
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 23 May 2013
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 4 - 2013 | https://doi.org/10.3389/fmicb.2013.00135
This article is part of the Research Topic Emerging antimicrobial resistance mechanisms in animal and zoonotic pathogens View all 21 articles
A commentary has been posted on this article:
Commentary on genetic mechanisms of antimicrobial resistance in bacteria from U.S. food animals: ESBLs are here
The prevalence of antimicrobial resistance (AR) in bacteria isolated from U.S. food animals has increased over the last several decades as have concerns of AR foodborne zoonotic human infections. Resistance mechanisms identified in U.S. animal isolates of Salmonella enterica included resistance to aminoglycosides (e.g., alleles of aacC, aadA, aadB, ant, aphA, and StrAB), β-lactams (e.g., blaCMY−2, TEM−1, PSE−1), chloramphenicol (e.g., floR, cmlA, cat1, cat2), folate pathway inhibitors (e.g., alleles of sul and dfr), and tetracycline [e.g., alleles of tet(A), (B), (C), (D), (G), and tetR]. In the U.S., multi-drug resistance (MDR) mechanisms in Salmonella animal isolates were associated with integrons, or mobile genetic elements (MGEs) such as IncA/C plasmids which can be transferred among bacteria. It is thought that AR Salmonella originates in food animals and is transmitted through food to humans. However, some AR Salmonella isolated from humans in the U.S. have different AR elements than those isolated from food animals, suggesting a different etiology for some AR human infections. The AR mechanisms identified in isolates from outside the U.S. are also predominantly different. For example the extended spectrum β-lactamases (ESBLs) are found in human and animal isolates globally; however, in the U.S., ESBLs thus far have only been found in human and not food animal isolates. Commensal bacteria in animals including Escherichia coli and Enterococcus spp. may be reservoirs for AR mechanisms. Many of the AR genes and MGEs found in E. coli isolated from U.S. animals are similar to those found in Salmonella. Enterococcus spp. isolated from animals frequently carry MGEs with AR genes, including resistances to aminoglycosides (e.g., alleles of aac, ant, and aph), macrolides [e.g., erm(A), erm(B), and msrC], and tetracyclines [e.g., tet(K), (L), (M), (O), (S)]. Continuing investigations are required to help understand and mitigate the impact of AR bacteria on human and animal health.
Antimicrobial compounds have been used to treat bacterial infections since the middle of the twentieth century. These compounds were highly successful in treating various diseases and were widely used in both human and veterinary medicine. However, resistance to these compounds was detected in target pathogens only a few years after initiation of therapeutic use in humans (Alanis, 2005). The selective pressure created by the use of antimicrobials was identified as a driving force behind the emergence of resistance which was genetically encoded, inherited by subsequent progeny of the resistant pathogens, and in some cases could be transferred horizontally even to distantly related bacteria [as reviewed by Linton (1977)]. Because of their efficacy in treating and preventing disease, antimicrobials were used widely in food animal husbandry, and were also found to promote the growth of some animals when fed to the animals at sub-therapeutic levels (Aarestrup and Wegener, 1999; Mathew et al., 2007). Over the past few decades, the number of human infections caused by antimicrobial resistant bacteria increased which can make treatment of these diseases more difficult [reviewed by Alanis (2005)]. Because of the genetic nature of resistance and the ability to select for resistant organisms through the use of antimicrobials, using these compounds in animals was considered a potential source of antimicrobial resistant bacteria which could be transmitted to humans (Aarestrup, 2005; Aarestrup et al., 2008; Shryock and Richwine, 2010).
Research into the impact of antimicrobial use in animals on human health has focused on determining the prevalence of resistance in bacteria isolated from animals and from human infections (Tollefson et al., 1998; Aarestrup, 1999, 2004; Bager et al., 1999a; McEwen and Fedorka-Cray, 2002). These studies were then followed by characterization of the mechanisms leading to resistance and determining if resistance found in human infections were likely to have first occurred in animals followed by transmission to humans. To achieve these goals, antimicrobial resistance (AR) monitoring programs have been initiated globally. In the U.S., the Centers for Disease Control and Prevention (CDC), the Food and Drug Administration (FDA), and the U.S. Department of Agriculture (USDA) established the National Antimicrobial Resistance Monitoring System (NARMS) to monitor changes in antimicrobial susceptibilities of zoonotic pathogens from human and animal clinical specimens, from healthy farm animals, and from raw product of food-producing animals at slaughter and processing (http://www.fda.gov/AnimalVeterinary/SafetyHealth
/AntimicrobialResistance/NationalAntimicrobialResistance
MonitoringSystem/default.htm; Tollefson et al., 1998, 1999). Similar programs were developed in Canada, Mexico, the European Union, and in other locations (Martel et al., 2000; Barton et al., 2003; Aarestrup, 2004; Biedenbach et al., 2006; Hammerum et al., 2007). This information is shared globally to detect the emergence of resistance and the dissemination of antimicrobial resistant foodborne pathogens (http://www.who.int/foodborne_disease/resistance/agisar/en/index.html). Many of these programs also include epidemiological, microbiological, and molecular biology research projects to improve our understanding of the data collected.
In addition to determining the prevalence of resistant bacteria, research associated with the surveillance programs focuses on determining the mechanism leading to resistance. Typically, mechanisms of AR fall into three categories: (1) inactivation of the antimicrobial, (2) efflux or changes in permeability or transport of the antimicrobial, or (3) modification or replacement of the antimicrobial target (McDermott et al., 2003; Walsh, 2003; Boerlin and Reid-Smith, 2008; Foley and Lynne, 2008). Resistances are genetically encoded and can vary from mutations in endogenous genes, to horizontally acquired foreign resistance genes carried by mobile genetic elements (MGEs) like plasmids. Both point mutations and horizontally acquired genes can encode all three categories of resistance. Point mutations in a promoter or operator can result in the overexpression of endogenous genes such as an antimicrobial inactivation enzyme like the AmpC β-lactamase gene, or an efflux system like the mar locus (Van et al., 2000; Siu et al., 2003; Tracz et al., 2005). Point mutations in genes encoding antimicrobial targets can result in a resistant target, such as mutations to the gyrase gene leading to the expression of a fluoroquinolone-resistant gyrase enzyme (Eaves et al., 2004; Hopkins et al., 2005). Exogenous resistance genes encoded on plasmids, integrons, phage, and transposons can be horizontally transmitted by transformation, conjugation, or transduction and these foreign genes can encode all three mechanisms of resistance. This includes genes encoding enzymes that inactivate the antimicrobial, such as β-lactamases that cleave the four membered ring in β-lactams, genes which encode efflux systems like tet(A), genes encoding a modified version of the enzyme that is the target of the antimicrobial, such as dfrA, or genes encoding an enzyme that modifies the antimicrobial target like a ribosomal RNA methylase, such as ermB (Carattoli, 2001, 2009; Boerlin and Reid-Smith, 2008; Ajiboye et al., 2009). Analysis of these resistance mechanisms can then be used to determine the genetic relationship between resistances found in isolates from animals and humans. Because of the diversity of genetic elements that lead to AR, it may be possible to determine if resistances seen in bacterial isolates from human infections are closely related to those found in animal isolates, thus identifying animal sources of resistant bacteria in human infections that can be targeted in order to reduce human disease (Bager et al., 1999b; Aarestrup, 2000b; Boerlin, 2004).
When first detected, AR in bacteria was relatively rare and new antimicrobial compounds were discovered or developed that were not susceptible to the resistance mechanisms that had arose (Alanis, 2005). However, resistance mechanisms to new antimicrobials can develop, or existing ones can emerge due to selective pressure from their use, leading to increasing resistance in human and animal isolates over time (Swaminathan et al., 2006; Frye and Fedorka-Cray, 2007; Gilbert et al., 2007; Frye et al., 2008, 2011). Increasing infections with antimicrobial resistant bacteria is also accompanied by a decrease in efficacy of treatment with these compounds (Alanis, 2005; Stoycheva and Murdjeva, 2006; Walsh and Fanning, 2008; Ajiboye et al., 2009). In addition, infections caused by antimicrobial resistant bacteria have also been shown to result in increased morbidity and mortality in humans and animals (Mundy et al., 2000; Alanis, 2005; Foley and Lynne, 2008; Ajiboye et al., 2009; Gebreyes et al., 2009; Huehn et al., 2010). The consequences of antimicrobial use are also caused by prophylactic treatment of animals to prevent infection or antimicrobials employed as growth promoters. Concerns about selective pressure caused by the utilization of sub-therapeutic growth promoting antimicrobials have led to precautionary restrictions and bans on these applications (DuPont and Steele, 1987; Aarestrup and Seyfarth, 2000; Aarestrup et al., 2001; Anthony et al., 2001; White et al., 2002, 2004; Stokes et al., 2008). In some cases these bans have appeared to result in increased animal illnesses and increased therapeutic use of some antimicrobials in animal husbandry, thus confounding the evaluation of these practices in preventing resistance and protecting human and animal health (DuPont and Steele, 1987; Aarestrup and Seyfarth, 2000; Aarestrup et al., 2001; Anthony et al., 2001; White et al., 2002, 2004; Stokes et al., 2008).
While the proportions of resistant bacteria have fluctuated from year to year, the percentage of antimicrobial resistant bacteria seems to be increasing as well as the fraction of bacteria that are multi-drug resistant (MDR; Devasia et al., 2005; Alcaine et al., 2007; Johnson et al., 2011a). MDR bacteria are troublesome because it is possible for a pathogen to be resistant to all of the antimicrobial compounds that are used to treat the infections it causes (Alanis, 2005). In addition, horizontally transferred mechanisms of resistance are often physically linked on genetic loci such as plasmids, integrons, and transposons (Poole and Crippen, 2009; Douard et al., 2010; Frye et al., 2011; Glenn et al., 2011, 2012; Lindsey et al., 2011a). These genetic elements often encode MDR and are capable of being transferred to sensitive bacteria, rendering a new bacterial host MDR through a single transfer event (Zhao et al., 2003; Poole and Crippen, 2009; Frye et al., 2011; Lindsey et al., 2011a).
Salmonella enterica is a ubiquitous pathogen which can infect many host species and cause different diseases (D'Aoust, 1997; Lavigne and Blanc-Potard, 2008). Hosts can range from companion animals to food animals and humans, and disease symptoms can range from self-limiting gastroenteritis to invasive systemic infections with a high mortality rate (Lavigne and Blanc-Potard, 2008). Some serovars of S. enterica can survive in a wide variety of hosts and cause different diseases such as Salmonella Typhimurium, which causes no symptoms in adult poultry, can cause gastroenteritis in humans, or cause highly invasive systemic enteric fever in mice (Goldberg and Rubin, 1988; Chan et al., 2003; Lavigne and Blanc-Potard, 2008). Other serovars are host specific like Salmonella Typhi which is an obligate human pathogen that causes invasive Typhoid fever (Bhan et al., 2005), which is extremely rare in the U.S., but can be endemic in other parts of the world. Most other serovars of Salmonella usually cause only gastroenteritis in humans while some other serovars can potentially cause both gastroenteritis and invasive infections in humans. Salmonella is usually transmitted by contaminated food with contamination originating from animal products or feces, contaminated processing equipment, or contaminated food handlers (Goldberg and Rubin, 1988). Additionally, Salmonella is sometimes antimicrobial resistant which can result in difficulty treating infections (Fluit, 2005; Stoycheva and Murdjeva, 2006). Because of the impact on human health, zoonotic transmission, and ability to acquire AR, Salmonella has been chosen as the sentinel organism for foodborne disease and for AR monitoring (Tollefson et al., 1998, 1999; White et al., 2001, 2002).
In the U.S., Salmonella is estimated to cause over one million human infections each year (Scallan et al., 2011). Most of these infections result in gastroenteritis that resolves after a few days; however, some infections can be chronic or invasive, especially in the very young, the old, and groups of people with compromised immune systems (Alcaine et al., 2007). In these cases Salmonella infections may require antimicrobial treatment to prevent further morbidity or mortality (Alcaine et al., 2007). First line antibiotic treatment in the U.S. is typically a fluoroquinolone-like ciprofloxacin or a third generation cephalosporin β-lactam such as ceftriaxone, and folic acid pathway inhibitors are also available (Mandal, 1990; Guerrant et al., 2001; Hohmann, 2001; Habib, 2004; Parry and Threlfall, 2008). However, in children and pregnant women, treatment is usually limited to β-lactams due to fluoroquinolone's interference with cartilage formation; therefore resistance to β-lactams is a considerable concern in Salmonella (Parry and Threlfall, 2008). In cases where infection is caused by Salmonella resistant to first line treatments, alternative second line antimicrobials may be used, such as aminoglycosides, or folic acid pathway inhibitors like sulfisoxazole or sulfamethoxazole with or without trimethoprim (Guerrant et al., 2001). In MDR Salmonella infections, the last line treatments are usually the aminoglycoside, amikacin or the carbapenems, imipenem or meropenem which are administered intravenously. Due to observed increases in morbidity and mortality in antimicrobial resistant infections, it has been suggested that resistant Salmonella are more virulent than sensitive strains (Mundy et al., 2000; Alanis, 2005; Foley and Lynne, 2008; Ajiboye et al., 2009; Gebreyes et al., 2009; Huehn et al., 2010). However, research into this has been inconclusive, and some studies have demonstrated that resistance to some antimicrobials such as fluoroquinolones actually reduces virulence in Salmonella (O'Regan et al., 2010).
Antimicrobial resistant Salmonella have been isolated globally from human infections, clinically ill animals, healthy food animals, food animal products, and fresh produce. In the U.S., since 1997, NARMS has collected data on AR in Salmonella isolated from humans, and animals, including chickens, turkeys, cattle, swine, and their retail meat products. The percentage of resistant isolates from these sources has been presented in NARMS reports for each year from 1997 to 2009 (http://www.fda.gov/AnimalVeterinary/SafetyHealth/AntimicrobialResistance
/NationalAntimicrobialResistanceMonitoringSystem/default.htm). Antimicrobial compounds used for susceptibility testing in Salmonella are listed in Table 1. Resistance to all of these antimicrobials was detected in isolates from humans during 1997–2009; however, only 26 human isolates were resistant to the first line treatment ciprofloxacin and only two human isolates were resistant to amikacin making it a possible treatment option (Table 1). However, substantial resistance to ceftriaxone, which is also often used as a first line treatment for salmonellosis, was detected in humans and increased from 1997 (0.5%) to 2003 (4.4%) (http://www.fda.gov/AnimalVeterinary/SafetyHealth/Antimicrobial
Resistance/NationalAntimicrobialResistanceMonitoringSystem/default.htm) and has remained fairly steady from 2004 to 2009 (average of 3.1%; Table 1) (Frye and Fedorka-Cray, 2007; Frye et al., 2008). A similar trend was observed for animal isolates, especially from cattle where resistance to ceftriaxone increased from 0% in 1997 to 21.6% in 2003 (Frye and Fedorka-Cray, 2007; Frye et al., 2008) and then leveled-off. Resistance to other antimicrobials varied between human and animal isolates, and especially by animal source of the isolates. In human isolates, resistance to tetracycline averaged 16.9%; in animal isolates resistance averaged 34.9% for all animals, and over 50% in turkey and swine isolates (Table 1). Sulfamethoxazole/sulfisoxazole resistance in human isolates was 15.3% and in animal isolates it was 19.9% for all sources. Resistance to streptomycin was also found in human and animal isolates at 14.4% in humans and 26.3% for all animals. Similarly, ampicillin resistance was 14.3% in human isolates and 16.0% in animal isolates. Resistance to chloramphenicol was detected in human (8.8%) and animal (7.3%) isolates; however, this drug is not usually used to treat human infections in the U.S. Levels of resistance to gentamicin was detected at 2.1% in isolates from humans and 6.5% from animals, and resistance to combined folate pathway inhibitors, trimethoprim-sulfamethoxazole, were 2.0% in human isolates and 1.4% in animal isolates, making it also suitable as a treatment in the U.S. (Guerrant et al., 2001).
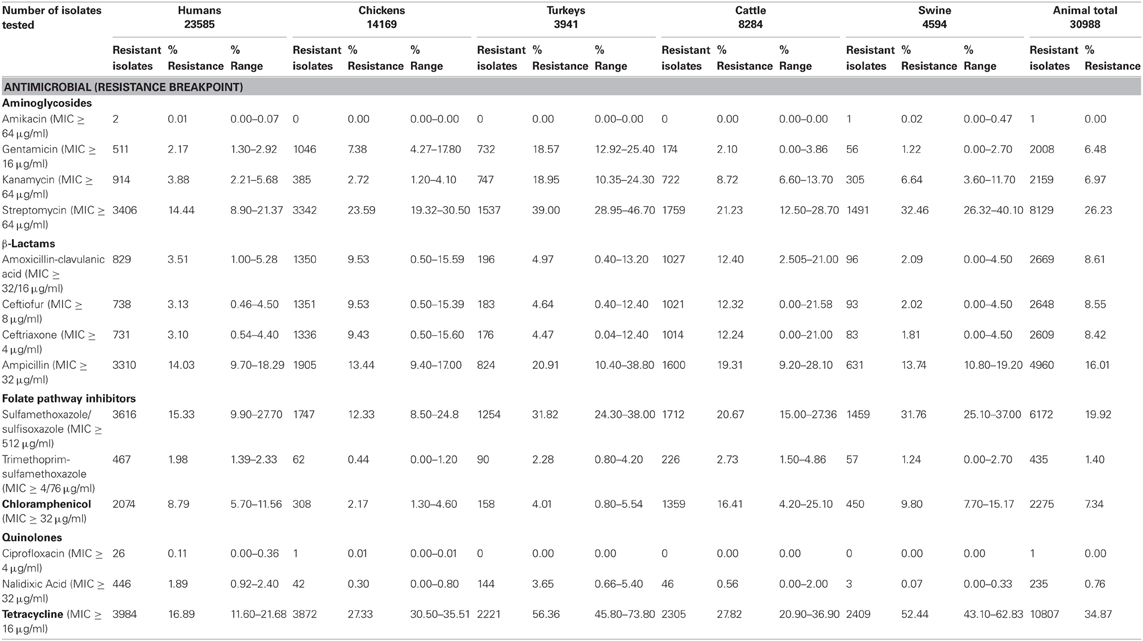
Table 1. Resistance to antimicrobials in Salmonella enterica isolated from human and animal sources by NARMS 1997–2009.
Aminoglycoside antimicrobials were first introduced into clinical application in the middle and last half of the twentieth century primarily to treat severe infections caused by Gram-negative bacteria in animals (Maurin and Raoult, 2001; Schwarz and Chaslus-Dancla, 2001; Schwarz et al., 2001). Their use in treatment of infections in food animals is limited due to both their toxic nature and the persistence of residual antimicrobial in the tissue of the animals. In swine, aminoglycosides including gentamicin, neomycin, or streptomycin have been used to treat intestinal diseases such as scours in weanling pigs and swine dysentery (Maurin and Raoult, 2001; Schwarz and Chaslus-Dancla, 2001). Gentamicin is also used to prevent or treat Salmonella or E. coli infections in poults. In some European countries, neomycin is used in combination with the macrolide antimicrobial, lincomycin, for treatment of mastitis in dairy cattle caused by E. coli and Staphylococcus aureus (De Oliveira et al., 2000). The synergistic effect of an aminoglycoside antimicrobial with an antimicrobial that targets the cell wall of enterococci such as a β-lactam like ampicillin or penicillin are also used in human medicine to treat enterococcal infections (Arias and Murray, 2012).
The aminoglycosides function by binding to the 30S ribosomal subunit inhibiting protein translation. Salmonella resistance to aminoglycosides is usually an enzymatic modification of the compound; however, in other bacteria, active efflux of the compound or enzymatic modification of the 16S rRNA subunit to prevent the aminoglycoside from binding to its ribosomal target can lead to resistance. Mechanisms of aminoglycoside resistances in U.S. Salmonella animal isolates are primarily due to acetyltransferases, phosphotransferases, and nucleotidyltransferases which modify and inactivate the aminoglycoside (Shaw et al., 1993; Ramirez and Tolmasky, 2010). There are two systems of nomenclature for these genes and some researchers have also used additional name modifications to indicate new alleles resulting in a complex set of names, many of which are synonymous. These have been expertly reviewed in several papers as well as their prevalence in Salmonella throughout the world (Ramirez and Tolmasky, 2010).
The aminoglycoside acetyltransferases are usually named aac, followed by a numeral in parentheses to designate the target of their enzymatic activity on the aminoglycoside molecule [e.g., aacC(3′)]. This can then be followed by a roman numeral to indicate the resistance phenotype, and then a letter to indicate the allele or variant of the gene. The aac genes found in U.S. Salmonella isolates can confer resistance to gentamicin, tobramycin, and kanamycin. Aminoglycoside phosphotransferases confer resistance to kanamycin and neomycin, and are usually named aph. These genes also have a designation of the location they modify on the antibiotic [e.g., aph(3′)], and some aph genes also have other names, such as strA and strB which encode resistance to streptomycin. Aminoglycoside nucleotidyltransferases can confer resistance to gentamicin, tobramycin, or streptomycin and include aad and ant groups of genes that can also have extensions to indicate the target of the enzyme. The most common genes reported are listed in Table 2, and include variants of aac, aad, aph, and str genes. Specific alleles of aminoglycoside resistance genes detected in several studies of NARMS U.S. food animal isolates have included aac(3′), aac(6′), aadA, aadA1, aadA2, aadA12, aphAI, aph(3′)-Ii-iv, strA, and strB (Frye and Fedorka-Cray, 2007; Frye et al., 2008, 2011; Glenn et al., 2011; Lindsey et al., 2011a).
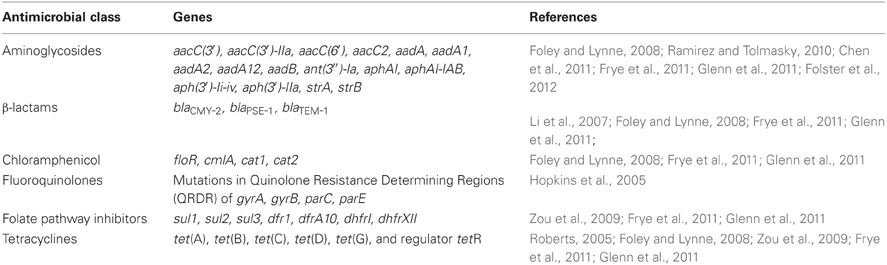
Table 2. Antimicrobial resistance genes found in Salmonella enterica isolated from U.S. food animals.
Penicillin was one of the first β-lactams developed for clinical use in humans, and was also one of the first antibiotics to which bacteria became resistant. The β-lactams prevent synthesis and maintenance of the peptidoglycan component of the bacterial cell wall by mimicking one of the building blocks used by enzymes to construct peptidoglycan (Prescott, 2000c,d,g). The β-lactams have a unique four membered “β-lactam” ring that when acted upon by enzymes that build the cell wall, forms an irreversible bond to the enzyme, inactivating it and preventing the enzyme from completing cell wall synthesis. These enzymes are also known as penicillin-binding proteins (PBP). Most resistance to β-lactams is conferred by β-lactamases that enzymatically cleave the β-lactam ring and prevent it from bonding to and inactivating cell wall enzymes (Prescott, 2000c,d,g). Because of this, new β-lactams were synthesized through modification of the chemical groups around the β-lactam ring to produce β-lactams that are resistant to the β-lactamases; other modifications also improved their activity on specific bacteria or accessibility to certain infection sites. These include modified penicillins such as methicillin and oxacillin; the cephalosporins like cephalothin, cefoxitin, ceftriaxone, and cefipime, which are 1st through 4th generation cephalosporins, respectively; and the carbapenems such as imipenem and meropenem (Prescott, 2000c,d,g). In response to the selective pressure created by these new antibiotics, mutations in β-lactamase genes have also created enzymes that can digest these later generation β-lactams. Some important groups of these are the extended spectrum β-lactamases (ESBLs) (Bush, 2008), cephalosporinases (Arlet et al., 2006), and carbapenemases (Miriagou et al., 2010). However, some β-lactamases can also be inactivated by β-lactamase inhibitors, like clavulanic acid, which bind irreversibly to particular β-lactamases, thus allowing the β-lactam to work when the β-lactam and β-lactamase inhibitor are used as a combined treatment, such as Augmentin (ampicillin/clavulanic acid; Prescott, 2000g). Other resistance mechanisms include genes that encode modified PBPs that have a low affinity for β-lactams and are not inactivated by them or that use different building blocks to construct the cell wall. Efflux of the β-lactam or modification of porins (e.g., ompF and ompC) is also a resistance mechanism to β-lactams. Often these different mechanisms are found in the same bacterium, resulting in high level β-lactam resistance (Batchelor et al., 2005).
Most of the β-lactam resistance in Salmonella is encoded by horizontally acquired β-lactamases (Table 2); however, many other bacteria have an intrinsic β-lactamase, such as ampC found in E. coli (Table 3) (Siu et al., 2003). In Salmonella isolated from U.S. animals, the most prevalent β-lactamase genes are blaTEM−1 and blaPSE−1 (a.k.a. blaCARB2) encoding resistance to ampicillin, and blaCMY−2 that encodes resistance to ampicillin, 1st, 2nd, and 3rd generation cephalosporins and is also resistant to β-lactamase inhibitors such as those found in Augmentin (Table 2) (Frye and Fedorka-Cray, 2007; Frye et al., 2008, 2011; Glenn et al., 2011; Lindsey et al., 2011a). In contrast, other β-lactamases have been detected globally including blaTEM, blaCTX−M, blaIMP, blaVIM, blaKPC, blaSHV, and blaOXA and variants of these genes may encode ESBL or carbapenemase activity (Falagas and Karageorgopoulos, 2009). Of particular concern may be the global emergence of the blaNDM−1 metallo-β-lactamase that confers resistance to carbapenems which are often the last line of defense in Gram negative infections (Walsh, 2010). Recently this gene was found in Salmonella isolated from a human in the U.S. after travel to India (Walsh, 2010; Savard et al., 2011). NARMS U.S. food animal isolates of Salmonella collected from 1999 to 2011 have been screened by both phenotypic and genotypic assays for ESBLs and none have been detected in isolates from U.S. animals by these or any other published studies [(Frye and Fedorka-Cray, 2007; Frye et al., 2008) and Fedorka-Cray, pers. communication]. The β-lactam resistance genes identified in NARMS animal isolates have been limited to mostly blaTEM−1, blaPSE−1, and blaCMY−2 (Table 2) (Frye and Fedorka-Cray, 2007; Frye et al., 2008, 2011; Glenn et al., 2011; Lindsey et al., 2011a).
Chloramphenicol and related compounds such as florphenicol inhibit protein synthesis by binding to the 50S ribosomal subunit. In the developed world, chloramphenicol is virtually irrelevant clinically and has been banned in the U.S. and other countries for use in humans or food animals due to its potential toxic effects on humans. In most of the developing world, its use is also limited by high levels of resistance likely due to the low-cost of the antimicrobial and unregulated, widespread over use. It is primarily used for treatment of systemic salmonellosis caused by bacteria that are resistant to other drugs of choice (Prescott, 2000e). Chloramphenicol has also been used for eye infections and sparingly for treatment of infections caused by bacterial anaerobes. Most resistance mechanisms are efflux pumps such as floR and cmlA, as well as inactivating enzymes such as chloramphenicol acetyltransferase, cat1 (Prescott, 2000b,e). Resistance in Salmonella and other bacteria isolated from animals is often seen by these mechanisms. Mechanisms of phenicol resistance found in U.S. NARMS animal isolates have been floR, cmlA, and cat1 (Table 2). In addition, the chloramphenicol resistance gene floR is often found in the class I integron located in Salmonella Genomic Island 1(SGI-1) (Frye et al., 2011; Glenn et al., 2011, 2012).
Quinolones and fluoroquinolones are a synthetic group of antimicrobials used in food animals to combat various infectious agents (Hopkins et al., 2005). Introduced into use over two decades ago, they have broad-spectrum activity coupled with low toxicity and other pharmacokinetic characteristics which make them attractive antimicrobials for use in food animals. A number of fluoroquinolones have been used in food animals including enrofloxacin, difloxacin, marbofloxacin, orbifloxacin, and sarafloxacin (Hopkins et al., 2005). Enrofloxacin and danofloxacin are both useful for treatment of respiratory track disease in cattle; enrofloxacin and sarafloxacin were approved in the mid-1990's for treatment of chickens and turkeys with E. coli infections (Endtz et al., 1991; Nelson et al., 2007). Although valuable for treatment of disease in food animals, fluoroquinolones are also used in human medicine for the treatment of Salmonella, E. coli, and other bacterial infections. Bacteria resistant to fluoroquinolones used in animals could also be resistant to fluoroquinolones used in human medicine due to the shared mechanism of action of these drugs (Nelson et al., 2007). In Europe, after the introduction of enrofloxacin in food animals, ciprofloxacin-resistant Campylobacter jejuni were detected in food animals and humans and were suspected to have been transmitted through food; this led to EU withdrawal of use of these compounds (Endtz et al., 1991; Nelson et al., 2007). In the U.S., fluoroquinolone use in poultry is banned and use in cattle is limited to non-lactating heifers under 20 months of age for treating bovine respiratory disease in cattle.
Quinolones and fluoroquinolones bind to and prevent DNA processing enzymes such as topoisomerases from assisting in DNA replication and maintenance. Mechanisms found in Salmonella and E. coli have been recently reviewed (Hopkins et al., 2005). Most resistances to these compounds are due to mutations within the genes that encode the enzymes such as gyrA, gyrB, parC, and parE. Most of these mutations occur in the quinolone resistance determining region (QRDR) which is a conserved site in these enzymes targeted by these antimicrobials. Resistance to nalidixic acid and then to fluoroquinolones builds in a stepwise process of mutations in the QRDR region producing an enzyme with a target region that quinolones cannot bind to (Chen et al., 2007). Resistance to nalidixic acid and fluoroquinolones like ciprofloxacin has been found in human isolates of bacteria globally. However, animal isolates of Salmonella in the U.S. have very low levels of resistance while the close relative E. coli has higher levels of resistance; for example, only a handful of Salmonella isolated from animals by NARMS were resistant to ciprofloxacin (Tankson et al., 2006). Studies have shown that Salmonella resistant to ciprofloxacin also had a growth defect in vitro and in vivo, while E. coli does not (O'Regan et al., 2010). This may be responsible for the low levels of ciprofloxacin resistance seen in Salmonella. Other resistance mechanisms have also been identified including the qnr efflux system, and an aminoglycoside acetyltransferase, aac(6′)-Ib, which can modify and inactivate ciprofloxacin (Cavaco et al., 2007a,b; Cavaco and Aarestrup, 2009). While these mechanisms have been found in E. coli, they are rare or undetected in U.S. Salmonella isolates (Tables 2, 3) (Frye and Fedorka-Cray, 2007; Frye et al., 2008, 2011; Glenn et al., 2011, 2012; Lindsey et al., 2011a).
These are compounds that compete for substrates of the essential folic acid pathway in bacteria at two different steps, the sulfonamides, which inhibit DHPS (dihydropteroate synthase) and trimethoprim, which inhibit DHFR (dihydrofolate reductase). Sulfonamides are one of the oldest classes of antimicrobials introduced into use more than 80 years ago (Skold, 2001). Both sulfonamides and trimethoprim act on the folic acid pathway in bacteria by interfering with the production of dihydrofolic acid. They have been used extensively in food animals as growth promoters in swine and for treatment of diseases such as colibacillosis in swine and coccidiosis in poultry (Prescott, 2000b,h). Sulfonamides are bacteriostatic when used alone or bacteriocidal when used in combination (trimethoprim-sulfamethoxazole; Walsh, 2003).
Resistance to both of these antimicrobials occurs by acquisition of genes encoding enzymes that do not bind these compounds. These include the sul genes, sul1, sul2, and sul3, which encode an insensitive DHPS enzyme and are found in Salmonella globally; however, in the U.S. most of the resistance is due to either sul1 or sul2 (note that sometimes roman numerals are used to designate these genes; Prescott, 2000h). Resistance to trimethoprim is by DHFR encoding genes, either dhfr or dfr, both of which have been found in Salmonella animal isolates in the U.S. (Prescott, 2000h; Frye et al., 2011; Glenn et al., 2011). More than 30 dfr genes have been identified and are presently divided into two families (White and Rawlinson, 2001). Type A dihydrofolate reductase genes encode proteins between 157 and 187 amino acid residues, while type B dihydrofolate reductase genes encode proteins of ~78 residues (White and Rawlinson, 2001). Sulfonamides and trimethoprim are often used in combination to treat Salmonella infections that are resistant to other antimicrobials (Hohmann, 2001). Currently, resistance to these combined compounds is relatively rare in the U.S. as reported by NARMS and is a good second line treatment for salmonellosis (Table 1). Genes that have been identified in studies of NARMS Salmonella animal isolates include: sul1 (a.k.a. sulI), sul2 (a.k.a. sulII), dfr1, dfrA, dfrAI, dhf, and dhfrI (Table 2) (Zou et al., 2009; Frye et al., 2011; Glenn et al., 2011).
Tetracycline has been used in food animals to combat vector-borne infections such as borreliosis, erlichiosis, rickettsiosis, and tularemia as well as other infections including pneumonia, brucellosis, and listeriosis (Roberts, 1996, 2002, 2005). In food animals, tetracycline or doxycycline are used mainly in treatment of respiratory infections (Mathers et al., 2011). Tetracyclines (chlortetracycline or oxytetracycline) have also been used for growth promotion and to promote feed efficiency in cattle, swine, and poultry. Tetracycline targets the 30S subunit of the bacterial ribosome binding to the ribosome and inhibiting protein synthesis. Resistance mechanisms include efflux, modification of the rRNA target, and inactivation of the compound. However, in Salmonella, active efflux systems are most commonly observed and include tet(A), tet(B), tet(C), tet(D), tet(G), and tet(H). In U.S. Salmonella animal isolates, tet(A), tet(B), tet(C), tet(D), and tet(G) are most often detected and are usually accompanied by the tetR regulator (Frye and Fedorka-Cray, 2007; Frye et al., 2008, 2011; Zou et al., 2009; Glenn et al., 2011; Lindsey et al., 2011a). As reported by NARMS, tetracycline resistance is 16.9% in human isolates and 34.9% in animal isolates of Salmonella in the U.S. (Table 1). Tetracycline resistance has been linked to use over a long time in humans, in animals, and as growth promoters in animals (Witte, 2000; Chopra and Roberts, 2001; Jones and Ricke, 2003). The tetracycline resistance genes found most often in studies of NARMS Salmonella animal isolates are tet(A), tet(B), tet(G), and tetR (Table 2) (Frye and Fedorka-Cray, 2007; Frye et al., 2008, 2011; Glenn et al., 2011; Lindsey et al., 2011a).
AR genes are often arranged in cassette-like genetic elements, which may include multiple resistances and be associated with integrons or MGEs that can facilitate the expression of these genes as well as their movement both within a bacterium's genome and horizontally between bacteria (Carattoli, 2001, 2003, 2008, 2009; Blake et al., 2003; White et al., 2003; Lindsey et al., 2011a; Douard et al., 2010; Folster et al., 2011; Frye et al., 2011; Glenn et al., 2011, 2012; Johnson et al., 2011b; Lindsey et al., 2009). These cassette-like elements may be associated with insertion sequences (IS elements), integrons, and MGEs like transposons, bacteriophage, and others. These cassettes may also be located on plasmids, which are self-replicating, extra chromosomal DNA elements that can also encode machinery for DNA transfer between cells such as conjugation systems (Carattoli, 2003, 2009). On a global level, Salmonella harbor a wide variety of these elements (Carattoli, 2003, 2009). These vary greatly with the location or host source of the isolates and often reflect the environment to which they are exposed (Carattoli, 2008, 2009). In Salmonella isolated from U.S. farm animals, the most prevalent genetic elements identified to date are plasmids and integrons (White et al., 2003; Lindsey et al., 2009, 2011a; Folster et al., 2011; Frye et al., 2011; Glenn et al., 2011, 2012; Johnson et al., 2011b).
Plasmids are categorized by incompatibility groups, or Inc types, as only one plasmid of the same type can be stably maintained during cell division; thus two plasmids of the same Inc type are “incompatible.” Recently replicon typing has been accepted as an improved plasmid typing method using specific PCR assays (Carattoli et al., 2005, 2006). Replicon typing further divides Inc groups into sub-groups of replicons based on their specific DNA sequence. Therefore, replicon type is not necessarily interchangeable with Inc type (Carattoli et al., 2005, 2006). Studies of antimicrobial resistant, including MDR, Salmonella have determined that specific plasmid replicon types are associated with resistance, source host, and geographic origin (Osborn and Boltner, 2002; Su et al., 2004; Carattoli, 2008, 2009; Lindsey et al., 2009, 2011a,b). In U.S. food animals, isolates harbor multiple replicon types, including IncFIB, IncFIIA, IncHI1, IncHI2, IncI1, IncA/C, and IncP (Lindsey et al., 2009, 2011a; Glenn et al., 2011). Because IncA/C plasmids are wide spread in U.S. isolates and often carry MDR, they have been well-studied (Welch et al., 2007, 2009; Fricke et al., 2009; Lindsey et al., 2009, 2011a; Glenn et al., 2011).
Integrons are also associated with AR. These are inserted into the host bacterium's genome or plasmids, have an integrase gene, have cassettes and promoters bounded by IS elements, can mobilize their cassettes, and may have a variable number of genes inserted into the cassettes, including AR genes. While there are also several kinds of integrons detected in Salmonella isolated from animals in the U.S., the most prevalent is IntI1 and its variants, which often encode several AR genes and consequently may confer MDR phenotypes (White et al., 2003; Ebner et al., 2004; Gebreyes et al., 2004; Evershed et al., 2009; Krauland et al., 2009; Frye et al., 2011; Glenn et al., 2011; Lindsey et al., 2011a).
The development and dissemination of MDR in Salmonella is a major concern and the IntI1 integrons and the IncA/C plasmids play a major role in MDR found in isolates from U.S. food animals (Carattoli, 2001; Carattoli et al., 2002a,b; Welch et al., 2007, 2009; Carattoli, 2008, 2009; Fricke et al., 2009; Douard et al., 2010; Mataseje et al., 2010; Frye et al., 2011; Glenn et al., 2011; Lindsey et al., 2011a). Salmonella enterica serovar Typhimurium Definitive Phage Type 104 (DT104) was a MDR Salmonella that affected humans and animals worldwide in the mid 1990's (Wall et al., 1994; Akkina et al., 1999; Bolton et al., 1999; Briggs and Fratamico, 1999; Baggesen et al., 2000; Allen et al., 2001). It was first detected in Europe in the 1990's and was eventually found in U.S. swine and humans (Wall et al., 1994; Akkina et al., 1999; Bolton et al., 1999; Briggs and Fratamico, 1999; Baggesen et al., 2000; Allen et al., 2001). DT104 was reported as penta-resistant to ampicillin, chloramphenicol, streptomycin, sulfamethoxazole, and tetracycline (ACSSuT); however, isolates were subsequently detected with different phenotypes. The resistance genes usually associated with DT104 are blaPSE−1, floR, aadA2, sulI, dfrA10, tet(G), and tetR located on an IntI1integron (Boyd et al., 2001; Ebner et al., 2004; Mulvey et al., 2006). IntI1 is site specific and usually integrates into the genome resulting in Salmonella Genomic Island 1 (SGI1) a wide spread MDR carrying MGE (Ebner et al., 2004; Mulvey et al., 2006). In the U.S., penta-resistant Salmonella Typhimurium DT104 is found primarily in swine but has also been isolated from other animal sources including chickens, turkeys, and cattle. These integrons can also be found in other serovars of Salmonella and other species of bacteria, and can encode different resistance genes for multiple antimicrobials, resulting in many variants of IntI1 and SGI1 (Mulvey et al., 2006).
Recently, MDR Salmonella enterica serovar Newport was isolated from animals and humans with a MDR-AmpC phenotype which is similar to the ACSSuT DT104 phenotype, but also includes resistance to 3rd generation cephalosporins and sometimes additional antimicrobials (Gupta et al., 2003; Varma et al., 2006; Lindsey et al., 2009, 2011a; Glenn et al., 2011). As described previously, the β-lactam often used to treat human Salmonella infections is the 3rd generation cephalosporin, ceftriaxone, and resistance to ceftriaxone increased from 1998 to 2003 in humans and animals (Table 1). Several studies have suggested that this increase was associated with the use of ceftiofur, which is similar to ceftriaxone, in dairy cattle to treat mastitis and diarrhea (Clegg et al., 1983; Zhao et al., 2003; Cobbold et al., 2006; Lindsey et al., 2009; Glenn et al., 2011). This use may have exerted selective pressure causing MDR S. enterica serovar Newport to increase in prevalence. However, direct evidence for increased prevalence has not been reported (Cobbold et al., 2006; Call et al., 2008; Glenn et al., 2011). Ceftriaxone and ceftiofur resistance in MDR-AmpC isolates was determined to be encoded by the blaCMY−2 β-lactamase gene and was located on an IncA/C plasmid found in S. Newport isolated from U.S. humans and cattle (Gupta et al., 2003; Varma et al., 2006). These IncA/C plasmids were demonstrated to often be mobile, found in many serovars other than S. Newport, and also encoded resistance to multiple antimicrobials including other β-lactams, aminoglycosides, chloramphenicol, folic acid pathway inhibitors, and tetracycline (Fricke et al., 2009; Lindsey et al., 2009; Welch et al., 2009; Douard et al., 2010; Folster et al., 2011; Glenn et al., 2011; Lindsey et al., 2011a). Genes encoding the resistance mechanisms found in U.S. animal isolates often include blaCMY−2, aacC, aadA, aphA1, strB/A, floR, cat, sulI, dhfr1, tet(A), and tetR (Table 2) (Glenn et al., 2011). Several studies have shown that the resistance cassettes found in the IncA/C plasmids are similar to those in SGI1, and that these genes were likely previously transferred to the SGI1integron from an IncA/C plasmid in the distant past, prior to human influences (Fricke et al., 2009; Lindsey et al., 2009; Welch et al., 2009; Douard et al., 2010; Folster et al., 2011; Glenn et al., 2011; Lindsey et al., 2011a). It has also been demonstrated that IncA/C MDR plasmids can mobilize SGI1 in trans (Douard et al., 2010).
Interestingly, throughout most of the rest of the world, resistance to 3rd generation cephalosporins in Salmonella is often encoded by ESBLs, which are mutations of several different lineages of β-lactamases (Bradford, 2001; Paterson, 2006). The ESBLs can be found in Salmonella isolated from both animals and humans in Europe, Asia, and South America (Bradford, 2001; Nijssen et al., 2004; Hasman et al., 2005; Yates and Amyes, 2005; Biedenbach et al., 2006; Carattoli, 2008). Salmonella with ESBLs are infrequently isolated from humans in the U.S.; however, they have not been detected in U.S. food animals (Mulvey et al., 2003; Frye and Fedorka-Cray, 2007; Bush, 2008; Frye et al., 2008). In addition to intI1 and IncA/C plasmids, other vehicles for resistance in the U.S. include many other plasmids such as IncF, IncH, IncI, and IncP which have also been associated with AR (Lindsey et al., 2009; Folster et al., 2010, 2011; Glenn et al., 2011). For example, the IncI1 plasmids are also known to carry the blaCMY−2 gene; however, they do not usually carry MDR, are found mostly in Salmonella serovar Heidelberg, and are associated with chickens and turkeys rather than cattle (Folster et al., 2010, 2012).
Outbreaks of Salmonella in the U.S. have been associated with food originating from animal sources such as chicken, eggs, beef, and ground turkey (Scallan et al., 2011). However, in the past few decades there has been an increasing number of Salmonella outbreaks that have been associated with unusual sources, such as tomatoes, peanut butter, black and red pepper, and spinach (Boxrud et al., 2010). These outbreaks indicate that Salmonella infection is not always associated with food animal products such as meat. It is unclear if non-meat sources of Salmonella infections have increased or if public health agency programs such as Food Net, Pulse Net, and NARMS have simply enabled detection and tracing of these outbreaks to their sources (Boxrud, 2010; Boxrud et al., 2010). Few outbreaks have been associated with antimicrobial resistant Salmonella; however, an exception to this was a 2011 outbreak connected with ground turkey (Folster et al., 2012). A strain of Salmonella Heidelberg resistant to ampicillin, gentamicin, streptomycin, and tetracycline, encoded by blaTEM−1, aac(3)-IIa, aadA1, ant(3″)-Ia, and tet(A) on a IncI1 plasmid was detected (Folster et al., 2012).
The wide variety of outbreak sources and variability in genetics causing AR indicates resistance in Salmonella is multifaceted, and the trend of unusual food sources and antimicrobial resistant outbreaks may become more prevalent in the future. Additionally, it is also likely that AR elements found mostly outside the U.S. will eventually be found in U.S. food animals. For example, ESBLs have already been found in U.S. human infections and it can be expected to eventually find them in the animal population (Bush, 2008). Surveillance and research of AR in Salmonella is a global priority and an ongoing effort will need to continue to follow these trends and identify the mechanisms leading to resistance. Data collected by these studies will improve our understanding of resistance, how it develops, how it spreads, and what can be done to prevent infections in humans and animals.
Much less studied and understood is the influence exerted by non-pathogens on the acquisition, transfer, and persistence of AR in pathogens sharing the same environment. The intestinal tract of humans and animals are inhabited predominately by Gram-negative and Gram-positive anaerobes such as Bacteroides and Bifidobacterium. However, many other genera of bacteria including enterococci and Escherichia coli are also present. Transient pathogenic colonizers such as Streptococcus and Neisseria are also present (Salyers et al., 1995). Both indirect and direct evidence exists that AR can be transferred between different genera in the intestinal tract (McConnell et al., 1991; Salyers et al., 1995; Netherwood et al., 1999; Avrain et al., 2004). Furthermore, total numbers and types of bacteria in the intestinal tract can be altered by antimicrobial treatment which may influence the transfer of resistance (McConnell et al., 1991). E. coli and enterococci are two commensals of particular interest because they are ubiquitous in animals and the environment and are used most often in bacterial source tracking (Scott et al., 2002; Hassan et al., 2007). They are also known to harbor resistance genes on plasmids, transposons, and integrons (Johnson and Nolan, 2009; Hegstad et al., 2010).
The recognition of E. coli as a principal commensal intestinal bacteria and the relative ease of isolating E. coli has led to its adoption as a sentinel organism for fecal contamination, particularly as applied to water safety. Those strains which ferment lactose were included in a “coliform bacteria” group of enterobacteria, and coliform enumeration is commonly used to indicate the microbiological safety of food products and water (Blood and Curtis, 1995). However, this simple method cannot be used to identify enteric pathogens nor provide the population data necessary for identifying sources of contamination (Leclerc et al., 2001).
Research has shown that the enteropathogenic E. coli (EPEC) are a virulent subgroup possessing a complex array of disease-producing genetic factors absent from commensal strains (Spears et al., 2006). In the 1980's, a pathogenic variant of E. coli was recognized as the etiologic agent of potentially fatal hemorrhagic intestinal and hemolytic uremic illnesses (Paton and Paton, 1998; Karch et al., 1999). These toxigenic E. coli have acquired Shiga-like toxin genes (Sandvig, 2001; Schmidt, 2001) as a key component of their virulence profile and are termed Shiga toxin-producing E. coli (STEC). The STEC were found to kill Vero cells in culture which led to the name verocytotoxigenic E. coli (VTEC); they are also categorized as enterohemorrhagic E. coli (EHEC). While EPEC and STEC strains share a number of characteristics, the STEC are not simply EPEC that generate a cytolytic toxin (Kaper, 2005). The EPEC appear to be exclusively human pathogens and remain a significant cause of infant diarrheal disease, especially in developing countries. In contrast, the STEC are niche-specific gut commensals in certain animals, particularly ruminants.
The STEC serotype O157:H7 from cattle has become the paradigm for this pathotype (Rasmussen and Casey, 2001; Renter and Sargeant, 2002), but other significant serogroups are emerging (Bettelheim, 2007). Controlling outbreaks of human disease due to contamination of food products by STEC sources is an important food safety issue (Olsvik et al., 1991; Mainil and Daube, 2005; Erickson and Doyle, 2007).
In food animals, E. coli has been documented as the etiological agent of many different diseases (Prescott, 2000b). In cattle, E. coli infections of the gastrointestinal tract, skin, the genital and urinary tract, the musculoskeletal system, the central nervous system, and the cardiovascular system have been reported (Prescott, 2000b). Some infections such as septicemia and meningitis are often fatal. Colibacillosis in both swine and poultry is also a major cause of mortality in these two groups of food animals (Prescott, 2000b).
Since all types of E. coli acquire AR and are commonly associated with many different animal and environmental sources, they are typically included in AR surveillance studies (Erb et al., 2007). Resistance to aminoglycosides, cephalosporins, fluoroquinolones, and sulfonamide antimicrobials is widespread in E. coli isolated from animals and humans and may compromise treatment efficacy since drugs of these classes are frequently used to treat Gram-negative infections (Hammerum and Heuer, 2009). In particular, the prevalence of resistance to clinically important 3rd and 4th generation cephalosporins mediated by extended-spectrum β-lactamases (ESBLs) has serious implications for human and veterinary medicine (Shah et al., 2004a,b; Paterson and Bonomo, 2005; Li et al., 2007).
Multi-drug resistance in E. coli is also a concern (Hawkey and Jones, 2009). Resistance in E. coli is often mediated by plasmids that encode AR genes (Carattoli, 2009; Johnson et al., 2011a). These extra-chromosomal MGEs are readily acquired by E. coli thereby conferring resistance to one or more antimicrobials. The potential threat to human health from multiple drug resistant pathogenic E. coli such as STEC acquired from food products may pose a serious yet underestimated food safety risk (Rasko et al., 2011). It is therefore essential to improve our limited knowledge of the basis for the increasing drug resistance of commensal and pathogenic E. coli. Since resistance plasmids can be transferred between strains of the same species or different genera (such as Salmonella or Klebsiella), such exchanges may serve to establish reservoirs of resistance genes in animals and the environment (Su et al., 2008). Furthermore, it should also be noted that there are differences in resistance levels of E. coli (including MDR strains) depending on the age of the host. Past studies have shown that younger animals such as cows and swine harbor more resistant E. coli than older animals (Langlois et al., 1988; Khachatryan et al., 2004; Berge et al., 2005). It has been suggested that this observation could be due to a higher fitness of the resistant isolates in the intestine of the animals or that the intestine of the younger animals may be more readily colonized by the resistant isolates as compared to the older animals (Langlois et al., 1988; Khachatryan et al., 2004; Berge et al., 2005). Many of the same resistance phenotypes and genotypes presented for Salmonella in the previous section are also present in E. coli; therefore, AR in E. coli will focus on resistance and resistance genes of specific interest in E. coli (Table 3).
Since E. coli induced diarrhea often occurs in food animals, several aminoglycosides are used in treatment including neomycin for calves, gentamicin in swine, and apramycin in both calves and swine (Prescott, 2000a). Gentamicin is also used for treating septicemia in cattle. Resistance to aminoglycosides is mediated in E. coli by genes from all three classes of aminoglycoside modifying enzymes [AAC, ANT, and APH; (Ramirez and Tolmasky, 2010)]. Several acetyltransferases including type II 3-N-aceyltransferase, type IV 3-N-aceyltransferase, and a type VI 3-N-aceyltransferase have all been described in E. coli isolated from animals and are located on plasmids (Hedges and Shannon, 1984; Chaslus-Dancla et al., 1987; Johnson et al., 2006a,b, 2011a). Type IV 3-N-aceyltransferase was first detected in bovine E. coli in France in 1984 and conferred resistance to gentamicin and apramycin. Type II 3-N-aceyltransferase was identified a short time later also from bovine isolates. This enzyme was different from the previously described type IV 3-N-aceyltransferase which only mediated resistance to gentamicin. A 241-kb plasmid was detected in avian pathogenic E. coli isolates (APEC) in 2006 which encoded resistance to gentamicin via aac(3)-VI and streptomycin via aadA (an adenyltransferase), and additionally conferred resistance to potassium tellurite, silver nitrate, copper sulfate, tetracycline, benzylkonium chloride, and sulfisoxazole (Johnson et al., 2006b). The dissemination of these genes is most likely due to their location on plasmids most of which are capable of mobilization within different E. coli strains and to other bacteria as well (Johnson et al., 2006b).
β-lactam and cephem antimicrobials are well-tolerated and present few side-effects in food animals, which make them useful for treatment of disease (Prescott, 2000c,d,g). They have been used to treat E. coli infections in poults as well as diarrhea caused by E. coli in both neonatal calves and swine. Use of these antimicrobials in food animals has been controversial because they are also very useful in human medicine and represent some of the most critically important antimicrobials used in humans. Resistance to β-lactams and cephems in E. coli isolated from U.S. animals has been reported (Li et al., 2007). As reported by NARMS, resistance to β-lactams in E. coli isolated from chickens has remained below 26% between the years 2000–2009 (http://www.ars.usda.gov/SP2UserFiles/Place/66120508/NARMS/NARMS2009/Table3C.pdf). Resistance to amoxicillin-clavulanic acid, cefoxitin, ceftriaxone, and ceftiofur for 2009 was 12.4, 11.4, 11.5, and 9.5%, respectively. Resistance to ampicillin ranged from 20% in year 2000 to 19.8% in year 2009 with the highest resistance reported in year 2006 at 25.6%.
Resistance genes detected in E. coli isolated from animals are not markedly different from those reported in Salmonella from animals (see previous section, Tables 2, 3) with one exception. E. coli harbors an endogenous ampC gene located on the chromosome, whereas this gene is absent in Salmonella (Li et al., 2007). Expression of the “classic” AmpC β-lactamase is not inducible due to the absence of a regulatory gene and is produced at low-levels in the wild-type strains. In contrast, hyper-producers of AmpC β-lactamase have been detected that confer resistance to ampicillin and some cephalosporins (Li et al., 2007). Several factors that influence hyper-production of β-lactamase have been described and many of the hyper-producers have mutations in the promoter region of ampC (Siu et al., 2003). A possible homolog to the “classic” ampC in E. coli has recently been identified in S. Typhimurium, but only shares 11% identity with the “classic” ampC β-lactamase (McClelland et al., 2001). The ability of this gene to confer functional β-lactam resistance in S. Typhimurium has not been demonstrated.
Because they have been in use for a long time, resistance to sulfonamides is widespread. Over 50% of E. coli isolated from poultry samples by the NARMS program was resistant to sulfisoxazole in year 2009. Conversely, resistance to the combination drug trimethoprim-sulfamethoxazole in poultry E. coli was only 7% in year 2009 (http://www.ars.usda.gov/SP2UserFiles/Place/66120508/NARMS/NARMS2009/Table3C.pdf). Resistance to the sulfonamides can be conferred by chromosomal mutations or by resistance genes, sul1, sul2, and sul3, which results in antimicrobial-resistant variants of the target enzymes as described in the Salmonella section and Tables 2, 3 (Skold, 2001). Widespread dissemination of the resistance genes by plasmids, integrons, or insertion elements is evidenced by detection of sulfonamide resistance in E. coli in humans and food animals in both the U.S. and Europe (Hammerum and Heuer, 2009; Frye et al., 2011; Johnson et al., 2011a; Lindsey et al., 2011b; Glenn et al., 2012). Several mechanisms of resistance to trimethoprim have been identified, including efflux pumps, impaired drug penetration, and mutation in the target enzymes (Skold, 2001). But, as seen for sulfonamide resistance, trimethoprim resistance genes (dfr) located on plasmids or transposons-encoding variants to dihydrofolate reductase are predominant.
Chloramphenicol is banned for use in U.S. food animals, and resistance is typically low (less than 5% for years 2000–2009 in E. coli from poultry from the NARMS program). Several resistance genes have been described that confer resistance to chloramphenicol (White et al., 2000). The plasmid-mediated chloramphenicol acetyltransferase (cat) gene is the most common mechanism (Table 3) (Glenn et al., 2012). A non-enzymatic resistance gene encoding an efflux system, cmlA, has also been identified in E. coli plasmids. A third chloramphenicol resistance gene, flo, is also plasmid-encoded and confers resistance to chloramphenicol and florfenicol, a fluorinated structural analog of thiamphenicol and chloramphenicol. In the U.S., florfenicol was approved for use in cattle in 1996 for treatment of respiratory disease (Keyes et al., 2000). Florfenicol resistance has since been detected in clinically ill cattle and chickens in the U.S. (Keyes et al., 2000; White et al., 2000).
Mechanisms of fluoroquinolone resistance in E. coli and Salmonella have been reviewed (Hopkins et al., 2005). Resistance to fluoroquinolones can be due to decreased permeability of the antimicrobial to the cell, efflux pumps, or mutations in DNA gyrase or topoisomerase genes' QRDR motifs (Hopkins et al., 2005). E. coli differs from Salmonella in that it readily mutates to become resistant under selective pressure, and will maintain the mutations and resistance without selective pressure as there is no apparent fitness cost associated with maintaining resistance in E. coli (Bagel et al., 1999; Giraud et al., 1999, 2003; Hopkins et al., 2005). Plasmids harboring quinolone resistance genes (qnrA, qnrB, and qnrS) have also been described, but have not been detected in isolates from U.S. animals. Enrofloxacin and sarafloxacin were approved for use in poultry production in 1995 and 1996, respectively, but this approval was withdrawn in 2005 (Iovine and Blaser, 2004a,b). However, monitoring showed that only 26 of 14,398 (0.18%) E. coli isolated from chickens by NARMS from 2000 to 2009 were ciprofloxacin resistant and genetic screening did not detect any known horizontally exchanged quinolone resistance genes (http://www.ars.usda.gov/SP2UserFiles/Place/66120508/NARMS/NARMS2009/Table3C.pdf).
Tetracyclines have been used for decades in both human and animal medicine and resistance genes are easily acquired. During years 2000–2009, the lowest percentage of resistance among E. coli from poultry was 40.2% in 2007 with the highest level of resistance recorded at 68.4% in year 2000 (http://www.ars.usda.gov/SP2UserFiles/Place/66120508/NARMS/NARMS2009/Table3C.pdf). As described for Salmonella, tetracycline acts by inhibiting protein synthesis in the bacterial cell. Although the most common acquired mechanisms of tetracycline resistance are efflux pumps and ribosomal protection, the efflux pumps are more prevalent in E. coli (Roberts, 2002). To date, nine genes have been identified in E. coli isolates that encode proteins for active efflux of tetracycline and/or its derivatives. These include tet(A), tet(B), tet(C), tet(D), tet(E), tet(G), tet(J), tet(L), and tet(Y) (Roberts, 2002, 2005). Resistance genes identified in E. coli isolated from chickens in the U.S. are shown in Table 3 (Frye et al., 2011; Glenn et al., 2012). Two genes which encode proteins for ribosomal protection, tet(M) and tet(W), have also been detected in E. coli. However, the tet(M) gene may not have a major role in conferring tetracycline resistance in E. coli as it has been reported to only confer low-levels of resistance to tetracycline (Roberts, 2002).
Although primarily defined as a commensal organism, Enterococcus is a Gram-positive bacterium that has an additional role as an opportunistic pathogen causing infections in both humans and animals (Martone, 1998; Cetinkaya et al., 2000; Kuhn et al., 2000). Enterococci are a common inhabitant of the intestinal tract of humans and animals, but have also been isolated from vegetation, soil, water, and food (Niemi et al., 1993; Svec and Sedlacek, 1999; Muller et al., 2001; Giraffa, 2002). Their presence in the digestive tracts of humans and animals coupled with the available methods that exist for molecular typing of the group of bacteria make them useful as an indicator of fecal contamination (Svec and Sedlacek, 1999; Scott et al., 2005; Layton et al., 2010).
An opportunistic pathogen, enterococci are only second to staphylococci as a leading cause of nosocomial infections, accounting for ~12% of hospital-associated infections yearly in the U.S. (Hidron et al., 2008). The majority of infections are caused by two enterococcal species, Enterococcus faecium and E. faecalis (Huycke et al., 1998). The enterococci have been implicated in a number of human clinical diseases including endocarditis, bacteremia, and urinary tract infections (Jett et al., 1994; Huycke et al., 1998). The majority of enterococcal infections are associated with devices used in hospital settings such as central-lines and catheters, but they are also a common cause of surgical site infections (Hidron et al., 2008). Complicating treatment of enterococcal nosocomial infections is the tendency of the bacterium to harbor AR genes conferring resistance to antimicrobials, such as vancomycin, used to treat enterococcal infections. Furthermore, enterococci are also able to transfer AR genes and some virulence factors to other members of the intestinal microflora, including pathogenic bacteria which increase the risk of resistant nosocomial pathogens (Murray, 1990; Chow et al., 1993; Wirth, 1994; Hancock and Gilmore, 2000).
In addition to causing infections in humans, enterococci have been indicated in infections in animals including food animals such as poultry and cattle (Martone, 1998; Cetinkaya et al., 2000; Kuhn et al., 2000). In poultry, enterococcal species may change over time in the chicken gut and enterococcal infections in poultry can be caused by any of the species that are commonly found in the intestines of the birds; although infections in poultry are sporadic, they can be lethal. Pulmonary hypertension syndrome, amyloid athropathy, bacteremia, encephalomalacia, neurological disorders, and endocarditis have all been described in poultry associated with infection by E. faecalis, E. durans, and E. hirae (Randall et al., 1993; McNamee and King, 1996; Tankson et al., 2001; Steentjes et al., 2002). In dairy cattle, enterococci are primarily associated with bovine mastitis although enterococcal-induced diarrhea in calves has also been reported (Rogers et al., 1992; Madsen et al., 2000). Staphylococci are a major cause of bovine mastitis, but enterococci were implicated in 2–20% of cases where an etiological agent has been identified (Poutrel and Ryniewicz, 1984; Aarestrup et al., 1995; Sobiraj et al., 1997). The route of transmission of enterococci in bovine mastitis is most likely from the environment to the animal as animal to animal infections have not been reported (Rossitto et al., 2002).
While their role as an opportunistic nosocomial pathogen has been well-documented, their ability to cause food-borne illnesses remains largely unknown. While enterococci have been reported to cause diarrhea in animals, this has not been proven in humans. In humans, vomiting and headaches indicative of food intoxication are believed to be caused by the ingestion of fermented food containing enterococci which have produced biogenic amines (Tham et al., 1990; Gardin et al., 2001; Giraffa, 2002). The safety of using enterococci in food production has not been determined as they may be both beneficial as well as detrimental in food processing. In fermented foods, enterococci are essential in manufacturing fermented milk products such as cheeses due to the specific biochemical traits that they possess. Alternatively, they may also be indicative of food spoilage for fermented meats or unsanitary conditions in other food industries (Giraffa, 2002; Foulquie Moreno et al., 2006). Determination of innate traits of the enterococci such as AR and virulence need to be addressed before the safety of using enterococci in food production can be determined.
Intrinsic resistance to antimicrobial agents used in hospital settings is a common characteristic of enterococci compared to other bacteria primarily found there (Facklam et al., 2002; Malani et al., 2002). Enterococcal infections caused by antimicrobial resistant isolates, including MDR isolates, are more serious and difficult to treat than those caused by susceptible isolates. Some enterococcal species, particularly E. faecium, are inherently resistant to some penicillins; and in the past few years, they have also shown increased resistance to vancomycin, cephalosporins, and aminoglycosides in nosocomial infections (Arias et al., 2010). Vancomycin is often considered the last treatment available in serious, MDR infections in humans (Wilson et al., 1995; Marshall et al., 1998; Boneca and Chiosis, 2003). Newer drugs including daptomycin, linezolid, Quinupristin/Dalfopristin, and tigecycline have been developed recently to combat infections caused by Gram-positive bacteria and appear to be promising in the treatment of infections caused by enterococci (Swaney et al., 1998; Projan, 2000; Hancock, 2005; Shoemaker et al., 2006).
Resistance of enterococci in food animals in the U.S. is very similar to what has been described of enterococci isolated from nosocomial infections. Resistance to aminoglycosides, lincosamides, macrolides, nitrofurans, penicillins, quinolones, streptogramins, tetracycline, and rarely vancomycin has been described in poultry, swine, and cattle (Table 4) (Jackson, 2004a,b, 2005, 2011; Jackson et al., 2007; Donabedian et al., 2010; Hammerum et al., 2010; Frye et al., 2011; Marshall and Levy, 2011). With the exception of daptomycin, resistance to the newer antimicrobials (linezolid and tigecycline) has not been detected in food animals (Jackson, 2011).
According to available data from NARMS, resistance in enterococci from chickens to gentamicin, kanamycin, and streptomycin ranged from 18.6 to 22.3, 29.2 to 32.1, and 14.3 to 20.7%, respectively (http://www.ars.usda.gov/SP2UserFiles/Place/66120508/NARMS/percent_resistance/ENT-RSummary.pdf). Acquired resistance to aminoglycosides is mediated by enzymes categorized as acetyltransferases, adenyltransferases, or phosphotransferases which modify the antimicrobial and is described above (Chow, 2000). Seven aminoglycoside resistance genes have been detected in enterococci from food animals in both the U.S. and Europe (Table 4). Of those, the bifunctional gene aac(6′)-Ie-aph(2″)-Ia, is considered the most important in enterococci as it confers resistance to all aminoglycosides used to treat infections including gentamicin, kanamycin, and streptomycin (Aarestrup, 2000a; Chow, 2000). Alleles of aph(2″) including aph(2″)-Ib, aph(2″)-Ic, and aph(2″)-Id confer high-level resistance to gentamicin (MIC ≥ 500 μg/ml). Another allele, aph(2″)-Ie, was only recently detected in 2006 in E. casseliflavus isolated from a hospitalized patient in China (Chen et al., 2006; Watanabe et al., 2009). The deduced amino acid sequence of aph(2″)-Ie was almost 94% identical to the amino acid of aph(2″)-Id. Prevalence of these aph(2″) alleles in enterococci from both humans and animals remains largely undetermined as some reports suggest that they are present in large numbers of enterococci from humans while data from other studies indicate that they are not as widespread in animals (Chow, 2000; Jackson, 2004b, 2005). These discrepancies may be explained by the recent discovery of most of the genes conferring high-level resistance to gentamicin (Chow et al., 1997; Chow, 2000; Chen et al., 2006). It is possible that while they may exist, they may not yet be as widely disseminated as aac(6′)-Ie-aph(2″)-Ia.
Compared to gentamicin, fewer aminoglycoside resistance genes have been described that confer resistance to kanamycin and streptomycin. Kanamycin-resistance can be mediated by either aph(3″)-IIIa or aac(6′)-Ii both of which have been detected in humans and animals in the U.S. (Chow, 2000; Jackson, 2004b, 2005). The high prevalence of aac(6′)-Ii in E. faecium is most likely due to its presence on the chromosome of all E. faecium isolates in both humans and animals. Similarly, intrinsic resistance to streptomycin also exists in enterococci, but it is not mediated by a foreign gene; ribosomal mutations can occur rendering streptomycin resistance levels ≤100 μg/ml (Chow, 2000). Acquired resistance to streptomycin is encoded by ant(3″)-Ia (aadA) or ant(6′)-Ia (aadE). Presently, studies have been performed that have detected multiple aminoglycoside resistance genes in enterococci from human and animal sources (Chow, 2000; Jackson, 2004b, 2005). The number of aminoglycoside resistance genes detected to date suggests that this trend of multiple aminoglycoside resistance will continue.
Macrolide, lincosomide, and streptogramin antimicrobials act by binding to the ribosome and preventing protein synthesis (Roberts, 2004). Macrolides and lincosamides are not used to treat human enterococcal infections; they are used in treatment of other bacterial infections and may be substituted in place of other antimicrobials due to allergic reactions (Clermont and Horaud, 1990). In food animals, both macrolides and lincosamides are used to treat infections or as growth promoters (Prescott, 2000f). Lincosamides are used in combination with an aminoglycoside to treat mastitis in dairy cattle in some European countries (De Oliveira et al., 2000). Similarly, lincosamides were used to treat mastitis in dairy cattle on farms in the U.S. (Jackson, 2011). In swine, both lincomycin and the macrolide tylosin are used to treat swine arthritis, ileitis, erysipelas, pneumonia, and swine dysentery; lincomycin is also used to prevent swine dysentery by medicating herds via their drinking water (Prescott, 2000b). Tylosin is also used as medication in the feed of swine in the U.S. to improve feed efficiency and increase weight gain; this practice was banned by the European Union in 1999 when antimicrobials were no longer used as growth promoters (Aarestrup et al., 2001). The ban on growth promoters in Europe also ended the use of virginiamycin, a streptogramin antimicrobial. Virginiamycin has been used in animal production as a growth promoter in the U.S. for decades; it is used in both poultry and swine to increase feed efficiency and promote growth (McEwen and Fedorka-Cray, 2002). More recently, a human analogue of virginiamycin, Synercid® (Quinupristin/Dalfopristin-Q/D) a combination of streptogramin A and B antimicrobials, was approved for use in treatment of vancomycin-resistant enterococci (VRE) in humans (Hancock, 2005). Although virginiamycin has only been used in animals and Q/D in humans, cross-resistance is of particular concern as streptogramin resistance genes can confer resistance to both antimicrobials.
Resistance to macrolides in human clinical enterococci and enterococci from animal sources in Europe as well as the U.S. has been reported (Collignon et al., 2009). Acquired resistance to the macrolides can be due to alteration of the antimicrobial, pumping of the antimicrobial from the cell, or modification of the target. The most common mechanism appears to be target modification by erythromycin resistance methylase (erm) genes, primarily erm(B) (Mlynarczyk et al., 2010). The erm(B) gene confers cross resistance to macrolide, lincosamide, and streptogramin type B antimicrobials, which is characteristic of the macrolide-lincosamide-streptogramin type B (MLSB) phenotype (Roberts et al., 1999).
Other macrolide resistance genes which have been detected in the enterococci include erm(A) and msrC, which is described as an ATP-binding transporter belonging to the efflux pump family of genes (Roberts et al., 1999; Werner et al., 2001; Roberts, 2004; Schwaiger and Bauer, 2008). For lincomycin resistance, lnu(B) is the only resistance gene described thus far for Enterococcus. The first lincomycin resistance gene, lnu(A), was originally described in the staphylococci; to our knowledge lnu(B) has not been detected in enterococci from food animals, but it has been detected in enterococci isolated from companion animals (Jackson et al., 2009).
For streptogramin resistance, resistance to the A component is the sole requirement for resistance to both components of the streptogramin A and B combination (Rende-Fournier et al., 1993; Soltani et al., 2000). Higher levels of resistance to streptogramins have been reported as a result of the presence of resistance genes to each single component. Resistance to the B component (quinupristin) in enterococci is mediated by erm genes and msrC described above. Resistance to the A component (dalfopristin) in the enterococci is mediated by streptogramin A acetyltransferases encoded by vat(B), vat(D), vat(E), or vat(G), ATP-binding transporters encoded by vga(B), or hydrolases encoded by vgb(A) (Hammerum et al., 1998; Jensen et al., 1998; Werner and Witte, 1999; Hershberger, 2004; Roberts, 2004; Jung et al., 2010). The presence of resistance genes to streptogramin A and B antimicrobials differs slightly between Europe and the U.S. Previously, erm(B), msrC, vat(D), and vat(E) had all been identified in enterococci from humans, animals, and the environment in Europe; three of the four genes [erm(B), msrC, and vat(E)] had also been identified in enterococci in the U.S. (Hershberger, 2004). The fourth gene, vat(D), was only recently found in E. faecium and E. hirae isolated from chicken carcass rinsates in the U.S. (Jackson et al., 2007). Two newly identified streptogramin resistance genes, vat(G) and vga(D), were also recently characterized in E. faecium isolated from the stool of a healthy human from Korea (Jung et al., 2010). Both genes were detected together on the same plasmid, but have yet to be detected in the U.S. Two additional streptogramin A resistance genes, vat(B) and vga(B), encoding an acetyltransferase and ATP binding transporter, respectively, were identified in E. gallinarum from chicken carcass rinsates in 2008 in the U.S. (Jackson et al., 2008). Both vat(B) and vga(B) were originally described in a clinical isolate of S. aureus from France (Allignet and El Solh, 1995, 1997). Although those genes were located on a plasmid in both S. aureus and E. gallinarum, plasmid pIP1633 from S. aureus is conjugal whereas the E. gallinarum plasmid containing vat(B) and vga(B) is not (Jackson et al., 2008). This was the first report of these genes in Enterococcus worldwide. Additional resistance mechanisms to the streptogramin antimicrobials may exist although they have yet to be identified and characterized (McDermott et al., 2005). Streptogramin resistance in E. faecalis is typically not reported due the presence of an intrinsic E. faecalis species-specific gene, lsa (lincosamides-streptogramin A; Singh et al., 2002).
Tetracycline or the semisynthetic derivative, doxycycline, is used in food animals mainly in treatment of respiratory infections (Mathers et al., 2011). Doxycycline has been used rarely to treat VRE infections in humans possibly due to the high numbers of antimicrobial resistant clinical isolates (Landman and Quale, 1997; Matsumura and Simor, 1998). Overall, tetracycline resistance in enterococci from both humans and animals is widespread and has been previously reviewed (Roberts, 2005). Resistance to tetracycline in enterococci is largely due to ribosomal protection or efflux of the antimicrobial. The most common tetracycline resistance gene is tet(M) which encodes proteins for ribosomal protection. The location of this gene in enterococci includes the chromosome, conjugal transposons, Tn916, as well as conjugal plasmids all of which may account for its prevalence. Two additional genes, tet(O) and tet(S) also confer resistance to tetracycline via ribosomal protection and have been detected in enterococci from food animals (Aarestrup et al., 2002).
The tet(L) gene is the most frequently detected tetracycline efflux gene in the enterococci (Bentorcha et al., 1991; Platteeuw et al., 1995). Like tet(M), it has also been localized on the chromosome and plasmids in enterococci. A second tetracycline efflux gene, tet(K), has also been described in Enterococcus (Roberts, 2005; Fard et al., 2011). Previously, a sixth tetracycline resistance gene, tet(U) was detected in E. faecium, but recent reports suggest that tet(U) does not confer resistance to tetracycline in enterococci, but may instead be part of a gene encoding a replication initiator protein (Caryl et al., 2012). New tetracycline derivatives, glycylcyclines, have recently been developed; the first of these is tigecycline (Projan, 2000). For enterococcal infections, tigecycline has been approved for treatment of complicated skin and skin structure infections and complicated intra-abdominal infections caused by vancomycin-susceptible E. faecalis. The safety and efficacy of this drug is still being evaluated as reports suggest that there may be an increased risk of mortality when using tigecycline (Yahav et al., 2011). Tigecycline is not approved for use in food animals.
Glycopeptides such as vancomycin bind to peptidoglycan cell wall components and inhibit further synthesis of the bacterial cell wall resulting in their antimicrobial effect. In the U.S., neither vancomycin nor the glycopeptide-related compound, avoparcin, has been approved for use in food animals. Until the European ban on use of growth promoters in food animals, avoparcin was used in European countries for growth promotion (Aarestrup and Seyfarth, 2000; Aarestrup et al., 2000, 2001). The differences in the use of glycopeptide antimicrobials in food animal production in the two regions most likely account for the differences in glycopeptide resistance observed in food animals, but not in humans. In Europe, vancomycin resistance has been found in humans, animals, and the environment (Werner et al., 2008). In contrast, vancomycin resistance in the U.S. was confined to humans where VRE was a leading cause of MDR healthcare-associated infections (Hidron et al., 2008). In a study of healthcare-associated infections conducted by the U.S. CDC, the percentage of VRE in the U.S. healthcare system (33%) was higher than that in Europe and the rest of North America (range 13–28%; Hidron et al., 2008). More specific, the percentages of vancomycin-resistant E. faecium pathogenic isolates were also higher in the U.S. (80%) compared to France (0.8%) and Italy (24%). E. faecium is more likely than other enterococcal species to harbor vancomycin resistance (Donabedian et al., 2010).
The mechanisms conferring resistance to vancomycin have been previously reviewed (Courvalin, 2006). Briefly, the predominant vancomycin-resistance gene, vanA, is an acquired resistance gene and confers resistance to vancomycin and teicoplanin. This gene encodes an enzyme that offers an alternative pathway for peptidoglycan cell wall synthesis that circumvents the obstruction created by glycopeptide antimicrobials bound to the cell wall components. The vanB gene is also acquired, but confers resistance to vancomycin only. Both genes have an inducible phenotype and can be located on the chromosome or plasmids. Conversely, vanC (vanC1, C2, or C3), an intrinsic gene localized to the chromosome in either E. casseliflavus, E. gallinarum, or E. flavescens, mediates lower levels of resistance (MIC 2–32 μg/ml) to vancomycin only. Several new vancomycin-resistance genes have recently been identified. These include vanD, vanE, vanG, vanL, vanM, and vanN (Courvalin, 2006; Boyd et al., 2008; Xu et al., 2010; Lebreton et al., 2011). Both vanD and vanM encode D-Ala-D-Lac ligase while vanE, vanG, vanL, and vanN encode D-Ala-D-Ser ligase. In addition to the modified target, all of the genes identified to date can be distinguished from each other based upon a number of characteristics including whether they are acquired or intrinsic, the level of resistance to vancomycin and/or teicoplanin, the expression of the resistance (constitutive or inducible), the location of the resistance operon, and the ability of the genes to transfer to other enterococci (Courvalin, 2006). Although the van gene cluster organization of vanM is most similar to that of vanD, the two genes are characteristically different. While vanD confers intermediate resistance to vancomycin and teicoplanin, is located on the chromosome and is not transferable by conjugation, vanM confers high-level resistance to both vancomycin and teicoplanin, is located on a plasmid, and is transferable (Courvalin, 2006; Xu et al., 2010; Nilsson, 2012). Vancomycin genes vanE, vanG, and vanL confer low level resistance to vancomycin and susceptibility to teicoplanin (Courvalin, 2006; Xu et al., 2010; Nilsson, 2012). All three genes have inducible resistance and are located on the chromosome, but are not mobile. The newest vancomycin-resistance gene, vanN, confers resistance to vancomycin only and is the only D-Ala-D-Ser ligase gene that is transferable by conjugation (Lebreton et al., 2011). Until recently, vancomycin-resistance in food animals had not been observed in the U.S.; the first report of vancomycin-resistance in food animals in the U.S. was published in 2010 in which vancomycin-resistant E. faecium (MIC ≥ 256 μg/ml) were isolated from swine in Michigan (Donabedian et al., 2010). Those isolates contained vanA located on the Tn1546 transposon as previously described for enterococci containing vanA. The origin of vanA in the Michigan swine samples remains undetermined. No other vancomycin-resistance genes have been identified in the U.S.
Globally, resistance to antimicrobials appears to be increasing in commensal and environmental isolates as well as in pathogens including foodborne pathogens. This may cause difficulty in treating human and animal infections in the future and merits further surveillance and analyses. Salmonella isolated from human and animal infections and also from healthy animals in the U.S. have acquired additional resistance to antimicrobials since the 1990's. Resistance to some antimicrobials appears to have moderated in recent years; however, monitoring will need to be maintained to determine if this trend will continue. In addition, resistance to a number of antimicrobials has also been detected in commensal bacteria and opportunistic pathogens, E. coli and Enterococcus, isolated from food animals in the U.S. There is also evidence that these and other commensal organisms may serve as reservoirs for AR genes and may transfer these to Salmonella and other pathogens. Compounding resistance concerns, international travel and trade makes it likely that resistance mechanism found in other areas of the world may eventually be found in the U.S., as seen for the emergence of ESBLs or vancomycin resistance. The recent identification of the blaMDM−1 gene in Salmonella isolated from a human in the U.S. presents the possibility of a virtually untreatable Salmonella infection and underscores these concerns (Savard et al., 2011). Research is ongoing to determine the sources of AR in animals and humans, and the results from these studies will enable an understanding of the complex events behind AR. Analysis of these results may aid in the development of practices that will prevent resistance or slow its spread and thus reduce its impact on human and animal health.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to thank Mark Englen for helpful discussions on the topics of this review. Funding provided by U.S. Department of Agriculture, Agricultural Research Service, project number: 6612-32000-006-00. Note: The mention of trade names or commercial products in this manuscript is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Aarestrup, F. M. (1999). Association between the consumption of antimicrobial agents in animal husbandry and the occurrence of resistant bacteria among food animals. Int. J. Antimicrob. Agents 12, 279–285.
Aarestrup, F. M. (2000a). Comparison of antimicrobial resistance phenotypes and resistance genes in Enterococcus faecalis and Enterococcus faecium from humans in the community, broilers, and pigs in Denmark. Diagn. Microbiol. Infect. Dis. 37, 127–137.
Aarestrup, F. M. (2000b). Occurrence, selection and spread of resistance to antimicrobial agents used for growth promotion for food animals in Denmark. APMIS Suppl. 101, 1–48.
Aarestrup, F. M. (2004). Monitoring of antimicrobial resistance among food animals: principles and limitations. J. Vet. Med. B Infect. Dis. Vet. Public Health 51, 380–388.
Aarestrup, F. M. (2005). Veterinary drug usage and antimicrobial resistance in bacteria of animal origin. Basic Clin. Pharmacol. Toxicol. 96, 271–281.
Aarestrup, F. M., Dangler, C. A., and Sordillo, L. M. (1995). Prevalence of coagulase gene polymorphism in Staphylococcus aureus isolates causing bovine mastitis. Can. J. Vet. Res. 59, 124–128.
Aarestrup, F. M., Hasman, H., Jensen, L. B., Moreno, M., Herrero, I. A., Dominguez, L., et al. (2002). Antimicrobial resistance among enterococci from pigs in three European countries. Appl. Environ. Microbiol. 68, 4127–4129.
Aarestrup, F. M., Kruse, H., Tast, E., Hammerum, A. M., and Jensen, L. B. (2000). Associations between the use of antimicrobial agents for growth promotion and the occurrence of resistance among Enterococcus faecium from broilers and pigs in Denmark, Finland, and Norway. Microb. Drug Resist. 6, 63–70.
Aarestrup, F. M., and Seyfarth, A. M. (2000). Effect of intervention on the occurrence of antimicrobial resistance. Acta Vet. Scand. Suppl. 93, 99–102.
Aarestrup, F. M., Seyfarth, A. M., Emborg, H. D., Pedersen, K., Hendriksen, R. S., and Bager, F. (2001). Effect of abolishment of the use of antimicrobial agents for growth promotion on occurrence of antimicrobial resistance in fecal enterococci from food animals in Denmark. Antimicrob. Agents Chemother. 45, 2054–2059.
Aarestrup, F. M., and Wegener, H. C. (1999). The effects of antibiotic usage in food animals on the development of antimicrobial resistance of importance for humans in Campylobacter and Escherichia coli. Microbes Infect. 1, 639–644.
Aarestrup, F. M., Wegener, H. C., and Collignon, P. (2008). Resistance in bacteria of the food chain: epidemiology and control strategies. Expert Rev. Anti Infect. Ther. 6, 733–750.
Ajiboye, R. M., Solberg, O. D., Lee, B. M., Raphael, E., Debroy, C., and Riley, L. W. (2009). Global spread of mobile antimicrobial drug resistance determinants in human and animal Escherichia coli and Salmonella strains causing community-acquired infections. Clin. Infect. Dis. 49, 365–371.
Akkina, J. E., Hogue, A. T., Angulo, F. J., Johnson, R., Petersen, K. E., Saini, P. K., et al. (1999). Epidemiologic aspects, control, and importance of multiple-drug resistant Salmonella Typhimurium DT104 in the United States. J. Am. Vet. Med. Assoc. 214, 790–798.
Alanis, A. J. (2005). Resistance to antibiotics: are we in the post-antibiotic era? Arch. Med. Res. 36, 697–705.
Alcaine, S. D., Warnick, L. D., and Wiedmann, M. (2007). Antimicrobial resistance in nontyphoidal Salmonella. J. Food Prot. 70, 780–790.
Allen, C. A., Fedorka-Cray, P. J., Vazquez-Torres, A., Suyemoto, M., Altier, C., Ryder, L. R., et al. (2001). In vitro and in vivo assessment of Salmonella enterica serovar Typhimurium DT104 virulence. Infect. Immun. 69, 4673–4677.
Allignet, J., and El Solh, N. (1995). Diversity among the gram-positive acetyltransferases inactivating streptogramin A and structurally related compounds and characterization of a new staphylococcal determinant, vatB. Antimicrob. Agents Chemother. 39, 2027–2036.
Allignet, J., and El Solh, N. (1997). Characterization of a new staphylococcal gene, vgaB, encoding a putative ABC transporter conferring resistance to streptogramin A and related compounds. Gene 202, 133–138.
Anthony, F., Acar, J., Franklin, A., Gupta, R., Nicholls, T., Tamura, Y., et al. (2001). Antimicrobial resistance: responsible and prudent use of antimicrobial agents in veterinary medicine. Rev. Sci. Tech. 20, 829–839.
Arias, C. A., Contreras, G. A., and Murray, B. E. (2010). Management of multidrug-resistant enterococcal infections. Clin. Microbiol. Infect. 16, 555–562.
Arias, C. A., and Murray, B. E. (2012). The rise of the Enterococcus: beyond vancomycin resistance. Nat. Rev. Microbiol. 10, 266–278.
Arlet, G., Barrett, T. J., Butaye, P., Cloeckaert, A., Mulvey, M. R., and White, D. G. (2006). Salmonella resistant to extended-spectrum cephalosporins: prevalence and epidemiology. Microbes Infect. 8, 1945–1954.
Avrain, L., Vernozy-Rozand, C., and Kempf, I. (2004). Evidence for natural horizontal transfer of tetO gene between Campylobacter jejuni strains in chickens. J. Appl. Microbiol. 97, 134–140.
Bagel, S., Hullen, V., Wiedemann, B., and Heisig, P. (1999). Impact of gyrA and parC mutations on quinolone resistance, doubling time, and supercoiling degree of Escherichia coli. Antimicrob. Agents Chemother. 43, 868–875.
Bager, F., Aarestrup, F. M., Jensen, N. E., Madsen, M., Meyling, A., and Wegener, H. C. (1999a). Design of a system for monitoring antimicrobial resistance in pathogenic, zoonotic and indicator bacteria from food animals. Acta Vet. Scand. Suppl. 92, 77–86.
Bager, F., Aarestrup, F. M., Madsen, M., and Wegener, H. C. (1999b). Glycopeptide resistance in Enterococcus faecium from broilers and pigs following discontinued use of avoparcin. Microb. Drug Resist. 5, 53–56.
Baggesen, D. L., Sandvang, D., and Aarestrup, F. M. (2000). Characterization of Salmonella enterica serovar Typhimurium DT104 isolated from Denmark and comparison with isolates from Europe and the United States. J. Clin. Microbiol. 38, 1581–1586.
Barton, M. D., Pratt, R., and Hart, W. S. (2003). Antibiotic resistance in animals. Commun. Dis. Intell. 27(Suppl.), S121–S126.
Batchelor, M., Threlfall, E. J., and Liebana, E. (2005). Cephalosporin resistance among animal-associated Enterobacteria: a current perspective. Expert Rev. Anti Infect. Ther. 3, 403–417.
Bentorcha, F., De, C. G., and Horaud, T. (1991). Tetracycline resistance heterogeneity in Enterococcus faecium. Antimicrob. Agents Chemother. 35, 808–812.
Berge, A. C., Atwill, E. R., and Sischo, W. M. (2005). Animal and farm influences on the dynamics of antibiotic resistance in faecal Escherichia coli in young dairy calves. Prev. Vet. Med. 69, 25–38.
Bettelheim, K. A. (2007). The non-O157 shiga-toxigenic (verocytotoxigenic) Escherichia coli; under-rated pathogens. Crit. Rev. Microbiol. 33, 67–87.
Bhan, M. K., Bahl, R., and Bhatnagar, S. (2005). Typhoid and paratyphoid fever. Lancet 366, 749–762.
Biedenbach, D. J., Toleman, M., Walsh, T. R., and Jones, R. N. (2006). Analysis of Salmonella spp. with resistance to extended-spectrum cephalosporins and fluoroquinolones isolated in North America and Latin America: report from the SENTRY Antimicrobial Surveillance Program (1997–2004). Diagn. Microbiol. Infect. Dis. 54, 13–21.
Bischoff, K. M., White, D. G., McDermott, P. F., Zhao, S., Gaines, S., Maurer, J. J., et al. (2002). Characterization of chloramphenicol resistance in beta-hemolytic Escherichia coli associated with diarrhea in neonatal swine. J. Clin. Microbiol. 40, 389–394.
Blake, D. P., Hillman, K., Fenlon, D. R., and Low, J. C. (2003). Transfer of antibiotic resistance between commensal and pathogenic members of the Enterobacteriaceae under ileal conditions. J. Appl. Microbiol. 95, 428–436.
Blood, R. M., and Curtis, G. D. (1995). Media for ‘total’ Enterobacteriaceae, coliforms and Escherichia coli. Int. J. Food Microbiol. 26, 93–115.
Boerlin, P. (2004). Molecular epidemiology of antimicrobial resistance in veterinary medicine: where do we go? Anim. Health Res. Rev. 5, 95–102.
Boerlin, P., and Reid-Smith, R. J. (2008). Antimicrobial resistance: its emergence and transmission. Anim. Health Res. Rev. 9, 115–126.
Bolton, L. F., Kelley, L. C., Lee, M. D., Fedorka-Cray, P. J., and Maurer, J. J. (1999). Detection of multidrug-resistant Salmonella enterica serotype typhimurium DT104 based on a gene which confers cross-resistance to florfenicol and chloramphenicol. J. Clin. Microbiol. 37, 1348–1351.
Boneca, I. G., and Chiosis, G. (2003). Vancomycin resistance: occurrence, mechanisms and strategies to combat it. Expert Opin. Ther. Targets 7, 311–328.
Boxrud, D. (2010). Advances in subtyping methods of foodborne disease pathogens. Curr. Opin. Biotechnol. 21, 137–141.
Boxrud, D., Monson, T., Stiles, T., and Besser, J. (2010). The role, challenges, and support of pulsenet laboratories in detecting foodborne disease outbreaks. Public Health Rep. 125(Suppl. 2), 57–62.
Boyd, D., Peters, G. A., Cloeckaert, A., Boumedine, K. S., Chaslus-Dancla, E., Imberechts, H., et al. (2001). Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J. Bacteriol. 183, 5725–5732.
Boyd, D. A., Willey, B. M., Fawcett, D., Gillani, N., and Mulvey, M. R. (2008). Molecular characterization of Enterococcus faecalis N06-0364 with low-level vancomycin resistance harboring a novel D-Ala-D-Ser gene cluster, vanL. Antimicrob. Agents Chemother. 52, 2667–2672.
Bradford, P. A. (2001). Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14, 933–951.
Briggs, C. E., and Fratamico, P. M. (1999). Molecular characterization of an antibiotic resistance gene cluster of Salmonella typhimurium DT104. Antimicrob. Agents Chemother. 43, 846–849.
Bryan, A., Shapir, N., and Sadowsky, M. J. (2004). Frequency and distribution of tetracycline resistance genes in genetically diverse, nonselected, and nonclinical Escherichia coli strains isolated from diverse human and animal sources. Appl. Environ. Microbiol. 70, 2503–2507.
Bush, K. (2008). Extended-spectrum beta-lactamases in North America, 1987–2006. Clin. Microbiol. Infect. 14(Suppl. 1), 134–143.
Call, D. R., Davis, M. A., and Sawant, A. A. (2008). Antimicrobial resistance in beef and dairy cattle production. Anim. Health Res. Rev. 9, 159–167.
Carattoli, A. (2001). Importance of integrons in the diffusion of resistance. Vet. Res. 32, 243–259.
Carattoli, A. (2003). Plasmid-mediated antimicrobial resistance in Salmonella enterica. Curr. Issues Mol. Biol. 5, 113–122.
Carattoli, A. (2008). Animal reservoirs for extended spectrum beta-lactamase producers. Clin. Microbiol. Infect. 14(Suppl. 1), 117–123.
Carattoli, A. (2009). Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 53, 2227–2238.
Carattoli, A., Bertini, A., Villa, L., Falbo, V., Hopkins, K. L., and Threlfall, E. J. (2005). Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63, 219–228.
Carattoli, A., Filetici, E., Villa, L., Dionisi, A. M., Ricci, A., and Luzzi, I. (2002a). Antibiotic resistance genes and Salmonella genomic island 1 in Salmonella enterica serovar Typhimurium isolated in Italy. Antimicrob. Agents Chemother. 46, 2821–2828.
Carattoli, A., Tosini, F., Giles, W. P., Rupp, M. E., Hinrichs, S. H., Angulo, F. J., et al. (2002b). Characterization of plasmids carrying CMY-2 from expanded-spectrum cephalosporin-resistant Salmonella strains isolated in the United States between 1996 and 1998. Antimicrob. Agents Chemother. 46, 1269–1272.
Carattoli, A., Miriagou, V., Bertini, A., Loli, A., Colinon, C., Villa, L., et al. (2006). Replicon typing of plasmids encoding resistance to newer beta-lactams. Emerg. Infect. Dis. 12, 1145–1148.
Caryl, J. A., Cox, G., Trimble, S., and O'Neill, A. J. (2012). “tet(U)” is not a tetracycline resistance determinant. Antimicrob. Agents Chemother. 56, 3378–3379.
Cavaco, L. M., and Aarestrup, F. M. (2009). Evaluation of quinolones for use in detection of determinants of acquired quinolone resistance, including the new transmissible resistance mechanisms qnrA, qnrB, qnrS, and aac(6′)Ib-cr, in Escherichia coli and Salmonella enterica and determinations of wild-type distributions. J. Clin. Microbiol. 47, 2751–2758.
Cavaco, L. M., Hansen, D. S., Friis-Moller, A., Aarestrup, F. M., Hasman, H., and Frimodt-Moller, N. (2007a). First detection of plasmid-mediated quinolone resistance (qnrA and qnrS) in Escherichia coli strains isolated from humans in Scandinavia. J. Antimicrob. Chemother. 59, 804–805.
Cavaco, L. M., Hendriksen, R. S., and Aarestrup, F. M. (2007b). Plasmid-mediated quinolone resistance determinant qnrS1 detected in Salmonella enterica serovar Corvallis strains isolated in Denmark and Thailand. J. Antimicrob. Chemother. 60, 704–706.
Cavaco, L. M., Korsgaard, H., Sorensen, G., and Aarestrup, F. M. (2008). Plasmid-mediated quinolone resistance due to qnrB5 and qnrS1 genes in Salmonella enterica serovars Newport, Hadar and Saintpaul isolated from turkey meat in Denmark. J. Antimicrob. Chemother. 62, 632–634.
Cetinkaya, Y., Falk, P., and Mayhall, C. G. (2000). Vancomycin-resistant enterococci. Clin. Microbiol. Rev. 13, 686–707.
Chan, K., Baker, S., Kim, C. C., Detweiler, C. S., Dougan, G., and Falkow, S. (2003). Genomic Comparison of Salmonella enterica serovars and Salmonella bongori by use of an S. enterica serovar Typhimurium DNA microarray. J. Bacteriol. 185, 553–563.
Chaslus-Dancla, E., Gerbaud, G., Martel, J. L., Lagorce, M., Lafont, J. P., and Courvalin, P. (1987). Detection of a second mechanism of resistance to gentamicin in animal strains of Escherichia coli. Antimicrob. Agents Chemother. 31, 1274–1277.
Chen, C. Y., Strobaugh, T. P. Jr., Lindsey, R. L., Frye, J. G., and Uhlich, G. (2011). Sequence analysis of a group of low molecular-weight plasmids carrying multiple IS903 elements flanking a kanamycin resistance aph gene in Salmonella enterica serovars. Plasmid 65, 246–252.
Chen, S., Cui, S., McDermott, P. F., Zhao, S., White, D. G., Paulsen, I., et al. (2007). Contribution of target gene mutations and efflux to decreased susceptibility of Salmonella enterica serovar typhimurium to fluoroquinolones and other antimicrobials. Antimicrob. Agents Chemother. 51, 535–542.
Chen, Y. G., Qu, T. T., Yu, Y. S., Zhou, J. Y., and Li, L. J. (2006). Insertion sequence ISEcp1-like element connected with a novel aph(2”) allele [aph(2”)-Ie] conferring high-level gentamicin resistance and a novel streptomycin adenylyltransferase gene in Enterococcus. J. Med. Microbiol. 55, 1521–1525.
Chopra, I., and Roberts, M. (2001). Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65, 232–260.
Chow, J. W., Thal, L. A., Perri, M. B., Vazquez, J. A., Donabedian, S. M., Clewell, D. B., et al. (1993). Plasmid-associated hemolysin and aggregation substance production contribute to virulence in experimental enterococcal endocarditis. Antimicrob. Agents Chemother. 37, 2474–2477.
Chow, J. W., Zervos, M. J., Lerner, S. A., Thal, L. A., Donabedian, S. M., Jaworski, D. D., et al. (1997). A novel gentamicin resistance gene in Enterococcus. Antimicrob. Agents Chemother. 41, 511–514.
Clegg, F. G., Chiejina, S. N., Duncan, A. L., Kay, R. N., and Wray, C. (1983). Outbreaks of Salmonella newport infection in dairy herds and their relationship to management and contamination of the environment. Vet. Rec. 112, 580–584.
Clermont, D., and Horaud, T. (1990). Identification of chromosomal antibiotic resistance genes in Streptococcus anginosus (“S. milleri”). Antimicrob. Agents Chemother. 34, 1685–1690.
Cobbold, R. N., Rice, D. H., Davis, M. A., Besser, T. E., and Hancock, D. D. (2006). Long-term persistence of multi-drug-resistant Salmonella enterica serovar Newport in two dairy herds. J. Am. Vet. Med. Assoc. 228, 585–591.
Collignon, P., Powers, J. H., Chiller, T. M., idara-Kane, A., and Aarestrup, F. M. (2009). World Health Organization ranking of antimicrobials according to their importance in human medicine: a critical step for developing risk management strategies for the use of antimicrobials in food production animals. Clin. Infect. Dis. 49, 132–141.
Courvalin, P. (2006). Vancomycin resistance in gram-positive cocci. Clin. Infect. Dis. 42(Suppl. 1), S25–S34.
D'Aoust, J. Y. (1997). “Salmonella species,” in Food Microbiology: Fundamentals and Frontiers, eds M. P. Doyle, L. R. Beauchat, and T. J. Montville (Washington, DC: ASM Press), 129–158.
De Oliveira, A. P., Watts, J. L., Salmon, S. A., and Aarestrup, F. M. (2000). Antimicrobial susceptibility of Staphylococcus aureus isolated from bovine mastitis in Europe and the United States. J. Dairy Sci. 83, 855–862.
Devasia, R. A., Varma, J. K., Whichard, J., Gettner, S., Cronquist, A. B., Hurd, S., et al. (2005). Antimicrobial use and outcomes in patients with multidrug-resistant and pansusceptible Salmonella Newport infections, 2002–2003. Microb. Drug Resist. 11, 371–377.
Donabedian, S. M., Perri, M. B., Abdujamilova, N., Gordoncillo, M. J., Naqvi, A., Reyes, K. C., et al. (2010). Characterization of vancomycin-resistant Enterococcus faecium isolated from swine in three Michigan counties. J. Clin. Microbiol. 48, 4156–4160.
Douard, G., Praud, K., Cloeckaert, A., and Doublet, B. (2010). The Salmonella genomic island 1 is specifically mobilized in trans by the IncA/C multidrug resistance plasmid family. PLoS ONE 5:e15302. doi: 10.1371/journal.pone.0015302
DuPont, H. L., and Steele, J. H. (1987). Use of antimicrobial agents in animal feeds: implications for human health. Rev. Infect. Dis. 9, 447–460.
Eaves, D. J., Randall, L., Gray, D. T., Buckley, A., Woodward, M. J., White, A. P., et al. (2004). Prevalence of mutations within the quinolone resistance-determining region of gyrA, gyrB, parC, and parE and association with antibiotic resistance in quinolone-resistant Salmonella enterica. Antimicrob. Agents Chemother. 48, 4012–4015.
Ebner, P., Garner, K., and Mathew, A. (2004). Class 1 integrons in various Salmonella enterica serovars isolated from animals and identification of genomic island SGI1 in Salmonella enterica var. Meleagridis. J. Antimicrob. Chemother. 53, 1004–1009.
Endtz, H. P., Ruijs, G. J., van, K. B., Jansen, W. H., van der, R. T., and Mouton, R. P. (1991). Quinolone resistance in campylobacter isolated from man and poultry following the introduction of fluoroquinolones in veterinary medicine. J. Antimicrob. Chemother. 27, 199–208.
Erb, A., Sturmer, T., Marre, R., and Brenner, H. (2007). Prevalence of antibiotic resistance in Escherichia coli: overview of geographical, temporal, and methodological variations. Eur. J. Clin. Microbiol. Infect. Dis. 26, 83–90.
Erickson, M. C., and Doyle, M. P. (2007). Food as a vehicle for transmission of Shiga toxin-producing Escherichia coli. J. Food Prot. 70, 2426–2449.
Evershed, N. J., Levings, R. S., Wilson, N. L., Djordjevic, S. P., and Hall, R. M. (2009). Unusual class 1 integron-associated gene cassette configuration found in IncA/C plasmids from Salmonella enterica. Antimicrob. Agents Chemother. 53, 2640–2642.
Facklam, R. R., Carvalho, M. G. S., and Teixeiram, L. M. (2002). “History, taxonomy, biochemical characteristics, and antibiotic susceptibility testing of enterococci,” in The Enterococci: Pathogenesis, Molecular Biology, and Antibiotic Resistance, eds M. S. Gilmore, D. B. Clewell, P. Courvalin, G. M. Dunny, B. E. Murray, and L. B. Rice (Washington, DC: ASM Press), 1–54.
Falagas, M. E., and Karageorgopoulos, D. E. (2009). Extended-spectrum beta-lactamase-producing organ-isms. J. Hosp. Infect. 73, 345–354.
Fard, R. M., Heuzenroeder, M. W., and Barton, M. D. (2011). Antimicrobial and heavy metal resistance in commensal enterococci isolated from pigs. Vet. Microbiol. 148, 276–282.
Fluit, A. C. (2005). Towards more virulent and antibiotic-resistant Salmonella? FEMS Immunol. Med. Microbiol. 43, 1–11.
Foley, S. L., and Lynne, A. M. (2008). Food animal-associated Salmonella challenges: pathogenicity and antimicrobial resistance. J. Anim. Sci. 86, E173–E187.
Folster, J. P., Pecic, G., Bolcen, S., Theobald, L., Hise, K., Carattoli, A., et al. (2010). Characterization of extended-spectrum cephalosporin-resistant Salmonella enterica serovar Heidelberg isolated from humans in the United States. Foodborne Pathog. Dis. 7, 181–187.
Folster, J. P., Pecic, G., McCullough, A., Rickert, R., and Whichard, J. M. (2011). Characterization of bla(CMY)-encoding plasmids among Salmonella isolated in the United States in 2007. Foodborne Pathog. Dis. 8, 1289–1294.
Folster, J. P., Pecic, G., Rickert, R., Taylor, J., Zhao, S., Fedorka-Cray, P. J., et al. (2012). Characterization of multidrug-resistant Salmonella enterica serovar heidelberg from a ground turkey-associated outbreak in the United States in 2011. Antimicrob. Agents Chemother. 56, 3465–3466.
Foulquie Moreno, M. R., Sarantinopoulos, P., Tsakalidou, E., and De Vuyst, L. (2006). The role and application of enterococci in food and health. Int. J. Food Microbiol. 106, 1–24.
Fricke, W. F., Welch, T. J., McDermott, P. F., Mammel, M. K., LeClerc, J. E., White, D. G., et al. (2009). Comparative genomics of the IncA/C multidrug resistance plasmid family. J. Bacteriol. 191, 4750–4757.
Frye, J. G., and Fedorka-Cray, P. J. (2007). Prevalence, distribution and characterisation of ceftiofur resistance in Salmonella enterica isolated from animals in the USA from 1999 to 2003. Int. J. Antimicrob. Agents 30, 134–142.
Frye, J. G., Fedorka-Cray, P. J., Jackson, C. R., and Rose, M. (2008). Analysis of Salmonella enterica with reduced susceptibility to the third-generation cephalosporin ceftriaxone isolated from U.S. cattle during 2000–2004. Microb. Drug Resist. 14, 251–258.
Frye, J. G., Lindsey, R. L., Meinersmann, R. J., Berrang, M. E., Jackson, C. R., Englen, M. D., et al. (2011). Related antimicrobial resistance genes detected in different bacterial species co-isolated from swine fecal samples. Foodborne Pathog. Dis. 8, 663–679.
Gardin, F., Martuscelli, M., Caruso, M. C., Galgano, F., Crudele, M. A., Favati, F., et al. (2001). Effects of pH, temperature and NaCl concentration on the growth kinetics, proteolytic activity and biogenic amine production of Enterococcus faecalis. Int. J. Food Microbiol. 64, 105–117.
Gebreyes, W. A., Thakur, S., Davies, P. R., Funk, J. A., and Altier, C. (2004). Trends in antimicrobial resistance, phage types and integrons among Salmonella serotypes from pigs, 1997–2000. J. Antimicrob. Chemother. 53, 997–1003.
Gebreyes, W. A., Thakur, S., Dorr, P., Tadesse, D. A., Post, K., and Wolf, L. (2009). Occurrence of spvA virulence gene and clinical significance for multidrug-resistant Salmonella strains. J. Clin. Microbiol. 47, 777–780.
Gilbert, J. M., White, D. G., and McDermott, P. F. (2007). The US national antimicrobial resistance monitoring system. Future Microbiol. 2, 493–500.
Giraud, E., Brisabois, A., Martel, J. L., and Chaslus-Dancla, E. (1999). Comparative studies of mutations in animal isolates and experimental in vitro- and in vivo-selected mutants of Salmonella spp. suggest a counterselection of highly fluoroquinolone-resistant strains in the field. Antimicrob. Agents Chemother. 43, 2131–2137.
Giraud, E., Cloeckaert, A., Baucheron, S., Mouline, C., and Chaslus-Dancla, E. (2003). Fitness cost of fluoroquinolone resistance in Salmonella enterica serovar Typhimurium. J. Med. Microbiol. 52, 697–703.
Glenn, L. M., Englen, M. D., Lindsey, R. L., Frank, J. F., Turpin, J. E., Berrang, M. E., et al. (2012). Analysis of antimicrobial resistance genes detected in multiple-drug-resistant Escherichia coli isolates from broiler chicken carcasses. Microb. Drug Resist. 18, 453–463.
Glenn, L. M., Lindsey, R. L., Frank, J. F., Meinersmann, R. J., Englen, M. D., Fedorka-Cray, P. J., et al. (2011). Analysis of antimicrobial resistance genes detected in multidrug-resistant Salmonella enterica serovar Typhimurium isolated from food animals. Microb. Drug Resist. 17, 407–418.
Goldberg, M. B., and Rubin, R. H. (1988). The spectrum of Salmonella infection. Infect. Dis. Clin. North Am. 2, 571–598.
Graves, A. K., Hagedorn, C., Teetor, A., Mahal, M., Booth, A. M., and Reneau, R. B. Jr. (2002). Antibiotic resistance profiles to determine sources of fecal contamination in a rural Virginia watershed. J. Environ. Qual. 31, 1300–1308.
Guerrant, R. L., Van, G. T., Steiner, T. S., Thielman, N. M., Slutsker, L., Tauxe, R. V., et al. (2001). Practice guidelines for the management of infectious diarrhea. Clin. Infect. Dis. 32, 331–351.
Gupta, A., Fontana, J., Crowe, C., Bolstorff, B., Stout, A., Van Duyne, S., et al. (2003). Emergence of multidrug-resistant Salmonella enterica serotype Newport infections resistant to expanded-spectrum cephalosporins in the United States. J. Infect. Dis. 188, 1707–1716.
Habib, A. G. (2004). A clinical audit of presentation and outcome of Salmonella septicaemia. Ann. Acad. Med. Singapore 33, 749–753.
Hammerum, A. M., and Heuer, O. E. (2009). Human health hazards from antimicrobial-resistant Escherichia coli of animal origin. Clin. Infect. Dis. 48, 916–921.
Hammerum, A. M., Heuer, O. E., Emborg, H. D., Bagger-Skjot, L., Jensen, V. F., Rogues, A. M., et al. (2007). Danish integrated antimicrobial resistance monitoring and research program. Emerg. Infect. Dis. 13, 1632–1639.
Hammerum, A. M., Jensen, L. B., and Aarestrup, F. M. (1998). Detection of the satA gene and transferability of virginiamycin resistance in Enterococcus faecium from food-animals. FEMS Microbiol. Lett. 168, 145–151.
Hammerum, A. M., Lester, C. H., and Heuer, O. E. (2010). Antimicrobial-resistant enterococci in animals and meat: a human health hazard? Foodborne Pathog. Dis. 7, 1137–1146.
Hancock, L., and Gilmore, M. S. (2000). “Pathogenicity of enterococci,” in Gram-Positive Pathogens, eds V. A. Fiscetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (Washington, DC: ASM Press), 251–257.
Hancock, R. E. (2005). Mechanisms of action of newer antibiotics for Gram-positive pathogens. Lancet Infect. Dis. 5, 209–218.
Hasman, H., Mevius, D., Veldman, K., Olesen, I., and Aarestrup, F. M. (2005). beta-Lactamases among extended-spectrum beta-lactamase (ESBL)-resistant Salmonella from poultry, poultry products and human patients in The Netherlands. J. Antimicrob. Chemother. 56, 115–121.
Hassan, W. M., Ellender, R. D., and Wang, S. Y. (2007). Fidelity of bacterial source tracking: Escherichia coli vs Enterococcus spp and minimizing assignment of isolates from nonlibrary sources. J. Appl. Microbiol. 102, 591–598.
Hawkey, P. M., and Jones, A. M. (2009). The changing epidemiology of resistance. J. Antimicrob. Chemother. 64(Suppl. 1), i3–i10.
Hedges, R. W., and Shannon, K. P. (1984). Resistance to apramycin in Escherichia coli isolated from animals: detection of a novel aminoglycoside-modifying enzyme. J. Gen. Microbiol. 130, 473–482.
Hegstad, K., Mikalsen, T., Coque, T. M., Werner, G., and Sundsfjord, A. (2010). Mobile genetic elements and their contribution to the emergence of antimicrobial resistant Enterococcus faecalis and Enterococcus faecium. Clin. Microbiol. Infect. 16, 541–554.
Hershberger, E. (2004). Quinupristin-dalfopristin resistance in gram-positive bacteria: mechanism of resistance and epidemiology. Clin. Infect. Dis. 38, 92–98.
Hidron, A. I., Edwards, J. R., Patel, J., Horan, T. C., Sievert, D. M., Pollock, D. A., et al. (2008). NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infect. Control Hosp. Epidemiol. 29, 996–1011.
Hopkins, K. L., Davies, R. H., and Threlfall, E. J. (2005). Mechanisms of quinolone resistance in Escherichia coli and Salmonella: recent developments. Int. J. Antimicrob. Agents 25, 358–373.
Huehn, S., La Ragione, R. M., Anjum, M., Saunders, M., Woodward, M. J., Bunge, C., et al. (2010). Virulotyping and antimicrobial resistance typing of Salmonella enterica serovars relevant to human health in Europe. Foodborne Pathog. Dis. 7, 523–535.
Huycke, M. M., Sahm, D. F., and Gilmore, M. S. (1998). Multiple-drug resistant enterococci: the nature of the problem and an agenda for the future. Emerg. Infect. Dis. 4, 239–249.
Iovine, N. M., and Blaser, M. J. (2004a). Antibiotics in animal feed and spread of resistant Campylobacter from poultry to humans. Emerg. Infect. Dis. 10, 1158–1159.
Iovine, N. M., and Blaser, M. J. (2004b). Antimicrobial resistance in Campylobacter. Emerg. Infect. Dis. 10, 1346.
Jackson, C. R. (2004a). Effects of tylosin use on erythromycin resistance in enterococci isolated from swine. Appl. Environ. Microbiol. 70, 4205–4210.
Jackson, C. R. (2004b). Genetic relatedness of high-level aminoglycoside-resistant enterococci isolated from poultry carcasses. Avian Dis. 48, 100–107.
Jackson, C. R. (2005). High-level aminoglycoside resistant enterococci isolated from swine. Epidemiol. Infect. 133, 367–371.
Jackson, C. R. (2011). Prevalence, species distribution and antimicrobial resistance of enterococci isolated from US dairy cattle. Lett. Appl. Microbiol. 52, 41–48.
Jackson, C. R., Fedorka-Cray, P. J., Barrett, J. B., Hiott, L. M., and Woodley, T. A. (2007). Prevalence of streptogramin resistance in enterococci from animals: identification of vatD from animal sources in the USA. Int. J. Antimicrob. Agents 30, 60–66.
Jackson, C. R., Fedorka-Cray, P. J., Barrett, J. B., Hiott, L. M., and Woodley, T. A. (2008). First report of vatB and vgaB from Enterococcus gallinarum in the USA. Int. J. Antimicrob. Agents 31, 175–176.
Jackson, C. R., Fedorka-Cray, P. J., Davis, J. A., Barrett, J. B., and Frye, J. G. (2009). Prevalence, species distribution and antimicrobial resistance of enterococci isolated from dogs and cats in the United States. J. Appl. Microbiol. 107, 1269–1278.
Jensen, L. B., Hammerum, A. M., Aerestrup, F. M., van den Bogaard, A. E., and Stobberingh, E. E. (1998). Occurrence of satA and vgb genes in streptogramin-resistant Enterococcus faecium isolates of animal and human origins in the Netherlands. Antimicrob. Agents Chemother. 42, 3330–3331.
Jett, B. D., Huycke, M. M., and Gilmore, M. S. (1994). Virulence of enterococci. Clin. Microbiol. Rev. 7, 462–478.
Johnson, T. J., Logue, C. M., Johnson, J. R., Kuskowski, M. A., Sherwood, J. S., Barnes, H. J., et al. (2011a). Associations between multidrug resistance, plasmid content, and virulence potential among extraintestinal pathogenic and commensal Escherichia Coli from humans and poultry. Foodborne Pathog. Dis. 9, 37–46.
Johnson, T. J., Shepard, S. M., Rivet, B., Danzeisen, J. L., and Carattoli, A. (2011b). Comparative genomics and phylogeny of the IncI1 plasmids: a common plasmid type among porcine enterotoxigenic Esc-herichia coli. Plasmid 66, 144–151.
Johnson, T. J., and Nolan, L. K. (2009). Pathogenomics of the virulence plasmids of Escherichia coli. Microbiol. Mol. Biol. Rev. 73, 750–774.
Johnson, T. J., Siek, K. E., Johnson, S. J., and Nolan, L. K. (2006a). DNA sequence of a ColV plasmid and prevalence of selected plasmid-encoded virulence genes among avian Escherichia coli strains. J. Bacteriol. 188, 745–758.
Johnson, T. J., Wannemeuhler, Y. M., Scaccianoce, J. A., Johnson, S. J., and Nolan, L. K. (2006b). Complete DNA sequence, comparative genomics, and prevalence of an IncHI2 plasmid occurring among extraintestinal pathogenic Escherichia coli isolates. Antimicrob. Agents Chemother. 50, 3929–3933.
Jones, F. T., and Ricke, S. C. (2003). Observations on the history of the development of antimicrobials and their use in poultry feeds. Poult. Sci. 82, 613–617.
Jung, Y. H., Shin, E. S., Kim, O., Yoo, J. S., Lee, K. M., Yoo, J. I., et al. (2010). Characterization of two newly identified genes, vgaD and vatH, [corrected] conferring resistance to streptogramin A in Enterococcus faecium. Antimicrob. Agents Chemother. 54, 4744–4749.
Karch, H., Bielaszewska, M., Bitzan, M., and Schmidt, H. (1999). Epidemiology and diagnosis of Shiga toxin-producing Escherichia coli infections. Diagn. Microbiol. Infect. Dis. 34, 229–243.
Keyes, K., Hudson, C., Maurer, J. J., Thayer, S., White, D. G., and Lee, M. D. (2000). Detection of florfenicol resistance genes in Escherichia coli isolated from sick chickens. Antimicrob. Agents Chemother. 44, 421–424.
Khachatryan, A. R., Hancock, D. D., Besser, T. E., and Call, D. R. (2004). Role of calf-adapted Escherichia coli in maintenance of antimicrobial drug resistance in dairy calves. Appl. Environ. Microbiol. 70, 752–757.
Krauland, M. G., Marsh, J. W., Paterson, D. L., and Harrison, L. H. (2009). Integron-mediated multidrug resistance in a global collection of nontyphoidal Salmonella enterica isolates. Emerg. Infect. Dis. 15, 388–396.
Kuhn, I., Iversen, A., Burman, L. G., Olsson-Liljequist, B., Franklin, A., Finn, M., et al. (2000). Epidemiology and ecology of enterococci, with special reference to antibiotic resistant strains, in animals, humans and the environment: example of an ongoing project within the European research programme. Int. J. Antimicrob. Agents 14, 337–342.
Landman, D., and Quale, J. M. (1997). Management of infections due to resistant enterococci: a review of therapeutic options. J. Antimicrob. Chemother. 40, 161–170.
Langlois, B. E., Dawson, K. A., Leak, I., and Aaron, D. K. (1988). Effect of age and housing location on antibiotic resistance of fecal coliforms from pigs in a non-antibiotic-exposed herd. Appl. Environ. Microbiol. 54, 1341–1344.
Lavigne, J. P., and Blanc-Potard, A. B. (2008). Molecular evolution of Salmonella enterica serovar Typhimurium and pathogenic Escherichia coli: from pathogenesis to therapeutics. Infect. Genet. Evol. 8, 217–226.
Layton, B. A., Walters, S. P., Lam, L. H., and Boehm, A. B. (2010). Enterococcus species distribution among human and animal hosts using multiplex PCR. J. Appl. Microbiol. 109, 539–547.
Lebreton, F., Depardieu, F., Bourdon, N., Fines-Guyon, M., Berger, P., Camiade, S., et al. (2011). D-Ala-d-Ser VanN-type transferable vancomycin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 55, 4606–4612.
Leclerc, H., Mossel, D. A., Edberg, S. C., and Struijk, C. B. (2001). Advances in the bacteriology of the coliform group: their suitability as markers of microbial water safety. Annu. Rev. Microbiol. 55, 201–234.
Li, X. Z., Mehrotra, M., Ghimire, S., and Adewoye, L. (2007). Beta-lactam resistance and beta-lactamases in bacteria of animal origin. Vet. Microbiol. 121, 197–214.
Lindsey, R. L., Fedorka-Cray, P. J., Frye, J. G., and Meinersmann, R. J. (2009). Inc A/C plasmids are prevalent in multidrug-resistant Salmonella enterica isolates. Appl. Environ. Microbiol. 75, 1908–1915.
Lindsey, R. L., Frye, J. G., Fedorka-Cray, P. J., and Meinersmann, R. J. (2011a). Microarray-based analysis of IncA/C plasmid-associated genes from multidrug-resistant Salmonella enterica. Appl. Environ. Microbiol. 77, 6991–6999.
Lindsey, R. L., Frye, J. G., Thitaram, S. N., Meinersmann, R. J., Fedorka-Cray, P. J., and Englen, M. D. (2011b). Characterization of multidrug-resistant Escherichia coli by antimicrobial resistance profiles, plasmid replicon typing, and pulsed-field gel electrophoresis. Microb. Drug Resist. 17, 157–163.
Linton, A. H. (1977). Antibiotic resistance: the present situation reviewed. Vet. Rec. 100, 354–360.
Madsen, L., Aarestrup, F. M., and Olsen, J. E. (2000). Characterisation of streptomycin resistance determinants in Danish isolates of Salmonella Typhimurium. Vet. Microbiol. 75, 73–82.
Mainil, J. G., and Daube, G. (2005). Verotoxigenic Escherichia coli from animals, humans and foods: who's who? J. Appl. Microbiol. 98, 1332–1344.
Malani, P. N., Kauffman, C. A., and Zervos, M. J. (2002). “Enterococcal disease, epidemiology, and treatment,” in The Enterococci: Pathogenesis, Molecular Biology, and Antibiotic Resistance, eds M. S. Gilmore, D. B. Clewell, P. Courvalin, G. M. Dunny, B. E. Murray, and L. B. Rice (Washington, DC: ASM Press), 385–408.
Marshall, B. M., and Levy, S. B. (2011). Food animals and antimicrobials: impacts on human health. Clin. Microbiol. Rev. 24, 718–733.
Marshall, C. G., Lessard, I. A., Park, I., and Wright, G. D. (1998). Glycopeptide antibiotic resistance genes in glycopeptide-producing organisms. Antimicrob. Agents Chemother. 42, 2215–2220.
Martel, J. L., Tardy, F., Brisabois, A., Lailler, R., Coudert, M., and Chaslus-Dancla, E. (2000). The French antibiotic resistance monitoring programs. Int. J. Antimicrob. Agents 14, 275–283.
Martone, W. J. (1998). Spread of vancomycin-resistant enterococci: why did it happen in the United States? Infect. Control Hosp. Epidemiol. 19, 539–545.
Mataseje, L. F., Baudry, P. J., Zhanel, G. G., Morck, D. W., Read, R. R., Louie, M., et al. (2010). Comparison of CMY-2 plasmids isolated from human, animal, and environmental Escherichia coli and Salmonella spp. from Canada. Diagn. Microbiol. Infect. Dis. 67, 387–391.
Mathers, J. J., Flick, S. C., and Cox, L. A. Jr. (2011). Longer-duration uses of tetracyclines and penicillins in U.S. food-producing animals: indications and microbiologic effects. Environ. Int. 37, 991–1004.
Mathew, A. G., Cissell, R., and Liamthong, S. (2007). Antibiotic resistance in bacteria associated with food animals: a United States perspective of livestock production. Foodborne Pathog. Dis. 4, 115–133.
Matsumura, S., and Simor, A. E. (1998). Treatment of endocarditis due to vancomycin-resistant Enterococcus faecium with quinupristin/dalfopristin, doxycycline, and rifampin: a synergistic drug combination. Clin. Infect. Dis. 27, 1554–1556.
Maurin, M., and Raoult, D. (2001). Use of aminoglycosides in treatment of infections due to intracellular bacteria. Antimicrob. Agents Chemother. 45, 2977–2986.
McClelland, M., Sanderson, K. E., Spieth, J., Clifton, S. W., Latreille, P., Courtney, L., et al. (2001). Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413, 852–856.
McConnell, M. A., Mercer, A. A., and Tannock, G. W. (1991). Transfer of plasmid pAM?1 between members of the normal microflora inhabiting the murine digestive tract and modification of the plasmid in a Lactobacillus reuteri host. Microbial Ecol. Health Dis. 4, 343–355.
McDermott, P. F., Cullen, P., Hubert, S. K., McDermott, S. D., Bartholomew, M., Simjee, S., et al. (2005). Changes in antimicrobial susceptibility of native Enterococcus faecium in chickens fed virginiamycin 29. Appl. Environ. Microbiol. 71, 4986–4991.
McDermott, P. F., Walker, R. D., and White, D. G. (2003). Antimicrobials: modes of action and mechanisms of resistance. Int. J. Toxicol. 22, 135–143.
McEwen, S. A., and Fedorka-Cray, P. J. (2002). Antimicrobial use and resistance in animals. Clin. Infect. Dis. 34(Suppl. 3) S93-S106.
McNamee, P. T., and King, D. C. (1996). Endocarditis in broiler breeder rearers due to Enterococcus hirae. Vet. Rec. 138, 240.
Miriagou, V., Cornaglia, G., Edelstein, M., Galani, I., Giske, C. G., Gniadkowski, M., et al. (2010). Acquired carbapenemases in Gram-negative bacterial pathogens: detection and surveillance issues. Clin. Microbiol. Infect. 16, 112–122.
Mlynarczyk, B., Mlynarczyk, A., Kmera-Muszynska, M., Majewski, S., and Mlynarczyk, G. (2010). Mechanisms of resistance to antimicrobial drugs in pathogenic Gram-positive cocci. Mini Rev. Med. Chem. 10, 928–937.
Muller, T., Ulrich, A., Ott, E. M., and Muller, M. (2001). Identification of plant-associated enterococci. J. Appl. Microbiol. 91, 268–278.
Mulvey, M. R., Boyd, D. A., Olson, A. B., Doublet, B., and Cloeckaert, A. (2006). The genetics of Salmonella genomic island 1. Microbes Infect. 8, 1915–1922.
Mulvey, M. R., Soule, G., Boyd, D., Demczuk, W., and Ahmed, R. (2003). Characterization of the first extended-spectrum beta-lactamase-producing Salmonella isolate identified in Canada. J. Clin. Microbiol. 41, 460–462.
Mundy, L. M., Sahm, D. F., and Gilmore, M. (2000). Relationships between enterococcal virulence and antimicrobial resistance. Clin. Microbiol. Rev. 13, 513–522.
Nelson, J. M., Chiller, T. M., Powers, J. H., and Angulo, F. J. (2007). Fluoroquinolone-resistant Campylobacter species and the withdrawal of fluoroquinolones from use in poultry: a public health success story. Clin. Infect. Dis. 44, 977–980.
Netherwood, T., Bowden, R., Harrison, P., O'Donnell, A. G., Parker, D. S., and Gilbert, H. J. (1999). Gene transfer in the gastrointestinal tract. Appl. Environ. Microbiol. 65, 5139–5141.
Niemi, R. M., Niemela, S. I., Bamford, D. H., Hantula, J., Hyvarinen, T., Forsten, T., et al. (1993). Presumptive fecal streptococci in environmental samples characterized by one-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Appl. Environ. Microbiol. 59, 2190–2196.
Nijssen, S., Florijn, A., Bonten, M. J., Schmitz, F. J., Verhoef, J., and Fluit, A. C. (2004). Beta-lactam susceptibilities and prevalence of ESBL-producing isolates among more than 5000 European Enterobacteriaceae isolates. Int. J. Antimicrob. Agents 24, 585–591.
Nilsson, O. (2012). Vancomycin resistant enterococci in farm animals - occurrence and importance. Infect. Ecol. Epidemiol. 2:16959. doi: 10.3402/iee.v2i0.16959
O'Regan, E., Quinn, T., Frye, J. G., Pages, J. M., Porwollik, S., Fedorka-Cray, P. J., et al. (2010). Fitness costs and stability of a high-level ciprofloxacin resistance phenotype in Salmonella enterica serotype enteritidis: reduced infectivity associated with decreased expression of Salmonella pathogenicity island 1 genes. Antimicrob. Agents. Chemother. 54, 367–374.
Olsvik, O., Wasteson, Y., Lund, A., and Hornes, E. (1991). Pathogenic Escherichia coli found in food. Int. J. Food Microbiol. 12, 103–113.
Osborn, A. M., and Boltner, D. (2002). When phage, plasmids, and transposons collide: genomic islands, and conjugative- and mobilizable-transposons as a mosaic continuum. Plasmid 48, 202–212.
Parry, C. M., and Threlfall, E. J. (2008). Antimicrobial resistance in typhoidal and nontyphoidal salmonellae. Curr. Opin. Infect. Dis. 21, 531–538.
Paterson, D. L. (2006). Resistance in gram-negative bacteria: Enterobacteriaceae. Am. J. Infect. Control 34: S20–S28.
Paterson, D. L., and Bonomo, R. A. (2005). Extended-spectrum beta-lactamases: a clinical update. Clin. Microbiol. Rev. 18, 657–686.
Paton, J. C., and Paton, A. W. (1998). Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 11, 450–479.
Platteeuw, C., Michiels, F., Joos, H., Seurinck, J., and de Vos, W. M. (1995). Characterization and heterologous expression of the tetL gene and identification of iso-ISS1 elements from Enterococcus faecalis plasmid pJH1. Gene 160, 89–93.
Poole, T., and Crippen, T. (2009). Conjugative plasmid transfer between Salmonella enterica Newport and Escherichia coli within the gastrointestinal tract of the lesser mealworm beetle, Alphitobius diaperinus (Coleoptera: Tenebrionidae). Poult. Sci. 88, 1553–1558.
Poutrel, B., and Ryniewicz, H. Z. (1984). Evaluation of the API 20 Strep system for species identification of streptococci isolated from bovine mastitis. J. Clin. Microbiol. 19, 213–214.
Prescott, J. F. (2000a). “Aminoglycosides and aminocyclitols,” in Antimicrobial Therapy in Veterinary Medicine, eds J. F. Prescott, J. D. Baggot, and R. D. Walker (Ames, IA: Iowa State University Press), 191–228.
Prescott, J. F. (2000b). Antimicrobial Therapy in Veterinary Medicine. Ames, IA: Iowa State University Press.
Prescott, J. F. (2000c). “Beta-lactam antibiotics: cephalosporins and cephamycins,” in Antimicrobial Therapy in Veterinary Medicine, eds J. F. Prescott, J. D. Baggot, and R. D. Walker (Ames, IA: Iowa State University Press), 134–159.
Prescott, J. F. (2000d). “Beta-lactam antibiotics: penam penicillins,” in Antimicrobial Therapy in Veterinary Medicine, eds J. F. Prescott, J. D. Baggot, and R. D. Walker (Ames, IA: Iowa State University Press), 105–133.
Prescott, J. F. (2000e). “Chloramphenicol, thiamphenicol, and flofenicol,” in Antimicrobial Therapy in Veterinary Medicine, eds J. F. Prescott, J. D. Baggot, and R. D. Walker (Ames, IA: Iowa State University Press), 263–274.
Prescott, J. F. (2000f). “Lincosamides, macrolides, and pleuromutilins,” in Antimicrobial Therapy in Veterinary Medicine, eds J. F. Prescott, J. D. Baggot, and R. D. Walker (Ames, IA: Iowa State University Press), 229–262.
Prescott, J. F. (2000g). “Other Beta-lactam Antibiotics: beta-lactamase inhibitors, carbapenems, and monobactams,” in Antimicrobial Therapy in Veterinary Medicine. eds J. F. Prescott, J. D. Baggot, and R. D. Walker (Ames, IA: Iowa State University Press), 160–176.
Prescott, J. F. (2000h). “Sulfonamides, diaminopyrimidines, and their combinations,” in Antimicrobial Therapy in Veterinary Medicine, eds J. F. Prescott, J. D. Baggot, and R. D. Walker (Ames, IA: Iowa State University Press), 290–314.
Projan, S. J. (2000). Preclinical pharmacology of GAR-936, a novel glycylcycline antibacterial agent. Pharmacotherapy 20, 219S–223S.
Ramirez, M. S., and Tolmasky, M. E. (2010). Aminoglycoside modifying enzymes. Drug Resist. Updat. 13, 151–171.
Randall, C. J., Wood, A. M., and MacKenzie, G. (1993). Encephalomalacia in first-week chicks. Vet. Rec. 132, 419.
Rasko, D. A., Webster, D. R., Sahl, J. W., Bashir, A., Boisen, N., Scheutz, F., et al. (2011). Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N. Engl. J. Med. 365, 709–717.
Rasmussen, M. A., and Casey, T. A. (2001). Environmental and food safety aspects of Escherichia coli O157:H7 infections in cattle. Crit. Rev. Microbiol. 27, 57–73.
Rende-Fournier, R., Leclercq, R., Galimand, M., Duval, J., and Courvalin, P. (1993). Identification of the satA gene encoding a streptogramin A acetyltransferase in Enterococcus faecium BM4145. Antimicrob. Agents Chemother. 37, 2119–2125.
Renter, D. G., and Sargeant, J. M. (2002). Enterohemorrhagic Escherichia coli O157: epidemiology and ecology in bovine production environments. Anim. Health Res. Rev. 3, 83–94.
Roberts, M. C. (1996). Tetracycline resistance determinants: mechanisms of action, regulation of expression, genetic mobility, and distribution. FEMS Microbiol. Rev. 19, 1–24.
Roberts, M. C. (2002). Resistance to tetracycline, macrolide-lincosamide-streptogramin, trimet-hoprim, and sulfonamide drug classes. Mol. Biotechnol. 20, 261–283.
Roberts, M. C. (2004). Resistance to macrolide, lincosamide, streptogramin, ketolide, and oxazolidinone antibiotics. Mol. Biotechnol. 28, 47–62.
Roberts, M. C. (2005). Update on acquired tetracycline resistance genes. FEMS Microbiol. Lett. 245, 195–203.
Roberts, M. C., Sutcliffe, J., Courvalin, P., Jensen, L. B., Rood, J., and Seppala, H. (1999). Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resis-tance determinants. Antimicrob. Agents Chemother. 43, 2823–2830.
Rogers, D. G., Zeman, D. H., and Erickson, E. D. (1992). Diarrhea associated with Enterococcus durans in calves. J. Vet. Diagn. Invest. 4, 471–472.
Rossitto, P. V., Ruiz, L., Kikuchi, Y., Glenn, K., Luiz, K., Watts, J. L., et al. (2002). Antibiotic susceptibility patterns for environmental streptococci isolated from bovine mastitis in central California dairies. J. Dairy Sci. 85, 132–138.
Salyers, A. A., Shoemaker, N. B., Stevens, A. M., and Li, L. Y. (1995). Conjugative transposons: an unusual and diverse set of integrated gene transfer elements. Microbiol. Rev. 59, 579–590.
Savard, P., Gopinath, R., Zhu, W., Kitchel, B., Rasheed, J. K., Tekle, T., et al. (2011). First NDM-positive Salmonella sp. strain identified in the United States. Antimicrob. Agents Chemother. 55, 5957–5958.
Scallan, E., Hoekstra, R. M., Angulo, F. J., Tauxe, R. V., Widdowson, M. A., Roy, S. L., et al. (2011). Foodborne illness acquired in the United States–major pathogens. Emerg. Infect. Dis. 17, 7–15.
Schwaiger, K., and Bauer, J. (2008). Detection of the erythromycin rRNA methylase gene erm(A) in Enterococcus faecalis. Antimicrob. Agents Chemother. 52, 2994–2995.
Schwarz, S., and Chaslus-Dancla, E. (2001). Use of antimicrobials in veterinary medicine and mechanisms of resistance. Vet. Res. 32, 201–225.
Schwarz, S., Kehrenberg, C., and Walsh, T. R. (2001). Use of antimicrobial agents in veterinary medicine and food animal production. Int. J. Antimicrob. Agents 17, 431–437.
Scott, T. M., Jenkins, T. M., Lukasik, J., and Rose, J. B. (2005). Potential use of a host associated molecular marker in Enterococcus faecium as an index of human fecal pollution. Environ. Sci. Technol. 39, 283–287.
Scott, T. M., Rose, J. B., Jenkins, T. M., Farrah, S. R., and Lukasik, J. (2002). Microbial source tracking: current methodology and future directions. Appl. Environ. Microbiol. 68, 5796–5803.
Shah, A. A., Hasan, F., Ahmed, S., and Hameed, A. (2004a). Characteristics, epidemiology and clinical importance of emerging strains of Gram-negative bacilli producing extended-spectrum beta-lactamases. Res. Microbiol. 155, 409–421.
Shah, A. A., Hasan, F., Ahmed, S., and Hameed, A. (2004b). Extended-spectrum beta-lactamases (ESBLs): characterization, epidemiology and detection. Crit. Rev. Microbiol. 30, 25–32.
Shaw, K. J., Rather, P. N., Hare, R. S., and Miller, G. H. (1993). Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 57, 138–163.
Shoemaker, D. M., Simou, J., and Roland, W. E. (2006). A review of daptomycin for injection (Cubicin) in the treatment of complicated skin and skin structure infections. Ther. Clin. Risk Manag. 2, 169–174.
Shryock, T. R., and Richwine, A. (2010). The interface between veterinary and human antibiotic use. Ann. N.Y. Acad. Sci. 1213, 92–105.
Singh, K. V., Weinstock, G. M., and Murray, B. E. (2002). An Enterococcus faecalis ABC homologue (Lsa) is required for the resistance of this species to clindamycin and quinupristin-dalfopristin. Antimicrob. Agents Chemother. 46, 1845–1850.
Siu, L. K., Lu, P. L., Chen, J. Y., Lin, F. M., and Chang, S. C. (2003). High-level expression of ampC beta-lactamase due to insertion of nucleotides between -10 and -35 promoter sequences in Escherichia coli clinical isolates: cases not responsive to extended-spectrum-cephalosporin treatment. Antimicrob. Agents Chemother. 47, 2138–2144.
Sobiraj, A., Kron, A., Schollmeyer, U., and Failing, K. (1997). Federal investigations on the distribution and in vitro resistance of udder pathogenic bacteria in the milk of cows with subclinical mastitis. Tierarztl. Prax. 25, 108–115.
Soltani, M., Beighton, D., Philpott-Howard, J., and Woodford, N. (2000). Mechanisms of resistance to quinupristin-dalfopristin among isolates of Enterococcus faecium from animals, raw meat, and hospital patients in Western Europe. Antimicrob. Agents Chemother. 44, 433–436.
Spears, K. J., Roe, A. J., and Gally, D. L. (2006). A comparison of enteropathogenic and enterohaemorrhagic Escherichia coli pathogenesis. FEMS Microbiol. Lett. 255, 187–202.
Steentjes, A., Veldman, K. T., Mevius, D. J., and Landman, W. J. (2002). Molecular epidemiology of unilateral amyloid arthropathy in broiler breeders associated with Enterococcus faecalis. Avian Pathol. 31, 31–39.
Stokes, D. J., Kelly, A. F., Gould, S. W., Cassar, C. A., and Fielder, M. D. (2008). The withdrawal of antimicrobial treatment as a mechanism for defeating resistant microorganisms. FEMS Immunol. Med. Microbiol. 53, 300–305.
Stoycheva, M. V., and Murdjeva, M. A. (2006). Antimicrobial therapy of salmonelloses–current state and perspectives. Folia Med. (Plovdiv) 48, 5–10.
Su, L. H., Chiu, C. H., Chu, C., and Ou, J. T. (2004). Antimicrobial resistance in nontyphoid Salmonella serotypes: a global challenge. Clin. Infect. Dis. 39, 546–551.
Su, L. H., Chu, C., Cloeckaert, A., and Chiu, C. H. (2008). An epidemic of plasmids? Dissemination of extended-spectrum cephalosporinases among Salmonella and other Enterobacteriaceae. FEMS Immunol. Med. Microbiol. 52, 155–168.
Svec, P., and Sedlacek, I. (1999). Occurrence of Enterococcus spp. in waters. Folia Microbiol. (Praha) 44, 3–10.
Swaminathan, B., Barrett, T. J., and Fields, P. (2006). Surveillance for human Salmonella infections in the United States. J. AOAC Int. 89, 553–559.
Swaney, S. M., Aoki, H., Ganoza, M. C., and Shinabarger, D. L. (1998). The oxazolidinone linezolid inhibits initiation of protein synthesis in bacteria. Antimicrob. Agents Chemother. 42, 3251–3255.
Tankson, J. D., Fedorka-Cray, P. J., Jackson, C. R., and Headrick, M. (2006). Genetic relatedness of a rarely isolated Salmonella: Salmonella enterica serotype Niakhar from NARMS animal isolates. J. Antimicrob. Chemother. 57, 190–198.
Tankson, J. D., Thaxton, J. P., and Vizzier-Thaxton, Y. (2001). Pulmonary hypertension syndrome in broilers caused by Enterococcus faecalis. Infect. Immun. 69, 6318–6322.
Tham, W., Karp, G., and nielsson-Tham, M. L. (1990). Histamine formation by enterococci in goat cheese. Int. J. Food Microbiol. 11, 225–229.
Tollefson, L., Angulo, F. J., and Fedorka-Cray, P. J. (1998). National surveillance for antibiotic resistance in zoonotic enteric pathogens. Vet. Clin. North Am. Food Anim. Pract. 14, 141–150.
Tollefson, L., Fedorka-Cray, P. J., and Angulo, F. J. (1999). Public health aspects of antibiotic resistance monitoring in the USA. Acta Vet. Scand. Suppl. 92, 67–75.
Tracz, D. M., Boyd, D. A., Bryden, L., Hizon, R., Giercke, S., Van, C. P., et al. (2005). Increase in ampC promoter strength due to mutations and deletion of the attenuator in a clinical isolate of cefoxitin-resistant Escherichia coli as determined by RT-PCR. J. Antimicrob. Chemother. 55, 768–772.
Van, B. F., Balzi, E., and Tulkens, P. M. (2000). Antibiotic efflux pumps. Biochem. Pharmacol. 60, 457–470.
Varma, J. K., Marcus, R., Stenzel, S. A., Hanna, S. S., Gettner, S., Anderson, B. J., et al. (2006). Highly resistant Salmonella Newport-MDRAmpC transmitted through the domestic US food supply: a FoodNet case-control study of sporadic Salmonella Newport infections, 2002–2003. J. Infect. Dis. 194, 222–230.
Wall, P. G., Morgan, D., Lamden, K., Ryan, M., Griffin, M., Threlfall, E. J., et al. (1994). A case control study of infection with an epidemic strain of multiresistant Salmonella typhimurium DT104 in England and Wales. Commun. Dis. Rep. CDR Rev. 4: R130–R135.
Walsh, C., and Fanning, S. (2008). Antimicrobial resistance in foodborne pathogens–a cause for concern? Curr. Drug Targets 9, 808–815.
Walsh, T. R. (2010). Emerging carbapenemases: a global perspective. Int. J. Antimicrob. Agents 36(Suppl. 3), S8–S14.
Watanabe, S., Kobayashi, N., Quinones, D., Nagashima, S., Uehara, N., and Watanabe, N. (2009). Genetic diversity of enterococci harboring the high-level gentamicin resistance gene aac(6′)-Ie-aph(2″)-Ia or aph(2”)-Ie in a Japanese hospital. Microb. Drug Resist. 15, 185–194.
Welch, T. J., Evenhuis, J., White, D. G., McDermott, P. F., Harbottle, H., Miller, R. A., et al. (2009). IncA/C plasmid-mediated florfenicol resistance in the catfish pathogen Edwardsiella ictaluri. Antimicrob. Agents Chemother. 53, 845–846.
Welch, T. J., Fricke, W. F., McDermott, P. F., White, D. G., Rosso, M. L., Rasko, D. A., et al. (2007). Multiple antimicrobial resistance in plague: an emerging public health risk. PLoS ONE 2:e309. doi: 10.1371/journal.pone.0000309
Werner, G., Coque, T. M., Hammerum, A. M., Hope, R., Hryniewicz, W., Johnson, A., et al. (2008). Emergence and spread of vancomycin resistance among enterococci in Europe. Euro. Surveill. 13, 19046.
Werner, G., Hildebrandt, B., and Witte, W. (2001). The newly described msrC gene is not equally distributed among all isolates of Enterococcus faecium. Antimicrob. Agents Chemother. 45, 3672–3673.
Werner, G., and Witte, W. (1999). Characterization of a new enterococcal gene, satG, encoding a putative acetyltransferase conferring resistance to streptogramin A compounds 1. Antimicrob. Agents Chemother. 43, 1813–1814.
White, D. G., Acar, J., Anthony, F., Franklin, A., Gupta, R., Nicholls, T., et al. (2001). Antimicrobial resistance: standardisation and harmonisation of laboratory methodologies for the detection and quantification of antimicrobial resistance. Rev. Sci. Tech. 20, 849–858.
White, D. G., Hudson, C., Maurer, J. J., Ayers, S., Zhao, S., Lee, M. D., et al. (2000). Characterization of chloramphenicol and florfenicol resistance in Escherichia coli associated with bovine diarrhea. J. Clin. Microbiol. 38, 4593–4598.
White, D. G., Zhao, S., McDermott, P. F., Ayers, S., Friedman, S., Sherwood, J., et al. (2003). Characterization of integron mediated antimicrobial resistance in Salmonella isolated from diseased swine. Can. J. Vet. Res. 67, 39–47.
White, D. G., Zhao, S., Simjee, S., Wagner, D. D., and McDermott, P. F. (2002). Antimicrobial resistance of foodborne pathogens. Microbes Infect. 4, 405–412.
White, D. G., Zhao, S., Singh, R., and McDermott, P. F. (2004). Antimicrobial resistance among gram-negative foodborne bacterial pathogens associated with foods of animal origin. Foodborne Pathog. Dis. 1, 137–152.
White, P. A., and Rawlinson, W. D. (2001). Current status of the aadA and dfr gene cassette families. J. Antimicrob. Chemother. 47, 495–496.
Wilson, W. R., Karchmer, A. W., Dajani, A. S., Taubert, K. A., Bayer, A., Kaye, D., et al. (1995). Antibiotic treatment of adults with infective endocarditis due to streptococci, enterococci, staphylococci, and HACEK microorganisms. American Heart Association. JAMA 274, 1706–1713.
Wirth, R. (1994). The sex pheromone system of Enterococcus faecalis. More than just a plasmid-collection mechanism? Eur. J. Biochem. 222, 235–246.
Witte, W. (2000). Selective pressure by antibiotic use in livestock. Int. J. Antimicrob. Agents 16(Suppl. 1), S19–S24.
Xu, X., Lin, D., Yan, G., Ye, X., Wu, S., Guo, Y., et al. (2010). vanM, a new glycopeptide resistance gene cluster found in Enterococcus faecium. Antimicrob. Agents Chemother. 54, 4643–4647.
Yahav, D., Lador, A., Paul, M., and Leibovici, L. (2011). Efficacy and safety of tigecycline: a systematic review and meta-analysis. J. Antimicrob. Chemother. 66, 1963–1971.
Yates, C., and Amyes, S. (2005). Extended-spectrum beta-lactamases in non-typhoidal Salmonella spp. isolated in the UK are now a reality: why the late arrival? J. Antimicrob. Chemother. 56, 262–264.
Zhao, S., Qaiyumi, S., Friedman, S., Singh, R., Foley, S. L., White, D. G., et al. (2003). Characterization of Salmonella enterica serotype newport isolated from humans and food animals. J. Clin. Microbiol. 41, 5366–5371.
Keywords: antimicrobial, resistant, multidrug-resistance, animals, foodborne, Salmonella, E. coli, Enterococcus
Citation: Frye JG and Jackson CR (2013) Genetic mechanisms of antimicrobial resistance identified in Salmonella enterica, Escherichia coli, and Enteroccocus spp. isolated from U.S. food animals. Front. Microbiol. 4:135. doi: 10.3389/fmicb.2013.00135
Received: 04 January 2013; Accepted: 07 May 2013;
Published online: 23 May 2013.
Edited by:
Axel Cloeckaert, Institut National de la Recherche Agronomique, FranceReviewed by:
Patrick R. Butaye, Ghent University, BelgiumCopyright © 2013 Frye and Jackson. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Jonathan G. Frye, Bacterial Epidemiology and Antimicrobial Resistance Research Unit, Agricultural Research Service, U.S. Department of Agriculture, 950 College Station Rd., Athens, GA 30605, USA. e-mail:am9uYXRoYW4uZnJ5ZUBhcnMudXNkYS5nb3Y=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.