
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 09 April 2015
Sec. Food Microbiology
Volume 6 - 2015 | https://doi.org/10.3389/fmicb.2015.00270
This article is part of the Research Topic Ecology, virulence and detection of pathogenic and pandemic Vibrio parahaemolyticus View all 13 articles
 Oscar Escalante-Maldonado1
Oscar Escalante-Maldonado1 Ahmad Y. Kayali1
Ahmad Y. Kayali1 Wataru Yamazaki2
Wataru Yamazaki2 Varaporn Vuddhakul3
Varaporn Vuddhakul3 Yoshitsugu Nakaguchi4†
Yoshitsugu Nakaguchi4† Mitsuaki Nishibuchi4*
Mitsuaki Nishibuchi4*
Vibrio parahaemolyticus is a marine microorganism that can cause seafood-borne gastroenteritis in humans. The infection can be spread and has become a pandemic through the international trade of contaminated seafood. Strains carrying the tdh gene encoding the thermostable direct hemolysin (TDH) and/or the trh gene encoding the TDH-related hemolysin (TRH) are considered to be pathogenic with the former gene being the most frequently found in clinical strains. However, their distribution frequency in environmental isolates is below 1%. Thus, very sensitive methods are required for detection and quantitation of tdh+ strains in seafood. We previously reported a method to detect and quantify tdh+ V. parahaemolyticus in seafood. This method consists of three components: the most-probable-number (MPN), the immunomagnetic separation (IMS) targeting all established K antigens, and the loop-mediated isothermal amplification (LAMP) targeting the tdh gene. However, this method faces regional issues in tropical zones of the world. Technicians have difficulties in securing dependable reagents in high-temperature climates where we found MPN underestimation in samples having tdh+ strains as well as other microorganisms present at high concentrations. In the present study, we solved the underestimation problem associated with the salt polymyxin broth enrichment for the MPN component and with the immunomagnetic bead-target association for the IMS component. We also improved the supply and maintenance of the dependable reagents by introducing a dried reagent system to the LAMP component. The modified method is specific, sensitive, quick and easy and applicable regardless of the concentrations of tdh+ V. parahaemolyticus. Therefore, we conclude this modified method is useful in world tropical, sub-tropical, and temperate zones.
Vibrio parahaemolyticus inhabits estuarine and marine environments (Joseph et al., 1982). This bacterium thrives in high-temperature environments and thus it is prevalent in tropical areas year around and is found at lower concentrations only in summer in temperate regions.
Vibrio parahaemolyticus is the major cause of seafood-borne infections in the world (Raghunath, 2015). This bacterium can cause gastroenteritis in humans only when it propagates in the harvested seafood to the number exceeding the infectious dose when consumed by humans without proper cooking (Okuda et al., 1997a). A large number of V. parahaemolyticus cells distributed in the eutrophic coastal environments may accumulate in the digestive tract of molluscan bivalves because they filter-feed (Nishibuchi and Kaper, 1995; Manas et al., 2014). Therefore, molluscan shellfish accumulates V. parahaemolyticus at high concentrations and very frequently cause infection. However, not all strains are considered pathogenic. Only those possessing and expressing the gene (tdh) encoding the thermostable direct hemolysin (TDH) and/or the gene (trh) encoding the TDH-related hemolysin (TRH) are considered pathogenic (Nishibuchi and Kaper, 1985). Most clinical isolates of V. parahaemolyticus carry the tdh and trh genes, either alone or in combination; however, distribution of these genes in environmental isolates is usually low (1–2%; Nishibuchi and Kaper, 1995; Yamazaki et al., 2010) although some workers reported extremely frequent detection (up to 48%; Rodriguez-Castro et al., 2010; Gutierrez West Casandra et al., 2013).
The concentration of pathogenic V. parahaemolyticus can exceed a detectable level all the year around in the tropical zone but only exists at lower levels in the summer season in temperate zones. To not under-report pathogenic V. parahaemolyticus, sensitive methods have been devised for shellfish examination. These include the use of alkaline peptone water (APW) and salt polymyxin broth (SPB) as selective media in an enrichment procedure (Hara-Kudo et al., 2003); the immunomagnetic separation (IMS) technique targeting the K6 antigen shown to be useful in selective isolation of the O3:K6 pandemic strain of V. parahaemolyticus (Okuda et al., 1997b; Vuddhakul et al., 2000); and loop-mediated isothermal amplification (LAMP) reported to be more sensitive than conventional PCR (Yamazaki et al., 2010). Based on these reports, our group recently developed an most-probable-number (MPN) procedure for enumeration of tdh+ V. parahaemolyticus in shellfish samples where a PickPen-assisted IMS technique (hereinafter abbreviated simply as IMS) targeting as many as 69 established K antigens and a LAMP assay targeting the tdh gene were incorporated (Tanaka et al., 2014; hereinafter referred to as MPN-IMS-LAMP).
Experiments in southern Thailand show the MPN-IMS-LAMP performed well in general in detection and quantitation of tdh+ V. parahaemolyticus in shellfish products (Tanaka et al., 2014). However, there was a problem of underestimating MPN values because the study was conducted in a tropical environment where the total microbial population including target and non-target organisms is generally large; and, in such an environment, it is difficult to properly detect tdh+ V. parahaemolyticus (Vuddhakul et al., 2006). Overgrowth of non-target organism(s) and a failure in the IMS and the SPB enrichment are most likely responsible for the underestimation problem.
PickPen, an eight-channel intrasolution magnetic particle separation device enables a straight forward 96-well plate-based IMS procedure was successfully applied to increase the sensitivity and specificity of Escherichia coli O157:H7 detection in food (Nou et al., 2006). The IMS consists of two steps: the incubation of immunomagnetic beads (IMBs) with enriched culture containing the target bacterium and others (hereinafter referred to as IMB Incubation); and the washing step using the PickPen (hereinafter referred to as PickPen Operation) to remove non-targeted microbial population. The E. coli O157:H7 study demonstrated that constant shaking during the IMB Incubation could increase the efficiency of IMS (Nou et al., 2006). Our previous method employed incubation with intermittent mixing but not constant shaking (Tanaka et al., 2014). We also noticed loss of IMB during PickPen Operation suggesting improvement of this step in the IMS component. In this study we adopted the IMB Incubation with constant shaking and examined whether increased PickPen Operation time could further improve the efficiency of IMS.
Loop-mediated isothermal amplification allows one-step detection of gene amplification at a single temperature (Notomi et al., 2000) and it has been reported that LAMP is more simple and sensitive than the currently popular conventional PCR methods targeting the tdh and trh genes (Yamazaki et al., 2010). The conventional liquid LAMP reagent is practically inconvenient because refrigerated environment is recommended during storage, transportation, and operation; whereas the dried form does not require a refrigerated environment and therefore it can be used to detect pathogenic microorganisms even in tropical countries (Boehme et al., 2007; Mitarai et al., 2011). If the dried LAMP reagent is applicable to detection of the pathogenic genes of V. parahaemolyticus, it would be very useful for examination of seafood in various parts of the world.
In the present study, we attempted to improve the previously reported MPN-IMS-LAMP for quantitation of tdh+ V. parahaemolyticus. We evaluated the improvement by applying a series of modifications in the three components of the protocol: MPN, IMS, and LAMP. First, we evaluated two important factors (IMB incubation and PickPen Operation time) in IMS and we determined the benefits of the dried LAMP reagents for seafood analysis. Then, we compared the MPN values at three steps (after APW incubation, after IMS application, and after SPB enrichment). Finally, based on the results we present a new recommended MPN-IMS-LAMP method where the high sensitivity, MPN accuracy, and shellfish analysis applicability was determined.
Immunomagnetic bead was prepared as previously described (Tanaka et al., 2014). In brief, magnetic beads were coated with antibodies partially purified with ammonium sulfate precipitation from polyvalent K antisera groups I–IX and monovalent anti-K70 to -K75 antibodies using commercially available V. parahaemolyticus K antisera from Denka Seiken Co., Ltd., Tokyo, Japan.
The shellfish sample was shucked and homogenized in a plastic bag. A three-tubes MPN dilution series was prepared as described in the U.S. Food and Drug Administration’s Bacteriological Analytical Manual (DePaola and Kaysner, 2004) with slight modifications (shown schematically in Figure 1).Briefly, a 25 g portion of the homogenate was added to 225 ml of APW (Nissui Pharmaceutical Co., Ltd., Tokyo, Japan). For determination of the MPN, the shellfish homogenate was diluted 10-fold as follows. Ten-ml of shellfish homogenate in APW was transferred to an empty tube, and subsequent 10-fold dilutions were prepared by transferring 1 ml aliquot to the tube containing 9 ml of APW in triplicate. The tubes were incubated at 37°C for 6 h. The modified IMS was applied as described below (IMS subsection). The 1 ml suspension in PBS resulting from IMS was used for DNA template preparation followed by LAMP assay using dried reagent as described below. Finally, MPN values were determined.
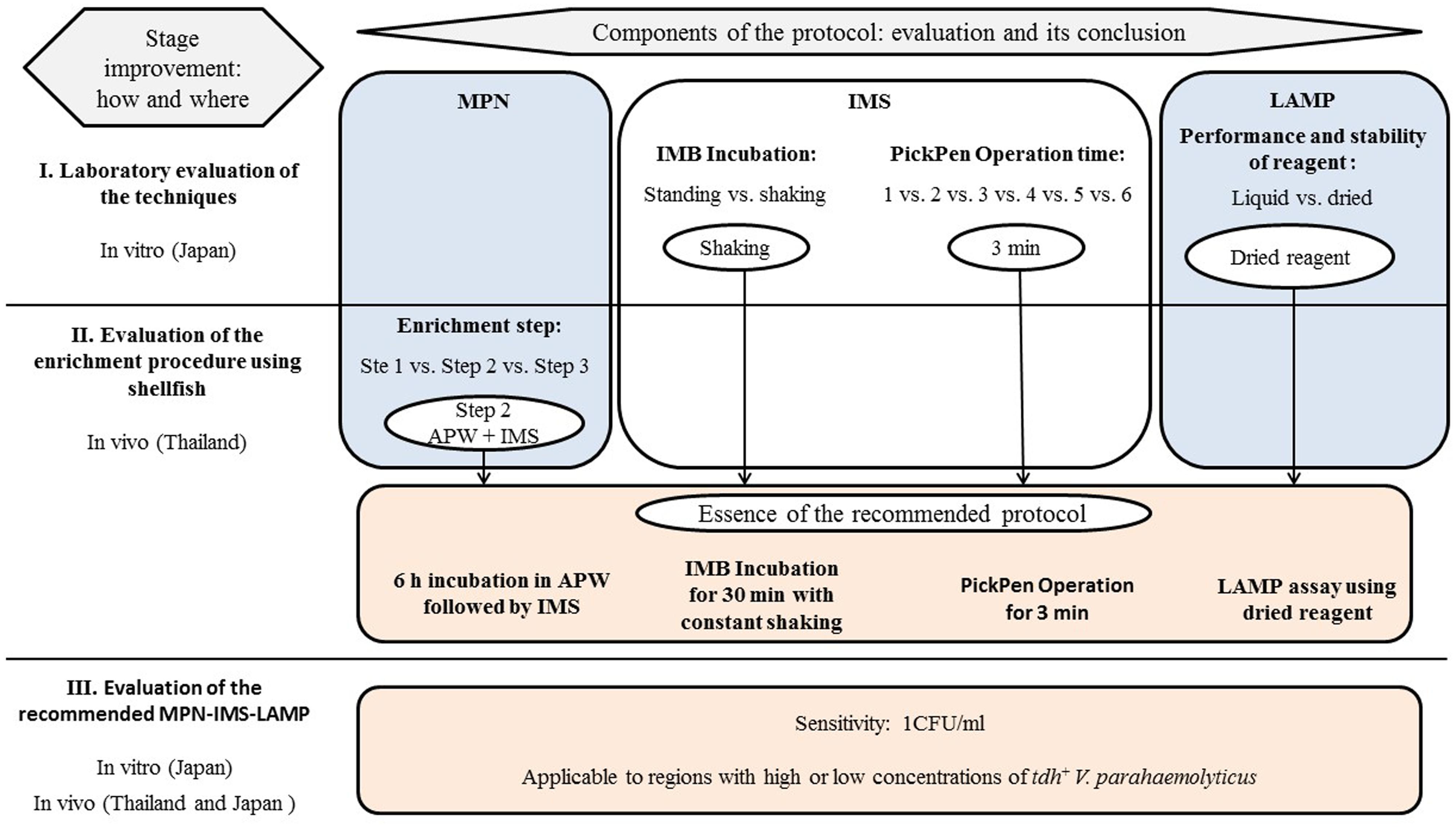
FIGURE 1. MPN-IMS-LAMP protocol for quantitation of tdh+ V. parahaemolyticus recommended in this study and the scheme of the comparative experiments used in this study.
Immunomagnetic separation was performed as previously described (Nou et al., 2006; Tanaka et al., 2014) with modifications. Briefly, 1-ml aliquots of each culture were transferred to a well in a 96-well (2-ml capacity) titer plate. The cultures were mixed with 25 μl of the IMB were incubated on a shaker (140 rpm) at room temperature for 30 min. The subsequent bead washing and bead suspension steps were performed without changing the tips. The beads were captured with PickPen (BioControl Systems, Bellevue, WA, USA) by gently stirring the cultures with an up-and-down motion for 3 min. The captured beads were then washed twice by releasing into and recapturing from 1 ml of phosphate buffered saline (SIGMA-Aldrich Co., St. Louis, MO, USA) and were finally suspended in 1 ml of the same buffer.
A 1-ml aliquot of a test culture was centrifuged at 15,000 rpm for 1 min, and the supernatant was discarded. The pellet was washed and suspended in 1 ml 0.85% NaCl solution, heated at 100°C for 10 min, and immediately cooled on ice for 5 min. After centrifugation at 15,000 rpm for 5 min, the supernatant was transferred to a new tube and was stored at -20°C until used.
The microtubes containing the dried reagent were taken from a commercially available kit for detection of Influenza virus (Loopamp Type A Influenza detection kit, Eiken Chemical Co., Ltd., Tokyo, Japan). The microtubes were transported, stored, and rehydrated at room temperature. Five μl of DNA template solution, 1.3 μl of tdh LAMP primer set (Yamazaki et al., 2010) and 23.7 μl of distilled water were added to make a final volume of 30 μl per reaction were added to the microtube. The cap was tightly secured and the microtube was inverted for 3 min in order to rehydrate the reagent which is located in the cap of the tube. The tube was heated at 65°C for 1 h (reaction) and at 80°C for 5 min (enzyme inactivation). Results of the reaction was judged using the visual system by the color change from brown to green in the reaction solution.
Vibrio parahaemolyticus O3:K6 strain VP81 originally isolated from a fecal sample of a patient with diarrhea in India (Matsumoto et al., 2000) was used in this study. The strain was grown in 5 ml Luria broth (SIGMA-Aldrich Co.) at 37°C with shaking at 160 rpm for 18 h. Serial 10-fold dilutions of the culture were made in sterile saline. Based on our preliminary experiment (data not shown), to best evaluate the effect of the PickPen Operation time, a concentration of 103 CFU/ml was used for this experiment. Six sets of 25 μl of IMB mixed with the diluted culture were prepared and incubated for 30 min with shaking as explained above. The IMB in each set was then washed with different PickPen Operation times (1, 2, 3, 4, 5, or 6 min). After the IMB capture, the remaining supernatant as well as the diluted culture without IMS treatment were plated onto thiosulfate citrate bile salt sucrose agar (TCBS, Eiken Chemical Co.) and incubated at 37°C for 24 h and colonies were counted. The capture efficiency (CE) was calculated using the following equation: CE (%) = (Co-Ca)/Co x 100%, where Co is the total CFU/ml present in the sample, and Ca is the CFU/ml not bound to IMB (Zeng et al., 2014).
To compare the performance between liquid reagent (DNA amplification kit, Eiken Chemical Co.) and the dried reagent (contained in the microtubes provided in the Loopamp Type A Influenza detection kit), the DNA templates prepared from 14 V. parahaemolyticus reference strains were used. LAMP reaction with liquid reagent was performed according to the manufacture’s instruction. LAMP reaction with dried reagent is described above. The LAMP primer sets for the detection of tdh, trh1 and trh2 genes were used to conduct four LAMP assays targeting the tdh, trh1, trh2 and tdh plus trh2 genes as previously reported (Yamazaki et al., 2010). We judged the results using a turbidimetric system 1 h after the beginning of the reaction using the Loopamp EXIA LA-320A (Eiken Chemical Co.). Results were judged using the visual system after 1 h by a change in the color of the reaction solution. Finally, to assess the utility of the dried LAMP reagent in tropical countries, we tested its stability at high-temperatures. The reagents were exposed to temperatures of 30, 40, 50, and 60°C for 15 days; and after, a tdh LAMP assay using a standard tdh+ V. parahaemolyticus strain was performed (Yamazaki et al., 2010).
Twenty eight samples of shellfish harvested in Thailand consisting of 12 bloody clams (Anadara granosa), 12 hard clams (Meretrix lusoria), and four green mussels (Perna viridis) were purchased at a local morning market in Hat Yai, Thailand, during April and May 2014. The shellfish samples were transported to the Prince of Songkla University, Hat Yai, Thailand at room temperature (∼30°C) and were processed within 1 h of purchase. Sixteen samples of shellfish harvested in Japan consisting of 12 Japanese littlenecks (Ruditapes philippinarum) and four hard clams were obtained at Osaka Municipal Wholesale Market, Osaka, Japan during August and September 2014. These shellfish samples were transported to the laboratory at Kyoto University at room temperature (∼25°C) and processed within 2 h of purchase.
A minor modification was employed in the experiments in southern Thailand for comparison. Samples were processed as described above after up to 6 h incubation in APW at 37°C (Figure 1, Step 1). After IMS application (Figure 1, Step 2), 500 μl of 1-ml bead suspension was inoculated into 4 ml of SPB (Nissui Pharmaceutical Co., Ltd.) and was incubated at 37°C for 18 h (Figure 1, Step 3). One-milliliter aliquots from each culture tube at Steps 1 and 3 and 500 μl of the bead suspension at Step 2 were used for DNA template preparation. LAMP using the liquid reagent was employed in these experiments.
Two-hundred-fifty ml of the Japanese littleneck sample suspension in APW was prepared by inoculation of tdh+ V. parahaemolyticus VP81 in the mid log phase at known concentrations (0.1, 1.0, 10.0 CFU/ml). These VP81 suspensions were examined for MPN of tdh+ V. parahaemolyticus using the protocol recommended in this study.
All experiments were performed in triplicate. The means and standard deviations of all collected data were calculated for every triplicate group. A student’s t-test was used for statistical analysis between two groups. A p-value ≤ 0.05 was considered statistically significant.
To improve the MPN-IMS-LAMP previously reported (Tanaka et al., 2014), we evaluated the components of the protocol in two stages to propose the recommended procedure for quantitation of tdh+ V. parahaemolyticus and evaluated the recommended procedure as schematically shown in Figure 2.
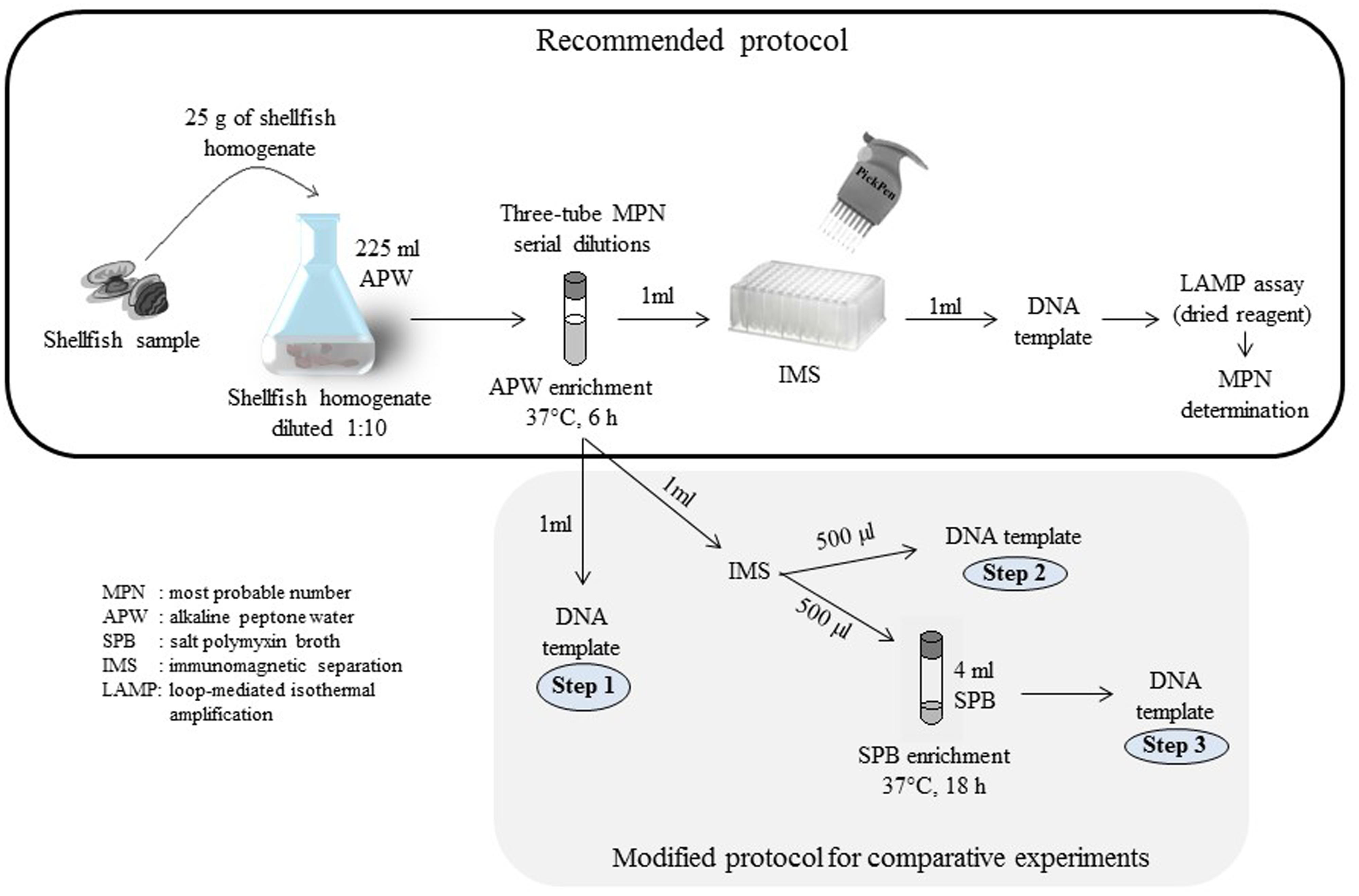
FIGURE 2. Schematic representation of the strategy used to improve the method for quantitation of tdh+ Vibrio parahaemolyticus.
As explained in the introduction, we noticed loss of IMB during PickPen Operation suggesting improvement of this step. Therefore, we evaluated whether increasing the time during the washing step of IMB helped to minimize the loss of IMB and thus improve the CE of IMB. We used the student’s t-test to compare the different PickPen Operation times (1–6 min) and to evaluate the significance of increasing time (Figure 3). The CE value gradually increased with increase in the PickPen Operation time until it reached 5 min; in particular, the significant increase in CE was apparent after an PickPen Operation time of 3 min.
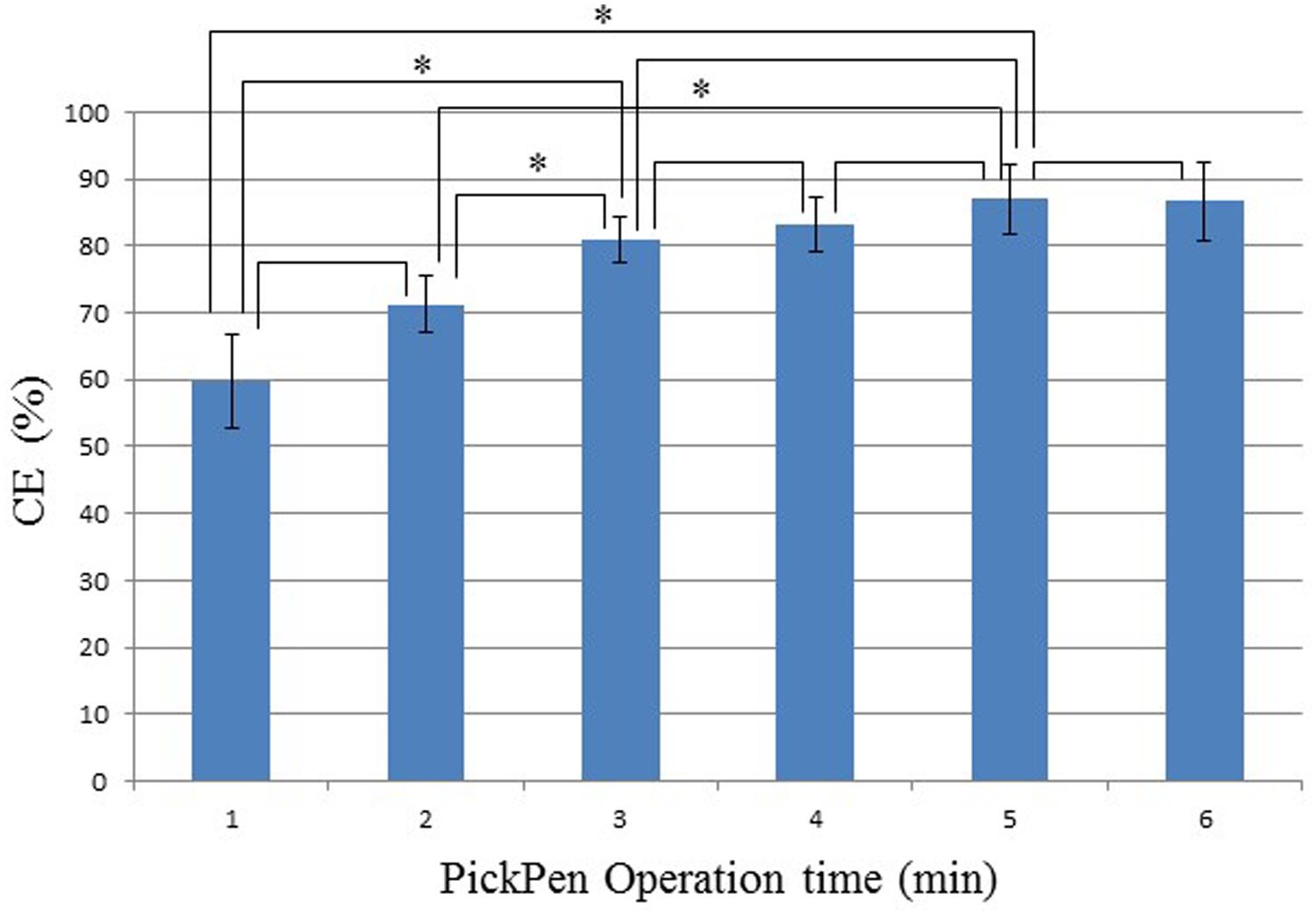
FIGURE 3. Effect of PickPen Operation time on the capture efficiency (CE) of V. parahaemolyticus. CE (%) = (Co-Ca)/Co x 100%, where Co is the total CFU/ml present in the sample, and Ca is the CFU/ml not bound to IMB (Zeng et al., 2014). The pairs of the samples varying in PickPen Operation time that were compared for the difference in CE (%) using the Student’s t-test are indicated; *pairs showing a significant difference (p= 0.05).
To evaluate whether easy-to-use dried reagent can replace the standard liquid reagent we compared the two methods using 14 reference strains. The method using the liquid reagent or using dried reagent were equally sensitive and specific when the results were judged using the turbidimetric system (Table 1). In addition, when the product of the reaction was judged by eye, the results using the dried reagent were clearer, due to the color change of the rehydrated reagent from brown to green, than that using the liquid reagent (Figure 4A). Furthermore, it was more difficult to judge by eye the results of trh (trh1 and trh2) detection than tdh detection using the liquid reagent. Conversely, judgment of color by eye of the results for all these genes was equally easy when the dried reagent was used (data not shown). Further, the positive results in the tdh LAMP assays obtained after the dried LAMP reagent was exposed to different temperatures (30, 40, 50, and 60°C), showed the dried reagent was equally stable at all of these temperatures during the examination period (15 days).

TABLE 1. Comparison of liquid reagent and dried reagent for detection of the tdh, trh1 and trh2 genes in Vibrio parahaemolyticus using the turbidimetric system.
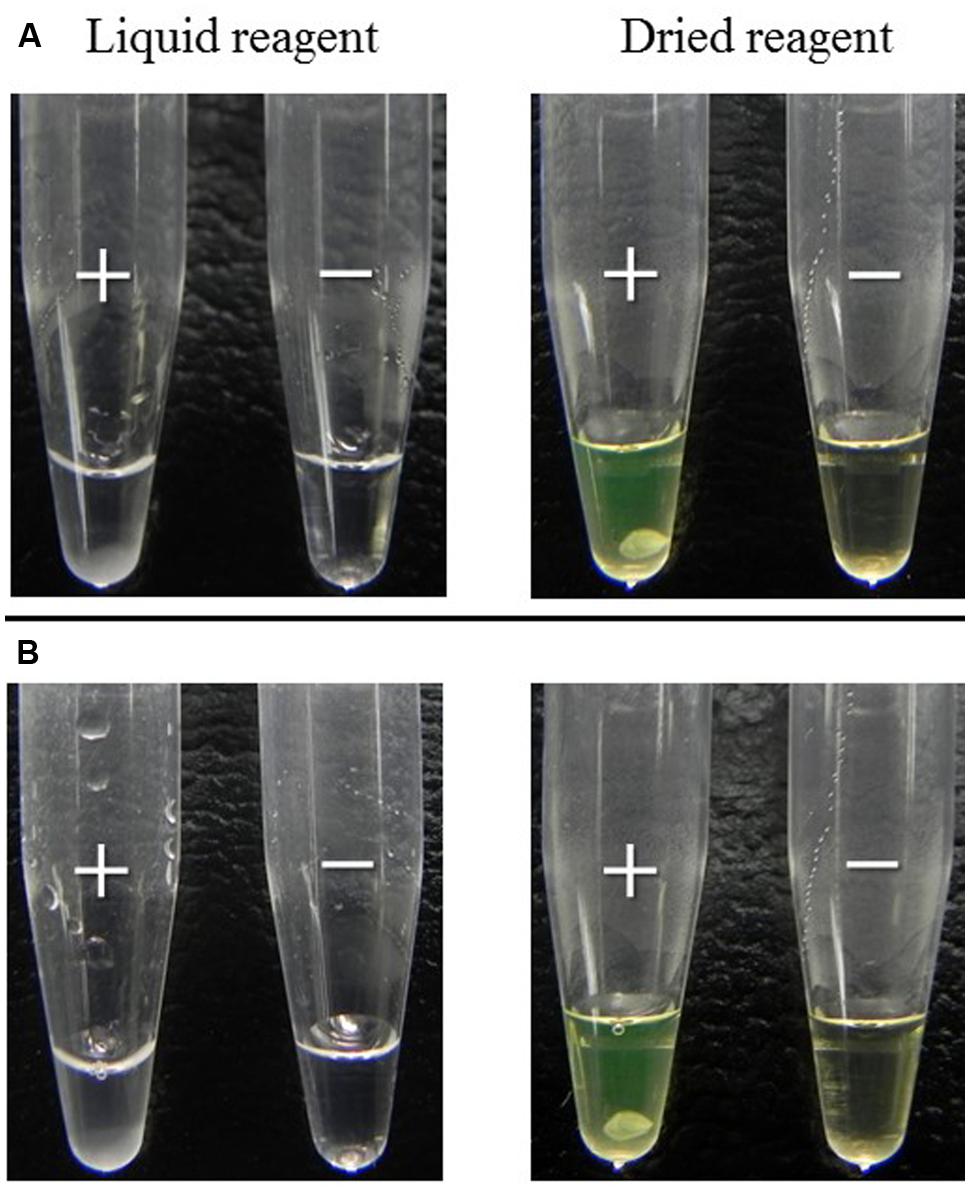
FIGURE 4. Comparison of the LAMP results between liquid and dried reagents. DNA template obtained using the boiling method from: (A) the pure culture of control strains examined for LAMP component at Stage I (Figure 2); (B) retail Thai shellfish samples examined at Stage III (Figure 2) by the recommended MPN-IMS-LAMP protocol (Figure 1). Positive (+) and negative (-) LAMP reaction directly judged by eye.
We explored the possibility of finding a factor affecting the protocol in the enrichment step. We compared the MPN values after each of the three treatments (Figure 1, Steps 1–3) in the new protocol including the above modifications. Twenty-eight shellfish samples purchased in southern Thailand were examined for the MPN of tdh+ V. parahaemolyticus as described above (Table 2). The data clearly shows tdh+ V. parahaemolyticus is prevalent and the concentration is very high in these samples, with the log MPN/10 g value ranging from 1.0 to 5.4. Hard clams gave higher log MPN than the other two shellfish. Regardless of the kind of the shellfish, the values in Step 3 were lower than those of the other two steps. When the MPN values of Steps 1 and 2 are compared, three samples (Table 2, sample no. 8–10) were higher in Step 1, two samples (Table 2, sample no. 1 and 2) were higher for Step 2 and 23 samples were the same. The average MPN values for all samples between Steps 1 and 2 did not differ significantly.
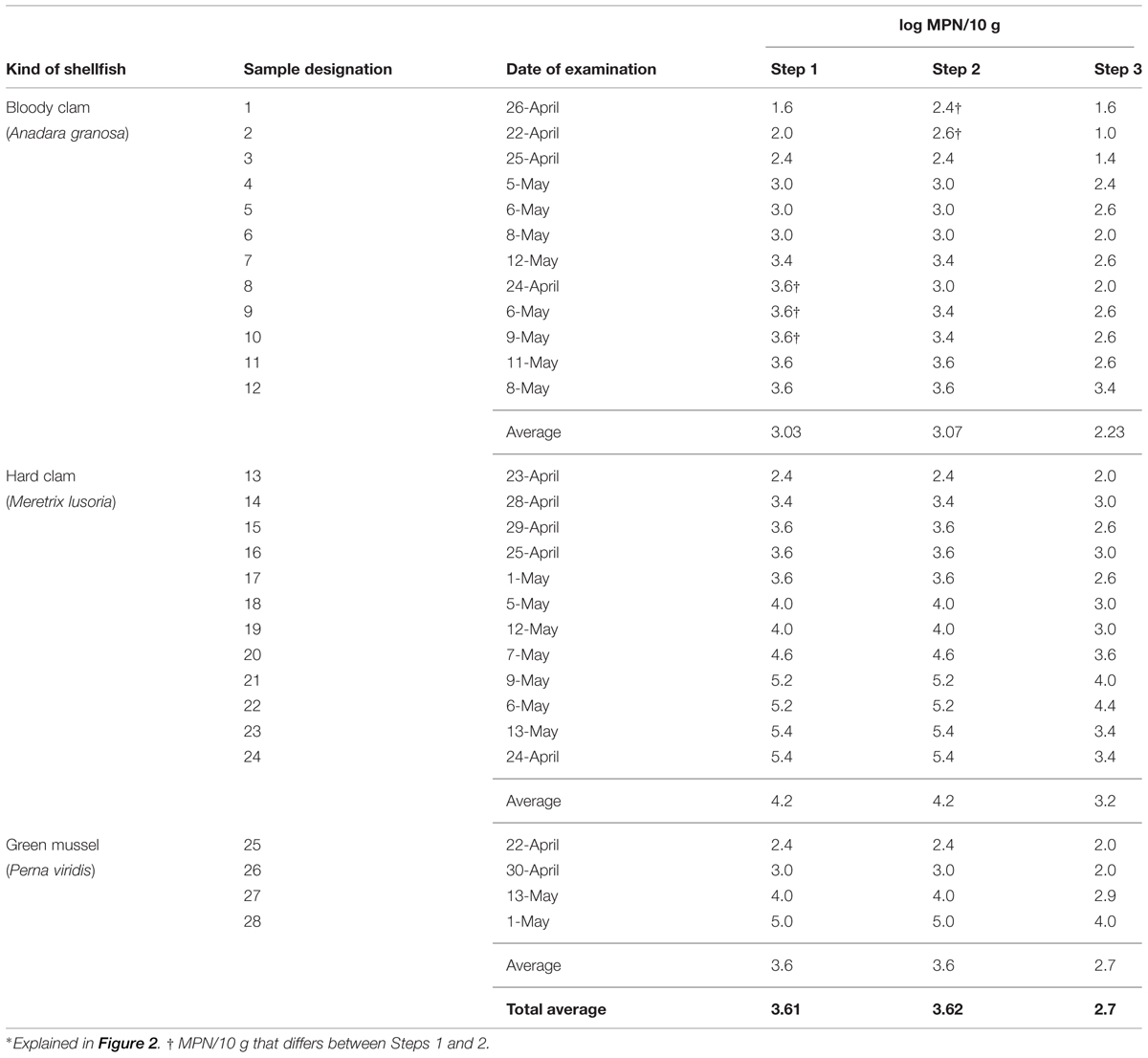
TABLE 2. Comparison of the MPN values of tdh+ V. parahaemolyticus obtained at the three Steps of the modified protocol∗.
Shellfish homogenates artificially contaminated with a reference strain (tdh+) at known concentrations were examined by the recommended protocol. Positive tubes in the three-tubes MPN format were 0-0-0, 3-0-0, and 3-3-0, for the homogenates contaminated at 0.1, 1.0 and 10.0 CFU/ml, respectively. The sensitivity of the recommended MPN-IMS-LAMP was therefore determined to be 1 CFU/ml. Based on the 3-tubes MPN format results, the MPN values obtained were <0.03 (range: <0.005–0.09) MPN/ml, 0.23 (range: 0.04–1.2) MPN/ml, and 2.4 (range: 0.36–13.0) MPN/ml, respectively.
Twenty-eight Thai samples were examined using liquid LAMP reagent in Stage II. Three (Table 2, sample designations 4, 21, and 25) of the 28 samples representing each shellfish group were also examined using dried LAMP reagent as in the new recommended protocol. The MPN values obtained at this stage were exactly the same, indicating the dried reagent had the same sensitivity as the liquid reagent as judged by the turbidimetric system. However, we confirmed that judgment by eye is easier when dried reagent is applied (Figure 4B).
Sixteen Japanese shellfish samples were also examined by the recommended new protocol (Table 3). The tdh gene was not detected in 14 of the 16 samples, with the MPN values being below the detection limit (<3 MPN/10 g). Two of the samples were positive for the tdh gene but the MPN values (15 and 23 MPN/10 g) were much lower than those obtained with Thai samples.
Our international research group attempted to develop an easy and sensitive method for quantitative detection of tdh+ V. parahaemolyticus in molluscan shellfish that can be applied in any part of the world; being practical and feasible even in resources-limited and/or tropical countries. We previously reported an MPN-IMS-LAMP method that was shown to be more sensitive than the MPN-PCR-based method (Hara-Kudo et al., 2003; Jones et al., 2012; Tanaka et al., 2014). However, a problem of under-estimation of the MPN values for some samples in a tropical environment was found as well as the technical practical inconvenience of the liquid LAMP reagent. In the current study, we improved the previously reported method using a series of technical modifications in the MPN and IMS components (Figure 2). In addition, we replaced the liquid reagent by the dried reagent in the LAMP component. As a result, much of the technical problems were solved (Tables 2 and 3; Figure 4).
While our study was in progress, a LAMP (targeting the tlh gene) and IMS (using nanoparticules targeting flagella) detection method for V. parahaemolyticus was reported (Zeng et al., 2014). However, CODEX 2011 recommends quantitative detection of pathogenic rather than total V. parahaemolyticus for risk assessment of V. parahaemolyticus in seafood (Food and Agriculture Organization World Health Organization of the United Nations [FAO/WHO], 2011). Along this line, our IMS method screens for clinically important V. parahaemolyticus by targeting all established K antigens and our LAMP method targets the tdh gene. Though, primer sets for trh1 and trh2 are available, variation in the trh gene sequence is widely observed in environmental strains is of major concern (Kishishita et al., 1992). A new trh LAMP primer set to overcome this issue is currently under development (Escalante-Maldonado and Nishibuchi, unpublished data).
The IMS component is an essential part of our protocol, therefore we needed to improve the efficiency of the IMS performance. In vitro experiments in Stage I confirmed that IMB incubation with constant shaking (Nou et al., 2006) for 30 min (Tanaka et al., 2014) enhances IMB-target association (data not shown). Figure 3 shows that increasing the PickPen Operation time up to 3 min (1–3 and 2–3 min) increased CE value significantly. Increase in operation time after 3 min showed no significance difference in CE value where further extension of PickPen Operation time increases the workload. Taken together, we judged 3 min is the most effective PickPen Operation time. In vivo experiments conducted in Thailand employing the new protocol confirmed that these two modifications are a valuable contribution to the IMS performance (Table 2). The MPN values obtained after IMS application in this study are higher than the under-estimated MPN values reported in our previous study where similar shellfish samples were examined (Tanaka et al., 2014).
Another very valuable contribution to our recommended protocol is the application of the dried LAMP reagent. We utilized the dried reagent included in the commercially available kit for detection of two other pathogens. Results of the comparative experiments in Stage I indicate that both dried and liquid reagents are equally sensitive and specific for the LAMP assays for V. parahaemolyticus regardless of the primer set (Table 1). Evaluation of the stability of the dried reagent at different temperatures (30–60°C) confirmed the dried reagent is very stable even at high-temperatures during the examination period, corroborating its utility even in tropical coastal areas of the world. Likewise, the dried reagent was proven useful because its transportation and storage do not necessarily require refrigerated or frozen conditions as confirmed during experiments in southern Thailand in Stage III.
Prevalence of infection by tdh+ V. parahaemolyticus was reported previously in southern Thailand. This prevalence is due to consumption of under-cooked molluscan shellfish, which is very common in southern Thailand (Laohaprertthisan et al., 2003). A risk assessment study conducted in southern Thailand reported low concentration of tdh+ V. parahaemolyticus in the bloody clam sold in the evening market (Yamamoto et al., 2008). However, pre-incubation of shellfish samples prior to examination may allow growth of tdh+ V. parahaemolyticus to a detectable level (Hara-Kudo et al., 2003; Yamamoto et al., 2008). Our previous study indicated that molluscan shellfish kept overnight and sold at the morning market contained relatively high levels of tdh+ V. parahaemolyticus (Tanaka et al., 2014). Under this condition, the under-estimation problem of the MPN values was raised. The current study examining similar shellfish samples solved the under-estimation problem. Comparative experiments in Stage II showed the problem is due at least in-part to the addition of the SPB enrichment step (Figure 1, Step 3). Among the MPN values observed at the three different steps, those of Step 3 were lower for all seafood samples examined (Table 2). SPB enrichment probably supported preferentially the growth of competing bacterial population rather than that of tdh+ V. parahaemolyticus.
The method yielding the highest MPN values is presumed to be most sensitive. We therefore compared the values obtained at Steps 1 and 2. The averages of the MPN values for these two steps were indistinguishable (Table 2). Difference in MPN values between Step 1 and Step 2 were observed only with five of 12 bloody clam samples. Two samples require special attention. They showed lower MPN values at Step 1 which were close to the detection limit. Application of IMS could avoid under-estimation of these values and assure the detection of the target.
Modifications in the three components of the protocol (Figure 2) were critical to improve the MPN-IMS-LAMP. The under-estimation problem was solved by modifying the IMS and excluding the SPB from the enrichment step. Introduction of the dried LAMP reagent made the method quicker, easier and allows its use at high environmental temperatures. We therefore recommend the modified MPN-IMS-LAMP for detection and quantitation of tdh+ V. parahaemolyticus as a universal method useful in tropical, subtropical, and temperate zones of the world.
Conceived and designed the experiments: OE-M and MN. Performed the experiments and analyzed data: OE-M and AK. Technical assistance: WY, VV, and YN. Interpretated and wrote the paper: OE-M and MN.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are grateful to Kensuke Ozawa, Fumio Gondaira, and Akira Oshima for valuable comments and technical assistance; and to Yohko Takeda, Jetnapang Kongrueng, Natthawan Sermwittayawong, and Pharanai Sukhumungoon and their graduate students for technical assistance. This research was supported, in-part, by Kakenhi Grant-in-Aid for Scientific Research (KAKENHI 24249038) and from the Japan Society for the Promotion of Sciences and a fund for Research on international cooperation in medical science, Research on global health issues, Health and Labor Science Research Grants, the Ministry of Health, Labor, Welfare of Japan (H26-KOKUI-SITEI-001), a fund from the GSS Program, a Kyoto University Program for Leading Graduate Schools, supported by the JSPS and MIXT, and a fund from Yakult Honsha Co. Ltd., Japan.
Boehme, C. C., Nabeta, P., Henostroza, G., Raqib, R., Rahim, Z., Gerhardt, M., et al. (2007). Operational feasibility of using loop-mediated isothermal amplification for diagnosis of pulmonary tuberculosis in microscopy centers of developing countries. J. Clin. Microbiol. 45, 1936–1940. doi: 10.1128/JCM.02352-06
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
DePaola, A., and Kaysner, C. A. (2004). Vibrio/Bacteriological Analytical Manual Online, 9th Edn. Available at: http://www.fda.gov/Food/FoodScienceResearch/LaboratoryMethods/ucm070830.htm
Food and Agriculture Organization World Health Organization of the United Nations [FAO/WHO]. (2011). Risk Assessment of Vibrio parahaemolyticus in Seafood. Microbiol. Risk Assessment Series: 16. Available at: http://www.who.int/foodsafety/publications/micro/MRA_16_JEMRA.pdf
Gutierrez West Casandra, K., Klein Savannah, L., and Lovell Charles, R. (2013). High frequency of virulence factor genes tdh, trh, and tlh in Vibrio parahaemolyticus strains isolated from a pristine estuary. Appl. Environ. Microbiol. 79, 72247–72252. doi: 10.1128/AEM.03792-12
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hara-Kudo, Y., Sugiyama, K., Nishibuchi, M., Chowdhury, A., Yatsuyanagi, J., Ohtomo, Y., et al. (2003). Prevalence of pandemic thermostable direct hemolysin-producing Vibrio parahaemolyticus O3:K6 in seafood and the coastal environment in Japan. Appl. Environ. Microbiol. 69, 3883–3891. doi: 10.1128/AEM.69.7.3883-3891.2003
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Jones J. L., Hara-Kudo, Y., Krantz, J. A., Benner, R. A., Smith, A. B. Jr., Dambaugh T. R., et al. (2012). Comparison of molecular detection methods for Vibrio parahaemolyticus and Vibrio vulnificus. Food Microbiol. 30, 105–111. doi: 10.1016/j.fm.2011.12.011
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Joseph, S. W. R., Colwell, R., and Kaper, J. B. (1982). Vibrio parahaemolyticus and related halophilic vibrios. Crit. Rev. Microbiol. 10, 77–124. doi: 10.3109/10408418209113506
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kishishita, M., Matsuoka, N., Kumagai, K., Yamasaki, S., Takeda, Y., and Nishibuchi, M. (1992). Sequence variation in the thermostable direct hemolysin-related hemolysin (trh) gene of Vibrio parahaemolyticus. Appl. Environ. Microbiol. 58, 2449–2457.
Laohaprertthisan, V., Chowdhury, A., Kongmuang, U., Kalnauwakul, S., Ishibashi, M., Matsumoto, C., et al. (2003). Prevalence and serodiversity of the pandemic clone among the clinical strains of Vibrio parahaemolyticus isolated in southern Thailand. Epidemiol. Infect. 130, 395–406.
Manas, H., Mimouni, R., Chaouqy, N., Hamadi, F., and Martinez-Urtaza, J. (2014). Ocurrence of Vibrio and Salmonella species in mussels (Mytilus galloprovincialis) collected along the Moroccan Atlantic coast. J. SringerPlus 3, 265. doi: 10.1186/2193-1801-3-265
Matsumoto, C., Okuda, J., Ishibashi, M., Iwanaga, M., Garg, P., Rammamurthy, T., et al. (2000). Pandemic spread of an O3:K6 clone of Vibrio parahaemolyticus and emergence of related strains evidenced by arbitrarily primed PCR and toxRS sequence analysis. J. Clin. Microbiol. 38, 578–585.
Mitarai, S., Okumura, M., Toyota, E., Yoshiyama, T., Aono, A., Sejimo, A., et al. (2011). Evaluation of a simple loop-mediated isothermal amplification test kit for the diagnosis of tuberculosis. Int. J. Tuberc. Lung Dis. 15, 1211–1217. doi: 10.5588/ijtld.10.0629
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Nishibuchi, M., and Kaper, J. B. (1985). Nucleotide sequence of the thermostable direct hemolysin gene of Vibrio parahaemolyticus. J. Bacteriol. 162, 558–564.
Nishibuchi, M., and Kaper, J. B. (1995). Thermostable direct hemolysin gene of Vibrio parahaemolyticus: a virulence gene acquired by a marine bacterium. Infect. Immun. 63, 2093–2099.
Notomi, T., Okayama, H., Masubuchi, H., Yonekawa, T., Watanabe, K., Amino, N., et al. (2000). Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:E63. doi: 10.1093/nar/28.12.e63
Nou, X., Arthur, T. M., Bosilevac, J. M., Brichta-Harhay, D. M., Guerini, M. N., Kalchayanand, N., et al. (2006). Improvement of immunomagnetic separation for Escherichia coli O157:H7 detection by the PickPen magnetic particle separation device. J. Food Prot. 69, 2870–2874.
Okuda, J., Ishibashi, M., Abbott, S. L., Janda, J. M., and Nishibuchi, M. (1997a). Analysis of the thermostable direct hemolysin (tdh) gene and the tdh-related hemolysin (trh) genes in urease-positive strains of Vibrio parahaemolyticus isolated on the West Coast of the United States. J. Clin. Microbiol. 35, 1965–1971.
Okuda, J., Ishibashi, M., Hayakawa, E., Nishino, T., Takeda, Y., Mukhopadhyay A. K., et al. (1997b). Emergence of a unique O3:K6 clone of Vibrio parahaemolyticus in Calcutta, India, and isolation of strains from the same clonal group from Southeast Asian travelers arriving in Japan. J. Clin. Microbiol. 35, 3150–3155.
Raghunath, P. (2015). Roles of thermostable direct hemolysin (TDH) and TDH-related hemolysin (TRH) in Vibrio parahaemolyticus. Front. Microbiol. 5:805. doi: 10.3389/fmicb.2014.00805
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Rodriguez-Castro, A., Ansede-Bermejo, J., Blanco-Abad, V., Varela-Pet, J., Garcia-Martin, O., and Martinez-Urtaza, J. (2010). Prevalence and genetic diversity of pathogenic populations of Vibrio parahaemolyticus in coastal waters of Galicia, Spain. Environ. Microbiol. Rep. 2, 58–66. doi: 10.1111/j.1758-2229.2009.00064.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tanaka, N., Iwade, Y., Yamazaki, W., Gondaira, F., Vuddhakul, V., Nakaguchi, Y., et al. (2014). Most-probable-number loop-mediated isothermal amplification-based procedure enhanced with K antigen-specific immunomagnetic separation for quantifying tdh+ Vibrio parahaemolyticus in Molluscan Shellfish. J. Food Prot. 77, 1078–1085. doi: 10.4315/0362-028X.JFP-13-536
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Vuddhakul, V., Chowdhury, A., Laohaprertthisan, V., Pungrasamee, P., Patararungrong, N., Thianmontri, P., et al. (2000). Isolation of Vibrio parahaemolyticus strains belonging to a pandemic O3:K6 clone from environmental and clinical sources in Thailand. Appl. Environ. Microbiol. 66, 2685–2689. doi: 10.1128/AEM.66.6.2685-2689.2000
Vuddhakul, V., Soboon, S., Sunnghiran, W., Kaewpiboon, S., Chowdhury, A., Ishibashi, M., et al. (2006). Distribution of virulent and pandemic strains of Vibrio parahaemolyticus in three molluscan shellfish species (Meretrix meretrix, Perna viridis, and Anadara granosa) and their association with Foodborne disease in southern Thailand. J. Food Prot. 69, 2615–2620.
Yamamoto, A., Iwahori, J., Vuddhakul, V., Charernjiratragulc, W., Vose, D., Osaka, K., et al. (2008). Quantitative modeling for risk assessment of Vibrio parahaemolyticus in bloody clams in southern Thailand. Int. J. Food Microbiol. 124, 70–78. doi: 10.1016/j.ijfoodmicro.2008.02.021
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Yamazaki, W., Kumeda, Y., Misawa, N., Nakaguchi, Y., and Nishibuchi, M. (2010). Development of a loop-mediated isothermal amplification assay for sensitive and rapid detection of the tdh and trh genes of Vibrio parahaemolyticus and related Vibrio species. Appl. Environ. Microbiol. 76, 820–828. doi: 10.1128/AEM.02284-09
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zeng, J., Wei, H., Zhang, L., Liu, X., Zhang, H., Cheng, J., et al. (2014). Rapid detection of Vibrio parahaemolyticus in raw oysters using immunomagnetic separation combined with loop-mediated isothermal amplification. J. Food Microbiol. 174, 123–128. doi: 10.1016/j.ijfoodmicro.2014.01.004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Keywords: Vibrio parahaemolyticus, most-probable-number, immunomagnetic separation, loop-mediated isothermal amplification, K antigen
Citation: Escalante-Maldonado O, Kayali AY, Yamazaki W, Vuddhakul V, Nakaguchi Y and Nishibuchi M (2015) Improvement of the quantitation method for the tdh+ Vibrio parahaemolyticus in molluscan shellfish based on most-probable-number, immunomagnetic separation, and loop-mediated isothermal amplification. Front. Microbiol. 6:270. doi: 10.3389/fmicb.2015.00270
Received: 10 January 2015; Accepted: 18 March 2015;
Published online: 09 April 2015.
Edited by:
Iddya Karunasagar, Food Safety Consultant, IndiaReviewed by:
Jaime Martinez-Urtaza, University of Bath, UKCopyright © 2015 Escalante-Maldonado, Kayali, Yamazaki, Vuddhakul, Nakaguchi and Nishibuchi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mitsuaki Nishibuchi, Division of Human-Nature Dynamics, Center for Southeast Asian Studies, Kyoto University, 46 Shimoadachi-cho, Yoshida, Sakyo-ku, Kyoto 606-8501, JapanbmlzaWJ1dGlAY3NlYXMua3lvdG8tdS5hYy5qcA==
†Present address: Yoshitsugu Nakaguchi, Department of Food Science, Faculty of Bioresources and Environmental Sciences, Ishikawa Prefectural University, Nonoichi, Japan
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.