- Eco-Systems Biology Group, Luxembourg Centre for Systems Biomedicine, University of Luxembourg, Esch-sur-Alzette, Luxembourg
Biological wastewater treatment plants (BWWTPs) based on the activated sludge (AS) process have dramatically improved worldwide water sanitation despite increased urbanization and industrialization. However, current AS-based operations are considered economically and environmentally unsustainable. In this Perspective, we discuss our current understanding of microbial populations and their metabolic transformations in AS-based BWWTPs in view of developing more sustainable processes in the future. In particular, much has been learned over the course of the past 25 years about specialized microorganisms, which could be more comprehensively leveraged to recover energy and/or nutrients from wastewater streams. To achieve this, we propose a bottom-up design approach, focused around the concept of a “wastewater biorefinery column”, which would rely on the engineering of distinct ecological niches into a BWWTP in order to guarantee the targeted enrichment of specific organismal groups which in turn will allow the harvest of high-value resources from wastewater. This concept could be seen as a possible grand challenge to microbial ecologists and engineers alike at the centenary of the discovery of the AS process.
Modern society is placing considerable strain on essential resources including water and energy (e.g., fossil fuels). Human interferences in biogeochemical cycles has already substantially altered the structure and function of atmospheric, aquatic, and terrestrial ecosystems (Vitousek, 1994). This is projected to worsen over the coming decades, as current estimates predict the human population to grow to more than 9 billion by 2050 (United Nations, 2013). In particular, waste residues may become pollutants if care is not given to their discharge into the environment. Thus, apart from public health considerations, systematic waste management is a central premise of sustainable development.
One hundred years ago, Ardern and Lockett (1914) described the process of activated sludge (AS) for biological remediation of sewage, based on heterotrophic microbial biomass either assimilating or oxidizing dissolved organic matter in influent wastewater. Following an aeration phase, the suspended biomass is separated from the treated wastewater by gravity filtration with a subsequent recycling of the majority of the sludge which is termed activated. To date, numerous variations of the original AS-based biological wastewater treatment plants (BWWTPs) are being operated world-wide including for enhanced biological phosphorus removal (Gujer et al., 1995), nitrification-denitrification (Henze et al., 1999), and anaerobic oxidation of ammonium (Van Dongen et al., 2001) together with anaerobic digestion of excess sludge (Grady et al., 2011). However, given the need for aeration as well as moving biomass between treatment tanks, AS-based BWWTPs consume considerable amounts of fossil fuel-derived energy resulting in considerable anthropogenic greenhouse gas emissions. In addition, the processes themselves also generate potent greenhouse gases such as CH4 (El-Fadel and Massoud, 2001) and N2O (Kampschreur et al., 2008). In this light, the current reliance on AS-based processes is environmentally unsustainable.
Although the organic and inorganic composition of wastewater depends on the influent (either municipal or industrial), it is estimated that the amount of chemical energy contained within wastewater is at least 10-fold higher than the amount of energy currently used to treat it (Shizas and Bagley, 2004; Heidrich et al., 2010). To date, energy recovery yields from anaerobic digestion of excess AS or from microbial fuel cells (MFCs) are significantly lower than the actual chemical energy contained within wastewater (Pham et al., 2006). Therefore, future sustainable strategies to reclaim energy from wastewater should not only reduce our dependence on fossil fuels but could also meet our demands of daily resources such as plastics and fertilizers.
In this Perspective, we introduce the concept of a “wastewater biorefinery column”, an approach for potentially leveraging the existing microbiological and biochemical knowledge of AS-based processes in a bottom-up design approach. This concept is based on the engineering of distinct ecological niches into future wastewater treatment processes, which may in turn allow the targeted enrichment of distinct organismal groups and the comprehensive recovery of energy and biotechnologically relevant molecules.
The Current Microbiological and Biochemical Knowledge of as-Based Processes
Microbial communities of BWWTPs have for a long time been viewed as “black boxes”, as their structure and function have remained largely unknown. The majority of early studies on the microbiology of BWWTPs involved classical isolation techniques (Prakasam and Dondero, 1967) and standard light microscopy for morphological identification of specific bacterial groups, e.g., filamentous bacteria (Eikelboom, 1975; Figure 1A). Using classical culture-dependent microbiological techniques, Acinetobacter spp. was implicated in phosphorus removal (Fuhs and Chen, 1975), Nitrosospira spp. was considered to be the key ammonium oxidizer (Prosser, 1989), Nitrobacter spp. (Henze et al., 1997) to be the dominating nitrite oxidizer and Hypomicrobium spp. to be a key denitrifier (Timmermans and Van Haute, 1983).
Intense research based on the retrieval of 16S rRNA gene sequences from BWWTPs in the last decades have ruled out the involvement of Acinetobacter spp. as a major player in phosphorus removal (Wagner et al., 1994; Kämpfer et al., 1996; Bond et al., 1995; Santos et al., 1999). So far, an uncultured and unclassified genus belonging to the Beta-proteobacteria, named Candidatus Accumulibacter phosphatis (CAp), has been shown to be a dominating phosphorus accumulating organism (PAO) in laboratory-scale reactors (Hesselmann et al., 1999; Crocetti et al., 2000) as well as in full-scale BWWTPs (Zilles et al., 2002; Wong et al., 2005). Moreover, subsequent molecular investigations have identified glycogen accumulating organisms (GAOs; Mino et al., 1995) as major competitors of CAp in anaerobic/aerobic sludge cycling. This includes a novel group belonging to Gamma-proteobacteria named as Candidatus Competibacter phosphatis (Crocetti et al., 2002) and other groups belonging to Alpha-proteobacteria (Wong et al., 2004).
Regarding the cycling of nitrogen, new key players identified using molecular approaches included diverse populations of ammonium oxidizers (Juretschko et al., 1998; Purkhold et al., 2000; Daims et al., 2001), Nitrospira-like microorganisms as dominant nitrite oxidizers (Juretschko et al., 1998; Daims et al., 2001; Dionisi et al., 2002; Maixner et al., 2006; Spieck et al., 2006) and denitrifiers belonging to the genera Aquaspirillum, Azoarcus, Thauera, and PAOs such as CAp (Kong et al., 2004; Hesselsoe et al., 2005; Thomsen et al., 2007). More recently, archaeal members of the Thaumarchaeota were also found to be capable of catalyzing the aerobic oxidation of ammonium (Park et al., 2006). The complexity of nitrogen cycling in BWWTPs was further brought to the forefront when distinct microorganisms having novel physiological properties, i.e., being able to carry out anaerobic ammonium oxidation (anammox) were identified (Mulder et al., 1995). These organisms belong to the phylum Planctomycetes (Strous et al., 1999) and have been putatively named “Candidatus Kuenenia stuttgartiensis” (Schmid et al., 2000).
Numerous investigations into the ecophysiology of microorganisms in BWWTPs were carried out using fluorescence in situ hybridization coupled with microautoradiography (MAR-FISH), which allowed direct visualization of specific microorganisms and qualitatively link specific carbon (C), nitrogen (N), and phosphorus (P) substrate transformations (Nielsen and Nielsen, 2002; Wagner et al., 2006). MAR-FISH allowed for example the study of competition between PAOs and GAOs for carbon substrates and allowed the formulation of models (Oehmen et al., 2010). During anaerobic conditions, PAOs assimilate organic carbon substrates, e.g., volatile fatty acids and store them as polyhydroxyalkanoates (PHAs; Kong et al., 2004). When exposed to aerobic conditions, PAOs oxidize PHAs, which provides energy for polyphosphate accumulation, leading to P removal from the wastewater by biomass retrieval. GAOs compete with PAOs for volatile fatty acids under anaerobic conditions for PHA synthesis (Bengtsson et al., 2008) and do not accumulate polyphosphate under aerobic conditions but rather use energy for glycogen accumulation (Crocetti et al., 2002; Kong et al., 2006). Further information on the intra-cellular accumulation of PHA or polyphosphate indicating storage and/or cycling of these polymers was confirmed by combining MAR-FISH with staining methods (Crocetti et al., 2000; Wong et al., 2004). Other MAR-FISH studies demonstrated the physiological potential of lipid accumulating organisms (LAOs), such as Candidatus Microthrix parvicella to take up and store lipid substrates under anaerobic conditions and highlighted a competitive advantage of these filamentous bacteria compared to other organisms unable to assimilate these energy-rich substrates anaerobically (Andreasen and Nielsen, 1998; Nielsen et al., 2002).
Remarkably, advances in high-resolution molecular approaches, so called “meta-omics”, are now allowing concrete linkages to be drawn between microbial community compositions and functions. For example, metagenomic studies have unraveled some of the enigmatic microbial phenotypic traits by identifying candidate genes for ladderane biosynthesis and biological hydrazine metabolism in Candidatus Kuenenia stuttgartiensis (Strous et al., 2006). Full-scale BWWTP metagenomic studies have underlined the need for more reference genomes of key species (Albertsen et al., 2011), which can be used as a template to interpret in situ data as illustrated by recent laboratory studies (Kristiansen et al., 2013; McIlroy et al., 2013). In this context, metagenomic or single cell sequencing data may be fruitfully employed to infer media formulation and growth conditions for organisms of interest.
While metagenomic studies have greatly contributed to the clarification of the functional capabilities of specific populations of interest, metaproteomics (Wilmes et al., 2008), metatranscriptomics (Yu and Zhang, 2012), and the combination of these two approaches (Haroon et al., 2013) have furthered our knowledge about the actual expression of relevant genes involved in key processes under different environmental conditions such as alternating anaerobic/aerobic phases. Thus, integrated “omics” over space and time (Muller et al., 2013) in combination with recorded physico-chemical parameters will allow reconstruction of the ecological networks and detailed definition of organismal niches (Figure 1B). Such knowledge may then be used to identify key determinants of overall microbial community structure and function, which in turn may be harnessed for comprehensive reclamation of energy and biotechnologically relevant products from wastewater.
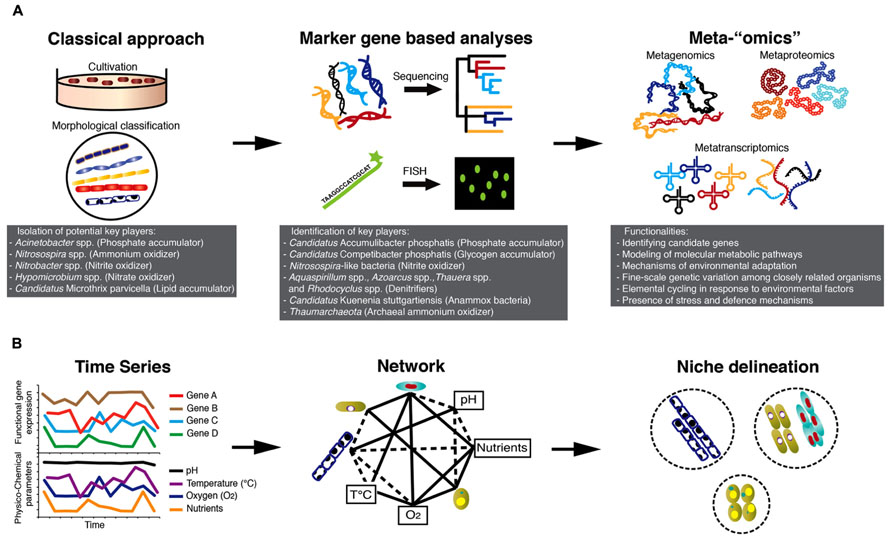
FIGURE 1. Technology-driven understanding of the microbiology of AS-based processes. (A) Advances in microbial ecology methods starting with cultivation-based approaches leading to community “omics” allowing the unveiling of the microbial “black box” of AS-based processes. (B) Integrated high-throughput “omics” together with recorded physico-chemical parameters of BWWTPs hold great promise to define niches of individual community members and their elemental transformation capabilities. The combined information may be used to engineer niches of specific microbial communities for subsequent energy/nutrient recovery.
Into the Future: Using the Intelligence of Wastewater Microbes to Recover the Hidden Treasure
Given the numerous organismal groups which have now been identified, as well as the short-term prospect of additional unprecedented data from multi-omic analyses, the time is ripe at the centenary of the AS-process to think about formulation of new biological wastewater treatment processes. In particular, we can start to consider bottom-up design approaches rather than the top-down approaches promulgated thus far.
Here, we introduce the concept of a “wastewater biorefinery column” (Figure 2), based on a hypothetical bottom-up design approach, which takes into account the detailed knowledge of how specific microorganisms behave in situ and accumulate different storage compounds of interest over changing BWWTPs environmental conditions. Consequently, the engineering of specific niches would allow the harvest of high-value resources from wastewater. Such niche engineering may be achieved for example by (1) establishing distinct substrate gradients within the entire resource space of wastewater thereby exploiting the individual organismal niche breadths; (2) manipulation of the vertical distribution of key microorganisms by exploiting their respective settling velocities which in turn may be influenced by microbial/floc size and intracellular storage compounds; (3) harvesting dominant members for energy and/or nutrient reclamation. In laboratory-scale reactors, pronounced organismal enrichments have already been obtained for organisms of interest, in particular PAOs and GAOs (Winkler et al., 2011a), highlighting the feasibility of enriching these organisms by providing them with appropriate environmental conditions. With the advent of advanced meta-omic approaches, we will be able to define the niches of the respective organismal groups much more precisely, thereby allowing these to be engineering into future systems. Given the heterogeneous and dynamic composition of wastewater, niches may have to be continually adjusted. Such niche fine-tuning could be based on feedback from microbial fuel cells acting as biosensors (Di Lorenzo et al., 2009), which would allow the continuous monitoring of organic and inorganic composition of influents.
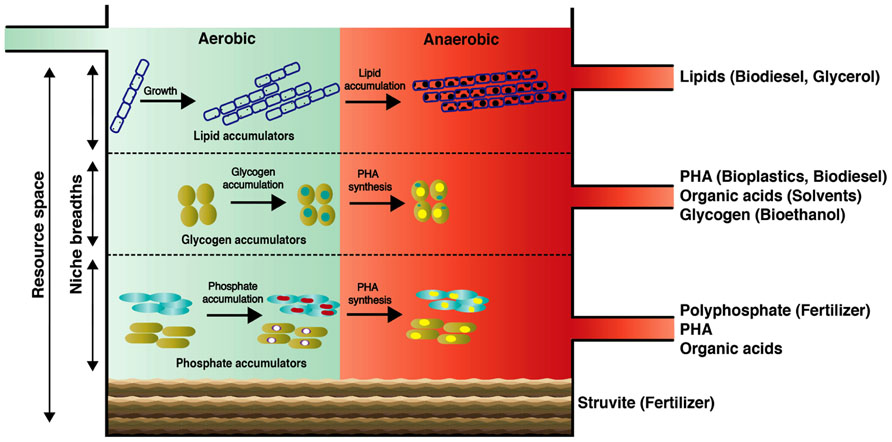
FIGURE 2. Conceptual scheme of the proposed “wastewater biorefinery column” concept for dedicated resource recovery through engineered organismal niches. Our proposed concept may serve as a future grand challenge for microbial ecologists and engineers to tackle current AS process marking the centenary of its discovery.
Among the renewable sources of energy currently being explored, biodiesel holds significant promise as a potential alternative to partially replace petroleum-based fuels. At present, up to 85% of the overall biodiesel production cost is associated with the feedstocks (Knothe et al., 2005; Mondala et al., 2009). Consequently, biomolecules from wastewater of immediate bioenergy interest are saponifiable lipids since they can be easily transformed into biodiesel (Fukuda et al., 2001; Chisti, 2007; Mondala et al., 2009). In municipal wastewater, lipids can represent up to 41% of the total organic pools (Raunkjae, 1994), with a vast majority being triacylglycerides (TAGs) and a minor part free long chain fatty acids (Quemeneur and Marty, 1994). Importantly, the long chain fatty acids comprising TAGs within wastewater sludge are predominantly in the range of C14–C18 which are ideal for the production of methyl esters (Dufreche et al., 2007; Mondala et al., 2009). Due to their hydrophobicity, wastewater lipids are usually sorbed onto particles and not readily extractable (Dueholm et al., 2000). However, LAOs excrete extracellular lipases, which allow hydrolysis of lipids and efficient bacterial assimilation. Following our “wastewater biorefinery column” concept, the buildup of filamentous LAO biomass could be favored at the top of the column (Figure 2). Given the high levels of lipid accumulation by filamentous organisms and straightforward production of biodiesel from this lipid-rich biomass, a large potential exists in wastewater biodiesel production as its synthesis is economically viable (Knothe et al., 2005). Interestingly, a byproduct of TAG-derived biodiesel production is glycerol, which can be furthered used to produce PHA (Mothes et al., 2007), thereby leading to a complete and high-value valorization of the wastewater TAG fraction.
PHA synthesis naturally occurs within the microbial biomass of AS. Wastewater-derived PHAs are currently being used to synthesize biodegradable bioplastics on an industrial scale (Chen, 2009; Morgan-Sagastume et al., 2010) and they exhibit similar thermomechanical properties compared to chemically synthesized polypropylenes (Curran, 1996). Additionally, PHAs can be chemically transformed into the biofuel hydroxybutyrate methyl ester by acid catalyzed hydrolysis (Zhang et al., 2009). Therefore, wastewater-derived PHAs represent a suitable renewable resource for plastic production, as resource expenses in the entire PHA production chain can account up to 50% of the total production costs (Choi and Lee, 1999). The microbial accumulation of PHAs from wastewater can be very rapid (ca. 5 h) and pronounced (PHAs can constitute up to 77% of cell dry weight (Jiang et al., 2012). Both PAOs and GAOs are known to accumulate PHAs under alternating anaerobic/aerobic conditions. Given tailored environmental conditions, targeted enrichment of these organisms would therefore allow reclamation of PHA and fermentation products along with polyphosphate or glycogen (see below). PAOs in full-scale BWWTPs can contribute up to 35% of the total bacterial biomass (Oehmen et al., 2010) and laboratory reactor studies suggest that PAOs exhibit fast settling rates when compared to GAOs because of their differing cell densities (Vlaeminck et al., 2010; Winkler et al., 2011b; Volcke et al., 2012). These properties could be harnessed for separating both organismal groups and carrying out targeted resource reclamation (Figure 2). Through the establishment of density gradients, e.g., through manipulation of settling times, it could therefore be envisaged that GAOs could occupy the middle layers followed by PAOs in the lower section of the “wastewater biorefinery column” (Figure 2).
Although poorly studied in AS-based BWWTPs, fermentative bacteria contribute to the hydrolysis of complex organic macromolecules into low-molecular-weight substrates providing energy and carbon sources to other microbes. Produced molecules include industrially relevant commodities such as propionic acid, lactic acid, acetic acid, and formic acid (Kong et al., 2008). Interestingly, the production of alcohols and/or organic acids using fermentative treatment phases could be combined with biodiesel and bioplastic production since both processes require organic solvents. Most importantly, harvesting glycogen from GAO biomass could also be used for bioethanol production, an important biofuel (Hahn-Hägerdal et al., 2006).
Renewable fertilizer production using nutrient recovery from wastewater (primarily N and P) could cover up to 30% of the current agricultural fertilizer demand (Verstraete et al., 2009). A global estimate suggests that fertilizer production consumes up to 1.2% of the world’s energy (out of which 92.5% for N and 3% for P), contributing about 1.2% of the total anthropogenic greenhouse gas emissions (Kongshaug, 1998). In particular, PAO-rich biomass holds great promise as fertilizer. A major limitation of its current use as an agricultural fertilizer is that the biomass is typically rich in heavy metals (Veeken and Hamelers, 1999). However, there is increasing evidence that wastewater derived adsorbents (e.g., blast furnace slag) possesses high heavy metal adsorption capacities and offers a low-cost alternative treatment of metal-contaminated wastewater (Kurniawan et al., 2006) prior to further processing of the wastewater.
Apart from PAO-enriched biomass, struvite (MgNH4PO4·6H2O), a commonly occurring mineral found as precipitates in BWWTPs (Rawn et al., 1939) is a commercially produced fertilizer (Mavinic et al., 2007). Estimates suggest that within BWWTPs, 100 m3 of wastewater can yield up to 1 kg of struvite (Shu et al., 2006). The low solubility and the presence of high abundances of N and P in struvite is advantageous as a fertilizer since it reduces nutrient run-off in turn limiting eutrophication in receiving water bodies (Shu et al., 2006; El Diwani et al., 2007). Presence of low concentrations of suspended solids and high concentrations of ammonium and phosphate enhances struvite production (Doyle and Parsons, 2002). Thus, accelerating struvite production by retaining high levels of these ions in the wastewater while continuously reclaiming biomass fractions, e.g., from fast-growing populations could be another aim to be achieved in the “wastewater biorefinery column.”
In the context of N recovery for fertilizer production, nitrate accumulating organisms which are known to occur in varied habitats (McHatton et al., 1996; Schulz et al., 1999) may also be of pronounced interest. Although nitrate accumulators in AS-processes have so far not yet been described, they could represent a significant N source for fertilizer production. A selective enrichment strategy (Spieck et al., 2006) may provide a plausible approach to identify potential nitrate accumulators within full-scale AS-based BWWTPs.
In the past few decades, increasing environmental concerns have triggered the formulation and development of new strategies for energy and nutrient recovery from wastewater through AS-based processes (Verstraete et al., 2009). However, the yields in terms of recovery of either energy and/or biotechnological resources have so far been limited. Our proposed concept of a “wastewater biorefinery column” would use the existing and future wealth of information concerning the genetic repertoire of microorganisms and their metabolic transformations for sustainable production of bioenergy, bioplastics and fertilizers. However, in order to have this come to fruition, it is essential that we first obtain detailed descriptions of the niches of the individual community members. Once such knowledge has been obtained, wastewater treatment processes should be (re-)engineered taking into account the individual organismal niches using bottom-up design approaches rather the top-down strategies pursued so far. The optimization of processes may involve an iterative bottom-up design approach based on a discovery-driven planning approach (Muller et al., 2013) which would involve systematic omic analyses based on which ecological niches may be fine-tuned and, thus, the process continually tweaked to guarantee optimal resource recovery. We still have a long way to go to bring this vision to fruition but it may represent a grand challenge for microbial ecologists and engineers to tackle at the centenary of the discovery of the AS process.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was funded by a Luxembourg National Research Fund (FNR) ATTRACT program grant to Paul Wilmes (ATTRACT/A09/03) as well as postdoctoral grants Aide à la Formation Recherche (AFR) to Abdul R. Sheik (PDR-2013-1/5748561) and Emilie E. L. Muller (PDR-2011-1/SR). We thank reviewers for their valuable suggestions to improve this manuscript.
References
Albertsen, M., Hansen, L. B. S., Saunders, A. M., Nielsen, P. H., and Nielsen, K. L. (2011). A metagenome of a full-scale microbial community carrying out enhanced biological phosphorus removal. ISME J. 6, 1094–1106. doi: 10.1038/ismej.2011.176
Andreasen, M., and Nielsen, P. (1998). In situ characterization of substrate uptake by Microthrix parvicella using microautoradiography. Water Sci. Technol. 37, 19–26. doi: 10.1016/S0273-1223(98)00079-1
Ardern, E., and Lockett, W. T. (1914). Experiments on the oxidation of sewage without the aid of filters. J. Soc. Chem. Ind. 33, 523–539. doi: 10.1002/jctb.5000331005
Bengtsson, S., Werker, A., and Welander, T. (2008). Production of polyhydroxyalkanoates by glycogen accumulating organisms treating a paper mill wastewater. Water Sci. Technol. 58, 323–330. doi: 10.2166/wst.2008.381
Bond, P. L., Hugenholtz, P., Keller, J., and Blackall, L. L. (1995). Bacterial community structures of phosphate-removing and non-phosphate-removing activated sludges from sequencing batch reactors. Appl. Environ. Microbiol. 61, 1910–1916.
Chen, G. Q. (2009). A microbial polyhydroxyalkanoates (PHA) based bio-and materials industry. Chem. Soc. Rev. 38, 2434–2446. doi: 10.1039/b812677c
Chisti, Y. (2007). Biodiesel from microalgae. Biotechnol. Adv. 25, 294–306. doi: 10.1016/j.biotechadv.2007.02.001
Choi, J., and Lee, S. Y. (1999). Factors affecting the economics of polyhydroxyalkanoate production by bacterial fermentation. Appl. Microbiol. Biotechnol. 51, 13–21. doi: 10.1007/s002530051357
Crocetti, G. R., Banfield, J. F., Keller, J., Bond, P. L., and Blackall, L. L. (2002). Glycogen-accumulating organisms in laboratory-scale and full-scale wastewater treatment processes. Microbiology 148, 3353–3364.
Crocetti, G. R., Hugenholtz, P., Bond, P. L., Schuler, A., Keller, J., Jenkins, D., et al. (2000). Identification of polyphosphate-accumulating organisms and design of 16S rRNA-directed probes for their detection and quantitation. Appl. Environ. Microbiol. 66, 1175–1182. doi: 10.1128/AEM.66.3.1175-1182.2000
Curran, M. A. (1996). Environmental life-cycle assessment. Int. J. Life Cycle Assess. 1, 179. doi: 10.1007/BF02978949
Daims, H., Nielsen, J. L., Nielsen, P. H., Schleifer, K.-H., and Wagner, M. (2001). In situ characterization of Nitrospira-like nitrite-oxidizing bacteria active in wastewater treatment plants. Appl. Environ. Microbiol. 67, 5273–5284. doi: 10.1128/AEM.67.11.5273-5284.2001
Dionisi, H. M., Layton, A. C., Harms, G., Gregory, I. R., Robinson, K. G., and Sayler, G. S. (2002). Quantification of Nitrosomonas oligotropha-like ammonia-oxidizing bacteria and Nitrospira spp. from full-scale wastewater treatment plants by competitive PCR. Appl. Environ. Microbiol. 68, 245–253. doi: 10.1128/AEM.68.1.245-253.2002
Di Lorenzo, M., Curtis, T. P., Head, I. M., and Scott, K. (2009). A single-chamber microbial fuel cell as a biosensor for wastewaters. Water Res. 43, 3145–3154. doi: 10.1016/j.watres.2009.01.005
Doyle, J. D., and Parsons, S. A. (2002). Struvite formation, control and recovery. Water Res. 36, 3925–3940. doi: 10.1016/S0043-1354(02)00126-4
Dueholm, T., Andreasen, K., and Nielsen, P. (2000). Transformation of lipids in activated sludge. Water Sci. Technol. 43, 165–172.
Dufreche, S., Hernandez, R., and French, T. (2007). Extraction of lipids from municipal wastewater plant microorganisms for production of biodiesel. J. Am. Oil Chem. Soc. 84, 181–187. doi: 10.1007/s11746-006-1022-4
Eikelboom, D. H. (1975). Filamentous organisms observed in activated sludge. Water Res. 9, 365–388. doi: 10.1016/0043-1354(75)90182-7
El Diwani, G., El Rafie, S., El Ibiari, N. N., and El-Aila, H. I. (2007). Recovery of ammonia nitrogen from industrial wastewater treatment as struvite slow releasing fertilizer. Desalination 214, 200–214. doi: 10.1016/j.desal.2006.08.019
El-Fadel, M., and Massoud, M. (2001). Methane emissions from wastewater management. Environ. Pollut. 114, 177–185. doi: 10.1016/S0269-7491(00)00222-0
Fuhs, G. W., and Chen, M. (1975). Microbiological basis of phosphate removal in the activated sludge process for the treatment of wastewater. Microb. Ecol. 2, 119–138. doi: 10.1007/BF02010434
Fukuda, H., Kondo, A., and Noda, H. (2001). Biodiesel fuel production by transesterification of oils. J. Biosci. Bioeng. 92, 405–416. doi: 10.1016/S1389-1723(01)80288-7
Grady, C. P. L. Jr., Daigger, G. T., Love, N. G., Filipe, C. D. M., and Leslie Grady, C. P. (2011). Biological Wastewater Treatment. London: IWA Publishing.
Gujer, W., Henze, M., Mino, T., Matsuo, T., Wentzel, M. C., and Marais, G. V. R. (1995). The activated sludge model No. 2: Biological phosphorus removal. Water Sci. Technol. 31, 1–11. doi: 10.1016/0273-1223(95)00175-M
Hahn-Hägerdal, B., Galbe, M., Gorwa-Grauslund, M. F., Liden, G., and Zacchi, G. (2006). Bio-ethanol–the fuel of tomorrow from the residues of today. Trends Biotechnol. 24, 549–556. doi: 10.1016/j.tibtech.2006.10.004
Haroon, M. F., Hu, S., Shi, Y., Imelfort, M., Keller, J., Hugenholtz, P., et al. (2013). Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 500, 567–570. doi: 10.1038/nature12375
Heidrich, E. S., Curtis, T. P., and Dolfing, J. (2010). Determination of the internal chemical energy of wastewater. Environ. Sci. Technol. 45, 827–832. doi: 10.1021/es103058w
Henze, M., Gujer, W., Mino, T., Matsuo, T., Wentzel, M. C., and Van Loosdrecht, M. C. M. (1999). Activated sludge model no. 2d, ASM2d. Water Sci. Technol. 39, 165–182. doi: 10.1016/S0273-1223(98)00829-4
Henze, M., Harremoës, P., Cour, J. J. L., and Arvin, E. eds. (1997). Wastewater Treatment. Biological and Chemical Processes, 2nd Edn. Berlin: Springer-Verlag. doi: 10.1007/978-3-662-22605-6
Hesselmann, R. P. X., Werlen, C., Hahn, D., van der Meer, J. R., and Zehnder, A. J. B. (1999). Enrichment, phylogenetic analysis and detection of a bacterium that performs enhanced biological phosphate removal in activated sludge. Syst. Appl. Microbiol. 22, 454–465. doi: 10.1016/S0723-2020(99)80055-1
Hesselsoe, M., Nielsen, J. L., Roslev, P., and Nielsen, P. H. (2005). Isotope labeling and microautoradiography of active heterotrophic bacteria on the basis of assimilation of 14CO2. Appl. Environ. Microbiol. 71, 646–655. doi: 10.1128/AEM.71.2.646-655.2005
Jiang, Y., Marang, L., Tamis, J., Van Loosdrecht, M. C. M., Dijkman, H., and Kleerebezem, R. (2012). Waste to resource: Converting paper mill wastewater to bioplastic. Water Res. 46, 5517–5530. doi: 10.1016/j.watres.2012.07.028
Juretschko, S., Timmermann, G., Schmid, M., Schleifer, K.-H., Pommerening-Röser, A., Koops, H.-P., et al. (1998). Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl. Environ. Microbiol. 64, 3042–3051.
Kämpfer, P., Erhart, R., Beimfohr, C., Böhringer, J., Wagner, M., and Amann, R. (1996). Characterization of bacterial communities from activated sludge: culture-dependent numerical identification versus in situ identification using group-and genus-specific rRNA-targeted oligonucleotide probes. Microb. Ecol. 32, 101–121. doi: 10.1007/BF00185883
Kampschreur, M. J., van der Star, W. R. L., Wielders, H. A., Mulder, J. W., Jetten, M. S. M., and Van Loosdrecht, M. C. M. (2008). Dynamics of nitric oxide and nitrous oxide emission during full-scale reject water treatment. Water Res. 42, 812–826. doi: 10.1016/j.watres.2007.08.022
Knothe, G., Van Gerpen, J. H., and Krahl, J. (2005). The Biodiesel Handbook. Champaign, IL: AOCS press. doi: 10.1201/9781439822357
Kong, Y., Nielsen, J. L., and Nielsen, P. H. (2004). Microautoradiographic study of Rhodocyclus-related polyphosphate-accumulating bacteria in full-scale enhanced biological phosphorus removal plants. Appl. Environ. Microbiol. 70, 5383–5390. doi: 10.1128/AEM.70.9.5383-5390.2004
Kong, Y., Xia, Y., Nielsen, J. L., and Nielsen, P. H. (2006). Ecophysiology of a group of uncultured gammaproteobacterial glycogen-accumulating organisms in full-scale enhanced biological phosphorus removal wastewater treatment plants. Environ. Microbiol. 8, 479–489. doi: 10.1111/j.1462-2920.2005.00914.x
Kong, Y., Xia, Y., and Nielsen, P. H. (2008). Activity and identity of fermenting microorganisms in full-scale biological nutrient removing wastewater treatment plants. Environ. Microbiol. 10, 2008–2019. doi: 10.1111/j.1462-2920.2008.01617.x
Kongshaug, G. (1998). “Energy consumption and greenhouse gas emissions in fertilizer production,” in IFA Technical Conference Marrakech. Available at: http://www.fertilizer.org/HomePage/LIBRARY/Publication-database.html/Energy-Consumption-and-Greenhouse-Gas-Emissions-in-Fertilizer-Production.html
Kristiansen, R., Nguyen, H. T. T., Saunders, A. M., Nielsen, J. L., Wimmer, R., Le, V. Q., et al. (2013). A metabolic model for members of the genus Tetrasphaera involved in enhanced biological phosphorus removal. ISME J. 7, 543–554. doi: 10.1038/ismej.2012.136
Kurniawan, T. A., Chan, G., Lo, W., and Babel, S. (2006). Comparisons of low-cost adsorbents for treating wastewaters laden with heavy metals. Sci. Total Environ. 366, 409–426. doi: 10.1016/j.scitotenv.2005.10.001
Maixner, F., Noguera, D. R., Anneser, B., Stoecker, K., Wegl, G., Wagner, M., et al. (2006). Nitrite concentration influences the population structure of Nitrospira – like bacteria. Environ. Microbiol. 8, 1487–1495. doi: 10.1111/j.1462-2920.2006.01033.x
Mavinic, D. S., Koch, F. A., Huang, H., and Lo, K. V. (2007). Phosphorus recovery from anaerobic digester supernatants using a pilot-scale struvite crystallization process. J. Environ. Eng. Sci. 6, 561–571. doi: 10.1139/S07-007
McHatton, S. C., Barry, J. P., Jannasch, H. W., and Nelson, D. C. (1996). High nitrate concentrations in vacuolate, autotrophic marine Beggiatoa spp. Appl. Environ. Microbiol. 62, 954–958.
McIlroy, S. J., Kristiansen, R., Albertsen, M., Michael Karst, S., Rossetti, S., Lund Nielsen, J., et al. (2013). Metabolic model for the filamentous “Candidatus Microthrix parvicella” based on genomic and metagenomic analyses. ISME J. 7, 1161–1172. doi: 10.1038/ismej.2013.6
Mino, T., Liu, W.-T., Kurisu, F., and Matsuo, T. (1995). Modelling glycogen storage and denitrification capability of microorganisms in enhanced biological phosphate removal processes. Water Sci. Technol. 31, 25–34. doi: 10.1016/0273-1223(95)00177-O
Mondala, A., Liang, K., Toghiani, H., Hernandez, R., and French, T. (2009). Biodiesel production by in situ transesterification of municipal primary and secondary sludges. Bioresour. Technol. 100, 1203–1210. doi: 10.1016/j.biortech.2008.08.020
Morgan-Sagastume, F., Karlsson, A., Johansson, P., Pratt, S., Boon, N., Lant, P., et al. (2010). Production of polyhydroxyalkanoates in open, mixed cultures from a waste sludge stream containing high levels of soluble organics, nitrogen and phosphorus. Water Res. 44, 5196–5211. doi: 10.1016/j.watres.2010.06.043
Mothes, G., Schnorpfeil, C., and Ackermann, J. (2007). Production of PHB from crude glycerol. Eng. Life Sci. 7, 475–479. doi: 10.1002/elsc.200620210
Mulder, A., Graaf, A. A., Robertson, L. A., and Kuenen, J. G. (1995). Anaerobic ammonium oxidation discovered in a denitrifying fluidized bed reactor. FEMS Microbiol. Ecol. 16, 177–184. doi: 10.1111/j.1574-6941.1995.tb00281.x
Muller, E. E. L., Glaab, E., May, P., Vlassis, N., and Wilmes, P. (2013). Condensing the omics fog of microbial communities. Trends Microbiol. 21, 325–333. doi: 10.1016/j.tim.2013.04.009
Nielsen, J., and Nielsen, P. (2002). Quantification of functional groups in activated sludge by microautoradiography. Water Sci. Technol. 46, 389–395.
Nielsen, P., Roslev, P., Dueholm, T., and Nielsen, J. (2002). Microthrix parvicella, a specialized lipid consumer in anaerobic-aerobic activated sludge plants. Water Sci. Technol. 46, 73–80.
Oehmen, A., Carvalho, G., Lopez-Vazquez, C. M., Van Loosdrecht, M. C. M., and Reis, M. A. M. (2010). Incorporating microbial ecology into the metabolic modelling of polyphosphate accumulating organisms and glycogen accumulating organisms. Water Res. 44, 4992–5004. doi: 10.1016/j.watres.2010.06.071
Park, H.-D., Wells, G. F., Bae, H., Criddle, C. S., and Francis, C. A. (2006). Occurrence of ammonia-oxidizing archaea in wastewater treatment plant bioreactors. Appl. Environ. Microbiol. 72, 5643–5647. doi: 10.1128/AEM.00402-06
Pham, T. H., Rabaey, K., Aelterman, P., Clauwaert, P., De Schamphelaire, L., Boon, N., et al. (2006). Microbial fuel cells in relation to conventional anaerobic digestion technology. Eng. Life Sci. 6, 285–292. doi: 10.1002/elsc.200620121
Prakasam, T. B. S., and Dondero, N. C. (1967). Aerobic heterotrophic bacterial populations of sewage and activated sludge I. Enumeration. Appl. Microbiol. 15, 461–467.
Purkhold, U., Pommerening-Röser, A., Juretschko, S., Schmid, M. C., Koops, H.-P., and Wagner, M. (2000). Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl. Environ. Microbiol. 66, 5368–5382. doi: 10.1128/AEM.66.12.5368-5382.2000
Quemeneur, M., and Marty, Y. (1994). Fatty acids and sterols in domestic wastewaters. Water Res. 28, 1217–1226. doi: 10.1016/0043-1354(94)90210-0
Raunkjær, K., Hvitved-Jacobsen, T., and Nielsen, P. H. (1994). Measurement of pools of protein, carbohydrate and lipid in domestic wastewater. Water Res. 28, 251–262. doi: 10.1016/0043-1354(94)90261-5
Rawn, A. M., Banta, A. P., and Pomeroy, R. (1939). Multiple-stage sewage sludge digestion. Trans. Am. Soc. Civ. Eng. 104, 93–119.
Santos, M. M., Lemos, P. C., Reis, M. A. M., and Santos, H. (1999). Glucose metabolism and kinetics of phosphorus removal by the fermentative bacterium Microlunatus phosphovorus. Appl. Environ. Microbiol. 65, 3920–3928.
Schmid, M., Twachtmann, U., Klein, M., Strous, M., Juretschko, S., Jetten, M., et al. (2000). Molecular evidence for genus level diversity of bacteria capable of catalyzing anaerobic ammonium oxidation. Syst. Appl. Microbiol. 23, 93–106. doi: 10.1016/S0723-2020(00)80050-8
Schulz, H. N., Brinkhoff, T., Ferdelman, T. G., Mariné, M. H., Teske, A., and Jørgensen, B. B. (1999). Dense populations of a giant sulfur bacterium in Namibian shelf sediments. Science 284, 493–495. doi: 10.1126/science.284.5413.493
Shizas, I., and Bagley, D. M. (2004). Experimental determination of energy content of unknown organics in municipal wastewater streams. J. Energy Eng. 130, 45–53. doi: 10.1061/(ASCE)0733-9402(2004)130:2(45)
Shu, L., Schneider, P., Jegatheesan, V., and Johnson, J. (2006). An economic evaluation of phosphorus recovery as struvite from digester supernatant. Bioresour. Technol. 97, 2211–2216. doi: 10.1016/j.biortech.2005.11.005
Spieck, E., Hartwig, C., McCormack, I., Maixner, F., Wagner, M., Lipski, A., et al. (2006). Selective enrichment and molecular characterization of a previously uncultured Nitrospira – like bacterium from activated sludge. Environ. Microbiol. 8, 405–415. doi: 10.1111/j.1462-2920.2005.00905.x
Strous, M., Fuerst, J. A., Kramer, E. H. M., Logemann, S., Muyzer, G., van de Pas-Schoonen, K. T., et al. (1999). Missing lithotroph identified as new planctomycete. Nature 400, 446–449. doi: 10.1038/22749
Strous, M., Pelletier, E., Mangenot, S., Rattei, T., Lehner, A., Taylor, M. W., et al. (2006). Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature 440, 790–794. doi: 10.1038/nature04647
Thomsen, T. R., Kong, Y., and Nielsen, P. H. (2007). Ecophysiology of abundant denitrifying bacteria in activated sludge. FEMS Microbiol. Ecol. 60, 370–382. doi: 10.1111/j.1574-6941.2007.00309.x
Timmermans, P., and Van Haute, A. (1983). Denitrification with methanol: fundamental study of the growth and denitrification capacity of Hyphomicrobium sp. Water Res. 17, 1249–1255. doi: 10.1016/0043-1354(83)90249-X
United Nations, Department of Economic and Social Affairs, Population Division (2013). World Population Prospects: The 2012 Revision, Highlights and Advance Tables. ESA/P/WP.228
Van Dongen, U., Jetten, M., and Van Loosdrecht, M. C. M. (2001). The SHARON-Anammox process for treatment of ammonium rich wastewater. Water Sci. Technol. 44, 153–160.
Veeken, A. H. M., and Hamelers, H. V. M. (1999). Removal of heavy metals from sewage sludge by extraction with organic acids. Water Sci. Technol. 40, 129–136. doi: 10.1016/S0273-1223(99)00373-X
Verstraete, W., Van de Caveye, P., and Diamantis, V. (2009). Maximum use of resources present in domestic “used water.” Bioresour. Technol. 100, 5537–5545. doi: 10.1016/j.biortech.2009.05.047
Vitousek, P. M. (1994). Beyond global warming: ecology and global change. Ecology 75, 1861–1876. doi: 10.2307/1941591
Vlaeminck, S. E., Terada, A., Smets, B. F., De Clippeleir, H., Schaubroeck, T., Bolca, S., et al. (2010). Aggregate size and architecture determine microbial activity balance for one-stage partial nitritation and anammox. Appl. Environ. Microbiol. 76, 900–909. doi: 10.1128/AEM.02337-09
Volcke, E. I. P., Picioreanu, C., De Baets, B., and van Loosdrecht, M. C. M. (2012). The granule size distribution in an anammox-based granular sludge reactor affects the conversion – implications for modeling. Biotechnol. Bioeng. 109, 1629–1636. doi: 10.1002/bit.24443
Wagner, M., Erhart, R., Manz, W., Amann, R., Lemmer, H., Wedi, D., et al. (1994). Development of an rRNA-targeted oligonucleotide probe specific for the genus Acinetobacter and its application for in situ monitoring in activated sludge. Appl. Environ. Microbiol. 60, 792–800.
Wagner, M., Nielsen, P. H., Loy, A., Nielsen, J. L., and Daims, H. (2006). Linking microbial community structure with function: fluorescence in situ hybridization-microautoradiography and isotope arrays. Curr. Opin. Biotechnol. 17, 83–91. doi: 10.1016/j.copbio.2005.12.006
Wilmes, P., Andersson, A. F., Lefsrud, M. G., Wexler, M., Shah, M., Zhang, B., et al. (2008). Community proteogenomics highlights microbial strain-variant protein expression within activated sludge performing enhanced biological phosphorus removal. ISME J. 2, 853–864. doi: 10.1038/ismej.2008.38
Winkler, M. K. H., Bassin, J. P., Kleerebezem, R., De Bruin, L. M. M., Van den Brand, T. P. H., and Van Loosdrecht, M. C. M. (2011a). Selective sludge removal in a segregated aerobic granular biomass system as a strategy to control PAO–GAO competition at high temperatures. Water Res. 45, 3291–3299. doi: 10.1016/j.watres.2011.03.024
Winkler, M. K. H., Kleerebezem, R., Kuenen, J. G., Yang, J., and Van Loosdrecht, M. C. M. (2011b). Segregation of biomass in cyclic anaerobic/aerobic granular sludge allows the enrichment of anaerobic ammonium oxidizing bacteria at low temperatures. Environ. Sci. Technol. 45, 7330–7337. doi: 10.1021/es201388t
Wong, M.-T., Mino, T., Seviour, R. J., Onuki, M., and Liu, W.-T. (2005). In situ identification and characterization of the microbial community structure of full-scale enhanced biological phosphorous removal plants in Japan. Water Res. 39, 2901–2914. doi: 10.1016/j.watres.2005.05.015
Wong, M.-T., Tan, F. M., Ng, W. J., and Liu, W.-T. (2004). Identification and occurrence of tetrad-forming Alphaproteobacteria in anaerobic–aerobic activated sludge processes. Microbiology 150, 3741–3748. doi: 10.1099/mic.0.27291-0
Yu, K., and Zhang, T. (2012). Metagenomic and metatranscriptomic analysis of microbial community structure and gene expression of activated sludge. PLoS ONE 7:e38183. doi: 10.1371/journal.pone.0038183
Zhang, X., Luo, R., Wang, Z., Deng, Y., and Chen, G.-Q. (2009). Application of (R)-3-hydroxyalkanoate methyl esters derived from microbial polyhydroxyalkanoates as novel biofuels. Biomacromolecules 10, 707–711. doi: 10.1021/bm801424e
Keywords: activated sludge, wastewater microbial diversity and function, integrated omics, niche engineering, sustainability and renewable resources, energy reclamation – biodiesel and bioethanol, nutrient recovery and fertilizers, wastewater biorefinery column concept
Citation: Sheik AR, Muller EEL and Wilmes P (2014) A hundred years of activated sludge: time for a rethink. Front. Microbiol. 5:47. doi: 10.3389/fmicb.2014.00047
Received: 28 November 2013; Accepted: 22 January 2014;
Published online: 03 March 2014.
Edited by:
Kevin Bradley Clark, Veterans Affairs Greater Los Angeles Healthcare System, USAReviewed by:
Xiao Zhang, Washington State University, USAPer Halkjær Nielsen, Aalborg University, Denmark
Copyright © 2014 Sheik, Muller and Wilmes. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Abdul R. Sheik and Paul Wilmes, Eco-Systems Biology Group, Luxembourg Centre for Systems Biomedicine, University of Luxembourg, Campus Belval, 7, avenue des Hauts-Fourneaux, L-4362 Esch-sur-Alzette, Luxembourg e-mail:YWJkdWwuc2hlaWtAdW5pLmx1;cGF1bC53aWxtZXNAdW5pLmx1
 Abdul R. Sheik
Abdul R. Sheik Emilie E. L. Muller
Emilie E. L. Muller Paul Wilmes
Paul Wilmes