- DOE Great Lakes Bioenergy Research Center, University of Wisconsin-Madison, Madison, WI, USA
Lignocellulosic hydrolysate (LCH) inhibitors are a large class of bioactive molecules that arise from pretreatment, hydrolysis, and fermentation of plant biomass. These diverse compounds reduce lignocellulosic biofuel yields by inhibiting cellular processes and diverting energy into cellular responses. LCH inhibitors present one of the most significant challenges to efficient biofuel production by microbes. Development of new strains that lessen the effects of LCH inhibitors is an economically favorable strategy relative to expensive detoxification methods that also can reduce sugar content in deconstructed biomass. Systems biology analyses and metabolic modeling combined with directed evolution and synthetic biology are successful strategies for biocatalyst development, and methods that leverage state-of-the-art tools are needed to overcome inhibitors more completely. This perspective considers the energetic costs of LCH inhibitors and technologies that can be used to overcome their drain on conversion efficiency. We suggest academic and commercial research groups could benefit by sharing data on LCH inhibitors and implementing “translational biofuel research.”
Introduction
Lignocellulosic biofuels offer the promise of sustainable, domestically produced fuels with favorable carbon balances. Fast-growing grasses like Miscanthus and agricultural residues provide fermentable sugars at lower energy and fertilizer costs than grains (Schmer et al., 2008), making them preferable feedstocks for advanced biofuels. Cellulosic ethanol is an obvious next-generation biofuel to implement given its production and delivery infrastructures are compatible with existing fuels.
Central to the success of cellulosic ethanol is efficient conversion of biomass-derived sugars to ethanol by microbes such as Saccharomyces cerevisiae, Escherichia coli, and Zymomonas mobilis (Alper and Stephanopoulos, 2009; Lau et al., 2010; Yang et al., 2010a). Under optimal conditions, these microbes are powerful ethanologens; however, lignocellulosic hydrolysates (LCH) and industrial scale fermentation tanks are not optimal conditions. Thermal, osmotic, and ethanol stresses are just some of the environmental factors that inhibit fermentation and reduce yield (Attfield, 1997; Gibson et al., 2007; Jin et al., 2013). Industrial microbes are pushed to the limits of stress tolerance to make biofuel production energetically favorable.
Although environmental stressors limit yields in present day ethanol facilities, cellulosic biomass conversion comes with new challenges. Specifically, LCH inhibitors, a group of small, bioactive molecules can significantly reduce conversion efficiency. LCH inhibitors such as aliphatic acids, furans, and phenolics are released or condensed from cellulose and hemicellulose during pretreatment and hydrolysis (Larsson et al., 1999, 2000; Yang et al., 2010a); however, chemical residues from newer hydrolysis strategies and synergies with biofuel end products (ethanol, isobutanol) are less well studied. Removal of these inhibitors can be expensive and may reduce titers of fermentable sugars; some estimates suggest that detoxification can remove up to 26% of total fermentable sugars (Larsson et al., 1999). Thus, a preferred strategy is to develop microbial strains with properties that minimize the effects of LCH inhibitors on biofuel yields.
With commercially available industrial stains that are robust to thermal and ethanol stress (e.g., Ethanol Red, Fermentis, Milwaukee, WI, USA), recent attention has been directed to overcoming the challenge of LCH inhibitors. These compounds are ubiquitous in hydrolysates, and their abundance and composition depends on pretreatment (Chundawat et al., 2010), feedstock (Klinke et al., 2004; Almeida et al., 2007), and seasonality (Bunnell et al., 2013; Greenhalf et al., 2013). Given their chemical diversity, these compounds can target many cellular processes. LCH inhibitors can also generate a substantial cellular energy drain. Cells have evolved to detoxify, excrete inhibitors, or repair the resultant cellular damage fast enough to reproduce. However, evolved coping mechanisms may also negatively affect the efficiency of conversion by competing for cellular resources (Bellissimi et al., 2009; Miller et al., 2009). Although it is in the microbe's best interest to use its resources to limit the effects of LCH inhibitors and maintain cellular viability, this may be reducing biofuel production. In this perspective, we consider the diversity and cellular costs of LCH inhibitors from traditional and novel pretreatment and hydrolysis strategies, describe new technologies and their application to strain development, and finally identify key needs of the cellulosic biofuel community that will empower “translational biofuel research” to take discoveries quickly to industrial scale.
Diversity of Fermentation Inhibitors
Prior to microbial conversion of lignocellulosic sugars into biofuel, biomass must be deconstructed into monomeric sugars by enzymatic or chemical hydrolysis. This hydrolysis step is often preceded by a pretreatment step that expands the plant fibers and allows cellulolytic enzymes access to the polysaccharide matrices. The resulting hydrolysates are complex, ill-defined mixtures that include sugars and a diversity of bioactive molecules (Table 1). Small acids and phenolic compounds are released from cellulose and hemicelluloses during hydrolysis and furans arise from the dehydration of pentose and hexose monomers (Klinke et al., 2004). Pretreatments such as acid hydrolysis, steam explosion or NH3 expansion each impart their own “profile” of LCH inhibitors. For example, AFEX (Ammonia Fiber EXpansion) uses high-pressure/temperature ammonia to alter the cellulose matrix to allow hydrolysis by cellulases (Lau and Dale, 2009), and this produces amide versions of inhibitors (e.g., feruloyl amide from ferulic acid) with potentially new biological properties (Chundawat et al., 2010).
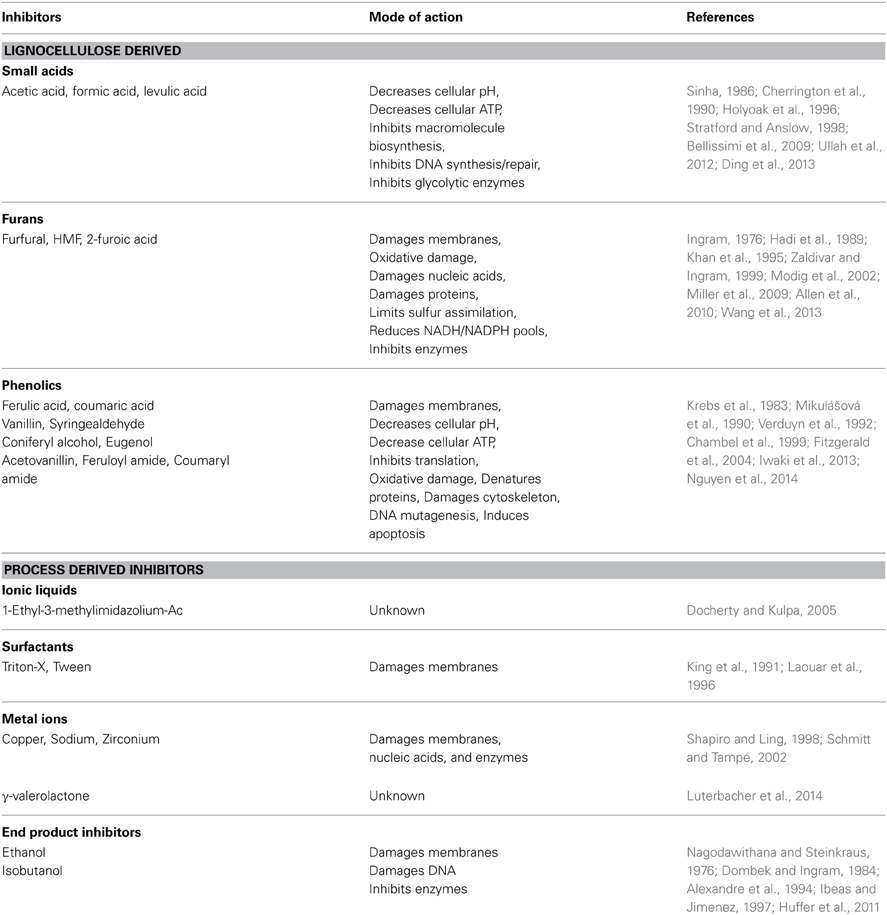
Table 1. Classes of lignocellulosic hydrolysate (LCH) inhibitors and their described modes of toxicity.
Besides these common inhibitors, residual pretreatment chemicals may complicate fermentation. Ionic liquids are pretreatment and hydrolysis solvents, but are toxic to many microorganisms (Docherty and Kulpa, 2005; Ouellet et al., 2011). Alkaline hydrogen peroxide (AHP) pretreatment limits the production of furans; however this method introduces significant amounts of Na+ from NaOH, which can cause osmotic stress (Sato et al., 2014). Copper(II) 2,2′-bipyridine is a catalyst that enhances AHP pretreatment by reducing the H2O2 requirement (Li et al., 2013), but copper is toxic to most microbes. Next-generation pre-treatments and hydrolysis methods like γ-valerolactone (Luterbacher et al., 2014), surfactants (Sindhu et al., 2013), zirconium phosphate catalysts (Gliozzi et al., 2014), and other incipient hydrolysis technologies may imbue hydrolysates with novel toxicities and synergisms with common inhibitors.
Biofuel end-products themselves are inhibitory. Ethanol can directly damage cellular membranes, DNA, as well as inhibit enzymes (Nagodawithana and Steinkraus, 1976; Dombek and Ingram, 1984; Alexandre et al., 1994; Ibeas and Jimenez, 1997; Huffer et al., 2011). The ethanologens S. cerevisiae and Z. mobilis are not immune to ethanol toxicity at high concentrations (Carmona-Gutierrez et al., 2012; Yang et al., 2013). Advanced biofuels like isobutanol are toxic at significantly lower concentrations than ethanol (Brynildsen and Liao, 2009; Atsumi et al., 2010; Huffer et al., 2011; Minty et al., 2011). Inhibition by end products has been an area of research interest (Baez et al., 2011; McEwen and Atsumi, 2012; Zingaro et al., 2013), and ethanol tolerance is a pre-requisite for all industrial yeast.
Effects of inhibitors at a minimum can be additive, but an even greater concern is the potential for synergy between LCH inhibitors and fermentation condition, including high osmolarity and absence of O2. Some studies have described synergies between acetic acid, furfural, and phenolics in yeast (Oliva et al., 2006; Ding et al., 2011), but a comprehensive evaluation of synergisms between compounds and conditions on both growth rate and fermentation will be essential. Such assessment will be a massive undertaking that will also require defined synthetic hydrolysate media to permit meaningful definition of minimum inhibitory concentrations (MICs) of the individual inhibitors, and how this value will change in combination with other LCH inhibitors (synergy/antagonism) and fermentation conditions. Nevertheless, documenting interactions between inhibitors on sugar conversion is crucial to prioritizing future research for improved biofuel microbes.
Small Molecule Inhibitors Deplete Cellular Resources
LCH inhibitors directly affect biofuel yield as well as the production rate, which can extend fermentation time and result in higher operating costs. In simplest terms, these inhibitors affect conversion efficiency by depleting cellular energy resources (e.g., ATP, NADH, NADPH; Figure 1). Some inhibitors can act broadly and damage key enzymes of fermentation pathways (Modig et al., 2002). The coping mechanisms available to the biofuel microbes fall into 4 main categories: (i) detoxification, (ii) efflux, (iii) repair, or (iv) tolerance. The first three are of most concern given their effects on cellular energy and resources.
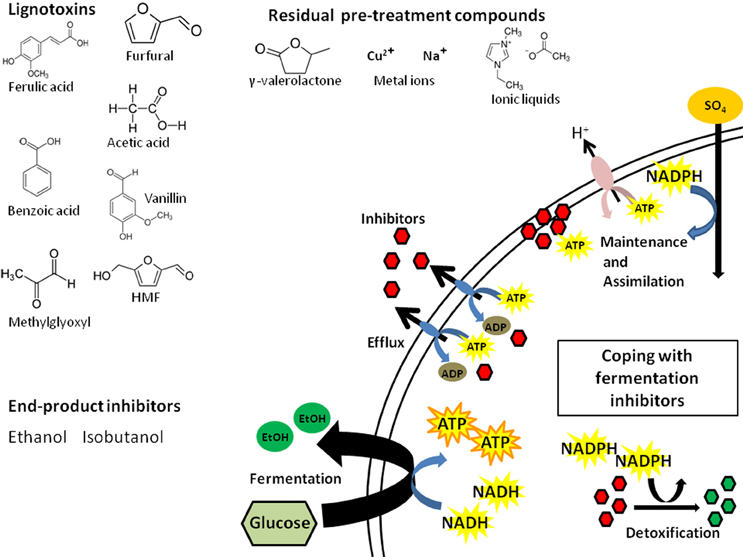
Figure 1. Inhibitor classes and the cellular energy consequences of LCH inhibitors. Presented are examples from three main classes of inhibitors and the ways cells can cope with these: efflux via pumps, detoxification via enzymes, and repair of the damage caused by the compounds. Each coping strategy comes at the expense of cellular energy that is diverted from the stores used to maintain cell integrity by chemiosmotic exchange and assimilation as well as energy required for fermentation of sugars to fuel.
Detoxification and efflux are the most well characterized mechanisms of inhibitor tolerance in microbes, with deep literature not only from biofuel research but also in the medical literature from a wealth of antibiotic and pharmaceutical research. Detoxification is the major route of tolerance for aldehydes in both bacteria and yeast. Reduction of compounds like furfural and vanillin to less toxic alcohols by NADH/NADPH dependent reductases occurs in ethanol fermentation using S. cerevisiae and E. coli (Jarboe, 2011). Oxidoreductase expression significantly increases in yeast and E. coli in the presence of aldehydes (Liu et al., 2008; Wang et al., 2011), depleting cellular NADH/NADPH. This results in inhibition of NADPH-dependent processes (e.g., assimilation of sulfur) leading ultimately to slower conversion of sugars (Miller et al., 2009). Interestingly, yields can be increased by disabling the detoxification pathway, suggesting that tolerance may be more energetically efficient than detoxification (Wang et al., 2013). Alternatively, changing the source of reducing equivalent for aldehyde detox from NADPH to NADH also can improve biofuel yield (Wang et al., 2013).
Efflux is mediated by ATP-dependent trans-membrane pumps that selectively or non-selectively pump out toxic compounds usually at the cost of 1 ATP per molecule (Shapiro and Ling, 1998; Schmitt and Tampé, 2002). The yeast S. cerevisiae has 29 different ATP-binding cassette (ABC) efflux transporters (Decottignies and Goffeau, 1997) and 5% of the E. coli genome is composed of genes with ABC-transporter domains, many involved in efflux (Linton and Higgins, 1998). In both yeast and E. coli, expression of transporters increases in response to LCH inhibitors (Schüller et al., 2004; Lee et al., 2012; Schwalbach et al., 2012). The yeast weak acid response is mediated by increased expression of ABC-transporters (e.g., Pdr1p and Pdr5p) (Schüller et al., 2004; Pereira Rangel et al., 2010). As long as cells are exposed to LCH inhibitors, a significant fraction of cellular ATP will be diverted to efflux pumps. Of particular importance for overall efficiency of LCH conversion, ATP-depletion may have a disproportionate effect on xylose conversion compared to glucose conversion because xylose produces less cellular energy per molecule transported (Matsushika et al., 2013). Bellissimi et al. (2009) found that acetic acid could specifically inhibit xylose fermentation, but that this effect could be reversed with glucose addition. The authors posit that ATP generated from xylose fermentation cannot match ATP depletion from the weak acid response and efflux/proton pumps used to maintain cellular pH. Inhibitors that require ATP-dependent coping mechanisms can directly reduce xylose conversion. Some inhibitors can be particularly draining, affecting both NADPH and ATP pools. Ask et al. (2013) found that furfural and HMF not only reduced cellular NAPDH in S. cerevisiae, but also elicited increased expression of ATP-dependent efflux pumps Pdr5p and Yor1p. This suggests that coping with furans requires both NADPH dependent detoxification and ATP-dependent efflux.
An unanswered question is whether the ATP-dependent action of efflux pumps has a net positive (by inhibitor removal) or net negative (by energy consumption) effect on biofuel production. Although the answer may vary by inhibitor, because some inhibitors like aldehydes are more damaging to cells than others, a general test of the positive or negative consequences of efflux pumps for biofuel yield will help advance strategies for biofuel microbe design.
The cellular energy costs of maintenance and repair are more difficult to quantify but could account for significant energy loss. If cells can repair the damage caused by fermentation inhibitors quickly, then fermentation may proceed. Inhibitors can acidify cells (Verduyn et al., 1992), damage cellular membranes (Russell, 1992), DNA (Allen et al., 2010), and individual proteins (Modig et al., 2002). Repairing structures requires biogenesis; this comes at the expense of ATP, NADPH, carbon, and nitrogen. Maintaining pH is mediated by ATP-dependent proton pump Pma1p; and ATP cost under acidic conditions is the primary cause of reduced cellular growth (Verduyn et al., 1992; Ullah et al., 2012). Biogenesis requires NADPH-dependent assimilation of nutrients like sulfur, which is drained by repair enzymes. Phenolics and furans can damage membranes, requiring more energy to maintain the proton gradient required for basic metabolism (Ding et al., 2012; Schwalbach et al., 2012; Stratford et al., 2013). Growth and sugar conversion will be slowed as resources are diverted to maintenance and repair. Given that hydrolysates contain a mixture of inhibitors with diverse modes of action requiring all of these coping mechanisms simultaneously, the energy drain from fermentation inhibitors is truly death by a thousand cuts.
Adapting to the Changing Landscape of LCH Inhibitors
Strain development is an economical route to deal with LCH inhibitors. Resistance to the suite of inhibitors requires complex response of many cellular systems, and as such is not easily conferred by engineering of individual genes. Moreover, inhibitor pools can vary between hydrolysate preparations, thus even the most robust strains in ammonia-pretreated hydrolysate may wither in dilute acid pretreated hydrolysates. It is unlikely that one strain will be optimal for all conditions. The reality is that microbial strains will need to be tailored to specific hydrolysates through engineering and directed evolution. Accelerating this process is crucial to making new cellulosic technologies industrially viable.
The tools of systems biology can give a detailed view of the microbial stress response (Jozefczuk et al., 2010; Lee et al., 2011). Transcriptomic, proteomic, and metabolomic responses to inhibitors can be tracked in detail, and this “multiomic” approach can give high-resolution insight to the global cellular consequences (Miller et al., 2009; Yang et al., 2010b; Schwalbach et al., 2012; Skerker et al., 2013; Yang et al., 2013). Advanced techniques such as ribosome profiling can give a view into relationships between transcription and protein abundance in the presence of LCH inhibitors (Ingolia et al., 2009; Brar et al., 2012). Metabolic and flux-balance models are valuable in determining energy balances within cells (Varma and Palsson, 1994; Fiaux et al., 2003; Jin and Jeffries, 2004; Herrgård et al., 2008; Taymaz-Nikerel et al., 2010). These models combined with a systems biology view of protein and gene expression can be used to identify key energetic bottlenecks as targets for engineering. Recently, Wei et al. (2013) demonstrated an elegant way to overcome the redox cofactor imbalance in yeast designed to ferment xylose by engineering acetate metabolism from E. coli into S. cerevisiae. The authors combined an acetate utilization pathway that consumes NADH with a xylose utilization pathway that produces NADH to overcome the redox imbalance of engineered xylose fermentation. The resultant strain has both better xylose conversion and the ability to detoxify acetate (Wei et al., 2013). Detailed accounting of ATP and NAD(P)H in the presence of LCH inhibitors and industrial conditions will be necessary to disentangle and understand points for rational engineering of microbial catalysts.
Model biofuel microbes like S. cerevisiae and E. coli benefit from well-developed suites of functional genomics resources, such as deletion mutant or overexpression collections (Giaever et al., 2002; Baba et al., 2006; Kitagawa et al., 2006). These tools have revealed effects of some inhibitors such as furfural (Gorsich et al., 2006), vanillin (Endo et al., 2008; Iwaki et al., 2013), and acetic acid (Mira et al., 2010). Genome-wide mutant collections also allow powerful studies of inhibitors via “chemical genomics” (Giaever et al., 2004; Parsons et al., 2006; Ho et al., 2011). This new tool in the multiomic arsenal, when combined with the information in genetic interaction networks (Butland et al., 2008; Costanzo et al., 2010), can allow precise predictions of the cellular targets of fermentation inhibitors. Recently, Skerker et al. (2013) used a chemical genomics approach to discover a previously undescribed inhibitor in acid pretreated hydrolysate, methyl glyoxal (MG), and identified mutations that confer MG resistance. Chemical genomics can be used to identify the chemical biological signatures within hydrolysates and mutations conferring resistance, but more broadly can serve as a “biological fingerprinting” technique for hydrolysate to identify variation in production, and as a method to benchmark the biological properties of novel hydrolysates. Resources such as the MoBY-ORF collections (Ho et al., 2009; Magtanong et al., 2011), which are barcoded plasmids collections carrying nearly all S. cerevisiae used to assess the effects of increased gene dose and gene complementation, could be used with industrial, wild, and engineered yeast to identify genetic interactions within diverse yeast strains. Combined with traditional selection for resistance and directed evolution, systems biology tools offer great potential for strain development.
Further, new tools are now available that can accelerate strain development. Genome editing and optimization techniques such as CRISPR/Cas9 and MAGE allows rapid, detailed genome editing in both bacteria and eukaryotes (Wang et al., 2012; Cong et al., 2013; DiCarlo et al., 2013; Gilbert et al., 2013). Next steps in strain development will be tuning genomes to inhibitor tolerance by parallel disruption or activation of inhibitor responsive genes using CRISPR/Cas9-based systems. Additionally, large-scale gene synthesis can be used to identify genes that not only aid xylose utilization, but also confer inhibitor tolerance. Systems biology tools for biocatalyst development are coalescing to a pipeline that can keep pace with the changing landscape of fermentation inhibitors, allowing for the rapid tailoring of ethanologens with robust and efficient sugar to biofuel conversion from any new hydrolysate.
Necessary Tools to Meet a Common Goal
Much like the interface between commercial and academic drug discovery communities, applying next-generation biofuel technologies will require cooperative translational research. Academic biofuel communities are developing advanced system biology techniques whereas the commercial community excels at scale-up commercialization. However, similar to drug discovery, these two groups are often isolated in their research. In both cases, public-private partnerships such as the NIH translational medicine initiatives (Zerhouni, 2003) and NCERC industrial partnerships (http://www.siue.edu/ethanolresearch/), as well as shared computational resources like Kbase (http://www.kbase.us/) can help bridge the divide.
How can researchers and funding agencies best enhance collaboration? A key advance would be greater data sharing about LCH inhibitors. Diverse hydrolysates and their respective inhibitors are major variables in the field of cellulosic biofuel production, but detailed information on specific hydrolysate compositions is not broadly available. Comprehensive efforts to identify all major LCH components and inhibitors across feedstocks and hydrolysis treatments are needed and will require a central repository of LCH data that includes data on feedstocks, pretreatment, hydrolysis, nutrients, and inhibitor. Chemical genomic profiling could be used to generate “biological fingerprints” of hydrolysates to allow comparisons among hydrolysates and identify the biological effects of LCH components by standard analytical methods. A central resource would allow researchers to compare the composition and biological fingerprints of new hydrolysates with existing knowledge about tolerant microbes for further strain development. The DOE's Systems Biology Portal, KBase, offers the best outlet for community resources, and could serve as the authority of the response of biocatalysts to LCH inhibitors with open-source, community-developed analytical tools for chemical genomics datasets.
Conclusions
LCH inhibitors are major barriers for cellulosic biofuels. The cellular energy costs of coping with these compounds are a significant drain on the already thin margins of biofuel production. However, the increasingly powerful tools of systems biology can be used to gain a detailed understanding of the cellular consequences of individual and mixtures of fermentation inhibitors, which will serve as a basis for rational engineering of customizable microbes. The biofuel research community would benefit from shared computational and database resources that can improve communication between the academic and commercial sides of biofuels.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
All authors are funded by the DOE Great Lakes Bioenergy Research Center (DOE BER Office of Science DE-FC02-07ER64494).
References
Alexandre, H., Berlot, J. P., and Charpentier, C. (1994). Effect of ethanol on membrane fluidity of protoplasts from Saccharomyces cerevisiae and Kloeckera apiculata grown with or without ethanol, measured by fluorescence anisotropy. Biotechnol. Tech. 8, 295–300. doi: 10.1007/BF02428970
Allen, S. A., Clark, W., McCaffery, J. M., Cai, Z., Lanctot, A., Slininger, P. J., et al. (2010). Furfural induces reactive oxygen species accumulation and cellular damage in Saccharomyces cerevisiae. Biotechnol. Biofuels 3, 2. doi: 10.1186/1754-6834-3-2
Almeida, J. R., Modig, T., Petersson, A., Hähn-Hägerdal, B., Lidén, G., and Gorwa-Grauslund, M. F. (2007). Increased tolerance and conversion of inhibitors in lignocellulosic hydrolysates by Saccharomyces cerevisiae. J. Chem. Technol. Biotechnol. 82, 340–349. doi: 10.1002/jctb.1676
Alper, H., and Stephanopoulos, G. (2009). Engineering for biofuels: exploiting innate microbial capacity or importing biosynthetic potential? Nat. Rev. Microbiol. 7, 715–723. doi: 10.1038/nrmicro2186
Ask, M., Bettiga, M., Mapelli, V., and Olsson, L. (2013). The influence of HMF and furfural on redox-balance and energy-state of xylose-utilizing Saccharomyces cerevisiae. Biotechnol. Biofuels 6, 22. doi: 10.1186/1754-6834-6-22
Atsumi, S., Wu, T.-Y., Machado, I. M. P., Huang, W.-C., Chen, P.-Y., Pellegrini, M., et al. (2010). Evolution, genomic analysis, and reconstruction of isobutanol tolerance in Escherichia coli. Mol. Syst. Biol. 6, 449. doi: 10.1038/msb.2010.98
Attfield, P. V. (1997). Stress tolerance: the key to effective strains of industrial baker's yeast. Nat. Biotechnol. 15, 1351–1357. doi: 10.1038/nbt1297-1351
Baba, T., Ara, T., Hasegawa, M., Takai, Y., Okumura, Y., Baba, M., et al. (2006). Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2. doi: 10.1038/msb4100050
Baez, A., Cho, K.-M., and Liao, J. C. (2011). High-flux isobutanol production using engineered Escherichia coli: a bioreactor study with in situ product removal. Appl. Microbiol. Biotechnol. 90, 1681–1690. doi: 10.1007/s00253-011-3173-y
Bellissimi, E., Van Dijken, J. P., Pronk, J. T., and Van Maris, A. J. A. (2009). Effects of acetic acid on the kinetics of xylose fermentation by an engineered, xylose-isomerase-based Saccharomyces cerevisiae strain. FEMS Yeast Res. 9, 358–364. doi: 10.1111/j.1567-1364.2009.00487.x
Brar, G. A., Yassour, M., Friedman, N., Regev, A., Ingolia, N. T., and Weissman, J. S. (2012). High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science 335, 552–557. doi: 10.1126/science.1215110
Brynildsen, M. P., and Liao, J. C. (2009). An integrated network approach identifies the isobutanol response network of Escherichia coli. Mol. Syst. Biol. 5, 277. doi: 10.1038/msb.2009.34
Bunnell, K., Rich, A., Luckett, C., Wang, Y.-J., Martin, E., and Carrier, D. J. (2013). Plant maturity effects on the physicochemical properties and dilute acid hydrolysis of switchgrass (Panicum virgatum, L.) hemicelluloses. ACS Sustain. Chem. Eng. 1, 649–654. doi: 10.1021/sc4000175
Butland, G., Babu, M., Diaz-Mejia, J. J., Bohdana, F., Phanse, S., Gold, B., et al. (2008). eSGA: E. coli synthetic genetic array analysis. Nat. Methods 5, 789–795. doi: 10.1038/nmeth.1239
Carmona-Gutierrez, D., Sommer, C., Andryushkova, A., Kroemer, G., and Madeo, F. (2012). A higher spirit: avoiding yeast suicide during alcoholic fermentation. Cell Death Differ. 19, 913–914. doi: 10.1038/cdd.2012.31
Chambel, A., Viegas, C. A., and Sá-Correia, I. (1999). Effect of cinnamic acid on the growth and on plasma membrane H+–ATPase activity of Saccharomyces cerevisiae. Int. J. Food Microbiol. 50, 173–179. doi: 10.1016/S0168-1605(99)00100-2
Cherrington, C. A., Hinton, M., and Chopra, I. (1990). Effect of short-chain organic acids on macromolecular synthesis in Escherichia coli. J. Appl. Bacteriol. 68, 69–74. doi: 10.1111/j.1365-2672.1990.tb02550.x
Chundawat, S. P. S., Vismeh, R., Sharma, L. N., Humpula, J. F., da Costa Sousa, L., Chambliss, C. K., et al. (2010). Multifaceted characterization of cell wall decomposition products formed during ammonia fiber expansion (AFEX) and dilute acid based pretreatments. Bioresour. Technol. 101, 8429–8438. doi: 10.1016/j.biortech.2010.06.027
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. doi: 10.1126/science.1231143
Costanzo, M., Baryshnikova, A., Bellay, J., Kim, Y., Spear, E. D., Sevier, C. S., et al. (2010). The genetic landscape of a cell. Science 327, 425–431. doi: 10.1126/science.1180823
Decottignies, A., and Goffeau, A. (1997). Complete inventory of the yeast ABC proteins. Nat. Genet. 15, 137–145. doi: 10.1038/ng0297-137
DiCarlo, J. E., Norville, J. E., Mali, P., Rios, X., Aach, J., and Church, G. M. (2013). Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 41, 4336–4343. doi: 10.1093/nar/gkt135
Ding, J., Bierma, J., Smith, M. R., Poliner, E., Wolfe, C., Hadduck, A. N., et al. (2013). Acetic acid inhibits nutrient uptake in Saccharomyces cerevisiae: auxotrophy confounds the use of yeast deletion libraries for strain improvement. Appl. Microbiol. Biotechnol. 97, 7405–7416. doi: 10.1007/s00253-013-5071-y
Ding, M.-Z., Wang, X., Yang, Y., and Yuan, Y.-J. (2011). Metabolomic study of interactive effects of phenol, furfural, and acetic acid on Saccharomyces cerevisiae. OMICS J. Integr. Biol. 15, 647–653. doi: 10.1089/omi.2011.0003
Ding, M.-Z., Wang, X., Yang, Y., and Yuan, Y.-J. (2012). Comparative metabolic profiling of parental and inhibitors-tolerant yeasts during lignocellulosic ethanol fermentation. Metabolomics 8, 232–243. doi: 10.1007/s11306-011-0303-6
Docherty, K. M., and Kulpa, C. F. Jr. (2005). Toxicity and antimicrobial activity of imidazolium and pyridinium ionic liquids. Green Chem. 7, 185–189. doi: 10.1039/B419172B
Dombek, K. M., and Ingram, L. O. (1984). Effects of ethanol on the Escherichia coli plasma membrane. J. Bacteriol. 157, 233–239.
Endo, A., Nakamura, T., Ando, A., Tokuyasu, K., and Shima, J. (2008). Genome-wide screening of the genes required for tolerance to vanillin, which is a potential inhibitor of bioethanol fermentation, in Saccharomyces cerevisiae. Biotechnol. Biofuels 1, 3. doi: 10.1186/1754-6834-1-3
Fiaux, J., Çakar, Z. P., Sonderegger, M., Wüthrich, K., Szyperski, T., and Sauer, U. (2003). Metabolic-flux profiling of the yeasts Saccharomyces cerevisiae and Pichia stipitis. Eukaryot. Cell 2, 170–180. doi: 10.1128/EC.2.1.170-180.2003
Fitzgerald, D. J., Stratford, M., Gasson, M. J., Ueckert, J., Bos, A., and Narbad, A. (2004). Mode of antimicrobial action of vanillin against Escherichia coli, Lactobacillus plantarum and Listeria innocua. J. Appl. Microbiol. 97, 104–113. doi: 10.1111/j.1365-2672.2004.02275.x
Giaever, G., Chu, A. M., Ni, L., Connelly, C., Riles, L., Véronneau, S., et al. (2002). Functional profiling of the Saccharomyces cerevisiae genome. Nature 418, 387–391. doi: 10.1038/nature00935
Giaever, G., Flaherty, P., Kumm, J., Proctor, M., Nislow, C., Jaramillo, D. F., et al. (2004). Chemogenomic profiling: identifying the functional interactions of small molecules in yeast. Proc. Natl. Acad. Sci. U.S.A. 101, 793–798. doi: 10.1073/pnas.0307490100
Gibson, B. R., Lawrence, S. J., Leclaire, J. P. R., Powell, C. D., and Smart, K. A. (2007). Yeast responses to stresses associated with industrial brewery handling. FEMS Microbiol. Rev. 31, 535–569. doi: 10.1111/j.1574-6976.2007.00076.x
Gilbert, L. A., Larson, M. H., Morsut, L., Liu, Z., Brar, G. A., Torres, S. E., et al. (2013). CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451. doi: 10.1016/j.cell.2013.06.044
Gliozzi, G., Innorta, A., Mancini, A., Bortolo, R., Perego, C., Ricci, M., et al. (2014). Zr/P/O catalyst for the direct acid chemo-hydrolysis of non-pretreated microcrystalline cellulose and softwood sawdust. Appl. Catal. B Environ. 145, 24–33. doi: 10.1016/j.apcatb.2012.12.035
Gorsich, S. W., Dien, B. S., Nichols, N. N., Slininger, P. J., Liu, Z. L., and Skory, C. D. (2006). Tolerance to furfural-induced stress is associated with pentose phosphate pathway genes ZWF1, GND1, RPE1, and TKL1 in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 71, 339–349. doi: 10.1007/s00253-005-0142-3
Greenhalf, C. E., Nowakowski, D. J., Yates, N., Shield, I., and Bridgwater, A. V. (2013). The influence of harvest and storage on the properties of and fast pyrolysis products from Miscanthus x giganteus. Biomass Bioenergy 56, 247–259. doi: 10.1016/j.biombioe.2013.05.007
Hadi, S. M., Shahabuddin, and Rehman, A. (1989). Specificity of the interaction of furfural with DNA. Mutat. Res. Lett. 225, 101–106. doi: 10.1016/0165-7992(89)90125-5
Herrgård, M. J., Swainston, N., Dobson, P., Dunn, W. B., Arga, K. Y., Arvas, M., et al. (2008). A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology. Nat. Biotechnol. 26, 1155–1160. doi: 10.1038/nbt1492
Ho, C. H., Magtanong, L., Barker, S. L., Gresham, D., Nishimura, S., Natarajan, P., et al. (2009). A molecular barcoded yeast ORF library enables mode-of-action analysis of bioactive compounds. Nat. Biotechnol. 27, 369–377. doi: 10.1038/nbt.1534
Ho, C. H., Piotrowski, J., Dixon, S. J., Baryshnikova, A., Costanzo, M., and Boone, C. (2011). Combining functional genomics and chemical biology to identify targets of bioactive compounds. Curr. Opin. Chem. Biol. 15, 66–78. doi: 10.1016/j.cbpa.2010.10.023
Holyoak, C. D., Stratford, M., McMullin, Z., Cole, M. B., Crimmins, K., Brown, A. J., et al. (1996). Activity of the plasma membrane H(+)-ATPase and optimal glycolytic flux are required for rapid adaptation and growth of Saccharomyces cerevisiae in the presence of the weak-acid preservative sorbic acid. Appl. Environ. Microbiol. 62, 3158–3164.
Huffer, S., Clark, M. E., Ning, J. C., Blanch, H. W., and Clark, D. S. (2011). Role of alcohols in growth, lipid composition, and membrane fluidity of yeasts, bacteria, and archaea. Appl. Environ. Microbiol. 77, 6400–6408. doi: 10.1128/AEM.00694-11
Ibeas, J. I., and Jimenez, J. (1997). Mitochondrial DNA loss caused by ethanol in Saccharomyces flor yeasts. Appl. Environ. Microbiol. 63, 7–12.
Ingolia, N. T., Ghaemmaghami, S., Newman, J. R. S., and Weissman, J. S. (2009). Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223. doi: 10.1126/science.1168978
Iwaki, A., Ohnuki, S., Suga, Y., Izawa, S., and Ohya, Y. (2013). Vanillin inhibits translation and induces messenger ribonucleoprotein (mRNP) granule formation in Saccharomyces cerevisiae: application and validation of high-content, image-based profiling. PloS ONE 8:e61748. doi: 10.1371/journal.pone.0061748
Jarboe, L. R. (2011). YqhD: a broad-substrate range aldehyde reductase with various applications in production of biorenewable fuels and chemicals. Appl. Microbiol. Biotechnol. 89, 249–257. doi: 10.1007/s00253-010-2912-9
Jin, M., Sarks, C., Gunawan, C., Bice, B. D., Simonett, S. P., Avanasi Narasimhan, R., et al. (2013). Phenotypic selection of a wild Saccharomyces cerevisiae strain for simultaneous saccharification and co-fermentation of AFEX™ pretreated corn stover. Biotechnol. Biofuels 6, 108. doi: 10.1186/1754-6834-6-108
Jin, Y.-S., and Jeffries, T. W. (2004). Stoichiometric network constraints on xylose metabolism by recombinant Saccharomyces cerevisiae. Metab. Eng. 6, 229–238. doi: 10.1016/j.ymben.2003.11.006
Jozefczuk, S., Klie, S., Catchpole, G., Szymanski, J., Cuadros-Inostroza, A., Steinhauser, D., et al. (2010). Metabolomic and transcriptomic stress response of Escherichia coli. Mol. Syst. Biol. 6:364. doi: 10.1038/msb.2010.18
Khan, Q. A., Shamsi, F. A., and Hadi, S. M. (1995). Mutagenicity of furfural in plasmid DNA. Cancer Lett. 89, 95–99. doi: 10.1016/0304-3835(95)90163-9
King, A. T., Davey, M. R., Mellor, I. R., Mulligan, B. J., and Lowe, K. C. (1991). Surfactant effects on yeast cells. Enzyme Microb. Technol. 13, 148–153. doi: 10.1016/0141-0229(91)90171-6
Kitagawa, M., Ara, T., Arifuzzaman, M., Ioka-Nakamichi, T., Inamoto, E., Toyonaga, H., et al. (2006). Complete set of ORF clones of Escherichia coli ASKA library (A Complete Set of E. coli K-12 ORF Archive): unique resources for biological research. DNA Res. 12, 291–299. doi: 10.1093/dnares/dsi012
Klinke, H. B., Thomsen, A. B., and Ahring, B. K. (2004). Inhibition of ethanol-producing yeast and bacteria by degradation products produced during pre-treatment of biomass. Appl. Microbiol. Biotechnol. 66, 10–26. doi: 10.1007/s00253-004-1642-2
Krebs, H. A., Wiggins, D., Stubbs, M., Sols, A., and Bedoya, F. (1983). Studies on the mechanism of the antifungal action of benzoate. Biochem. J. 214, 657–663.
Laouar, L., Lowe, K. C., and Mulligan, B. J. (1996). Yeast responses to nonionic surfactants. Enzyme Microb. Technol. 18, 433–438. doi: 10.1016/0141-0229(95)00122-0
Larsson, S., Quintana-Sáinz, A., Reimann, A., Nilvebrant, N. O., and Jönsson, L. J. (2000). Influence of lignocellulose-derived aromatic compounds on oxygen-limited growth and ethanolic fermentation by Saccharomyces cerevisiae. Appl. Biochem. Biotechnol. 84–86, 617–632. doi: 10.1385/ABAB:84-86:1-9:617
Larsson, S., Reimann, A., Nilvebrant, N.-O., and Jönsson, L. J. (1999). Comparison of different methods for the detoxification of lignocellulose hydrolyzates of spruce. Appl. Biochem. Biotechnol. 77, 91–103. doi: 10.1385/ABAB:77:1-3:91
Lau, M. W., and Dale, B. E. (2009). Cellulosic ethanol production from AFEX-treated corn stover using Saccharomyces cerevisiae 424A(LNH-ST). Proc. Natl. Acad. Sci. U.S.A. 106, 1368–1373. doi: 10.1073/pnas.0812364106
Lau, M. W., Gunawan, C., Balan, V., and Dale, B. E. (2010). Comparing the fermentation performance of Escherichia coli KO11, Saccharomyces cerevisiae 424A(LNH-ST) and Zymomonas mobilis AX101 for cellulosic ethanol production. Biotechnol. Biofuels 3, 11. doi: 10.1186/1754-6834-3-11
Lee, M. V., Topper, S. E., Hubler, S. L., Hose, J., Wenger, C. D., Coon, J. J., et al. (2011). A dynamic model of proteome changes reveals new roles for transcript alteration in yeast. Mol. Syst. Biol. 7:514. doi: 10.1038/msb.2011.48
Lee, S., Nam, D., Jung, J. Y., Oh, M.-K., Sang, B.-I., and Mitchell, R. J. (2012). Identification of Escherichia coli biomarkers responsive to various lignin-hydrolysate compounds. Bioresour. Technol. 114, 450–456. doi: 10.1016/j.biortech.2012.02.085
Li, Z., Chen, C. H., Liu, T., Mathrubootham, V., Hegg, E. L., and Hodge, D. B. (2013). Catalysis with Cu(II) (bpy) improves alkaline hydrogen peroxide pretreatment. Biotechnol. Bioeng. 110, 1078–1086. doi: 10.1002/bit.24793
Linton, K. J., and Higgins, C. F. (1998). The Escherichia coli ATP-binding cassette (ABC) proteins. Mol. Microbiol. 28, 5–13. doi: 10.1046/j.1365-2958.1998.00764.x
Liu, Z. L., Moon, J., Andersh, B. J., Slininger, P. J., and Weber, S. (2008). Multiple gene-mediated NAD(P)H-dependent aldehyde reduction is a mechanism of in situ detoxification of furfural and 5-hydroxymethylfurfural by Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 81, 743–753. doi: 10.1007/s00253-008-1702-0
Luterbacher, J. S., Rand, J. M., Alonso, D. M., Han, J., Youngquist, J. T., Maravelias, C. T., et al. (2014). Nonenzymatic sugar production from biomass using biomass-derived γ-valerolactone. Science 343, 277–280. doi: 10.1126/science.1246748
Magtanong, L., Ho, C. H., Barker, S. L., Jiao, W., Baryshnikova, A., Bahr, S., et al. (2011). Dosage suppression genetic interaction networks enhance functional wiring diagrams of the cell. Nat. Biotechnol. 29, 505–511. doi: 10.1038/nbt.1855
Matsushika, A., Nagashima, A., Goshima, T., and Hoshino, T. (2013). Fermentation of xylose causes inefficient metabolic state due to carbon/energy starvation and reduced glycolytic flux in recombinant industrial Saccharomyces cerevisiae. PloS ONE 8:e69005. doi: 10.1371/journal.pone.0069005
McEwen, J. T., and Atsumi, S. (2012). Alternative biofuel production in non-natural hosts. Curr. Opin. Biotechnol. 23, 744–750. doi: 10.1016/j.copbio.2011.12.019
Mikulášová, M., Vodnımath, Š., and Pekarovièová, A. (1990). Influence of phenolics on biomass production by Candida utilis and Candida albicans. Biomass 23, 149–154. doi: 10.1016/0144-4565(90)90032-F
Miller, E. N., Jarboe, L. R., Turner, P. C., Pharkya, P., Yomano, L. P., York, S. W., et al. (2009). Furfural inhibits growth by limiting sulfur assimilation in ethanologenic Escherichia coli strain LY180. Appl. Environ. Microbiol. 75, 6132–6141. doi: 10.1128/AEM.01187-09
Minty, J. J., Lesnefsky, A. A., Lin, F., Chen, Y., Zaroff, T. A., Veloso, A. B., et al. (2011). Evolution combined with genomic study elucidates genetic bases of isobutanol tolerance in Escherichia coli. Microb. Cell Fact. 10, 18. doi: 10.1186/1475-2859-10-18
Mira, N. P., Palma, M., Guerreiro, J. F., and Sa-Correia, I. (2010). Genome-wide identification of Saccharomyces cerevisiae genes required for tolerance to acetic acid. Microb. Cell Fact. 9, 79. doi: 10.1186/1475-2859-9-79
Modig, T., Lidén, G., and Taherzadeh, M. J. (2002). Inhibition effects of furfural on alcohol dehydrogenase, aldehyde dehydrogenase and pyruvate dehydrogenase. Biochem. J. 363, 769–776. doi: 10.1042/0264-6021:3630769
Nagodawithana, T. W., and Steinkraus, K. H. (1976). Influence of the rate of ethanol production and accumulation on the viability of Saccharomyces cerevisiae in “rapid fermentation”. Appl. Environ. Microbiol. 31, 158–162.
Nguyen, T. T. M., Iwaki, A., Ohya, Y., and Izawa, S. (2014). Vanillin causes the activation of Yap1 and mitochondrial fragmentation in Saccharomyces cerevisiae. J. Biosci. Bioeng. 117, 33–38. doi: 10.1016/j.jbiosc.2013.06.008
Oliva, J. M., Negro, M. J., Sáez, F., Ballesteros, I., Manzanares, P., González, A., et al. (2006). Effects of acetic acid, furfural and catechol combinations on ethanol fermentation of Kluyveromyces marxianus. Process Biochem. 41, 1223–1228. doi: 10.1016/j.procbio.2005.12.003
Ouellet, M., Datta, S., Dibble, D. C., Tamrakar, P. R., Benke, P. I., Li, C., et al. (2011). Impact of ionic liquid pretreated plant biomass on Saccharomyces cerevisiae growth and biofuel production. Green Chem. 13, 2743. doi: 10.1039/c1gc15327g
Parsons, A., Lopez, A., Givoni, I., Williams, D., Gray, C., Porter, J., et al. (2006). Exploring the mode-of-action of bioactive compounds by chemical-genetic profiling in yeast. Cell 126, 611–625. doi: 10.1016/j.cell.2006.06.040
Pereira Rangel, L., Fritzen, M., Yunes, R. A., Leal, P. C., Creczynski-Pasa, T. B., and Ferreira-Pereira, A. (2010). Inhibitory effects of gallic acid ester derivatives on Saccharomyces cerevisiae multidrug resistance protein Pdr5p. FEMS Yeast Res. 10, 244–251. doi: 10.1111/j.1567-1364.2010.00603.x
Russell, J. B. (1992). Another explanation for the toxicity of fermentation acids at low pH: anion accumulation versus uncoupling. J. Appl. Bacteriol. 73, 363–370. doi: 10.1111/j.1365-2672.1992.tb04990.x
Sato, T. K., Liu, T., Parreiras, L. S., Williams, D. L., Wohlbach, D. J., Bice, B. D., et al. (2014). Harnessing genetic diversity in Saccharomyces cerevisiae for improved fermentation of xylose in hydrolysates of alkaline hydrogen peroxide pretreated biomass. Appl. Environ. Microbiol. 80, 540–554. doi: 10.1128/AEM.01885-13
Schmer, M. R., Vogel, K. P., Mitchell, R. B., and Perrin, R. K. (2008). Net energy of cellulosic ethanol from switchgrass. Proc. Natl. Acad. Sci. U.S.A. 105, 464–469. doi: 10.1073/pnas.0704767105
Schmitt, L., and Tampé, R. (2002). Structure and mechanism of ABC transporters. Curr. Opin. Struct. Biol. 12, 754–760. doi: 10.1016/S0959-440X(02)00399-8
Schüller, C., Mamnun, Y. M., Mollapour, M., Krapf, G., Schuster, M., Bauer, B. E., et al. (2004). Global phenotypic analysis and transcriptional profiling defines the weak acid stress response regulon in Saccharomyces cerevisiae. Mol. Biol. Cell 15, 706–720. doi: 10.1091/mbc.E03-05-0322
Schwalbach, M. S., Keating, D. H., Tremaine, M., Marner, W. D., Zhang, Y., Bothfeld, W., et al. (2012). Complex physiology and compound stress responses during fermentation of alkali-pretreated corn stover hydrolysate by an Escherichia coli ethanologen. Appl. Environ. Microbiol. 78, 3442–3457. doi: 10.1128/AEM.07329-11
Shapiro, A. B., and Ling, V. (1998). Transport of LDS-751 from the cytoplasmic leaflet of the plasma membrane by the rhodamine-123-selective site of P-glycoprotein. Eur. J. Biochem. FEBS 254, 181–188. doi: 10.1046/j.1432-1327.1998.2540181.x
Sindhu, R., Kuttiraja, M., Elizabeth Preeti, V., Vani, S., Sukumaran, R. K., and Binod, P. (2013). A novel surfactant-assisted ultrasound pretreatment of sugarcane tops for improved enzymatic release of sugars. Bioresour. Technol. 135, 67–72. doi: 10.1016/j.biortech.2012.09.050
Sinha, R. P. (1986). Toxicity of organic acids for repair-deficient strains of Escherichia coli. Appl. Environ. Microbiol. 51, 1364–1366.
Skerker, J. M., Leon, D., Price, M. N., Mar, J. S., Tarjan, D. R., Wetmore, K. M., et al. (2013). Dissecting a complex chemical stress: chemogenomic profiling of plant hydrolysates. Mol. Syst. Biol. 9, 674. doi: 10.1038/msb.2013.30
Stratford, M., and Anslow, P. A. (1998). Evidence that sorbic acid does not inhibit yeast as a classic “weak acid preservative.” Lett. Appl. Microbiol. 27, 203–206. doi: 10.1046/j.1472-765X.1998.00424.x
Stratford, M., Nebe-von-Caron, G., Steels, H., Novodvorska, M., Ueckert, J., and Archer, D. B. (2013). Weak-acid preservatives: pH and proton movements in the yeast Saccharomyces cerevisiae. Int. J. Food Microbiol. 161, 164–171. doi: 10.1016/j.ijfoodmicro.2012.12.013
Taymaz-Nikerel, H., Borujeni, A. E., Verheijen, P. J. T., Heijnen, J. J., and van Gulik, W. M. (2010). Genome-derived minimal metabolic models for Escherichia coli MG1655 with estimated in vivo respiratory ATP stoichiometry. Biotechnol. Bioeng. 107, 369–381. doi: 10.1002/bit.22802
Ullah, A., Orij, R., Brul, S., and Smits, G. J. (2012). Quantitative analysis of the modes of growth inhibition by weak organic acids in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 78, 8377–8387. doi: 10.1128/AEM.02126-12
Varma, A., and Palsson, B. O. (1994). Stoichiometric flux balance models quantitatively predict growth and metabolic by-product secretion in wild-type Escherichia coli W3110. Appl. Environ. Microbiol. 60, 3724–3731.
Verduyn, C., Postma, E., Scheffers, W. A., and Van Dijken, J. P. (1992). Effect of benzoic acid on metabolic fluxes in yeasts: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast 8, 501–517. doi: 10.1002/yea.320080703
Wang, H. H., Kim, H., Cong, L., Jeong, J., Bang, D., and Church, G. M. (2012). Genome-scale promoter engineering by coselection MAGE. Nat. Methods 9, 591–593. doi: 10.1038/nmeth.1971
Wang, X., Miller, E. N., Yomano, L. P., Zhang, X., Shanmugam, K. T., and Ingram, L. O. (2011). Increased furfural tolerance due to overexpression of NADH-Dependent oxidoreductase FucO in Escherichia coli strains engineered for the production of ethanol and lactate. Appl. Environ. Microbiol. 77, 5132–5140. doi: 10.1128/AEM.05008-11
Wang, X., Yomano, L. P., Lee, J. Y., York, S. W., Zheng, H., Mullinnix, M. T., et al. (2013). Engineering furfural tolerance in Escherichia coli improves the fermentation of lignocellulosic sugars into renewable chemicals. Proc. Natl. Acad. Sci. U.S.A. 110, 4021–4026. doi: 10.1073/pnas.1217958110
Wei, N., Quarterman, J., Kim, S. R., Cate, J. H. D., and Jin, Y.-S. (2013). Enhanced biofuel production through coupled acetic acid and xylose consumption by engineered yeast. Nat. Commun. 4:2580. doi: 10.1038/ncomms3580
Yang, S., Land, M. L., Klingeman, D. M., Pelletier, D. A., Lu, T.-Y. S., Martin, S. L., et al. (2010a). Paradigm for industrial strain improvement identifies sodium acetate tolerance loci in Zymomonas mobilis and Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 107, 10395–10400. doi: 10.1073/pnas.0914506107
Yang, S., Pan, C., Tschaplinski, T. J., Hurst, G. B., Engle, N. L., Zhou, W., et al. (2013). Systems biology analysis of Zymomonas mobilis ZM4 ethanol stress responses. PloS ONE 8:e68886. doi: 10.1371/journal.pone.0068886
Yang, S., Pelletier, D. A., Lu, T.-Y. S., and Brown, S. D. (2010b). The Zymomonas mobilis regulator hfq contributes to tolerance against multiple lignocellulosic pretreatment inhibitors. BMC Microbiol. 10:135. doi: 10.1186/1471-2180-10-135
Zaldivar, J., and Ingram, L. O. (1999). Effect of organic acids on the growth and fermentation of ethanologenic Escherichia coli LY01. Biotechnol. Bioeng. 66, 203–210. doi: 10.1002/(SICI)1097-0290(1999)66:4<203::AID-BIT1>3.0.CO;2-#
Keywords: cellulosic biofuels, lignocellulosic hydrolysate inhibitors, systems biology, chemical genomics, metabolic modeling, ethanologens
Citation: Piotrowski JS, Zhang Y, Bates DM, Keating DH, Sato TK, Ong IM and Landick R (2014) Death by a thousand cuts: the challenges and diverse landscape of lignocellulosic hydrolysate inhibitors. Front. Microbiol. 5:90. doi: 10.3389/fmicb.2014.00090
Received: 16 November 2013; Accepted: 18 February 2014;
Published online: 14 March 2014.
Edited by:
Shihui (Shane) Yang, National Renewable Energy Laboratory, USAReviewed by:
Shiyong Peng, National Institute of Health, USAQiang J. Fei, National Renewable Energy Lab, USA
Copyright © 2014 Piotrowski, Zhang, Bates, Keating, Sato, Ong and Landick. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jeff S. Piotrowski, DOE Great Lakes Bioenergy Research Center, University of Wisconsin-Madison, 1552 University Ave., WEI4152, Madison, WI 53726, USA e-mail:anBpb3Ryb3dza2lAd2lzYy5lZHU=
 Jeff S. Piotrowski
Jeff S. Piotrowski Yaoping Zhang
Yaoping Zhang Donna M. Bates
Donna M. Bates David H. Keating
David H. Keating Trey K. Sato
Trey K. Sato