- Agricultural Bioinformatics Research Unit, Graduate School of Agricultural and Life Sciences, University of Tokyo, Tokyo, Japan
In bacteria and archaea, genome size and guanine–cytosine (GC) content are correlated (Bentley and Parkhill, 2004; Musto et al., 2006; Mitchell, 2007; Suzuki et al., 2008; Guo et al., 2009). These parameters show greater correlation in bacteria (Pearson's correlation coefficient r = 0.46) than in archaea (r = 0.195) (Nishida, 2012a). The GC content in bacteria varies widely from 13.5% in “Candidatus Zinderia insecticola” (McCutcheon and Moran, 2010) to 74.9% in Anaeromyxobacter dehalogenans (Thomas et al., 2008). Although the GC content is similar among closely related bacteria, sometimes, the GC content is similar in phylogenetically distant bacteria. The distribution of GC content in bacterial genomes differs from a Gaussian distribution with multiple peaks.
Bacterial chromosome organization is mediated by nucleoid-associated proteins (NAPs) (Wang et al., 2011). The specificity of NAP–DNA binding is determined by the differences in the GC content in specific regions of the DNA (Lucchini et al., 2006; Navarre et al., 2006; Castang et al., 2008; Smits and Grossman, 2010; Yun et al., 2010; Gordon et al., 2011). For example, the Salmonella NAP specifically binds to DNA regions with low GC content and inhibits expression of the genes present in these regions (Lucchini et al., 2006; Navarre et al., 2006). NAPs vary among bacteria (Ali et al., 2012). In addition, the NAP genes are located in the plasmid as well as in the chromosome, suggesting that these genes have been distributed via plasmids (Takeda et al., 2011). I hypothesize that the GC content distribution may be related to the variation in bacterial NAPs (Nishida, 2012b, 2013). However, the correlation between the genome size and GC content in bacteria is poorly understood.
The obligate host-associated bacteria contain short genomes with low GC content (Mira et al., 2002; Moran, 2002; McCutcheon and Moran, 2012). Insertion sequence elements play an important role in the genome reduction of the host-associated bacteria (Song et al., 2010), whose small population size and asexual mode of reproduction lead to reduction of the genome size. In addition, deletion of the genes involved in DNA repair may contribute to a GC-poor genome (Moran et al., 2008). However, genome size reduction is not limited to the obligate host-associated bacteria (Nilsson et al., 2005). Generally, bacteria show a bias toward genomic deletions than insertions (Mira et al., 2001). Thus, bacteria must acquire additional genes to adapt to different environments.
Some bacterial lineages, for example, Actinobacteria, have maintained long genomes with high GC content. Plasmids (and viruses) have played an important role in additional gene uptake into chromosomes (Davison, 1999; Sørensen et al., 2005; Harrison and Brockhurst, 2012). Occasionally, the plasmid DNA gets integrated into the host chromosomal DNA (Harrison and Brockhurst, 2012). In addition, viral DNA occasionally remains in the chromosome as a prophage. Horizontally transferred DNA, plasmid DNA, and virus DNA have lower GC content than host chromosome DNAs (Rocha and Danchin, 2002). In a previous study, I compared the GC content across 953 pairs of bacterial chromosomes and plasmids. Among the 953 pairs, 746 (78.3%) pairs showed <10% difference in the GC content of the plasmid DNA and the host chromosomal DNA (Nishida, 2012a). Probably, most bacteria are unable to maintain and regulate plasmids that show very different GC content from their own. However, why most bacteria have not acquired DNAs with GC content higher than that of their own chromosome, but have acquired DNAs with lower GC content is not clear.
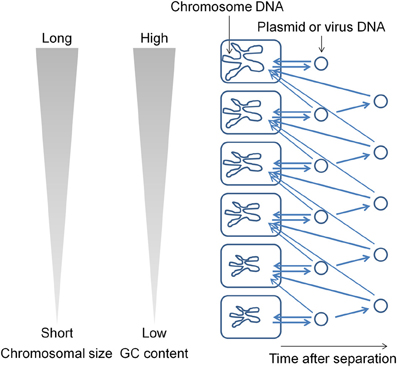
During evolution, DNA base mutations occurred intracellulaly and not in extracellular environments. The DNA polymerase components that are involved in DNA replication directly influence the base composition of the genome (Zhao et al., 2007; Wu et al., 2012). Variations in the bacterial genome DNA sequences cannot be fully explained on the basis of neutral mutations alone (Sueoka, 1988). In bacterial genomes, mutations from GC to adenosine–thymine (AT) are more common than mutations from AT to GC (Lind and Andersson, 2008; Hershberg and Petrov, 2010; Hildebrand et al., 2010; Rocha and Feil, 2010). I hypothesize that plasmids (and viruses) have been generated from the chromosome (Frontiers Research Topics, “Evolution and function of bacterial and archaeal genome sequences”). The host bacterial genome DNA has undergone a series of changes during evolution to become AT rich. On the other hand, the GC content in a plasmid that is in an extracellular environment would not change. Such a plasmid will not be accepted by the original host bacterium because most bacteria appear unable to acquire DNAs with GC content higher than that in their own chromosome. Such plasmids are transferred to bacteria that contain genomes with GC content higher than that in the plasmid (Figure 1). This natural system may be effective for obtaining useful genetic information (DNA fragments) from phylogenetically distant bacteria. I propose that the genome size and GC content in bacteria are correlated because genetic information has been transferred from AT-rich chromosomes to GC-rich chromosomes during evolution. If most bacteria could acquire DNAs with GC content higher than that in their own chromosome in addition to the lower GC content DNAs, then the flow of DNA fragments would not be biased and the genome size and GC content would not show a correlation. The results of previous studies indicate that bacteria contain a system (or systems) to generate and maintain GC content differences in the chromosomal DNA (e.g., Lawrence and Ochman, 1997). In order to maintain GC content, bacteria should estimate the DNA base composition of the transferred DNA fragments.
References
Ali, S. S., Xia, B., Liu, J., and Navarre, W. W. (2012). Silencing of foreign DNA in bacteria. Curr. Opin. Microbiol. 15, 175–181.
Bentley, S. D., and Parkhill, J. (2004). Comparative genomic structure of prokaryotes. Ann. Rev. Genet. 38, 771–792.
Castang, S., McManus, H. R., Turner, K. H., and Dove, S. L. (2008). H-NS family members function coordinately in an opportunistic pathogen. Proc. Natl. Acad. Sci. U.S.A. 105, 18947–18952.
Gordon, B. R. G., Li, Y., Cote, A., Weirauch, M. T., Ding, P., Hughes, T. R., et al. (2011). Structural basis for recognition of AT-rich DNA by unrelated exnogeneic silencing proteins. Proc. Natl. Acad. Sci. U.S.A. 108, 10690–10695.
Guo, F. B., Lin, H., and Huang, J. (2009). A plot of G + C content against sequence length of 640 bacterial chromosomes shows the points are widely scattered in the upper triangular area. Chromosome Res. 17, 359–364.
Harrison, E., and Brockhurst, M. A. (2012). Plasmid-mediated horizontal gene transfer is a coevolutionary process. Trends Microbiol. 20, 262–267.
Hershberg, R., and Petrov, D. A. (2010). Evidence that mutation is universally biased towards AT in bacteria. PLoS Genet. 6:e1001115. doi: 10.1371/journal.pgen.1001115
Hildebrand, F., Meyer, A., and Eyre-Walker, A. (2010). Evidence of selection upon genomic GC-content in bacteria. PLoS Genet. 6:e1001107. doi: 10.1371/journal.pgen.1001107
Lawrence, J. G., and Ochman, H. (1997). Amelioration of bacterial genomes: rates of change and exchange. J. Mol. Evol. 44, 383–397.
Lind, P. A., and Andersson, D. I. (2008). Whole-genome mutational biases in bacteria. Proc. Natl. Acad. Sci. U.S.A. 105, 17878–17883.
Lucchini, S., Rowley, G., Goldberg, M. D., Hurd, D., Harrison, M., and Hinton, J. C. (2006). H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2:e81. doi: 10.1371/journal.ppat.0020081
McCutcheon, J. P., and Moran, N. A. (2010). Functional convergence in reduced genomes of bacterial symbionts spanning 200 My of evolution. Genome Biol. Evol. 2, 708–718.
McCutcheon, J. P., and Moran, N. A. (2012). Extreme genome reduction in symbiotic bacteria. Nat. Rev. Microbiol. 10, 13–26.
Mira, A., Klasson, L., and Andersson, S. G. (2002). Microbial genome evolution: sources of variability. Curr. Opin. Microbiol. 5, 506–512.
Mira, A., Ochman, H., and Moran, N. A. (2001). Deletional bias and the evolution of bacterial genomes. Trends Genet. 17, 589–596.
Mitchell, D. (2007). GC content and genome length in Chargaff compliant genomes. Biochem. Biophys. Res. Commun. 353, 207–210.
Moran, N. A. (2002). Microbial minimalism: genome reduction in bacterial pathogens. Cell 108, 583–586.
Moran, N. A., McCutcheon, J. P., and Nakabachi, A. (2008). Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 42, 165–190.
Musto, H., Naya, H., Zavala, A., Romero, H., Alvarez-Valín, F., and Bernardi, G. (2006). Genomic GC level, optimal growth temperature, and genome size in prokaryotes. Biochem. Biophys. Res. Commun. 347, 1–3.
Navarre, W. W., Porwollik, S., Wang, Y., McClelland, M., Rosen, H., Libby, S. J., et al. (2006). Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313, 236–238.
Nilsson, A. I., Koskiniemi, S., Eriksson, S., Kugelberg, E., Hinton, J. C. D., and Andersson, D. I. (2005). Bacterial genome size reduction by experimental evolution. Proc. Natl. Acad. Sci. U.S.A. 102, 12112–12116.
Nishida, H. (2012a). Comparative analyses of base composition, DNA sizes, and dinucleotide frequency profiles in archaeal and bacterial chromosomes and plasmids. Int. J. Evol. Biol. 2012, 342482.
Nishida, H. (2013). Genome DNA sequence variation, evolution, and function in bacteria and archaea. Curr. Issues Mol. Biol. 15, 19–24.
Rocha, E. P., and Danchin, A. (2002). Base composition bias might result from competition for metabolic resources. Trends Genet. 18, 291–294.
Rocha, E. P., and Feil, E. J. (2010). Mutational patterns cannot explain genome composition: are there any neutral sites in the genomes of bacteria? PLoS Genet. 6:e1001104. doi: 10.1371/journal.pgen.1001104
Smits, W. K., and Grossman, A. D. (2010). The transcriptional regulator Rok binds A+T-rich DNA and is involved in repression of a mobile genetic element in Bacillus subtilis. PLoS Genet. 6:e1001207. doi: 10.1371/journal.pgen.1001207
Song, H., Hwang, J., Yi, H., Ulrich, R. L., Yu, Y., Nierman, W. C., et al. (2010). The early stage of bacterial genome-reductive evolution in the host. PLoS Pathog. 6:e1000922. doi: 10.1371/journal.ppat.1000922
Sørensen, S. J., Bailey, M., Hansen, L. H., Kroer, N., and Wuertz, S. (2005). Studying plasmid horizontal transfer in situ: a critical review. Nat. Rev. Microbiol. 3, 700–710.
Sueoka, N. (1988). Directional mutation pressure and neutral molecular evolution. Proc. Natl. Acad. Sci. U.S.A. 85, 2653–2657.
Suzuki, H., Sota, M., Brown, C. J., and Top, E. M. (2008). Using Mahalanobis distance to compare genomic signatures between bacterial plasmids and chromosomes. Nucleic Acids Res. 36, e147.
Takeda, T., Yun, C.-S., Shintani, M., Yamane, H., and Nojiri, H. (2011). Distribution of genes encoding nucleoid-associated protein homologs in plasmids. Int. J. Evol. Biol. 2011, 685015.
Thomas, S. H., Wagner, R. D., Arakaki, A. K., Skolnick, J., Kirby, J. R., Shimkets, L. J., et al. (2008). The mosaic genome of Anaeromyxobacter dehalogenans strain 2CP-C suggests an aerobic common ancestor to the delta-Proteobacteria. PLoS ONE 3:e2103. doi: 10.1371/journal.pone.0002103
Wang, W., Li, G.-W., Chen, C., Xie, X. S., and Zhuang, X. (2011). Chromosome organization by a nucleoid-associated protein in live bacteria. Science 333, 1445–1449.
Wu, H., Zhang, Z., Hu, S., and Yu, J. (2012). On the molecular mechanism of GC content variation among eubacterial genomes. Biol. Direct 7, 2.
Yun, C. S., Suzuki, C., Naito, K., Takeda, T., Takahashi, Y., Sai, F., et al. (2010). Pmr, a histone-like protein H1 (H-NS) family protein encoded by the lncP-7 plasmid pCAR1, is a key global regulator that alters host function. J. Bacteriol. 192, 4720–4731.
Citation: Nishida H (2012) Evolution of genome base composition and genome size in bacteria. Front. Microbio. 3:420. doi: 10.3389/fmicb.2012.00420
Received: 19 October 2012; Accepted: 21 November 2012;
Published online: 06 December 2012.
Edited by:
Patrick S. G. Chain, Lawrence Livermore National Laboratory, USAReviewed by:
Patrick S. G. Chain, Lawrence Livermore National Laboratory, USACopyright © 2012 Nishida. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: ahn@mail.ecc.u-tokyo.ac.jp