- 1Alimentary Pharmabiotic Centre and Department of Microbiology, Bioscience Institute, National University of Ireland, Cork, Ireland
- 2Laboratory of Probiogenomics, Department of Life Sciences, University of Parma, Parma, Italy
- 3Department of Food, Environmental and Nutritional Sciences, University of Milan, Milan, Italy
Bifidobacteria are considered dominant and for this reason key members of the human gut microbiota, particularly during the first one to two years following birth. A substantial proportion of the bifidobacterial population in the intestine of infants belong to the Bifidobacterium bifidum taxon, whose members have been shown to display remarkable physiological and genetic features involving adhesion to epithelia, as well as utilization of host-derived glycans. Here, we reviewed the current knowledge on the genetic features and associated adaptations of B. bifidum to the human gut.
General Features of the Genus Bifidobacterium
The genus Bifidobacterium, a member of the Bifidobacteriaceae family, belongs to the Actinobacteria phylum (Stackebrandt and Tindall, 2000). Bifidobacteria are Gram-positive microorganisms with a high G+C DNA content, which were first isolated from feces of a breast-fed infant by Tissier in 1899, and then named Bacillus bifidus (Tissier, 1900). However, because of their morphological and physiological features, which are similar to those of lactobacilli, they were classified as members of the genus Lactobacillus for most of the 20th century and only starting from 1974 have been recognized as a separate genus (Buchanan, 2009). Currently, the genus Bifidobacterium is comprised of 48 different taxa, 40 of which have been isolated from the gastro-intestinal tract (GIT) contents of mammals, birds, or insects, while the remaining eight from sewage and fermented milk (Ventura et al., 2007a, 2009b, 2014). In addition, two bifidobacterial taxa, i.e., Bifidobacterium crudilactis and Bifidobacterium mongoliense, were isolated from raw milk cheeses (St Marcellin, Vercors area, France) (Delcenserie et al., 2013). Taking their different ecological niches into account and combining this information with a comparative analysis of their 16S rRNA sequences, as well as with other housekeeping genes (clpC, dnaJ, xfp, dnaB, rpoC, and purF), the various Bifidobacterium taxa can be clustered into six different phylogenetic groups, designated as the Bifidobacterium adolescentis-, Bifidobacterium asteroides-, Bifidobacterium boum-, Bifidobacterium longum-, Bifidobacterium pullorum-, and Bifidobacterium pseudolongum-phylogenetic groups (Ventura et al., 2007a). In this context, the B. bifidum species was shown not to fit in any of the above mentioned phylogenetic groups, thus suggesting the existence of unique and specific genetic features.
Bifidobacterial Ecology
Bifidobacteria have been isolated from six different ecological niches. Bifidobacteria are widely distributed among animals whose offspring enjoy parental care, such as mammals, birds, social insects. Therefore, the reason for their ecological distribution may be due to direct transmission of bifidobacterial cells from mother/carer to offspring. Though bifidobacteria are commonly found in the animal gut, these microorganisms have also been found in three other ecological niches: (iv) human blood (Bifidobacterium scardovii), (v) sewage (e.g., Bifidobacterium minimum and Bifidobacterium thermacidophilum), and (vi) food products (e.g., Bifidobacterium animalis subsp. lactis). These atypical ecological niches are rather different from that of the GIT, and it is plausible that the identification of bifidobacteria in these environments is the consequence of contaminations from GIT (Ventura et al., 2007b).
Notably, bifidobacteria that belong to the species Bifidobacterium animalis, Bifidobacterium adolescentis, Bifidobacterium dentium, and Bifidobacterium catenulatum display a more cosmopolitan lifestyle (Lamendella et al., 2008).
Bifidobacterium bifidum, Bifidobacterium breve, and Bifidobacterium longum are specifically identified in the human gut and have been shown to represent part of the dominant bacterial members of the gut microbiota of breast-fed infants (Turroni et al., 2009a, 2012).
Bifidobacterial Population in the Human Gut
Bifidobacteria quickly colonize the intestine of infants during the first weeks of life due to selection by breast or formula milk, as confirmed by metagenomic analyses (Roger et al., 2010; Koenig et al., 2011). In breast-fed infants, B. breve is the dominant species, followed by B. bifidum and B. longum subsp. infantis (Turroni et al., 2012; Milani et al., 2013).
The fecal microbiota of infants is characterized by high levels of bifidobacteria (Harmsen et al., 2000; Turroni et al., 2012). The level of abundance of bifidobacteria within the human gut decreases with age, although ecological analyses based on FISH and metagenomic studies have estimated that their presence in the adult colon is around 4.3 ± 4.4% of fecal microbes (Eckburg et al., 2005; Mueller et al., 2006). In adult feces, B. adolescentis and B. catenulatum species are commonly detected, followed by B. longum. Recently, a study involving the isolation of bifidobacteria from human intestinal mucosal samples and fecal samples on selective media (i.e., by a culture-dependent method), followed by the sequencing of the 16S rRNA internal transcribed spacer (ITS) regions of individual isolates allowed a description of the biodiversity of the bifidobacterial population present in the human gut (Turroni et al., 2009a). This study identified the most abundant bifidobacterial species present in the human gut, being represented by B. longum, B. pseudolongum, B. animalis subsp. lactis, B. adolescentis, B. bifidum, B. pseudocatenulatum, and B. breve. Furthermore, this analysis highlighted that the distribution of bifidobacteria present in various human subjects underline both an inter-subject, and an intra-subject variability, as also confirmed through real-time quantitative polymerase chain reaction (qPCR) analyses of fecal samples from healthy adults (Guglielmetti et al., 2013).
Turroni et al. (2009b) evaluated the bifidobacterial composition of the human intestine by a microbiomic approach, through the analyses of five colonic mucosal samples from healthy adults. This work showed how each subject possesses a specific population of colonic bifidobacteria that is in agreement with the large inter-variability of the whole intestinal microbiota previously described (Eckburg et al., 2005; Palmer et al., 2007). These samples were dominated by 16S rRNA gene sequences closely related to the B. pseudolongum phylogenetic groups (74.5%), followed by members of the B. longum (17%) and B. adolescentis (8.5%) phylogenetic groups. Moreover, this culture-independent approach led to the identification of many novel bifidobacterial 16S rRNA gene sequences, which are presumed to represent as yet undefined novel bifidobacterial species.
As mentioned above, B. bifidum is among the first colonizers of the human gut, reaching high numbers in the infant gut, but also detected at low levels in adults (Turroni et al., 2012). Particularly, the analyses by qPCR of fecal specimens from healthy adults revealed the presence of B. bifidum in 76% of the analyzed samples (n = 82), with a mean log(10) number of cells per g of feces (±SD) of 6.5 ± 1.4, whereas the mean concentration of total bifidobacteria in the same samples was 8.6 ± 1.2 (Guglielmetti et al., 2013). Therefore, B. bifidum species is a frequent member of the intestinal bifidobacterial population in healthy adults.
Genomics Insights into the B. bifidum Taxon
Due to the availability of novel whole-genome sequencing approaches, research in molecular microbiology, in particular that related to pathogens, has undergone dramatic changes during the last decade. In recent times, genome-decoding efforts have also been directed towards gut commensals and probiotic bacteria such as members of the genus Bifidobacterium (Schell et al., 2002; Lee et al., 2008; Sela et al., 2008; Barrangou et al., 2009; Turroni et al., 2010; Bottacini et al., 2011, 2012; O’Connell Motherway et al., 2011). In 2009, a genomics-based discipline, named probiogenomics, was established, which aims to provide insights into the diversity and evolution of beneficial gut commensals, and to reveal the molecular basis for their adaptation and interaction with the mammalian gut (Ventura et al., 2009a). Thanks to these probiogenomics efforts, we have significantly expanded our understanding of the biology of gut microorganisms, such as bifidobacteria, and we have generated a large amount of data on metabolic capabilities, genetics, and phylogeny of these bacteria. Within the genus Bifidobacterium, just eight members of the B. bifidum species have had their genome sequenced out of 23 currently publicly available complete bifidobacterial genome sequences (NCBI source). Notably, of these eight, only three genome sequences are complete, while the remaining five B. bifidum genome sequences are still fragmented in multiple contigs. The genome size of a B. bifidum taxon ranges from 2.14 to 2.28 Mb, whereas such a genome displays a GC content of about 62%, which is in line with the average values described for genomes of members of the genus Bifidobacterium (Ventura et al., 2007b, 2012; Turroni et al., 2013b). The currently NCBI-deposited reference genome of the B. bifidum species belongs to the infant stool isolate PRL2010 (Turroni et al., 2010), which was sequenced and published in 2010. The B. bifidum PRL2010 genome is also similar (89% identity at nucleotide level) to that of B. longum subsp. infantis ATCC15697 (Sela et al., 2008), even though they belong to two distinct bifidobacterial phylogenetic clusters, perhaps reflecting the fact that they share a common ecological niche (Ventura et al., 2007a). A functional classification of the genes present in the B. bifidum genomes according to the Cluster of Orthologous Genes (COG) families allowed the identification of a large proportion (>10%) of genes assigned to the COG family of carbohydrate metabolism and transport, including genes predicted to be involved in mucin metabolism (see below).
In silico analyses of the B. bifidum PRL2010 chromosome identified candidate genes displaying a deviant G+C content and for this reason possibly acquired through Horizontal Gene Transfer (HGT), collectively referred to as the mobilome. The predicted PRL2010 mobilome includes a prophage-like element Bbif-1 (Ventura et al., 2010) and two loci encompassing type I Restriction/Modification (R/M) systems as well as a type III R/M system (Turroni et al., 2010). Moreover, the predicted mobilome encompasses a 19 kb DNA region, which appears to represent an integrated plasmid.
The evaluation of genome variability within members of the B. bifidum species was assayed by Comparative Genomic Hybridization (CGH) experiments and the use of B. bifidum PRL2010-based microarrays (Turroni et al., 2010). In this way genes from the sequenced B. bifidum PRL2010 strain were assessed for their presence or absence in the genomes of a set of seven B. bifidum strains, which included isolates from various infant fecal samples as well as the neotype of B. bifidum species. Interestingly, among the variable regions of the CGH map there are those predicted as the mobilome of PRL2010 as well as other genes predicted to be involved in bacterium–environment interaction, such as the genes specifying sortase-dependent pili (Turroni et al., 2010). In addition, analysis of the B. bifidum PRL2010 chromosome revealed novel insights into the metabolic strategies followed by this strain to degrade host-derived glycans, and in particular mucin-associated carbohydrates.
The Genomics of Mucin Breakdown in B. bifidum
Mucin represents the main component of the mucus gel layer that is covering the epithelial surface of the GIT (Podolsky, 1985). The main carbohydrate monomers found in mucin include N-acetylglucosamine, N-acetylgalactosamine, fucose, and galactose, which are sometimes linked to sialic acid and sulfate groups (Forstner et al., 1995). In addition, mucin contains salts, lipids, and many proteins, such as growth factors, lysozyme, defensins, immunoglobulins, trefoil factors, and several intestinal proteins (Johansson et al., 2008).
Among members of the genus Bifidobacterium, the ability to degrade mucin is a peculiar property of the B. bifidum species, which can hydrolyse the glycosydic bonds of mucin (Turroni et al., 2010, 2011) and utilize it as the sole carbon source (Guglielmetti et al., 2009).
In silico analyses of the genome sequences of PRL2010 revealed a relatively small set of genes dedicated to carbohydrate metabolism, which predominantly specified glycosyl hydrolases (GH), compared to other bifidobacteria (Turroni et al., 2011). Remarkably, about 60% of the identified GH-encoding enzymes from B. bifidum PRL2010 are predicted to be involved in the degradation of mucin-derived oligosaccharides, most of which are uniquely present in the B. bifidum chromosome relative to other currently available bifidobacterial genomes. Furthermore, according to the Carbohydrate Active Enzymes (CAZy) system (Cantarel et al., 2009), the B. bifidum PRL2010 genome is predicted to encode members of two carbohydrate-binding module (CBM) families, CBM32 and CBM51, that are suggested to bind to carbohydrate residues encountered in the mucin core structure. Notably, in bifidobacteria the genetic information corresponding to predicted CBM32 and CBM51 members were only detected in the genomes of B. bifidum (Turroni et al., 2011).
Additional insights into the behavior of B. bifidum PRL2010 to utilize mucin were obtained from functional genomics approaches, such as whole proteome profiling as well as transcriptomic investigations (Turroni et al., 2010). Several of the enzymes encoded by B. bifidum PRL2010 involved in mucin metabolism encompass extracellular enzymes, such as putative exo-α-sialidases, as well as a predicted 1,2-α-L-fucosidase and 1,3/4-α-L-fucosidase, and a putative cell wall-anchored endo-α-N-acetylgalactosaminidase (Ashida et al., 2008, 2009; Kiyohara et al., 2012). Additional PRL2010 encoded enzymes that are believed to be involved in mucin breakdown include four N-acetyl-β-hexosaminidases, and four β-galactosidases. The mucin-catabolic phenotype of PRL2010 is further facilitated by the presence of carbohydrate transporters belonging to various families, such as the ATP-binding cassette (ABC-type), phosphoenolpyruvate phosphotransferase system (PEP-PTS) and major facilitator superfamily (MFS).
The genome of B. bifidum PRL2010 encompasses a DNA region spanning eight genes, which encode enzymes for the breakdown of galacto-N-biose that forms one of the core structures of mucin-oligosaccharides. In this context it is believed that B. bifidum PRL2010 accesses mucin-derived galacto-N-biose by the action of extracellular enzymes like the exo-α-sialidases and 1,2-α-L-/α-1,3/4-fucosidases, which perform de-sialidation and de-fucosylation, respectively, of mucin-derived oligosaccharides to facilitate further breakdown by the action of other enzymes encoded by PRL2010 such as the lacto-N-biosidase and endo-α-N-acetylgalactosaminidase.
Comparative genomics analyses involving all other currently available B. bifidum genomes revealed a high conservation of the predicted genetic arsenal involved in mucin breakdown (Turroni et al., 2011). Thus, it is tempting to conclude that mucin metabolism in PRL2010 is a genetic feature of most if not all members of the B. bifidum species rather than of a unique strain. Mucin degradation, which is expected to reduce the mucin layer and consequently reduce the protective barrier covering the intestinal mucosa, is generally considered as an undesirable event. However, one may also consider breakdown as an evolved “host-settler mechanism.” In fact, mucin production in the GIT normally initiates only several months after birth and reaches its mature level at about 12 months (Hooper et al., 1999). Interestingly, mucin breakdown activity as operated by B. bifidum could trigger the secretion of additional colonic mucin, thus increasing the thickness of the total amount of mucus layer covering the gut and so reinforcing the epithelial barrier function, which constitutes an important feature especially in those subjects affected by irritable bowel syndrome (Caballero-Franco et al., 2007). The capacity to efficiently use mucus is a typical feature also of Akkermansia muciniphila, a human intestinal species that has been associated with healthy intestines and disease prevention (Guglielmetti et al., 2008b).
Pili Production By B. bifidum as Key Host–Microbe Effector Molecules
Non-flagellar appendages decorating the microbial cell surface were identified in bacteria in the early 1950s and since then the molecular data about their assembly, composition, and function has greatly expanded, especially for pathogens (Telford et al., 2006). In this context, these extracellular structures are considered crucial in the initial establishment of pathogens inside the host and are consequently considered key effector molecules in pathogenesis. However, their identification in bifidobacteria was only very recently established (Foroni et al., 2011; O’Connell Motherway et al., 2011). It has been shown that bifidobacterial genomes belonging to B. bifidum, B. longum subsp. longum, B. adolescentis, B. dentium, B. animalis subsp. Lactis, and B. breve contain one to seven predicted sortase-dependent pilus gene loci, each of which are predicted to encode one major pilin subunit (represented by FimAPRL2010 or FimPPRL2010 for the pil2PRL2010 and pil3PRL2010 clusters, respectively) plus a minor pilin subunit (represented by FimBPRL2010 and FimQPRL2010 for the pil2PRL2010 and pil3PRL2010 clusters, respectively), as well as a so-called sortase, a protein specifically dedicated to covalently assemble these pilin subunits (Foroni et al., 2011).
Very recently, the four B. bifidum genome sequences that are currently publicly available have been screened for sortase-dependent pili leading to the identification of three loci (Turroni et al., 2013a). Of these three identified loci, only two were shown to be genetically intact whereas the third appeared to be non-functional due to a frameshift within the coding region of the gene encoding major pilus subunit (Turroni et al., 2013a). When FimAPRL2010 was compared to FimA homologs encoded by other B. bifidum strains, their amino acid sequences were shown to display much higher variability compared to the FimP homologs (Turroni et al., 2013a). Furthermore, FimAPRL2010 includes a CnaB-type domain that is known to serve as a stalk in binding to components of the Extra Cellular Matrix proteins of the host, such as fibronectin, collagen types I to XV, and laminin (Deivanayagam et al., 2000).
Transcriptomic investigations performed on B. bifidum PRL2010 upon colonization of mice as well as upon contact with human cell lines, demonstrated a clear transcriptional up-regulation of those genes encompassing two sortase-dependent pili, named pil2 and pil3 (Turroni et al., 2013a). Heterologous expression of the pilus-encoding genes corresponding to Pil2 and Pil3 in the non-piliated, Gram-positive host Lactococcus lactis showed that both types of pili are modulating the adhesion to human enterocytes through extracellular matrix (ECM) proteins and bacterial aggregation. ECM deglycosylation provoked a dramatic reduction in PRL2010 pili-mediated binding ability compared to untreated ECM (Turroni et al., 2013a), pointing that N- and/or O-linked glycoproteins are involved in adhesion of PRL2010 pili to ECM. Furthermore, carbohydrate binding competition experiments demonstrated that mannose and fucose act as potential receptors for Pil2 of B. bifidum PRL2010 in a fashion that is reminiscent of that previously described for other enteric bacteria (Farfan et al., 2011), whereas the putative binding partners for Pil3 appear to encompass a larger set of carbohydrates (Turroni et al., 2013a).
Recombinant piliated L. lactis cells were also shown to evoke a higher tumor necrosis factor alpha (TNFα) response during murine colonization compared to their non-piliated parent, indicating that B. bifidum PRL2010 sortase-dependent pili not only contribute to adhesion but also display immunomodulatory activity (Turroni et al., 2013a). Triggering TNFα production by pili synthetized by B. bifidum PRL2010 may represent an intriguing feature of this species as one of the first colonizers of the human gut (Turroni et al., 2012). In this context, it is worth mentioning that cytokines belonging to the TNFα superfamily are not only linked to the occurrence of inflammatory diseases (Yasutake et al., 1999), but also exert a major role in the rejection of tumors and the response to infections (Wajant et al., 2003; Lebeer et al., 2010). Furthermore, the induction of TNFα may be crucial for the initiation of cross-talk among immune cells without provoking any inflammation or detrimental effects (Galdeano et al., 2007).
Other Host Response Effector Molecules Encoded by B. bifidum
Recently, the murine lytic enzyme TgaA encoded by B. bifidum MIMBb75 has been molecularly characterized (Guglielmetti et al., 2014b). This peptidoglycan-degrading enzyme contains two active domains, i.e., a lytic murine transglycosylase and a cysteine histidine-dependent amidohydrolase/peptidase (CHAP) domain and was demonstrated to exert immunomodulatory effects (Guglielmetti et al., 2014a). The TgA-encoding gene does not appear to be widely distributed among the currently available B. bifidum genomes and thus represents an example of a strain-dependent gene (Guglielmetti et al., 2014b).
Other B. bifidum proteins involved in host interaction are represented by the surface lipoprotein BopA, which was originally described to be involved in adhesion to intestinal epithelium (Guglielmetti et al., 2008b; Gleinser et al., 2012). However, recently the role of BopA in the adhesion of B. bifidum was reassessed and, in contrast to what was published earlier, the strong adhesion of B. bifidum to epithelial cell lines is mainly BopA-independent (Kainulainen et al., 2013).
Within the surface proteins encoded by B. bifidum strains responsible of adhesion to the human intestine, the transaldolase Tal of B. bifidum A8 has been also proposed (Gonzalez-Rodriguez et al., 2012). Notably, such protein has been shown in modulating the adhesion to mucin as well as to promote bacterial aggregation, thus could act as a key colonization factor in driving the establishment of B. bifidum cells in the human gut.
Immune Response of B. bifidum Strains
Members of the B. bifidum species have been claimed to exert an important role in the evolution and maturation of the immune system of the host, which is still undeveloped at birth (Lopez et al., 2011). The interaction of B. bifidum with the host immune system has been assayed by investigating the impact of B. bifidum Z9 in combination with a second human gut commensal, Lactobacillus acidophilus, on the transcriptome of dendritic cells (DCs) (Weiss et al., 2010). This study highlighted that B. bifidum Z9 down-regulates the expression of genes involved to the adaptive immune system in murine DCs. Such findings corroborated other studies based on in vitro assays and involving various strains belonging to different Bifidobacterium species, which display a clear and distinct induction of cytokine profile by bifidobacteria. In particular, it was shown that B. bifidum strains, in contrast to representatives of other bifidobacterial species, provoked a significantly increased production of the IL-17 cytokine (Turroni et al., 2010; Lopez et al., 2011). The observation that B. bifidum strains induce an immune response affecting Treg/TH17 plasticity (Turroni et al., 2010; Lopez et al., 2011) leads to hypothesize that such commensal bacteria have a key role in mucosal tolerance, as also suggested by the demonstration that B. bifidum, differently from several other species of the genus, possesses the ability to induce IL-2 secretion by DCs (Guglielmetti et al., 2014a). In particular, it was demonstrated that the cell-surface-exposed molecule Tga of B. bifidum MIMBb75 is capable alone through its C-terminal CHAP domain of inducing DCs activation and IL-2 production (Guglielmetti et al., 2014b).
Recently, the host response triggered by the presence of B. bifidum PRL2010 cells was investigated thanks to a high-throughput gene expression technology and by utilizing both an in vitro cell line model as well as a murine model (Turroni et al., 2014a). Notably, the overall host-response scenario driven by B. bifidum PRL2010 cells can be described as a pro-inflammatory response priming the immune system, yet at the same time attenuating the pro-inflammatory response by down-regulation of certain chemokines, heat shock proteins (HSP) as well as stimulating the up-regulation of defensin and tight junction genes. In addition, results from ELISA experiments displayed that exposure to B. bifidum PRL2010 triggers the synthesis of IL-6 and IL-8 cytokines, presumably through NF-κb activation (Turroni et al., 2014a).
Other in vivo observations involving B. bifidum strains have been performed under chemically-induced diseases such as colitis. In this context, administration of B. bifidum strain S17 to mice with colitis was shown to suppress intestinal inflammation with a significant reduction in histology scores and the levels of pro-inflammatory cytokines interleukin IL-1β, IL-6, keratinocyte-derived chemokine and the inflammatory markers cyclooxygenase and myeloperoxidase (Philippe et al., 2011).
B. bifidum Strains as Potential Health-Promoting Candidates
Various strains of the B. bifidum species have been reported to exert health benefits to their human host, including antibacterial activities against pathogens such as Helicobacter pylori (Shirasawa et al., 2010; Chenoll et al., 2011), reduction of apoptosis in the intestinal epithelium of infants suffering from necrotizing enterocolitis (Khailova et al., 2010), modulation of the host-immune system (Fu et al., 2010; Philippe et al., 2011), and alleviation of anti-inflammatory activities associated with certain chronic large bowel dysfunctions (Mouni et al., 2009; Guglielmetti et al., 2011). In addition, B. bifidum together with other bifidobacterial species like B. breve and B. longum subsp. infantis are considered important for the establishment of a well-balanced, autochthonous intestinal microbiota in newborns (Tabbers et al., 2011). However, in order to exert a potential health-promoting activity in the human gut, bacteria need to reach this compartment in a viable form, while they should also be able to persist within the intestine. In this context, many B. bifidum strains have been shown to possess a strong adhesion phenotype to human epithelial intestinal cell monolayers (Caco-2 and HT29) (Guglielmetti et al., 2009; Serafini et al., 2013) and in a few cases, such as for B. bifidum PRL2010 and MIMBb75, were demonstrated to survive under gastrointestinal challenges (Serafini et al., 2013) and colonize the intestine impacting on the resident microbial communities at various intestinal loci (Singh et al., 2013). Another interesting phenotype displayed by probiotic bacteria is their displacement and competition against pathogens. Interestingly, in vitro trials based on HT29 monolayer involving B. bifidum PRL2010 cells displayed a clear inhibition of adhesion of pathogenic bacteria such as Escherichia coli and Cronobacter sakazakii (Serafini et al., 2013).
A health-promoting microorganism needs to be administered to its human host within a specific matrix in order to assure survival and biological functionality of its cells (e.g., producing probiotic molecules). Recently, kefir and kefiran were shown to affect the transcriptome of B. bifidum PRL2010 causing increased transcription of genes involved in the metabolism of dietary glycans as well as genes acting as host–microbe effector molecules such as pili (Serafini et al., 2014). Thus, the use of kefir and perhaps other (fermented) food products may be considered as a valuable means for the administration of B. bifidum cells to humans and may represent an effective food matrix to pre-adapt bifidobacterial cells to the host in order to enhance probiotic efficacy.
Industrial Use of B. bifidum
Due to their health-promoting activities, much effort has been invested in the incorporation of bifidobacteria into probiotic food, supplements, and pharmaceutical preparations. By regulatory definition, microbial cells must be alive in a sufficient number in order to define a product as probiotic. In this perspective, the commercial use of B. bifidum as a probiotic has been limited by the fact that members of this species are particularly sensitive to stresses such as acidity and, in particular, oxygen (Jayamanne and Adams, 2006). Strategies to preserve probiotic cell viability are available and involve, for instance, microencapsulation (Zhang et al., 2013), and the addition of prebiotic molecules to the formulation (Guglielmetti et al., 2008a). However, these strategies do not overcome the problem of the limited biomass yields generally obtained in industrial fermentations due to the intrinsic stress sensitivity of B. bifidum, resulting in a strong increase of production costs. In this context, an aspect of particular importance is represented by strain “domestication.” In fact, once a bifidobacterial strain is isolated from its natural environment, its intrinsic sensitivity to oxygen decreases slowly and progressively during the numerous subcultivations under laboratory conditions. In this direction, an example is represented by strain B. bifidum MIMBb75, which was isolated about twelve years ago from a fecal sample of a healthy adult. Although originally very recalcitrant to laboratory cultivation, after hundreds of subculturings in aerobic atmosphere followed by incubation in a gas-pack with Anaerocult A, this strain drastically improved its ability to resist oxidative and other laboratory stresses (Guglielmetti S., personal communication). Thus, the selective pressure of a laboratory environment induced physiological changes that allowed the employment of strain B. bifidum MIMBb75 at industrial level; this strain, in fact, is now commercially available in a pharmaceutical probiotic product consisting of an encapsulated formulation, in which MIMBb75 cells can maintain a viability of at least 109 CFU per capsule during the entire shelf-life of the product (Guglielmetti et al., 2011; Guglielmetti S., personal communication).
Conclusion
During the last 5–10 years research in bifidobacteria has blossomed (Ventura et al., 2012, 2014; Turroni et al., 2014b). In this context, genomic investigations of bifidobacteria have revealed genetic repertoires that are considered crucial for conveying the typical saccharolytic phenotype of these bacteria, and which are worthy of detailed investigation for their potential roles in colonization of the human gut and their dependence on our diet. However, the discovery of the specific functional contribution to the host by each member of the bifidobacterial population in the human gut is still at its infancy. Furthermore, very little is known about the cross-talk that is believed to occur between individual members of the bifidobacterial microbiota (Egan et al., 2014) and with other members of the gut microbiota (Sonnenburg et al., 2006). The genetic data retrieved from the genome analysis of B. bifidum suggests the existence of various molecules that are responsible for specific health-promoting activities exerted by members of this bifidobacterial taxon (Figure 1). Future work needs to be carried out in order to better understand how these genetic features are exploited in the human gut through the use of high throughput metagenomic and post-genomic approaches. Nonetheless, currently available experimental data already supports the notion that B. bifidum represents a highly interesting bacterial species that is able to benefit human health in the prevention and treatment of gastrointestinal dysfunctions.
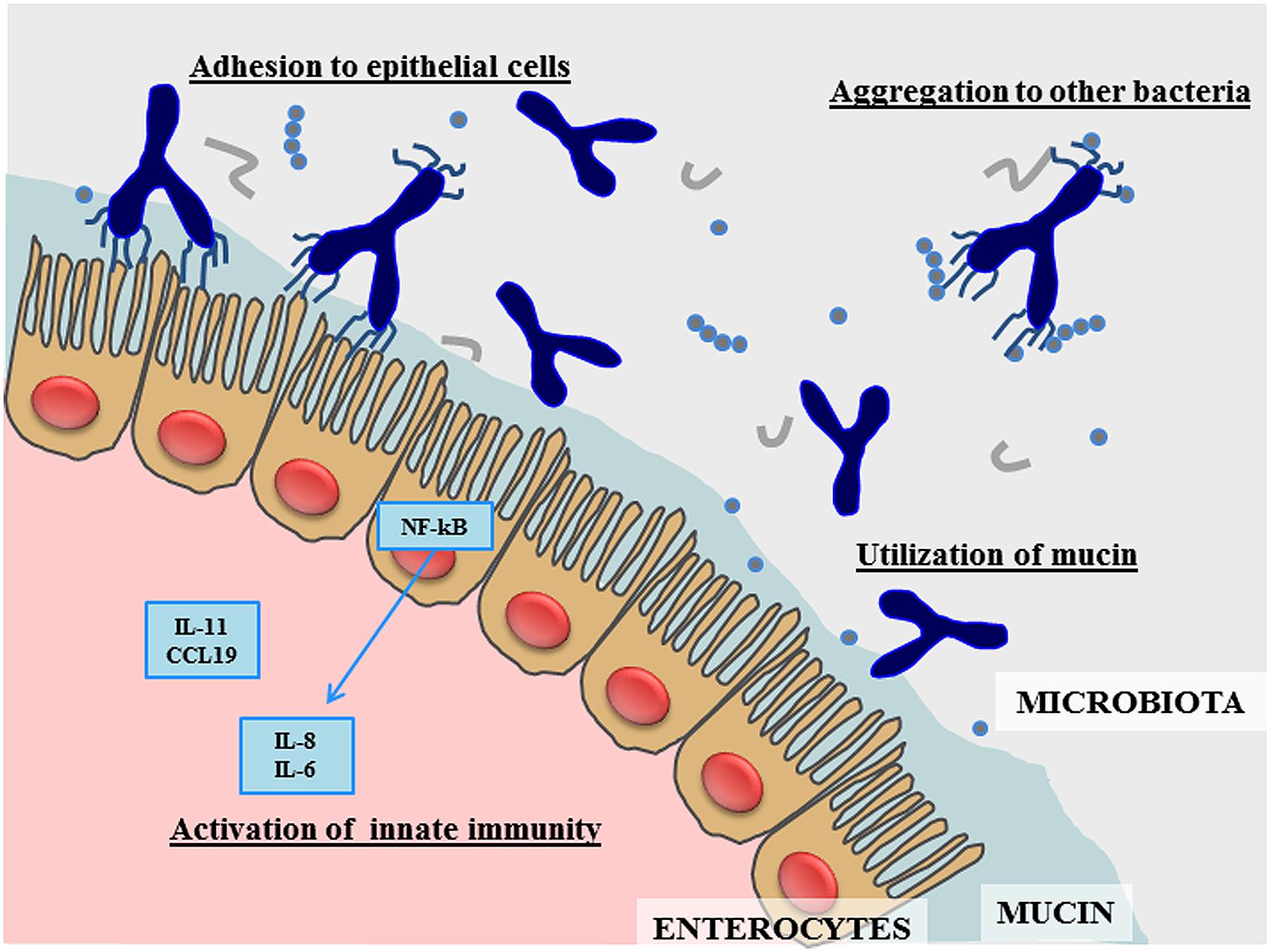
FIGURE 1. Schematic representation of the main effects produced by Bifidobacterium bifidum in the human gut.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank GenProbio srl for the financial support of the Laboratory of Probiogenomics. This publication has emanated from research conducted with the financial support of Science Foundation Ireland (SFI) under Grant Numbers 07/CE/B1368 and SFI/12/RC/2273.
References
Ashida, H., Maki, R., Ozawa, H., Tani, Y., Kiyohara, M., Fujita, M.,et al. (2008). Characterization of two different endo-alpha-N-acetylgalactosaminidases from probiotic and pathogenic enterobacteria, Bifidobacterium longum and Clostridium perfringens. Glycobiology 18, 727–734. doi: 10.1093/glycob/cwn053
Ashida, H., Miyake, A., Kiyohara, M., Wada, J., Yoshida, E., Kumagai, H.,et al. (2009). Two distinct alpha-L-fucosidases from Bifidobacterium bifidum are essential for the utilization of fucosylated milk oligosaccharides and glycoconjugates. Glycobiology 19, 1010–1017. doi: 10.1093/glycob/cwp082
Barrangou, R., Briczinski, E. P., Traeger, L. L., Loquasto, J. R., Richards, M., Horvath, P.,et al. (2009). Comparison of the complete genome sequences of Bifidobacterium animalis subsp. lactis DSM 10140 and Bl-04. J. Bacteriol. 191, 4144–4151. doi: 10.1128/JB.00155-09
Bottacini, F., Dal Bello, F., Turroni, F., Milani, C., Duranti, S., Foroni, E.,et al. (2011). Complete genome sequence of Bifidobacterium animalis subsp. lactis BLC1. J. Bacteriol. 193, 6387–6388. doi: 10.1128/JB.06079-11
Bottacini, F., Milani, C., Turroni, F., Sanchez, B., Foroni, E., Duranti, S.,et al. (2012). Bifidobacterium asteroides PRL2011 genome analysis reveals clues for colonization of the insect gut. PLoS ONE 7:e44229. doi: 10.1371/journal.pone.0044229
Buchanan, R. E. (2009). Bergey’s Manual of Systematic Bacteriology, ed. N. E. Gibbons. New York: Springer.
Caballero-Franco, C., Keller, K., De Simone, C., and Chadee, K. (2007). The VSL#3 probiotic formula induces mucin gene expression and secretion in colonic epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 292, G315–G322. doi: 10.1152/ajpgi.00265.2006
Cantarel, B. L., Coutinho, P. M., Rancurel, C., Bernard, T., Lombard, V., and Henrissat, B. (2009). The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D238. doi: 10.1093/nar/gkn663
Chenoll, E., Casinos, B., Bataller, E., Astals, P., Echevarria, J., Iglesias, J. R.,et al. (2011). Novel probiotic Bifidobacterium bifidum CECT 7366 strain active against the pathogenic bacterium Helicobacter pylori. Appl. Environ. Microbiol. 77, 1335–1343. doi: 10.1128/AEM.01820-10
Deivanayagam, C. C., Rich, R. L., Carson, M., Owens, R. T., Danthuluri, S., Bice, T.,et al. (2000). Novel fold and assembly of the repetitive B region of the Staphylococcus aureus collagen-binding surface protein. Structure 8, 67–78. doi: 10.1016/S0969-2126(00)00081-2
Delcenserie, V., Taminiau, B., Gavini, F., De Schaetzen, M. A., Cleenwerck, I., Theves, M.,et al. (2013). Detection and characterization of Bifidobacterium crudilactis and B. mongoliense able to grow during the manufacturing process of French raw milk cheeses. BMC Microbiol. 13:239. doi: 10.1186/1471-2180-13-239
Eckburg, P. B., Bik, E. M., Bernstein, C. N., Purdom, E., Dethlefsen, L., Sargent, M.,et al. (2005). Diversity of the human intestinal microbial flora. Science 308, 1635–1638. doi: 10.1126/science.1110591
Egan, M., O’connell Motherway, M., Ventura, M., and Van Sinderen, D. (2014). Metabolism of sialic acid by Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 80, 4414–4426. doi: 10.1128/AEM.01114-14
Farfan, M. J., Cantero, L., Vidal, R., Botkin, D. J., and Torres, A. G. (2011). Long polar fimbriae of enterohemorrhagic Escherichia coli O157:H7 bind to extracellular matrix proteins. Infect. Immun. 79, 3744–3750. doi: 10.1128/IAI.05317-11
Foroni, E., Serafini, F., Amidani, D., Turroni, F., He, F., Bottacini, F.,et al. (2011). Genetic analysis and morphological identification of pilus-like structures in members of the genus Bifidobacterium. Microb. Cell Fact. 10(Suppl. 1), S16. doi: 10.1186/1475-2859-10-S1-S16
Forstner, J. F., Oliver., M. G., and Sylvester, F. A. (1995). Production, Structure and Biologic Relevance of Gastrointestinal Mucins. New York: Raven Press.
Fu, Y. R., Yi, Z. J., Pei, J. L., and Guan, S. (2010). Effects of Bifidobacterium bifidum on adaptive immune senescence in aging mice. Microbiol. Immunol. 54, 578–583. doi: 10.1111/j.1348-0421.2010.00255.x
Galdeano, C. M., De Moreno De Leblanc, A., Vinderola, G., Bonet, M. E., and Perdigon, G. (2007). Proposed model: mechanisms of immunomodulation induced by probiotic bacteria. Clin. Vaccine Immunol. 14, 485–492. doi: 10.1128/CVI.00406-06
Gleinser, M., Grimm, V., Zhurina, D., Yuan, J., and Riedel, C. U. (2012). Improved adhesive properties of recombinant bifidobacteria expressing the Bifidobacterium bifidum-specific lipoprotein BopA. Microb. Cell Fact. 11, 80. doi: 10.1186/1475-2859-11-80s
Gonzalez-Rodriguez, I., Sanchez, B., Ruiz, L., Turroni, F., Ventura, M., Ruas-Madiedo, P.,et al. (2012). Role of extracellular transaldolase from Bifidobacterium bifidum in mucin adhesion and aggregation. Appl. Environ. Microbiol. 78, 3992–3998. doi: 10.1128/AEM.08024-11
Guglielmetti, S., Balzaretti, S., Taverniti, V., Miriani, M., Milani, C., Scarafoni, A.,et al. (2014a). Characterization of TgaA, a VirB1-like component belonging to a putative type IV secretion system of Bifidobacterium bifidum MIMBb75. Appl. Environ. Microbiol. doi: 10.1128/AEM.01413-14 [Epub ahead of print].
Guglielmetti, S., Zanoni, I., Balzaretti, S., Miriani, M., Taverniti, V., De Noni, I.,et al. (2014b). The murein lytic enzyme TgaA of Bifidobacterium bifidum MIMBb75 modulates dendritic cell maturation through its CHAP amidase domain. Appl. Environ. Microbiol. doi: 10.1128/AEM.00761-14 [Epub ahead of print].
Guglielmetti, S., Ciranna, A., Mora, D., Parini, C., and Karp, M. (2008a). Construction, characterization and exemplificative application of bioluminescent Bifidobacterium longum biovar longum. Int. J. Food Microbiol. 124, 285–290. doi: 10.1016/j.ijfoodmicro.2008.03.033
Guglielmetti, S., Tamagnini, I., Mora, D., Minuzzo, M., Scarafoni, A., Arioli, S.,et al. (2008b). Implication of an outer surface lipoprotein in adhesion of Bifidobacterium bifidum to Caco-2 cells. Appl. Environ. Microbiol. 74, 4695–4702. doi: 10.1128/AEM.00124-08
Guglielmetti, S., Fracassetti, D., Taverniti, V., Del Bo, C., Vendrame, S., Klimis-Zacas, D.,et al. (2013). Differential modulation of human intestinal Bifidobacterium populations after consumption of a wild blueberry (Vaccinium angustifolium) drink. J. Agric. Food Chem. 61, 8134–8140. doi: 10.1021/jf402495k
Guglielmetti, S., Mora, D., Gschwender, M., and Popp, K. (2011). Randomised clinical trial: Bifidobacterium bifidum MIMBb75 significantly alleviates irritable bowel syndrome and improves quality of life – a double-blind, placebo-controlled study. Aliment. Pharmacol. Ther. 33, 1123–1132. doi: 10.1111/j.1365-2036.2011.04633.x
Guglielmetti, S., Tamagnini, I., Minuzzo, M., Arioli, S., Parini, C., Comelli, E.,et al. (2009). Study of the adhesion of Bifidobacterium bifidum MIMBb75 to human intestinal cell lines. Curr. Microbiol. 59, 167–172. doi: 10.1007/s00284-009-9415-x
Harmsen, H. J., Wildeboer-Veloo, A. C., Raangs, G. C., Wagendorp, A. A., Klijn, N., Bindels, J. G.,et al. (2000). Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J. Pediatr. Gastroenterol. Nutr. 30, 61–67. doi: 10.1097/00005176-200001000-00019
Hooper, L. V., Xu, J., Falk, P. G., Midtvedt, T., and Gordon, J. I. (1999). A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc. Natl. Acad. Sci. U.S.A. 96, 9833–9838. doi: 10.1073/pnas.96.17.9833
Jayamanne, V. S., and Adams, M. R. (2006). Determination of survival, identity and stress resistance of probiotic bifidobacteria in bio-yoghurts. Lett. Appl. Microbiol. 42, 189–194. doi: 10.1111/j.1472-765X.2006.01843.x
Johansson, M. E., Phillipson, M., Petersson, J., Velcich, A., Holm, L., and Hansson, G. C. (2008). The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc. Natl. Acad. Sci. U.S.A. 105, 15064–15069. doi: 10.1073/pnas.0803124105
Kainulainen, V., Reunanen, J., Hiippala, K., Guglielmetti, S., Vesterlund, S., Palva, A.,et al. (2013). BopA does not have a major role in the adhesion of Bifidobacterium bifidum to intestinal epithelial cells, extracellular matrix proteins, and mucus. Appl. Environ. Microbiol. 79, 6989–6997. doi: 10.1128/AEM.01993-13
Khailova, L., Mount Patrick, S. K., Arganbright, K. M., Halpern, M. D., Kinouchi, T., and Dvorak, B. (2010). Bifidobacterium bifidum reduces apoptosis in the intestinal epithelium in necrotizing enterocolitis. Am. J. Physiol. Gastrointest. Liver Physiol. 299, G1118–G1127. doi: 10.1152/ajpgi.00131.2010
Kiyohara, M., Nakatomi, T., Kurihara, S., Fushinobu, S., Suzuki, H., Tanaka, T.,et al. (2012). Alpha-N-acetylgalactosaminidase from infant-associated bifidobacteria belonging to novel glycoside hydrolase family 129 is implicated in alternative mucin degradation pathway. J. Biol. Chem. 287, 693–700. doi: 10.1074/jbc.M111.277384
Koenig, J. E., Spor, A., Scalfone, N., Fricker, A. D., Stombaugh, J., Knight, R.,et al. (2011). Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. U.S.A. 108(Suppl. 1), 4578–4585. doi: 10.1073/pnas.1000081107
Lamendella, R., Santo Domingo, J. W., Kelty, C., and Oerther, D. B. (2008). Bifidobacteria in feces and environmental waters. Appl. Environ. Microbiol. 74, 575–584. doi: 10.1128/AEM.01221-07
Lebeer, S., Vanderleyden, J., and De Keersmaecker, S. C. (2010). Host interactions of probiotic bacterial surface molecules: comparison with commensals and pathogens. Nat. Rev. Microbiol. 8, 171–184. doi: 10.1038/nrmicro2297
Lee, J. H., Karamychev, V. N., Kozyavkin, S. A., Mills, D., Pavlov, A. R., Pavlova, N. V.,et al. (2008). Comparative genomic analysis of the gut bacterium Bifidobacterium longum reveals loci susceptible to deletion during pure culture growth. BMC Genomics 9:247. doi: 10.1186/1471-2164-9-247
Lopez, P., Gonzalez-Rodriguez, I., Gueimonde, M., Margolles, A., and Suarez, A. (2011). Immune response to Bifidobacterium bifidum strains support Treg/Th17 plasticity. PLoS ONE 6:e24776. doi: 10.1371/journal.pone.0024776
Milani, C., Hevia, A., Foroni, E., Duranti, S., Turroni, F., Lugli, G. A.,et al. (2013). Assessing the fecal microbiota: an optimized ion torrent 16S rRNA gene-based analysis protocol. PLoS ONE 8:e68739. doi: 10.1371/journal.pone.0068739
Mouni, F., Aissi, E., Hernandez, J., Gorocica, P., Bouquelet, S., Zenteno, E.,et al. (2009). Effect of Bifidobacterium bifidum DSM 20082 cytoplasmic fraction on human immune cells. Immunol. Invest. 38, 104–115. doi: 10.1080/08820130802608303
Mueller, S., Saunier, K., Hanisch, C., Norin, E., Alm, L., Midtvedt, T.,et al. (2006). Differences in fecal microbiota in different European study populations in relation to age, gender, and country: a cross-sectional study. Appl. Environ. Microbiol. 72, 1027–1033. doi: 10.1128/AEM.72.2.1027-1033.2006
O’Connell Motherway, M., Zomer, A., Leahy, S. C., Reunanen, J., Bottacini, F., Claesson, M. J.,et al. (2011). Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc. Natl. Acad. Sci. U.S.A. 108, 11217–11222. doi: 10.1073/pnas.1105380108
Palmer, C., Bik, E. M., Digiulio, D. B., Relman, D. A., and Brown, P. O. (2007). Development of the human infant intestinal microbiota. PLoS Biol. 5:e177. doi: 10.1371/journal.pbio.0050177
Philippe, D., Heupel, E., Blum-Sperisen, S., and Riedel, C. U. (2011). Treatment with Bifidobacterium bifidum 17 partially protects mice from Th1-driven inflammation in a chemically induced model of colitis. Int. J. Food Microbiol. 149, 45–49. doi: 10.1016/j.ijfoodmicro.2010.12.020
Podolsky, D. K. (1985). Oligosaccharide structures of isolated human colonic mucin species. J. Biol. Chem. 260, 15510–15515.
Roger, L. C., Costabile, A., Holland, D. T., Hoyles, L., and Mccartney, A. L. (2010). Examination of faecal Bifidobacterium populations in breast- and formula-fed infants during the first 18 months of life. Microbiology 156, 3329–3341. doi: 10.1099/mic.0.043224-0
Schell, M. A., Karmirantzou, M., Snel, B., Vilanova, D., Berger, B., Pessi, G.,et al. (2002). The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. U.S.A. 99, 14422–14427. doi: 10.1073/pnas.212527599212527599
Sela, D. A., Chapman, J., Adeuya, A., Kim, J. H., Chen, F., Whitehead, T. R.,et al. (2008). The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome.Proc. Natl. Acad. Sci. U.S.A. 105, 18964–18969. doi: 10.1073/pnas.0809 584105
Serafini, F., Strati, F., Ruas-Madiedo, P., Turroni, F., Foroni, E., Duranti, S.,et al. (2013). Evaluation of adhesion properties and antibacterial activities of the infant gut commensal Bifidobacterium bifidum PRL2010. Anaerobe 21, 9–17. doi: 10.1016/j.anaerobe.2013.03.003
Serafini, F., Turroni, F., Ruas-Madiedo, P., Lugli, G. A., Milani, C., Duranti, S.,et al. (2014). Kefir fermented milk and kefiran promote growth of Bifidobacterium bifidum PRL2010 and modulate its gene expression. Int. J. Food Microbiol. 178, 50–59. doi: 10.1016/j.ijfoodmicro.2014.02.024
Shirasawa, Y., Shibahara-Sone, H., Iino, T., and Ishikawa, F. (2010). Bifidobacterium bifidum BF-1 suppresses Helicobacter pylori-induced genes in human epithelial cells. J. Dairy Sci. 93, 4526–4534. doi: 10.3168/jds.2010-3274
Singh, N., Arioli, S., Wang, A., Villa, C. R., Jahani, R., Song, Y. S.,et al. (2013). Impact of Bifidobacterium bifidum MIMBb75 on mouse intestinal microorganisms. FEMS Microbiol. Ecol. 85, 369–375. doi: 10.1111/1574-6941.12124
Sonnenburg, J. L., Chen, C. T., and Gordon, J. I. (2006). Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host. PLoS Biol. 4:e413. doi: 10.1371/journal.pbio.0040413
Stackebrandt, E., and Tindall, B. J. (2000). Appreciating microbial diversity: rediscovering the importance of isolation and characterization of microorganisms. Environ. Microbiol. 2, 9–10.
Tabbers, M. M., De Milliano, I., Roseboom, M. G., and Benninga, M. A. (2011). Is Bifidobacterium breve effective in the treatment of childhood constipation? Results from a pilot study. Nutr. J. 10:19. doi: 10.1186/1475-2891-10-19
Telford, J. L., Barocchi, M. A., Margarit, I., Rappuoli, R., and Grandi, G. (2006). Pili in Gram-positive pathogens. Nat. Rev. Microbiol. 4, 509–519. doi: 10.1038/nrmicro1443
Tissier. (1900). Recherchers Sur La Flora Intestinale Normale et Pathologique Du Nourisson. Ph.D. thesis, University of Paris, Paris.
Turroni, F., Bottacini, F., Foroni, E., Mulder, I., Kim, J. H., Zomer, A.,et al. (2010). Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc. Natl. Acad. Sci. U.S.A. 107, 19514–19519. doi: 10.1073/pnas.1011100107
Turroni, F., Foroni, E., Pizzetti, P., Giubellini, V., Ribbera, A., Merusi, P.,et al. (2009a). Exploring the diversity of the bifidobacterial population in the human intestinal tract. Appl. Environ. Microbiol. 75, 1534–1545. doi: 10.1128/AEM.02216-08
Turroni, F., Marchesi, J. R., Foroni, E., Gueimonde, M., Shanahan, F., Margolles, A.,et al. (2009b). Microbiomic analysis of the bifidobacterial population in the human distal gut. ISME J. 3, 745–751. doi: 10.1038/ismej.2009.19
Turroni, F., Milani, C., Van Sinderen, D., and Ventura, M. (2011). Genetic strategies for mucin metabolism in Bifidobacterium bifidum PRL2010: an example of possible human–microbe co-evolution. Gut Microbes 2, 183–189. doi: 10.4161/gmic.2.3.16105
Turroni, F., Peano, C., Pass, D. A., Foroni, E., Severgnini, M., Claesson, M. J.,et al. (2012). Diversity of bifidobacteria within the infant gut microbiota. PLoS ONE 7:e36957. doi: 10.1371/journal.pone.0036957
Turroni, F., Serafini, F., Foroni, E., Duranti, S., O’connell Motherway, M., Taverniti, V.,et al. (2013a). Role of sortase-dependent pili of Bifidobacterium bifidum PRL2010 in modulating bacterium–host interactions. Proc. Natl. Acad. Sci. U.S.A. 110, 11151–11156. doi: 10.1073/pnas.1303897110
Turroni, F., Ventura, M., Butto, L. F., Duranti, S., O’toole, P. W., Motherway, M. O.,et al. (2013b). Molecular dialogue between the human gut microbiota and the host: a Lactobacillus and Bifidobacterium perspective. Cell. Mol. Life Sci. 71, 183–203. doi: 10.1007/s00018-013-1318-0
Turroni, F., Taverniti, V., Ruas-Madiedo, P., Duranti, S., Guglielmetti, S., Lugli, G. A.,et al. (2014a). Bifidobacterium bifidum PRL2010 modulates the host innate immune response. Appl. Environ. Microbiol. 80, 730–740. doi: 10.1128/AEM.03313-13
Turroni, F., Ventura, M., Butto, L. F., Duranti, S., O′Toole, P. W., Motherway, M. O.,et al. (2014b). Molecular dialogue between the human gut microbiota and the host: a Lactobacillus and Bifidobacterium perspective. Cell. Mol. Life. Sci. 71, 183–203. doi: 10.1007/s00018-013-1318-0
Ventura, M., Canchaya, C., Fitzgerald, G. F., Gupta, R. S., and Van Sinderen, D. (2007a). Genomics as a means to understand bacterial phylogeny and ecological adaptation: the case of bifidobacteria. Antonie Van Leeuwenhoek 91, 351–372. doi: 10.1007/s10482-006-9122-6
Ventura, M., Canchaya, C., Tauch, A., Chandra, G., Fitzgerald, G. F., Chater, K. F.,et al. (2007b). Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 71, 495–548. doi: 10.1128/MMBR.00005-07
Ventura, M., O’flaherty, S., Claesson, M. J., Turroni, F., Klaenhammer, T. R., Van Sinderen, D.,et al. (2009a). Genome-scale analyses of health-promoting bacteria: probiogenomics. Nat. Rev. Microbiol. 7, 61–71. doi: 10.1038/nrmicro2047
Ventura, M., Turroni, F., Canchaya, C., Vaughan, E. E., O’toole, P. W., and Van Sinderen, D. (2009b). Microbial diversity in the human intestine and novel insights from metagenomics. Front. Biosci. (Landmark Ed.) 14:3214–3221. doi: 10.2741/3445
Ventura, M., Turroni, F., Foroni, E., Duranti, S., Giubellini, V., Bottacini, F.,et al. (2010). Analyses of bifidobacterial prophage-like sequences. Antonie Van Leeuwenhoek 98, 39–50. doi: 10.1007/s10482-010-9426-4
Ventura, M., Turroni, F., Lugli, G. A., and Van Sinderen, D. (2014). Bifidobacteria and humans: our special friends, from ecological to genomics perspectives. J. Sci. Food Agric. 94, 163–168. doi: 10.1002/jsfa.6356
Ventura, M., Turroni, F., Motherway, M. O., Macsharry, J., and Van Sinderen, D. (2012). Host-microbe interactions that facilitate gut colonization by commensal bifidobacteria. Trends Microbiol. 20, 467–476. doi: 10.1016/j.tim.2012.07.002
Wajant, H., Pfizenmaier, K., and Scheurich, P. (2003). Tumor necrosis factor signaling. Cell Death Differ. 10, 45–65. doi: 10.1038/sj.cdd.4401189
Weiss, G., Rasmussen, S., Nielsen Fink, L., Jarmer, H., Nohr Nielsen, B., and Frokiaer, H. (2010). Bifidobacterium bifidum actively changes the gene expression profile induced by Lactobacillus acidophilus in murine dendritic cells. PLoS ONE 5:e11065. doi: 10.1371/journal.pone.0011065
Yasutake, N., Matsuzaki, T., Kimura, K., Hashimoto, S., Yokokura, T., and Yoshikai, Y. (1999). The role of tumor necrosis factor (TNF)-alpha in the antitumor effect of intrapleural injection of Lactobacillus casei strain Shirota in mice. Med. Microbiol. Immunol. 188, 9–14. doi: 10.1007/s004300050099
Keywords: Bifidobacterium bifidum, bifidobacteria, probiotics, genomics, microbiota
Citation: Turroni F, Duranti S, Bottacini F, Guglielmetti S, Van Sinderen D and Ventura M (2014) Bifidobacterium bifidum as an example of a specialized human gut commensal. Front. Microbiol. 5:437. doi: 10.3389/fmicb.2014.00437
Received: 12 June 2014; Accepted: 01 August 2014;
Published online: 21 August 2014.
Edited by:
Maria De Angelis, University of Bari Aldo Moro, ItalyReviewed by:
Stella Maris Reginensi Rivera, Universidad de la República Oriental del Uruguay, UruguayDiego Mora, Università degli Studi di Milano, Italy
Copyright © 2014 Turroni, Duranti, Bottacini, Guglielmetti, Van Sinderen and Ventura. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marco Ventura, Laboratory of Probiogenomics, Department of Life Sciences, University of Parma, Parco Area delle Scienze 11a, 43124 Parma, Italy e-mail:bWFyY28udmVudHVyYUB1bmlwci5pdA==
 Francesca Turroni1
Francesca Turroni1 Simone Guglielmetti
Simone Guglielmetti Douwe Van Sinderen
Douwe Van Sinderen Marco Ventura
Marco Ventura