- 1Plant Research International, Wageningen University and Research Centre, Wageningen, Netherlands
- 2Applied Plant Research, Wageningen University and Research Centre, Lisse, Netherlands
- 3Food and Biobased Research, Wageningen University and Research Centre, Wageningen, Netherlands
- 4Central Veterinary Institute, Wageningen University and Research Centre, Lelystad, Netherlands
Disease incidences related to Escherichia coli and Salmonella enterica infections by consumption of (fresh) vegetables, sprouts, and occasionally fruits made clear that these pathogens are not only transmitted to humans via the “classical” routes of meat, eggs, and dairy products, but also can be transmitted to humans via plants or products derived from plants. Nowadays, it is of major concern that these human pathogens, especially the ones belonging to the taxonomical family of Enterobacteriaceae, become adapted to environmental habitats without losing their virulence to humans. Adaptation to the plant environment would lead to longer persistence in plants, increasing their chances on transmission to humans via consumption of plant-derived food. One of the mechanisms of adaptation to the plant environment in human pathogens, proposed in this paper, is horizontal transfer of genes from different microbial communities present in the arable ecosystem, like the ones originating from soil, animal digestive track systems (manure), water and plants themselves. Genes that would confer better adaptation to the phytosphere might be genes involved in plant colonization, stress resistance and nutrient acquisition and utilization. Because human pathogenic enterics often were prone to genetic exchanges via phages and conjugative plasmids, it was postulated that these genetic elements may be hold key responsible for horizontal gene transfers between human pathogens and indigenous microbes in agroproduction systems. In analogy to zoonosis, we coin the term phytonosis for a human pathogen that is transmitted via plants and not exclusively via animals.
Introduction
The Plant Environment as a Habitat for Human Pathogens
Agricultural plants have become a source for human pathogens, especially the emerging ones belonging to the group of Shiga toxin-producing Escherichia coli (STEC) strains (Feng and Reddy, 2013). The threat of human pathogens in freshly consumable products of plant origin became apparent during the outbreak caused by Escherichia coli O104:H4 in Germany and France in 2011, where almost 4000 persons became infected leading to 54 casualties and over 900 incidences of hemolytic uremic syndrome (HUS; Karch et al., 2011; Beutin and Martin, 2012). The most likely transmission route of the pathogen to consumable products was remarkable as the source was fenugreek seeds that were transported from Egypt to Rotterdam harbor, the Netherlands, approximately 17 months before appearance of the first disease incidences in Hamburg and surrounding area (Karch et al., 2011). Although the pathogen was neither found in Fenugreek sprouts, nor in the seeds themselves, epidemiological facts revealed that the pathogen must have been associated with seeds over a relatively long period of time. The questions rise on how a human pathogen can persist as a viable entity in a hostile environment for such a long period and why similar observations had not been made before. Must this be considered as the first incidence of a disease outbreak caused by a human pathogenic bacterium that was adapted to the plant environment?
To address this question, it must be referred to the nature of the causative agent that is atypical for pathogenic Escherichia coli strains commonly occurring in Europe and the USA (Scheutz et al., 2011; Beutin and Martin, 2012). The Escherichia coli O104: H4 outbreak strain was an entero-aggregative Escherichia coli strain that does not have animals, which is often the case for other Escherichia coli O type strains, but instead only humans as major reservoir (Wieler et al., 2011). Outbreaks caused by this type of pathogen are rare in Western societies, whereas those caused by Escherichia coli O157:H7 and Salmonella enterica are more common (Table 1). This indicates that particular features in these human pathogens already exist, extending their life-time in the plant environment. The question is whether these features were intrinsic to particular subsets of human pathogens or were recently gained, e.g., via horizontal gene transfer. The strain causing the outbreak in the Hamburg area must be considered as a highly virulent pathogen and it must have acquired its virulence and antibiotic resistance (extended spectrum beta-lactamase, ESBL) traits via horizontal gene transfer events like transduction (phage infection) and conjugation (plasmid transfer; Mellmann et al., 2011; Muniesa et al., 2012). Acquisition of new virulence traits is one aspect in the evolution of a new pathogen, but selection pressure is another, and the outbreak strain must have been evolved by increasing its virulence in humans side-by-side with improvement of its ecological competence in plants. The threat of this development is the emergence of new types of highly virulent human bacterial pathogens that are fully adapted to life near, or may be inside agricultural plants.
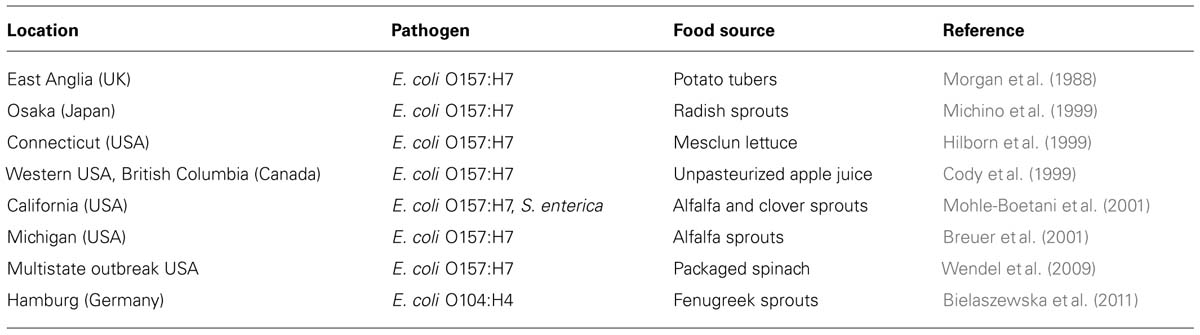
TABLE 1. Examples of large disease outbreaks resulting from contamination of vegetables and sprouts with human pathogenic bacteria.
Bacteria Associated with Multiple Habitats
The agricultural plant environment is an environment where microbial communities of at least four different ecosystems may come together, namely those from soil, plants, farm animal digestive track systems (manure), and fresh water sources (irrigation). Agricultural plant production thus must be considered as a cross road of communities originating from these different sources. The microbial community compositions in these systems substantially differ from each other, but representatives of the different communities may, at least temporarily, coexist with micro-organisms typically associated with plants. However, much is still unknown about the microbial composition and functioning of these microbes in the different environments. Some remarkable correspondence in the taxonomy of human disease-causing bacteria can be found with typical soil/rhizosphere bacterial species. Clearest examples are the so called cross-domain pathogens belonging to the genera of Burkhoderia (e.g., Burkhoderia cepacia) and Pseudomonas (e.g., Pseudomonas aeruginosa; Barak and Schroeder, 2012; Kumar et al., 2013). These are opportunistic pathogens in humans and are commonly found in plant and soil habitats. Distinction of pathogenic from non-pathogenic types (of which some can be excellent plant growth promoters) of both genera is often difficult to accomplish. Pseudomonas fluorescens is a bacterial species that is commonly found near plant roots, however, representatives of this species were also shown to be associated with patients suffering from Crohn’s disease (Wei et al., 2002; Eckburg and Relman, 2007). A representative of Pseudomonas veronii, another typical rhizosphere-borne bacterial species was found to be associated with human intestinal inflammatory pseudotumour formation (Cheuk et al., 2000). Typical plant pathogens belonging to the genus of Pantoea (Pantoea ananatis and P. agglomerans) and Erwinia (Erwinia tasmaniensis) were also demonstrated to cause diseases in humans (Cruz et al., 2007; Shin et al., 2008; Coutinho and Venter, 2009). Taxonomical commonalities exist between human pathogens and species known to thrive in soil habitats. Recently, a new taxa belonging to a hitherto uncultured group of bacteria named Verrucomicrobia subdivision 1 was shown to be competent in the rhizospheres of different plant species (Nunes da Rocha et al., 2011), whereas a representative of the same group (Akkermansia muciniphila) had been found as a commensal species living in the intestinal track system of humans (Derrien et al., 2004), where it deemed to play a role in degradation of mucus and health support of humans. This illustrates that the taxonomical diversity of cross domain species are high and up to date still rather unexplored. Important proposition in this paper is that the chance on genetic exchanges via horizontal transfer will be highest between cross-domain species commonly occurring in different ecosystems and coming into physical contact with each other near or within plants. Human pathogens may acquire genes from plant-associated bacteria (Szmolka and Nagy, 2013), leading to new phenotypes of increased persistence in plants or their propagative materials, higher resistance to stresses or broader spectra in acquisition or utilization of available nutrients. These traits will make human pathogens fitter for survival in the plant environment.
Especially, the rhizosphere and phyllosphere of plants must be considered as hot spots of microbial life because of the nutrients leaking from growing roots and leaves. Plant surfaces are important reservoirs for (enteric) opportunistic human pathogens (Berg et al., 2005; Holden et al., 2009). Escherichia coli belong to the Enterobacteriaceae, and species belonging to this taxonomical class can be found frequently as mutualistic, commensal, or plant pathogenic species in plants. Species belonging to the genera of Enterobacter, Serratia, Klebsiella, Erwinia, Pantoea, and Pectobacterium are typical plant-associated bacteria and if gene transfer events with plant-invasive human pathogens, like ones belonging to Escherichia coli and S. enterica, will occur then expectedly it will be highest with these species. Prophages and plasmids in plant-associated Enterobacteriaceae are extrachromosomal gene pools that, thus far, remained inaccessible for human pathogens. On the other hand harmless bacteria living as commensals in plant environments may pick up mobile elements from plant-invasive human pathogenic species to become human pathogens (Szmolka and Nagy, 2013). The crucial difference between a relatively harmless bacterium and a dangerous human pathogen can rely on presence or absence of a single mobile genetic element (Frost et al., 2005). It, thus, might be possible that harmless bacterial species living as commensals in plants acquire traits making them virulent to humans although these type of incidences has not been reported to occur in plants to date.
Practices common in agriculture make it possible to bring gene pools from different ecosystems together and making these accessible for commensal and human pathogenic species, leading to acquisition of new traits and possibly to emergence of new human pathogens (Figure 1). Phytonoses (singular phytonosis), occurring from human infecting agents transmitted by plants (van der Riet, 1997), in analogy to zoonoses, which are diseases transmitted by animals, can be coined as a new term for the group of diseases caused by human pathogens, viz. Escherichia coli and S. enterica, that are transmitted via consumption of fresh produce.
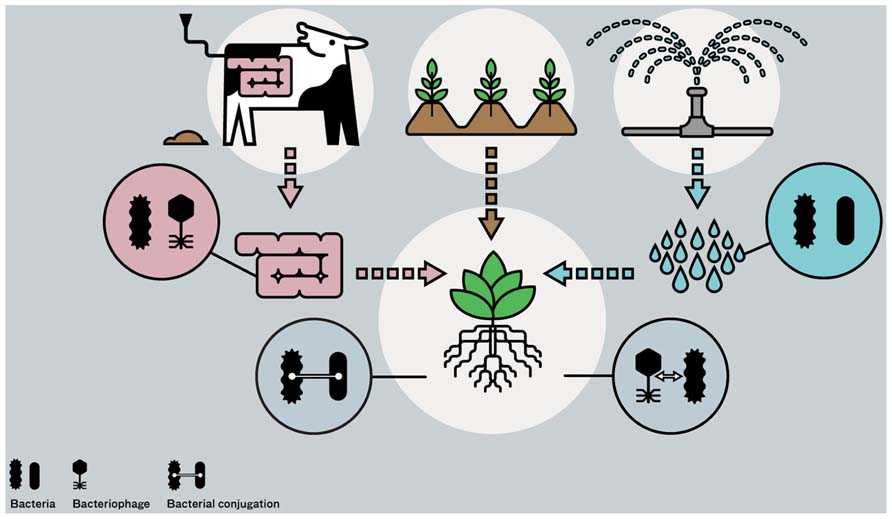
FIGURE 1. Bacterial communities from different habitats, i.e., from cattle intestinal track system via manure, surface water via irrigation, and from soil and plants, all coming together at plant growth in arable production systems. Four magnifications in the figure depict origin of different bacterial groups and their phages, and the possible occurrences of gene transfer near or inside plants, either via conjugation or via transduction.
The Arable Plant Environment as Cross Road of Different Ecosystems
The Animal Digestive Track System as Major Reservoir for Escherichia coli O157:H7 and Other Human Pathogenic Enterics
Shiga toxin-producing Escherichia coli serotypes, the most commonly found human pathogens in plant-derived products, pose serious threats to human health. The question often raised in different studies is why these strains, and especially the ones of Escherichia coli O157:H7, are so often associated with plants (Table 1). Escherichia coli O157:H7 live as commensal species in the digestive track system of ruminants without causing any visible symptoms to the host (Wells et al., 1991; Chapman et al., 1993). The bacterium is excreted via feces and can survive in manure for over a long period of time. The most important STEC serotypes causing disease in humans are O157:H7, O26:H11, O103:H2, O145:H28, and O111:H8 (Pearce et al., 2004, 2006). In North America, Japan and Europe, O157:H7 is the type causing most disease incidences in humans and this is the reason why most research is focussed on this serotype. However, other serotypes like O26, O103, and O145 were regularly found in cattle from Scotland and USA (Wells et al., 1991; Wieler et al., 1996; Pearce et al., 2004, 2006; Shaw et al., 2004). Shiga toxin-producing O104:H4 or other entero-aggregative Escherichia coli strains were not found in cattle manure from Northern Germany during the Hamburg outbreak in 2011, indicating that cattle is not a major reservoir for the entero-aggregative Escherichia coli pathotype, to which the Escherichia coli O104:H4 outbreak strain belong to (Wieler et al., 2011).
The occurrence of Escherichia coli O157:H7 is more common in the intestinal track systems of cattle than in that of other ruminants and farm animals (Karmali et al., 2010; Ferens and Hovde, 2011 and references therein). The distribution of the pathogen in cattle feces is heterogeneous and occasionally numbers can be very high in excrements of particular individuals within the herd, so called super shedders (Ferens and Hovde, 2011). Epidemiological research on Escherichia coli O157:H7 infections in livestock in slaughterhouses in the Netherlands revealed prevalence of 10.6% (n = 540) in adult cattle, 0.5% (n = 397) in veal calves, 3.8% (n = 52) in sheep, 4.1% (n = 49) in lambs, 1.4% (n = 145) in pigs, 0% (n = 501) in chicken and 1.3% (n = 459) in turkey (Heuvelink et al., 1998a, 1999). Highest incidences in serotype O157:H7 contamination was thus found in adult cattle, but this serotype was also present in carcasses of other animals, with the exception of chicken. Examination of 10 dairy farms in the Netherlands, of which five were demonstrated to be serotype O157:H7 positive in their cattle, resulted in seven farms where at least one serotype O157:H7-positive individual was identified to be present (Heuvelink et al., 1998a). Within-herd prevalence on these seven farms varied between 0.8 and 22.4%. Surveillance of Escherichia coli O157:H7 in pooled fecal samples over 1051 dairy herds in the years 1997–2005 revealed on average prevalence of 8.0% (variation between 6.4 and 9.6%; Berends et al., 2008). In the same study surveillance of two distinct types of veal herds (so called pink and white veal herds) revealed clear differences between both types, i.e., 107 positive of 269 examined pink herds (39.8%) and 10 positive of 661 examined white herds (1.5%; Berends et al., 2008). The reason for this difference in infection prevalence was not further investigated, but may be related to difference in age before slaughtering, which is 35 weeks for pink veal and 25 weeks for white veal. Interestingly, screening of Dutch dairy farms on the basis of presence of Shiga toxin genes revealed much higher prevalence (80%) than via screening on serotype O157:H7 determinants (Franz et al., 2007). This would indicate that Shiga toxin genes, which are considered to be major virulence determinants in enterohemorrhagic Escherichia coli (EHEC), are not only present in serotype O157:H7, but can also be present in other serotypes as well. One important observation in the longitudinal study done in the Netherlands was the fact that the number of serotype O157:H7 in positive cattle herds shifted during the season showing maximum peaks in the second half of the summer period, whereas this serotype was undetectable in samples collected in wintertime (Heuvelink et al., 1998b; Bouwknegt et al., 2004; Schouten et al., 2004, 2005; Valkenburgh et al., 2007). Such a fluctuation during the season was also observed in other countries (Hancock et al., 1994; Chapman et al., 1997; Conedera et al., 2001; Barkocy-Gallagher et al., 2003; Milnes et al., 2009). This would indicate that fluctuations in serotype O157:H7, and possibly also in other serotypes, exist in cattle manure, which has a consequence in the control of serotype O157:H7 contamination of vegetable plants grown in manure-amended soils. Depending on the period in the season, contamination with pathogenic Escherichia coli serotypes expectedly will vary.
It was hypothesized that Escherichia coli O157:H7 is better adapted to circumstances prevailing in habitats outside of the digestive track of cattle than other non-Shiga-toxin producing Escherichia coli serotypes (Durso et al., 2004). This study revealed that a higher incidence of competitive strains were found among Escherichia coli O157:H7 than over non-Shiga toxin producing serotypes, but the non-Shiga toxin types were able to utilize a broader spectrum of tested substrates than Escherichia coli O157:H7. There were no other factors that could explain better fitness of Escherichia coli O157:H7 over non-Shiga toxin producing serotypes in alternative habitats, in spite of their relatively larger genome sizes that might be indicative for better adaptation to multiple habitats. Other, unknown factors therefore must be held responsible for survival of Escherichia coli O157:H7 outside the intestinal track systems of cattle.
The most likely route of contamination from cattle to plants is via manure through soil to plant roots, but other routes may exist like irrigation water (Erickson et al., 2010), flies (Talley et al., 2009) and equipment used by field workers (Johnston et al., 2009). Different transmission routes of Escherichia coli O157:H7 to arable plants may exist and remarkable is the rather long-term persistence of these strains in plant and soil ecosystems. It seems that at least some of the STEC strains and strains of other human pathogens like Salmonella, Campylobacter, and Listeria are better adapted to the circumstances prevailing in plants and soils than ever thought before.
Transmission Routes of Human Pathogens in Plant Production Fields
One of the first reported disease outbreaks caused by Escherichia coli O157:H7 and associated with consumption of vegetables was from potatoes in the UK (Morgan et al., 1988; Table 1). In this particular case, it was potato tubers that were suspected to be contaminated with Escherichia coli O157:H7 most likely originating from cattle manure and applied at potato production in the field. Because potatoes are not eaten raw, it was postulated that the transmission route of the pathogen was not via potato consumption itself, but rather from hand-to-mouth transfer at food preparation and handling of potatoes. From this case it became clear that the pathogen was able to persist over longer periods of time in soil near or at the surface of potato tubers. Potato lots containing the pathogen were not traceable anymore, so it is unknown at which densities and locations on, or may be inside tubers the human pathogen was present (Morgan et al., 1988). Concerning internalization, invasiveness of the pathogen would depend on specific enzymes like endoglucanases to create openings in plant cells, like plant pathogens do, or they would enter plants via natural openings (stomata and hydathodes) or woundings caused by feeding insects or nematodes or via co-infections with plant pathogens or soil-indigenous micro-organisms. The underground route of Escherichia coli O157:H7 and other human pathogenic enterics to plants will be discussed below as it is often hypothesized, but not always accepted as a realistic transmission route to plants.
Survival of human pathogens in manure-amended soils
The underground route of transmission of Escherichia coli O157:H7 to plants can be plausible if the survival time of the pathogen in manure-amended soil is long enough to allow successful colonization of plants. Under experimental circumstances, Escherichia coli O157:H7, introduced into autoclaved soils and mixed at different ratios with manure, was able to survive for more than 226 days at 15 and 21°C (Jiang et al., 2002). Lowering the temperature to 5°C led to a reduction in survival time to down to 35 days. Introduction into non-autoclaved manure-amended soil reduced survival to a maximal time of 193 days at 15 and 21°C, but there was no reduction in survival time at 5°C. This indicates that temperature is an important factor directly affecting survival time of the pathogen in soil, but also indirectly by influencing the activities of microbial populations resident in soil that antagonizes the invasive human pathogen. Higher average temperatures and higher oscillation amplitudes resulted in faster decline of both populations (Semenov et al., 2007). However, lower temperatures, between 10 and 15°C, commonly occur in temperate regions where incidences with Escherichia coli O157:H7 outbreaks are highest. Soil horizons where plant roots grow are often more constant in temperature than topsoil horizons and therefore, invasion of human pathogens to deeper soil layers would favor longer-term persistence in soil and thus increasing their chances on contact with plant roots. The effect of manure type and application to soil on percolation to deeper soil layers of the same Escherichia coli O157:H7 and S. enterica serovar Typhimurium strains were investigated in another study (Semenov et al., 2009). Both strains percolated to deeper layers (maximal studied depth was 40 cm) within a few hours after introduction, independent of manure type, and application of slurry resulted in percolation of both pathogens to greater depths than of solid manure, whereas the mode of application (spreading over soil surface versus injection into topsoil) had smaller effects on percolation depths of both pathogens. Percolation to deeper soil layers in fields may have two consequences for transmission to crop plants: one, that survival time may be increased because of the lower and more constant temperature present in deeper soil layers, and two, ground water may become contaminated which can be used for irrigation of crop plants (Brennan et al., 2010).
Available nutrients in manure and soil also were demonstrated to play important roles in survival of Escherichia coli O157:H7 and S. enterica serovar Typhimurium strains (Franz and Van Bruggen, 2008). Survival of Escherichia coli O157:H7 across 36 different manure-amended soils was investigated and it was shown that survival increased when slurry or chemical fertilizer (relatively rich in available C and/or N) were applied to soils in comparison with applications of farmyard manure or compost (relatively poor in available C and N). Therefore, it was concluded that circumstances characterized to be oligotrophic for bacteria decreased Escherichia coli O157:H7 survival in manure-amended soils. Manure type itself is also another factor influencing survival of human pathogens in manure-amended soils. Manure type depends on the diets that cows had received and the effect of roughage of the diet on the fate of introduced Escherichia coli O157:H7 and S. enterica serovar Typhimurium strains in manure was investigated (Franz et al., 2005). Population decline of introduced Escherichia coli O157:H7 was fastest in manure from cows fed with the roughest diet type (straw). The effect of roughage of the diet on S. enterica serovar Typhimurium survival in manure was less clear than for Escherichia coli O157:H7, although decline rates of both pathogens were fastest in manures of cows fed with the roughest diet. Fiber content and pH of manure were the best explanatory factors for decline rates of both pathogens.
The nutrient status in manure-amended soils is a factor that plays a direct role in survival of Escherichia coli O157:H7 and other human pathogens. However, indirectly, the nutrient level also may play a role in the diversity and evenness of microbial populations that are indigenous to manure-amended soil that might compete with invasive human pathogens (Semenov et al., 2008; Van Overbeek et al., 2010). Soil bacterial diversity as an experimental variable indeed was shown to be negatively correlated with longevity in survival of an introduced Escherichia coli O157:H7 strain, representing a species invasive to soil (van Elsas et al., 2012). These data all together illustrate the importance of high microbial diversity to reduce survival time of human pathogens as species invasive to soils.
Finally, survival time is dependent on strain differences, even within the same species. Remarkable differences in survival time between different Escherichia coli O157:H7 strains was observed among 18 different strains of which eight originated from animal, one from food, and nine from human sources (Franz et al., 2011). Survival time in manure-amended soils of the nine human strains were longer than of the other nine strains. Principle component analyses on metabolic profiles of all 18 strains revealed separate clustering of the human strains from the others and the ability of the nine human strains to oxidize propionic acid, α-ketobutyric acid and α-hydroxybutyric acid appeared to be discriminative for this particular environmental group. It was therefore concluded that phenotypic diversity found among different Escherichia coli O157:H7 strains may explain observed differences in survival time in manure-amended soils. Differences in survival time among different strains of Escherichia coli O157:H7 would indicate that some strains are better adapted to circumstances prevailing in natural habitats, like soils, than others. Presence or absence of important virulence factors in all 18 Escherichia coli O157:H7 strains appeared not to play any role in survival time in manure-amended soils. This is in line with observations made in a study done on different stx1, stx2, both stx and eae (intimin) gene deletion mutants made from Escherichia coli O157:H7 EDL933 strain in soils differing in texture (loamy sand, sandy loam, silty clay; Ma et al., 2011). There are thus no indications that virulence to humans has an effect on Escherichia coli O157:H7 survival time in (manure-amended) soils. However, mutation in an important gene involved in stress regulation, rpoS, had an effect on survival of Escherichia coli O157:H7, leading to reduced persistence in manure-amended soil (van Hoek et al., 2013).
Plant colonization by human pathogens
Presence of human pathogens in contaminated soil at high cell densities may lead to plant internalization. This was demonstrated in lettuce plants grown in soil amended with cow manure containing high doses (106 and 108 CFU/g soil) of a GFP-marked Escherichia coli O157:H7 strain (Solomon et al., 2002). The introduced strain was retrieved from lettuce seedlings even after 10 min treatments with HgCl2, indicating that internalization of plant tissue by the pathogen had occurred. Confocal microscopy on lettuce leafs colonized by the GFP-marked Escherichia coli O157:H7 strain revealed the presence of fluorescent aggregates in intercellular spaces. There was no evidence for intracellular colonization by Escherichia coli O157:H7, but the presence of the pathogens inside plant tissue of the edible parts of the plant already indicate that there is a risk upon consumption of fresh produce, because cells cannot easily be removed or inactivated by washing and disinfection procedures. Adherence and internalization of plant tissue are consequences when human pathogens come into contact with plants.
Longer-term survival of human pathogens in manure-amended soils ultimately will lead to lower inoculum levels in soils and thus to lower risks of crop plants to become contaminated. The question that arises is whether human pathogens are still capable to contaminate plants after longer periods of residence in soils. A mixture (1:1) of S. enterica (serovars Baildon and Enteritidis) strains was detectable to up to 6 weeks after introduction into soil (Barak and Liang, 2008). Sowing tomato seeds at weekly intervals after inoculation of this mixture revealed the presence of the pathogen mixture in the rhizoplane and phylosphere of plants for up to 7 weeks after inoculation. Inoculation of soils with Escherichia coli O157:H7 via three different composted manure types and irrigation water revealed the presence of the pathogen strain in lettuce and parsley plants for up to, respectively, 77 and 177 days after transplanting (Islam et al., 2004a). Using the same carriers, S. enterica serovar Typhimurium was detectable in radish and carrots after, respectively 84 and 203 days after sowing of seeds into soils (Islam et al., 2004b). No clear effects of the carrier types for the Escherichia coli O157:H7 and S. enterica serovar Typhimurium inoculants were found in these studies, however, longevity of survival differed among different tested plant species, indicating that there was an effect of plant type on survival of both pathogen strains. Escherichia coli O157:H7 and S. enterica serovar Typhimurium strains inoculated at levels of 107 CFU per g manure-amended soil were retrieved from the rhizosphere (Ongeng et al., 2011a) and internal compartments (Ongeng et al., 2011b) of transplanted cabbage (Brassica oleracea) plants grown under tropical field conditions. These two studies indicated that the presence of plant roots may extend survival time of human pathogens in soils and that internalization of crop plants may occur following successful rhizosphere colonization.
Long-term persistence in soil may lead to nutrient starvation of human pathogens in soil. These pathogens may gear to forms that are more resistant against stresses, although they also may become metabolically arrested, entering the so called viable-but non-culturable (VBNC) state as was demonstrated for another gammaproteobacterial species, P. fluorescens (Van Overbeek et al., 1995). Cells of the Escherichia coli O104:H4 Hamburg-outbreak strain (Table 1) imposed to copper and low temperature as stress factors became VBNC as demonstrated by viability staining of non-culturable cells (Aurass et al., 2011). However, non-culturability most likely did not affect virulence, as genes responsible for virulence appeared to remain intact. This to the contrary to a typical soil-borne plant pathogenic bacterium, Ralstonia solanacearum biovar 2, that also became VBNC upon treatment at low temperature, but these cells apparently lost their virulence upon injection into host (tomato) plants, where these cells still were capable to multiply, demonstrating that they were resuscitated from the VBNC state (van Overbeek et al., 2004). Exposure of S. enterica serovar Typhimurium LT2 to cold stress (5 h at 5°C) resulted in higher resistance to acid (pH 4.0) stress (Shah et al., 2013). Persistence of S. enterica serovar Typhimurium DT104 in soil and lettuce plants resulted in better survival in simulated gastric fluid than cells freshly grown in culture, however, these cells were less capable to attach and invade epithelial cells (Oliveira et al., 2011). Observed effects slightly differed between the two studied strains (one originating from pig carcass and the other from lettuce plants), but illustrates that physiological aspects in cells of human pathogens can play a role in adaptation to circumstances prevailing in the phytosphere, possibly leading to increased transmission to humans upon consumption.
Presence of plant roots in soil play an important role in the entrance of human pathogens into plants. During root colonization, human pathogens must successfully compete with rhizosphere-indigenous micro-organisms for available nutrients. Iron is an essential element for bacteria and a limiting factor for growth of bacteria in the rhizosphere. Mutations in a precursor in siderophore production, or in siderophore production itself, in S. enterica serovar Typhimurium resulted in a significant lower colonization of alfalfa roots (Hao et al., 2012). Siderophore production in S. enterica is essential for root colonization, as it is typical for rhizosphere bacteria like P. fluorescens and R. solanacearum. Siderophore production is a colonization fitness factor for S. enterica (Hao et al., 2012) and this would indicate that the pathogen is well-adapted to circumstances prevailing in the rhizosphere.
However, human pathogens contaminating vegetable plants do not necessarily originate from soil, but may originate from seeds, as seeds were supposed to be the main contamination route for the Escherichia coli O104:H4 outbreak strain to fenugreek sprouts at the Hamburg outbreak incidence in 2011. S. enterica and Escherichia coli O157:H7 were both shown to persist on lettuce seeds for 2 years and still capable to colonize young lettuce plants (van der Linden et al., 2013). Of both human pathogens, S. enterica was shown to be the best survivor on lettuce seeds. These examples show that contamination of arable plants can occur via different routes and that long-term persistence in soil or on dry seeds both can be important factors in successful colonization of plants.
Proposed mechanisms of plant internalization by human pathogens
Internalization of plants by human pathogens may result from passive or active processes. Cells of Escherichia coli O157:H7 “Sakai” strain, introduced to spinach and lettuce plants under experimental conditions using a high inoculum dose of 2 × 107 CFU per ml, were shown to be present inside root tissue of both plant species (Wright et al., 2013), indicating that this human pathogen potentially can enter these plants by itself. It may be assumed that human pathogens do not possess the same specific traits, like cell wall degrading (pectinolytic) enzymes present in plant pathogens that are required for efficient invasion of plants. The presence of plant pathogens at the same locations on the plant surface where human pathogens reside would facilitate entrance of human pathogens into plants as was proposed in Teplitski et al. (2009, 2011). Under practical circumstances in agronomic systems, other soil species including plant pathogens indeed appeared to play important roles in spread, colonization, and internalization of plants by human pathogens. The bacterivorous nematode Caenorhabditis elegans was demonstrated to transport S. enterica serovar Newport, initially introduced into manure, through manure-amended soil to lettuce, strawberry and carrot plants (Kenney et al., 2006). Xanthomonas campestris pathovar vesicatoria facilitated a mixture of S. enterica strains to colonize the tomato phytosphere (Barak and Liang, 2008). Soft rot caused by Pectobacterium chrysanthemi in postharvest lettuce resulted in higher density levels of Escherichia coli O157:H7 in lettuce leafs (Brandl, 2008) and coinoculation of S. enterica serovar Typhimurium with the plant pathogen Dickeya dadantii under experimental conditions in lettuce and cilantro leaves resulted in higher densities of S. enterica serovar Typhimurium in leafs of both plant species than by inoculation with the human pathogenic strain alone (Goudeau et al., 2013). However, the presence of the root knot nematode Meloidogyne hapla in soil with bioluminescent-labeled Escherichia coli O157:H7 cells did not result in colonization of the areal parts of young spinach plants (Hora et al., 2005). Mechanic wounding of roots and coinoculation of leafs with the Escherichia coli O157:H7 strain and Pseudomonas syringae also did not result in colonization of the aerial parts of spinach plants, indicating that not all plant pathogens are facilitating entrance of human pathogens into, and/or spread through plants. Other species living in soil, like protozoa and fungi, may play an important role in the spread of S. enterica through soil and colonization of plants (Brandl et al., 2013) and it was demonstrated that S. enterica also can live inside protozoa cells (Jacobsen and Bech, 2012, and references therein). All together, these studies indicate that presence of plant pathogens and other species living in soil can play different roles in colonization and internalization of plants by human pathogens. Interesting within this aspect is that Enterobacter sp. 638, an endophyte in poplar tree, is capable to degrade pectate, facilitating this strain to colonize spaces between plant cells (Taghavi et al., 2010). Considering its close taxonomical relatedness with Escherichia coli and S. enterica, it emphasizes the fact that specific traits required for plant colonization must have been acquired by particular Enterobacteriaceae during evolution near or inside plants.
Enterobacteriaceae are common inhabitants of the phytosphere. To this taxonomical family belong important plant pathogens like Erwinia, Pantoea, and Pectobacterium species (Holden et al., 2009; Teplitski et al., 2009, 2011). Of more importance are genera whose species are beneficial to plants and pathogenic to animals, such as the ones belonging to Serratia and Klebsiella (Berg, 2000; Tan et al., 2001; Dong et al., 2003; Tyler and Triplett, 2008; Holden et al., 2009). Plant and animal-associated enteric species share genes that encode for important virulence traits such as attachment, plant colonization and internalization, biofilm formation and cell invasion (Tyler and Triplett, 2008; Holden et al., 2009; Teplitski et al., 2011). The taxonomic relatedness between human pathogens and plant-associated species may explain why at least some of the human pathogenic strains can colonize plants so well. Ancestors of both groups diversified along evolution, but parts of their genomes, so called cores, remained the same and possibly some of the orthologous genes shared by both groups may be involved in plant interactions (Holden et al., 2009).
Attachment to plant surfaces is an important feature related with settlement of human pathogens to, or inside plants. Constituents of the extracellular matrix of human pathogens like curli fibers, cellulose and lipopolysaccharide capsule (O antigen) were important for attachment to, and colonization of alfalfa sprouts by S. enterica serovar Newport (Barak et al., 2007) and spinach leafs by Escherichia coli O157:H7 (Macarisin et al., 2012). Possible mutations in regulatory genes responsible for curli and/or cellulose fiber production and leading to the non-rdar morphotype (defective in the formation of red dry and rough colonies on Congo red agar plates; Römling, 2005) resulted in better fitness of S. enterica serovar Typhimurium inside tomato fruits (Gu et al., 2011; Zaragoza et al., 2012). The same phenotype in S. enterica was less competitive in growth medium in comparison with its near isogenic wildtype strain (Zaragoza et al., 2012). Particular subsets of strains of human pathogens appear to possess features enabling them to attach to plant surfaces.
Entrance of human pathogens via aerial plant parts like woundings or natural openings such as hydathodes or stomata is realistic. A GFP-labeled S. enterica serovar Typhimurium strain was attracted to the stomatal openings of iceberg lettuce leafs under the influence of light, where it entered the stomatal cavity (Kroupitski et al., 2009). This strain was able to circumvent the stoma-based innate immune system. Internalization of Escherichia coli O157:H7 via stomata was also demonstrated in spinach leafs (Saldaña et al., 2011). Reduced cell numbers of a type 3 secretion defective mutant of this strain was found in the stomatal opening, in comparison with the wild type strain, indicating that type 3 secretion must play an important role in internal colonization of plants. Transfer of the locus of enterocyte effacement (LEE) pathogenicity island, containing type 3 secretion effector genes, of the Escherichia coli O157:H7 strain into a non-pathogenic Escherichia coli K12 strain resulted in improved colonization of derived strain. Both S. enterica and Escherichia coli O157:H7 possess mechanisms to actively enter the apoplast, thereby circumventing plant host immunity responses. Colonization of the interior parts of Roman lettuce by S. enterica serovar Dublin strain evoked an upshift in the expression of stress-related plant genes (Klerks et al., 2007b). Of the five different S. enterica serovars tested (Dublin, Typhimurium, Enteritidis, Newport and Montevideo), the strain of serovar Dublin was demonstrated to be the best colonizer of Roman lettuce, indicating the existence of differences in plant colonizing traits among different S. enterica serovars (Klerks et al., 2007a). S. enterica serovar Dublin strain was attracted to sugar-like carbon sources present in root exudates of lettuce and these compounds were responsible for induction of different genes amongst which genes that are regulators of the type 3 secretion system (Klerks et al., 2007a). S. enterica serovars actively colonize plants and expression of genes under type 3 secretion control also appeared to play an important role in the suppression of the plant immune system (Schikora et al., 2012).
Once inside plants, human pathogens may colonize plants locally, but also may systemically spread through plants by making use of the vascular tissues present in plants and needed for the transport of water and inorganic substances to leafs (xylem) or photosynthates to roots (phloem; McCully, 2001; Warriner et al., 2003). Five different S. enterica serovar strains, applied to plants via inoculation of the flowers, were later found to be present in tomato fruit by Guo et al. (2001). Systemic transport to aerial parts of tomato plants occurred upon growth in hydroponic solution containing the five different S. enterica serovar strains at levels of between 104 and 105 cell per ml (Guo et al., 2002). From the last study there are clear indications that transport inside tomato plants to developing leafs and branches must have occurred. Presence of S. enterica serovar Newport and Escherichia coli O157:H7 strains in chaff and seed of Arabidopsis plants was demonstrated when plants were grown under gnotobiotic circumstances and in autoclaved and non-autoclaved soils (Cooley et al., 2003). All together, the last two studies are indicative for systemic transport of human pathogens through vascular tissue, but no conclusive data could be provided yet. Vascular transport may occur in low quantities, but numbers may be too low to be detectable with common technologies (Warriner and Namvar, 2010).
In conclusion, it is likely to assume that features in particular subsets of Escherichia coli and S. enterica groups exist, enabling them to persist near or inside plants over extended periods in time. Genes in human pathogens involved in persistence in plants may be orthologs, but it cannot be ruled out that at least some of these genes were also recently gained via horizontal gene transfer, as was demonstrated to be the case for the Escherichia coli O104:H4 outbreak strain in Hamburg and surrounding area in 2011. In modern agriculture, practices are applied to optimize crop yields by irrigation, fertilization, pest, and disease control and maintenance of soil quality. This requires input from different sources, and bacterial communities from plants themselves, soils, intestinal track systems of farm animals (manure), water reservoirs (for sprinkling and irrigation of plants) and agricultural runoff water (Jacobsen and Bech, 2012) all come together at plant production (Figure 1). Human pathogens or their phages (examples are provided later) were found to be present in all four ecosystems, potentially bringing these into contact with indigenous bacteria near or inside plants. The consequences of these contacts can be exchanges in genetic material and the likelihood on occurrence of genetic exchanges between human pathogens and plant-associated bacteria and potential risks resulting from these will be discussed in the following section.
Horizontal Gene Transfer between Human Pathogens and their (Pro) Phages in the Phytosphere
Historical Occurrences of Gene Exchanges in Two Major Human Pathogens, Escherichia coli Serotypes O104:H4 and O157:H7
Gene transfer can occur when bacterial cells physically meet within the same habitat (Toth et al., 2006). Proposed vehicles for transmission of genomic islands are plasmids, (pro) phages and conjugative transposons (Juhas et al., 2009). Acquisition of new phenotypic traits will occur when human pathogens reside outside the human host. The plant-soil ecosystem is such an environment where these species can occur and thus it is likely to assume that auxiliary traits will be acquired from the microbial communities indigenous to this ecosystem.
Escherichia coli strain O104:H4, the outbreak strain in Germany and France in 2011, substantially differed from Escherichia coli O157:H7 outbreak strains (Bielaszewska et al., 2011; Eppinger et al., 2011; Mellmann et al., 2011). Some characteristics of the outbreak strain were: (i) that it belonged to the pathotype of enteroaggregative Escherichia coli and not to that of enterohemorrhagic Escherichia coli, (ii) that it did not produce intimin (encoded by the eae gene located on the LEE) but instead Iha adhesin and (iii) that it only produced Shiga toxin 2. The strong adherence of the outbreak strain to the intestinal epithelium in humans in combination with high tolerance to acid (to survive passage through the stomach) and Shiga toxin production were believed to be the main responsible factors for the occurrence of high incidence in HUS in patients, stressing the risks for humans by blends of virulence factors that can occur among human pathogens (Bielaszewska et al., 2011). These differences can be explained by differences in the nature of both serotypes, but also in genomic changes resulting from gene insertions and deletions. The LEE in serotype O157:H7 strains is a conserved pathogenicity island containing genes that are coding for chaperone and effector proteins, belonging to the type 3 secretion system, and responsible for the attaching and effacing lesions in the human large intestine. Intimine is a protein that is involved in cellular attachment of O157:H7serotype strains and the gene coding for this protein (eae) was not present in serotype O104:H4 as well as in other entero-aggregative Escherichia coli strains.
Escherichia coli O157:H7 strains commonly possess a virulence plasmid (pO157) and a conjugal plasmid (pEC4115; Eppinger et al., 2011), whereas in both serotypes, O104:H4 and O157:H7, stx genes are located on lambdoid prophages integrated in the wrbA gene (Mellmann et al., 2011). The genome of the serotype O157 Sakai strain possessed a total of 18 prophages, of which two contained the Shiga toxin genes stx1 and stx2 (Asadulghani et al., 2009). Despite the fact that these prophages contained multiple mutations, they were still functional, indicating that these genetic elements were able to propagate and to recombine with other genetic elements. The roles of phages that did not carry stx genes were inferred from complete genome sequence and comparative genome analysis of Escherichia coli strains (Ogura et al., 2008; Iguchi et al., 2009). It revealed that virulence genes on exchangeable effector loci were present that code for non-LEE and LEE effectors belonging to the type 3 secretion system. Some of these genes harbor homology to effector proteins of plant pathogens, strengthening the idea of a common evolutionary plan for the type 3 secretion system.
An evolutionary model of the serotype O104:H4 outbreak strain was constructed, based on available whole genome sequence data from a proposed ancestor strain, another Escherichia coli O104:H4 strain from an outbreak in 2001 (Mellmann et al., 2011). Interesting to note is that the German Escherichia coli O104:H4 outbreak strain of 2011 must have acquired the plasmid containing the aggregative adherence fimbriae type I (AAF/I) locus, but lost a plasmid containing the AAF/III locus and gained a plasmid with the gene encoding CTX-M-15 ESBL.
Virulence traits were gained, likely via horizontal gene transfer, and it is still under debate whether plasmid or phage exchanges between human pathogens and commensal species will occur under changing environmental circumstances. In a model for genome evolution proposed by Dini-Andreote et al. (2012), it was postulated that bacterial genomes will tend to expand under situations of shifting environmental conditions and that genomes shrink under stable environmental conditions. Translating this model for our study, it means that genomes of pathogens that occupy single habitats, like inside human bodies, will shrink whereas the ones that are exposed to multiple habitats will tend to increase in size, which will be the case for environmental strains. Proposed increase in genome size may occur by acquisition of new genes via gene transfer events. The “pan-genome” encompasses all genes that are present among strains originating from different environments, but still belonging the same taxonomic group (Dini-Andreote et al., 2012). It is characteristic for genomes of enterohemorrhagic species that stretches of housekeeping genes, forming the core genome, are punctuated by gene islands, forming the flexible genome (Mellmann et al., 2009). Genes belonging to the flexible genome most easily will be exchanged between bacteria of the same species, but also between bacteria of sometimes entirely different taxonomic groups. The flexible gene pool will consist of gene clusters, so called genomic islands that are responsible for auxiliary traits. The formation of genomic islands in bacterial genomes will facilitate the transfer of new phenotypic traits to recipients, resulting in “quantum leaps” in the adaptation of receiving strains to new environments (Juhas et al., 2009).
Horizontal Gene Transfer by Bacteriophages
Bacteriophages play a role in the virulence of EHEC strains by conferring genes involved in toxin production and type 3 secretion. Bacteriophages are recognized to be the main contributors to the transmission of virulence determinants, via transduction and lysogenic conversion, between bacterial strains. This process of lateral gene transfer is a significant factor in the evolution of bacteria. DNA of bacteriophage origin often comprises 10–20% of a bacterial genome, and approximately two-third of gammaproteobacterial and low-GC Gram-positive species harbor intact bacteriophages or their remnants in their genomes (Davidson et al., 1990; Saye et al., 1990; Kidambi et al., 1994; Boyd et al., 2000; Perna et al., 2001; Wagner and Waldor, 2002; Canchaya et al., 2003a,b; Casjens, 2003; Toth et al., 2003). A recent metagenomics survey on the virome of the bovine rumen showed that 64% of known virus genomes are from bacteriophages (Berg Miller et al., 2012). In particular, prophages were shown to originate from bacterial hosts within the taxonomical groups of Firmicutes (68 ± 1%), Proteobacteria (18 ± 1%) and Bacteriodetes (8 ± 1%). Comparison of the bovine rumen virome with a selected set of mobile elements from microbial genomes of the bovine rumen revealed a similarity of 80 ± 19%. The vast majority (79 ± 19%) was similar to prophages, demonstrating the importance of this type of mobile element within this ecosystem.
Shiga toxins, encoded by stx1 and stx 2 genes, are located on temperate bacteriophages and low or avirulent forms of Escherichia coli can be converted to pathogenic strains upon infection with these phages. Bacterial conversion by phages from non-pathogenic to pathogenic forms occur more often, for example in Corynebacterium diphtheria and in different Salmonella species (Saunders et al., 2001). Bacterial phages are responsible for horizontal transfer of virulence determinants between different bacterial species. Bacteriophage Φ24B, isolated from an enterohemorrhagic Escherichia coli O157 strain and carrying the stx2 gene, was able to infect commensal and pathogenic Escherichia coli and Shigella strains from different sources (James et al., 2001). This indicates that the host-range of this bacteriophage is broader, conferring new virulence traits toward different species among the group of Enterobacteriaceae. Expression of Shiga toxin genes are controlled by lambdoid prophage cI repressor (Mauro and Koudelka, 2011). Shiga toxin gene expression is stimulated upon cleavage of cI by RecA, and on its turn RecA expression can be stimulated by the presence of reactive oxygen species released from eukaryotic predators like Tetrahymena thermophila (Lainhart et al., 2009; Mauro and Koudelka, 2011). Shiga toxin production is a virulence determinant whose contribution to bacterial fitness is obscure. As discussed before, knock out mutants in Shiga toxin production did not survive better or worse in soils and plants. A question that still remains open is the role of Shiga toxin production in bacterial survival in the environment. Shiga toxin production may be a defense mechanism against eukaryotic unicellular predators, suggesting that humans may not be the prime targets for Shiga-toxin producing Escherichia coli strains.
A total of 15 phages carrying Shiga toxin stx2 genes, purified from environmental water samples, were shown to belong to three different virus families: Podoviridae (9), Myoviridae (6) and Siphidoviridae, the family to which phage lambda belong (2; Rooks et al., 2012). Environmental water thus can be a source of stx-carying phages and transduction with this type of phages may increase virulence levels in Escherichia coli strains. A Siphidoviridae lytic phage targeting the plant pathogenic bacterium Pectobacterium carotovorum subsp. carotovorum, My1, was recently sequenced (Lee et al., 2012). Although no stx genes were found in the genome of this phage, it illustrates the possibility that lytic phages released from plant-pathogenic bacteria can infect plant-invasive Escherichia coli strains. Broad-host range lytic phages have been characterized, like phage ΦOT8 from treated sewage effluent, that was able to infect and tranduce resistance and prodigiosin marker genes from Serratia sp. ATCC 39006 to Pantoea agglomerans (Evans et al., 2010). Phages isolated from soil and trees from Ontario, Canada (Gill et al., 2003) and from apple and pear orchard soils in Switzerland (Born et al., 2011) were able to infect the plant pathogenic bacterium Erwinia amylovora. All bacteriophages obtained (42 from the study of Gill et al., 2003 and 24 of the one of Born et al., 2011) belonged to Podoviridae or Myoviridae. Commonalities in genome sequence were found between two Myoviridae phages from both studies, ΦEa21-4 and M7, and both were considered to be broad-host range phages infecting three different Erwinia species and Pantoea agglomerans (Born et al., 2011). The same phage type (ΦEa21-4) showed a remarkable similarity at a protein level of 56% with Salmonella phage Felix O1 (Lehman et al., 2009). Major difference between phages ΦEa21-4 and Felix O1 was the presence of the nadV homolog encoding a nicotinamide-scavenging enzyme that may supplement nicotinic acid in E. amylovora (Lehman et al., 2009). Typical ΦEa21-4-like phages may be cosmopolites able to infect multiple species within the class of Enterobacteriaceae. In potential, this type of phages could also be able to horizontally transfer genes from and to human pathogens upon invasion into the phytosphere.
Horizontal Gene Transfer by Conjugal Plasmids
Virulence traits are present on different plasmids in Escherichia coli (Johnson and Nolan, 2009). Commonalities exist in pathogenicity determinants among plant and animal Enterobacteriaceae (Toth et al., 2006). Therefore, conjugal plasmids can play pivoting roles in horizontal transmission of virulence factors located on pathogenicity islands in plant-soil ecosystems.
The Hamburg serotype O104:H4 outbreak strain contained TEM-1 and CTX-M-15 beta lactamase genes on an incompatibility group I (IncI) type plasmid (Mellmann et al., 2011). Extended spectrum beta lactamase phenotypes have been found in many enteric species over the world, and CTX-M15 was found in Escherichia coli, Klebsiella pneumonia, Enterobacter aerogenes and S. marcescens (Bonnet, 2004 and references therein). Further, CTX-M class of beta lactamase genes were found in Erwinia persicina (Vimont et al., 2002) and in P. aeruginosa and S. maltophilia (Naiemi et al., 2005). IncI-type plasmids are narrow host-range plasmids and were not transferrable between donor and recipient Escherichia coli strains in nutrient-amended soil (Pukall et al., 1996). In that study, broad-host range plasmids belonging to incompatibility groups IncN, IncP1, IncW3, and IncQ were transferred to the Escherichia coli recipient strain and it was hypothesized that size and flexibility of the pili covering the cellular surface of donor cells would play an important role in the frequency of plasmid transfer in soil (Pukall et al., 1996; Ghigo, 2001). IncP-type of plasmids are self-transmissible plasmids, whereas IncQ types are not, however, both were considered to be the major vectors in horizontal transfer of antibiotic resistances in natural environments (Heuer et al., 2012; Popowska and Krawczyk-Balska, 2013).
Virulence factors in human pathogens are often located on plasmids. Tellurite (TeO3) resistance is a trait that is commonly found among different human pathogens including Escherichia coli O157:H7 and O46:H-, and were located on a pathogenicity island (Taylor, 1999). Tellurite resistance genes in enteric bacteria, located on IncHI2 and IncHII plasmids, showed remarkable homology with tellurite resistance genes located on plasmid R478 from S. marcescens (Taylor et al., 2002). Tellurite resistance determinants were also found on a IncP2 plasmid from P. aeruginosa, but these genes were not related with the ones found in Escherichia coli O157:H7 (Hou and Taylor, 1994). Iron acquisition genes of Yersinia pathogens (Y. pestis, Y. pseudotuberculosis, Y. enetrocolitica) is located on the Yersinia high pathogenicity island (HPI) and this HPI was found among different species of Enterobacteriaceae, including Escherichia coli, Citrobacter diversus, different Klebsiella species, non-I serotypes of S. enterica, but not in S. enterica serovars Enteritidis and Typhimurium and in EHEC (Bach et al., 2000; Carniel, 2001). Mobility of the HPI among enteric species must occur, although in Y. pseudotuberculosis excision was shown to be a rare event (frequency of about 10-9; Carniel, 2001). An HPI found in Escherichia coli showed strong (99%) identity with HPI present in Y. pestis, indicating that gene transfer between Y. pestis and Escherichia coli must have occurred in the past (Schubert et al., 2004). Mobilization of HPI in the bacterial chromosome is a phage-mediated process and P4-phage-related integrase and excision genes were found to be present in HPI (Benedek and Schubert, 2007). However, promiscuous mobile plasmids play important roles as shuttle vectors in the horizontal transfer of HPI (Antonenka et al., 2005; Schubert et al., 2009) and other pathogenicity islands in Escherichia coli (Schneider et al., 2011) and in environmental and other pathogenic micro-organisms (Dobrindt et al., 2004). Upon introduction of the self-transmissible plasmid RP4 into Y. pseudotuberculosis, a cointegrate between this plasmid and HPI was formed and transferred to a recipient strain (Antonenka et al., 2005). Trapping of HPI into the cointegrate was based on precise excision of the element from the chromosome and the frequency in occurrence of this event was estimated at 1 on 106 cells. Precise excision did not always occur, and occasionally regions flanking the HPI were also integrated and transferred to recipients (Schubert et al., 2009). Salmonella genomic island 1 (SGI1), conferring multiple antibiotic resistances to the bacterial host, was shown to be excised from the chromosome by a lambdoid integrase (Doublet et al., 2005). However, the circular form of SGI1 was not transferred to recipient strains by itself under experimental circumstances. A helper IncC plasmid (R55) was needed for the transfer of SGI1 from different Salmonella donor strains to Escherichia coli K12 as recipient (Doublet et al., 2005). A screening over 902 S. enterica serovar types from poultry revealed a higher incidence of a ColV type plasmid among a clonal type of serovar Kentucky than over other serovar types (Johnson et al., 2010). The ColV plasmid conferred virulence and fitness traits toward its host and strains carrying the plasmid better colonized the chicken cecum and successfully competed with the indigenous microbial community in there. This illustrates the ecological advantage that may be gained by human pathogens via transfer of genes located on mobile elements like plasmids. The genes present on plasmids in human pathogens may come from different sources as demonstrated by the full sequence of the large virulence plasmid, pO157, from Escherichia coli O157:H7 (Burland et al., 1998). A strong resemblance between genes located on this plasmid was found with ToxA and ToxB toxin protein genes from Clostridium difficile. Genes of archeal origin (presumably from Methanobacterium thermoautotrophicum and Halobacterium species) were proposed to be present in the genome of Escherichia coli O157:H7 strain EDL933 (Faguy, 2003). The last would indicate that cross-domain gene transfer events would have occurred in the past between Escherichia coli and Archeae species.
Besides transfer of genes belonging to the flexible genome (mostly located on genomic or pathogenicity islands), also genes of the core genome can be transferred like household and metabolic genes. Among these genes can be ones that improve fitness of the recipient strain under environmental circumstances like present in the rhizosphere. Transfer of metabolic genes involved in oxidation or fermentation of small carbohydrates like sugars, acids, and amino acids may improve fitness of receiving strains in the rhizosphere where these compounds are exuded by plant roots. Genes responsible for L-sorbose transport and metabolism in Escherichia coli was also found in K. pneumonia (Wehmeier et al., 1992) and genes involved in glucose fermentation were transferred via genetic elements among different Salmonella species (Wohlhieter et al., 1975; Bartlett and Trust, 1980). Only a few strains of Salmonella and Escherichia coli can ferment glucose and acquisition these genes can be important for adaptation to new environments. Transfer of sucrose fermenting genes was found to be facilitated by conjugative transposons (Pembroke et al., 2002).
Under experimental circumstances, transfer of an environmental (wastewater treatment plant) self-transmissible and broad-host-range (IncP1β) plasmid from P. putida to different S. enterica serovar and Escherichia coli O157:H7 strains was demonstrated (Van Meervenne et al., 2012). Upon bacterial mating between donor and recipient strains, transconjugants were found at most, but not all occasions, indicating that human pathogens are not equally susceptible for horizontal gene transfer events under applied conditions. Bacterial gene transfer between introduced plasmid donor and/or recipient strains was demonstrated at different places in the phytosphere, such as in the rhizosphere (Lilley and Bailey, 1997), the phylloplane (Normander et al., 1998) and inside plants between endophytes (Taghavi et al., 2005). Transfer of two plasmids from P. putida recipients to indigenous bacteria associated with ready-to-eat alfalfa sprouts was demonstrated (Mølbak et al., 2003). In this study, transconjugants were found among different Pseudomonas and Erwinia species. Mobilizable plasmids from indigenous bacteria of piggery manure were obtained by introduction of Escherichia coli and P. putida strains, acting as recipients for exogenous plasmid isolation, in samples from different ecosystems (Smalla et al., 2000). Identities of plasmids selected from transconjugants revealed that they were all IncQ like. IncQ and IncP, IncN, and IncW type plasmids carrying gentamycin (Heuer et al., 2002) or streptomycin (van Overbeek et al., 2002) resistance determinants were obtained by exogenous plasmid isolations from bulk and rhizosphere soils, manure, sludge, and seawater samples. These data demonstrate the omnipresence of mobilisable (like IncQ type) and sometimes self-transmissible (like IncP type) plasmids in a wide variety of ecosystems, including the ones that are relevant for agriculture. Remarkable was the high abundance of IncQ-type of plasmids obtained by exogenous plasmid isolation, because these plasmids need a helper plasmid from another incompatibility group for transfer. Transfer of IncQ plasmids would require three parents for mating and it was believed that these type of matings would occur less frequently in soils (Pukall et al., 1996).
Cryptic plasmids can play important roles in horizontal transfer of IncQ plasmids in natural ecosystems. A cryptic plasmid, pIPO2, was found in the rhizosphere of wheat and this plasmid was able to mobilize IncQ plasmids to recipient strains (Van Elsas et al., 1998). PCR detection based on pIPO2 revealed that this plasmid was present in 7 of 10 tested rhizosphere DNA samples, in 2 of 12 bulk soil DNA samples, but not in any of the single DNA samples from manure, seawater and wastewater (Tauch et al., 2002). Sequence comparisons made between pIPO2 and plasmid pTER331 from the soil bacterium Collimonas fungivorans revealed a high similarity between both plasmids (Tauch et al., 2002; Mela et al., 2008). Cryptic plasmids belonging to the pIPO2-type thus are common in the rhizosphere where they are supposed to be responsible for the mobilization and retromobilization of plasmids. In principle, the roles of these type of plasmids in horizontal gene transmission in arable plant environments have not been explored so much.
Among the species that are typical for the plant-soil ecosystem are plant pathogenic and endophytic bacteria and different plasmid types were found among species belonging to alpha, beta and gammaproteobacteria (Vivian et al., 2001). Whole-genome sequencing of the poplar endophyte Enterobacter sp. 638 revealed the presence of important adhesion and colonization genes on plasmid pENT638-1 (Taghavi et al., 2010), explaining the excellent colonization of strain 638 in poplar trees. This plasmid is thus of ecological relevance and supportive for the endophytic lifestyle of this strain. In E. amylovora, a plant pathogen commonly causing diseases in a wide variety of (rosaceous) plants, plasmids were found in different strains of this species and DNA sequences of three plasmids were annotated. Plasmid pEA29 from E. amylovora is a non-self-transmissible plasmid that stably replicates within its host (McGhee and Jones, 2000). On this plasmid auxiliary virulence and streptomycin resistance genes were found. DNA sequences of two other plasmids from E. amylovora pEU30 and pEL60, respectively, revealed strong similarities with conjugal transfer genes found in Pseudomonas syringae pathovar syringae and with pCTX-M3 plasmid of Citrobacter freundii (Foster et al., 2004). Plasmid pEL60 was an IncL/M type of plasmid and contained genes that are responsible for DNA repair and was proposed to increase environmental fitness of the host. The close resemblance of this plasmid with a plasmid from an animal (mouse) pathogenic species revealed that animal and plant pathogenic enteric species might share the same gene pool (Foster et al., 2004). In two strains of another plant pathogenic bacterial species, P. chrysanthemi, a gene was found that was immunologically related with the intimin-encoding gene eae commonly found among EHEC strains (Duarté et al., 2000). The two strains were able to kill human colon carcinoma cells, whereas a mutant defective in type 3 secretion lost the capacity to kill these cells. The combination of type 3 secretion and intimin is important for attachment to human cells and demonstrates a high conservation of virulence genes among plant and human pathogens (Duarté et al., 2000). Among strains of Klebsiella pneumoniae are human pathogens and (commensal) endophytes. Genome comparisons between a human pathogenic and an endophytic strain of this species revealed that there was a higher abundance of genes supposed to be responsible for survival under circumstances present inside plants like polysaccharide degradation, transport, protection against oxidative agents and nitrogen fixation (Fouts et al., 2008). In the same study, the endophytic strain was virulent in a mouse model test system, although virulence was milder in comparisons with the human pathogenic strain. Pseudomonas aeruginosa is a bacterial species that harbors plant as well as (opportunistic) human pathogenic strains and it was found that virulence mechanisms necessary for infection of evolutionary different hosts (vertebrate and invertebrate animal and plant species) were conserved (Rahme et al., 2000). Mutations in the genes coding for exotoxin A (toxA), responsible for protein synthesis inhibition in mammalian cells, phospholipase C (plcC), phospholipid degradation in eukaryotic cells, and gacA, transcriptional activator of genes responsible for production of extracellular compounds, lead to reduced virulence both in Arabidopsis and mouse test systems. Other genes responsible for multihost pathogenesis were involved in synthesis of membrane derived oligosaccharides, quorum sensing, trans-membrane export of proteins, multidrug efflux, phenazine-1-carboxylate production, motility, and surface attachment (Rahme et al., 2000). A strain of the non-pathogenic species Wolinella succinogenes, originating from the intestinal track system of cattle, contained many homologs of known virulence genes (Baar et al., 2003). W. succinogenes is a epsilon proteobacterium closely related with human pathogenic species Campylobacter jejuni and Helicobacter pylori. Next to pathogenicity gene homologs, genes proposed to be important for survival in soil and plants, including nitrogen fixation genes, were found (Baar et al., 2003). Conservation of genes that are known to be responsible for pathogenesis in humans thus are found among entirely different plant and soil bacterial species that have human pathogens as close relatives. This indicates that the plant-soil ecosystem is a source of genes that may increase fitness of many human pathogens besides Escherichia coli and S. enterica that can reside in the phytosphere.
In Conclusion
Horizontal gene transfer, either mediated by phages or by conjugal plasmids, can play a role in the acquisition of new phenotypes in human pathogens present in one of the four ecosystems relevant for arable plant production, i.e., soil, plant, animal (cattle) gut, and fresh water systems (Figure 1). Acquired phenotypes are related to resistance, virulence and ecological competence. All human pathogens had been prone to gene transfer events in the past and examples of recent and multiple gene transfer events were demonstrated in Escherichia coli serotypes O104:H4 and O157:H7. Serotype O157:H7 and some of the other O serovar types are zoonotic pathogens, which is not the case for serotype O104:H4 (entero-aggregative Escherichia coli), which is believed to be transmitted via humans only. From epidemiological studies it became clear that the Hamburg Escherichia coli O104:H4 outbreak was not transmitted via humans, but most likely via seeds. It is clear that human pathogens passing these four ecosystems might acquire mobile genetic elements, supplying them additional virulence genes and/or making them better adapted to their new environment. The combination of increased resistance to antibiotics and higher virulence levels in commensal or low-pathogenic bacteria is a major threat for human health (Szmolka and Nagy, 2013). Another hotspot boosting horizontal gene transfer thus might be, next to the mammalian gut system (Stecher et al., 2012), the rhizosphere. Studies going on from the 80-ties of the last century revealed that gene transfer via conjugation occur in the rhizosphere. New pathogenic types thus may arise at agricultural practice that are adapted to life near or inside plants; the so called phytonic types.
Products derived from plants are considered to be healthy and microbiologically safe by consumers. The public is unaware of eventual microbiological risks related to consumption of fresh produce (Beutin and Martin, 2012) and therefore emergence of phytonic human pathogens deserves attention. Appropriate and timely detection of emerging pathogens in plants and food derived from plants is of importance, but will be complicated by the flexibility of their genomes. Appearance of new features, encrypted in genomes of STEC, S. enterica and other human pathogenic strains, that determine fitness in plants and virulence to humans, like genes from plasmids and bacteriophage sequences, should be monitored frequently (Mellmann et al., 2011; Noguera et al., 2011a,b; Laing et al., 2012). Novel, rapid, innovative high throughput detection technologies will facilitate screenings for these regions in genomes of human pathogenic isolates from clinical and environmental samples.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This literature overview was conducted within the framework program e-H@C HUPAction financed by EU Interreg IV A, German-Dutch cross border collaboration (North Rhine Westphalia and Gelderland regions) and the Dutch Ministry of Economic affairs under the top sector initiative for horticulture and starting materials (“kennisbasis programma,” KB-12-006.01-001).
References
Antonenka, U., Nolting, C., Heesemann, J., and Rakin, A. (2005). Horizontal transfer of Yersinia high-pathogenicity island by the conjugative RP4 attB target-presenting shuttle plasmid. Mol. Microbiol. 57, 727–734. doi: 10.1111/j.1365-2958.2005.04722.x
Asadulghani, M., Ogura, Y., Ooka, T., Itoh, T., Sawaguchi, A., Iguchi, A., et al. (2009). The defective prophage pool of Escherichia coli O157: prophage–prophage interactions potentiate horizontal transfer of virulence determinants. PLoS Pathog. 5:e1000408. doi: 10.1371/journal.ppat.1000408
Aurass, P., Prager, R., and Flieger, A. (2011). EHEC/EAEC O104:H4 strain linked with the 2011 German outbreak of haemolytic uremic syndrome enters into the viable but non-culturable state in response to various stresses and resuscitates upon stress relief. Environ. Microbiol. 13, 3139–3148. doi: 10.1111/j.1462-2920.2011.02604.x
Baar, C., Eppinger, M., Raddatz, G., Simon, J., Lanz, C., Klimmek, O., et al. (2003). Complete genome sequence and analysis of Wolinella succinogenes. Proc. Natl Acad Sci. U.S.A. 100, 11690–11695. doi: 10.1073/pnas.1932838100
Bach, S., de Almeida, A., and Carniel, E. (2000). The Yersinia high-pathogenicity island is present in different members of the family Enterobacteriaceae. FEMS Microbiol. Lett. 183, 289–294. doi: 10.1111/j.1574-6968.2000.tb08973.x
Barak, J. D., Jahn, C. E., Gibson, D. L., and Charkowski, A. O. (2007). The role of cellulose and O-antigen capsule in the colonization of plants by Salmonella enterica. Mol. Plant Microbe Interact. 20, 1083–1091. doi: 10.1094/MPMI-20-9-1083
Barak, J. D., and Liang, A. S. (2008). Role of soil, crop debris, and a plant pathogen in Salmonella enterica contamination of tomato plants. PLoS ONE 3:e1657. doi: 10.1371/journal.pone.0001657
Barak, J. D., and Schroeder, B. K. (2012). Interrelationships of food safety and plant pathology: the life cycle of human pathogens on plants. Annu. Rev. Phytopathol. 50, 241–266. doi: 10.1146/annurev-phyto-081211-172936
Barkocy-Gallagher, G. A., Arthur, T. M., Rivera-Betancourt, M., Nou, X., Shackelford, S. D., Wheeler, T. L., et al. (2003). Seasonal prevalence of Shiga toxin-producing Escherichia coli, including O157:H7 and non-O157 serotypes, and Salmonella in commercial beef processing plants. J. Food Prot. 66, 1978–1986.
Bartlett, K. H., and Trust, T. J. (1980). Plasmid-specified sucrose fermentation in Salmonella arizonae. 121, 255–257. doi: 10.1099/00221287-121-1-255
Benedek, O., and Schubert, S. (2007). Mobility of the yersinia high-pathogenicity Island (HPI): transfer mechanisms of pathogenicity islands (PAIS) revisited (a review). Acta Microbiol. Immunol. Hung. 54, 89–105. doi: 10.1556/AMicr.54.2007.2.1
Berends, I. M. G. A., Graat, E. A. M., Swart, W. A. J. M., Weber, M. F., van de Giessen, A. W., Lam, T. J. G. M., et al. (2008). Prevalence of VTEC O157 in dairy and veal herds and risk factors for veal herds. Prev. Vet. Med. 87, 301–310. doi: 10.1016/j.prevetmed.2008.05.004
Berg, G. (2000). Diversity of antifungal and plant-associated Serratia plymuthica strains. J. Appl. Microbiol. 88, 952–960. doi: 10.1046/j.1365-2672.2000.01064.x
Berg, G., Eberl, L., and Hartmann, A. (2005). The rhizosphere as a reservoir for opportunistic human pathogenic bacteria. Environ. Microbiol. 7, 1673–1685. doi: 10.1111/j.1462-2920.2005.00891.x
Berg Miller, M. E., Yeoman, C. J., Chia, N., Tringe, S. G., Angly, F. E., Edwards, R. A., et al. (2012). Phage-bacteria relationships and CRISPR elements revealed by a metagenomic survey of the rumen microbiome. Environ. microbiol. 14, 207–227. doi: 10.1111/j.1462-2920.2011.02593.x
Beutin, L., and Martin, A. (2012). Outbreak of Shiga toxin-producing Escherichia coli (STEC) O104:H4 infection in Germany causes a paradigm shift with regard to human pathogenicity of STEC strains. J. Food Prot. 75, 408–418. doi: 10.4315/0362-028X.JFP-11-452
Bielaszewska, M., Mellmann, A., Zhang, W., Kock, R., Fruth, A., Bauwens, A., et al. (2011). Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: a microbiological study. Lancet Infect. Dis. 11, 671–676. doi: 10.1016/S1473-3099(11)70165-7
Bonnet, R. (2004). Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 48, 1–14. doi: 10.1128/AAC.48.1.1-14.2004
Born, Y., Fieseler, L., Marazzi, J., Lurz, R., Duffy, B., and Loessner, M. J. (2011). Novel virulent and broad-host-range Erwinia amylovora bacteriophages reveal a high degree of mosaicism and a relationship to Enterobacteriaceae phages. Appl. Environ. Microbiol. 77, 5945–5954. doi: 10.1128/AEM.03022-10
Bouwknegt, M., Dam-Deisz, W. D. C., Wannet, W. J. B., van Pelt, W., Visser, G., and van de Giessen, A. W. (2004). Surveillance of Zoonotic Bacteria in Farm Animals in the Netherlands: Results from January 1998 Until December 2002. RIVM report 330050001/2004.
Boyd, E. F., Moyer, K. E., Shi, L., and Waldor, M. K. (2000). Infectious CTXPhi and the vibrio pathogenicity island prophage in Vibrio mimicus: evidence for recent horizontal transfer between V. mimicus and V. cholerae. Infect. Immun. 68, 1507–1513. doi: 10.1128/IAI.68.3.1507-1513.2000
Brandl, M. T. (2008). Plant lesions promote the rapid multiplication of Escherichia coli O157:H7 on postharvest lettuce. Appl. Environ. Microbiol. 74, 5285–5289. doi: 10.1128/AEM.01073-08
Brandl, M. T., Clayton, E. C., and Teplitski, M. (2013). Salmonella interactions with plants and their associated microbiota. Phytopathology 103, 316–325. doi: 10.1094/PHYTO-11-12-0295-RVW
Brennan, F. P., O’Flaherty, V., Kramers, G., Grant, J., and Richards, K. G. (2010). Long-term persistence and leaching of Escherichia coli in temperate maritime soils. Appl. Environ. Microbiol. 76, 1449–1455. doi: 10.1128/AEM.02335-09
Breuer, T., Benkel, D. H., Shapiro, R. L., Hall, W. N., Winnett, M. M., and Linn, M. J. (2001). A multistate outbreak of Escherichia coli O157:H7 infections linked to alfalfa sprouts grown from contaminated seeds. Emerg. Infect. Dis. 7, 977–982. doi: 10.3201/eid0706.010609
Burland, V., Shao, Y., Perna, N. T., Plunkett, G., Sofia, H. J., and Blattner, F. R. (1998). The complete DNA sequence and analysis of the large virulence plasmid of Escherichia coli O157:H7. Nucleic Acids Res. 26, 4196–4204. doi: 10.1093/nar/26.18.4196
Canchaya, C., Fournous, G., Chibani-Chennoufi, S., Dillmann, M. L., and Brussow, H. (2003a). Phage as agents of lateral gene transfer. Curr. Opin. Microbiol. 6, 417–424. doi: 10.1016/S1369-5274(03)00086-9
Canchaya, C., Proux, C., Fournous, G., Bruttin, A., and Brussow, H. (2003b). Prophage genomics. Microbiol. Mol. Biol. Rev. 67, 238–276. doi: 10.1128/MMBR.67.2.238-276.2003
Carniel, E. (2001). The Yersinia high-pathogenicity island: an iron-uptake island. Microbes Infect. 3, 561–569. doi: 10.1016/S1286-4579(01)01412-5
Casjens, S. (2003). Prophages and bacterial genomics: what have we learned so far? Mol. Microbiol. 49, 277–300. doi: 10.1046/j.1365-2958.2003.03580.x
Chapman, P. A., Siddons, C. A., Gerdan Malo, A. T., and Harkin, M. A. (1997). A 1-year study of Escherichia coli O157 in cattle, sheep, pigs and poultry. Epidemiol. Infect. 119, 245–250. doi: 10.1017/S0950268897007826
Chapman, P. A., Siddons, C. A., Wright, D. J., Norman, P., Fox, J., and Crick, E. (1993). Cattle as a possible source of verocytotoxin-producing Escherichia coli O157 infections in man. Epidemiol. Infect. 111, 439–447. doi: 10.1017/S0950268800057162
Cheuk, W., Woo, P. C. Y., Yuen, K. Y., Yu, P. H., and Chan, J. K. C. (2000). Intestinal inflammatory pseudotumour with regional lymph node involvement: identification of a new bacterium as the etiological agent. J. Pathol. 192, 289–292. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH767>3.0.CO;2-F
Cody, S. H., Glynn, M. K., Farrar, J. A., Cairns, K. L., Griffin, P. M., and Kobayashi, J. (1999). An outbreak of Escherichia coli O157:H7 infection from unpasteurized commercial apple juice. Ann. Int. Med. 130, 202–209. doi: 10.7326/0003-4819-130-3-199902020-00005
Conedera, G., Chapman, P. A., Marangon, S., Tisato, E., Dalvit, P., and Zuin, A. (2001). A field survey of Escherichia coli O157 ecology on a cattle farm in Italy. Int. J. Food Microbiol. 66, 85–93. doi: 10.1016/S0168-1605(00)00489-X
Cooley, M. B., Miller, W. G., and Mandrell, R. E. (2003). Colonization of Arabidopsis thaliana with Salmonella enterica and enterohemorrhagic Escherichia coli O157:H7 and competition by Enterobacter asburiae. Appl. Environ. Microbiol. 69, 4915–4926. doi: 10.1128/AEM.69.8.4915-4926.2003
Coutinho, T. H., and Venter, S. N. (2009). Pathogen profile. Pantoea ananatis: an unconventional plant pathogen. Mol. Plant Pathol. 10, 325–335. doi: 10.1111/j.1364-3703.2009.00542.x
Cruz, A. T., Cazacu, A. C., and Allen, C. H. (2007). Pantoea agglomerans, a plant pathogen causing human disease. J. Clin. Microbiol. 45, 1989–1992. doi: 10.1128/JCM.00632-07
Davidson, B. E., Powell, I. B., and Hillier, A. J. (1990). Temperate bacteriophages and lysogeny in lactic acid bacteria. FEMS Microbiol. Rev. 7, 79–90. doi: 10.1111/j.1574-6968.1990.tb04880.x
Derrien, M., Vaughan, E. E., Plugge, C. M., and De Vos, W. M. (2004). Akkermansia muciniphila gen. Nov., sp. Nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 50, 1965–1972.
Dini-Andreote, F., Andreote, F. D., Araujo, W. L., Trevors, J. T., and van Elsas, J. D. (2012). Bacterial genomes: habitat specificity and uncharted organisms. Microb. Ecol. 64, 1–7. doi: 10.1007/s00248-012-0017-y
Dobrindt, U., Hochhut, B., Hentschel, U., and Hacker, J. (2004). Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2, 414–424. doi: 10.1038/nrmicro884
Dong, Y., Iniguez, A. L., Ahmer, B. M., and Triplett, E. W. (2003). Kinetics and strain specificity of rhizosphere and endophytic colonization by enteric bacteria on seedlings of Medicago sativa and Medicago truncatula. Appl. Environ. Microbiol. 69, 1783–1790. doi: 10.1128/AEM.69.3.1783-1790.2003
Doublet, B., Boyd, D., Mulvey, M. R., and Cloeckaert, A. (2005). The Salmonella genomic island 1 is an integrative mobilizable element. Mol. Microbiol. 55, 1911–1924. doi: 10.1111/j.1365-2958.2005.04520.x
Duarté, X., Anderson, C. T., Grimson, M., Barabote, R. D., Strauss, R. E., Gollahon, L. S., et al. (2000). Erwinia chrysanthemi strains cause death of human gastrointestinal cells in culture and express an intimin-like protein. FEMS Microbiol. Lett. 190, 81–86. doi: 10.1016/S0378-1097(00)00325-6
Durso, L. M., Smith, D., and Hutkins, R. W. (2004). Measurements of fitness and competition in commensal Escherichia coli and E. coli O157:H7 strains. Appl. Environ. Microbiol. 70, 6466–6472. doi: 10.1128/AEM.70.11.6466-6472.2004
Eckburg, P. B., and Relman, D. A. (2007). The role of microbes in Crohn’s disease. Clin. Infect. Dis. 44, 256–262. doi: 10.1086/510385
Eppinger, M., Mannel, M. K., Leclerck, J. E., Ravel, J., and Cebula, T. A. (2011). Genomic anatomy of Escherichia coli O157:H7 outbreaks. Proc. Natl. Acad. Sci. U.S.A. 108, 20142–20147. doi: 10.1073/pnas.1107176108
Erickson, M. C., Webb, C. C., Diaz-Perez, J. C., Phatak, S. C., Silvoy, J. J., Davey, L., et al. (2010). Surface and internalized Escherichia coli O157:H7 on field-grown spinach and lettuce treated with spray-contaminated irrigation water. J. Food Prot. 73, 1023–1029.
Evans, T. J., Crow, M. A., Williamson, N. R., Orme, W., Thomson, N. R., Komitopoulou, E., et al. (2010). Characterization of a broad-host-range flagellum-dependent phage that mediates high-efficiency generalized transduction in, and between, Serratia and Pantoea. Microbiology 156, 240–247. doi: 10.1099/mic.0.032797-0
Faguy, D. M. (2003). Lateral gene transfer (LGT) between Archaea and Escherichia coli is a contributor to the emergence of novel infectious disease. BMC Infect. Dis. 3:13. doi: 10.1186/1471-2334-3-13
Feng, P. C. H., and Reddy, S. (2013). Prevalences of Shiga toxin subtypes and selected other virulence factors among Shiga-toxigenic Escherichia coli strains isolated from fresh produce. Appl. Environ. Microbiol. 79, 6917–6923. doi: 10.1128/AEM.02455-13
Ferens, W. A., and Hovde, C. J. (2011). Escherichia coli O157:H7: animal reservoir and sources of human infection. Foodborne Pathog. Dis. 8, 465–487. doi: 10.1089/fpd.2010.0673
Foster, G. C., McGhee, G. C., Jones, A. L., and Sundin, G. W. (2004). Nucleotide sequences, genetic organization, and distribution of pEU30 and pEL60 from Erwinia amylovora. Appl. Environ. Microbiol. 70, 7539–7544. doi: 10.1128/AEM.70.12.7539-7544.2004
Fouts, D. E., Tyler, H. L., DeBoy, R. T., Daugherty, S., Ren, Q., Badger, J. H., et al. (2008). Complete genome sequence of the N2-fixing broad host range endophyte Klebsiella pneumoniae 342 and virulence predictions verified in mice. PLoS Genet. 4:e1000141. doi: 10.1371/journal.pgen.1000141
Franz, E., Klerks, M. M., De Vos, O. J., Termorshuizen, A. J., and Van Bruggen, A. H. C. (2007). Prevalence of Shiga toxin-producing Escherichia coli stx1, stx2, eaeA, and rfbE genes and survival of E. coli O157:H7 in manure from organic and low-input conventional dairy farms. Appl. Environ. Microbiol. 73, 2180–2190. doi: 10.1128/AEM.01950-06
Franz, E., and Van Bruggen, A. H. C. (2008). Ecology of E. coli O157:H7 and Salmonella enterica in the primary vegetable production chain. Crit. Rev. Microbiol. 34, 143–161. doi: 10.1080/10408410802357432
Franz, E., Van Diepeningen, A. D., De Vos, O. J., and Van Bruggen, A. H. C. (2005). Effects of cattle feeding regimen and soil management type on the fate of Escherichia coli O157:H7 and Salmonella enterica Serovar Typhimurium in manure, manure-amended soil, and lettuce. Appl. Environ. Microbiol. 71, 6165–6174. doi: 10.1128/AEM.71.10.6165-6174.2005
Franz, E., van Hoek, A. H., Bouw, E., and Aarts, H. J. (2011). Variability of Escherichia coli O157 strain survival in manure-amended soil in relation to strain origin, virulence profile, and carbon nutrition profile. Appl. Environ. Microbiol. 77, 8088–8096. doi: 10.1128/AEM.00745-11
Frost, L. S., Leplae, R., Summers, A. O., and Toussaint, A. (2005). Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3, 722–732. doi: 10.1038/nrmicro1235
Ghigo, J.-M. (2001). Natural conjugative plasmids induce bacterial biofilm development. Lett. Nat. 412, 442–445. doi: 10.1038/35086581
Gill, J. J., Svircev, A. M., Smith, R., and Castle, A. J. (2003). Bacteriophages of Erwinia amylovora. Appl. Environ. Microbiol. 69, 2133–2138. doi: 10.1128/AEM.69.4.2133-2138.2003
Goudeau, D. M., Parker, C. T., Zhou, Y., Sela, S., Kroupitski, Y., and Brandl, M. T. (2013). The Salmonella transcriptome in lettuce and cilantro soft rot reveals a niche overlap with the animal host intestine. Appl. Environ. Microbiol. 79, 250–262. doi: 10.1128/AEM.02290-12
Gu, G., Hu, J., Cevallos-Cevallos, J. M., Richardson, S. M., Bartz, J. A., and Van Bruggen, A. H. (2011). Internal colonization of Salmonella enterica serovar Typhimurium in tomato plants. PLoS ONE 6:e27340. doi: 10.1371/journal.pone.0027340
Guo, X., Chen, J., Brackett, R. E., and Beuchat, L. R. (2001). Survival of salmonellae on and in tomato plants from the time of inoculation at flowering and early stages of fruit development through fruit ripening. Appl. Environ. Microbiol. 67, 4760–4764. doi: 10.1128/AEM.67.10.4760-4764.2001
Guo, X., Van Iersel, M. W., Chen, J., Brackett, R. E., and Beuchat, L. R. (2002). Evidence of association of salmonellae with tomato plants grown hydroponically in inoculated nutrient solution. Appl. Environ. Microbiol. 68, 3639–3643. doi: 10.1128/AEM.68.7.3639-3643.2002
Hancock, D. D., Besser, T. E., Kinsel, M. L., Tarr, P. I., Rice, D. H., and Paros, M. G. (1994). The prevalence of Escherichia coli O157.H7 in dairy and beef cattle in Washington State. Epidemiol. Infect. 113, 199–207. doi: 10.1017/S0950268800051633
Hao, L. Y., Willis, D. K., Andrews-Polymenis, H., McClelland, M., and Barak, J. D. (2012). Requirement of siderophore biosynthesis for plant colonization by Salmonella enterica. Appl. Environ. Microbiol. 78, 4561–4570. doi: 10.1128/AEM.07867-11
Heuer, H., Binh, C. T. T., Jechalke, S., Kopmann, C., Zimmerling, U., Krögerrecklenfort, E., et al. (2012). Inc-P1ε plasmids are important vectors of antibiotic resistance genes in agricultural systems: diversification driven by class 1 integron gene cassettes. Front. Microbiol. 3:e2. doi: 10.3389/fmicb.2012.00002
Heuer, H., Krögerrecklenfort, E., Wellington, E. M., Egan, S., Van Elsas, J. D., Van Overbeek L., et al. (2002). Gentamicin resistance genes in environmental bacteria: prevalence and transfer. FEMS Microbiol. Ecol. 42, 289–302. doi: 10.1111/j.1574-6941.2002.tb01019.x
Heuvelink, A. E., van den Biggelaar, F., de Boer, E., Herbes, R. G., Melchers, W. J. G., Huis in ’t Veld, J. H. J., et al. (1998a). Isolation and characterization of verocytotoxin-producing Escherichia coli O157 strains from Dutch cattle and sheep. J. Clin. Microbiol. 36, 878–882.
Heuvelink, A. E., Van den Biggelaar, F. L. A. M., Zwartkruis-Nahuis, J. T. M., Herbes, R. G., Huyben, R., Nagelkerke, N., et al. (1998b). Occurrence of verocytotoxin-producing Escherichia coli O157 on Dutch dairy farms. J. Clin. Microbiol. 36, 3480–3487.
Heuvelink, A. E., Zwartkruis-Nahuis, J. T. M., van den Biggelaar, F., van Leeuwen, W. J., and de Boer, E. (1999). Isolation and characterization of verocytotoxin-producing Escherichia coli O157 from slaughter pigs and poultry. Int. J. Food Microbiol. 52, 67–75. doi: 10.1016/S0168-1605(99)00119-1
Hilborn, E. D., Mermin, J. H., Mshar, P. A., Hadler, J. L., Voetsch, A., Wojtkunski, C., et al. (1999). A multistate outbreak of Escherichia coli O157:H7 infections associated with consumption of mesclun lettuce. Arch. Int. Med. 159,1758–1764. doi: 10.1001/archinte.159.15.1758
Holden, N., Pritchard, L., and Toth, I. (2009). Colonization outwith the colon: plants as an alternative environmental reservoir for human pathogenic enterobacteria. FEMS Microbiol. Rev. 33, 689–703. doi: 10.1111/j.1574-6976.2008.00153.x
Hora, R., Warriner, K., Shelp, B. J., and Griffiths, M. W. (2005). Internalization of Escherichia coli O157:H7 following biological and mechanical disruption of growing spinach plants. J. Food Prot. 68, 2506–2509.
Hou, Y., and Taylor, D. E. (1994). Incidence of tellurite resistance determinants among plasmids of different incompatibility groups. Plasmid 32, 306–311. doi: 10.1006/plas.1994.1069
Iguchi, A., Thomson, N. R., Ogura, Y., Saunders, D., Ooka, T., Henderson, I. R., et al. (2009). Complete genome sequence and comparative genome analysis of enteropathogenic Escherichia coli O127:H6 strain E2348/69. J. Bacteriol. 191, 347–354. doi: 10.1128/JB.01238-08
Islam, M., Doyle, M. P., Phatak, S. C., Millner, P., and Jiang, X. (2004a). Persistence of enterohemorrhagic Escherichia coli O157:H7 in soil and on leaf lettuce and parsley grown in fields treated with contaminated manure composts or irrigation water. J. Food Prot. 67, 1365–1370.
Islam, M., Morgan, J., Doyle, M. P., Phatak, S. C., Millner, P., and Jiang, X. (2004b). Fate of Salmonella enterica serovar Typhimurium on carrots and radishes grown in fields treated with contaminated manure composts or irrigation water. Appl. Environ. Microbiol. 70, 2497–2502. doi: 10.1128/AEM.70.4.2497-2502.2004
Jacobsen, C. S., and Bech, T. B. (2012). Soil survival of Salmonella and transfer to freshwater and fresh produce. Food Res. Int. 45, 557–566. doi: 10.1016/j.foodres.2011.07.026
James, C. E., Stanley, K. N., Allison, H. E., Flint, H. J., Stewart, C. S., Sharp, R. J., et al. (2001). Lytic and lysogenic infection of diverse Escherichia coli and Shigella strains with a verocytotoxigenic bacteriophage. Appl. Environ. Microbiol.67, 4335–4337. doi: 10.1128/AEM.67.9.4335-4337.2001
Jiang, X., Morgan, J., and Doyle, M. P. (2002). Fate of Escherichia coli O157:H7 in manure-amended soil. Appl. Environ. Microbiol. 68, 2605–2609. doi: 10.1128/AEM.68.5.2605-2609.2002
Johnson, T. J., and Nolan, L. K. (2009). Pathoge nomics of the virulence plasmids of Escherichia coli. Microbiol. Mol. Biol. Rev. 73, 750–774. doi: 10.1128/MMBR.00015-09
Johnson, T. J., Thorsness, J. L., Anderson, C. P., Lynne, A. M., Foley, S. L., Han, J., et al. (2010). Horizontal gene transfer of a ColV plasmid has resulted in a dominant avian clonal type of Salmonella enterica serovar Kentucky. PLoS ONE 5:e15524. doi: 10.1371/journal.pone.0015524
Johnston, M. A., Harrison, M. A., and Morrow, R. A. (2009). Microbial antagonists of Escherichia coli O157:H7 on fresh-cut lettuce and spinach. J. Food Prot. 72, 1569–1575.
Juhas, M., van der Meer, J. R., Gaillard, M., Harding, R. M., Hood, D. W., and Crook, D. W. (2009). Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol. Rev. 33, 376–393. doi: 10.1111/j.1574-6976.2008.00136.x
Karch, H., Denamus, E., Dobrindt, U., Finlay, B. B., Hengge, R., Johannes, L., et al. (2011). The enemy within us: lessons from the 2011 European Escherichia coli O104:H4 outbreak. EMBO Mol. Med. 4, 841–848. doi: 10.1002/emmm.201201662
Karmali, M. A., Gannon, V., and Sargeant, J. M. (2010). Verocytotoxin-producing Escherichia coli (VTEC). Vet. Microbiol. 140, 360–370. doi: 10.1016/j.vetmic.2009.04.011
Kenney, S. J., Anderson, G. L., Williams, P. L., Millner, P. D., and Beuchat, L. R. (2006). Migration of Caenorhabditis elegans to manure and manure compost and potential for transport of Salmonella newport to fruits and vegetables. Int. J. Food Microbiol. 106, 61–68. doi: 10.1016/j.ijfoodmicro.2005.05.011
Klerks, M. M., Franz, E., Van Gent-Pelzer, M., Zijlstra, C., and Van Bruggen, A. H. (2007a). Differential interaction of Salmonella enterica serovars with lettuce cultivars and plant-microbe factors influencing the colonization efficiency. ISME J. 1, 620–631. doi: 10.1038/ismej.2007.82
Klerks, M. M., van Gent-Pelzer, M., Franz, E., Zijlstra,C., and Van Bruggen, A. H. (2007b). Physiological and molecular responses of Lactuca sativa to colonization by Salmonella enterica serovar Dublin. Appl. Environ. Microbiol. 73, 4905–4914. doi: 10.1128/AEM.02522-06
Kidambi, S. P., Ripp, S., and Miller, R. V. (1994). Evidence for phage-mediated gene transfer among Pseudomonas aeruginosa strains on the phylloplane. Appl. Environ. Microbiol. 60, 496–500.
Kroupitski, Y., Golberg, D., Belausov, E., Pinto, R., Swartzberg, D., Granot, D., et al. (2009). Internalization of Salmonella enterica in leaves is induced by light and involves chemotaxis and penetration through open stomata. Appl. Environ. Microbiol. 75, 6076–6086. doi: 10.1128/AEM.01084-09
Kumar, A., Munder, A., Aravind, R., Eapen, S. J., Tümmler, B., and Raaijmakers, J. M. (2013). Friend or foe: genetic and functional characterization of plant endophytic Pseudomonas aeruginosa. Environ. Microbiol. 15, 764–779. doi: 10.1111/1462-2920.12031
Laing, C. R., Zhang, Y., Gilmour, M. W., Allen, V., Johnson, R., Thomas, J. E., et al. (2012). A comparison of Shiga-toxin 2 bacteriophage from classical enterohemorrhagic Escherichia coli serotypes and the German E. coli O104:H4 outbreak strain. PLoS ONE 7:e37362. doi: 10.1371/journal.pone.0037362
Lainhart, W., Stolfa, G., and Koudelka, G. B. (2009). Shiga toxin as a bacterial defense against a eukaryotic predator, Tetrahymena thermophila. J. Bacteriol. 191, 5116–5122. doi: 10.1128/JB.00508-09
Lee, D. H., Lee, J.-H., Shin, H., Ji, S., Roh, E., Jung, K., et al. (2012). Complete genome sequence of Pectobacterium carotovorum subsp. carotovorum bacteriophage My1. J. Virol. 86, 11410–11411. doi: 10.1128/JVI.01987-12
Lehman, S. M., Kropinski, A. M., Castle, A. J., and Svircev, A. M. (2009). Complete genome of the broad-host-range Erwinia amylovora phage ΦEa21-4 and its relationship to Salmonella phage Felix O1. Appl. Environ. Microbiol. 75, 2139–2147. doi: 10.1128/AEM.02352-08
Lilley, A. K., and Bailey, M. J. (1997). The acquisition of indigenous plasmids by a genetically marked pseudomonad population colonizing the sugar beet phytosphere is related to local environmental conditions. Appl. Environ. Microbiol 63, 1577–1583.
Ma, J., Ibekwe, A. M., Yi, X., Wang, X. H., Yamazaki, A., Crowley, D. E., et al. (2011). Persistence of Escherichia coli O157:H7 and its mutants in soils. PLoS ONE 6:e23191. doi: 10.1371/journal.pone.0023191
Macarisin, D., Patel, J., Bauchan, G., Giron, J. A., and Sharma, V. K. (2012). Role of curli and cellulose expression in adherence of Escherichia coli O157:H7 to spinach leaves. Foodborne Pathog. Dis. 9, 160–167. doi: 10.1089/fpd.2011.1020
Mauro, S. A., and Koudelka, G. B. (2011). Shiga toxin: expression, distribution, and its role in the environment. Toxins 3, 608–625. doi: 10.3390/toxins3060608
McCully, M. E. (2001). Niches for bacterial endophytes in crop plants: a plant biologist’s view. Aust. J. Plant Physiol. 28, 983–990.
McGhee, G. C., and Jones, A. L. (2000). Complete nucleotide sequence of ubiquitous plasmid pEA29 from Erwinia amylovora strain Ea88: gene organization and intraspecies variation. Appl. Environ. Microbiol. 66, 4897–4907. doi: 10.1128/AEM.66.11.4897-4907.2000
Mela, F., Fritsche, K., Boersma, H., Van Elsas, J. D., Bartels, D., Meyer, F., et al. (2008). Comparative genomics of the pIPO2/pSB102 family of environmental plasmids: sequence, evolution, and ecology of pTer331 isolated from Collimonas fungivorans Ter331. FEMS Microbiol. Ecol. 66, 45–62. doi: 10.1111/j.1574-6941.2008.00472.x
Mellmann, A., Bielaszewska, M., and Karch, H. (2009). Intrahost genome alterations in enterohemorrhagic Escherichia coli. Gastroenterology 136, 1925–1938. doi: 10.1053/j.gastro.2008.12.072
Mellmann, A., Harmsen, D., Cummings, C. A., Zentz, E. B., Leopold, S. R., Rico, A., et al. (2011). Prospective genomic characterisation of the German Enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology”. PLoS ONE 6:e22751. doi: 10.1371/journal.pone.0022751
Michino, H., Araki, K., Minami, S., Takaya, S., Sakai, N., Miyazaki, M., et al. (1999). Massive outbreak of Escherichia coli O157:H7 infection in schoolchildren in Sakai City, Japan, associated with consumption of white radish sprouts. Am. J. Epidemiol. 150, 787–796. doi: 10.1093/oxfordjournals.aje.a010082
Milnes, A. S., Sayers, A. R., Stewart, I., Clifton-Hadley, F. A., Davies, R. H., Newell, D. G., et al. (2009). Factors related to the carriage of Verocytotoxigenic E. coli, Salmonella, thermophilic Campylobacter and Yersinia enterocolitica in cattle, sheep and pigs at slaughter. Epidemiol. Infect. 137, 1135–1148. doi: 10.1017/S095026880900199X
Mohle-Boetani, J. C., Farrar, J. A., Werner, S. B., Minassian, D., Bryant, R., Abbott, S., et al. (2001). Escherichia coli O157 and Salmonella infections associated with sprouts in California, 1996–1998. Ann. Int. Med. 135, 239–247. doi: 10.7326/0003-4819-135-4-200108210-00008
Mølbak, L., Licht, T. R., Kvist, T., Kroer, N., and Andersen, S. R. (2003). Plasmid transfer from Pseudomonas putida to the indigenous bacteria on alfalfa sprouts: characterization, direct quantification, and in situ location of transconjugant cells. Appl. Environ. Microbiol. 69, 5536–5542. doi: 10.1128/AEM.69.9.5536-5542.2003
Morgan, G. M., Newman, C., Palmer, S. R., Allen, J. B., Shepherd, W., Rampling, A. M., et al. (1988). First recognized community outbreak of haemorrhagic colitis due to verotoxin-producing Escherichia coli O 157:H7 in the UK. Epidemiol. Infect. 101, 83–91. doi: 10.1017/S0950268800029241
Muniesa, M., Hammerl, J. A., Hertwig,S., Appel, B., and Brüssow, H. (2012). Shiga toxin-producing Escherichia coli O104:H4: a new challenge for microbiology. Appl. Environ. Microbiol. 78, 4065–4073. doi: 10.1128/AEM.00217-12
Naiemi, N. A., Duim, B., Savelkoul, P. H. M., Spanjaard, L., De Jonge, E., Bart, A., et al. (2005). Widespread transfer of resistance genes between bacterial species in an intensive care unit: implications for hospital epidemiology. J. Clin. Microbiol. 43, 4862–4864. doi: 10.1128/JCM.43.9.4862-4864.2005
Noguera, P., Posthuma-Trumpie, G., Van Tuil, M., Van der Wal, F., De Boer, A., Moers, A., et al. (2011a). Carbon nanoparticles in lateral flow methods to detect genes encoding virulence factors of Shiga toxin-producing Escherichia coli. Anal. Bioanal. Chem. 399, 831–838. doi: 10.1007/s00216-010-4334-z
Noguera, P., Posthuma-Trumpie, G., Van Tuil, M., Van der Wal, F., De Boer, A., Moers, A., et al. (2011b). Carbon nanoparticles as detection labels in antibody microarrays, detection of genes encoding virulence factors in Shiga toxin-producing Escherichia coli. Anal. Chem. 83, 8531–8536. doi: 10.1021/ac201823v
Normander, B., Christensen, B. B., Molin, S., and Kroer, N. (1998). Effect of bacterial distribution and activity on conjugal gene transfer on the phylloplane of the bush bean (Phaseolus vulgaris). Appl. Environ. Microbiol. 64, 1902–1909.
Nunes da Rocha, U., Van Elsas, J. D., and Van Overbeek, L. S. (2011). Verrucomicrobia subdivision 1 strains display a difference in colonization in the colonization of the leek (Allium porrum) rhizosphere. FEMS Microbiol. Ecol. 78, 297–303. doi: 10.1111/j.1574-6941.2011.01156.x
Ogura, Y., Abe, H., Katsura,K., Kurokawa, K., Asadulghani, M., Iguchiet, A., et al. (2008). Systematic identification and sequence analysis of the genomic islands of the enteropathogenic Escherichia coli strain B171-8 by the combined use of whole-genome PCR scanning and fosmid mapping. J. Bacteriol. 190, 6948–6960. doi: 10.1128/JB.00625-08
Oliveira, M., Wijnands, L., Abadias, M., Aarts, H., and Franz, E. (2011). Pathogenic potential of Salmonella Typhimurium DT104 following sequential passage through soil, packaged fresh-cut lettuce and a model gastro-intestinal tract. Int. J. Food Microbiol. 148, 149–155.
Ongeng, D., Muyanja, C., Ryckeboer, J., Geeraerd, A. H., and Springael, D. (2011a). Rhizosphere effect on survival of Escherichia coli O157:H7 and Salmonella enterica serovar Typhimurium in manure-amended soil during cabbage (Brassica oleracea) cultivation under tropical field conditions in Sub-Saharan Africa. Int. J. Food Microbiol. 149, 133–142. doi: 10.1016/j.ijfoodmicro.2011.06.009
Ongeng, D., Vasquez, G. A., Muyanja, C., Ryckeboer, J., Geeraerd, A. H., and Springael, D. (2011b). Transfer and internalisation of Escherichia coli O157:H7 and Salmonella enterica serovar Typhimurium in cabbage cultivated on contaminated manure-amended soil under tropical field conditions in Sub-Saharan Africa. Int. J. Food Microbiol. 145, 301–310. doi: 10.1016/j.ijfoodmicro.2011.01.018
Pearce, M. C., Evans, J., McKendrick, I. J., Smith, A. W., Knight, H. I., Mellor, D. J., et al. (2006). Prevalence and virulence factors of Escherichia coli serogroups O26, O103, O111, and O145 shed by cattle in Scotland. Appl. Environ. Microbiol. 72, 653–659. doi: 10.1128/AEM.72.1.653-659.2006
Pearce, M. C., Jenkins, C., Vali, L., Smith, A. W., Knight, H. I., Cheasty, T., et al. (2004). Temporal shedding patterns and virulence factors of Escherichia coli serogroups O26, O103, O111, O145, and O157 in a cohort of beef calves and their dams. Appl. Environ. Microbiol. 70, 1708–1716. doi: 10.1128/AEM.70.3.1708-1716.2004
Pembroke, J. T., MacMahon, C., and McGrath, B. (2002). The role of conjugative transposons in the Enterobacteriaceae. Cell. Mol. Life Sci. 59, 2055–2064. doi: 10.1007/s000180200005
Perna, N. T., Plunkett, G., Burland., V., Mau, B., Glasner, J. D., Rose, D. J., et al. (2001). Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409, 529–533. doi: 10.1038/35054089
Popowska, M., and Krawczyk-Balska, A. (2013). Broad-host-range IncP-1 plasmids and their resistance potential. Front. Microbiol. 4:e44. doi: 10.3389/fmicb.2013.00044
Pukall, R., Tschäpe, H., and Smalla, K. (1996). Monitoring the spread of broad host and narrow host range plasmids in soil microcosms. FEMS Microbiol. Lett. 20, 53–66. doi: 10.1111/j.1574-6941.1996.tb00304.x
Rahme, L. G., Ausubel, F. M., Cao, H., Drenkard, E., Goumnerov, B. C., Lau, G. W., et al. (2000). Plants and animals share functionally common bacterial virulence factors. Proc. Natl. Acad. Sci. U.S.A. 97, 8815–8821. doi: 10.1073/pnas.97.16.8815
Römling, U. (2005). Characterization of the rdar morphotype, a multicellular behaviour in Enterobacteriaceae. Cell. Mol. Life Sci. 62, 1234–1246. doi: 10.1007/s00018-005-4557-x
Rooks, D. J., Libberton, B., Woodward, M. J., Allison, H. E., and McCarthy, A. J. (2012). Development and application of a method for the purification of free Shiga-toxigenic bacteriophage from environmental samples. J. Microbiol. Methods 91, 240–245. doi: 10.1016/j.mimet.2012.08.017
Saldaña, Z., Sanchez, E., Xicohtencatl-Cortes, J., Puente, J. L., and Giron, J. A. (2011). Surface structures involved in plant stomata and leaf colonization by Shiga-toxigenic Escherichia coli O157:H7. Front. Microbiol. 2:e119. doi: 10.3389/fmicb.2011.00119
Saunders, J. R., Allison, H., James, C. E., McCarthy, A. J., and Sharp, R. (2001). Phage-mediated transfer of virulence genes. J. Chem. Technol. Biotechnol. 76, 662–666. doi: 10.1002/jctb.437
Saye, D. J., Ogunseitan, O. A., Sayler, G. S., and Miller, R. V. (1990). Transduction of linked chromosomal genes between Pseudomonas aeruginosa strains during incubation in situ in a freshwater habitat. Appl. Environ. Microbiol. 56, 140–145.
Scheutz, F., Møller Nielsen, E. M., Frimodt-Møller, J., Boisen, N., Morabito, S., Tozzoli R., et al. (2011). Characteristics of the enteroaggregative Shiga toxin/verotoxin-producing Escherichia coli O104:H4 strain causing the outbreak of haemolytic uraemic syndrome in Germany, May to June 2011. Euro surveill. 16, 19889. www.eurosurveillance.org.
Schikora, A., Garcia, A. V., and Hirt, H. (2012). Plants as alternative hosts for Salmonella. Trends Plant Sci. 17, 245–249. doi: 10.1016/j.tplants.2012.03.007
Schneider, G., Dobrindt, U., Middendorf, B., Hochhut, B., Szijarto, V., Emody, L., et al. (2011). Mobilisation and remobilisation of a large archetypal pathogenicity island of uropathogenic Escherichia coli in vitro support the role of conjugation for horizontal transfer of genomic islands. BMC Microbiol. 11:210. doi: 10.1186/1471-2180-11-210
Schouten, J. M., Bouwknegt, M., Van De Giessen, A. W., Frankena, K., De Jong, M. C. M., and Graat, E. A. M. (2004). Prevalence estimation and risk factors for Escherichia coli O157 on Dutch dairy farms. Prev. Vet. Med. 64, 49–61. doi: 10.1016/j.prevetmed.2004.03.004
Schouten, J. M., Van de Giessen, A. W., Frankena, K., De Jong, M. C. M., and Graat, E. A. M. (2005) Escherichia coli O157 prevalence in Dutch poultry, pig finishing and veal herds and risk factors in Dutch veal herds. Prev. Vet. Med. 70, 1–15. doi: 10.1016/j.prevetmed.2004.12.010
Schubert, S., Darlu, P., Clermont, O., Wieser, A., Magistro, G., Hoffmann, C., et al. (2009). Role of intraspecies recombination in the spread of pathogenicity islands within the Escherichia coli species. PLoS Pathog. 5:e1000257. doi: 10.1371/journal.ppat.1000257
Schubert, S., Dufke, S., Sorsa, J., and Heesemann, J. (2004). A novel integrative and conjugative element (ICE) of Escherichia coli: the putative progenitor of the Yersinia high-pathogenicity island. Mol. Microbiol. 51, 837–848. doi: 10.1046/j.1365-2958.2003.03870.x
Semenov, A. V., Franz, E., Van Overbeek, L., Termorshuizen, A. J., and Van Bruggen, A. H. (2008). Estimating the stability of Escherichia coli O157:H7 survival in manure-amended soils with different management histories. Environ. Microbiol. 10, 1450–1459. doi: 10.1111/j.1462-2920.2007.01558.x
Semenov, A. V., Van Bruggen, A. H., Van Overbeek, L., Termorshuizen, A. J., and Semenov, A. M. (2007). Influence of temperature fluctuations on Escherichia coli O157:H7 and Salmonella enterica serovar Typhimurium in cow manure. FEMS Microbiol. Ecol. 60, 419–428. doi: 10.1111/j.1574-6941.2007.00306.x
Semenov, A. V., Van Overbeek, L., and Van Bruggen, A. H. (2009). Percolation and survival of Escherichia coli O157:H7 and Salmonella enterica serovar Typhimurium in soil amended with contaminated dairy manure or slurry. Appl. Environ. Microbiol. 75, 3206–3215. doi: 10.1128/AEM.01791-08
Shah, J., Desai, P. T., Chen, D., Stevens, J. R., and Weimer, B. C. (2013). Preadaptation to cold stress in Salmonella enterica serovar typhimurium increases survival during subsequent acid stress. Appl. Environ. Microbiol. 79, 7281–7289. doi: 10.1128/AEM.02621-13
Shaw, D. J., Jenkins, C., Pearce, M. C., Cheasty, T., Gunn, G. J., Dougan, G., et al. (2004). Shedding patterns of verocytotoxin-producing Escherichia coli strains in a cohort of calves and their dams on a Scottish beef farm. Appl. Environ. Microbiol. 70, 7456–7465. doi: 10.1128/AEM.70.12.7456-7465.2004
Shin, S. Y., Young Lee, M., Song, J.-H., and Ko, K. S. (2008). New Erwinia-like organism causing cervical lymphadenitis. J. Clin. Microbiol. 46, 3156–3158. doi: 10.1128/JCM.00716-08
Smalla, K., Heuer, H., Gotz, A., Niemeyer, D., Krögerrecklenfort, E., and Tietze, E. (2000). Exogenous isolation of antibiotic resistance plasmids from piggery manure slurries reveals a high prevalence and diversity of IncQ-like plasmids. Appl. Environ. Microbiol. 66, 4854–4862. doi: 10.1128/AEM.66.11.4854-4862.2000
Solomon, E. B., Yaron, S., and Matthews, K. R. (2002). Transmission of Escherichia coli O157:H7 from contaminated manure and irrigation water to lettuce plant tissue and its subsequent internalization. Appl. Environ. Microbiol. 68, 397–400. doi: 10.1128/AEM.68.1.397-400.2002
Stecher, B., Denzler, R., Maier, L., Bernet, F., Sanders, M. J., and Pickard, D. J., et al. (2012). Gut inflammation can boost horizontal gene transfer between pathogenic and commensal Enterobactericaceae. Proc. Natl. Acad. Sci. U.S.A. 109, 1269–1274. doi: 10.1073/pnas.1113246109
Szmolka, A., and Nagy, B. (2013). Multidrug resistant commensal Escherichia coli in animals and its impact in public health. Front. Microbiol. 4:e258. doi: 10.3389/fmicb.2013.00258
Taghavi, S., Barac, T., Greenberg, B., Borremans, B., Vangronsveld, J., and Van der Lelie, D. (2005). Horizontal gene transfer to endogenous endophytic bacteria from poplar improves phytoremediation of toluene. Appl. Environ. Microbiol. 71, 8500–8505. doi: 10.1128/AEM.71.12.8500-8505.2005
Taghavi, S., Van der Lelie, D., Hoffman, A., Zhang, Y.-B., Walla, M. D., Vangronsveld, J., et al. (2010). Genome sequence of the plant growth promoting endophytic bacterium Enterobacter sp. 638. PLoS Genet. 6:e1000943. doi: 10.1371/journal.pgen.1000943
Talley, J. L., Wayadande, A. C., Wasala, L. P., Gerry, A. C., Fletcher, J., DeSilva, U., et al. (2009). Association of Escherichia coli O157:H7 with filth flies (Muscidae and Calliphoridae) captured in leafy greens fields and experimental transmission of E. coli O157:H7 to spinach leaves by house flies (Diptera: Muscidae). J. Food Prot. 72,1547–1552.
Tan, Z., Hurek, T., Gyaneshwar, P., Ladha, J. K., and Reinhold-Hurek, B. (2001). Novel endophytes of rice form a taxonomically distinct subgroup of Serratia marcescens. Syst. Appl. Microbiol. 24, 245–251. doi: 10.1078/0723-2020-00002
Tauch, A., Schneiker, S., Selbitschka, W., Puhler, A., van Overbeek, L. S., Smalla, K., et al. (2002). The complete nucleotide sequence and environmental distribution of the cryptic, conjugative, broad-host-range plasmid pIPO2 isolated from bacteria of the wheat rhizosphere. Microbiology 148, 1637–1653.
Taylor, D. E. (1999). Bacterial tellurite resistance. Trends Microbiol. 7, 111–115. doi: 10.1016/S0966-842X(99)01454-7
Taylor, D. E., Rooker, M., Keelan, M., Ng, L. K., Martin, I., Perna, N. T., et al. (2002). Genomic variability of O islands encoding tellurite resistance in enterohemorrhagic Escherichia coli O157:H7 isolates. J. Bacteriol. 184, 4690–4698. doi: 10.1128/JB.184.17.4690-4698.2002
Teplitski, M., Barak, J. D., and Schneider, K. R. (2009). Human enteric pathogens in produce: un-answered ecological questions with direct implications for food safety. Curr. Opin. Biotechnol. 20, 166–171. doi: 10.1016/j.copbio.2009.03.002
Teplitski, M., Warriner, K., Bartz J., and Schneider K. R. (2011). Untangling metabolic and communication networks: interactions of enterics with phytobacteria and their implications in produce safety. Trends Microbiol. 19, 121–127. doi: 10.1016/j.tim.2010.11.007
Toth, I., Schmidt, H., Dow, H., Malik, A., Oswald, E., and Nagy, B. (2003). Transduction of porcine enteropathogenic Escherichia coli with a derivative of a Shiga toxin 2-encoding bacteriophage in a porcine ligated ileal loop system. Appl. Environ. Microbiol. 69, 7242–7247. doi: 10.1128/AEM.69.12.7242-7247.2003
Toth, I. K., Pritchard, L., and Birch, P. R. (2006). Comparative genomics reveals what makes an enterobacterial plant pathogen. Annu. Rev. Phytopathol. 44, 305–336. doi: 10.1146/annurev.phyto.44.070505.143444
Tyler, H. L., and Triplett, E. W. (2008). Plants as a habitat for beneficial and/or human pathogenic bacteria. Annu. Rev. Phytopathol. 46, 53–73. doi: 10.1146/annurev.phyto.011708.103102
Valkenburgh, S., van Oosterom, R., Stenvers, O., Aalten, M., Braks, M., Schimmer, B., et al. (2007). Zoonoses and zoonotic agents in humans, food, animals and feed in the Netherlands 2003–2006. RIVM Report 330152001, 140.
van der Linden, I., Cottyn, B., Uyttendaele, M., Vlaemynck, G., Maes, M., and Heyndrickx, M. (2013). Long-term survival of Escherichia coli O157:H7 and Salmonella enterica on butterhead lettuce seeds, and their subsequent survival and growth on the seedlings. Int. J. Food Microbiol. 161, 214–219. doi: 10.1016/j.ijfoodmicro.2012.12.015
van der Riet, F. S. (1997). Diseases of plants transmissible between plants and man (phytonoses) exist. Med. Hypotheses 49, 359–361. doi: 10.1016/S0306-9877(97)90202-4
van Elsas, J. D., Chiurazzi, M., Mallon, C. A., Elhottova, D., Kristufek, V., and Salles, J. F. (2012). Microbial diversity determines the invasion of soil by a bacterial pathogen. Proc. Natl. Acad. Sci. U.S.A. 109, 1159–1164. doi: 10.1073/pnas.1109326109
Van Elsas, J. D., Gardener, B. B., Wolters, A. C., and Smit, E. (1998). Isolation, characterization, and transfer of cryptic gene-mobilizing plasmids in the wheat rhizosphere. Appl. Environ. Microbiol. 64, 880–889.
van Hoek, A. H., Aarts, H. J., Bouw, E., Van Overbeek, W. M., and Franz, E. (2013). The role of rpoS in Escherichia coli O157 manure-amended soil survival and distribution of allelic variations among bovine, food and clinical isolates. FEMS Microbiol. Lett. 338, 18–23. doi: 10.1111/1574-6968.12024
Van Meervenne, E., Van Coillie, E., Kerckhof, F. M., Devlieghere, F., Herman, L., De Gelder, L. S., et al. (2012). Strain-specific transfer of antibiotic resistance from an environmental plasmid to foodborne pathogens. J. Biomed. Biotechnol. 2012, e834598. doi: 10.1155/2012/834598
van Overbeek, L. S., Bergervoet, J. H. W., Jacobs, F., and Van Elsas, J. D. (2004). The low-temperature-induced viable-but-nonculturable state affects the virulence of Ralstonia solanacearum biovar 2. Phytopathology 94, 463–469. doi: 10.1094/PHYTO.2004.94.5.463
Van Overbeek, L. S., Eberl, L., Givskov, M., Molin, S., and Van Elsas, J. D. (1995). Survival of, and induced stress resistance in, carbon-starved Pseudomonas fluorescens cells residing in soil. Appl. Environ. Microbiol. 61, 4202–4208.
Van Overbeek, L. S., Franz, E., Semenov, A. V., De Vos, O. J., and Van Bruggen, A. H. C. (2010). The effect of the native bacterial community structure on the predictability of E. coli O157:H7 survival in manure-amended soil. Lett. Appl. Microbiol. 50, 425–430. doi: 10.1111/j.1472-765X.2010.02817.x
van Overbeek, L. S., Wellington, E. M., Egan, S., Smalla, K., Heuer, H., Collard, J. M., et al. (2002). Prevalence of streptomycin-resistance genes in bacterial populations in European habitats. FEMS Microbiol. Ecol. 42, 277–288. doi: 10.1111/j.1574-6941.2002.tb01018.x
Vimont, S., Poirel, L., Naas, T., and Nordmann, P. (2002). Identification of a chromosome-borne expanded-spectrum class A β-lactamase from Erwinia persicina. Antimicrob. Agents Chemother. 46, 3401–3405. doi: 10.1128/AAC.46.11.3401-3405.2002
Vivian, A., Murillo, J., and Jackson, R. W. (2001).The roles of plasmids in phytopathogenic bacteria: mobile arsenals? Microbiology 147, 763–780.
Wagner, P., and Waldor, M. K. (2002). Bacteriophage control of bacterial virulence. Infect. Immun. 70, 3985–3993. doi: 10.1128/IAI.70.8.3985-3993.2002
Warriner, K., Ibrahim, F., Dickinson, M., Wright, C., and Waites, W. M. (2003). Internalization of human pathogens within growing salad vegetables. Biotechnol. Genet. Eng. Rev. 20, 117–134. doi: 10.1080/02648725.2003.10648040
Warriner, K., and Namvar, A. (2010). The tricks learnt by human enteric pathogens from phytopathogens to persist within the plant environment. Curr. Opin. Biotechnol. 21, 131–136. doi: 10.1016/j.copbio.2010.01.004
Wehmeier, U. F., Nobelmann, B., and Lengeler, J. W. (1992). Cloning of the Escherichia coli sor genes for L-sorbose transport and metabolism and physical mapping of the genes near metH and iclR. J. Bacteriol. 174, 7784–7790. doi: 10.1111/j.1365-2958.1990.tb02067.x
Wei, B., Huang, T., Dalwadi, H., Sutton, C. L., Bruckner, D., and Braun, J. (2002). Pseudomonas fluorescens encodes the Crohn’s disease-associated I2 sequence and T-cell superantigen. Infect. Immun. 70, 6567–6575. doi: 10.1128/IAI.70.12.6567-6575.2002
Wells, J. G., Shipman, L. D., Greene, K. D., Sowers, E. G., Green, J. H., Cameron, D. N., et al. (1991). Isolation of Escherichia coli serotype O157:H7 and other Shiga-like-toxin-producing E. coli from dairy cattle. J. Clin. Microbiol. 29, 985–989.
Wendel, A. M., Johnson, D. H., Sharapov, U., Grant, J., Archer, J. R., Monson, T., et al. (2009). Multistate outbreak of Escherichia coli O157:H7 infection associated with consumption of packaged spinach, August-September 2006: the Wisconsin investigation. Clin. Infect. Dis. 15, 1079–1086. doi: 10.1086/597399
Wieler, L. H., Semmler, T., Eichhorn, I., Antao, E. M., Kinnemann, B., Geue, L., et al. (2011). No evidence of the Shiga toxin-producing E. coli O104:H4 outbreak strain or enteroaggregative E. coli (EAEC) found in cattle faeces in northern Germany, the hotspot of the 2011 HUS outbreak area. Gut Pathog. 3, 17. doi: 10.1186/1757-4749-3-17
Wieler, L. H., Vieler, E., Erpenstein, C., Schlapp, T., Steinruck, H., and Bauerfeind, R. (1996). Shiga toxin-producing Escherichia coli strains from bovines: association of adhesion with carriage of eae and other genes. J. Clin. Microbiol. 34, 2980–2984.
Wohlhieter, J. A., Lazere, J. R., Snellings, N. J., Johnson, E. M., Synenki, R. M., and Baron, L. S. (1975). Characterization of transmissible genetic elements from sucrose-fermenting Salmonella strains. J. Bacteriol. 122, 401–406.
Wright, K. M., Chapman, S., McGeachy, K., Humphris, S., Campbell, E., Toth, I. K., et al. (2013). The endophytic lifestyle of Escherichia coli O157:H7: quantification and internal localization in roots. Phytopathology 103, 333–340. doi: 10.1094/PHYTO-08-12-0209-FI
Keywords: EHEC, emerging pathogens, phytonoses, gene transfer, rhizosphere, Enterobacteriaceae
Citation: van Overbeek LS, van Doorn J, Wichers JH, van Amerongen A, van Roermund HJW and Willemsen PTJ (2014) The arable ecosystem as battleground for emergence of new human pathogens. Front. Microbiol. 5:104. doi: 10.3389/fmicb.2014.00104
Received: 18 November 2013; Accepted: 27 February 2014;
Published online: 20 March 2014.
Edited by:
Gabriele Berg, Graz University of Technology, AustriaReviewed by:
Eelco Franz, Centre for Infectious Disease Control, NetherlandsPaolina Garbeva, Netherlands Institute of Ecology, Netherlands
Copyright © 2014 van Overbeek, van Doorn, Wichers, van Amerongen, van Roermund and Willemsen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Leonard S. van Overbeek, Plant Research International, Wageningen University and Research Centre, Droevendaalsesteeg 1, 6708 PB, Wageningen, Netherlands e-mail:bC5zLnZhbm92ZXJiZWVrQHd1ci5ubA==
†Present address: Joop van Doorn, Enza Zaden Seed Operations BV, Enhuizen, Netherlands
 Leonard S. van Overbeek
Leonard S. van Overbeek Joop van Doorn
Joop van Doorn Jan H. Wichers
Jan H. Wichers Aart van Amerongen
Aart van Amerongen Herman J. W. van Roermund
Herman J. W. van Roermund Peter T. J. Willemsen
Peter T. J. Willemsen