
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
OPINION article
Front. Microbiol. , 10 July 2013
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 4 - 2013 | https://doi.org/10.3389/fmicb.2013.00192
This article is part of the Research Topic Low-dose antibiotics: current status and outlook for the future View all 20 articles
In a recent publication by Leski et al. (2013), the authors reported the occurrence of multidrug-resistant tet(X)-containing bacterial strains in a hospital in Sierra Leone. Among 52 clinical isolates, 11 (21%) have been confirmed tet(X)-positive. All the positive strains have been isolated from urinary tract infections and identified as Enterobacter cloacae, Comamonas testosteroni, Escherichia coli, Klebsiella pneumoniae, Delftia acidovorans, Enterobacter sp., and other members of Enterobacteriaceae and Pseudomonadaceae (Leski et al., 2013).
The need for careful monitoring of tet(X) dissemination is dictated by the fact that the enzyme encoded by the gene, a flavin-dependent monooxygenase, is capable of degrading almost all tetracyclines, including the third-generation tetracycline, tigecycline (the minocycline derivative 9-tert-butyl-glycylamido-minocycline) (Yang et al., 2004; Moore et al., 2005). The US FDA approved tigecycline in 2005, and its use in the EU was authorized in 2006. Its use is approved for complicated skin and intra-abdominal infections as well as community-acquired pneumonia (http://www.accessdata.fda.gov/drugsatfda_docs/label/2010/021821s021lbl.pdf). The antibiotic is very efficient in treatment of a number of infections, including those resistant to the first- and second-generation tetracyclines (Bertrand and Dowzicky, 2012). Despite being considered as a drug of last resort, its use is steadily increasing, at least in the US (Huttner et al., 2012).
Although tigecycline resistance has not been tested at the time of isolation (Leski et al., 2013), the high frequency of tet(X) encountered in clinical samples signifies a worrying trend. In the previous analysis of the occurrence and phylogeny of the tet(X) genes it has been established that these genes can be detected in environmental DNAs and isolates as well as commensal bacteria (Aminov, 2009). Further studies have not spotted any expansion beyond these ecological niches. The presence of tet(X) has been detected in the human gut bacteria (de Vries et al., 2011), intestinal Bacteroides strains (Bartha et al., 2011), sewage treatment plants (Zhang and Zhang, 2011), and an oxytetracycline production wastewater treatment system (Liu et al., 2012). But now tet(X) is detected in a variety of clinical isolates and accepted human pathogens (Leski et al., 2013). The tet(X) sequences from this study have been added to the previous dataset (Aminov, 2009), and the phylogenetic tree has been recomputed (Figure 1). It is not surprising to see a tight clustering, with a 100% bootstrap support, of the tet(X) sequences from Enterobacteriaceae bacterium SL1 and Delftia sp. SL20 with the known tet(X) genes, given the high similarity of sequences within the cluster that exceeds 99%.
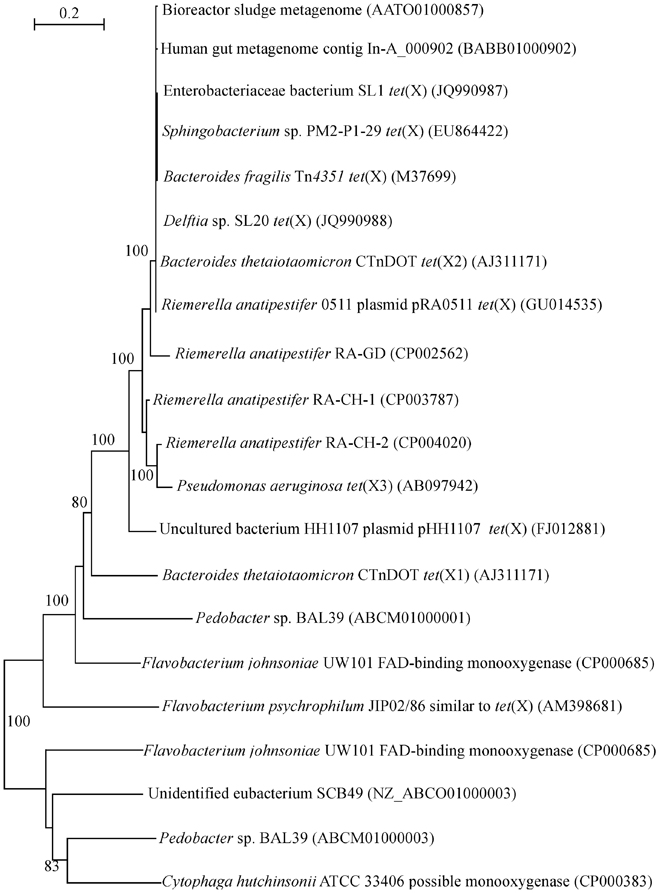
Figure 1. Neighbor-joining tree of the tet(X) and flavin monooxygenase-encoding genes. Numbers above each node show the percentage of tree configurations that occurred during 1000 bootstrap trials. The scale bar is in fixed nucleotide substitutions per sequence position. GenBank accession numbers of nucleotide sequences used in this analysis are given in parenthesis.
It is important to note here that there is no access to tigecycline (Tygacil®, Pfizer Inc.) in the hospital where tet(X)-positive samples were collected nor it is available through the independent pharmacies and hospital dispensaries operating in the area (Leski et al., 2013). Still, 87% of pharmacies dispense the “older” tetracyclines without prescription. As the authors suggest, this selective pressure of continuous application of tetracyclines may serve to maintain and spread tet(X) and other tetracycline resistance genes into pathogenic microbiota. Also, the probability of co-selection cannot be ruled out. The authors indicated the presence of mobile genetic elements in some isolates, and 10 out of 11 isolates appeared to be harboring multidrug resistance determinants.
In animal production systems, the penetration of tet(X) into the pathogens happened earlier. This can be demonstrated with the example of Riemerella anatipestifer, a causative agent of septicaemia anserum exsudativa (Segers et al., 1993). Septicaemia leads to major economic losses in duck production (Ryll et al., 2001; Sarver et al., 2005) but it also affects other bird species (Sandhu and Rimler, 1997; Hess et al., 2013). The R. anatipestifer strain, resistant to ampicillin, chloramphenicol, gentamicin, amikacin, tetracycline, nalidixic acid, and trimethoprim/sulfamethoxazole, was isolated in 2005 from waterfowl in Taiwan (Chen et al., 2010). It carries pRA0511 plasmid, which, in addition to two chloramphenicol acetyltransferases and a multi-drug ABC transporter permease/ATPase, also encodes TetX. The gene sequence has been incorporated into the existing dataset (Aminov, 2009) and recomputed (Figure 1). Similar to the genes from human pathogens, the gene from the poultry pathogen is confidently grouped into the tet(X) cluster. Three genomic sequences of R. anatipestifer, published (Yuan et al., 2011) or available as database entries (GenBank accession numbers CP003787 and CP004020), also carry chromosomally encoded genes similar to tet(X) (Figure 1). Interestingly, four other strains of R. anatipestifer, for which genome sequences are available (Mavromatis et al., 2011; Zhou et al., 2011; Wang et al., 2012; Yuan et al., 2013), have not yet acquired tet(X). No information regarding antibiotic use practices at sampling sites where R. anatipestifer strains have been isolated is available in the cited publications.
It seems that the use of even ‘older’ antibiotics may contribute to the resistance to newer antibiotics. There is no access to the third-generation tetracycline, tigecycline (Tygacil®, Pfizer Inc.), in the areas sampled in Sierra Leone (Leski et al., 2013). It is also highly unlikely that this expensive new antibiotic is used in duck production, most likely these are the first-generation tetracyclines. Thus the conclusion is that the selective pressure by older antibiotics drives the resistance to a newer antibiotic and contributes to the dissemination of this resistance to pathogens.
The flavoprotein monooxygenase group of enzymes is found in many metabolic pathways involved in the region-specific hydroxylation of organic substrates in all three domains of life (Harayama et al., 1992). Based on sequence similarity and 3D structural data, the enzymes are divided into six classes (van Berkel et al., 2006). Class A enzymes, to which TetX belongs, are generally involved in the degradation of phenolic compounds by ortho- or para-hydroxylation of the aromatic ring (Moonen et al., 2002).
Bacteria that carry these genes are omnipresent and can be encountered in a variety of ecosystems, including soil, aquatic ecosystems, and intestinal tract; some are opportunistic pathogens. Accordingly, the range of biochemical reactions performed by this class of enzymes is quite broad, and they may play an important role in the global carbon and nitrogen cycles (Chen et al., 2011; Wang and Shao, 2012). Interestingly, the range of metabolic activities expressed by these enzymes also includes the modification of many antibiotics. Besides the tetracylines discussed here, this range is extended to such structurally different antibiotics as rifampin (Andersen et al., 1997), mithramycin (Prado et al., 1999), griseorhodin (Li and Piel, 2002), chromomycin (Menendez et al., 2004), and auricin (Novakova et al., 2005).
The genetic context of flavin monooxygenase genes has been discussed earlier (Aminov, 2009). In brief, the majority of the genes analysed is almost uniformly associated with mobile genetic elements, including the plasmid-encoded tet(X) discussed here (Chen et al., 2010). The genes in this class are also highly incongruent with taxonomic positioning suggesting horizontal gene transfer events. They are also subject to frequent duplication events, which are partially illustrated here with the paralogous genes from Flavobacterium johnsoniae UW101 and Pedobacter sp. BAL39 (Figure 1).
The case of flavin monooxygenases is a vivid example demonstrating enormous adaptability of bacteria: they can freely move their protective armours amongst a variety of ecological compartments in response to yet another challenge, this time inflicted by humans in the form of antibiotic selective pressure. The global microbiota has been dealing with environmental challenges for billions of years to become sophisticated genetic engineers moving genes around with ease (Aminov, 2011). Combined with the readily available massive metabolic resources of the environmental metagenome, the microbiota seem capable of countering any kind of environmental or anthropogenic assault.
We are living in a fascinating era with technological advancements that allow us to see almost instantaneously the evolutionary events leading to the emergence of novel pathogens armed with resistance mechanisms against the most advanced antibiotics that we have been able to design. We should not underestimate the enormous genetic flexibility and the vast metabolic capabilities of the environmental microbiota. Based on our technical capabilities and knowledge acquired during the antibiotic era (Aminov, 2010), we have to make every effort, at every level possible, to preserve the power of antibiotics. Taking a bystander position in this situation is not acceptable.
Aminov, R. I. (2009). The role of antibiotics and antibiotic resistance in nature. Environ. Microbiol. 11, 2970–2988. doi: 10.1111/j.1462-2920.2009.01972.x
Aminov, R. I. (2010). A brief history of the antibiotic era: lessons learned and challenges for the future. Front. Microbiol. 1:134. doi: 10.3389/fmicb.2010.00134
Aminov, R. I. (2011). Horizontal gene exchange in environmental microbiota. Front. Microbiol. 2:158. doi: 10.3389/fmicb.2011.00158
Andersen, S. J., Quan, S., Gowan, B., and Dabbs, E. R. (1997). Monooxygenase-like sequence of a Rhodococcus equi gene conferring increased resistance to rifampin by inactivating this antibiotic. Antimicrob. Agents Chemother. 41, 218–221.
Bartha, N. A., Sóki, J., Urbán, E., and Nagy, E. (2011). Investigation of the prevalence of tetQ, tetX and tetX1 genes in Bacteroides strains with elevated tigecycline minimum inhibitory concentrations. Int. J. Antimicrob. Agents 38, 522–525. doi: 10.1016/j.ijantimicag.2011.07.010
Bertrand, X., and Dowzicky, M. J. (2012). Antimicrobial susceptibility among gram-negative isolates collected from intensive care units in North America, Europe, the Asia-Pacific Rim, Latin America, the Middle East, and Africa between 2004 and 2009 as part of the Tigecycline Evaluation and Surveillance Trial. Clin. Ther. 34, 124–137. doi: 10.1016/j.clinthera.2011.11.023
Chen, Y., Patel, N. A., Crombie, A., Scrivens, J. H., and Murrell, J. C. (2011). Bacterial flavin-containing monooxygenase is trimethylamine monooxygenase. Proc. Natl. Acad. Sci. U.S.A. 108, 17791–17796. doi: 10.1073/pnas.1112928108
Chen, Y. P., Tsao, M. Y., Lee, S. H., Chou, C. H., and Tsai, H. J. (2010). Prevalence and molecular characterization of chloramphenicol resistance in Riemerella anatipestifer isolated from ducks and geese in Taiwan. Avian Pathol. 39, 333–338. doi: 10.1080/03079457.2010.507761
de Vries, L. E., Vallès, Y., Agersø, Y., Vaishampayan, P. A., García-Montaner, A., Kuehl, J. V., et al. (2011). The gut as reservoir of antibiotic resistance: microbial diversity of tetracycline resistance in mother and infant. PLoS ONE 6:e21644. doi: 10.1371/journal.pone.0021644
Harayama, S., Kok, M., and Neidle, E. L. (1992). Functional and evolutionary relationships among diverse oxygenases. Annu. Rev. Microbiol. 46, 565–601. doi: 10.1146/annurev.mi.46.100192.003025
Hess, C., Enichlmayr, H., Jandreski-Cvetkovic, D., Liebhart, D., Bilic, I., and Hess, M. (2013). Riemerella anatipestifer outbreaks in commercial goose flocks and identification of isolates by MALDI-TOF mass spectrometry. Avian Pathol. 42, 151–156. doi: 10.1080/03079457.2013.775401
Huttner, B., Jones, M., Rubin, M. A., Neuhauser, M. M., Gundlapalli, A., and Samore, M. (2012). Drugs of last resort? The use of polymyxins and tigecycline at US Veterans Affairs medical centers, 2005–2010. PLoS ONE 7:e36649. doi: 10.1371/journal.pone.0036649
Leski, T. A., Bangura, U., Jimmy, D. H., Ansumana, R., Lizewski, S. E., Stenger, D. A., et al. (2013). Multidrug-resistant tet(X)-containing hospital isolates in Sierra Leone. Int. J. Antimicrob. Agents 42, 83–86. doi: 10.1016/j.ijantimicag.2013.04.014
Li, A., and Piel, J. (2002). A gene cluster from a marine Streptomyces encoding the biosynthesis of the aromatic spiroketal polyketide griseorhodin A. Chem. Biol. 9, 1017–10126. doi: 10.1016/S1074-5521(02)00223-5
Liu, M., Zhang, Y., Yang, M., Tian, Z., Ren, L., and Zhang, S. (2012). Abundance and distribution of tetracycline resistance genes and mobile elements in an oxytetracycline production wastewater treatment system. Environ. Sci. Technol. 46, 7551–7557. doi: 10.1021/es301145m
Mavromatis, K., Lu, M., Misra, M., Lapidus, A., Nolan, M., Lucas, S., et al. (2011). Complete genome sequence of Riemerella anatipestifer type strain (ATCC 11845). Stand. Genomic Sci. 4, 144–153. doi: 10.4056/sigs.1553865
Menendez, N., Nur-e-Alam, M., Brana, A. F., Rohr, J., Salas, J. A., and Mendez, C. (2004). Biosynthesis of the antitumor chromomycin A3 in Streptomyces griseus: analysis of the gene cluster and rational design of novel chromomycin analogs. Chem. Biol. 1, 21–32.
Moonen, M. J. H., Fraaije, M. W., Rietjens, I. M. C. M., Laane, C., and van Berkel, W. J. H. (2002). Flavoenzyme-catalyzed oxygenations and oxidations of phenolic compounds. Adv. Synth. Catal. 344, 1–13.
Moore, I. F., Hughes, D. W., and Wright, G. D. (2005). Tigecycline is modified by the flavin-dependent monooxygenase TetX. Biochemistry 44, 11829–111835. doi: 10.1021/bi0506066
Novakova, R., Homerova, D., Feckova, L., and Kormanec, J. (2005). Characterization of a regulatory gene essential for the production of the angucycline-like polyketide antibiotic auricin in Streptomyces aureofaciens CCM 3239. Microbiology 151, 2693–2706. doi: 10.1099/mic.0.28019-0
Prado, L., Fernandez, E., Weissbach, U., Blanco, G., Quiros, L. M., Brana, A. F., et al. (1999). Oxidative cleavage of premithramycin B is one of the last steps in the biosynthesis of the antitumor drug mithramycin. Chem. Biol. 6, 19–30. doi: 10.1016/S1074-5521(99)80017-9
Ryll, M., Christensen, H., Bisgaard, M., Christensen, J. P., Hinz, K. H., and Kohler, B. (2001). Studies on the prevalence of Riemerella anatipestifer in the upper respiratory tract of clinically healthy ducklings and characterization of untypable strains. J. Vet. Med. B Infect. Dis. Vet. Public Health 48, 537–546. doi: 10.1046/j.1439-0450.2001.00471.x
Sandhu, T. S., and Rimler, R. B. (1997). “Riemerella anatipestifer infection,” in Diseases of Poultry, 10th Edn., eds B. W. Calnek, H. J. Barnes, H. J. Beard, L. R. McDougald, and Y. M. Saif (Ames, IA: Iowa State University Press), 161–166.
Sarver, C. F., Morishita, T. Y., and Nersessian, B. (2005). The effect of route of inoculation and challenge dosage on Riemerella anatipestifer infection in Pekin ducks (Anas platyrhynchos). Avian Dis. 49, 104–107. doi: 10.1637/7248-073004R1
Segers, P., Mannheim, W., Vancanneyt, M., De Brandt, K., Hinz, K. H., Kersters, K., et al. (1993). Riemerella anatipestifer gen. nov., comb. nov., the causative agent of septicemia anserum exsudativa, and its phylogenetic affiliation within the Flavobacterium-Cytophaga rRNA homology group. Int. J. Syst. Bacteriol. 43, 768–776. doi: 10.1099/00207713-43-4-768
van Berkel, W. J., Kamerbeek, N. M., and Fraaije, M. W. (2006). Flavoprotein monooxygenases, a diverse class of oxidative biocatalysts. J. Biotechnol. 124, 670–689. doi: 10.1016/j.jbiotec.2006.03.044
Wang, W., and Shao, Z. (2012). Diversity of flavin-binding monooxygenase genes (almA) in marine bacteria capable of degradation long-chain alkanes. FEMS Microbiol. Ecol. 80, 523–533. doi: 10.1111/j.1574-6941.2012.01322.x
Wang, X., Zhu, D., Wang, M., Cheng, A., Jia, R., Zhou, Y., et al. (2012). Complete genome sequence of Riemerella anatipestifer reference strain. J. Bacteriol. 194, 3270–3271. doi: 10.1128/JB.00366-12
Yang, W., Moore, I. F., Koteva, K. P., Bareich, D. C., Hughes, D. W., and Wright, G. D. (2004). TetX is a flavin-dependent monooxygenase conferring resistance to tetracycline antibiotics. J. Biol. Chem. 279, 52346–52352. doi: 10.1074/jbc.M409573200
Yuan, J., Liu, W., Sun, M., Song, S., Cai, J., and Hu, S. (2011). Complete genome sequence of the pathogenic bacterium Riemerella anatipestifer strain RA-GD. J. Bacteriol. 193, 2896–2897. doi: 10.1128/JB.00301-11
Yuan, J., Li, L., Sun, M., Dong, J., and Hu, Q. (2013). Genome sequence of avirulent Riemerella anatipestifer strain RA-SG. Genome Announc. 1:e0021812. doi: 10.1128/genomeA.00218-12
Zhang, X. X., and Zhang, T. (2011). Occurrence, abundance, and diversity of tetracycline resistance genes in 15 sewage treatment plants across China and other global locations. Environ. Sci. Technol. 45, 2598–2604. doi: 10.1021/es103672x
Citation: Aminov RI (2013) Evolution in action: dissemination of tet(X) into pathogenic microbiota. Front. Microbiol. 4:192. doi: 10.3389/fmicb.2013.00192
Received: 22 June 2013; Accepted: 23 June 2013;
Published online: 10 July 2013.
Edited by:
Kunihiko Nishino, Osaka University, JapanCopyright © 2013 Aminov. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence:cnVzdGFtLmFtaW5vdkB1d2ltb25hLmVkdS5qbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.