- 1Laboratory of Veterinary Public Health, Department of Veterinary Medicine, Joint Faculty of Veterinary Medicine, Kagoshima University, Kagoshima, Japan
- 2Department of Veterinary Public Health, Faculty of Veterinary Medicine, Zagazig University, Zagazig, Egypt
- 3Safety Research Team, National Institute of Animal Health, Tsukuba, Japan
Epidemiologic surveillance study was conducted in southern Japan to determine the antimicrobial resistance phenotypes and characterize the β-lactamase genes and the plasmids harboring these genes in Salmonella enterica serovar Infantis (S. Infantis) isolates from broilers. Between January, 2007 and December, 2008, a total of 1,472 fecal samples were collected and examined at the Laboratory of Veterinary Public Health, Kagoshima University, Japan. In 93 (6.3%) isolates recovered, 33 (35.5%) isolates showed resistance to cefotaxime, an extended-spectrum cephalosporin (ESC), conferred by TEM-20, TEM-52 and CTX-M-25 extended-spectrum β-lactamases (ESBLs). In addition to ESC-resistance, eight (8.6%) isolates exhibited resistance to cefoxitin mediated by CMY-2 AmpC β-lactamase. Plasmid analysis and polymerase chain reaction replicon typing revealed the blaTEM-20 and blaCMY-2 genes were associated with IncP plasmids, blaTEM-52 was linked with a non-typable plasmid and blaCTX-M-25 was carried by an IncA/C plasmid. Non-β-lactam resistance to streptomycin, sulfamethoxazole, and oxytetracycline encoded by the aadA1, sul1, and tet(A) genes, respectively, was found in 86 (92.5%) isolates. Resistance to kanamycin and ofloxacin was exhibited in 12 (12.9%) and 11 (11.8%) isolates, respectively, the former was mediated by aphA1-Iab. These data indicate that S. Infantis isolates producing ESBLs and AmpC β-lactamase have spread among broiler farms in Japan. These data demonstrated that the incidence of ESC-resistant S. Infantis carrying blaTEM-52 remarkably increased and S. Infantis strains harboring blaCMY-2, blaTEM-20, or blaCTX-M-25 genes emerged from broilers in Japan for the first time in 2007 and 2008.
Introduction
Non-typhoidal Salmonella enterica serovars (Salmonella) are a major cause of bacterial food-borne diseases world-wide (Majowicz et al., 2010). The food poisoning statistics in Japan show that bacterial food poisoning patients during the year 2008 numbered at 10,331 and Salmonella were the leading etiological agents accounting for 24.7% of the cases [http://idsc.nih.go.jp/iasr/29/342/graph/t3421.gif]. Poultry products are important vehicles in the transmission, and have been incriminated in several Salmonella outbreaks (Kimura et al., 2004; Chittick et al., 2006). Since the late 1990s, Salmonella enterica serovar Infantis (S. Infantis) has been the commonest serovar of Salmonella isolated from both broiler flocks and retail chicken meat in Japan (Asai et al., 2007b; Iwabuchi et al., 2011).
The emergence of multidrug-resistant Salmonella has become a serious global health problem because antimicrobial treatment is lifesaving for invasive infections, particularly in neonates of <1 year of age. Preventive antimicrobial treatment is also generally given to patients suffering from immunosuppressive or other predisposing conditions (Hohmann, 2001). The options of first-line therapy for Salmonella infection include ampicillin (AMP), sulfamethoxazole-trimethoprim, fluoroquinolones, and extended-spectrum cephalosporins (ESCs). ESCs are among the preferred drugs because of resistance to the other aforementioned drugs is relatively frequent in Salmonella isolates (Hohmann, 2001). At present, ESC-resistance in Salmonella is mainly attributed to the acquisition of plasmid-mediated extended-spectrum β-lactamases (ESBLs) and AmpC β-lactamases (Bonnet, 2004; Arlet et al., 2006).
Salmonella carrying the blaCMY-2 gene were recently recovered from bovine and porcine salmonellosis cases (Dahshan et al., 2010; Sugawara et al., 2011). Besides, S. Infantis isolates harboring the blaTEM-52 gene were reported from broilers for the first time in the year 2004 (Shahada et al., 2010a). Thus, a major concern has been increased prevalence of resistance to ESCs noted in S. Infantis isolates. This study was conducted to determine the antimicrobial resistance phenotypes and characterize the β-lactamase genes and the plasmids harboring these genes in S. Infantis isolated from broiler flocks.
Materials and Methods
Bacterial Isolates
Between January, 2007 and December, 2008, a total of 1,472 cecal specimens derived from 92 broiler flocks (ca. 10,000 birds per flock) were collected from a poultry processing plant located in the southern part of Japan. Usually, 16 samples per flock were randomly selected fortnightly. The isolation, identification and serotyping of S. Infantis isolates were performed at the Laboratory of Veterinary Public Health, Kagoshima University, Japan as previously described (Shahada et al., 2008).
Determination of Minimum Inhibitory Concentrations
Antimicrobial susceptibility testing was assayed by the agar dilution method on Mueller-Hinton (MH) agar (Oxoid Ltd., Basingstoke, Hampshire, England) plates according to the National Committee for Clinical and Laboratory Standards guidelines (National Committee for Clinical Laboratory Standards, 2001). S. Infantis isolates were tested for sensitivities to AMP, cefotaxime (CTX), cefoxitin (FOX), chloramphenicol (CHL), streptomycin (STR), sulfamethoxazole (SUL), oxytetracycline (TET), kanamycin (KAN), and ofloxacin (OFX). The MIC range was set at 0.125–512 μg/ml for all tested antimicrobial agents. The MIC breakpoints were interpreted according to the new criteria established by the Clinical and Laboratory Standards Institute (2012). Escherichia coli (E. coli) ATCC 25922 and Staphylococcus aureus ATCC29213 were used as quality control strains.
Double-Disk Synergy Assay
The double-disk synergy testing was conducted to screen for ESBLs and AmpC β-lactamases as previously described (Shahada et al., 2010a). This test was performed as a standard Kirby-Bauer disk diffusion assay on MH agar (Oxoid) plates.
Detection of Resistance Determinants
All DNA templates were prepared using the InstaGene Matrix kit (Bio-Rad Laboratories, Hercules, CA, USA). Detection of resistance genes was performed by polymerase chain reaction (PCR). The amplification reactions were carried out using primers and conditions as previously described (Dahshan et al., 2010; Shahada et al., 2010b). Briefly, the targets were as follows: blaTEM, blaOXA, blaPSE, blaSHV, blaCTX-M-1 group, blaCTX-M-2 group, blaCTX-M-25, and Toho-1 encoding for penicillinases; blaCIT, blaCMY-2, blaDHA, blaFOX, blaMOX, blaACC, and blaEBC encoding for cephalosporinases; tet(A), tet(B), and tet(G) mediating tetracycline efflux proteins; aadA1 and aadA2 encoding for resistance to STR and spectinomycin; sul1 conferring resistance to SUL; and aphA1-Iab encoding for resistance to KAN.
When β-lactamase-encoding genes were positive following PCR amplification, obtained products were directly sequenced using a BioDye Terminator version 3.1 Ready Reaction sequencing kit and ABI 3100 automated DNA sequencer (Applied Biosystems, Foster City, CA, USA). The DNA alignments and deduced amino acid sequences were examined using the BLAST program (National Center for Biotechnology Information, USA).
Plasmid Analysis
All S. Infantis isolates were examined for the carriage of plasmids by employing the alkaline lysis method as previously described (Kado and Liu, 1981). The molecular size of plasmids was determined by using the standard Salmonella enterica serovar Choleraesuis ATCC 7001 (50 kbp) and S. Typhimurium DT104 strain 300-98 (90 kbp). The plasmid size was estimated by graphing the molecular size of standard strains versus the distance traveled from the wells using logistic graph paper. Conjugation experiments were performed as described previously (Shahada et al., 2010a). In brief, S. Infantis donor isolates and rifampicin-resistant E. coli DH5α recipient derivatives were used for conjugal mating. Conjugants were selected onto deoxycholate hydrogen sulfide lactose agar containing 128 μg/ml rifampicin and 128 μg/ml AMP, followed by antimicrobial testing and the detection of transferred resistance genes as above. Plasmids were characterized by the PCR-based replicon typing method as described previously (Carattoli et al., 2005) to detect the plasmid types: IncI1, IncA/C, IncHI1, IncHI2, IncN, IncX, IncW, IncY, IncP, IncT, IncFIIs, IncL/M, IncFIA, IncFIB, IncFIC, IncFrepB, IncK/B, and IncB/O.
Results
Antimicrobial Resistance
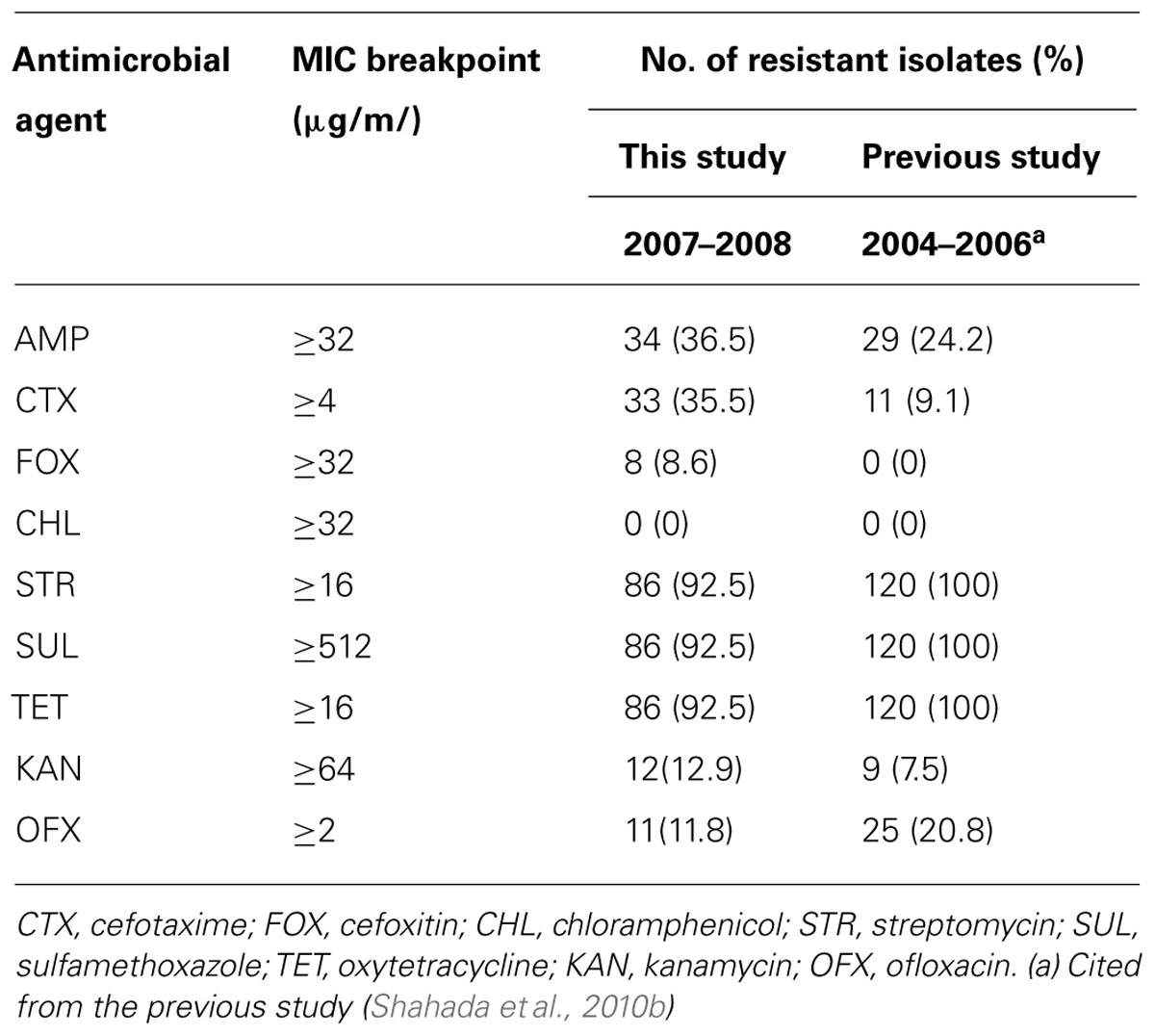
S. Infantis isolates were recovered in 54 (58.7%) of 92 flocks surveyed and 93 (6.3%) of 1,472 samples examined. Antimicrobial susceptibility profiles of the isolates are summarized in Table 1. Of 93 S. Infantis isolates recovered, 34 (36.5%) exhibited resistance to AMP (MIC, 256 to >512 μg/ml), 33 (35.5%) showed resistance to CTX (MIC, 4-256 μg/ml) and eight (8.6%) demonstrated resistance to FOX (MIC, 32–128 μg/ml). From the 93 isolates and with regard to non-β-lactam antibiotics, 86 (92.5%) isolates exhibited resistance to STR (MIC, 16–512 μg/ml, conferred by aadA1), SUL (MIC >512 μg/ml, conferred by sul1) and TET (MIC, 64–512 μg/ml, conferred by tetA); 12 (12.9%) isolates showed resistance to KAN (MIC,≥512 μg/ml, conferred by aphA1); and 11 (11.8%) isolates demonstrated resistance to OFX (MIC, 2–8 μg/ml). All 93 isolates were fully susceptible to CHL. On the basis of resistance patterns, 11 phenotypes (A to K) were demonstrated as depicted in Table 2. The most frequent pattern was STR/SUL/TET (43 isolates) followed by AMP/CTX/STR/SUL/TET (14 isolates).
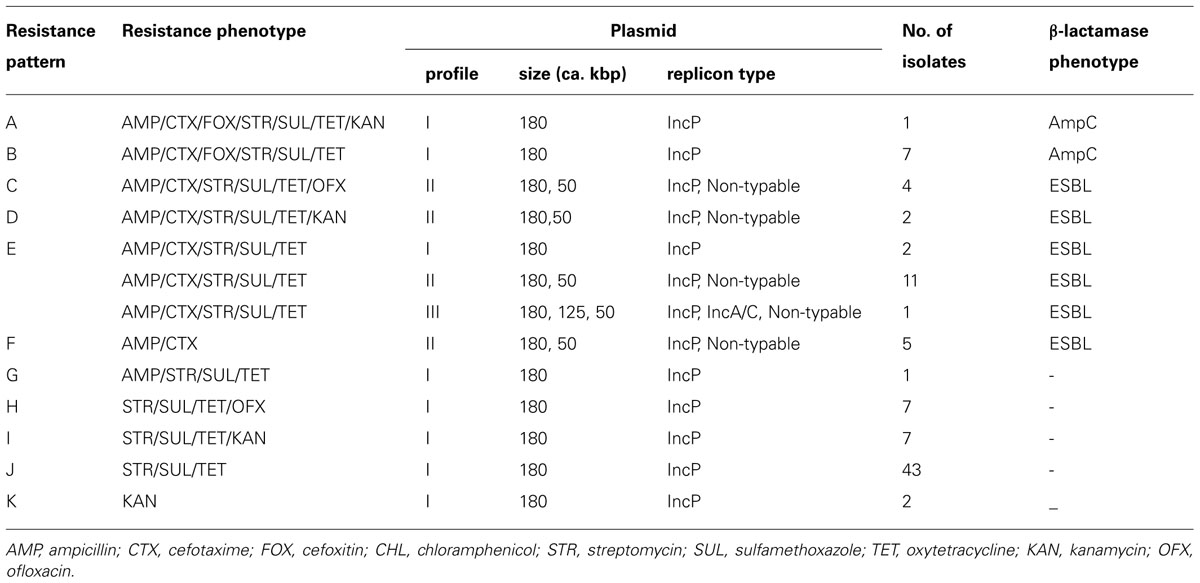
TABLE 2. Distribution of resistance phenotypes, plasmid profiles and β-lactamase phenotypes in Salmonella Infantis isolates
ESBL and AMPC Characterization
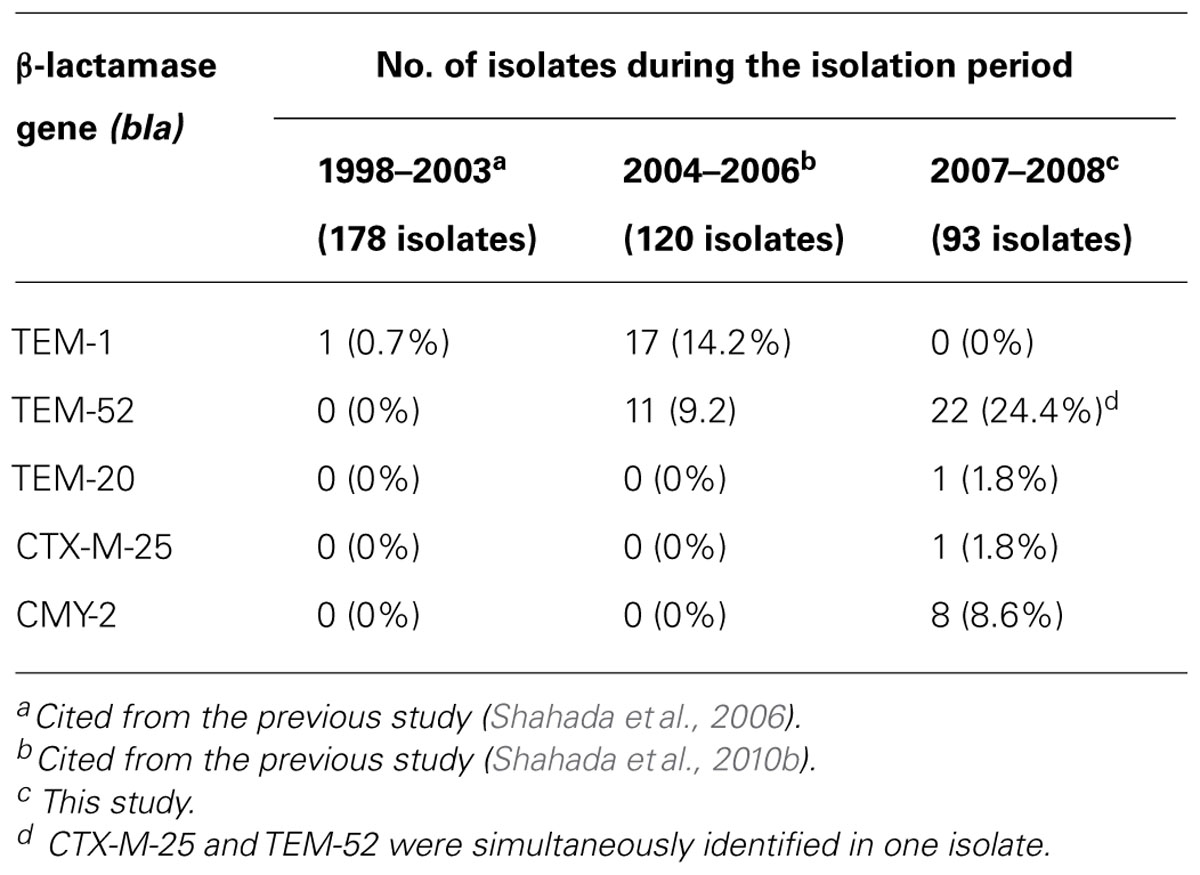
Double-disk synergy testing revealed 25 ESBL- and eight AmpC β-lactamase-producing S. Infantis isolates. TEM-type ESBL was identified in 23 (24.7%) isolates, CTX-M-type ESBL was demonstrated in one isolate and CMY-type AmpC β-lactamase was detected in eight isolates. Nucleotide sequencing of PCR products confirmed that the amplicons were blaTEM-52 (22 isolates), blaTEM-20 (one isolate), blaCTX-25 (one isolate in combination with blaTEM-52), and blaCMY-2 (8 isolates). Of 54 S. Infantis-positive flocks, As indicated in Table 3, S. Infantis isolates harboring blaTEM-1 were not detected during the present study; but, they were found during 1998–2003 (one isolate) and 2004–2006 (17 isolates). On the other hand, isolates harboring blaTEM-52 were identified during 2004–2006 (11 isolates) and 2007–2008 (22 isolates). Isolates carrying the blaTEM-20, blaCTX-25, and blaCMY-2 genes were demonstrated during 2007–2008 study period.
Plasmid Characterization
Three plasmid profiles were defined among S. Infantis isolates. Seventy isolates harbored ca. 180-kbp plasmids (profile I), 22 isolates carried two plasmids of ca. 50 and 180 kbp (profile II), and one isolate possessed three plasmids of ca. 50, 125, and 180 kbp (profile III; Figure 1). Isolates exhibiting resistance to ESCs displayed diverse profiles: Of 34 ESC-resistant S. Infantis isolates, 22 belonged to profile II, 11 were classified into profile I and one isolate was categorized into profile III (Table 2). PCR-based replicon typing revealed three kinds of plasmids. Large-size plasmids (ca.180 kbp) were typed as IncP, intermediate-size plasmid (ca. 125 kbp) was typed as IncA/C and small plasmids (ca. 50 kbp) were non-typable. Large-, intermediate- and small-size plasmids were self-transmissible to E. coli recipient strain by conjugation. The blaTEM-20 and blaCMY-2 genes were associated with large IncP plasmids, blaCTX-M-25 was affiliated with intermediate IncA/C plasmid and the blaTEM-52 gene was linked to small non-typable plasmids.
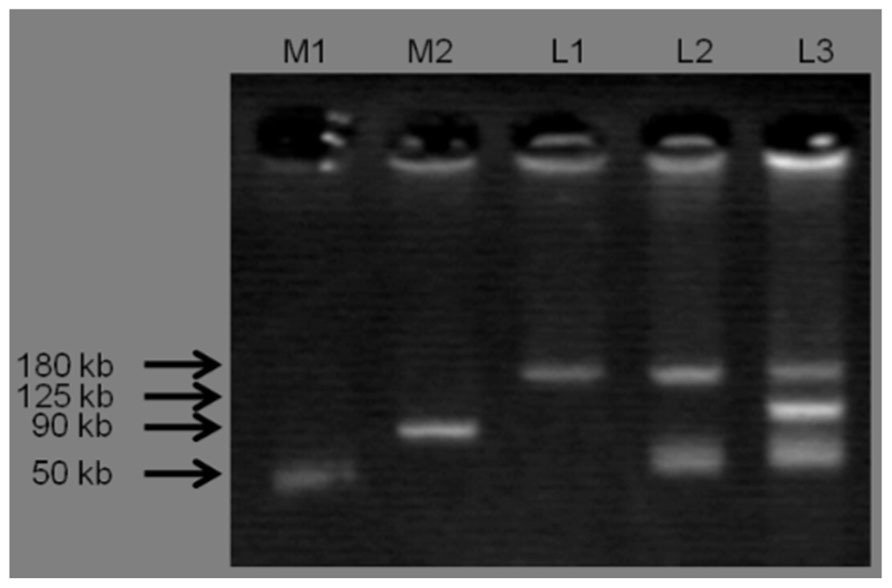
FIGURE 1. Three plasmid profiles obtained from S. Infantis isolates. Lane M1 and M2 shows standard sizes of plasmids extracted from S. Choleraesuis ATCC 7001 (50 kbp) and S. Typhimurium DT104 strain 300-98 (90 kbp), respectively. L1: IncP (ca. 180 kbp), L2: IncP (ca. 180 kbp) and Untypable plasmid (ca. 50 kbp), L3: IncP (ca. 180 kbp), IncA/C (ca. 125 kbp), and Untypable plasmid (ca. 50 kbp).
Discussion
The results obtained in this study show that resistance to ESCs has increased to 35.5% from the previous 9.2% reported during an earlier investigation (Shahada et al., 2010b). Other new findings include the detection of S. Infantis isolates harboring the blaTEM-20, blaCTX-M-25, and blaCMY-2 genes. Demonstration of these resistance traits in S. Infantis serovar is a rare phenomenon. This is the first report describing these mechanisms of ESC-resistance exhibited by S. Infantis isolates derived from broilers. Previous reports indicate that β-lactam resistance in broilers was once mediated by blaTEM-1, a narrow-spectrum β-lactamase gene that was demonstrated in one S. Infantis isolate recovered during 1998–2003 (Shahada et al., 2006) and 14.2% of the isolates obtained in 2004–2006 (Shahada et al., 2010a). Besides, during the period of 2004–2006, resistance to ESCs mediated by blaTEM-52 was reported for the first time from the broiler industry in Japan (Shahada et al., 2010a).
In the present study, we identified one S. Infantis isolate harboring the blaTEM-20 gene while other isolates had blaTEM-52. It’s worth noting that the wild-type blaTEM-1 gene was not detected during this study suggesting the likelihood of occurrence of point mutations which led to the emergence of observed variants, blaTEM-20 and blaTEM-52. This hypothesis is supported by deduced amino acid sequence analysis which revealed that the blaTEM-20 gene differs from blaTEM-1 by two substitutions, Met182→Thr and Gly238→Ser; whereas the blaTEM-52 gene differs from blaTEM-1 by three substitutions, Glu104 Lys, Met182→Thr, and Gly238→Ser (Arlet et al., 1999; Weill et al., 2004). This phenomenon has the implications for bacterial adaptation mechanism because blaTEM-1 seems to have lost its fitness as a potential resistance trait necessary for survival of Salmonella. Thus, point mutations have taken place as the evolutionary adaptation mechanism crucial for successful colonization of the broiler chicken intestinal tract.
In Japan, CTX-M-type ESBL-producing Enterobacteriaceae are important nosocomial infectious agents raising considerable concern in the public health community. Similarly, the detection of CTX-M-2 and CTX-M-25 ESBL-producing E. coli isolates from chickens affected with colibacillosis has raised concerns in the veterinary public health community (Kojima et al., 2005; Asai et al., 2011). We report for the first time one S. Infantis isolate harboring blaCTX-M-25 on IncA/C plasmid and blaTEM-52 on non-typable plasmid. Because blaCTX-M-25 was initially demonstrated in E. coli (Asai et al., 2011) a comprehensive molecular study is required to determine the likely source and elucidate mechanisms involved in collecting antimicrobial resistance traits and mobilizing them across taxonomical borders.
To date, several reports have described the emergence of AmpC β-lactamase-producing Salmonella derived from farm animals affected with salmonellosis. One of the studies involving Salmonella enterica serovar Typhimurium (S. Typhimurium) identified blaCMY-2 associated with self-transmissible IncI1-Iγ and A/C plasmids (Sugawara et al., 2011). Another study revealed a novel chromosomally integrated multi-drug resistance genomic island harboring blaCMY-2 among clonally related S. Typhimurium isolates (Shahada et al., 2011). Consequently, the detection of blaCMY-2 in several S. Infantis isolates from broilers poses another challenge in the veterinary public health community. Carriers of blaCMY-2 harbored IncP plasmids initially demonstrated in Pseudomonas bacteria (Shintani et al., 2011).
Antimicrobial susceptibility data indicate a gradual decrease of OFX resistance to 11.8 from 20.8% previously reported (Shahada et al., 2010b). Fluoroquinolones (e.g., OFX) are broad-spectrum antimicrobial agents widely used in clinical medicine. Emergence of fluoroquinolone resistance was attributed to overuse of this group of drugs in domestic animals either therapeutically or for the purpose of growth promotion (Asai et al., 2007a). Since 1991, the Japanese Ministry of Agriculture, Forestry and Fisheries (JMAFF) approved this class of antimicrobials in veterinary medicine for therapeutic purposes and prohibited its use as feed additives. Fluoroquinolones might have been prescribed prudently among antimicrobial agents used in broiler farms in Japan. This likely has contributed to the decline of resistance to OFX observed in the present study.
Taken together, these results show increased resistance against ESCs mediated by both ESBLs and AmpC β-lactamases. It seems these resistance traits have spread among farm animals in Japan while their likely source remains largely unknown. The probability that resistance to ESCs may continue to spread among members of the Enterobacteriaceae family poses another public health challenge because ESBLs and AmpC β-lactamases limit the effectiveness of cephalosporin therapy.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
This study was partly supported by a Grants-in-Aid for Scientific Research (No.23580426) from the Japan Society for the Promotion of Science.
References
Arlet, G., Barrett, T. J., Butaye, P., Cloeckaert, A., Mulvey, M. R., and White, D. G. (2006). Salmonella resistant to extended-spectrum cephalosporins: prevalence and epidemiology. Microbes Infect. 8, 1945–1954.
Arlet, G., Goussard, S., Courvalin, P., and Philippon, A. (1999). Sequences of the genes for the TEM-20, TEM-21, TEM-22, and TEM-29 extended-spectrum beta-lactamases. Antimicrob. Agents Chemother. 43, 969–971.
Asai, T., Harada, K., Ishihara, K., Kojima, A., Sameshima, T., Tamura, Y., et al. (2007a). Association of antimicrobial resistance in Campylobacter from food-producing animals with antimicrobial use on farms. Jpn. J. Infect. Dis. 60, 290–294.
Asai, T., Ishihara, K., Harada, K., Kojima, A., Tamura, Y., Sato, S., et al. (2007b). Long-term prevalence of antimicrobial-resistant Salmonella enterica subspecies enterica serovar Infantis in the broiler chicken industry in Japan. Microbiol. Immunol. 51, 111–115.
Asai, T., Masani, K., Sato, C., Hiki, M., Usui, M., Baba, K., et al. (2011). Phylogenetic groups and cephalosporin resistance genes of Escherichia coli from diseased food-producing animals in Japan. Acta Vet. Scand. 53, 52.
Bonnet, R. (2004). Growing group of extended-spectrum beta-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 48, 1–14.
Carattoli, A., Bertini, A., Villa, L., Falbo, V., Hopkins, K. L., and Threlfall, E. J. (2005). Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63, 219–228.
Chittick, P., Sulka, A., Tauxe, R. V., and Fry, A. M. (2006). A summary of national reports of foodborne outbreaks of Salmonella Heidelberg infections in the United States: clues for disease prevention. J. Food Prot. 69, 1150–1153.
Clinical and Laboratory Standards Institute. (2012). Performance Standards for Antimicrobial Susceptibility Testing: 22th Information Supplement M100-S22, Wayne, PA.
Dahshan, H., Chuma, T., Shahada, F., Akiba, M., Fujimoto, H., Akasaka, K., et al. (2010). Characterization of antibiotic resistance and the emergence of AmpC-producing Salmonella Infantis from pigs. J. Vet. Med. Sci. 72, 1437–1442.
Iwabuchi, E., Yamamoto, S., Endo, Y., Ochiai, T., and Hirai, K. (2011). Prevalence of Salmonella isolates and antimicrobial resistance patterns in chicken meat throughout Japan. J. Food Prot. 74, 270–273.
Kado, C. I., and Liu, S. T. (1981). Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145, 1365–1373.
Kimura, A. C., Reddy, V., Marcus, R., Cieslak, P. R., Mohle-Boetani, J. C., Kassenborg, H. D., et al. (2004). Chicken consumption is a newly identified risk factor for sporadic Salmonella enterica serotype Enteritidis infections in the United States: a case-control study in FoodNet sites. Clin. Infect. Dis. 38, S244–S252.
Kojima, A., Ishii, Y., Ishihara, K., Esaki, H., Asai, T., Oda, C., et al. (2005). Extended-spectrum-beta-lactamase-producing Escherichia coli strains isolated from farm animals from 1999 to 2002: report from the Japanese Veterinary Antimicrobial Resistance Monitoring Program. Antimicrob. Agents Chemother. 49, 3533–3537.
Majowicz, S. E., Musto, J., Scallan, E., Angulo, F. J., Kirk, M., O’Brien, S. J., et al. (2010). The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis. 50, 882–889.
National Committee for Clinical Laboratory Standards. (2001). Performance Standards for Antimicrobial Disk and Dilution Susceptibility Test for Bacteria Isolated from Animals-Second Edition: Approved Standard M31-A2, Wayne, PA.
Shahada, F., Chuma, T., Tobata, T., Okamoto, K., Sueyoshi, M., and Takase, K. (2006). Molecular epidemiology of antimicrobial resistance among Salmonella enterica serovar Infantis from poultry in Kagoshima, Japan. Int. J. Antimicrob. Agents 28, 302–307.
Shahada, F., Chuma, T., Okamoto, K., and Sueyoshi, M. (2008). Temporal distribution and genetic fingerprinting of Salmonella in broiler flocks from southern Japan. Poult. Sci. 87, 968–972.
Shahada, F., Chuma, T., Dahshan, H., Akiba, M., Sueyoshi, M., and Okamoto, K. (2010a). Detection and characterization of extended-spectrum beta-lactamase (TEM-52)-producing Salmonella serotype Infantis from broilers in Japan. Foodborne Pathog. Dis. 7, 515–521.
Shahada, F., Sugiyama, H., Chuma, T., Sueyoshi, M., and Okamoto, K. (2010b). Genetic analysis of multi-drug resistance and the clonal dissemination of beta-lactam resistance in Salmonella Infantis isolated from broilers. Vet. Microbiol. 140, 136–141.
Shahada, F., Sekizuka, T., Kuroda, M., Kusumoto, M., Ohishi, D., Matsumoto, A., et al. (2011). Characterization of Salmonella enterica serovar Typhimurium isolates harboring a chromosomally encoded CMY-2 beta-lactamase gene located on a multidrug resistance genomic island. Antimicrob. Agents Chemother. 55, 4114–4121.
Shintani, M., Tokumaru, H., Takahashi, Y., Miyakoshi, M., Yamane, H., Nishida, H., et al. (2011). Alterations of RNA maps of IncP-7 plasmid pCAR1 in various Pseudomonas bacteria. Plasmid 66, 85–92.
Sugawara, M., Komori, J., Kawakami, M., Izumiya, H., Watanabe, H., and Akiba, M. (2011). Molecular and phenotypic characteristics of CMY-2 beta-lactamase-producing Salmonella enterica serovar Typhimurium isolated from cattle in Japan. J. Vet. Med. Sci. 73, 345–349.
Keywords: antimicrobial resistance, β-lactamase gene, broiler, extended-spectrum cephalosporin,Salmonella enterica serovar Infantis, plasmids
Citation: Chuma T, Miyasako D, Dahshan H, Takayama T, Nakamoto Y, Shahada F, Akiba M and Okamoto K (2013) Chronological change of resistance to β-lactams in Salmonella enterica serovar Infantis isolated from broilers in Japan. Front. Microbiol. 4:113. doi:10.3389/fmicb.2013.00113
Received: 05 January 2013; Paper pending published: 20 March 2013;
Accepted: 23 April 2013; Published online: 21 May 2013.
Edited by:
Kunihiko Nishino, Osaka University, JapanReviewed by:
Bruno Gonzalez-Zorn, Universidad Complutense de Madrid, SpainBéla Nagy, Hungarian Academy of Sciences, Hungary
Copyright: © 2013 Chuma, Miyasako, Dahshan, Takayama, Nakamoto, Shahada, Akiba and Okamoto. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Takehisa Chuma, Laboratory of Veterinary Public Health, Department of Veterinary Medicine, Faculty of Agriculture, Kagoshima University, 1-21-24 Korimoto, Kagoshima 890-0065, Japan. e-mail:Y2h1bWFAdmV0LmthZ29zaGltYS11LmFjLmpw

 Daisuke Miyasako1
Daisuke Miyasako1