
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol., 08 September 2015
Sec. Plant Pathogen Interactions
Volume 6 - 2015 | https://doi.org/10.3389/fmicb.2015.00945
This article is part of the Research TopicSignaling in the phytomicrobiomeView all 12 articles
Although signal exchange between legumes and their rhizobia is among the best-known examples of this biological process, most of the more characterized data comes from just a few legume species and environmental stresses. Although a relative wealth of information is available for some model legumes and some of the major pulses such as soybean, little is known about tropical legumes. This relative disparity in current knowledge is also apparent in the research on the effects of environmental stress on signal exchange; cool-climate stresses, such as low-soil temperature, comprise a relatively large body of research, whereas high-temperature stresses and drought are not nearly as well understood. Both tropical legumes and their environmental stress-induced effects are increasingly important due to global population growth (the demand for protein), climate change (increasing temperatures and more extreme climate behavior), and urbanization (and thus heavy metals). This knowledge gap for both legumes and their environmental stresses is compounded because whereas most temperate legume-rhizobia symbioses are relatively specific and cultivated under relatively stable environments, the converse is true for tropical legumes, which tend to be promiscuous, and grow in highly variable conditions. This review will clarify some of this missing information and highlight fields in which further research would benefit our current knowledge.
Biological nitrogen fixation is one of the main biological cycles worldwide (Canfield et al., 2010) and is estimated to contribute close to half (Herder et al., 2010) of the world’s biologically available nitrogen. Most of that fixed nitrogen comes from the legume-rhizobia symbiosis, which is based on a very large and constantly changing group of bacteria generically called rhizobia, including Allorhizobium, Aminobacter, Azorhizobium, Bradyrhizobium, Devosia, Ensifer (Sinorhizobium), Mesorhizobium, Methylobacterium, Microvirga, Ochrobactrum, Phyllobacterium, Rhizobium, and Shinella among the α-Proteobacteria; Burkholderia, Cupriavidus, and Herbaspirillum among the β-Proteobacteria (Vinuesa, 2015); and at least one Pseudomonas sp. from the γ-Proteobacteria (Shiraishi et al., 2010). This usage of rhizobia as a catch-all name has been challenged recently because it was based initially on the Rhizobium genus (then the Rhizobiaceae family), whereas we now know that at least three classes of the Proteobacteria include at least one genus with this capability. In contrast, this well-recognized term has been used extensively and, as such, is used throughout this review.
This symbiosis begins with an elaborate signal exchange process that is among the best studied between bacteria and plants (Hirsch and Fujishige, 2012). Initially, the legume root releases exudate compounds such as sugars, amino acids, several classes of proteins classes (De-la-Peña et al., 2008, 2010; Badri and Vivanco, 2009; Badri et al., 2009), and flavonoids, and phenolic compounds (Broughton et al., 2003), such as flavone, flavonones, isoflavones, and betains (Cooper, 2007). These compounds induce chemoostatic reactions from the bacteria and act as nodulation gene inducers (Hirsch and Fujishige, 2012; Ryu et al., 2012).
These compounds may act as weak or strong inducers, whereas others are inhibitors or have no effect on nodulation (Mulligan and Long, 1985; Firmin et al., 1986; Peters et al., 1986; Redmond et al., 1986; Hartwig et al., 1989, 1990; Hungria et al., 1992; Bolaños-Vásquez and Werner, 1997; Begum et al., 2001; Mabood et al., 2006; Subramanian et al., 2007).
Which compounds, or class of compounds, induce nodulation the strongest varies among symbiotic pairs. For common beans (Phaseolus vulgaris), the strongest inducers are genistein-3-O-glucoside, eriodictyol, naringenin, daidzein, genistein, and coumesterol (Hungria et al., 1991a; Dakora et al., 1993b); this plant also releases other classes of compounds such as anthocyanidins, flavonols, isoflavonoids, and flavones (Hungria et al., 1992). For soybeans (Glycine max), the most effective plant-to-bacteria signal has been variously found to be an isoflavone (Subramanian et al., 2006), jasmonic acid and its derivatives (Mabood and Smith, 2005), or genistein (Zhang and Smith, 1995).
After the nodulation genes are activated, the rhizobia release nod factors, lipochitooligosaccharides specific to each symbiotic association that are sufficient to activate nodule organogenesis at least under some conditions, and these factors may induce cellular modifications associated with early rhizobial root infection (Oldroyd and Downie, 2004; Cooper, 2007; Jones et al., 2007). In addition to the nod factors, several other bacterial compounds affect several stages of the interaction, including exopolysaccharides (EPS), lipopolysaccharides, K-antigen polysaccharides, cyclic β-glucan, high-molecular-weight neutral polysaccharides (glucomannan), and gel-forming polysaccharides (Fraysse et al., 2003; Laus et al., 2006; Downie, 2010; Janczarek, 2011).
The complex signal exchange between plant and bacterial partners in symbiosis is also a key component of symbiotic specificity, which varies from highly specific to highly promiscuous. For example, although Sinorhizobium sp. NGR234 nodulates 232 legume species from 112 distantly related genera, with varying efficacy, some strains of Rhizobium leguminosarum bv. viciae do not nodulate pea (Pisum sativum) cultivars from different origins (Ovtsyna et al., 1998; Masson-Boivin et al., 2009).
The lack of effective signal exchange between legumes and bacteria precludes symbiosis establishment for incompatible partners, but in some situations, nodules may be formed in which the rhizobia do not enter, are not liberated from the infection thread, or do not fix nitrogen (Miller et al., 2007). This lack of recognition may occur even after the initial signal exchange. For example, R. leguminosarum bv. trifolii (Rlt) strain ICC105 does not fix nitrogen with white clover (Trifolium repens), whereas this strain is effective when paired with Caucasian clover (T. ambiguum). According to Miller et al. (2007), this difference is due to a region between the nifH gene and the fixA promoter that is differentially activated when in symbiosis with the two Trifolium species. It is not clear if this difference is due to positive or negative regulation by a specific plant signal, nor is it clear how NifA activity is regulated (Miller et al., 2007).
The combination of a vast range of compounds secreted by both plants and bacteria is one of the main characteristics of this symbiotic compatibility. Because the first step is exudation by the plant, this step may be considered the most important one. These exudates are continuously secreted into the rhizosphere, but both the number and concentration of these compounds increases when compatible bacteria are detected by the plant (Zaat et al., 1989; Dakora et al., 1993a,b; Hassan and Mathesius, 2012).
These plant-bacteria signals activate three main groups of nodulation genes in the bacteria: the common nodABC genes that are present in almost all rhizobia (the exception being some photosynthetic bradyrhizobia and some Burkholderia, Giraud et al., 2007) and are required to produce the basic structure of the nod factors; host-specific nod genes that are linked to specific modifications of the basic nod factor structure that allows for symbiotic specificity, such as nodEF, nodG, nodH, nodPQ, and nodRL; and regulatory genes that are linked to the activation and transcription of both the common and specific nod genes (Horvath et al., 1986; Göttfert et al., 1990; Lerouge et al., 1990; Sanjuan et al., 1994; Moulin et al., 2001; Schlaman et al., 2006).
Nod factor perception is mediated by Nod factor receptors (NFRs), which are serine/threonine kinases that are located in the plasma membrane and that contain LysM motifs in their extracellular domains (Limpens et al., 2003; Madsen et al., 2003; Radutoiu et al., 2003; Arrighi et al., 2006). These NFRs correspond to the Nod factor structure and act as host determinants for symbiotic specificity. This specificity was shown by the transfer of Lj-NFR1 and Lj-NFR5 to Medicago truncatula, which enabled nodulation by the Lotus japonicus symbiont Mesorhizobium loti (Radutoiu et al., 2007); the specificity of two Lotus species is the function of a single amino acid residue in one of the LysM domains of Lj-NFR5 (Radutoiu et al., 2007).
In addition to Nod factors, some rhizobia secrete proteins that are involved in nodulation via a type III secretion system (T3SS; Fauvart and Michiels, 2008; Deakin and Broughton, 2009). These proteins, called nodulation outer proteins (Nops), are believed to contribute to legume immune response suppression or to modulate root cell cytoskeletal rearrangement during nodule development (Bartsev et al., 2004; Skorpil et al., 2005; Soto et al., 2009). The nopP and nopL genes are found in Rhizobium sp. NGR234, Sinorhizobium fredii and Bradyrhizobium japonicum and are absent in pathogenic bacteria (Deakin and Broughton, 2009). In Rhizobium sp. NGR234, these genes are required for the nodulation of the tropical legumes Tephrosia vogelii and Flemingia congesta (Marie et al., 2003; Skorpil et al., 2005). Moreover, the nodulation of Vigna unguiculata by S. fredii is also affected by Nop proteins injected by S. fredii in a T3SS-dependent fashion (Schechter et al., 2010), but further studies on their effects on host specificity are still necessary.
Exopolysaccharides, bacterial cellular wall constituents, are also known to have important effects on symbiosis. For example, a defect on the EPS surface may induce failures both in the early and late stages of symbiosis, such as those observed in strains of S. meliloti presenting normal nodules in some ecotypes of M. truncatula but defective nodules in others, and this pattern may be transferred by a change in the EPS biosynthesis locus (Simsek et al., 2007). Because M. loti EPS mutants result in nonfunctional nodules in L. leucocephala but functional ones in L. pedunculatus, the EPS surface has also been linked to specificity in the nitrogen fixing phase (Hotter and Scott, 1991), as demonstrated by a B. japonicum exoB mutant fixing nitrogen in G. max but not in G. soja (Parniske et al., 1994) or some R. leguminosarum LPS mutants fixing nitrogen in peas (Pisum sativum), whereas other mutants do not (Kannenberg et al., 1992).
One point that deserves attention is the almost complete lack of literature on this signal exchange in tropical legumes, which are typically more promiscuous than temperate ones. Because of this knowledge gap, it is not known how the degree of promiscuity of a legume affects the signal exchange process because with the exception of Phaseolus, the best-studied legumes are all generally considered to nodulate with a few species or genera at the most (Michiels et al., 1998; Martínez-Romero, 2003; Rodríguez-Navarro et al., 2011; Rufini et al., 2013). A synthesis of a large portion of the literature identifying seed or root exudate compounds with known nod-gene activating properties (Table 1) indicates that more promiscuous (or less-selective) legumes may exhibit a broader range of these compounds, as per a comparison between P. vulgaris and G. max, which are less and more selective, respectively, for the rhizobial partner of the symbiosis. In contrast, the only paper we could find on V. unguiculata identifies only three compounds, although it has a very broad range of rhizobial partners. One further puzzle is that genistein is a known inducer for G. max, P. vulgaris, and V. unguiculata, although the rhizobia of these three species are not identical.
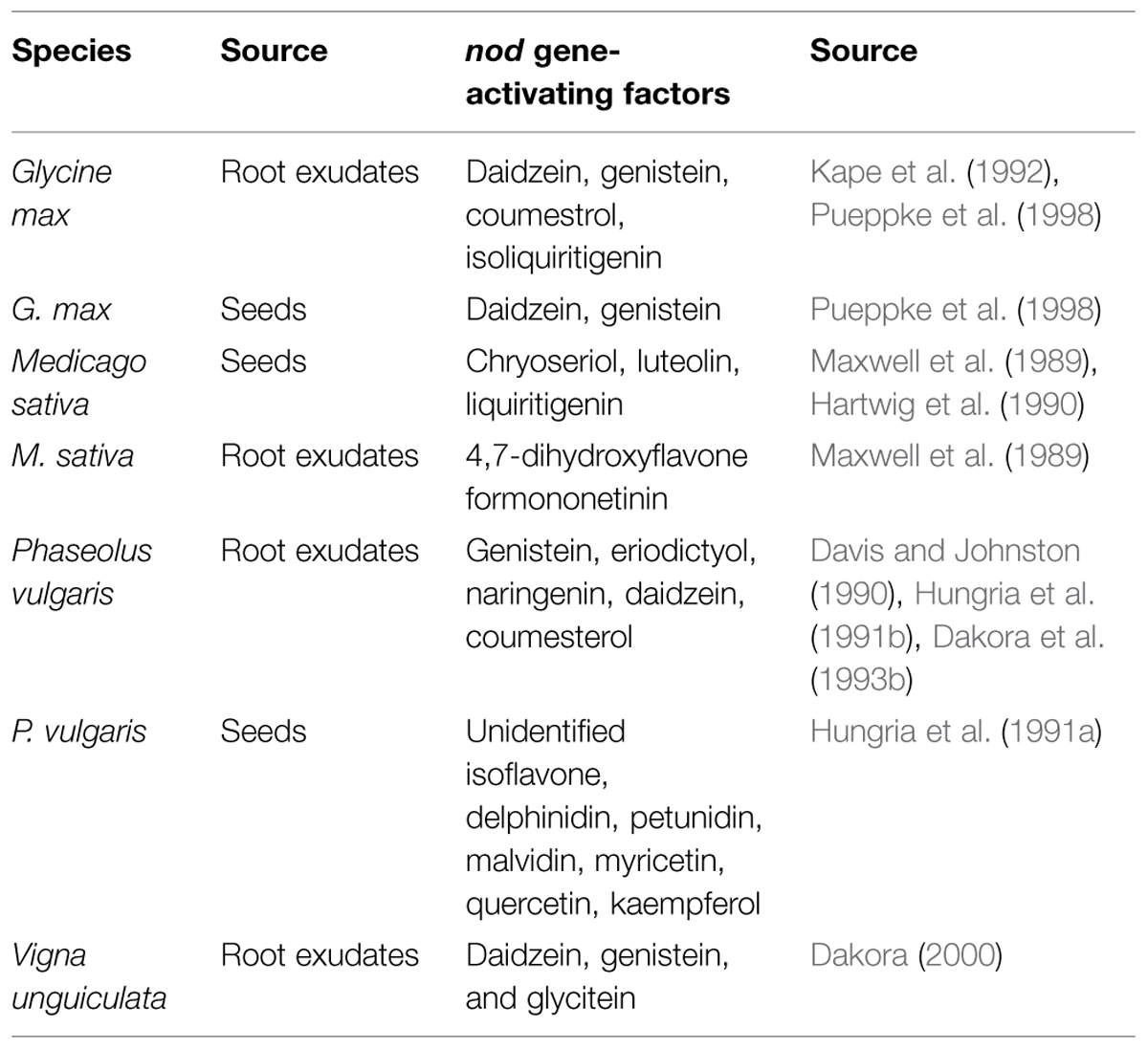
TABLE 1. Seed and root exudate compounds with known nod gene-activating factors, from legumes with broad or narrow ranges of symbiotic compatibility.
A lack of depth in the literature on this topic leads to ambiguity in how to relate legume promiscuity (or specificiticity) with the signal exchange process, although this relationship is expected to exist due to the specific nature of this exchange. Thus, this relationship might be an interesting line of future research; a better understanding of this relationship may lead to biotechnological approaches to enhance or reduce the compatibility profile of a given legume similarly, to soybean breeding for broad bacterial compatibility in Africa (Gwata et al., 2005).
Although the interaction between environmental stresses and legume-rhizobia signal exchange has been investigated, as will be discussed, these studies have also centered on temperate climate pulses, and their stresses. Much work is still needed to understand how the signal exchange process of other legumes is affected by their more typical stresses.
Much research has examined low root zone temperatures and their effects on signal exchange and nodulation, particularly in soybeans, but little is known about the effects of high root zone temperatures.
Low root zone temperatures inhibit the synthesis and secretion of plant-to-bacteria signals, as shown in G. max, in which the root exudation of genistein is strongly reduced below 17.5°C (Zhang and Smith, 1994, 1996a; Zhang et al., 1995; Pan and Smith, 1998). Low root zone temperatures also reduce nod factor synthesis and/or excretion in R. leguminosarum bv. trifolii (McKay and Djordjevic, 1993) and B. japonicum (Zhang et al., 2002). The molecular basis of this effect indicates that the T3SS gene cluster was progressively activated as temperatures increased, whereas the nod genes were rapidly induced at 15°C (Wei et al., 2010). Genistein has been proposed to induce this gene cluster through a regulatory cascade involving NodD1 and NodW (Krause et al., 2002).
These signal exchange effects combine to delay nodulation onset (Pan and Smith, 1998) and reduce the nodule growth rate, leading to smaller nodules (Lira Junior et al., 2005). Further confirmation that these stresses are directly linked to signal exchange is that the exogenous application of genistein is sufficient to mitigate a delay in nodulation under environmental conditions in which the root system temperature is below this threshold and the shoot is above it (Zhang and Smith, 1995, 1997; Pan et al., 1997). This mitigation is stronger for lower soil temperatures or stronger stresses (Zhang and Smith, 1996b).
Although salinity is known to affect Nod factor production by R. tropici CIAT 899 in the presence of apigenin (Estévez et al., 2009), there are indications that high salt concentrations may induce nod genes even in the absence of flavonoid inducers (Guasch-Vidal et al., 2012).
However, increased salinity reduces Nod factor production by S. arboris, which nodulates Acacia and Prosopis, both of which are legume trees tolerant to salt stresses (Penttinen et al., 2013). Similar effects were found for R. tropici and R. etli, which nodulate P. vulgaris (Dardanelli et al., 2012).
Similarly, to what is observed at low soil temperatures, as previously described, some of the salinity effects may be reduced if the bacteria are pre-incubated with their respective legume signals, such as genistein for B. japonicum (Miransari and Smith, 2009) or hesperetin and apigenin for R. tibeticum (Abd-Alla et al., 2013).
Soil pH affects symbiosis in several ways, including signal exchange (Hungria and Vargas, 2000). For example, both G. max and P. vulgaris isoflavonoid exudation from roots were reduced when the pH was lowered from 5.8 to 4.5 (Hungria and Stacey, 1997), and some nodulation genes, including nodA, are inactivated by reducing the pH in R. leguminosarum bv. trifolii (Richardson et al., 1988a,b). The production and excretion of Nod factors were also reduced in acidic soils (McKay and Djordjevic, 1993).
Another effect is a change in the profile of the Nod factors secreted by R. tropici CIAT 899, which is tolerant to acid conditions. A total of 52 different molecules were produced under an acidic pH and 29 at a neutral pH; only 15 are common to both conditions (Moron et al., 2005). This phenomenon might be linked to the reduction in nodC expression by the Arachis hypogaea bacterial symbionts under acidic conditions (Angelini et al., 2003).
In contrast to what is observed for low soil temperatures and salinity, the addition of flavonoids did not reduce the effects of low pH on acid-sensitive or acid-tolerant A. hypogaea (Angelini et al., 2003), which was apparently due to increased flavonoid uptake and toxicity.
Low pH also activates a systemic, shoot-controlled, and GmNARK-dependent (Nodulation Autoregulation Receptor Kinase) mechanism that negatively regulates initial nodule development in soybeans (Lin et al., 2012), as confirmed by the reduced expression of the GmENOD40b, GmNIN-2b, GmRIC1, GmRabA2, and cytochrome P450 genes, which are critical to early nodulation stages.
The legume-rhizobia symbiosis demands high levels of iron due to its inclusion in the compositions of leghemoglobin, nitrogenase, and cytochromes (Brear et al., 2013). Iron deficiency effects vary between legume species and may include altered nodule initiation, as seen in Lupinus angustifolius L. (Tang et al., 1990), or late development, as seen in peanuts (A. hypogaea), common beans (P. vulgaris), and soybeans (O’Hara et al., 1988; Soerensen et al., 1988; Slatni et al., 2011).
Iron absorption regulation by rhizobia in culture media has been extensively researched, and iron-responsive transcription regulators such as IrrA and RirA and the genes they control under iron deficiency and sufficiency have been determined (Viguier et al., 2005; Todd et al., 2006). Several of these genes encode siderophore production, heme biosynthesis, and transporters, such as the ferric siderophore ATP-binding cassette (ABC)-related genes.
Under iron-limiting conditions, free-living rhizobia express TonB-dependent receptors after activation by an iron regulator (Small et al., 2009), although bacteriod active siderophore transport is not necessary for symbiosis (Chang et al., 2007; Small et al., 2009). Mutations in ABC transporters, TonB-dependent receptors and TonB do not affect symbiosis establishment (Lynch et al., 2001; Nienaber et al., 2001), suggesting that bacteriods do not require a high affinity for siderophore absorption to obtain iron during symbiosis (Brear et al., 2013), although S. meliloti strains deficient in the siderophore absorption system exhibited lower nodule occupation rates under iron-deficient conditions than the corresponding wild types (Battistoni et al., 2002).
N2 fixation has a high energy cost, and P deficiency is an important restriction for legume production, particularly in the low-P soils of most tropical regions (Sulieman and Tran, 2015). Organic phosphates are the main source to sustain nodule symbiotic activities (Li et al., 2012), and several genes involved in recycling P are up-regulated under low-P conditions (Hernandez et al., 2009), particularly those encoding acid phosphatases (Maougal et al., 2014; Zhang et al., 2014).
Generally, the specific activity of acid phosphatases in nodules strongly increases when P supply is reduced in the growth medium but is stable when P supply is high (Araujo et al., 2008). The expression of several genes of the purple acid phosphatase GmPAP family was highly induced in soybean nodules under low-P availability (Li et al., 2012); the expression of phytate and phosphoenol pyruvate phosphatase was also increased in nodules under these conditions (Araujo et al., 2008; Bargaz et al., 2012). Acid phosphatases may have multiple functions, such as carbon metabolism, nodule permeability for O2 diffusion, and oxidative stress attenuation (Sulieman and Tran, 2015), which makes their study both more challenging and necessary.
The current literature lacks information on the effects of either drought or flooding on legume-rhizobia signal exchange, although both situations are well known to reduce nodulation and nitrogen fixation (Arayangkoon et al., 1990; Marcar et al., 1991; Purwantari et al., 1995; Hatimi, 1999). Thus, further research is necessary on this topic. Nodule formation ceases completely under sufficiently long or severe drought conditions, and nitrogenase and nodule respiratory activities are also strongly diminished in soybeans and common beans (Gerosa-Ramos et al., 2003). In alfalfa, such nitrogenase activity reduction has been linked to diminished bacteriod metabolic capability and oxidative damage to nodule cell components (Naya et al., 2007).
At the other extreme, several legumes are highly sensitive to water-logged conditions, with nodule development and function being more impaired than infection. Some of these effects, including nitrogenase activity, may be even stronger than observed for drought conditions. This phenomenon appears to be mostly linked to reduced O2 availability (Andres et al., 2012).
Although the literature contains little information on the effects of pesticides and heavy metals on signal exchange, some in vitro work with 30 different pesticides and environmental contaminants showed that S. meliloti NodD was affected, delaying nodulation, and reducing biological nitrogen fixation by M. sativa (Fox et al., 2001, 2004). M. sativa and G. max fungicide-treated seeds also exhibited reduced nod gene activity for their respective partners (Andrés et al., 1998).
More recently, it has been shown that R. alamii, an EPS producer, modulates its metabolism in response to cadmium (Schue et al., 2011) through the activation of biofilm formation, both in the wild type and in EPS-deficient mutants, which may reduce the effects of this heavy metal.
Although signal exchange between legumes and their bacterial symbionts is a well-studied process, much still needs to be clarified, particularly in relation to tropical legumes, which have been barely studied, and environmental effects other than low soil temperature.
Under at least some conditions, a delay in nodulation onset and, therefore, biological nitrogen fixation may be reduced by the exogenous supply of the appropriate legume signal. Because current predictions indicate a probable reduction in global agricultural season lengths, this phenomenon should receive increased attention.
Another field that deserves more attention is the study of signal exchange with non-traditional rhizobia, such as Burkholderia and Cupriavidus, and its effects on the plant host, for which no literature was found.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the Brazilian Funding Agencies CAPES, CNPq, and FACEPE for their funding through several different projects, including a Research Fellowship for the first author, a Post-Doctoral Fellowship for the second author, and joint research funding for all three authors.
Abd-Alla, M. H., El-Enany, A.-W. E., Bagy, M. K., and Bashandy, S. R. (2013). Alleviating the inhibitory effect of salinity stress on nod gene expression in Rhizobium tibeticum – fenugreek (Trigonella foenum graecum) symbiosis by isoflavonoids treatment. J. Plant Int. 9, 275–284. doi: 10.1080/17429145.2013.824622
Andrés, J. A., Correa, N. S., and Rosas, S. B. (1998). Alfalfa and soybean seed and root exudates treated with thiram inhibit the expression of rhizobia nodulation genes. Phyton Int. J. Exp. Bot. 62, 47–53.
Andres, J. A., Rovera, M., Guiñazú, L. B., Pastor, N. A., and Rosas, S. B. (2012). “Interactions between legumes and rhizobia under stress conditions,” in Bacteria in Agrobiology: Stress Management, ed. K. M. Dinesh (Berlin: Springer-Verlag), 77–94. doi: 10.1007/978-3-642-23465-1-5
Angelini, J., Castro, S., and Fabra, A. (2003). Alterations in root colonization and nodC gene induction in the peanut-rhizobia interaction under acidic conditions. Plant Physiol. Biochem. 41, 289–294. doi: 10.1016/s0981-9428(03)00021-24
Araujo, A. P., Plassard, C., and Drevon, J. J. (2008). Phosphatase and phytase activities in nodules of common bean genotypes at different levels of phosphorus supply. Plant Soil 312, 129–138. doi: 10.1007/s11104-008-9595-3
Arayangkoon, T., Schomber, H. H., and Weaver, R. W. (1990). Nodulation and N2 fixation of guar at high root temperature. Plant Soil 126, 209–213. doi: 10.1007/BF00012824
Arrighi, J. F., Barre, A., Ben Amor, B., Bersoult, A., Soriano, L. C., Mirabella, R., et al. (2006). The Medicago truncatula lysine motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 142, 265–279. doi: 10.1104/pp.106.084657
Badri, D. V., and Vivanco, J. M. (2009). Regulation and function of root exudates. Plant Cell Environ. 32, 666–681. doi: 10.1111/j.1365-3040.2009.01926.x
Badri, D. V., Weir, T. L., Van Der Lelie, D., and Vivanco, J. M. (2009). Rhizosphere chemical dialogues: plant-microbe interactions. Curr. Opin. Biotechnol. 20, 642–650. doi: 10.1016/j.copbio.2009.09.014
Bargaz, A., Ghoulam, C., Amenc, L., Lazali, M., Faghire, M., Abadie, J., et al. (2012). A phosphoenol pyruvate phosphatase transcript is induced in the root nodule cortex of Phaseolus vulgaris under conditions of phosphorus deficiency. J. Exp. Bot. 63, 4723–4730. doi: 10.1093/jxb/ers151
Bartsev, A. V., Deakin, W. J., Boukli, N. M., McAlvin, C. B., Stacey, G., Malnoe, P., et al. (2004). NopL, an effector protein of Rhizobium sp. NGR234, thwarts activation of plant defense reactions. Plant Physiol. 134, 871–879. doi: 10.1104/pp.103.031740
Battistoni, F., Platero, R., Noya, F., Arias, A., and Fabiano, E. (2002). Intracellular Fe content influences nodulation competitiveness of Sinorhizobium meliloti strains as inocula of alfalfa. Soil Biol. Biochem. 34, 593–597. doi: 10.1016/S00380717(01)00215-2
Begum, A. A., Leibovitch, S., Migner, P., and Zhang, F. (2001). Specific flavonoids induced nod gene expression and pre-activated nod genes of Rhizobium leguminosarum increased pea (Pisum sativum L.) and lentil (Lens culinaris L.) nodulation in controlled growth chamber environments. J. Exp. Bot. 52, 1537–1543. doi: 10.1093/jexbot/52.360.1537
Bolaños-Vásquez, M. C., and Werner, D. (1997). Effects of Rhizobium tropici, R. etli, and R. leguminosarum bv. phaseoli on nod Gene-Inducing Flavonoids in Root Exudates of Phaseolus vulgaris. Mol. Plant Microbe Int. 10, 339–346. doi: 10.1094/MPMI.1997.10.3.339
Brear, E. M., Day, D. A., and Smith, P. M. C. (2013). Iron: an essential micronutrient for the legume-rhizobium symbiosis. Front. Plant Sci. 4:359. doi: 10.3389/fpls.2013.00359
Broughton, W. J., Zhang, F., Perret, X., and Staehelin, C. (2003). Signals exchanged between legumes and Rhizobium : agricultural uses and perspectives. Plant Soil 252, 129–137. doi: 10.1023/a:1024179717780
Canfield, D. E., Glazer, A. N., and Falkowski, P. G. (2010). The evolution and future of earth’s nitrogen cycle. Science 330, 192–196. doi: 10.1126/science.1186120
Chang, W. S., Franck, W. L., Cytryn, E., Jeong, S., Joshi, T., Emerich, D. W., et al. (2007). An oligonucleotide microarray resource for transcriptional profiling of Bradyrhizobium japonicum. Mol. Plant Microbe Int. 20, 1298–1307. doi: 10.1094/mpmi-20-10-1298
Cooper, J. E. (2007). Early interactions between legumes and rhizobia: disclosing complexity in a molecular dialogue. J. Appl. Microbiol. 103, 1355–1365. doi: 10.1111/j.1365-2672.2007.03366.x
Dakora, F. D. (2000). Commonality of root nodulation signals and nitrogen assimilation in tropical grain legumes belonging to the tribe Phaseoleae. Funct. Plant Biol. 27, 885–892. doi: 10.1071/pp00015
Dakora, F. D., Joseph, C. M., and Phillips, D. A. (1993a). Alfalfa (Medicago sativa L.) root exudates contain isoflavonoids in the presence of Rhizobium meliloti. Plant Physiol. 101, 819–824. doi: 10.1071/pp00015
Dakora, F. D., Joseph, C. M., and Phillips, D. A. (1993b). Common bean root exudates contain elevated levels of daidzein and coumesterol in response to Rhizobium inoculation. Mol. Plant Microbe Int. 6, 665–668. doi: 10.1094/mpmi-6-665
Dardanelli, M. S., De Córdoba, F. J. F., Estévez, J., Contreras, R., Cubo, M. T., Rodríguez-Carvajal, M. T., et al. (2012). Changes in flavonoids secreted by Phaseolus vulgaris roots in the presence of salt and the plant growth-promoting rhizobacterium Chryseobacterium balustinum. Appl. Soil Ecol. 57, 31–38. doi: 10.1016/j.apsoil.2012.01.005
Davis, E. O., and Johnston, A. W. B. (1990). Regulatory function of the three nodD gens of Rhizobium leguminosarum biovar phaseoli. Mol. Microbiol. 4, 933–941. doi: 10.1111/j.1365-2958.1990.tb00666.x
Deakin, W. J., and Broughton, W. J. (2009). Symbiotic use of pathogenic strategies: rhizobial protein secretion systems. Nat. Rev. Microbiol. 7, 312–320. doi: 10.1038/nrmicro2091
De-la-Peña, C., Badri, D. V., Lei, Z., Watson, B. S., Brandão, M. M., Silva-Filho, M. C., et al. (2010). Root secretion of defense-related proteins is development-dependent and correlated with flowering time. J. Biol. Chem. 285, 30654–30665. doi: 10.1074/jbc.M110.119040
De-la-Peña, C., Lei, Z., Watson, B. S., Sumner, L. W., and Vivanco, J. M. (2008). Root-Microbe Communication through Protein Secretion. J. Biol. Chem. 283, 25247–25255. doi: 10.1074/jbc.M801967200
Downie, J. A. (2010). The roles of extracellular proteins, polysaccharides and signals in the interactions of rhizobia with legume roots. FEMS Microbiol. Rev. 34, 150–170. doi: 10.1111/j.1574-6976.2009.00205.x
Estévez, J., Dardanelli, M. S., Megías, M., and Rodríguez-Navarro, D. N. (2009). Symbiotic performance of common bean and soybean co-inoculated with rhizobia and Chryseobacterium balustinum Aur9 under moderate saline conditions. Symbiosis 49, 29–36. doi: 10.1007/s13199-009-0008-z
Fauvart, M., and Michiels, J. (2008). Rhizobial secreted proteins as determinants of host specificity in the rhizobium-legume symbiosis. FEMS Microbiol. Lett. 285, 1–9. doi: 10.1111/j.1574-6968.2008.01254.x
Firmin, J. L., Wilson, K. E., Rossen, L., and Johnston, W. B. (1986). Flavonoid activation of nodulation genes in Rhizobium reversed by other compounds present in plants. Nature 324, 90–92. doi: 10.1038/324090a0
Fox, J. E., Starcevic, M., Jones, P. E., Burow, M. E., and Mclachlan, J. A. (2004). Phytoestrogen signaling and symbiotic gene activation are disrupted by endocrine-disrupting chemicals. Environ. Health Perspect. 112, 672–677. doi: 10.1289/ehp.6456
Fox, J. E., Starcevic, M., Kow, K. Y., Burow, M. E., and Mclachlan, J. A. (2001). Nitrogen fixation. Endocrine disrupters and flavonoid signalling. Nature 413, 128–129. doi: 10.1038/35093163
Fraysse, N., Couderc, F., and Poinsot, V. (2003). Surface polysaccharide involvement in establishing the rhizobium–legume symbiosis. Eur. J. Biochem. 270, 1365–1380. doi: 10.1046/j.1432-1033.2003.03492.x
Gerosa-Ramos, M. L., Parsons, R., Sprent, J. I., and James, E. K. (2003). Effect of water stress on nitrogen fixation and nodule structure of common bean. Pesq. Agropec. Bras. 38, 339–347. doi: 10.1590/S0100-204X2003000300002
Giraud, E., Moulin, L., Vallenet, D., Barbe, V., Cytryn, E., Avarre, J. C., et al. (2007). Legume symbiosis: absence of Nod genes in photosynthetic bradyrhizobia. Science 316, 1307–1312. doi: 10.1126/science.1139548
Göttfert, M., Grob, P., and Hennecke, H. (1990). Proposed regulatory pathway encoded by the nodV and nodW genes, determinants of host specificity in Bradyrhizobium japonicum. Proc. Natl. Acad. Sci. U.S.A. 87, 2680–2684. doi: 10.1073/pnas.87.7.2680
Guasch-Vidal, B., Estévez, J., Dardanelli, M. S., Soria-Díaz, M. E., De Córdoba, F. F., Balog, C. I. A., et al. (2012). High NaCl concentrations induce the nod genes of Rhizobium tropici CIAT899 in the absence of flavonoid inducers. Mol. Plant Microbe Interact. 26, 451–460. doi: 10.1094/MPMI-09-12-0213-R
Gwata, E. T., Wofford, D. S., Boote, K. J., Blount, A. R., and Pfahler, P. L. (2005). Inheritance of promiscuous nodulation in soybean. Crop Sci. 45, 635–638. doi: 10.2135/cropsci2005.0635
Hartwig, U. A., Maxwell, C. A., Joseph, C. M., and Phillips, D. A. (1989). Interactions among flavonoid nod gene inducers released from alfalfa seeds and roots. Plant Physiol. 91, 1138–1142. doi: 10.1104/pp.91.3.1138
Hartwig, U. A., Maxwell, C. A., Joseph, C. M., and Phillips, D. A. (1990). Chrysoeriol and luteolin released from alfalfa seeds induce nod genes in Rhizobium meliloti. Plant Physiol. 92, 116–122. doi: 10.1104/pp.92.1.116
Hassan, S., and Mathesius, U. (2012). The role of flavonoids in root–rhizosphere signalling: opportunities and challenges for improving plant–microbe interactions. J. Exp. Bot. doi: 10.1093/jxb/err430
Hatimi, A. (1999). Effect of salinity on the association between root symbionts and Acacia cyanophylla Lind: growth and nutrition. Plant Soil 216, 93–101. doi: 10.1023/A:1004745707277
Herder, G. D., Van Isterdael, G., Beeckman, T., and De Smet, I. (2010). The roots of a new green revolution. Trends Plant Sci. 15, 600–607. doi: 10.1016/j.tplants.2010.08.009
Hernandez, G., Valdes-Lopez, O., Ramirez, M., Goffard, N., Weiller, G., Aparicio-Fabre, R., et al. (2009). Global changes in the transcript and metabolic profiles during symbiotic nitrogen fixation in phosphorus-stressed common bean plants. Plant Physiol. 151, 1221–1238. doi: 10.1104/pp.109.900306
Hirsch, A. M., and Fujishige, N. A. (2012). “Molecular signals and receptors: communication between nitrogen-fixing bacteria and their plant hosts,” in Biocommunication of Plants, eds G. Witzany and F. Baluška (Berlin: Springer Berlin Heidelberg), 255–280.
Horvath, B., Kondorosi, E., John, M., Schmidt, J., Torok, I., Gyorgypal, Z., et al. (1986). Organization, structure and symbiotic function of Rhizobium meliloti nodulation genes determining host specificity for alfalfa. Cell 46, 335–343. doi: 10.1016/0092-8674(86)90654-90659
Hotter, G. S., and Scott, D. B. (1991). Exopolysaccharide mutants of Rhizobium loti are fully effective on a determinate nodulating host but are ineffective on an indeterminate nodulating host. J. Bacteriol. 173, 851–859.
Hungria, M., Johnston, A. W., and Phillips, D. A. (1992). Effects of flavonoids released naturally from bean (Phaseolus vulgaris) on nodD-regulated gene transcription in Rhizobium leguminosarum bv. phaseoli. Mol. Plant Microbe Int. 5, 199–203. doi: 10.1094/mpmi-5-199
Hungria, M., Joseph, C. M., and Phillips, D. A. (1991a). Anthocyanidins and flavonols, major nod gene inducer from seeds of a black-seeded common bean (Phaseolus vulgaris L.). Plant Physiol. 97, 758. doi: 10.1104/pp.97.2.751
Hungria, M., Joseph, C. M., and Phillips, D. A. (1991b). Rhizobium nod gene inducers exuded naturally from roots of common bean (Phaseolus vulgaris L.). Plant Physiol. 97, 759–764. doi: 10.1104/pp.97.2.759
Hungria, M., and Stacey, G. (1997). Molecular signals exchange between host plants and rhizobia - basic aspects and potential application in agriculture. Soil Biol. Biochem. 29, 819–830. doi: 10.1016/s0038-0717(96)00239-238
Hungria, M., and Vargas, M. A. T. (2000). Environmental factors affecting N2 fixation in grain legumes in the tropics, with an emphasis on Brazil. Field Crops Res. 65, 151–164. doi: 10.1016/s0378-4290(99)00084-82
Janczarek, M. (2011). Environmental signals and regulatory pathways that influence exopolysaccharide production in rhizobia. Int. J. Mol. Sci. 12, 7898–7933. doi: 10.3390/ijms12117898
Jones, K. M., Kobayashi, H., Davies, B. W., Taga, M. E., and Walker, G. C. (2007). How rhizobial symbionts invade plants: the Sinorhizobium -Medicago model. Nat. Rev. Microbiol. 5, 619–633. doi: 10.1038/nrmicro1705
Kannenberg, E. L., Rathbun, E. A., and Brewin, N. J. (1992). Molecular dissection of structure and function in the lipopolysaccharide of Rhizobium leguminosarum strain 3841 using monoclonal antibodies and genetic analysis. Mol. Microbiol. 6, 2477–2487. doi: 10.1111/j.1365-2958.1992.tb01424.x
Kape, R., Parniske, M., Brandt, S., and Werner, D. (1992). Isoliquiritigenin, a stron nod gene- and glyceollin resistance-inducing flavonoid from soybean root exudate. Appl. Environ. Microbiol. 58, 1705–1710.
Krause, A., Doerfel, A., and Gottfert, M. (2002). Mutational and transcriptional analysis of the type III secretion system of Bradyrhizobium japonicum. Mol. Plant Microbe Int. 15, 1228–1235. doi: 10.1094/mpmi.2002.15.12.1228
Laus, M. C., Logman, T. J., Lamers, G. E., Van Brussel, A. A. N., Carlson, R. W., and Kijne, J. W. (2006). A novel polar surface polysaccharide from Rhizobium leguminosarum binds host plant lectin. Mol. Microbiol. 59, 1704–1713. doi: 10.1111/j.1365-2958.2006.05057.x
Lerouge, P., Roche, P., Faucher, C., Maillet, F., Truchet, G., Prome, J. C., et al. (1990). Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature 344, 781–784. doi: 10.1038/344781a0
Li, C., Gui, S., Yang, T., Wang, X., and Liao, H. (2012). Identification of soybean purple acid phosphatase genes and their expression responses to phosphorus availability and symbiosis. Ann. Bot. 109, 275–285. doi: 10.1093/aob/mcr246
Limpens, E., Franken, C., Smit, P., Willemse, J., Bisseling, T., and Geurts, R. (2003). LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302, 630–633. doi: 10.1126/science.1090074
Lin, M. H., Gresshoff, P. M., and Ferguson, B. J. (2012). Systemic regulation of soybean nodulation by acidic growth conditions. Plant Physiol. 160, 2028–2039. doi: 10.1104/pp.112.204149
Lira Junior, M. A., Lima, A. S. T., Arruda, J. R. F., and Smith, D. L. (2005). Effect of root temperature on nodule development of bean, lentil and pea. Soil Biol. Biochem. 37, 235–239. doi: 10.1016/j.soilbio.2004.07.032
Lynch, D., O’Brien, J., Welch, T., Clarke, P., Cuiv, P. O., Crosa, J. H., et al. (2001). Genetic organization of the region encoding regulation, biosynthesis, and transport of rhizobactin 1021, a siderophore produced by Sinorhizobium meliloti. J. Bacteriol. 183, 2576–2585. doi: 10.1128/jb.183.8.2576-2585.2001
Mabood, F., and Smith, D. L. (2005). Pre-incubation of Bradyrhizobium japonicum with jasmonates accelerates nodulation and nitrogen fixation in soybean (Glycine max) at optimal and suboptimal root zone temperatures. Physiol. Plant. 125, 311–323. doi: 10.1111/j.1399-3054.2005.00559.x
Mabood, F., Souleimanov, A., Khan, W., and Smith, D. L. (2006). Jasmonates induce Nod factor production by Bradyrhizobium japonicum. Plant Physiol. Biochem. 44, 759–765. doi: 10.1016/j.plaphy.2006.10.025
Madsen, E. B., Madsen, L. H., Radutoiu, S., Olbryt, M., Rakwalska, M., Szczyglowski, K., et al. (2003). A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425, 637–640. doi: 10.1038/nature02045
Maougal, R. T., Bargaz, A., Sahel, C., Amenc, L., Djekoun, A., Plassard, C., et al. (2014). Localization of the Bacillus subtilis beta-propeller phytase transcripts in nodulated roots of Phaseolus vulgaris supplied with phytate. Planta 239, 901–908. doi: 10.1007/s00425-013-2023-9
Marcar, N. E., Dart, P., and Sweeney, C. (1991). Effect of root-zone salinity on growth and chemical composition of Acacia ampliceps B. R. Maslin, A. auriculiformis A. Cunn ex Benth and A. mangium Wild at two nitrogen levels. New Phytol. 119, 567–573. doi: 10.1111/j.1469-8137.1991.tb01049.x
Marie, C., Deakin, W. J., Viprey, V., Kopcinska, J., Golinowski, W., Krishnan, H. B., et al. (2003). Characterization of Nops, nodulation outer proteins, secreted via the type III secretion system of NGR234. Mol. Plant Microbe Interact. 16, 743–751. doi: 10.1094/MPMI.2003.16.9.743
Martínez-Romero, E. (2003). Diversity of rhizobium-Phaseolus vulgaris symbiosis: overview and perspectives. Plant Soil 252, 11–23. doi: 10.1023/a:1024199013926
Masson-Boivin, C., Giraud, E., Perret, X., and Batut, J. (2009). Establishing nitrogen-fixing symbiosis with legumes: how many rhizobium recipes? Trends Microbiol. 17, 458–466. doi: 10.1016/j.tim.2009.07.004
Maxwell, C. A., Hartwig, U. A., and Joseph, C. M. (1989). A chalcone and two related flavonoids released from alfalfa roots induce nod genes of Rhizobium meliloti. Plant Physiol. 91, 842–847. doi: 10.1104/pp.91.3.842
McKay, I. A., and Djordjevic, M. A. (1993). Production and excretion of Nod metabolites by Rhizobium leguminosarum bv. trifolii are disrupted by the same enviromental factors that reduce nodulation in the field. Appl. Environ. Microbiol. 59, 3385–3392.
Michiels, J., Dombrecht, B., Vermeiren, N., Xi, C., Luyten, E., and Vanderleyden, J. (1998). Phaseolus vulgaris is a non-selective host for nodulation. FEMS Microbiol. Ecol. 26, 193–205. doi: 10.1016/s0168-6496(98)00035-x
Miller, S. H., Elliot, R. M., Sullivan, J. T., and Ronson, C. W. (2007). Host-specific regulation of symbiotic nitrogen fixation in Rhizobium leguminosarum biovar trifolii. Microbiology 153, 3184–3195. doi: 10.1099/mic.0.2007/006924-0
Miransari, M., and Smith, D. L. (2009). Alleviating salt stress on soybean (Glycine max (L.) Merr.) - Bradyrhizobium japonicum symbiosis, using signal molecule genistein. Eur. J. Soil Biol. 45, 146–152. doi: 10.1016/j.ejsobi.2008.11.002
Moron, B., Soria-Diaz, M. E., Ault, J., Verroios, G., Noreen, S., Rodriguez-Navarro, D. N., et al. (2005). Low pH changes the profile of nodulation factors produced by Rhizobium tropici CIAT899. Chem. Biol. 12, 1029–1040. doi: 10.1016/j.chembiol.2005.06.014
Moulin, L., Munive, A., Dreyfus, B., and Boivin-Masson, C. (2001). Nodulation of legumes by members of the b-subclass of Proteobacteria. Nature 411, 948–950. doi: 10.1038/35082070
Mulligan, J. T., and Long, S. R. (1985). Induction of Rhizobium meliloti nodC expression by plant exudate requires nodD. Proc. Natl. Acad. Sci. U.S.A. 82, 6609–6613. doi: 10.1073/pnas.82.19.6609
Naya, L., Ladrera, R., Ramos, J., Gonzalez, E., Arrese-Igor, C., Minchin, F. R., et al. (2007). The response of carbon metabolism and antioxidant defenses of alfafa nodules to drought stress and to the subsequente recovery of plants. Plant Physiol. 144, 1104–1114. doi: 10.1104/pp.107.099648
Nienaber, A., Hennecke, H., and Fischer, H. M. (2001). Discovery of a haem uptake system in the soil bacterium Bradyrhizobium japonicum. Mol. Microbiol. 41, 787–800. doi: 10.1046/j.1365-2958.2001.02555.x
O’Hara, G. W., Dilworth, M. J., Boonkerd, N., and Parkpian, P. (1988). Iron-deficiency specifically limits nodule development in peanut inoculated with Bradyrhizobium sp. New Phytol. 108, 51–57. doi: 10.1111/j.1469-8137.1988.tb00203.x
Oldroyd, G. E. D., and Downie, J. A. (2004). Calcium, kinases and nodulation signalling in legumes. Nat. Rev. Mol. Cell Biol. 5, 566–576. doi: 10.1038/nrm1424
Ovtsyna, A. O., Geurts, R., Bisseling, T., Lugtenberg, B. J. J., Tikhonovich, I. A., and Spaink, H. P. (1998). Restriction of host range by the sym2 allele of Afghan pea is nonspecific for the type of modification at the reducing terminus of nodulation signals. Mol. Plant Microbe Interact. 11, 418–422. doi: 10.1094/mpmi.1998.11.5.418
Pan, B., and Smith, D. L. (1998). Genistein and daidzein concentrations and contents in seedling roots of three soybean cultivars grown under three root zone temperatures. J. Agronomy Crop Sci. 180, 77–82. doi: 10.1111/j.1439-037x.1998.tb00374.x
Pan, B., Zhang, F., and Smith, D. L. (1997). “Genistein addition to the rhizosphere of soybean (Glycine max L. Merr.) at the onset of nitrogen fixation increases overall nodulation and nitrogen fixation,” in Abstracts. 77th Annual Conference of the Agricultural Institute of Canada, ed. A. I. C. Program Committee (Truro: Agricultural Institute of Canada), 26–27.
Parniske, M., Schmidt, P. E., Kosch, K., and Muller, P. (1994). Plant defense responses of host plants with determinate nodules induced by EPS-defective exoB mutants of Bradyrhizobium japonicum. Mol. Plant Microbe Interact. 7, 631–638. doi: 10.1094/MPMI-7-0631
Penttinen, P., Rasanen, L. A., Lortet, G., and Lindstrom, K. (2013). Stable isotope labelling reveals that NaCl stress decreases the production of Ensifer (Sinorhizobium) arboris lipochitooligosaccharide signalling molecules. FEMS Microbiol. Lett. 349, 117–126. doi: 10.1111/1574-6968.12303
Peters, N. K., Frost, J. W., and Long, S. R. (1986). A plant flavone, luteolin, induces expression of Rhizobium meliloti nodulation genes. Science 233, 977–980. doi: 10.1126/science.3738520
Pueppke, S. G., Bolaños-V squez, M. C., Werner, D., Bec-Fert, M. P., Prom, J. C., and Krishnan, H. B. (1998). Release of flavonoids by the soybean sultivars McCall and Peking and their perception as signals by the nitrogen-fixing symbiont Sinorhizobium fredii. Plant Physiol. 117, 599–608. doi: 10.1104/pp.117.2.599
Purwantari, N. D., Date, R. A., and Dart, P. J. (1995). Nodulation and N2 by Calliandra calothyrsus and Sesbania sesban grown at differente root temperatures. Soil Biol. Biochem. 27, 421–425. doi: 10.1016/0038-0717(95)98613-S
Radutoiu, S., Madsen, L. H., Madsen, E. B., Felle, H. H., Umehara, Y., Grønlund, M., et al. (2003). Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425, 585–592. doi: 10.1038/nature02039
Radutoiu, S., Madsen, L. H., Madsen, E. B., Jurkiewicz, A., Fukai, E., Quistgaard, E. M., et al. (2007). LysM domains mediate lipochitin-oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO J. 26, 3923–3935. doi: 10.1038/sj.emboj.7601826
Redmond, J. W., Batley, M., Innes, R. W., Kuempel, P. L., Djordjevic, M. A., and Rolfe, B. G. (1986). “Flavones induce expression of the nodulation genes in Rhizobium,” in Recognition in Microbe-Plant Symbiotic and Pathogenic Interactions, ed. B. Lugtenberg (Berlin: Springer-Verlag), 115–121.
Richardson, A. E., Djordjevic, M. A., Rolfe, B. G., and Simpson, R. J. (1988a). Effects of pH, Ca and Al on the exudation from clover seedling of compounds that induce the expression of nodulation genes in Rhizobium trifolii. Plant Soil 109, 37–47. doi: 10.1007/bf02197578
Richardson, A. E., Simpson, R. J., Djordjevic, M. A., and Rolfe, B. G. (1988b). Expression of nodulation genes in Rhizobium leguminosarum biovar trifolii is affected by low pH and by Ca and Al ions. Appl. Environ. Microbiol. 54, 2541–2548.
Rodrí guez-Navarro, D. N., Margaret Oliver, I., Albareda Contreras, M., and Ruiz-Sainz, J. E. (2011). Soybean interactions with soil microbes, agronomical and molecular aspects. Agronomy Sustainable Dev. 31, 173–190. doi: 10.1051/agro/2010023
Rufini, M., Silva, M. A., Avelar Ferreira, P. A., Souza Cassetari, A., Lima Soares, B., Andrade, M. J., et al. (2013). Symbiotic efficiency and identification of rhizobia that nodulate cowpea in a Rhodic Eutrudox. Biol. Fertility Soils 50, 115–122. doi: 10.1007/s00374-013-0832-834
Ryu, H., Cho, H., Choi, D., and Hwang, I. (2012). Plant hormonal regulation of nitrogen-fixing nodule organogenesis. Mol. Cells 34, 117–126. doi: 10.1007/s10059-012-0131-131
Sanjuan, J., Grob, P., Göttfert, M., Hennecke, H., and Stacey, G. (1994). NodW is essential for full expression of the common nodulation genes in Bradyrhizobium japonicum. Mol. Plant Microbe Interact. 7, 364–369. doi: 10.1094/mpmi-7-0364
Schechter, L. M., Guenther, J., Olcay, E. A., Jang, S., and Krishnan, H. B. (2010). Translocation of NopP by Sinorhizobium fredii USDA257 into Vigna unguiculata root nodules. Appl. Environ. Microbiol. 76, 3758–3761. doi: 10.1128/AEM.03122-09
Schlaman, H. R., Olsthoorn, M. M., Harteveld, M., Dorner, L., Djordjevic, M. A., Thomas-Oates, J. E., et al. (2006). The production of species-specific highly unsaturated fatty acyl-containing LCOs from Rhizobium leguminosarum bv. trifolii is stringently regulated by nodD and involves the nodRL genes. Mol. Plant Microbe Interact. 19, 215–226. doi: 10.1094/mpmi-19-0215
Schue, M., Fekete, A., Ortet, P., Brutesco, C., Heulin, T., Schmitt-Kopplin, P., et al. (2011). Modulation of metabolism and switching to biofilm prevail over exopolysaccharide production in the response of Rhizobium alamii to cadmium. PLoS ONE 6:e26771. doi: 10.1371/journal.pone.0026771
Shiraishi, A., Matsushita, N., and Hougetsu, T. (2010). Nodulation in black locust by the Gammaproteobacteria Pseudomonas sp. and the Betaproteobacteria Burkholderia sp. Syst. Appl. Microbiol. 33, 269–274. doi: 10.1016/j.syapm.2010.04.005
Simsek, S., Ojanen-Reuhs, T., Stephens, S. B., and Reuhs, B. L. (2007). Strain-ecotype specificity in Sinorhizobium meliloti -Medicago truncatula symbiosis is correlated to succinoglycan oligosaccharide structure. J. Bacteriol. 189, 7733–7740. doi: 10.1128/JB.00739-07
Skorpil, P., Saad, M. M., Boukli, N. M., Kobayashi, H., Ares-Orpel, F., Broughton, W. J., et al. (2005). NopP, a phosphorylated effector of Rhizobium sp. strain NGR234, is a major determinant of nodulation of the tropical legumes Flemingia congesta and Tephrosia vogelii. Mol. Microbiol. 57, 1304–1317. doi: 10.1111/j.1365-2958.2005.04768.x
Slatni, T., Vigani, G., Salah, I. B., Kouas, S., Dell’Orto, M., Gouia, H., et al. (2011). Metabolic changes of iron uptake in N2-fixing common bean nodules during iron deficiency. Plant Sci. 181, 151–158. doi: 10.1016/j.plantsci.2011.04.015
Small, S. K., Puri, S., Sangwan, I., and O’brian, M. R. (2009). Positive control of ferric siderophore receptor gene expression by the Irr protein in Bradyrhizobium japonicum. J. Bacteriol. 191, 1361–1368. doi: 10.1128/jb.01571-08
Soerensen, K. U., Terry, R. E., Jolley, V. D., Brown, J. C., and Vargas, M. E. (1988). The interaction of iron-stress response and root nodules in iron efficient and inefficient soybeans. J. Plant Nutr. 11, 853–862. doi: 10.1080/01904168809363850
Soto, M. J., Domínguez-Ferreras, A., Pérez-Mendoza, D., Sanjuán, J., and Olivares, J. (2009). Mutualism versus pathogenesis: the give-and-take in plant-bacteria interactions. Cell Microbiol. 11, 381–388. doi: 10.1111/j.1462-5822.2009.01282.x
Subramanian, S., Stacey, G., and Yu, O. (2006). Endogenous isoflavones are essential for the establishment of symbiosis between soybean and Bradyrhizobium japonicum. Plant J. 48, 261–273. doi: 10.1111/j.1365-313x.2006.02874.x
Subramanian, S., Stacey, G., and Yu, O. (2007). Distinct, crucial roles of flavonoids during legume nodulation. Trends Plant Sci. 12, 282–285. doi: 10.1016/j.tplants.2007.06.006
Sulieman, S., and Tran, L. S. P. (2015). Phosphorus homeostasis in legume nodules as an adaptive strategy to phosphorus deficiency. Plant Sci. 239, 36–43. doi: 10.1016/j.plantsci.2015.06.018
Tang, C., Robson, A. D., and Dilworth, M. J. (1990). The role of iron in nodulation and nitrogen fixation in Lupinus angustifolius L. New Phytol. 114, 173–182. doi: 10.1111/j.1469-8137.1990.tb00388.x
Todd, J. D., Sawers, G., Rodionov, D. A., and Johnston, A. W. B. (2006). The Rhizobium leguminosarum regulator IrrA affects the transcription of a wide range of genes in response to Fe availability. Mol. Genet. Genomics 275, 564–577. doi: 10.1007/s00438006-0155-y
Viguier, C., Cuiv, P. O., Clarke, P., and O’connell, M. (2005). RirA is the iron response regulator of the rhizobactin 1021 biosynthesis and transport genes in Sinorhizobium meliloti 2011. FEMS Microbiol. Lett. 246, 235–242. doi: 10.1016/j.femsle.2005.04.012
Vinuesa, P. (2015). Rhizobial Taxonomy Up-to-Date [Online]. Mexico: Universidad Nacional Autonoma de Mexico. [accessed 01 April, 2015].
Wei, M., Takeshima, K., Yokoyama, T., Minamisawa, K., Mitsui, H., Itakura, M., et al. (2010). Temperature-dependent expression of type III secretion system genes and its regulation in Bradyrhizobium japonicum. Mol. Plant Microbe Int. 23, 628–637. doi: 10.1094/mpmi-23-5-0628
Zaat, S. A., Schripsema, J., Wijffelman, C. A., Van Brussel, A. A., and Lugtenberg, B. J. (1989). Analysis of the major inducers of the Rhizobium nodA promoter from Vicia sativa root exudate and their activity with different nodD genes. Plant Mol. Biol. 13, 175–188. doi: 10.1007/bf00016136
Zhang, F., Lynch, D. H., and Smith, D. L. (1995). Impact of low root temperatures in soybean [Glycine max (L.) Merr.] on nodulation and nitrogen fixation. Exp. Appl. Bot. 35, 279–285. doi: 10.1016/0098-8472(95)00017-17
Zhang, F., and Smith, D. L. (1994). Effects of low root zone temperatures on the early stages of symbiosis establishment between soybean [Glycine max (L.) Merr.] and Bradyrhizobium japonicum. J. Exp. Bot. 45, 1467–1473. doi: 10.1093/jxb/45.10.1467
Zhang, F., and Smith, D. L. (1995). Preincubation of Bradyrhizobium japonicum with genistein accelerates nodule development of soybean at suboptimal root zone temperatures. Plant Physiol. 108, 961–968.
Zhang, F., and Smith, D. L. (1996a). Genistein accumulation in soybean (Glycine max [L.] Merr.) root systems under suboptimal root zone temperature. J. Exp. Bot. 47, 785–792. doi: 10.1093/jxb/47.6.785
Zhang, F., and Smith, D. L. (1996b). Inoculation of soybean (Glycine max (L.) Merr.) with genistein-preincubated Bradyrhizobium japonicum or genistein directly applied into soil increases soybean protein and dry matter yield under short season conditions. Plant Soil 179, 233–241. doi: 10.1007/bf00009333
Zhang, F., and Smith, D. L. (1997). Application of genistein to inocula and soil to overcome low spring soil temperature inhibition of soybean nodulation and nitrogen fixation. Plant Soil 192, 141–151. doi: 10.1023/a:1004284727885
Zhang, H., Prithiviraj, B., Souleimanov, A., D′aoust, F., Charles, T. C., Driscoll, B. T., et al. (2002). The effect of temperature and genistein concentration on lipo-chitooligosaccharide (LCO) production by wild-type and mutant strains of Bradyrhizobium japonicum. Soil Biol. Biochem. 34, 1175–1180. doi: 10.1016/s0038-0717(02)00054-58
Keywords: tropical legumes, broad spectrum, soil acidity, soil temperature, salinity
Citation: Lira MA Jr., Nascimento LRS and Fracetto GGM (2015) Legume-rhizobia signal exchange: promiscuity and environmental effects. Front. Microbiol. 6:945. doi: 10.3389/fmicb.2015.00945
Received: 10 April 2015; Accepted: 27 August 2015;
Published: 08 September 2015.
Edited by:
Etienne Yergeau, National Research Council Canada, CanadaReviewed by:
Oswaldo Valdes-Lopez, National Autonomous University of Mexico, MexicoCopyright © 2015 Lira, Nascimento and Fracetto. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mario A. Lira Jr., Agronomy Department, Federal Rural University of Pernambuco, Recife 52061160, Brazil,bWFyaW9saXJhanVuaW9yQGdtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.