- 1Food Safety and Enteric Pathogens Research Unit, National Animal Disease Center, Agricultural Research Service, United States Department of Agriculture, Ames, IA, USA
- 2Agroecosystems Management Research Unit, National Laboratory for Agriculture and the Environment, Agricultural Research Service, United States Department of Agriculture, Ames, IA, USA
Salmonella enterica serovar Typhimurium is one of the most common serovars isolated from humans and livestock, and over 35% of these isolates are resistant to three or more antibiotics. Multidrug-resistant (MDR) Salmonella is a public health concern as it is associated with increased morbidity in patients compared to antibiotic sensitive strains, though it is unknown how the antibiotic resistant isolates lead to a more severe infection. Cellular invasion is temporally regulated in Salmonella and normally occurs during late-log and stationary growth. However, our previous work determined that a 30 min exposure to a sub-inhibitory concentration of tetracycline can induce the full invasion phenotype during early-log growth in certain MDR S. Typhimurium isolates. The current study examined whether sub-inhibitory concentrations of other antibiotics could also induce the invasiveness in the same set of isolates. Ampicillin and streptomycin had no effect on invasion, but certain concentrations of chloramphenicol were found to induce invasion in a subset of isolates. Two of the isolates induced by chloramphenicol were also inducible by tetracycline. RNA-seq analyses demonstrated that chloramphenicol and tetracycline both down-regulated motility gene expression, while up-regulating genes associated with attachment, invasion, and intracellular survival. Eleven fimbrial operons were up-regulated, which is notable as only three fimbrial operons were thought to be inducible in culture; six of these up-regulated operons have been reported to play a role in Salmonella persistence in mice. Overall, these data show that the normal progression of the genetic pathways that regulate invasion can be expedited to occur within 30 min due to antibiotic exposure. This altered invasion process due to antibiotics may play a role in the increased intensity and duration of infection observed in patients with MDR Salmonella.
Introduction
Salmonella is a highly prevalent bacterial food-borne disease. Annually in the United States, human salmonellosis is estimated to cause over 1 million cases, 19,000 hospitalizations, 350 deaths, and $2.6 billion in associated costs (Economic Research Service, 2009; Scallan et al., 2011). Salmonella enterica serovar Typhimurium is one of the most commonly isolated salmonellae in both humans and livestock, and there is a high incidence of antibiotic resistance among these isolates. According to a 10-year average from the National Antimicrobial Resistance Monitoring System (NARMS), isolates resistant to three or more antibiotics are frequent among S. Typhimurium from humans (35%), chickens (41%), cattle (63%), and swine (78%) (CDC, 2012). The NARMS report also lists that the four most common antibiotics to which human isolates of S. Typhimurium are resistant are tetracycline (33% of isolates), ampicillin (31%), streptomycin (31%), and chloramphenicol (24%). Resistance to these antibiotics is mediated by antibiotic-specific mechanisms that can be acquired via horizontal transfer (tetA, floR, strA, etc.). Multidrug-resistant (MDR) Salmonella has emerged as an important food safety concern and is associated with increased morbidity in humans compared to antibiotic sensitive strains (Molbak, 2005; Varma et al., 2005). However, little is known why MDR Salmonella is more virulent as it is not due to an intrinsically higher capacity to invade eukaryotic cells in vitro; in fact, MDR Salmonella has been reported to be less invasive in tissue culture experiments compared to antibiotic-sensitive strains (Carlson et al., 2000). The increased virulence capacity may be triggered by effectors in the environment, the host, or both.
A temporal progression of regulated expression occurs in Salmonella for genes associated with virulence and cellular invasion (Ibarra et al., 2010; Saini et al., 2010; Fabrega and Vila, 2013). During early-log growth, Salmonella express flagella and are motile, but have limited-to-no invasive capabilities. As late-log and stationary phases are reached, motility is down-regulated, and genes associated with attachment, invasion, and intracellular survival are up-regulated. Attachment is mediated by fimbriae, and S. Typhimurium encodes up to 13 fimbrial operons (McClelland et al., 2001). Invasion genes are located in Salmonella Pathogenicity Island 1 (SPI-1), which encodes a type three secretion system (TTSS) and effector molecules that promote cellular entry (Lostroh and Lee, 2001). SPI-2 also encodes a TTSS and effector molecules, but these and the genes expressed in SPI-3 promote Salmonella intracellular survival (Kuhle and Hensel, 2004).
Antibiotics can affect the regulation of cellular processes in bacteria (Goh et al., 2002; Kuroda et al., 2007; Shen et al., 2008; Deneve et al., 2009; Morita et al., 2014). For example, florfenicol can induce adherence of Staphylococcus aureus (Blickwede et al., 2005), and tetracycline can increase TTSS expression and cytotoxicity in Pseudomonas aeruginosa (Linares et al., 2006). However, the majority of bacteria in such studies are resistant to antibiotics due to intrinsic factors (such as mutations) or efflux pumps that work against a suite of antimicrobials. Antibiotic-specific genes often provide a much higher level of resistance compared to the non-specific antimicrobial pumps and can allow growth at an antibiotic concentration that might otherwise be inhibitory to sensitive bacteria. We previously characterized the effect different sub-inhibitory concentrations of the antibiotic tetracycline had on the invasiveness of eight MDR S. Typhimurium isolates that encoded tetracycline-specific resistance efflux pumps (Brunelle et al., 2013). We found that a subset of the isolates had an induced invasion phenotype during early-log growth phase in response to 16 μg/ml of tetracycline. The induced invasiveness was not observed with lower concentrations of tetracycline during early-log growth (1 and 4 μg/ml), nor did tetracycline enhance invasion in any isolate during late-log growth at any antibiotic concentration tested. The goals of the current study were to establish if ampicillin, chloramphenicol, or streptomycin can also induce invasion in the same isolates, and to assess transcriptional changes for genes associated with the virulence functional groups (motility, SPI-1, SPI-2, SPI-3, and attachment) due to antibiotic exposure.
Materials and Methods
Isolates
Eight isolates of Salmonella enterica serovar Typhimurium phage types DT104 (530, 290, 360) and DT193 (1434, 5317, 752, 1306, 4584) originally from cattle, and previously tested for differences in invasiveness after exposure to tetracycline (Brunelle et al., 2013), were selected for study. These isolates were cultured on solid media with antibiotic concentrations based on CLSI breakpoints, and it was identified that they are resistant to ampicillin (32 μg/ml), chloramphenicol (32 μg/ml), streptomycin (64 μg/ml), and tetracycline (16 μg/ml), but are sensitive to gentamicin (16 μg/ml).
Characterization of Antibiotic Resistance Genes
Primers (Table 1) were used to amplify and detect the most common resistance genes in Salmonella for the following antibiotics: ampicillin (blaCMY, blaOXA, blaPSE, blaTEM, and blaSHV), chloramphenicol (cat, cml, and floR), and streptomycin (aadA2, strA, and strB). Tetracycline resistance genes in these isolates (tetA, B, C, D, and G) were characterized previously (Brunelle et al., 2013). DNA was obtained by boiling a single colony from each isolate in 30 μl water. Each 25 μL PCR reaction contained 1.5 μl DNA, 1.5 units of Taq polymerase (Promega), 1x PCR buffer with 1.5 mM MgCl2, 1 mM each dNTP, and 0.8 μM of each primer. Amplification conditions were: 94°C for 1 min; 35 cycles of 94°C for 30 s, 56°C for 30 s, 72°C for 30 s; 72°C for 2 min; 4°C hold. Amplifications were done in duplicate and products were visualized on 2% NuSieve agarose gels (Cambrex, Rockland, ME).
Characterization of Antibiotic Concentrations that Inhibit Growth
The antibiotic concentration that inhibited growth in broth culture for each isolate was determined. Overnight cultures were diluted 1:200 in LB and grown to early-log phase (OD600 = 0.15) at 37°C with agitation before adding serial 2-fold dilutions of ampicillin, chloramphenicol, or streptomycin (0, 2-512 μg/ml). All conditions were done in triplicate. Cultures were continuously shaken at 37°C, and growth measurements (OD600) were taken every hour for 24 h using a Bioscreen C instrument (Growth Curves LTD, Raisio, Finland). Growth curves on serial dilutions of tetracycline were established previously (Brunelle et al., 2013).
Culture Conditions
For each experiment, Salmonella was plated on Lennox L (LB) agar plates (Invitrogen, Carlsbad, CA), and a single colony was selected and grown in LB broth with agitation for 6 h at 37°C. The 6 h culture was diluted 1:1000 in fresh LB broth and grown with agitation at 37°C overnight. The overnight culture was diluted 1:200 in fresh LB broth and divided into 16 × 100 mm glass tubes. Cultures were grown to early-log (OD600 = 0.15) with agitation at 37°C before antibiotic addition. An aliquot was taken for RNA analysis from each culture and placed in RNAProtect (QIAGEN, Germantown, MD). Ampicillin, chloramphenicol, and streptomycin were added to separate tubes for a final concentration of 0 (control), 16, 32, 64, and 128 μg/ml, and these were incubated with agitation at 37°C for 30 min; tetracycline was assayed at only 0 and 16 μg/ml. Aliquots for RNA analysis were taken from each bacterial culture and placed in RNAProtect. An additional aliquot was taken from each culture for a cell culture invasion assay. All experiments were performed four separate times.
Salmonella Invasion Assays
The aliquots of Salmonella cultures taken following the 30 min incubation with and without antibiotics were centrifuged at 16,000 × g for 2 min, and the pellets were re-suspended in fresh LB broth to remove the antibiotic. Invasion assays were performed with technical replicates for each biological replicate using a gentamicin protection assay in HEp-2 cells at a multiplicity of infection of ~40 as previously described (Elsinghorst, 1994). Percent invasion was calculated by dividing the CFU/ml recovered by the CFU/ml originally added to the cells. The significance of the differences in invasion was determined by a one-way repeated measures ANOVA with Dunnett's post-test to assess pair-wise differences between the no-antibiotic control and the other sample conditions using GraphPad Prism 5 (San Diego, CA). Differences were considered significant if P < 0.05. Each isolate has an intrinsically different invasion rate when grown without antibiotics; therefore the invasion data for each isolate was normalized to its respective control for graphical representation.
Real-Time PCR Assays
RNA was isolated using the RNeasy Mini Kit (QIAGEN, Germantown, MD), and genomic DNA was removed by the Turbo DNase DNA-free kit (Ambion, Austin, TX) according to the product directions. Total RNA was quantitated on a Nanodrop ND-1000 spectrophotometer (Thermo Scientific, Wilmington, DE). Reverse transcription was carried out by the Applied Biosystems High capacity cDNA reverse transcription kit on total RNA with random primers (Life Technologies, Grand Island, NY), and two technical replicates were performed for each of the three biological replicates. Real-Time PCR was performed in a Bio-Rad CFX96 Real-Time PCR Detection System (BioRad Laboratories, Hercules, CA) using the SYBR Green Master Mix (Applied Biosystems, Foster City, CA). Primer sets were used to evaluate the 16S rRNA, hilA, floR, tetA, tetC, tetD, and tetG transcripts (Table 1). For control assays, reverse transcriptase was not added to parallel mixtures for each sample. The amplification cycle conditions were as follows: 95°C for 10 min; 40 cycles of 95°C for 15 s, 55°C for 30 s, 72°C for 30 s; melting curve analysis from 65°C to 95°C. Raw fluorescence data was analyzed with the LinRegPCR software, and amplification efficiencies and cycle threshold (CT) values were determined by linear regression in a Window of Linearity for each primer set (Ramakers et al., 2003). Expression differences were calculated using the CT value and efficiency for each locus by the Pfaffl method (Pfaffl, 2001), where the 16S gene was the reference gene and the pre-antibiotic sample was the control condition for each isolate at each antibiotic concentration. Values were log2 transformed, and GraphPad Prism 5 was used to perform a one-way repeated measures ANOVA with Dunnett's post-test to assess pair-wise differences between the no-antibiotic control and the other sample conditions. Differences were considered significant if P < 0.05.
RNA Transcriptome Sequencing
Isolates 1434, 5317, 752, 4584, and 530 were selected for RNA-seq analysis to identify changes in virulence gene expression due to tetracycline (0 and 16 μg/ml) and chloramphenicol (0, 32, and 64 μg/ml) exposure. RNA from three biological replicates of each isolate and condition were used for sequencing. Quality and integrity of the RNA was assessed with the 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). The RiboZero kit was used to deplete the ribosomal RNA (rRNA) sequences according to the manufacturer's instructions (Epicentre, Madison, WI). The 2100 Bioanalyzer was used to verify removal of the rRNA. Libraries were constructed using TruSeq RNA Sample prep kit and were sequenced on a HiSeq 2500 employing a 100-cycle run (Illumina Inc., San Diego, CA) at the Iowa State University DNA core facility.
RNA-Seq Data Analyses
CLC Genomics Workbench v7.0 was used to import the Illumina sequence data. Failed reads were removed prior to analysis. The S. Typhimurium SL1344 genome [GenBank: FQ312003] and three plasmids [GenBank: HE654724-26], as well as a MDR plasmid from S. Heidelberg [GenBank JN983048], were used as references for mapping with the following parameters: 2 maximum mismatches, 90% minimum length fraction, 80% minimum similarity fraction, and 10 maximum hits per read. Differences in gene expression were calculated using EdgeR (total count filter cut-off = 5.0) with False Discovery Rate (FDR) corrected P-values (Robinson et al., 2010). Differences were considered significant if P < 0.05. Fold-changes for RNA-seq data described in the study are the weighted differences between groups based on counts per million calculated in EdgeR. Functional categories for genes associated with motility, attachment, SPI-1, SPI-2, and SPI-3 were manually assigned.
Electron Microscopy
Salmonella isolate 1434 was grown in LB to early-log growth phase followed by a 30 min antibiotic exposure to 64 μg/ml chloramphenicol, 16 μg/ml tetracycline, or no antibiotic (control) as described above. One ml of each culture was centrifuged at 1000 × g for 5 min, the supernatant was removed, and the pellet was re-suspended in 0.5 ml of 0.5% methyl cellulose solution in water (Mitani and Iino, 1968). Samples were prepared for negative stain examination by electron microscopy following previously described procedures (Doane and Anderson, 1987). Each sample was negatively stained with 2% phosphotungstic acid (pH 7.0) and examined with a FEI Tecnai G2 BioTWIN electron microscope (FEI Co., Hillsboro, OR) at the National Animal Disease Center Core Facility.
Results and Discussion
Our previous work investigated eight MDR S. Typhimurium isolates that were resistant to ampicillin, chloramphenicol, streptomycin, and tetracycline and demonstrated that a sub-inhibitory concentration of tetracycline induced the full invasive phenotype in a subset of these isolates during early-log growth phase (Brunelle et al., 2013). In order to determine if other antibiotics could induce invasiveness in the same eight isolates, the current study used invasion assays to test their response to sub-inhibitory concentrations of ampicillin, chloramphenicol, and streptomycin. Real-time PCR and RNA-seq were used to assess expression changes of genes associated with antibiotic resistance, as well as with the temporal regulation of the invasion phenotype.
Characterization of Antibiotic Resistance
PCR was used to identify the presence of commonly encoded MDR Salmonella genes that mediate resistance to ampicillin (blaCMY, blaOXA, blaPSE, blaTEM, and blaSHV), chloramphenicol (cat, cml, and floR), and streptomycin (aadA2, strA, and strB). With the exception of blaTEM and blaOXA, all genes were present in some combination in the eight isolates (Table 2). The tetracycline resistance genes in these isolates were established previously (Brunelle et al., 2013). As expected, blaPSE, aadA2, and tetG were identified in all three DT104 isolates, but not in any of the DT193 isolates, as these resistance genes are typically chromosomally encoded on the Salmonella Genomic Island 1 (SGI-1) in DT104 (Boyd et al., 2001). Of note is that 4584 was the only isolate to lack the floR gene, as well as the only one to encode the cat gene.
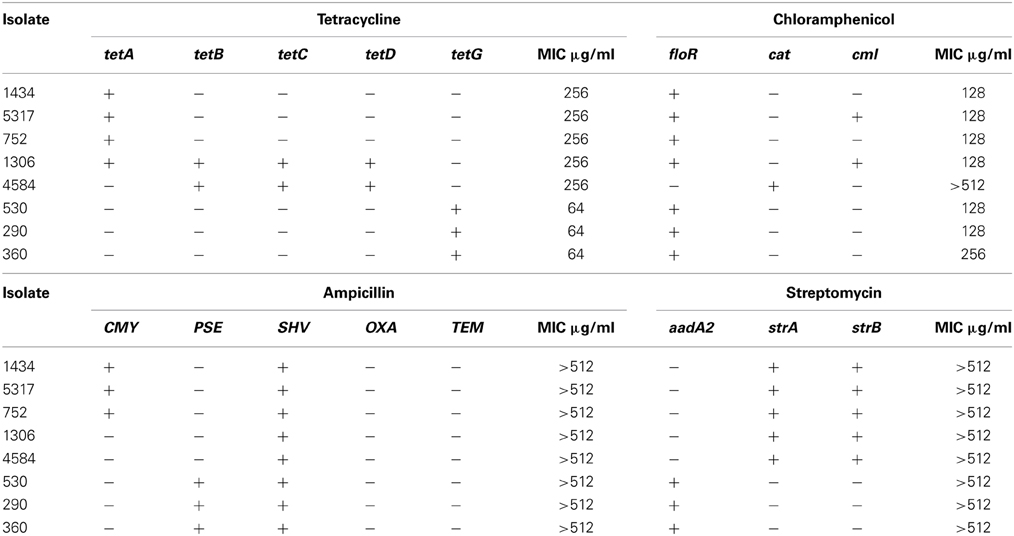
The effect different concentrations (0, 2-512 μg/ml) of ampicillin, chloramphenicol, and streptomycin had on the growth of each isolate was determined (Table 2). For chloramphenicol, growth was inhibited at 128 μg/ml for six of the isolates, including isolate 1434 (Figure 1), and at 256 μg/ml for isolate 360 (Table 2). Isolate 4584 grew at 512 μg/ml, which is likely because it encodes the antibiotic-modifying enzyme chloramphenicol acetyltransferase (cat) instead of the efflux pumps floR or cml (Schwarz et al., 2004). The ampicillin and streptomycin resistance genes in the isolates encode modifying enzymes, and neither antibiotic inhibited growth at any of the concentrations that were tested.
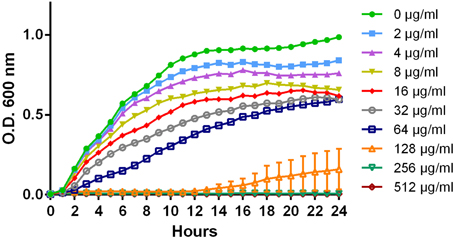
Figure 1. Growth curve of multidrug-resistant S. Typhimurium isolate 1434 exposed to chloramphenicol. Serial two-fold dilutions of chloramphenicol (0, 2-512 μg/ml) were added at OD600 = 0.15 to the eight isolates in the study to assess the effect chloramphenicol exposure had on growth. The growth curve of isolate 1434 is representative of the other isolates, with the exception of 4584.
Chloramphenicol Induces Invasion in Three Isolates
Based on the growth data, 0, 16, 32, 64, and 128 μg/ml of ampicillin, chloramphenicol, and streptomycin were chosen as the sub-inhibitory concentrations to test the effect each antibiotic had on invasion during early-log growth phase of the eight isolates. Chloramphenicol induced a significant increase in invasiveness at 32 or 64 μg/ml compared to the no-antibiotic control for three DT193 isolates (1434, 5317, and 1306; Figure 2). The chloramphenicol-induced invasion rates during early-log are similar to the maximum natural invasion rates for each isolate during late-log, which parallels the results observed for tetracycline (Brunelle et al., 2013) (data not shown). Isolates 1434 and 5317 were previously shown to have the invasion phenotype induced after exposure to 16 μg/ml tetracycline for 30 min during early-log growth. Several isolates had a significant decrease in cellular invasion at 64 μg/ml (752 and 530) and 128 μg/ml (752, 530, and 360) of chloramphenicol. Neither ampicillin nor streptomycin had an effect on the invasiveness of any of the isolates at any concentration tested (data not shown). Therefore, invasion was induced by two different antibiotics in a subset of isolates that utilize efflux pump-mediated resistance (floR and tetA), but not in the isolates that use enzyme-mediated resistance (cat, all ampicillin and streptomycin genes). However, several isolates that utilize TetA, TetG, or FloR efflux pumps did not have the induced invasion phenotype.
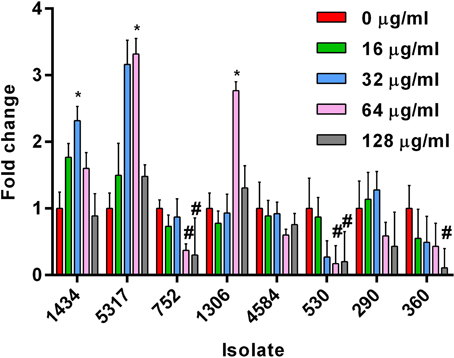
Figure 2. Changes in cellular invasiveness after chloramphenicol exposure in S. Typhimurium isolates during early-log growth. Invasion assays were performed using HEp-2 cells. S. Typhimurium isolates grown to early-log phase and exposed to chloramphenicol (0, 16, 32, 64, or 128 μg/ml) for 30 min. Changes in invasion were normalized to the control dose (0 μg/ml) for each isolate. The “*” indicates a significant increase based on the pre-normalized data, while “#” denotes a significant decrease (P < 0.05).
Real-Time PCR Analyses
Real-time PCR was used to assess expression changes of hilA, floR, and the tet genes in all the isolates in response to the 30 min chloramphenicol exposure.
Differential expression of the hilA gene
The HilA protein is the master regulator of SPI-1, and increases or decreases in expression of the hilA gene typically have a corresponding increase or decrease on invasion (Lee et al., 1992). However, increases in hilA expression due to chloramphenicol (Figure 3A) did not always correlate with increased invasiveness (Figure 2); this is highlighted in isolate 530 as 16, 32, and 64 μg/ml of chloramphenicol significantly increased hilA expression without any corresponding changes on invasion. This lack of congruence between increased hilA gene expression and increased invasiveness was previously observed when the isolates were exposed to tetracycline (Brunelle et al., 2013). No changes in hilA expression were detected due to either ampicillin or streptomycin exposure in any of the isolates, which is consistent with the invasion data (data not shown).
Figure 3. Gene expression changes in S. Typhimurium at early-log growth after chloramphenicol exposure. Real-time gene expression assays were performed on eight MDR S. Typhimurium isolates grown to early-log phase and exposed to different concentrations of chloramphenicol (0, 16, 32, and 64 μg/ml) for 30 min. The “*” denotes a significant change in expression over the 30 min compared to the control (pre-incubation) (P < 0.05). (A) Gene expression changes in the hilA gene. (B) Gene expression changes in the chloramphenicol-resistance floR gene (not present in isolate 4584). (C) Gene expression differences in the tetracycline-resistance genes in the eight isolates due to chloramphenicol exposure.
Differential expression of the floR gene
An increase in the expression of floR was significant at all concentrations of chloramphenicol in the seven isolates encoding the gene (Figure 3B). These data indicate that there is no direct association between the up-regulation of floR and the induced invasiveness in isolates 1434, 5317, and 1306. Therefore, floR may be necessary, but is not sufficient for the observed effect. This parallels previous findings that the increased expression of tet genes due to tetracycline exposure was not directly responsible for tetracycline-induced invasion (Brunelle et al., 2013).
Differential expression of the tetA,C,D,G genes
Expression changes of the tetracycline resistance genes tetA, tetC, tetD, and tetG were also analyzed in response to chloramphenicol (Figure 3C); tetB expression with or without antibiotic exposure was undetectable, and therefore was not included in the figure. Interestingly, tetA, C, D, and G were found to be up-regulated due to exposure to various concentrations of chloramphenicol (and in the absence of tetracycline) in most of the isolates. Only isolate 4584, which encodes tetC, tetD, and cat but lacks tetA and floR, did not have a change in expression of its tet genes. In contrast, the tetC and tetD genes in isolate 1306 were up-regulated at 64 μg/ml of chloramphenicol. Isolate 1306 encodes both floR and tetA, and it may be the presence of one or both of these two genes that is needed to co-regulate tetC and tetD expression after exposure to chloramphenicol. Additionally, floR is frequently adjacent to tetA in DT193 and tetG in DT104. Although these genes are in opposite orientations and would not be in the same operon, it could be the proximity of these genes that allows chloramphenicol to induce the tet genes independently of tetracycline. PCR confirmed the order and orientation of the floR and tetA/G genes in the isolates.
RNA-Seq Identifies Differential Regulation of the Virulence-Associated Functional Groups Due to Antibiotic Exposure
In order to assess if the early induction of the full invasive phenotype by chloramphenicol and tetracycline coincided with an acceleration of the temporally regulated virulence pathways, RNA-seq was used to evaluate differences in the expression of genes in the functional groups required for invasion. Five isolates were chosen to study: isolates 1434 and 5317 because they both have an increase in invasion after exposure to tetracycline and chloramphenicol, and both encode floR and tetA; isolates 752 and 530 because they do not have increased invasiveness due to antibiotic exposure, but encode floR and either tetA (752) or tetG (530); and isolate 4584 as it did not have a change in its invasion phenotype and lacks floR, tetA, and tetG. Genes associated with motility, invasion (SPI-1), intracellular survival (SPI-2 and SPI-3), and attachment were examined in isolates exposed separately to no antibiotic (control), 16 μg/ml of tetracycline, and 32 and 64 μg/ml of chloramphenicol for 30 min during early-log growth. The results for the samples exposed to 32 and 64 μg/ml of chloramphenicol were highly similar, so only the 64 μg/ml of chloramphenicol data are presented. Except for SPI-3, the data for isolate 4584 is not included below as there were no significant changes in any of the genes among the other functional groups.
Motility
Fifty-four genes associated with motility were assessed, and the majority of these were found to be down-regulated in isolates 1434, 5317, 752, and 530 when exposed to chloramphenicol and tetracycline (Figure 4, Table 3). This is highlighted by the flgA-J and fliE-N genes that encode proteins for the basal body, flagellar hook, and flagellar motor switch. The 10 flg and the 10 fli genes are all significantly down-regulated an average of 18.5-fold and 12.8-fold, respectively, in each of the four isolates in response to both antibiotics. The genes that encoded the master regulators of motility, flhCD, were not differentially regulated except in isolate 752 where their expression increased due to chloramphenicol. However, the fliA gene that encodes a σ 28 factor and controls regulation of the late-flagellar genes (Chilcott and Hughes, 2000) was significantly down-regulated in most of the samples (the exception was isolate 5317 exposed to chloramphenicol that was down-regulated, but had a P = 0.054). Motility genes are typically up-regulated during early-log growth, and are then down-regulated during the mid- to late-growth phase as the invasion genes are up-regulated (Saini et al., 2010). In this study, we observed that tetracycline and chloramphenicol down-regulate the motility genes in Salmonella during early-log growth.
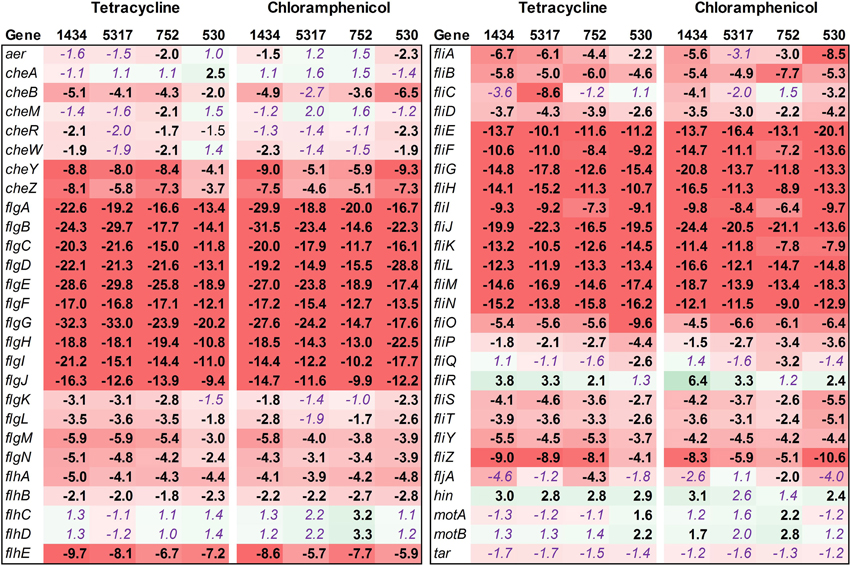
Figure 4. RNA-seq analysis of genes associated with motility. RNA-seq identified the fold-change in expression of genes associated with motility due to exposure to the antibiotics tetracycline (16 μg/ml) or chloramphenicol (64 μg/ml) when compared to the no-antibiotic control. Numbers in bold are significantly different (FDR-adjusted P < 0.05), while numbers in purple italics are not significant. Green denotes increased gene expression, and red indicates decreased gene expression; the intensity of the color highlights greater change.
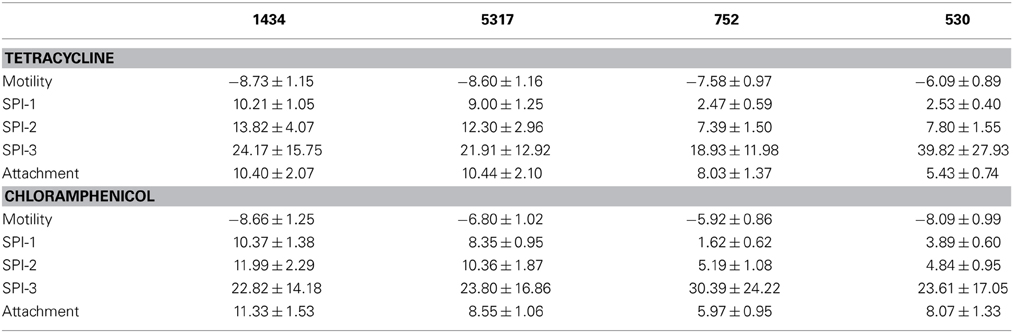
Table 3. Mean and Standard Error of the Mean of all gene expression differences within each functional group after antibiotic exposure for isolates 1434, 5317, 752, and 530.
SPI-1
Thirty-seven SPI-1 genes related to invasion were examined, which included those genes for SPI-1 regulation (hil), type-three secretion apparatus (sip, spa), and effector molecules (inv, prg) (Figure 5). While many of the SPI-1 genes are up-regulated in the four isolates, there is a striking difference between the two isolates associated with the induced invasion phenotype (1434 and 5317) and the other two isolates (752 and 530) (Table 3). This is denoted by the differences in the average fold-change between the two sets of isolates for all of the inv (12.4 ± 0.9 vs. 2.4 ± 0.3), prg (4.8 ± 0.7 vs. 0.7 ± 0.5), sip (10.5 ± 1.5 vs. 3.5 ± 0.7), and spa genes (16.0 ± 2.1 vs. 5.9 ± 0.8). Changes in hilA expression are consistent with the real-time data where 1434, 5317, and 530 had significant differences, but again the average fold-change for 1434 and 5317 was much higher than 530 (12.2 ± 1.1 vs. 3.3 ± 0.7, respectively). Thus, tetracycline and chloramphenicol can increase transcription of SPI-1 genes, but there may be a minimum threshold of induction that is necessary before the invasive phenotype is also induced.
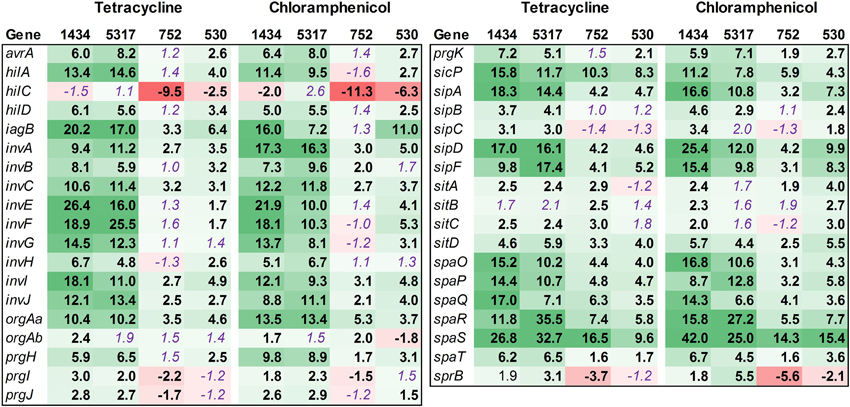
Figure 5. RNA-seq analysis of SPI-1 genes. RNA-seq identified the fold-change in expression of SPI-1 genes due to exposure to the antibiotics tetracycline (16 μg/ml) or chloramphenicol (64 μg/ml) when compared to the no-antibiotic control. Numbers in bold are significantly different (FDR-adjusted P < 0.05), while numbers in purple italics are not significant. Green denotes increased gene expression, and red indicates decreased gene expression; the intensity of the color highlights greater change.
SPI-2
SPI-2 encodes a TTSS that is required for intracellular replication and survival, and of the 40 genes examined in this functional group, the majority were up-regulated (Figure 6). The bias in up-regulation that appeared in SPI-1 for isolates 1434 and 5317 was also present in SPI-2 (Table 3). Several genes encoding effector molecules, such as sifA, pipB, and sseJ, were up-regulated between 6.4 and 51.5-fold for the four isolates exposed to either antibiotic. Only two genes were significantly down-regulated, ttrR and ttrS, and these encode a two-component regulatory system for reducing tetrathionate.
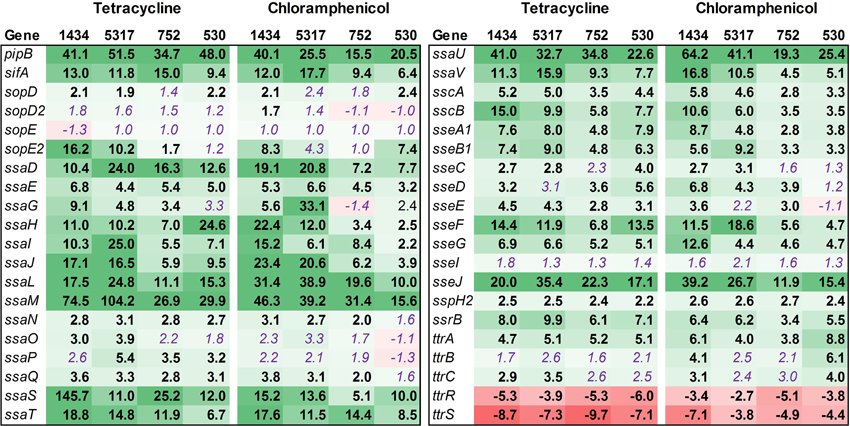
Figure 6. RNA-seq analysis of SPI-2 genes. RNA-seq identified the fold-change in expression of SPI-2 genes due to exposure to the antibiotics tetracycline (16 μg/ml) or chloramphenicol (64 μg/ml) when compared to the no-antibiotic control. Numbers in bold are significantly different (FDR-adjusted P < 0.05), while numbers in purple italics are not significant. Green denotes increased gene expression, and red indicates decreased gene expression; the intensity of the color highlights greater change.
SPI-3
The SPI-3 region has few genes, but these play a role in virulence and intracellular survival in monocytes. Most of the SPI-3 genes were up-regulated (Figure 7), including mgtBC. MgtB is used by Salmonella to transport magnesium into the bacterium in magnesium-deficient environments such as an intracellular environment. The tetracycline and chloramphenicol efflux pumps are major facilitator superfamily antiporters that export an antibiotic:proton complex outside of the cell (Chopra and Roberts, 2001; Braibant et al., 2005). It is known that tetracycline molecules bind to magnesium, and the export of this complex would therefore decrease the internal concentration of the ion. It would be beneficial for Salmonella to up-regulate this system in order to continue to export the antibiotic. The mgtB and mgtC genes are up-regulated in isolate 4584 exposed to tetracycline but not chloramphenicol, and these are the only virulence-associated genes examined in this study to be differentially regulated in this isolate.
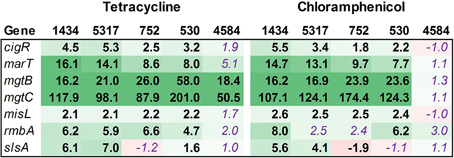
Figure 7. RNA-seq analysis of SPI-3 genes. RNA-seq identified the fold-change in expression of SPI-3 genes due to exposure to the antibiotics tetracycline (16 μg/ml) or chloramphenicol (64 μg/ml) when compared to the no-antibiotic control. Numbers in bold are significantly different (FDR-adjusted P < 0.05), while numbers in purple italics are not significant. Green denotes increased gene expression, and red indicates decreased gene expression; the intensity of the color highlights greater change.
Attachment
Salmonella Typhimurium isolates encode up to 13 fimbrial operons (bcf, csg, fim, lpf, pef, saf, stb, stc, std, stf, sth, sti, and stj). These genes, as well as other genes related to attachment, were evaluated (Figure 8). Overall, most of the fimbrial operons were up-regulated in each isolate due to antibiotic exposure. However, there were a few notable differences among the isolates. Most of the pef genes were up-regulated in 530 (DT104) whereas the three DT193 isolates (1434, 5317, and 752) lacked this operon. Isolates 1434 and 5317 had a greater level of fim gene expression compared to isolates 752 and 530. The fim genes encode the Type I fimbriae, which are important for Salmonella entry into epithelial cells (Ernst et al., 1990; Baumler et al., 1996; Wilson et al., 2000). The increased fim expression in isolates 1434 and 5317 may also play a role in the induced invasion phenotype observed for these two isolates.
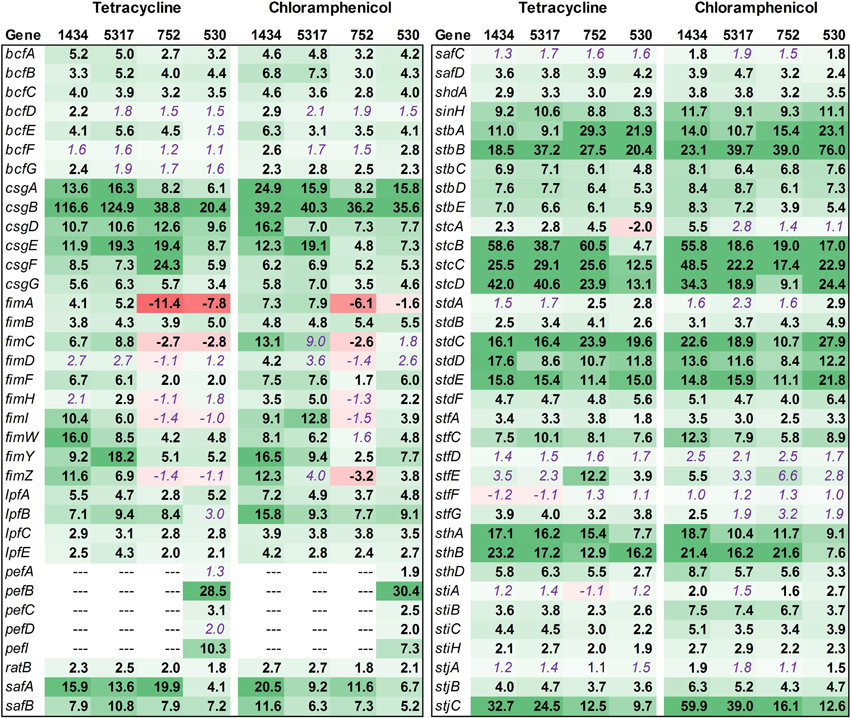
Figure 8. RNA-seq analysis of genes associated with attachment. RNA-seq identified the fold-change in expression of genes associated with attachment due to exposure to the antibiotics tetracycline (16 μg/ml) or chloramphenicol (64 μg/ml) when compared to the no-antibiotic control. Numbers in bold are significantly different (FDR-adjusted P < 0.05), while numbers in purple italics are not significant. Green denotes increased gene expression, and red indicates decreased gene expression; the intensity of the color highlights greater change.
Previously, only three fimbrial operons (csg, fim, and pef) had been shown to be expressed in culture, and each requires specific growth conditions (Humphries et al., 2003). Typically, expression of csg is dependent on growth for over 24 h on solid medium, while fim expression occurs in the stationary phase for Salmonella grown in liquid medium. The pef genes are encoded on a plasmid and require liquid medium and a low pH to be expressed in culture. The expression of the remaining fimbrial genes has only been observed in vivo (Nicholson and Baumler, 2001; Humphries et al., 2003). However, in the current study, tetracycline and chloramphenicol induced the expression of genes in up to 11 different fimbrial operons in isolates 1434, 5317, 752, and 530. Also, the fimbrial genes in the bcf, lpf, stb, stc, std, and sth operons have been shown to be associated with persistence in mice (Weening et al., 2005); as all six of these operons can be induced by the two antibiotics, this could be one factor that contributes to the increased morbidity observed in MDR Salmonella patients that have been treated with antibiotics (Molbak, 2005).
Electron Microscopy Demonstrates Decreased Flagella and Increased Agglutination Due to Antibiotic Exposure
Electron microscopy was used to visualize isolate 1434 in early-log phase treated with 64 μg/ml chloramphenicol compared to a no antibiotic control (Figure 9). Salmonella that were not exposed to chloramphenicol (control) had many flagella (Figure 9A) and were dispersed (Figure 9B). The chloramphenicol-treated Salmonella had few-to-no flagella (Figure 9C) and had increased bacterial agglutination compared to the untreated sample (Figure 9D). These figures are representative of other regions of the samples that were also visualized. Isolate 1434 exposed to 16 μg/ml tetracycline had the same result (data not shown). The decreased flagella and increased agglutination are consistent with the RNA-seq data that demonstrates decreased motility and increased attachment gene expression.
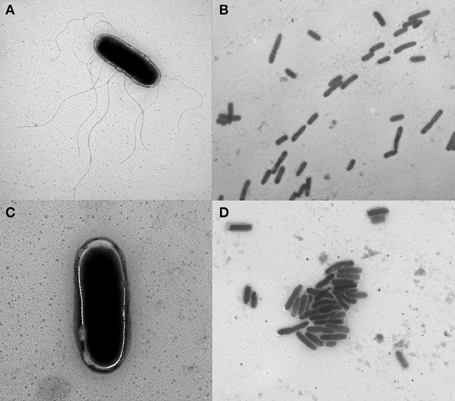
Figure 9. Electron micrographs of Salmonella with and without chloramphenicol exposure. S. Typhimurium isolate 1434 grown to early-log phase (OD600 = 0.15) and exposed to (A,B) 0 μg/ml or (C,D) 64 μg/ml chloramphenicol for 30 min. Isolates exposed to chloramphenicol had fewer flagella and increased bacterial agglutination compared to the control (no antibiotic) condition. These figures are representative of other regions of the samples that were also visualized.
Concluding Remarks
Tetracyclines are approved for growth promotion and therapeutic use in swine, cattle, and poultry in the U.S.; it was reported that over 4.6 million kg of tetracyclines were sold for use in livestock in a single year (USGAO, 2011). While tetracycline usage may directly select for resistant Salmonella encoding tet genes, it may also indirectly select for other resistance genes (such as floR) and virulence factors that are encoded on the same plasmid. For example, Hradecka et al. reported the Salmonella virulence plasmid can recombine with an antibiotic resistance plasmid, ensuring that the plasmid and its content can be selected based on either virulence or antibiotics (Hradecka et al., 2008). Because of the widespread use of antibiotics in humans and livestock, it is important to understand the effects these antibiotics have on MDR Salmonella.
Tetracycline and chloramphenicol exposure of MDR Salmonella in our transcriptomic data indicate that the regulation of the invasive phenotype in Salmonella, which normally takes over 8 h to induce, can be accelerated to occur in 30 min. While gene expression data cannot assess protein levels or post-transcriptional interactions of each individual gene that have an impact on the invasion phenotype, the data suggests there is a coordinated regulation of the regulons involved in attachment, motility, and invasion due to chloramphenicol and tetracycline. Furthermore, the attachment regulon appears to extend beyond the fim operon to include other fimbrial operons that would not normally be expressed in culture. Chloramphenicol and tetracycline may be acting on a complex stimulon with multiple interacting regulons in a system that is currently uncharacterized in culture.
The induced invasion phenotype observed may be directly activated by the antibiotic response pathway that is stimulated to express the efflux pump after antibiotic exposure. It may also be an indirect response due to the changes in the cellular environment due to the presence of the antibiotic and potential changes in H+ and Mg2+ concentrations (Yamaguchi et al., 1990). For instance, the response may be due to the “tetracycline transport cycle” in resistant cells where extracellular tetracycline is protonated and diffuses into the bacterial cell in a neutral form. Once inside the cell, the tetracycline is predicted to dissociate from the proton due the higher intracellular pH. The intracellular concentration of Mg may facilitate the formation of a tetracycline-Mg chelate complex that can then be exported by the TetA antiporter with the simultaneous import of a proton. In this model, the export of tetracycline is electrically neutral (Mg+ exported while H+ imported) but there is a net gain of one proton from the initial diffusion of tetracycline into the bacterial cell. Therefore, the internal pH of the bacteria may shift due to the accumulation of protons with the decreased pH conditions triggering a secondary response that activates the invasion genes. Interestingly, mgtBC present in SPI-3 is up-regulated by both tetracycline and chloramphenicol exposure. The MgtC protein inhibits the bacterium's own F1F0 ATP synthase that couples proton translocation across the bacterial membrane with ATP synthesis/hydrolysis (Lee et al., 2013). The intracellular pH is lower and ATP levels higher for an mgtC mutant compared to wild-type S. Typhimurium suggesting that intracellular pH or ATP/ADP levels could influence the gene expression seen in our investigation. Because the induced invasion phenotype does not occur in all the MDR isolates tested, there is likely a genetic component that is involved. Additional investigations using genomics and proteomics will provide greater insights into the effect antibiotics have on these MDR Salmonella isolates.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Briony Atkinson for her outstanding technical assistance, as well as Judith Stasko for her fantastic electron microscopy work. We would also like to thank Dr. Tracy Nicholson and Dr. Thomas Casey for their critical review of the manuscript. This research was supported by USDA, ARS CRIS funds. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendations or endorsement by the US Department of Agriculture. USDA is an equal opportunity provider and employer.
Abbreviations
MDR, multidrug-resistant; FDR, false discovery rate; SPI, Salmonella pathogenicity island; TTSS, type three secretion system.
References
Baumler, A. J., Tsolis, R. M., and Heffron, F. (1996). Contribution of fimbrial operons to attachment to and invasion of epithelial cell lines by Salmonella typhimurium. Infect. Immun. 64, 1862–1865.
Blickwede, M., Goethe, R., Wolz, C., Valentin-Weigand, P., and Schwarz, S. (2005). Molecular basis of florfenicol-induced increase in adherence of Staphylococcus aureus strain Newman. J. Antimicrob. Chemother. 56, 315–323. doi: 10.1093/jac/dki233
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Boyd, D., Peters, G. A., Cloeckaert, A., Boumedine, K. S., Chaslus-Dancla, E., Imberechts, H., et al. (2001). Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J. Bacteriol. 183, 5725–5732. doi: 10.1128/JB.183.19.5725-5732.2001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Braibant, M., Chevalier, J., Chaslus-Dancla, E., Pages, J. M., and Cloeckaert, A. (2005). Structural and functional study of the phenicol-specific efflux pump FloR belonging to the major facilitator superfamily. Antimicrob. Agents Chemother. 49, 2965–2971. doi: 10.1128/AAC.49.7.2965-2971.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brunelle, B. W., Bearson, S. M., and Bearson, B. L. (2013). Tetracycline accelerates the temporally-regulated invasion response in specific isolates of multidrug-resistant Salmonella enterica serovar Typhimurium. BMC Microbiol. 13:202. doi: 10.1186/1471-2180-13-202
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brunelle, B. W., Bearson, S. M. D., and Bearson, B. L. (2011). Salmonella enterica serovar Typhimurium DT104 invasion is not enhanced by sub-inhibitory concentrations of the antibiotic florfenicol. Vet. Sci. Technol. 2:104. doi: 10.4172/2157-7579.1000104
Carlson, S. A., Browning, M., Ferris, K. E., and Jones, B. D. (2000). Identification of diminished tissue culture invasiveness among multiple antibiotic resistant Salmonella typhimurium DT104. Microb. Pathog. 28, 37–44. doi: 10.1006/mpat.1999.0322
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
CDC. (2012). National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): Human Isolates Final Report, 2010. Atlanta: U.S. Department of Health and Human Services, CDC.
Chang, H. R., Loo, L. H., Jeyaseelan, K., Earnest, L., and Stackebrandt, E. (1997). Phylogenetic relationships of Salmonella typhi and Salmonella typhimurium based on 16S rRNA sequence analysis. Int. J. Syst. Bacteriol. 47, 1253–1254. doi: 10.1099/00207713-47-4-1253
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chilcott, G. S., and Hughes, K. T. (2000). Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64, 694–708. doi: 10.1128/MMBR.64.4.694-708.2000
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chopra, I., and Roberts, M. (2001). Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65, 232–260. doi: 10.1128/MMBR.65.2.232-260.2001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Deneve, C., Bouttier, S., Dupuy, B., Barbut, F., Collignon, A., and Janoir, C. (2009). Effects of subinhibitory concentrations of antibiotics on colonization factor expression by moxifloxacin-susceptible and moxifloxacin-resistant Clostridium difficile strains. Antimicrob. Agents Chemother. 53, 5155–5162. doi: 10.1128/AAC.00532-09
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Doane, F. W., and Anderson, N. (1987). Electron Microscopy in Diagnostic Virology: a Practical Guide and Atlas. Cambridge, UK: Cambridge University Press.
Eckert, C., Gautier, V., Saladin-Allard, M., Hidri, N., Verdet, C., Ould-Hocine, Z., et al. (2004). Dissemination of CTX-M-type beta-lactamases among clinical isolates of Enterobacteriaceae in Paris, France. Antimicrob. Agents Chemother. 48, 1249–1255. doi: 10.1128/AAC.48.4.1249-1255.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Economic Research Service. (2009). Foodborne Illness Cost Calculator: Salmonella. Washington, DC: United States Department of Agriculture.
Elsinghorst, E. A. (1994). Measurement of invasion by gentamicin resistance. Meth. Enzymol. 236, 405–420. doi: 10.1016/0076-6879(94)36030-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ernst, R. K., Dombroski, D. M., and Merrick, J. M. (1990). Anaerobiosis, type 1 fimbriae, and growth phase are factors that affect invasion of HEp-2 cells by Salmonella typhimurium. Infect. Immun. 58, 2014–2016.
Fabrega, A., and Vila, J. (2013). Salmonella enterica serovar Typhimurium skills to succeed in the host: virulence and regulation. Clin. Microbiol. Rev. 26, 308–341. doi: 10.1128/CMR.00066-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Goh, E. B., Yim, G., Tsui, W., McClure, J., Surette, M. G., and Davies, J. (2002). Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics. Proc. Natl. Acad. Sci. U.S.A. 99, 17025–17030. doi: 10.1073/pnas.252607699
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Golding, G. R., Olson, A. B., Doublet, B., Cloeckaert, A., Christianson, S., Graham, M. R., et al. (2007). The effect of the Salmonella genomic island 1 on in vitro global gene expression in Salmonella enterica serovar Typhimurium LT2. Microbes Infect. 9, 21–27. doi: 10.1016/j.micinf.2006.10.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hradecka, H., Karasova, D., and Rychlik, I. (2008). Characterization of Salmonella enterica serovar Typhimurium conjugative plasmids transferring resistance to antibiotics and their interaction with the virulence plasmid. J. Antimicrob. Chemother. 62, 938–941. doi: 10.1093/jac/dkn286
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Humphries, A. D., Raffatellu, M., Winter, S., Weening, E. H., Kingsley, R. A., Droleskey, R., et al. (2003). The use of flow cytometry to detect expression of subunits encoded by 11 Salmonella enterica serotype Typhimurium fimbrial operons. Mol. Microbiol. 48, 1357–1376. doi: 10.1046/j.1365-2958.2003.03507.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ibarra, J. A., Knodler, L. A., Sturdevant, D. E., Virtaneva, K., Carmody, A. B., Fischer, E. R., et al. (2010). Induction of Salmonella pathogenicity island 1 under different growth conditions can affect Salmonella-host cell interactions in vitro. Microbiology 156, 1120–1133. doi: 10.1099/mic.0.032896-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kuhle, V., and Hensel, M. (2004). Cellular microbiology of intracellular Salmonella enterica: functions of the type III secretion system encoded by Salmonella pathogenicity island 2. Cell. Mol. Life Sci. 61, 2812–2826. doi: 10.1007/s00018-004-4248-z
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kuroda, H., Kuroda, M., Cui, L., and Hiramatsu, K. (2007). Subinhibitory concentrations of beta-lactam induce haemolytic activity in Staphylococcus aureus through the SaeRS two-component system. FEMS Microbiol. Lett. 268, 98–105. doi: 10.1111/j.1574-6968.2006.00568.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lee, C. A., Jones, B. D., and Falkow, S. (1992). Identification of a Salmonella typhimurium invasion locus by selection for hyperinvasive mutants. Proc. Natl. Acad. Sci. U.S.A. 89, 1847–1851. doi: 10.1073/pnas.89.5.1847
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lee, E. J., Pontes, M. H., and Groisman, E. A. (2013). A bacterial virulence protein promotes pathogenicity by inhibiting the bacterium's own F1Fo ATP synthase. Cell 154, 146–156. doi: 10.1016/j.cell.2013.06.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Linares, J. F., Gustafsson, I., Baquero, F., and Martinez, J. L. (2006). Antibiotics as intermicrobial signaling agents instead of weapons. Proc. Natl. Acad. Sci. U.S.A. 103, 19484–19489. doi: 10.1073/pnas.0608949103
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lostroh, C. P., and Lee, C. A. (2001). The Salmonella pathogenicity island-1 type III secretion system. Microbes Infect. 3, 1281–1291. doi: 10.1016/S1286-4579(01)01488-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
McClelland, M., Sanderson, K. E., Spieth, J., Clifton, S. W., Latreille, P., Courtney, L., et al. (2001). Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413, 852–856. doi: 10.1038/35101614
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mitani, M., and Iino, T. (1968). Electron microscopy of Salmonella flagella in methylcellulose solution. J. Gen. Microbiol. 50, 459–464. doi: 10.1099/00221287-50-3-459
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Molbak, K. (2005). Human health consequences of antimicrobial drug-resistant Salmonella and other foodborne pathogens. Clin. Infect. Dis. 41, 1613–1620. doi: 10.1086/497599
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Morita, Y., Tomida, J., and Kawamura, Y. (2014). Responses of Pseudomonas aeruginosa to antimicrobials. Front. Microbiol. 4:422. doi: 10.3389/fmicb.2013.00422
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nicholson, T. L., and Baumler, A. J. (2001). Salmonella enterica serotype typhimurium elicits cross-immunity against a Salmonella enterica serotype enteritidis strain expressing LP fimbriae from the lac promoter. Infect. Immun. 69, 204–212. doi: 10.1128/IAI.69.1.204-212.2001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ramakers, C., Ruijter, J. M., Deprez, R. H., and Moorman, A. F. (2003). Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339, 62–66. doi: 10.1016/S0304-3940(02)01423-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Saini, S., Slauch, J. M., Aldridge, P. D., and Rao, C. V. (2010). Role of cross talk in regulating the dynamic expression of the flagellar Salmonella pathogenicity island 1 and type 1 fimbrial genes. J. Bacteriol. 192, 5767–5777. doi: 10.1128/JB.00624-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Scallan, E., Griffin, P. M., Angulo, F. J., Tauxe, R. V., and Hoekstra, R. M. (2011). Foodborne illness acquired in the United States–unspecified agents. Emerging Infect. Dis. 17, 16–22. doi: 10.3201/eid1701.091101p2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schwarz, S., Kehrenberg, C., Doublet, B., and Cloeckaert, A. (2004). Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol. Rev. 28, 519–542. doi: 10.1016/j.femsre.2004.04.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shen, L., Shi, Y., Zhang, D., Wei, J., Surette, M. G., and Duan, K. (2008). Modulation of secreted virulence factor genes by subinhibitory concentrations of antibiotics in Pseudomonas aeruginosa. J. Microbiol. 46, 441–447. doi: 10.1007/s12275-008-0054-x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
USGAO. (2011). Agencies Have Made Limited Progress Addressing Antibiotic Use in Animals. US Government Accountability Office. Available online at: http://www.gao.gov/products/GAO-11–801
Varma, J. K., Greene, K. D., Ovitt, J., Barrett, T. J., Medalla, F., and Angulo, F. J. (2005). Hospitalization and antimicrobial resistance in Salmonella outbreaks, 1984-2002. Emerging Infect. Dis. 11, 943–946. doi: 10.3201/eid1106.041231
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Weening, E. H., Barker, J. D., Laarakker, M. C., Humphries, A. D., Tsolis, R. M., and Baumler, A. J. (2005). The Salmonella enterica serotype Typhimurium lpf, bcf, stb, stc, std, and sth fimbrial operons are required for intestinal persistence in mice. Infect. Immun. 73, 3358–3366. doi: 10.1128/IAI.73.6.3358-3366.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Weill, F. X., Guesnier, F., Guibert, V., Timinouni, M., Demartin, M., Polomack, L., et al. (2006). Multidrug resistance in Salmonella enterica serotype Typhimurium from humans in France (1993 to 2003). J. Clin. Microbiol. 44, 700–708. doi: 10.1128/JCM.44.3.700-708.2006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Weir, E. K., Martin, L. C., Poppe, C., Coombes, B. K., and Boerlin, P. (2008). Subinhibitory concentrations of tetracycline affect virulence gene expression in a multi-resistant Salmonella enterica subsp. enterica serovar Typhimurium DT104. Microbes Infect. 10, 901–907. doi: 10.1016/j.micinf.2008.05.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wilson, R. L., Elthon, J., Clegg, S., and Jones, B. D. (2000). Salmonella enterica serovars gallinarum and pullorum expressing Salmonella enterica serovar typhimurium type 1 fimbriae exhibit increased invasiveness for mammalian cells. Infect. Immun. 68, 4782–4785. doi: 10.1128/IAI.68.8.4782-4785.2000
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: antibiotics, attachment, chloramphenicol, fimbriae, invasion, motility, Salmonella, tetracycline
Citation: Brunelle BW, Bearson BL and Bearson SMD (2015) Chloramphenicol and tetracycline decrease motility and increase invasion and attachment gene expression in specific isolates of multidrug-resistant Salmonella enterica serovar Typhimurium. Front. Microbiol. 5:801. doi: 10.3389/fmicb.2014.00801
Received: 15 September 2014; Accepted: 28 December 2014;
Published online: 30 January 2015.
Edited by:
Kunihiko Nishino, Osaka University, JapanReviewed by:
Yuji Morita, Aichi Gakuin University, JapanMasato Akiba, National Institute of Animal Health, Japan
Copyright © 2015 Brunelle, Bearson and Bearson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Brian W. Brunelle, Food Safety and Enteric Pathogens Research Unit, National Animal Disease Center, Agricultural Research Service, United States Department of Agriculture, 1920 Dayton Ave., Ames, IA 50010, USA e-mail: brian.brunelle@ars.usda.gov
 Brian W. Brunelle
Brian W. Brunelle Bradley L. Bearson
Bradley L. Bearson Shawn M. D. Bearson
Shawn M. D. Bearson