- 1The Infectious Diseases Research Laboratory, Sheba Medical Center, Tel-Hashomer, Israel
- 2Bernhard Nocht Institute for Tropical Medicine, Hamburg, Germany
- 3Institute for Experimental Medicine, Christian Albrechts University Kiel, Kiel, Germany
- 4Research Center Borstel, Borstel, Germany
Human infections by the bacterial pathogen Salmonella enterica represent major disease burdens worldwide. This highly ubiquitous species consists of more than 2600 different serovars that can be divided into typhoidal and non-typhoidal Salmonella (NTS) serovars. Despite their genetic similarity, these two groups elicit very different diseases and distinct immune responses in humans. Comparative analyses of the genomes of multiple Salmonella serovars have begun to explain the basis of the variation in disease manifestations. Recent advances in modeling both enteric fever and intestinal gastroenteritis in mice will facilitate investigation into both the bacterial- and host-mediated mechanisms involved in salmonelloses. Understanding the genetic and molecular mechanisms responsible for differences in disease outcome will augment our understanding of Salmonella pathogenesis, host immunity, and the molecular basis of host specificity. This review outlines the differences in epidemiology, clinical manifestations, and the human immune response to typhoidal and NTS infections and summarizes the current thinking on why these differences might exist.
Introduction
Salmonella enterica is a highly diverse Gram negative bacterial species containing more than 2600 different serovars differentiated by their antigenic presentation. Various serovars are characterized by their host specificity or by the clinical syndrome they cause ranging from asymptomatic carriage to invasive systemic disease. Most S. enterica serovars associated with diseases in humans and other warm blooded animals belong to subspecies I consisting of both typhoidal and non-typhoidal serovars. Several excellent recent reviews have highlighted different aspects of invasive salmonellosis (De Jong et al., 2012; Feasey et al., 2012), discussed the mechanisms behind host restriction (Baumler and Fang, 2013), and detailed salmonelloses in immunocompromised individuals (Gordon, 2008; Maclennan, 2014). Here, we will discuss how typhoidal and non-typhoidal serovars differ in epidemiology, clinical manifestations, and the immune response they trigger in humans.
Epidemiology
While many non-typhoidal Salmonella (NTS) serovars such as Typhimurium and Enteritidis are generalist pathogens with broad host specificity, a few S. enterica serovars including Typhi, Sendai, and Paratyphi A, B, or C are highly adapted to the human host that is used as their exclusive reservoir. These specialist pathogens, collectively referred to as typhoidal Salmonella serovars, are the causative agents of enteric fever (also known as typhoid or paratyphoid fever if caused by serovar Typhi or Paratyphi, respectively). Enteric fever is an invasive, life-threatening, systemic disease with an estimated global annual burden of over 27 million cases, resulting in more than 200,000 deaths (Crump et al., 2004; Buckle et al., 2012). Enteric fever is endemic in the developing world in regions that lack clean water and adequate sanitation, facilitating the spread of these pathogens via the fecal-oral route. In recent years, for unknown reasons, the incidence of infections with serovar Paratyphi A is on the rise and in some regions of the globe, particularly in South–East Asia, this serovar is accountable for up to 50% of all enteric fever cases (Ochiai et al., 2005; Meltzer and Schwartz, 2010).
In contrast to typhoid fever which is common in the developing world, NTS salmonelloses occur worldwide. There are an estimated 93.8 million cases of gastroenteritis due to NTS infection each year, resulting in approximately 155,000 deaths (Majowicz et al., 2010). Despite global morbidity, mortality due to NTS infection is primarily restricted to the developing world. In addition to contaminated animal-derived food products such as poultry, eggs, and dairy products, NTS transmission can result from person to person contact or from contact with pets such as cats, dogs, rodents, reptiles, or amphibians (Hohmann, 2001; Mermin et al., 2004; Braden, 2006; Haeusler and Curtis, 2013). Another important source of infection is consumption of contaminated produce especially sprouts, tomatoes, fruits, peanuts, and spinach which have all been associated with recent outbreaks (Berger et al., 2009, 2010; Barton Behravesh et al., 2011; Cavallaro et al., 2011; Jackson et al., 2013; Bayer et al., 2014).
While normally NTS infections in humans induces gastroenteritis, in up to 5% of NTS cases, bacteria cause an invasive, extra-intestinal disease leading to bacteremia and focal systemic infections, henceforth referred to as invasive NTS (iNTS; Mandal and Brennand, 1988). Interestingly, various NTS serovars (e.g., Typhimurium, Dublin, Choleraesuis, 9,12:l,v:-) tend to have more potential to cause extraintestinal infections than others. This implies there is a genetic basis for the emergence iNTS disease; however, these differences are still not understood (Wilkins and Roberts, 1988; Marzel et al., 2014). In Sub-Saharan Africa, iNTS is a major cause of bacteremia in adults and children, with an estimated annual incidence of 175–388 cases per 100,000 children and 2000–7500 cases per 100,000 HIV-infected adults. Especially S. Typhimurium sequence type (ST) 313 is associated with invasive disease. Startlingly, in 20–25% of cases, invasive infection results in the death of the patients. Other major risk factors for invasive disease in addition to HIV are co-infection with malaria and malnutrition (Feasey et al., 2012; Maclennan, 2014).
Clinical Manifestations
Enteric fever caused by typhoidal serovars differs dramatically from the gastroenteritis normally associated with NTS. Infections caused by different typhoidal serovars (e.g., Typhi and Paratyphi A) cannot be distinguished by clinical presentation (Meltzer et al., 2005; Patel et al., 2010). The average incubation period for typhoidal serovars is 14 days with symptoms persisting for up to 3 weeks (Olsen et al., 2003; Wangdi et al., 2012). Patients most typically present with a gradual onset of sustained fever (39–40°C). Other frequent symptoms include chills, abdominal pain, hepatosplenomegaly, rash (rose spots), nausea, anorexia, diarrhea or constipation, headache, and a dry cough (Stuart and Pullen, 1946). In contrast to enteric fever, individuals infected with NTS have self-limiting, acute gastroenteritis and watery diarrhea. Nausea, vomiting, abdominal pain, and fever are also common symptoms (McGovern and Slavutin, 1979). With NTS infection, symptoms appear 6–12 h after the ingestion of the pathogen and clinical symptoms last less than 10 days (Glynn and Palmer, 1992). In the case of iNTS infections, which are often associated with patients with immunodeficiency, disease more closely resembles enteric fever in that patients often suffer from high fever, hepatosplenomegaly, and have respiratory complications with intestinal symptoms often being absent.
Both typhoidal and NTS serovars initially adhere to and invade the intestinal epithelium of the small intestine (Liu et al., 1988). Unlike NTS infection, infection by typhoidal serovars does not induce a high inflammatory response during the initial invasion of the intestinal mucosa (Sprinz et al., 1966; Kraus et al., 1999; Nguyen et al., 2004). Minimal intestinal inflammation during enteric fever is correlated with negligible neutrophil transmigration across the intestinal epithelium in contrast to massive neutrophil recruitment during intestinal inflammation caused by NTS serovars (McCormick et al., 1995). In immunocompetent patients, NTS gastroenteritis is self-limiting, with infection being confined to the terminal ileum and colon. In the case of typhoidal salmonellae, after passing the intestinal mucosa, bacteria gain access to underlying lymphoid tissues and multiply intracellularly within mononuclear phagocytes. Infection quickly becomes systemic with spreading of the pathogen from the intestine to the mesenteric lymph nodes, liver, spleen, bone marrow, and gallbladder. Secondary infection of typhoidal organisms to the small bowel can occur via secretion in the bile through the enterohepatic cycle (Gordon, 2008). The absence of robust intestinal inflammation and the lack of neutrophil transmigration are thought to facilitate the invasion of typhoidal serovars into the deeper tissues of the gut and its dissemination to systemic sites (House et al., 2001).
Interestingly, up to 10% of convalescing, untreated patients continue to shed S. Typhi in their stool for up to three months after infection (Parry et al., 2002). One to four percent of individuals infected with S. Typhi become asymptomatic, chronic carriers that continue to excrete 106–1010 S. Typhi bacteria per gram of feces for more than 12 months. The role of such chronic carriers in disease transmission was notoriously demonstrated by the case of Mary Mallon (Typhoid Mary). During her work at different households as a cook in the New York City area in the early 20th century, Mary Mallon infected between 26 and 54 people (Marr, 1999). Another example of an asymptomatic S. Typhi carrier was “Mr. N” who worked as a cowman and milker in South–East England and was responsible for a 207 case outbreak of typhoid fever, which peaked in 1899 but continued until 1909 (Mortimer, 1999). The suspected site of persistence of S. Typhi in carriers is the gallbladder and gallstones are thought to be an important risk factor for developing chronic carriage (Levine et al., 1982) as they are conducive for biofilm formation which protects bacteria from antimicrobial compounds and the host immune system. Long-term carriage of S. Paratyphi has received much less attention and is currently less characterized than S. Typhi, but a recent study in Nepal suggests a similar rate of persistence for serovars Typhi and Paratyphi A in endemic regions (Khatri et al., 2009; Dongol et al., 2012).
Long-term carriage of NTS has not been described. However, even though symptoms usually last only for a few days, adults excrete Salmonella on average for 1 month after infection and children under the age of 5 years shed bacteria in their feces for an average of 7 weeks (Buchwald and Blaser, 1984; Hohmann, 2001). Interestingly, several studies have shown that treatment with antibiotics can prolong shedding of NTS bacteria (Aserkoff and Bennett, 1969; Murase et al., 2000), although these findings are controversial (Dryden et al., 1996; Hohmann, 2001). In comparison to NTS serovars, the long-term persistence of typhoidal serovars in humans suggests an enhanced ability of these pathogens to evade the human immune system (Raffatellu et al., 2008b).
Human Immune Response
Infection in humans by NTS serovars induces a strong Th1 response with high levels of IFN-γ, IL-18, IL-12, IL-15, TNF-α, and IL-10 detected in serum from patients (Mizuno et al., 2003; Stoycheva and Murdjeva, 2005). Expression of several chemokines is also induced upon NTS infection, which leads to the recruitment and activation of macrophages and dendritic cells, and a significant influx of neutrophils into the intestinal lumen, which is a hallmark of NTS gastroenteritis. The fact that typhoidal serovars are not typically associated with acute diarrhea or a strong influx of neutrophils into the intestinal lumen (Sprinz et al., 1966; Kraus et al., 1999; Nguyen et al., 2004) suggests that their initial interaction with the human gut mucosa is less inflammatory than that of NTS serovars.
Recent studies have shown that patients with inherited deficiency of the IL-12/IL-23 system (IL-12p40/IL-12Rβ1) are highly susceptible to NTS infections, but not to S. Typhi or S. Paratyphi infections, even though some of these patients live in endemic areas (MacLennan et al., 2004; Van de Vosse and Ottenhoff, 2006). These observations support the possibility that different inflammatory pathways may be involved in NTS vs. typhoidal infections including a distinct role for the IL-12 pathway. This idea is further supported by additional epidemiological observations indicating that invasive infections caused by NTS, but not by typhoidal serovars, are often associated with immunocompromised adults, in particular individuals infected with HIV (Gordon, 2008; MacLennan and Levine, 2013). This implies that certain immune responses, malfunctioning during HIV infection, are required for the immune defense against systemic infection of NTS, but not against typhoidal serovars.
The immune response to infection with typhoidal serovars is complex and involves both humoral and cell-mediated immune responses (Sztein, 2007). Clinical studies that examined the immune response of patients infected with S. Typhi showed a significant CD4 and CD8 T cell response to specific S. Typhi antigens during typhoid fever, with elevated levels of IFN-γ during the acute phase of the disease (Butler et al., 1993; Sheikh et al., 2011). Transcriptome analysis of peripheral blood mononuclear cells (PBMCs) from patients with acute typhoid fever also demonstrated up-regulation of the genes from the IFN-γ pathway compared to healthy individuals (Thompson et al., 2009). Induction of other cytokines in response to S. Typhi infection include IL-6 and IL-8 which are secreted into the serum during the acute phase of typhoid fever (Butler et al., 1993; Keuter et al., 1994; Gasem et al., 2003). PBMCs from immunized volunteers orally vaccinated with an attenuated S. Typhi vaccine secrete Th1 cytokines including IFN-γ, TNF-α, and IL-10, following sensitization with a number of S. Typhi antigens including flagella (Wahid et al., 2007). Collectively, these findings indicate that the human immune response to S. Typhi infection is predominantly Th1-associated.
Given that typhoidal serovars do not typically illicit septic shock, in contrast to many other Gram-negative pathogens that induce bacteremia and leukopenia (Pohan, 2004; Tsolis et al., 2008; Gal-Mor et al., 2012), suggests a restrained immune response mediated by these pathogens in the human host. This view is consistent with the clinical observation that serum levels of pyrogenic cytokines IL-1β and TNF-α are relatively low in patients with typhoid fever compared to the levels found in patients with sepsis caused by other Gram-negative pathogens. In fact, IL-1β and TNF-α production by PBMCs has been shown to be suppressed during the acute phase of typhoid fever (Butler et al., 1978; Girardin et al., 1988; Keuter et al., 1994; Gasem et al., 2003).
Despite the increasing prevalence of S. Paratyphi A in endemic regions, the immune response to S. Paratyphi infection is much less characterized than the one to S. Typhi. A recent study done in our group examined the circulating cytokine profile of healthy Israeli travelers that became infected with S. Paratyphi A during an outbreak in Nepal. Comparison of 16 cytokines demonstrated considerable (more than 10-fold) increase in the serum concentration of IFN-γ, but only a moderate elevation in the concentration of IL-6, IL-8, IL-10, and TNF-α between convalescence and the peak time of clinical presentation (Gal-Mor et al., 2012). These results suggest that the prominent IFN-γ and the moderate TNF-α, IL-6, and IL-8 responses are common to both typhoid and paratyphoid fever. Interestingly, no changes in IL-12 serum concentrations were detected during the acute phase of the disease (Gal-Mor et al., 2012), in contrast to its induction seen during gastroenteritis caused by NTS serovars (Stoycheva and Murdjeva, 2005).
Current Therapies and Vaccines
Antibiotic therapy can prolong the duration of excretion of NTS and therefore is only recommended for people with severe illness, invasive disease, or for certain risk groups including infants, the elderly, and immunocompromised individuals. Enteric fever, on the other hand is always immediately treated with antibiotics. In the 1990s, physicians moved away the first-line antibiotics chloramphenicol, ampicillin, and cotrimoxazole due to widespread resistance amongst S. enterica serovars. Since then, fluoroquinolones (like ciprofloxacin) have been the primary treatment for salmonelloses, as this class of drug is particularly effective against intracellular Gram-negative bacteria. While there is increasing resistance to fluoroquinolones, new fluoroquinolones like gatifloxacin hold promise. Third generation cephalosporins are often the second-line treatment to treat salmonelloses. In addition, azithromycin is relatively new drug with activity against both nalidixic acid resistant and multidrug resistant (MDR) strains (Hohmann, 2001; Arjyal and Pandit, 2008).
Multidrug-resistance is an increasing problem in S. enterica serovars. Resistance to multiple antibiotics is especially common in serovars Typhimurium and Newport and multidrug-resistant strains are also linked to more severe disease outcome (Krueger et al., 2014). Notably, many strains of S. Typhimurium Definitive Type (DT) 104, which have caused multiple outbreaks since the 1990s, are resistant to ampicillin, chloramphenicol, streptomycin, sulphonamides, and tetracycline (Mather et al., 2013). Moreover, new resistant strains of S. enterica are continuously emerging worldwide. For example, an MDR strain of serovar Infantis now accounts for up to 35% of the NTS infections in Israel (Gal-Mor et al., 2010; Aviv et al., 2014). Additional examples are the emergence of resistant strains of serovars Virchow (Weill et al., 2004) and Heidelberg (Dutil et al., 2010). Similarly, many iNTS strains are resistant against ampicillin, chloramphenicol, kanamycin, streptomycin, trimethoprim, and cotrimoxazole (Gordon, 2008; Kingsley et al., 2009; Msefula et al., 2012). Therefore, there is a high need to (i) prevent further resistance development through the prudent use of antibiotics, (ii) improve measures that prevent spread of MDR strains, and (iii) discover new therapies for salmonelloses. Interestingly, the re-emergence of chloramphenicol sensitive strains in areas where resistance was previously prevalent suggests that cycling or rotation of antibiotics could also be an effective strategy to deal with antibiotic resistance, rendering older antibiotics useful once again (Abel Zur Wiesch et al., 2014).
Three types of vaccines against S. Typhi are currently commercially available, but unfortunately, there is still not a single licensed vaccine available against S. Paratyphi A, with very little, if any, cross-protection provided by the available S. Typhi vaccines. Vaccination strategies against typhoid fever including a description of ongoing trials were recently reviewed in detail (Waddington et al., 2014). The currently licensed S. Typhi vaccines include (i) a killed whole cell parenteral vaccine (Engels et al., 1998), (ii) a live attenuated oral vaccine, designated Ty21a (Germanier and Fuer, 1975) and, (iii) a Vi polysaccharide capsule-based vaccine (Tacket et al., 1986). There are vaccines against NTS serovars Enteritidis and Typhimurium which are effective in poultry (Desin et al., 2013). However, there are no vaccines available for NTS in humans or other animal reservoirs such as cattle or pigs. This represents a significant limitation in the existing prevention strategies. Understanding the host specificity determinants of S. enterica serovars will aid in future therapeutic and vaccine development.
Why do Typhoidal and NTS Serovars Elicit Such Different Host Immune Responses?
How do pathogens so similar, belonging to the same subspecies (S. enterica ssp. I), with >96% DNA sequence identity between shared genes (McClelland et al., 2001) induce such different clinical manifestations and immune responses in humans? Despite significant advances in the field, this question is still far from being answered. Understanding the genetic and molecular mechanisms responsible for differences in disease outcome will aid in our understanding of Salmonella pathogenesis, host immunity, and the molecular basis of host specificity (Table 1).
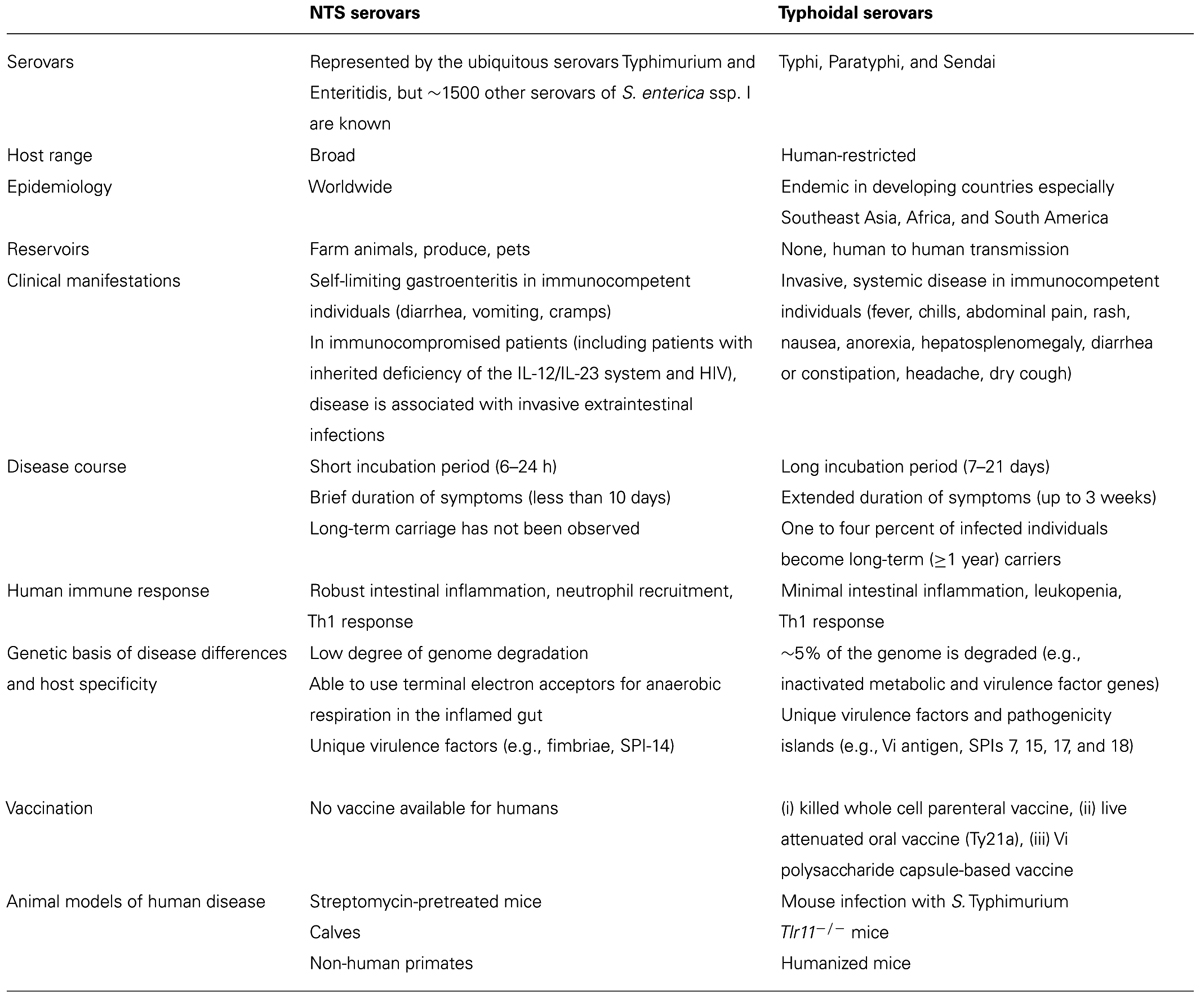
TABLE 1. Summary of the differences between NTS and typhoidal serovars associated with disease in humans.
In vitro tissue culture studies suggest that S. Typhi induces restrained inflammatory responses that do not trigger a pro-inflammatory response via TLR5. Similarly, polarized human colonic epithelial (T84) cells infected with S. Typhi induce significantly lower levels of the neutrophil chemoattractant IL-8 compared to S. Typhimurium infection (Raffatellu et al., 2005). Raffatellu et al. (2008b) have therefore postulated that S. Typhi expresses unique virulence factors that allow this pathogen to overcome the innate immune response in the intestinal mucosa resulting in the absence of neutrophil infiltration and inflammatory diarrhea. One of the current hypotheses in the field suggests that the polysaccharide capsular antigen Vi in S. Typhi enables this pathogen to resist phagocytosis and complement killing (Robbins and Robbins, 1984) and masks access to pattern recognition molecules, resulting in less IL-8 production (Raffatellu et al., 2005), limited neutrophil influx, and thereby reduced small bowel inflammation (Sharma and Qadri, 2004; Wilson et al., 2008). The role of the Vi antigen regulator TviA, and its putative contribution to S. Typhi’s ability to evade the immune system have been recently reviewed (Wangdi et al., 2012). Nevertheless, since the Vi capsule is largely restricted to serovar Typhi and is absent from serovars Paratyphi A and Sendai, it cannot explain why the clinical manifestations of these other typhoidal serovars differ from that of NTS. Furthermore, the fact that Vi-negative mutants of S. Typhi are still able to cause a typhoid-like illness in human volunteers (Zhang et al., 2008), suggests that additional mechanisms are involved (Figure 1).
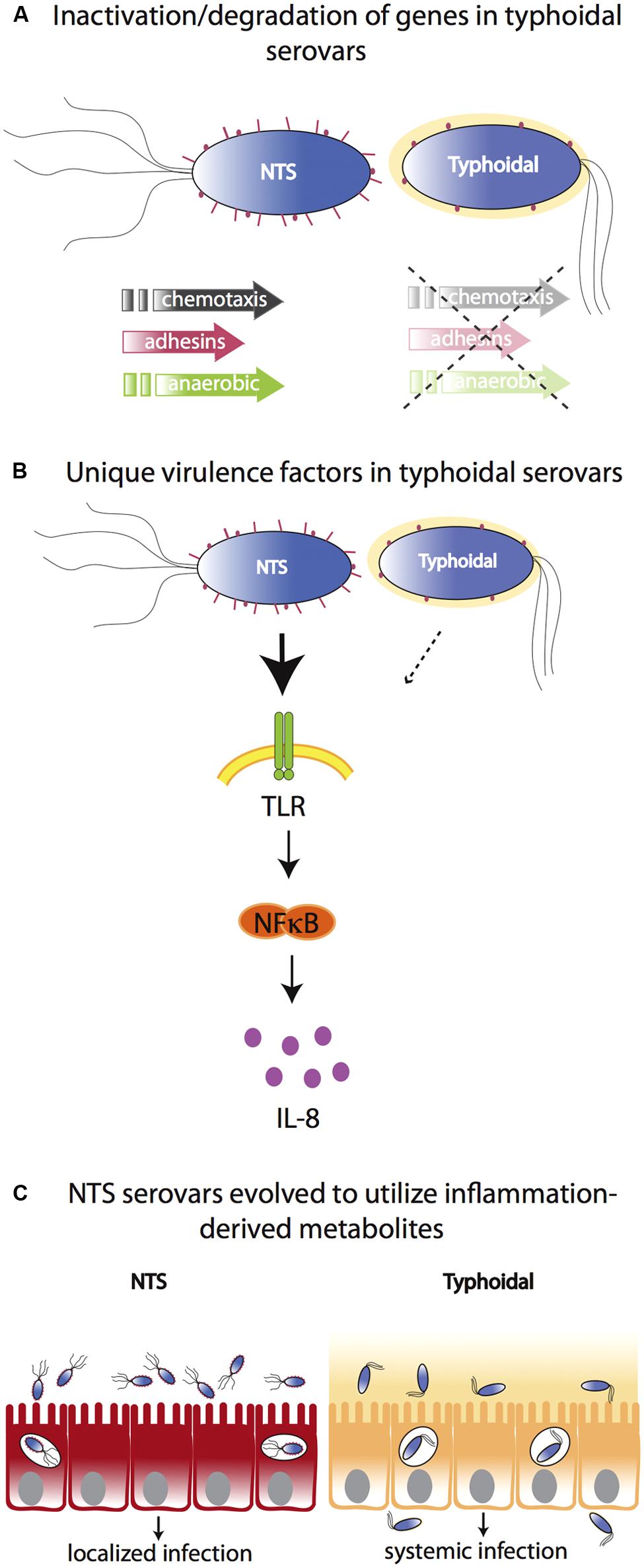
FIGURE 1. Molecular bases for differences between typhoidal and NTS serovars. (A) Typhoidal serovars possess several inactive/degraded genes compared to NTS serovars such as genes for chemotaxis, adhesion, and anaerobic metabolism. (B) Both typhoidal and NTS serovars possess unique virulence factors. For example, some S. Typhi strains express Vi capsule that reduces TLR-dependent IL-8 production in the intestinal mucosa. However, while the Vi capsule plays a role in typhoid fever manifestation, it is not necessary as it is absent from other typhoidal serovars and Vi-negative mutants of S. Typhi are still able to cause a typhoid-like illness in humans. (C) In contrast to typhoidal serovars, NTS cause severe intestinal inflammation. NTS serovars have evolved to utilize inflammation-derived metabolites (e.g., nitrate and tetrathionate), thereby enhancing their growth in the inflamed intestine. Typhoidal serovars have lost the ability to benefit from inflammation-derived metabolites and disseminate to systemic sites to a much greater extent.
Of the approximately 4400 S. Typhi and S. Paratyphi A genes, about 200 are inactivated or functionally disrupted, while most of their homologs in S. Typhimurium are intact. Many of the degraded genes found in the genomes of the typhoid serovars are involved in motility and chemotaxis or encode for type 3 secretion system effectors, fimbriae, or adhesins that play a role in Salmonella pathogenicity (McClelland et al., 2004). Furthermore, Salmonella pathogenicity island (SPI)-7 (encoding the Vi antigen), SPI-15, SPI-17, and SPI-18 are present in the genome of S. Typhi, but not in the genome of S. Typhimurium, while SPI-14, present in S. Typhimurium, is absent from the genome of typhoidal serovars (Sabbagh et al., 2010). Therefore, it is highly possible that differences in virulence and colonization factor composition affect host–pathogen interactions and disease outcome in humans. This notion has recently been demonstrated by the expression of the S. Typhimurium effector, GtgE, in S. Typhi. When secreted into host cells, GtgE proteolytically degrades Rab29 and confers the ability of S. Typhi to survive and replicate within macrophages and in tissues from mice, a normally non-permissive host (Spano and Galan, 2012).
Recent evidence suggests that NTS serovars have evolved to flourish in the inflamed gut environment and use inflammation to outcompete microbiota (Stecher et al., 2007; Thiennimitr et al., 2011). It has been proposed that typhoidal strains may have lost this ability and therefore have evolved to not induce inflammation in the gut but rather thrive systemically. For example, a by-product of the acute intestinal inflammation triggered by S. Typhimurium and other NTS serovars is the generation of the terminal electron acceptors nitrate and tetrathionate in the lumen of the inflamed gut. These compounds can be used by S. Typhimurium and other NTS serovars to outcompete the fermenting gut microbes that are unable to utilize these electron acceptors (Winter et al., 2010). In another recent report, Nuccio and Baumler (2014) have identified a network of 469 genes involved in central anaerobic metabolic pathways that are intact in NTS, but are decayed in the genome of typhoid serovars. Some of these degraded genes include the ethanolamine utilization pathway (eut genes) as well as the vitamin B12 biosynthesis pathway (cbi and cob genes) required for ethanolamine utilization (Nuccio and Baumler, 2014). These pathways are hypothesized to enable NTS to utilize inflammation-derived nutrients to outcompete other gut microbes.
Collectively, a substantial degree of metabolic and virulence gene degradation exists in the genomes of typhoidal serovars which may explain the restricted host-tropism of these pathogens and may also provide at least a partial explanation as to why typhoidal and NTS-infections induce such different clinical presentations and immune responses in humans.
Animal Models
Animal Models of Non-Typhoidal Salmonelloses
There are several animal models used to model human gastroenteritis caused by NTS. The model which most resembles human disease is arguably infection of non-human primates (Kent et al., 1966; Rout et al., 1974). Rhesus macacques are especially useful for investigating co-infection with simian immunodeficiency virus. For example, infection of SIV-infected macacques with S. Typhimurium results in a blunted immune response and invasive bacterial disease similar to what is seen in HIV-infected patients (Raffatellu et al., 2008a). Furthermore, this model is useful for testing the efficacy and safety of potential live Salmonella vaccines for HIV infected people (Ault et al., 2013). However, the use of primates is limited by ethical concerns, cost, and the inability for genetic manipulation. Infection of calves with S. Typhimurium results in similar pathology to humans. Furthermore, S. Typhimurium is a natural pathogen of cattle and beef is a common reservoir for human infection (Santos et al., 2001; Costa et al., 2012). Data from the calf model have provided valuable insights into host–Salmonella interaction. However, this model is also restricted by cost and the limited possibility for genetic manipulation of the host.
Due to the low cost, ease of housing/handling, and genetic manipulation possible, mouse models are the most widely used animal models to study bacterial disease. NTS infection of mice does not mimic gastroenteritis as seen in humans but results in a typhoid-like systemic disease. However, after pretreatment of mice with antibiotics such as streptomycin or kanamycin, S. Typhimurium can overcome the “colonization resistance” presented by the natural microbiota and thus efficiently colonize the cecum and colon. In the now widely used streptomycin pretreatment model, NTS infection has been shown to lead to overt inflammation characterized by transmural inflammation including epithelial destruction, infiltration of inflammatory cells into the mucosa, formation of crypt abscesses, submucosal edema, and hyperplasia (Barthel et al., 2003; Hapfelmeier and Hardt, 2005; Sekirov et al., 2008; Woo et al., 2008). This model is now being exploited by many research groups to dissect both the bacterial- and host-mediated mechanisms involved in intestinal inflammation induction by NTS.
Animal Models of Enteric Fever
S. Typhi, S. Paratyphi, and S. Sendai are human-restricted pathogens. Historically, attempts at eliciting enteric fever in animal models by infection with S. Typhi have proven to be rather inadequate. Chimpanzees infected with S. Typhi develop a mild disease that resembles enteric fever, but only when infected with a very high dose of 1 × 1011 CFU (Edsall et al., 1960). Another model for S. Typhi consists of inoculating mice intraperitoneally with S. Typhi suspended in hog gastric mucin (Pasetti et al., 2003). However, this model has not been found to correlate well with human enteric fever and with the expected attenuation of key Salmonella virulence regulators, such as PhoP (Baker et al., 1997).
Therefore until recently, due to the lack of suitable animal models, much of our understanding of enteric fever had been extrapolated from S. Typhimurium infection in mice. Mice infected with S. Typhimurium display minimal intestinal pathology but become systemically colonized as seen in humans with enteric fever. This model also allows for investigation of gallbladder colonization which is most likely the niche for chronic S. Typhi carriage in humans (Menendez et al., 2009; Gonzalez-Escobedo et al., 2013). Susceptible (Slc11a1-/-, also known as Nramp1) mouse strains have been widely used but also resistant (Slc11a1+/+) mice have proven useful. Mice with a wild-type Slc11a1 gene (e.g., 129Sv, DBA) are relatively resistant to high doses of S. Typhimurium and have been particularly useful to investigate chronic infection, carriage (Lawley et al., 2006; Monack et al., 2004), and transmission (Lawley et al., 2008; Gopinath et al., 2012; Monack, 2012). In general, infection of mice with NTS has provided invaluable insight into the role of specific virulence factors in host invasion, dissemination, and transmission and although the murine inflammatory response to NTS in some ways resembles the human response to typhoidal serovars (Santos et al., 2001), conclusions from this model regarding the relevance to human typhoid disease must be carefully inferred.
In recent years, more sophisticated mouse models have been developed to study S. Typhi infection. Mathur et al. (2012) have shown that Salmonella flagellin is recognized in the mouse intestine by Toll-like receptor 11 (TLR11), which is absent from humans. Tlr11 knockout mice are severely attenuated in innate epithelial responses to S. Typhi (and S. Typhimurium) and exhibit significant systemic infection following oral administration (Mathur et al., 2012; Shi et al., 2012). It will be exciting to see if this model can also be used for infection with S. Paratyphi.
Another promising novel model is the use of humanized mice whereby immunodeficient mice (either Rag2-/- Il2rg-/- or NOD⋅Cg-Prkdcscid Il2rg-/-) lacking murine T, B, and NK cells are engrafted with human CD34+ hematopoietic stem cells (Shultz et al., 2007). These chimeric mice contain human immune cells including B cells, CD4+ and CD8+ T cells, NK cells, monocytes, and myeloid and plasmacytoid dendritic cells. Such humanized mice facilitate S. Typhi replication in the liver, spleen, and gallbladder and allow long-term persistence to be modeled (Song et al., 2010; Firoz Mian et al., 2011). In addition, infection results in a progressive, lethal infection within two to three days with inflammatory cytokine responses resembling human typhoid (Libby et al., 2010). These models suggest that the presence of human immune cells is prerequisite for systemic infection and in vivo replication of S. Typhi in the mouse. Although these humanized mice have proven informative to the study of S. Typhi infection, they are expensive and labor-intensive models and (so far) not widely used. Another limitation of such models is that they are subject to considerable inconsistency as a result of the genetic heterogeneity of donors and the variable degree of engraftment (Libby et al., 2010; Mian et al., 2011).
Perspectives
In-depth comparative analyses of the genomes of Salmonella serovars have begun to explain the basis for the variation seen in disease manifestations; however, this is still far from being fully understood. An interesting question in this regard is whether there is a genetic basis for the emergence of iNTS strains and why some NTS serovars (e.g., Typhimurium, Dublin, Choleraesuis, Schwarzengrund) tend to cause more invasive disease than others. In addition, the mechanisms by which co-infections (e.g., with Plasmodium falciparum, HIV) contribute to the increased risk of iNTS bacteremia must be further investigated. From the perspective of the host response, one unanswered question is whether there are unique immune responses to different typhoidal strains (e.g., Typhi vs. Paratyphi). And lastly, a fast-developing area of research that has already had implications on our understanding of salmonelloses is that of the role of the microbiota in disease outcome (see review by Santos in this issue). In the case of gastrointestinal pathogens, the influence of the host microbiota on pathogenesis, host immunity, and disease progression can no longer be overlooked.
Exploitation of the recent advances in modeling typhoid and NTS infection in mice is likely to provide novel insights into how these serovars are able to cause such different diseases. Opportunities remain, however, in the development of “next generation” humanized mouse models with enhanced human cell engraftment and function. These models hold much promise as they allow one to study the pathogenesis of human-restricted serovars, as well as to test the efficacy of therapeutic agents and experimental vaccines. Understanding the genetic and molecular mechanisms responsible for differences in disease outcome will aid in our understanding of Salmonella pathogenesis, host immunity, and the molecular basis of host specificity. Together, this information may be applied to control Salmonella infection, with specific determinants being targeted for therapeutic and vaccine development.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Work in the lab of OGM is funded by grant number 249241 from the European Community’s Seventh Framework program (PF7/2007–2013) and by a joint grant (1096-39.11/2010) from the German Israel Foundation for Scientific Research and Development to Ohad Gal-Mor and Guntram A. Grassl. Work in the lab of Guntram A. Grassl is also funded by the DFG Excellence Cluster “Inflammation at Interfaces” EXC306 and a DFG Priority Program grant (GR 2666/5-1).
References
Abel Zur Wiesch, P., Kouyos, R., Abel, S., Viechtbauer, W., and Bonhoeffer, S. (2014). Cycling empirical antibiotic therapy in hospitals: meta-analysis and models. PLoS Pathog. 10:e1004225. doi: 10.1371/journal.ppat.1004225
Aserkoff, B., and Bennett, J. V. (1969). Effect of antibiotic therapy in acute salmonellosis on the fecal excretion of Salmonellae. N. Engl. J. Med. 281, 636–640. doi: 10.1056/NEJM196909182811202
Ault, A., Tennant, S. M., Gorres, J. P., Eckhaus, M., Sandler, N. G., Roque, A.,et al. (2013). Safety and tolerability of a live oral Salmonella Typhimurium vaccine candidate in SIV-infected nonhuman primates. Vaccine 31, 5879–5888. doi: 10.1016/j.vaccine.2013.09.041
Aviv, G., Tsyba, K., Steck, N., Salmon-Divon, M., Cornelius, A., Rahav, G.,et al. (2014). A unique megaplasmid contributes to stress tolerance and pathogenicity of an emergent Salmonella enterica serovar Infantis strain. Environ. Microbiol. 16, 977–994. doi: 10.1111/1462-2920.12351
Baker, S. J., Daniels, C., and Morona, R. (1997). PhoP/Q regulated genes in Salmonella Typhi identification of melittin sensitive mutants. Microb. Pathog. 22, 165–179. doi: 10.1006/mpat.1996.0099
Barthel, M., Hapfelmeier, S., Quintanilla-Martinez, L., Kremer, M., Rohde, M., Hogardt, M.,et al. (2003). Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect. Immun. 71, 2839–2858. doi: 10.1128/IAI.71.5.2839-2858.2003
Barton Behravesh, C., Mody, R. K., Jungk, J., Gaul, L., Redd, J. T., Chen, S.,et al. (2011). 2008 outbreak of Salmonella Saintpaul infections associated with raw produce. N. Engl. J. Med. 364, 918–927. doi: 10.1056/NEJMoa1005741
Baumler, A., and Fang, F. C. (2013). Host specificity of bacterial pathogens. Cold Spring Harb. Perspect. Med. 3, 1–19. doi: 10.1101/cshperspect.a010041
Bayer, C., Bernard, H., Prager, R., Rabsch, W., Hiller, P., Malorny, B.,et al. (2014). An outbreak of Salmonella Newport associated with mung bean sprouts in Germany and the Netherlands, October to November 2011. Euro Surveill. 19. doi: 10.2807/1560-7917.ES2014.19.1.20665
Berger, C. N., Shaw, R. K., Brown, D. J., Mather, H., Clare, S., Dougan, G.,et al. (2009). Interaction of Salmonella enterica with basil and other salad leaves. ISME J. 3, 261–265. doi: 10.1038/ismej.2008.95
Berger, C. N., Sodha, S. V., Shaw, R. K., Griffin, P. M., Pink, D., Hand, P.,et al. (2010). Fresh fruit and vegetables as vehicles for the transmission of human pathogens. Environ. Microbiol. 12, 2385–2397. doi: 10.1111/j.1462-2920.2010.02297.x
Braden, C. R. (2006). Salmonella enterica serotype Enteritidis and eggs: a national epidemic in the United States. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 43, 512–517. doi: 10.1086/505973
Buchwald, D. S., and Blaser, M. J. (1984). A review of human salmonellosis: II. Duration of excretion following infection with nontyphi Salmonella. Rev. Infect. Dis. 6, 345–356. doi: 10.1093/clinids/6.3.345
Buckle, G. C., Walker, C. L., and Black, R. E. (2012). Typhoid fever and paratyphoid fever: systematic review to estimate global morbidity and mortality for 2010. J. Glob. Health 2:010401. doi: 10.7189/jogh.02.010401
Butler, T., Bell, W. R., Levin, J., Linh, N. N., and Arnold, K. (1978). Typhoid fever. Studies of blood coagulation, bacteremia, and endotoxemia. Arch. Intern. Med. 138, 407–410. doi: 10.1001/archinte.1978.03630270047018
Butler, T., Ho, M., Acharya, G., Tiwari, M., and Gallati, H. (1993). Interleukin-6, gamma interferon, and tumor necrosis factor receptors in typhoid fever related to outcome of antimicrobial therapy. Antimicrob. Agents Chemother. 37, 2418–2421. doi: 10.1128/AAC.37.11.2418
Cavallaro, E., Date, K., Medus, C., Meyer, S., Miller, B., Kim, C.,et al. (2011). Salmonella Typhimurium infections associated with peanut products. N. Engl. J. Med. 365, 601–610. doi: 10.1056/NEJMoa1011208
Costa, L. F., Paixão, T. A., Tsolis, R. M., Bäumler, A. J., and Santos, R. L. (2012). Salmonellosis in cattle: advantages of being an experimental model. Res. Vet. Sci. 93, 1–6. doi: 10.1016/j.rvsc.2012.03.002
Crump, J. A., Luby, S. P., and Mintz, E. D. (2004). The global burden of typhoid fever. Bull. World Health Organ. 82, 346–353.
De Jong, H. K., Parry, C. M., van der Poll, T., and Wiersinga, W. J. (2012). Host–pathogen interaction in invasive salmonellosis. PLoS Pathog. 8:e1002933. doi: 10.1371/journal.ppat.1002933
Desin, T. S., Köster, W., and Potter, A. A. (2013). Salmonella vaccines in poultry: past, present and future. Expert Rev. Vaccines 12, 87–96. doi: 10.1586/erv.12.138
Dongol, S., Thompson, C. N., Clare, S., Nga, T. V., Duy, P. T., Karkey, A.,et al. (2012). The microbiological and clinical characteristics of invasive Salmonella in gallbladders from cholecystectomy patients in Kathmandu, Nepal. PLoS ONE 7:e47342. doi: 10.1371/journal.pone.0047342
Dryden, M. S., Gabb, R. J., and Wright, S. K. (1996). Empirical treatment of severe acute community-acquired gastroenteritis with ciprofloxacin. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 22, 1019–1025. doi: 10.1093/clinids/22.6.1019
Dutil, L., Irwin, R., Finley, R., Ng, L. K., Avery, B., Boerlin, P.,et al. (2010). Ceftiofur resistance in Salmonella enterica serovar Heidelberg from chicken meat and humans, Canada. Emerg. Infect. Dis. 16, 48–54. doi: 10.3201/eid1601.090729
Edsall, G., Gaines, S., Landy, M., Tigertt, W. D., Sprinz, H., Trapani, R. J.,et al. (1960). Studies on infection and immunity in experimental typhoid fever. I. Typhoid fever in chimpanzees orally infected with Salmonella typhosa. J. Exp. Med. 112, 143–166. doi: 10.1084/jem.112.1.143
Engels, E. A., Falagas, M. E., Lau, J., and Bennish, M. L. (1998). Typhoid fever vaccines: a meta-analysis of studies on efficacy and toxicity. Br. Med. J. 316, 110–116. doi: 10.1136/bmj.316.7125.110
Feasey, N. A., Dougan, G., Kingsley, R. A., Heyderman, R. S., and Gordon, M. A. (2012). Invasive non-typhoidal salmonella disease: an emerging and neglected tropical disease in Africa. Lancet 379, 2489–2499. doi: 10.1016/S0140-6736(11)61752-2
Firoz Mian, M., Pek, E. A., Chenoweth, M. J., and Ashkar, A. A. (2011). Humanized mice are susceptible to Salmonella Typhi infection. Cell. Mol. Immunol. 8, 83–87. doi: 10.1038/cmi.2010.52
Gal-Mor, O., Suez, J., Elhadad, D., Porwollik, S., Leshem, E., Valinsky, L.,et al. (2012). Molecular and cellular characterization of a Salmonella enterica serovar Paratyphi a outbreak strain and the human immune response to infection. Clin. Vaccine Immunol. 19, 146–156. doi: 10.1128/CVI.05468-11
Gal-Mor, O., Valinsky, L., Weinberger, M., Guy, S., Jaffe, J., Schorr, Y. I.,et al. (2010). Multidrug-resistant Salmonella enterica serovar Infantis, Israel. Emerg. Infect. Dis. 16, 1754–1757. doi: 10.3201/eid1611.100100
Gasem, M. H., Keuter, M., Dolmans, W. M., Van Der Ven-Jongekrijg, J., Djokomoeljanto, R., and Van Der Meer, J. W. (2003). Persistence of Salmonellae in blood and bone marrow: randomized controlled trial comparing ciprofloxacin and chloramphenicol treatments against enteric fever. Antimicrob. Agents Chemother. 47, 1727–1731. doi: 10.1128/AAC.47.5.1727-1731.2003
Germanier, R., and Fuer, E. (1975). Isolation and characterization of Gal E mutant Ty 21a of Salmonella Typhi: a candidate strain for a live, oral typhoid vaccine. J. Infect. Dis. 131, 553–558. doi: 10.1093/infdis/131.5.553
Girardin, E., Grau, G. E., Dayer, J. M., Roux-Lombard, P., and Lambert, P. H. (1988). Tumor necrosis factor and interleukin-1 in the serum of children with severe infectious purpura. N. Engl. J. Med. 319, 397–400. doi: 10.1056/NEJM198808183190703
Glynn, J. R., and Palmer, S. R. (1992). Incubation period, severity of disease, and infecting dose: evidence from a Salmonella outbreak. Am. J. Epidemiol. 136, 1369–1377.
Gonzalez-Escobedo, G., La Perle, K. M. D., and Gunn, J. S. (2013). Histopathological analysis of Salmonella chronic carriage in the mouse hepatopancreatobiliary system. PloS ONE 8:e84058. doi: 10.1371/journal.pone.0084058
Gopinath, S., Carden, S., and Monack, D. (2012). Shedding light on Salmonella carriers. Trends Microbiol. 20, 320–327. doi: 10.1016/j.tim.2012.04.004
Gordon, M. A. (2008). Salmonella infections in immunocompromised adults. J. Infect. 56, 413–422. doi: 10.1016/j.jinf.2008.03.012
Haeusler, G. M., and Curtis, N. (2013). Non-typhoidal Salmonella in children: microbiology, epidemiology and treatment. Adv. Exp. Med. Biol. 764, 13–26. doi: 10.1007/978-1-4614-4726-9_2
Hapfelmeier, S., and Hardt, W. D. (2005). A mouse model for S. Typhimurium-induced enterocolitis. Trends Microbiol. 13, 497–503. doi: 10.1016/j.tim.2005.08.008
Hohmann, E. L. (2001). Nontyphoidal salmonellosis. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 32, 263–269. doi: 10.1086/318457
House, D., Wain, J., Ho, V. A., Diep, T. S., Chinh, N. T., Bay, P. V.,et al. (2001). Serology of typhoid fever in an area of endemicity and its relevance to diagnosis. J. Clin. Microbiol. 39, 1002–1007. doi: 10.1128/JCM.39.3.1002-1007.2001
Jackson, B. R., Griffin, P. M., Cole, D., Walsh, K. A., and Chai, S. J. (2013). Outbreak-associated Salmonella enterica serotypes and food commodities, United States, 1998–2008. Emerg. Infect. Dis. 19, 1239–1244. doi: 10.3201/eid1908.121511
Kent, T. H., Formal, S. B., and Labrec, E. H. (1966). Salmonella gastroenteritis in rhesus monkeys. Arch. Pathol. 82, 272–279.
Keuter, M., Dharmana, E., Gasem, M. H., van der Ven-Jongekrijg, J., Djokomoeljanto, R., Dolmans, W. M.,et al. (1994). Patterns of proinflammatory cytokines and inhibitors during typhoid fever. J. Infect. Dis. 169, 1306–1311. doi: 10.1093/infdis/169.6.1306
Khatri, N. S., Maskey, P., Poudel, S., Jaiswal, V. K., Karkey, A., Koirala, S.,et al. (2009). Gallbladder carriage of Salmonella Paratyphi A may be an important factor in the increasing incidence of this infection in South Asia. Ann. Intern. Med. 150, 567–568. doi: 10.7326/0003-4819-150-8-200904210-00017
Kingsley, R. A., Msefula, C. L., Thomson, N. R., Kariuki, S., Holt, K. E., Gordon, M. A.,et al. (2009). Epidemic multiple drug resistant Salmonella Typhimurium causing invasive disease in sub-Saharan Africa have a distinct genotype. Genome Res. 19, 2279–2287. doi: 10.1101/gr.091017.109
Kraus, M. D., Amatya, B., and Kimula, Y. (1999). Histopathology of typhoid enteritis: morphologic and immunophenotypic findings. Mod. Pathol. 12, 949–955.
Krueger, A. L., Greene, S. A., Barzilay, E. J., Henao, O., Vugia, D., Hanna, S.,et al. (2014). Clinical outcomes of nalidixic acid, ceftriaxone, and multidrug-resistant nontyphoidal Salmonella infections compared with pansusceptible infections in FoodNet sites, 2006–2008. Foodborne Pathog. Dis. 11, 335–341. doi: 10.1089/fpd.2013.1642
Lawley, T. D., Bouley, D. M., Hoy, Y. E., Gerke, C., Relman, D. A., and Monack, D. M. (2008). Host transmission of Salmonella enterica serovar Typhimurium is controlled by virulence factors and indigenous intestinal microbiota. Infect Immun. 76, 403–416. doi: 10.1128/IAI.01189-07
Lawley, T. D., Chan, K., Thompson, L. J., Kim, C. C., Govoni, G. R., and Monack, D. M. (2006). Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLoS Pathog. 2:e11. doi: 10.1371/journal.ppat.0020011
Levine, M. M., Black, R. E., and Lanata, C. (1982). Precise estimation of the numbers of chronic carriers of Salmonella Typhi in Santiago, Chile, an endemic area. J. Infect. Dis. 146, 724–726. doi: 10.1093/infdis/146.6.724
Libby, S. J., Brehm, M. A., Greiner, D. L., Shultz, L. D., McClelland, M., Smith, K. D.,et al. (2010). Humanized nonobese diabetic-scid IL2rgammanull mice are susceptible to lethal Salmonella Typhi infection. Proc. Natl. Acad. Sci. U.S.A. 107, 15589–15594. doi: 10.1073/pnas.1005566107
Liu, S. L., Ezaki, T., Miura, H., Matsui, K., and Yabuuchi, E. (1988). Intact motility as a Salmonella Typhi invasion-related factor. Infect. Immun. 56,1967–1973.
Maclennan, C. A. (2014). Out of Africa: links between invasive nontyphoidal Salmonella disease, typhoid fever, and malaria. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 58, 648–650. doi: 10.1093/cid/cit803
MacLennan, C., Fieschi, C., Lammas, D. A., Picard, C., Dorman, S. E., Sanal, O.,et al. (2004). Interleukin (IL)-12 and IL-23 are key cytokines for immunity against Salmonella in humans. J. Infect. Dis. 190, 1755–1757. doi: 10.1086/425021
MacLennan, C. A., and Levine, M. M. (2013). Invasive nontyphoidal Salmonella disease in Africa: current status. Expert Rev. Anti. Infect. Ther. 11, 443–446. doi: 10.1586/eri.13.27
Majowicz, S. E., Musto, J., Scallan, E., Angulo, F. J., Kirk, M., O’Brien, S. J.,et al. (2010). International collaboration on enteric disease “Burden of Illness” studies. The global burden of nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 50, 882–889. doi: 10.1086/650733
Mandal, B. K., and Brennand, J. (1988). Bacteraemia in salmonellosis: a 15 year retrospective study from a regional infectious diseases unit. Br. Med. J. 297, 1242–1243. doi: 10.1136/bmj.297.6658.1242
Marzel, A., Desai, P. T., Nissan, I., Schorr, Y. I., Suez, J., Valinsky, L.,et al. (2014). Integrative analysis of salmonellosis in Israel, 1995–2012 reveals association of serovar 9,12:l,v: with extraintestinal infections, dissemination of endemic S. Typhimurium DT104 biotypes and a severe under-reporting of outbreaks. J. Clin. Microbiol. 52, 2078–2088. doi: 10.1128/JCM.00399-14
Mather, A. E., Reid, S. W. J., Maskell, D. J., Parkhill, J., Fookes, M. C., Harris, S. R.,et al. (2013). Distinguishable epidemics within different hosts of the multidrug resistant zoonotic pathogen Salmonella Typhimurium DT104. Science 341, 1514–1517. doi: 10.1126/science.1240578
Mathur, R., Oh, H., Zhang, D., Park, S.-G., Seo, J., Koblansky, A.,et al. (2012). Mouse model of Salmonella Typhi infection. Cell 151, 590–602. doi: 10.1016/j.cell.2012.08.042
McClelland, M., Sanderson, K. E., Clifton, S. W., Latreille, P., Porwollik, S., Sabo, A.,et al. (2004). Comparison of genome degradation in Paratyphi A and Typhi, human-restricted serovars of Salmonella enterica that cause typhoid. Nat. Genet. 36, 1268–1274. doi: 10.1038/ng1470
McClelland, M., Sanderson, K. E., Spieth, J., Clifton, S. W., Latreille, P., Courtney, L.,et al. (2001). Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413, 852–856. doi: 10.1038/35101614
McCormick, B. A., Miller, S. I., Carnes, D., and Madara, J. L. (1995). Transepithelial signaling to neutrophils by salmonellae: a novel virulence mechanism for gastroenteritis. Infect. Immun. 63, 2302–2309.
McGovern, V. J., and Slavutin, L. J. (1979). Pathology of Salmonella colitis. Am. J. Surg. Pathol. 3, 483–490. doi: 10.1097/00000478-197912000-00001
Meltzer, E., Sadik, C., and Schwartz, E. (2005). Enteric fever in Israeli travelers: a nationwide study. J. Travel. Med. 12, 275–281. doi: 10.2310/7060.2005.12507
Meltzer, E., and Schwartz, E. (2010). Enteric fever: a travel medicine oriented view. Curr. Opin. Infect. Dis. 23, 432–437. doi: 10.1097/QCO.0b013e32833c7ca1
Menendez, A., Arena, E. T., Guttman, J. A., Thorson, L., Vallance, B. A., Vogl, W.,et al. (2009). Salmonella infection of gallbladder epithelial cells drives local inflammation and injury in a model of acute typhoid fever. J. Infect. Dis. 200, 1703–1713. doi: 10.1086/646608
Mermin, J., Hutwagner, L., Vugia, D., Shallow, S., Daily, P., Bender, J.,et al. (2004). Emerging Infections Program FoodNet Working Group. Reptiles, amphibians, and human Salmonella infection: a population-based, case-control study. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 38(Suppl. 3), S253–S261. doi: 10.1086/381594
Mian, M. F., Pek, E. A., Chenoweth, M. J., Coombes, B. K., and Ashkar, A. A. (2011). Humanized mice for Salmonella Typhi infection: new tools for an old problem. Virulence 2, 248–252. doi: 10.4161/viru.2.3.16133
Mizuno, Y., Takada, H., Nomura, A., Jin, C. H., Hattori, H., Ihara, K.,et al. (2003). Th1 and Th1-inducing cytokines in Salmonella infection. Clin. Exp. Immunol. 131, 111–117. doi: 10.1046/j.1365-2249.2003.02060.x
Monack, D. M. (2012). Salmonella persistence and transmission strategies. Curr. Opin. Microbiol. 15, 100–107. doi: 10.1016/j.mib.2011.10.013
Monack, D. M., Bouley, D. M., and Falkow, S. (2004). Salmonella Typhimurium persists within macrophages in the mesenteric lymph nodes of chronically infected Nramp1+/+ mice and can be reactivated by IFNgamma neutralization. J. Exp. Med. 199, 231–241. doi: 10.1084/jem.20031319
Mortimer, P. P. (1999). Mr N the milker, and Dr Koch’s concept of the healthy carrier. Lancet 353, 1354–1356. doi: 10.1016/S0140-6736(98)11315-6
Msefula, C. L., Kingsley, R. A., Gordon, M. A., Molyneux, E., Molyneux, M. E., MacLennan, C. A.,et al. (2012). Genotypic homogeneity of multidrug resistant S. Typhimurium infecting distinct adult and childhood susceptibility groups in Blantyre, Malawi. PloS ONE 7:e42085. doi: 10.1371/journal.pone.0042085
Murase, T., Yamada, M., Muto, T., Matsushima, A., and Yamai, S. (2000). Fecal excretion of Salmonella enterica Serovar Typhimurium following a Food-Borne outbreak. J. Clin. Microbiol. 38, 3495–3497.
Nguyen, Q. C., Everest, P., Tran, T. K., House, D., Murch, S., Parry, C.,et al. (2004). A clinical, microbiological, and pathological study of intestinal perforation associated with typhoid fever. Clin. Infect. Dis. 39, 61–67. doi: 10.1086/421555
Nuccio, S. P., and Baumler, A. J. (2014). Comparative analysis of Salmonella genomes identifies a metabolic network for escalating growth in the inflamed gut. MBio 5:e00929-14. doi: 10.1128/mBio.00929-14
Ochiai, R. L., Wang, X., von Seidlein, L., Yang, J., Bhutta, Z. A., Bhattacharya, S. K.,et al. (2005). Salmonella Paratyphi a rates, Asia. Emerg. Infect. Dis. 11, 1764–1766. doi: 10.3201/eid1111.050168
Olsen, S. J., Bleasdale, S. C., Magnano, A. R., Landrigan, C., Holland, B. H., Tauxe, R. V.,et al. (2003). Outbreaks of typhoid fever in the United States, 1960–99. Epidemiol. Infect. 130, 13–21. doi: 10.1017/S0950268802007598
Parry, C. M., Hien, T. T., Dougan, G., White, N. J., and Farrar, J. J. (2002). Typhoid fever. N. Engl. J. Med. 347, 1770–1782. doi: 10.1056/NEJMra020201
Pasetti, M. F., Levine, M. M., and Sztein, M. B. (2003). Animal models paving the way for clinical trials of attenuated Salmonella enterica serovar Typhi live oral vaccines and live vectors. Vaccine 21, 401–418. doi: 10.1016/S0264-410X(02)00472-3
Patel, T. A., Armstrong, M., Morris-Jones, S. D., Wright, S. G., and Doherty, T. (2010). Imported enteric fever: case series from the hospital for tropical diseases, London, United Kingdom. Am. J. Trop. Med. Hyg. 82, 1121–1126. doi: 10.4269/ajtmh.2010.10-0007
Pohan, H. T. (2004). Clinical and laboratory manifestations of typhoid fever at Persahabatan Hospital, Jakarta. Acta Med. Indones. 36, 78–83.
Raffatellu, M., Chessa, D., Wilson, R. P., Dusold, R., Rubino, S., and Baumler, A. J. (2005). The Vi capsular antigen of Salmonella enterica serotype Typhi reduces Toll-like receptor-dependent interleukin-8 expression in the intestinal mucosa. Infect. Immun. 73, 3367–3374. doi: 10.1128/IAI.73.6.3367-3374.2005
Raffatellu, M., Santos, R. L., Verhoeven, D. E., George, M. D., Wilson, R. P., Winter, S. E.,et al. (2008a). Simian immunodeficiency virus-induced mucosal interleukin-17 deficiency promotes Salmonella dissemination from the gut. Nat. Med. 14, 421–428. doi: 10.1038/nm1743
Raffatellu, M., Wilson, R. P., Winter, S. E., and Baumler, A. J. (2008b). Clinical pathogenesis of typhoid fever. J. Infect. Dev. Ctries. 2, 260–266.
Robbins, J. D., and Robbins, J. B. (1984). Reexamination of the protective role of the capsular polysaccharide (Vi antigen) of Salmonella Typhi. J. Infect. Dis. 150, 436–449. doi: 10.1093/infdis/150.3.436
Rout, W. R., Formal, S. B., Dammin, G. J., and Giannella, R. A. (1974). Pathophysiology of Salmonella diarrhea in the Rhesus monkey: intestinal transport, morphological and bacteriological studies. Gastroenterology 67, 59–70.
Sabbagh, S. C., Forest, C. G., Lepage, C., Leclerc, J. M., and Daigle, F. (2010). So similar, yet so different: uncovering distinctive features in the genomes of Salmonella enterica serovars Typhimurium and Typhi. FEMS Microbiol. Lett. 305, 1–13. doi: 10.1111/j.1574-6968.2010.01904.x
Santos, R. L., Zhang, S., Tsolis, R. M., Kingsley, R. A., Adams, L. G., and Bäumler, A. J. (2001). Animal models of Salmonella infections: enteritis versus typhoid fever. Microbes Infect. Inst. Pasteur 3, 1335–1344. doi: 10.1016/S1286-4579(01)01495-2
Sekirov, I., Tam, N. M., Jogova, M., Robertson, M. L., Li, Y., Lupp, C.,et al. (2008). Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect. Immun. 76, 4726–4736. doi: 10.1128/IAI.00319-08
Sharma, A., and Qadri, A. (2004). Vi polysaccharide of Salmonella Typhi targets the prohibitin family of molecules in intestinal epithelial cells and suppresses early inflammatory responses. Proc. Natl. Acad. Sci. U.S.A. 101, 17492–17497. doi: 10.1073/pnas.0407536101
Sheikh, A., Khanam, F., Sayeed, M. A., Rahman, T., Pacek, M., Hu, Y.,et al. (2011). Interferon-gamma and proliferation responses to Salmonella enterica serotype Typhi proteins in patients with S. Typhi bacteremia in Dhaka, Bangladesh. PLoS Negl. Trop. Dis. 5:e1193. doi: 10.1371/journal.pntd.0001193
Shi, Z., Cai, Z., Yu, J., Zhang, T., Zhao, S., Smeds, E.,et al. (2012). Toll-like receptor 11 (TLR11) prevents Salmonella penetration into the murine Peyer patches. J. Biol. Chem. 287, 43417–43423. doi: 10.1074/jbc.M112.411009
Shultz, L. D., Ishikawa, F., and Greiner, D. L. (2007). Humanized mice in translational biomedical research. Nat. Rev. Immunol. 7, 118–130. doi: 10.1038/nri2017
Song, J., Willinger, T., Rongvaux, A., Eynon, E. E., Stevens, S., Manz, M. G.,et al. (2010). A mouse model for the human pathogen Salmonella Typhi. Cell Host Microbe 8, 369–376. doi: 10.1016/j.chom.2010.09.003
Spano, S., and Galan, J. E. (2012). A Rab32-dependent pathway contributes to Salmonella Typhi host restriction. Science 338, 960–963. doi: 10.1126/science.1229224
Sprinz, H., Gangarosa, E. J., Williams, M., Hornick, R. B., and Woodward, T. E. (1966). Histopathology of the upper small intestines in typhoid fever. Biopsy study of experimental disease in man. Am. J. Dig. Dis. 11, 615–624. doi: 10.1007/BF02233509
Stecher, B., Robbiani, R., Walker, A. W., Westendorf, A. M., Barthel, M., Kremer, M.,et al. (2007). Salmonella enterica serovar Typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 5:2177–2189. doi: 10.1371/journal.pbio.0050244
Stoycheva, M., and Murdjeva, M. (2005). Serum levels of interferon-gamma, interleukin-12, tumour necrosis factor-alpha, and interleukin-10, and bacterial clearance in patients with gastroenteric Salmonella infection. Scand. J. Infect. Dis. 37, 11–14. doi: 10.1080/00365540410026068
Stuart, B. M., and Pullen, R. L. (1946). Typhoid: clinical analysis of 360 cases. Arch. Intern. Med. (Chic.) 78, 629–661. doi: 10.1001/archinte.1946.00220060002001
Sztein, M. B. (2007). Cell-mediated immunity and antibody responses elicited by attenuated Salmonella enterica serovar Typhi strains used as live oral vaccines in humans. Clin. Infect. Dis. 45(Suppl. 1), S15–S19. doi: 10.1086/518140
Tacket, C. O., Ferreccio, C., Robbins, J. B., Tsai, C. M., Schulz, D., Cadoz, M.,et al. (1986). Safety and immunogenicity of two Salmonella Typhi Vi capsular polysaccharide vaccines. J. Infect. Dis. 154, 342–345. doi: 10.1093/infdis/154.2.342
Thiennimitr, P., Winter, S. E., Winter, M. G., Xavier, M. N., Tolstikov, V., Huseby, D. L.,et al. (2011). Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc. Natl. Acad. Sci. U.S.A. 108, 17480–17485. doi: 10.1073/pnas.1107857108
Thompson, L. J., Dunstan, S. J., Dolecek, C., Perkins, T., House, D., Dougan, G.,et al. (2009). Transcriptional response in the peripheral blood of patients infected with Salmonella enterica serovar Typhi. Proc. Natl. Acad. Sci. U.S.A. 106, 22433–22438. doi: 10.1073/pnas.0912386106
Tsolis, R. M., Young, G. M., Solnick, J. V., and Baumler, A. J. (2008). From bench to bedside: stealth of enteroinvasive pathogens. Nat. Rev. Microbiol. 6, 883–892. doi: 10.1038/nrmicro2012
Van de Vosse, E., and Ottenhoff, T. H. (2006). Human host genetic factors in mycobacterial and Salmonella infection: lessons from single gene disorders in IL-12/IL-23-dependent signaling that affect innate and adaptive immunity. Microbes Infect. 8, 1167–1173. doi: 10.1016/j.micinf.2005.10.032
Waddington, C. S., Darton, T. C., and Pollard, A. J. (2014). The challenge of enteric fever. J. Infect. 68(Suppl. 1), S38–S50. doi: 10.1016/j.jinf.2013.09.013
Wahid, R., Salerno-Goncalves, R., Tacket, C. O., Levine, M. M., and Sztein, M. B. (2007). Cell-mediated immune responses in humans after immunization with one or two doses of oral live attenuated typhoid vaccine CVD 909. Vaccine 25, 1416–1425. doi: 10.1016/j.vaccine.2006.10.040
Wangdi, T., Winter, S. E., and Baumler, A. J. (2012). Typhoid fever: “you can’t hit what you can’t see.” Gut Microbes 3, 88–92. doi: 10.4161/gmic.18602
Weill, F.-X., Lailler, R., Praud, K., Kérouanton, A., Fabre, L., Brisabois, A.,et al. (2004). Emergence of extended-spectrum-beta-lactamase (CTX-M-9)-producing multiresistant strains of Salmonella enterica serotype Virchow in poultry and humans in France. J. Clin. Microbiol. 42, 5767–5773. doi: 10.1128/JCM.42.12.5767-5773.2004
Wilkins, E. G., and Roberts, C. (1988). Extraintestinal salmonellosis. Epidemiol. Infect. 100, 361–368. doi: 10.1017/S095026880006711X
Wilson, R. P., Raffatellu, M., Chessa, D., Winter, S. E., Tükel, C., and Bäumler, A. J. (2008). The Vi-capsule prevents Toll-like receptor 4 recognition of Salmonella. Cell Microbiol. 10, 876–890. doi: 10.1111/j.1462-5822.2007.01090.x
Winter, S. E., Thiennimitr, P., Winter, M. G., Butler, B. P., Huseby, D. L., Crawford, R. W.,et al. (2010). Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467, 426–429. doi: 10.1038/nature09415
Woo, H., Okamoto, S., Guiney, D., Gunn, J. S., and Fierer, J. (2008). A model of Salmonella colitis with features of diarrhea in SLC11A1 wild-type mice. PloS ONE 3:e1603. doi: 10.1371/journal.pone.0001603
Keywords: Salmonella enterica, typhoid, enteric fever, NTS, salmonellosis, gastroenteritis
Citation: Gal-Mor O, Boyle EC and Grassl GA (2014) Same species, different diseases: how and why typhoidal and non-typhoidal Salmonella enterica serovars differ. Front. Microbiol. 5:391. doi: 10.3389/fmicb.2014.00391
Received: 23 May 2014; Paper pending published: 20 June 2014;
Accepted: 12 July 2014; Published online: 04 August 2014.
Edited by:
Constantino López-Macías, Mexican Social Security Institute, Mexico and University of Oxford, UKReviewed by:
Laurel L. Lenz, National Jewish Health, USARanjit Kumar, University of Alabama at Birmingham, USA
Copyright © 2014 Gal-Mor, Boyle and Grassl. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guntram A. Grassl, Institute for Experimental Medicine, Christian Albrechts University Kiel, Kiel, Germany; Research Center Borstel, Parkallee 29, 23845 Borstel, Germany e-mail: g.grassl@iem.uni-kiel.de
 Ohad Gal-Mor
Ohad Gal-Mor Erin C. Boyle
Erin C. Boyle Guntram A. Grassl
Guntram A. Grassl