
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 14 April 2015
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 6 - 2015 | https://doi.org/10.3389/fmicb.2015.00294
This article is part of the Research TopicEmerging Enterobacteriaceae infections: antibiotic resistance and novel treatment optionsView all 8 articles
 Zhenhong Chen1†
Zhenhong Chen1† Hongxia Li1†
Hongxia Li1† Jiao Feng2
Jiao Feng2 Yuxue Li3
Yuxue Li3 Xin Chen4
Xin Chen4 Xuemin Guo4
Xuemin Guo4 Weijun Chen5
Weijun Chen5 Li Wang1
Li Wang1 Lei Lin2
Lei Lin2 Huiying Yang2
Huiying Yang2 Wenhui Yang2
Wenhui Yang2 Jie Wang2
Jie Wang2 Dongsheng Zhou2*
Dongsheng Zhou2* Changting Liu1*
Changting Liu1* Zhe Yin2*
Zhe Yin2*A carbapenem-nonsusceptible Enterobacter aerogenes strain named 3-SP was isolated from a human case of pneumonia in a Chinese teaching hospital. NDM-1 carbapenemase is produced by a pNDM-BJ01-like conjugative plasmid designated p3SP-NDM to account for carbapenem resistance of 3-SP. p3SP-NDM was fully sequenced and compared with all publically available pNDM-BJ01-like plasmids. The genetic differences between p3SP-NDM and pNDM-BJ01 include only 18 single nucleotide polymorphisms, a 1 bp deletion and a 706 bp deletion. p3SP-NDM and pNDM-BJ01 harbor an identical Tn125 element organized as ISAba125, blaNDM−1, bleMBL, ΔtrpF, dsbC, cutA, ΔgroES, groEL, ISCR27, and ISAba125. The blaNDM−1 surrounding regions in these pNDM-BJ01-like plasmids have a conserved linear organization ISAba14-aphA6-Tn125-unknown IS, with considerable genetic differences identified within or immediately downstream of Tn125. All reported pNDM-BJ01-like plasmids are exclusively found in Acinetobacter, whereas this is the first report of identification of a pNDM-BJ01-like plasmid in Enterobacteriaceae.
Enterobacter aerogenes is a Gram-negative bacterium widely found in the human gastrointestinal tract and in the environment, and generally non-pathogenic to healthy humans. Since 1990s, E. aerogenes has become an important opportunistic pathogen commonly affecting those with weakened immune systems to cause hospital-acquired infections such as pneumonia, bacteremia, urinary tract infection, surgical site infection, and meningitis (Georghiou et al., 1995; Davin-Regli et al., 1996; De Gheldre et al., 1997; Jalaluddin et al., 1998; Ronveaux et al., 1999).
E. aerogenes strains isolated from hospitalized patients generally exhibit high resistance to commonly used broad-spectrum antibiotics; in particular, the use of carbapenems imipenem and meropenem as the first-line antimicrobial agents for treating serious or refractory infections has led to considerable increase in prevalence of carbapenem-resistant E. aerogenes (De Gheldre et al., 1997; Chen et al., 2008; Lavigne et al., 2013). Carbapenem resistance of E. aerogenes is usually a result of production of plasmid-encoding carbapenemases such as KPC (Chen et al., 2014; Kuai et al., 2014; Luo et al., 2014; Qin et al., 2014), IMP (Biendo et al., 2008; Ding et al., 2014), VIM (Biendo et al., 2008; Souli et al., 2008) and NDM (Ho et al., 2012), decreased membrane permeability (due to altered porin expression or efflux pump overexpression) together with production of AmpC-type cephalosporinase or extended-spectrum β-lactamase (ESBL) (Bornet et al., 2003; Lavigne et al., 2012, 2013), and lipopolysaccharide modification (Leying et al., 1991).
The 47.27 Kb plasmid pNDM-BJ01 is isolated from a clinical A. lwoffii strain in China in 2010 (Hu et al., 2012) and it cannot be assigned into any known incompatibility group. In this study, phenotypic and biochemical experiments combined with plasmid sequencing and comparative genomics analyses disclose that production of NDM-1 by a pNDM-BJ01-like conjugative plasmid p3SP-NDM accounts for carbapenem resistance of a clinical E. aerogenes isolate recovered from a human case of pneumonia in a Chinese teaching hospital.
The use of human specimens and all related experimental protocols were approved by the Committee on Human Research of indicated institutions and carried out in accordance with the approved guidelines, and moreover the informed consent was obtained from indicated patient. All the bacterial strains in this study were subjective for species identification by BioMérieux VITEK 2, Bruker MALDI Biotyper, and 16s rRNA gene sequencing. For determination of 16S rRNA gene sequence, the almost complete coding region of 16S rRNA gene was amplified by PCR with the universal primers 27f (AGAGTTTGATCCTGGCTCAG) and 1492r (TACCTTGTTACGACTT) (Frank et al., 2008). The major carbapenemase and ESBL genes as listed in Table S1 were subjected to PCR detection. All PCR amplicons were sequenced on ABI 3730 Sequencer with the same primers for PCR.
Plasmid conjugal transfer experiments were carried out with the rifampin-resistant E. coli EC600 being used as recipient and the blaNDM-positive strain 3-SP as donor. 3 ml of overnight culture of each of donor and recipient bacteria were mixed together, harvested, and resuspended in 80 μl of Brain Heart Infusion (BHI) broth (BD Biosciences). The mixture was spotted on a 1 cm2 filter membrane that was placed on BHI agar (BD Biosciences) plate, and then incubated for mating at 37°C for 12–18 h. Bacteria were washed from filter membrane and spotted on Muller-Hinton (MH) agar (BD Biosciences) plate containing 750 μg/ml rifampin and 200 μg/ml ampicillin for selection of blaNDM-positive E. coli transconjugants.
To prepare competent cells for plasmid electroporation, 200 ml of overnight culture of E. coli DH10B in Super Optimal Broth (SOB) at an optical density (OD600) of 0.4–0.6 was washed three times with electroporation buffer (0.5 M mannitol and 10% glycerol) and concentrated into a final volume of 2 ml. 1 μg of plasmid DNA, which was isolated from 3-SP with QIAGEN Plasmid Midi Kit, were mixed with 100 μl of competent cells for electroporation at 25 μF, 200 Ω, and 2.5 Kv. The resulting cells were suspended in 500 μl of SOB, and an appropriate aliquot was spotted on SOB agar plate containing 200 μg/ml ampicillin for selection of blaNDM-positive E. coli electroporants.
Bacterial genomic DNA was prepared in agarose plugs and digested with S1 nuclease (Takara). The linearized plasmids and partially digested genomic DNA were separated through the CHEF-Mapper XA PFGE system (Bio-Rad). The DNA fragments were stained with ethidium bromide (EtBr), transferred to a Hybond N+ membrane (GE Amersham Biosciences) and hybridized with a DIG-labeled probe specific to blaNDM (Rasheed et al., 2013). Probe labeling and signal detection were carried out with DIG high primer DNA labeling and detection starter kit II according to the manufacturer's instructions (Roche Diagnostics).
Activity of class A/B/D carbapenemases was determined by CarbaNP test (Dortet et al., 2012) with modifications. Overnight bacterial cell culture in MH broth was diluted 1:100 into 3 ml of fresh MH broth, and bacteria were allowed to grow at 37°C with shaking at 200 rpm to reach an OD600 of 1.0–1.4. If required, ampicillin was used at 200 μg/ml. Bacterial cells were harvested from 2 ml of the above culture, and washed twice with 20 mM Tris-HCl (pH 7.8). Cell pellets were resuspended in 500 μl of 20 mM Tris-HCl (pH 7.8), and lysed by soniation, followed by centrifugation at 10,000 ×g at 4 °C for 5 min. 50 μl of the supernatant (the enzymatic bacterial suspension) were mixed with 50 μl of substrate I to V, respectively, followed by incubation at 37°C for a maximum of 2 h. Substrate I: 0.054% phenol red plus 0.1 mM ZnSO4 (pH 7.8). Substrate II: 0.054% phenol red plus 0.1 mM ZnSO4 (pH 7.8), and 0.6 mg/μl imipenem. Substrate III: 0.054% phenol red plus 0.1 mM ZnSO4 (pH 7.8), 0.6 mg/μl mg imipenem, and 0.8 mg/μl tazobactam. Substrate IV: 0.054% phenol red plus 0.1 mM ZnSO4 (pH 7.8), 0.6 mg/μl mg imipenem, and 3 mM EDTA (pH 7.8). Substrate V: 0.054% phenol red plus 0.1 mM ZnSO4 (pH 7.8), 0.6 mg/μl mg imipenem, 0.8 mg/μl tazobactam, and 3 mM EDTA (pH 7.8).
The MIC values of indicated bacterial strains were tested by using VITEK 2 according to manufacturer's instructions, and antimicrobial susceptibility was judged by Clinical and Laboratory Standards Institute (CLSI) standard.
The chromosome DNA-free plasmid DNA was isolated from the cell cultures of indicated E. coli transconjugant using a Qiagen large construct kit, and then sequenced by using whole-genome shotgun strategy in combination with Illumina HiSeq 2500 sequencing technology. The contigs were assembled with Velvet, and the gaps were filled through combinatorial PCR and Sanger Sequencing on ABI 3730 Sequencer. The genes were predicted with GeneMarkS and further annotated by BLASTP against UniPort and NR databases.
The complete sequence of plasmid p3SP-NDM was submitted to GenBank under accession number KP900015.
In June 2012, an 86-year-old male with cough and fever visited a teaching hospital in Xi'an city of China. The patient had underlying sequelae of cerebral hemorrhage, and complained of recurrent pulmonary infection. The patient received oral administration with cefradine for a week, but his symptoms did not improve. The patient was subsequently hospitalized, and chest X-ray examination confirmed presence of bilateral pulmonary infection and he was accordingly diagnosed to have pneumonia. The sputum specimens were sampled on the same day of admission. On the next day, round bacterial colonies were observed after cultivation of sputum on MH agar, and the bacterial isolate designated 3-SP was identified as E. aerogenes by VITEK 2, Bruker MALDI Biotyper, and 16s rRNA gene sequencing. The antimicrobial susceptibility test using VITEK 2 indicated 3-SP was resistant to multiple β-lactam antibiotics including imipenem and meropenem but remained susceptible to fluoroquinolones. The patient accordingly received intravenous administration with levofloxacin, and his symptoms associated with pulmonary infection disappeared and he was discharged after 10 days of antimicrobial treatment.
PCR detection of the major ESBL and carbapenemase genes (Table S1) indicated presence of only blaNDM in E. aerogenes 3-SP (Figure S1), which was confirmed by PCR amplicon sequencing. A blaNDM-positive E. coli EC600 transconjugant named 3-SP-NDM-EC600 and a blaNDM-positive E. coli DH10B electroporant designated 3-SP-NDM-DH10B were obtained. The S1-PFGE/southern hybridization assay detected a ~48 kb plasmid in each of 3-SP, 3-SP-NDM-EC600 and 3-SP-NDM-DH10B, which could hybridize with a blaNDM-specific probe (Figure S2). The modified CarbaNP test showed that 3-SP, 3-SP-NDM-EC600, and 3-SP-NDM-DH10B had Ambler class B carbapenemase activity (Figure S3). The antibiotic susceptibility test showed that 3-SP, 3-SP-NDM-EC600, and 3-SP-NDM-DH10B were highly resistant to all the penicillin, β-lactamase, monobactam, cephalosporin, and carbapenem drugs tested, but remained to be susceptible to fluoroquinolones, furanes, aminoglycosides, and sulfanilamide tested (Table 1). The above results indicated that 3-SP contained a conjugative NDM-encoding plasmid (designated p3SP-NDM), which accounted for carbapenem resistance of 3-SP and could be transferred into and mobilized in E. coli recipients.
Plasmid DNA was isolated from 3-SP-NDM-EC600, and the whole genome sequence of p3SP-NDM was determined to 46,570 bp in length with a 137 fold coverage, forming a circular DNA sequence with a total of 45 open reading frames annotated (Figure 1).
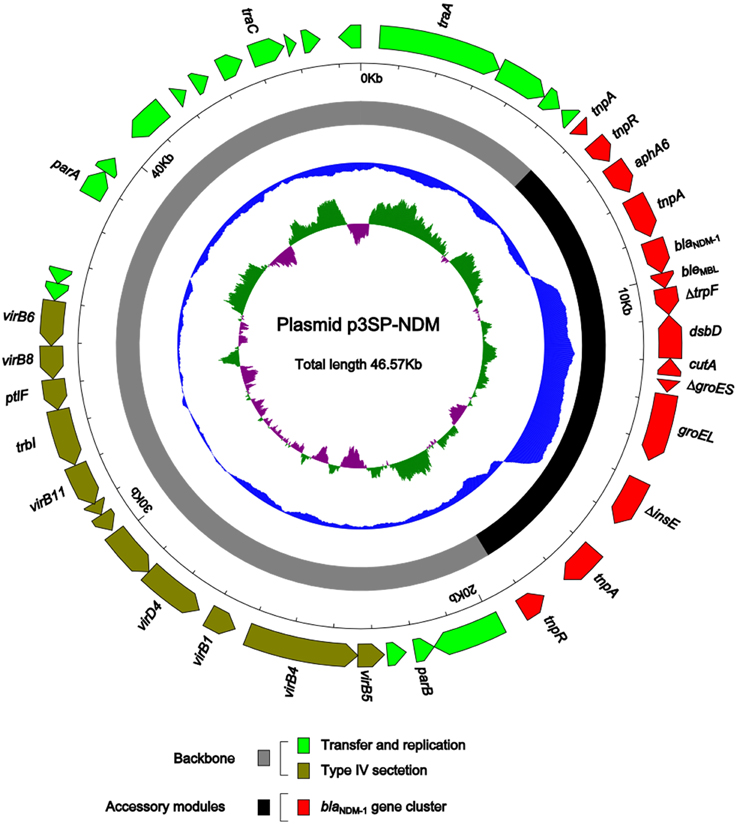
Figure 1. Schematic map of p3SP-NDM. Genes are denoted by arrows and colored based on gene function classification. The innermost circle presents GC-Skew [(G − C)/(G + C)] with a window size of 500 bp and a step size of 20 bp. The blue circle presents GC content. Shown also are backbone and accessory module regions. All the gene organization figures in this work were drawn by using the Inkscape software (https://inkscape.org/).
p3SP-NDM is highly similar to pNDM-BJ01 with genetic differences including only 18 single nucleotide polymorphisms and an 1 bp deletion (Table S2) and a 706 bp deletion (Figure S3, see also below).
Linear structural comparison (Figure 2) was performed with whole genome sequences of p3SP-NDM, pNDM-BJ01 and all the six additional pNDM-BJ01-like plasmids pNDM-BJ02 (Hu et al., 2012), pNDM-40-1 (Jones et al., 2014), pNDM-AB (Zhang et al., 2013), pNDM-Iz4b (KJ547696), pAbNDM-1 (JN377410), and pM131_NDM1 (JX072963) (collected from GenBank on November 20, 2014). p3SP-NDM and pNDM-Iz4b essentially had the same genomic organization.
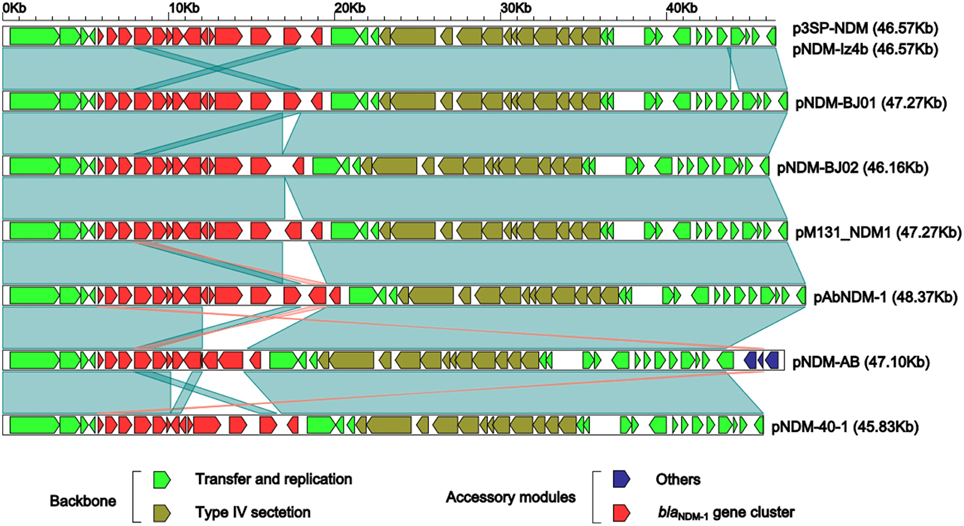
Figure 2. Linear comparison of sequenced plasmids. Genes are denoted by arrows and colored based on gene function classification. Dark green shading regions denote regions of homology (>99% nucleotide similarity).
The above eight plasmids contain a highly conserved backbone composed of two separate regions of plasmid replication/transfer and one region of type VI section system, with only one structural polymorphism that a 706 bp fragment (nucleotide position 43,861–44,566 in pNDM-BJ01; located within the plasmid replication/transfer region and contains only one annotated gene encoding hypothetical protein) is deleted from p3SP-NDM and pNDM-Iz4b relative to all the other plasmids (Figure 2).
As for accessory modules (Figure 2), each of these eight plasmids contains a blaNDM−1 gene cluster located around nucleotide position 5685; in addition, pNDM-AB harbors an additional 3.5 Kb accessory region, which is located around nucleotide position 5570 and composed of an ISAba14 element and a gene encoding type I restriction-modification system methyl transferase subunit.
The blaNDM−1 gene clusters from the above eight plasmids show a conserved linear organization ISAba14-aphA6-Tn125-unknown IS, and the ISAba14-aphA6 and unknown IS fragments are essentially identical structurally in these plasmids while structural differences occur within or immediately downstream of the composite transposon Tn125 (Figure 3). pNDM-BJ01 and p3SP-NDM contain the prototype Tn125, which is sequentially organized as ISAba125, blaNDM−1, bleMBL (bleomycin resistance), ΔtrpF, dsbC, cutA, ΔgroES, groEL, ISCR27, and ISAba125 (Figure 3); Tn125 is inserted into a site downstream of aphA6 (aminoglycoside resistance), which is evidenced by presence of GTT direct repeats at both ends, and the two copies of ISAba125 likely target blaNDM−1 surrounding sequences to promote formation and transposition of Tn125 (Poirel et al., 2012).
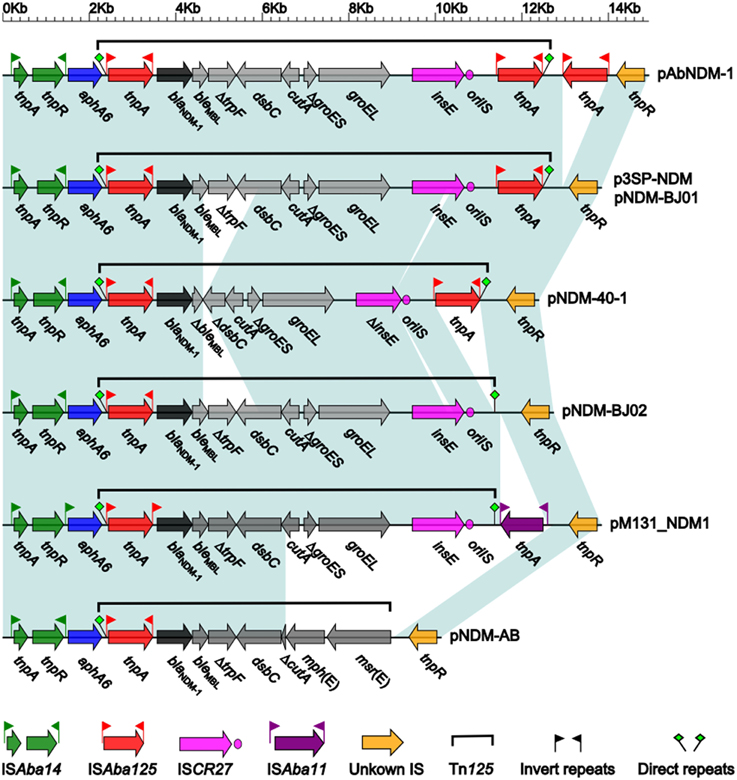
Figure 3. Linear comparison of blaNDM−1 gene clusters. Genes are denoted by arrows and colored. Dark green shading regions denote regions of homology (>99% nucleotide similarity).
Compared to the counterpart in pNDM-BJ01/p3SP-NDM, Tn125 in pNDM-40-1 (Figure 3) is a truncated version with two deletions: a 1298 bp deletion within original bleMBL-ΔtrpF-dsbC to generate ΔbleMBL-ΔdsbC, and a 150 bp deletion within ISCR27 (Jones et al., 2014). A third copy of ISAba125 or an intact ISAba11 element is inserted immediately downstream of the intact Tn125 of pAbNDM-1 or pM131_NDM1, respectively, while the downstream copy of ISAba125 is deleted from Tn125 of pNDM-BJ02 (Figure 3). As for Tn125 in pNDM-AB, cutA-ΔgroES-groEL-ISCR27 observed in pNDM-BJ01 is replaced by ΔcutA-mph(E)- msr(E), leading to absence of downstream GTT direct repeat (Zhang et al., 2013); by contrast, GTT direct repeats are intact in all other seven plasmids (Figure 3). The mph(E) and msr(E) genes confer macrolide/triamilide resistance (Michael et al., 2012).
NDM, initially identified in Klebsiella pneumoniae in 2009, is a metallo-β-lactamase (MBL) capable of hydrolyzing almost all clinically used β-lactams (Tiwari and Moganty, 2013), and the blaNDM genes have been found in a large collection of Gram-negative bacteria of clinical, environmental and animal origins, especially including Acinetobacter, Enterobacteriaceae, and Pseudomonas (Nordmann et al., 2011; Johnson and Woodford, 2013; Dortet et al., 2014). Fourteen NDM variants have been described, differing by several amino acid changes, and a few of them have been tested for their enzymatic kinetics, which denotes that amino acid substitution is a major source of MBL activity extension (Nordmann et al., 2012; Tada et al., 2013). Nevertheless, a systematic characterization of enzymatic kinetics of all the identified NDM variants is needed.
Intact ISAba125 has never been found in bacterial species other than Acinetobacter, and thus ISAba125 ought to originate from Acinetobacter. blaNDM−1 is most likely generated in an Acinetobacter background by a fusion event between aphA6 and an ancestral metallo-β-lactamase gene (Poirel et al., 2012; Toleman et al., 2012; Zong and Zhang, 2013). Insertion of various derivates of blaNDM−1-carrying Tn125 have been found within Acinetobacter chromosomes (Pfeifer et al., 2011; Bonnin et al., 2012; Partridge and Iredell, 2012; Poirel et al., 2012) and plasmids (Hu et al., 2012; Partridge and Iredell, 2012; Zhang et al., 2013; Zong and Zhang, 2013; Jones et al., 2014) at different locations, and moreover Tn125 derivates also represent plasmid-borne blaNDM−1 contexts in Enterobacteriaceae (Sekizuka et al., 2011; Mcgann et al., 2012; Partridge and Iredell, 2012; Fiett et al., 2014; Mataseje et al., 2014). These indicate emergency of blaNDM−1in Acinetobacter and then dissemination among Enterobacteriaceae. In addition, the upstream copy of ISAba125, either intact or interrupted by other mobile elements, of Tn125 provides blaNDM with a strong promoter to drive high-level production of NDM enzymes (Poirel et al., 2011; Toleman et al., 2012).
At the time of writing this paper, there are at least eight additional pNDM-BJ01-like plasmids have been deposited in GenBank. All the above plasmids are exclusively found in Acinetobacter species including A. lwoffii, A. baumannii, A. ereziniae, A. pittii, and an unidentified Acinetobacter species from China, India, and Pakistan. This is the first report of identification of a pNDM-BJ01-like plasmid in Enterobacteriaceae, indicating spread of pNDM-BJ01-like plasmids from Acinetobacter to Enterobacteriaceae.
There is only one preliminary report describing detection of blaNDM in E. aerogenes, and this strain harbors a ~50 Kb blaNDM−1-encoding plasmid and is recovered from the stool sample of a 1-year-old infant with cough and intermittent fever in Hunan Province of China (Ho et al., 2012). This work presents extended evidence that NDM-1 is produced by a conjugative 46.57 Kb plasmid p3SP-NDM, and accounts for carbapenem resistance of clinical E. aerogenes; phenotypic and biochemical experiments combined with plasmid sequencing and comparative genomics analyses give a deeper understanding of antibiotic resistance mechanism of this NDM-1-producing E. aerogenes strain.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work is funded by National Key Program for Infectious Disease of China (2013ZX10004216), National Basic Research Program of China (2014CB744400), Program of Manned Spaceflight (040203), and National Natural Science Foundation of China (81350020, and 81373077).
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fmicb.2015.00294/abstract
Figure S1. PCR detection of bla genes. The major ESBL and carbapenemase genes (Table S1) were screened by PCR with strains 3-SP, 3-SP-NDM-EC600, 3-SP-NDM-DH10B, ATCC BAA-2146 (a NDM-1-producing reference strain of K. pneumoniae, Rasheed et al., 2013), EC600, and DH10B. Of all the bla genes detected, only blaNDM was shown to be present in each of 3-SP, 3-SP-NDM-EC600, and 3-SP-NDM-DH10B.
Figure S2. S1-PFGE/Southern blot. The S1-digested genomic DNA samples was analyzed on an EtBr-stained PFGE gel (A), and then subjected to Southern blot hybridization with a DIG-labeled probe specific to blaNDM−1(B).
Figure S3. Detection of carbapenemase activity. In the presence of any carbapenemase, relevant carbapenems are hydrolyzed and transformed into its carboxylic form, thus leading to a pH decrease which is detected by a color change of phenol red solution (red to yellow–orange). Ambler class A carbapenemases are, at least partially, inhibited by tazobactam, whereas class B carbapenemases (metallo-ß-lactamases) are inhibited by divalent cation chelators such as EDTA. There is no available chemical inhibitor for class D carbapenemases. In this study, the blaNDM-psitive strains 3-SP, 3-SP-NDM-EC600, 3-SP-NDM-DH10B, and ATCC BAA-2146 had class B carbapenemase activity. As expected, E. coli EC600 and DH10B had no carbapenemase activity.
Biendo, M., Canarelli, B., Thomas, D., Rousseau, F., Hamdad, F., Adjide, C., et al. (2008). Successive emergence of extended-spectrum beta-lactamase-producing and carbapenemase-producing Enterobacter aerogenes isolates in a university hospital. J. Clin. Microbiol. 46, 1037–1044. doi: 10.1128/JCM.00197-07
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bonnin, R. A., Poirel, L., Naas, T., Pirs, M., Seme, K., Schrenzel, J., et al. (2012). Dissemination of New Delhi metallo-beta-lactamase-1-producing Acinetobacter baumannii in Europe. Clin. Microbiol. Infect. 18, E362–E365. doi: 10.1111/j.1469-0691.2012.03928.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bornet, C., Chollet, R., Mallea, M., Chevalier, J., Davin-Regli, A., Pages, J. M., et al. (2003). Imipenem and expression of multidrug efflux pump in Enterobacter aerogenes. Biochem. Biophys. Res. Commun. 301, 985–990. doi: 10.1016/S0006-291X(03)00074-3
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chen, L., Chavda, K. D., Melano, R. G., Jacobs, M. R., Koll, B., Hong, T., et al. (2014). Comparative genomic analysis of KPC-encoding pKpQIL-like plasmids and their distribution in New Jersey and New York Hospitals. Antimicrob. Agents Chemother. 58, 2871–2877. doi: 10.1128/AAC.00120-14
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chen, Y. G., Zhang, Y., Yu, Y. S., Qu, T. T., Wei, Z. Q., Shen, P., et al. (2008). In vivo development of carbapenem resistance in clinical isolates of Enterobacter aerogenes producing multiple beta-lactamases. Int. J. Antimicrob. Agents 32, 302–307. doi: 10.1016/j.ijantimicag.2008.02.014
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Davin-Regli, A., Saux, P., Bollet, C., Gouin, F., and De Micco, P. (1996). Investigation of outbreaks of Enterobacter aerogenes colonisation and infection in intensive care units by random amplification of polymorphic DNA. J. Med. Microbiol. 44, 89–98. doi: 10.1099/00222615-44-2-89
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
De Gheldre, Y., Maes, N., Rost, F., De Ryck, R., Clevenbergh, P., Vincent, J. L., et al. (1997). Molecular epidemiology of an outbreak of multidrug-resistant Enterobacter aerogenes infections and in vivo emergence of imipenem resistance. J. Clin. Microbiol. 35, 152–160.
Ding, B., Hu, F., Yang, Y., Guo, Q., Huang, J., and Wang, M. (2014). Four carbapenem-resistant gram-negative species carrying distinct carbapenemases in a single patient. J. Clin. Microbiol. 53, 1031–1033. doi: 10.1128/JCM.03623-14
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dortet, L., Poirel, L., and Nordmann, P. (2012). Rapid identification of carbapenemase types in Enterobacteriaceae and Pseudomonas spp. by using a biochemical test. Antimicrob. Agents Chemother. 56, 6437–6440. doi: 10.1128/AAC.01395-12
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dortet, L., Poirel, L., and Nordmann, P. (2014). Worldwide dissemination of the NDM-type carbapenemases in Gram-negative bacteria. Biomed Res. Int. 2014:249856. doi: 10.1155/2014/249856
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fiett, J., Baraniak, A., Izdebski, R., Sitkiewicz, I., Zabicka, D., Meler, A., et al. (2014). The first NDM metallo-beta-lactamase-producing Enterobacteriaceae isolate in Poland: evolution of IncFII-type plasmids carrying the bla(NDM-1) gene. Antimicrob. Agents Chemother. 58, 1203–1207. doi: 10.1128/AAC.01197-13
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Frank, J. A., Reich, C. I., Sharma, S., Weisbaum, J. S., Wilson, B. A., and Olsen, G. J. (2008). Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl. Environ. Microbiol. 74, 2461–2470. doi: 10.1128/AEM.02272-07
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Georghiou, P. R., Hamill, R. J., Wright, C. E., Versalovic, J., Koeuth, T., Watson, D. A., et al. (1995). Molecular epidemiology of infections due to Enterobacter aerogenes: identification of hospital outbreak-associated strains by molecular techniques. Clin. Infect. Dis. 20, 84–94. doi: 10.1093/clinids/20.1.84
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ho, P. L., Li, Z., Lai, E. L., Chiu, S. S., and Cheng, V. C. (2012). Emergence of NDM-1-producing Enterobacteriaceae in China. J. Antimicrob. Chemother. 67, 1553–1555. doi: 10.1093/jac/dks095
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hu, H., Hu, Y., Pan, Y., Liang, H., Wang, H., Wang, X., et al. (2012). Novel plasmid and its variant harboring both a bla(NDM-1) gene and type IV secretion system in clinical isolates of Acinetobacter lwoffii. Antimicrob. Agents Chemother. 56, 1698–1702. doi: 10.1128/AAC.06199-11
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Jalaluddin, S., Devaster, J. M., Scheen, R., Gerard, M., and Butzler, J. P. (1998). Molecular epidemiological study of nosocomial Enterobacter aerogenes isolates in a Belgian hospital. J. Clin. Microbiol. 36, 1846–1852.
Johnson, A. P., and Woodford, N. (2013). Global spread of antibiotic resistance: the example of New Delhi metallo-beta-lactamase (NDM)-mediated carbapenem resistance. J. Med. Microbiol. 62, 499–513. doi: 10.1099/jmm.0.052555-0
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Jones, L. S., Carvalho, M. J., Toleman, M. A., White, P. L., Connor, T. R., Mushtaq, A., et al. (2014). Characterisation of plasmids in extensively drug-resistant (XDR) Acinetobacter species from India and Pakistan. Antimicrob. Agents Chemother. 59, 923–929. doi: 10.1128/AAC.00117-12
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kuai, S., Shao, H., Huang, L., Pei, H., Lu, Z., Wang, W., et al. (2014). KPC-2 carbapenemase and DHA-1 AmpC determinants carried on the same plasmid in Enterobacter aerogenes. J. Med. Microbiol. 63, 367–370. doi: 10.1099/jmm.0.054627-0
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lavigne, J. P., Sotto, A., Nicolas-Chanoine, M. H., Bouziges, N., Bourg, G., Davin-Regli, A., et al. (2012). Membrane permeability, a pivotal function involved in antibiotic resistance and virulence in Enterobacter aerogenes clinical isolates. Clin. Microbiol. Infect. 18, 539–545. doi: 10.1111/j.1469-0691.2011.03607.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lavigne, J. P., Sotto, A., Nicolas-Chanoine, M. H., Bouziges, N., Pages, J. M., and Davin-Regli, A. (2013). An adaptive response of Enterobacter aerogenes to imipenem: regulation of porin balance in clinical isolates. Int. J. Antimicrob. Agents 41, 130–136. doi: 10.1016/j.ijantimicag.2012.10.010
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Leying, H., Cullmann, W., and Dick, W. (1991). Carbapenem resistance in Enterobacter aerogenes is due to lipopolysaccharide alterations. Chemotherapy 37, 106–113. doi: 10.1159/000238841
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Luo, Y., Yang, J., Ye, L., Guo, L., Zhao, Q., Chen, R., et al. (2014). Characterization of KPC-2-producing Escherichia coli, Citrobacter freundii, Enterobacter cloacae, Enterobacter aerogenes, and Klebsiella oxytoca isolates from a Chinese Hospital. Microb. Drug Resist. 20, 264–269. doi: 10.1089/mdr.2013.0150
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Mataseje, L. F., Boyd, D. A., Lefebvre, B., Bryce, E., Embree, J., Gravel, D., et al. (2014). Complete sequences of a novel blaNDM-1-harbouring plasmid from Providencia rettgeri and an FII-type plasmid from Klebsiella pneumoniae identified in Canada. J. Antimicrob. Chemother. 69, 637–642. doi: 10.1093/jac/dkt445
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Mcgann, P., Hang, J., Clifford, R. J., Yang, Y., Kwak, Y. I., Kuschner, R. A., et al. (2012). Complete sequence of a novel 178-kilobase plasmid carrying bla(NDM-1) in a Providencia stuartii strain isolated in Afghanistan. Antimicrob. Agents Chemother. 56, 1673–1679. doi: 10.1128/AAC.05604-11
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Michael, G. B., Eidam, C., Kadlec, K., Meyer, K., Sweeney, M. T., Murray, R. W., et al. (2012). Increased MICs of gamithromycin and tildipirosin in the presence of the genes erm(42) and msr(E)-mph(E) for bovine Pasteurella multocida and Mannheimia haemolytica. J. Antimicrob. Chemother. 67, 1555–1557. doi: 10.1093/jac/dks076
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Nordmann, P., Boulanger, A. E., and Poirel, L. (2012). NDM-4 metallo-beta-lactamase with increased carbapenemase activity from Escherichia coli. Antimicrob. Agents Chemother. 56, 2184–2186. doi: 10.1128/AAC.05961-11
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Nordmann, P., Poirel, L., Walsh, T. R., and Livermore, D. M. (2011). The emerging NDM carbapenemases. Trends Microbiol. 19, 588–595. doi: 10.1016/j.tim.2011.09.005
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Partridge, S. R., and Iredell, J. R. (2012). Genetic contexts of blaNDM-1. Antimicrob. Agents Chemother. 56, 6065–6067; author reply 6071. doi: 10.1128/AAC.00117-12
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Pfeifer, Y., Wilharm, G., Zander, E., Wichelhaus, T. A., Gottig, S., Hunfeld, K. P., et al. (2011). Molecular characterization of blaNDM-1 in an Acinetobacter baumannii strain isolated in Germany in 2007. J. Antimicrob. Chemother. 66, 1998–2001. doi: 10.1093/jac/dkr256
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Poirel, L., Bonnin, R. A., Boulanger, A., Schrenzel, J., Kaase, M., and Nordmann, P. (2012). Tn125-related acquisition of blaNDM-like genes in Acinetobacter baumannii. Antimicrob. Agents Chemother. 56, 1087–1089. doi: 10.1128/AAC.05620-11
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Poirel, L., Dortet, L., Bernabeu, S., and Nordmann, P. (2011). Genetic Features of blaNDM-1-Positive Enterobacteriaceae. Antimicrob. Agents Chemother. 55, 5403–5407. doi: 10.1128/AAC.00585-11
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Qin, X., Yang, Y., Hu, F., and Zhu, D. (2014). Hospital clonal dissemination of Enterobacter aerogenes producing carbapenemase KPC-2 in a Chinese teaching hospital. J. Med. Microbiol. 63, 222–228. doi: 10.1099/jmm.0.064865-0
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Rasheed, J. K., Kitchel, B., Zhu, W., Anderson, K. F., Clark, N. C., Ferraro, M. J., et al. (2013). New Delhi metallo-beta-lactamase-producing Enterobacteriaceae, United States. Emerging Infect. Dis. 19, 870–878. doi: 10.3201/eid1906.121515
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ronveaux, O., Gheldre, Y., Glupczynski, Y., Struelens, M., and Mol, P. (1999). Emergence of Enterobacter aerogenes as a major antibiotic-resistant nosocomial pathogen in Belgian hospitals. Clin. Microbiol. Infect. 5, 622–627. doi: 10.1111/j.1469-0691.1999.tb00419.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sekizuka, T., Matsui, M., Yamane, K., Takeuchi, F., Ohnishi, M., Hishinuma, A., et al. (2011). Complete sequencing of the bla(NDM-1)-positive IncA/C plasmid from Escherichia coli ST38 isolate suggests a possible origin from plant pathogens. PLoS ONE 6:e25334. doi: 10.1371/journal.pone.0025334
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Souli, M., Kontopidou, F. V., Papadomichelakis, E., Galani, I., Armaganidis, A., and Giamarellou, H. (2008). Clinical experience of serious infections caused by Enterobacteriaceae producing VIM-1 metallo-beta-lactamase in a Greek University Hospital. Clin. Infect. Dis. 46, 847–854. doi: 10.1086/528719
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tada, T., Miyoshi-Akiyama, T., Dahal, R. K., Sah, M. K., Ohara, H., Kirikae, T., et al. (2013). NDM-8 metallo-beta-lactamase in a multidrug-resistant Escherichia coli strain isolated in Nepal. Antimicrob. Agents Chemother. 57, 2394–2396. doi: 10.1128/AAC.02553-12
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tiwari, V., and Moganty, R. R. (2013). Structural studies on New Delhi Metallo-beta-lactamase (NDM-2) suggest old beta-lactam, penicillin to be better antibiotic for NDM-2-harbouring Acinetobacter baumanni. J. Biomol. Struct. Dyn. 31, 591–601. doi: 10.1080/07391102.2012.706075
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Toleman, M. A., Spencer, J., Jones, L., and Walsh, T. R. (2012). blaNDM-1 is a chimera likely constructed in Acinetobacter baumannii. Antimicrob. Agents Chemother. 56, 2773–2776. doi: 10.1128/AAC.06297-11
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhang, W. J., Lu, Z., Schwarz, S., Zhang, R. M., Wang, X. M., Si, W., et al. (2013). Complete sequence of the bla(NDM-1)-carrying plasmid pNDM-AB from Acinetobacter baumannii of food animal origin. J. Antimicrob. Chemother. 68, 1681–1682. doi: 10.1093/jac/dkt066
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zong, Z., and Zhang, X. (2013). blaNDM-1-carrying Acinetobacter johnsonii detected in hospital sewage. J. Antimicrob. Chemother. 68, 1007–1010. doi: 10.1093/jac/dks505
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Keywords: Enterobacter aerogenes, NDM-1, Plasmid, p3SP-NDM
Citation: Chen Z, Li H, Feng J, Li Y, Chen X, Guo X, Chen W, Wang L, Lin L, Yang H, Yang W, Wang J, Zhou D, Liu C and Yin Z (2015) NDM-1 encoded by a pNDM-BJ01-like plasmid p3SP-NDM in clinical Enterobacter aerogenes. Front. Microbiol. 6:294. doi: 10.3389/fmicb.2015.00294
Received: 28 January 2015; Accepted: 25 March 2015;
Published: 14 April 2015.
Edited by:
Ghassan M. Matar, American University of Beirut, LebanonReviewed by:
Benjamin Andrew Evans, University of Edinburgh, UKCopyright © 2015 Chen, Li, Feng, Li, Chen, Guo, Chen, Wang, Lin, Yang, Yang, Wang, Zhou, Liu and Yin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dongsheng Zhou and Zhe Yin, State Key Laboratory of Pathogen and Biosecurity, Beijing Institute of Microbiology and Epidemiology, No. 20, Dongdajie, Fengtai, Beijing 100853, ChinaZG9uZ3NoZW5nemhvdTE5NzdAZ21haWwuY29t;amVycnk5eWluQDE2My5jb20=;
Changting Liu, Nanlou Respiratory Diseases Department, Chinese People's Liberation Army General Hospital, No. 28, Fuxing Road, Haidian, Beijing 100071, ChinabGl1Y2hhbmd0QGdtYWlsLmNvbQ==
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.