
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 21 May 2015
Sec. Plant Pathogen Interactions
Volume 6 - 2015 | https://doi.org/10.3389/fmicb.2015.00387
This article is part of the Research Topic Unravelling Plant–Microbe Interactions: from ecology to mechanisms View all 47 articles
Paenibacillus polymyxa is a common soil bacterium with broad range of practical applications. An important group of secondary metabolites in P. polymyxa are non-ribosomal peptide and polyketide derived metabolites (NRPs/PKs). Modular non-ribosomal peptide synthetases catalyze main steps in the biosynthesis of the complex secondary metabolites. Here we report on the inactivation of an A26 Sfp-type 4'-phosphopantetheinyl transferase (Sfp-type PPTase). The inactivation of the gene resulted in loss of NRPs/PKs production. In contrast to the former Bacillus spp. model the mutant strain compared to wild type showed greatly enhanced biofilm formation ability. A26Δsfp biofilm promotion is directly mediated by NRPs/PKs, as exogenous addition of the wild type metabolite extracts restores its biofilm formation level. Wheat inoculation with bacteria that had lost their Sfp-type PPTase gene resulted in two times higher plant survival and about three times increased biomass under severe drought stress compared to wild type. Challenges with P. polymyxa genetic manipulation are discussed.
The global agricultural system is experiencing profound changes as a result of anthropogenic pressures. The ever-increasing human population (more than 9 billion by 2050), together with the impacts of climate change, is shaping what we eat, and how much, more than ever. A key challenge for plant growth is global water shortage, limiting crop yields already today in more than 70% (Foley et al., 2011; Smol, 2012). Against this background, the global food system will have to improve its resource use efficiency and environmental performance significantly to ensure the sustainability of global food production and consumption. The microbial world is a large unexplored reservoir of biodiversity which exists at diverse and sometimes extreme ecological niches. Paenibacillus polymyxa is a bacterium widely used in agriculture, industry, and environmental remediation because it has multiple functions (Choi et al., 2009; Timmusk et al., 2009, 2014; Timmusk and Nevo, 2011). Recently it was shown that the species has very high metabolic diversity which results in great differences in the bacterial potential for bio-technological applications (Timmusk et al., 2014). P. polymyxa strains from the harsh South Facing Slope (SFS) in comparison to the moderate North Facing Slope (NFS) at “Evolution Canyon,” Israel show huge differences in their metabolism, drought tolerance enhancement and biocontrol ability (Timmusk et al., 2005, 2011; Kim and Timmusk, 2013). Our P. polymyxa strain A26 is isolated from the stressful SFS and has been shown capable of moderate drought stress tolerance enhancement (Timmusk et al., 2011, 2014).
An important pool of the bioactive compounds of great interest for biotechnology are non-ribosomal peptides/polyketides (NRPs/PKs). The spectrum of application of both classes of compounds is large. NRPs are produced by non-ribosomal peptide synthetases (NRPS) and PKs by polyketide synthetases (PKS). Both are very diverse families of natural products with an extremely broad range of biological activities that include adaptation to unfavorable environments, and communication or competition with other microorganism in their natural habitat (Lambalot et al., 1996; Chen et al., 2009; Sunbul et al., 2009b; Powell et al., 2007; Timmusk and Nevo, 2011). The diverse structure of the compounds can be explained by how the NRPs and PKs molecules are synthesized. They are produced as secondary metabolites by microorganisms, by the consecutive condensation of amino acids. The process is not limited to the 20 protein amino acids. Around 500 different monomers, including monoproteinogenic amino acids, fatty acids, and α-hydroxy acids, have been identified as building blocks for NRPs and PKs (de Bruijn et al., 2007). The building blocks contribute to the structural versatility of the compounds and are likely to contribute substantially to the observed biological effect. Despite the enormous chemical diversity in non-ribosomal peptides/polyketides, NRPS and PKS and hybrid PKS-NRPSs share a common point of regulation (Sunbul et al., 2009a,b). These enzymes require activation by phosphopantetheinyl transferases (PPTases) (Beld et al., 2014; Bunet et al., 2014). Bacterial PPTases are divided into two groups based on their sequence homologies and substrate spectra. The members of the first group are associated with primary metabolism and catalyze the activation of the fatty acid acyl carrier domains (Beld et al., 2014; Bunet et al., 2014). The second group prototype is a PPTase Sfp which activates peptidyl carrier protein domains in Bacillus subtilis (Quadri et al., 1998; Beld et al., 2014; Bunet et al., 2014). This group of enzymes has been shown to exhibit broad spectrum activity. During the compound assembly, the biosynthesis intermediates are attached to carrier protein domains of these megaenzymes via a phosphopantetheinyl arm. The Sfp-type PPTases transfer 4′-phosphopantetheinyl (Ppant) groups from CoA to conserved serine residues on peptidyl carrier protein (PCP) and acyl carrier protein (ACP) domains (Sunbul et al., 2009a,b; Beld et al., 2014; Bunet et al., 2014). The post-translational modification of the PCP and ACP domains with Ppant as catalyzed by Sfp-type 4′-phosphopantetheinyl transferase is crucial for the activation of NRPS and PKS (Sunbul et al., 2009a,b; Beld et al., 2014; Bunet et al., 2014). The virulence of various pathogens is dependent on non-ribosomal peptide or polyketide production and their Sfp-type PPTases represent gate keepers to pathogenicity. Hence recently the enzyme has been highlighted as a potential target for antibacterial drug development in the medical industry as well as a means of fighting agricultural pathogens (Chalut et al., 2006; Leblanc et al., 2012; Zheng and Burr, 2013). Inhibition of the enzyme has been shown to reduce the pathogen's growth and virulence (Chalut et al., 2006; Leblanc et al., 2012; Zheng and Burr, 2013; Beld et al., 2014; Foley et al., 2014).
In the frame of the current work, a P. polymyxa A26 mutant with an inactivated Sfp-type PPTase gene (A26Δsfp) was created. The mutant, which is incapable of producing the enzymatically active 4′-phosphopantetheinyl transferase, in turn results in a P. polymyxa A26 mutant strain lacking enzymatically active NRPS and PKS. In contrast to earlier reports on bacterial reduced growth caused by an Sfp-type PPTase mutation, A26Δsfp was significantly enhanced in biofilm production. The A26Δsfp strain had the effect of increasing plant dry weight during normal watering conditions and in particular when the plant was exposed to drought stress. To our knowledge the fact that a bacterial Sfp-type PPTase mutation can promote biofilm formation and enhance the benefits of bacterial interaction with eukaryotes has not been reported earlier.
P. polymyxa A26 was isolated from the “Evolution Canyon” SFS as earlier described (Timmusk et al., 2011). P. polymyxa E681 was provided by SH Park (Ryu et al., 2005). Bacillus subtilis 3610 and B. subtilis sfp::mls (DS3337) were provided by DB Kearns (McLoon et al., 2011). The primers for this experiment are described in Table S1. Escherichia coli DH5alpha cells (Invitrogen) acted as host for recombinant plasmids and were cultivated at 37°C on LB agar. P. polymyxa A26 and E681 were grown in half strength Tryptic Soy Broth (TSB) (Difco) at 30°C. Brain Heart Infusion medium (Difco) containing 10% sucrose was used for the transformation of P. polymyxa A26 and E681. The extraction of plasmid and chromosomal DNA was performed according to standard procedures (Harwood and Cutting, 1990). For antibiotic selection the media were supplemented with 5 μg/ml chloramphenicol (Cm), 1 μg/ml ampicillin (Ap), final concentration.
To analyse the role of A26 Sfp-type PPTase in A26 non-ribosomal peptide production, the Sfp-type PPTase gene was inactivated (Figure 1) (Datsenko and Wanner, 2000; Kim and Timmusk, 2013). As the gene homologous recombination was not successful in A26 the initial gene replacement was performed in the P. polymyxa E681 strain. The gene replacement vector was constructed by cloning a chloramphenicol resistance marker gene (cmr) into the pGEM7Z/Cm EcoRI-BamHI site. Then the flanking regions of the Sfp-type PPTase gene in A26 were amplified by PCR from the A26 chromosomal DNA using the sfp FF and sfp FR primers or the sfp RF and sfp RR primers resulting in upstream and downstream fragments. The upstream fragment was cloned into the pGEM7Z/Cm vector at the NsiI/BamHI site and the downstream fragment at the XbaI/ApaI restriction site (Figure 1). The gene replacement pGEM7Z/Cm-sfp plasmid was introduced into E681. Homologous recombination was confirmed by PCR and sequencing and resulted in the replacement of E681 Sfp-type PPTase gene. Then the E68Δsfp chromosomal DNA was extracted and transferred to A26. The PCR and sequencing conformation analysis was repeated on genomic DNA extracted from A26Δsfp.
Transformation of P. polymyxa was performed by electroporation. In order to prepare competent cells, a single P. polymyxa E681 or A26 colony was picked from 1/2 TSA solid medium and cultured in BHIS broth (Harwood and Cutting, 1990) at 30°C and 200 rpm for 15 h. Two milliliters of the pre-culture was inoculated into 200 mL of BHIS and cultured at 30°C, 200 rpm. When the cells reached an OD600 of 0.5, the bacterial growth was interrupted by placing the culture on ice for 10 min and centrifuged at 5000 × g for 10 min at 4°C. After washing the cells twice with cold SM buffer (Harwood and Cutting, 1990), the competent cells were resuspended in SM buffer. Electroporation was performed using a Gene Pulser (Bio-Rad Laboratories, Richmond, Calif.) as described earlier (Kim and Timmusk, 2013). For P. polymyxa A26 transformation, 20 μg of chromosomal DNA was used instead of plasmid. Transformants were selected on Cm selection plates and cassette insertion was confirmed by PCR and sequencing.
For complementation the Sfp-type PPTase gene was amplified from A26 using primers A26 sfp LF and A26 sfp LR. The PCR product was purified, cleaved with EcoRI and BamHI, and cloned into pHPS9 (BGSC, Ohio), an E. coli/Bacillus shuttle vector, via the NdeI and BamHI restriction sites. The product, i.e. the pHPS9-sfp vector, was transformed into E. coli, purified using QIAprep Spin Miniprep Kit (Qiagen) confirmed by PCR and sequencing and finally transformed into A26Δsfp as described above. The transformation resulted in complemented mutant strain (A26Δsfp/pHPS9-sfp).
Carbohydrate metabolism shown to be characteristic for P. polymyxa species was studied using the BioMerieux API50CH system following the instructions provided by the manufacturer. Additional biochemical identification tests were performed using the BioMerieux API20E system following the instructions provided by the manufacturer.
The antagonistic ability of P. polymyxa A26 wild type, mutant and complemented strains was detected against Fusarium graminearum on potato dextrose agar (PDA) plates for 3 days at 30°C. A26 and A26Δsfp lipopeptide extraction was performed as described by Li et al. (2007). Fusaricidin and polymyxin bioactivity assays have been previously described (Choi et al., 2008, 2009).
Metabolite extraction and antimicrobial activity analysis for LC-MS was conducted as previously described by Choi et al. (2008). The final methanol extract was evaporated and dissolved in 2 ml water. The concentrated samples were further purified on a C18 column (Sili-Cycle Inc. Quebeck, CA) and eluted with a water/methanol gradient. Active eluted fractions were used for LC-MS. LC-MS was performed using a high pressure liquid chromatography system (Thermo Electron Co, USA). The samples were injected into reverse phase columns and analyzed in a mixed solvent of water and acetonitrile containing 0.1% formic acid (0.2 ml/min).
Lipopeptide extraction for MALDI-TOF MS was performed as previously described by Li et al. (2007). Bruker Ultraflex II mass spectrometer using alpha-hydroxycinnamic acid (alpha-HCCA) was used as matrix and spectra were visualized with the mMass software (Niedermeyer and Strohalm, 2012).
Winter wheat (cv. Stava) seeds were sterilized with 5% chlorine solution. A26 and A26Δsfp strains were grown in TSB at 28°C overnight. Culture density was confirmed by colony forming analysis. Inoculation was performed by watering 7 day old plantlets with bacterial solutions containing 107 bacteria. Wheat seeds were grown in pots filled with sand mixed with 10% greenhouse potting mix soil in a growth cabinet at 24/16°C (day/night) temperature, and 16 h photoperiod at 250 μmol m2 s−1. The soil moisture was adjusted to 75% of water holding capacity. Soil moisture (12.5% of soil dry weight) was kept constant during the first 10 days of seedling growth. In 10 days after germination drought stress was induced by stopping watering. Soil volumetric water content was evaluated using 5TE soil moisture sensors (Decagon Devices, Inc.). Root and shoot dry weights were determined after 14 days of drought stress. Plant survival was calculated daily after stress application using 32 stressed plants that were randomly selected and divided into two groups with 16 plants each. Plants were watered and allowed to recover for 4 days. The plants recovering were counted as survived plants. The survived and recovered plants were harvested, washed and dried. For estimates of roots with adhering soil, 12 plantlets per treatment were sampled. Roots with adhering soil (RAS) were carefully separated from bulk sand and sand soil mix by shaking. Soil and root dry mass (RT) was recorded after drying the samples at 105°C, and the RAS/RT ratio was calculated. Root hair length and density were evaluated using 12 plants. Plantlets were carefully separated from soil by shaking. After the loose soil separation the roots of plantlets were washed in distilled water and left to dry in Petri dishes containing 5 ml of water. The other set of plants was homogenized and used for bacterial identification and quantification. Water use efficiency (WUE) was calculated as total dry mass/total water usage. Plant relative water content (RWC) after inoculation with A26 and A26Δsfp was estimated as described elsewhere using the formula RWC (%) = [(W-DW)/TW-DW)] × 100. W is sample fresh weight, TW sample turgid weight and DW sample dry weight. Eight independent experiments were performed.
To study A26 Sfp-type PPTase mediated metabolite direct effect on wheat root growth the metabolite extraction of A26 and A26Δsfp was performed as described by Li et al. (2007). Fusaricidin and polymyxin bioactivity assays were conducted as described earlier (Choi et al., 2008, 2009). Active fractions were evaporated to dryness and taken with saline to initial culture volumes. Wheat seeds were germinated for 24 h, transformed to culture vessels in water for 48 h, immersed in A26 and A26Δsfp metabolite extracts for 12 h at 25°C, rinsed briefly with water and placed in saline for a further 96 h. Then dry weights of the lipopeptide-treated roots were estimated and roots were prepared for scanning electron microscopy (SEM). To compare the metabolite effect with the effect of A26 and A26Δsfp the bacterial cultures were grown in TSB o/n and diluted to 107/ml. Wheat seeds were germinated for 24 h, transformed to culture vessels in water for 48 h, inoculated with A26 and A26Δsfp cultures for 2 h at 25°C, rinsed briefly with water and placed in saline for a further 96 h. Dry weights of the lipopeptide-treated roots were estimated and roots were prepared for SEM.
The other set of A26, A26Δsfp and their metabolite treated roots were placed on sand soil left to grow for 5 more days in controlled environment as described above. Soil moisture content was kept constant. Seven day old seedlings drought stress was induced by stopping watering.
Commercial antibiotics polymyxin B or polymyxin E sulfate salts (Sigma-Aldrich) were used. Seeds were treated with three concentrations of the antibiotics (0.3, 2.5, and 7.5 μg/ml) and left to grow in sand soil in controlled environment and constant soil moisture. Seven day old seedlings were exposed to drought stress by stopping watering. Dry weight experiments were performed after 6 days. Each value represents the mean of three experiments.
Leaf samples for enzyme activity determination were taken after 8 days from drought-treated and well-watered plants. Plant tissue was mixed with 10 ml extraction buffer as described by Knöerzer et al. (1996). The mixture was centrifuged at 14,000 rpm (Eppendorf, 5415C) for 10 min at 5°C, and the supernatant was used to determine protein content and activity of key antioxidant enzymes. Monodehydroascorbate reductase (MDAR) activity was determined following the decrease in light absorbance at 340 nm due to NADH oxidation as described by Hossain et al. (1984). Glutathione reductase (GR) activity was determined by increase in absorbance at 412 nm according to Smith et al. (1989). Superoxide dismutase (SOD) activity was determined by reduction in light absorbance at 490 nm using an Oxiselect SOD activity assay kit (Cell Biolabs, San Diego, CA, USA) according to manufacturer's instructions. Catalase (CAT) activity was measured by reduction in light absorbance at 520 nm, using an OxiselectTM CAT activity assay kit (Cell Biolabs). For CAT and SOD, enzyme activities were determined per gram of fresh mass (FM). Ultimately, the enzyme activities were normalized with respect to the activities in well-watered plants. The corresponding activities were 20 nmol (mg FM)−1 min−1 for MDHAR, 25 nmol (mg FM)−1 min−1 for GR, 0.45 μmol (g FM)−1 min−1 for SOD and 53 μmol (mg FM)−1 for CAT.
The assay was performed based on pellicle weights as described by Beauregard et al. (2013). Briefly, cells were cultured from 1 day old colonies resuspended in 3 ml potato dextrose broth (PDB). After 2 h the cells were diluted 1:100 in 3 ml PDB. The dilution was repeated two more times. After the last dilution, cells were harvested at OD600 < 0.5 and adjusted to a final OD600 of 0.3. The assays were performed in 24 well plates. Pre-weighed PELCO prep-eze individual wells with a mesh bottom (opening size 420 uM) (Ted Pella) were put in the wells to which 1 ml medium and 14 ul of cells were added. Plates were incubated at 30°C for 96 h to allow pellicles to develop. Individual wells were then removed, dried and weighed. Each value represents the mean of three experiments.
To study the Sfp-type PPTase mediated secondary metabolite effect on A26, A26Δsfp, and A26Δsfp/pHPS9-sfp biofilm formation the metabolite extraction was performed as described by Li et al. (2007). Fusaricidin and polymyxin bioactivity assays were conducted as described earlier (Choi et al., 2008, 2009). Active fractions were evaporated to dryness and taken with saline to initial culture volumes. Single colony selection plates were supplemented with the filter-sterilized metabolite extracts. Biofilm formation assays were performed as described above with exception that all growth media were supplemented with A26 or A26Δsfp filter-sterilized metabolite extracts.
Wheat seedlings were inoculated as described above with A26, A26Δsfp, and A26Δsfp/pHPS9-sfp. Another set of roots was prepared as described under culture vessel assay. Scanning electron microscopy (SEM) was performed as described earlier by Timmusk et al. (2005). Briefly, 10 day old plant root tips were fixed in glutaraldehyde or in a solution containing formaldehyde. The samples were dehydrated using a graded ethanol series and critical point dried in CO2. The pressure was decreased very slowly to prevent tissue damage. Samples were mounted on stubs and shadowed with gold (22 nm) before they were viewed with a Philips Autoscan SEM. All images were computer processed.
For the plant root hair assay, wheat seedlings were inoculated as described above with A26 A26Δsfp, and A26Δsfp/pHPS9-sfp. Ten day old plantlets were carefully separated from soil by shaking, and roots were washed in distilled water and left to dry in Petri dishes containing 5 ml of water. The dried root system was evaluated using a Zeiss LSM 710 microscope.
Experiments were repeated three times to confirm reproducibility. Replicated data were analyzed by ANOVA, and all treatment effects were considered significant at a conservative level of significance of P ≤ 0.01.
The analysis of A26 genome shows that it contains a single Sfp-type PPTase encoding gene (ppa26_1634). The gene shares 97, 92 and 91% homology with P. polymyxa E681, SC2 and M1 PPTase genes, respectively and contains three conserved sequence motives described by Lambalot et al. (1996).
The suicide vector was constructed as described on Figure 1. The initial gene replacement was performed in the P. polymyxa E681 strain. The chromosomal DNA was isolated from E681 and transformed into A26. After the confirmations by PCR and sequencing, the culture extracts were isolated from wild type strain, mutant strain and complemented strain. To date the only characterized non-ribosomal peptides in P. polymyxa are fusaricidins and polymyxins. As the P. polymyxa A26 sequence shares 96% homology with the well-studied P. polymyxa E681 sequence, comparative in silico prediction of P. polymyxa A26 NRPS based on E681 data and peptide structures according to NRPS architecture was performed using NRPS prediction program, ClustalW and MegAlign (Choi et al., 2008, 2009; Royer et al., 2013). The analysis confirms our LC-MS data and suggests that A26 is able to produce fusaricidins of molecular weights 883, 897, 911, 931, 947, and 961 Da and a polymyxin of molecular weight 1094 Da (Figure 2 and Figure S1). The extracts from the wild type, A26Δsfp and complemented strain cultures were isolated, tested for bioactivity and analyzed by MALDI-TOF MS. Neither of the non-ribosomal peptides was detected in A26Δsfp extracts (Figure 2). To prove that the phenotypic changes were due to Sfp-type PPTase replacement, the gene was cloned into the pHPS9 vector and transformed into A26 to complement the inactivated gene. In the complemented strain the wild type phenotype was fully restored (Figure 2). The mutant, wild type and complemented strains were tested on agar plates against a pathogen F. graminearum. A typical example is shown in Figure 2A.
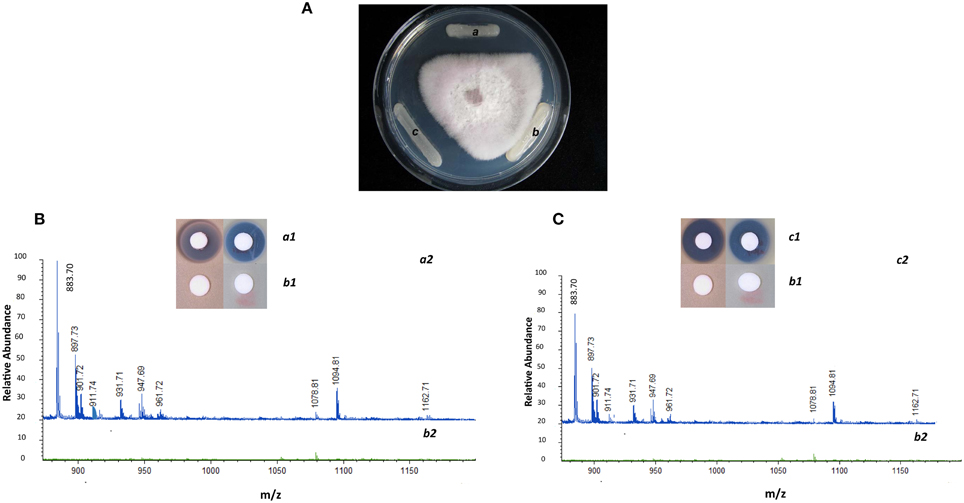
Figure 2. Phenotypic and chemical analysis of P. polymyxa A26 Sfp-type PPTase mutant (A26Δsfp). (A) Inhibitory effect of wild type A26 (a), A26Δsfp (b) and complemented strain A26Δsfp/pHPS9-sfp (c) against F. graminearum. Note that the zone of antagonism observed with wild type has disappeared with mutant and is fully restored with complemented strain. (B) Antimicrobial activities of fusaricidin, polymyxin and MALDI-TOF MS analysis of culture extracts of wild type A26 (a1 and a2, respectively; blue line) and A26Δsfp (b1 and b2; green line). Note that the fusaricidins (molecular weights 883, 897, 911, 931, 947, and 961 Da) and polymyxin (molecular weight 1094 Da) produced in wild type are eradicated in A26Δsfp culture extracts. (C) Antimicrobial activities of fusaricidin, polymyxin and MALDI-TOF MS analysis of complemented strain A26Δsfp/pHPS9-sfp culture extracts (c1 and c2; blue line) is compared to mutant A26Δsfp (b1 and b2; green line). Note that synthesis of fusaricidins and polymyxin is fully restored by A26Δsfp strain complemented with plasmid pHPS9-sfp.
Various assays were used to evaluate biofilm formation of the wild type, A26Δsfp and complemented strain. Enhanced slime production and changed colony morphology were observed on plate assays PDA agar plates (Figure 3A). The deletion of the Sfp-type PPTase gene resulted in about 40% higher biofilm formation based on pellicle weight assay (Figure 3A). Complementation restored the biofilm formation rates to wild type level (Figure 3A). A26 and the other P. polymyxa strain E681 Sfp-type PPTase mutants were almost identical in their colony morphology but in E681Δsfp the enhancement of biofilm formation (30–35%) was slightly smaller than in A26Δsfp (40%). For comparison B. subtilis 3610 and its Sfp mutant were also studied for their biofilm formation. In contrast to P. polymyxa strains, B. subtilis 3610 Sfp mutation significantly impaired its colony morphology and biofilm formation. B. subtilis Sfp mutant impaired its pellicle weight by 60% (Figure 3A). SEM images of wheat root tips colonized with wild type, A26Δsfp and complemented strain confirmed the results observed on colony morphology plate assays and biofilm formation pellicle assays. Significantly more porous extracellular matrix is formed by A26Δsfp (Figure 3B). Complementation successfully restores the extracellular matrix formation to the wild type level (Figure 3B).
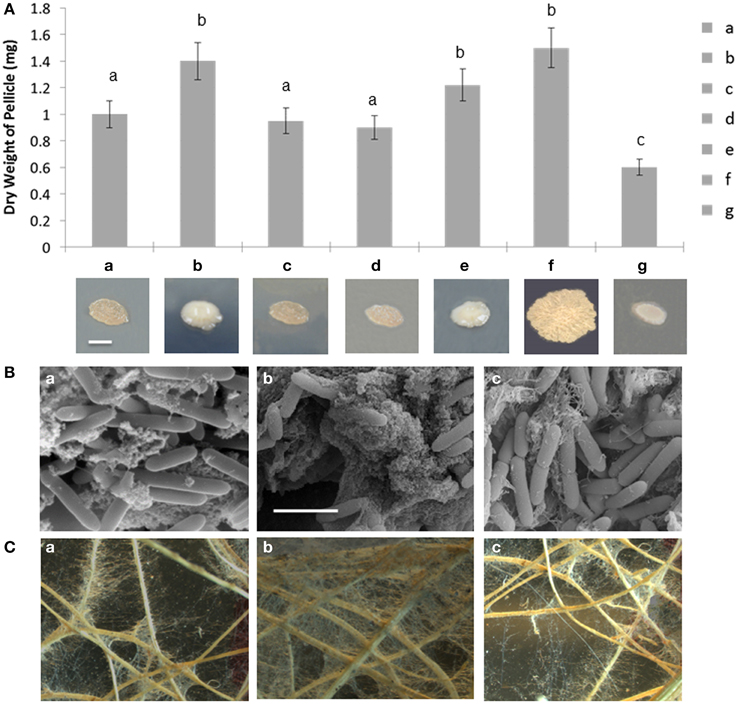
Figure 3. Biofilm and root hair formation analysis of P. polymyxa Sfp-type PPTase mutants. (A) In vitro biofilm formation of A26 (a), A26Δsfp (b), A26Δsfp/pHPS9-sfp (c), E681 (d), E681Δsfp (e), compared to B. subtilis 3610 (f), and 3610Δsfp (g). Colony phenotypes of the strains are shown. Colonies were grown on PDA agar for 4 days at 30°C. The scale bar represents 2 mm. (B) Scanning electron microscopic images of A26 (a), A26Δsfp (b), A26Δsfp/pHPS9-sfp (c) inoculated wheat roots. Significantly more biofilm compared to A26 is formed on the roots inoculated with A26Δsfp; complementation of the strain with pHPS9-sfp restores the wild type biofilm formation level. The scale bar represents 3. (C) Light microscopic images of biofilm and root hair formation on wheat roots inoculated A26 (a), A26Δsfp (b), and A26Δsfp/pHPS9-sfp (c). Note that compared to A26 significantly more root hair and biofilm are formed on wheat roots inoculated with A26Δsfp. Complementation of A26Δsfp restores the wild type of root hair and biofilm formation levels.
In order to study if the A26 Sfp-type PPTase mediated NRPs/PKs are directly involved in the observed biofilm promotion, additional weight-based biofilm formation assays were performed. Metabolite extracts from A26 and A26Δsfp were exogenously added to A26, A26Δsfp and its complemented strain A26Δsfp pHPS9-sfp (Table 2). A26 Sfp-type PPTase mutant biofilm formation level was restored with the external addition of the A26 metabolites (Table 2). At the same time there was no significant effect of any other metabolite extract treatment on any tested strain (Table 2).
Another assay was performed with the A26 and A26Δsfp inoculated plant roots grown in sand, washed and left to dry in 5 ml water on Petri plates. Biofilm formation was observed to significantly enhance root hair growth of A26Δsfp inoculated plants (Figure 3C). Additional quantitative estimation of biofilm formation was performed based on amount of soil attached to roots (Table 1). Two times more soil was attached to the wheat seedling roots inoculated with A26Δsfp (Table 1).
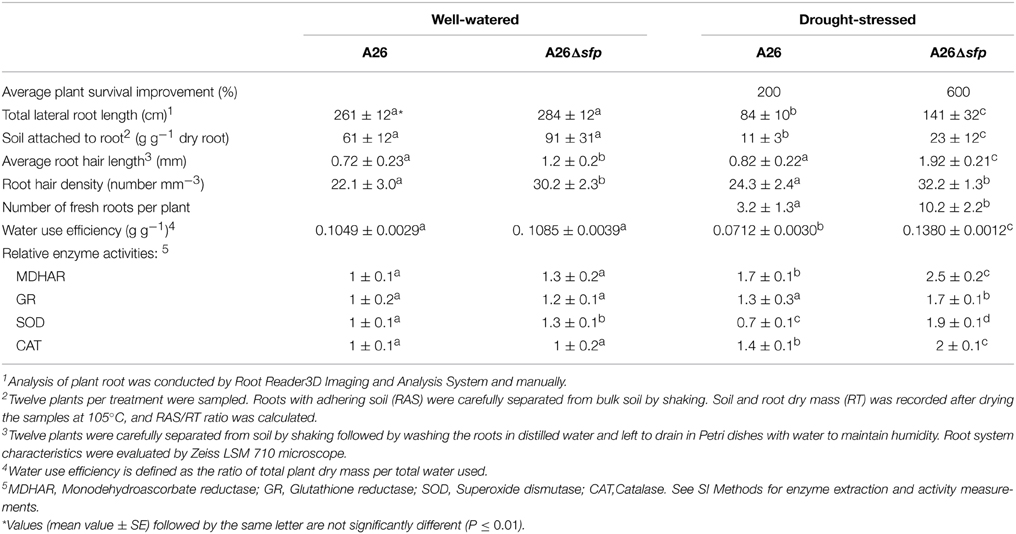
Table 1. Comparative effect of priming by P. polymyxa A26 and A26Δsfp on wheat (Triticum aestivum L. cv. Stava) average (±SD) growth characteristics, water use efficiency and antioxidant enzyme activities.
The metabolism of the wild type, mutated and complemented strains of P. polymyxa was studied for the species characteristic metabolic traits using the BioMerieux API50CH system. Similar metabolic traits were confirmed of all three strains. Additional biochemical identification tests were performed using the BioMerieux API20E system. Acetoin production was confirmed to be produced by the wild-type, mutant and complemented strain.
Comparative effects of the wild type A26 and A26Δsfp on wheat (Triticum aestivum cv. Stava) growth characteristics water use efficiency and relative water content were studied. Initially the colonization ability of the strains was evaluated using PCR assays. The mutation did not influence the A26 root colonization and 105 - 106 cells of wild type as well as mutant were detected in plant roots. The mutant significantly increases seed germination, root hair length, density, amount of soil attached to roots and plant water use efficiency (Table 1). One hundred percent of the A26Δsfp-primed seeds germinated under normal and stress conditions. A26Δsfp increases root length and root hair length and density. A26Δsfp inoculation resulted in 4.5 and 2.5 times improvements in root hair length and density, respectively. This is about twice the improvements obtained with the wild-type gene. The detailed analysis of plant root systems reveals that plant lateral root length was improved about 150 % by A26Δsfp (Table 1). The inoculation with A26Δsfp results in 1.6 times improved water use efficiency compared to the wild type and plant relative water content is significantly improved (Table 1 and Figure S2).
Antioxidant enzymes play important role in reactive oxygen species scavenging and plant drought tolerance enhancement. Hence the activities of monodehydroascorbate reductase (MDHAR), glutathione reductase (GR), catalase (CAT), and superoxide dismutase (SOD) were studied after 8 days in drought-stressed and well-watered plants. Enzyme activities were normalized to the well-watered seedlings enzyme activities. The relative activity of MDHAR was increased by A26 and A26Δsfp colonization (Table 1). Activity of GR was slightly enhanced by A26, and significantly enhanced by A26Δsfp. Both SOD and CAT activities were increased by A26Δsfp under drought stress (Table 1).
The comparative effects of inoculation with A26 or A26Δsfp on the survival and dry weight of winter wheat (cv. Stava) plants grown in sand with 10% greenhouse soil and exposed to drought stress was investigated. Dry weights were estimated after 14 days and survival after 14 days of drought exposure and 4 days recovery watering. A26Δsfp improved plant dry weight under drought stress about 6 times, which is 3 times improvement over wild type (Figure 4A). A26Δsfp primed plant survival was 4 times higher than controls after 14 days of drought stress and 4 days of recovery watering (Figure 4B). This is twice the increase obtained with the wild type.
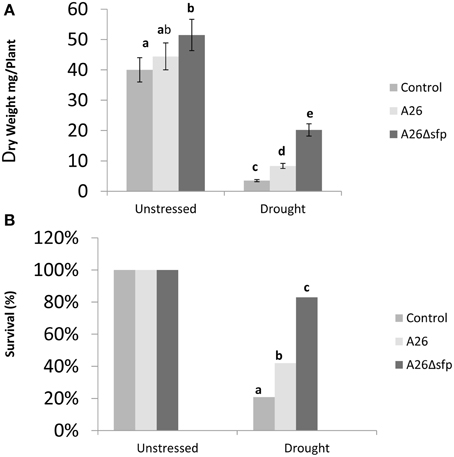
Figure 4. P. polymyxa A26 and its Sfp-type PPTase mutant (A26Δsfp) wheat drought tolerance enhancement analysis. Effect of A26 and A26Δsfp inoculation on wheat dry mass (A) and survival (B).
Wild type A26 and A26Δsfp bacteria and their metabolite extracts were studied for biological activity on wheat root growth in water vessels. A26 and its extracts caused damage and reduced root growth compared to A26Δsfp bacteria and their metabolite extracts (Figure 5). Both bacterial root counts were 109/cells per g root after 96 h incubation in saline. Wheat seedlings inoculated with A26 or A26Δsfp or treated with the respective metabolite extracts, were grown in sand and exposed to drought stress. 4–5% of wild type culture or metabolite treated survived the 5 day stress (Figure 5). Forty to forty five percent of wheat seedling treated with A26 or A26Δsfp bacteria or metabolites survived 5 days of drought stress (Figure 5).
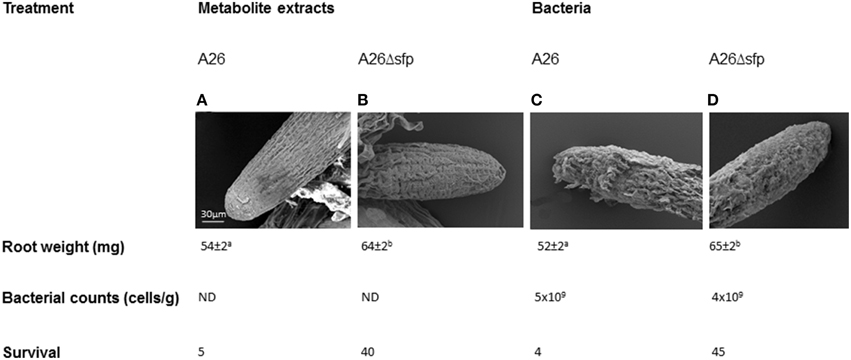
Figure 5. Direct effect of A26 Sfp-type PPTase mediated metabolites on wheat root. Scanning electron microscopic images, dry mass, root bacterial counts and survival of A26 (A), A26Δsfp metabolite extract treated roots compared to (B), A26 (C), and A26Δsfp bacterial culture (D) treated roots.
Three concentrations of the antibiotics polymyxin B and E (0.3, 2.5, and 7.5 μg/ml) were used to study direct effects of lipopeptides on the growth of wheat seedlings. Dry weights of the plants treated with polymyxin B and E at the two higher concentrations (2.5 and 7.5 μg/ml) were significantly reduced under both the well watered and under drought conditions (Figure 6). All concentrations of both antibiotics had significant negative effect on plants under drought stress (Figure 6). Owning to germination impairment by polymyxin E only lowest concentration of polymyxin E was studied for its effect on the dry weight of shoots and roots.
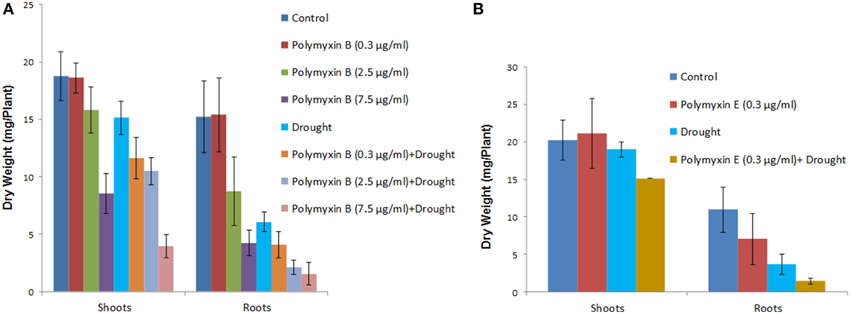
Figure 6. Direct effect of polymyxin B and polymyxin E on wheat root. Three concentrations of the antibiotics (0.3, 2.5 and 7.5 μg/ml) were used in the study. Effect of polymyxin B (A) and polymyxin E (B) on shoot and root dry weight under well watered regime and drought stress. Note that due to germination impairment by polymyxin E only lowest concentration of polymyxin E treated plant dry weights of shoots and roots are shown.
Here we report on the inactivation of the Sfp-type PPTase gene of P. polymyxa A26. Recently we published a simplified method for P. polymyxa genetic manipulation (Kim and Timmusk, 2013). Despite the success of direct manipulation of the well-studied laboratory strain E681, we were not able to carry out Sfp-type PPTase gene homologous recombination in the natural isolate A26. This further confirms the diverse nature of P. polymyxa. Some of the bacterial natural isolates are recalcitrant to any kind of genetic manipulation (Timmusk et al., 2005). Hence the initial gene replacement was performed in the P. polymyxa E681 strain (Figure 1). The chromosomal DNA was isolated from E681 and transformed into A26. Bioactivity and plate antagonism assays, and MALDI-TOF MS analysis, confirm the loss of fusaricidins and polymyxin production by the mutant and successful restoration by the complemented strain (Figure 2). The results indicate that, similar to non-ribosomal peptide/polyketide synthesis in various beneficial and pathogenic bacteria, P. polymyxa A26 is dependent on the presence of a single functional Sfp-type 4′-phosphopantetheinyl transferase identified in its genome (Nakano et al., 1988; Chen et al., 2009; Leblanc et al., 2012).
The wild type, mutant and complemented strain metabolism was studied using BioMerieux® systems and no metabolic differences were detected. The major phenotypic difference of the mutant strain is its enhanced biofilm production (Figure 3 and Table 1). B. subtilis strain 3610, which is genetically almost identical to B. subtilis 168, was used in the study. It is generally known that the Sfp mutation impairs Bacillus spp. biofilm formation and for that reason root colonization is also impaired (Bais et al., 2004; Angelini et al., 2009; Chen et al., 2009; Dietel et al., 2013; Mielich-Suss and Lopez, 2014; Zeriouh et al., 2014). Using the weight-based quantification assay we showed that B. subtilis 3610 Sfp mutant biofilm formation was about 60% impaired. At the same time biofilm formation in the A26 Sfp-type PPTase mutant was increased about 40% (Figure 3). The A26Δsfp biofilm formation enhancement is confirmed in vitro on plate assays and liquid assays as well as in planta on wheat roots using scanning electron and light microscopy and indirectly by soil attached to roots (Figure 3 and Table 1). In order to confirm the A26 biofilm promotion by the mutation the assays were performed with another isolate E681Δsfp. A significant, but slightly smaller, increase in biofilm formation compared to A26Δsfp was observed (Figure 3). The link between lipopeptide antibiotic production and biofilm formation was suggested by Li et al. who observed increased slime production of the P. polymyxa PKB1 lipopeptide mutant (Li et al., 2007).
The biofilm increase of P. polymyxa Sfp-type PPTase mutants is in contrast to the Bacillus spp. model (Angelini et al., 2009; Lopez et al., 2009, 2010a,b; Mielich-Suss and Lopez, 2014; Zeriouh et al., 2014). The PPTase Sfp was first identified in 1988 by Nakamo (Nakano et al., 1988). Since then the enzyme has been studied in various organisms but B. subtilis is the organism where biofilm formation and related Sfp gene involvement is understood in molecular detail (Branda et al., 2001; Chu et al., 2008; Lopez et al., 2009, 2010a,b; McLoon et al., 2011; Beauregard et al., 2013; Vlamakis et al., 2013). In accordance with the model surfactin induces potassium leakage, which stimulates the activity of membrane protein kinase, kin C. Kin C activates the master regulator for biofilm formation. The stimulation may be triggered by a variety of natural products that cause cell membrane potassium leakage (Lopez et al., 2009). In contrast to B. subtilis P. polymyxa is a bacterium that is famously hard to manipulate. It is only during recent years that technologies have been discovered that somewhat simplify genetic manipulation of the species and allow mechanistic studies (Kim and Timmusk, 2013). Although P. polymyxa is one of the best rhizosphere biofilm formers, the mechanism of its biofilm formation has not been studied owning to difficulties to work with the species. An examination of the A26 genome indicates that no surfactins are encoded, whereas genes coding for polymyxins, fusaricidins as well as quite a number of potentially new non-ribosomal lipopeptides/antibiotics are present. The enzymes, NRPS and PKS required for the synthesis, are post-translationally modified by a single Sfp-type PPTase. The A26 Sfp-type PPTase inactivation eliminates the secondary metabolites (Figure 2). In order to study if the compounds are directly involved in the observed biofilm promotion, additional weight-based biofilm formation assays were performed. Metabolite extracts from A26 and A26Δsfp were exogenously added to A26, A26Δsfp and its complemented strain (Table 2). A26 Sfp-type PPTase mutant biofilm formation level was restored with the A26 metabolite complementation and there was no effect with any other metabolite exogenous addition (Table 2). This confirms that the Sfp-type PPTase- mediated A26 secondary metabolites are directly involved in its biofilm formation. Studies are directed now to identify the key regions in the respective synthetase gene clusters, and perform knockouts of all known and potential lipopeptide candidates.

Table 2. In vitro biofilm formation of A26, A26Δsfp, and A26Δsfp pHPS9-sfp complemented with A26 and A26Δsfp metabolite extracts.
Our studies show that the A26 Sfp-type PPTase deficient mutant greatly enhances wheat drought tolerance (Figure 4). The increase is confirmed by survival, dry mass, relative water content water use efficiency and antioxidant enzyme analyses (Figure 4, Figure S2, and Table 1). The discovery opens new areas for biotechnology in the changing world as wheat is the primary staple food of mankind. It is generally believed that bacterial capacity to form biofilms on the root is required for the strain colonization and beneficial effect (Bais et al., 2004; Timmusk et al., 2005, 2009, 2014; Haggag and Timmusk, 2008; Timmusk and Nevo, 2011; Chen et al., 2013; Dietel et al., 2013; Garcia-Gutierrez et al., 2013; Zeriouh et al., 2014). It is tempting to speculate that biofilm formation facilitates the A26Δsfp colonization and that the drought tolerance enhancement is linked to the improved colonization. However, we showed that the colonization by the wild type mutant and complemented mutant did not differ significantly. Can plants benefit from a bacterial biofilm? Bacterial biofilms are comprised of cells and extracellular matrix and form layers around a root hair (Figure 3). The dense biofilm matrix limits diffusion of ACC deaminase and biologically active compounds secreted by bacteria, and these are therefore concentrated for plant uptake (Table 1). It is suggested that B. subtilis biofilm may be generated in response to antimicrobials produced by other microorganisms and may thus constitute a defense mechanism to protect B. subtilis from the action of antibiotics in natural settings (Lopez et al., 2009). P. polymyxa is generally considered as a great biofilm forming biocontrol agent which owning to its unique antibiotic spectra is even able to form single species root biofilms under natural conditions (Timmusk et al., 2005; Timmusk and Nevo, 2011; Timmusk et al., 2011). Hence, A26Δsfp enhanced tight extracellular matrix around roots may contribute to observed drought tolerance enhancement without being its primary cause.
A26 and its Sfp-type PPTase mutant metabolism was studied using BioMerieux® system. No metabolic differences were detected between the strains. It is confirmed in our MALDI-TOF MS that no lipopeptide antibiotics were produced by A26Δsfp in contrast to A26 (Figure 2). Is there any direct effect of P. polymyxa Sfp-type PPTase mediated secondary metabolites on the host plant? To study the effect under gnotobiotic conditions, germinated wheat seeds were exposed to high inoculum densities of A26, A26Δsfp or their lipopeptide extracts and synthetic polymyxins. A26 and its extracts reduced root growth, caused damage to the root tip and synthetic polymyxins had negative effect on seedlings drought tolerance (Figures 5, 6). Lipopeptides, which are amphiphilic molecules with an amino or hydroxy-fatty acid integrated into a peptide moiety, interact with the biological membranes and at some concentrations may induce cell leakage and death (Ongena et al., 2007; Zeriouh et al., 2011). We have previously reported that plant growth promoting P. polymyxa strains may cause mild negative effects on plant root tips which induce plant systemic resistance against Erwinia carotovora (Timmusk and Wagner, 1999; Timmusk et al., 2005). Formerly it has been suggested that microbial hydrolytic enzymes and auxins may be responsible for the deleterious effects (Timmusk and Wagner, 1999; Timmusk et al., 2005; Ludwig-Muller, 2015). The results here however support that NRP/PK-origin compounds produced by P. polymyxa may be the primary reason for its temporary mild deleterious influence (Figure 5). Even though several studies have speculated on rhizobacterially produced lipopeptide role in plant induced systemic resistance, the first evidence was provided only in 2007 (Ongena et al., 2007; Ongena and Jacques, 2008).
P. polymyxa A26 which is isolated from SFS, EC in the northern part of Israel is known to be a biofilm-forming ACC deaminase-containing phosphorus-solubilizing biocontrol bacterium capable of moderate plant drought tolerance enhancement (Timmusk et al., 2011, 2014). There are certainly more scenarios behind A26Δsfp enhanced drought tolerance but our results indicate that the bacterial Sfp-type PPTase- mediated compound restriction contributes to the observed drought tolerance enhancement. The high inoculum densities of P. polymyxa used in our gnotobiotic experiment (Figure 5) don't occur in natural settings (Timmusk et al., 2005). Yet we show that the bacterial lipopeptide production might be the reason for temporary negative effects which induce plant systemic resistance but affect negatively plant drought tolerance. Salicylic acid related defense pathway activation during the host plant interaction has been shown by various plant-beneficial microorganisms, indicating that temporary mild negative effects under particular environmental conditions may be widely spread among plant growth promoting microbes (Phi et al., 2010; Alonso-Ramirez et al., 2014). Lipopeptide antibiotics have been successfully employed in the treatment of various pathogens since 1950s. The bulk of the work has focused on classifying modes of their action. How much of the lipopeptides are produced by A26 under natural conditions? Which of its Sfp-type PPTase-mediated compounds are involved in direct antagonism and induced systemic resistance? It is clear that the physiological concentration of any biologically active compound varies considerably depending on the dose and on all manners of parameters of treated host. More studies need to be performed on the exciting area of the A26 lipopeptide production under natural conditions. Cell type specific transcriptional regulation probing should reveal exactly how wheat root cells are influenced by A26 and A26Δsfp priming. An important challenge for the future will be to determine which of the Sfp-type PPTase- mediated compounds contribute to the A26Δsfp strain drought tolerance enhancement and biofilm promotion.
Rhizobacterial ability to enhance plant drought stress tolerance was serendipitously discovered in 1999 by Timmusk and Wagner in an attempt to study the soil bacteria for plant induced resistance and nitrogen fixation ability (Timmusk and Wagner, 1999). Recently the discovery was updated with an in principle new technology showing that the bacteria from harsh environments are more likely to be efficient in enhancing host plant drought tolerance (Timmusk et al., 2014). Here we show that the Sfp-type PPTase gene in P. polymyxa is a gate-keeper for the bacterial drought tolerance enhancement.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Prof. S-H Park is gratefully acknowledged for providing E681 strain and help with initial stages of work. Prof. D. Kearns is thanked for NCIB3610 and DS3337 strains. We are indebted to D. Clapham for valuable discussions and critical reading of the manuscript. Financial support for the study was provided by the European Commission through FP7:247669, the Swedish Research Council Formas (2009-243), Vetenskapsrådet (2012-6217), Carl Tryggers Stiftelse for vetenskaplig forskning (CTS09:385), Stiftelsen Oscar och Lili Lamms Minne (DO2009-0052) (to ST).
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00387/abstract
Table S1. Primers used in this study.
Figure S1. Analysis of P. polymyxa A26 fusaricidins synthesis. (A) LC analysis of A26 culture supernatants. (B) MS data for the fusaricidins produced by A26.
Figure S2. Relative water content (RWC) of P. polymyxa A26Δsfp, A26 and untreated wheat under drought and well watered regime.
Alonso-Ramirez, A., Poveda, J., Martin, I., Hermosa, R., Monte, E., and Nicolas, C. (2014). Salicylic acid prevents Trichoderma harzianum from entering the vascular system of roots. Mol. Plant Pathol. 15, 823–831. doi: 10.1111/mpp.12141
Angelini, T. E., Roper, M., Kolter, R., Weitz, D. A., and Brenner, M. P. (2009). Bacillus subtilis spreads by surfing on waves of surfactant. Proc. Natl. Acad. Sci. U.S.A. 106, 18109–18113. doi: 10.1073/pnas.0905890106
Bais, H. P., Fall, R., and Vivanco, J. M. (2004). Biocontrol of Bacillus subtilis against infection of Arabidopsis roots by Pseudomonas syringae is facilitated by biofilm formation and surfactin production. Plant Physiol. 134, 307–319. doi: 10.1104/pp.103.028712
Beauregard, P. B., Chai, Y., Vlamakis, H., Losick, R., and Kolter, R. (2013). Bacillus subtilis biofilm induction by plant polysaccharides. Proc. Natl. Acad. Sci. U.S.A. 110: E1621–1630. doi: 10.1073/pnas.1218984110
Beld, J., Sonnenschein, E. C., Vickery, C. R., Noel, J. P., and Burkart, M. D. (2014). The phosphopantetheinyl transferases: catalysis of a post-translational modification crucial for life. Nat. Prod. Rep. 31, 61–108. doi: 10.1039/C3NP70054B
Branda, S. S., Gonzalez-Pastor, J. E., Ben-Yehuda, S., Losick, R., and Kolter, R. (2001). Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 98, 11621–11626. doi: 10.1073/pnas.191384198
Bunet, R., Riclea, R., Laureti, L., Hotel, L., Paris, C., Girardet, J. M., et al. (2014). A single Sfp-type phosphopantetheinyl transferase plays a major role in the biosynthesis of PKS and NRPS derived metabolites in Streptomyces ambofaciens ATCC23877. PLoS ONE 9:e87607. doi: 10.1371/journal.pone.0087607
Chalut, C., Botella, L., de Sousa-D'Auria, C., Houssin, C., and Guilhot, C. (2006). The nonredundant roles of two 4′-phosphopantetheinyl transferases in vital processes of Mycobacteria. Proc. Natl. Acad. Sci. U.S.A. 103, 8511–8516. doi: 10.1073/pnas.0511129103
Chen, X. H., Koumoutsi, A., Scholz, R., and Borriss, R. (2009). More than anticipated - production of antibiotics and other secondary metabolites by Bacillus amyloliquefaciens FZB42. J. Mol. Microbiol. Biotechnol. 16, 14–24. doi: 10.1159/000142891
Chen, Y., Yan, F., Chai, Y., Liu, H., Kolter, R., Losick, R., et al. (2013). Biocontrol of tomato wilt disease by Bacillus subtilis isolates from natural environments depends on conserved genes mediating biofilm formation. Environ. Microbiol. 15, 848–864. doi: 10.1111/j.1462-2920.2012.02860.x
Choi, S. K., Park, S. Y., Kim, R., Kim, S. B., Lee, C. H., Kim, J. F., et al. (2009). Identification of a polymyxin synthetase gene cluster of Paenibacillus polymyxa and heterologous expression of the gene in Bacillus subtilis. J. Bacteriol. 191, 3350–3358. doi: 10.1128/JB.01728-08
Choi, S. K., Park, S. Y., Kim, R., Lee, C. H., Kim, J. F., and Park, S. H. (2008). Identification and functional analysis of the fusaricidin biosynthetic gene of Paenibacillus polymyxa E681. Biochem. Biophys. Res. Commun. 365, 89–95. doi: 10.1016/j.bbrc.2007.10.147
Chu, F., Kearns, D. B., McLoon, A., Chai, Y., Kolter, R., and Losick, R. (2008). A novel regulatory protein governing biofilm formation in Bacillus subtilis. Mol. Microbiol. 68, 1117–1127. doi: 10.1111/j.1365-2958.2008.06201.x
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645. doi: 10.1073/pnas.120163297
de Bruijn, I., de Kock, M. J., Yang, M., de Waard, P., van Beek, T. A., and Raaijmakers, J. M. (2007). Genome-based discovery, structure prediction and functional analysis of cyclic lipopeptide antibiotics in Pseudomonas species. Mol. Microbiol. 63, 417–428. doi: 10.1111/j.1365-2958.2006.05525.x
Dietel, K., Beator, B., Budiharjo, A., Fan, B., and Borriss, R. (2013). Bacterial traints involved in colonization of Arabidopsis thaliana roots by Bacillus amyloliquefaciens FZB42. Plant Pathol. J. 29, 59–66. doi: 10.5423/PPJ.OA.10.2012.0155
Foley, J. A., Ramankutty, N., Brauman, K. A., Cassidy, E. S., Gerber, J. S., Johnston, M., et al. (2011). Solutions for a cultivated planet. Nature 478, 337–342. doi: 10.1038/nature10452
Foley, T. L., Rai, G., Yasgar, A., Daniel, T., Baker, H. L., Attene-Ramos, M., et al. (2014). 4-(3-Chloro-5-(trifluoromethyl)pyridin-2-yl)-N-(4-methoxypyridin-2-yl)piperazine- 1-carbothioamide (ML267), a potent inhibitor of bacterial phosphopantetheinyl transferase that attenuates secondary metabolism and thwarts bacterial growth. J. Med. Chem. 57, 1063–1078. doi: 10.1021/jm401752p
Garcia-Gutierrez, L., Zeriouh, H., Romero, D., Cubero, J., de Vicente, A., and Perez-Garcia, A. (2013). The antagonistic strain Bacillus subtilis UMAF6639 also confers protection to melon plants against cucurbit powdery mildew by activation of jasmonate- and salicylic acid-dependent defence responses. Microb. Biotechnol. 6, 264–274. doi: 10.1111/1751-7915.12028
Haggag, W. M., and Timmusk, S. (2008). Colonization of peanut roots by biofilm-forming Paenibacillus polymyxa initiates biocontrol against crown rot disease. J. Appl. Microbiol. 104, 961–969. doi: 10.1111/j.1365-2672.2007.03611.x
Harwood, C., and Cutting, S. (1990). Molecular Biological Methods for Bacillus. New York, NY: John Wiley & Sons.
Hossain, M., Nakano, Y., and Asada, K. (1984). Mono dehydro ascorbate reductase EC-1.6.5.4 in spinach chloroplasts and its participation in regeneration of ascorbate for scavenging hydrogen per oxide. Plant Cell Physiol. 25, 385–396.
Kim, S.-B., and Timmusk, S. (2013). A simplified method for Paenibacillus polymyxa gene knockout and insertional screening. PLoS ONE 8:e68092. doi: 10.1371/journal.pone.0068092
Knöerzer, O., Durner, J., and Boeger, P. (1996). Alterations in the antioxidative system of suspension-cultured soybean cells (Glycine max) induced by oxidative stress. Physiol. Plant 97, 388–396. doi: 10.1034/j.1399-3054.1996.970225.x
Lambalot, R. H., Gehring, A. M., Flugel, R. S., Zuber, P., LaCelle, M., Marahiel, M. A., et al. (1996). A new enzyme superfamily - the phosphopantetheinyl transferases. Chem. Biol. 3, 923–936. doi: 10.1016/S1074-5521(96)90181-7
Leblanc, C., Prudhomme, T., Tabouret, G., Ray, A., Burbaud, S., Cabantous, S., et al. (2012). 4′-Phosphopantetheinyl transferase PptT, a new drug target required for Mycobacterium tuberculosis growth and persistence in vivo. PLoS Pathog. 8:e1003097. doi: 10.1371/journal.ppat.1003097
Li, J., Beatty, P. K., Shah, S., and Jensen, S. E. (2007). Use of PCR-targeted mutagenesis to disrupt production of fusaricidin-type antifungal antibiotics in Paenibacillus polymyxa. Appl. Environ. Microbiol. 73, 3480–3489. doi: 10.1128/AEM.02662-06
Lopez, D., Fischbach, M. A., Chu, F., Losick, R., and Kolter, R. (2009). Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 106, 280–285. doi: 10.1073/pnas.0810940106
Lopez, D., Gontang, E. A., and Kolter, R. (2010a). Potassium sensing histidine kinase in Bacillus subtilis. Meth. Enzymol. 471, 229–251. doi: 10.1016/S0076-6879(10)71013-2
Lopez, D., Vlamakis, H., and Kolter, R. (2010b). Biofilms. Cold Spring Harb. Perspect. Biol. 2: a000398. doi: 10.1101/cshperspect.a000398
Ludwig-Muller, J. (2015). Bacteria and fungi controlling plant growth by manipulating auxin: balance between development and defense. J. Plant Physiol. 172, 4–12. doi: 10.1016/j.jplph.2014.01.002
McLoon, A. L., Guttenplan, S. B., Kearns, D. B., Kolter, R., and Losick, R. (2011). Tracing the domestication of a biofilm-forming bacterium. J. Bacteriol. 193, 2027–2034. doi: 10.1128/JB.01542-10
Mielich-Suss, B., and Lopez, D. (2014). Molecular mechanisms involved in Bacillus subtilis biofilm formation. Environ. Microbiol. 17, 555–565. doi: 10.1111/1462-2920.12527
Nakano, M. M., Marahiel, M. A., and Zuber, P. (1988). Identification of a genetic locus required for biosynthesis of the lipopeptide antibiotic surfactin in Bacillus subtilis. J. Bacteriol. 170, 5662–5668.
Niedermeyer, T. H., and Strohalm, M. (2012). mMass as a software tool for the annotation of cyclic peptide tandem mass spectra. PLoS ONE 7:e44913. doi: 10.1371/journal.pone.0044913
Ongena, M., and Jacques, P. (2008). Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 16, 115–125. doi: 10.1016/j.tim.2007.12.009
Ongena, M., Jourdan, E., Adam, A., Paquot, M., Brans, A., Joris, B., et al. (2007). Surfactin and fengycin lipopeptides of Bacillus subtilis as elicitors of induced systemic resistance in plants. Environ. Microbiol. 9, 1084–1090. doi: 10.1111/j.1462-2920.2006.01202.x
Phi, Q. T., Park, Y. M., Seul, K. J., Ryu, C. M., Park, S. H., Kim, J. G., et al. (2010). Assessment of root-associated Paenibacillus polymyxa groups on growth promotion and induced systemic resistance in pepper. J. Microbiol. Biotechnol. 20, 1605–1613.
Powell, A., Borg, M., Amir-Heidari, B., Neary, J. M., Thirlway, J., Wilkinson, B., et al. (2007). Engineered biosynthesis of nonribosomal lipopeptides with modified fatty acid side chains. J. Am. Chem. Soc. 129, 15182–15191. doi: 10.1021/ja074331o
Quadri, L. E., Weinreb, P. H., Lei, M., Nakano, M. M., Zuber, P., and Walsh, C. T. (1998). Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. Biochemistry 37, 1585–1595. doi: 10.1021/bi9719861
Royer, M., Koebnik, R., Marguerettaz, M., Barbe, V., Robin, G. P., Brin, C., et al. (2013). Genome mining reveals the genus Xanthomonas to be a promising reservoir for new bioactive non-ribosomally synthesized peptides. BMC Genomics 14:658. doi: 10.1186/1471-2164-14-658
Ryu, C.-M., Kim, J., Choi, O., Park, S.-Y., Park, S.-H., and Park, C.-S. (2005). Nature of a root-associated Paenibacillus polymyxa from fieldgrown winter barley in Korea. J. Microbiol. Biotechnol. 15, 984–991.
Smith, I., Vierheller, T., and Thorne, C. (1989). Properties and functions of glutathione reductase in plants. Physiol. Plant 77, 449–456. doi: 10.1111/j.1399-3054.1989.tb05666.x
Sunbul, M., Marshall, N. J., Zou, Y., Zhang, K., and Yin, J. (2009a). Catalytic turnover-based phage selection for engineering the substrate specificity of Sfp phosphopantetheinyl transferase. J. Mol. Biol. 387, 883–898. doi: 10.1016/j.jmb.2009.02.010
Sunbul, M., Zhang, K., and Yin, J. (2009b). Chapter 10 using phosphopantetheinyl transferases for enzyme posttranslational activation, site specific protein labeling and identification of natural product biosynthetic gene clusters from bacterial genomes. Meth. Enzymol. 458, 255–275. doi: 10.1016/S0076-6879(09)04810-1
Timmusk, S., El Daim, I., Copolovici, L., Kannaste, A., Behers, L., Nevo, E., et al. (2014). Drought-tolerance of wheat improved by rhizosphere bacteria from harsh environments: enhanced biomass production and reduced emissions of stress volatiles. PLoS ONE 9:e96086. doi: 10.1371/journal.pone.0096086
Timmusk, S., Grantcharova, N., and Wagner, E. G. (2005). Paenibacillus polymyxa invades plant roots and forms biofilms. Appl. Environ. Microbiol. 71, 7292–7300. doi: 10.1128/AEM.71.11.7292-7300.2005
Timmusk, S., and Nevo, E. (2011). “Plant root associated biofilms,” in Bacteria in Agrobiology, Vol. 3: Plant Nutrient Management, ed D. K. Maheshwari (Berlin: Springer Verlag), 285–300.
Timmusk, S., Paalme, V., Pavlicek, T., Bergquist, J., Vangala, A., Danilas, T., et al. (2011). Bacterial distribution in the rhizosphere of wild barley under contrasting microclimates. PLoS ONE 6:e17968. doi: 10.1371/journal.pone.0017968
Timmusk, S., van West, P., Gow, N. A., and Huffstutler, R. P. (2009). Paenibacillus polymyxa antagonizes oomycete plant pathogens Phytophthora palmivora and Pythium aphanidermatum. J. Appl. Microbiol. 106, 1473–1481. doi: 10.1111/j.1365-2672.2009.04123.x
Timmusk, S., and Wagner, E. G. (1999). The plant-growth-promoting rhizobacterium Paenibacillus polymyxa induces changes in Arabidopsis thaliana gene expression: a possible connection between biotic and abiotic stress responses. Mol. Plant Microbe Interact. 12, 951–959. doi: 10.1094/MPMI.1999.12.11.951
Vlamakis, H., Chai, Y., Beauregard, P., Losick, R., and Kolter, R. (2013). Sticking together: building a biofilm the Bacillus subtilis way. Nat. Rev. Microbiol. 11, 157–168. doi: 10.1038/nrmicro2960
Zeriouh, H., de Vicente, A., Perez-Garcia, A., and Romero, D. (2014). Surfactin triggers biofilm formation of Bacillus subtilis in melon phylloplane and contributes to the biocontrol activity. Environ. Microbiol. 16, 2196–2211. doi: 10.1111/1462-2920.12271
Zeriouh, H., Romero, D., Garcia-Gutierrez, L., Cazorla, F. M., de Vicente, A., and Perez-Garcia, A. (2011). The iturin-like lipopeptides are essential components in the biological control arsenal of Bacillus subtilis against bacterial diseases of cucurbits. Mol. Plant Microbe Interact. 24, 1540–1552. doi: 10.1094/MPMI-06-11-0162
Keywords: Sfp-type PPTase, Paenibacillus polymyxa, Evolution Canyon, rhizobacterial biofilm, plant drought tolerance, natural isolate genetic manipulation
Citation: Timmusk S, Kim S-B, Nevo E, Abd El Daim I, Ek B, Bergquist J and Behers L (2015) Sfp-type PPTase inactivation promotes bacterial biofilm formation and ability to enhance wheat drought tolerance. Front. Microbiol. 6:387. doi: 10.3389/fmicb.2015.00387
Received: 16 March 2015; Accepted: 15 April 2015;
Published: 21 May 2015.
Edited by:
Brigitte Mauch-Mani, Université de Neuchâtel, SwitzerlandReviewed by:
Choong-Min Ryu, Korea Research Institute of Bioscience and Biotechnology, South KoreaCopyright © 2015 Timmusk, Kim, Nevo, Abd El Daim, Ek, Bergquist and Behers. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Salme Timmusk, Department of Forest Mycology and Plant Pathology, Uppsala BioCenter, Swedish University of Agricultural Sciences, PO Box 7026, SE-75007 Uppsala, Sweden,c2FsbWUudGltbXVza0BzbHUuc2U=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.