
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 03 March 2015
Sec. Evolutionary and Genomic Microbiology
Volume 6 - 2015 | https://doi.org/10.3389/fmicb.2015.00163
This article is part of the Research Topic Mobilomes of bacterial communities in different habitats View all 14 articles
 Sven Jechalke1*
Sven Jechalke1* Melanie Broszat2,3
Melanie Broszat2,3 Friederike Lang4
Friederike Lang4 Christina Siebe5
Christina Siebe5 Kornelia Smalla1
Kornelia Smalla1 Elisabeth Grohmann2,3*
Elisabeth Grohmann2,3*Long-term irrigation with untreated wastewater can lead to an accumulation of antibiotic substances and antibiotic resistance genes in soil. However, little is known so far about effects of wastewater, applied for decades, on the abundance of IncP-1 plasmids and class 1 integrons which may contribute to the accumulation and spread of resistance genes in the environment, and their correlation with heavy metal concentrations. Therefore, a chronosequence of soils that were irrigated with wastewater from 0 to 100 years was sampled in the Mezquital Valley in Mexico in the dry season. The total community DNA was extracted and the absolute and relative abundance (relative to 16S rRNA genes) of antibiotic resistance genes (tet(W), tet(Q), aadA), class 1 integrons (intI1), quaternary ammonium compound resistance genes (qacE+qacEΔ1) and IncP-1 plasmids (korB) were quantified by real-time PCR. Except for intI1 and qacE+qacEΔ1 the abundances of selected genes were below the detection limit in non-irrigated soil. Confirming the results of a previous study, the absolute abundance of 16S rRNA genes in the samples increased significantly over time (linear regression model, p < 0.05) suggesting an increase in bacterial biomass due to repeated irrigation with wastewater. Correspondingly, all tested antibiotic resistance genes as well as intI1 and korB significantly increased in abundance over the period of 100 years of irrigation. In parallel, concentrations of the heavy metals Zn, Cu, Pb, Ni, and Cr significantly increased. However, no significant positive correlations were observed between the relative abundance of selected genes and years of irrigation, indicating no enrichment in the soil bacterial community due to repeated wastewater irrigation or due to a potential co-selection by increasing concentrations of heavy metals.
Wastewater irrigation is a widely used practice worldwide, especially in arid and semiarid regions, to alleviate water shortages in agriculture (Siebe and Cifuentes, 1995; Jimenez and Chávez, 2004; Elifantz et al., 2011; Frenk et al., 2014). More than 20 million ha of land are estimated to be irrigated with wastewater globally, and particularly in developing countries the number of people consuming produce irrigated with poorly or non-treated water is increasing (Amoah et al., 2007; Raschid-Sally and Priyantha, 2008). Since wastewater irrigation provides nutrients that improve plant growth, reduces the need for fertilizer application, and increases the productivity of soils with poor fertility, it is expected to expand further (Gatica and Cytryn, 2013). An example is the Mezquital Valley, where untreated wastewater mixed with surface run-off released from the Mexico City Metropolitan area, located 80 km south of the valley, has been used since more than 100 years (Raschid-Sally and Priyantha, 2008; Dalkmann et al., 2012).
However, besides high concentrations of organic matter, wastewater typically contains large amounts of pollutants including detergents, heavy metals, pharmaceuticals including antibiotics, as well as pathogenic and antibiotic resistant bacteria carrying resistance determinants, class 1 integrons and mobile genetic elements (MGEs) (Baquero et al., 2008; Levantesi et al., 2010; Moura et al., 2010; Chávez et al., 2011; Malik and Aleem, 2011; Bruchmann et al., 2013; Rizzo et al., 2013; Manzetti and Ghisi, 2014). The coexistence of pathogens, antibiotic resistance genes, antibiotics, and heavy metals raises concerns about antibiotic resistance genes being mobilized, propagated, and ultimately transferred to bacteria that are pathogenic to humans (Baquero et al., 2009; Canton, 2009; Wright, 2010). Previous studies showed an increase of pharmaceuticals (Kinney et al., 2006; Ternes et al., 2007; Chen et al., 2011; Tamtam et al., 2011; Dalkmann et al., 2012), resistance determinants, and antibiotic resistant bacteria in soils due to wastewater-irrigation (Dalkmann et al., 2012; Broszat et al., 2014; Chen et al., 2014).
In soils from the Mezquital Valley, long-term wastewater irrigation led to an increase in the abundance of sul genes, encoding resistance toward sulfonamides, and an accumulation of antibiotics. The sulfonamide resistance gene sul1 is often associated with the qacEΔ1 gene on class 1 integrons, especially in clinical settings (Wellington et al., 2013; Gillings, 2014). However, class 1 integrons are also widely disseminated in environmental settings such as soil, sewage sludge and animal slurries, and are able to acquire, exchange, and express genes embedded in gene cassettes, which can include resistance genes for almost all antibiotic families (Moura et al., 2010; Gaze et al., 2011; Stalder et al., 2012; Gillings, 2014; Jechalke et al., 2014b). For example, it was observed that class 1 integrons from manure and manured soil frequently carried aadA gene cassettes, conferring resistance toward streptomycin and spectinomycin (Binh et al., 2009; Heuer et al., 2012). Exposure to antibiotics can up-regulate the intI1 expression by triggering the SOS response and ultimately increase gene cassette recombination rates (Cambray et al., 2011; Hocquet et al., 2012) and might also lead to co-selection of antibiotic resistance, e.g., by heavy metals (Rosewarne et al., 2010). Furthermore, class 1 integrons are often located on MGEs such as transposons and plasmids, e.g., of the IncP-1ε incompatibility group, facilitating their transfer and spread within bacterial communities (Stokes and Gillings, 2011; Heuer et al., 2012; Gillings, 2014). Hence, due to their ability to foster bacterial adaptation to environmental perturbation, class 1 integrons might be used as a universal marker for selective pressure in the environment. However, knowledge of long-term effects of wastewater application containing antibiotics and heavy metals on the abundance of class 1 integrons, related antibiotic resistance genes and MGEs is scarce.
An interesting study site to explore such long-term effects of human impact is located in the Mezquital Valley, where long-term untreated wastewater irrigation for up to 100 years has increased soil organic matter contents as well as nitrogen and phosphorous concentrations up to 2-fold (Siebe, 1998), which in turn resulted in an increase of total soil microbial biomass (Friedel et al., 2000). Also total heavy metal contents including Zn, Cu, Cd, Pb, and Cr as well as bioavailable concentrations of Zn, Cu, and Cd increased with increasing duration of irrigation between 3- and 9-fold in relation to non-irrigated soils (Gutiérrez-Ruiz et al., 1995; Siebe, 1995; Siebe and Cifuentes, 1995). In the present study, the effects of long-term wastewater application on the abundance of IncP-1 plasmids, class 1 integrons and their typically associated resistance genes (qacEΔ1, aadA), as well as municipal wastewater associated resistance genes tet(W) and tet(Q) (Auerbach et al., 2007; Storteboom et al., 2010; Burch et al., 2014) were analyzed in soil from Mezquital Valley. Additionally, concentrations of heavy metals and other inorganic substances were determined and correlated with the duration of irrigation and with the abundance of the selected genes.
The sites of the irrigation chronosequence and the soil sampling were described by Dalkmann et al. (2012). Briefly, sites were selected on behalf of duration of irrigation with untreated wastewater (0, 1.5, 3, 6, 8, 85, and 100 years of irrigation) and soil type. Soils in the Mezquital Valley have been classified as Leptosols, Vertisols, and Phaeozems (Siebe, 1998). In all selected fields water is provided by the same main distribution channel, and each field is irrigated about 10–12 times every year by overflow. Each single irrigation event lasts 6 to 24 h according to the field size. The discharges from Mexico City are dominated by domestic sewage (65%), followed by discharges of the service sector (20%) and the industry (15%). The travel distance to the valley is 80 km, along which the sewage is partly homogenized and large suspended particles and floating materials are removed by sedimentation or retained by bar screens (Gutiérrez-Ruiz et al., 1995). Although water quality is variable in time, we assume that frequent and long lasting irrigations produce similar pollutant loads in all fields over time.
All fields analyzed here were sampled during the dry season. Each individual field was subdivided into four parcels, two on the wastewater inflow side and two on the wastewater outflow side of the field. Samples were taken from each of the parcels by combining 12 subsamples taken with an auger at a depth of 0–30 cm. Soil samples were transported in plastic bags at 4°C and stored at −21°C until further processing.
Total community (TC-) DNA was extracted from 0.5 g soil using the NucleoSpin® Soil Kit according to the manufacturer's protocol (Macherey-Nagel, Düren, Germany). DNA extracted from the four parcels was combined and target genes were quantified in triplicate by quantitative real-time PCR 5′-nuclease assays (qPCR) in a CFX96 real-time PCR detection system (Bio-Rad, Hercules, CA) as previously described for korB of IncP-1 plasmids (Jechalke et al., 2013), class 1 integron integrase gene intI1 (Barraud et al., 2010), quaternary ammonium compound resistance genes qacE and the qacEΔ1 variant (Jechalke et al., 2014b), tetracycline resistance gene tet(W) (Smith et al., 2004), and the streptomycin resistance gene aadA (Walsh et al., 2011), which was frequently found on class 1 integrons (Moura et al., 2007; Schlüter et al., 2007; Binh et al., 2009; Gaze et al., 2011). The 16S rRNA gene (rrn) copies were quantified using the primers BACT1369F and PROK1492R and the probe TM1389F (Suzuki et al., 2000). To adjust for differences in bacterial DNA and amplification efficiency between samples, target numbers of the respective genes were divided by the rrn copy numbers and the results were log transformed. The Pearson product-moment correlations between target gene copy number/relative abundance and years of irrigation were tested using the CORR procedure of the SAS statistical package (p < 0.05; SAS 9.3; SAS Institute Inc., Cary, NC). Multiple comparisons of means were performed using the GLIMMIX procedure (Tukey test, p < 0.05; SAS 9.3).
Soil samples were air-dried (<40°C), sieved to pass a 2 mm nylon mesh, and homogenized using a disc mill with agate stone beakers. Afterwards, 200 mg of homogenized and dried (105°C) samples were placed in clean Teflon vessels and 4 ml of HNO3 (65%, Suprapur, Merck KGaA, Darmstadt, Germany) and 2 ml of H2O2 (35%, Suprapur, Merck KGaA) were added. After 12 h of pre-hydrolysis vessels were locked and subjected to 30 min of microwave treatment (MLS 1200 mega microwave, MLS GmbH, Leutkirch, Germany) with a maximum power of 600 W and a maximum temperature of 110°C. After digestion, samples were quantitatively transferred to volumetric flasks, filled up to 100 ml, and afterwards stored in polythene containers. Analyses of total concentrations of Zn, Cu, Cd, Pb, Ni, Cr, Mn, S, and P were performed by Inductive Coupled Plasma Atomic Emission Spectroscopy (ICP-AES, site-on plasma; Spectro CIROS CCD, SPECTRO Analytical Instruments, Kleve, Germany) from soil samples of years 0, 1.5, 3, 6, 8, 85, and 100. All samples were digested and analyzed in duplicate and concentrations were expressed on dry weight basis.
Potentially bioaccessible concentrations of Zn, Cu, and Cd were analyzed in 1 M NH4NO3 extracts (soil:solution ratio 1:2.5) by graphite furnace atomic absorption spectroscopy (Schöning and Brümmer, 2008) from samples of years 0, 3, 8, 85, and 100 (see above). The soil pH values from samples of all years were measured potentiometrically in a suspension of 0.01 M CaCl2 solution (soil:solution ratio 1:2.5 wt/vol), and the electric conductivities in suspensions with distilled water (soil:water ratio 1:2.5 wt/vol). Total organic carbon (TOC) was quantified in a CHNS/O analyzer (Perkin Elmer 2400 Series II, PerkinElmer, Waltham, MA, USA). Bioaccessible P in samples of years 0, 8, 85, and 100 was quantified colorimetrically in 1 M NaHCO3 extracts (Schlichting et al., 1995).
The Pearson product-moment correlations between the concentrations of metals, target genes and the years of irrigation were tested using the CORR procedure of the SAS statistical package (p < 0.05; SAS 9.3; SAS Institute Inc., Cary, NC).
The abundance of rrn copies per gram soil (dry weight) was quantified and revealed similar results as observed in the previous study by Dalkmann et al. (2012) for the same samples. A significant positive correlation was observed between the rrn copy number and years of irrigation (0–100 years, p < 0.0001, Pearson correlation coefficient 0.75; 1.5–100 years, p < 0.0001, Pearson correlation coefficient 0.79), which coincides with previous findings of an increased microbial biomass in long-term irrigated soils (Friedel et al., 2000). Samples including the period of 1.5–100 years were selected for the analysis of target genes (Figure 1) due to a low abundance in the samples not irrigated with wastewater (0 years), which was predominantly below detection limit of the qPCR method (between log 2–3 gene copies/g dry soil). Significant correlations between gene abundance (per gram dry soil) and years of irrigation were observed for intI1, qacE+qacEΔ1, aadA, korB, and tet(W), while no significant correlation was observed for tet(Q) (Table 1). On the contrary, regarding the abundance of genes relative to rrn copies, no significant correlations were found between years of irrigation and intI1, qacE+qacEΔ1, aadA, korB, and tet(W), while a significant negative correlation was found for tet(Q). The abundance of qacE and qacEΔ1 genes in irrigated soils on average was three times higher than of intI1 (Tukey test, p < 0.05), while the gene abundances of korB, intI1, and aadA were not significantly different. The mean abundances of tet(W) and tet(Q) were about 1 and 2 orders of magnitude lower than the abundance of korB, intI1, and aadA, respectively.
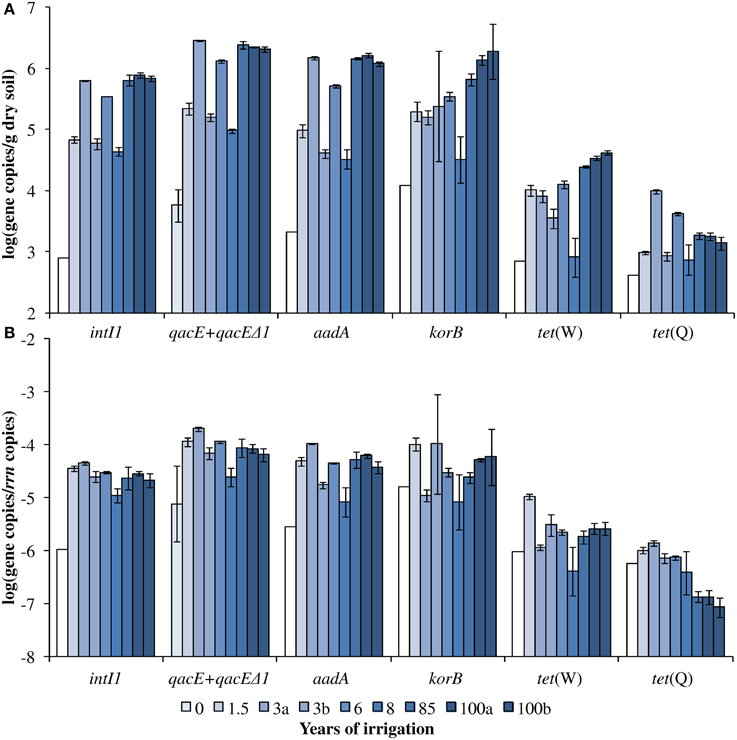
Figure 1. Dry season (A) absolute and (B) relative gene abundance over the period of 100 years of irrigation. Bars and error bars indicate means and respective standard deviations of three technical replicates of one DNA extract from one field, respectively. Fields irrigated for the same period of time are differentiated by small letters (a, b). Except for qacE+qacEΔ1 absolute and relative abundance (indicated by light blue bars), the year 0 values indicate the detection limits for the respective genes (white bars). The detection limits for the relative abundances were calculated based on the mean 16S rRNA gene copy number of year 0 and the detection limits of the respective genes.
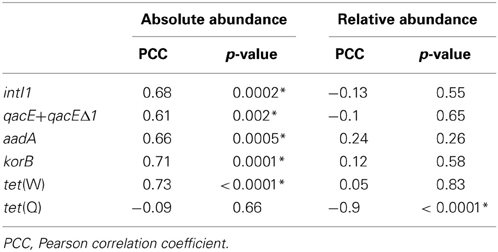
Table 1. Correlation analysis of absolute (log(gene copies/g dry soil)) and relative (log(gene copies/rrn copies)) gene abundances in soil samples from the dry season and the duration of wastewater irrigation (1.5–100 years of irrigation; Pearson correlation, p < 0.05; significance indicated by asterisk).
Total (acid extractable) and potentially bioaccessible (NH4NO3-extractable) concentrations of selected metals were analyzed in dry season samples and ranged from 0 until 0.8 mg/g soil (Table 2, Tables S1, S2). Additionally, total concentrations of P and S in soil samples increased with the length of irrigation.
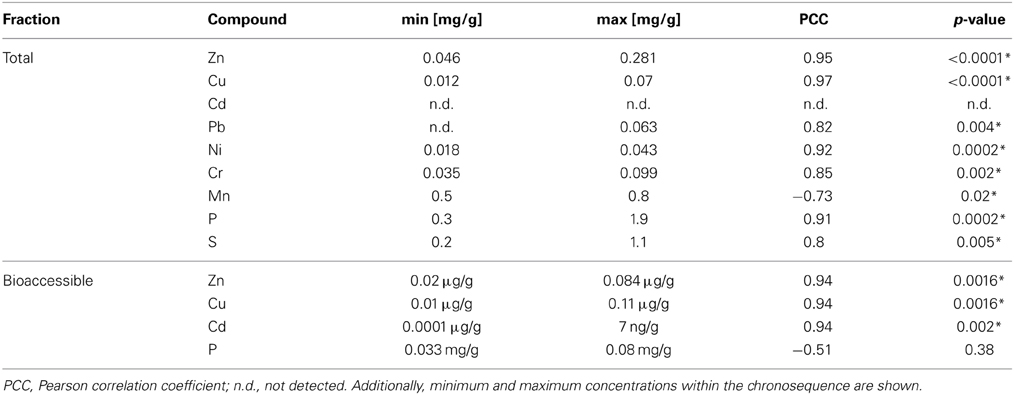
Table 2. Correlation of metals and inorganic compounds with the duration of wastewater irrigation (0–100 years of irrigation; Pearson correlation, p < 0.05; significance indicated by asterisk).
Total soil organic carbon contents also increased significantly with the duration of irrigation (0–100 years irrigation, p = 0.0013, Pearson correlation coefficient 0.89), while soil pH, electrical conductivity, and plant available P contents are not significantly correlated with the years of irrigation.
Absolute and relative abundances of genes intI1, qacE+qacEΔ1, aadA, korB, tet(W), and tet(Q) were correlated with total concentrations of heavy metals Zn, Cu, Pb, Ni, Cr, and Mn as well as with total concentrations of P and S (Table 3). While for the relative abundance of genes only tet(Q) showed significant negative correlations to Zn, Cu, Pb, Ni, Cr, and P, absolute abundances of korB and tet(W) showed positive correlations to Zn, Cu, Pb, Ni, Cr (only korB), P, and S, as well as negative correlations to Mn (Table 3). The concentration of sulfur was positively correlated with absolute abundances of intI1, qacE+qacEΔ1, aadA, korB, and tet(W), but not with tet(Q).
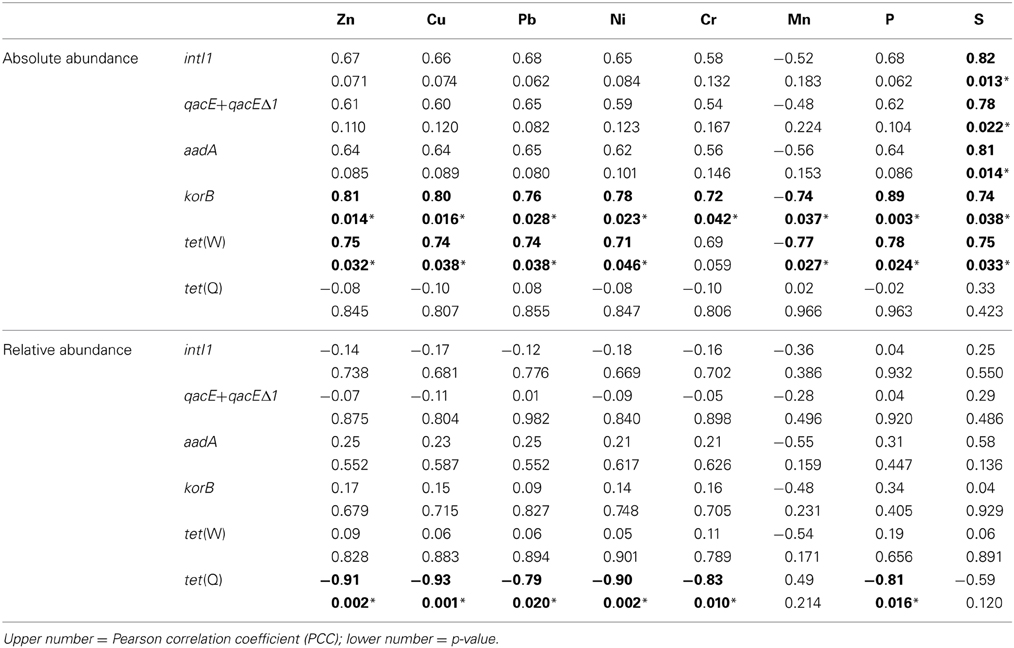
Table 3. Correlation of absolute (log(gene copies/g soil)) and relative abundance (log(gene copies/rrn copies)) of target genes with concentrations of heavy metals, S, and P (1.5–100 years of irrigation; Pearson correlation, p < 0.05; significance indicated in bold and by asterisk).
Correlations between absolute and relative abundances of genes intI1, qacE+qacEΔ1, aadA, korB, tet(W), and tet(Q) and potentially bioaccessible concentrations of heavy metals Zn, Cu, and Cd, as well as with soil electrical conductivity, pH, and TOC were also analyzed (Table 4). Similar to the total concentrations, only the relative abundance of tet(Q) showed a significant correlation to bioaccessible concentrations of Zn, Cu, and Cd, but also to TOC. For the absolute abundance of genes only korB and tet(W) were correlated to the bioaccessible concentrations of Cu. Except for tet(Q) all absolute abundances of tested genes were positively correlated to TOC concentrations. The electrical conductivity showed a significant positive correlation to tet(Q) absolute abundance only. No significant correlations were observed between gene abundances and soil pH.
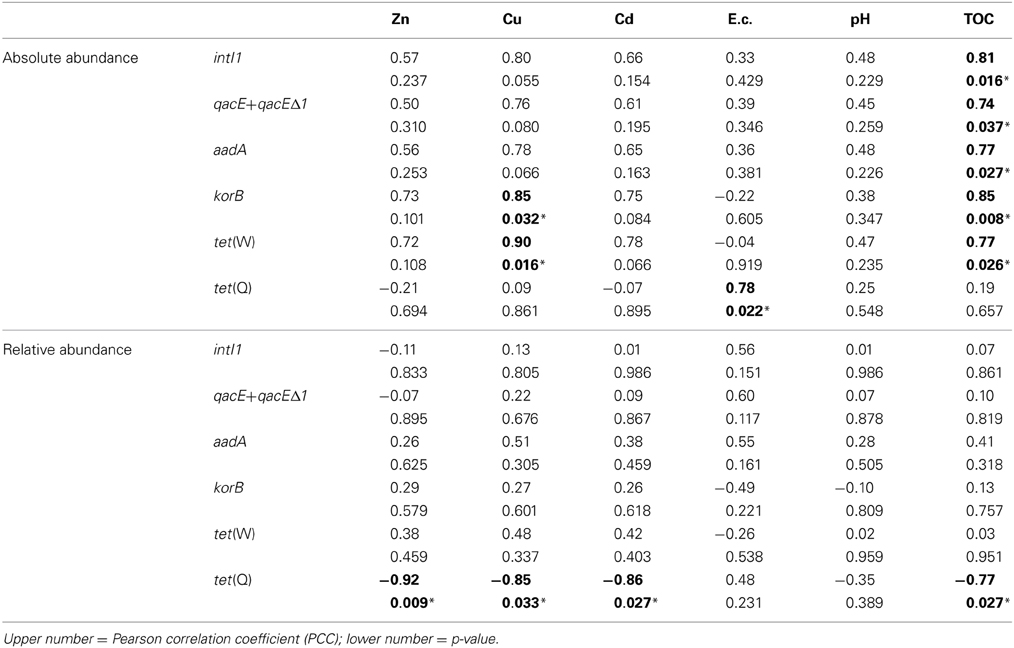
Table 4. Correlation of absolute (log(gene copies/g soil)) and relative abundance (log(gene copies/rrn copies)) of target genes with bioaccessible concentrations of heavy metals as well as with electrical conductivity (E.c.), pH and total organic carbon (TOC) (1.5–100 years of irrigation; Pearson correlation, p < 0.05; significance indicated in bold and by asterisk).
Wastewater typically contains a diverse mixture of pharmaceuticals, pathogenic bacteria, antibiotic resistant bacteria, resistance genes, and heavy metals, which can reach and affect the environment and might pose a risk for human health when wastewater is not properly treated or directly applied as fertilizer (Moura et al., 2010; Bouki et al., 2013; Norton-Brandao et al., 2013; Rivera-Utrilla et al., 2013). In the Mezquital Valley, results from a cross-sectional survey done 20 years ago revealed that intestinal helminth infections represent the highest risk associated with exposure to wastewater irrigation (Blumenthal et al., 1991; Cifuentes et al., 1991). However, no recent epidemiological data exist on the prevalence of disease in the region with respect to wastewater irrigation. Also, little is known worldwide about long-term effects of wastewater application on the accumulation of heavy metals and effects on the abundance of resistance genes and associated MGE in agricultural soil. Therefore, a chronosequence of soils was sampled from the Mezquital Valley that was irrigated with wastewater from zero up to 100 years.
A previous study using the same samples had demonstrated that irrigation with wastewater over several decades led to an accumulation of several pharmaceuticals in soil, such as ciprofloxacin, sulfamethoxazole, and carbamazepine (Dalkmann et al., 2012). At the same time, long-term irrigation with waste-water led to a reduced sorption of sulfamethoxazole, while the sorption of ciprofloxacin was not affected (Dalkmann et al., 2014). Furthermore, wastewater application was correlated with an increase in organic carbon, total microbial biomass and activity accompanied by an increase in absolute numbers of sul1 and sul2 genes, conferring resistance to sulfonamide antibiotics such as sulfamethoxazole (Siebe and Fischer, 1996; Friedel et al., 2000; Dalkmann et al., 2012). However, the abundance of sul genes relative to 16S rRNA genes did not correlate with the duration of irrigation (Dalkmann et al., 2012), indicating no enrichment of sulfonamide resistant populations within the soil bacterial community.
In this study we observed, besides the increase in absolute abundance of rrn copies, which was already demonstrated by Dalkmann et al. (2012) using the same soil samples, an increase in absolute abundance of intI1, qacE+qacEΔ1, aadA, korB, and tet(W) genes with the duration of irrigation (Table 1). No significant increase was observed for tet(Q) which was detected in very low abundance. Additionally, confirming the results of previous studies (Siebe, 1994; Siebe and Cifuentes, 1995; Guedron et al., 2014), we could show that the concentrations of the heavy metals Zn, Cu, Pb, Ni, and Cr increased with the duration of irrigation, while Mn decreased and Cd was below detection limit (Table 2). The decrease of Mn in the soils is coherent with the frequent flooding with large amounts of water, which induces temporal reducing conditions in the soil under which Mn is mobilized and lixiviated with the percolating water as Mn2+ (Siebe and Fischer, 1996). Additionally, NH4NO3-extractable concentrations of Zn, Cu, and Cd, which are considered to represent the bioaccessible fraction due to the gentle extraction, increased with the duration of irrigation (Siebe, 1994).
Similar to antibiotics, which interact with the soil solid phase in sorption and desorption reactions controlling their biotransformation and biological effects (Jechalke et al., 2014a), metals are not readily degraded and thus can affect bacterial populations for extended periods (Stepanauskas et al., 2005). Genes encoding metal resistance determinants are frequently found on MGEs such as transposons and plasmids, which also carry integrons and antibiotic resistance genes (Baker-Austin et al., 2006). Thus, the exposure of bacterial communities to metals can select for metal-resistant strains but also indirectly for bacteria resistant to unrelated toxicants, such as antibiotics, by co-selection processes (Stepanauskas et al., 2005; Baker-Austin et al., 2006). Consequently, a number of studies comparing contaminated and reference sites observed direct or indirect associations between the presence of metals and elevated antibiotic resistance and abundance of class 1 integrons (Baker-Austin et al., 2006; Wright et al., 2008; Rosewarne et al., 2010). Class 1 integrons, which are able to acquire, exchange, and accumulate resistance genes embedded in gene cassettes (Gillings, 2014; Jechalke et al., 2014b), were frequently found on plasmids of the IncP-1ε group (Heuer et al., 2012). These plasmids are able to efficiently transfer and replicate in a broad range of hosts and are widely spread in clinical and environmental settings, such as agricultural soil and wastewater, where they might be important vectors of antibiotic and heavy metal resistance genes (Schlüter et al., 2007; Sen et al., 2011; Heuer et al., 2012; Popowska and Krawczyk-Balska, 2013).
However, in this study no significant positive correlations between relative abundance of resistance genes, class 1 integrons, IncP-1 plasmids and years of irrigation were observed, indicating no enrichment in the soil bacterial community. Similarly, no positive correlations were observed between the relative abundance of genes and total or bioaccessible concentrations of heavy metals or total and CaCl2 extracted concentrations of antibiotics (obtained from Supporting Information of Dalkmann et al., 2012, data not shown). Nevertheless, except for qacE+qacEΔ1 the absolute and relative abundances of the detected resistance genes, class 1 integrons and IncP-1 plasmids were below the detection limit in the soils from rain-fed fields, i.e., soils never irrigated with wastewater, but were detectable in irrigated soils, pointing to the wastewater irrigation as a common source of these genes in soil. The large absolute numbers of intI1, qacE+qacEΔ1, and aadA genes in soils irrigated for 3 and 6 years could eventually be due to applications of cattle manure to these fields, since both fields show a relatively large TOC content (more similar to that of long-term wastewater-irrigated fields, Table S2). Duration of irrigation is for sure not longer than 3 and 6 years, since the wastewater distribution channel was constructed recently. However, it cannot be excluded that a few farmers applied manure irregularly on rain-fed fields before wastewater was available as fertilizer.
The abundances of tet(W) and tet(Q) were below detection limit in non-irrigated soils, which is in agreement with a previous study reporting low abundances of tet(W) and tet(Q) in agricultural soils without manure fertilization (Kyselková et al., 2013), and still close to the detection limit after irrigation, which might indicate a low concentration of these genes and their hosts in the irrigation water.
Positive correlations were observed between absolute abundances of korB, tet(W), and the bioaccessible concentration of Cu (Table 4), which further supports a common source of IncP-1 plasmids, tetracycline resistance genes, and this heavy metal. Correlations between absolute abundances of korB, tet(W), and Zn were significant for the total concentrations of this heavy metal but not for its bioaccessible fraction, which might indicate a less relevant association compared to Cu.
Not much data are available regarding minimum co-selective concentrations (MCCs) of heavy metals in soil, that potentially drive co-selection of antibiotic resistance (Seiler and Berendonk, 2012). In a field study of Berg et al. (2005), a concentration of 116.7 mg Cu/kg soil was associated with an increase of the absolute number of antibiotic resistant bacteria. The maximum concentration of 70 mg acid extractable Cu/kg soil detected in the Mexican soils was smaller than the one observed by Berg and colleagues, which might explain that no enrichment of resistance genes, IncP-1 plasmids and class 1 integrons occurred in the soil bacterial community over the period of 100 years of irrigation.
MCCs for Zn ranged from 19.61 μg/L in water samples to 22.75 mg/kg and 46.1 mg/kg in manure and sediment samples, respectively (Seiler and Berendonk, 2012), which is in the range of 46–281 mg Zn/kg detected in the Mexican soil samples but even lower than the 800 μg/L measured in wastewater samples from the Mezquital Valley (Guedron et al., 2014). This might indicate that higher concentrations of Zn are necessary in soil to co-select for antibiotic resistance. In accordance with this, Knapp et al. (2011) did not observe an increased abundance of antibiotic resistance genes in correlation with elevated Zn concentrations in archived Scottish soils, and similar to the results of the present study no positive correlations were found between metal concentrations and tet(Q) relative abundance. However, similar to the present study, Knapp et al. (2011) found correlations between Ni and Cu concentrations and tet(W) relative abundance.
Confirming a previous study by Gaze et al. (2011) for pig slurry, but in contrast to a study by Jechalke et al. (2014b) for bulk soil and rhizosphere, the abundance of qacE and qacEΔ1 in irrigated Mexican soils were about three times higher than of intI1 (Tukey test, p < 0.05), suggesting that more than one copy of these genes was associated with class 1 integrons or that these genes are carried by other integron classes, MGEs, or located chromosomally. The mean abundances of korB, intI1, and aadA genes were not significantly different, which might suggest that class 1 integrons and the streptomycin resistance gene aadA were associated with IncP-1 plasmids.
Besides changes in metal concentrations, a significant increase was also observed for S and P concentrations along with extended duration of irrigation. Although Shi et al. (2013) could show by a modeling approach that soil concentrations of S and P affected bacterial Cu resistance, in this study positive correlations between concentrations of S and P and antibiotic resistance levels were only observed for the absolute but not for the relative abundance, suggesting that there was no co-selection of antibiotic resistance genes at present S and P concentrations.
In summary, wastewater irrigation in Mezquital Valley, Mexico, over a period of 100 years led to an increase in bacterial biomass in soil and, accordingly, to an increase in absolute abundance of intI1, qacE+qacEΔ1, aadA, korB, and tet(W) genes. In parallel, concentrations of Zn, Cu, Pb, Ni, Cr, P, and S significantly increased. However, no significant positive correlations were observed between the relative abundance of selected genes and years of irrigation or concentration of heavy metals, indicating no enrichment in the soil bacterial community due to wastewater irrigation or due to a potential selection by increasing concentrations of heavy metals and antibiotic compounds. Nevertheless, the increase in gene copy numbers per gram of soil along with extended duration of irrigation might lead to a higher exposure of field crops and farmers to resistant bacteria, resistance genes and MGEs. Further epidemiological surveys are needed to address if the increased presence of antibiotic resistance genes in the soils of this region poses a health risk for the inhabitants of the region and the consumers of the crops.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
SJ was funded by the Federal Environment Agency (Umweltbundesamt) (FKZ 3713 63 402). EG and MB are grateful to the Deutsche Forschungsgemeinschaft (DFG, grants GR1792/4-1 and GR1792/4-2) for support, CS is grateful to the Mexican Consejo Nacional de Ciencia y Tecnología (CONACYT, grants CB 83767 and I 443 0110-193-10). Mario Cayetano and Kumiko Shimada analyzed the bioaccessible heavy metal contents. We thank Ilse-Marie Jungkurth for proofreading the manuscript.
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fmicb.2015.00163/abstract
Amoah, P., Drechsel, P., Abaidoo, R. C., and Henseler, M. (2007). Irrigated urban vegetable production in Ghana: microbiological contamination in farms and markets and associated consumer risk groups. J. Water Health 5, 455–466. doi: 10.2166/wh.2007.041
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Auerbach, E. A., Seyfried, E. E., and McMahon, K. D. (2007). Tetracycline resistance genes in activated sludge wastewater treatment plants. Water Res. 41, 1143–1151. doi: 10.1016/j.watres.2006.11.045
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Baker-Austin, C., Wright, M. S., Stepanauskas, R., and McArthur, J. V. (2006). Co-selection of antibiotic and metal resistance. Trends Microbiol. 14, 176–182. doi: 10.1016/j.tim.2006.02.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Baquero, F., Alvarez-Ortega, C., and Martinez, J. L. (2009). Ecology and evolution of antibiotic resistance. Environ. Microbiol. Rep. 1, 469–476. doi: 10.1111/j.1758-2229.2009.00053.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Baquero, F., Martinez, J. L., and Canton, R. (2008). Antibiotics and antibiotic resistance in water environments. Curr. Opin. Biotechnol. 19, 260–265. doi: 10.1016/j.copbio.2008.05.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Barraud, O., Baclet, M. C., Denis, F., and Ploy, M. C. (2010). Quantitative multiplex real-time PCR for detecting class 1, 2 and 3 integrons. J. Antimicrob. Chemother. 65, 1642–1645. doi: 10.1093/jac/dkq167
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Berg, J., Tom-Petersen, A., and Nybroe, O. (2005). Copper amendment of agricultural soil selects for bacterial antibiotic resistance in the field. Lett. Appl. Microbiol. 40, 146–151. doi: 10.1111/j.1472-765X.2004.01650.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Binh, C. T. T., Heuer, H., Kaupenjohann, M., and Smalla, K. (2009). Diverse aadA gene cassettes on class 1 integrons introduced into soil via spread manure. Res. Microbiol. 160, 427–433. doi: 10.1016/j.resmic.2009.06.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Blumenthal, U. J., Abisudjak, B., Cifuentes, E., Bennett, S., and Ruiz-Palacios, G. (1991). Recent epidemiological studies to test microbiological quality guidelines for wastewater use in agriculture and aquaculture. Public Health Rev. 19, 237–242.
Bouki, C., Venieri, D., and Diamadopoulos, E. (2013). Detection and fate of antibiotic resistant bacteria in wastewater treatment plants: a review. Ecotoxicol. Environ. Saf. 91, 1–9. doi: 10.1016/j.ecoenv.2013.01.016
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Broszat, M., Nacke, H., Blasi, R., Siebe, C., Huebner, J., Daniel, R., et al. (2014). Wastewater irrigation increases abundance of potentially harmful Gammaproteobacteria in soils from Mezquital Valley, Mexico. Appl. Environ. Microbiol. 80, 5282–5291. doi: 10.1128/AEM.01295-14
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bruchmann, J., Kirchen, S., and Schwartz, T. (2013). Sub-inhibitory concentrations of antibiotics and wastewater influencing biofilm formation and gene expression of multi-resistant Pseudomonas aeruginosa wastewater isolates. Environ. Sci. Pollut. Res. 20, 3539–3549. doi: 10.1007/s11356-013-1521-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Burch, T. R., Sadowsky, M. J., and LaPara, T. M. (2014). Fate of antibiotic resistance genes and class 1 integrons in soil microcosms following the application of treated residual municipal wastewater solids. Environ. Sci. Technol. 48, 5620–5627. doi: 10.1021/es501098g
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cambray, G., Sanchez-Alberola, N., Campoy, S., Guerin, E., Da Re, S., Gonzalez-Zorn, B., et al. (2011). Prevalence of SOS-mediated control of integron integrase expression as an adaptive trait of chromosomal and mobile integrons. Mob. DNA 2:6 doi: 10.1186/1759-8753-2-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Canton, R. (2009). Antibiotic resistance genes from the environment: a perspective through newly identified antibiotic resistance mechanisms in the clinical setting. Clin. Microbiol. Infect. 15, 20–25. doi: 10.1111/j.1469-0691.2008.02679.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chávez, A., Maya, C., Gibson, R., and Jiménez, B. (2011). The removal of microorganisms and organic micropollutants from wastewater during infiltration to aquifers after irrigation of farmland in the Tula Valley, Mexico. Environ. Pollut. 159, 1354–1362. doi: 10.1016/j.envpol.2011.01.008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chen, C., Li, J., Chen, P., Ding, R., Zhang, P. F., and Li, X. (2014). Occurrence of antibiotics and antibiotic resistances in soils from wastewater irrigation areas in Beijing and Tianjin, China. Environ. Pollut. 193, 94–101. doi: 10.1016/j.envpol.2014.06.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chen, F., Ying, G. G., Kong, L. X., Wang, L., Zhao, J. L., Zhou, L. J., et al. (2011). Distribution and accumulation of endocrine-disrupting chemicals and pharmaceuticals in wastewater irrigated soils in Hebei, China. Environ. Pollut. 159, 1490–1498. doi: 10.1016/j.envpol.2011.03.016
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cifuentes, E., Blumenthal, U., Ruiz-Palacios, G., and Bennett, S. (1991). Health impact evaluation of wastewater use in Mexico. Public Health Rev. 19, 243–250.
Dalkmann, P., Broszat, M., Siebe, C., Willaschek, E., Sakinc, T., Huebner, J., et al. (2012). Accumulation of pharmaceuticals, Enterococcus, and resistance genes in soils irrigated with wastewater for zero to 100 years in central Mexico. PLoS ONE 7:e45397. doi: 10.1371/journal.pone.0045397
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dalkmann, P., Willaschek, E., Schiedung, H., Bornemann, L., Siebe, C., and Siemens, J. (2014). Long-term wastewater irrigation reduces sulfamethoxazole sorption, but not ciprofloxacin binding in Mexican soils. J. Environ. Qual. 43, 964–970. doi: 10.2134/jeq2013.11.0473
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Elifantz, H., Kautsky, L., Mor-Yosef, M., Tarchitzky, J., Bar-Tal, A., Chen, Y. N., et al. (2011). Microbial activity and organic matter dynamics during 4 years of irrigation with treated wastewater. Microb. Ecol. 62, 973–981. doi: 10.1007/s00248-011-9867-y
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Frenk, S., Hadar, Y., and Minz, D. (2014). Resilience of soil bacterial community to irrigation with water of different qualities under Mediterranean climate. Environ. Microbiol. 16, 559–569. doi: 10.1111/1462-2920.12183
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Friedel, J. K., Langer, T., Siebe, C., and Stahr, K. (2000). Effects of long-term waste water irrigation on soil organic matter, soil microbial biomass and its activities in central Mexico. Biol. Fertil. Soils 31, 414–421. doi: 10.1007/s003749900188
Gatica, J., and Cytryn, E. (2013). Impact of treated wastewater irrigation on antibiotic resistance in the soil microbiome. Environ. Sci. Pollut. Res. 20, 3529–3538. doi: 10.1007/s11356-013-1505-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gaze, W. H., Zhang, L. H., Abdouslam, N. A., Hawkey, P. M., Calvo-Bado, L., Royle, J., et al. (2011). Impacts of anthropogenic activity on the ecology of class 1 integrons and integron-associated genes in the environment. ISME J. 5, 1253–1261. doi: 10.1038/ismej.2011.15
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gillings, M. R. (2014). Integrons: past, present, and future. Microbiol. Mol. Biol. Rev. 78, 257–277. doi: 10.1128/MMBR.00056-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Guedron, S., Duwig, C., Lucia Prado, B., Point, D., Giovana Flores, M., and Siebe, C. (2014). (Methyl) mercury, arsenic, and lead contamination of the world's largest wastewater irrigation system: the Mezquital Valley (Hidalgo State-Mexico). Water Air Soil Poll. 225, 2045. doi: 10.1007/s11270-014-2045-3
Gutiérrez-Ruiz, M. E., Siebe, C., and Sommer, I. (1995). Effects of land application of waste water from Mexico City on soil fertility and heavy metal accumulation: a bibliographical review. Environ. Rev. 3, 318–330. doi: 10.1139/a95-017
Heuer, H., Binh, C. T. T., Jechalke, S., Kopmann, C., Zimmerling, U., Krögerrecklenfort, E., et al. (2012). IncP-1ε plasmids are important vectors of antibiotic resistance genes in agricultural systems: diversification driven by class 1 integron gene cassettes. Front. Microbiol. 3:2. doi: 10.3389/fmicb.2012.00002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hocquet, D., Llanes, C., Thouverez, M., Kulasekara, H. D., Bertrand, X., Plesiat, P., et al. (2012). Evidence for induction of integron-based antibiotic resistance by the SOS response in a clinical setting. PLoS Pathog. 8:e1002778. doi: 10.1371/journal.ppat.1002778
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jechalke, S., Dealtry, S., Smalla, K., and Heuer, H. (2013). Quantification of IncP-1 plasmid prevalence in environmental samples. Appl. Environ. Microbiol. 79, 1410–1413. doi: 10.1128/AEM.03728-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jechalke, S., Heuer, H., Siemens, J., Amelung, W., and Smalla, K. (2014a). Fate and effects of veterinary antibiotics in soil. Trends Microbiol. 22, 536–545. doi: 10.1016/j.tim.2014.05.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jechalke, S., Schreiter, S., Wolters, B., Dealtry, S., Heuer, H., and Smalla, K. (2014b). Widespread dissemination of class 1 integron components in soils and related ecosystems as revealed by cultivation-independent analysis. Front. Microbiol. 4:420. doi: 10.3389/fmicb.2013.00420
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jimenez, B., and Chávez, A. (2004). Quality assessment of an aquifer recharged with wastewater for its potential use as drinking source: “El Mezquital Valley” case. Water Sci. Technol. 50, 269–276.
Kinney, C. A., Furlong, E. T., Werner, S. L., and Cahill, J. D. (2006). Presence and distribution of wastewater-derived pharmaceuticals in soil irrigated with reclaimed water. Environ. Toxicol. Chem. 25, 317–326. doi: 10.1897/05-187R.1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Knapp, C. W., McCluskey, S. M., Singh, B. K., Campbell, C. D., Hudson, G., and Graham, D. W. (2011). Antibiotic resistance gene abundances correlate with metal and geochemical conditions in archived Scottish soils. PLoS ONE 6:e27300. doi: 10.1371/journal.pone.0027300
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kyselková, M., Jirout, J., Chronakova, A., Vrchotova, N., Bradley, R., Schmitt, H., et al. (2013). Cow excrements enhance the occurrence of tetracycline resistance genes in soil regardless of their oxytetracycline content. Chemosphere 93, 2413–2418. doi: 10.1016/j.chemosphere.2013.08.058
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Levantesi, C., La Mantia, R., Masciopinto, C., Bockelmann, U., Ayuso-Gabella, M. N., Salgot, M., et al. (2010). Quantification of pathogenic microorganisms and microbial indicators in three wastewater reclamation and managed aquifer recharge facilities in Europe. Sci. Total Environ. 408, 4923–4930. doi: 10.1016/j.scitotenv.2010.07.042
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Malik, A., and Aleem, A. (2011). Incidence of metal and antibiotic resistance in Pseudomonas spp. from the river water, agricultural soil irrigated with wastewater and groundwater. Environ. Monit. Assess. 178, 293–308. doi: 10.1007/s10661-010-1690-2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Manzetti, S., and Ghisi, R. (2014). The environmental release and fate of antibiotics. Mar. Pollut. Bull. 79, 7–15. doi: 10.1016/j.marpolbul.2014.01.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Moura, A., Henriques, I., Ribeiro, R., and Correia, A. (2007). Prevalence and characterization of integrons from bacteria isolated from a slaughterhouse wastewater treatment plant. J. Antimicrob. Chemother. 60, 1243–1250. doi: 10.1093/jac/dkm340
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Moura, A., Henriques, I., Smalla, K., and Correia, A. (2010). Wastewater bacterial communities bring together broad-host range plasmids, integrons and a wide diversity of uncharacterized gene cassettes. Res. Microbiol. 161, 58–66. doi: 10.1016/j.resmic.2009.11.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Norton-Brandao, D., Scherrenberg, S. M., and van Lier, J. B. (2013). Reclamation of used urban waters for irrigation purposes - A review of treatment technologies. J. Environ. Manag. 122, 85–98. doi: 10.1016/j.jenvman.2013.03.012
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Popowska, M., and Krawczyk-Balska, A. (2013). Broad-host-range IncP-1 plasmids and their resistance potential. Front. Microbiol. 4:44. doi: 10.3389/fmicb.2013.00044
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Raschid-Sally, L., and Priyantha, J. (2008). Drivers and Characteristics of Wastewater Agriculture in Developing Countries: Results from a Global Assessment. Research Report 127, Colombo: International Water Management Institute.
Rivera-Utrilla, J., Sanchez-Polo, M., Ferro-Garcia, M. A., Prados-Joya, G., and Ocampo-Perez, R. (2013). Pharmaceuticals as emerging contaminants and their removal from water. A review. Chemosphere 93, 1268–1287. doi: 10.1016/j.chemosphere.2013.07.059
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rizzo, L., Manaia, C., Merlin, C., Schwartz, T., Dagot, C., Ploy, M. C., et al. (2013). Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: a review. Sci. Total Environ. 447, 345–360. doi: 10.1016/j.scitotenv.2013.01.032
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rosewarne, C. P., Pettigrove, V., Stokes, H. W., and Parsons, Y. M. (2010). Class 1 integrons in benthic bacterial communities: abundance, association with Tn402-like transposition modules and evidence for coselection with heavy-metal resistance. FEMS Microbiol. Ecol. 72, 35–46. doi: 10.1111/j.1574-6941.2009.00823.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schlüter, A., Szczepanowski, R., Pühler, A., and Top, E. M. (2007). Genomics of IncP-1 antibiotic resistance plasmids isolated from wastewater treatment plants provides evidence for a widely accessible drug resistance gene pool. FEMS Microbiol. Rev. 31, 449–477. doi: 10.1111/j.1574-6976.2007.00074.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schöning, A., and Brümmer, G. W. (2008). Extraction of mobile element fractions in forest soils using ammonium nitrate and ammonium chloride. J. Plant Nutr. Soil Sci. 171, 392–398. doi: 10.1002/jpln.200625169
Seiler, C., and Berendonk, T. U. (2012). Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Front. Microbiol. 3:399. doi: 10.3389/fmicb.2012.00399
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sen, D., Van der Auwera, G. A., Rogers, L. M., Thomas, C. M., Brown, C. J., and Top, E. M. (2011). Broad-host-range plasmids from agricultural soils have IncP-1 backbones with diverse accessory genes. Appl. Environ. Microbiol. 77, 7975–7983. doi: 10.1128/AEM.05439-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shi, Z., Cao, Z., Qin, D., Zhu, W., Wang, Q., Li, M., et al. (2013). Correlation models between environmental factors and bacterial resistance to antimony and copper. PLoS ONE 8:e78533. doi: 10.1371/journal.pone.0078533
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Siebe, C. (1994). Accumulación y disponibilidad de metales pesados en suelos regados con aguas residuales en el distrito de riego 03, Tula, Hidalgo, México. Revista International de Contaminación Ambiental 10, 15–21.
Siebe, C. (1995). Heavy metal availability to plants in soils irrigated with wastewater from Mexico City. Water Sci. Technol. 32, 29–34. doi: 10.1016/0273-1223(96)00135-7
Siebe, C. (1998). Nutrient inputs to soils and their uptake by alfalfa through long-term irrigation with untreated sewage effluent in Mexico. Soil Use Manag. 14, 119–122. doi: 10.1111/j.1475-2743.1998.tb00628.x
Siebe, C., and Cifuentes, E. (1995). Environmental impact of wastewater irrigation in central Mexico: an overview. Int. J. Environ. Heal. R. 5, 161–173. doi: 10.1080/09603129509356845
Siebe, C., and Fischer, W. R. (1996). Effect of long-term irrigation with untreated sewage effluents on soil properties and heavy metal adsorption of Leptosols and Vertisols in Central Mexico. Z. Pflanzenernähr. Bodenk. 159, 357–364. doi: 10.1002/jpln.1996.3581590408
Smith, M. S., Yang, R. K., Knapp, C. W., Niu, Y. F., Peak, N., Hanfelt, M. M., et al. (2004). Quantification of tetracycline resistance genes in feedlot lagoons by real-time PCR. Appl. Environ. Microbiol. 70, 7372–7377. doi: 10.1128/AEM.70.12.7372-7377.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Stalder, T., Barraud, O., Casellas, M., Dagot, C., and Ploy, M.-C. (2012). Integron involvement in environmental spread of antibiotic resistance. Front. Microbiol. 3:119. doi: 10.3389/fmicb.2012.00119
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Stepanauskas, R., Glenn, T. C., Jagoe, C. H., Tuckfield, R. C., Lindell, A. H., and McArthur, J. V. (2005). Elevated microbial tolerance to metals and antibiotics in metal-contaminated industrial environments. Environ. Sci. Technol. 39, 3671–3678. doi: 10.1021/es048468f
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Stokes, H. W., and Gillings, M. R. (2011). Gene flow, mobile genetic elements and the recruitment of antibiotic resistance genes into Gram-negative pathogens. FEMS Microbiol. Rev. 35, 790–819. doi: 10.1111/j.1574-6976.2011.00273.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Storteboom, H., Arabi, M., Davis, J. G., Crimi, B., and Pruden, A. (2010). Identification of antibiotic-resistance-gene molecular signatures suitable as tracers of pristine river, urban, and agricultural sources. Environ. Sci. Technol. 44, 1947–1953. doi: 10.1021/es902893f
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Suzuki, M. T., Taylor, L. T., and DeLong, E. F. (2000). Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 66, 4605–4614. doi: 10.1128/AEM.66.11.4605-4614.2000
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tamtam, F., van Oort, F., Le Bot, B., Dinh, T., Mompelat, S., Chevreuil, M., et al. (2011). Assessing the fate of antibiotic contaminants in metal contaminated soils four years after cessation of long-term waste water irrigation. Sci. Total Environ. 409, 540–547. doi: 10.1016/j.scitotenv.2010.10.033
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ternes, T. A., Bonerz, M., Herrmann, N., Teiser, B., and Andersen, H. R. (2007). Irrigation of treated wastewater in Braunschweig, Germany: an option to remove pharmaceuticals and musk fragrances. Chemosphere 66, 894–904. doi: 10.1016/j.chemosphere.2006.06.035
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Walsh, F., Ingenfeld, A., Zampicolli, M., Hilber-Bodmer, M., Frey, J. E., and Duffy, B. (2011). Real-time PCR methods for quantitative monitoring of streptomycin and tetracycline resistance genes in agricultural ecosystems. J. Microbiol. Methods 86, 150–155. doi: 10.1016/j.mimet.2011.04.011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wellington, E. M. H., Boxall, A. B. A., Cross, P., Feil, E. J., Gaze, W. H., Hawkey, P. M., et al. (2013). The role of the natural environment in the emergence of antibiotic resistance in Gram-negative bacteria. Lancet Infect. Dis. 13, 155–165. doi: 10.1016/S1473-3099(12)70317-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wright, G. D. (2010). Antibiotic resistance in the environment: a link to the clinic? Curr. Opin. Microbiol. 13, 589–594. doi: 10.1016/j.mib.2010.08.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wright, M. S., Baker-Austin, C., Lindell, A. H., Stepanauskas, R., Stokes, H. W., and McArthur, J. V. (2008). Influence of industrial contamination on mobile genetic elements: class 1 integron abundance and gene cassette structure in aquatic bacterial communities. ISME J. 2, 417–428. doi: 10.1038/ismej.2008.8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: wastewater irrigation, IncP-1 plasmids, class 1 integrons, quaternary ammonium compound resistance, tetracycline resistance, aminoglycoside resistance
Citation: Jechalke S, Broszat M, Lang F, Siebe C, Smalla K and Grohmann E (2015) Effects of 100 years wastewater irrigation on resistance genes, class 1 integrons and IncP-1 plasmids in Mexican soil. Front. Microbiol. 6:163. doi: 10.3389/fmicb.2015.00163
Received: 31 October 2014; Accepted: 12 February 2015;
Published online: 03 March 2015.
Edited by:
Eva M. Top, University of Idaho, USAReviewed by:
Thibault Stalder, Univerité de Limoges, FranceCopyright © 2015 Jechalke, Broszat, Lang, Siebe, Smalla and Grohmann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sven Jechalke, Institute for Epidemiology and Pathogen Diagnostics, Julius Kühn-Institut – Federal Research Centre for Cultivated Plants (JKI), Messeweg 11-12, 38104 Braunschweig, Germany e-mail:c3Zlbi5qZWNoYWxrZUBqa2kuYnVuZC5kZQ==;
Elisabeth Grohmann, Department of Infectious Diseases, University Hospital Freiburg, Hugstetter Straße 55, 79106 Freiburg, Germany e-mail:ZWxpc2FiZXRoLmdyb2htYW5uQGdvb2dsZW1haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.