- 1Department of Crop and Soil Science, Oregon State University, Corvallis, OR, USA
- 2Department of Microbiology, Oregon State University, Corvallis, OR, USA
We examine and discuss literature targeted at identifying “active” subpopulations of soil microbial communities with regard to the factors that affect the balance between mineralization and immobilization/assimilation of N. Whereas a large fraction (≥50%) of soil microbial biomass can immediately respire exogenous substrates, it remains unclear what percentage of both bacterial and fungal populations are capable of expressing their growth potential. The factors controlling the relative amounts of respiratorily responsive biomass versus growth-active biomass will impact the balance between N mineralization and N immobilization. Stable isotope probing of de novo DNA synthesis, and pyrosequence analyses of rRNA:rDNA ratios in soils have identified both numerically dominant and rare microbial taxa showing greatest growth potential. The relative growth responses of numerically dominant or rare members of a soil community could influence the amount of N immobilized into biomass during a “growth” event. Recent studies have used selective antibiotics targeted at protein synthesis to measure the relative contributions of fungi and bacteria to ammonification and
Introduction
The soil N cycle consists of several N pools and inter-connected transformations (Figure 1). Microbial biomass is central to the processes involved in producing and immobilizing inorganic N. Furthermore, microbial biomass contributes directly to the pool of soil organic N through its death and turnover. The placement of soil microbial diversity and dynamics into context with biological functions associated with the N cycle still remains a grand challenge (Myrold and Bottomley, 2008; Strickland et al., 2009; McGuire and Treseder, 2010). Conservation of soil N depends on the maintenance of a balance between the rate of depolymerization of organic N, the portion of it that is mineralized to
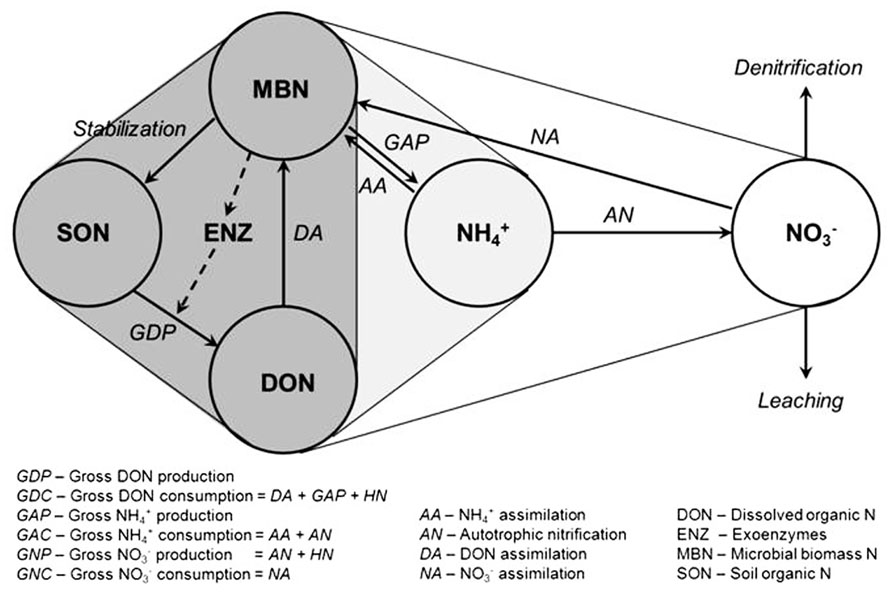
FIGURE 1. Diagram of the microbial N cycle in aerobic soil. Major pools of N are shown in circles, major fluxes by solid arrows, and the dashed arrow represents the production of exoenzymes (e.g., proteases) for the depolymerization of soil organic N. Pools and fluxes in dark gray relate to turnover of dissolved organic N, those in light gray relate to turnover of
How Does Gross Mineralization of N (Nmin) and Gross Immobilization of N (Nassim) Occur in the Same Soil Volume?
For >50 year soil scientists have measured fluxes of
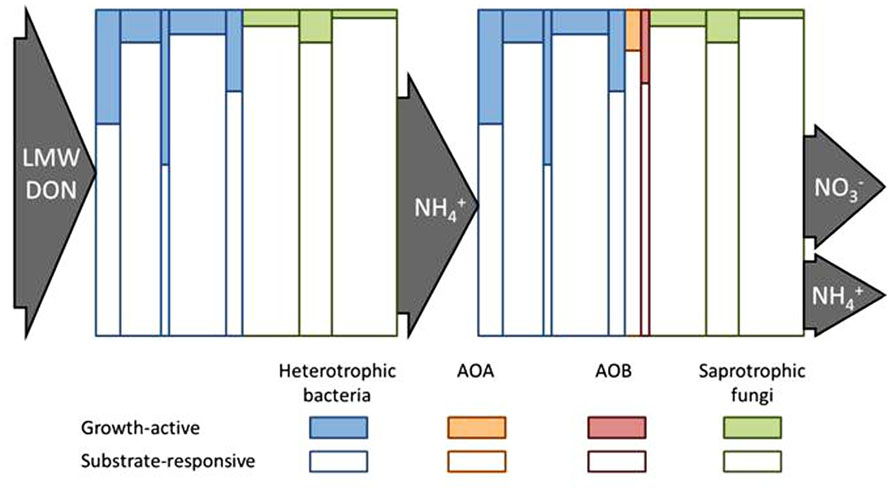
FIGURE 2. Conceptual diagram to illustrate how the fate of soil N pools might be controlled by the growth-active fraction (GAF) of the substrate-responsive population (SRP) of microorganisms. The heights of the individual vertical columns represent the SRP of different taxa and their widths represent the relative sizes of each taxa as part of the whole microbial community. The shaded portion of each column represents the GAF of each SRP taxon. The left side depicts that low molecular weight (LMW) dissolved organic N (DON) is taken up and metabolized by the SRP but N is only assimilated by the GAF. The balance between
Substrate-Responsive Population: Growth Ready or not?
Whereas it is well accepted that the vast majority (>80%) of soil microbes probably reside in some state of dormancy, several studies have shown that at least one-half of soil microbial populations are respiratorily active (Jones and Lennon, 2010; Lennon and Jones, 2011; Hobbie and Hobbie, 2012). Furthermore, it is well known that soil respiration responds rapidly (within 1–3 h) to the addition of exogenous C substrates, including amino acids (Jones and Murphy, 2007; Jones et al., 2009). This response is the basis of the well-established substrate-induced respiration (SIR) assay used to measure soil microbial biomass (Anderson and Domsch, 1978). In SIR the rule-of-thumb is that 40 μg of respiratorily active soil biomass C generates a respiratory response of ~0.48 μg CO2-C/h. With this relationship in mind, a recent series of papers by Rousk et al. (2008, 2009, 2010b) focused on factors influencing the relative contributions of fungi and bacteria to soil activities. They determined that substrate-induced soil microbial biomass ranged between 100 and 200 μg microbial biomass C/g soil which would support a substrate-induced rate of respiration of ~1.3–2.6 μg CO2-C/g soil/h or (~30–60 μg CO2-C/g soil/day). The bacterial and fungal growth potentials of the same soils were measured by following 3H-leucine incorporation into hot trichloroacetic acid (TCA) precipitable material (presumed to be protein), and 14C acetate incorporation into ergosterol, respectively. The rates of leucine assimilation ranged between 10 and 100 pmol/g soil/h. We have attempted to extrapolate from these data to obtain a rate of N assimilation into protein. We used the following correction factors/assumptions, that ~50% of assimilated leucine is incorporated into protein, and that only 10% of the total soil bacterial population is extracted and represented in the soil slurry assay (Baath, 1994, 1998; Rousk, personal communication). We further assumed that leucine incorporation into protein tracks total protein synthesis, that leucine represents ~8–10% of microbial cell protein (Kirchman et al., 1985; Neidhardt et al., 1990), and that the average N content of protein is 16%. These rates of leucine incorporation extrapolate to ~0.28–2.8 μg N assimilated/g soil/day. In the case of fungal growth potential, the rates of acetate incorporation into ergosterol (C28H44O, mw = 396) ranged between 10 and 80 pmol acetate incorporated/g soil/h, equivalent to ~0.7–5.7 pmol ergosterol synthesized/g soil/h (assuming a minimum of 14 acetate molecules required per molecule of ergosterol), and ~ 6.8–54.4 ng ergosterol synthesized/g soil/day. Using the authors’ biomass conversion factor (5 ng ergosterol/1 μg fungal biomass), the ergosterol synthesis rate extrapolates to 1.3–10.9 μg fungal biomass formed/g soil/day. Assuming that fungal biomass is 45% C and has a C:N of 10:1 this growth rate is equivalent to ~0.06–0.48 μg N immobilized/g soil/day. From the sum of the leucine/ergosterol assays the rate of N assimilation ranges from 0.34 to 3.4 μg N/g/day. These values fall within the range of
Important unanswered questions are: (1) to what extent does the dormant, substrate-responsive microbial population actually process DON and contribute to Nmin? (2) How quickly does a substrate-responsive population transition into a N assimilatory sink and shift the Nmin:Nassim balance? (3) How well does the leucine plus acetate assimilatory subpopulations of bacteria and fungi detected in the Rousk et al. (2008, 2009, 2010b) studies represent the
What Controls the Outcome of Competition for
It has been shown that the extent of
Identification of “Potentially Growth-Active” Subpopulations by Targeting Nucleic Acid Synthesis
We have raised the issue of why it might be important to delineate between substrate-responsive and growth-active subpopulations and hypothesized that the latter are most likely candidates for N immobilizing activity [see How Does Gross Mineralization of N (Nmin) and Gross Immobilization of N (Nassim) Occur in the Same Soil Volume?, Substrate-Responsive Population: Growth Ready or Not?, and What Controls the Outcome of Competition for
BrdU-Labeled DNA
McMahon et al. (2011) used the thymidine analog, 5-bromo-3-deoxyuridine (BrdU), to identify the growth responsive subpopulation in Arctic tundra shrub and tussock associated soils at different times of the year. BrdU was added to soil along with a small quantity of various monomeric substrates (100 μg C/g soil of either glucose, glutamate, or vanillin) sufficient to “act as a tracer of extant microbial activity” and yet insufficient to stimulate overall CO2 production. Winter and summer soils were incubated at -2°C for 28 days, and at 4°C for 2 days, respectively. After incubation DNA was extracted, BrdU-labeled DNA recovered by immunocapture, and 16S rRNA genes PCR amplified and analyzed with the T-RFLP approach. None of the BrdU-labeled terminal restriction fragments (T-RFs) recovered from winter samples of shrub soil matched up with the T-RFs representing the total winter community suggesting that “winter growth-active” organisms were minor components of the community. Although there were more T-RF overlaps between the active and total communities of the summer shrub soil samples, many “active” T-RFs in the summer samples did not overlap with the total summer community. Because the balance between N cycling activities of tundra soils shifts between winter (net N mineralization dominant) and summer (net N immobilization dominant), this study serves to illustrate the importance of determining the amounts of growth that occur in the growth-active subpopulations during the winter and summer periods. This will impact the amount of N immobilized and the overall balance of the N cycle.
18O-Labeled DNA
Aanderud and Lennon (2011) targeted the growth-active members of a soil population carrying out DNA synthesis after rewetting a soil with 18O-labeled H2O. Labeled H2O was added to field dry soil to raise water content from 0.05 to 0.25 g H2O/g. Samples were incubated for 72 h, DNA was extracted, and “heavy” DNA separated from “light” DNA by gradient density centrifugation. Pyrosequencing was used to compare the composition of 16S rRNA gene sequences recovered from the light DNA at time zero with that of heavy DNA extracted after 72 h of incubation. The contributions of some phyla to the heavy DNA fraction increased by about 10% (Alpha-, Beta-, and Gamma-proteobacteria), whereas the contribution of other taxa declined (Chloroflexi, Delta-proteobacteria) implying that selective growth had occurred. Because Alpha-proteobacteria are generally more abundant than Gamma-proteobacteria in bulk soils (Lauber et al., 2009; Rousk et al., 2010a) the similar relative population increases during the rewet, implies that the amount of biomass increase and N immobilized by Alpha-proteobacteria might be greater than by Gamma-proteobacteria.
Relative Quantities of rRNA Versus rRNA Gene Sequences
Baldrian et al. (2011) extracted DNA and RNA from both the litter and humus layers of a spruce forest soil and used a pyrosequencing approach to compare the relative amounts of 16S rRNA and 16S rRNA gene sequences of bacteria, and compared the relative amounts of RNA and DNA sequences of the ITS1–5.8S–ITS2 region of fungi. Whereas several abundant fungal OTUs identified from DNA sequences were also highly enriched in RNA sequences, a substantial percentage of fungal OTUs (18%) were only found in the RNA community, implying that less abundant fungi might be potentially growth-active. This idea was further supported by the most abundant “cellobiohydrolase” (cbh) transcripts originating from less abundant fungi.
Identifying the Relative Contributions of Different Microbial Groups to N Cycle Activities Using Selective Protein Synthesis Inhibitors
Several studies have attempted to tease apart the relative contributions of different groups of soil microorganisms to N cycling activities by targeting protein synthesis (Castaldi and Smith, 1998; Laughlin and Stevens, 2002; Tungaraza et al., 2003; Castaldi, 2005; Myrold and Posavatz, 2007; Boyle et al., 2008; Rousk et al., 2008, 2009, 2010b). In this section, we highlight two published papers where protein synthesis inhibitory antibiotics were used to differentiate (a) the contributions of bacteria and fungi to
Fungal and Bacterial Contributions to N Cycling Based Upon Use of Antibiotics and 15N Isotope Pool Dilution Measurements
Boyle et al. (2008) compared the effects of the bacterial and fungal protein synthesis inhibitors, bronopol and cycloheximide on gross rates of N cycling in forest soils using the 15N isotope pool dilution method. Bronopol completely stopped
The Relative Contributions of Ammonia-Oxidizing Archaea and Bacteria to Soil Nitrification Potentials
Although the ability of AOA to oxidize NH3 and grow autotrophically in soil has been well established (Offre et al., 2009; Zhang et al., 2010; Verhamme et al., 2011), and that AOA dominate
Concluding Remarks
Over the past 25 years considerable effort has been spent refining our understanding of how the physical and chemical properties of the soil environment interact with microbial communities to maintain an overall balance between the mineralization and immobilization of soil N. During the past 10 years considerable information has been generated on the overall diversity and composition of soil microbial communities. In this review, we have selected a few recent publications that are focused upon the activities of subpopulations of soil microbes and discussed how the implications of this work may lead to a better understanding of N cycling. Clearly, this mini-review is not meant to be all inclusive, but we hope that the readers’ attention has been drawn to “soil phenomena” into which they might “dig” and “unearth” an increased level of understanding about the physiology and growth response behaviors of soil subpopulations and how they influence the balance between mineralization, nitrification, and immobilization of soil N.
Conflict of Interest Statement
The authors declare that the findings cited from their own research in this minireview was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The concepts and ideas presented in this mini-review are based upon published work of our own laboratory, of our professional colleagues, and by the thoughts of two anonymous reviewers. Our own research was funded by grants from NSF IGERT, USDA CSREES, USDA NIFA, and USDOE.
References
Aanderud, Z. T., and Lennon, J. T. (2011). Validation of heavy-water stable isotope probing for the characterization of rapidly responding soil bacteria. Appl. Environ. Microbiol. 77, 4589–4596.
Allison, S. D., and Vitousek, P. M. (2005). Responses of extracellular enzymes to simple and complex nutrient inputs. Soil Biol. Biochem. 37, 937–944.
Allison, S. D., Wallenstein, M. D., and Bradford, M. A. (2010). Soil-carbon response to warming dependent on microbial physiology. Nat. Geosci. 3, 336–340.
Anderson, J. P. E., and Domsch, K. H. (1978). Physiological method for quantitative measurement of microbial biomass in soils. Soil Biol. Biochem. 10, 215–221.
Baath, E. (1994). Measurement of protein synthesis by soil bacterial assemblages with the leucine incorporation technique. Biol. Fertil. Soils 17, 147–153.
Baath, E. (1998). Growth rates of bacterial communities in soils at varying pH: a comparison of the thymidine and leucine incorporation techniques. Microb. Ecol. 36, 316–327.
Baldrian, P., Kolarik, M., Stursova, M., Kopecky, J., Valaskova, V., Vetrovsky, T., et al. (2011). Active and total microbial communities in forest soil are largely different and highly stratified during decomposition. ISME J. 6, 248–258.
Bollmann, A., Schmidt, I., Saunders, A. M., and Nicolaisen, M. H. (2005). Influence of starvation on potential ammonia-oxidizing activity and amoA mRNA levels of Nitrosospira briensis. Appl. Environ. Microbiol. 71, 1276–1282.
Booth, M. S., Stark, J. M., and Rastetter, E. (2005). Controls on nitrogen cycling in terrestrial ecosystems: a synthetic analysis of literature data. Ecol. Monogr. 75, 139–157.
Boyle, S. A., Yarwood, R. R., Bottomley, P. J., and Myrold, D. D. (2008). Bacterial and fungal contributions to soil nitrogen cycling under Douglas fir and red alder at two sites in Oregon. Soil Biol. Biochem. 40, 443–451.
Burger, M., and Jackson, L. E. (2003). Microbial immobilization of ammonium and nitrate in relation to ammonification and nitrification rates in organic and conventional cropping systems. Soil Biol. Biochem. 35, 29–36.
Castaldi, S. (2005). Cycloheximide inhibition of peptone-induced nitrate production across a soil moisture gradient. Biol. Fertil. Soils 41, 288–290.
Castaldi, S., and Smith, K. A. (1998). Effect of cycloheximide an N2O and NO3-production in a forest and an agricultural soil. Biol. Fertil. Soils 27, 27–34.
Chen, J., and Stark, J. M. (2000). Plant species effects and carbon and nitrogen cycling in a sagebrush-crested wheatgrass soil. Soil Biol. Biochem. 32, 47–57.
Davidson, E. A., Hart, S. C., Shanks, C. A., and Firestone, M. K. (1991). Measuring gross nitrogen mineralization, immobilization, and nitrification by N-15 isotopic pool dilution in intact soil cores. J. Soil Sci. 42, 335–349.
Dworkin, J., and Shah, I. M. (2010). Exit from dormancy in microbial organisms. Nat. Rev. Microbiol. 8, 890–896.
Geisseler, D., and Horwath, W. R. (2008). Regulation of extracellular protease activity in soil in response to different sources and concentrations of nitrogen and carbon. Soil Biol. Biochem. 40, 3040–3048.
Hart, S. C., Nason, G. E., Myrold, D. D., and Perry, D. A. (1994). Dynamics of gross nitrogen transformations in an old-growth forest – the carbon connection. Ecology 75, 880–891.
Hobbie, J. E., and Hobbie, E. A. (2012). Amino acid cycling in plankton and soil microbes studied with radioisotopes: measured amino acids in soil do not reflect bioavailability. Biogeochemistry 107, 339–360.
Jiang, Q. Q., and Bakken, L. R. (1999). Comparison of Nitrosospira strains isolated from terrestrial environments. FEMS Microbiol. Ecol. 30, 171–186.
Jones, D. L., Kielland, K., Sinclair, F. L., Dahlgren, R. A., Newsham, K. K., Farrar, J. F., et al. (2009). Soil organic nitrogen mineralization across a global latitudinal gradient. Glob. Biogeochem. Cycles 23, GB1016.
Jones, D. L., and Murphy, D. V. (2007). Microbial response time to sugar and amino acid additions to soil. Soil Biol. Biochem. 39, 2178–2182.
Jones, S. E., and Lennon, J. T. (2010). Dormancy contributes to the maintenance of microbial diversity. Proc. Natl. Acad. Sci. U.S.A. 107, 5881–5886.
Kirchman, D., Knees, E., and Hodson, R. (1985). Leucine incorporation and its potential as a measure of protein synthesis by bacteria in natural aquatic systems. Appl. Environ. Microbiol. 49, 599–607.
Kirkham, D., and Bartholomew, W. V. (1954). Equations for following nutrient transformations in soil, utilizing tracer data. Proc. Soil Sci. Soc. U.S.A. 18, 33–34.
Kleiner, D. (1981). The transport of NH3 and
Lauber, C. L., Hamady, M., Knight, R., and Fierer, N. (2009). Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 75, 5111–5120.
Laughlin, R. J., and Stevens, R. J. (2002). Evidence for fungal dominance of denitrification and codenitrification in a grassland soil. Soil Sci. Soc. Am. J. 66, 1540–1548.
Lehtovirta-Morley, L. E., Stoecker, K., Vilcinskas, A., Prosser, J. I., and Nicol, G. W. (2011). Cultivation of an obligate acidophilic ammonia oxidizer from a nitrifying acid soil. Proc. Natl. Acad. Sci. U.S.A. 108, 15892–15897.
Lennon, J. T., and Jones, S. E. (2011). Microbial seed banks: the ecological and evolutionary implications of dormancy. Nat. Rev. Microbiol. 9, 119–130.
Levicnik-Hofferle, S., Nicol, G. W., Ausec, L., Mandic-Mulec, I., and Prosser, J. I. (2012). Stimulation of thaumarchaeal ammonia oxidation by ammonia derived from organic nitrogen but not added inorganic nitrogen. FEMS Microbiol. Ecol. 80, 114–123.
Manzoni, S., Porporato, A., and Schimel, J. P. (2008). Soil heterogeneity in lumped mineralization-immobilization models. Soil Biol. Biochem. 40, 1137–1148.
Martens-Habbena, W., Berube, P. M., Urakawa, H., De La Torre, J. R., and Stahl, D. A. (2009). Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461, 976–979.
McGuire, K. L., and Treseder, K. K. (2010). Microbial communities and their relevance for ecosystem models: decomposition as a case study. Soil Biol. Biochem. 42, 529–535.
McMahon, S. K., Wallenstein, M. D., and Schimel, J. P. (2011). A cross-seasonal comparison of active and total bacterial community composition in Arctic tundra soil using bromodeoxyuridine labeling. Soil Biol. Biochem. 43, 287–295.
Murphy, D. V., Recous, S., Stockdale, E. A., Fillery, I. R. P., Jensen, L. S., Hatch, D. J., et al. (2003). Gross nitrogen fluxes in soil: theory, measurement and application of 15N pool dilution techniques. Adv. Agron. 79, 69–118.
Myrold, D. D., and Bottomley, P. J. (2008). “Nitrogen mineralization and immobilization,” in Nitrogen in Agricultural Systems, eds J. S. Schepers and W. R. Raun (Madison, WI: ASA-CSSA-SSSA), 153–168.
Myrold, D. D., and Posavatz, N. R. (2007). Potential importance of bacteria and fungi in nitrate assimilation in soil. Soil Biol. Biochem. 39, 1737–1743.
Neidhardt, F. C., Ingraham, J. L., and Schaechter, M. (1990). Physiology of the Bacterial Cell: A Molecular Approach. Sunderland, MA: Sinauer Associates, Inc.
Offre, P., Prosser, J. I., and Nicol, G. W. (2009). Growth of ammonia-oxidizing archaea in soil microcosms is inhibited by acetylene. FEMS Microbiol. Ecol. 70, 99–108.
Reay, D. S., Nedwell, D. B., Priddle, J., and Ellis-Evans, J. C. (1999). Temperature dependence of inorganic nitrogen uptake: reduced affinity for nitrate at suboptimal temperatures in both algae and bacteria. Appl. Environ. Microbiol. 65, 2577–2584.
Rinnan, R., and Baath, E. (2009). Differential utilization of carbon substrates by bacteria and fungi in tundra soil. Appl. Environ. Microbiol. 75, 3611–3620.
Rousk, J., Baath, E., Brookes, P. C., Lauber, C. L., Lozupone, C., Caporaso, J. G., et al. (2010a). Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 4, 1340–1351.
Rousk, J., Brookes, P. C., and Baath, E. (2010b). Investigating the mechanisms for the opposing pH relationships of fungal and bacterial growth in soil. Soil Biol. Biochem. 42, 926–934.
Rousk, J., Brookes, P. C., and Baath, E. (2009). Contrasting soil pH effects on fungal and bacterial growth suggest functional redundancy in carbon mineralization. Appl. Environ. Microbiol. 75, 1589–1596.
Rousk, J., Demoling, L. A., Bahr, A., and Baath, E. (2008). Examining the fungal and bacterial niche overlap using selective inhibitors in soil. FEMS Microbiol. Ecol. 63, 350–358.
Schimel, J., Balser, T. C., and Wallenstein, M. (2007). Microbial stress–response physiology and its implications for ecosystem function. Ecology 88, 1386–1394.
Schimel, J. P., and Bennett, J. (2004). Nitrogen mineralization: challenges of a changing paradigm. Ecology 85, 591–602.
Schimel, J. P., and Hattenschwiler, S. (2007). Nitrogen transfer between decomposing leaves of different N status. Soil Biol. Biochem. 39, 1428–1436.
Sims, G. K., and Wander, M. M. (2002). Proteolytic activity under nitrogen or sulfur limitation. Appl. Soil Ecol. 19, 217–221.
Stockdale, E. A., Hatch, D. J., Murphy, D. V., Ledgard, S. F., and Watson, C. J. (2002). Verifying the nitrification to immobilisation ratio (N/I) as a key determinant of potential nitrate loss in grassland and arable soils. Agronomie 22, 831–838.
Stopnisek, N., Gubry-Rangin, C., Hofferle, S., Nicol, G. W., Mandic-Mulec, I., and Prosser, J. I. (2010). Thaumarchaeal ammonia oxidation in an acidic forest peat soil is not influenced by ammonium amendment. Appl. Environ. Microbiol. 76, 7626–7634.
Strickland, M. S., Lauber, C., Fierer, N., and Bradford, M. A. (2009). Testing the functional significance of microbial community composition. Ecology 90, 441–451.
Taylor, A. E., and Bottomley, P. J. (2006). Nitrite production by Nitrosomonas europaea and Nitrosospira sp. AV in soils at different solution concentrations of ammonium. Soil Biol. Biochem. 38, 828–836.
Taylor, A. E., Zeglin, L., Wanzek, T. A., Myrold, D. D., and Bottomley, P. J. (2012). Dynamics of ammonia-oxidizing archaea and bacteria populations and contributions to soil nitrification potentials. ISME J. doi: 10.1038/ismej.2012.51 [Epub ahead of print].
Taylor, A. E., Zeglin, L. H., Dooley, S., Myrold, D. D., and Bottomley, P. J. (2010). Evidence for different contributions of archaea and bacteria to the ammonia-oxidizing potential of diverse Oregon soils. Appl. Environ. Microbiol. 76, 7691–7698.
Tourna, M., Stieglmeier, M., Spang, A., Konneke, M., Schintlmeister, A., Urich, T., et al. (2011). Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc. Natl. Acad. Sci. U.S.A. 108, 8420–8425.
Tungaraza, C., Brion, N., Rousseau, V., Baeyens, W., and Goeyens, L. (2003). Influence of bacterial activities on nitrogen uptake rates determined by the application of antibiotics. Oceanologia 45, 473–489.
Verhamme, D. T., Prosser, J. I., and Nicol, G. W. (2011). Ammonia concentration determines differential growth of ammonia-oxidising archaea and bacteria in soil microcosms. ISME J. 5, 1067–1071.
Keywords: ammonium consumption, dominant and rare taxa, growth active subpopulations, N immobilization, N mineralization, nitrification, nitrogen cycling, substrate-responsive subpopulations
Citation: Bottomley PJ, Taylor AE and Myrold DD (2012) A consideration of the relative contributions of different microbial subpopulations to the soil N cycle. Front. Microbio. 3:373. doi: 10.3389/fmicb.2012.00373
Received: 02 April 2012; Paper pending published: 17 July 2012;
Accepted: 28 September 2012; Published online: 23 October 2012.
Edited by:
Graeme W. Nicol, University of Aberdeen, UKReviewed by:
Graeme W. Nicol, University of Aberdeen, UKAngela Kent, University of Illinois at Urbana-Champaign, USA
Copyright: © 2012 Bottomley, Taylor and Myrold. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Peter J. Bottomley, Department of Microbiology, Oregon State University, Nash Hall, Room 220, Corvallis, OR 97331, USA. e-mail:cGV0ZXIuYm90dG9tbGV5QG9yZWdvbnN0YXRlLmVkdQ==


