
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 20 December 2013
Sec. Plant Pathogen Interactions
Volume 4 - 2013 | https://doi.org/10.3389/fmicb.2013.00400
This article is part of the Research TopicThe plant microbiome and its importance for plant and human healthView all 19 articles
Past medicinal plant research primarily focused on bioactive phytochemicals, however, the focus is currently shifting due to the recognition that a significant number of phytotherapeutic compounds are actually produced by associated microbes or through interaction with their host. Medicinal plants provide an enormous bioresource of potential use in modern medicine and agriculture, yet their microbiome is largely unknown. The objective of this review is (i) to introduce novel insights into the plant microbiome with a focus on medicinal plants, (ii) to provide details about plant- and microbe-derived ingredients of medicinal plants, and (iii) to discuss possibilities for plant growth promotion and plant protection for commercial cultivation of medicinal plants. In addition, we also present a case study performed both to analyse the microbiome of three medicinal plants (Matricaria chamomilla L., Calendula officinalis L., and Solanum distichum Schumach. and Thonn.) cultivated on organically managed Egyptian desert farm and to develop biological control strategies. The soil microbiome of the desert ecosystem was comprised of a high abundance of Gram-positive bacteria of prime importance for pathogen suppression under arid soil conditions. For all three plants, we observed a clearly plant-specific selection of the microbes as well as highly specific diazotrophic communities that overall identify plant species as important drivers in structural and functional diversity. Lastly, native Bacillus spec. div. strains were able to promote plant growth and elevate the plants’ flavonoid production. These results underline the numerous links between the plant-associated microbiome and the plant metabolome.
All plant-associated microenvironments are colonized in high abundances by microorganisms, especially the nutrient-rich rhizosphere, the soil area influenced by plant roots, hosts a plethora of microbes that are of central importance for plant nutrition, health, and quality (Hiltner, 1904 in Hartmann et al., 2008; Berg, 2009; Mendes et al., 2011, 2013). The rhizosphere can host up to 1011 microbial cells per gram root with more than 30,000 different prokaryotic species (Berendsen et al., 2012). These rhizospheric microorganisms from a highly diverse reservoir of soil microbes are attracted by the rhizosphere’s plant root secretions and other rhizodeposits (Compant et al., 2010), and driven via the compositional variability of these exudates (Bais et al., 2006; Doornbos et al., 2012). Each plant species harbors a specific rhizosphere microbiome dependent of the present soil community (Smalla et al., 2001). Besides plant species, the composition and diversity of microbial rhizosphere communities is shaped by soil type and pedoclimate, plant health and developmental stage, climate and season, pesticide treatments, grazers and animals, and several other biotic and abiotic factors (Singh and Mukerji, 2006; Berg and Smalla, 2009; Barnard et al., 2013). Some genera are ubiquitous and can be found distributed over the entire plant, such as the well-known plant-associated genera Bacillus and Pseudomonas (Berg et al., 2011). However, a high degree of specificity for each microenvironment was also observed via comparison of microbial colonization patterns of different microhabitats (Berg et al., 2005b; Fürnkranz et al., 2012; Köberl et al., 2013). Certain plant-associated microorganisms including beneficials and pathogens are also passed down from previous generations via the seed (Hardoim et al., 2012; Hirsch and Mauchline, 2012), and likewise a transmission between plants through pollen grains was recently observed (Fürnkranz et al., 2012). Interestingly, the phylogenetically oldest land plants, mosses, transfer a highly diverse core microbiome of primarily potential beneficial bacteria from the sporophyte to the gametophyte and vice versa (Bragina et al., 2012).
Medicinal plants harbor a distinctive microbiome due to their unique and structurally divergent bioactive secondary metabolites that are most likely responsible for the high specificity of the associated microorganisms (Qi et al., 2012). The analyses of several Chinese medicinal plant microbiomes showed interesting results (Ainsliaea henryi Diels, Dioscorea opposita, Potentilla discolor Bge, Stellera chamaejasme L., Ophiopogon japonicus (Thunb) Ker-Gawl., Juncus effusus L. var. decipiens Buchen., Rhizoma arisaematis, and others; Li et al., 2008; Zhao et al., 2011), as each of them hosted a specific actinobacterial community and showed a remarkably high and diverse rhizospheric and endophytic colonization with Actinobacteria featuring both antimicrobial and antitumor properties (Zhao et al., 2012). These Gram-positive and often spore-forming bacteria are promising biological control agents (BCAs), such as the genus Streptomyces that is a known and unique source of novel antibiotics (Goodfellow and Fiedler, 2010; Niraula et al., 2010; Nachtigall et al., 2011; Raaijmakers and Mazzola, 2012).
Plants contain numerous different biologically active compounds, and plant-derived medicines have been part of traditional healthcare in most parts of the world for thousands of years. Traditional Chinese medicine (TCM), phytotherapeutic knowledge from the Mayans, the aboriginal medicine of Australia, and several other cultures comprise a huge spectrum of natural remedies that can be exploited as sources for new and effective therapeutic agents. Still widely practiced in the modern era, TCM supplies ethnopharmaceutical knowledge on over 5,000 plant species used for the treatment of numerous diseases and has already provided the basis for the discovery of many modern drugs, such as anticancer agents (Miller et al., 2012a, b). In general, natural products play a highly considerable role in the drug discovery and development process, as about 26% of the new chemical entities introduced into the market worldwide from 1981 to 2010 were either natural products or those derived directly therefrom, reaching a high of 50% in 2010 (Newman and Cragg, 2012). In the past, medicinal plant research focused primarily on their ingredients, however, recently the focus has shifted to include the structure and function of several medicinal plant microbiomes. Surprisingly, not only were the plants themselves able to produce substances with phytotherapeutic properties, but their associated microbes, in particular endophytes, could as well (Table 1). Currently, research continues to show that a significant number of natural products are actually produced by microbes and/or microbial interactions with the host from whence they were isolated (Gunatilaka, 2006), and for several medicinal plants it is presumed that the plant-associated microbiome, especially the complex community of the endomicrobiome, is directly or indirectly involved in the production of bioactive phytochemicals. Presently, however, only a small subset of potential microbial strains could be definitively attributed to phytotherapeutic properties (Strobel and Daisy, 2003; Strobel et al., 2004; Chandra, 2012; Miller et al., 2012a,b), and their relative contribution to the recognized valuable bioactivity of medicinal plants is not clear as of yet.
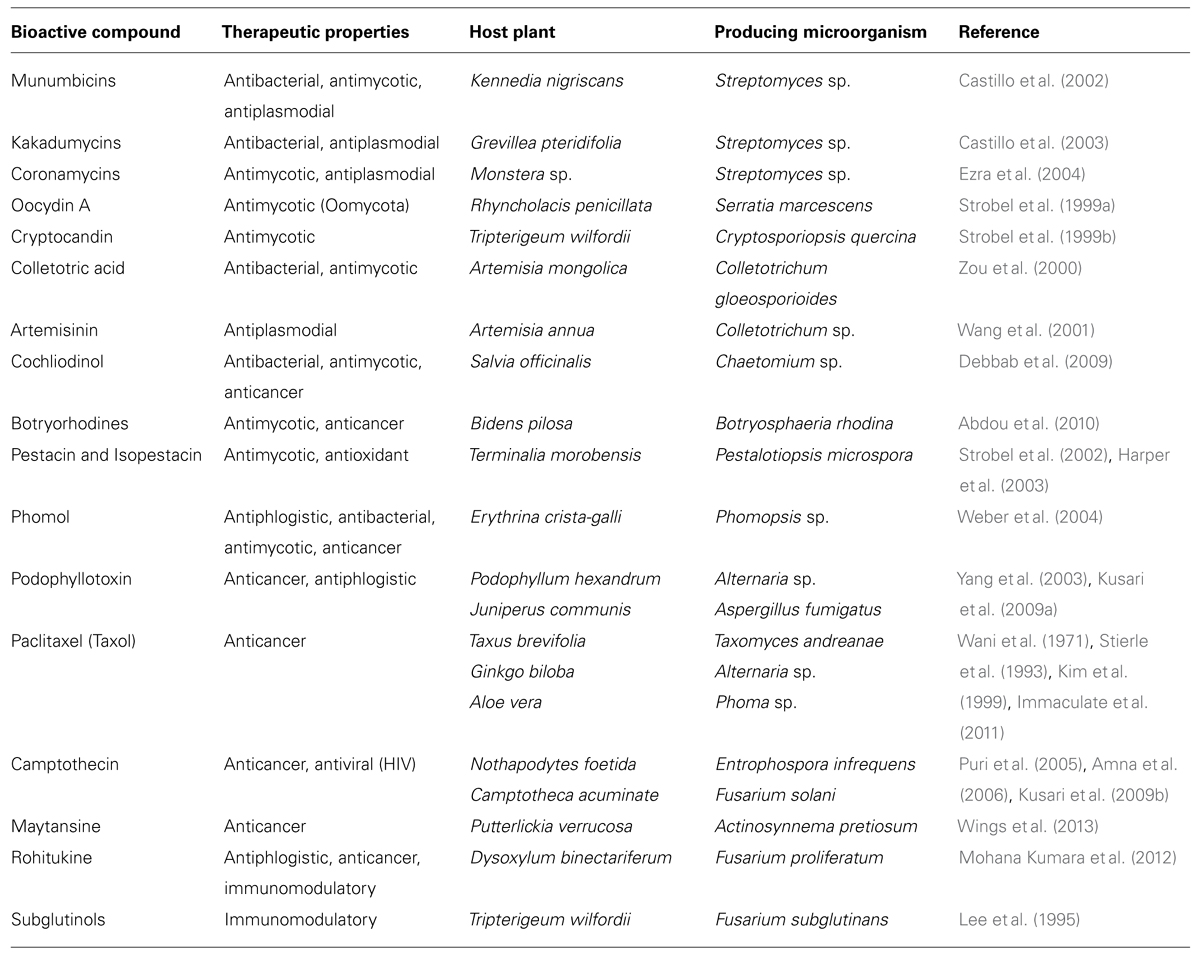
TABLE 1. Examples for bioactive phytometabolites where microorganisms are involved in their production.
In regards to the alarming incidence of antibiotic resistance in bacteria with medical relevancy, medicinal plants with antibacterial properties are of central importance as bioresources for novel active metabolites (Palombo and Semple, 2001). Likewise, there is an increasing need for more and better antimycotics to treat those with weakened immune systems who are more prone to developing fungal infections, such as from the AIDS epidemic, cancer therapy, or organ transplants (Strobel and Daisy, 2003; Strobel et al., 2004). For centuries, several phytotherapeutics have also been known for their antiphlogistic features, yet despite the progress within medical research, chronic inflammatory diseases such as asthma, arthritis, and rheumatism remain one of the world’s leading health problems (Li et al., 2003). Hypertension is another critical issue for human health and is a primary risk factor for stroke, heart disease, and renal failure. Many herbal remedies as well as foods, however, are known and effective folk medicines in the prevention and/or treatment of high blood pressure (Abdel-Aziz et al., 2011). Hence, nature must still harbor plenty of currently unknown active agents that may serve as leads and scaffolds for the development of desperately needed efficacious drugs for a multitude of diseases (Newman and Cragg, 2012). Today, globalization has also had an impact on the use of medicinal plants and has proven beneficial in allowing greater access to these medicines for people all across the globe. For example, TCM plants are very popular in Europe, whereas the traditional German chamomile is primarily produced in Egypt. Growth, quality, and health of the medicinal plants are highly influenced and controlled by their microbiota through microbial metabolisms and host interactions.
Several rhizospheric microbes interact beneficially via different mechanisms with their host plant. They can have a direct plant growth promoting effect based on improved nutrient acquisition or hormonal stimulation, or indirectly affect the plant health by suppression of phytopathogens (Berg, 2009; Lugtenberg and Kamilova, 2009). Biofertilizers are microbes that supply the plant with nutrients, for example symbiotic root-nodulating rhizobia are the most prominent among the nitrogen-fixing microorganisms. Other microbial biofertilizers, such as mycorrhizal fungi and several rhizobacteria, are able to solubilise plant-available phosphate from either organic or inorganic bound phosphate (Lugtenberg et al., 2002). Microbes that hormonally promote plant growth are termed phytostimulators, and the phytohormone auxin, for instance, produced by fluorescent pseudomonads is one of the best understood examples (Kamilova et al., 2006; Khare and Arora, 2010). Various rhizobacteria, including for example Burkholderia cepacia, Staphylococcus epidermidis, and strains of the Bacillus subtilis group, stimulate plant growth by the emission of volatile organic compounds (VOCs; Ryu et al., 2003; Kai et al., 2007; Effmert et al., 2012; Bitas et al., 2013). VOCs are low molecular weight molecules (<300 Da) that have high vapor pressures and are therefore able to diffuse over long distances through the porous structure of the soil and through water-filled pores (Kai et al., 2007; Insam and Seewald, 2010). Indirectly, the plant growth can be promoted via biological control of phytopathogens. Pathogen growth can be inhibited by antibiotics or VOCs, toxins, biosurfactants, or extracellular cell wall-degrading enzymes, but microbial antagonism can also occur via degradation of pathogenicity factors like toxins, or simply by the competition for nutrients, minerals, or colonization sites (Berg, 2009). Another possible way to reduce the activity of pathogenic microorganisms is the activation of the plant defense mechanisms, or the so called induced systemic resistance (ISR) triggered by certain non-pathogenic rhizobacteria. Flagella, lipopolysaccharides, siderophores, VOCs, and several other bacterial components are thought to be involved in activating the non-pathogenic rhizobacteria-mediated ISR signaling pathway (van Loon et al., 1998; Lugtenberg and Kamilova, 2009).
Biological control of plant pathogens as well as plant growth promotion with microorganisms has been intensively studied over the past decades and is becoming a realistic alternative to chemical pesticides and fertilizers in sustainable agriculture (Weller, 2007). Several microbial inoculants have already been successfully commercialized (Berg, 2009, 2013), but a specific biological control strategy for medicinal plants, which are increasingly affected by different soil-borne phytopathogens, has not been available until now. While specific biocontrol agents for medicinal plants are needed, their associated microbiomes with outstanding metabolic activities also provide a promising source for novel BCAs.
Traditional medicinal plants are often consumed raw, such as berries or other edible fruits, or in dried form as herbal brews or teas. Therefore, it is of crucial importance that any potentially harmful effect of associated microorganisms or of an applied biocontrol agent on human health be avoided completely. Recently, for instance, bacterial strains closely related to Stenotrophomonas maltophilia and Rhodococcus sp. were isolated from the roots of oregano (Origanum vulgare L.) cultivated in a sub-Himalayan region (Bafana, 2013). Similarly, Ochrobactrum and Rhodococcus were also detected on the studied medicinal plants in Egypt (Matricaria chamomilla L., Calendula officinalis L., and Solanum distichum Schumach. and Thonn.; Köberl et al., 2011). Among several others, these bacterial genera are known for their ambiguous interactions with eukaryotic hosts whereby the mechanisms responsible for plant growth promotion are similar to those also responsible for opportunistic infections in humans and animals (Berg et al., 2005a). In addition to the suppression of phytopathogens, antagonistic activity against potentially harmful human pathogens should also be considered in the biocontrol strategy.
Conversely, ethanolic extracts from the Chinese medicinal plants Mallotus yunnanensis Pax et. Hoffm., Schima sinensis (Hemsl. et. Wils) Airy-Shaw., Garcinia morella Desr., Evodia daneillii (Benn) Hemsl., Meliosma squamulata Hance., Skimmia arborescens Anders., and Brandisia hancei Hook. f. were determined as highly active against the clinical pathogens Staphylococcus aureus, Escherichia coli, Pseudomonas aeruginosa, and Candida albicans which corresponds to their traditional applications in skin and other infections (Zuo et al., 2012). Promising antimicrobial activities against human multi-drug-resistant pathogens have been observed for Mexican medicinal plants as well (Jacobo-Salcedo Mdel et al., 2011). As previously discussed for phytotherapeutic properties, the suppression of human pathogens can also be frequently attributed to medicinal plant-associated microbes and their secondary metabolites (Miller et al., 2012b; Mousa and Raizada, 2013).
In conclusion, medicinal plants should be considered as meta-organisms that comprise both the plant themselves and their microbiome. As meta-organisms, they are a largely untapped and enormous bioresource for bioactive compounds and microorganisms of potential use in modern medicine, agriculture, and pharmaceutical industry. As such, more research is necessary to exploit this immense reservoir for mankind.
In comparison to soils of humid areas, the soil microbiome of the Egyptian desert farm Sekem was comprised of a high abundance of Gram-positive, spore-forming bacteria primarily of the Firmicutes branch with 37% of the total bacterial soil community as revealed through a pyrosequencing-based amplicon sequencing approach (Köberl et al., 2011). However, a global soil community analysis including 32 libraries of 16S rRNA and 16S rRNA gene libraries from a variety of soils reported Firmicutes contribute a mean of only 2% in the total bacterial soil community (Janssen, 2006). Bacillus and Paenibacillus play the key role in explaining this remarkably high abundance of Firmicutes in the investigated desert agro-ecosystem. These drought-resistant genera are of prime importance for pathogen suppression under arid conditions as nearly all isolated antagonists with activity against soil-borne phytopathogenic fungi could be affiliated to this taxonomic group. This is in direct contrast to humid soils, where primarily Gram-negative bacteria like Pseudomonas are responsible for the indigenous antagonistic potential (Berg et al., 2005b; Haas and Défago, 2005; Costa et al., 2006; Zachow et al., 2008). A significantly higher proportion of Firmicutes and antifungal isolates were observed in field soil from the Egyptian farm than in the surrounding desert soil uninfluenced by human activities. In general, the total bacterial soil microbiome of the anthropogenic ecosystem exhibited a higher diversity and better ecosystem function for plant health in comparison to the natural desert soil (Figure 1). Due to the long-term agricultural use of the desert and the associated increasing occurrence of plant pathogens, the indigenous antagonistic potential in soil was almost twice as high as in the uncultivated desert soil. However, the diversity of antagonistic bacteria was lower and highly dominated by isolates of the Bacillus subtilis group. The most efficient antagonists from the native desert soil belonged to Streptomyces, and Bacillus and Paenibacillus species were the most frequently isolated antagonists from all investigated arid habitats including both desert and agriculturally used soil, as well as from the rhizosphere and endorhiza of three different species of medicinal plants cultivated on the desert farm (Matricaria chamomilla L., Calendula officinalis L., and Solanum distichum Schumach. and Thonn.). None of the plants are native to Egypt, and therefore were exposed to a previously unencountered microbiome. Interestingly, despite a clearly plant-specific selection of the associated bacterial microbiome, indigenous Bacillus and Paenibacillus strains of native desert soil with promising antagonistic properties against a wide range of soil-borne phytopathogens were enriched in all investigated plant roots. Conversely, several extremophilic bacterial groups, such as Acidimicrobium, Rubellimicrobium, and Deinococcus–Thermus decreased or completely disappeared from soil after agricultural use (Köberl et al., 2011).
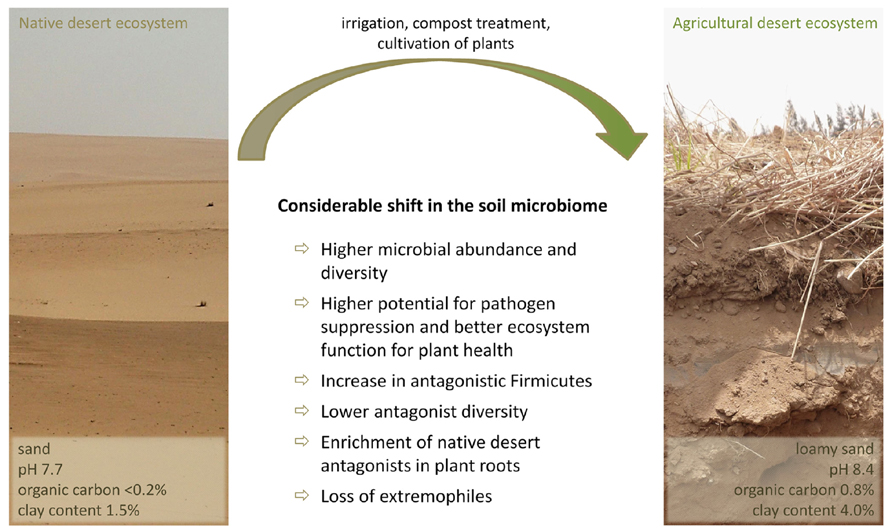
FIGURE 1. Community shift in the soil microbiome after long-term agricultural use of the desert under organic management.
Nitrogen is an essential macronutrient for plants and one of the most yield-limiting factors in agricultural production systems throughout the world (Bhattacharjee et al., 2008; Orr et al., 2011). To gain insight into the indigenous community of diazotrophic plant growth promoting microorganisms that inhabit desert agro-ecosystems, community profiles of the nifH gene encoding the nitrogenase reductase subunit were assessed. A broad diversity and high abundance of diazotrophs were detected in all investigated habitats, thus underlining their importance in native and agricultural desert ecosystems. Due to watering and cultivation of desert soil, a considerable shift toward a higher abundance and diversity was also observed for the nitrogen-fixing community. Phylogenetic analyses distinguishing between the major nifH gene types (Zehr et al., 2003; Gaby and Buckley, 2012) revealed that all NifH sequences from soil libraries were affiliated with the canonical nifH clusters I (conventional molybdenum nitrogenases) and III (molybdenum nitrogenases from anaerobes), while no sequences of alternative nitrogenases (cluster II) and nifH paralogs (clusters IV and V) were found. In general, the diazotrophic soil microbiota was highly dominated by NifH sequences related to Alphaproteobacteria. Each investigated medicinal plant cultivated on the desert farm harbored a specific root-associated diazotrophic microbiome. The rhizosphere inhabitants of Matricaria chamomilla (Figure 2) and Calendula officinalis were similar and both dominated by potential root-nodulating rhizobia acquired mainly from soil. Conversely, the rhizosphere of Solanum distichum was colonized in higher abundances by free-living nitrogen fixers most likely transmitted between plants as they were undetectable in soils. Although well-known for taxonomic community structure (Berg and Smalla, 2009; Bulgarelli et al., 2012), this high degree of plant-specificity identified plants as important drivers for functional diversity as well (Köberl, 2013). The total bacterial and fungal communities also revealed similar colonization patterns between the medicinal plants Matricaria chamomilla and Calendula officinalis compared to Solanum distichum (Köberl et al., 2013). This effect may have been intensified as a result of the close relationship between Matricaria chamomilla and Calendula officinalis who both belong to the Asteraceae family and therefore produce more similar bioactive metabolites. Furthermore, both Matricaria chamomilla and Calendula officinalis are annual herbal medicinal plants, while Solanum distichum is a perennial plant thus providing a longer timeframe to specifically select a stable associated microbiome.
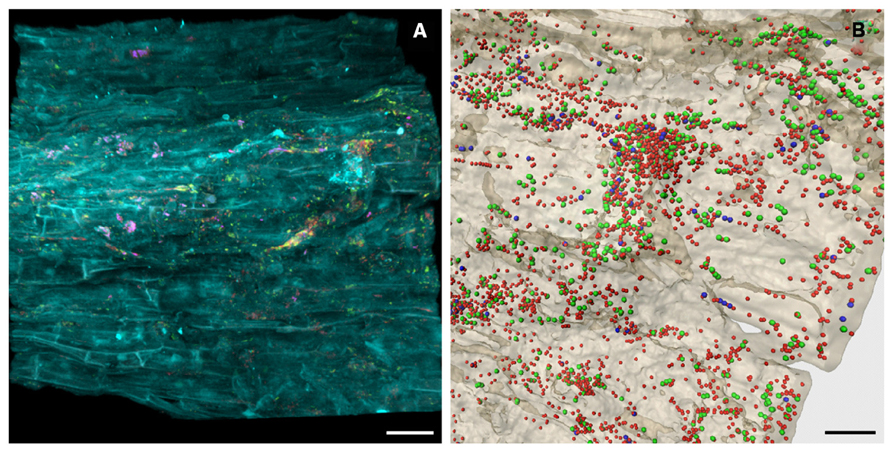
FIGURE 2. In situ visualization of the bacterial root colonization of Matricaria chamomilla. Volume rendering (A) and three-dimensional reconstruction model (B) of confocal laser scanning microscopy stacks. (A) yellow = Alphaproteobacteria, pink = Betaproteobacteria, red = other eubacteria, cyan = root tissue, scale bar = 30 μm. (B) green = Alphaproteobacteria, blue = Betaproteobacteria, red = other eubacteria, beige = root tissue, scale bar = 15 μm.
In contrast to the highly specific bacterial communities associated with cultivated medicinal plants, fungal communities were less discriminative and characterized primarily by potential pathogens. Phytopathogenic species Fusarium, Verticillium, and several others were frequently identified, and, apart from Rhizoctonia, were the main soil-borne pathogens on the investigated desert farm that caused high yield losses on a wide host range of economically important crops, including the medicinal plants. To biologically control these soil-borne diseases, different desert habitats were screened for potential BCAs adapted to the unique and arid conditions of desert farming. Due to this high content of potential plant pathogens in the fungal community, the selection of antagonists was focused on the indigenous bacterial microbiome. An in vitro screening of 1,212 bacterial isolates linked with the comprehensive ecological data resulted in an antagonist collection of 45 genotypically different antifungal strains. In a hierarchical evaluation including their antifungal properties against Verticillium dahliae, Rhizoctonia solani, and Fusarium culmorum in addition to their antagonistic activity against the soil-borne plant pathogenic bacterium Ralstonia solanacearum and the nematode Meloidogyne incognita, three promising drought- and heat-resistant biocontrol candidates were selected: Streptomyces subrutilus Wb2n-11 isolated from desert soil in Sinai, Bacillus subtilis subsp. subtilis Co1-6 obtained from the rhizosphere of Calendula officinalis, and Paenibacillus polymyxa Mc5Re-14 isolated from the endorhiza of Matricaria chamomilla. Each belongs to risk group 1 and poses no risk for humans or the environment. These three potential BCAs have already shown promising in vitro plant growth promoting activities and stress tolerances; Bacillus subtilis Co1-6 exhibited high drought and salt resistance, protease and glucanase activity, and the production of siderophores, Paenibacillus polymyxa Mc5Re-14 had a lower tolerance to abiotic stresses in comparison to the Bacillus strain, but also tested positive for siderophores and glucanase activity, and the desert bacterium Streptomyces subrutilus Wb2n-11 showed hydrolytic degradation of chitin and glucan. All of them produced antibiotics against the nematode Meloidogyne incognita, however, their antibacterial activities were highly specific. While Bacillus subtilis and Streptomyces subrutilus exhibited antagonistic suppression of the plant pathogen Ralstonia solanacearum, only the Paenibacillus isolate was active against the opportunistic human pathogen Escherichia coli (Köberl et al., 2013).
These three autochthonous Gram-positive strains were selected for ad planta evaluation in the field under desert farming conditions in comparison to three allochthonous Gram-negative strains already known for their beneficial plant–microbe interactions in humid soils: Pseudomonas fluorescens L13-6-12 isolated from the rhizosphere of potato (Solanum tuberosum), Stenotrophomonas rhizophila P69 from oilseed rape (Brassica napus) rhizosphere, and Serratia plymuthica 3Re4-18 from the endorhiza of potato (Lottmann and Berg, 2001; Wolf et al., 2002; Kai et al., 2007; Zachow et al., 2010; Alavi et al., 2013). The first results revealed that priming chamomile seedlings with the autochthonous strains not only showed a stabilizing effect on plant performance, but Bacillus subtilis Co1-6 and Paenibacillus polymyxa Mc5Re-14 were also able to further elevate the plants’ flavonoid production. Higher contents of the bioactive compounds apigenin-7-O-glucoside and apigenin, which belong to the major flavonoids of chamomile florets (Kato et al., 2008; Srivastava and Gupta, 2009), were measured in blossoms of plants treated with the two Bacillales strains compared to blossoms of other treatments and uninoculated control plants (Schmidt et al., 2013). These findings demonstrate that a targeted bacterial treatment could influence the metabolic activity of the plant, and therefore represent one of the many poorly understood links between the structure and metabolic profile of the plant-associated microbiome and the plant metabolome.
Conceived and designed the experiments: Gabriele Berg, Rudolf Bauer, Elshahat M. Ramadan and Martina Köberl. Performed the experiments: Martina Köberl and Ruth Schmidt. Analyzed the data: Martina Köberl, Ruth Schmidt and Gabriele Berg. Contributed reagents/materials/analysis tools: Gabriele Berg. Wrote the paper: Martina Köberl and Gabriele Berg.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by the EU-Egypt Innovation Fund (RDI C2/S1/190) and the EC (Tempus IV JP-00241-2008). We wish to give many thanks to Ibrahim Abouleish and his family (Cairo), without whom the project would not have been possible. Furthermore, we would like to thank Massimiliano Cardinale and Henry Müller (Graz) for their relevant practical and theoretical support, and Margaret R. Starcher (Washington/Graz) is gratefully acknowledged for critically reading the manuscript.
Abdel-Aziz, H., Fawzy, N., Ismail, A. I., and El-Askary, H. (2011). Toxicological studies on a standardized extract of Solanum indicum ssp. distichum. Food Chem. Toxicol. 49, 903–909. doi: 10.1016/j.fct.2010.11.048
Abdou, R., Scherlach, K., Dahse, H. M., Sattler, I., and Hertweck, C. (2010). Botryorhodines A-D, antifungal and cytotoxic depsidones from Botryosphaeria rhodina, an endophyte of the medicinal plant Bidens pilosa. Phytochemistry 71, 110–116. doi: 10.1016/j.phytochem.2009.09.024
Alavi, P., Starcher, M. R., Zachow, C., Müller, H., and Berg, G. (2013). Root-microbe systems: the effect and mode of interaction of stress protecting agent (SPA) Stenotrophomonas rhizophila DSM14405(T.). Front. Plant Sci. 4:141. doi: 10.3389/fpls.2013.00141
Amna, T., Puri, S. C., Verma, V., Sharma, J. P., Khajuria, R. K., Musarrat, J., et al. (2006). Bioreactor studies on the endophytic fungus Entrophospora infrequens for the production of an anticancer alkaloid camptothecin. Can. J. Microbiol. 52, 189–196. doi: 10.1139/W05-122
Bafana, A. (2013). Diversity and metabolic potential of culturable root-associated bacteria from Origanum vulgare in sub-Himalayan region. World J. Microbiol. Biotechnol. 29, 63–74. doi: 10.1007/s11274-012-1158-3
Bais, H. P., Weir, T. L., Perry, L. G., Gilroy, S., and Vivanco, J. M. (2006). The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 57, 233–266. doi: 10.1146/annurev.arplant.57.032905.105159
Barnard, R. L., Osborne, C. A., and Firestone, M. K. (2013). Responses of soil bacterial and fungal communities to extreme desiccation and rewetting. ISME J. 7, 2229–2241. doi: 10.1038/ismej.2013.104
Berendsen, R. L., Pieterse, C. M., and Bakker, P. A. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Berg, G., Eberl, L., and Hartmann, A. (2005a). The rhizosphere as a reservoir for opportunistic human pathogenic bacteria. Environ. Microbiol. 7, 1673–1685. doi: 10.1111/j.1462-2920.2005.00891.x
Berg, G., Krechel, A., Ditz, M., Sikora, R. A., Ulrich, A., and Hallmann, J. (2005b). Endophytic and ectophytic potato-associated bacterial communities differ in structure and antagonistic function against plant pathogenic fungi. FEMS Microbiol. Ecol. 51, 215–229. doi: 10.1016/j.femsec.2004.08.006
Berg, G. (2009). Plant-microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture. Appl. Microbiol. Biotechnol. 84, 11–18. doi: 10.1007/s00253-009-2092-7
Berg, G., and Smalla, K. (2009). Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 68, 1–13. doi: 10.1111/j.1574-6941.2009.00654.x
Berg, G., Zachow, C., Cardinale, M., and Müller, H. (2011). "Ecology and human pathogenicity of plant-associated bacteria," in Regulation of Biological Control Agents, ed. R. U. Ehlers (Dordrecht: Springer), 175–189. doi: 10.1007/978-90-481-3664-3_8
Berg, G., Zachow, C., Müller, H., Philipps, J., and Tilcher, R. (2013). Next-generation bio-products sowing the seeds of success for sustainable agriculture. Agronomy 3, 648–656. doi: 10.3390/agronomy3040648
Bhattacharjee, R. B., Singh, A., and Mukhopadhyay, S. N. (2008). Use of nitrogen-fixing bacteria as biofertiliser for non-legumes: prospects and challenges. Appl. Microbiol. Biotechnol. 80, 199–209. doi: 10.1007/s00253-008-1567-2
Bitas, V., Kim, H. S., Bennett, J. W., and Kang, S. (2013). Sniffing on microbes: diverse roles of microbial volatile organic compounds in plant health. Mol. Plant Microbe Interact. 26, 835–843. doi: 10.1094/MPMI-10-12-0249-CR
Bragina, A., Berg, C., Cardinale, M., Shcherbakov, A., Chebotar, V., and Berg, G. (2012). Sphagnum mosses harbour highly specific bacterial diversity during their whole lifecycle. ISME J. 6, 802–813. doi: 10.1038/ismej.2011.151
Bulgarelli, D., Rott, M., Schlaeppi, K., Ver Loren van Themaat, E., Ahmadinejad, N., Assenza, F., et al. (2012). Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488, 91–95. doi: 10.1038/nature11336
Castillo, U., Strobel, G. A., Ford, E. J., Hess, W. M., Porter, H., Jensen, J. B., et al. (2002). Munumbicins, wide-spectrum antibiotics produced by Streptomyces NRRL 30562, endophytic on Kennedia nigriscans. Microbiology 148, 2675–2685.
Castillo, U., Harper, J. K., Strobel, G. A., Sears, J., Alesi, K., Ford, E., et al. (2003). Kakadumycins, novel antibiotics from Streptomyces sp. NRRL 30566, an endophyte of Grevillea pteridifolia. FEMS Microbiol. Lett. 224, 183–190. doi: 10.1016/S0378-1097(03)00426-9
Chandra, S. (2012). Endophytic fungi: novel sources of anticancer lead molecules. Appl. Microbiol. Biotechnol. 95, 47–59. doi: 10.1007/s00253-012-4128-7
Compant, S., Clément, C., and Sessitsch, A. (2010). Plant growth-promoting bacteria in the rhizo- and endosphere of plants: their role, colonization, mechanisms involved and prospects for utilization. Soil Biol. Biochem. 42, 669–678. doi: 10.1016/j.soilbio.2009.11.024
Costa, R., Gomes, N. C., Peixoto, R. S., Rumjanek, N., Berg, G., Mendonça-Hagler, L. C., et al. (2006). Diversity and antagonistic potential of Pseudomonas spp. associated to the rhizosphere of maize grown in a subtropical organic farm. Soil Biol. Biochem. 38, 2434–2447. doi: 10.1016/j.soilbio.2006.03.003
Debbab, A., Aly, A. H., Edrada-Ebel, R. A., Müller, W. E., Mosaddak, M., Hakiki, A., et al. (2009). Bioactive secondary metabolites from the endophytic fungus Chaetomium sp. isolated from Salvia officinalis growing in Morocco. Biotechnol. Agron. Soc. Environ. 13, 229–234.
Doornbos, R. F., van Loon, L. C., and Bakker, P. A. (2012). Impact of root exudates and plant defense signaling on bacterial communities in the rhizosphere. Agron. Sustain. Dev. 32, 227–243. doi: 10.1007/s13593-011-0028-y
Effmert, U., Kalderás, J., Warnke, R., and Piechulla, B. (2012). Volatile mediated interactions between bacteria and fungi in the soil. J. Chem. Ecol. 38, 665–703. doi: 10.1007/s10886-012-0135-5
Ezra, D., Castillo, U. F., Strobel, G. A., Hess, W. M., Porter, H., Jensen, J. B., et al. (2004). Coronamycins, peptide antibiotics produced by a verticillate Streptomyces sp. (MSU-2110) endophytic on Monstera sp. Microbiology 150, 785–793. doi: 10.1099/mic.0.26645-0
Fürnkranz, M., Lukesch, B., Müller, H., Huss, H., Grube, M., and Berg, G. (2012). Microbial diversity inside pumpkins: microhabitat-specific communities display a high antagonistic potential against phytopathogens. Microb. Ecol. 63, 418–428. doi: 10.1007/s00248-011-9942-4
Gaby, J. C., and Buckley, D. H. (2012). A comprehensive evaluation of PCR primers to amplify the nifH gene of nitrogenase. PLoS ONE 7:e42149. doi: 10.1371/journal.pone.0042149
Goodfellow, M., and Fiedler, H. P. (2010). A guide to successful bioprospecting: informed by actinobacterial systematics. Antonie Van Leeuwenhoek 98, 119–142. doi: 10.1007/s10482-010-9460-2
Gunatilaka, A. A. (2006). Natural products from plant-associated microorganisms: distribution, structural diversity, bioactivity, and implications of their occurrence. J. Nat. Prod. 69, 509–526. doi: 10.1021/np058128n
Haas, D., and Défago, G. (2005). Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 3, 307–319. doi: 10.1038/nrmicro1129
Hardoim, P. R., Hardoim, C. C., van Overbeek, L. S., and van Elsas, J. D. (2012). Dynamics of seed-borne rice endophytes on early plant growth stages. PLoS ONE 7:e30438. doi: 10.1371/journal.pone.0030438
Harper, J. K., Arif, A. M., Ford, E. J., Strobel, G. A., Porco, J. A., Tomer, D. P., et al. (2003). Pestacin: a 1,3-dihydro isobenzofuran from Pestalotiopsis microspora possessing antioxidant and antimycotic activities. Tetrahedron 59, 2471–2476. doi: 10.1016/S0040-4020(03)00255-2
Hartmann, A., Rothballer, M., and Schmid, M. (2008). Lorenz Hiltner, a pioneer in rhizosphere microbial ecology and soil bacteriology research. Plant Soil 312, 7–14. doi: 10.1007/s11104-007-9514-z
Hirsch, P. R., and Mauchline, T. H. (2012). Who’s who in the plant root microbiome? Nat. Biotechnol. 30, 961–962. doi: 10.1038/nbt.2387
Immaculate, N. R., Kumar, M., Srimathi, S., Muthumary, J., and Kalaichelvan, P. T. (2011). Isolation of Phoma species from Aloe vera: an endophyte and screening the fungus for taxol production. World J. Sci. Technol. 1, 23–31.
Insam, H., and Seewald, M. S. (2010). Volatile organic compounds (VOCs) in soils. Biol. Fertil. Soils 46, 199–213. doi: 10.1007/s00374-010-0442-3
Jacobo-Salcedo Mdel, R., Alonso-Castro, A. J., Salazar-Olivo, L. A., Carranza-Alvarez, C., González-Espíndola, L. A., Domínguez, F., et al. (2011). Antimicrobial and cytotoxic effects of Mexican medicinal plants. Nat. Prod. Commun. 6, 1925–1928.
Janssen, P. H. (2006). Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 72, 1719–1728. doi: 10.1128/AEM.72.3.1719-1728.2006
Kai, M., Effmert, U., Berg, G., and Piechulla, B. (2007). Volatiles of bacterial antagonists inhibit mycelial growth of the plant pathogen Rhizoctonia solani. Arch. Microbiol. 187, 351–360. doi: 10.1007/s00203-006-0199-0
Kamilova, F., Kravchenko, L. V., Shaposhnikov, A. I., Azarova, T., Makarova, N., and Lugtenberg, B. (2006). Organic acids, sugars, and L-tryptophane in exudates of vegetables growing on stonewool and their effects on activities of rhizosphere bacteria. Mol. Plant Microbe Interact. 19, 250–256. doi: 10.1094/MPMI-19-0250
Kato, A., Minoshima, Y., Yamamoto, J., Adachi, I., Watson, A. A., and Nash, R. J. (2008). Protective effects of dietary chamomile tea on diabetic complications. J. Agric. Food Chem. 56, 8206–8211. doi: 10.1021/jf8014365
Khare, E., and Arora, N. K. (2010). Effect of indole-3-acetic acid (IAA) produced by Pseudomonas aeruginosa in suppression of charcoal rot disease of chickpea. Curr. Microbiol. 61, 64–68. doi: 10.1007/s00284-009-9577-6
Kim, S. U., Strobel, G. A., and Ford, E. (1999). Screening of taxol-producing endophytic fungi from Ginkgo biloba and Taxus cuspidata in Korea. Agric. Chem. Biotechnol. 42, 97–99. doi: 10.1042/BA20080110
Köberl, M., Müller, H., Ramadan, E. M., and Berg, G. (2011). Desert farming benefits from microbial potential in arid soils and promotes diversity and plant health. PLoS ONE 6:e24452. doi: 10.1371/journal.pone.0024452
Köberl, M. (2013). The Microbiome of Medicinal Plants and its Potential for Biocontrol and Promotion of Plant Growth and Quality. Doctoral thesis, Graz University of Technology, Graz.
Köberl, M., Ramadan, E. M., Adam, M., Cardinale, M., Hallmann, J., Heuer, H., et al. (2013). Bacillus and Streptomyces were selected as broad-spectrum antagonists against soilborne pathogens from arid areas in Egypt. FEMS Microbiol. Lett. 342, 168–178. doi: 10.1111/1574-6968.12089
Kusari, S., Lamshöft, M., and Spiteller, M. (2009a). Aspergillus fumigatus Fresenius, an endophytic fungus from Juniperus communis L. Horstmann as a novel source of the anticancer pro-drug deoxypodophyllotoxin. J. Appl. Microbiol. 107, 1019–1030. doi: 10.1111/j.1365-2672.2009.04285.x
Kusari, S., Zühlke, S., and Spiteller, M. (2009b). An endophytic fungus from Camptotheca acuminata that produces camptothecin and analogues. J. Nat. Prod. 72, 2–7. doi: 10.1021/np800455b
Lee, J. C., Lobkovsky, E., Pliam, N. B., Strobel, G., and Clardy, J. (1995). Subglutinols A and B: immunosuppressive compounds from the endophytic fungus Fusarium subglutinans. J. Org. Chem. 60, 7076–7077. doi: 10.1021/jo00127a001
Li, J., Zhao, G. Z., Chen, H. H., Wang, H. B., Qin, S., Zhu, W. Y., et al. (2008). Antitumour and antimicrobial activities of endophytic streptomycetes from pharmaceutical plants in rainforest. Lett. Appl. Microbiol. 47, 574–580. doi: 10.1111/j.1472-765X.2008.02470.x
Li, R. W., Myers, S. P., Leach, D. N., Lin, G. D., and Leach, G. (2003). A cross-cultural study: anti-inflammatory activity of Australian and Chinese plants. J. Ethnopharmacol. 85, 25–32. doi: 10.1016/S0378-8741(02)00336-7
Lottmann, J., and Berg, G. (2001). Phenotypic and genotypic characterization of antagonistic bacteria associated with roots of transgenic and non-transgenic potato plants. Microbiol. Res. 156, 75–82. doi: 10.1078/0944-5013-00086
Lugtenberg, B., Chin-A-Woeng, T. F., and Bloemberg, G. V. (2002). Microbe-plant interactions: principles and mechanisms. Antonie Van Leeuwenhoek 81, 373–383. doi: 10.1023/A:1020596903142
Lugtenberg, B., and Kamilova, F. (2009). Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 63, 541–556. doi: 10.1146/annurev.micro.62.081307.162918
Mendes, R., Kruijt, M., de Bruijn, I., Dekkers, E., van der Voort, M., Schneider, J. H., et al. (2011). Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332, 1097–1100. doi: 10.1126/science.1203980
Mendes, R., Garbeva, P., and Raaijmakers, J. M. (2013). The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 37, 634–663. doi: 10.1111/1574-6976.12028
Miller, K. I., Qing, C., Sze, D. M., and Neilan, B. A. (2012a). Investigation of the biosynthetic potential of endophytes in traditional Chinese anticancer herbs. PLoS ONE 7:e35953. doi: 10.1371/journal.pone.0035953
Miller, K. I., Qing, C., Sze, D. M., Roufogalis, B. D., and Neilan, B. A. (2012b). Culturable endophytes of medicinal plants and the genetic basis for their bioactivity. Microb. Ecol. 64, 431–449. doi: 10.1007/s00248-012-0044-8
Mohana Kumara, P., Zuehlke, S., Priti, V., Ramesha, B. T., Shweta, S., Ravikanth, G., et al. (2012). Fusarium proliferatum, an endophytic fungus from Dysoxylum binectariferum Hook.f, produces rohitukine, a chromane alkaloid possessing anti-cancer activity. Antonie Van Leeuwenhoek 101, 323–329. doi: 10.1007/s10482-011-9638-2
Mousa, W. K., and Raizada, M. N. (2013). The diversity of anti-microbial secondary metabolites produced by fungal endophytes: an interdisciplinary perspective. Front. Microbiol. 4:65. doi: 10.3389/fmicb.2013.00065
Nachtigall, J., Kulik, A., Helaly, S., Bull, A. T., Goodfellow, M., Asenjo, J. A., et al. (2011). Atacamycins A-C, 22-membered antitumor macrolactones produced by Streptomyces sp. C38. J. Antibiot. (Tokyo) 264, 775–780. doi: 10.1038/ja.2011.96
Newman, D. J., and Cragg, G. M. (2012). Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Nat. Prod. 75, 311–335. doi: 10.1021/np200906s
Niraula, N. P., Kim, S. H., Sohng, J. K., and Kim, E. S. (2010). Biotechnological doxorubicin production: pathway and regulation engineering of strains for enhanced production. Appl. Microbiol. Biotechnol. 87, 1187–1194. doi: 10.1007/s00253-010-2675-3
Orr, C. H., James, A., Leifert, C., Cooper, J. M., and Cummings, S. P. (2011). Diversity and activity of free-living nitrogen-fixing bacteria and total bacteria in organic and conventionally managed soils. Appl. Environ. Microbiol. 77, 911–919. doi: 10.1128/AEM.01250-10
Palombo, E. A., and Semple, S. J. (2001). Antibacterial activity of traditional Australian medicinal plants. J. Ethnopharmacol. 77, 151–157. doi: 10.1016/S0378-8741(01)00290-2
Puri, S. C., Verma, V., Amna, T., Qazi, G. N., and Spiteller, M. (2005). An endophytic fungus from Nothapodytes foetida that produces camptothecin. J. Nat. Prod. 68, 1717–1719. doi: 10.1021/np0502802
Qi, X., Wang, E., Xing, M., Zhao, W., and Chen, X. (2012). Rhizosphere and non-rhizosphere bacterial community composition of the wild medicinal plant Rumex patientia. World J. Microbiol. Biotechnol. 28, 2257–2265. doi: 10.1007/s11274-012-1033-2
Raaijmakers, J. M., and Mazzola, M. (2012). Diversity and natural functions of antibiotics produced by beneficial and plant pathogenic bacteria. Annu. Rev. Phytopathol. 50, 403–424. doi: 10.1146/annurev-phyto-081211-172908
Ryu, C. M., Farag, M. A., Hu, C. H., Reddy, M. S., Wei, H. X., Paré, P. W., et al. (2003). Bacterial volatiles promote growth in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 100, 4927–4932. doi: 10.1073/pnas.0730845100
Schmidt, R., Köberl, M., Mostafa, A., Ramadan, E. M., Monschein, M., Jensen, K. B., et al. (2013). Effects of bacterial inoculants on the indigenous microbiome and secondary metabolites of chamomile plants. Front. Microbiol. (in press).
Singh, G., and Mukerji, K. G. (2006). “Root exudates as determinant of rhizospheric microbial biodiversity,” in Microbial Activity in the Rhizosphere, eds K. G. Mukerji, C. Manoharachary, and J. Singh (Berlin/Heidelberg: Springer), 39–53.
Smalla, K., Wieland, G., Buchner, A., Zock, A., Parzy, J., Kaiser, S., et al. (2001). Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed. Appl. Envrion. Microbiol. 67, 4742–4751. doi: 10.1128/AEM.67.10.4742-4751.2001 PMCid:PMC93227
Srivastava, J. K., and Gupta, S. (2009). Extraction, characterization, stability and biological activity of flavonoids isolated from chamomile flowers. Mol. Cell. Pharmacol. 1, 138. doi: 10.4255/mcpharmacol.09.18
Stierle, A., Strobel, G., and Stierle, D. (1993). Taxol and taxane production by Taxomyces andreanae, an endophytic fungus of Pacific yew. Science 260, 214–216. doi: 10.1126/science.8097061
Strobel, G., Li, J. Y., Sugawara, F., Koshino, H., Harper, J., and Hess, W. M. (1999a). Oocydin A, a chlorinated macrocyclic lactone with potent anti-oomycete activity from Serratia marcescens. Microbiology 145, 3557–3564.
Strobel, G., Miller, R. V., Martinez-Miller, C., Condron, M. M., Teplow, D. B., and Hess, W. M. (1999b). Cryptocandin, a potent antimycotic from the endophytic fungus Cryptosporiopsis cf. quercina. Microbiology 145, 1919–1926. doi: 10.1099/13500872-145-8-1919
Strobel, G., Ford, E., Worapong, J., Harper, J. K., Arif, A. M., Grant, D. M., et al. (2002). Isopestacin, an isobenzofuranone from Pestalotiopsis microspora, possessing antifungal and antioxidant activities. Phytochemistry 60, 179–183. doi: 10.1016/S0031-9422(02)00062-6
Strobel, G., and Daisy, B. (2003). Bioprospecting for microbial endophytes and their natural products. Microbiol. Mol. Biol. Rev. 67, 491–502. doi: 10.1128/MMBR.67.4.491-502.2003
Strobel, G., Daisy, B., Castillo, U., and Harper, J. (2004). Natural products from endophytic microorganisms. J. Nat. Prod. 67, 257–268. doi: 10.1021/np030397v
van Loon, L. C., Bakker, P. A., and Pieterse, C. M. (1998). Systemic resistance induced by rhizosphere bacteria. Annu. Rev. Phytopathol. 36, 453–483. doi: 10.1146/annurev.phyto.36.1.453
Wang, J. W., Zhang, Z., and Tan, R. X. (2001). Stimulation of artemisinin production in Artemisia annua hairy roots by the elicitor from the endophytic Colletotrichum sp. Biotechnol. Lett. 23, 857–860. doi: 10.1023/A:1010535001943
Wani, M. C., Taylor, H. L., Wall, M. E., Coggon, P., and McPhail, A. T. (1971). Plant antitumor agents. VI. The isolation and structure of taxol, a novel antileukemic and antitumor agent from Taxus brevifolia. J. Am. Chem. Soc. 93, 2325–2327. doi: 10.1021/ja00738a045
Weber, D., Sterner, O., Anke, T., Gorzalczancy, S., Martino, V., and Acevedo, C. (2004). Phomol, a new antiinflammatory metabolite from an endophyte of the medicinal plant Erythrina crista-galli. J. Antibiot. (Tokyo) 57, 559–563. doi: 10.7164/antibiotics.57.559
Weller, D. M. (2007). Pseudomonas biocontrol agents of soilborne pathogens: looking back over 30 years. Phytopathology 97, 250–256. doi: 10.1094/PHYTO-97-2-0250
Wings, S., Müller, H., Berg, G., Lamshöft, M., and Leistner, E. (2013). A study of the bacterial community in the root system of the maytansine containing plant Putterlickia verrucosa. Phytochemistry 91, 158–164. doi: 10.1016/j.phytochem.2012.06.016
Wolf, A., Fritze, A., Hagemann, M., and Berg, G. (2002). Stenotrophomonas rhizophila sp. nov., a novel plant-associated bacterium with antifungal properties. Int. J. Syst. Evol. Microbiol. 52, 1937–1944. doi: 10.1099/ijs.0.02135-0
Yang, X., Guo, S., Zhang, L., and Shao, H. (2003). Selection of producing podophyllotoxin endophytic fungi from podophyllin plant. Nat. Prod. Res. Dev. 15, 419–422.
Zachow, C., Tilcher, R., and Berg, G. (2008). Sugar beet-associated bacterial and fungal communities show a high indigenous antagonistic potential against plant pathogens. Microb. Ecol. 55, 119–129. doi: 10.1007/s00248-007- 9257-7
Zachow, C., Fatehi, J., Cardinale, M., Tilcher, R., and Berg, G. (2010). Strain-specific colonization pattern of Rhizoctonia antagonists in the root system of sugar beet. FEMS Microbiol. Ecol. 74, 124–135. doi: 10.1111/j.1574-6941.2010. 00930.x
Zehr, J. P., Jenkins, B. D., Short, S. M., and Steward, G. F. (2003). Nitrogenase gene diversity and microbial community structure: a cross-system comparison. Environ. Microbiol. 5, 539–554. doi: 10.1046/j.1462-2920.2003.00451.x
Zhao, K., Penttinen, P., Guan, T., Xiao, J., Chen, Q., Xu, J., et al. (2011). The diversity and anti-microbial activity of endophytic actinomycetes isolated from medicinal plants in Panxi plateau, China. Curr. Microbiol. 62, 182–190. doi: 10.1007/s00284-010-9685-3
Zhao, K., Penttinen, P., Chen, Q., Guan, T., Lindström, K., Ao, X., et al. (2012). The rhizospheres of traditional medicinal plants in Panxi, China, host a diverse selection of actinobacteria with antimicrobial properties. Appl. Microbiol. Biotechnol. 94, 1321–1335. doi: 10.1007/s00253-011-3862-6
Zou, W. X., Meng, J. C., Lu, H., Chen, G. X., Shi, G. X., Zhang, T. Y., et al. (2000). Metabolites of Colletotrichum gloeosporioides, an endophytic fungus in Artemisia mongolica. J. Nat. Prod. 63, 1529–1530. doi: 10.1021/np000204t
Keywords: biocontrol, desert farming, medicinal plants, microbial communities, organic agriculture, soil-borne pathogens
Citation: Köberl M, Schmidt R, Ramadan EM, Bauer R and Berg G (2013) The microbiome of medicinal plants: diversity and importance for plant growth, quality, and health. Front. Microbiol. 4:400. doi: 10.3389/fmicb.2013.00400
Received: 30 September 2013; Paper pending published: 04 November 2013;
Accepted: 05 December 2013; Published online: 20 December 2013.
Edited by:
Martin Grube, Karl-Franzens-University Graz, AustriaReviewed by:
Franz Narberhaus, Ruhr University Bochum, GermanyCopyright © 2013 Köberl, Schmidt, Ramadan, Bauer and Berg. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Martina Köberl, Institute for Environmental Biotechnology, Graz University of Technology, Petersgasse 12/I, 8010 Graz, Austria e-mail:bWFydGluYS5rb2ViZXJsQHR1Z3Jhei5hdA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.