- 1Institute of Clinical Medicine, Schsool of Medicine, National Yang-Ming University, Taipei, Taiwan
- 2National Institute of Infectious Diseases and Vaccinology, National Health Research Institutes, Taipei, Taiwan
- 3Division of Infectious Diseases, Taipei Veterans General Hospital, Taipei, Taiwan
- 4Emergency Department, Taipei Veterans General Hospital, Taipei, Taiwan
- 5Department of Internal Medicine, Mackay Memorial Hospital, Hsin-Chu, Taiwan
- 6Department of Molecular Medicine and Institute of Life Science, National Tsing Hua University, Hsin-Chu, Taiwan
The sulbactam resistance rate in Acinetobacter baumannii has increased worldwide. Previous reports have shown that the β-lactamase blaTEM-1 confers resistance to sulbactam in A. baumannii. The purpose of this study was to examine whether other β-lactamases, including the Acinetobacter-derived cephalosporinase (ADC), OXA-23, OXA-24/72, and OXA-58 families, also contribute to sulbactam resistance in A. baumannii. The correlation between these β-lactamases and the sulbactam minimal inhibitory concentration (MIC) was determined using A. baumannii clinical isolates from diverse clonality, which were collected in a nationwide surveillance program from 2002 to 2010 in Taiwan. A possible association between the genetic structure of ISAba1-blaADC-30 and sulbactam resistance was observed because this genetic structure was detected in 97% of sulbactam-resistant strains compared with 10% of sulbactam-susceptible strains. Transformation of ISAba1-blaADC-30 into susceptible strains increased the sulbactam MIC from 2 to 32 μg/ml, which required blaADC-30 overexpression using an upstream promoter in ISAba1. Flow cytometry showed that ADC-30 production increased in response to sulbactam, ticarcillin, and ceftazidime treatment. This effect was regulated at the RNA level but not by an increase in the blaADC-30 gene copy number as indicated by quantitative PCR. Purified ADC-30 decreased the inhibitory zone created by sulbactam or ceftazidime, similarly to TEM-1. In conclusion, ADC-30 overexpression conferred resistance to sulbactam in diverse clinical A. baumannii isolates.
Introduction
Acinetobacter baumannii causes various nosocomial infections, and the prevalence of multidrug-resistant (MDR) A. baumannii has been increasing in different countries. This bacterium has intrinsic resistance to multiple drugs and can gain resistance mechanisms from other species (Peleg et al., 2008). The SENTRY program documented non-susceptibility to carbapenems, the last resort of drugs for the treatment of MDR A. baumannii, increased from 34.5% in 2006 to 59.8% in 2009 worldwide (Gales et al., 2011). In Taiwan, the rate of multidrug resistance in Acinetobacter spp. also increased from 1.3% in 2002 to 41.0% in 2010 (Kuo et al., 2012). In severely ill patients, infections with MDR isolates have been associated with high mortality due to the absence of appropriate or effective treatment options (Peleg et al., 2008). Combination therapies or new drugs such as antimicrobial peptides or silver nanoparticles have been proposed as novel modalities to treat MDR A. baumannii (Peleg et al., 2008; Tiwari et al., 2014)
Sulbactam is a β-lactamase inhibitor that is typically combined with penicillins because sulbactam lacks antimicrobial activity against most bacterial species (Adnan et al., 2013). However, sulbactam has demonstrated bacteriostatic or bactericidal effects against A. baumannii (Corbella et al., 1998). Combination treatment with sulbactam and carbapenems has shown promising in vivo and in vitro synergistic effects against MDR A. baumannii (Wolff et al., 1999; Ko et al., 2004; Song et al., 2007), and clinical success has been reported (Karageorgopoulos and Falagas, 2008). The addition of sulbactam to other antibiotics has been proposed in the treatment of MDR A. baumannii; however, the resistance rate to ampicillin/sulbactam in Acinetobacter spp. has increased to approximately 60% in certain area (Kuo et al., 2012).
Until recently, the mechanism underlying sulbactam resistance in A. baumannii was less commonly studied. In 2013, Krizova and colleagues demonstrated that the β-lactamase TEM-1 contributes to sulbactam resistance (Krizova et al., 2013), which led us to examine whether other selected β-lactamases found in A. baumannii, including the Acinetobacter-derived cephalosporinase (ADC), OXA-23, OXA-24/72, and OXA-58 families, also contribute to sulbactam resistance. Using clinical isolates collected from a Taiwanese surveillance program, we aimed to identify the β-lactamases associated with sulbactam resistance and to test the role of these β-lactamases in sulbactam resistance in A. baumannii.
Materials and Methods
Association of Selected β-Lactamases with Sulbactam Resistance in A. baumannii
A. baumannii clinical isolates were randomly selected from the Taiwan Surveillance of Antimicrobial Resistance (TSAR) program, which contains 1640 Acinetobacter isolates collected from 2002 to 2010 (Kuo et al., 2012). A. baumannii was identified at the species level using multiplex PCR targeting the specific 16-23S rDNA intergenic spacer region (Chen et al., 2007). Pulsed-field gel electrophoresis was performed to determine clonality as previously described (Kuo et al., 2013). Isolates with similarity of >80% was designated as a single clone. The sulbactam-resistant and sulbactam-susceptible isolates were randomly selected and subjected to PCR testing for the presence of genes encoding ADC, OXA-23, OXA-24/72, and OXA-58 β-lactamases (Table S1). The PCR program (Krizova et al., 2013; Kuo et al., 2013) for genes encoding OXA was as followed: 94°C for 1 min, and 30 cycles of 25 s at 94°C, 40 s at 52°C and 50 s at 72°C; for PCR of blaADC, 30 cycles of 60 s at 94°C, 60 s at 58°C, and 120 s at 72°C. GoTaq Flexi DNA polymerase (Promega, Madison, WI) was used for PCR assays performed in the GeneAmp PCR System 2700 (Applied Biosystems, Foster City, CA). Amplified DNA product was resolved by electrophoresis in agarose 2% w/v gels, stained with ethidium bromide, and purified according to the manufacturer's instruction (Geneaid Biotech Ltd, Taipei, Taiwan).
Transformation of Plasmids Carrying Different β-Lactamase Genes
Representative β-lactamase genes and their associated promoters, including blaTEM−1, blaADC and blaOXA−23 with their upstream insertion sequence ISAba1 (ISAba1-blaADC and ISAba1-blaOXA−23, respectively), blaOXA−24/72, and blaOXA−58 with its upstream ISAba3 that was truncated with IS1008 (IS1008–ΔISAba3-blaOXA−58) (Chen et al., 2010), were PCR-amplified using the forward and reverse primers shown in Table S1. The PCR products were amplified with a proofreading DNA polymerase (Phusion High-Fidelity DNA Polymerase, Finnzymes, Espoo, Finland), cloned into the pCRII-TOPO vector (Invitrogen, Carlsbad, CA, USA) and subjected to sequencing (Mission Biotech, Taipei, Taiwan). The digested fragments were cloned into the XbaI and XhoI sites of the Escherichia coli-A. baumannii shuttle vector pYMAb2 (Kuo et al., 2013), which contains a kanamycin-resistant determinant. The fragment was cloned in-frame with a polyhistidine (His) tag, causing the resulting protein to be His-tagged. The recombinant plasmid and a control plasmid (pYMAb2 without β-lactamase genes) were transformed into the kanamycin-susceptible A. baumannii strain ATCC15151. ATCC15151 already contained blaOXA−51; therefore, blaOXA−51was not included in the experiment. Electroporation was performed with a gene pulser electroporator (Bio-Rad, Hercules, CA, USA) and 2-mm electrode gap cuvettes (Kuo et al., 2013). Transformants were selected based on kanamycin resistance, and sequencing was performed to confirm the presence of each β-lactamase gene.
Antimicrobial Susceptibility
Minimal inhibitory concentrations (MICs) of sulbactam, ceftazidime, ampicillin, imipenem, meropenem, and ticarcillin were determined by the agar dilution method according to the guidelines provided by the Clinical and Laboratory Standards Institute (CLSI) (Clinical and Laboratory Standards Institute, 2012). Sulbactam susceptibility parameters were adopted from the previous CLSI guidelines in which sulbactam MICs of less than 4 or more than 16 μg/ml were defined as susceptible or resistant, respectively.
Immunofluorescent Staining and Enumeration by Flow Cytometry for Protein Expression
Immunofluorescent staining was performed as previously described with several modifications (Moe et al., 1999). Specifically, bacterial cultures were diluted in phosphate buffered saline (PBS) to ~108 CFU/mL, and 0.5-ml samples were transferred into 1.5-ml Eppendorf microtubes. The samples were centrifuged at 5000 × g for 5 min, washed with 0.1% NaN3 PBS, and centrifuged for 5 min. The resulting bacterial pellet was resuspended in fixative (4% paraformaldehyde in PBS) for 20 min. The fixed samples were washed twice with quenching solution (100 mM NaCl, 50 mM Tris-HCl, pH 8.0) and resuspended in permeable buffer (1% Triton X-100, 0.1% NaN3 in PBS) for 5 min. After centrifugation, the samples were resuspended in 500 μL blocking buffer (1% BSA and 0.1% NaN3 in PBS).
To identify His-tagged ADC-30 expressed by ATCC15151 (pYMAb2::ISAba1-blaADC−30), each 100-μL sample was incubated with 2 μL of mouse anti-His-6-tag antibody (Sigma-Aldrich, St. Louis, MO, USA) at 4°C for 2 h. After washing by blocking buffer, the samples were stained for 1 h at 4°C with 5 μL of phycoerythrin (PE)-conjugated anti-mouse IgG antibody (Sigma-Aldrich). The stained samples were spun at 5000 × g for 5 min, and the cell pellet was resuspended in 500 μL of 1% paraformaldehyde buffer and stored at 4°C overnight. Cytometry samples were resuspended in PBS and analyzed in a flow cytometer system with wavelength of 575 nm (FACScanto II, BD Biosciences, San Jose, CA, USA).
Quantitative PCR (qPCR) to Determine the blaADC−30 Gene Copy Number after Challenging with Different Antimicrobials
ATCC15151 (pYMAb2::ISAba1-blaADC−30) strains at mid-log phase were incubated in Luria-Bertani (LB) broth with different antimicrobial agents (25% of MIC) for 6 h. The blaADC−30 copy number in these bacteria was estimated by qPCR using primers targeting blaADC−30, and the housekeeping gene, recA, was used as an internal control. Each qPCR reaction contained a total volume of 10 μL with 2 ng of genomic DNA as template, 100 nmol/L of each primer, and 1 × SYBR Green® PCR Master Mix (Applied Biosystems, Carlsbad, CA, USA) with ROX™ (Kapa Biosystems, Woburn, MA, USA). The relative blaADC−30 copy number in the bacteria treated with different antimicrobial agents was normalized to the number found in the bacteria treated with LB broth without antimicrobial agents. The qPCR conditions included 2 min at 50°C (UNG activation), 10 min at 95°C, followed by 45 cycles of 15 s at 95°C and 1 min at 60°C. At the end, a dissociation stage was added: 15 s at 95°C, 15 s at 60°C, and 15 s at 95°C. All experiments were conducted using the ABI 7500 Fast Real-time PCR system (Applied Biosystems, Inc., Carlsbad, CA, USA) and were performed in triplicate.
Quantitative Reverse Transcription PCR (qRT-PCR) to Assess mRNA Expression after Challenging with Different Antimicrobials
After incubation with different antimicrobial agents, the ADC-30 mRNA levels in ATCC15151 (pYMAb2::ISAba1-blaADC−30) were compared using qRT-PCR (Chen et al., 2010). Briefly, around 2 μg of RNA was extracted with RNAprotect Bacteria Reagent and an RNeasy mini-kit (Qiagen, Valencia, CA, USA). Residual genomic DNA was removed using RNase-free DNase. The RNA was reverse transcribed into single-stranded cDNA with random hexamers and Moloney Murine Leukemia Virus reverse transcriptase (Epicenter, Madison, WI, USA). The cDNAs were subsequently quantified by real-time PCR amplification with conditions mentioned above. Expression level results were standardized to the transcription levels of rpoB gene for each strain, but relatively to the culture in LB (2 delta–delta Ct method). Negative controls without reverse transcription were performed to detect DNA contamination in the purified RNA.
ADC-30 Purification
His-tagged ADC-30 (in which the stop codon was deleted, and the proteins were fused with 6 His amino acids) expressed by ATCC15151 (pYMAb2::ISAba1-blaADC−30) was purified with Ni-NTA Superflow column (Qiagen). Briefly, the bacteria equal to ~107 CFU/ml were centrifuged, resuspended in lysis buffer and sonicated. The lysate was diluted in binding buffer (25 mM Tris, 150 mM NaCl, 10 mM imidazole, pH 7.5) and loaded onto the column. The column was washed with five column volumes of wash buffer, and the protein was eluted with five column volumes of elution buffer (25 mM Tris, 150 mM NaCl, 300 mM imidazole). The protein solution was then dialyzed and concentrated by ultrafiltration on a 10 KDa-cutoff Amicon membrane (Millipore). The purity was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting as greater than 95%.
SDS-PAGE and Western Blotting to Detect Purified β-Lactamases
For SDS-PAGE and Western blotting (Blair et al., 2009), the purified protein was first separated by SDS-PAGE on 12% acrylamide gel using Mini-PROTEAN system (Bio-Rad) and transferred to a nitrocellulose membrane (PerkinElmer, Boston, MA, USA) in transfer buffer (25 mM Tris-HCl, 190 mM glycine, 20% methanol, 0.1% SDS, pH 8.3) at 350 mA for 1 h. The membranes were blocked with 10% non-fat dried milk in Tris-bufferred saline with 0.5% Tween-20. After hybridization with anti-His antibodies (Sigma-Aldrich) and peroxidase-conjugated goat anti-mouse secondary antibodies (Millipore, Temecula, CA, USA), the band was visualized with an ECL Western blot kit (PerkinElmer, Boston, MA, USA).
Bioassays to Confirm the Activity of Purified ADC-30 against Ceftazidime and Sulbactam
The first bioassay involved the mixture of 10 μL of purified ADC-30 or extracts of sulbactam- resistant or -susceptible strains with 10 μL of ceftazidime for 30 min at 37°C. Each 20-μL mixture was loaded onto a blank disk (Becton Dickinson, Sparks, MD, USA) that was placed in an agar plate containing a lawn of ceftazidime-susceptible A. baumannii ATCC 15151. Inhibitory zones were measured after incubating the plates overnight at 37°C.
The second bioassay conducted was similar to the modified Hodge test for carbapenemase detection. A 0.5 McFarland standard suspension of the sulbactam-susceptible A. baumannii ATCC 15151 strain was diluted to 1:10 and inoculated on an Mueller-Hinton agar plate for routine disk diffusion test. The 30-μg sulbactam disk was placed in the center of the plate, and the purified ADC-30, TEM-1, or PBS samples were drawn in a straight line out from the edge of the disk. The phenotype was evaluated after an overnight incubation at 37°C.
Results and Discussion
Detection of Selected β-Lactamases in Clinical A. baumannii Isolates using a Nationwide Surveillance System
Of the 30 sulbactam-resistant isolates tested, 14 (47%) were positive for blaTEM−1, and 29 (97%) possessed blaADC with ISAba1 upstream (ISAba1-blaADC). The PFGE shown in Figure 1 depicts the strain diversity. Based on clonality, 19 isolates belonging to different clones and carrying ISAba1-blaADC were sent for sequencing. The blaADC sequences of all 19 strains were consistent with the blaADC−30 sequence. ISAba1-blaOXA−23, blaOXA−24/72 and IS1008–ΔISAba3-blaOXA−58 were present in16, 6, and 0 isolates, respectively. In 10 sulbactam-susceptible isolates, one possessed ISAba1-blaADC−30. None of the isolates possessed blaTEM−1. Therefore, in addition to TEM-1(Krizova et al., 2013), ISAba1-blaADC−30 may play a role in providing A. baumannii with sulbactam resistance.
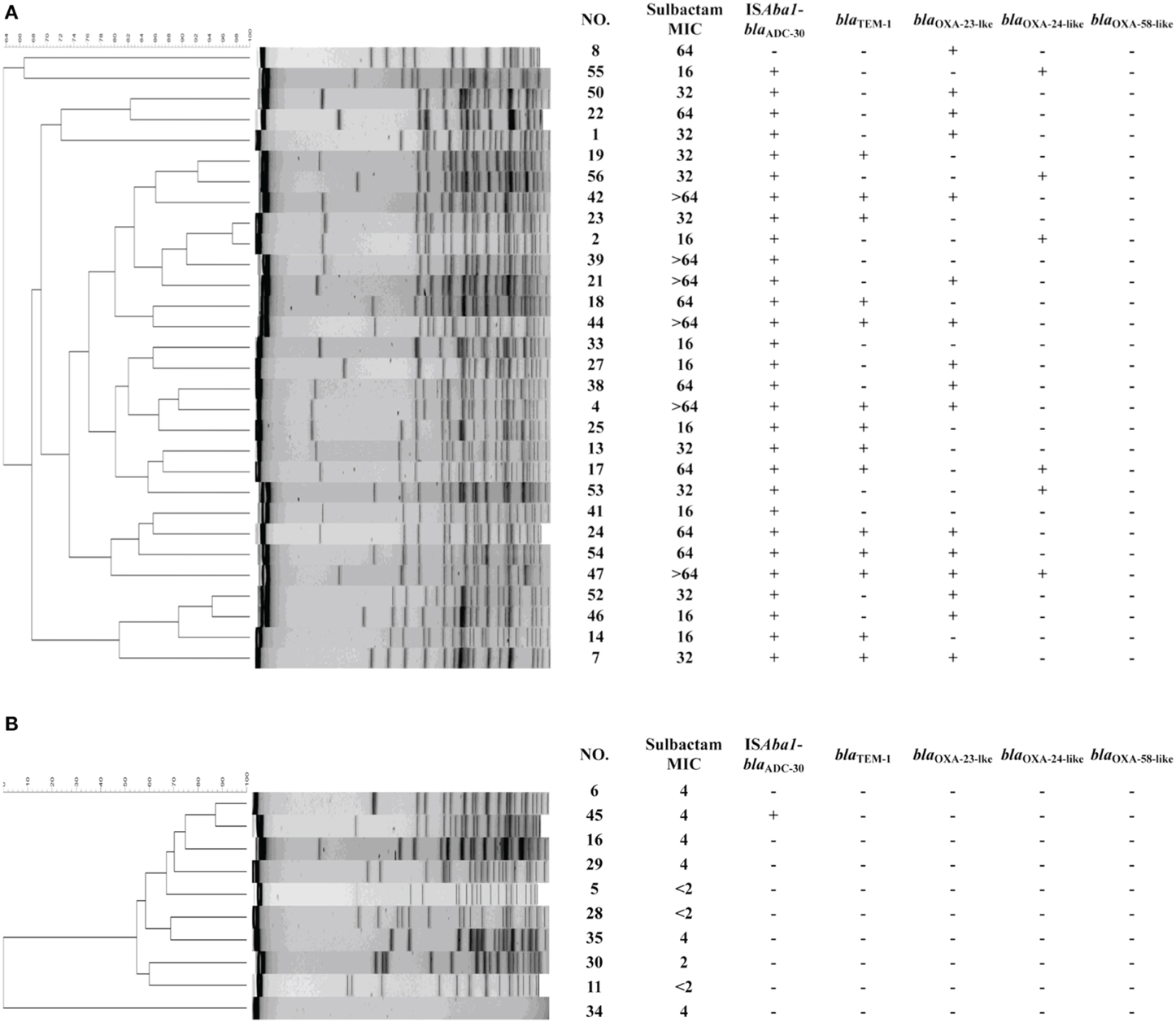
Figure 1. Molecular characteristics of randomly selected Acinetobacter baumannii from the Taiwan Surveillance of Antimicrobial Resistance (TSAR) program, 2002–2010. The results of pulsed-field gel electrophoresis are shown, followed by the minimal inhibitory concentrations (MICs), and the presence of ISAba1-blaADC-30, blaTEM-1, ISAba1-blaOXA-23-lke, blaOXA-24-like, and blaOXA-58-like in the (A) sulbactam-resistant strains and (B) sulbactam-susceptible strains.
The presence of one resistant strain without ISAba1-blaADC nor blaTEM−1 indicates that another mechanism may be involved in sulbactam resistance. Various combinations of resistance mechanisms, including β-lactamase overexpression, the up-regulation of the efflux pump and the inactivation or down-regulation of porin, are often required for the development of β-lactam resistance in Pseudomonas aeruginosa and A. baumannii (Quale et al., 2006; Peleg et al., 2008; Tiwari et al., 2012; Tiwari and Moganty, 2014), which may explain the presence of ISAba1-blaADC−30 in one susceptible strain. However, the high rate of ISAba1-blaADC−30 in resistant strains and its low rate in susceptible strains (97% vs. 10%, Chi-square test p < 0.001) indicate the importance of ISAba1-blaADC−30 for the development of sulbactam resistance.
Contribution of ADC-30 and ISAba1 to Sulbactam Resistance
To further confirm the association observed in the epidemiological survey, ISAba1-blaADC−30, blaTEM−1,ISAba1-blaOXA−23, blaOXA−24/72 (and its promoter), and IS1008–ΔISAba3-blaOXA−58 were cloned and transformed into a sulbactam-susceptible reference strain, respectively. Shuttle vectors were also transformed, and changes in the MICs were measured (Table 1).
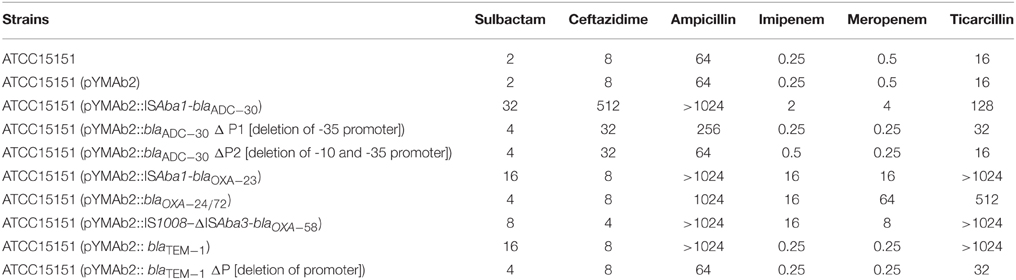
Table 1. Minimal inhibitory concentrations (μg/ml) of Acinetobacter baumannii reference strain and different transformants.
ATCC15151 (pYMAb2::ISAba1-blaADC−30) exhibited the highest increases of sulbactam MIC (16-fold). Other β-lactamases, including OXA-23, OXA-72, and OXA-58, contributed to the increase in the sulbactam MICs, although at lower levels. ISAba1-blaADC−30 was used in the study due to the high level of sulbactam resistance and high prevalence of ISAba1-blaADC−30, which were comparable to the sulbactam resistance levels and prevalence of blaTEM−1.
The regulatory mechanism for ADC expression in Acinetobacter spp. may be different from the mechanism in many Enterobacteriaceae because Acinetobacter spp. lack the AmpR gene (Jacoby, 2009). The presence of ISAba1 upstream of blaAmpC is essential for ceftazidime resistance due to the AmpC overexpression (Heritier et al., 2006). To confirm the role of the promoter located within ISAba1 in mediating sulbactam resistance, plasmids harboring blaADC−30 without upstream -35 (within ISAba1) or -35/-10 promoters were transformed into ATCC15151. The MICs increased by only 2-fold compared with the control (Table 1). Therefore, as previous studies have indicated (Heritier et al., 2006), these promoters in the ISAba1 are required for ADC-mediated sulbactam resistance. Similarly, deletion of the promoter upstream of blaTEM−1 decreased the sulbactam MIC.
ADC-30 Production in Response to Treatment with Sulbactam and Other Substrates
The production of β-lactamase usually increases in response to its substrates; therefore, we asked whether the addition of sulbactam increases ADC-30 production. Flow cytometry (Figure 2) showed that ADC-30 protein expression increased significantly after the addition of sulbactam or ceftazidime compared with the control. ADC-30 production was also induced by ticarcillin but not by ciprofloxacin or imipenem. The results indicate that the co-selection of sulbactam resistance by other antimicrobial agents occurs.
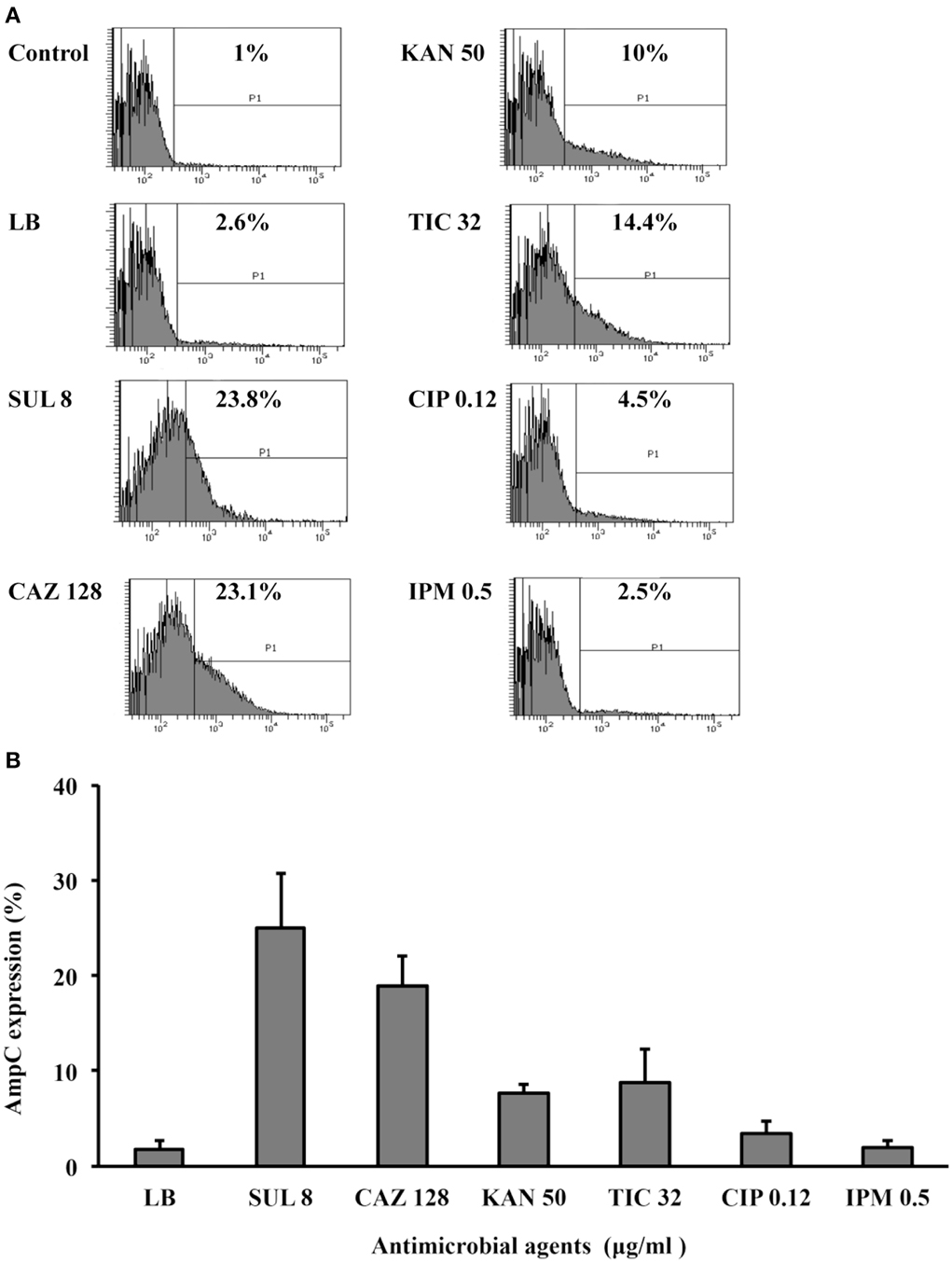
Figure 2. ADC-30 production in response to treatment with sulbactam and other substrates. (A) Flow cytometry showed that the ADC-30 protein expression level increased in response to its substrates (ceftazidime, sulbactam, and ticarcillin) compared with the level observed in bacteria cultured in Luria-Bertani (LB) broth without antimicrobial agents. The tests were performed in triplicate; however, only one of the representative experiments is shown. (B) Quantified values of experiments performed in triplicate are shown in the bar graph. LB broth supplemented with kanamycin was used as a positive control because pYMAb2 also carries a kanamycin resistance determinant. CAZ, ceftazidime; CIP, ciprofloxacin; IPM, imipenem; KAN, kanamycin; SUL, sulbactam.
The qRT-PCR results (Figure 3) showed that ADC-30 mRNA expression increased in response to its substrates (ceftazidime and sulbactam) but not in response to other antimicrobial agents (ciprofloxacin). Carbapenemase expression has been related to the high blaOXA−58 plasmid copy number (Bertini et al., 2007; Chen et al., 2008). Therefore, we asked whether the increased ADC-30 protein and mRNA expression levels in the presence of sulbactam or ceftazidime are attributed to an increase in the blaADC−30 copy number. The qPCR results (Figure 3) showed no differences in the blaADC−30 copy numbers of cells treated with sulbactam or ceftazidime compared with the negative controls. In contrast, the gene copy number increased in response to kanamycin. The results indicate that the increase in ADC-30 was regulated at the RNA level.
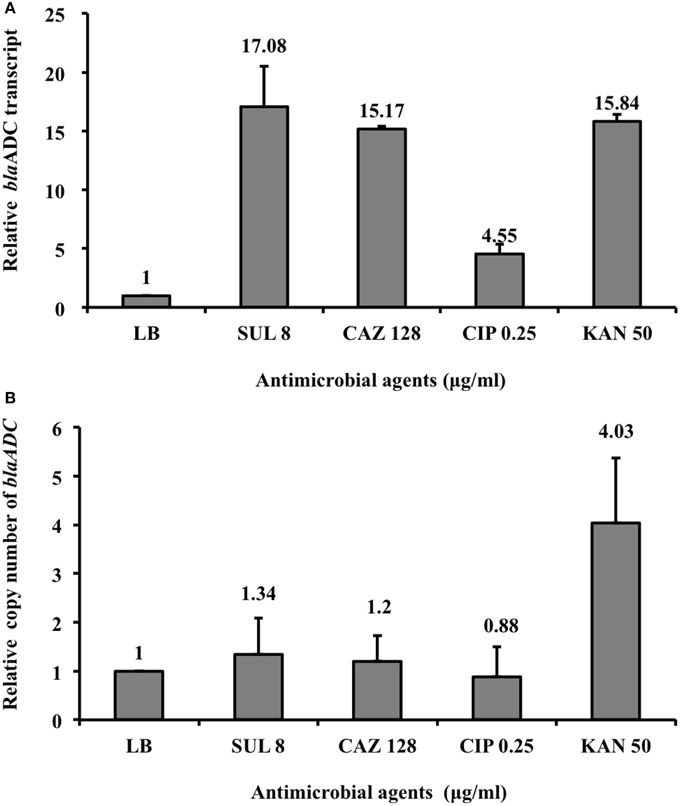
Figure 3. BlaADC-30 mRNA expression level and gene copy number in response to treatment with sulbactam and other antimicrobial agents. (A) The blaADC-30 mRNA expression level increased after treatment with sulbactam and ceftazidime. ATCC15151 (pYMAb2::ISAba1-blaADC-30) treated with ciprofloxacin or without antimicrobial agents were used as negative controls. (B) The blaADC-30 gene copy number did not differ, regardless of the antimicrobial agents added. The mRNA expression level increased in response to kanamycin because of the increased number of plasmids (pYMAb2) carrying the kanamycin-resistant gene. The test was performed in triplicate. CAZ, ceftazidime; CIP, ciprofloxacin; KAN, kanamycin; LB, Luria-Bertani broth; SUL, sulbactam.
How the addition of sulbactam increased ADC-30 protein and mRNA expression level in A. baumannii is unknown. The induction of AmpC by β-lactams has been commonly described in P. aeruginosa and many Enterobacteriaceae (Jacoby, 2009). After the treatment of β-lactams, altered peptidoglycan synthesis leads to increased expression of AmpC through AmpG–AmpR–AmpC pathway (Zeng and Lin, 2013). Clavulanate, another β-lactamase inhibitor with structure similar to β-lactams, also induces the expression of AmpC in many Enterobacteriaceae (Drawz and Bonomo, 2010). Although sulbactam may interfere the wall synthesis by interacting with penicillin-binding protein 1 and 3 (Penwell et al., 2015), lack of AmpR in A. baumannii indicated other mechanisms, rather than AmpG–AmpR–AmpC pathway, are responsible. The two-component system has been proposed to be involved in the induction of AmpC and other chromosomally encoded β-lactamases (Jacoby, 2009; Zeng and Lin, 2013). However, the mechanism regarding the induction of ADC in response to sulbactam in A. baumannii requires further investigation.
Contribution of Purified ADC-30 to Sulbactam Resistance
ADC-30 was purified (Figure S1), and the addition of purified ADC-30 to the disk decreased the inhibitory zone caused by ceftazidime (Figure 4A), confirming the activity of the purified protein. The bioassay resembling the modified Hodge test showed that purified ADC-30 promoted the inward growth of sulbactam-susceptible A. baumannii similarly to purified TEM-1 (Figure 4B). Therefore, ADC-30 may directly interact with sulbactam and confer sulbactam resistance.
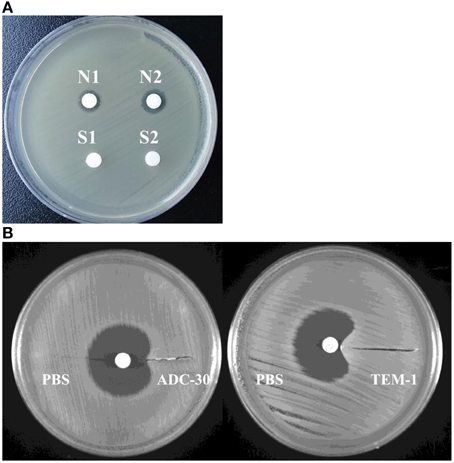
Figure 4. Activity of ADC-30 against ceftazidime and sulbactam. (A) ADC-30 remained active against ceftazidime in Acinetobacter baumannii after purification. Disks containing ceftazidime and different samples were placed in an agar plate containing a lawn of ceftazidime-susceptible A. baumannii ATCC 15151. Crude extract of the ATCC15151 (pYMAb2::ISAba1-blaADC-30) strain and purified ADC-30 (denoted as S1 and S2 in the lower filed) promoted the growth of the A. baumannii ATCC 15151 strain against disks loaded with ceftazidime. Controls included N1 (phosphate buffered saline) and N2 [crude extract of ATCC15151 (pYMAb2)], with which the ceftazidime created an inhibitory zone. (B) The bioassay resembling modified the Hodge test showed that purified ADC-30 and TEM-1 both promoted A. baumannii ATCC 15151 growth against sulbactam. PBS, phosphate buffered salinex.
In conclusion, blaADC−30 overexpression contributes to sulbactam resistance in A. baumannii, which is prevalent in clinical isolates of different clones in Taiwan over a long period of time. The resistance mechanisms are induced at the mRNA and protein levels by other antimicrobial agents in addition to sulbactam, supporting the cautious use of these antibiotics to avoid the selection of sulbactam-resistant A. baumannii isolates.
Funding
This work was supported by grants from the National Health Research Institute and the National Science Council (MOST 103-2314-B-400 -020 -MY2 and 101-2314-B-010-027-MY3). The sponsors did not have a role in the study design, in the collection, analysis, or interpretation of data, in the writing of the report, or in the decision to submit the article for publication.
Ethical Approval
This study was approved by the Institutional Review Board of National Health Research Institutes.
Conflict of Interest Statement
Te-Li Chen is a medical advisor of TTY Biopharm. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We express our sincere appreciation to the 26 hospitals that participated in the Taiwan Surveillance of Antimicrobial Resistance (TSAR) program.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00231/abstract
References
Adnan, S., Paterson, D. L., Lipman, J., and Roberts, J. A. (2013). Ampicillin/sulbactam: its potential use in treating infections in critically ill patients. Int. J. Antimicrob. Agents 42, 384–389. doi: 10.1016/j.ijantimicag.2013.07.012
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bertini, A., Poirel, L., Bernabeu, S., Fortini, D., Villa, L., Nordmann, P., et al. (2007). Multicopy blaOXA-58 gene as a source of high-level resistance to carbapenems in Acinetobacter baumannii. Antimicrob. Agents Chemother. 51, 2324–2328. doi: 10.1128/AAC.01502-06
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Blair, J. M., La Ragione, R. M., Woodward, M. J., and Piddock, L. J. (2009). Periplasmic adaptor protein AcrA has a distinct role in the antibiotic resistance and virulence of Salmonella enterica serovar Typhimurium. J. Antimicrob. Chemother. 64, 965–972. doi: 10.1093/jac/dkp311
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chen, T. L., Chang, W. C., Kuo, S. C., Lee, Y. T., Chen, C. P., Siu, L. K., et al. (2010). Contribution of a plasmid-borne blaOXA-58 gene with its hybrid promoter provided by IS1006 and an ISAba3-like element to beta-lactam resistance in Acinetobacter genomic species 13TU. Antimicrob. Agents Chemother. 54, 3107–3112. doi: 10.1128/AAC.00128-10
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chen, T. L., Siu, L. K., Wu, R. C., Shaio, M. F., Huang, L. Y., Fung, C. P., et al. (2007). Comparison of one-tube multiplex PCR, automated ribotyping and intergenic spacer (ITS) sequencing for rapid identification of Acinetobacter baumannii. Clin. Microbiol. Infect. 13, 801–806. doi: 10.1111/j.1469-0691.2007.01744.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chen, T. L., Wu, R. C., Shaio, M. F., Fung, C. P., and Cho, W. L. (2008). Acquisition of a plasmid-borne blaOXA-58 gene with an upstream IS1008 insertion conferring a high level of carbapenem resistance to Acinetobacter baumannii. Antimicrob. Agents Chemother. 52, 2573–2580. doi: 10.1128/AAC.00393-08
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Clinical Laboratory Standards Institute. (2012). Performance Standards for Antimicrobial Susceptibility Testing. Twenty-Second Informational Supplement. CLSI Document M100-S22. Wayne, PA: CLSI.
Corbella, X., Ariza, J., Ardanuy, C., Vuelta, M., Tubau, F., Sora, M., et al. (1998). Efficacy of sulbactam alone and in combination with ampicillin in nosocomial infections caused by multiresistant Acinetobacter baumannii. J. Antimicrob. Chemother. 42, 793–802. doi: 10.1093/jac/42.6.793
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Drawz, S. M., and Bonomo, R. A. (2010). Three decades of beta-lactamase inhibitors. Clin. Microbiol. Rev. 23, 160–201. doi: 10.1128/CMR.00037-09
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gales, A. C., Jones, R. N., and Sader, H. S. (2011). Contemporary activity of colistin and polymyxin B against a worldwide collection of Gram-negative pathogens: results from the SENTRY Antimicrobial Surveillance Program (2006-09). J. Antimicrob. Chemother. 66, 2070–2074. doi: 10.1093/jac/dkr239
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Heritier, C., Poirel, L., and Nordmann, P. (2006). Cephalosporinase over-expression resulting from insertion of ISAba1 in Acinetobacter baumannii. Clin. Microbiol. Infect. 12, 123–130. doi: 10.1111/j.1469-0691.2005.01320.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Jacoby, G. A. (2009). AmpC beta-lactamases. Clin. Microbiol. Rev. 22, 161–182. doi: 10.1128/CMR.00036-08
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Karageorgopoulos, D. E., and Falagas, M. E. (2008). Current control and treatment of multidrug-resistant Acinetobacter baumannii infections. Lancet Infect. Dis. 8, 751–762. doi: 10.1016/S1473-3099(08)70279-2
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ko, W. C., Lee, H. C., Chiang, S. R., Yan, J. J., Wu, J. J., Lu, C. L., et al. (2004). In vitro and in vivo activity of meropenem and sulbactam against a multidrug-resistant Acinetobacter baumannii strain. J. Antimicrob. Chemother. 53, 393–395. doi: 10.1093/jac/dkh080
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Krizova, L., Poirel, L., Nordmann, P., and Nemec, A. (2013). TEM-1 beta-lactamase as a source of resistance to sulbactam in clinical strains of Acinetobacter baumannii. J. Antimicrob. Chemother. 68, 2786–2791. doi: 10.1093/jac/dkt275
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kuo, S. C., Chang, S. C., Wang, H. Y., Lai, J. F., Chen, P. C., Shiau, Y. R., et al. (2012). Emergence of extensively drug-resistant Acinetobacter baumannii complex over 10 years: nationwide data from the Taiwan Surveillance of Antimicrobial Resistance (TSAR) program. BMC Infect. Dis. 12:200. doi: 10.1186/1471-2334-12-200
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kuo, S. C., Yang, S. P., Lee, Y. T., Chuang, H. C., Chen, C. P., Chang, C. L., et al. (2013). Dissemination of imipenem-resistant Acinetobacter baumannii with new plasmid-borne blaOXA-72 in Taiwan. BMC Infect. Dis. 13:319. doi: 10.1186/1471-2334-13-319
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Moe, G. R., Tan, S., and Granoff, D. M. (1999). Differences in surface expression of NspA among Neisseria meningitidis group B strains. Infect. Immun. 67, 5664–5675.
Peleg, A. Y., Seifert, H., and Paterson, D. L. (2008). Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21, 538–582. doi: 10.1128/CMR.00058-07
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Penwell, W. A., Shapiro, A. B., Giacobbe, R. A., Gu, R. F., Gao, N., Thresher, J., et al. (2015). The molecular mechanisms of sulbactam antibacterial activity and resistance determinants in Acinetobacter baumannii. Antimicrob. Agents Chemother. 59, 1680–1689. doi: 10.1128/AAC.04808-14
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Quale, J., Bratu, S., Gupta, J., and Landman, D. (2006). Interplay of efflux system, ampC, and oprD expression in carbapenem resistance of Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 50, 1633–1641. doi: 10.1128/AAC.50.5.1633-1641.2006
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Song, J. Y., Kee, S. Y., Hwang, I. S., Seo, Y. B., Jeong, H. W., Kim, W. J., et al. (2007). In vitro activities of carbapenem/sulbactam combination, colistin, colistin/rifampicin combination and tigecycline against carbapenem-resistant Acinetobacter baumannii. J. Antimicrob. Chemother. 60, 317–322. doi: 10.1093/jac/dkm136
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tiwari, V., Kapil, A., and Moganty, R. R. (2012). Carbapenem-hydrolyzing oxacillinase in high resistant strains of Acinetobacter baumannii isolated from India. Microb. Pathog. 53, 81–86. doi: 10.1016/j.micpath.2012.05.004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tiwari, V., Khokar, M. K., Tiwari, M., Barala, S., and Kumar, M. (2014). Anti-bacterial activity of polyvinyl pyrrolidone capped silver nanoparticles on the carbapenem resistant strain of Acinetobacter baumannii. J. Nanomed. Nanotechnol. 5:1000246. doi: 10.4172/2157-7439.1000246
Tiwari, V., and Moganty, R. R. (2014). Conformational stability of OXA-51 beta-lactamase explains its role in carbapenem resistance of Acinetobacter baumannii. J. Biomol. Struct. Dyn. 32, 1406–1420. doi: 10.1080/07391102.2013.819789
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wolff, M., Joly-Guillou, M. L., Farinotti, R., and Carbon, C. (1999). In vivo efficacies of combinations of beta-lactams, beta-lactamase inhibitors, and rifampin against Acinetobacter baumannii in a mouse pneumonia model. Antimicrob. Agents Chemother. 43, 1406–1411.
Zeng, X., and Lin, J. (2013). Beta-lactamase induction and cell wall metabolism in Gram-negative bacteria. Front. Microbiol. 4:128. doi: 10.3389/fmicb.2013.00128
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Keywords: sulbactam, mechanisms of resistance, Acinetobacter baumannii, transformation, Acinetobacter-derived cephalosporinase (ADC)
Citation: Kuo S-C, Lee Y-T, Yang Lauderdale T-L, Huang W-C, Chuang M-F, Chen C-P, Su S-C, Lee K-R and Chen T-L (2015) Contribution of Acinetobacter-derived cephalosporinase-30 to sulbactam resistance in Acinetobacter baumannii. Front. Microbiol. 6:231. doi: 10.3389/fmicb.2015.00231
Received: 25 December 2014; Accepted: 09 March 2015;
Published: 25 March 2015.
Edited by:
Jose L. Martinez, Centro Nacional de Biotecnología, SpainReviewed by:
Agnese Lupo, University of Bern, SwitzerlandVishvanath Tiwari, Central University of Rajasthan, India
Copyright © 2015 Kuo, Lee, Yang Lauderdale, Huang, Chuang, Chen, Su, Lee and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kuan-Rong Lee, Department of Molecular Medicine and Institute of Life Science, National Tsing Hua University, No.101, Section 2, Kuang-Fu Road, Hsin-chu 30013, Taiwan kuan.r.lee@gmail.com;
Te-Li Chen, Institute of Clinical Medicine, School of Medicine, National Yang-Ming University, No. 155, Sec. 2, Linong Street, Taipei 112, Taiwan tecklayyy@gmail.com
 Shu-Chen Kuo
Shu-Chen Kuo Yi-Tzu Lee1,4
Yi-Tzu Lee1,4 Te-Li Chen
Te-Li Chen