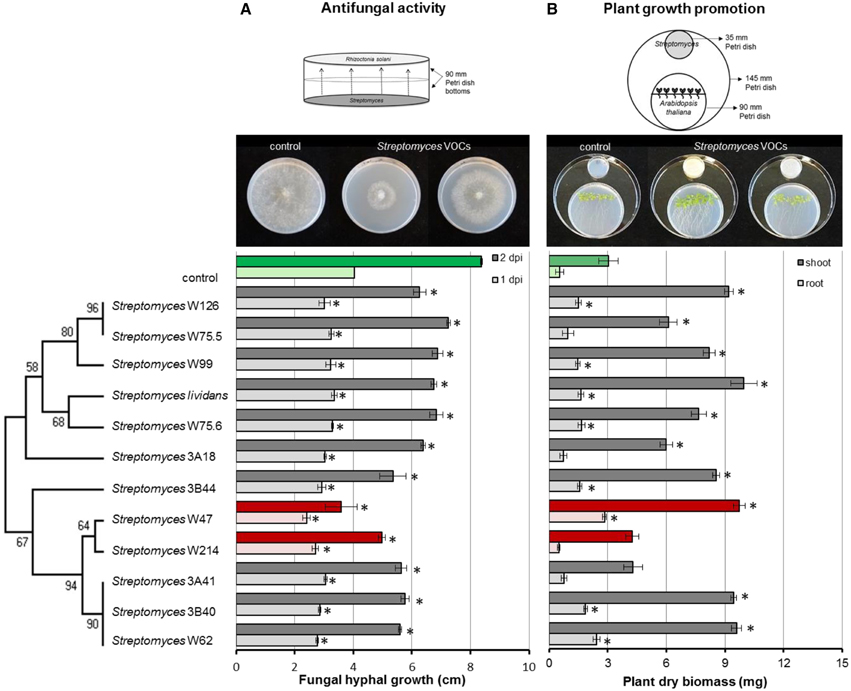
Figure 6. Inhibition of fungal growth after 1 and 2 days of exposure to Streptomyces VOCs (A) and growth of Arabidopsis thaliana seedlings after 2 weeks of exposure to Streptomyces VOCs (B). The controls are displayed in green and isolates with the strongest antifungal activity in red. Bars represent standard errors of the mean of 3 independent biological replicates. Asterisks indicate a statistical difference as compared to controls (exposed to medium only) using Student's t-Test (p < 0.05, n = 3). Pictures of antifungal activity and plant growth promotion were made after 3 and 14 days of exposure, respectively.
Additionally, we tested whether Streptomyces VOCs could promote plant growth. To that end, we exposed 7-day-old A. thaliana seedlings to VOCs from each of the isolates and determined root and shoot biomass. After 2 weeks of exposure to Streptomyces VOCs, no negative effects on plant growth were observed. Ten out of 12 isolates significantly increased shoot biomass, and 8 significantly increased root biomass compared to the control (Figure 6B). S. lividans 1326, and Streptomyces strains W47 and W62 led to the largest increase in plant biomass, whereas Streptomyces strains W214 and 3A41 did not increase shoot and root biomass.
Identification of Streptomyces VOCs Contributing to Antifungal Activity
Since Streptomyces strains W47 and W214 are phylogenetically closely related and both showed strong antifungal activity, these isolates were selected to identify VOCs with activity against R. solani. Screening of VOCs with potential antifungal activity was computed with One-way ANOVA [p < 0.05; with false discovery rate (FDR) correction] and a fold change >2 using the peak intensity of VOCs from W214/control and W47/control. For the selection of VOCs for in vitro antifungal activity, three criteria were used: (1) match factor and reverse match factor higher than 850, (2) reliable annotation based on retention indices and, (3) availability of pure (synthetic) reference compounds.
A comparison of the VOC profiles of Streptomyces strains W47 and W214 with the control (medium only) pinpointed VOCs potentially involved in antifungal activity (Figure 7A). A total of 96 VOCs were shared between these two isolates; 65 and 7 VOCs were unique for Streptomyces strains W47 and W214, respectively (Figures 7A,B). Since both Streptomyces strains W47 and W214 showed antifungal activity, we looked into the VOCs detected for both strains. We selected five common VOCs (methyl butanoate, methyl 2-methylpentanoate, methyl 3-methylpentanoate, 1,3,5-trichloro-2-methoxy benzene, and 3-octanone) which could be reliably annotated based on RI and mass spectral similarity and which were commercially available as authentic reference standards. The identity of these compounds was verified by analyzing pure standards by the GC-MS and comparing their mass spectra and RI with those of the VOCs detected for Streptomyces strains W47 and W214. Subsequently, different concentrations of these five VOCs were used to test their inhibitory effect on hyphal growth of R. solani (Figure 7C). The VOC 1,3,5-trichloro-2-methoxy benzene completely inhibited radial hyphal growth of R. solani at concentrations of 1 M and 100 mM (Figure 7D). Exposure to this VOC led to melanization of R. solani hyphae (Figure 7E). The VOC methyl 2-methylpentanoate reduced fungal growth by 47 and 25% after 1 and 2 days of exposure, respectively. Additionally, a mix of the 5 synthetic VOCs, each at a final concentration of 200 mM, inhibited hyphal growth by 58 and 42% after 1 and 2 days of exposure, respectively.
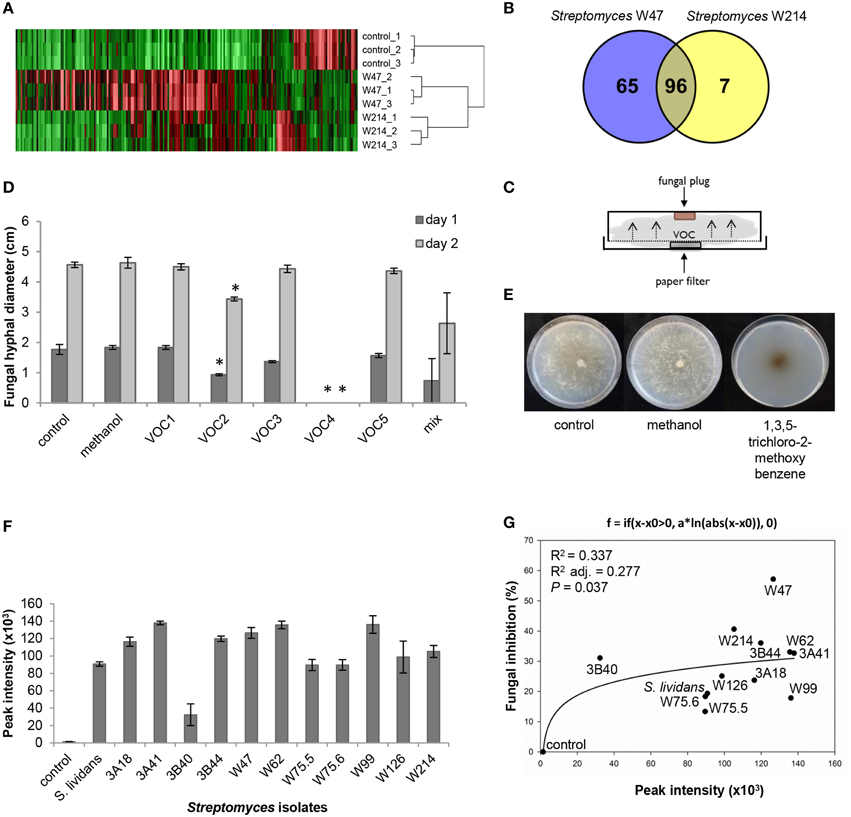
Figure 7. (A) VOC profiles of Streptomyces strains W47 and W214 compared to control (medium only). (B) Venn diagram for common and unique VOCs produced by Streptomyces strains W47 and W214. (C) Experimental set-up for in vitro antifungal activity assay with synthetic VOCs. (D) In vitro antifungal activity with synthetic VOCs at 1 M [control, methanol, VOC1 (methyl butanoate), VOC2 (methyl 2-methylpentanoate), VOC3 (methyl 3-methylpentanoate), VOC4 (1,3,5-trichloro-2-methoxy benzene), VOC5 (3-octanone)]. Methanol was used to dilute all VOCs. Bars represent standard errors of the mean of 3 independent replicates. Asterisks indicate statistical differences compared to control according to Student's t-Test (p < 0.05, n = 3). (E) Fungal growth after exposure to 1,3,5-trichloro-2-methoxy benzene. (F) Abundance of 1,3,5-trichloro-2-methoxy benzene produced by different Streptomyces isolates based on GC-MS peak intensities. (G) Nonlinear relationship between fungal growth inhibition and abundance of 1,3,5-trichloro-2-methoxy benzene.
To further determine if the antifungal VOC 1,3,5-trichloro-2-methoxy benzene is typically found for Streptomyces isolates that inhibit hyphal growth of R. solani, we determined the relative amounts of this VOC produced by each of the 12 Streptomyces isolates tested in this study. The results show that production of this VOC is widespread among the 12 Streptomyces isolates. Moreover, a positive nonlinear correlation was found between the percentage of hyphal growth inhibition and the abundance (peak intensity) of 1,3,5-trichloro-2-methoxy benzene detected for the 12 isolates (Figures 7F,G).
Discussion
The production of VOCs by microorganisms is known for several decades. Only recently an increasing number of studies reported on the chemical diversity and possible functions of this group of microbial compounds (Schmidt et al., 2015). In comparison to plant VOCs, knowledge about the natural functions of microbial VOCs is still limited (Bitas et al., 2013). Here we studied the diversity and activities of VOCs produced by different streptomycetes from a Rhizoctonia-suppressive soil.
VOC profiling has been extensively used for food flavoring and aroma as well as indicators of fungal growth in buildings and in post-harvest management (Morath et al., 2012). More recently, VOC chemotyping allowed not only to identify species- and strain-specific VOCs but also to study soil microbial activity and shifts in microbial community compositions (McNeal and Herbert, 2009; Müller et al., 2013; Trefz et al., 2013). We showed that VOC profiling can be used for chemotyping different streptomycetes. Most of the 381 VOCs detected for the different streptomycetes from the Rhizoctonia-suppressive soil were found to be specific for some isolates whereas fewer VOCs were found to be commonly produced by all isolates. The best known VOCs from streptomycetes are 2-methylisoborneol (MIB) and trans-1,10-dimethyl-trans-9-decalol (geosmin) which are responsible for the characteristic musty or earthy smell of moist soils (Gerber, 1968; Jiang et al., 2007). Our results also show that these VOCs are widely produced by Streptomyces isolates from the rhizosphere of sugar beet plants grown in Rhizoctonia-suppressive soil. Geosmin was detected for all isolates, whereas MIB was detected for eight isolates. Members of the Streptomyces genus differ greatly in their morphology, physiology, and biochemical characteristics (Anderson and Wellington, 2001). Taxonomic delineation of this genus remains complex and leads to over- or under-classified groups. Current approaches for classification of Streptomyces as well as other prokaryotes rely on genetic and phenotypic traits, mainly on 16S rRNA gene sequences. This molecular marker, however, is not always sufficient to discriminate between closely related species and between strains of a given species (Girard et al., 2013). We showed that concatenation of atpD, recA, and 16S rRNA gene sequences displayed a better phylogenetic delineation of the different streptomycetes than 16S rRNA gene sequences alone, although closely related isolates could not be distinguished. We revealed that VOC profiling allowed discrimination of Streptomyces isolates that are phylogenetically close but phenotypically different, such as Streptomyces strains W75.5/W126 and W47/W214.
The genus Streptomyces is well-known for the production of several antifungal and antiviral compounds and accounts for 80% of the currently available antibiotic compounds (Watve et al., 2001). Streptomyces also produces VOCs which reduce the incidence and/or the severity of several plant diseases caused by fungi and cause morphological abnormalities in different fungi (Moore-Landecker and Stotzky, 1973; Wan et al., 2008; Boukaew et al., 2013; Wang et al., 2013; Wu et al., 2015). VOCs produced by the streptomycetes tested here exhibited antifungal and plant growth promoting properties. Several isolates inhibited hyphal growth, with Streptomyces strains W47 and W214 showing the strongest inhibitory effect. Given that these streptomycetes were obtained from a Rhizoctonia-suppressive soil suggests that VOCs may contribute to disease suppressiveness. This suggestion needs to be further investigated in situ but fits well with one of the initial hypotheses of Lockwood (Lockwood, 1977) for the potential role of microbial VOCs in soil fungistasis. To provide more conclusive proof of the role of these Streptomyces VOCs in disease suppression in the soil ecosystem, specific soil bioassays are needed where the VOC producers and the pathogen are physically separated. However, there are several technical limitations to accomplish this. First, the strains used here are rhizospheric bacteria that need to be positioned in their ecological context (the rhizosphere) to provide meaningful results. Given the need for the localization of the Streptomyces strains in the rhizosphere where also the pathogen colonizes and infects, it has not been possible yet to physically separate the Streptomyces strains from the fungal pathogen. This is due in part to the prolific growth of this particular fungus. The physical separation in situ is needed to exclude a possible role of mechanisms other than VOCs. An alternative approach would be to generate site-directed mutants of the Streptomyces strains that do not produce one or more of the specific VOCs identified in this study. Comparison of the activity of these mutants with their wildtype strains would then more conclusively resolve the role of specific VOCs in disease suppression in situ. For this alternative approach, however, we have not yet been able to generate mutants as many environmental Streptomyces species/strains are not or very difficult to access for genetic modification.
Several studies have described antifungal activity by bacterial VOCs, however, few have identified single or blends of VOCs responsible for the antifungal activity (Kai et al., 2007; Wang et al., 2013). For Pseudomonas, six VOCs (cyclohexanal, decanal, 2-ethyl 1-hexanol, nonanal, benzothiazole, and dimethyl trisulfide) were found to inhibit mycelial growth and sclerotial germination of Sclerotinia sclerotiorum at tested volumes of 100 and 150 μl (Fernando et al., 2005). Regarding VOCs produced by Streptomyces species, butanone (methyl vinyl ketone) and dimethyl disulfide were described to inhibit the spore germination in Cladosporium cladosporioides and mycelial growth of Fusarium moniliforme, respectively (Herrington et al., 1987; Wang et al., 2013). Here we showed that two out of five VOCs detected for Streptomyces strains W47 and W214 (methyl 2-methylpentanoate and 1,3,5-trichloro-2-methoxy benzene) as well as the mix of these VOCs exhibited antifungal activity, albeit at high concentrations. The VOC 1,3,5-trichloro-2-methoxy benzene completely inhibited fungal growth and caused melanization of the fungal hyphae. 1,3,5-Trichloro-2-methoxy benzene is also known as 2,4,6-trichloroanisole (TCA) and causes off-flavor in wine, coffee and water (Spadone et al., 1990; Jensen et al., 1994). Anisole produced by S. albulus has recently been described for activity against S. sclerotiorum and F. oxysporum (Wu et al., 2015). Derivatives of anisole have been described to be produced by bacteria and fungi (Mauriello et al., 2004; Blom et al., 2011), but no function has been ascribed to this specific VOC yet. To our knowledge, this is the first time that 1,3,5-trichloro-2-methoxy benzene is described for its antifungal activity. The VOC methyl 2-methylpentanoate, which also exhibited antifungal activity, is known for other streptomycetes, but also for this VOC no specific function has been described so far (Wilkins and Scholler, 2009; Dickschat et al., 2011). For both 1,3,5-trichloro-2-methoxy benzene and methyl 2-methylpentanoate, the concentrations needed to inhibit fungal growth were high. However, in the experimental setup used here, we do not know how much of the applied VOCs actually contact the fungal hyphae, which part of the fungal hyphae are the most VOC sensitive and how long VOC exposure is necessary to exert the antifungal activity. These aspects will be subject of future studies. Also, the identification of Streptomyces VOCs involved in plant growth promotion was not further pursued in this study but a possible candidate is acetoin (3-hydroxy-2-butanone) which was detected for several isolates tested here. Acetoin and 2,3-butanediol were the first bacterial VOCs described for their role in plant growth promotion (Ryu et al., 2003). More recently, other VOCs have been identified for their role in plant growth promotion such as indole, 1-hexanol, pentadecane, 13-tetradecadien-1-ol, 2-butanone, and 2-methyl-n-1-tridecene (Blom et al., 2011; Park et al., 2015). Plant growth-promoting effects can also be, at least partially, due to CO2 accumulation as products of microbial metabolism when using closed Petri dishes (Kai and Piechulla, 2009). In the experimental set-up used in our study, however, CO2 appears to have only a minor role since two isolates (3A41 and W214) out of the 12 tested isolates did not promote shoot and root growth, and two isolates (3A18 and W75.5) did not promote root growth.
In conclusion, VOCs produced by rhizosphere-associated streptomycetes are chemically diverse and display antifungal and plant growth-promoting properties. Hence, VOC profiling can provide a new resource of novel metabolites and biochemical pathways involved in antifungal activity and plant growth promotion by streptomycetes. We identified two VOCs with antifungal activity, but it remains to be determined whether these compounds are produced in situ at the biologically relevant concentrations. Our work further demonstrated the utility of VOC profiling for the characterization of streptomycetes, providing an additional tool for phylogenetic delineation of closely related strains.
Author Contributions
VC designed and performed the experiments and drafted the manuscript. GV and HZ assisted with the isolation of the Actinobacteria. VJC assisted with the molecular characterization of the Streptomyces isolates. VC, RM, and DE analyzed the GC-MS data. JR supervised the work and assisted with the experimental design and writing. All authors revised the manuscript and approved submission.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Jacques Davies for assistance with the GC-MS. This manuscript is publication number 5937 of Netherlands Institute of Ecology (NIOO-KNAW). The authors also acknowledge funding support from the Netherlands Organization for Scientific Research (NWO) and the Consortium of Improved Plant Yield which is part of the Netherlands Genomics Initiative (NGI).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.01081
References
Anderson, A. S., and Wellington, E. M. H. (2001). The taxonomy of Streptomyces and related genera. Int. J. Syst. Evol. Microbiol. 51, 797–814. doi: 10.1099/00207713-51-3-797
Bailly, A., and Weisskopf, L. (2012). The modulating effect of bacterial volatiles on plant growth: current knowledge and future challenges. Plant Signal. Behav. 7, 79–85. doi: 10.4161/psb.7.1.18418
Bitas, V., Kim, H. S., Bennett, J. W., and Kang, S. (2013). Sniffing on microbes: diverse roles of microbial volatile organic compounds in plant health. Mol. Plant Microbe Interact. 26, 835–843. doi: 10.1094/MPMI-10-12-0249-CR
Blom, D., Fabbri, C., Connor, E. C., Schiestl, F. P., Klauser, D. R., Boller, T., et al. (2011). Production of plant growth modulating volatiles is widespread among rhizosphere bacteria and strongly depends on culture conditions. Environ. Microbiol. 13, 3047–3058. doi: 10.1111/j.1462-2920.2011.02582.x
Boukaew, S., Plubrukam, A., and Prasertsan, P. (2013). Effect of volatile substances from Streptomyces philanthi RM-1-138 on growth of Rhizoctonia solani on rice leaf. BioControl 58, 471–482. doi: 10.1007/s10526-013-9510-6
Chapelle, E., Mendes, R., Bakker, P. A., and Raaijmakers, J. M. (2015). Fungal invasion of the rhizosphere microbiome. ISME J. doi: 10.1038/ismej.2015.82. [Epub ahead of print].
Claessen, D., Rozen, D. E., Kuipers, O. P., Søgaard-Andersen, L., and van Wezel, G. P. (2014). Bacterial solutions to multicellularity: a tale of biofilms, filaments and fruiting bodies. Nat. Rev. Microbiol. 12, 115–124. doi: 10.1038/nrmicro3178
Cruz-Morales, P., Vijgenboom, E., Iruegas-Bocardo, F., Girard, G., Yáńez-Guerra, L. A., Ramos-Aboites, H. E., et al. (2013). The genome sequence of Streptomyces lividans 66 reveals a novel tRNA-dependent peptide biosynthetic system within a metal-related genomic island. Genome Biol. Evol. 5, 1165–1175. doi: 10.1093/gbe/evt082
Deangelis, K. M., Brodie, E. L., Desantis, T. Z., Andersen, G. L., Lindow, S. E., and Firestone, M. K. (2009). Selective progressive response of soil microbial community to wild oat roots. ISME J. 3, 168–178. doi: 10.1038/ismej.2008.103
Dickschat, J. S., Bruns, H., and Riclea, R. (2011). Novel fatty acid methyl esters from the actinomycete Micromonospora aurantiaca. Beilstein J. Org. Chem. 7, 1697–1712. doi: 10.3762/bjoc.7.200
Effmert, U., Kalderás, J., Warnke, R., and Piechulla, B. (2012). Volatile mediated interactions between bacteria and fungi in the soil. J. Chem. Ecol. 38, 665–703. doi: 10.1007/s10886-012-0135-5
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791.
Fernando, W. G. D., Ramarathnam, R., Krishnamoorthy, A. S., and Savchuk, S. C. (2005). Identification and use of potential bacterial organic antifungal volatiles in biocontrol. Soil Biol. Biochem. 37, 955–964. doi: 10.1016/j.soilbio.2004.10.021
Garbeva, P., Hordijk, C., Gerards, S., and de Boer, W. (2014). Volatiles produced by the mycophagous soil bacterium Collimonas. FEMS Microbiol. Ecol. 87, 639–649. doi: 10.1111/1574-6941.12252
Gerber, N. N. (1968). Geosmin, from microorganisms, is -1, 10-dimethyl-9-decalol. Tetrahedron Lett. 25, 2971–2974. doi: 10.1016/S0040-4039(00)89625-2
Girard, G., Traag, B. A., Sangal, V., Mascini, N., Hoskisson, P. A., Goodfellow, M., et al. (2013). A novel taxonomic marker that discriminates between morphologically complex actinomycetes. Open Biol. 3:130073. doi: 10.1098/rsob.130073
Goodfellow, M. (2012). “Phylum XXVI. Actinobacteria phyl. nov.,” in Bergey's Manual of Systematic Bacteriology, 2nd Edn., eds M. Goodfellow, P. Kämpfer, H.-J. Busse, M. E. Trujillo, K.-I. Suzuki, W. Ludwig, and W. B. Whitman (New York, NY: Springer), 1–2083.
Guo, Y., Zheng, W., Rong, X., and Huang, Y. (2008). A multilocus phylogeny of the Streptomyces griseus 16S rRNA gene clade: use of multilocus sequence analysis for streptomycete systematics. Int. J. Syst. Evol. Microbiol. 58, 149–159. doi: 10.1099/ijs.0.65224-0
Hagai, E., Dvora, R., Havkin-Blank, T., Zelinger, E., Porat, Z., Schulz, S., et al. (2014). Surface-motility induction, attraction and hitchhiking between bacterial species promote dispersal on solid surfaces. ISME J. 8, 1147–1151. doi: 10.1038/ismej.2013.218
Herrington, P. R., Crakz, J. T., and Sheridan, J. E. (1987). Methyl vinyl ketone: a volatile fungistatic inhibitor from Streptomyces griseoruber. Soil Bid. Biochem. 19, 509–512. doi: 10.1016/0038-0717(87)90092-7
Hopwood, D. A. (2007). Streptomyces in Nature and Medicine: The Antibiotic Makers. New York, NY: Oxford University Press.
Hopwood, D. A., Kieser, T., Wright, H. M., and Bibb, M. J. (1983). Plasmids, recombination and chromosome mapping in Streptomyces lividans 66. J. Gen. Microbiol. 129, 2257–2269. doi: 10.1099/00221287-129-7-2257
Hornby, D. (1983). Suppressive soils. Annu. Rev. Phytopatol. 21, 65–85. doi: 10.1146/annurev.py.21.090183.000433
Jensen, S. E., Goatcher, L. J., Perley, T., Kenefick, S., and Hrudey, S. E. (1994). Actinomycetes as a factor in odour problems affecting drinking water from the North Saskatchewan River. Water Res. 28, 1393–1401. doi: 10.1016/0043-1354(94)90306-9
Jiang, J., He, X., and Cane, D. E. (2007). Biosynthesis of the earthy odorant geosmin by a bifunctional Streptomyces coelicolor enzyme. Nat. Chem. Biol. 3, 711–715. doi: 10.1038/nchembio.2007.29
Kai, M., Effmert, U., Berg, G., and Piechulla, B. (2007). Volatiles of bacterial antagonists inhibit mycelial growth of the plant pathogen Rhizoctonia solani. Arch. Microbiol. 187, 351–360. doi: 10.1007/s00203-006-0199-0
Kai, M., Haustein, M., Molina, F., Petri, A., Scholz, B., and Piechulla, B. (2009). Bacterial volatiles and their action potential. Appl. Microbiol. Biotechnol. 81, 1001–1012. doi: 10.1007/s00253-008-1760-3
Kai, M., and Piechulla, B. (2009). Plant growth promotion due to rhizobacterial volatiles—an effect of CO2? FEBS Lett. 583, 3473–3477. doi: 10.1016/j.febslet.2009.09.053
Kim, Y.-C., Glick, B. R., Bashan, Y., and Ryu, C.-M. (2012). “Enhancement of plant drought tolerance by microbes,” in Plant Responses to Drought Stress, ed R. Aroca (Berlin; Heidelberg: Springer-Verlag), 383–413.
Labeda, D. P., Goodfellow, M., Brown, R., Ward, A. C., Lanoot, B., Vanncanneyt, M., et al. (2012). Phylogenetic study of the species within the family Streptomycetaceae. Antonie Van Leeuwenhoek 101, 73–104. doi: 10.1007/s10482-011-9656-0
Letunic, I., and Bork, P. (2011). Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 39, W475–W478. doi: 10.1093/nar/gkr201
Lockwood, J. L. (1977). Fungistasis in soils. Biol. Rev. 52, 1–43. doi: 10.1111/j.1469-185X.1977.tb01344.x
Lommen, A., and Kools, H. J. (2012). MetAlign 3.0: performance enhancement by efficient use of advances in computer hardware. Metabolomics 8, 719–726. doi: 10.1007/s11306-011-0369-1
Mauriello, G., Marino, R., D'Auria, M., Cerone, G., and Rana, G. L. (2004). Determination of volatile organic compounds from truffles via SPME–GC–MS. J. Chromatogr. Sci. 42, 299–305. doi: 10.1093/chromsci/42.6.299
McDonald, D., Price, M. N., Goodrich, J., Nawrocki, E. P., Desantis, T. Z., Probst, A., et al. (2012). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6, 610–618. doi: 10.1038/ismej.2011.139
McNeal, K. S., and Herbert, B. E. (2009). Volatile organic metabolites as indicators of soil microbial activity and community composition shifts. Soil Sci. Am. J. 73, 579. doi: 10.2136/sssaj2007.0245
Mendes, R., Kruijt, M., de Bruijn, I., Dekkers, E., van der Voort, M., Schneider, J. H., et al. (2011). Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332, 1097–1100. doi: 10.1126/science.1203980
Moore-Landecker, E., and Stotzky, G. (1973). Morphological abnormalities of fungi induced by volatile microbial metabolites. Mycologia 65, 519–530. doi: 10.2307/3758256
Morath, S. U., Hung, R., and Bennett, J. W. (2012). Fungal volatile organic compounds: a review with emphasis on their biotechnological potential. Fungal Biol. Rev. 26, 73–83. doi: 10.1016/j.fbr.2012.07.001
Müller, A., Faubert, P., Hagen, M., Zu Castell, W., Polle, A., Schnitzler, J. P., et al. (2013). Volatile profiles of fungi—chemotyping of species and ecological functions. Fungal Genet. Biol. 54, 25–33. doi: 10.1016/j.fgb.2013.02.005
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Park, Y. S., Dutta, S., Ann, M., Raaijmakers, J. M., and Park, K. (2015). Promotion of plant growth by Pseudomonas fluorescens strain SS101 via novel volatile organic compounds. Biochem. Biophys. Res. Commun. 461, 361–365. doi: 10.1016/j.bbrc.2015.04.039
Piechulla, B., and Degenhardt, J. (2014). The emerging importance of microbial volatile organic compounds. Plant Cell Environ. 37, 811–812. doi: 10.1111/pce.12254
Ryu, C. M., Farag, M. A., Hu, C. H., Reddy, M. S., Kloepper, J. W., and Pare, P. W. (2004). Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol. 134, 1017–1026. doi: 10.1104/pp.103.026583
Ryu, C. M., Farag, M. A., Hu, C. H., Reddy, M. S., Wei, H. X., Pare, P. W., et al. (2003). Bacterial volatiles promote growth in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 100, 4927–4932. doi: 10.1073/pnas.0730845100
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Schmidt, R., Cordovez, V., de Boer, W., Raaijmakers, J., and Garbeva, P. (2015). Volatile affairs in microbial interactions. ISME J. doi: 10.1038/ismej.2015.42. [Epub ahead of print].
Schulz, S., and Dickschat, J. S. (2007). Bacterial volatiles: the smell of small organisms. Nat. Prod. Rep. 24, 814–842. doi: 10.1039/b507392h
Spadone, J. C., Takeoka, G., and Liardon, R. (1990). Analytical investigation of rio off-flavor in green coffee. J. Agric. Food Chem. 38, 226–233. doi: 10.1021/jf00091a050
Stotzky, G., and Schenck, S. (1976). Volatile organic compounds and microorganisms. CRC Crit. Rev. Microbiol. 4, 333–382. doi: 10.3109/10408417609102303
Strehmel, N., Hummel, J., Erban, A., Strassburg, K., and Kopka, J. (2008). Retention index thresholds for compound matching in GC-MS metabolite profiling. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 871, 182–190. doi: 10.1016/j.jchromb.2008.04.042
Tamura, K., and Nei, M. (1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10, 1073–1095.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tikunov, Y. M., Laptenok, S., Hall, R. D., Bovy, A., and de Vos, R. C. (2012). MSClust: a tool for unsupervised mass spectra extraction of chromatography-mass spectrometry ion-wise aligned data. Metabolomics 8, 714–718. doi: 10.1007/s11306-011-0368-2
Trefz, P., Koehler, H., Klepik, K., Moebius, P., Reinhold, P., Schubert, J. K., et al. (2013). Volatile emissions from Mycobacterium avium subsp. paratuberculosis mirror bacterial growth and enable distinction of different strains. PLoS ONE 8:e76868. doi: 10.1371/journal.pone.0076868
van de Mortel, J. E., de Vos, R. C., Dekkers, E., Pineda, A., Guillod, L., Bouwmeester, K., et al. (2012). Metabolic and transcriptomic changes induced in Arabidopsis by the rhizobacterium Pseudomonas fluorescens SS101. Plant Physiol. 160, 2173–2188. doi: 10.1104/pp.112.207324
van Wezel, G. P., McKenzie, N. L., and Nodwell, J. R. (2009). Chapter 5. Applying the genetics of secondary metabolism in model actinomycetes to the discovery of new antibiotics. Methods Enzymol. 458, 117–141. doi: 10.1016/S0076-6879(09)04805-8
Verhulst, N. O., Beijleveld, H., Knols, B. G., Takken, W., Schraa, G., Bouwmeester, H. J., et al. (2009). Cultured skin microbiota attracts malaria mosquitoes. Malar. J. 8:302. doi: 10.1186/1475-2875-8-302
Vespermann, A., Kai, M., and Piechulla, B. (2007). Rhizobacterial volatiles affect the growth of fungi and Arabidopsis thaliana. Appl. Environ. Microbiol. 73, 5639–5641. doi: 10.1128/AEM.01078-07
Wan, M., Li, G., Zhang, J., Jiang, D., and Huang, H.-C. (2008). Effect of volatile substances of Streptomyces platensis F-1 on control of plant fungal diseases. Biol. Control 46, 552–559. doi: 10.1016/j.biocontrol.2008.05.015
Wang, Z., Wang, C., Li, F., Li, Z., Chen, M., Wang, Y., et al. (2013). Fumigant activity of volatiles from Streptomyces alboflavus TD-1 against Fusarium moniliforme Sheldon. J. Microbiol. 51, 477–483. doi: 10.1007/s12275-013-2586-y
Watve, M. G., Tickoo, R., Jog, M. M., and Bhole, B. D. (2001). How many antibiotics are produced by the genus Streptomyces? Arch. Microbiol. 176, 386–390. doi: 10.1007/s002030100345
Weller, D. M., Raaijmakers, J. M., Gardener, B. B., and Thomashow, L. S. (2002). Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu. Rev. Phytopathol. 40, 309–348. doi: 10.1146/annurev.phyto.40.030402.110010
Wilkins, K., and Scholler, C. (2009). Volatalie organic metabolites from selected streptomyces strains. Actinomycetologica 23, 27–33. doi: 10.3209/saj.SAJ230202
Wu, Y., Yuan, J., E, Y., Raza, W., Shen, Q., and Huang, Q. (2015). Effects of volatile organic compounds from Streptomyces albulus NJZJSA2 on growth of two fungal pathogens. J. Basic Microbiol. 55, 1104–1117. doi: 10.1002/jobm.201400906
Keywords: Actinobacteria, SPME-GC-MS, antifungal activity, plant growth promotion, suppressive soil
Citation: Cordovez V, Carrion VJ, Etalo DW, Mumm R, Zhu H, van Wezel GP and Raaijmakers JM (2015) Diversity and functions of volatile organic compounds produced by Streptomyces from a disease-suppressive soil. Front. Microbiol. 6:1081. doi: 10.3389/fmicb.2015.01081
Received: 27 July 2015; Accepted: 22 September 2015;
Published: 09 October 2015.
Edited by:
Stéphane Hacquard, Max Planck Institute for Plant Breeding Research, GermanyReviewed by:
Mika Tapio Tarkka, Helmholtz Centre for Environmental Research - UFZ, GermanyTomislav Cernava, Graz University of Technology, Austria
Copyright © 2015 Cordovez, Carrion, Etalo, Mumm, Zhu, van Wezel and Raaijmakers. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jos M. Raaijmakers, Department of Microbial Ecology, Netherlands Institute of Ecology (NIOO-KNAW), Droevendaalsesteeg 10, 6708 PB Wageningen, Netherlands,ai5yYWFpam1ha2Vyc0BuaW9vLmtuYXcubmw=
 Viviane Cordovez
Viviane Cordovez Victor J. Carrion
Victor J. Carrion Desalegn W. Etalo
Desalegn W. Etalo Roland Mumm
Roland Mumm Hua Zhu5
Hua Zhu5 Gilles P. van Wezel
Gilles P. van Wezel Jos M. Raaijmakers
Jos M. Raaijmakers