- 1Jeffrey Cheah School of Medicine and Health Sciences, Monash University Malaysia, Selangor Darul Ehsan, Malaysia
- 2School of Science, Monash University Malaysia, Selangor Darul Ehsan, Malaysia
- 3UKM Medical Molecular Biology Institute (UMBI), UKM Medical Centre, Bandar Tun Razak, Kuala Lumpur, Malaysia
- 4Division of Genetics and Molecular Biology, Institute of Biological Sciences, Faculty of Science, University of Malaya, Kuala Lumpur, Malaysia
The incidence of foodborne diseases has increased over the years and resulted in major public health problem globally. Foodborne pathogens can be found in various foods and it is important to detect foodborne pathogens to provide safe food supply and to prevent foodborne diseases. The conventional methods used to detect foodborne pathogen are time consuming and laborious. Hence, a variety of methods have been developed for rapid detection of foodborne pathogens as it is required in many food analyses. Rapid detection methods can be categorized into nucleic acid-based, biosensor-based and immunological-based methods. This review emphasizes on the principles and application of recent rapid methods for the detection of foodborne bacterial pathogens. Detection methods included are simple polymerase chain reaction (PCR), multiplex PCR, real-time PCR, nucleic acid sequence-based amplification (NASBA), loop-mediated isothermal amplification (LAMP) and oligonucleotide DNA microarray which classified as nucleic acid-based methods; optical, electrochemical and mass-based biosensors which classified as biosensor-based methods; enzyme-linked immunosorbent assay (ELISA) and lateral flow immunoassay which classified as immunological-based methods. In general, rapid detection methods are generally time-efficient, sensitive, specific and labor-saving. The developments of rapid detection methods are vital in prevention and treatment of foodborne diseases.
Introduction
Foodborne diseases have become a major public health problem worldwide due to the significantly increased incidence of foodborne diseases over the last 20 years (Oliver et al., 2005). Although it is difficult to estimate the global incidence of foodborne diseases as some of the cases are under-reported especially in developing countries, but the increased incidence of foodborne diseases were reported in many parts of the world (Van de Venter, 2000). For instance, the outbreak of foodborne disease in Taiwan increased rapidly from 121 in 1995 to 177 in 1996 and since then the incidence keep rising (Chiou et al., 2000). According to report from Centers for Diseases Control and Prevention (CDC), approximately 48 million people in the United States get ill, 128000 people are hospitalized and 3000 people die annually due to foodborne diseases despite United States has the safest food supplies in the world (Oliver et al., 2005; Centers for Disease Control and Prevention, 2011). In addition, about a quarter of the population is at a higher risk for foodborne diseases nowadays (Oliver et al., 2005).
Generally, foodborne diseases are caused by the consumption of food or water contaminated with pathogens or their toxins. Pathogens that caused foodborne diseases are often referred as foodborne pathogens and they include bacteria, viruses, fungi and parasites (Zhao et al., 2014). There are 31 identified foodborne pathogens in the United State and it is estimated that viruses are the primary causes of illnesses whereas bacteria are the primary causes of hospitalizations and deaths (Scallan et al., 2011). The common foodborne pathogens which are responsible for most of the foodborne disease outbreaks are Listeria monocytogenes, Escherichia coli O157:H7, Staphylococcus aureus, Salmonella enterica, Bacillus cereus, Vibrio spp., Campylobacter jejuni, Clostridium perfringens, and Shiga toxin-producing Escherichia coli (STEC) (Oliver et al., 2005; Scallan et al., 2011; Zhao et al., 2014).
The increasing amounts of street foods and the increasing demand for minimally processed ready-to-eat products have begun to concern public health agencies on food safety assurance (Lee et al., 2014). Foodborne pathogens are present in various foods such as fruits, vegetables and ready-to-eat products which are consumed without any further treatment (Chung et al., 2010; Lee et al., 2014). This may lead to foodborne diseases if food safety issues are not taken into consideration. Also, foodborne diseases are often associated with the consumption of raw or undercooked foods such as seafood, meat and poultry (Wingstrand et al., 2006; Rosec et al., 2012; Omurtag et al., 2013). It is essential to analyze the food for the presence of foodborne pathogens in order to ensure a safe food supply and to minimize the occurrence of foodborne diseases.
The conventional methods for detecting the foodborne bacterial pathogens present in food are based on culturing the microorganisms on agar plates followed by standard biochemical identifications (Mandal et al., 2011). Conventional methods are usually inexpensive and simple but these methods can be time consuming as they depend on the ability of the microorganisms to grow in different culture media such as pre-enrichment media, selective enrichment media and selective plating media. Usually conventional methods require 2 to 3 days for preliminary identification and more than a week for confirmation of the species of the pathogens (Zhao et al., 2014). Conventional methods are laborious as they require the preparation of culture media, inoculation of plates and colony counting (Mandal et al., 2011). Furthermore, conventional methods may be limited by their low sensitivity (Lee et al., 2014). False negative results may occur due to viable but non-culturable (VBNC) pathogens. The failure to detect foodborne pathogens would increase the transmission risk of pathogens.
Recently, different rapid methods with high sensitivity and specificity have been developed to overcome the limitations of conventional methods for the detection and identification of foodborne pathogens. Furthermore, researchers are still developing novel methods with improvements in terms of rapidity, sensitivity, specificity and suitability for in situ analysis and distinction of the viable cell (Zhao et al., 2014). Rapid detection methods are important, particularly in food industry, as they are able to detect the presence of pathogens in raw and processed foods immediately. Rapid methods are also sensitive enough to detect pathogens that present in low numbers in the food. Sensitivity is important because a single pathogen present in food has the risk to cause infection. Rapid methods are more time-efficient, labor-saving and able to reduce human errors (Mandal et al., 2011). Nevertheless, each of the rapid method has its own advantages and limitations. Generally, rapid detection methods are categorized into nucleic acid-based, biosensor-based and immunological-based methods (Zhao et al., 2014). This review examines these recent rapid detection methods and their applications in foodborne bacterial pathogens detection and along with their advantages and limitations.
Nucleic Acid-Based Methods
Nucleic acid-based methods operate by detecting specific DNA or RNA sequences in the target pathogen. This is done by hybridizing the target nucleic acid sequence to a synthetic oligonucleotide (probes or primers) which is complementary to the target sequence (Zhao et al., 2014). There are many bacterial pathogens such as Clostridium botulinum, Vibrio cholerae, Staphylococcus aureus, and Escherichia coli O157 which produced toxins that caused foodborne diseases (Singh et al., 2001; Akbulut et al., 2004; Fusco et al., 2011; Son et al., 2014). The toxin-related genes in these pathogens can be detected by nucleic acid-based methods (Zhao et al., 2014). Furthermore pathogens showing ambiguous phenotypic characteristics such as hippurate negative Campylobacter jejuni strains can be identified and confirmed by nucleic acid-based methods (Adzitey et al., 2012). Nucleic-acid based methods detect the specific genes in the target pathogens, therefore preventing ambiguous or wrongly interpreted results. The recent nucleic acid-based methods described are simple polymerase chain reaction (PCR), multiplex polymerase chain reaction (mPCR), real-time/quantitative polymerase chain reaction (qPCR), nucleic acid sequence-based amplification (NASBA), loop-mediated isothermal amplification (LAMP) and microarray technology.
Polymerase Chain Reaction (PCR)
One of the most commonly used molecular-based method for the detection of foodborne bacterial pathogens is polymerase chain reaction (PCR). PCR was invented about 30 years ago and it allows the detection of a single bacterial pathogen that present in food by detecting a specific target DNA sequence (Velusamy et al., 2010). PCR operates by amplifying a specific target DNA sequence in a cyclic three steps process (Mandal et al., 2011). Firstly, the target double-stranded DNA is denatured into single-stranded DNA at high temperature. Then, two single-stranded synthetic oligonucleotides or specific primers which are the forward and reverse primer will anneal to the DNA strands. This is followed by the polymerization process whereby the primers complementary to the single-stranded DNA are extended with the presence of deoxyribonucleotides and a thermostable DNA polymerase. The PCR amplification products are visualized on electrophoresis gel as bands by staining with ethidium bromide (Zhao et al., 2014). PCR have been used in the detection of numerous foodborne pathogens like Listeria monocytogenes, Escherichia coli O157:H7, Staphylococcus aureus, Campylobacter jejuni, Salmonella spp. and Shigella spp. (Cheah et al., 2008; Lee et al., 2008; Alves et al., 2012; Chiang et al., 2012; Zhou et al., 2013).
Multiplex PCR (mPCR)
Multiplex PCR offers a more rapid detection as compared to simple PCR through the simultaneous amplification of multiple gene targets. The basic principle of mPCR is similar to conventional PCR. However, several sets of specific primers are used in mPCR assay whereas only one set of specific primers are used in conventional PCR assay. Primer design is very important for the development of mPCR, as the primer sets should have similar annealing temperature in order to produce a successful mPCR assay (Zhao et al., 2014). Besides, the concentration of primers is also important in mPCR. This is because interaction may occur between the multiple primer sets in mPCR that results in primer dimers, thus, the concentration of primers may need to be adjusted to ensure the production of reliable PCR products (Zhao et al., 2014). Other important factors for a successful mPCR assay include the PCR buffer concentrations, the balance between magnesium chloride and deoxynucleotide concentrations, the quantities of DNA template, cycling temperatures and Taq DNA polymerase (Markoulatos et al., 2002; Cheah et al., 2008; Khoo et al., 2009).
Previously, mPCR was used to detect around two to three pathogens only. Now, mPCR is more advanced and it can detect up to five or more pathogens simultaneously. Chen et al. (2012) had carried out mPCR for the simultaneous detection of Salmonella Enteritidis, Staphylococcus aureus, Shigella flexneri, Listeria monocytogenes, and Escherichia coli O157:H7 using five pairs of primers targeting invasion protein (invA), 16S rDNA, invasion plasmid antigen H (ipaH), listeriolysin O (hlyA) and intimin (eaeA) gene, respectively. Ryu et al. (2013) had developed a novel mPCR assay which is able to detect and discriminate six Listeria species simultaneously in one tube with high accuracy. The Listeria species that were successfully identified are Listeria monocytogenes, Listeria grayi, Listeria ivanovii, Listeria innocua, Listeria welshimeri, and Listeria seeligeri. The mPCR detection limit was 7.58 × 104 copies for mixed genomic DNA.
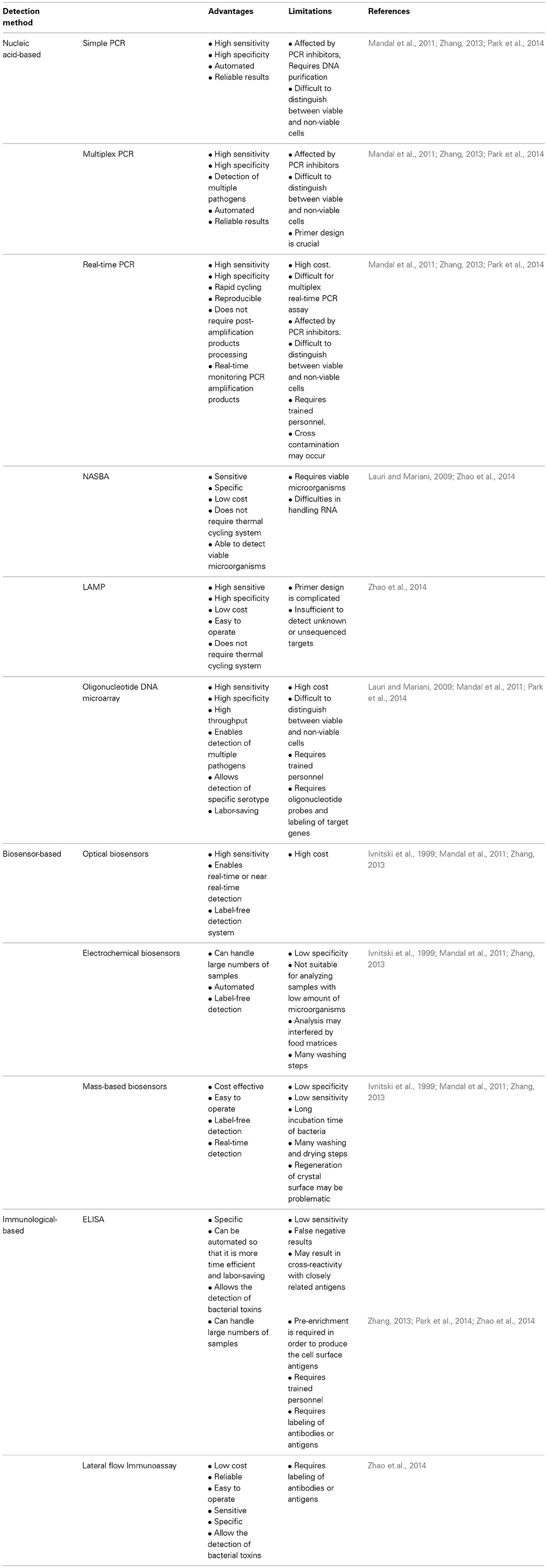
Further improvements of mPCR include the development of a novel GeXP-PCR by Zhou et al. (2013) for the simultaneous detection of six foodborne bacterial pathogens: Salmonella enterica, Listeria monocytogenes, Staphylococcus aureus, Escherichia coli O157:H7, Shigella spp. and Campylobacter jejuni. GenomeLab Gene Expression Profiler (GeXP) genetic analysis system allows high-throughput and detection of multiple pathogens in a single reaction. The analytical procedure involves primer design, PCR amplification and capillary electrophoretic separation of PCR products instead of agarose gel electrophoresis. The GeXP multiplex PCR amplification involves the use of chimeric primers, universal primers and capillary electrophoretic separation of PCR products instead of agarose gel electrophoresis. The chimeric primers contain a gene-specific sequence with a universal tag at the 5′end and they are used to produce amplicons with universal tags. Then, the universal primers which contain the same sequence as the universal tags used in the chimeric primers will drive the remaining PCR reactions. The forward universal primer is covalently labeled with a fluorescent dye at the 5′ end and it is used for the detection during capillary electrophoresis (Zhou et al., 2013). This method was found to be more sensitive and suitable for high-throughput analysis. The detection limit of GeXP-PCR for Salmonella, Listeria monocytogenes, Staphylococcus aureus, Escherichia coli O157:H7, Shigella spp. and Campylobacter jejuni was 420, 310, 270, 93, 85, and 66 CFU/mL respectively. Additional examples of the application of mPCR for the detection of foodborne pathogens in various food matrices are listed in Table 1.
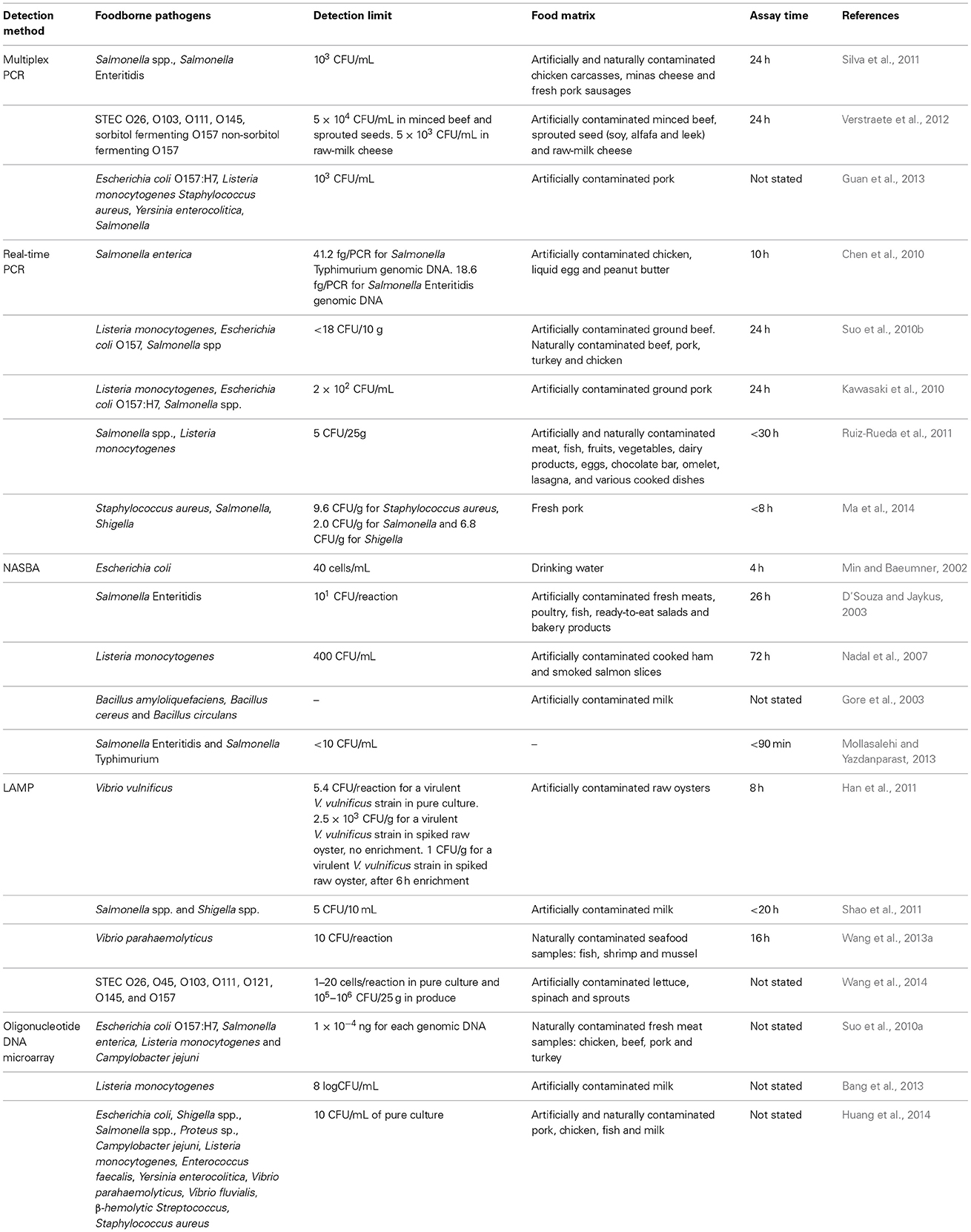
Table 1. Examples of the application of nucleic acid-based methods for the detection of various foodborne pathogens present in food samples.
Real-Time or Quantitative PCR (qPCR)
Real-time PCR or quantitative PCR is different from simple PCR whereby it does not require agarose gel electrophoresis for the detection of PCR products. This method is able to monitor the PCR products formation continuously in the entire reaction by measuring the fluorescent signal produced by specific dual-labeled probes or intercalating dyes. The fluorescence intensity is proportional to the amount of PCR amplicons (Omiccioli et al., 2009; Zhao et al., 2014).
Several fluorescent systems have been developed for qPCR and the most commonly used fluorescent systems include SYBR green, TaqMan probes and molecular beacons. SYBR green is a double-stranded DNA (dsDNA)-binding fluorescent dye (Hein et al., 2001). This non-sequence-specific intercalating dye emits little fluorescence and the fluorescence signal is enhanced when bound to the minor groove of the DNA double helix (Fukushima et al., 2003; Levin, 2005; Singh et al., 2009).
TaqMan probes and molecular beacons are the common alternatives to SYBR green. TaqMan probes, also known as double-dye probes, are oligonucleotides that contain a fluorophore as the reporter dye at the 5′-end and the quenching dye at the 3′-end (Hein et al., 2001; Levin, 2005). The reporter dye and the quenching dye are close to each other and this prevent the emitted fluorescence of the reporter (Levin, 2005). TaqMan probe is complementary to a specific nucleotide sequence in one of the strands of amplicon internal to both primers and the system depends on the 5′-3′ exonuclease activity of Taq DNA polymerase that cleaves the probe and separates both dyes in order to generate the fluorophore signal (Patel et al., 2006).
Molecular beacons are oligonucleotide probes with hairpin/stem-and-loop configuration, in which the sequence complementary to a target sequence is present in the loop portion and the stem is produced by annealing of two complementary arm sequences. A reporter dye is attached to one end of the probe and a quencher dye is attached to the other end. Both dyes are in close proximity which maintained by the hybrid stem, hence, no fluorescence is produced (Leone et al., 1998; Chen et al., 2000; Levin, 2005; Patel et al., 2006). Molecular beacon produces fluorescence signal upon hybridization of the probe to its complementary nucleotide sequence in the amplicon. During hybridization, the probe undergoes spontaneous conformational change that separates the two dyes and this allow fluorescence to occur (Leone et al., 1998; Chen et al., 2000; Liming and Bhagwat, 2004; Levin, 2005).
The detection of Salmonella in fresh-cut fruits and vegetables by molecular beacon qPCR targeting the invasion associated gene (iagA) was first reported by Liming and Bhagwat (2004). The detection limit was approximately 4 CFU/25 g of produce after enrichment. Tyagi et al. (2009) developed a sensitive and highly reproducible SYBR green qPCR assay for the detection of pathogenic tdh-positive Vibrio parahaemolyticus in tropical shellfish. The detection limit was 102 CFU/ml for shrimp homogenates spiked with pure culture. Other advances in qPCR include the development of TaqMan and SYBR Green qPCR to identify and quantitatively detect Staphylococcus aureus strains that harbor the enterotoxin gene cluster (egc) in raw milk. Approximately 1 × 103 CFU/mL Staphylococcus aureus was detected by SYBR green qPCR whereas 1 × 104 CFU/mL Staphylococcus aureus was detected by TaqMan qPCR in raw milk (Fusco et al., 2011).
Furthermore, multiplex qPCR assay is also developed for the detection and quantification of multiple foodborne pathogens. Fratamico et al. (2011) performed a study to detect Shiga toxin-producing Escherichia coli serogroups O26, O45, O103, O111, O121, and O145 in ground beef using multiplex qPCR with primers which target Shiga toxins (stx1 and stx2), intimin (eae) and wzx genes. Taqman probe with different combinations of fluorophores were used to detect the product of multiplex qPCR with detection limit around 50 CFU per PCR. Additionally, a simultaneous detection of Vibrio parahaemolyticus, Vibrio cholerae, and Vibrio vulnificus by multiplex qPCR using primers that target vmrA, zot and vuuA genes respectively was developed by Kim et al. (2012). The multiplex qPCR products was detected and quantitated by using SYBR green. Hu et al. (2014) conducted a study to detect eight foodborne bacterial pathogens which include Salmonella enterica subsp. enterica, Listeria monocytogenes, Escherichia coli O157, Vibrio parahaemolyticus, Vibrio vulnificus, Campylobacter jejuni, Enterobacter sakazakii and Shigella spp. by using molecular beacon based multiplex qPCR targeting ssaR, hlyA, rfbE, toxR, vvh, gyrA, 16SrRNA and ipaH genes respectively. The detection limits of the assay ranged from 1.3 × 103–1.6 × 104 CFU/g stool.
Among the discussed fluorescent systems for qPCR, SYBR green is simple and less costly as compared to TaqMan probes or molecular beacons (Fukushima et al., 2003; Levin, 2005). Besides, the advantage of using SYBR green is that DNA melting curve can be generated after PCR along with the calculation of the Tm value of the amplified products. This is required in order to differentiate the target products from the primer dimer formation. The primer dimer products have lower Tm values as compared to the amplicons (Levin, 2005). SYBR green lacks specificity and it binds to all double-stranded DNA, thus, it can be used to detect any PCR product (Levin, 2005; Madani et al., 2005). However, SYBR green dye will bind to other non-specific reaction products which include primer dimers (Madani et al., 2005). As for TaqMan probes and molecular beacons, they are sequence specific probes and they only bind to their target sequence, hence, primer dimers will not be detected (Levin, 2005). There are some studies have shown that TaqMan-based qPCR is more sensitive compared to SYBR green or molecular beacons-based qPCR (Hein et al., 2001; Klerks et al., 2004). Nevertheless, the sensitivity of PCR-based method is mainly affected by primer specificity, primer sequence and annealing temperature, rather than the choice of detection probe (Klerks et al., 2004). Overall, qPCR is more sensitive than conventional PCR and it minimizes the risk of cross-contamination (Omiccioli et al., 2009). More examples of the detection of foodborne bacterial pathogens by qPCR are presented in Table 1.
Conventional PCR and multiplex PCR that require agarose gel analysis for the detection of PCR products are laborious and time-consuming, thus, not suitable for high-throughput analysis and difficult to automate (Patel et al., 2006; Zhou et al., 2013). As for qPCR, post-PCR processing is not required. This allows low risk of cross-contamination, high-throughput analysis and automation (Fricker et al., 2007). The advantages of qPCR have led to the development of various commercial qPCR kits for the detection of foodborne pathogens such as Salmonella, Listeria monocytogenes, Escherichia coli O157:H7 and Campylobacter (Maurer, 2011).
The examples of commercial qPCR kits for the detection of Salmonella include Salmonella BAX™ PCR (DuPont Qualicon), AnDiaTec® Salmonella real time PCR Kit (AnDia Tec), Probelia™ Salmonella sp. (Sanofi Diagnostics Pasteur), and TaqMan™ Salmonella detection kit (PE Applied Biosystems) (Maciorowski, 2005; Maurer, 2011; Park et al., 2014). Kimura et al. (1999) evaluated the TaqMan™ Salmonella detection kit for the detection of Salmonella in shrimp and meat. The detection limit was 120 CFU/mL in pure culture. Wan et al. (2000) evaluated the Probelia™ Salmonella sp. PCR amplification and detection kits for the detection of Salmonella agona in artificially contaminated milk powder and ricotta cheese. The detection limit was approximately 8–79 CFU/mL. Besides, Margot et al. (2013) evaluated seven commercial qPCR kits for the detection of Salmonella which include BAX® system Q7 real-time PCR (DuPont Qualicon) and other commercial qPCR kit such as ADIAFOOD® Salmonella (AES chemunex), BIOTECON foodproof Salmonella Detection Kit (Biotecon Diagnostics), BioControl Assurance GDS® TM Salmonella (BioControl), Genedisc® Shiga Toxic E. colii and Salmonella spp. (Pall GeneDisc® Technologies), BioRad iQ-Check Salmonella 2 (Biorad) and MicroSeq® Salmonella spp. Detection Kit (Applied Biosystems). A total of 49 Salmonella strains were included in this study and correctly identified by all the evaluated systems.
The commercial qPCR kits available for the detection of Listeria monocytogenes are BAX® Listeria monocytogenes Detection System (DuPont Qualicon), TaqMan® Listeria monocytogenes Detection Kit (Applied Biosystems), ADIAFOOD rapid pathogen detection system for Listeria monocytogenes (AES Chemunex), Probelia® Listeria monocytogenes PCR system (Biorad), LightCycler® Listeria monocytogenes Detection Kit (Roche/Biotecon) and GeneVision® Rapid Pathogen Detection System for Listeria monocytogenes (Janzten et al., 2006; Maurer, 2011; Traunšek et al., 2011). The BAX® Listeria monocytogenes Detection System is adopted by the USDA (2012) as a screening test for L. monocytogenes. Besides, Wan et al. (2003) conducted a study to compare the Probelia® Listeria monocytogenes PCR system and the International Organization for Standardization (ISO) method 11290–1 for the detection of L. monocytogenes in salmon samples. The results indicated that Probelia® Listeria monocytogenes PCR system is as good as the ISO method and the detection limit was approximately 20 CFU/mL broth culture for pure culture of L. monocytogenes.
There are many more commercial qPCR kits that are available for the detection of other foodborne bacterial pathogens such as Escherichia coli and Campylobacter. For instance, Escherichia coli O157 BAX® PCR (DuPont Qualicon), ADIAFOOD rapid pathogen detection system for E. coli O157 and E. coli O157:H7 (AES Chemunex) and ADIAFOOD Campylobacter JCL qPCR commercial kit (AES Chemunex) for the detection of C. jejuni, C. coli and C. lari (Maurer, 2011; Vencia et al., 2014).
Nucleic Acid Sequence-Based Amplification (NASBA)
NASBA is developed by Compton (1991) in the early 90s and it operates by amplifying nucleic acids under isothermal conditions, unlike PCR which requires thermocycling system. NASBA is normally used for the amplification of RNA whereby the single-stranded RNA template is converted into complementary DNA (cDNA) by the reverse transcriptase during the reaction. NASBA reaction occurs at around 41°C, involving two specific primers and three enzymes: avian myeloblastosis virus (AMV) reverse transcriptase, T7 RNA polymerase and RNase H. The NASBA amplicons can be detected by agarose gel electrophoresis (Leone et al., 1998; Zhao et al., 2014).
The post-NASBA product detection methods such as agarose gel electrophoresis or enzyme-linked gel assay is considered labor-intensive and not cost-effective. This leads to the development of a novel real-time NASBA which uses fluorescently labeled probes which are molecular beacons to detect the single-stranded RNA amplicons, thus, producing a homogenous NASBA assay (Leone et al., 1998). Real-time NASBA has been used for the detection of various foodborne pathogens such as Salmonella enterica, Vibrio cholerae, Staphylococcus aureus, Campylobacter jejuni, and Campylobacter coli (Simpkins et al., 2000; Churruca et al., 2007; Fykse et al., 2007; O'Grady et al., 2009). In addition, real-time NASBA is able to detect viable microorganisms that present in food samples through mRNA amplification and the detection of RNA targets will indicate the presence of viable cells (Simpkins et al., 2000). Real-time NASBA has been used to distinguish viable from non-viable bacterial cells and RNase treatment is usually required to degrade target mRNA from dead cells before nucleic acid extraction or treating the samples with RNase-free DNase is required prior to performing the NASBA assay (Blais et al., 1997; Nadal et al., 2007; Dwivedi and Jaykus, 2011).
NASBA offers high-throughput analysis and it has been commercialized as kits. There are several commercial NASBA kits manufactured by Life Sciences, KIT Biomedical Research, Gen-Probe and bioMérieux (Gracias and McKillip, 2007). However, the commercial NASBA kit that is used for the detection of foodborne bacterial pathogens is mainly the Nuclisens EasyQ® Basic Kit (bioMérieux). Nuclisens EasyQ® Basic Kit can be used for the detection and identification of Listeria monocytogenes, Salmonella enterica and Vibrio cholerae (D'Souza and Jaykus, 2003; Fykse et al., 2007; Nadal et al., 2007).
Loop-Mediated Isothermal Amplification (LAMP)
LAMP is a novel nucleic acid amplification method developed by Notomi et al. (2000) which provides a rapid, sensitivity and specific detection of foodborne pathogens. LAMP is based on auto-cycling strand displacement DNA synthesis carried out by Bst DNA polymerase large fragment under isothermal conditions between 59°C and 65°C for 60 min. In LAMP, four primers comprising two inner primers and two outer primers are used to target six specific regions of target DNA. Cauliflower-like DNA structures bearing multiple loops as well as stem-loop DNAs of different sizes are the final products of LAMP. Large amount of amplicons can be produced by LAMP within 60 min which is usually 103-fold or higher as compared to simple PCR. The LAMP amplicons can be detected by agarose gel electrophoresis or SYBR Green I dye (Wang et al., 2008; Zhao et al., 2014).
In the field of foodborne pathogen detection, LAMP was used for the first time to detect stxA2 gene in Escherichia coli O157:H7 (Maruyama et al., 2003). Since then, LAMP has been used for the detection of various foodborne pathogens due to its rapidity and sensitivity. LAMP is proven to be more specific and sensitive as compared to PCR assays for the detection of foodborne pathogens (Hara-Kudo et al., 2005; Wang et al., 2008; Yamazaki et al., 2008). This is because LAMP uses four primers targeting six specific regions and it provides rapid amplification, greater yield of amplification products and lower detection limits than PCR assays (Hara-Kudo et al., 2005; Xu et al., 2012). Till date, commercial LAMP kits are available for the detection of Listeria, Salmonella, Campylobacter, Legionella, and verotoxin-producing Escherichia coli (Mori and Notomi, 2009). For example, the Loompamp detection kit (Eiken Chemical) is commercially available for the detection of foodborne pathogens such as Salmonella enterica (Ohtsuka et al., 2005), Shigella (Song et al., 2005), enteroinvasive Escherichia coli (Song et al., 2005), verotoxigenic Escherichia coli O157 and O26 (Hara-Kudo et al., 2008) and Campylobacter (Yamazaki et al., 2009).
Besides, different types of LAMP assays have been developed for the detection of foodborne pathogens. For instance, multiplex LAMP, reverse-transcription LAMP, real-time LAMP and in situ LAMP (Chen et al., 2008; Han and Ge, 2010; Shao et al., 2011; Ye et al., 2011). The availability of real-time monitoring of LAMP amplification products by the presence of turbidity or fluorescence eliminates the need for staining with ethidium bromide and gel electrophoresis. Therefore, this allows high-throughput analysis along with its high sensitivity and specificity (Yang et al., 2010). More examples of the application of NASBA and LAMP in food samples are shown in Table 1.
Oligonucleotide DNA Microarray
The recent progress in multi-gene detection technology includes the microarray technology (Call et al., 2001). Microarrays were originally used for the study of gene expression, but oligonucleotide DNA microarray has been widely used in the field of foodborne pathogen detection. Microarrays are made up of glass slides or chips coated with up to hundreds of specific oligonucleotide probes and these probes are chemically synthesized short sequences range from 25 to 80 bp (Severgnini et al., 2011). Each oligonucleotide probe is able to target a specific part of a gene sequence. In this method, the sample nucleic acid fragments (DNA, mRNA or cDNA) are labeled with fluorescent dye, and then they are denaturated to generate single-stranded fragments. These fragments will hybridize to the array through binding to their corresponding oligonucleotide probes. The results are obtained through the visualization of the fluorescence signal produced from the probe-sample complex. The fluorescence intensity is proportional to the concentration of each labeled nucleic acid fragment (Lauri and Mariani, 2009).
Li et al. (2006) had presented the first report of detection of pathogenic Shigella and Escherichia coli serotypes by oligonucleotide DNA microarray. The detection of a specific serotype can be crucial especially for Escherichia coli, as this pathogen has different serotypes with different level of pathogenicity that ranges from harmless strain Escherichia coli K-12 to deadly strain Escherichia coli O157:H7 (Lauri and Mariani, 2009). Moreover, Wang et al. (2007) developed a microarray assay which allows the detection and identification of 22 foodborne pathogens. Some examples of these pathogens are Staphylococcus aureus, Listeria monocytogenes, Vibrio parahaemolyticus, Vibrio cholerae, Campylobacter jejuni, Clostridium perfringens, Shigella spp. Salmonella spp., and Bacillus cereus. Other studies that involved the application of oligonucleotide DNA microarray for the detection of foodborne pathogens are presented in Table 1.
DNA microarrays are commercially available but most of them are designed for gene expression analysis studies (Rasooly and Herold, 2008). The commercial in situ-synthesized arrays are high-density microarrays where short oligonucleotide probes range from 20 to 25 bp are synthesized directly on the surface of the microarray. In addition, multiple probes per target are included for higher sensitivity, specificity and accuracy. These high density microarrays require special manufacturing and they are relatively high in cost (Rasooly and Herold, 2008; Severgnini et al., 2011). There are various commercially available DNA microarrays manufactured by Affymetrix, Roche NimbleGen and Agilent Technologies (Severgnini et al., 2011).
Most of the commercial microarrays are not desirable for specialized application such as food microbial analysis or diagnostic laboratory because low to medium density array will serve as the ideal microarray platform that can provide reliable results without involving the use of complicated equipments and data management (Rasooly and Herold, 2008; Severgnini et al., 2011). In this case, custom microarrays are available from the Department of Bioresources at Seibersdorf and other organizations. Custom microarrays are sensitive, specific and less expensive than commercial microarrays (Mothershed and Whitney, 2006; Severgnini et al., 2011). Nevertheless, low-density microarray is commercially available. For instance, StaphyChips® developed by Affymetrix and in collaboration with Advanced Array Technology (ATT, Eppendorf Array Technologies). StaphyChips® is able to detect a total of 15 Staphylococcus species, including methicillin-resistant Staphylococcus aureus (MRSA) (Mothershed and Whitney, 2006).
In general, DNA oligonucleotide microarray allows simultaneous identification of multiple foodborne bacterial pathogens. Therefore, it is capable of high-throughput analysis and it also has the potential to be automated (Al-Khaldi, 2002; De Boer and López, 2012; Zhou et al., 2013).
Biosensor-Based Methods
Biosensor is an analytical device that consists of two main elements: a bioreceptor and a transducer. The bioreceptor responsible for recognizing the target analyte can either be a:
(1) Biological material: enzymes, antibodies, nucleic acids and cell receptors, or
(2) Biologically derived material: aptamers and recombinant antibodies, or
(3) Biomimic: imprinted polymers and synthetic catalysts.
The transducer that converts the biological interactions into a measurable electrical signal can be optical, electrochemical, mass-based, thermometric, micromechanical or magnetic (Velusamy et al., 2010; Zhao et al., 2014).
Biosensors are easy to operate and they do not require sample pre-enrichment, unlike nucleic-acid based methods and immunological methods which require sample pre-enrichment for concentrating the pathogens before detection (Singh et al., 2013). The recent biosensors that commonly used for the detection of foodborne pathogens are optical, electrochemical and mass-based biosensors (Zhang, 2013; Zhao et al., 2014). The examples of the application of different types of biosensors in foodborne pathogen detection are given in Table 2.
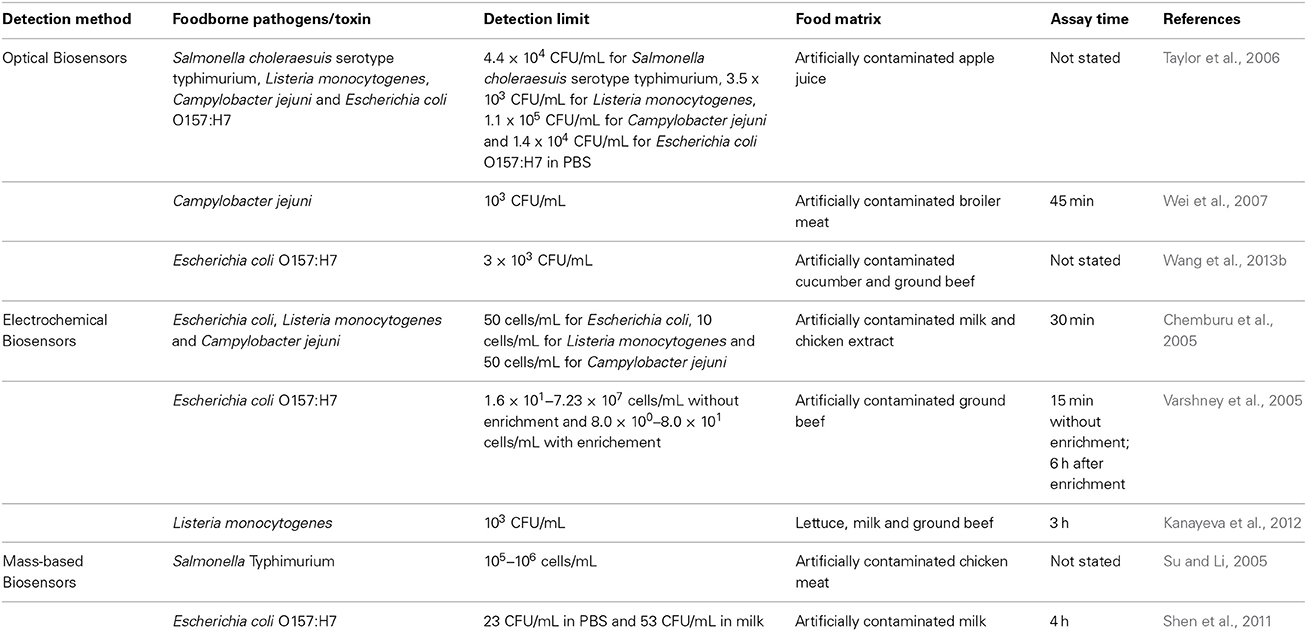
Table 2. Examples of the application of biosensor-based methods for the detection of various foodborne pathogens present in food samples.
Optical Biosensors
The most commonly used optical biosensor for the detection of foodborne pathogen is surface plasmon resonance (SPR) biosensor due to their sensitivity. SPR employs reflectance spectroscopy for the pathogen detection (Velusamy et al., 2010). In SPR, bioreceptors are immobilized on the surface of a thin metal. The electromagnetic radiation of a certain wavelength interacts with the electron cloud of the thin metal and produces a strong resonance. When the pathogen binds to the metal surface, this interaction alters its refractive index which results in the change of wavelength required for electron resonance (Zhang, 2013; Zhao et al., 2014).
Commercial optical biosensors using SPR techniques such as SPREETA biosensor and BIACORE 3000 biosensor are currently available for the detection of foodborne pathogens. Waswa et al. (2007) used SPREETA biosensor for the detection of E. coli O157:H7 in milk, apple juice and ground beef. The detection limit was around 102–103 CFU/mL. Furthermore, Salmonella Enteritidis and Salmonella Typhimurium were successfully detected by SPREETA biosensor (Son et al., 2007; Lan et al., 2008). Besides, Listeria monocytogenes was successfully detected by BIACORE 3000 biosensor with detection limit of 1 × 105 cells/mL (Leonard et al., 2004). Salmonella group B, D, and E, Escherichia coli O157:H7 and Salmonella Enteritidis were also successfully detected by BIACORE biosensor (Bokken et al., 2003; Waswa et al., 2006; Wang et al., 2011).
The commercially available biosensors for the detection of foodborne pathogens are mostly optical-based biosensors. The commercial biosensors offer varying degrees of automation (Leonard et al., 2003). The commercialization of biosensors is slower than other rapid methods due to several factors such as cost consideration, quality assurance, stability issues, sensitivity issues and instrumentation design. There are difficulties in the methods of producing inexpensive and reliable sensors, the storage of biosensors, the stabilization of biosensors, methods of sensor calibration and total integration of the sensor system (Velasco-Garcia and Mottram, 2003).
Electrochemical Biosensors
Electrochemical biosensors are further classified into several types such as amperometric, impedimetric, potentiometric, and conductometric according to the measurement of changes in current, impedance, voltage and conductance respectively, which caused by antigen-bioreceptor interactions (Zhang, 2013).
Many researchers had reported the successful detection of foodborne pathogens by electrochemical biosensors. For example, Pal et al. (2008) successfully detected Bacillus cereus present in alfalfa sprouts, strawberries, lettuce, tomatoes, fried rice and cooked corn by a direct-charge transfer conductometric biosensor. The detection limit of this method was around 35–88 CFU/mL. Amperometric magnetoimmunosensor was used for the detection of Staphylococcus aureus, the detection limit of this assay was 1 CFU/mL and the analysis time was 2 h (de Ávila et al., 2012). Munoz-Berbel et al. (2008) had described the use of impedimetric spectroscopy for the detection and quantification of Escherichia coli, the detection limit was found to be 101–107 CFU/mL. Ercole et al. (2003) had reported the successful detection of Escherichia coli in vegetable food by using antibody-based potentiometric biosensor with detection limit of 10 cells/mL.
Mass-Based Biosensors
Mass-based or mass-sensitive biosensors operate based on the detection of small changes in mass. Mass-based biosensors involve the use of piezoelectric crystal which will vibrate at a certain frequency when induced by an electrical signal of a certain frequency. The bioreceptors (e.g., antibodies) for the detection of pathogens (e.g., antigens) are immobilized on this crystal. Once the target antigens bind to the antibodies immobilized on the crystal, this will cause a measurable change in the vibrational frequency of the crystal which correlates with the added mass on the crystal surface. There are two major types of mass-based biosensors which are the bulk acoustic wave resonators (BAW) or quartz crystal microbalance (QCM) and surface acoustic wave resonators (SAW) (Velusamy et al., 2010; Zhang, 2013).
However, the application of mass-based biosensors in the field of foodborne pathogen detection is generally lesser than electrochemical and optical biosensors (Velusamy et al., 2010). The use of piezoelectric immunosensor for the detection of Salmonella Enteritidis with detection limit of 1 × 105 cells/mL (Si et al., 2001) and the detection of Escherichia coli with detection limit of 106–109 CFU/mL (Pohanka et al., 2007). Detection of toxigenic Escherichia coli O157:H7 by using a SAW biosensor was reported by Berkenpas et al. (2006). The QCM immunosensor was employed by Vaughan et al. (2001) for the detection of Listeria monocytogenes and the detection limit was 1 × 107 cells/mL. Moreover, Liu et al. (2007) employed QCM immunosensor for the detection of Escherichia coli O157:H7 and the detection limit was 102 CFU/mL with detection time of less than 1.5 h.
Immunological-Based Methods
The detection of foodborne pathogens by immunological-based methods is based on antibody-antigen interactions, whereby a particular antibody will bind to its specific antigen. The binding strength of a particular antibody to its antigen determines the sensitivity and specificity of immunological-based methods. Immunological-based methods involve the use of polyclonal and monoclonal antibodies (Zhao et al., 2014). Enzyme-linked immunosorbent assay (ELISA) and lateral flow immunoassay is among the immunological-based methods which recently used for the detection of foodborne pathogens.
Enzyme-Linked Immunosorbent Assay (ELISA)
ELISA is one of the most commonly used immunological methods for the detection of foodborne pathogens. Sandwich ELISA is the most effective form of ELISA whereby it involves two antibodies (Zhao et al., 2014). The primary antibody is usually immobilized onto the walls of the microtiter plate wells. The target antigen like bacterial cells or bacterial toxins from the food sample binds to the immobilized primary antibody and the remaining unbound antigens are removed. After that, an enzyme-conjugated secondary antibody is added and it will bind to the antigen and the remaining unbound antibodies are removed. The complex consisting antigen sandwiched between two antibodies is formed and it can be detected by adding a colorless substrate which will be converted into a colored form in the presence of the enzyme (Zhang, 2013). There are different types of enzymes can be used in ELISA, some of the most commonly used enzymes include horseradish peroxidase (HRP), alkaline phosphatase and beta-galactosidase (Yeni et al., 2014).
Many studies have been performed using the sandwich ELISA for rapid detection of foodborne pathogens. For example, Kumar et al. (2011) performed the detection of pathogenic Vibrio parahaemolyticus in seafood with sandwich ELISA, using monoclonal antibodies against the TDH-related hemolysin (TRH) of pathogenic Vibrio parahaemolyticus. The detection limit of this assay was 103 cells of pathogenic Vibrio parahaemolyticus. Commercial ELISA test kit such as BIOLINE Salmonella ELISA Test is also available for the detection of Salmonella in food products. The detection limit of this test kit was 1 CFU/25 g sample with minimum four of the 20 food matrixes tested (Bolton et al., 2000). ELISA is also commonly used for the detection of toxins present in foods such as Clostridium perfringens α, β, and ε toxin, staphylococcal enteroxins A, B, C, and E, botulinum toxins and Escherichia coli enterotoxins (Aschfalk and Mülller, 2002; Zhao et al., 2014).
Recently, high-throughput and automated ELISA systems such as VIDAS (BioMerieux) and Assurance EIA (BioControl) are available for the detection of foodborne pathogens (Glynn et al., 2006). VITEK immunodiagnostic assay system (VIDAS) is system that performs entire ELISA procedure automatically. It utilizes enzyme-linked fluorescent immunoassay (ELFA) which is similar to ELISA, but it is a more sensitive fluorescent immunoassay for reporting the results. Generally, this system can complete an assay in 45 min to 2 h which also depends on the test kit. The VIDAS system involves the use of reagent strip and a plastic tube known as solid phase receptacle (SPR). A liquid sample of an enriched sample is placed in the reagent strip that contains all the required reagents in a ready-to-use format. The SPR serves as the pipette and the solid phase for the assay. The instrument will perform ELFA by automatically transferring the sample to the SPR that coated with antibodies in its interior wall in order to capture the target pathogen or toxin. The SPR is then automatically transferred to a series of wells that contain enzyme-conjugated secondary antibodies and enzymes in a sequence manner. Once the assay is completed, the result will be automatically analyzed by the instrument and interpreted as positive or negative (Vaz-Velho et al., 2000; Fung, 2002).
Several studies applied VIDAS for the detection of Salmonella in pork sample, fruits and vegetables (Vieira-Pinto et al., 2007; Gómez-Govea et al., 2012), Listeria monocytogenes in fish samples, beef, pork, fruits, and vegetables (Vaz-Velho et al., 2000; Meyer et al., 2011; Gómez-Govea et al., 2012), Escherichia coli O157:H7 in Minas Frescal cheese, fruits, and vegetables (Gómez-Govea et al., 2012; Carvalho et al., 2014), Campylobacter spp. in fruits and vegetables (Gómez-Govea et al., 2012), and staphylococcal enterotoxin in raw milk cheese (Cremonesi et al., 2007).
Assurance Enzyme Immunoassay (EIA) is a commercial ELISA kit that allows automation and high-throughput testing (Fung, 2002). The Assurance EIA test kits for Salmonella, Escherichia coli, O157:H7, Listeria monocytogenes, and Campylobacter are currently available (Fung, 2002). Assurance EIA test kit for the detection of Escherichia coli O157:H7 in raw and cooked beef was used in the study conducted by Feldsine et al. (2002). Besides, Stewart et al. (2001) performed the detection of Salmonella in alfalfa sprouts by Assurance EIA test kit and Taha et al. (2010) performed the detection of Salmonella in chicken meat by Assurance EIA test kit.
Lateral Flow Immunoassay
ELISA offers a sensitive and accurate detection of foodborne pathogens. However, the operation of ELISA requires specialized equipment and trained personnel (Zhao et al., 2014). Hence, other immunological detection methods which are rapid, cheap, simple and reliable are required. Lateral flow immunoassays such as dipstick and immunochromatographic strips have been developed for rapid on-site detection of foodborne pathogens. Lateral flow immunoassay device is made up of four sections which are arranged orderly on a plastic backing, with sample pad starting at the bottom, followed by conjugate pad, nitrocellulose membrane and then absorbent pad. The sample fluid will migrate along the four sections of lateral flow immunoassay via capillary action. The sample fluid encounters and mixes with the conjugate, which can be antibody or antigen labeled by a color particle, at the conjugate pad and then pass through the lines in the nitrocellulose membrane that immobilized with antibody or antigen. The color particle can bind to the antibody or antigen immobilized at test line depending on the analytes present in the sample. The color can be visualized approximately two to 10 min after the addition of sample. There are two basic formats of lateral flow immunoassays: competitive assay which used to test analytes with single epitope and sandwich assay which used to test analytes with several epitopes (Ngom et al., 2010; Zhao et al., 2014).
The detection of foodborne pathogens by lateral flow immunoassay employs labels such as monodisperse latex, colloidal gold, carbon and fluorescent tags (Zhao et al., 2014). Immunochromatographic strip developed by Jung et al. (2005) to detect Escherichia coli O157 in enriched samples had used colloidal gold particles as label. This study had showed that the detection limit for Escherichia coli O157 without enrichment was 1.8 × 105 CFU/mL and after enrichment was 1.8 CFU/mL. Niu et al. (2014) employed an immunochromatographic test strip based on sandwich format with colloidal gold as label for the detection of Staphylococcus aureus. The detection limit for Staphylococcus aureus the study was 103 CFU/mL. Moreover, Xu et al. (2013) developed a novel immunochromatographic strip test which based on sandwich format with fluorescent microspheres as label for the detection of Campylobacter jejuni. The detection limit of this test was 106 CFU/mL. Lateral flow immonuassay is also used for detection of other foodborne bacterial pathogens such as Listeria spp. and Salmonella (Kim et al., 2007; Shukla et al., 2011). It can also be used to detect toxins which may cause foodborne diseases such as brevetoxins and staphylococcal enterotoxin B (Zhou et al., 2009; Rong-Hwa et al., 2010).
Commercial immunochromatographic test strips are also available for the detection of foodborne bacterial pathogens. For instance, Reveal® test kits (Neogen) for Listeria, Salmonella and Escherichia coli O157, VIP® GOLD™ (BioControl Systems) for Listeria, Salmonella, and Escherichia coli and DuPont™ Lateral Flow System (DuPont Qualicon) for Listeria (Fung, 2002; Shukla et al., 2011; Cho and Irudayaraj, 2013; Leem et al., 2014). Lateral flow immunoassays are simple and fast, but they are designed for individual tests rather than high-throughput screening (De Boer and López, 2012). More examples of the application of immunological-based methods for the detection of foodborne pathogens in different food matrices are presented in Table 3.
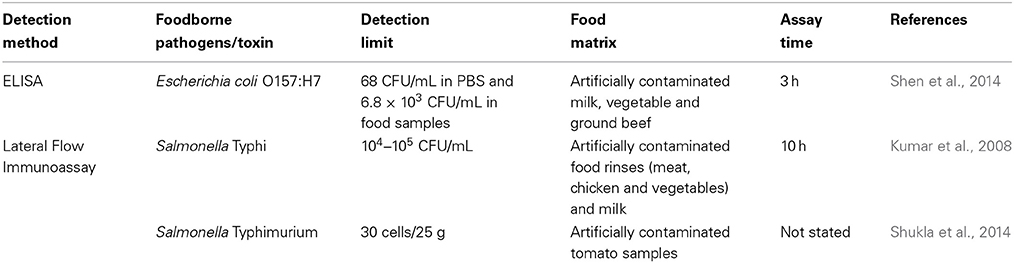
Table 3. Examples of the application of immunological-based methods for the detection of various foodborne pathogens present in food samples.
Advantages and Limitations of Rapid Methods
Rapid methods provide various advantages for the detection of foodborne pathogens, however, they also have several limitations as summarized in Table 4.
Conclusion
Conventional methods for the detection of foodborne pathogens which based on culturing the microorganisms are selective, but they can be time-consuming and laborious. Hence, various rapid detection methods have been developed in order to overcome the limitations of conventional detection methods. Rapid methods are important for the rapid detection of foodborne pathogens in food products to prevent outbreaks of foodborne diseases and the spread of foodborne pathogens. Rapid detection methods are generally more sensitive, specific, time-efficient, labor-saving, and reliable than conventional methods.
Nucleic acid-based methods such as PCR, mPCR, qPCR, and DNA microarray have high sensitivity and they are widely used for the detection of foodborne pathogens, but these methods require trained personnel and specialized instruments. Alternative nucleic acid-based methods such as NASBA and LAMP are available for the detection of foodborne pathogens and their toxins. NASBA and LAMP are relatively sensitive, specific and cost efficient. They do not require thermocycling system therefore they are useful especially in low resource settings. Furthermore, numerous biosensors-based methods have recently emerged and employed in the field of foodborne pathogen detection due to their rapidness and cost effectiveness. Biosensors-based methods are easy to operate and they do not require trained personnel, furthermore the techniques can be used for the detection of foodborne pathogens without sample pre-enrichment. However, improvement in food matrixes detection is still needed for these methods for on-site detection. Immunological-based methods such as ELISA and lateral flow immunoassay are also used for the detection of foodborne bacterial pathogens and their toxins. Immunological methods work best in the absence of interfering molecules in the samples such as non-targeted cells, DNA or proteins. Combination of several rapid methods for the detection of a particular foodborne pathogen is also possible as the use of only one detection method may not be sufficient to confirm the detected pathogen. Further studies on the effect of different combinations of rapid methods for foodborne pathogen detection are required in order to develop the most effective and accurate detection method.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the University of Malaya for High Impact Research Grant (UM-MOHE HIR Nature Microbiome Grant No. H-50001-A000027) to Dr. Chan Kok-Gan and External Industry Grant (Biotek Abadi - Vote No. GBA-808138) awarded to Dr. Lee Learn-Han.
References
Adzitey, F., Rusul, G., Huda, N., Cogan, T., and Corry, J. (2012). Prevalence, antibiotic resistance and RAPD typing of Campylobacter species isolated from ducks, their rearing and processing environments in Penang, Malaysia. Int. J. Food Microbiol. 154, 197–205. doi: 10.1016/j.ijfoodmicro.2012.01.006
Akbulut, D., Grant, K. A., and McLauchlin, J. (2004). Development and application of real-time PCR assays to detect fragments of the Clostridium botulinum types A, B, and E neurotoxin genes for investigation of human foodborne and infant botulism. Foodborne Pathog. Dis. 1, 247–257. doi: 10.1089/fpd.2004.1.247
Al-Khaldi, S. F. (2002). DNA microarray technology used for studying foodborne pathogens and microbial habitats: minireview. J. AOAC Int. 85, 906–910.
Alves, J., Marques, V. V., Pereira, L. F. P., Hirooka, E. Y., and Moreira de Oliveira, T. C. R. (2012). Multiplex PCR for the detection of Campylobacter spp. and Salmonella spp. in chicken meat. J. Food Safety 32, 345–350. doi: 10.1111/j.1745-4565.2012.00386.x
Aschfalk, A., and Mülller, W. (2002). Clostridium perfringens toxin types from wild caught Atlantic cod (Gadus morhua L.)., determined by PCR and ELISA. Can. J. Microbiol. 48, 365–368. doi: 10.1139/w02-015
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bang, J., Beuchat, L. R., Song, H., Gu, M. B., Chang, H. I., Kim, H. S., et al. (2013). Development of random genomic DNA microarray for the detection and identification of Listeria monocytogenes in milk. Int. J. Food Microbiol. 161, 134–141. doi: 10.1016/j.ijfoodmicro.2012.11.023
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Berkenpas, E., Millard, P., and da Cunha, M. P. (2006). Detection of Escherichia coli O157:H7 with langasite pure shear horizontal surface acoustic wave sensors. Biosens. Bioelectron. 21, 2255–2262. doi: 10.1016/j.bios.2005.11.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Blais, B. W., Turner, G., Sooknanan, R., and Malek, L. T. (1997). A nucleic acid sequence-based amplification system for detection of Listeria monocytogenes hlyA sequences. Appl. Eviron. Microbiol. 63, 310–313.
Bokken, G. C. A. M., Corbee, R. J., van Knapen, F., and Bergwerff, A. A. (2003). Immunochemical detection of Salmonella group B, D and E using an optical surface plasmon resonance biosensor. FEMS Microbiol. Lett. 222, 75–82. doi: 10.1016/S0378-1097(03)00250-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bolton, F. J., Fritz, E., and Poynton, S. (2000). Rapid enzyme-linked immunoassay for the detection of Salmonella in food and feed products: performance testing program. J. AOAC Int. 83, 299–304.
Call, D. R., Brockman, F. J., and Chandler, D. P. (2001). Detecting and genotyping Escherichia coli O157:H7 using multiplexed PCR and nucleic acid microarrays. Int. J. Food Microbiol. 67, 71–80. doi: 10.1016/S0168-1605(01)00437-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Carvalho, R. N., Oliveira, A. N., Mesquita, A. J., Rezende, C. S. M., Mesquita, A. Q., and Romero, R. A. M. (2014). PCR and ELISA (VIDAS ECO O157®) Escherichia coli O157:H7 identification in Minas Frescal cheese commercialized in Goiânia, GO. Braz. J. Microbiol. 45, 7–10. doi: 10.1590/S1517-83822014000100002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Centers for Disease Control and Prevention (CDC). (2011). CDC Estimates of Foodborne Illness in the United States. Available online at: http://www.cdc.gov/foodborneburden/PDFs/FACTSHEET_A_FINDINGS_updated4-13.pdf (Accessed 15 June 2014).
Cheah, Y. K., Noorzaleha, A. S., Lee, L. H., Radu, S., Sukardi, S., and Sim, J. H. (2008). Comparison of PCR fingerprinting techniques for the discrimination of Salmonella enterica subsp. enterica serovar Weltevreden isolated from indigenous vegetables in Malaysia. World J. Microbiol. Biotechnol. 24, 327–335. doi: 10.1007/s11274-007-9474-8
Chemburu, S., Wilkins, E., and Abdel-Hamid, I. (2005). Detection of pathogenic bacteria in food samples using highly-dispersed carbon particles. Biosens. Bioelectron. 21, 491–499. doi: 10.1016/j.bios.2004.11.025
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chen, H. T., Zhang, J., Sun, D. H., Ma, L. N., Liu, X. T., Cai, X. P., et al. (2008). Development of reverse transcription loop-mediated isothermal amplification for rapid detection of H9 avian influenza virus. J. Virol. Methods 151, 200–203. doi: 10.1016/j.jviromet.2008.05.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chen, J., Tang, J., Liu, J., Cai, Z., and Bai, X. (2012). Development and evaluation of a multiplex PCR for simultaneous detection of five foodborne pathogens. J. Appl. Microbiol. 112, 823–830. doi: 10.1111/j.1365-2672.2012.05240.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chen, J., Zhang, L., Paoli, G. C., Shi, C., Tu, S. I., and Shi, X. (2010). A real-time PCR method for the detection of Salmonella enterica from food using a target sequence identified by comparative genomic analysis. Int. J. Food Microbiol. 137, 168–174. doi: 10.1016/j.ijfoodmicro.2009.12.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chen, W., Martinez, G., and Mulchandani, A. (2000). Molecular beacons: a real-time polymerase chain reaction assay for detecting Salmonella. Anal. Biochem. 280, 166–172. doi: 10.1006/abio.2000.4518
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chiang, Y. C., Tsen, H. Y., Chen, H. Y., Chang, Y. H., Lin, C. K., Chen, C. Y., et al. (2012). Multiplex PCR and a chromogenic DNA macroarray for the detection of Listeria monocytogenes, Staphylococcus aureus, Streptococcus agalactiae, Enterobacter sakazakii, Escherichia coli O157:H7, Vibrio parahaemolyticus, Salmonella spp. and Pseudomonas fluorescens in milk and meat samples. J. Microbiol. Meth. 88, 110–116. doi: 10.1016/j.mimet.2011.10.021
Chiou, C. S., Hsu, S. Y., Chiu, S. I., Wang, T. K., and Chao, C. S. (2000). Vibrio parahaemolyticus Serovar O3:K6 as cause of unusually high incidence of food-borne disease outbreaks in Taiwan from 1996 to 1999. J. Clin. Microbiol. 38, 4621–4625.
Cho, H. H., and Irudayaraj, J. (2013). Lateral-flow enzyme immunoconcentration for rapid detection of Listeria monocytogenes. Anal. Bioanal. Chem. 405, 3313–3319. doi: 10.1007/s00216-013-6742-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chung, M. S., Kim, C. M., and Ha, S. D. (2010). Detection and enumeration of microorganisms in ready-to-eat foods, ready-to-cook foods and fresh-cut produce in Korea. J. Food Safety 30, 480–489. doi: 10.1111/j.1745-4565.2010.00221.x
Churruca, E., Girbau, C., Martinez, I., Mateo, E., Alonso, R., and Fernandez-Astorga, A. (2007). Detection of Campylobacter jejuni and Campylobacter coli in chicken meat samples by real-time nucleic acid sequence-based amplification with molecular beacons. Int. J. Food Microbiol. 117, 85–90. doi: 10.1016/j.ijfoodmicro.2007.02.007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Compton, J. (1991). Nucleic acid sequence-based amplification. Nature 350, 91–92. doi: 10.1038/350091a0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cremonesi, P., Perez, G., Pisoni, G., Moroni, P., Morandi, S., Luzzana, M., et al. (2007). Detection of enterotoxigenic Staphylococcus aureus isolates in raw milk cheese. Lett. Appl. Microbiol. 45, 586–591. doi: 10.1111/j.1472-765X.2007.02231.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
de Ávila, B. E. F., Pedrero, M., Campuzano, S., Escamilla-Gómez, V., and Pingarrón, J. M. (2012). Sensitive and rapid amperometric magnetoimmunosensor for the determination of Staphylococcus aureus. Anal. Bioanal. Chem. 403, 917–925. doi: 10.1007/s00216-012-5738-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
De Boer, S. H., and López, M. M. (2012). New grower-friendly methods for plant pathogen monitoring. Annu. Rev. Phytopathol. 50, 197–218. doi: 10.1146/annurev-phyto-081211-172942
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
D'Souza, D. H., and Jaykus, L. A. (2003). Nucleic acid sequence based amplification for the rapid and sensitive detection of Salmonella enterica from foods. J. Appl. Microbiol. 95, 1343–1350. doi: 10.1046/j.1365-2672.2003.02106.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dwivedi, H. P., and Jaykus, L. A. (2011). Detection of pathogens in foods: the current state-of-the-art and future directions. Crit. Rev. Microbiol. 37, 40–63. doi: 10.3109/1040841X.2010.506430
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ercole, C., Del Gallo, M., Mosiello, L., Baccella, S., and Lepidi, A. (2003). Escherichia coli detection in vegetable food by a potentiometric biosensor. Sensors and Actuat. B-Chem. 91, 163–168. doi: 10.1016/S0925-4005(03)00083-2
Feldsine, P. T., Kerr, D. E., Leung, S. C., Lienau, A. H., Miller., S. M., and Mui, L. A. (2002). Assurance® enzyme immunoassay eight hour method for detection of enterohemorrhagic Escherichia coli O157: H7 in raw and cooked Beef. J. AOAC Int. 85, 1037–1044.
Fratamico, P. M., Bagi, L. K., Cray Jr, W. C., Narang, N., Yan, X., Medina, M., et al. (2011). Detection by multiplex real-time polymerase chain reaction assays and isolation of Shiga toxin-producing Escherichia coli serogroups O26, O45, O103, O111, O121 and O145 in ground beef. Foodborne Pathog. Dis. 8, 601–607. doi: 10.1089/fpd.2010.0773
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fricker, M., Messelhäuber, U., Busch, U., Scherer, S., and Ehling-Schulz, M. (2007). Diagnostic real-time PCR assays for the detection of emetic Bacillus cereus strains in foods and recent food-borne outbreaks. Appl. Environ. Microbiol. 73, 1892–1898. doi: 10.1128/AEM.02219-06
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fukushima, H., Tsunomori, Y., and Seki, R. (2003). Duplex real-time SYBR green PCR assays for detection of 17 species of food- or waterborne pathogens in stools. J. Clin. Microbiol. 41, 5134–5146. doi: 10.1128/JCM.41.11.5134-5146.2003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fung, D. Y. C. (2002). Rapid methods and automation in microbiology. Compr. Rev. Food Sci. F 1, 3–22. doi: 10.1111/j.1541-4337.2002.tb00003.x
Fusco, V., Quero, G. M., Morea, M., Blaiotta, G., and Visconti, A. (2011). Rapid and reliable identification of Staphylococcus aureus harbouring the enterotoxin gene cluster (egc). and quantitative detection in raw milk by real time PCR. Int. J. Food Microbiol. 144, 528–537. doi: 10.1016/j.ijfoodmicro.2010.11.016
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fykse, E. M., Skogan, G., Davies, W., Olsen, J. S., and Blatny, J. M. (2007). Detection of Vibrio cholerae by real-time nucleic acid sequence-based amplification. Appl. Environ. Microbiol. 73, 1457–1466. doi: 10.1128/AEM.01635-06
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Glynn, B., Lahiff, S., Wernecke, M., Barry, T., Smith, T. J., and Maher, M. (2006). Current and emerging molecular diagnostic technologies applicable to bacterial food safety. Int. J. Dairy Technol. 59, 126–139. doi: 10.1111/j.1471-0307.2006.00253.x
Gómez-Govea, M., Solís-Soto, L., Heredia, N., García, S., Moreno, G., Tovar, O., et al. (2012). Analysis of microbial contamination levels of fruits and vegetables at retail in Monterrey, Mexico. J. Food Agric. Environ. 10, 152–156.
Gore, H. M., Wakeman, C. A., Hull, R. M., and McKillip, J. L. (2003). Real-time molecular beacon NASBA reveals hblC expression from Bacillus spp. in milk. Biochem. Biophys. Res. Commun. 311, 386–390. doi: 10.1016/j.bbrc.2003.10.016
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gracias, K. S., and McKillip, J. L. (2007). Nucleic acid sequence-based amplification (NASBA) in molecular bacteriology: a procedural guide. J. Rapid Meth. Aut. Mic. 15, 295–309. doi: 10.1111/j.1745-4581.2007.00099.x
Guan, Z. P., Jiang, Y., Gao, F., Zhang, L., Zhou, G. H., and Guan, Z. J. (2013). Rapid and simultaneous analysis of five foodborne pathogenic bacteria using multiplex PCR. Eur. Food Res. Technol. 237, 627–637. doi: 10.1007/s00217-013-2039-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Han, F., and Ge, B. (2010). Quantitative detection of Vibrio vulnificus in raw oysters by real-time-loop-mediated isothermal amplification. Int. J. Food Microbiol. 142, 60–66. doi: 10.1016/j.ijfoodmicro.2010.05.029
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Han, F., Wang, F., and Ge, B. (2011). Detecting potentially virulent Vibrio vulnificus strains in raw oysters by quantitative loop-mediated isothermal amplification. Appl. Environ. Microbiol. 77, 2589–2595. doi: 10.1128/AEM.02992-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hara-Kudo, Y., Konishi, N., Ohtsuka, K., Hiramatsu, R., Tanaka, H., Konuma, H., et al. (2008). Detection of verotoxigenic Escherichia coli O157 and O26 in food by plating methods and LAMP method: a collaborative study. Int. J. Food Microbiol. 122, 156–161. doi: 10.1016/j.ijfoodmicro.2007.11.078
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hara-Kudo, Y., Yoshino, M., Kojima, T., and Ikedo, M. (2005). Loop-mediated isothermal amplification for the rapid detection of Salmonella. FEMS Microbiol. Lett. 253, 155–161. doi: 10.1016/j.femsle.2005.09.032
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hein, I., Lehner, A., Rieck, P., Klein, K., Brandl, E., and Wagner, M. (2001). Comparison of different approaches to quantify Staphylococcus aureus cells by real-time quantitative PCR and application of this technique for examination of cheese. Appl. Environ. Microbiol. 67, 3122–3126. doi: 10.1128/AEM.67.7.3122-3126.2001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hu, Q., Lyu, D., Shi, X., Jiang, Y., Lin, Y., Li, Y., et al. (2014). A modified molecular beacons-based multiplex real-time PCR assay for simultaneous detection of eight foodborne pathogens in a single reaction and its application. Foodborne Pathog. Dis. 11, 207–214. doi: 10.1089/fpd.2013.1607
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Huang, A., Qiu, Z., Jin, M., Shen, Z., Chen, Z., Wang, X., et al. (2014). High-throughput detection of food-borne pathogenic bacteria using oligonucleotide microarray with quantum dots as fluorescent labels. Int. J. Food Microbiol. 185, 27–32. doi: 10.1016/j.ijfoodmicro.2014.05.012
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ivnitski, D., Abdel-Hamid, I., Atanasov, P., and Wilkins, E. (1999). Biosensors for detection of pathogenic bacteria. Biosens. Bioelectron. 14, 599–624. doi: 10.1016/S0956-5663(99)00039-1
Janzten, M. M., Navas, J., Corujo, A., Moreno, R., López, V., and Martínez-Suárez, J. V. (2006). Review. Specific detection of Listeria monocytogenes in foods using commercial methods: from chromogenic media to real-time PCR. Span. J. Agric. Res. 4, 235–247. doi: 10.5424/sjar/2006043-198
Jung, B. Y., Jung, S. C., and Kweon, C. H. (2005). Development of a rapid immunochromatographic strip for the detection of Escherichia coli O157. J. Food Prot. 68, 2140–2143.
Kanayeva, D. A., Wang, R., Rhoads, D., Erf, G. F., Slavik, M. F., Tung, S., et al. (2012). Efficient separation and sensitive detection of Listeria monocytogenes using an impedance immunosensor based on magnetic nanoparticles, a microfluidic chip, and an interdigitated microelectrode. J. Food Prot. 75, 1951–1959. doi: 10.4315/0362-028X.JFP-11-516
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kawasaki, S., Fratamico, P. M., Horikoshi, N., Okada, Y., Takeshita, K., Sameshima, T., et al. (2010). Multiplex real-time polymerase chain reaction assay for simultaneous detection and quantification of Salmonella species, Listeria monocytogenes, and Escherichia coli O157:H7 in ground pork samples. Foodborne Pathog. Dis. 7, 549–554. doi: 10.1089/fpd.2009.0465
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Khoo, C. H., Cheah, Y. K., Lee, L. H., Sim, J. H., Noorzaleha, A. S., Sidik, M. S., et al. (2009). Virulotyping of Salmonella enterica subsp. enterica isolated from indigenous vegetables and poultry meat in Malaysia using multiplex-PCR. Antonie Van Leeuwenhoek 96, 441–457. doi: 10.1007/s10482-009-9358-z
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kim, H. J., Lee, H. J., Lee, K. H., and Cho, J. C. (2012). Simultaneous detection of pathogenic Vibrio species using multiplex real-time PCR. Food Cont. 23, 491–498. doi: 10.1016/j.foodcont.2011.08.019
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kim, S. H., Kim, J. Y., Han, W., Jung, B. Y., Chuong, P. D., and Joo, H. (2007). Development and evaluation of an immunochromatographic assay for Listeria spp. in pork and milk. Food Sci. Biotechnol. 16, 515–519.
Kimura, B., Kawasaki, S., Fujii, T., Kusunoki, J., Itoh, T., and Flood, S. J. A. (1999). Evaluation of TaqMan PCR assay for detecting Salmonella in raw meat and shrimp. J. Food Prot. 4, 307–425.
Klerks, M. M., Zijlstra, C., and van Bruggen, A. H. C. (2004). Comparison of real-time PCR methods for detection of Salmonella enterica and Escherichia coli O157:H7, and introduction of a general internal amplification control. J. Microbiol. Meth. 59, 337–349. doi: 10.1016/j.mimet.2004.07.011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kumar, B. K., Raghunath, P., Devegowda, D., Deekshit., V. K., Venugopal, M. N., Karunasagar, I., et al. (2011). Development of monoclonal antibody based sandwich ELISA for the rapid detection of pathogenic Vibrio parahaemolyticus in seafood. Int. J. Food Microbiol. 145, 244–249. doi: 10.1016/j.ijfoodmicro.2010.12.030
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kumar, S., Balakrishna, K., and Batra, H. V. (2008). Enrichment-ELISA for detection of Salmonella typhi from food and water samples. Biomed. Environ. Sci. 21, 137–143. doi: 10.1016/S0895-3988(08)60019-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lan, Y. B., Wang, S. Z., Yin, Y. G., Hoffmann, W. C., and Zheng, X. Z. (2008). Using a surface plasmon resonance biosensor for rapid detection of Salmonella Typhimurium in chicken carcass. J. Bionic. Eng. 5, 239–246. doi: 10.1016/S1672-6529(08)60030-X
Lauri, A., and Mariani, P. O. (2009). Potentials and limitations of molecular diagnostic methods in food safety. Genes Nutr. 4, 1–12. doi: 10.1007/s12263-008-0106-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lee, L. H., Cheah, Y. K., Noorzaleha, A. S., Sabrina, S., Sim, J. H., Khoo, C. H., et al. (2008). Analysis of Salmonella Agona and Salmonella Weltevreden in Malaysia by PCR fingerprinting and antibiotic resistance profiling. Antonie Van Leeuwenhoek 94, 377–387. doi: 10.1007/s10482-008-9254-y
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lee, N., Kwon, K. Y., Oh, S. K., Chang, H. J., Chun, H. S., and Choi, S. W. (2014). A multiplex PCR assay for simultaneous detection of Escherichia coli O157:H7, Bacillus cereus, Vibrio parahaemolyticus, Salmonella spp., Listeria monocytogenes, and Staphylococcus aureus in Korea ready-to-eat food. Foodborne Pathog. Dis. 11, 574–580. doi: 10.1089/fpd.2013.1638
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Leem, H., Shukla, S., Song, X., Heu, S., and Kim, M. (2014). An efficient liposome-based immunochromatographic strip assay for the sensitive detection of Salmonella typhimurium in pure culture. J. Food Safety 34, 239–248. doi: 10.1111/jfs.12119
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Leonard, P., Hearty, S., Brennan, J., Dunne, L., Quinn, J., Chakraborty, T., et al. (2003). Advances in biosensors for detection of pathogens in food and water. Enzyme Microb. Technol. 32, 3–13. doi: 10.1016/S0141-0229(02)00232-6
Leonard, P., Hearty, S., Quinn, J., and O'Kennedy, R. (2004). A generic approach for the detection of whole Listeria monocytogenes cells in contaminated samples using surface plasmon resonance. Biosens. Bioelectron. 19, 1331–1335. doi: 10.1016/j.bios.2003.11.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Leone, G., van Schijndel, H., van Gemen, B., Kramer, F. R., and Schoen, C. D. (1998). Molecular beacon probes combined with amplification by NASBA enable homogeneous, real-time detection of RNA. Nucleic Acids Res. 26, 2150–2155. doi: 10.1093/nar/26.9.2150
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Levin, R. E. (2005). The application of real-time PCR to food and agricultural systems. A review. Food Biotechnol. 18, 97–133. doi: 10.1081/FBT-120030386
Li, Y., Liu, D., Cao, B., Han, W., Liu, Y., Liu, F., et al. (2006). Development of a serotype-specific DNA microarray for identification of some Shigella and pathogenic Escherichia coli strains. J. Clin. Microbiol. 44, 4376–4383. doi: 10.1128/JCM.01389-06
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Liming, S. H., and Bhagwat, A. A. (2004). Application of a molecular beacon-real-time PCR technology to detect Salmonella species contaminating fruits and vegetables. Int. J. Food Microbiol. 95, 177–187. doi: 10.1016/j.ijfoodmicro.2004.02.013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Liu, F., Li, Y., Su, X. L., Slavik, M. F., Ying, Y., and Wang, J. (2007). QCM immunosensor with nanoparticle amplification for detection of Escherichia coli O157:H7. Sens. Instrum. Food Qual. Saf. 1, 161–168. doi: 10.1007/s11694-007-9021-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ma, K., Deng, Y., Bai, Y., Xu, D., Chen, E., Wu, H., et al. (2014). Rapid and simultaneous detection of Salmonella, Shigella, and Staphylococcus aureus in fresh pork using a multiplex real-time PCR assay based on immunomagnetic separation. Food Cont. 42, 87–93. doi: 10.1016/j.foodcont.2014.01.042
Maciorowski, K. G. (2005). Polymerase chain reaction detection of foodborne Salmonella spp. in animal feeds. Crit. Rev. Microbiol. 31, 45–53. doi: 10.1080/10408410590912970
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Madani, M., Subbotin, S. A., and Moens, M. (2005). Quantitative detection of the potato cyst nematode, Globodera pallida, and the beet cyst nematode, Heterodera schachtii, using real-time PCR with SYBR green I dye. Mol. Cell. Probes 19, 81–86. doi: 10.1016/j.mcp.2004.09.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mandal, P. K., Biswas, A. K., Choi, K., and Pal, U. K. (2011). Methods for rapid detection of foodborne pathogens: an overview. Am. J. Food. Technol. 6, 87–102. doi: 10.3923/ajft.2011.87.102
Margot, H., Stephan, R., Guarino, S., Jagadeesan, B., Chilton, D., O'Mahony, E., et al. (2013). Inclusivity, exclusivity and limit of detection of commercially available real-time PCR assays for the detection of Salmonella. Int. J. Food Microbiol. 165, 221–226. doi: 10.1016/j.ijfoodmicro.2013.05.012
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Markoulatos, P., Siafakas, N., and Moncany, M. (2002). Multiplex polymerase chain reaction: a practical approach. J. Clin. Lab. Anal. 16, 47–51. doi: 10.1002/jcla.2058
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Maruyama, F., Kenzaka, T., Yamaguchi, N., Tani, K., and Nasu, M. (2003). Detection of bacteria carrying the stx2 gene by in situ loop-mediated isothermal amplification. Appl. Environ. Microbiol. 69, 5023–5028. doi: 10.1128/AEM.69.8.5023-5028.2003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Maurer, J. J. (2011). Rapid detection and limitations of molecular techniques. Annu. Rev. Food Sci. Technol. 2, 259–279. doi: 10.1146/annurev.food.080708.100730
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Meyer, C., Fredriksson-Ahomaa, M., Sperner, B., and Märtlbauer, E. (2011). Detection of Listeria monocytogenes in pork and beef using the VIDAS® LMO2 automated enzyme linked immunoassay method. Meat Sci. 88, 594–596. doi: 10.1016/j.meatsci.2011.01.035
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Min, J., and Baeumner, A. J. (2002). Highly sensitive and specific detection of viable Escherichia coli in drinking water. Anal. Biochem. 303, 186–193. doi: 10.1006/abio.2002.5593
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mollasalehi, H., and Yazdanparast, R. (2013). Development and evaluation of a novel nucleic acid sequence-based amplification method using one specific primer and one degenerate primer for simultaneous detection of Salmonella Enteritidis and Salmonella Typhimurium. Anal. Chim. Acta 770, 169–174. doi: 10.1016/j.aca.2013.01.053
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mori, Y., and Notomi, T. (2009). Loop-mediated isothermal amplification (LAMP).: a rapid, accurate, and cost-effective diagnostic method for infectious diseases. J. Infect. Chemother. 15, 62–69. doi: 10.1007/s10156-009-0669-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mothershed, E. A., and Whitney, A. M. (2006). Nucleic acid-based methods for the detection of bacterial pathogens: present and future considerations for clinical laboratory. Clin. Chim. Acta 363, 206–220. doi: 10.1016/j.cccn.2005.05.050
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Munoz-Berbel, X., Vigués, N., Jenkins, A. T. A., Mas, J., and Munoz, F. J. (2008). Impedimetric approach for quantifying low bacteria concentrations based on the changes produced in the electrode–solution interface during the pre-attachment stage. Biosens. Bioelectron. 23, 1540–1546. doi: 10.1016/j.bios.2008.01.007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nadal, A., Coll, A., Cook, N., and Pla, M. (2007). A molecular beacon-based real time NASBA assay for detection of Listeria monocytogenes in food products: role of target mRNA secondary structure on NASBA design. J. Microbiol. Meth. 68, 623–632. doi: 10.1016/j.mimet.2006.11.011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ngom, B., Guo, Y., Wang, X., and Bi, D. (2010). Development and application of lateral flow test strip technology for the detection of infectious agents and chemical contaminants: a review. Anal. Bioanal. Chem. 397, 1113–1135. doi: 10.1007/s00216-010-3661-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Niu, K., Zheng, X., Huang, C., Xu, K., Zhi, Y., Shen, H., et al. (2014). A colloidal gold nanoparticle-based immunochromatographic test strip for rapid and convenient detection of Staphylococcus aureus. J. Nanosci. Nanotechnol. 14, 5151–5156. doi: 10.1166/jnn.2014.8703
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Notomi, T., Okayama, H., Masubuchi, H., Yonekawa, T., Watanabe, K., Amino, N., et al. (2000). Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:e63. doi: 10.1093/nar/28.12.e63
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
O'Grady, J., Lacey, K., Glynn, B., Smith, T. J., Barry, T., and Maher, M. (2009). tmRNA-a novel high-copy-number RNA diagnostic target-its application for Staphylococcus aureus detection using real-time NASBA. FEMS Microbiol. Lett. 301, 218–223. doi: 10.1111/j.1574-6968.2009.01822.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ohtsuka, K., Yanagawa, K., Takatori, K., and Hara-Kudo, Y. (2005). Detection of Salmonella enterica in naturally contaminated liquid eggs by loop-mediated isothermal amplification, and characterization of Salmonella isolates. Appl. Environ. Microbiol. 71, 6730–6735. doi: 10.1128/AEM.71.11.6730-6735.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Oliver, S. P., Jayarao, B. M., and Almeida, R. A. (2005). Foodborne pathogens in milk and the dairy farm environment: food safety and public health implications. Foodborne Pathog. Dis. 2, 115–129. doi: 10.1089/fpd.2005.2.115
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Omiccioli, E., Amagliani, G., Brandi, G., and Magnani, M. (2009). A new platform for real-time PCR detection of Salmonella spp., Listeria monocytogenes and Escherichia coli O157 in milk. Food Microbiol. 26, 615–622. doi: 10.1016/j.fm.2009.04.008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Omurtag, I., Paulsen, P., Hilbert, F., and Smulders, F. J. M. (2013). The risk of transfer of foodborne bacterial hazards in Turkey through the consumption of meat; risk ranking of muscle foods with the potential to transfer Campylobacter spp. Food Security 5, 117–127. doi: 10.1007/s12571-012-0230-z
Pal, S., Ying, W., Alocilja, E. C., and Downes, F. P. (2008). Sensitivity and specificity performance of direct-charge transfer biosensor for detecting Bacillus cereus in selected food matrices. Biosyst. Eng. 99, 461–468. doi: 10.1016/j.biosystemseng.2007.11.015
Park, S. H., Aydin, M., Khatiwara, A., Dolan, M. C., Gilmore, D. F., Bouldin, J. L., et al. (2014). Current and emerging technologies for rapid detection and characterization of Salmonella in poultry and poultry products. Food Microbiol. 38, 250–262. doi: 10.1016/j.fm.2013.10.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Patel, J. R., Bhagwat, A. A., Sanglay, G. C., and Solomon, M. B. (2006). Rapid detection of Salmonella from hydrodynamic pressure-treated poultry using molecular beacon real-time PCR. Food Microbiol. 23, 39–46. doi: 10.1016/j.fm.2005.01.011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pohanka, M., Skládal, P., and Pavliš, O. (2007). Label−free piezoelectric immunosensor for rapid assay of Escherichia coli. J. Immunoassay Immunochem. 29, 70–79. doi: 10.1080/15321810701735120
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rasooly, A., and Herold, K. E. (2008). Food microbial pathogen detection and analysis using DNA microarray technologies. Foodborne Pathog. Dis. 5, 531–550. doi: 10.1089/fpd.2008.0119
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rong-Hwa, S., Shiao-Shek, T., Der-Jiang, C., and Yao-Wen, H. (2010). Gold nanoparticle-based lateral flow assay for detection of staphylococcal enterotoxin B. Food Chem. 118, 462–466. doi: 10.1016/j.foodchem.2009.04.106
Rosec, J. P., Causse, V., Cruz, B., Rauzier, J., and Carnat, L. (2012). The international standard ISO/TS 21872-1 to study the occurrence of total and pathogenic Vibrio parahaemolyticus and Vibrio cholerae in seafood: ITS improvement by use of a chromogenic medium and PCR. Int J. Food Microbiol. 157, 189–194. doi: 10.1016/j.ijfoodmicro.2012.04.026
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ruiz-Rueda, O., Soler, M., Calvó, L., and García-Gil, J. L. (2011). Multiplex real-time PCR for the simultaneous detection of Salmonella spp. and Listeria monocytogenes in food samples. Food Anal. Method. 4, 131–138. doi: 10.1007/s12161-010-9163-3
Ryu, J., Park, S. H., Yeom, Y. S., Shrivastav, A., Lee, S. H., Kim, Y. R., et al. (2013). Simultaneous detection of Listeria species isolated from meat processed foods using multiplex PCR. Food Cont. 32, 659–664. doi: 10.1016/j.foodcont.2013.01.048
Scallan, E., Hoekstra, R. M., Angulo, F. J., Tauxe, R. V., Widdowson, M. A., Roy, S. L., et al. (2011). Foodborne illness acquired in the United States- major pathogens. Emerg. Infect. Dis. 17, 7–15. doi: 10.3201/eid1701.P11101
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Severgnini, M., Cremonesi, P., Consolandi, C., De Bellis, G., and Castiglioni, B. (2011). Advances in DNA microarray technology for the detection of foodborne pathogens. Food Bioprocess Tech. 4, 936–953. doi: 10.1007/s11947-010-0430-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shao, Y., Zhu, S., Jin, C., and Chen, F. (2011). Development of multiplex loop-mediated isothermal amplification-RFLP (mLAMP-RFLP). to detect Salmonella spp. and Shigella spp. in milk. Int. J. Food Microbiol. 148, 75–79. doi: 10.1016/j.ijfoodmicro.2011.05.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shen, Z., Hou, N., Jin, M., Qiu, Z., Wang, J., Zhang, B., et al. (2014). A novel enzyme-linked immunosorbent assay for detection of Escherichia coli O157:H7 using immunomagnetic and beacon gold nanoparticles. Gut Pathog. 6, 14. doi: 10.1186/1757-4749-6-14
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shen, Z. Q., Wang, J. F., Qiu, Z. G., Jin, M., Wang, X. W., Chen, Z. L., et al. (2011). QCM immunosensor detection of Escherichia coli O157:H7 based on beacon immunomagnetic nanoparticles and catalytic growth of colloidal gold. Biosens. Bioelectron. 26, 3376–3381. doi: 10.1016/j.bios.2010.12.035
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shukla, S., Leem, H., and Kim, M. (2011). Development of a liposome-based immunochromatographic strip assay for the detection of Salmonella. Anal. Bioanal. Chem. 401, 2581–2590. doi: 10.1007/s00216-011-5327-2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shukla, S., Leem, H., Lee, J. S., and Kim, M. (2014). Immunochromatographic strip assay for the rapid and sensitive detection of Salmonella Typhimurium in artificially contaminated tomato samples. Can. J. Microbiol. 60, 399–406. doi: 10.1139/cjm-2014-0223
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Si, S. H., Li, X., Fung, Y. S., and Zhu, D. R. (2001). Rapid detection of Salmonella Enteritidis by piezoelectric immunosensor. Microchem. J. 68, 21–27. doi: 10.1016/S0026-265X(00)00167-3
Silva, D. S. P., Canato, T., Magnani, M., Alves, J., Hirooka, E. Y., and de Oliveira, T. C. R. M. (2011). Multiplex PCR for the simultaneous detection of Salmonella spp. and Salmonella Enteritidis in food. Int. J. Food Sci. Tech. 46, 1502–1507. doi: 10.1111/j.1365-2621.2011.02646.x
Simpkins, S. A., Chan, A. B., Hays, J., Popping, B., and Cook, N. (2000). An RNA transcription-based amplification technique (NASBA). for the detection of viable Salmonella enterica. Lett. Appl. Microbiol. 30, 75–79. doi: 10.1046/j.1472-765x.2000.00670.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Singh, A., Poshtiban, S., and Evoy, S. (2013). Recent advances in bacteriophage based biosensors for food-borne pathogen detection. Sensors 13, 1763–1786. doi: 10.3390/s130201763
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Singh, D. V., Matte, M. H., Matte, G. R., Jiang, S., Sabeena, F., Shukla, B. N., et al. (2001). Molecular analysis of Vibrio cholerae O1, O139, non-O1, and non-O139 strains: clonal relationships between clinical and environmental isolates. Appl. Environ. Microbiol. 67, 910–921. doi: 10.1128/AEM.67.2.910-921.2001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Singh, J., Batish, V. K., and Grover, S. (2009). A molecular beacon-based duplex real-time polymerase chain reaction assay for simultaneous detection of Escherichia coli O157:H7 and Listeria monocytogenes in milk and milk products. Foodborne Pathog. Dis. 6, 1195–1201. doi: 10.1089/fpd.2009.0310
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Son, I., Binet, R., Maounounen-Laasri, A., Lin, A., Hammack, T. S., and Kase, J. A. (2014). Detection of five Shiga toxin-producing Escherichia coli genes with multiplex PCR. Food Microbiol. 40, 31–40. doi: 10.1016/j.fm.2013.11.016
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Son, J. R., Kim, G., Kothapalli, A., Morgan, M. T., and Ess, D. (2007). Detection of Salmonella Enteritidis using a miniature optical surface plasmon resonance biosensor. J. Phys. 61, 1086–1090. doi: 10.1088/1742-6596/61/1/215
Song, T., Toma, C., Nakasone, N., and Iwanaga, M. (2005). Sensitive and rapid detection of Shigella and enteroinvasive Escherichia coli by a loop-mediated isothermal amplification method. FEMS Microbiol. Lett. 243, 259–263. doi: 10.1016/j.femsle.2004.12.014
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Stewart, D. S., Reineke, K. F., Ulaszek, J. M., and Tortorello, M. L. (2001). Growth of Salmonella during sprouting of alfalfa seeds associated with salmonellosis outbreaks. J. Food Prot. 64, 618–622.
Su, X. L., and Li, Y. (2005). A QCM immunosensor for Salmonella detection with simultaneous measurements of resonant frequency and motional resistance. Biosens. Bioelectron. 21, 840–848. doi: 10.1016/j.bios.2005.01.021
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Suo, B., He, Y., Paoli, G., Gehring, A., Tu, S. I., and Shi, X. (2010a). Development of an oligonucleotide-based microarray to detect multiple foodborne pathogens. Mol. Cell. Probes 24, 77–86. doi: 10.1016/j.mcp.2009.10.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Suo, B., He, Y., Tu, S. I., and Shi, X. (2010b). A multiplex real-time polymerase chain reaction for simultaneous detection of Salmonella spp., Escherichia coli O157, and Listeria monocytogenes in meat products. Foodborne Pathog. Dis. 7, 619–628. doi: 10.1089/fpd.2009.0430
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Taha, E. G., Mohamed, A., Srivastava, K. K., and Reddy, P. G. (2010). Rapid detection of Salmonella in chicken meat using immunomagnetic separation, CHROMagar, ELISA and real-time polymerase chain reaction (RT-PCR). Int. J. Poult. Sci. 9, 831–835. doi: 10.3923/ijps.2010.831.835
Taylor, A. D., Ladd, J., Yu, Q., Chen, S., Homola, J., and Jiang, S. (2006). Quantitative and simultaneous detection of four foodborne bacterial pathogens with a multi-channel SPR sensor. Biosens. Bioelectron. 22, 752–758. doi: 10.1016/j.bios.2006.03.012
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Traunšek, U., Toplak, N., Jeršek, B., Lapanje, A., Majstorović, T., and Kovač, M. (2011). Novel cost-efficient real-time PCR assays for detection and quantitation of Listeria monocytogenes. J. Microbiol. Meth. 85, 40–46. doi: 10.1016/j.mimet.2011.01.018
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tyagi, A., Saravanan, V., Karunasagar, I., and Karunasagar, I. (2009). Detection of Vibrio parahaemolyticus in tropical shellfish by SYBR green real-time PCR and evaluation of three enrichment media. Int. J. Food Microbiol. 129, 124–130. doi: 10.1016/j.ijfoodmicro.2008.11.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
USDA. (2012). FSIS Procedure for the Use of a Listeria monocytogenes Polymerase Chain Reaction (PCR) Screening Test. Effective June, 2012 MLG 8A.05. Available online at: http://www.fsis.usda.gov/wps/wcm/connect/1b6843c4-147f-4228-ae6a-c0993c0cf5fa/MLG-8A.pdf?MOD=AJPERES (Accessed 3 December 2014).
Van de Venter, T. (2000). Emerging food-borne diseases: a global responsibility. Food Nutr. Agr. 26, 4–13.
Velasco-Garcia, M., and Mottram, T. (2003). Biosensor technology addressing agricultural problems. Biosyst. Eng. 84, 1–12. doi: 10.1016/S1537-5110(02)00236-2
Yang, J. L., Ma, G. P., Yang, R., Yang, S. Q., Fu, L. Z., Cheng, A. C., et al. (2010). Simple and rapid detection of Salmonella serovar Enteritidis under field conditions by loop-mediated isothermal amplification. J. Appl. Microbiol. 109, 1715–1723. doi: 10.1111/j.1365-2672.2010.04800.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Varshney, M., Yang, L., Su, X. L., and Li, Y. (2005). Magnetic nanoparticle-antibody conjugates for the separation of Escherichia coli O157:H7 in ground beef. J. Food Prot. 68, 1804–1811.
Vaughan, R. D., O'Sullivan, C. K., and Guilbault, G. G. (2001). Development of a quartz crystal microbalance (QCM) immunosensor for the detection of Listeria monocytogenes. Enzyme Microb. Tech. 29, 635–638. doi: 10.1016/S0141-0229(01)00449-5
Vaz-Velho, M., Duarte, G., and Gibbs, P. (2000). Evaluation of mini-VIDAS rapid test for detection of Listeria monocytogenes from production lines of fresh to cold-smoked fish. J. Microbiol. Meth. 40, 147–151. doi: 10.1016/S0167-7012(00)00118-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Velusamy, V., Arshak, K., Korostynska, O., Oliwa, K., and Adley, C. (2010). An overview of foodborne pathogen detection: in the perspective of biosensors. Biotechnol. Adv. 28, 232–254. doi: 10.1016/j.biotechadv.2009.12.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vencia, W., Nogarol, C., Bianchi, D. M., Gallina, S., Zuccon, F., Adriano, D., et al. (2014). Validation according to ISO 16140:2003 of a commercial real-time PCR-based method for detecting Campylobacter jejuni, C. coli and C. lari in foods. Int. J. Food Microbiol. 177, 78–80. doi: 10.1016/j.ijfoodmicro.2014.02.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Verstraete, K., Robyn, J., Del-Favero, J., De Rijk, P., Joris, M. A., Herman, L., et al. (2012). Evaluation of a multiplex-PCR detection in combination with an isolation method for STEC O26, O103, O111, O145 and sorbitol fermenting O157 in food. Food Microbiol. 29, 49–55. doi: 10.1016/j.fm.2011.08.017
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vieira-Pinto, M., Oliveira, M., Bernardo, F., and Martins, C. (2007). Rapid detection of Salmonella sp. in pork samples using fluorescent in situ hybridization: a comparison with VIDAS®-SLM system and ISO 6579 cultural method. Arq. Bras. Med. Vet. Zootec. 59, 1388–1393. doi: 10.1590/S0102-09352007000600006
Wan, J., King, K., Craven, H., McAuley, C., Tan, S. E., and Coventry, M. J. (2000). Probelia™ PCR system for rapid detection of Salmonella in milk powder and ricotta cheese. Lett. Appl. Microbiol. 30, 267–271. doi: 10.1046/j.1472-765x.2000.00723.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wan, J., King, K., Forsyth, S., and Coventry, M. J. (2003). Detection of Listeria monocytogenes in salmon using the Probelia polymerase chain reaction system. J. Food Prot. 66, 436–440.
Wang, F., Yang, Q., Qu, Y., Meng, J., and Ge, B. (2014). Evaluation of a loop-mediated isothermal amplification suite for the rapid, reliable, and robust detection of shiga toxin-producing Escherichia coli in produce. Appl. Environ. Microbiol. 80, 2516–2525. doi: 10.1128/AEM.04203-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, L., Shi, L., Alam, M. J., Geng, Y., and Li, L. (2008). Specific and rapid detection of foodborne Salmonella by loop-mediated isothermal amplification method. Food Res. Int. 41, 69–74. doi: 10.1016/j.foodres.2007.09.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, L., Shi, L., Su, J., Ye, Y., and Zhong, Q. (2013a). Detection of Vibrio parahaemolyticus in food samples using in situ loop-mediated isothermal amplification method. Gene 515, 421–425. doi: 10.1016/j.gene.2012.12.039
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, X. W., Zhang, L., Jin, L. Q., Jin, M., Shen, Z. Q., An, S., et al. (2007). Development and application of an oligonucleotide microarray for the detection of food-borne bacterial pathogens. Appl. Microbiol. Biot. 76, 225–233. doi: 10.1007/s00253-007-0993-x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, Y., Ye, Z., Si, C., and Ying, Y. (2011). Subtractive inhibition assay for the detection of E. coli O157:H7 using surface plasmon resonance. Sensors 11, 2728–2739. doi: 10.3390/s110302728
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, Y., Ye, Z., Si, C., and Ying, Y. (2013b). Monitoring of Escherichia coli O157:H7 in food samples using lectin based surface plasmon resonance biosensor. Food Chem. 136, 1303–1308. doi: 10.1016/j.foodchem.2012.09.069
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Waswa, J., Irudayaraj, J., and DebRoy, C. (2007). Direct detection of E. coli O157:H7 in selected food systems by a surface plasmon resonance biosensor. LWT-Food Sci. Technol. 40, 187–192. doi: 10.1016/j.lwt.2005.11.001
Waswa, J. W., Debroy, C., and Irudayaraj, J. (2006). Rapid detection of Salmonella Enteritidis and Escherichia coli using surface plasmon resonance biosensor. J. Food Process Eng. 29, 373–385. doi: 10.1111/j.1745-4530.2006.00071.x
Wei, D., Oyarzabal, O. A., Huang, T. S., Balasubramanian, S., Sista, S., and Simonian, A. L. (2007). Development of a surface plasmon resonance biosensor for the identification of Campylobacter jejuni. J. Microbiol. Meth. 69, 78–85. doi: 10.1016/j.mimet.2006.12.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wingstrand, A., Neimann, J., Engberg, J., Nielsen, E. M., Gerner-Smidt, P., Wegener, H. C., et al. (2006). Fresh chicken as main risk factor for campylobacteriosis, Denmark. Emerg. Infect. Dis. 12, 280–284. doi: 10.3201/eid1202.050936
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Xu, D., Wu, X., Li, B., Li, P., Ming, X., Chen, T., et al. (2013). Rapid detection of Campylobacter jejuni using fluorescent microspheres as label for immunochromatographic strip test. Food Sci. Biotechnol. 22, 585–591. doi: 10.1007/s10068-013-0118-5
Xu, Z., Li, L., Chu, J., Peters, B. M., Harris, M. L., Li, B., et al. (2012). Development and application of loop-mediated isothermal amplification assays on rapid detection of various types of staphylococci strains. Food Res. Int. 47, 166–173. doi: 10.1016/j.foodres.2011.04.042
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yamazaki, W., Ishibashi, M., Kawahara, R., and Inoue, K. (2008). Development of a loop-mediated isothermal amplification assay for sensitive and rapid detection of Vibrio parahaemolyticus. BMC Microbiol. 8:163. doi: 10.1186/1471-2180-8-163
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yamazaki, W., Taguchi, M., Kawai, T., Kawatsu, K., Sakata, J., Inoue, K., et al. (2009). Comparison of loop-mediated isothermal amplification assay and conventional culture methods for detection of Campylobacter jejuni and Campylobacter coli in naturally contaminated chicken meat samples. Appl. Eviron. Microbiol. 75, 1597–1603. doi: 10.1128/AEM.02004-08
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ye, Y., Wang, B., Huang, F., Song, Y., Yan, H., Alam, M. J., et al. (2011). Application of in situ loop-mediated isothermal amplification method for detection of Salmonella in foods. Food Cont. 22, 438–444. doi: 10.1016/j.foodcont.2010.09.023
Yeni, F., Acar, S., Polat, O. G., Soyer, Y., and Alpas, H. (2014). Rapid and standardized methods for detection of foodborne pathogens and mycotoxins on fresh produce. Food Cont. 40, 359–367. doi: 10.1016/j.foodcont.2013.12.020
Zhang, G. (2013). Foodborne pathogenic bacteria detection: an evaluation of current and developing methods. Meducator 1, 15.
Zhao, X., Lin, C. W., Wang, J., and Oh, D. H. (2014). Advances in rapid detection methods for foodborne pathogens. J. Microbiol. Biotechn. 24, 297–312. doi: 10.4014/jmb.1310.10013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhou, B., Xiao, J., Liu, S., Yang, J., Wang, Y., Nie, F., et al. (2013). Simultaneous detection of six food-borne pathogens by multiplex PCR with GEXP analyzer. Food Cont. 32, 198–204. doi: 10.1016/j.foodcont.2012.11.044
Zhou, Y., Pan, F. G., Li, Y. S., Zhang, Y. Y., Zhang, J. H., Lu, S. Y., et al. (2009). Colloidal gold probe-based immunochromatographic assay for the rapid detection of brevetoxins in fishery product samples. Biosens. Bioelectron. 24, 2744–2747. doi: 10.1016/j.bios.2009.01.034
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: foodborne, pathogens, rapid, detection, PCR, NASBA, LAMP
Citation: Law JW-F, Ab Mutalib N-S, Chan K-G and Lee L-H (2015) Rapid methods for the detection of foodborne bacterial pathogens: principles, applications, advantages and limitations. Front. Microbiol. 5:770. doi: 10.3389/fmicb.2014.00770
Received: 21 October 2014; Accepted: 17 December 2014;
Published online: 12 January 2015.
Edited by:
Dongsheng Zhou, Beijing Institute of Microbiology and Epidemiology, ChinaReviewed by:
Kiiyukia Matthews Ciira, Jomo Kenyatta University of Agriculture and Technology, KenyaSheng Chen, Hong Kong Polytechnic University, China
Weili Liang, National Institute for Communicable Disease Control and Prevention, China
Copyright © 2015 Law, Ab Mutalib, Chan and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Learn-Han Lee, Jeffrey Cheah School of Medicine and Health Sciences, Monash University Malaysia, 46150 Bandar Sunway, Selangor Darul Ehsan, Malaysia e-mail:bGVlLmxlYXJuLmhhbkBtb25hc2guZWR1
 Jodi Woan-Fei Law
Jodi Woan-Fei Law Nurul-Syakima Ab Mutalib
Nurul-Syakima Ab Mutalib Kok-Gan Chan
Kok-Gan Chan Learn-Han Lee
Learn-Han Lee