
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 17 June 2014
Sec. Microbial Physiology and Metabolism
Volume 5 - 2014 | https://doi.org/10.3389/fmicb.2014.00275
This article is part of the Research Topic Oil Reservoir Microbiology View all 12 articles
We examined the potential for CH4 oxidation to be coupled with oxygen derived from the dissimilatory reduction of perchlorate, chlorate, or via chlorite (ClO−2) dismutation. Although dissimilatory reduction of ClO−4 and ClO−3 could be inferred from the accumulation of chloride ions either in spent media or in soil slurries prepared from exposed freshwater lake sediment, neither of these oxyanions evoked methane oxidation when added to either anaerobic mixed cultures or soil enriched in methanotrophs. In contrast, ClO−2 amendment elicited such activity. Methane (0.2 kPa) was completely removed within several days from the headspace of cell suspensions of Dechloromonas agitata CKB incubated with either Methylococcus capsulatus Bath or Methylomicrobium album BG8 in the presence of 5 mM ClO−2. We also observed complete removal of 0.2 kPa CH4 in bottles containing soil enriched in methanotrophs when co-incubated with D. agitata CKB and 10 mM ClO−2. However, to be effective these experiments required physical separation of soil from D. agitata CKB to allow for the partitioning of O2 liberated from chlorite dismutation into the shared headspace. Although a link between ClO−2 and CH4 consumption was established in soils and cultures, no upstream connection with either ClO−4 or ClO−3 was discerned. This result suggests that the release of O2 during enzymatic perchlorate reduction was negligible, and that the oxygen produced was unavailable to the aerobic methanotrophs.
In-situ production of CO2 by microbial activity is encouraged during enhanced oil recovery as a means of reducing oil viscosity and improving flow characteristics (Lazar et al., 2007; Youseff et al., 2009). Further, targeted growth of microbes and intentional precipitation of solid phase minerals can be applied to selectively decrease permeability and direct flow to enhance oil recovery (Jenneman et al., 1984; Zhu et al., 2013). These enhancements rely on the availability of appropriate electron acceptors to supply oxidant to microbes utilizing hydrocarbons or other reduced compounds as electron donors. Considerable attention has been paid thus far to the use of sulfate, nitrate or nitrite as electron acceptors in these applications (Youseff et al., 2009) with sulfate less favored because its reduction results in H2S and leads to oil souring (Gieg et al., 2011). There has been recent interest in the use of perchlorate or chlorate, together known as (per)chlorate as electron acceptors. However, little is known about the fate of (per)chlorate in anoxic environments like oil reservoirs. Here we examine the potential reaction of (per)chlorate and chlorite with the low molecular weight hydrocarbon methane.
Chemical reduction of perchlorate is generally quite slow (Urbansky, 2002). However, under anoxic conditions dissimilatory perchlorate reducing bacteria (DPRB) rapidly reduce (per)chlorate to form chlorite. Chlorite thus formed is further degraded by these bacteria using chlorite dismutase to produce Cl− and O2 (Rikken et al., 1996; Kostan et al., 2010; Mlynek et al., 2011). These microbial processes reduce (per)chlorate from both natural (Rao et al., 2007; Kounaves et al., 2010) and anthropogenic (Coates and Achenbach, 2004) sources. Degradation of intentionally added (per)chlorate is therefore likely in the proximity of hydrocarbon reservoirs given the abundance of suitable electron donors. There is a potential for O2 liberation during this process (see reaction 1 below) that may be used by aerobic bacteria to oxidize aromatic compounds (benzene, naphthalene, catechol) via oxygenase-dependent pathways in otherwise anoxic soils and sediments (Coates et al., 1998, 1999a; Coates and Achenbach, 2004; Weelink et al., 2007; Carlström et al., 2013). A similar phenomenon was noted that could link biological oxidation of arsenite to the reduction of chlorate ions, presumably also by liberation of O2 (Sun et al., 2010). This type of interaction has not been extended to the oxidation of low molecular weight hydrocarbons such as methane (CH4).
Methane is produced by geothermal and microbial processes in the Earth's crust (Martini et al., 1996) and in marine and terrestrial sediments (Cicerone and Oremland, 1988). Methane is removed photochemically in the atmosphere by reaction with hydroxyl radicals but the most important removal mechanism in aqueous and terrestrial environments is by the action of anaerobic and aerobic methane oxidizing microbes (Cicerone and Oremland, 1988; Boetius et al., 2000). Significant quantities of methane are associated with oil reservoirs (Jones et al., 2007; Gieg et al., 2008) hence we hypothesize that ClO−2 disproportionation, and by extension dissimilatory reduction of the upstream ClO−4 or ClO−3 ions, could be linked to aerobic CH4 oxidation by a biochemical release of O2:
Two other well-studied microbiological processes can achieve a net oxidation of CH4 under prevailing anaerobic conditions, (1) a reverse process of methanogenesis involving “ANME” archaea in syntrophy with bacterial sulfate- or sulfur-reduction (Hinrichs et al., 1999; Boetius et al., 2000; Milucka et al., 2012) and (2) nitrite-linked CH4 oxidation that putatively liberates O2 via NO dismutation as achieved by Methylomirabilis oxyfera (Ettwig et al., 2010). In our study we explored the potential for aerobic CH4 oxidizing bacteria to utilize oxygen produced by DPRB during (per)chlorate reduction and chlorite dismutation.
The DPRB Dechloromonas agitata CKB was grown at 30°C under N2 on 20 mM sodium acetate and 10 mM NaClO4 using phosphate buffer media (PBM) consisting of the following salts in solution (g/liter): Na2HPO4 (0.971), NaH2PO4 (0.379), NH4Cl (0.25) plus 10 ml/l vitamins and 10 ml/l mineral stock solution (Sun et al., 2009). Methylococcus capsulatus Bath, Methylosinus trichosporium OB3b, and Methylomicrobium album BG8, were grown and maintained at 30°C on air + 30 kPa CH4 using nitrate mineral salts media (NMS; Whittenbury et al., 1970). Cultures (1 l) for washed cell suspensions were harvested during late exponential phase, centrifuged (7000 × g), and washed twice with medium lacking substrates and vitamins. Final suspension volumes ranged from 5 to 150 ml. Cell concentrations at the start of incubations ranged from 1.8 × 108 cells ml−1 to 6.9 × 108 cells ml−1.
Soil from the seasonally exposed shoreline of Searsville Lake previously shown to harbor methanotrophic activity (Oremland and Culbertson, 1992) was air dried for two days at room temperature before sieving (<1 mm) to assure uniformity of soil particle size. Dried soil was stored for several weeks in stoppered 1 l glass flasks with air headspace and periodically augmented with 0.2 kPa CH4 after consumption had removed all of the previously added CH4 (4–6 days). Soil with thusly enhanced methanotrophic activity was used to determine CH4 uptake in studies with added ClO−4 and ClO−2 in the absence of O2.
Sediment slurries were prepared by adding 100 ml SeFr2 freshwater media (flushed with 20 kPa CO2/80 kPa N2; Miller et al., 2013) to 10 g Searsville Lake soil in N2 flushed serum bottles (160 ml). Slurry pH was adjusted to 7.1 using 1 ml of 1 M NaHCO3. Slurries were incubated under N2 headspace following periodic amendments with 1 to 2 mmoles acetate and 0.5 to 1 mmole ClO−4. Slurries were periodically sampled by syringe using 22 g needles and filtered through a 0.2 um Spin-X centrifuge tube. Acetate and ClO−4 amendments were made after both were depleted (usually several days to weeks) during which time copious quantities of CH4 were produced. Slurries with enhanced perchlorate reducing activity were used to determine methane uptake activity in the presence or absence of added O2.
Aliquots of washed cell suspension of D. agitata CKB were distributed into stoppered and N2 flushed 25 ml Balch tubes containing 10 ml PBM amended with 5 mM acetate and 10 mM NaClO4, NaClO3, or NaClO2. Initial cell densities were 1.8 × 108 cells per ml. The headspace was sampled over 7 days by syringe for CO2 and O2. Aqueous samples (0.3 ml) were collected by syringe for analysis of dissolved acetate and anions (Cl−, ClO−4, ClO−3, and ClO−2). A short-term (10 min) experiment was conducted to follow ClO−2 disproportionation. In this study, triplicate samples were sacrificed at pre-determined times. Activity was stopped by addition of 0.1 ml 4N NaOH before measurement of headspace O2. Aqueous samples were subsequently collected for analysis of dissolved anions (Cl− and ClO−2).
Mixtures (10 ml) of M. capsulatus Bath and D. agitata CKB were prepared by adding washed cell suspensions of the cultures together in N2 flushed Balch tubes (25 ml) sealed with butyl rubber stoppers. Methane (0.2 kPa) was introduced by syringe to all tubes and NaClO2 (5 mM) was added to 3 tubes at the start of the incubation which was conducted at 37°C. Headspace CH4 was monitored over 1 day. Single tubes were prepared without addition of ClO−2 or without one of the cultures (i.e., no D. agitata CKB or no M. capsulatus Bath) to act as negative controls. A tube containing only M. capsulatus Bath under an air headspace acted as a positive control.
Additional microcosms were prepared in serum bottles (37 or 67 ml) using washed cell suspensions of D. agitata CKB and M. trichosporium OB3b or M. album BG8. Inocula were either combined in bottles (5 ml each) in one aqueous phase or kept separate by placing methanotrophs (1 ml) inside an open-topped glass tube contained within the bottles before flushing with N2 and later adding D. agitata CKB (5 ml) by syringe to the bottom of the bottles. In this manner, the cultures were segregated but shared a common headspace. Methane (0.2 kPa) was introduced by syringe to all bottles and NaClO4, NaClO3, or NaClO2 (5 or 10 mM) was added aseptically to start the incubations which were conducted at 28°C. Headspace CH4 and aqueous anions were monitored over time. Controls were prepared without additions of ClO−2.
Washed cell suspensions (5 ml each) of D. agitata CKB and M. trichosporium OB3b were added together to N2 flushed serum bottles (13 ml). Radiolabeled 14CH4 (5 μCi; specific activity = 21 μCi/μmole) was added along with 1 kPa CH4 to the headspace of each bottle. Perchlorate (5 mM) was added to triplicate bottles and ClO−2 (5 mM) was added to a single bottle by syringe to start the incubation which was conducted at 30°C. Gas samples for analysis of 14CH4 and 14CO2 were collected by syringe. At the end of the incubation, samples were acidified using 0.5 ml of 1.2N HCl to cause dissolved inorganic carbon (DIC = HCO−3 + CO−23) to react to form CO2 gas which was partitioned into the headspace. The headspace was again sampled by syringe. Control incubations consisted of single bottles of M. trichosporium OB3b alone under an air headspace and D. agitata CKB alone under N2.
Slurry microcosms were prepared in N2 flushed serum bottles (57 ml) containing 5 g of dried Searsville Lake soil with enhanced methanotrophic activity to which 10 ml Searsville Lake sediment slurry with enhanced perchlorate reducing activity (above) was added. Half the bottles were maintained under N2 while half were flushed with air. Substrate (5 mM ClO−4 or ClO−2) was added by syringe followed by 0.5 ml CH4 (1 kPa). Incubations were conducted at 22°C. Headspace and liquid samples were collected by syringe over 9 days.
Soil microcosms were prepared in serum bottles (67 ml) using washed cell suspensions of D. agitata CKB and dried Searsville Lake soil which was enhanced in methanotrophic activity (above). Soil (2 g) was placed inside open-topped glass tubes contained within the bottles prior to sealing and flushing with N2. Subsequently, D. agitata CKB (10 ml) was added by syringe to the bottom of the bottles followed by aseptic addition of 5 mM acetate. The culture and the soil were thus segregated under a common headspace. Methane (0.1 kPa) was introduced by syringe to all bottles and 10 mM NaClO4, NaClO3, or NaClO−2 was added to D. agitata CKB to start the incubations. Incubations were conducted at 22°C. Headspace CH4 and CO2 and aqueous acetate and anions were monitored over 7 days.
Headspace O2 was determined by ECD-GC using a molecular sieve 5A column (3.2 mm O.D. × 2.4 m) operated at 75°C using hydrocarbon-free UHP N2 carrier. Background O2 was minimized by flushing syringes and needles with O2-free N2 prior to sampling. The detection limit was 0.05 mmol O2/L. Headspace CH4 and CO2 were determined by FID- and TCD-GC, respectively (Miller et al., 2013). Cell densities were determined by direct cell counting of liquid samples using acridine orange epi-fluorescence microscopy (Hobbie et al., 1977). Additional aqueous samples, including slurries, were filtered using Spin-X centrifuge filter tubes (0.2 μm; Corning Inc., Corning, NY) before determination of dissolved acetate by HPLC (Hoeft et al., 2004) or anions by IC (Miller et al., 2003). Dissolved ClO−4 was analyzed separately by suppressed conductivity IC using a Dionex ISC 1100 containing an AS16 analytical column (4 × 250 mm) and an AG16 guard column (4 × 50 mm) with 0.035 M NaOH eluent. Measurements of headspace 14CH4 and 14CO2 were made by gas proportional counting (Culbertson et al., 1981) following TCD-GC analysis of CH4 and CO2 with separation on a Hayesep D column (100/120; 3.2 mm O.D. × 4.8 m) using UHP He carrier.
The total amount of gas in each bottle or tube was calculated from the headspace concentration using Henry's Law and the volumes of gas and liquid present. The dimensionless Henry's Law constants (KH = CG/CL) used were 31.43 for O2, 29.46 for CH4 and 1.20 for CO2 and were not corrected for ionic strength.
Dissimilatory (per)chlorate reduction by D. agitata CKB resulted in conversion of 85–100 μmoles added ClO−4 or ClO−3 to Cl− in the presence of 50 μmoles added acetate (Figures 1A,B). Much less ClO−4 or ClO−3 (<15 μmoles) was consumed without added acetate and a corresponding lesser amount of Cl− was produced. These observations suggest endogenous metabolism of intrinsic electron donors such as glycogen or polyhydroxybutyrate (PHB). No activity was observed in killed controls or in incubations with media and chloroxyanions alone (data not shown). Biological reduction of ClO−4 or ClO−3 and consumption of acetate occurred over approximately 2 days. Chlorite dismutation was much more rapid. More than half of the 100 μmoles ClO−2 added was consumed and converted to Cl− before the initial sampling at T = 2 min (Figure 1C). An additional 20 μmoles ClO−2 were consumed over 7 days, however more than 20 μmoles ClO−2 remained unreacted at the end.
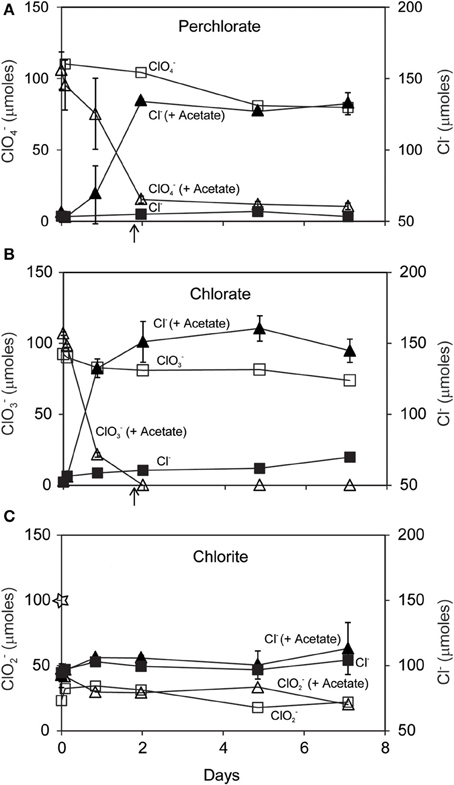
Figure 1. Time course of reaction of D. agitata CKB following addition of 10 mM ClO−4 (A), ClO−3 (B), or ClO−2 (C) showing consumption of added substrate (open symbols) and production of Cl− (closed symbols). Triangles symbolize incubations with added acetate (5 mM) while squares symbolize incubations without added acetate. Triangles represent the mean and range of duplicate samples. Absence of bars indicates that the error is smaller than the symbol size. Squares represent single samples. Arrows in (A) and (B) correspond to the time when 5 mM added acetate was completely consumed. The star in (C) corresponds to the initial amount of ClO−2 added.
Carbon dioxide (CO2) was the dominant gaseous product of dissimilatory reduction of ClO−4 or ClO−3 in the presence of acetate (Figures 2A,B). Slightly more CO2 was produced than could be accounted for by the added acetate. Little CO2 was produced without added acetate. Small amounts of O2 (up to 15 μmoles) were produced during incubations with ClO−4 or ClO−3. In contrast, ClO−2 dismutation resulted in substantial and rapid O2 production (Figure 2C) corresponding to release of >35% of the added ClO−2 within the first day. As expected, there was no effect of added acetate on disproportionation of ClO−2; however details of the early evolution of O2 were obscured by the coarse sampling schedule.
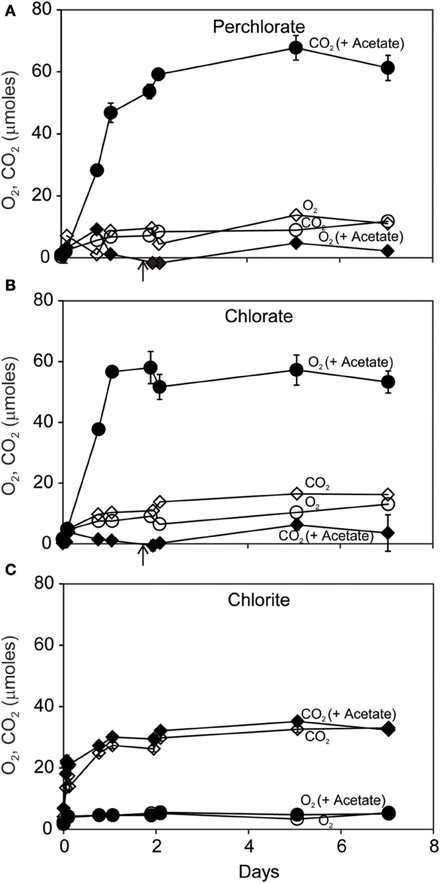
Figure 2. Time course of reaction of D. agitata CKB following addition of 10 mM ClO−4 (A), ClO−3 (B), or ClO−2 (C) showing production of gaseous products O2 (diamonds) and CO2 (circles), with (solid symbols), and without (open symbols) 5 mM acetate. Solid symbols represent the mean and range of duplicate samples. Absence of bars indicates that the error is smaller than the symbol size. Open symbols represent single samples. Arrows in (A) and (B) correspond to the time when 5 mM added acetate was completely consumed.
The pattern of early O2 production during ClO−2 dismutation was made clear in a subsequent short-term (10 min) experiment where 56 μmoles of both O2 and Cl− were produced during the consumption of 56 μmoles of ClO−2 (Figures 3A,B). A minor amount of CO2 (<1 μmole) was produced (Figure 3C). Nearly half (40 μmoles) of the added ClO−2 remained unreacted at the end of the experiment.
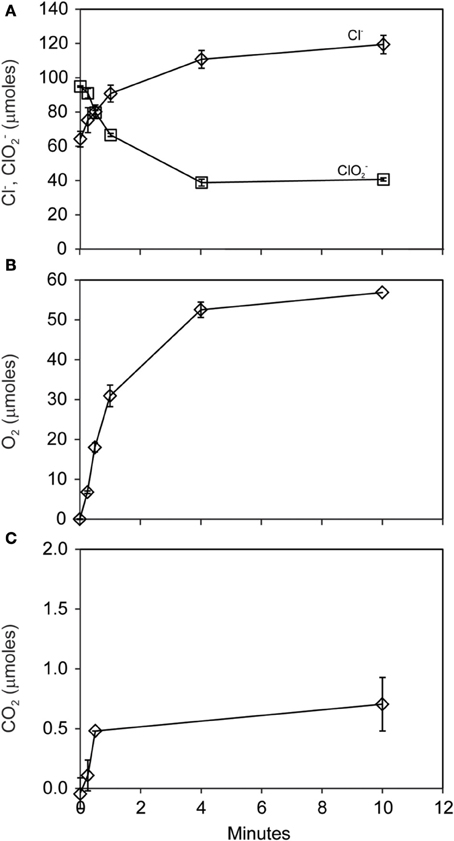
Figure 3. Short-term course of reaction of D. agitata CKB following addition of 10 mM ClO−2, showing loss of ClO−2 (squares) and production of Cl− (A) and production of O2 (B). A minor amount of CO2 was produced (C). Symbols represent the mean and standard deviation of triplicate measurements. Absence of bars indicates that the error is smaller than the symbol size.
Methane was oxidized by methanotrophic bacteria M. capsulatus Bath (Figure 4A) and M. album BG8 (Figure 4B) during the reaction of D. agitata CKB with ClO−2. No removal of CH4 was observed in mixed cell suspensions amended with ClO−4 or ClO−3 (i.e., no added ClO−2). Methane consumption occurred while the methanotrophs were in direct contact with up to 10 mM ClO−2. Methane removal during incubations of co-cultures of D. agitata CKB with M. capsulatus Bath at 37°C occurred within 1 day while removal of CH4 by M. album BG8 at 28°C occurred over 4 days. Similar rates of CH4 consumption were observed for these mixed cultures whether they were segregated or co-mingled (Figure 4B) indicating that direct contact of cells was not required for methane consumption to occur.
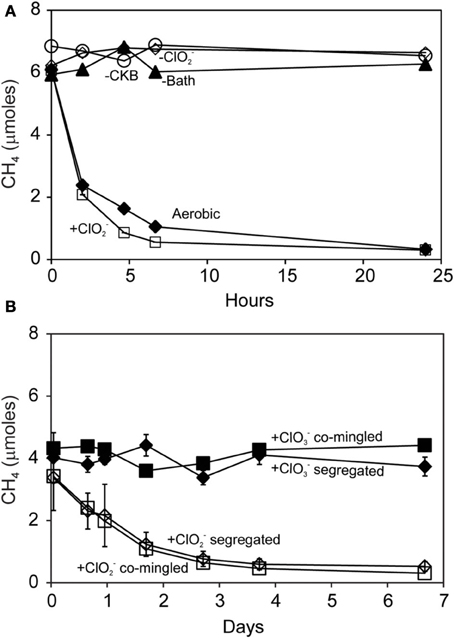
Figure 4. Methane uptake by mixed cultures of D. agitata CKB with M. capsulatus Bath (A) and M. album BG8 (B) during anaerobic incubations. Cultures in (A) were co-mingled and provided with 5 mM ClO−2 (open squares). Aerobic controls are also shown (solid diamonds). Negative controls were prepared by withholding M. capsulatus Bath (solid triangles), D. agitata CKB (open circles), or ClO2− (open diamonds). Cultures in (B) were either co-mingled (squares) or segregated under a common headspace (diamonds) and provided with either ClO−2 (open symbols) or ClO−3 (solid symbols). Symbols represent the mean and standard deviation of triplicate measurements. Absence of bars indicates that the error is smaller than the symbol size.
Strain M. trichosporium OB3b co-cultured with D. agitata CKB containing 5 mM acetate oxidized 14CH4 directly to 14CO2 (Figure 5). The rate of 14CH4 loss under anaerobic conditions with 5 mM added ClO−2 was similar to that under aerobic conditions. No loss of 14CH4 occurred in mixed cell suspensions amended with 5 mM ClO−4 in lieu of ClO−2 or in controls without methanotrophs. During the incubation, the product of methanotrophy (14CO2) was distributed about equally between liquid and gas phases. Roughly 20% of the added 14CH4 appeared as 14CO2 in the headspace after 20 h (0.8 days). Most of the 14CH4 added was recovered as 14CO2 (60–90% recovery after acidification).
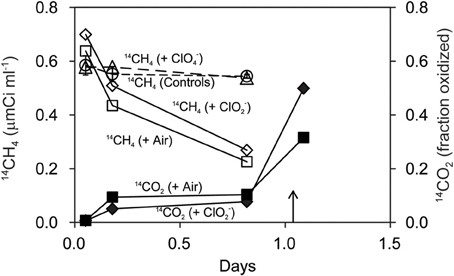
Figure 5. Oxidation of 14CH4 (open symbols) to 14CO2 (closed symbols) by M. trichosporium OB3b during anaerobic incubations with D. agitata CKB following addition of ClO−4 (triangles) and ClO−2 (diamonds). Triangles represent the mean and standard deviation of triplicate measurements of samples with ClO−4 added. All others were single bottles. Controls (no bacteria or no chloroxyanion added) were pooled and are shown as circles with error bars. Aerobic incubations with M. trichosporium OB3b alone are also shown (squares). Samples were acidified at the time indicated by the arrow.
Searsville Lake sediment slurries removed repeated pulses of added ClO−4 during anaerobic incubations using freshwater media with added acetate. Over a period of 1 month, 4 additions of 10 mM ClO−4 were removed in bottles with commensurate consumption of 4 additions of 5 mM added acetate (data not shown). These slurries, thus enhanced in ClO−4 reducing capacity, were used in separate incubations with added CH4 (Figure 6). Under aerobic conditions, incubations with or without additions of ClO−4 or ClO−2 (5 mM) completely consumed 30 μmoles CH4 within 5 days. However, no oxidation of CH4 occurred during anaerobic incubations of Searsville Lake sediment slurries either with or without additions of ClO−4 or ClO−2.
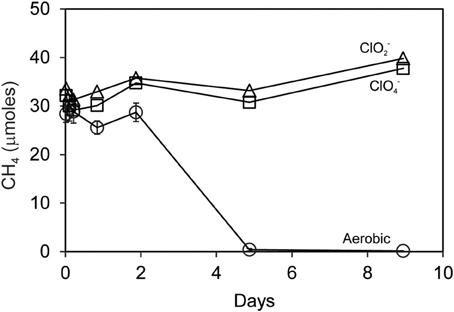
Figure 6. Methane uptake by Searsville Lake sediment slurries pre-adapted to reduce perchlorate using acetate as electron donor. Anaerobic incubations with 5 mM added ClO−4 (squares) or ClO−2 (triangles) showed no uptake. Aerobic incubations (circles) represent the mean and standard deviation of CH4 measurements in all bottles aerobic.
Methane oxidation was observed when Searsville Lake soil (previously enhanced in methanotrophic activity) was segregated from liquid cultures of D. agitata CKB. Methane was completely consumed over the next 5 days by soil methanotrophs during the reaction of DPRB with 10 mM ClO−2 (Figure 7). No CH4 loss was observed when DPRB were provided with either 10 mM ClO−4 or ClO−3.
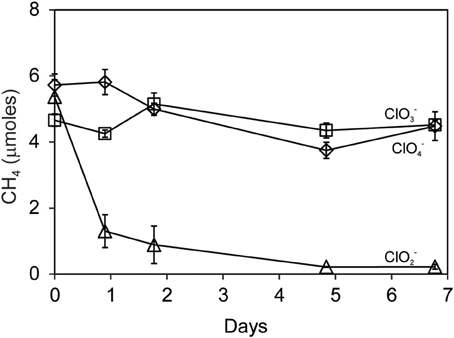
Figure 7. Methane uptake by Searsville Lake soils during anaerobic incubations with D. agitata CKB following addition of ClO−2 (triangles), ClO−4 (diamonds), or ClO−3 (squares). Symbols represent the mean and standard deviation of triplicate measurements. Absence of bars indicates that the error is smaller than the symbol size.
Few enzymes outside of photosystem II are capable of generating dioxygen in anoxic settings. In addition to chlorite dismutase (Cld), these include superoxide dismutase and catalase (McCord et al., 1971), and a putative nitric oxide dismutase (Ettwig et al., 2012). Chlorite dismutase has been purified from at least four DPRB (Mehboob et al., 2009) and is well studied (Coates et al., 1999b; Lee et al., 2008; Goblirsch et al., 2010, 2011). It is a heme enzyme which operates hyperselectively; its assumed sole function is to detoxify ClO−2. The present study is the first report of anaerobic methane oxidation linked to the combined presence of DPRB and ClO−2. We emphasize that this is a “cryptic” aerobic methane oxidation in that the organisms oxidizing methane are aerobic methanotrophs as opposed to anaerobic methanogenic archaea utilizing reverse methanogenesis to consume methane (Hinrichs et al., 1999). As such, this process is somewhat analogous to nitrite-dependent anaerobic methane oxidation as purportedly carried out by the mixed culture containing Methylomirabalis oxyfera (Ettwig et al., 2010). In our study, methanotrophs use O2 derived from disproportionation of ClO−2 by DPRB (reaction 1) to oxidize CH4 (reaction 3). This is in contrast to the mechanism proposed for M. oxyfera in which a single organism may use O2 from disproportionation of NO derived from NO2− (reaction 4) to oxidize CH4 (reaction 5):
Previous work by Coates et al. (1998, 1999a); Coates and Achenbach (2006) demonstrated a link between Cld activity and other aerobic hydrocarbon oxidizing bacteria during the degradation of benzene and naphthalene. In their experiments with pristine soil and hydrocarbon contaminated sediment, 14C-benzene was oxidized to 14CO2 over several days when provided with ClO−2 in the presence of washed cells of D. agitata CKB under anoxic conditions. Similarly, 14C-napthalene was rapidly oxidized to 14CO2 in the presence of washed cells of D. agitata CKB and Pseudomonas sp. strain JS150 (an aerobic hydrocarbon oxidizer) when provided with ClO−2.
Here we demonstrated that O2 released by the reaction of ClO−2 with pure cultures of DPRB could be utilized by a variety of methane oxidizing bacteria, including γ-Proteobacteria (M. capsulatus Bath and M. album BG8) and α-Proteobacteria (M. trichosporium OB3b) methanotrophs. Addition of 10 mM ClO−2 to DPRB resulted in only 40–60% recovery as O2 and Cl− (Figure 3). This may be attributed to a toxic effect of elevated ClO−2 or bleaching of the Cld enzyme (Streit and DuBois, 2008). Nonetheless, much of the available O2 was freely released during the reaction of ClO−2 with D. agitata CKB, consistent with localization of Cld in the periplasm of DPRB (O'Connor and Coates, 2002). We demonstrated that direct addition of 5 or 10 mM ClO−2 to mixed cultures of DPRB and methanotrophs did not inhibit the methanotrophs. We also showed that direct contact between the cells was not required as CH4 oxidation also occurred when the cells were contained in separate compartments under a common headspace. We further showed that 14CH4 was quantitatively oxidized to 14CO2 by the methanotrophs in culture. Our conclusion is that methane oxidizers utilized O2 provided by the dismutation of ClO−2 by DPRB.
We were unable to link methane oxidation to perchlorate or chlorate reduction. Small amounts of oxygen were produced when cultures of D. agitata CKB were amended with ClO−4 or ClO−3 (Figure 2), however methane was not consumed during co-culturing with methanotrophs (Figures 4, 6). This was previously observed for benzene and naphthalene (Coates et al., 1999b; Coates and Achenbach, 2006) and may be explained by O2 scavenging attributable to other processes, including activation of a terminal oxidase during (per)chlorate reduction (Rikken et al., 1996). It is also possible that slower kinetics of ClO−4 and ClO−3 reduction limits the production and subsequent dismutation of ClO−2 and therefore release of O2. Relief of this bottleneck could lead to more O2 being available to aerobic methanotrophs and stimulation of the unique process described herein.
In-situ oxidation of CH4 using O2 derived from chlorite dismutation may be useful in removing elevated levels of CH4 in subsurface environments. Chlorite is 104 times more soluble in water than O2 and could be easily and safely directed to the anaerobic zone (where methane may be present) during bioremediation. One example is enhanced oxidation of landfill methane without the use of forced air, reducing the risk of fire and explosion. The ability of methanotrophs and DPRB to function in separate compartments under a common headspace could be exploited at distal stages of oil development, for instance at well heads where unusable CH4 is typically flared off to reduce transportation costs or risk. In addition, production of CO2 formed during oxidation of CH4 may be viewed similarly to injected CO2 in efforts to dissolve and flush oil from developed petroleum reservoirs (Blunt et al., 1993). Further, bioclogging by cells and biocementation resulting from carbonate precipitation may be enhanced by the growth and activity of microbes capable of linking methane oxidation with (per)chlorate reduction. Bioclogging and biocementation are features of microbial enhanced hydrocarbon recovery most likely to be exploited by geotechnologists to direct hydrocarbon flow into more permeable substrates in order to enhance recovery. However, it must be noted that we observed no methane oxidation in water saturated soils exposed to both CH4 and ClO−2 (Figure 6) indicating that O2 derived from chlorite dismutation may face transport limitations in saturated anaerobic environments and may be consumed before reaching nearby CH4. Even under our unsaturated experimental conditions where liquid cultures and soil were segregated under a common headspace (Figure 7) the amount of soil present in each microcosm had to be optimized in order to balance methanotrophy with other soil O2 utilizing processes.
Oxidation of CH4 by the mechanism identified here may reduce the greenhouse impact of fugitive gases during hydrocarbon reservoir development and recovery because the global warming potential of CO2 is 25 times lower than CH4 on a 100 year time scale (IPCC 5th assessment, 2013). In addition, intermediates along the pathway of aerobic methane oxidation (e.g., methanol, formaldehyde, and formate) are themselves quite useful as chemical feedstocks for a myriad of industrial applications including biofuel production.
The Guest Associate Editor Hans Carlson declares that, despite having collaborated with author John Coates, the review process was handled objectively. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Jeremy Semrau and Jeongdai Im for supplying methanotrophic cultures. Nona Chiariello and Philippe Cohen provide access to Jasper Ridge Biological Preserve and Searsville Lake. Stacy Bennett assisted in the lab. Financial support was provided by USGS National Research Program and NASA Exobiology and Evolutionary Biology Program.
Blunt, M., Fayers, J. F., and Orr, F. M. Jr. (1993). Carbon dioxide in enhanced oil recovery. Energy Convers. Manag. 34, 1197–1204. doi: 10.1016/0196-8904(93)90069-M
Boetius, A., Ravenschlag, K., Schubert, C. J., Rickert, D., Widdel, F., Gieseke, A., et al. (2000). A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407, 623–626. doi: 10.1038/35036572
Carlström, C. I., Wang, O., Melnyk, R. A., Bauer, S., Lee, J., Engelbrekston, A., et al. (2013). Physiology and genomic description of the novel marine dissimilatory perchlorate reducing bacterium Arcobacter sp. strain CAB. MBio 4:e00217–e00213. doi: 10.1128/mBio.00217-13
Cicerone, R. J., and Oremland, R. S. (1988). Biogeochemical aspects of atmospheric methane. Global Biogeochem. Cycles 4, 299–327. doi: 10.1029/GB002i004p00299
Coates, J. D., and Achenbach, L. A. (2004). Microbial perchlorate reduction: rocket fueled metabolism. Nat. Rev. Microbiol. 2, 569–580. doi: 10.1038/nrmicro926
Coates, J. D., and Achenbach, L. A. (2006). “The microbiology of perchlorate reduction and its bioremediative application,” in Perchlorate: Environmental Occurrence, Interactions and Treatment, eds B. Gu and J. D. Coates (New York, NY: Springer), 279–295. doi: 10.1007/0-387-31113-0_12
Coates, J. D., Bruce, R. A., and Haddock, J. D. (1998). Anoxic bioremediation of hydrocarbons. Nature 396:730. doi: 10.1038/25470
Coates, J. D., Bruce, R. A., Patrick, J. A., and Achenbach, L. A. (1999a). Hydrocarbon bioremediative potential of (per)chlorate-reducing bacteria. Bioremed. J. 3, 323–334. doi: 10.1080/10889869991219415
Coates, J. D., Michaelidou, U., Bruce, R. A., O'Connor, S. M., Crespi, J. N., and Achenbach, L. A. (1999b). The ubiquity and diversity of dissimilatory (per)chlorate-reducing bacteria. Appl. Environ. Microbiol. 65, 5234–5241.
Culbertson, C. W., Zehnder, A. J. B., and Oremland, R. S. (1981). Anaerobic oxidation of acetylene by estuarine sediments and enrichment cultures. Appl. Environ. Microbiol. 41, 396–407.
Ettwig, K. F., Butler, M. K., Le Paslier, D., Pelletier, E., Mangenot, S., Kuypers, M. M., et al. (2010). Nitrite-driven anaerobic oxidation of methane by oxygenic bacteria. Nature 464, 543–548. doi: 10.1038/nature08883
Ettwig, K. F., Speth, D. R., Reimann, J., Wu, M. L., Jetten, M. S. M., and Keltjens, J. T. (2012). Bacterial oxygen production in the dark. Front. Microbiol. 3:273. doi: 10.3389/fmicb.2012.00273
Gieg, L. M., Duncan, K. E., and Suflita, J. M. (2008). Bioenergy production via microbial conversion of residual oil to natural gas. Appl. Environ. Microbiol. 74, 3022–3029. doi: 10.1128/AEM.00119-08
Gieg, L. M., Jack, T. R., and Foight, J. M. (2011). Biological souring and mitigation in oil reservoirs. Appl. Microbiol. Biotechnol. 92, 263–282. doi: 10.1007/s00253-011-3542-6
Goblirsch, B. R., Kurker, R. C., Streit, B. R., Wilmot, C. M., and DuBois, J. L. (2011). Chlorite dimutases, DyPs, and EfeB: 3 microbial heme enzyme families comprise the CDE structural superfamily. J. Mol. Biol. 408, 379–398. doi: 10.1016/j.jmb.2011.02.047
Goblirsch, B. R., Streit, B. R., DuBois, J. L., and Wilmot, C. M. (2010). Structural features promoting dioxygen production by Dechloromonas aromatica chlorite dismutase. J. Biol. Inorg. Chem. 15, 879–888. doi: 10.1007/s00775-010-0651-0
Hinrichs, K. U., Hayes, J. M., Sylva, S. P., Brewer, P. G., and DeLong, E. F. (1999). Methane-consuming archaebacteria in marine sediments. Nature 398, 802–805. doi: 10.1038/19751
Hobbie, J. E., Daley, R. L., and Jaspar, S. (1977). Use of Nuclepore filters for counting bacteria for fluorescence microscopy. Appl. Environ. Microbiol. 33, 1225–1228.
Hoeft, S. E., Kulp, T. R., Stolz, J. F., Hollibaugh, J. T., and Oremland, R. S. (2004). Dissimilatory arsenate reduction with sulfide as electron donor: experiments with Mono Lake water and isolation of strain MLMS-1, a chemoautotrophic arsenate respirer. Appl. Environ. Microbiol. 70, 2741–2747. doi: 10.1128/AEM.70.5.2741-2747.2004
IPCC Working group 1 contribution to the 5th assessment report. (2013). Climate Change 2013: The Physical Science Basis. Stockholm.
Jenneman, G. E., Knapp, R. M., McInerney, M. J., Menzie, D. E., and Revus, D. E. (1984). Experimental studies of in-situ microbial enhanced oil recovery. Soc. Petrol. Eng. J. 24, 35–37. doi: 10.2118/10789-PA
Jones, D. M., Head, I. M., Gray, N. D., Adams, J. J., Rowan, A. K., Aitken, C. M., et al. (2007). Crude-oil biodegradation via methanogenesis in subsurface petroleum reservoirs. Nature 451, 176–181. doi: 10.1038/nature06484
Kostan, J., Sjöblom, B., Maixner, F., Mlynek, G., Furtmüller, P. G., Obinger, C., et al. (2010). Structural and functional characterisation of the chlorite dismutase from the nitrite-oxidizing bacterium “Candidatus Nitrospira defluvii”: identification of a catalytically important amino acid residue. J. Struct. Biol. 172, 331–342. doi: 10.1016/j.jsb.2010.06.014
Kounaves, S. P., Stroble, S. T., Anderson, R. M., Moore, Q., Catling, D. C., Douglas, S., et al. (2010). Discovery of natural perchlorate in the Antarctic dry valleys and its global implications. Environ. Sci. Technol. 44, 2360–2364. doi: 10.1021/es9033606
Lazar, I., Petrisor, I. G., and Yen, T. F. (2007). Microbial Enhanced Oil Recovery. Petrol. Sci. Technol. 25, 1353–1366. doi: 10.1080/10916460701287714
Lee, A. Q., Streit, B. R., Zdilla, M. J., Abu-Omar, M. M., and DuBois, J. L. (2008). Mechanism of and exquisite selectivity for O-O bond formation by the heme-dependent chlorite dismutase. Proc. Nat. Acad. Sci. U.S.A. 105, 15654–15659. doi: 10.1073/pnas.0804279105
Martini, A. M., Budai, J. M., Walter, L. M., and Schoell, M. (1996). Microbial generation of economic accumulations of methane within a shallow organic-rich shale. Nature 383, 155–158. doi: 10.1038/383155a0
McCord, J. M., Keele, B. B. Jr., and Fridovich, I. (1971). An enzyme-based theory of obligate anaerobiosis: the physiological function of superoxide dismutase. Proc. Natl. Acad. Sci. U.S.A. 68, 1024–1027. doi: 10.1073/pnas.68.5.1024
Mehboob, F., Wolterink, A. F. M., Vermeulen, A. J., Jiang, B., Hagedoorn, P.-L., Stams, A. J. M., et al. (2009). Purification and characterization of a chlorite dismutase from Pseudomonas chloritidismutans. FEMS Microbiol. Lett. 293, 115–121. doi: 10.1111/j.1574-6968.2009.01517.x
Miller, L. G., Baesman, S. M., Kirshtein, J., Voytek, M. A., and Oremland, R. S. (2013). A biogeochemical and genetic survey of acetylene fermentation by environmental samples and bacterial isolates. Geomicrobiol. J. 30, 501–516. doi: 10.1080/01490451.2012.732662
Miller, L. G., Baesman, S. M., and Oremland, R. S. (2003). Bioreactors for removing methyl bromide following contained fumigations. Environ. Sci. Technol. 37, 1698–1704. doi: 10.1021/es026155j
Milucka, J., Ferdelman, T. G., Polerecky, L., Franzke, D., Wegener, G., Schmid, M., et al. (2012). Zero-valent sulfur is a key intermediate in marine methane oxidation. Nature 491, 541–546. doi: 10.1038/nature11656
Mlynek, G., Sjöblom, B., Kostan, J., Füreder, S., Maixner, F., Gysel, K., et al. (2011). Unexpected diversity of chlorite dismutases: a catalytically efficient dimeric enzyme from Nitrobacter winogradskyi. J. Bacteriol. 193, 2408–2417. doi: 10.1128/JB.01262-10
O'Connor, S. M., and Coates, J. D. (2002). Universal immunoprobe for (per)chlorate-reducing bacteria. Appl. Environ. Microbiol. 68, 3108–3113. doi: 10.1128/AEM.68.6.3108-3113.2002
Oremland, R. S., and Culbertson, C. W. (1992). Importance of methane oxidizing bacteria in the methane budget as revealed by the use of a specific inhibitor. Nature 356, 421–423. doi: 10.1038/356421a0
Rao, B., Anderson, T. A., Orris, G. J., Rainwater, K. A., Rajagopalan, S., Sandvig, R. M., et al. (2007). Widespread natural perchlorate in unsaturated zones of the southwest United States. Environ. Sci. Techol. 441, 4522–4528. doi: 10.1021/es062853i
Rikken, G. B., Kroon, A. G. M., and van Ginkel, C. G. (1996). Transformation of (per)chlorate into chloride by a newly isolated bacterium: reduction and dismutation. Appl. Microbiol. Biotechnol. 45, 420–426. doi: 10.1007/s002530050707
Streit, B. R., and DuBois, J. L. (2008). Chemical and steady-state kinetic analysis of a heterologously expressed heme enzyme dependent chlorite dismutase. Biochemistry 47, 5271–5280. doi: 10.1021/bi800163x
Sun, W., Sierra-Alvarez, R., Milner, L., and Field, J. A. (2010). Anaerobic oxidation of arsenite linked to chlorate reduction. Appl. Environ. Microbiol. 76, 6804–6811. doi: 10.1128/AEM.00734-10
Sun, Y., Gustavson, R. L., Ali, N., Weber, K. A., Westphal, L. L., and Coates, J. D. (2009). Behavioral response of dissimilatory perchlorate-reducing bacteria to different electron acceptors. Appl. Microbiol. Biotehnol. 84, 955–963. doi: 10.1007/s00253-009-2051-3
Urbansky, E. T. (2002). Perchlorate as an environmental contaminant. Environ. Sci. Poll. Res. 9, 187–192. doi: 10.1007/BF02987487
Weelink, S. A. B., Tan, N. C. G., ten Boreke, H., van Doesburg, W., Langenhoff, A. A. M., Gerritse, J., et al. (2007). Physiological and phylogenetic characterization of a stable benzene-degrading, chlorate-reducing microbial community. FEMS Microbiol. Ecol. 60, 312–321. doi: 10.1111/j.1574-6941.2007.00289.x
Whittenbury, R., Phillips, K. C., and Wilkinson, J. F. (1970). Enrichment, isolation and some properties of methane-utilizing bacteria. J. Gen. Microbiol. 61, 205–218. doi: 10.1099/00221287-61-2-205
Youseff, N., Elshahed, M. S., and McInerney, M. J. (2009). “Microbial processes in oil fields: culprits, problems, and opportunities,” in Advances in Applied Microbiology, ed A. I. Laskin, S. Sariaslani, and G. M. Gadd (Burlington: Academic Press, Elsevier Inc.) 141–251.
Keywords: chlorite, perchlorate, chlorate, methane, oxidation
Citation: Miller LG, Baesman SM, Carlström CI, Coates JD and Oremland RS (2014) Methane oxidation linked to chlorite dismutation. Front. Microbiol. 5:275. doi: 10.3389/fmicb.2014.00275
Received: 31 January 2014; Accepted: 19 May 2014;
Published online: 17 June 2014.
Edited by:
Hans Karl Carlson, University of California, Berkeley, USACopyright © 2014 Miller, Baesman, Carlström, Coates and Oremland. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Laurence G. Miller, United States Geological Survey, MS/480, 345 Middlefield Rd., Menlo Park, CA 94025, USA e-mail:bGdtaWxsZXJAdXNncy5nb3Y=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.