- 1Department of Animal Sciences, University of Florida, Gainesville, FL, USA
- 2Department of Animal Sciences, The Ohio State University, Columbus, OH, USA
Rumen microbes produce cellular protein inefficiently partly because they do not direct all ATP toward growth. They direct some ATP toward maintenance functions, as long-recognized, but they also direct ATP toward reserve carbohydrate synthesis and energy spilling (futile cycles that dissipate heat). Rumen microbes expend ATP by vacillating between (1) accumulation of reserve carbohydrate after feeding (during carbohydrate excess) and (2) mobilization of that carbohydrate thereafter (during carbohydrate limitation). Protozoa account for most accumulation of reserve carbohydrate, and in competition experiments, protozoa accumulated nearly 35-fold more reserve carbohydrate than bacteria. Some pure cultures of bacteria spill energy, but only recently have mixed rumen communities been recognized as capable of the same. When these communities were dosed glucose in vitro, energy spilling could account for nearly 40% of heat production. We suspect that cycling of glycogen (a major reserve carbohydrate) is a major mechanism of spilling; such cycling has already been observed in single-species cultures of protozoa and bacteria. Interconversions of short-chain fatty acids (SCFA) may also expend ATP and depress efficiency of microbial protein production. These interconversions may involve extensive cycling of intermediates, such as cycling of acetate during butyrate production in certain butyrivibrios. We speculate this cycling may expend ATP directly or indirectly. By further quantifying the impact of reserve carbohydrate accumulation, energy spilling, and SCFA interconversions on growth efficiency, we can improve prediction of microbial protein production and guide efforts to improve efficiency of microbial protein production in the rumen.
Introduction
Cattle and other ruminants can degrade fibrous feedstuffs owing to the consortium of bacteria, protozoa, fungi, and methanogens inhabiting their rumen and hindgut. This consortium ferments fiber and other feed components to short-chain fatty acids (SCFA) and, in the process, generates ATP that fuels microbial growth (synthesis of cellular protein in particular). This microbial protein supplies 60 to 85% of amino acids (AA) reaching the animal's small intestine (Storm et al., 1983). Maximizing efficiency of its production would consequently improve cattle productivity.
Production of microbial protein is inefficient because microbes do not direct all ATP toward growth. Rather, microbes can direct some ATP toward maintenance functions, synthesis of reserve carbohydrate, or energy spilling (futile cycles that dissipate heat). The impact of maintenance functions on growth efficiency has been recognized for decades and for both pure and mixed cultures (Pirt, 1965; Russell and Cook, 1995). Only recently, however, have reserve carbohydrate accumulation and energy spilling been accurately quantified in mixed rumen microbes such that their impact on efficiency can be considered (Hackmann et al., 2013a,b; Denton et al., 2015). We will review these recent advances in the context of other factors that depress efficiency of microbial protein production.
For many years, knowledge on the rumen microbiome steadily grew but was viewed myopically—from the context of a relatively few culturable isolates. Researchers used pure cultures or mixtures of a few pure cultures to establish niches, substrates used, growth factors, growth rates, fermentation end-products, and fundamental interactions between those cultures (Krause et al., 2013). Results from mixed cultures and from rumen-cannulated animals expanded those principles. In the past decade, however, the expansion of technology has greatly expanded our view of the rumen microbiome, sometimes from a hyperopic view—from the context of how to integrate extensive metagenomics data on the rumen microbiome with ruminant nutrition (Firkins and Yu, 2015). We will discuss how the growth efficiency of mixed ruminal microbes is affected by metabolic fluxes of anabolic and catabolic reactions, with emphasis on energy spilling.
Improving Efficiency of Microbial Growth
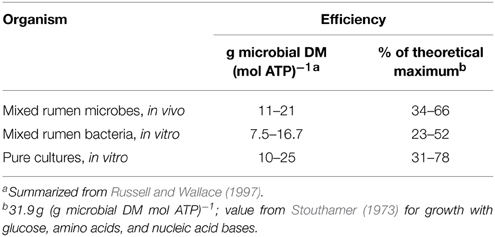
For more than 40 years, we have recognized that microbes grow (synthesize microbial protein) with far from perfect efficiency (Stouthamer, 1973). For mixed rumen microbes in vivo, actual growth efficiency ranges from only 1/3 to 2/3 of the theoretical maximum (as calculated from biochemical pathways) (Table 1). This implies that microbes spend as little as 1/3 of ATP on growth. Similar efficiencies are reported for mixed and pure cultures of bacteria in vitro (Table 1).
ATP not spent on growth is instead directed toward non-growth functions such as maintenance, energy spilling, and synthesis of reserve carbohydrate (Figure 1). Maintenance functions are those required for cellular “housekeeping” and include (1) re-synthesis of protein following intracellular turnover and (2) maintaining ion balances across the cell membrane (Russell and Cook, 1995). Motility is also a component of maintenance; it is a special case of maintaining ion balances because motility is driven by a proton or sodium motive force (Russell and Cook, 1995). Reserve carbohydrate synthesis refers to formation of glycogen and other compounds during energy excess (Preiss and Romeo, 1989). Although reserve carbohydrate can be mobilized later for growth (Figure 1) (Wilkinson, 1959), some ATP is irreversibly expended during synthesis. Energy spilling (e.g., futile cycling of ions or reserve carbohydrate; Russell and Cook, 1995; Portais and Delort, 2002; Russell, 2007b) refers to energy dissipated as heat when ATP exceeds needs for growth, maintenance functions, and reserve carbohydrate synthesis. It can be analogized to water spilling over the brim of an overfilled bucket (Figure S1). It is commonly a response to excess carbohydrate (Russell, 1998), as would occur when the ruminant is fed grain.
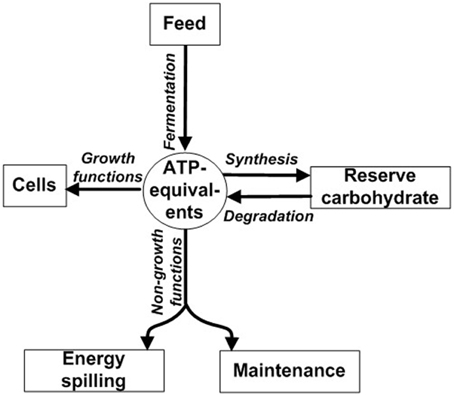
Figure 1. Partitioning of ATP energy toward growth functions, non-growth functions, and synthesis of reserve carbohydrate. ATP-equivalents can include ATP or ATP-yielding carbon compound (e.g., glucose). Modified from Russell and Wallace (1997) and Russell (2007a).
Maintenance functions have been long-recognized to be a sink for ATP energy and responsible for inefficient growth (Pirt, 1965). Maintenance energy becomes especially important when growth rates are low. Using the Pirt equation and values for mixed rumen bacteria in chemostats (Isaacson et al., 1975), Russell (2007a) calculated that maintenance energy would account for only 10% of total glucose consumption at the relatively high growth rate of 0.2 h−1. However, it would account for 31% total glucose consumption at the low growth rate of 0.05 h−1.
Because bacteria pass with digesta, their growth rate increases with increasing digesta passage rate in the rumen. Increasing passage rate by nutritional manipulation would be one strategy to decrease the relative impact of maintenance energy and improve growth efficiency. This is a facile strategy, however, because increasing passage rate, such as by grinding forage, decreases feed digestibility (Van Soest, 1994). A more defensible strategy to improving growth efficiency is to target other non-growth functions, such as energy spilling and reserve carbohydrate synthesis.
Occurrence of Energy Spilling in Pure Cultures
Although maintenance functions depress microbial growth efficiency, only energy spilling can explain very low efficiency during carbohydrate excess (and other growth-limiting conditions). Russell (1986) demonstrated energy spilling by pulse-dosing rumen bacterial cultures with glucose. Cultures fermented excess glucose rapidly, produced very little protein (growth efficiency approached 0), and dissipated (spilled) energy by producing heat. Van Kessel and Russell (1996) reported that mixed rumen bacteria fermented glucose 10-fold faster when spilling energy, implying that spilling could be a significant sink for ATP.
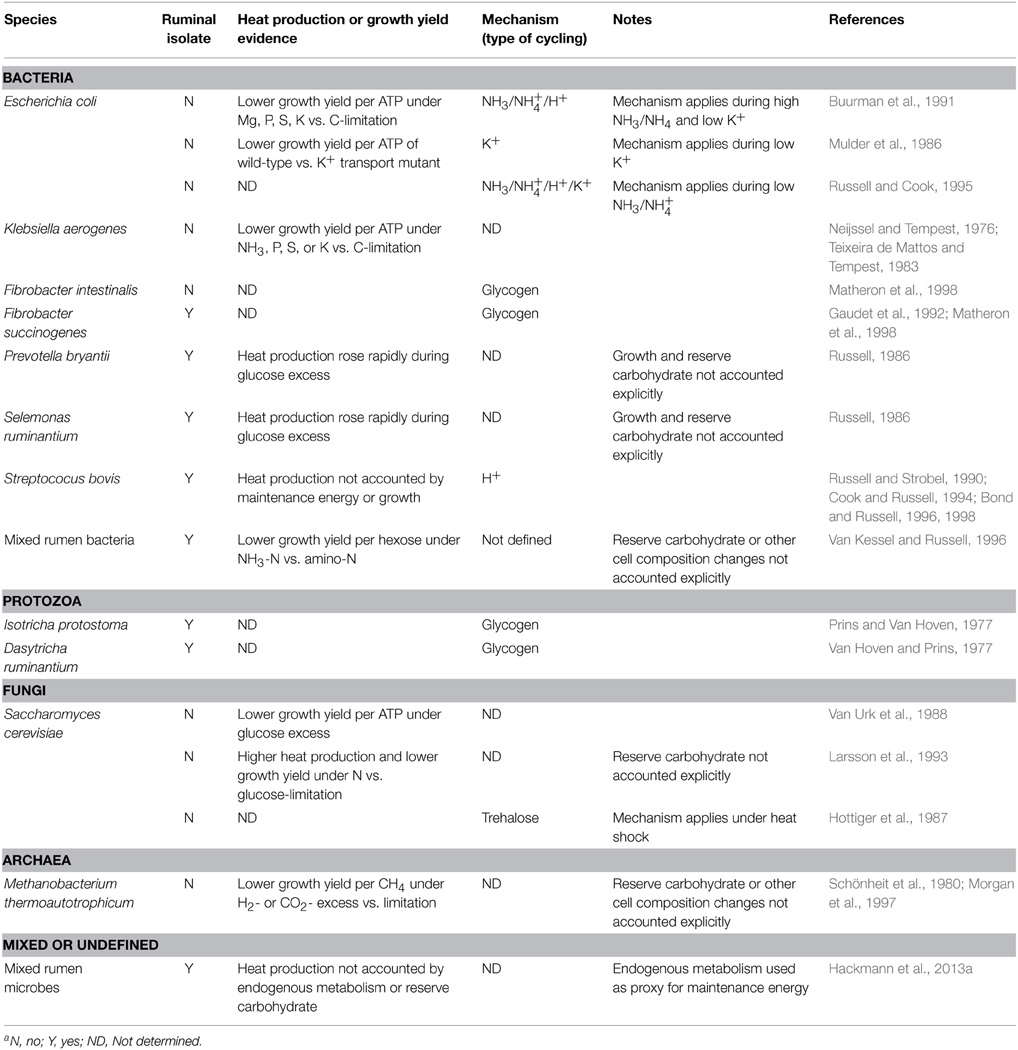
Spilling occurs in organisms across all three domains of life (Table 2). Spilling has been demonstrated extensively in a few rumen (Streptococcus bovis) and non-rumen (Escherichia coli, Klebsiella aerogenes) bacteria. Though evidence is less extensive, it also appears to occur in protozoa, fungi, and methanogens, suggesting broad importance. In almost all cases, single-species cultures, not mixed communities, have been studied.
Spilling can be evidenced by depressed growth efficiency or elevated heat production in response to excess carbohydrate (Table 2). Carbohydrate excesses have been generated by pulse dosing glucose or growing cells under limitation of an anabolic substrate (e.g., N, Mg, P, S, K). Spilling can also be evidenced in response to (1) ammonia-N replacing amino-N and (2) excess H2 or CO2 (for methanogens). When measuring changes in growth efficiency and heat production, one must account for any changes in maintenance energy, reserve carbohydrate, and growth (cf. Figure 1), though this is sometimes not done (Table 2).
The mechanism of spilling is by futile cycles of ions, glycogen, or trehalose (Table 2). The best-elucidated mechanism is for S. bovis, for which spilling occurs by futile cycling of protons. This cycling results from growth limitation and a cascade of biochemical events (Russell, 2002). Specifically, a growth limitation decreases use of ATP for protein synthesis, increasing fructose-1,6-bisphosphate, decreasing intracellular phosphate, increasing the absolute value of Gibbs energy of ATP hydrolysis, increasing activity of a proton-pumping ATPase, and decreasing membrane resistance to protons. The net result is heat, with no work done by the protons.
For E. coli, the mechanism of energy spilling is by cycling of K+ or NH+4 or a combination thereof, depending on the extracellular concentration of K+ and NH+4 (Table 2). For many other organisms, cycling of glycogen and trehalose may occur (Table 2). Such cycling implies energy spilling can occur, even though spilling often has not been measured directly (growth efficiency and heat production were often not determined; Table 2). Some authors have proposed that fructose-6-phosphate/fructose-1,6-bisphosphate (Otto, 1984) or other substrates (Newsholme et al., 1984) can be cycled. However, such cycling is difficult to establish under physiological conditions (Russell and Cook, 1995; Portais and Delort, 2002).
The biological rationale for spilling is not clear: why expend what can be conserved? Some have proposed spilling allows microbes to rapidly catabolize substrate and dissipate energy in order to (1) deprive competitors of energy or (2) hasten re-initiation of growth when a growth limitation is released (Tempest, 1978; Russell and Cook, 1995). Although all futile cycles dissipate energy and thus cause spilling, this spilling may be an unfortunate byproduct, not the primary function, of some futile cycles. Rather, the function of some futile cycles may be to sensitively control the net flux of metabolites (Newsholme and Crabtree, 1975). Regardless of function, energy spilling and futile cycles are wasteful from the perspective of growth efficiency, and their impact on growth of the rumen microbial community needs to be elucidated.
Occurrence of Energy Spilling in Mixed Cultures
Many pure cultures have been demonstrated to spill energy, but until recently, examples of mixed communities that spill energy were few (Table 2). In earlier studies, Van Kessel and Russell (1996) suggested that rumen bacteria spilled more energy when grown under ammonia-N vs. amino-N limitation. Their approach assumes constant cell composition, and they did not measure reserve carbohydrate. Some energy may have in fact been directed to reserve carbohydrate synthesis, not spilling. Chen et al. (2000) induced spilling in activated sludge by adding a protonophore, but spilling was not measured under more physiological conditions.
More recently, we quantified spilling for mixed rumen microbes (Hackmann et al., 2013a). When we washed cells with N-free buffer and dosed them with glucose, they consumed glucose rapidly, accumulated reserve carbohydrate, and did not grow (protein remained constant) (Figures 2A,B). When we dosed a moderate concentration of glucose (5 mM glucose), no spilling was detected, as nearly all heat production (93.7%) was accounted by reserve carbohydrate synthesis and endogenous metabolism (a proxy for maintenance energy) (Figure 2C). When we dosed a high concentration of glucose (20 mM), energy spilling was not detected immediately, but it accounted for a significant amount of heat production approximately 30 min after dosing (Figure 2D). Energy spilling accounted for as much as 38.7% of heat production in one incubation.
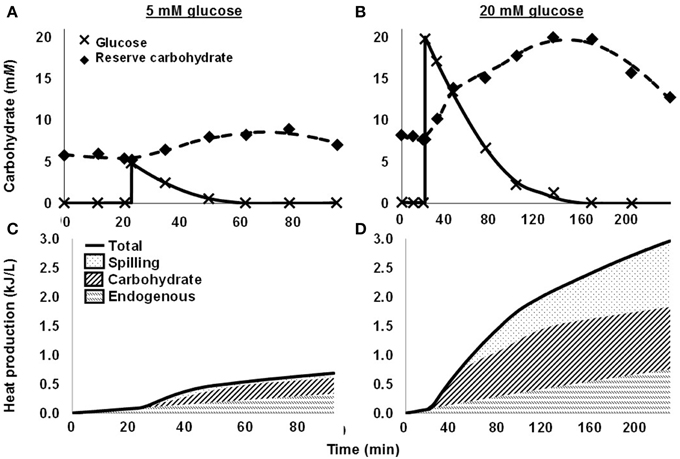
Figure 2. Energy spilling and other responses of mixed rumen microbes to pulse dose of glucose. (A,C) 5 mM glucose. (B,D) 20 mM glucose. (A,B) Glucose in media and reserve carbohydrate. Glucose was dosed at 20 min. (C,D) Heat production, including heat accounted by endogenous metabolism, synthesis of reserve carbohydrate, and energy spilling. Data are for 1 cow, and each glucose concentration represents a single experiment. Figure adapted from Hackmann et al. (2013a).
Identity and Occurrence of Reserve Carbohydrate
Rumen microbes can accumulate prodigious amounts of reserve carbohydrate. It can exceed 50% of cell weight for pure cultures of both rumen (Russell, 1998) and non-rumen (Preiss and Romeo, 1989) bacteria. Some protozoa (Isotrichidae) accumulate enough reserve carbohydrate to turn opaque (Williams and Coleman, 1992). Net accumulation of glucose into reserve carbohydrate can exceed 50% for mixed microbes (Hackmann et al., 2013a) and protozoa (Denton et al., 2015). The identity of this reserve carbohydrate is glycogen [glucan with (α1→4) and (α1→6) linkages] and appears ubiquitous across rumen bacteria, fungi, and protozoa (Table S1). Synthesis of these prodigious amounts of reserve carbohydrate irreversibly expends ATP, decreasing ATP available for protein synthesis (Figure 1). This lowers growth efficiency on a protein basis (g protein/mmol ATP), as it probably does on a dry matter basis (g DM/mmol ATP) (explained later).
Dynamics of Accumulation
In the rumen, reserve carbohydrate accumulates immediately after feeding (during carbohydrate excess), and then is mobilized thereafter (during carbohydrate limitation). This is observed for both rumen bacteria (Figure S2) and protozoa (Figure S3) (Jouany and Thiven, 1972; McAllan and Smith, 1974; Williams and Harfoot, 1976; Leedle et al., 1982). These dynamics are most dramatic for high-grain and low-N diets, which create large carbohydrate excesses after feeding (cf. basal diet + urea vs. basal diet in Figure S2).
For in vitro batch culture (Figures 2A,B), where conditions can be better defined, we observed that rumen microbes showed similar dynamics of accumulation and mobilization as in vivo (Figures S2, S3). When we washed mixed rumen microbes with N-free buffer and dosed glucose, reserve carbohydrate immediately accumulated (Figures 2A,B). At peak accumulation, microbes had incorporated 59.5% of glucose carbon in reserve carbohydrate when we dosed 5 mM glucose. The value was lower (52.6%) for 20 mM glucose. After glucose was exhausted, reserve carbohydrate quickly declined (Figures 2A,B).
In a subsequent batch culture study, we observed that protozoa, not bacteria, were responsible for most glycogen accumulation. In this study, we performed competition experiments in which mixtures of protozoa and bacteria were first dosed with glucose, and then at intervals the two groups were separated for glycogen analysis. When the mixtures were dosed with a moderate concentration of glucose (c. 5 mM), protozoa incorporated 58.7% of glucose carbon in reserve carbohydrate at time of peak reserve carbohydrate (Figure 3A). Bacteria had incorporated only 1.7%. When we dosed mixtures with a high concentration glucose (20 mM), the amounts incorporated were 21.4% for protozoa and 5.0% for bacteria, respectively (Figure 3B). Protozoa would thus appear the predominant group accumulating reserve carbohydrate. In sum, rumen microbes display a high capacity for accumulating and mobilizing reserve carbohydrate. As explained later, this can expend ATP and lower growth efficiency.
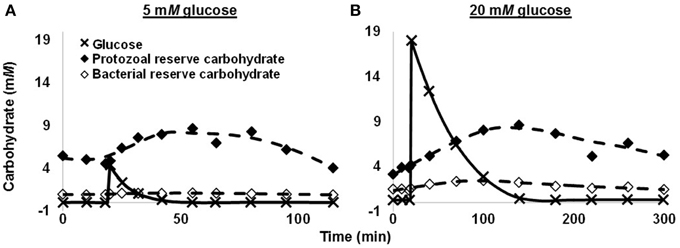
Figure 3. Glucose use and reserve carbohydrate accumulation of a mixture of rumen protozoa and bacteria in batch culture. (A) 5 mM glucose. (B) 20 mM glucose. Data are for 1 cow, and each glucose concentration represents a single experiment. Figure adapted from Denton et al. (2015).
Relation to Growth Efficiency
At first consideration, synthesis of reserve carbohydrate should not appear to depress growth efficiency; rather, it would seem to improve it. Glycogen, a common reserve carbohydrate, requires fewer ATP for synthesis than all other cellular macromolecules except lipid (Table 3). Simple arithmetic would suggest more dry matter could be formed when glycogen vs. most other macromolecules are synthesized—i.e., efficiency of growth on a dry matter basis (g DM/mmol ATP) would be higher.
Simple arithmetic does not consider that reserve carbohydrate accumulation is dynamic. As mentioned, rumen microbes vacillate between (1) reserve carbohydrate synthesis during carbohydrate excess and (2) degradation during carbohydrate limitation. The price paid for such vacillation is expenditure of ATP. For sequential synthesis and degradation of glycogen, 1 net ATP equivalent is expended per glucose (Figure S4). Synthesis of glycogen may cost few ATP compared to most other macromolecules (Table 3), but this low initial cost may be quickly outweighed by this sequential synthesis and degradation. Reserve carbohydrate synthesis could thus lower growth efficiency on a dry matter basis.
Reserve carbohydrate may have been historically overlooked in growth efficiency measurements because most experiments employ chemostats under steady-state conditions (see references in Russell and Cook, 1995). By design, the steady state input of substrate will prohibit vacillation between glycogen synthesis and degradation. Some experiments employ batch cultures (Russell and Cook, 1995), but they are usually terminated during exponential growth, before reserve carbohydrate degradation typically occurs.
Even when reserve carbohydrate would not lower growth efficiency on a DM basis (g DM/mmol ATP), it would always lower it on an N or protein basis (g N or protein/mmol ATP). No matter its course, reserve carbohydrate synthesis (2 ATP/glucose; Figure S4) would reduce ATP available for synthesis of protein and other N-containing macromolecules. This point is indirectly supported by the batch culture experiments of Hall (2011), in which destroying isotrichid protozoa by blending (1) decreased reserve carbohydrate accumulation by 43% and (2) increased growth efficiency on an N basis (g N/g soluble carbohydrate fermented) by 17%. In ruminant nutrition, growth efficiency is usually expressed in terms of protein or N, reflecting that microbial protein accounts for the majority of AA reaching the animal small intestine (Storm et al., 1983).
Although chemostats are more consistent with in vivo conditions than batch cultures because the former permit changes in dilution rate, such changes are simultaneous with increasing substrate supply. In contrast, increasing passage rate from the rumen should increase microbial growth rate, at least in part, independently from substrate supply (Dijkstra et al., 1998). As described in that report, increasing growth rate (decreasing division time) is projected to increase growth efficiency (g bacterial DM/g carbohydrate) and decrease the proportion of ATP used to support non-growth functions. Such models would be more accurate with better understanding of how much more reserve carbohydrate is accumulated under different dietary conditions.
Glycogen Cycling
On the surface, rumen microbes would seem to simply (1) synthesize reserve carbohydrate during carbohydrate excess and (2) degrade it during carbohydrate limitation. However, many rumen and non-rumen microbes have been shown to simultaneously synthesize and degrade (cycle) glycogen (Table 2) (Portais and Delort, 2002). This cycling expends ATP, just as sequential synthesis and degradation of glycogen do (Figure S4). It would thus depress growth efficiency.
Although we have discussed reserve carbohydrate synthesis and energy spilling independently, glycogen cycling would link these two functions because it is a form of energy spilling. Glycogen cycling has not yet been demonstrated for mixed rumen communities (Table 2), but we have speculated that it is the mechanism of spilling observed for mixed rumen microbes (Hackmann et al., 2013a).
Occurrence of Carbohydrate Excess in the Rumen
Both energy spilling and reserve carbohydrate synthesis occur primarily under carbohydrate excess. Carbohydrate is in greatest excess in the rumen for animals fed grain, particularly those transitioning to a high-grain diet, and also for animals fed high-sugar diets. For animals transitioning to a high grain diet, glucose can reach high concentrations [c. 5 mM (Ryan, 1964; Mackie et al., 1978)]. Even higher glucose concentrations (18 mM) have been reported for animals fed dextrose (Piwonka et al., 1994), and soluble sugar concentrations as high as 69 mM have been reported for animals fed beet pulp (Clapperton and Czerkawski, 1969). Concentrations in microenvironments (e.g., around starch granules) may also be high (Kajikawa et al., 1997).
For grain-fed animals, N availability can be low (NRC, 2000), also, and intensify carbohydrate excess (NRC, 2000). Availability of N can also be low for dairy rations with corn silage as the sole source of forage (Vandehaar, 2005). For animals in which N is chiefly in the form of ammonia, carbohydrate excess could be further intensified (Van Kessel and Russell, 1996) because rumen microbes grow far slower with ammonia-N than amino-N (Argyle and Baldwin, 1989; Van Kessel and Russell, 1996). Energy spilling and reserve carbohydrate synthesis would likely depress growth efficiency under these conditions, with spilling being more important at large carbohydrate excesses and reserve carbohydrate more important for smaller excesses (Hackmann et al., 2013a). Spilling has previously been suggested to account for low growth efficiency for high-concentrate diets (Clark et al., 1992).
For animals fed high-forage diets or adapted to grain, carbohydrate excess is relatively small, and glucose concentrations rarely exceed c. 2.5 mM (Kajikawa et al., 1997; Saleem et al., 2012). Energy spilling may play a minor role under these conditions, given that we did not detect spilling in batch cultures with glucose concentrations of 5 mM (Hackmann et al., 2013a). Reserve carbohydrate accumulation may still be important and decrease growth efficiency, however, given that reserve carbohydrate can still be detected even when cattle are provided low-quality grass diets (Van Kessel and Russell, 1997).
Other Factors Depressing Growth Efficiency
Other Responses to Excess Carbohydrate
Rumen microbes may respond to excess carbohydrate in ways other than spilling energy and synthesizing reserve carbohydrate. These other responses include reducing ATP yield by releasing metabolic intermediates (overflow metabolites) and shifting to catabolic pathways that yield less ATP (Russell, 1998). Responses are similar for non-rumen microbes (Tempest and Neijssel, 1984; Preiss and Romeo, 1989; Russell and Cook, 1995; Russell, 2007b).
Recycling of Microbial Protein
Recycling of microbial protein is another factor that depresses growth efficiency. As much as 50% of microbial protein is degraded to non-protein nitrogen in the rumen and recycled (Wells and Russell, 1996; Oldick et al., 2000). Protozoal predation, autolysis, and bacteriophages may all be causes (Wells and Russell, 1996). Most recycling has been thought to be mediated by protozoa predation, based on lysis of pure bacterial cultures in presence and absence of rumen fluid with protozoa (Wallace and McPherson, 1987). However, removing protozoa from the rumen was subsequently shown to have no effect on bacterial N recycling in vivo (Koenig et al., 2000). Firkins et al. (2007) reasoned that protozoa-mediated recycling of microbial protein is lessened with increasing passage rates by high-producing animals compared with some of the studies with low intakes or predictions based on measurements in vitro.
Exopolysaccharide Synthesis
In addition to synthesizing reserve carbohydrate, rumen bacteria can synthesize exopolysaccharides (Hobson and Macpherson, 1953, 1954; Costerton et al., 1974). One exopolysaccharide, dextran, is synthesized by S. bovis when given excess sucrose (Bailey and Oxford, 1958; Cheng et al., 1976), and it forms part of the “slime” observed in grain-fed animals with frothy bloat (Cheng et al., 1976). Up 80% of glucose in sucrose can be directed into its synthesis, but synthesis does not require ATP (Bailey, 1959; van Hijum et al., 2006), and there is no ATP cost of transport because synthesis occurs extracellularly (van Hijum et al., 2006). Thus, dextran formation would not depress growth efficiency on an ATP basis. Ruminococcus albus also forms an exopolysaccharide, but synthesis of this exopolysaccharide has been estimated to account for <4% of ATP of total used for synthesizing cell components (Weimer et al., 2006). Many other rumen bacteria form exopolysaccharides (Hobson and Macpherson, 1953, 1954; Costerton et al., 1974), but their formation has not been quantified, and their impact on growth efficiency remains unknown.
Cellodextrin Efflux
At least some cellulolytic bacteria expend ATP on cellodextrin efflux, which should depress their growth efficiency. The cellulolytic Fibrobacter spp. synthesize cellodextrins intracellularly, but these can be lost by extracellular efflux (Wells et al., 1995). The cell expends ~4/3 ATP from combined costs of synthesis (1 ATP) and active transport (~1/3 net ATP) (Figure S5). This ATP expended on cellodextrin synthesis would be recovered by non-cellulolytic bacteria after they take up the cellodextrin (assuming that transport is the exact opposite of efflux). Consequently, cellodextrin efflux should depress growth efficiency of some cellulolytics, not the microbial population as a whole.
Efflux of maltodextrins, not only cellodextrins, has been observed for F. succinogenes (Matulova et al., 2001; Nouaille et al., 2005). Maltodextrin efflux would be expected to expend ATP just as does cellodextrin efflux, but the exact expenditure is unknown because the pathway for maltodextrin synthesis is uncertain (cf. Matulova et al., 2001). Consequently, maltodextrin efflux likely depresses growth efficiency of some cellulolytics, but its exact impact on the cellulolytics and microbial population as a whole remains unknown.
Cross-feeding of cellodextrins is often depicted as being beneficial to the non-cellulolytics but also the cellulolytic populations by removing end-product inhibition of cellobiose on cellulases (Russell et al., 2009). As documented by those authors' model, increasing cellulolysis also diverts an increasing proportion of carbon toward cell growth and away from fermentation. However, if growth of the community is uncoupled by limitations of nitrogen or other growth factors, then an increasing proportion of carbon should be directed away from cell growth and toward SCFA, promoting energy spilling. In most studies measuring energy spilling, the medium was buffered. If total SCFA production was fast enough to decrease the ruminal pH below approximately 6.0, as can happen in the rumen, the proton gradient across the cell membrane could inhibit cellobiose transport by cellulolytics and thereby inhibit fiber degradation (Russell et al., 2009).
Cellobiose Hydrolysis or Transport
Russell (2002) described various mono or disaccharide transport mechanisms, including the phosphoenolpyruvate:phosphotransferase systems (PEP-PTS). Active transport also increases the ATP cost, but active transport of a disaccharide can have a decreased ATP charge if it is transported prior to hydrolysis into monosaccharides, and the ATP charge can be further decreased if disaccharide transport is coupled with a phosphorylase. Whether a phosphorylase or a PEP-phosphotransport system phosphorylates the sugar, this ATP charge is recovered by negating ATP required for a hexokinase reaction. Not all pure cultures were shown to express PEP-PTS transport of sugars (Martin, 1994). In mixed ruminal microbes, though, glucose or other sugars were assumed to be transported primarily by the PEP-PTS system (Kajikawa et al., 1997). Glucose is typically not fed to ruminants, but primarily is the product of cellulose and starch degradation. Some cellobiose was transported into mixed bacterial cells by a PEP-PTS but also by ATP-expending transporters (Kajikawa and Masaki, 1999). Increasing availability of maltose or maltodextrins for transport might lead to increased SCFA production and lower ruminal pH. Cellulolytic bacteria can be inhibited by pH < 6.0 through depressed adherence to cellulose, decreased cellobiose transport, or from inhibitory membrane gradients of anions or protons (Russell et al., 2009).
Genomics-based analyses have revealed a much more complicated mechanism in which genes are expressed as polysaccharide utilization loci (Wang et al., 2013). Di- or oligosaccharides from hydrolyzed cellulose can be transported in gram-negative F. succinogenes (Suen et al., 2011), gram-positive Ruminococcus flavefaciens (Flint et al., 2008), and from hydrolyzed hemicellulose in gram-positive Butyrivibrio proteoclasticus (Dunne et al., 2012), respectively, likely from a combination of ABC-transporters and those linked to phosphorolytic cleavage. Substrate source and availability probably regulates expression of many of these transporters (Bond et al., 2012). Based on metagenomics screening of cellulases and xylanases, many genes were novel, but a relatively high proportion were reputed transporters (Wang et al., 2013). There is likely periplasmic sequestration of oligosaccharides from cellulose (White et al., 2014), hemicellulose (Morgavi et al., 2013), and starch (Rosewarne et al., 2014). Therefore, the net ATP cost of di- and monosaccharide transport into cytosol is not fully known but probably varies with substrate availability.
Rapid growth decreased glycogen concentration of P. bryantii B14 (formerly P. ruminicola) (Lou et al., 1997b). However, with slower growth, maltose increased glycogen concentration more than when using glucose as substrate. When using maltose as substrate, maltose phosphorylase activity (which couples transport with phosphorylation of a glucose moiety) was increased and glycogen accumulated even when N was not limiting and growth rates increased. Similar results were detected when grown on cellobiose. In another study, growing P. bryantii on maltose or cellobiose increased the activity of UDP-glucose pyrophosphorylase and glycogen synthase compared with growth on sucrose or glucose (Lou et al., 1997a). Thus, transport and metabolism of maltose to glucose-1-phosphate was associated with glucose-1-phosphate polymerization into glycogen. Although poorly studied with mixed microbes, either gene expression of the reversible enzyme phosphoglucomutase or an accumulation of glucose-6-phosphate could help push synthesis of glycogen. UDP-glucose pyrophosphorylase (enzyme prior to glycogen synthase) was activated by fructose-1,6-phosphate in P. bryantii (Lou et al., 1997a). Pulse doses of glucose decreased metabolism of cellobiose and activity of cellobiose phosphorylase in P. bryantii, and vice versa (Lou et al., 1996). Thus, accumulation of disaccharides from rapid hydrolysis of cellulose or starch could stimulate glycogen synthesis in ruminal bacteria and thereby increase glycogen cycling as a means of energy spilling. Prevotella bryantii is well-known for energy spilling among cultivated prevotellas (Russell, 1992), although uncharacterized prevotellas often predominate in the rumen (Firkins and Yu, 2015).
Branched Short-chain Fatty Acids
The primary cellulolytics, most of which were characterized decades ago, have various requirements for growth factors such as branched chain SCFA and phenyl-substituted SCFA that are provided by secondary colonizers, which generally are much more proteolytic (Stewart et al., 1997). Despite the importance of branched chain SCFA required by isolates of cellulolytics, feeding these compounds in vivo primarily was associated with post-absorptive rather than ruminal responses (Andries et al., 1987). Given the importance of primary (or “keystone”) colonizers (Ze et al., 2013), such a lack of ruminal response can be reconciled with a broader view of how bacteria are stimulated by preformed AA to better balance anabolic and catabolic pathways during growth (Russell and Cook, 1995).
Peptides and Amino Acids vs. Ammonia-N
Although there seems to be little difference between AA and peptides for mixed cultures, many pure cultures of bacteria are stimulated by provision of small peptides rather than free AA (Wallace et al., 1997). For example, R. albus was shown to transport peptides but not AA (Kim et al., 2014). Peptides had a minor effect on this strain's growth rate, which was maximized at about 0.9/h. In contrast, S. bovis, which is known for rapid growth on starch or sugar, had growth rate of 0.9/h with NH3 that was stimulated to 1.6/h when AA were provided (Russell, 1993). In that study, incremental growth was synchronized with incremental boluses of glucose compared with a single dose, and the maximal growth available was particularly limited when glucose doses were incrementally staggered and when ammonia replaced AA. In contrast with pure cultures, when AA were provided with a mixture of carbohydrates as substrate, growth of mixed ruminal microbes was still stimulated by approximately 50% compared with providing NH3 even when the growth rates of the mixed cultures were lower (i.e., from 0.25–0.30 increased to 0.40–0.45/h) (Kajikawa et al., 2002). Because the latter stimulation is from growth rates below the threshold above which AA were expected to stimulate growth (Van Kessel and Russell, 1996), even growth of cellulolytics limited by rate of cellulolysis can be stimulated. In contrast with prior expectations, preformed AA now are considered as potentially stimulatory for consortia of microbes degrading fiber (Newbold, 1999).
Supply and Profile of Preformed Amino Acids
Although energy is required for bacteria to synthesize AA, this energy cost is small; when preformed AA are limiting, though, growth rate is slowed, and the balance of anabolic and catabolic rates leads to increased energy spilling (Russell and Cook, 1995). As depicted in Figure S6 (only for NH3 assimilation, not for transamination of other AA), Ala, Glu, and Gln are the primary AA formed from assimilation of ammonia in the rumen. Relative fluxes of these amination reactions depend on the Michaelis constant (Km) of ammonia for those enzymes but also based on transcription of ammonia-assimilating enzymes (Morrison and Mackie, 1997). Kim et al. (2014) documented an excellent example of transcriptional control of ammonia-assimilating enzymes in R. albus. Although bacteria can make most of their AA, gelatin (which has a poor profile of Leu and the aromatic AA) decreased growth rates of mixed bacteria (Van Kessel and Russell, 1996) and increased energy spilling.
Although AA are stimulatory to growth (Kajikawa et al., 2002), an imbalance of branched or aromatic AA was worse than deletion of the entire group of AA. Bacteria can partially control the flux of AA biosynthetic pathways (Figure S6), but congruent pathways likely antagonize availability of closely related AA when they are out of balance. Moreover, pathways for biosynthesis of AA intersect with central metabolic pathways used in fermentation. Most rumen bacteria lack a complete TCA cycle and must use what would be considered both forward and backward reactions of that cycle (if it was complete) to form α-ketoglutarate (Wallace et al., 1997). Thus, the alternating directional flux of this interrupted cycle must be able to provide the mix of intermediates for anabolism while intersecting with catabolic reactions to make ATP to drive anabolism. Because many of these intermediates are produced through dehydrogenases, the NADH/NAD ratio must be resolved with catabolic fermentation reactions. In support, the NADH/NAD ratio can regulate the deamination of reduced AA, in particular the branched chain AA, as this deamination produces NADH (Hino and Russell, 1985).
Part of the difficulty in assessing how AA profile affects growth efficiency lies in how various studies were done. For example, bolus doses of isotopically labeled AA or peptides mostly yielded catabolism for energy, except for Leu, Tyr, and Phe (Armstead and Ling, 1993; Atasoglu et al., 1999). Only deletion of Leu (not deletion of other AA) decreased bacterial growth (Atasoglu et al., 2003), but Atasoglu et al. (2004) noted that Ile, Phe, Lys, and (to a somewhat lesser effect) Leu were incorporated into bacterial protein to a greater extent than were other AA. These results are consistent in that the branched chain and aromatic AA have similar metabolism within their respective groups and are more limiting than other AA.
In some of these types of studies, the peptide or AA were dosed as both the primary nitrogen and carbon source. Most of the pure cultures of predominant saccharolytic bacteria with more moderate deaminative activity did not grow well when peptides or AA provided the sole substrate (Wallace et al., 1997). Those authors reasoned that proteolysis by saccharolytic bacteria might be more important in exposing carbohydrate. Compared with other studies, there was relatively high assimilation of preformed AA in the study of Atasoglu et al. (2004) because they dosed labeled AA concomitantly with carbohydrate substrate. Those authors also noted how preformed AA, after being taken up by cells, can be catabolized if not assimilated. Firkins et al. (2015) postulated that Met is incorporated into cellular protein but that surplus intracellular Met recycles intracellularly and likely exchanges with an extracellular pool. Secretion or leakage of AA is likely when AA are high relative to concentrations needed for protein synthesis or AA are imbalanced.
Some discrepancies among studies evaluating amount or profile of preformed peptides or AA for bacteria also might be a result of inadequate adjustment time upon removal of protozoa. Protozoa appear to contain high deaminase activity (Wallace et al., 1997). However, protozoa might excrete up to half of the degradation fragments from the bacterial protein it consumes (Hristov and Jouany, 2005). Little is known about the AA profile of excreted bacterial proteins (rich in cell wall proteins?), and the excreted peptides might also be mixed with excreted but active protozoal peptidases. Although clearly important, predation of bacteria and lysis rates of protozoa probably have been exacerbated under the in vitro conditions in which normal substrate was replaced with various bacterial strains to quantify bacterial predation by loss of planktonic bacterial counts (Diaz et al., 2014). The hyperammonia-producing bacteria make a significant contribution to the total deamination activity in the rumen but are in low numbers (Walker et al., 2005). Thus, effects on growth rate of this group would be masked in mixed cultures.
In studies that have separated protozoal and bacterial fractions, there often is synergistic action when these two groups are added together (Walker et al., 2005). Defaunation (removal of protozoa) consistently decreases ruminal ammonia concentration compared with faunated controls, with explanations typically assuming exclusion of protozoal proteolytic and deaminative enzymes (Hristov and Jouany, 2005). However, an alternative explanation is that defaunation should increase the abundance of bacteria most of which assimilate ammonia, whereas protozoa do not. Ruminal protozoa can limit efficiency of microbial protein synthesis in the rumen through predation of bacteria, but there is a large gap in studies with defaunated animals at production-level intakes (Firkins et al., 2006). Recent improvements in methods allow the extraction of metabolically active protozoa with minimal bacterial contamination (Denton et al., 2015), which is critical because protozoa can degrade endogenous protein (Forsberg et al., 1984).
Asynchrony and Primary and Secondary Colonizers
The rumen microbiome has received considerable attention to optimize fiber degradation and minimize problems with ruminal acidosis (Firkins and Yu, 2015). In contrast with many studies evaluating the synchrony of nutrients for pure cultures or simple communities, the rumen is far more complex. To maintain a balanced consortium, asynchrony of carbohydrate and nitrogen sources can have a profound rippling effect through entire communities of ruminal microbes.
Physical and environmental limitations alter the degradation and usage of carbohydrate by microbes in the rumen. Degradation rates of crystalline cellulose (Weimer, 1996) by cellulases are probably limited by surface area rather than enzymatic capacity (Fields et al., 2000) and therefore are much slower than the potential growth rates by cellulolytics on the resultant cellobiose or cellodextrins (Shi and Weimer, 1997). Secondary degraders cross-feed from the degradation products produced by primary degraders of starch (Cotta, 1992), cellulose (Russell, 1985), and hemicellulose (Cotta and Whitehead, 1998). Excessive degradation of carbohydrate can decrease cellulose degradation by primary degraders; it can do so by decreasing pH or leading to depletion of growth factors (Weimer, 1996; Mouriño et al., 2001).
Role of Short-chain Fatty Acid Interconversions in Mixed Ruminal Communities
The carbon used for substrate should be reconciled with carbon recovered as products, including SCFA and cellular growth. Many SCFA interconversions and usage for anabolism allow anaerobic bacteria to fill intermediates of metabolites. Exchanges of SCFA, especially between acetate and butyrate, are to be expected (Firkins et al., 2006). Therefore, researchers have used radio or stable isotopes to support the integration of anabolic and catabolic fluxes. Indeed, an estimated 28% of [2-13C]acetate infused into cattle was not recovered as absorbed acetate (Kristensen, 2001). Many of these exchanges allow microbes to reoxidize reducing equivalents and have little net effect on growth of the community. For human fecal bacteria, numerous interconversions are possible but with a major conversion of acetate to butyrate (Falony et al., 2006; Morrison et al., 2006). Unfortunately, there is much less known for ruminal bacteria.
Cycling of Acetate During Butyrate Production in Butyrivibrios
Exogenously derived acetate can be used in a cycle to produce butyrate from acetyl coA (Diez-Gonzalez et al., 1999). In that cycle (Figure S7A), exogenous acetate would not directly wind up in butyrate but would aid in reactions transferring coenzyme A. The butyrivibrios, which are the main characterized bacteria involved in biohydrogenation, cluster taxonomically by either high or low expression of butyrate kinase (Paillard et al., 2007). The cluster with high butyrate kinase activity (cf. Figure S7B) comprised the only stearate producers so far described, whereas the cluster with the lower butyrate kinase activity (cf. Figure S7A) could not complete biohydrogenation to stearate. That latter group expressed more butyryl coA-acetyl coA transferase (butyryl coA + acetate → butyrate + acetyl coA). Presumably, the acetyl coA would then produce acetyl-phosphate and then generate ATP as acetyl kinase yields acetate (Diez-Gonzalez et al., 1999), with the acetate then becoming available for another cycle. The two groups also can be characterized based on pyruvate flux. The group expressing butyryl kinase produces little lactate and produces more acetate than the other group. In contrast, the group expressing the butyryl coA-acetyl coA transferase enzyme increases lactate production with increasing concentration of fructose-1,6-phosphate (rapid glycolysis) but takes up considerable acetate. The increasing acetate uptake would indicate that some acetyl coA must be replenishing acetyl coA pools for butyrate production and perhaps other anabolic reactions such as fatty acid biosynthesis.
Although the butyrivibro group expressing the coA transferase was only inhibited by higher concentrations of linoleic acid, when one representative isolate was dosed with linoleic acid above the inhibition threshold, the various acyl coA pools and ATP production dramatically decreased (Maia et al., 2010). Based on that response, those authors proposed a metabolic inhibition rather than disrupted membrane function to explain the toxicity from linoleic acid. This cluster of butyrivibrios produced more lactate in pure cultures, and lactate exacerbated inhibition by linoleic acid (Paillard et al., 2007). A disrupted cycle involving acetate uptake and the butyryl coA-acetyl coA transferase reaction could help explain the depleted acetyl coA pools in the study of Maia et al. (2010).
Increased starch fermentability is well-known to shift biohydrogenation away from the trans-11 pathway used by most butyrivibrios and toward the trans-10 18:1 pathway used by as yet poorly characterized bacteria (Jenkins et al., 2008). To our knowledge, energy spilling has not received much attention with butyrivibrios. We note the commonality for fructose-1,6-bisphosphate accumulation to activate lactate dehydrogenase in both the lactate-producing butyrivibrios (Diez-Gonzalez et al., 1999) and for this stimulation of lactate production to coincide with energy spilling through proton cycling in S. bovis (Bond and Russell, 1998). Human gut bacteria closely related to Butyrivibrio fibrisolvens were projected to produce butyrate through a scheme projecting a proton motive force for subsequent ATP synthesis (Louis and Flint, 2009). A research question to be tested is whether or not the butyrivibrios (especially the stearate producers) might be inhibited by higher starch fermentability because of their inability to spill energy. In contrast, might the likely candidates to biohydrogenate linoleic acid through the trans-10 pathway (i.e., Propionibacterium, Streptococcus, and Lactobacillus; (Jenkins et al., 2008) be less inhibited by high starch fermentability and more equipped to spill energy.
If bioactive fatty acids accumulate enough to inhibit butyrivibrios, then methanogens also should be affected. Rather than sinking reducing equivalents into biohydrogenation, H2 is a more favorable and important sink (Jenkins et al., 2008). To our knowledge, detailed studies with polyunsaturated fatty acids have not been done. However, medium chain fatty acids inhibit methanogens by disrupting ion gradients (Zhou et al., 2013). In that report, despite the bolus doses of these fatty acids, many methanogens were stained still active. Firkins and Yu (2015) described the poor relationship between methane production and abundance of methanogens.
De novo Synthesis of Fatty Acids
Exogenous [2-13C]acetate was elongated to butyrate and a variety of longer chain fatty acids in bacterial samples (Kristensen, 2001). Notable recovery was in the odd and anteiso fatty acids needed for bacterial membranes and with minimal isotope recovery in palmitic and stearic acids. Although bacterial long chain fatty acid synthesis was suggested as a mechanism for aerobic bacteria to store excess energy (Bas et al., 2003), the main benefit might be in allowing acetate production to provide ATP in fermentation with subsequent usage of acetate in CoA transferase reactions that consume reducing equivalents while elongating fatty acids during de novo synthesis (Duncan et al., 2002). De novo fatty acids would clearly be an important sink for carbon diverted from fermentation but also for reducing equivalents derived by fermentation, so hydrogen and carbon recovery models should be reconsidering factors affecting fatty acid biosynthesis vs. fatty acid uptake.
Supplemental fat can improve efficiency of microbial protein synthesis either by inhibition of protozoal predation on bacteria or by alleviation of the need for de novo fatty acid synthesis (Hanigan et al., 2013), which would allow more diversion of carbon toward ATP-generating fermentation rather than anabolism. Ruminal bacteria are thought to lack desaturase enzymes and must rely on methylated long chain fatty acids to maintain membrane fluidity (Russell, 2002). As described in that source, branched chain SCFA are elongated in biosynthetic reactions and are therefore important growth factors for many bacteria. In contrast with bacteria, ruminal protozoa can take up more dietary fatty acids and rely less on biosynthesis (Karnati et al., 2009).
Shifts in Fermentation Pathways Corresponding with Growth Rate
Fermentation of lactate produced by another microbe is an important cross-feeding mechanism to help buffer against ruminal acidosis (Nagaraja and Titgemeyer, 2007). They explained that, when substrate increases, S. bovis shifts fermentation toward lactate to increase ATP yield per time while decreasing ATP yield per glucose fermented. With increasing lactate production, Selenomonas ruminantium produces propionate via succinate, and Megasphaera elsdenii produces propionate via acrylate; these species are regarded as among the most important of the characterized lactate-utilizing bacteria in the rumen (Nagaraja and Titgemeyer, 2007). Exogenously supplied lactate would be labeled as propionate in different ways, depending on the pathway by those two species (Counotte et al., 1983).
Studies have elucidated regulation of lactate metabolism in some pure cultures of bacteria, and enzymatic pathways likely depend on ATP status. The intracellular ATP concentration or some similar energy gauge apparently regulates lactate metabolism in S. ruminantium (Asanuma and Hino, 2001). Higher ATP was proposed to allosterically activate pyruvate kinase to stimulate lactate production (which would decrease ATP yield from glucose). In contrast, phosphoenolpyruvate (PEP) carboxykinase (PEPCK; PEP → OAA) would be induced with lower ATP concentration. Using PEPCK allows GTP synthesis and routes carbon to pyruvate through succinate. With the lactate producer S. bovis, high ATP concentration represses pyruvate formate lyase (converting pyruvate to acetate) but induces lactate dehydrogenase to produce lactate (Asanuma and Hino, 2002). Along with allosteric activation of lactate dehydrogenase (Bond and Russell, 1996) and energy spilling (Bond and Russell, 1998), S. bovis can greatly increase its rate of lactate production when glucose concentration increases.
Lactate fermentation in the rumen is more complicated because lactate is produced in both D and L stereoisomers (Nagaraja and Titgemeyer, 2007). Glucose concentration can influence the fermentation pathway of these stereoisomers (Weimer and Moen, 2013). In that latter study, a strain of M. elsdenii fermented lactate to acetate or propionate, but when the lactate was depleted, the strain fermented glucose to butyrate and valerate. Specifically, the strain fermented glucose to acetyl-CoA, then acetate and propionate were elongated to butyrate and valerate by their condensation with acetyl-CoA. CoA transferases appear to allow metabolic versatility in M. elsdenii (Prabhu et al., 2012).
Shifts in metabolism are not unique to lactate producers and consumers. In R. flavefaciens, the primary route of OAA formation seems to be via PEPCK, but pyruvate carboxylase activity (i.e., pyruvate → OAA) increased moderately with increasing growth rates, even though OAA derived this way would decrease ATP yield compared with pyruvate through PEPCK (Shi et al., 1997). Pyruvate is the route for acetate production (which yields more ATP than succinate considering ATP production by methanogens). Pyruvate carboxylase could also produce OAA as the precursor for Asp and several other AA (Figure S6). Thus, catabolic fermentation to yield ATP for anabolic reactions might be better balanced with catabolic fermentative routes by fluxing glucose-carbon through pyruvate.
Exogenous carbon dioxide is used and produced in propionate production through succinate (Mountfort and Roberton, 1978). Those authors also detected label from [2-14C]acetate in succinate. Shi et al. (1997) projected CO2 conversion to formate in a scheme to help R. flavefaciens resolve reducing equivalents during different phases of growth as affected by substrate supply.
Thermodynamic Control of Interconversions
Ungerfeld and Kohn (2006) have elaborated on SCFA interconversions on the basis of thermodynamic principles. These principles need better integration with relative abundance of various microbes, the metabolic pathways expressed by them, and their various approaches to handling asynchronous carbohydrate supply. Those authors discussed that interconversion of SCFA is much more thermodynamically likely for acetate than the more highly reduced propionate.
Multiple approaches have been used to inhibit methanogens to suppress enteric methane emissions from ruminant livestock operations (Hristov et al., 2013). However, many of these efforts were oversimplified because of reputed interacting dietary factors such as increasing forage digestibility and ruminal passage rate (Janssen, 2010). That author presented evidence that increasing aqueous H2 concentration would be associated with increasing methanogen growth rate but also decrease acetate production by the fermentative microbes based on thermodynamic principles. He acknowledged that the ratio of acetate to methane is not constant, but rather depends on whether H2 or propionate is formed along with acetate to balance reducing equivalents.
Interconversions of lactate and the SCFA could potentially influence various mechanistic models being derived to associate SCFA stoichiometry and methane production. For example, whether butyrate is produced through butyryl kinase or butyryl coA-acetyl coA transferase can vary the production of the intermediate lactate. If lactate is increased, there should be less of the intermediate H2 being produced in the first place and the lactate subsequently being fermented to propionate. If expressing butyryl coA-acetyl coA transferase does indeed have an advantage, probably through balancing reducing equivalents and ATP synthesis (Louis and Flint, 2009), then it might also allow a more efficient bacterial growth.
Conclusions
Microbial protein production is inefficient largely owing to maintenance functions, accumulation of reserve carbohydrate, and energy spilling. Reserve carbohydrate is accumulated primarily by protozoa and under even modest carbohydrate excesses, whereas energy spilling occurs under larger excesses. Future work needs to identify microbial groups and biochemical mechanisms for spilling. Interconversion of lactate and SCFA is another potential mechanism for microbes to better manage rates of catabolic and anabolic reactions or, conversely, might be associated with energy spilling.
Principles summarized in this review could improve prediction of microbial protein production by mechanistic models, guiding efforts to maximize efficiency of that production. Some models already represent energy spilling and reserve carbohydrate, but most model parameter values are simple constants or heuristic (not derived directly from data) (Dijkstra et al., 1992; Russell et al., 1992; Dijkstra, 1994; Baldwin, 1995; Hackmann and Spain, 2010). For example, most mechanistic models assume storage of reserve carbohydrate is a constant fraction of microbial biomass (Russell et al., 1992; Baldwin, 1995; Hackmann and Spain, 2010). This review suggests that in order to improve prediction of microbial protein, models need improved representation of energy spilling and reserve carbohydrate, and it also points out experimental data needed to achieve this better representation.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
TJH was supported by the USDA National Institute of Food and Agriculture, Hatch project FLA-ANS-005307 and Hatch/Multi-State project FLA-ANS-005304. JLF was supported by Agriculture and Food Research Initiative Competitive Grant no. 2012-67015-19437 from the USDA National Institute of Food and Agriculture and by state and federal funds appropriated to the Ohio Agricultural Research and Development Center, The Ohio State University.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00465/abstract
Figure S1. Bucket model of energy spilling. The large bucket represents the main pool of ATP-equivalents available to cell functions (maintenance, growth, reserve carbohydrate, energy spilling). The smaller bucket represents pool of ATP-equivalents in reserve carbohydrate, which can be stored from and mobilized to the main pool by pumps. Modified from Russell (2002).
Figure S2. Dynamics of reserve carbohydrate accumulation for mixed bacteria in the rumen. Diet was fed to calves once daily. Ingredient composition of basal diet 1.15 kg/d hay and straw and 1.26 kg/d flaked maize. Crude protein content was 2.56% DM for basal diet and between 6.56 and 8.32% for basal diet supplemented with urea. Reserve carbohydrate (“α-dextran glucose”) was measured as glucose detected by ion-exchange chromatography after hydrolysis in 0.5 N H2SO4 (100°C, 4 h). Figure redrawn from McAllan and Smith (1974).
Figure S3. Dynamics of carbohydrate accumulation for the protozoan Dasytricha ruminantium in the rumen. Diet was fed to sheep. Ingredient composition was 0.6 kg cubed molassed sugar-beet pulp and 0.3 kg chopped hay. Beet pulp was fed at 07.00 h and chopped hay fed at 16.00 h. Carbohydrate was measured by the phenol-sulfuric acid method after hydrolysis in 1 N NaOH (100°C, 5 min). Data from Williams and Harfoot (1976).
Figure S4. Equations for synthesis and degradation of glycogen.
Figure S5. Equations for cellodextrin synthesis and efflux. Na+ transport out of the cell is assumed to require 1/3 ATP (Russell, 2002). Cellobiose synthesis is shown, but synthesis of longer cellodextrins is also observed. After Wells et al. (1995).
Figure S6. Amino acid (AA) biosynthesis in various bacteria, primarily Escherichia coli. PEP, phosphoenolpyruvate; OAA, oxaloacetate; SAM, S-adenosyl methionine; and HMB, 2-hydroxy-4-(methylthio) butanoic acid. Redrawn from Morrison and Mackie (1997), with minor modifications (Paulus and Gray, 1967; Baldwin and Allison, 1983; Or-Rashid et al., 2001; Walker et al., 2005). Numerous reactions are combined in the solid arrows representing enzymatic reactions. Examples of feedback inhibition are indicated by (−) and dashed arrows were excerpted from studies with non-rumen bacteria, again primarily E. coli (Gottschalk, 1979; Kalcheva et al., 1994; Hindson, 2003; Lohkamp et al., 2004; Yang et al., 2005; Caldara et al., 2008; Ferla and Patrick, 2014). Most rumen bacteria lack a complete TCA cycle but can make α ketoglutarate by forward or backward reactions from OAA (Wallace et al., 1997).
Figure S7. Butyrate formation by two groups of ruminal Butyrivibrio species as proposed by Diez-Gonzalez et al. (1999) and further elaborated on by Paillard et al. (2007). (A) Butyrate formation by butyryl-CoA/acetate CoA transferase, phosphotransacetylase, and acetate kinase. (B) Butyrate formation by butyrate kinase. See those sources for enzymes. The group on the right expresses butyrate kinase but produces more acetate, consumes little acetate, and produces little lactate. This group fully biohydrogenates to stearate. The group on the left expresses butyryl coA-acetyl coA transferase, produces more butyrate and lactate and consumes more acetate. Dashed boxes denote carbon input, although glucose is generalized from disaccharide or other sugars entering the glycolysis pathway and even as a phosphorylated monosaccharide. ATP, adenosine triphosphate; CoA, coenzyme A; GTP, guanosine tri phosphate; OAA, oxaloacetate; P, phosphate; and PEP, phosphoenolpyruvate.
References
Andries, J. I., Buysse, F. X., de Brabander, D. L., and Cottyn, B. G. (1987). Isoacids in ruminant nutrition: their role in ruminal and intermediary metabolism and possible influences on performances—a review. Anim. Feed Sci. Technol. 18, 169–180. doi: 10.1016/0377-8401(87)90069-1
Argyle, J. L., and Baldwin, R. L. (1989). Effects of amino acids and peptides on rumen microbial growth yields. J. Dairy Sci. 72, 2017–2027. doi: 10.3168/jds.S0022-0302(89)79325-5
Armstead, I. P., and Ling, J. R. (1993). Variations in the uptake and metabolism of peptides and amino acids by mixed ruminal bacteria in vitro. Appl. Environ. Microbiol. 59, 3360–3366.
Asanuma, N., and Hino, T. (2001). Molecular characterization, enzyme properties and transcriptional regulation of phosphoenolpyruvate carboxykinase and pyruvate kinase in a ruminal bacterium, Selenomonas ruminantium. Microbiology 147, 681–690.
Asanuma, N., and Hino, T. (2002). Molecular characterization and xxpression of pyruvate formate-lyase-activating enzyme in a ruminal bacterium, Streptococcus bovis. Appl. Environ. Microbiol. 68, 3352–3357. doi: 10.1128/AEM.68.7.3352-3357.2002
Atasoglu, C., Guliye, A. Y., and Wallace, R. J. (2003). Use of a deletion approach to assess the amino acid requirements for optimum fermentation by mixed micro-organisms from the sheep rumen. Anim. Sci. 76, 147–153.
Atasoglu, C., Guliye, A. Y., and Wallace, R. J. (2004). Use of stable isotopes to measure de novo synthesis and turnover of amino acid-C and -N in mixed micro-organisms from the sheep rumen in vitro. Br. J. Nutr. 91, 253–262. doi: 10.1079/BJN20031040
Atasoglu, C., Valdés, C., Newbold, C. J., and Wallace, R. J. (1999). Influence of peptides and amino acids on fermentation rate and de novo synthesis of amino acids by mixed micro-organisms from the sheep rumen. Br. J. Nutr. 81, 307–314.
Bailey, R. W. (1959). Transglucosidase activity of rumen strains of Streptococcus bovis. 2. Isolation and properties of dextransucrase. Biochem. J. 72, 42–49.
Bailey, R. W., and Oxford, A. E. (1958). A quantitative study of the production of dextran from sucrose by rumen strains of Streptococcus bovis. J. Gen. Microbiol. 19, 130–145. doi: 10.1099/00221287-19-1-130
Bas, P., Archimède, H., Rouzeau, A., and Sauvant, D. (2003). Fatty acid composition of mixed-rumen bacteria: effect of concentration and type of forage. J. Dairy Sci. 86, 2940–2948. doi: 10.3168/jds.S0022-0302(03)73891-0
Bond, D. R., and Russell, J. B. (1996). A role for fructose 1,6-diphosphate in the ATPase-mediated energy-spilling reaction of Streptococcus bovis. Appl. Environ. Microbiol. 62, 2095–2099.
Bond, D. R., and Russell, J. B. (1998). Relationship between intracellular phosphate, proton motive force, and rate of nongrowth energy dissipation (energy spilling) in Streptococcus bovis JB1. Appl. Environ. Microbiol. 64, 976–981.
Bond, J. J., Dunne, J. C., Kwan, F. Y., Li, D., Zhang, K., Leahy, S. C., et al. (2012). Carbohydrate transporting membrane proteins of the rumen bacterium, Butyrivibrio proteoclasticus. J. Proteomics 75, 3138–3144. doi: 10.1016/j.jprot.2011.12.013
Buurman, E. T., Teixeira de Mattos, M. J., and Neijssel, O. M. (1991). Futile cycling of ammonium ions via the high affinity potassium uptake system (Kdp) of Escherichia coli. Arch. Microbiol. 155, 391–395. doi: 10.1007/BF00243460
Caldara, M., Dupont, G., Leroy, F., Goldbeter, A., De Vuyst, L., and Cunin, R. (2008). Arginine biosynthesis in Escherichia coli: experimental perturbation and mathematical modeling. J. Biol. Chem. 283, 6347–6358. doi: 10.1074/jbc.M705884200
Chen, G. H., Mo, H. K., Saby, S., Yip, W. K., and Liu, Y. (2000). Minimization of activated sludge production by chemically stimulated energy spilling. Water Sci. Tech. 42, 189–200.
Cheng, K. J., Hironaka, R., Jones, G. A., Nicas, T., and Costerton, J. W. (1976). Frothy feedlot bloat in cattle: production of extracellular polysaccharides and development of viscosity in cultures of Streptococcus bovis. Can. J. Microbiol. 22, 450–459. doi: 10.1139/m76-071
Clapperton, J. L., and Czerkawski, J. W. (1969). Methane production and soluble carbohydrates in the rumen of sheep in relation to the time of feeding and the effects of short-term intraruminal infusions of unsaturated fatty acids. Br. J. Nutr. 23, 813–826. doi: 10.1079/BJN19690092
Clark, J. H., Klusmeyer, T. H., and Cameron, M. R. (1992). Microbial protein synthesis and flows of nitrogen fractions to the duodenum of dairy cows. J. Dairy Sci. 75, 2304–2323. doi: 10.3168/jds.S0022-0302(92)77992-2
Cook, G. M., and Russell, J. B. (1994). Energy-spilling reactions of Streptococcus bovis and resistance of its membrane to proton conductance. Appl. Environ. Microbiol. 60, 1942–1948.
Costerton, J. W., Damgaard, H. N., and Cheng, K. J. (1974). Cell envelope morphology of rumen bacteria. J. Bacteriol. 118, 1132–1143.
Cotta, M. A. (1992). Interaction of ruminal bacteria in the production and utilization of maltooligosaccharides from starch. Appl. Environ. Microbiol. 58, 48–54.
Cotta, M. A., and Whitehead, T. R. (1998). Xylooligosaccharide utilization by the ruminal anaerobic bacterium Selenomonas ruminantium. Curr. Microbiol. 36, 183–189. doi: 10.1007/s002849900291
Counotte, G. H., Lankhorst, A., and Prins, R. A. (1983). Role of DL-lactic acid as an intermediate in rumen metabolism of dairy cows. J. Anim. Sci. 56, 1222–1235.
Denton, B. L., Diese, D. E., Firkins, J. L., and Hackmann, T. J. (2015). Accumulation of reserve carbohydrate by protozoa and bacteria in competition for glucose. Appl. Environ. Microbiol. 81, 1832–1838. doi: 10.1128/AEM.03736-14
Diaz, H. L., Barr, K. N., Godden, K. R., Plank, J. E., Zapata, I., Schappacher, A. N., et al. (2014). Eukaryotic inhibitors or activators elicit responses to chemosensory compounds by ruminal isotrichid and entodiniomorphid protozoa. J. Dairy Sci. 97, 2254–2269. doi: 10.3168/jds.2013-7698
Diez-Gonzalez, F., Bond, D. R., Jennings, E., and Russell, J. B. (1999). Alternative schemes of butyrate production in Butyrivibrio fibrisolvens and their relationship to acetate utilization, lactate production, and phylogeny. Arch. Microbiol. 171, 324–330. doi: 10.1007/s002030050717
Dijkstra, J. (1994). Simulation of the dynamics of protozoa in the rumen. Br. J. Nutr. 72, 679–699. doi: 10.1079/BJN19940071
Dijkstra, J., France, J., and Davies, D. R. (1998). Different mathematical approaches to estimating microbial protein supply in ruminants. J. Dairy Sci. 81, 3370–3384. doi: 10.3168/jds.S0022-0302(98)75902-8
Dijkstra, J., Neal, H. D., Beever, D. E., and France, J. (1992). Simulation of nutrient digestion, absorption and outflow in the rumen: model description. J. Nutr. 122, 2239–2256.
Duncan, S. H., Barcenilla, A., Stewart, C. S., Pryde, S. E., and Flint, H. J. (2002). Acetate utilization and butyryl coenzyme A (CoA):acetate-CoA transferase in butyrate-producing bacteria from the human large intestine. Appl. Environ. Microbiol. 68, 5186–5190. doi: 10.1128/AEM.68.10.5186-5190.2002
Dunne, J. C., Li, D., Kelly, W. J., Leahy, S. C., Bond, J. J., Attwood, G. T., et al. (2012). Extracellular polysaccharide-degrading proteome of Butyrivibrio proteoclasticus. J. Proteome Res. 11, 131–142. doi: 10.1021/pr200864j
Falony, G., Vlachou, A., Verbrugghe, K., and De Vuyst, L. (2006). Cross-feeding between Bifidobacterium longum BB536 and acetate-converting, butyrate-producing colon bacteria during growth on oligofructose. Appl. Environ. Microbiol. 72, 7835–7841. doi: 10.1128/AEM.01296-06
Ferla, M. P., and Patrick, W. M. (2014). Bacterial methionine biosynthesis. Microbiology 160, 1571–1584. doi: 10.1099/mic.0.077826-0
Fields, M. W., Mallik, S., and Russell, J. B. (2000). Fibrobacter succinogenes S85 ferments ball-milled cellulose as fast as cellobiose until cellulose surface area is limiting. Appl. Microbiol. Biotechnol. 54, 570–574. doi: 10.1007/s002530000426
Firkins, J. L., Fowler, C. M., Devillard, E., and Bequette, B. J. (2015). Kinetics of microbial methionine metabolism in continuous cultures administered different methionine sources. J. Dairy Sci. 98, 1178–1194. doi: 10.3168/jds.2014-8694
Firkins, J. L., Hristov, A. N., Hall, M. B., Varga, G. A., and St-Pierre, N. R. (2006). Integration of ruminal metabolism in dairy cattle. J. Dairy Sci. 89 (Suppl. 1), E31–E51. doi: 10.3168/jds.S0022-0302(06)72362-1
Firkins, J. L., and Yu, Z. (2015). How to use data on the rumen microbiome to improve our understanding of ruminant nutrition. J. Anim. Sci. 93, 1450–1470. doi: 10.2527/jas.2014-8754
Firkins, J. L., Yu, Z., and Morrison, M. (2007). Ruminal nitrogen metabolism: perspectives for integration of microbiology and nutrition for dairy. J. Dairy Sci. 90 (Suppl. 1), E1–E16. doi: 10.3168/jds.2006-518
Flint, H. J., Bayer, E. A., Rincon, M. T., Lamed, R., and White, B. A. (2008). Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat. Rev. Microbiol. 6, 121–131. doi: 10.1038/nrmicro1817
Forsberg, C. W., Lovelock, L. K., Krumholz, L., and Buchanan-Smith, J. G. (1984). Protease activities of rumen protozoa. Appl. Environ. Microbiol. 47, 101–110.
Gaudet, G., Forano, E., Dauphin, G., and Delort, A. M. (1992). Futile cycling of glycogen in Fibrobacter succinogenes as shown by in situ 1H-NMR and 13C-NMR investigation. Eur. J. Biochem. 207, 155–162. doi: 10.1111/j.1432-1033.1992.tb17032.x
Gottschalk, G. (1979). Bacterial Metabolism. New York, NY: Springer-Verlag. doi: 10.1007/978-1-4684-0465-4
Hackmann, T. J., Diese, L. E., and Firkins, J. L. (2013a). Quantifying the responses of mixed rumen microbes to excess carbohydrate. Appl. Environ. Microbiol. 79, 3786–3795. doi: 10.1128/AEM.00482-13
Hackmann, T. J., Keyser, B. L., and Firkins, J. L. (2013b). Evaluation of methods to detect changes in reserve carbohydrate for mixed rumen microbes. J. Microbiol. Methods 93, 284–291. doi: 10.1016/j.mimet.2013.03.025
Hackmann, T. J., and Spain, J. N. (2010). A mechanistic model for predicting intake of forage diets by ruminants. J. Anim. Sci. 88, 1108–1124. doi: 10.2527/jas.2008-1378
Hall, M. B. (2011). Isotrichid protozoa influence conversion of glucose to glycogen and other microbial products. J. Dairy Sci. 94, 4589–4602. doi: 10.3168/jds.2010-3878
Hanigan, M. D., Appuhamy, J. A., and Gregorini, P. (2013). Revised digestive parameter estimates for the Molly cow model. J. Dairy Sci. 96, 3867–3885. doi: 10.3168/jds.2012-6183
Hindson, V. J. (2003). Serine acetyltransferase of Escherichia coli: substrate specificity and feedback control by cysteine. Biochem. J. 375, 745–752. doi: 10.1042/BJ20030429
Hino, T., and Russell, J. B. (1985). Effect of reducing-equivalent disposal and NADH/NAD on deamination of amino acids by intact rumen microorganisms and their cell extracts. Appl. Environ. Microbiol. 50, 1368–1374.
Hobson, P. N., and Macpherson, M. J. (1953). Encapsulation in rumen bacterial fractions. Nature 171, 129–130. doi: 10.1038/171129b0
Hobson, P. N., and Macpherson, M. J. (1954). Some serological and chemical studies on materials extracted from an amylolytic streptococcus from the rumen of the sheep. Biochem. J. 57, 145–151.
Hottiger, T., Schmutz, P., and Wiemken, A. (1987). Heat-induced accumulation and futile cycling of trehalose in Saccharomyces cerevisiae. J. Bacteriol. 169, 5518–5522.
Hristov, A. N., and Jouany, J. P. (2005). “Factors affecting the efficiency of nitrogen utilization in the rumen,” in Nitrogen and Phosphorus Nutrition of Cattle: Reducing the Environmental Impact of Cattle Operations, eds E. Pfeffer and A. Hristov (Wallingford, UK: CABI), 117–166. doi: 10.1079/9780851990132.0117
Hristov, A. N., Oh, J., Firkins, J. L., Dijkstra, J., Kebreab, E., Waghorn, G., et al. (2013). Special topics–mitigation of methane and nitrous oxide emissions from animal operations: I. A review of enteric methane mitigation options. J. Anim. Sci. 91, 5045–5069. doi: 10.2527/jas.2013-6583
Isaacson, H. R., Hinds, F. C., Bryant, M. P., and Owens, F. N. (1975). Efficiency of energy utilization by mixed rumen bacteria in continuous culture. J. Dairy Sci. 58, 1645–1659. doi: 10.3168/jds.S0022-0302(75)84763-1
Janssen, P. H. (2010). Influence of hydrogen on rumen methane formation and fermentation balances through microbial growth kinetics and fermentation thermodynamics. Anim. Feed Sci. Tech. 160, 1–22. doi: 10.1016/j.anifeedsci.2010.07.002
Jenkins, T. C., Wallace, R. J., Moate, P. J., and Mosley, E. E. (2008). Board-invited review: recent advances in biohydrogenation of unsaturated fatty acids within the rumen microbial ecosystem. J. Anim. Sci. 86, 397–412. doi: 10.2527/jas.2007-0588
Jouany, J. P., and Thiven, P. (1972). Évolution postprandiale de la composition glucidique des coprs microbiens du rumen en foction de la nature des glucides du regime. Ann. Biol. Anim. Biochim. Biophys. 12, 673–677. doi: 10.1051/rnd:19720414
Kajikawa, H., Amari, M., and Masaki, S. (1997). Glucose transport by mixed ruminal bacteria from a cow. Appl. Environ. Microbiol. 63, 1847–1851.
Kajikawa, H., and Masaki, S. (1999). Cellobiose transport by mixed ruminal bacteria from a cow. Appl. Environ. Microbiol. 65, 2565–2569.
Kajikawa, H., Mitsumori, M., and Ohmomo, S. (2002). Stimulatory and inhibitory effects of protein amino acids on growth rate and efficiency of mixed ruminal bacteria. J. Dairy Sci. 85, 2015–2022. doi: 10.3168/jds.S0022-0302(02)74278-1
Kalcheva, E. O., Shanskaya, V. O., and Maliuta, S. S. (1994). Activities and regulation of the enzymes involved in the first and the third steps of the aspartate biosynthetic pathway in Enterococcus faecium. Arch. Microbiol. 161, 359–362. doi: 10.1007/BF00303593
Karnati, S. K., Sylvester, J. T., Ribeiro, C. V., Gilligan, L. E., and Firkins, J. L. (2009). Investigating unsaturated fat, monensin, or bromoethanesulfonate in continuous cultures retaining ruminal protozoa. I. Fermentation, biohydrogenation, and microbial protein synthesis. J. Dairy Sci. 92, 3849–3860. doi: 10.3168/jds.2008-1436
Kim, J. N., Henriksen, E. D., Cann, I. K., and Mackie, R. I. (2014). Nitrogen utilization and metabolism in Ruminococcus albus 8. Appl. Environ. Microbiol. 80, 3095–3102. doi: 10.1128/AEM.00029-14
Koenig, K. M., Newbold, C. J., McIntosh, F. M., and Rode, L. M. (2000). Effects of protozoa on bacterial nitrogen recycling in the rumen. J. Anim. Sci. 78, 2431–2445.
Krause, D. O., Nagaraja, T. G., Wright, A. D., and Callaway, T. R. (2013). Board-invited review: rumen microbiology: leading the way in microbial ecology. J. Anim. Sci. 91, 331–341. doi: 10.2527/jas.2012-5567
Kristensen, N. B. (2001). Rumen microbial sequestration of [2-13C]acetate in cattle. J. Anim. Sci. 79, 2491–2498.
Larsson, C., von Stockar, U., Marison, I., and Gustafsson, L. (1993). Growth and metabolism of Saccharomyces cerevisiae in chemostat cultures under carbon-, nitrogen-, or carbon- and nitrogen-limiting conditions. J. Bacteriol. 175, 4809–4816.
Leedle, J. A., Bryant, M. P., and Hespell, R. B. (1982). Diurnal variations in bacterial numbers and fluid parameters in ruminal contents of animals fed low- or high-forage diets. Appl. Environ. Microbiol. 44, 402–412.
Lohkamp, B., McDermott, G., Campbell, S. A., Coggins, J. R., and Lapthorn, A. J. (2004). The structure of Escherichia coli ATP-phosphoribosyltransferase: identification of substrate binding sites and mode of AMP inhibition. J. Mol. Biol. 336, 131–144. doi: 10.1016/j.jmb.2003.12.020
Lou, J., Dawson, K. A., and Strobel, H. J. (1996). Role of phosphorolytic cleavage in cellobiose and cellodextrin metabolism by the ruminal bacterium Prevotella ruminicola. Appl. Environ. Microbiol. 62, 1770–1773.
Lou, J., Dawson, K. A., and Strobel, H. J. (1997a). Glycogen biosynthesis via UDP-glucose in the ruminal bacterium Prevotella bryantii B1(4). Appl. Environ. Microbiol. 63, 4355–4359.
Lou, J., Dawson, K. A., and Strobel, H. J. (1997b). Glycogen formation by the ruminal bacterium Prevotella ruminicola. Appl. Environ. Microbiol. 63, 1483–1488.
Louis, P., and Flint, H. J. (2009). Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol. Lett. 294, 1–8. doi: 10.1111/j.1574-6968.2009.01514.x
Mackie, R., Gilchrist, F., Robberts, A., Hannah, P., and Schwartz, H. (1978). Microbiological and chemical changes in the rumen during the stepwise adaptation of sheep to high concentrate diets. J. Agric. Sci. 90, 241–254. doi: 10.1017/S0021859600055313
Maia, M. R., Chaudhary, L. C., Bestwick, C. S., Richardson, A. J., McKain, N., Larson, T. R., et al. (2010). Toxicity of unsaturated fatty acids to the biohydrogenating ruminal bacterium, Butyrivibrio fibrisolvens. BMC Microbiol. 10:52. doi: 10.1186/1471-2180-10-52
Martin, S. A. (1994). Nutrient transport by ruminal bacteria: a review. J. Anim. Sci. 72, 3019–3031.
Matheron, C., Delort, A. M., Gaudet, G., Forano, E., and Liptaj, T. (1998). 13C and 1H nuclear magnetic resonance study of glycogen futile cycling in strains of the genus Fibrobacter. Appl. Environ. Microbiol. 64, 74–81.
Matulova, M., Delort, A. M., Nouaille, R., Gaudet, G., and Forano, E. (2001). Concurrent maltodextrin and cellodextrin synthesis by Fibrobacter succinogenes S85 as identified by 2D NMR spectroscopy. Eur. J. Biochem. 268, 3907–3915. doi: 10.1046/j.1432-1327.2001.02300.x
McAllan, A. B., and Smith, R. H. (1974). Carbohydrate metabolism in the ruminant. Bacterial carbohydrates formed in the rumen and their contribution to digesta entering the duodenum. Br. J. Nutr. 31, 77–88. doi: 10.1079/BJN19740010
Morgan, R. M., Pihl, T. D., Nolling, J., and Reeve, J. N. (1997). Hydrogen regulation of growth, growth yields, and methane gene transcription in Methanobacterium thermoautotrophicum Δ H. J. Bacteriol. 179, 889–898.
Morgavi, D. P., Kelly, W. J., Janssen, P. H., and Attwood, G. T. (2013). Rumen microbial (meta)genomics and its application to ruminant production. Animal 7 (Suppl. 1), 184–201. doi: 10.1017/S1751731112000419
Morrison, D. J., Mackay, W. G., Edwards, C. A., Preston, T., Dodson, B., and Weaver, L. T. (2006). Butyrate production from oligofructose fermentation by the human faecal flora: what is the contribution of extracellular acetate and lactate? Br. J. Nutr. 96, 570–577. doi: 10.1079/BJN20061853
Morrison, M., and Mackie, R. I. (1997). “Biosynthesis of nitrogen-containing compounds,” in Gastrointestinal Ecosystems and Fermentations, eds R. I. Mackie and B. A. White (New York, NY: Chapman and Hall), 424–469.
Mountfort, D. O., and Roberton, A. M. (1978). Origins of fermentation products formed during growth of Bacteroides ruminicola on glucose. J. Gen. Microbiol. 106, 353–360. doi: 10.1099/00221287-106-2-353
Mouriño, F., Akkarawongsa, R., and Weimer, P. J. (2001). Initial pH as a determinant of cellulose digestion rate by mixed ruminal microorganisms in vitro. J. Dairy Sci. 84, 848–859. doi: 10.3168/jds.S0022-0302(01)74543-2
Mulder, M. M., Teixeira de Mattos, M. J., Postma, P. W., and van Dam, K. (1986). Energetic consequences of multiple K+ uptake systems in Escherichia coli. Biochim. Biophys. Acta 851, 223–228. doi: 10.1016/0005-2728(86)90129-5
Nagaraja, T. G., and Titgemeyer, E. C. (2007). Ruminal acidosis in beef cattle: the current microbiological and nutritional outlook. J. Dairy Sci. 90 (Suppl. 1), E17–E38. doi: 10.3168/jds.2006-478
Neijssel, O. M., and Tempest, D. W. (1976). Bioenergetic aspects of aerobic growth of Klebsiella aerogenes NCTC 418 in carbon-limited and carbon-sufficient chemostat culture. Arch. Microbiol. 107, 215–221. doi: 10.1007/BF00446843
Newsholme, E. A., and Crabtree, B. A. (1975). Substrate cycles in metabolic regulation and in heat generation. Biochem. Soc. Symp. 12, 61–109.
Newsholme, E., Challiss, R., and Crabtree, B. (1984). Substrate cycles: their role in improving sensitivity in metabolic control. Trends Biochem. Sci. 9, 277–280. doi: 10.1016/0968-0004(84)90165-8
Nouaille, R., Matulova, M., Delort, A. M., and Forano, E. (2005). Oligosaccharide synthesis in Fibrobacter succinogenes S85 and its modulation by the substrate. FEBS J. 272, 2416–2427. doi: 10.1111/j.1742-4658.2005.04662.x
Oldick, B. S., Firkins, J. L., and Kohn, R. A. (2000). Compartmental modeling with nitrogen-15 to determine effects of degree of fat saturation on intraruminal N recycling. J. Anim. Sci. 78, 2421–2430.
Or-Rashid, M. M., Onodera, R., and Wadud, S. (2001). Biosynthesis of methionine from homocysteine, cystathionine and homoserine plus cysteine by mixed rumen microorganisms in vitro. Appl. Microbiol. Biotechnol. 55, 758–764. doi: 10.1007/s002530100548
Otto, R. (1984). Uncoupling of growth and acid production in Streptococcus cremoris. Arch. Microbiol. 140, 225–230. doi: 10.1007/BF00454932
Paillard, D., McKain, N., Chaudhary, L. C., Walker, N. D., Pizette, F., Koppova, I., et al. (2007). Relation between phylogenetic position, lipid metabolism and butyrate production by different Butyrivibrio-like bacteria from the rumen. Antonie Van Leeuwenhoek 91, 417–422. doi: 10.1007/s10482-006-9121-7
Paulus, H., and Gray, E. (1967). Multivalent feedback inhibition of aspartokinase in Bacillus polymyxa. I. Kinetic studies. J. Biol. Chem. 242, 4980–4986.
Pirt, S. J. (1965). The maintenance energy of bacteria in growing cultures. Proc. R. Soc. Lond. B Biol. Sci. 163, 224–231. doi: 10.1098/rspb.1965.0069
Piwonka, E. J., Firkins, J. L., and Hull, B. L. (1994). Digestion in the rumen and total tract of forage-based diets with starch or dextrose supplements fed to Holstein heifers. J. Dairy Sci. 77, 1570–1579. doi: 10.3168/jds.S0022-0302(94)77099-5
Portais, J. C., and Delort, A. M. (2002). Carbohydrate cycling in micro-organisms: what can 13C-NMR tell us? FEMS Microbiol. Rev. 26, 375–402. doi: 10.1111/j.1574-6976.2002.tb00621.x
Prabhu, R., Altman, E., and Eiteman, M. A. (2012). Lactate and acrylate metabolism by Megasphaera elsdenii under batch and steady-state conditions. Appl. Environ. Microbiol. 78, 8564–8570. doi: 10.1128/AEM.02443-12
Preiss, J., and Romeo, T. (1989). Physiology, biochemistry and genetics of bacterial glycogen synthesis. Adv. Microb. Physiol. 30, 183–238. doi: 10.1016/S0065-2911(08)60113-7
Prins, R. A., and Van Hoven, W. (1977). Carbohydrate fermentation by the rumen ciliate Isotricha prostoma. Protistologica 13, 549–556.
Rosewarne, C. P., Pope, P. B., Cheung, J. L., and Morrison, M. (2014). Analysis of the bovine rumen microbiome reveals a diversity of Sus-like polysaccharide utilization loci from the bacterial phylum Bacteroidetes. J. Ind. Microbiol. Biotechnol. 41, 601–606. doi: 10.1007/s10295-013-1395-y
Russell, J. (2002). Rumen Microbiology and its Role in Ruminant Nutrition. Ithaca, NY: James B. Russell.
Russell, J. (2007a). Can the heat of ruminal fermentation be manipulated to decrease heat stress? Proc. Southwest Nutr. Conf. 22, 109–115.
Russell, J. B. (1985). Fermentation of cellodextrins by cellulolytic and noncellulolytic rumen bacteria. Appl. Environ. Microbiol. 49, 572–576.
Russell, J. B. (1986). Heat production by ruminal bacteria in continuous culture and its relationship to maintenance energy. J. Bacteriol. 168, 694–701.
Russell, J. B. (1992). Glucose toxicity and inability of Bacteroides ruminicola to regulate glucose transport and utilization. Appl. Environ. Microbiol. 58, 2040–2045.
Russell, J. B. (1993). Effect of amino acids on the heat production and growth efficiency of Streptococcus bovis: balance of anabolic and catabolic rates. Appl. Environ. Microbiol. 59, 1747–1751.
Russell, J. B. (1998). Strategies that ruminal bacteria use to handle excess carbohydrate. J. Anim. Sci. 76, 1955–1963.
Russell, J. B. (2007b). The energy spilling reactions of bacteria and other organisms. J. Mol. Microbiol. Biotechnol. 13, 1–11. doi: 10.1159/000103591
Russell, J. B., and Cook, G. M. (1995). Energetics of bacterial growth: balance of anabolic and catabolic reactions. Microbiol. Rev. 59, 48–62.
Russell, J. B., Muck, R. E., and Weimer, P. J. (2009). Quantitative analysis of cellulose degradation and growth of cellulolytic bacteria in the rumen. FEMS Microbiol. Ecol. 67, 183–197. doi: 10.1111/j.1574-6941.2008.00633.x
Russell, J. B., and Strobel, H. J. (1990). ATPase-dependent energy spilling by the ruminal bacterium, Streptococcus bovis. Arch. Microbiol. 153, 378–383. doi: 10.1007/BF00249009
Russell, J., O'Connor, J., Fox, D., Van Soest, P., and Sniffen, C. (1992). A net carbohydrate and protein system for evaluating cattle diets: I. Ruminal fermentation. J. Anim. Sci. 70, 3551–3561.
Russell, J., and Wallace, R. (1997). “Energy-yielding and energy-consuming reactions,” in The Rumen Microbial Ecosystem, 2nd Edn., eds P. N. Hobson and C. S. Stewart (New York, NY: Blackie Academic and Professional), 247–282.
Ryan, R. (1964). Concentrations of glucose and low-molecular-weight acids in the rumen of sheep changed gradually from a hay to a hay-plus-grain diet. Am. J. Vet. Res. 25, 653–659.
Saleem, F., Ametaj, B. N., Bouatra, S., Mandal, R., Zebeli, Q., Dunn, S. M., et al. (2012). A metabolomics approach to uncover the effects of grain diets on rumen health in dairy cows. J. Dairy Sci. 95, 6606–6623. doi: 10.3168/jds.2012-5403
Schönheit, P., Moll, J., and Thauer, R. K. (1980). Growth parameters (Ks, μmax, Ys) of Methanobacterium thermoautotrophicum. Arch. Microbiol. 127, 59–65. doi: 10.1007/BF00414356
Shi, Y., and Weimer, P. J. (1997). Competition for cellobiose among three predominant ruminal cellulolytic bacteria under substrate-excess and substrate-limited conditions. Appl. Environ. Microbiol. 63, 743–748.
Shi, Y., Weimer, P. J., and Ralph, J. (1997). Formation of formate and hydrogen, and flux of reducing equivalents and carbon in Ruminococcus flavefaciens FD-1. Antonie Van Leeuwenhoek 72, 101–109. doi: 10.1023/A:1000256221938
Stewart, C. S., Flint, H. J., and Bryant, M. P. (1997). “The rumen bacteria,” in The Rumen Microbial Ecosystem, 2nd Edn., eds P. N. Hobson and C. S. Stewart (New York, NY: Blackie Academic and Professional), 10–72. doi: 10.1007/978-94-009-1453-7_2
Storm, E., Orskov, E. R., and Smart, R. (1983). The nutritive value of rumen micro-organisms in ruminants. 2. The apparent digestibility and net utilization of microbial N for growing lambs. Br. J. Nutr. 50, 471–478. doi: 10.1079/BJN19830115
Stouthamer, A. H. (1973). A theoretical study on the amount of ATP required for synthesis of microbial cell material. Antonie Van Leeuwenhoek 39, 545–565. doi: 10.1007/BF02578899
Suen, G., Weimer, P. J., Stevenson, D. M., Aylward, F. O., Boyum, J., Deneke, J., et al. (2011). The complete genome sequence of Fibrobacter succinogenes S85 reveals a cellulolytic and metabolic specialist. PLoS ONE 6:e18814. doi: 10.1371/journal.pone.0018814
Teixeira de Mattos, M. J., and Tempest, D. W. (1983). Metabolic and energetic aspects of the growth of Klebsiella aerogenes NCTC 418 on glucose in anaerobic chemostat culture. Arch. Microbiol. 134, 80–85. doi: 10.1007/BF00429412
Tempest, D. (1978). The biochemical significance of microbial growth yields: a reassessment. Trends Biochem. Sci. 3, 180–184. doi: 10.1016/S0968-0004(78)91127-1
Tempest, D. W., and Neijssel, O. M. (1984). The status of YATP and maintenance energy as biologically interpretable phenomena. Annu. Rev. Microbiol. 38, 459–486. doi: 10.1146/annurev.mi.38.100184.002331
Ungerfeld, E. M., and Kohn, R. A. (2006). “The role of thermodynamics in the control of ruminal fermentation,” in Ruminant Physiology: Digestion, Metabolism and Impact of Nutrition on Gene Expression, Immunology and Stress, eds K. Sejrsen, T. Hvelplund, and M. O. Nielsen (Wageningen: Wageningen Academic Publishers), 55–86.
Vandehaar, M. J. (2005). “Dairy cattle,” in Basic Animal Nutrition and Feeding, eds W. G. Pond, D. C. Church, K. R. Pond, and P. A. Schoknecht (Hoboken, NJ: John Wiley and Sons), 413–437.
van Hijum, S. A., Kralj, S., Ozimek, L. K., Dijkhuizen, L., and van Geel-Schutten, I. G. (2006). Structure-function relationships of glucansucrase and fructansucrase enzymes from lactic acid bacteria. Microbiol. Mol. Biol. Rev. 70, 157–176. doi: 10.1128/MMBR.70.1.157-176.2006
Van Hoven, W., and Prins, R. A. (1977). Carbohydrate fermentation by the rumen ciliate Dasytricha ruminantium. Protistologica 13, 599–606.
Van Kessel, J., and Russell, J. (1997). The endogenous polysaccharide utilization rate of mixed ruminal bacteria and the effect of energy starvation on ruminal fermentation rates. J. Dairy Sci. 80, 2442–2448. doi: 10.3168/jds.S0022-0302(97)76196-4
Van Kessel, J. S., and Russell, J. B. (1996). The effect of amino nitrogen on the energetics of ruminal bacteria and its impact on energy spilling. J. Dairy Sci. 79, 1237–1243. doi: 10.3168/jds.S0022-0302(96)76476-7
Van Urk, H., Mark, P. R., Scheffers, W. A., and Van Dijken, J. P. (1988). Metabolic responses of Saccharomyces cerevisiae CBS 8066 and Candida utilis CBS 621 upon transition from glucose limitation to glucose excess. Yeast 4, 283–291. doi: 10.1002/yea.320040406
Walker, N. D., Newbold, C. J., and Wallace, R. J. (2005). “Nitrogen metabolism in the rumen,” in Nitrogen and Phosphorus Nutrition of Cattle, eds E. Pfeffer and A. Hristov (Cambridge, MA: CABI Publishing), 71–115. doi: 10.1079/9780851990132.0071
Wallace, R. J., and McPherson, C. A. (1987). Factors affecting the rate of breakdown of bacterial protein in rumen fluid. Br. J. Nutr. 58, 313–323. doi: 10.1079/BJN19870098
Wallace, R. J., Onodera, R., and Cotta, M. A. (1997). “Metabolism of nitrogen-containing compounds,” in The Rumen Microbial Ecosystem, 2nd Edn., eds P. N. Hobson and C. S. Stewart (New York, NY: Chapman and Hall), 283–328. doi: 10.1007/978-94-009-1453-7_7
Wang, L., Hatem, A., Catalyurek, U. V., Morrison, M., and Yu, Z. (2013). Metagenomic insights into the carbohydrate-active enzymes carried by the microorganisms adhering to solid digesta in the rumen of cows. PLoS ONE 8:e78507. doi: 10.1371/journal.pone.0078507
Weimer, P. J. (1996). Why don't ruminal bacteria digest cellulose faster? J. Dairy Sci. 79, 1496–1502. doi: 10.3168/jds.S0022-0302(96)76509-8
Weimer, P. J., and Moen, G. N. (2013). Quantitative analysis of growth and volatile fatty acid production by the anaerobic ruminal bacterium Megasphaera elsdenii T81. Appl. Microbiol. Biotechnol. 97, 4075–4081. doi: 10.1007/s00253-012-4645-4
Weimer, P. J., Price, N. P., Kroukamp, O., Joubert, L. M., Wolfaardt, G. M., and Van Zyl, W. H. (2006). Studies of the extracellular glycocalyx of the anaerobic cellulolytic bacterium Ruminococcus albus 7. Appl. Environ. Microbiol. 72, 7559–7566. doi: 10.1128/AEM.01632-06
Wells, J. E., and Russell, J. B. (1996). Why do many ruminal bacteria die and lyse so quickly? J. Dairy Sci. 79, 1487–1495. doi: 10.3168/jds.S0022-0302(96)76508-6
Wells, J. E., Russell, J. B., Shi, Y., and Weimer, P. J. (1995). Cellodextrin efflux by the cellulolytic ruminal bacterium Fibrobacter succinogenes and its potential role in the growth of nonadherent bacteria. Appl. Environ. Microbiol. 61, 1757–1762.
White, B. A., Lamed, R., Bayer, E. A., and Flint, H. J. (2014). Biomass utilization by gut microbiomes. Annu. Rev. Microbiol. 68, 279–296. doi: 10.1146/annurev-micro-092412-155618
Wilkinson, J. (1959). The problem of energy-storage compounds in bacteria. Exp. Cell Res. 7, 111–130. doi: 10.1016/0014-4827(59)90237-X
Williams, A., and Coleman, G. (1992). The Rumen Protozoa. New York, NY: Springer-Verlag. doi: 10.1007/978-1-4612-2776-2
Williams, A., and Harfoot, C. (1976). Factors affecting the uptake and metabolism of soluble carbohydrates by the rumen ciliate Dasytricha ruminantium isolated from ovine rumen contents by filtration. J. Gen. Microbiol. 96, 125. doi: 10.1099/00221287-96-1-125
Yang, C. R., Shapiro, B. E., Hung, S. P., Mjolsness, E. D., and Hatfield, G. W. (2005). A mathematical model for the branched chain amino acid biosynthetic pathways of Escherichia coli K12. J. Biol. Chem. 280, 11224–11232. doi: 10.1074/jbc.M411471200
Ze, X., Le Mougen, F., Duncan, S. H., Louis, P., and Flint, H. J. (2013). Some are more equal than others: the role of “keystone” species in the degradation of recalcitrant substrates. Gut Microbes 4, 236–240. doi: 10.4161/gmic.23998
Keywords: rumen microbiology, reserve carbohydrate, glycogen, energy spilling, short-chain fatty acids
Citation: Hackmann TJ and Firkins JL (2015) Maximizing efficiency of rumen microbial protein production. Front. Microbiol. 6:465. doi: 10.3389/fmicb.2015.00465
Received: 09 February 2015; Accepted: 28 April 2015;
Published: 15 May 2015.
Edited by:
Emilio M. Ungerfeld, Instituto de Investigaciones Agropecuarias - Carillanca, ChileReviewed by:
Evelyne Forano, Institut National de la Recherche Agronomique, FranceAmlan Kumar Patra, West Bengal University of Animal and Fishery Sciences, India
Copyright © 2015 Hackmann and Firkins. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Timothy J. Hackmann, Department of Animal Science, University of Florida, PO Box 110910, Gainesville, FL 32611, USA,dGhhY2ttYW5uQHVmbC5lZHU=
 Timothy J. Hackmann
Timothy J. Hackmann Jeffrey L. Firkins
Jeffrey L. Firkins