- Institute for Biomedical Sciences, Petit Science Center, Georgia State University, Atlanta, GA, USA
The paramyxovirus family includes major human and animal pathogens, including measles virus, mumps virus, and human respiratory syncytial virus (RSV), as well as the emerging zoonotic Hendra and Nipah viruses. In the U.S., RSV is the leading cause of infant hospitalizations due to viral infectious disease. Despite their clinical significance, effective drugs for the improved management of paramyxovirus disease are lacking. The development of novel anti-paramyxovirus therapeutics is therefore urgently needed. Paramyxoviruses contain RNA genomes of negative polarity, necessitating a virus-encoded RNA-dependent RNA polymerase (RdRp) complex for replication and transcription. Since an equivalent enzymatic activity is absent in host cells, the RdRp complex represents an attractive druggable target, although structure-guided drug development campaigns are hampered by the lack of high-resolution RdRp crystal structures. Here, we review the current structural and functional insight into the paramyxovirus polymerase complex in conjunction with an evaluation of the mechanism of activity and developmental status of available experimental RdRp inhibitors. Our assessment spotlights the importance of the RdRp complex as a premier target for therapeutic intervention and examines how high-resolution insight into the organization of the complex will pave the path toward the structure-guided design and optimization of much-needed next-generation paramyxovirus RdRp blockers.
Introduction
Paramyxoviruses are enveloped, non-segmented and single-stranded RNA viruses with negative genome polarity (NNRV) in the order Mononegavirales, which also includes the Bornaviridae, Filoviridae, and Rhabdoviridae families. The paramyxoviruses encompass major human and animal pathogens such as respiratory syncytial virus (RSV), measles virus (MeV), mumps virus (MuV), and Newcastle disease virus (NDV). The family is organized into two subfamilies, the Pneumovirinae and the Paramyxovirinae. While RSV belongs to the former subfamily, MeV, MuV, NDV, and the newly emerged Hendra and Nipah viruses are all part of the Paramyxovirinae.
All paramyxoviruses spread through the respiratory route and predominantly cause acute disease, and several members of the family are extremely contagious. For example, MeV is considered the most infectious viral pathogen identified to date (Kelly et al., 2002; Centers for Disease and Prevention, 2012a). Although vaccines are available for some paramyxoviruses, much-needed effective antiviral therapeutics for post-exposure prophylaxis and improved disease management are lacking. Moreover, vaccine prophylaxis against several clinically highly significant members of the family is still unavailable despite major past research efforts.
Respiratory syncytial virus, for instance, is the leading cause of infant mortality from viral respiratory disease and responsible for over 120,000 infant hospitalizations per year in the U.S. alone. Whereas clinical symptoms of paramyxovirus disease are frequently based on immunopathogenicity rather than directly virus induced (Hall et al., 1971; Auwaerter et al., 1999), in the case of RSV infection higher viral loads serve as a predictor of RSV lower respiratory tract infection in infected infants (DeVincenzo et al., 2005). Among hospitalized RSV-infected children less than 2 years of age, viral load on day three of hospitalization was also associated with a requirement for intensive care and respiratory failure (El Saleeby et al., 2011). These findings spotlight a window of opportunity for improved RSV disease management through therapeutics, but post-exposure prophylaxis may be the only viable indication against other clinically significant members of the family. For example, we propose that a combined prophylactic and post-exposure therapeutic anti-measles platform may be required to ultimately prevail in a prolonged endgame of gaining global measles control (Plemper and Snyder, 2009; Plemper and Hammond, 2014). Despite major educational efforts, herd immunity remains too low to interrupt endemic MeV transmission in large areas of Western Europe due to parental concerns against vaccination (Larson et al., 2011; Saint-Victor and Omer, 2013), and local pockets with low vaccination coverage increasingly sustain transmission of imported virus in the U.S. (Centers for Disease and Prevention, 2012b).
Executing essential and virus-specific enzymatic activities, the viral RdRp complex represents an attractive, albeit underexplored, target for therapeutics. This review will summarize current insight into the spatial organization and function of the paramyxovirus RdRp complex and assess candidate druggable targets within the complex based on the available structural information and experimental therapeutics.
Components of the RdRp Complex
The overall genome organization and fundamental principles for genome replication and transcription are conserved between different paramyxoviruses and, to some extend, all NNRV. Throughout the virus replication cycle, the genome exists as a unique ribonucleoprotein complex, the nucleocapsid (NC), which is composed of the genomic RNA completely sequestered by copies of the viral NC (N) protein. Only the NC can serve as a template for RNA synthesis by the RdRp complex, which consists of the viral large (L) and phospho-(P) proteins in addition to host co-factors. The L protein contains all enzymatic activities exercised by the complex, while P acts as an essential cofactor. The NC, P, and L core complex functions as both replicase and transcriptase. Although present in all paramyxoviruses, in most cases only homotypic N, P, and L combinations, in which each component is derived from the same paramyxovirus family member, are bioactive (Smallwood and Moyer, 2004; Dochow et al., 2012). Functional studies on N, P, and L have furthermore confirmed that each of the RdRp components can individually and differentially affect the processes of mRNA synthesis and genome replication (Perlman and Huang, 1973; Chen et al., 1997; Fearns et al., 1997; Hwang et al., 1999; Galloway and Wertz, 2008, 2009; Harouaka and Wertz, 2009).
Transcriptase Activity
Upon entry into the host cell, virion uncoating separates genome and viral envelope and releases the NC along with the attached RdRp into the cytoplasm. Once in the cytoplasm, encapsidated genomic RNA serves as the template for both transcription and replication. Leader (Le) and trailer (Tr) sequences are located at the 3′- and 5′-termini of the genome, respectively, and harbor the genomic and antigenomic promoter elements, (Figure 1). RSV contains a linear genomic promoter that spans the first 12 residues of the genome (Noton et al., 2014), while members of the paramyxovirinae subfamily contain bipartite promoters (Figure 2A; Pelet et al., 1996; Murphy et al., 1998; Tapparel et al., 1998). Encapsidation is essential for the assembly of a functional bipartite promoter, since distinct promoter elements are juxtaposed only in the helical NC. Consensus sequences that are involved in transcription initiation, polyadenylation, and transcription termination of individual genes are located at the beginning and end of each gene. In transcriptase mode, RdRp initiates synthesis of the first functional mRNA at the first gene-start consensus sequence. The nascent mRNA is capped and methylated by L, and then the full mRNA transcript is generated (Moyer and Banerjee, 1975; Ogino et al., 2005; Ogino and Banerjee, 2007). At the end of each open reading frame, RdRp recognizes a signal for non-templated polyadenylation, followed by release of the viral mRNA (Lamb and Parks, 2007). Next, the complex proceeds over the intergenic sequence and reinitiates transcription at the next downstream transcription start site. However, reinitation is only partially efficient, which results in a transcription gradient – the synthesis of progressively less of each viral mRNA as the RdRp advances along the template – that is characteristic for all members of the mononegavirales.
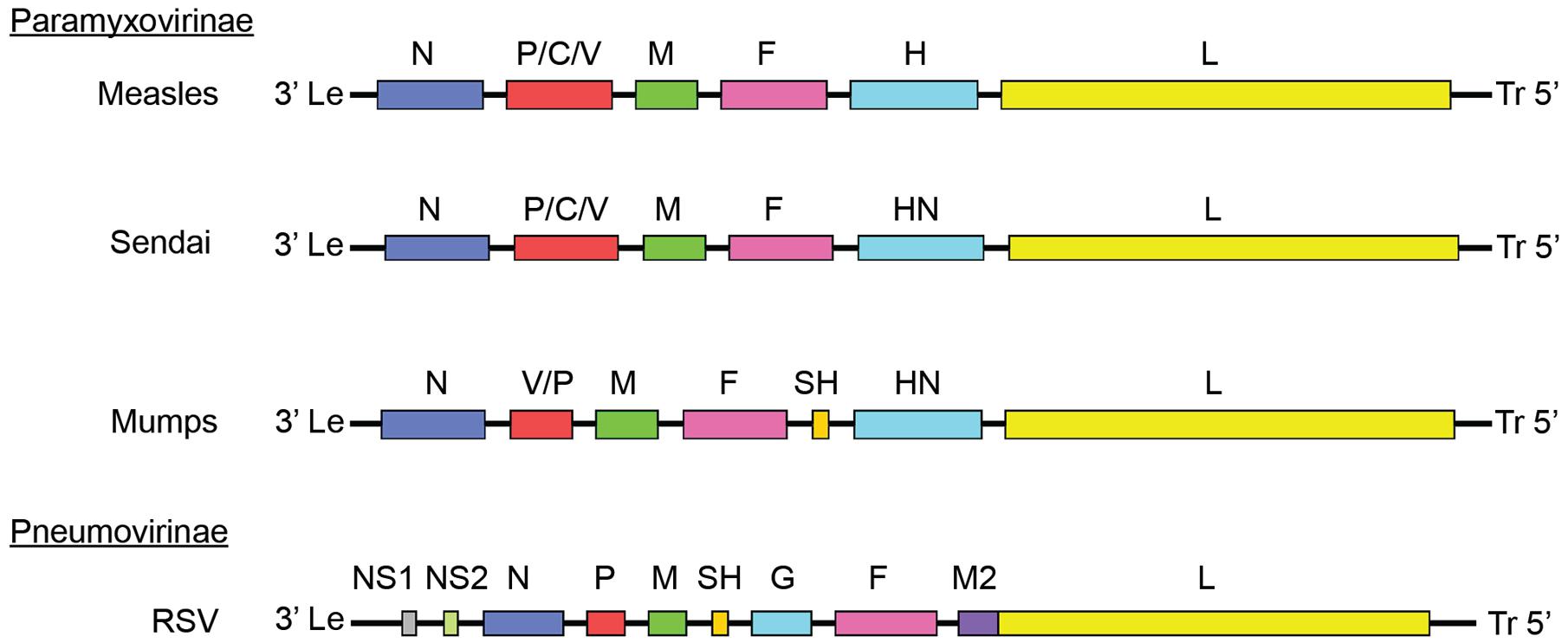
FIGURE 1. Paramyxoviridae genome organization. Genomes of different members of the Paramyxoviridae family. Members of the Paramyxovirinae subfamily include measles virus (MeV), mumps virus (MuV), and Sendai virus. Respiratory syncytial virus (RSV) is a member of the pneumovirinae subfamily. Genome organization and numbers of open reading frames differ between members of the family, but all paramyxoviruses use a core RdRp composed of the NC, P, and L.
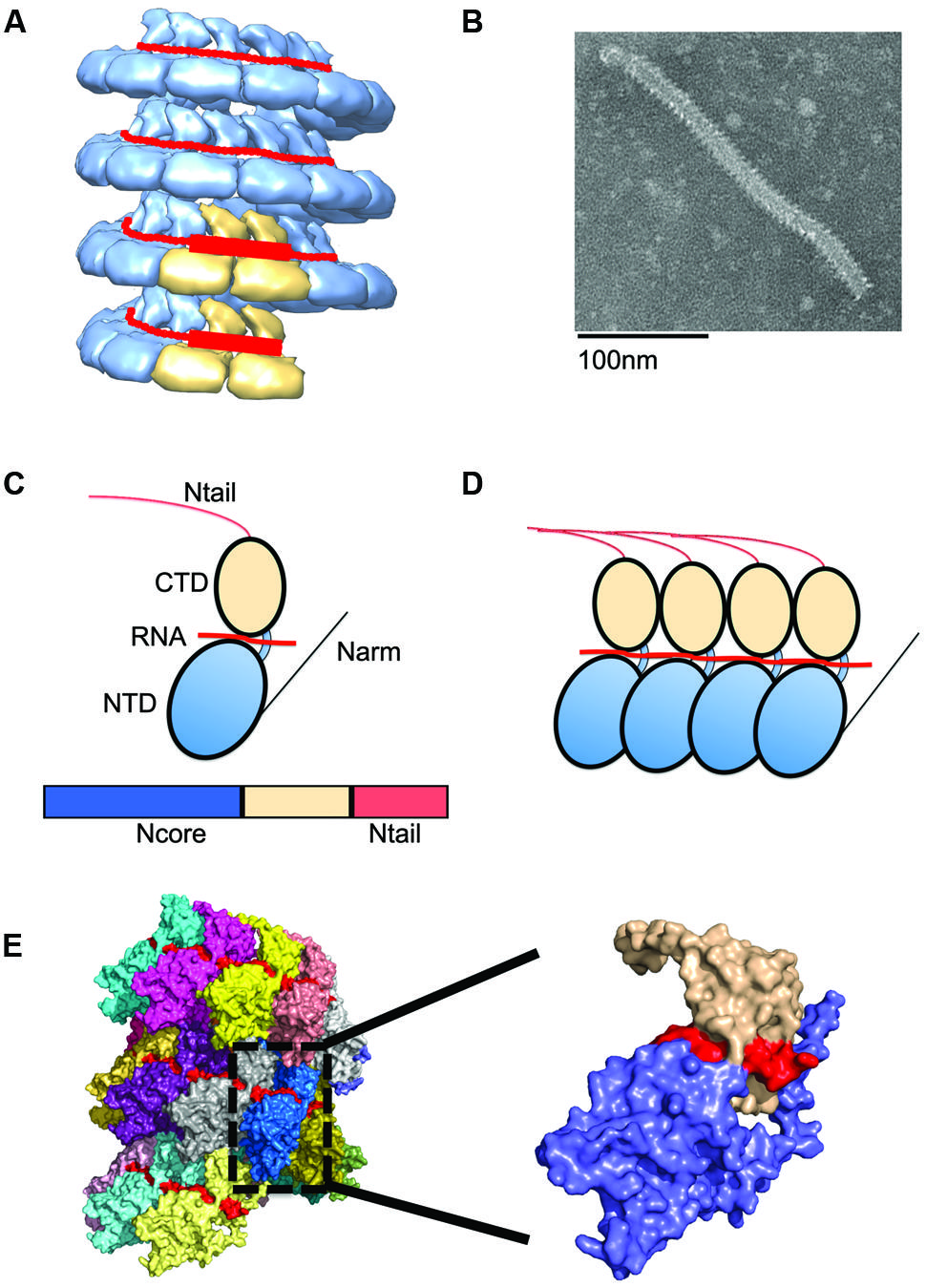
FIGURE 2. Nucleocapsid (NC) architecture. (A) Illustration of the bipartite promoter organization by example of the MuV NC (EMD-2630; Cox et al., 2014). N protein protomers encapsidating promoter regions in genomic RNA are highlighted in tan. (B) Typical ‘herring-bone’ structure of the paramyxovirus NC. (C) The paramyxovirus N protein is composed of two core domains (NCore), NTD (blue), and CTD (tan). A schematic of the N protein is shown below the cartoon model. For paramyxoviruses, the extreme C-terminus of N, NTail, extends out from the NC and interacts with P. (D) Extensions from these domains, N-arm and C-arm interact with neighboring N subunits in the helical NC. (D,E) N subunits assemble side by side along the genomic RNA. (E) The structure of the RSV NC showing the encapsidated RNA in red. PDB code 4BKK (Bakker et al., 2013). The insert shows an enlargement of a single N protein protomer, color coded as in (C).
Replicase Activity
Although transcription and replication use the same viral proteins, they are two distinct processes. While transcription by some NNRV RdRps can occur in vitro in the presence of NC, the correct salts, and ribonucleotides (Emerson and Wagner, 1972; Davis and Wertz, 1982), genome replication requires ongoing N protein synthesis, since the nascent genomic or antigenomic RNA is encapsidated concomitantly. In order to switch from transcription to replication, a sufficiently large amount of N protein must be available in order to encapsidate the newly synthesized genomes and antigenomes. In fact, in the case of the paramyxovirinae at least, the intracellular N protein pool serves as a major driver inducing the switch from initial transcription to replication (Baker and Moyer, 1988; Horikami et al., 1992). For the pneumovirinae, however, the available N protein pool alone is not responsible for the shift to replicase functionality, since increased levels of RSV N enhanced antigenome synthesis, but had no effect on transcription levels in RSV minireplicon experiments (Fearns et al., 1997). When in replicase mode, RdRps derived from either subfamily ignore all cis-acting signals, such as polyadenylation sites, to produce full-length genomic RNA copies.
The array of distinct enzymatic activities of the RdRp complex and the highly dynamic protein–protein and protein–RNA interactions that are required for bioactivity provide rich opportunity for therapeutic interference. As a basis for discussing individual druggable targets, we will illuminate the role of the viral protein components in RdRp complex assembly and function.
Nucleocapsid Protein
The paramyxovirus NC shows a characteristic herringbone structure in electron micrographs (Figure 2). Despite this defined appearance, the NC remains flexible with variations in pitch and helical symmetry parameter along its length, which may be required to allow the polymerase complex to access the encapsidated RNA without disassembling the helix (Heggeness et al., 1980; Egelman et al., 1989; Bhella et al., 2002, 2004).
N subunits in the NC are assembled side-by-side and parallel along the length of the RNA to form a highly unique protein–RNA complex, in which the viral RNA is entirely sequestered by the N protein (Figure 2; Tawar et al., 2009). Each N protomer is organized into an N-terminal (NTD) and C-terminal (CTD) core domain, which are connected through a hinge region (Figure 2). Both the NTD and CTD interact laterally with adjacent subunits. The RNA interaction site is positioned at the NTD/CTD interface, forming a basic surface groove into which the RNA threads belt-like along the outside of the NC (Figure 2). A crystal structure of the RSV N domains was recently solved and reveals parallel layers of RSV N:RNA rings (El Omari et al., 2011). The NTD and CTD of each N subunit have N- and C-terminal extensions, termed N-arm (residues 1–28) and C-arm (residues 360–375), respectively, which attach to neighboring N subunits (Tawar et al., 2009). Of these, the N-arm is considered to provide the main stabilizing lateral N–N interaction. However, weaker top–bottom interactions may likely exist between different rungs of the helical NC. Between layers, the RSV N subunits engage in weak contacts between the N-terminal domains of one layer and the C-terminal domains of the adjacent lower layer (El Omari et al., 2011). In the RSV N:RNA structure, the C-arm lies above the CTD, occupying space between consecutive turns of the helical NC. However, for other paramyxoviruses, the extreme C-terminus of N, called N-tail, is displayed on the exterior of the NC (Figure 3; Jensen et al., 2011; Communie et al., 2013b). Removal of the N-tail causes the NC to rigidify, rendering it more compact and biologically inactive (Schoehn et al., 2004).
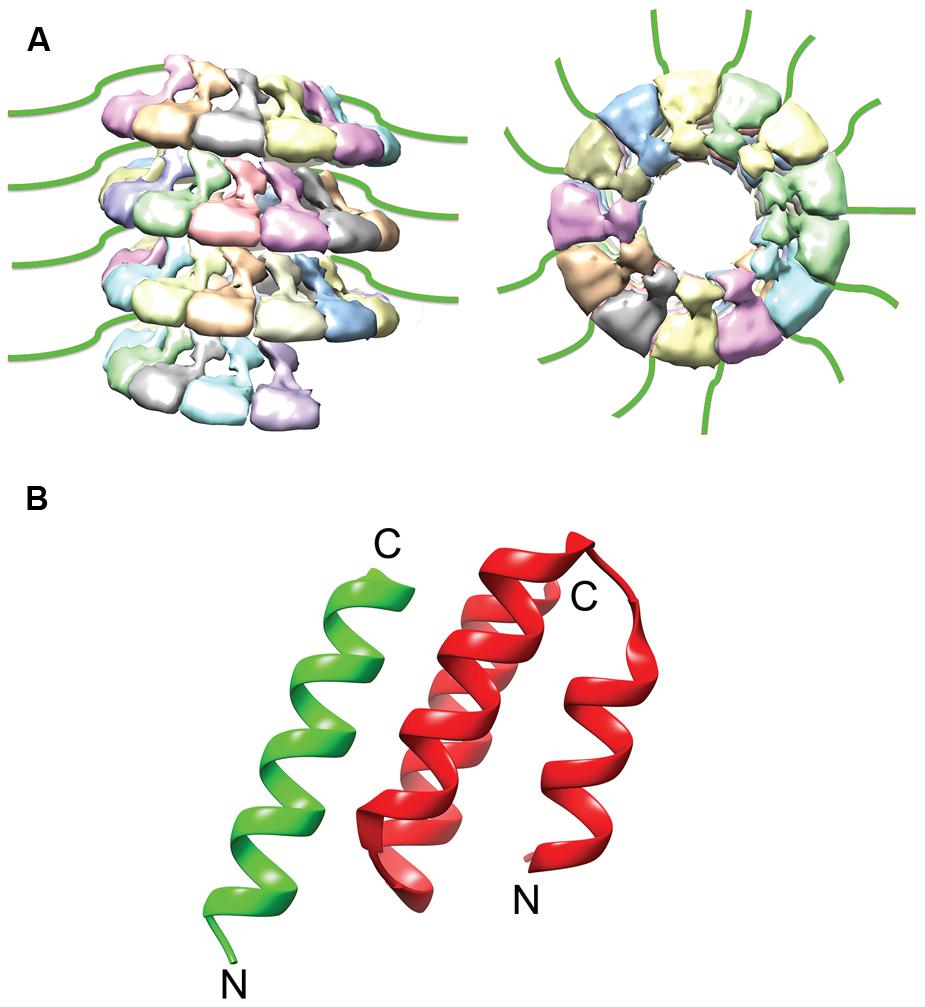
FIGURE 3. NTail and NTail–PCTD Interaction. (A) The extreme C-terminal region of N, NTail (green), is highly flexible and extends outward from the assembled NC (Jensen et al., 2011). (B) A short helix in NTail (green) is involved in binding the C-terminal NC binding domain of P (red). The N- and C- termini are labeled (PDB code 1T6O; Kingston et al., 2004a). The NC model was modified from the mumps NC structure (EMDataBank access code EMD-2630; Cox et al., 2014).
Phosphoprotein
The P protein lacks inherent catalytic activity, but is an essential co-factor of the RdRp complex. Although required for the replication of all NNRVs, paramyxovirus P proteins vary greatly in length and sequence (Figure 4; Tarbouriech et al., 2000b; Karlin et al., 2003; Ding et al., 2004, 2006; Mavrakis et al., 2004; Ivanov et al., 2010). P performs a dynamic range of different functions in the virus replication cycle. The protein is thought to properly position the L protein for RNA synthesis (Kingston et al., 2004a,b, 2008), interact with the NC template (Habchi et al., 2011; Longhi, 2012; Communie et al., 2013b; Cox et al., 2014), and chaperone newly synthesized, RNA-free N protein (N0) to the nascent viral RNA during replication (Mavrakis et al., 2006; Chen et al., 2007; Yabukarski et al., 2014). Reflecting these diverse tasks, P shows a modular organization of different functional domains separated by flexible linker regions (Tarbouriech et al., 2000a; Karlin et al., 2003; Blanchard et al., 2004; Llorente et al., 2006). Structures for individual domains of several NNRV P proteins have been solved previously (Figure 4; Tarbouriech et al., 2000b; Ding et al., 2004, 2006; Mavrakis et al., 2004; Ivanov et al., 2010), but the structure of a full-length paramyxovirus P has yet to be determined.
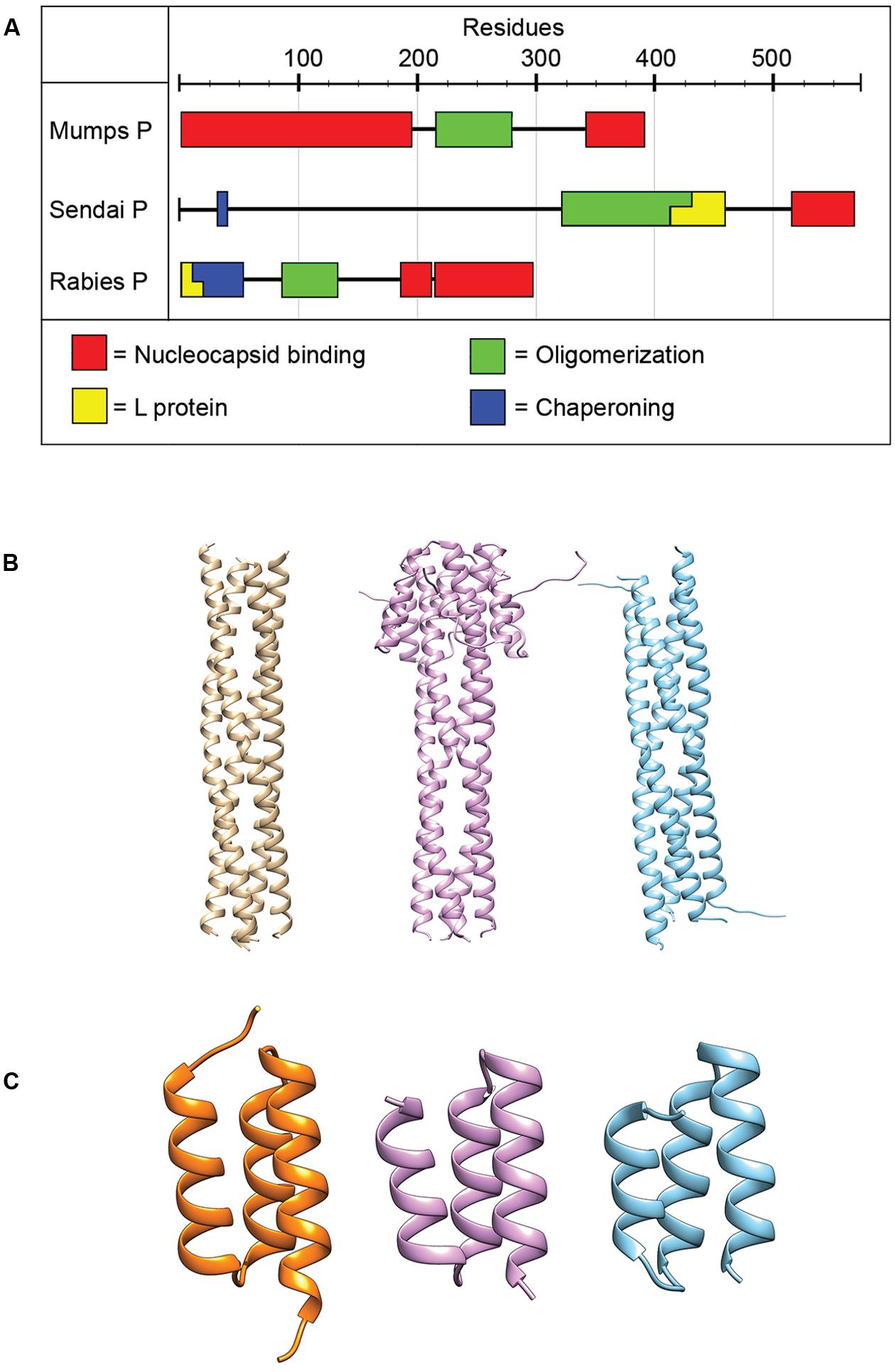
FIGURE 4. Phosphoprotein organization and structure. (A) The P proteins of NNRVs vary in length and domain organization. All NNRV P form oligomers. (B) The oligomerization state of all paramyxovirus P proteins is a tetramer. Tetrameric structures for NiV, MeV, and MuV are shown from left to right (PDB codes 4GJW, 4BHV, and 4EIJ for NiV, MeV, and MuV, respectively; Communie et al., 2013a; Cox et al., 2013; Bruhn et al., 2014). (C) The structures of the C-terminal NC binding domain of several paramyxovirus P have been solved and are highly conserved structurally. Structures shown from left to right are for HeV, MeV, and MuV. (PDB codes 4HEO, 3BBZ, and 1OKS, respectively; Johansson et al., 2003; Kingston et al., 2008; Communie et al., 2013b).
All NNRV P proteins contain a motif in their central region known as the oligomerization domain (Tarbouriech et al., 2000b; Ding et al., 2006; Gerard et al., 2009; Ivanov et al., 2010; Communie et al., 2013a; Cox et al., 2013; Bruhn et al., 2014). In addition, all NNRV P form homo-oligomers, but their lengths and oligomerization states vary (Tarbouriech et al., 2000b; Gerard et al., 2009; Communie et al., 2013a; Cox et al., 2013; Bruhn et al., 2014). However, the tetramer is considered to represent the physiological oligomer of paramyxovirus P proteins (Tarbouriech et al., 2000a,b; Cox et al., 2013; Bruhn et al., 2014). Proper P oligomerization is required for its role in both transcription and replication (Tarbouriech et al., 2000b; Kolakofsky et al., 2004; Chen et al., 2006), and structures of the oligomerization domains for several paramyxoviruses have been solved (Figure 4; Tarbouriech et al., 2000b; Communie et al., 2013a; Cox et al., 2013; Bruhn et al., 2014).
Since the L protein alone is unable to bind efficiently to the NC, a key function of P is to position the RdRp on the NC and ensure continued contact between the RdRp and the template as the complex progresses along the NC. According to the precedent set by vesicular stomatitis virus (VSV), an NNRV of the rhabdovirus family, after P binding to its NC, N-terminal L binding domains protrude outward and may serve as a latch to position L (Emerson and Schubert, 1987; Morin et al., 2012). Based off of previously solved crystal structures, it is possible that paramyxovirus P proteins bind L in a similar fashion (Tarbouriech et al., 2000a,b; Cox et al., 2013; Bruhn et al., 2014), since in all of these structures N-terminal domains are proposed to protrude outward.
In the absence of other viral proteins, N has a strong tendency to polymerize and to encapsidated non-viral cellular RNAs. To prevent non-productive N polymerization, P acts as a molecular chaperone and complexes RNA-free N0 forms in N0–P structures (Mavrakis et al., 2006; Chen et al., 2007; Leyrat et al., 2011; Yabukarski et al., 2014). In addition to blocking premature oligomerization, the N0–P complexes inhibit non-specific encapsidation of cellular RNA and keep N0 soluble (Masters and Banerjee, 1988; Chen et al., 2007; Leyrat et al., 2011; Yabukarski et al., 2014). Crystal structures for VSV and NiV N0–P complexes have been solved (Figure 5; Leyrat et al., 2011; Yabukarski et al., 2014). A comparison between the structures of the VSV and NiV N0–P complexes reveals a common mechanism of N0 chaperoning (Leyrat et al., 2011; Yabukarski et al., 2014). In both cases, the N-terminal N0-binding region of P prevents N polymerization by occupying the binding cavity for the N-arm and C-arm of adjacent N subunits. Proper encapsidation of the newly synthesized RNA genome requires the delivery of soluble RNA-free N0 to the site of RNA synthesis (Yabukarski et al., 2014). The N0–P complex can bind to the NC, but little is known about the reaction by which N0 is transferred from P to the RNA. Conceivably, an N0–P complex may bind to the NC template through the C-terminal NC binding domain of P, and the intrinsic flexibility of P may properly position and orient the N0 molecule within the replication complex and deliver it to the nascent RNA.
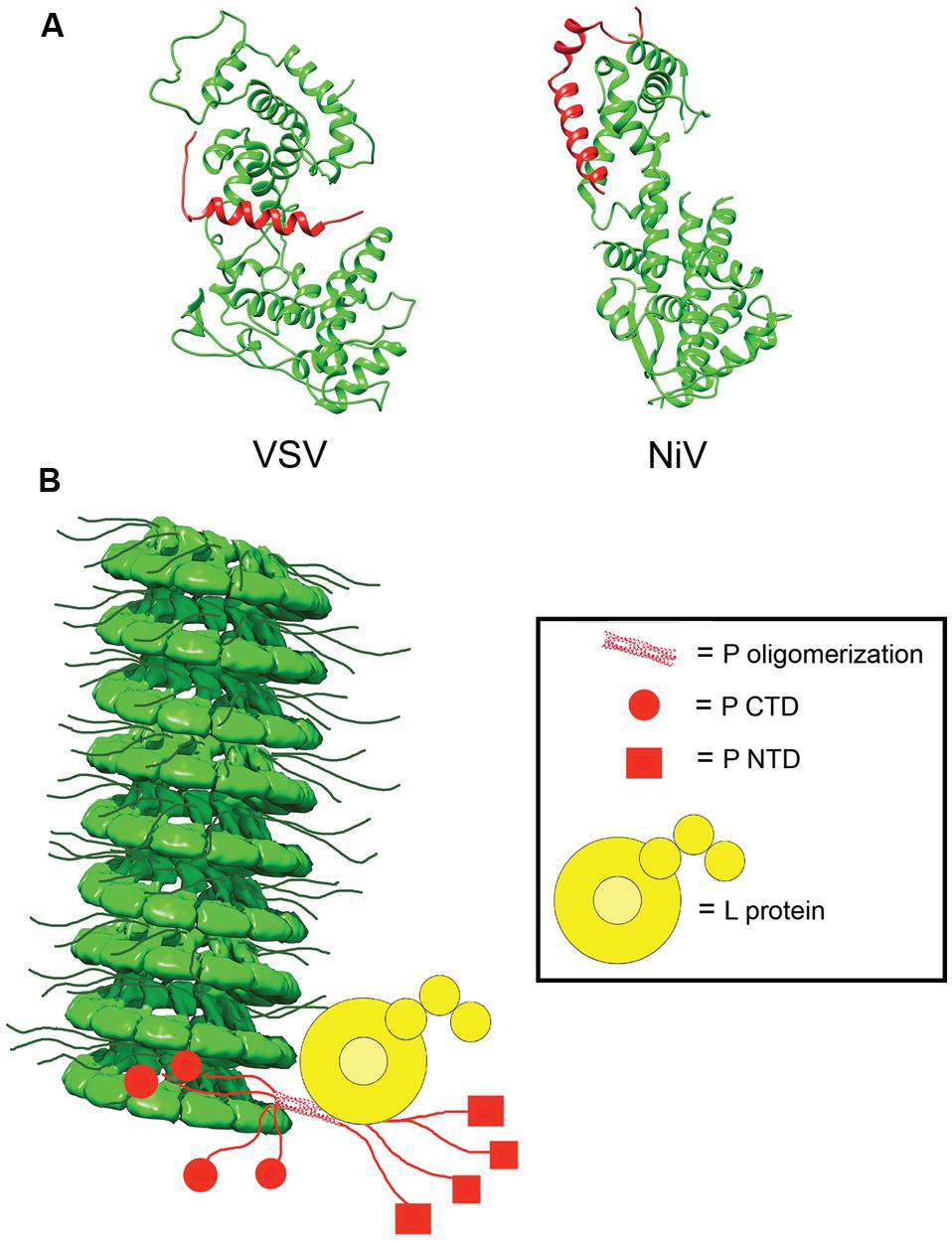
FIGURE 5. N0P and model of the RdRp complex. (A) Structures of the N0P complex of vesicular stomatitis virus (VSV) and NiV have been solved. P (red) acts to prevent premature encapsidation and oligomerization of N (green; PDB codes 3PMK and 4CO6 for VSV and NiV, respectively; Leyrat et al., 2011; Yabukarski et al., 2014). (B) Model of the interactions between N, P, and L in the viral RdRp complex. P anchors L to the NC via interactions in PCTD (red circle) and correctly positions the polymerase to begin synthesizing the viral RNA. The NC model was modified from the mumps NC structure (EMD-2630; Cox et al., 2014).
The interaction of P with the NC is mediated through a well-conserved nucleocapsid binding domain (NBD), which is located toward the C-terminal end of the P protein (Figure 4; Johansson et al., 2003; Kingston et al., 2004b). Structures of the NBDs of several paramyxoviruses complexed with their interacting domain in N have been solved (Figure 3; Gely et al., 2010; Habchi et al., 2011; Communie et al., 2013b). In the case of MeV, the P binding site is located near the C-terminus of the N protein, close to the end of the 125-residue N-tail domain (Kingston et al., 2004a). How the RdRp accesses the encapsidated RNA is unclear. One possibility is that a hinge movement of the NTD with respect to the CTD results in a transient opening of the groove and exposure of encapsidated nucleotides during RNA readout. In this model, the N protein acts as a helicase, dissociating the transient double-stranded RNA segment during procession of RNA synthesis along the genome (Tawar et al., 2009). The L protein, the P protein, or the L–P complex might be able to induce this conformational change (Cox et al., 2014). Physical movement of the polymerase along the NC during RNA synthesis has furthermore been hypothesized to involve the continuous attachment and release of the P NBD domains from its counterpart in the N tails (Kingston et al., 2004b), resulting in “cartwheeling” of the P–L complexes along the NC (Figure 5; Kingston et al., 2004b). In this model, the N-tail sections exposed on the outside of the NC are thought to serve as essential anchor points for recruitment of the polymerase complex (Curran et al., 1993; Kingston et al., 2004a,b).
Supporting this hypothesis, minireplicon reporter studies of truncated SeV and MeV N lacking the P binding domains in N suggested that N-tail truncated NCs cannot serve as a template for the RdRp, thus spotlighting a possible essential function for the N tail-P interaction in polymerase loading and/or advancement (Curran et al., 1993; Zhang et al., 2002). Strikingly, however, further truncation of the N-tail beyond the P interaction region largely restored template function of the NC in an MeV minigenome system (Krumm et al., 2013). This observation demonstrated that the P interaction with the N-tail is dispensable for initial productive loading of the RdRp onto the NC or subsequent advancement of the complex along the template (Krumm et al., 2013). Supported by RdRp activity experiments obtained with negative and positive sense replicon constructs, N-tail-independent RdRp loading appears not to be restricted to transcriptase configuration, but is also applicable to the replicase complex (Krumm et al., 2013). Interestingly, a recent characterization of the related MuV P protein revealed that its interaction with NC likewise does not depend on the N-tail but can be mediated by direct contacts between MuV P and the NTD core (Kingston et al., 2004a; Cox et al., 2013, 2014). Interestingly, this MuV P-NTD interaction would bring the associated L protein into close proximity of the encapsidated RNA. Taken together, these recent discoveries indicate that the initial tethering of the RdRp complex to the NC template is independent of the P and N tail interaction. Rather, cycles of N-tail to P binding and release may be necessary to stabilize the RdRp–NC complex as the polymerase progresses along the genome (Curran and Kolakofsky, 1999; Kolakofsky et al., 2004; Krumm et al., 2013).
Large Protein
The large (L) protein harbors the catalytic centers required for RNA synthesis, mRNA capping, and mRNA polyadenylation (Emerson and Yu, 1975; Hamaguchi et al., 1983; Gupta et al., 2002; Ogino et al., 2005). Bioinformatics analyses have identified six conserved domains (CR I to CR VI) in NNRV L proteins that are connected by variable linker regions (Figure 6; Poch et al., 1988, 1990; Svenda et al., 1997). However, the precise roles for each of these L domains in RdRp function are still largely unclear. CR I has been implicated in L oligomerization (Cevik et al., 2003, 2004; Smallwood and Moyer, 2004) and L–P interactions (Horikami et al., 1994; Holmes and Moyer, 2002; Cevik et al., 2003, 2004; Chattopadhyay and Shaila, 2004), CR III is involved in phosphodiester bond formation for RNA polymerization (Malur et al., 2002b), and CR VI contains methyltransferase activity (Poch et al., 1990; Ferron et al., 2002). A conserved GXXTnHR motif in CR V of VSV L is thought to mediate unusual capping of the viral mRNAs through transfer of 5′-monophosphate-mRNA onto GDP (Ogino and Banerjee, 2007; Li et al., 2008). However, paramyxovirus L proteins may possess traditional guanylyltransferase activity, since Rinderpest virus RdRp complexes reportedly form covalent guanosine monophosphate-L intermediates in vitro (Gopinath and Shaila, 2009). In addition, a conserved guanylyltransferase consensus motif required for transcriptase activity was identified in the C-terminal region of the L protein of human parainfluenza virus (HPIV) type 2 (Nishio et al., 2011).
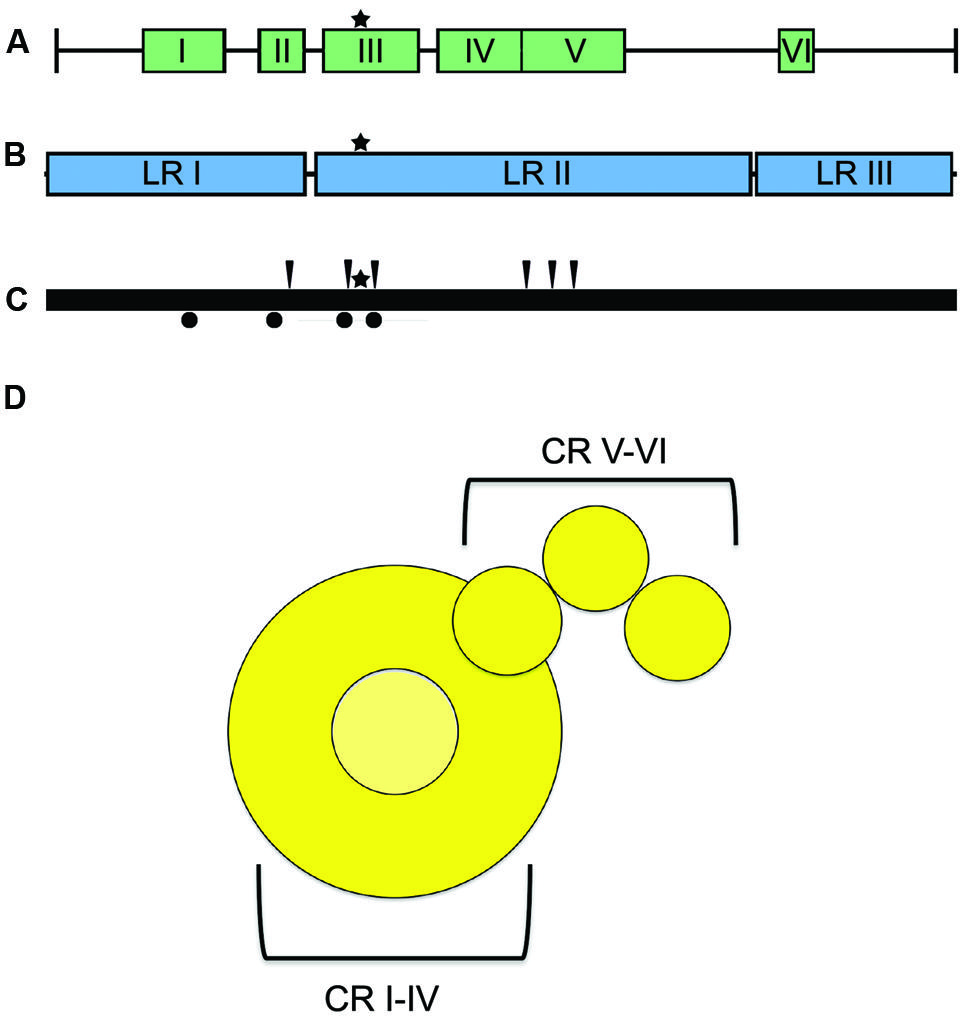
FIGURE 6. L protein domain organization and architecture. (A) The L protein is composed of six conserved regions (CR), I–VI, each containing separate functions (Sidhu et al., 1993). (B) Insertion analysis has shown that L can be further grouped into three large regions (LRI-III; Dochow et al., 2012). Star symbols mark the proposed position of key residues near the catalytic site for phosphodiester bond formation in CR III. (C) Positions of L mutations allowing for resistance to ERDRP-0519 in MeV (black arrows) and CDV (black circles) (Krumm et al., 2014). Mutations map to areas proximal to the active site for phosphodiester bond formation (black star) (D) Cartoon resembling the low-resolution organization of the VSV L protein revealed by electron microscopy analysis (Rahmeh et al., 2010). The cartoon representation is labeled with the proposed positions of the CR domains.
Consistent with L having a modular arrangement of functional domains, also studies of purified L proteins of NNRV by electron microscopy supported a linear organization of structural domains (Figure 6; Rahmeh et al., 2010). Analysis of the L protein of MeV has furthermore revealed that the protein can be split into distinct fragments that are capable of reconstituting RdRp bioactivity through trans-complementation (Duprex et al., 2002; Dochow et al., 2012). This study showed that MeV L is composed of at least two independently folding-competent domains. Consistent with these findings, sequence alignments of different morbillivirus L proteins had previously suggested two linker domains that separate three large regions (LR I to LR III; Figure 6; Mcllhatton et al., 1997; Duprex et al., 2002). Of these, LR I harbors CR I and II, LR II contains CR III-CR V, and LR III is considered to encompass the methyltransferase and, possibly, the recently proposed guanylyltransferase functions of CR VI. L proteins of MeV and rinderpest virus tolerated polypeptide insertions into the LR II/LR III but not the LR I/LR II junction (Duprex et al., 2002; Brown et al., 2005), consistent with at least a two-domain organization. Additional domain intersections may well exist in the paramyxovirus L protein.
In addition to the mandatory interaction with P, Sendai virus L was shown to exist as an oligomer in the RdRp complex (Smallwood et al., 2002). Homo-oligomerization was furthermore proposed for MeV and human parainfluenzavirus type 3 L proteins, and in all cases the L–L interaction domain was proposed to reside in the N-terminal region of the protein (Horikami et al., 1994; Chandrika et al., 1995; Holmes and Moyer, 2002; Malur et al., 2002a; Cevik et al., 2003; Smallwood and Moyer, 2004; Dochow et al., 2012). Although this finding spotlights that both the L–P and L–L interaction domains are located in N-terminal regions of the L protein, homo-oligomerization of MeV and SeV L is reportedly independent of P protein binding (Holmes and Moyer, 2002; Cevik et al., 2003; Dochow et al., 2012). The available information is limited, but the specificity for L–P binding apparently involves multiple non-consecutive amino acids that are distinct from those implemented in L–L interactions (Cevik et al., 2004).
Development of Antiviral Therapeutics
The dynamic interplay between the different viral protein components of the RdRp and the diverse enzymatic activities catalyzed by the L protein constitute an array of drug target candidates suitable for effective inhibition of virus replication. An inherent challenge of all pathogen-directed drug discovery campaigns is a narrow indication spectrum of the therapeutic candidate, limiting inhibitory activity to a specific member or, at best, a single genus within the paramyxovirus family. It may be possible to overcome this restriction by targeting a host cell-derived cofactor of the complex that is likewise indispensible for RdRp activity. For instance, the human translation elongation factor eEF1A is known to be required for VSV RdRp transcriptase activity (Das et al., 1998; Qanungo et al., 2004) and was recently shown to be critically involved also in RSV replication (Wei et al., 2014). A general requirement of eEF1A and/or additional host factors for paramyxovirus RdRp activity is possible, but direct therapeutic targeting of, for instance, eEF1A will likely be prohibited by its central role in host protein synthesis. While it may be hypothetically possible to reduce undesirable cytotoxicity through a campaign specifically designed to block a host cofactor-RdRp protein–protein interaction (PPI), we consider the development of pathogen-directed RdRp blockers more fruitful. Especially “open” high-throughput screening campaigns in search of RdRp inhibitors should yield pathogen-directed hits with higher propensity than compounds interfering with a cofactor-RdRp PPI.
In particular the L protein represents a rich target for drug discovery campaigns, due to its multidomain organization and the concentration of several essential enzymatic activities in a single protein. The L CR-V domain containing a guanylyltransferase domain responsible for 5′-cap formation (Li et al., 2008) is a case in point, since inhibiting the viral capping machinery using guanosine nucleotide analogs constitutes a proven antiviral approach (Lampio et al., 1999; Issur et al., 2011). Likewise, it may be possible to exploit the postulated S-adenosyl-L-methionine transferase domain responsible for 5′-cap methylation (Bujnicki and Rychlewski, 2002; Ferron et al., 2002; Ogino et al., 2005; Murphy and Grdzelishvili, 2009) in L CR-VI. S-adenosyl-L-homocysteine derivatives have been shown to selectively inhibit methyltransferase activity of dengue virus of the flavivirus family, setting an example for the therapeutic potential of antivirals targeting methyltransferase functions (Lim et al., 2011). The precedence established by the development of inhibitors of, for instance, HIV reverse transcriptase and Hepatitis C virus polymerase underscores the value of high-resolution structural information for the identification and optimization of hit structures, the molecular understanding of the mechanism of inhibitory activity, and, potentially, the proactive design of analogs with increased resilience against viral escape from inhibition (Nijhuis et al., 2009; Adams et al., 2010; Das et al., 2011; Halfon and Locarnini, 2011; Mayhoub, 2012; Lloyd et al., 2014). However, the paramyxovirus drug development field is hampered by the current lack of high-resolution structural information for any mononegavirales L protein. Overcoming this limitation will be a major milestone toward the development of next generation therapeutics.
An envisioned drug application including post-exposure prophylactic use affects the drug profile requested of a desirable anti-paramyxovirus therapeutic; a successful candidate must be safe and efficacious, amenable to cost-effective manufacture, ideally be shelf-stable at ambient temperature, and must be orally bioavailable. Of small-molecule chemical compounds, large molecule biologics, and peptidic biopharmaceutical as candidate drug classes, small-molecules are most suitable to fulfill these divergent demands (Ganellin et al., 2013). Two main classes of polymerase-targeted drugs are currently in clinical use for, among others, antiretroviral therapy, human cytomegalovirus therapy, and HCV therapy, competitive nucleotide/nucleoside substrate analogs and non-nucleoside allosteric inhibitors (Sun et al., 2007, 2008; Andrei et al., 2008; Brown, 2009; Krumm et al., 2011; Mercorelli et al., 2011). Table 1 provides an overview of some experimental drug candidates targeted against different paramyxovirus RdRps that represent these main classes and are currently under clinical consideration or were found efficacious in animal models of paramyxovirus disease. As discussed below, we consider it the most promising approach to combine, if available, a substrate analog with an allosteric inhibitor to maximize the prospect of capitalizing on drug combination synergies and in particular reduce the frequency of viral escape from inhibition and/or lower the fitness of escape variants with multiple resistance mutations.
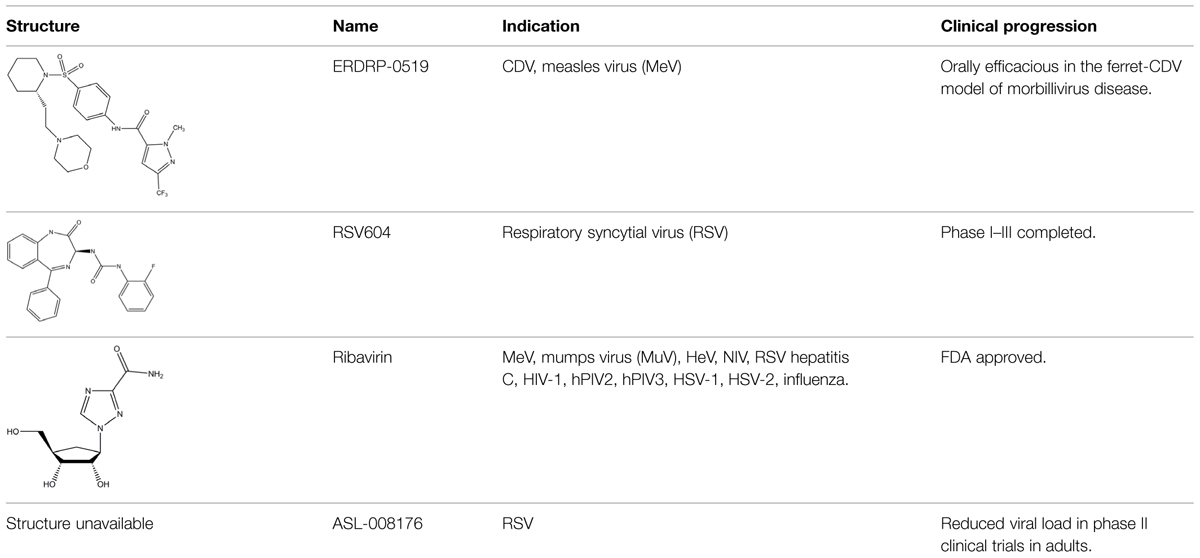
TABLE 1. Examples of substrate analog and allosteric paramyxovirus RNA-dependent RNA polymerase (RdRp) inhibitors that showed efficacy in animal models and/or were advanced to clinical trials.
Nucleoside and Nucleotide Analogs
Nucleoside analogs contain non-canonical bases that act as chain terminators after intracellular phosphorylation to the corresponding nucleotide and incorporation into the nascent chain (De Clercq and Neyts, 2009; Soriano et al., 2013) While nucleoside analogs have shown extreme clinical success, ribavirin is currently the only substrate analog licensed against a paramyxovirus disease, the treatment of RSV infection. The compound is a purine-analog capable of base-pairing with equal efficiency with either cytosine or uracil (Wright et al., 2005). Rather than acting as a chain terminator, the resulting hypermutation of the newly synthesized strand is considered to block virus replication through error catastrophe (Crotty and Andino, 2002). However, ribavirin efficacy against RSV is limited and severe adverse effects, in particular an increased risk of anemia and mitochondrial toxicity (Canonico, 1985; Gilbert and Knight, 1986; Huggins et al., 1991), undermine its clinical use for anti-RSV therapy. In contrast, ALS-008176, a recently presented novel nucleoside analog, is currently in phase II clinical trials for use against RSV infection (Devincenzo et al., 2014). In this trial, the compound ALS-008176 emerged as well tolerated and was capable of significantly reducing viral load in treated adults compared to the placebo control group, when treatment was initiated at the onset of infection. These data are highly encouraging, since they provide proof-of-concept for the clinical benefit of effective RSV inhibitors. In addition, Favipiravir (T-705), a nucleotide analog investigated for the treatment of several virus infections including influenza A, Ebola virus, and foot-and-mouth-disease virus (Furuta et al., 2002), showed activity against RSV in cell culture, albeit at prohibitively high concentrations for clinical use (Furuta et al., 2002, 2013). A novel nucleoside analog was recently reported as a screening hit emerging from a high-throughput anti-RSV campaign (Laganas et al., 2014). Remarkably, resistance mutations were characterized and mapped to the RSV P protein rather than the L polymerase, suggesting a novel mechanism of antiviral activity that is distinct from chain termination and error catastrophe.
Non-Nucleoside RdRp Inhibitors
Non-nucleoside allosteric inhibitors non-competitively block RdRp activity through docking into allosteric sites that are frequently located outside of the actual substrate binding site. Binding of an allosteric ligand can either indirectly alter the active site structurally through a long-range effect, rendering the enzyme catalytically inactive, or they may disrupt the formation of protein complexes required for correct enzymatic function. Examples of clinically approved allosteric polymerase inhibitors that served as primary medication in first-line highly active antiretroviral therapy are the first generation non-nucleoside RT inhibitors Nevirapine and Efavirenz (Basavapathruni and Anderson, 2007; de Bethune, 2010). However, the genetic barrier to resistance against these compounds is low and these drugs are unsuitable for monotherapy (Usach et al., 2013). Second-generation non-nucleoside RT inhibitors such as Etravirine and Rilpivirine show improved resistance profiles, allowing use of Etravirine in treatment-experienced patients containing multidrug-resistant HIV (de Bethune, 2010).
Analogous to the experience with non-nucleoside RT inhibitors, the paramyxovirus L protein should present an equally viable target for effective non-nucleoside therapeutics, in particular when used in combination with a nucleoside analog to prevent the induction of genetic drift in the endemic virus populations leading to the development of preexisting resistance.
We have recently developed and mechanistically characterized an allosteric morbillivirus RdRp inhibitor class that targets the L protein based on the experimental induction of escape mutants (Figure 6; White et al., 2007; Sun et al., 2008; Yoon et al., 2008, 2009; Krumm et al., 2011; Ndungu et al., 2012; Moore et al., 2013a,b). Specifically, resistance mutations clustered in L protein conserved domains of II, III, and IV (Yoon et al., 2009). Further development of this class yielded the clinical candidate ERDRP-0519, a well-tolerated orally efficacious pan-morbillivirus RdRp inhibitor that rendered normally lethal CDV disease in the ferret model clinically asymptomatic when administered in a post-exposure prophylactic regimen commencing at the onset of viremia (Krumm et al., 2014). Highly encouraging, all post-exposure-treated animals not only survived primary infection but mounted a robust immune response and were completely protected against a subsequent lethal CDV challenge infection (Krumm et al., 2014).
Currently at an early stage of development, several small molecule RSV inhibitors were shown to specifically block RdRp activity in cell culture and show high potential for lead development (Liuzzi et al., 2005; Laganas et al., 2014; Matharu et al., 2014; Tiong-Yip et al., 2014)
In addition to targeting the L protein directly, the paramyxovirus N protein also represents a potential target for viral therapeutics, as evidenced by the recently described RSV inhibitor RSV604 (Chapman et al., 2007). Resistance mutations to RSV604 were hypothesized to include residues involved in the interaction of N with the P–L complex (Chapman et al., 2007). Furthermore, locating resistance hot-spots in RSV N crystal structures revealed important candidate interaction sites, including the RNA binding cavity, the site of N-arm attachment, and the NTD region, which could all also be specifically targeted for the development of therapeutic treatments (Chapman et al., 2007; Tawar et al., 2009; El Omari et al., 2011). Clinical trials have shown that RSV604 was safe and well tolerated by healthy volunteers (Chapman et al., 2007; Marty, 2007; Chapman and Cockerill, 2011; Challa et al., 2014). The compound shows potent antiviral efficacy, using a unique mechanism of action, and is likewise orally bioavailable.
The resistance profile of RSV604 suggests that the compound could possibly interfere with critical PPIs required for RdRp activity. Considering the multitude of dynamic protein–protein contacts required for viral RNA synthesis, specifically targeting protein interfaces such as those between N and P, P and L, P and P, or L and L to block paramyxovirus RdRp represents a currently underexplored opportunity for therapeutic intervention that may hold high future promise.
Short-chain peptides have been explored as candidate inhibitors for a diverse panel of PPIs (DeLano et al., 2000; De Luca et al., 2011; Gavenonis et al., 2014), although poor intracellular availability and rapid proteolysis frequently limit therapeutic use. Small-molecules are more suitable to address these limitations, but until two decades ago, PPIs were essentially considered undruggable by synthetic molecules due to the large (typically 1,000–2000 A2) size and flat geometry of the typical PPI interface (Hwang et al., 2010). Subsequently, however, natural small molecule products such as rapamycin and cyclosporine spotlighted that only a subset of residues in small hot-spot areas confers most of the binding energy, making PPIs are amenable to small-molecule docking and interference (Arkin and Wells, 2004; Arkin et al., 2014). In recent years, over 40 PPIs were successfully subjected to small molecule targeting (Higueruelo et al., 2009; Basse et al., 2013; Labbe et al., 2013) and several candidate inhibitors were advanced to clinical testing (Arkin et al., 2014). The precedence set by these advanced PPI blockers demonstrates that PPIs most suitable for therapeutic intervention concentrate hot-spot residues in defined areas of less than 900 A2 and binding partners contain short primary sequences (Smith and Gestwicki, 2012; Basse et al., 2013). As our structural insight into the organization of the paramyxovirus RdRp complex and the geometry of the dynamic PPIs advances, well designed screening campaigns should commence with the structure-guided in silico evaluation of druggable candidate interfaces, followed by targeted in silico and/or high-throughput screens focused on identified suitable PPIs.
Conclusion
The high contagiousness of paramyxoviruses, the lack of vaccine protection against several clinically highly significant members of the family, and the deliberate decline of vaccination against other family members due to religious believes or concerns about vaccine safety create an urgent need for the development of efficacious paramyxovirus therapeutics. We believe that small-molecule antivirals are best suited to meet the stringent drug profile requested of a successful anti-paramyxovirus drug. The viral polymerase complex in particular presents a rich target for therapeutic interference through competitive substrate analogs and allosteric non-nucleoside inhibitors. The recent advance in the development of PPI inhibitors should furthermore open up the diverse RdRp protein interfaces to therapeutic interference, when more structural insight into the organization of the polymerase complex and its interaction with the NC template becomes available. Considering the challenges associated with rapidly emerging or preexisting viral resistance that we experience in influenza virus monotherapies, drug combination strategies should be explored and, if possible, implemented from the onset of anti-paramyxovirus therapy to reduce the frequency of inducing genetic drift in the endemic virus populations.
Conflict of Interest Statement
Richard K. Plemper is an inventor on PCT application No. PCT/US05/04565 paramyxovirus family inhibitors and methods of use.
Acknowledgments
We thank M. Luo for sharing EM images of MuV nucleocapsids. This work was supported, in part, by public health service grants AI071002 and HD 079327 from the NIH NIAID and NICHD, respectively, (to RKP).
References
Adams, J., Patel, N., Mankaryous, N., Tadros, M., and Miller, C. D. (2010). Nonnucleoside reverse transcriptase inhibitor resistance and the role of the second-generation agents. Ann. Pharmacother. 44, 157–165. doi: 10.1345/aph.1M359
Andrei, G., De Clercq, E., and Snoeck, R. (2008). Novel inhibitors of human CMV. Curr. Opin. Investig. Drugs 9, 132–145.
Arkin, M. R., Tang, Y., and Wells, J. A. (2014). Small-molecule inhibitors of protein-protein interactions: progressing toward the reality. Chem. Biol. 21, 1102–1114. doi: 10.1016/j.chembiol.2014.09.001
Arkin, M. R., and Wells, J. A. (2004). Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat. Rev. Drug Discov. 3, 301–317. doi: 10.1038/nrd1343
Auwaerter, P. G., Rota, P. A., Elkins, W. R., Adams, R. J., Delozier, T., Shi, Y., et al. (1999). Measles virus infection in rhesus macaques: altered immune responses and comparison of the virulence of six different virus strains. J. Infect. Dis. 180, 950–958. doi: 10.1086/314993
Baker, S. C., and Moyer, S. A. (1988). Encapsidation of Sendai virus genome RNAs by purified NP protein during in vitro replication. J. Virol. 62, 834–838.
Bakker, S. E., Duquerroy, S., Galloux, M., Loney, C., Conner, E., Eleouet, J. F., et al. (2013). The respiratory syncytial virus nucleoprotein-RNA complex forms a left-handed helical nucleocapsid. J. Gen. Virol. 94, 1734–1738. doi: 10.1099/vir.0.053025-0
Basavapathruni, A., and Anderson, K. S. (2007). Reverse transcription of the HIV-1 pandemic. FASEB J. 21, 3795–3808. doi: 10.1096/fj.07-8697rev
Basse, M. J., Betzi, S., Bourgeas, R., Bouzidi, S., Chetrit, B., Hamon, V., et al. (2013). 2P2Idb: a structural database dedicated to orthosteric modulation of protein-protein interactions. Nucleic Acids Res. 41, D824–D827. doi: 10.1093/nar/gks1002
Bhella, D., Ralph, A., Murphy, L. B., and Yeo, R. P. (2002). Significant differences in nucleocapsid morphology within the Paramyxoviridae. J. Gen. Virol. 83, 1831–1839.
Bhella, D., Ralph, A., and Yeo, R. P. (2004). Conformational flexibility in recombinant measles virus nucleocapsids visualised by cryo-negative stain electron microscopy and real-space helical reconstruction. J. Mol. Biol. 340, 319–331. doi: 10.1016/j.jmb.2004.05.015
Blanchard, L., Tarbouriech, N., Blackledge, M., Timmins, P., Burmeister, W. P., Ruigrok, R. W., et al. (2004). Structure and dynamics of the nucleocapsid-binding domain of the Sendai virus phosphoprotein in solution. Virology 319, 201–211. doi: 10.1016/j.virol.2003.10.029
Brown, D. D., Rima, B. K., Allen, I. V., Baron, M. D., Banyard, A. C., Barrett, T., et al. (2005). Rational attenuation of a morbillivirus by modulating the activity of the RNA-dependent RNA polymerase. J. Virol. 79, 14330–14338. doi: 10.1128/JVI.79.22.14330-14338.2005
Brown, N. A. (2009). Progress towards improving antiviral therapy for hepatitis C with hepatitis C virus polymerase inhibitors. Part I: nucleoside analogues. Expert Opin. Investig. Drugs 18, 709–725. doi: 10.1517/13543780902854194
Bruhn, J. F., Barnett, K. C., Bibby, J., Thomas, J. M., Keegan, R. M., Rigden, D. J., et al. (2014). Crystal structure of the nipah virus phosphoprotein tetramerization domain. J. Virol. 88, 758–762. doi: 10.1128/JVI.02294-13
Bujnicki, J. M., and Rychlewski, L. (2002). In silico identification, structure prediction and phylogenetic analysis of the 2’-O-ribose (cap 1) methyltransferase domain in the large structural protein of ssRNA negative-strand viruses. Protein Eng. 15, 101–108. doi: 10.1093/protein/15.2.101
Canonico, P. G. (1985). Efficacy, toxicology and clinical applications of ribavirin against virulent RNA viral infections. Antiviral Res. 5(Suppl. 1), 75–81. doi: 10.1016/S0166-3542(85)80011-5
Centers for Disease and Prevention. (2012a). Hospital-associated measles outbreak - Pennsylvania, March-April 2009. MMWR Morb. Mortal. Wkly. Rep. 61, 30–32.
Centers for Disease and Prevention. (2012b). Measles - United States, 2011. MMWR Morb. Mortal. Wkly. Rep. 61, 253–257.
Cevik, B., Holmes, D. E., Vrotsos, E., Feller, J. A., Smallwood, S., and Moyer, S. A. (2004). The phosphoprotein (P) and L binding sites reside in the N-terminus of the L subunit of the measles virus RNA polymerase. Virology 327, 297–306. doi: 10.1016/j.virol.2004.07.002
Cevik, B., Smallwood, S., and Moyer, S. A. (2003). The L-L oligomerization domain resides at the very N-terminus of the sendai virus L RNA polymerase protein. Virology 313, 525–536. doi: 10.1016/S0042-6822(03)00342-8
Challa, S., Scott, A. D., Yuzhakov, O., Zhou, Y., Tiong-Yip, C. L., Gao, N., et al. (2014). Mechanism of action for respiratory syncytial virus inhibitor RSV604. Antimicrob. Agents Chemother. 59, 1080–1087. doi: 10.1128/AAC.04119-14
Chandrika, R., Horikami, S. M., Smallwood, S., and Moyer, S. A. (1995). Mutations in conserved domain I of the Sendai virus L polymerase protein uncouple transcription and replication. Virology 213, 352–363. doi: 10.1006/viro.1995.0008
Chapman, J., Abbott, E., Alber, D. G., Baxter, R. C., Bithell, S. K., Henderson, E. A., et al. (2007). RSV604, a novel inhibitor of respiratory syncytial virus replication. Antimicrob. Agents Chemother. 51, 3346–3353. doi: 10.1128/AAC.00211-07
Chapman, J., and Cockerill, G. S. (2011). “Discovery and development of Rsv604,” in Antiviral Drugs: From Basic Discovery through Clinical Trials, ed. Kazmierski (Hoboken, NJ: John Wiley & Sons, Inc.), 367–382.
Chattopadhyay, A., and Shaila, M. S. (2004). Rinderpest virus RNA polymerase subunits: mapping of mutual interacting domains on the large protein L and phosphoprotein p. Virus Genes 28, 169–178. doi: 10.1023/B:VIRU.0000016855.25662.95
Chen, J. L., Das, T., and Banerjee, A. K. (1997). Phosphorylated states of vesicular stomatitis virus P protein in vitro and in vivo. Virology 228, 200–212. doi: 10.1006/viro.1996.8401
Chen, M., Ogino, T., and Banerjee, A. K. (2006). Mapping and functional role of the self-association domain of vesicular stomatitis virus phosphoprotein. J. Virol. 80, 9511–9518. doi: 10.1128/JVI.01035-06
Chen, M., Ogino, T., and Banerjee, A. K. (2007). Interaction of vesicular stomatitis virus P and N proteins: identification of two overlapping domains at the N terminus of P that are involved in N0-P complex formation and encapsidation of viral genome RNA. J. Virol. 81, 13478–13485. doi: 10.1128/JVI.01244-07
Communie, G., Crepin, T., Maurin, D., Jensen, M. R., Blackledge, M., and Ruigrok, R. W. (2013a). Structure of the tetramerization domain of measles virus phosphoprotein. J. Virol. 87, 7166–7169. doi: 10.1128/JVI.00487-13
Communie, G., Habchi, J., Yabukarski, F., Blocquel, D., Schneider, R., Tarbouriech, N., et al. (2013b). Atomic resolution description of the interaction between the nucleoprotein and phosphoprotein of Hendra virus. PLoS Pathog. 9:e1003631. doi: 10.1371/journal.ppat.1003631
Cox, R., Green, T. J., Purushotham, S., Deivanayagam, C., Bedwell, G. J., Prevelige, P. E., et al. (2013). Structural and functional characterization of the mumps virus phosphoprotein. J. Virol. 87, 7558–7568. doi: 10.1128/JVI.00653-13
Cox, R., Pickar, A., Qiu, S., Tsao, J., Rodenburg, C., Dokland, T., et al. (2014). Structural studies on the authentic mumps virus nucleocapsid showing uncoiling by the phosphoprotein. Proc. Natl. Acad. Sci. U.S.A. 111, 15208–15213. doi: 10.1073/pnas.1413268111
Crotty, S., and Andino, R. (2002). Implications of high RNA virus mutation rates: lethal mutagenesis and the antiviral drug ribavirin. Microbes Infect. 4, 1301–1307. doi: 10.1016/S1286-4579(02)00008-4
Curran, J., Homann, H., Buchholz, C., Rochat, S., Neubert, W., and Kolakofsky, D. (1993). The hypervariable C-terminal tail of the Sendai paramyxovirus nucleocapsid protein is required for template function but not for RNA encapsidation. J. Virol. 67, 4358–4364.
Curran, J., and Kolakofsky, D. (1999). Replication of paramyxoviruses. Adv. Virus Res. 54, 403–422. doi: 10.1016/S0065-3527(08)60373-5
Das, S. R., Hensley, S. E., David, A., Schmidt, L., Gibbs, J. S., Puigbo, P., et al. (2011). Fitness costs limit influenza A virus hemagglutinin glycosylation as an immune evasion strategy. Proc. Natl. Acad. Sci. U.S.A. 108, E1417–E1422. doi: 10.1073/pnas.1108754108
Das, T., Mathur, M., Gupta, A. K., Janssen, G. M., and Banerjee, A. K. (1998). RNA polymerase of vesicular stomatitis virus specifically associates with translation elongation factor-1 alphabetagamma for its activity. Proc. Natl. Acad. Sci. U.S.A. 95, 1449–1454. doi: 10.1073/pnas.95.4.1449
Davis, N. L., and Wertz, G. W. (1982). Synthesis of vesicular stomatitis virus negative-strand RNA in vitro: dependence on viral protein synthesis. J. Virol. 41, 821–832.
de Bethune, M. P. (2010). Non-nucleoside reverse transcriptase inhibitors (NNRTIs), their discovery, development, and use in the treatment of HIV-1 infection: a review of the last 20 years (1989-2009). Antiviral Res. 85, 75–90. doi: 10.1016/j.antiviral.2009.09.008
De Clercq, E., and Neyts, J. (2009). Antiviral agents acting as DNA or RNA chain terminators. Handb. Exp. Pharmacol. 189, 53–84. doi: 10.1007/978-3-540-79086-0_3
DeLano, W. L., Ultsch, M. H., De Vos, A. M., and Wells, J. A. (2000). Convergent solutions to binding at a protein-protein interface. Science 287, 1279–1283. doi: 10.1126/science.287.5456.1279
De Luca, L., Ferro, S., Morreale, F., De Grazia, S., and Chimirri, A. (2011). Inhibitors of the interactions between HIV-1 IN and the cofactor LEDGF/p75. ChemMedChem 6, 1184–1191. doi: 10.1002/cmdc.201100071
DeVincenzo, J. P., El Saleeby, C. M., and Bush, A. J. (2005). Respiratory syncytial virus load predicts disease severity in previously healthy infants. J. Infect. Dis. 191, 1861–1868. doi: 10.1086/430008
Devincenzo, J., Fathi, H., Mcclure, M., Westland, C., Chanda, S., Lambkin-Williams, R., et al. (2014). Treatment with oral ALS-008176, a nucleoside analog, rapidly reduces RSV viral load and clinical disease severity in a healthy volunteer challenge study. Open Forum Infect. Dis. 1, S66–S69. doi: 10.1093/ofid/ofu083
Ding, H., Green, T. J., Lu, S., and Luo, M. (2006). Crystal structure of the oligomerization domain of the phosphoprotein of vesicular stomatitis virus. J. Virol. 80, 2808–2814. doi: 10.1128/JVI.80.6.2808-2814.2006
Ding, H., Green, T. J., and Luo, M. (2004). Crystallization and preliminary X-ray analysis of a proteinase-K-resistant domain within the phosphoprotein of vesicular stomatitis virus (Indiana). Acta Crystallogr. D Biol. Crystallogr. 60, 2087–2090. doi: 10.1107/S0907444904024102
Dochow, M., Krumm, S. A., Crowe, J. E. Jr., Moore, M. L., and Plemper, R. K. (2012). Independent structural domains in the paramyxovirus polymerase protein. J. Biol. Chem. 28, 6878–6891. doi: 10.1074/jbc.M111.325258
Duprex, W. P., Collins, F. M., and Rima, B. K. (2002). Modulating the function of the measles virus RNA-dependent RNA polymerase by insertion of green fluorescent protein into the open reading frame. J. Virol. 76, 7322–7328. doi: 10.1128/JVI.76.14.7322-7328.2002
Egelman, E. H., Wu, S. S., Amrein, M., Portner, A., and Murti, G. (1989). The Sendai virus nucleocapsid exists in at least four different helical states. J. Virol. 63, 2233–2243.
El Omari, K., Dhaliwal, B., Ren, J., Abrescia, N. G., Lockyer, M., Powell, K. L., et al. (2011). Structures of respiratory syncytial virus nucleocapsid protein from two crystal forms: details of potential packing interactions in the native helical form. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 67, 1179–1183. doi: 10.1107/S1744309111029228
El Saleeby, C. M., Bush, A. J., Harrison, L. M., Aitken, J. A., and Devincenzo, J. P. (2011). Respiratory syncytial virus load, viral dynamics, and disease severity in previously healthy naturally infected children. J. Infect. Dis. 204, 996–1002. doi: 10.1093/infdis/jir494
Emerson, S. U., and Schubert, M. (1987). Location of the binding domains for the RNA polymerase L and the ribonucleocapsid template within different halves of the NS phosphoprotein of vesicular stomatitis virus. Proc. Natl. Acad. Sci. U.S.A. 84, 5655–5659. doi: 10.1073/pnas.84.16.5655
Emerson, S. U., and Wagner, R. R. (1972). Dissociation and reconstitution of the transcriptase and template activities of vesicular stomatitis B and T virions. J. Virol. 10, 297–309.
Emerson, S. U., and Yu, Y. (1975). Both NS and L proteins are required for in vitro RNA synthesis by vesicular stomatitis virus. J. Virol. 15, 1348–1356.
Fearns, R., Peeples, M. E., and Collins, P. L. (1997). Increased expression of the N protein of respiratory syncytial virus stimulates minigenome replication but does not alter the balance between the synthesis of mRNA and antigenome. Virology 236, 188–201. doi: 10.1006/viro.1997.8734
Ferron, F., Longhi, S., Henrissat, B., and Canard, B. (2002). Viral RNA-polymerases – a predicted 2’-O-ribose methyltransferase domain shared by all Mononegavirales. Trends Biochem. Sci. 27, 222–224. doi: 10.1016/S0968-0004(02)02091-1
Furuta, Y., Gowen, B. B., Takahashi, K., Shiraki, K., Smee, D. F., and Barnard, D. L. (2013). Favipiravir (T-705), a novel viral RNA polymerase inhibitor. Antiviral Res. 100, 446–454. doi: 10.1016/j.antiviral.2013.09.015
Furuta, Y., Takahashi, K., Fukuda, Y., Kuno, M., Kamiyama, T., Kozaki, K., et al. (2002). In vitro and in vivo activities of anti-influenza virus compound T-705. Antimicrob. Agents Chemother. 46, 977–981. doi: 10.1128/AAC.46.4.977-981.2002
Galloway, S. E., and Wertz, G. W. (2008). S-adenosyl homocysteine-induced hyperpolyadenylation of vesicular stomatitis virus mRNA requires the methyltransferase activity of L protein. J. Virol. 82, 12280–12290. doi: 10.1128/JVI.01225-08
Galloway, S. E., and Wertz, G. W. (2009). A temperature sensitive VSV identifies L protein residues that affect transcription but not replication. Virology 388, 286–293. doi: 10.1016/j.virol.2009.03.015
Ganellin, C. R., Jefferis, R., and Roberts, S. M. (eds). (2013). Introduction to Biological and Small Molecule Drug Research and Development: Theory and Case Studies. Oxford: Academic Press.
Gavenonis, J., Sheneman, B. A., Siegert, T. R., Eshelman, M. R., and Kritzer, J. A. (2014). Comprehensive analysis of loops at protein-protein interfaces for macrocycle design. Nat. Chem. Biol. 10, 716–722. doi: 10.1038/nchembio.1580
Gely, S., Lowry, D. F., Bernard, C., Jensen, M. R., Blackledge, M., Costanzo, S., et al. (2010). Solution structure of the C-terminal X domain of the measles virus phosphoprotein and interaction with the intrinsically disordered C-terminal domain of the nucleoprotein. J. Mol. Recognit. 23, 435–447. doi: 10.1002/jmr.1010
Gerard, F. C., Ribeiro Ede, A. Jr., Leyrat, C., Ivanov, I., Blondel, D., Longhi, S., et al. (2009). Modular organization of rabies virus phosphoprotein. J. Mol. Biol. 388, 978–996. doi: 10.1016/j.jmb.2009.03.061
Gilbert, B. E., and Knight, V. (1986). Biochemistry and clinical applications of ribavirin. Antimicrob. Agents Chemother. 30, 201–205. doi: 10.1128/AAC.30.2.201
Gopinath, M., and Shaila, M. S. (2009). RNA triphosphatase and guanylyl transferase activities are associated with the RNA polymerase protein L of rinderpest virus. J. Gen. Virol. 90, 1748–1756. doi: 10.1099/vir.0.010975-0
Gupta, A. K., Mathur, M., and Banerjee, A. K. (2002). Unique capping activity of the recombinant RNA polymerase (L) of vesicular stomatitis virus: association of cellular capping enzyme with the L protein. Biochem. Biophys. Res. Commun. 293, 264–268. doi: 10.1016/S0006-291X(02)00217-6
Habchi, J., Blangy, S., Mamelli, L., Jensen, M. R., Blackledge, M., Darbon, H., et al. (2011). Characterization of the interactions between the nucleoprotein and the phosphoprotein of Henipavirus. J. Biol. Chem. 286, 13583–13602. doi: 10.1074/jbc.M111.219857
Halfon, P., and Locarnini, S. (2011). Hepatitis C virus resistance to protease inhibitors. J. Hepatol. 55, 192–206. doi: 10.1016/j.jhep.2011.01.011
Hall, W. C., Kovatch, R. M., Herman, P. H., and Fox, J. G. (1971). Pathology of measles in rhesus monkeys. Vet. Pathol. 8, 307–319.
Hamaguchi, M., Yoshida, T., Nishikawa, K., Naruse, H., and Nagai, Y. (1983). Transcriptive complex of Newcastle disease virus. I. Both L and P proteins are required to constitute an active complex. Virology 128, 105–117. doi: 10.1016/0042-6822(83)90322-7
Harouaka, D., and Wertz, G. W. (2009). Mutations in the C-terminal loop of the nucleocapsid protein affect vesicular stomatitis virus RNA replication and transcription differentially. J. Virol. 83, 11429–11439. doi: 10.1128/JVI.00813-09
Heggeness, M. H., Scheid, A., and Choppin, P. W. (1980). Conformation of the helical nucleocapsids of paramyxoviruses and vesicular stomatitis virus: reversible coiling and uncoiling induced by changes in salt concentration. Proc. Natl. Acad. Sci. U.S.A. 77, 2631–2635. doi: 10.1073/pnas.77.5.2631
Higueruelo, A. P., Schreyer, A., Bickerton, G. R., Pitt, W. R., Groom, C. R., and Blundell, T. L. (2009). Atomic interactions and profile of small molecules disrupting protein-protein interfaces: the TIMBAL database. Chem. Biol. Drug Des. 74, 457–467. doi: 10.1111/j.1747-0285.2009.00889.x
Holmes, D. E., and Moyer, S. A. (2002). The phosphoprotein (P) binding site resides in the N terminus of the L polymerase subunit of sendai virus. J. Virol. 76, 3078–3083. doi: 10.1128/JVI.76.6.3078-3083.2002
Horikami, S. M., Curran, J., Kolakofsky, D., and Moyer, S. A. (1992). Complexes of Sendai virus NP-P and P-L proteins are required for defective interfering particle genome replication in vitro. J. Virol. 66, 4901–4908.
Horikami, S. M., Smallwood, S., Bankamp, B., and Moyer, S. A. (1994). An amino-proximal domain of the L protein binds to the P protein in the measles virus RNA polymerase complex. Virology 205, 540–545. doi: 10.1006/viro.1994.1676
Huggins, J. W., Hsiang, C. M., Cosgriff, T. M., Guang, M. Y., Smith, J. I., Wu, Z. O., et al. (1991). Prospective, double-blind, concurrent, placebo-controlled clinical trial of intravenous ribavirin therapy of hemorrhagic fever with renal syndrome. J. Infect. Dis. 164, 1119–1127. doi: 10.1093/infdis/164.6.1119
Hwang, H., Vreven, T., Janin, J., and Weng, Z. (2010). Protein-protein docking benchmark version 4.0. Proteins 78, 3111–3114. doi: 10.1002/prot.22830
Hwang, L. N., Englund, N., Das, T., Banerjee, A. K., and Pattnaik, A. K. (1999). Optimal replication activity of vesicular stomatitis virus RNA polymerase requires phosphorylation of a residue(s) at carboxy-terminal domain II of its accessory subunit, phosphoprotein P. J. Virol. 73, 5613–5620.
Issur, M., Picard-Jean, F., and Bisaillon, M. (2011). The RNA capping machinery as an anti-infective target. Wiley Interdiscip. Rev. RNA 2, 184–192. doi: 10.1002/wrna.43
Ivanov, I., Crepin, T., Jamin, M., and Ruigrok, R. W. (2010). Structure of the dimerization domain of the rabies virus phosphoprotein. J. Virol. 84, 3707–3710. doi: 10.1128/JVI.02557-09
Jensen, M. R., Communie, G., Ribeiro, E. A. Jr., Martinez, N., Desfosses, A., Salmon, L., et al. (2011). Intrinsic disorder in measles virus nucleocapsids. Proc. Natl. Acad. Sci. U.S.A. 108, 9839–9844. doi: 10.1073/pnas.1103270108
Johansson, K., Bourhis, J. M., Campanacci, V., Cambillau, C., Canard, B., and Longhi, S. (2003). Crystal structure of the measles virus phosphoprotein domain responsible for the induced folding of the C-terminal domain of the nucleoprotein. J. Biol. Chem. 278, 44567–44573. doi: 10.1074/jbc.M308745200
Karlin, D., Ferron, F., Canard, B., and Longhi, S. (2003). Structural disorder and modular organization in Paramyxovirinae N and P. J. Gen. Virol. 84, 3239–3252. doi: 10.1099/vir.0.19451-0
Kelly, H. A., Riddell, M. A., and Andrews, R. M. (2002). Measles transmission in healthcare settings in Australia. Med. J. Aust. 176, 50–51.
Kingston, R. L., Baase, W. A., and Gay, L. S. (2004a). Characterization of nucleocapsid binding by the measles virus and mumps virus phosphoproteins. J. Virol. 78, 8630–8640. doi: 10.1128/JVI.78.16.8630-8640.2004
Kingston, R. L., Hamel, D. J., Gay, L. S., Dahlquist, F. W., and Matthews, B. W. (2004b). Structural basis for the attachment of a paramyxoviral polymerase to its template. Proc. Natl. Acad. Sci. U.S.A. 101, 8301–8306. doi: 10.1073/pnas.0402690101
Kingston, R. L., Gay, L. S., Baase, W. S., and Matthews, B. W. (2008). Structure of the nucleocapsid-binding domain from the mumps virus polymerase; an example of protein folding induced by crystallization. J. Mol. Biol. 379, 719–731. doi: 10.1016/j.jmb.2007.12.080
Kolakofsky, D., Le Mercier, P., Iseni, F., and Garcin, D. (2004). Viral DNA polymerase scanning and the gymnastics of Sendai virus RNA synthesis. Virology 318, 463–473. doi: 10.1016/j.virol.2003.10.031
Krumm, S. A., Ndungu, J. M., Yoon, J. J., Dochow, M., Sun, A., Natchus, M., et al. (2011). Potent host-directed small-molecule inhibitors of myxovirus RNA-dependent RNA-polymerases. PLoS ONE 6:e20069. doi: 10.1371/journal.pone.0020069
Krumm, S. A., Takeda, M., and Plemper, R. K. (2013). The measles virus nucleocapsid protein tail domain is dispensable for viral polymerase recruitment and activity. J. Biol. Chem. 288, 29943–29953. doi: 10.1074/jbc.M113.503862
Krumm, S. A., Yan, D., Hovingh, E. S., Evers, T. J., Enkirch, T., Reddy, G. P., et al. (2014). An orally available, small-molecule polymerase inhibitor shows efficacy against a lethal morbillivirus infection in a large animal model. Sci. Transl. Med. 6, 232ra252. doi: 10.1126/scitranslmed.3008517
Labbe, C. M., Laconde, G., Kuenemann, M. A., Villoutreix, B. O., and Sperandio, O. (2013). iPPI-DB: a manually curated and interactive database of small non-peptide inhibitors of protein-protein interactions. Drug Discov. Today 18, 958–968. doi: 10.1016/j.drudis.2013.05.003
Laganas, V. A., Dunn, E. F., Mclaughlin, R. E., Tiong-Yip, C. L., Yuzhakov, O., Isabella, V. M., et al. (2014). Characterization of novel respiratory syncytial virus inhibitors identified by high throughput screen. Antiviral Res. 115C, 71–74.
Lamb, R. A., and Parks, G. D. (2007). “Paramyxoviridae: the viruses and their replication,” in Fields Virology, 5 Edn, eds D. M. Knipe and P. M. Howley (Philadelphia, PA: Wolters Kluwer/Lippincott Williams & Wilkins), 1449–1496.
Lampio, A., Ahola, T., Darzynkiewicz, E., Stepinski, J., Jankowska-Anyszka, M., and Kaariainen, L. (1999). Guanosine nucleotide analogs as inhibitors of alphavirus mRNA capping enzyme. Antiviral Res. 42, 35–46. doi: 10.1016/S0166-3542(99)00011-X
Larson, H. J., Cooper, L. Z., Eskola, J., Katz, S. L., and Ratzan, S. (2011). Addressing the vaccine confidence gap. Lancet 378, 526–535. doi: 10.1016/S0140-6736(11)60678-8
Leyrat, C., Yabukarski, F., Tarbouriech, N., Ribeiro, E. A. Jr., Jensen, M. R., Blackledge, M., et al. (2011). Structure of the vesicular stomatitis virus N-P complex. PLoS Pathog. 7:e1002248. doi: 10.1371/journal.ppat.1002248
Li, J., Rahmeh, A., Morelli, M., and Whelan, S. P. (2008). A conserved motif in region v of the large polymerase proteins of nonsegmented negative-sense RNA viruses that is essential for mRNA capping. J. Virol. 82, 775–784. doi: 10.1128/JVI.02107-07
Lim, S. V., Rahman, M. B., and Tejo, B. A. (2011). Structure-based and ligand-based virtual screening of novel methyltransferase inhibitors of the dengue virus. BMC Bioinformatics 12(Suppl. 13):S24. doi: 10.1186/1471-2105-12-S13-S24
Liuzzi, M., Mason, S. W., Cartier, M., Lawetz, C., Mccollum, R. S., Dansereau, N., et al. (2005). Inhibitors of respiratory syncytial virus replication target cotranscriptional mRNA guanylylation by viral RNA-dependent RNA polymerase. J. Virol. 79, 13105–13115. doi: 10.1128/JVI.79.20.13105-13115.2005
Llorente, M. T., Garcia-Barreno, B., Calero, M., Camafeita, E., Lopez, J. A., Longhi, S., et al. (2006). Structural analysis of the human respiratory syncytial virus phosphoprotein: characterization of an alpha-helical domain involved in oligomerization. J. Gen. Virol. 87, 159–169. doi: 10.1099/vir.0.81430-0
Lloyd, S. B., Kent, S. J., and Winnall, W. R. (2014). The high cost of fidelity. AIDS Res. Hum. Retroviruses 30, 8–16. doi: 10.1089/aid.2013.0153
Longhi, S. (2012). The measles virus N(TAIL)-XD complex: an illustrative example of fuzziness. Adv. Exp. Med. Biol. 725, 126–141. doi: 10.1007/978-1-4614-0659-4_8
Malur, A. G., Choudhary, S. K., De, B. P., and Banerjee, A. K. (2002a). Role of a highly conserved NH(2)-terminal domain of the human parainfluenza virus type 3 RNA polymerase. J. Virol. 76, 8101–8109. doi: 10.1128/JVI.76.16.8101-8109.2002
Malur, A. G., Gupta, N. K., De Bishnu, P., and Banerjee, A. K. (2002b). Analysis of the mutations in the active site of the RNA-dependent RNA polymerase of human parainfluenza virus type 3 (HPIV3). Gene Expr. 10, 93–100.
Marty, F. (2007). “A double-blind, randomized, placebo-controlled study to evaluate the safety and efficacy of RSV604 in adults with respiratory syncytial virus infection following stem cell transplantation,” in Proceedings of the IXth International Symposium on Respiratory Viral Infections, Hong Kong.
Masters, P. S., and Banerjee, A. K. (1988). Complex formation with vesicular stomatitis virus phosphoprotein NS prevents binding of nucleocapsid protein N to nonspecific RNA. J. Virol. 62, 2658–2664.
Matharu, D. S., Flaherty, D. P., Simpson, D. S., Schroeder, C. E., Chung, D., Yan, D., et al. (2014). Optimization of potent and selective quinazolinediones: inhibitors of respiratory syncytial virus that block RNA-dependent RNA-polymerase complex activity. J. Med. Chem. 57, 10314–10328. doi: 10.1021/jm500902x
Mavrakis, M., Mccarthy, A. A., Roche, S., Blondel, D., and Ruigrok, R. W. (2004). Structure and function of the C-terminal domain of the polymerase cofactor of rabies virus. J. Mol. Biol. 343, 819–831. doi: 10.1016/j.jmb.2004.08.071
Mavrakis, M., Mehouas, S., Real, E., Iseni, F., Blondel, D., Tordo, N., et al. (2006). Rabies virus chaperone: identification of the phosphoprotein peptide that keeps nucleoprotein soluble and free from non-specific RNA. Virology 349, 422–429. doi: 10.1016/j.virol.2006.01.030
Mayhoub, A. S. (2012). Hepatitis C RNA-dependent RNA polymerase inhibitors: a review of structure-activity and resistance relationships; different scaffolds and mutations. Bioorg. Med. Chem. 20, 3150–3161. doi: 10.1016/j.bmc.2012.03.049
Mcllhatton, M. A., Curran, M. D., and Rima, B. K. (1997). Nucleotide sequence analysis of the large (L) genes of phocine distemper virus and canine distemper virus (corrected sequence). J. Gen. Virol. 78(Pt 3), 571–576.
Mercorelli, B., Lembo, D., Palu, G., and Loregian, A. (2011). Early inhibitors of human cytomegalovirus: state-of-art and therapeutic perspectives. Pharmacol. Ther. 131, 309–329. doi: 10.1016/j.pharmthera.2011.04.007
Moore, T. W., Sana, K., Yan, D., Krumm, S. A., Thepchatri, P., Snyder, J. P., et al. (2013a). Synthesis and metabolic studies of host directed inhibitors for anti viral therapy. ACS Med. Chem. Lett. 4, 762–767. doi: 10.1021/ml400166b
Moore, T. W., Sana, K., Yan, D., Thepchatri, P., Ndungu, J. M., Saindane, M. T., et al. (2013b). Asymmetric synthesis of host-directed inhibitors of myxoviruses. Beilstein J. Org. Chem. 9, 197–203. doi: 10.3762/bjoc.9.23
Morin, B., Rahmeh, A. A., and Whelan, S. P. (2012). Mechanism of RNA synthesis initiation by the vesicular stomatitis virus polymerase. EMBO J. 31, 1320–1329. doi: 10.1038/emboj.2011.483
Moyer, S. A., and Banerjee, A. K. (1975). Messenger RNA species synthesized in vitro by the virion-associated RNA polymerase of vesicular stomatitis virus. Cell 4, 37–43. doi: 10.1016/0092-8674(75)90131-2
Murphy, A. M., and Grdzelishvili, V. Z. (2009). Identification of sendai virus L protein amino acid residues affecting viral mRNA cap methylation. J. Virol. 83, 1669–1681. doi: 10.1128/JVI.01438-08
Murphy, S. K., Ito, Y., and Parks, G. D. (1998). A functional antigenomic promoter for the paramyxovirus simian virus 5 requires proper spacing between an essential internal segment and the 3’ terminus. J. Virol. 72, 10–19.
Ndungu, J. M., Krumm, S. A., Yan, D., Arrendale, R. F., Reddy, G. P., Evers, T., et al. (2012). Non-nucleoside inhibitors of the measles virus RNA-dependent RNA polymerase: synthesis, structure-activity relationships, and pharmacokinetics. J. Med. Chem. 55, 4220–4230. doi: 10.1021/jm201699w
Nijhuis, M., Van Maarseveen, N. M., and Boucher, C. A. (2009). Antiviral resistance and impact on viral replication capacity: evolution of viruses under antiviral pressure occurs in three phases. Handb. Exp. Pharmacol. 189, 299–320. doi: 10.1007/978-3-540-79086-0_11
Nishio, M., Tsurudome, M., Garcin, D., Komada, H., Ito, M., Le Mercier, P., et al. (2011). Human parainfluenza virus type 2 L protein regions required for interaction with other viral proteins and mRNA capping. J. Virol. 85, 725–732. doi: 10.1128/JVI.01226-10
Noton, S. L., Aljabr, W., Hiscox, J. A., Matthews, D. A., and Fearns, R. (2014). Factors affecting de novo RNA synthesis and back-priming by the respiratory syncytial virus polymerase. Virology 462–463, 318–327. doi: 10.1016/j.virol.2014.05.032
Ogino, T., and Banerjee, A. K. (2007). Unconventional mechanism of mRNA capping by the RNA-dependent RNA polymerase of vesicular stomatitis virus. Mol. Cell 25, 85–97. doi: 10.1016/j.molcel.2006.11.013
Ogino, T., Kobayashi, M., Iwama, M., and Mizumoto, K. (2005). Sendai virus RNA-dependent RNA polymerase L protein catalyzes cap methylation of virus-specific mRNA. J. Biol. Chem. 280, 4429–4435. doi: 10.1074/jbc.M411167200
Pelet, T., Delenda, C., Gubbay, O., Garcin, D., and Kolakofsky, D. (1996). Partial characterization of a Sendai virus replication promoter and the rule of six. Virology 224, 405–414. doi: 10.1006/viro.1996.0547
Perlman, S. M., and Huang, A. S. (1973). RNA synthesis of vesicular stomatitis virus. V. Interactions between transcription and replication. J. Virol. 12, 1395–1400.
Plemper, R. K., and Hammond, A. L. (2014). Synergizing vaccinations with therapeutics for measles eradication. Expert Opin. Drug Discov. 9, 201–214. doi: 10.1517/17460441.2014.867324
Plemper, R. K., and Snyder, J. P. (2009). Measles control–can measles virus inhibitors make a difference? Curr. Opin. Investig. Drugs 10, 811–820.
Poch, O., Blumberg, B. M., Bougueleret, L., and Tordo, N. (1990). Sequence comparison of five polymerases (L proteins) of unsegmented negative-strand RNA viruses: theoretical assignment of functional domains. J. Gen. Virol. 71(Pt 5), 1153–1162. doi: 10.1099/0022-1317-71-5-1153
Poch, O., Tordo, N., and Keith, G. (1988). Sequence of the 3386 3’ nucleotides of the genome of the AVO1 strain rabies virus: structural similarities in the protein regions involved in transcription. Biochimie 70, 1019–1029. doi: 10.1016/0300-9084(88)90265-9
Qanungo, K. R., Shaji, D., Mathur, M., and Banerjee, A. K. (2004). Two RNA polymerase complexes from vesicular stomatitis virus-infected cells that carry out transcription and replication of genome RNA. Proc. Natl. Acad. Sci. U.S.A. 101, 5952–5957. doi: 10.1073/pnas.0401449101
Rahmeh, A. A., Schenk, A. D., Danek, E. I., Kranzusch, P. J., Liang, B., Walz, T., et al. (2010). Molecular architecture of the vesicular stomatitis virus RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 107, 20075–20080. doi: 10.1073/pnas.1013559107
Saint-Victor, D. S., and Omer, S. B. (2013). Vaccine refusal and the endgame: walking the last mile first. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368:20120148. doi: 10.1098/rstb.2012.0148
Schoehn, G., Mavrakis, M., Albertini, A., Wade, R., Hoenger, A., and Ruigrok, R. W. (2004). The 12 A structure of trypsin-treated measles virus N-RNA. J. Mol. Biol. 339, 301–312. doi: 10.1016/j.jmb.2004.03.073
Sidhu, M. S., Menonna, J. P., Cook, S. D., Dowling, P. C., and Udem, S. A. (1993). Canine distemper virus L gene: sequence and comparison with related viruses. Virology 193, 50–65. doi: 10.1006/viro.1993.1102
Smallwood, S., Cevik, B., and Moyer, S. A. (2002). Intragenic complementation and oligomerization of the L subunit of the sendai virus RNA polymerase. Virology 304, 235–245. doi: 10.1006/viro.2002.1720
Smallwood, S., and Moyer, S. A. (2004). The L polymerase protein of parainfluenza virus 3 forms an oligomer and can interact with the heterologous Sendai virus L, P and C proteins. Virology 318, 439–450. doi: 10.1016/j.virol.2003.09.045
Smith, M. C., and Gestwicki, J. E. (2012). Features of protein-protein interactions that translate into potent inhibitors: topology, surface area and affinity. Expert Rev. Mol. Med. 14:e16. doi: 10.1017/erm.2012.10
Soriano, V., Vispo, E., De Mendoza, C., Labarga, P., Fernandez-Montero, J. V., Poveda, E., et al. (2013). Hepatitis C therapy with HCV NS5B polymerase inhibitors. Expert Opin. Pharmacother. 14, 1161–1170. doi: 10.1517/14656566.2013.795543
Sun, A., Chandrakumar, N., Yoon, J. J., Plemper, R. K., and Snyder, J. P. (2007). Non-nucleoside inhibitors of the measles virus RNA-dependent RNA polymerase complex activity: synthesis and in vitro evaluation. Bioorg. Med. Chem. Lett. 17, 5199–5203. doi: 10.1016/j.bmcl.2007.06.084
Sun, A., Yoon, J. J., Yin, Y., Prussia, A., Yang, Y., Min, J., et al. (2008). Potent non-nucleoside inhibitors of the measles virus RNA-dependent RNA polymerase complex. J. Med. Chem. 51, 3731–3741. doi: 10.1021/jm701239a
Svenda, M., Berg, M., Moreno-Lopez, J., and Linne, T. (1997). Analysis of the large (L) protein gene of the porcine rubulavirus LPMV: identification of possible functional domains. Virus Res. 48, 57–70. doi: 10.1016/S0168-1702(96)01426-8
Tapparel, C., Maurice, D., and Roux, L. (1998). The activity of Sendai virus genomic and antigenomic promoters requires a second element past the leader template regions: a motif (GNNNNN)3 is essential for replication. J. Virol. 72, 3117–3128.
Tarbouriech, N., Curran, J., Ebel, C., Ruigrok, R. W., and Burmeister, W. P. (2000a). On the domain structure and the polymerization state of the sendai virus P protein. Virology 266, 99–109. doi: 10.1006/viro.1999.0066
Tarbouriech, N., Curran, J., Ruigrok, R. W., and Burmeister, W. P. (2000b). Tetrameric coiled coil domain of Sendai virus phosphoprotein. Nat. Struct. Biol. 7, 777–781. doi: 10.1038/79013
Tawar, R. G., Duquerroy, S., Vonrhein, C., Varela, P. F., Damier-Piolle, L., Castagne, N., et al. (2009). Crystal structure of a nucleocapsid-like nucleoprotein-RNA complex of respiratory syncytial virus. Science 326, 1279–1283. doi: 10.1126/science.1177634
Tiong-Yip, C. L., Aschenbrenner, L., Johnson, K. D., Mclaughlin, R. E., Fan, J., Challa, S., et al. (2014). Characterization of a respiratory syncytial virus L protein inhibitor. Antimicrob. Agents Chemother. 58, 3867–3873. doi: 10.1128/AAC.02540-14
Usach, I., Melis, V., and Peris, J. E. (2013). Non-nucleoside reverse transcriptase inhibitors: a review on pharmacokinetics, pharmacodynamics, safety and tolerability. J. Int. AIDS Soc. 16, 1–14. doi: 10.7448/IAS.16.1.18567
Wei, T., Li, D., Marcial, D., Khan, M., Lin, M. H., Snape, N., et al. (2014). The eukaryotic elongation factor 1A is critical for genome replication of the paramyxovirus respiratory syncytial virus. PLoS ONE 9:e114447. doi: 10.1371/journal.pone.0114447
White, L. K., Yoon, J. J., Lee, J. K., Sun, A., Du, Y., Fu, H., et al. (2007). Nonnucleoside inhibitor of measles virus RNA-dependent RNA polymerase complex activity. Antimicrob. Agents Chemother. 51, 2293–2303. doi: 10.1128/AAC.00289-07
Wright, P. J., Crameri, G., and Eaton, B. T. (2005). RNA synthesis during infection by Hendra virus: an examination by quantitative real-time PCR of RNA accumulation, the effect of ribavirin and the attenuation of transcription. Arch. Virol. 150, 521–532. doi: 10.1007/s00705-004-0417-5
Yabukarski, F., Lawrence, P., Tarbouriech, N., Bourhis, J. M., Delaforge, E., Jensen, M. R., et al. (2014). Structure of Nipah virus unassembled nucleoprotein in complex with its viral chaperone. Nat. Struct. Mol. Biol. 21:754. doi: 10.1038/nsmb.2868
Yoon, J. J., Chawla, D., Paal, T., Ndungu, M., Du, Y., Kurtkaya, S., et al. (2008). High-throughput screening-based identification of paramyxovirus inhibitors. J. Biomol. Screen. 13, 591–608. doi: 10.1177/1087057108321089
Yoon, J. J., Krumm, S. A., Ndungu, J. M., Hoffman, V., Bankamp, B., Rota, P. A., et al. (2009). Target analysis of the experimental measles therapeutic AS-136A. Antimicrob. Agents Chemother. 53, 3860–3870. doi: 10.1128/AAC.00503-09
Keywords: Paramyxovirus, RNA-dependent RNA polymerase, antiviral therapy, nucleoside analogs, allosteric inhibitor
Citation: Cox R and Plemper RK (2015) The paramyxovirus polymerase complex as a target for next-generation anti-paramyxovirus therapeutics. Front. Microbiol. 6:459. doi: 10.3389/fmicb.2015.00459
Received: 04 February 2015; Accepted: 27 April 2015;
Published online: 12 May 2015.
Edited by:
Ben Berkhout, University of Amsterdam, NetherlandsReviewed by:
Tatsuo Shioda, Osaka University, JapanLuis Menéndez Arias, Centro de Biología Molecular Severo Ochoa, Spain
Copyright © 2015 Cox and Plemper. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Richard K. Plemper, Institute for Biomedical Sciences, Petit Science Center, Georgia State University, 100 Piedmont Avenue, Ste 712, Atlanta, GA 30303-3222, USA,cnBsZW1wZXJAZ3N1LmVkdQ==
 Robert Cox
Robert Cox Richard K. Plemper
Richard K. Plemper