- 1Department of Synthetic Chemistry and Biological Chemistry, Graduate School of Engineering, Kyoto University, Katsura, Nishikyo-ku, Kyoto, Japan
- 2JST, CREST, Sanbancho, Chiyoda-ku, Tokyo, Japan
- 3Department of Biotechnology, College of Life Sciences, Ritsumeikan University, Noji-Higashi, Kusatsu, Shiga, Japan
- 4Department of Bioengineering, Graduate School of Bioscience and Biotechnology, Tokyo Institute of Technology, Nagatsuta, Midori-ku, Yokohama, Japan
This section provides an overview of the genetic systems developed in the Archaea. Genetic manipulation is possible in many members of the halophiles, methanogens, Sulfolobus, and Thermococcales. We describe the selection/counterselection principles utilized in each of these groups, which consist of antibiotics and their resistance markers, and auxotrophic host strains and complementary markers. The latter strategy utilizes techniques similar to those developed in yeast. However, Archaea are resistant to many of the antibiotics routinely used for selection in the Bacteria, and a number of strategies specific to the Archaea have been developed. In addition, examples utilizing the genetic systems developed for each group will be briefly described.
Introduction
Genetic manipulation, designated here as the ability to introduce, remove, or modify genes in a given organism, is a vital tool to study gene function. Deleting or overexpressing a gene may lead to phenotypic changes that provide valuable clues in determining the physiological role of the gene. Random mutagenesis and the isolation of mutant strains, followed by screening for genes that complement the mutations is a classical strategy to identify groups of genes that are involved in a particular biological function. Genetic manipulation can also be used to engineer cells to improve or introduce a desired function in a cell. The tools necessary for genetic manipulation have been developed in a wide variety of eukaryotes and bacteria, including the yeast Saccharomyces cerevisiae, the Gram-negative bacterium Escherichia coli and Gram-positive bacterium Bacillus subtilis, all of which have been subject to genome-wide gene disruption projects (Giaever et al., 2002; Kobayashi et al., 2003; Baba et al., 2006).
Compared to eukaryotes and bacteria, the development of genetic systems in Archaea is still at a modest stage. Many archaeal species have been found to be resistant against conventional antibiotics utilized for selection in bacterial genetic systems. In addition, many archaeal species can be regarded as extremophiles, preferring growth conditions that greatly differ to those of the mesophilic, aerobic model microbes such as S. cerevisiae, E. coli, and B. subtilis, which adds some difficulty to establish efficient screening methods. For example, when developing a system for hyperthermophilic archaea, the (thermo)stability of the compounds used for selection must also be taken into account, and establishing techniques necessary for growing colonies at high temperatures (and in many cases under an anaerobic environment) are necessary. These factors and others have hampered the development of archaeal genetic systems in the past, but the number of archaea with genetic systems is now increasing at a steady rate. Among the Crenarchaeotes, genetic manipulation is possible in a number of species in the genus Sulfolobus. In the Euryarchaeota, genetic systems have been developed in a number of halophiles, methanogens, and members of the Thermococcales. This section will give an overview of the genetic systems developed in these archaeal species, focusing on the principles applied for transformant selection (summarized in Table 1) and some examples of gene disruption that have led to a better understanding of gene function. An in-depth description of the individual organisms and detailed methodology, along with a historical account on the development of these systems, are available in the literature (Whitman et al., 1997; Tumbula and Whitman, 1999; Allers and Mevarech, 2005; Rother and Metcalf, 2005; Berkner and Lipps, 2008; Wagner et al., 2009; Buan et al., 2011; Leigh et al., 2011).
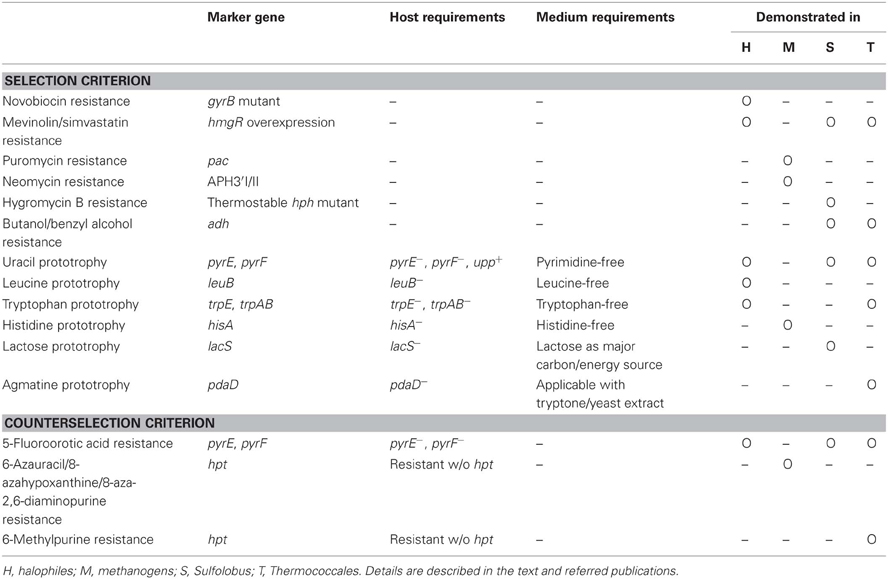
Table 1. A simple summary of the selection strategies employed for genetic manipulation in the Archaea.
Halophiles
Systems Based on Antibiotic Resistance
Genetic systems have mainly been developed in Halobacterium salinarum and Haloferax volcanii. Systems based on both antibiotic resistance markers and auxotrophic selectable markers have been established. In terms of systems based on antibiotic resistance, novobiocin, which inhibits DNA gyrase, and mevinolin/simvastatin, which inhibits 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase, are two antibiotics that have successfully been applied in halophiles belonging to the genera Haloferax and Halobacterium. DNA gyrase is a type II topoisomerase that introduces negative supercoils into DNA and whose function is essential for DNA synthesis. HMG-CoA reductase is one of the enzymes of the mevalonate pathway, which utilizes three molecules of acetyl-CoA to synthesize isopentenyl diphosphate (IPP) and its isomer dimethylallyl diphosphate (DMAPP) (Figure 1). IPP and DMAPP are precursors for isoprenoid compounds, which are particularly important for the archaea as their membrane lipids utilize isoprenoid chains. A gene that encodes a novobiocin-resistant DNA gyrase was isolated from Haloferax strain Aa2.2 and has been used as a selection marker in developing Hf. volcanii–E. coli shuttle vectors (Holmes and Dyall-Smith, 1990; Holmes et al., 1991, 1994). Furthermore in Hf. volcanii, shotgun cloning of DNA from spontaneous mevinolin-resistant strains led to the isolation of DNA fragments that could transform Hf. volcanii to mevinolin resistance, enabling the construction of shuttle vectors (Lam and Doolittle, 1989; Blaseio and Pfeifer, 1990). An examination of various Hf. volcanii mevinolin-resistant mutants have revealed that resistance is brought about by either gene amplification or up-promoter mutations, both resulting in enhanced and excess production of HMG-CoA reductase (Lam and Doolittle, 1992). Shuttle vectors such as pWL102 have been shown to also be applicable in transforming members of the genus Haloarcula (Cline and Doolittle, 1992). Additional shuttle vectors and gene disruption systems are now available in several members of the Haloarcula and their application has been demonstrated (Zhou et al., 2004; Ozawa et al., 2005; Tu et al., 2005).
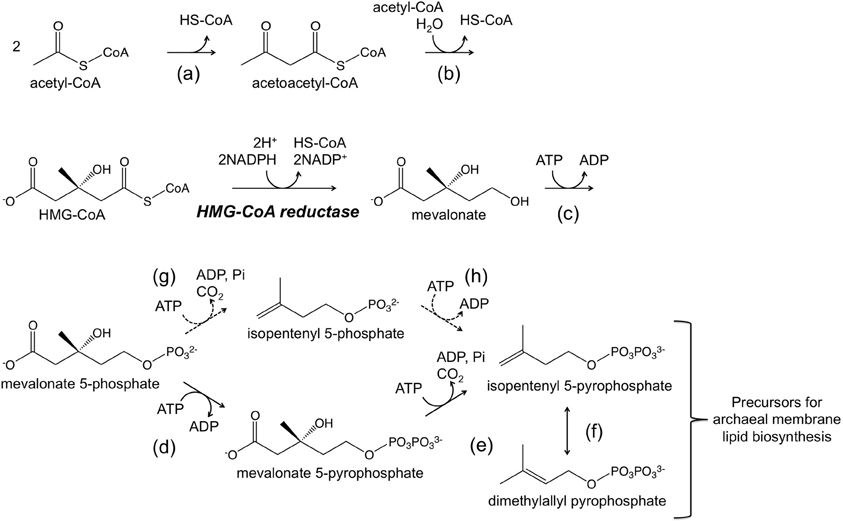
Figure 1. A schematic illustration of the mevalonate pathway. The dotted arrows indicate a possible route adopted in the Archaea (Grochowski et al., 2006; Matsumi et al., 2011). Enzymes catalyzing the reactions are (a) acetoacetyl-CoA thiolase, (b) HMG-CoA synthase, HMG-CoA reductase (written), (c) mevalonate kinase, (d) mevalonate-5-phosphate kinase, (e) mevalonate-5-pyrophosphate decarboxylase, (f) isopentenyl-5-pyrophosphate isomerase, (g) mevalonate-5-phosphate decarboxylase, and (h) isopentenyl-5-phosphate kinase.
Systems Based on Auxotrophic Selectable Markers
The ura3 (or pyrF) gene encoding orotidine-5′-monophosphate decarboxylase, an enzyme necessary for de novo pyrimidine biosynthesis, has been utilized as a selection marker in a number of halophilic archaea including Hb. salinarum NRC-1, Haloferax mediterranei, and Haloarcula hispanica (Liu et al., 2011). A host cell with a defect in ura3/pyrF can grow when uracil is added to the medium owing to the function of uracil phosphoribosyltransferase encoded by the upp gene. The ura3/pyrF system is especially convenient as it also allows counterselection. The addition of 5-fluoroorotic acid (5-FOA) to the medium prohibits growth of cells with an intact ura3/pyrF gene, as 5-FOA is converted to the toxic 5-fluorouridine 5′-phosphate and 5-fluorouracil (Boeke et al., 1984, 1987). These compounds inhibit DNA/RNA synthesis, with the latter known to inhibit thymidylate synthase, an enzyme necessary for thymidine synthesis. It is thus possible to specifically select cells that have lost a pyrF (or pyrE) gene by supplementing the medium with 5-FOA and a pyrimidine precursor such as uracil (Figure 2). In Hb. salinarum, a ura3/pyrF deletion strain was constructed using a mevinolin resistance marker, and the use of the ura3/pyrF gene as a counterselection marker has been extensively examined (Peck et al., 2000). Further improvements have enabled the use of ura3/pyrF as both a selection marker (uracil prototrophy) for initial plasmid integration, and as a counterselection marker (5-FOA resistance) for plasmid excision and gene deletion (Wang et al., 2004). The methodology has been successfully applied in disrupting and examining the arsenic resistance genes of this organism (Wang et al., 2004). The system has also been used to study the physiological roles of TATA binding proteins and transcription factor B proteins, whose genes are present in multiple copies on the genome (Coker and Dassarma, 2007). The methodology developed in Hb. salinarum can also be used in Hf. mediterranei and Ha. hispanica. The system was applied in deleting the phytoene synthase gene in both of these organisms (Liu et al., 2011).
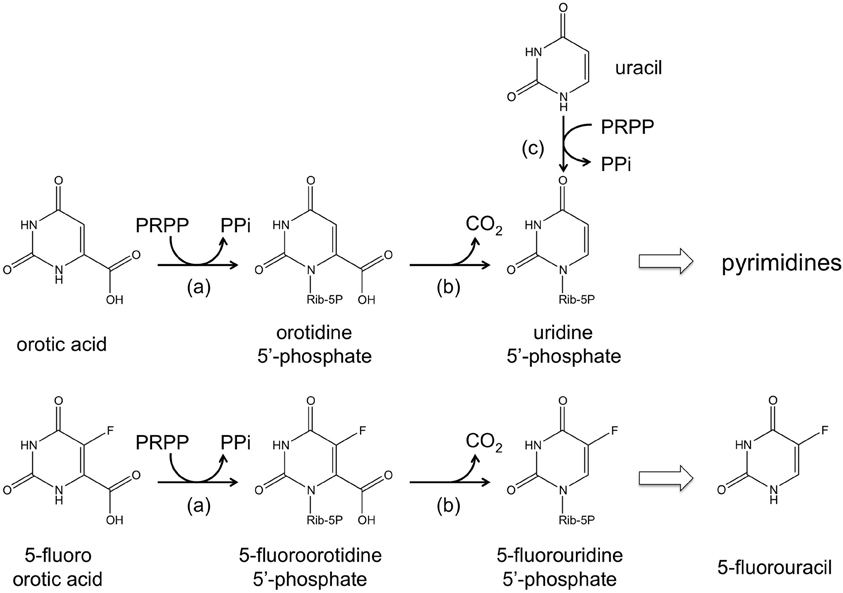
Figure 2. A schematic illustration of the reactions catalyzed by (a) orotate phosphoribosyltransferase (pyrE gene product), (b) orotidine- 5′-monophosphate decarboxylase (pyrF gene product), and (c) uracil phosphoribosyltransferase (upp gene product). The conversion from 5-fluoroorotic acid (5-FOA) to 5-fluorouridine 5′-phosphate and 5-fluorouracil is also shown.
Systems based on other auxotrophic selectable markers have been established in Hf. volcanii. The ura5 (or pyrE) gene encoding orotate phosphoribosyltransferase, responsible for the reaction preceding that of the ura3/pyrF product, has been demonstrated to be applicable as both a selection and counterselection marker (Bitan-Banin et al., 2003). Although two genes (pyrE1 and pyrE2) encoded proteins homologous with PyrE, pyrE2 was the gene actually involved in pyrimidine biosynthesis. A ΔpyrE2 strain was constructed and used as a host cell to disrupt the cmi4 gene of Hf. volcanii. Systems based on selection markers involved in amino acid biosynthesis have also been developed in Hf. volcanii (Allers et al., 2004). The leuB gene, encoding 3-isopropylmalate dehydrogenase in the leucine biosynthesis pathway, and the trpA gene that encodes one of the two subunits of tryptophan synthase have been used for selection based on leucine and tryptophan prototrophy, respectively. A convenient system for gene expression has also been developed in Hf. volcanii (Allers et al., 2010). Use of the tryptophanase promoter of Hf. volcanii (p.tna) promoter, which is induced by tryptophan, allows conditional overexpression of the target gene. The genetic background of the host strain has also been modified to facilitate the purification of His-tagged proteins and relieving the need to passage DNA through an E. coli dam mutant.
Application of the Genetic Systems in Halophiles
Gene manipulation is routinely performed in the halophiles and an overwhelming amount of genetic examinations has been reported in the literature (Leigh et al., 2011; Soppa, 2011). This most likely reflects the fact that genetic systems were developed at a relatively early stage in the halophiles and the versatility of the genetic systems themselves, along with the mesophilic and aerobic lifestyles of these organisms. To mention only several of the most recent studies, in Hb. salinarum, a gradual inducible gene expression system has been developed (Kixmüller and Greie, 2012). It relies on the promoter of the potassium uptake system operon (Pkdp), which responds to potassium cation concentrations in the medium. A workflow for genome-wide mapping of transcription factors from Hb. salinarum has also been reported (Wilbanks et al., 2012). Target genes such as those encoding the general transcription factor TfbD and the specific transcription factor Bat were modified to incorporate a hemagglutinin tag at the C-termini of the proteins, and the epitopes were used for chromatin immunoprecipitation coupled with high-throughput sequencing. Another study has examined the regulation of bacteriorhodopsin, particularly the relationship between bacterioopsin and retinal biosynthesis (Dummer et al., 2011). The results suggest that bacterioopsin accumulation promotes the production of its cofactor retinal by inhibiting bacterioruberin biosynthesis. In Hf. mediterranei, a genetic approach was applied in examining the functions of polyhydroxyalkanoate granule-associated proteins (Cai et al., 2012). The PhaP protein in this organism was found to act as the predominant structure protein on the PHA granules. In Hf. volcanii, a conserved archaeal gene with sequence similarity with a tRNA 3′-processing endonuclease has been studied biochemically and genetically, suggesting that in contrary to its annotation, the gene is involved in membrane transport (Fischer et al., 2012). Another study identifies the enzyme responsible for reduction of the ω-position isoprene of dolichol phosphate in Hf. volcanii (Naparstek et al., 2012). Single-stranded DNA-binding proteins have also been genetically examined. Five genes that encode proteins homologous to replication protein A (RPA) from Hf. volcanii (RpaA1A2, RpaB1B2, RpaC) were analyzed, revealing the essentiality of RpaC and the functional relationship among RPA proteins in this archaeon (Skowyra and Macneill, 2012). A metabolic study identified the enzymes responsible for fructose metabolism. Hf. volcanii adopts a bacteria-like phosphoenolpyruvate-dependent phosphotransferase system that generates fructose 1-phosphate, which is further converted to trioses via fructose-1-phosphate kinase and a Class II fructose-1,6-bisphosphate aldolase (Pickl et al., 2012).
Methanogens
Genetic systems have been developed in a number of species in the genera Methanococcus and Methanosarcina. DNA-mediated transformation was first demonstrated in Methanococcus voltae (Bertani and Baresi, 1987). Most systems in the methanogens rely on antibiotic resistance for selection. Puromycin and the puromycin transacetylase (pac) gene from the bacterium Streptomyces alboniger (Gernhardt et al., 1990) and its derivatives are often used as the antibiotic and the resistance marker gene, respectively, in methanogen genetics. Initial integration shuttle vectors using the pac gene were constructed and successfully used to transform M. voltae (Gernhardt et al., 1990; Patel et al., 1994) and Methanococcus maripaludis (Sandbeck and Leigh, 1991; Tumbula et al., 1994). Selection based on histidine auxotrophy/prototrophy using the hisA gene as a marker has also been demonstrated (Pfeifer et al., 1998). Counterselection methods have been developed (Moore and Leigh, 2005) based on the observation that M. maripaludis cells displaying growth were sensitive to the base analogs 6-azauracil and 8-azahypoxanthine (Bowen and Whitman, 1987; Ladapo and Whitman, 1990; Bowen et al., 1996; Kim and Whitman, 1999). Many of the principles and techniques developed in one methanogen have been shown to be applicable in other methanogen species.
In the Methanosarcina, efficient introduction of DNA is possible using liposome-mediated transformation (Metcalf et al., 1997). Replicating shuttle vectors were developed for Methanosarcina acetivorans and were also found to be applicable in a wide range of other Methanosarcina species including Methanosarcina barkeri, Methanosarcina mazei, and Methanosarcina thermophila. Gene disruption using the pac gene has been demonstrated in M. acetivorans and M. mazei. Markerless genetic exchange using the hypoxanthine phosphoribosyltransferase gene (hpt) was developed in M. acetivorans and also utilized in M. barkeri, with counterselection performed based on 8-aza-2,6-diaminopurine (8ADP) resistance (Pritchett et al., 2004; Rother and Metcalf, 2005; Welander and Metcalf, 2008; Buan et al., 2011). An in vivo transposon mutagenesis system has also been developed using a modified mariner-family transposable element originally derived from insect (Zhang et al., 2000).
Application of the Genetic Systems in Methanogens
Using these genetic systems, a number of genes in M. mazei Gö1, including those encoding the GlnK1 protein and archaeal histone, have been disrupted. glnK1 disruption revealed that GlnK1 is not directly involved in the transcriptional regulation of nitrogen assimilation/fixation genes, but does play a role in growth under nitrogen limiting conditions (Ehlers et al., 2005). Disruption of the histone gene was not lethal, but resulted in impaired growth on methanol and trimethylamine, and increased sensitivity to UV light. A broad genome-wide defect in gene transcription was also observed (Weidenbach et al., 2008). In M. acetivorans, the pylT gene encoding the tRNA for pyrrolysine was disrupted. The disruptant did not show growth defects when grown on methanol or acetate, but could not grow on methylamines, consistent with the fact that the methyltransferases from this organism that are involved in methylamine-dependent methanogenesis possess pyrrolysine (Mahapatra et al., 2006). A genetic approach was also used to distinguish the physiological roles of two gene clusters on the M. acetivorans genome encoding an archaeal A1A0-ATPase and a bacterial F1F0-ATPase. A mutant disrupted of the latter gene cluster did not display growth defects, and intracellular ATP levels were identical to those in wild-type cells, indicating that the F1F0-ATPase is dispensible for growth in M. acetivorans (Saum et al., 2009). The four studies introduced here have all utilized the pac gene for selection of the gene disruptants.
For M. voltae, protoplasts can efficiently be transformed by natural or electroporation-mediated uptake of exogenous DNA (Patel et al., 1994). Liposome-mediated transformation has also been applied (Heinicke et al., 2004; Chaban et al., 2009). Gene disruption has been demonstrated on the selenium-free Vhc and Frc hydrogenase genes in order to examine the individual roles of four hydrogenase gene clusters (Berghöfer and Klein, 1995). Four genes encoding the chromatin proteins histone (hstA, hstB), histone-like protein (hmvA), and an Alba homolog (AlbA) have been individually disrupted, revealing their involvement in regulation of gene expression (Heinicke et al., 2004). A genetic approach has also been taken to study post-translational protein modification. For example, two genes designated as aglC and aglK were shown to be necessary for proper N-glycosylation in this organism. It was suggested that the two genes are involved in the biosynthesis or transfer of diacetylated glucuronic acid within the glycan structure (Chaban et al., 2009).
In M. maripaludis, integration shuttle vectors and methods for auxotroph isolation (see above), and transposon insertion mutagenesis (Blank et al., 1995) and random insertional mutagenesis (Kim and Whitman, 1999) were developed at an early stage (Whitman et al., 1997; Tumbula and Whitman, 1999; Leigh et al., 2011). In addition to puromycin, M. maripaludis was found to be sensitive to neomycin, and the use of aminoglycoside phosphotransferase genes APH3′I and APH3′II as selectable markers has been demonstrated (Argyle et al., 1996). Transformation methods have been optimized and are performed via a polyethylene glycol-mediated method (Tumbula et al., 1994). A shuttle vector that replicates in both E. coli and M. maripaludis was constructed based on the plasmid pURB500 from this archaeon (Tumbula et al., 1997). Using the histone promoter from M. voltae, vectors for overexpression of endogenous and heterologous genes have been developed (Gardner and Whitman, 1999). A genetic approach has been taken to examine a wide variety of biological functions in M. maripaludis. The mechanisms and regulation of nitrogen fixation has been extensively examined. Repressor binding sites of nifH, encoding the nitrogenase reductase component of the nitrogenase complex, have been identified using a nifH promoter-lacZ in vivo reporter system (Cohen-Kupiec et al., 1997). A similar sequence was found upstream of the glutamine synthetase gene (glnA) and shown to function in repression. The repressor protein, NrpR, was identified, and its gene disruption, along with in vitro binding experiments, clearly demonstrated its function as a DNA-binding transcriptional repressor that regulates genes involved in nitrogen assimilation (Lie and Leigh, 2003). Further studies have revealed how NrpR binds to specific operator sequences and how it is released from DNA by 2-oxoglutarate binding (Lie et al., 2005, 2010). Furthermore, mechanisms governing posttranslation regulation, namely ammonia switchoff, of nitrogenase have also been examined in detail (Kessler and Leigh, 1999; Kessler et al., 2001; Dodsworth and Leigh, 2006). Genetics have also been utilized to study the energy-conserving hydrogenases in M. maripaludis. Gene disruption of one of the two membrane-bound hydrogenase complexes, Ehb, has revealed that the complex is involved in anabolic CO2 assimilation (Porat et al., 2006). Results of phenotypic analyses suggested that Ehb donates the electrons necessary for aromatic amino acid biosynthesis from aryl acids via the function of indolepyruvate oxidoreductase (Major et al., 2010). In addition to studies in M. voltae, N-glycosylation has also been examined in M. maripaludis. A putative acetyltransferase gene was subjected to gene disruption, and the mutant cells were found to produce flagellin proteins with sizes corresponding to proteins with defects in glycosylation. In addition to flagellar filament assembly, defects in pilus anchoring were also observed, indicating that flagellum and pilus assembly are linked in their post-translational modification mechanisms (Vandyke et al., 2008). Further studies have identified multiple genes that are necessary for piliation and have also led to the identification of the protein that corresponds to the major pilin monomer, a protein whose gene resides outside of the gene cluster that had been predicted to harbor most of the genes related to pilus formation (Ng et al., 2011). A number of recent studies have examined mechanisms of selenocysteine (Sec) biosynthesis in various strains of M. maripaludis (Stock et al., 2010, 2011; Hohn et al., 2011). A selenophosphate synthetase homolog (selD) in M. maripaludis S2 could not be deleted unless a bacterial selenophosphate synthetase gene was present in trans, whereas disruption of the corresponding gene in M. maripaludis JJ was possible. Further genetic examination on the latter strain indicated that selenophosphate is the selenium donor in this strain (Stock et al., 2010). In another strain M. maripaludis Mm900, which is related to S2, selD disruption was possible. Whereas the ability to grow on formate was abolished, hydrogenotrophic growth was unaffected (Hohn et al., 2011). Interestingly, disruption of genes encoding phosphoseryl-tRNASec kinase and phosphoseryl-tRNA:Sec-tRNA synthase was possible only when either selD was disrupted or if selenium-free hydrogenases were expressed. Detailed biochemical characterization of the gene disruption strains suggests a complex regulatory mechanism of Sec biosynthesis in M. maripaludis.
Sulfolobus
There are many natural genetic elements related to the crenarchaeal genus Sulfolobus, including viruses, cryptic plasmids, and transposons (Zillig et al., 1996, 1998; Prangishvili et al., 1998; Stedman et al., 2000). Several transformation systems based on these natural elements were developed in Sulfolobus solfataricus and Sulfolobus acidocaldarius at an early stage, followed by the establishment of gene manipulation systems based on useful selectable markers (Aagaard et al., 1996; Elferink et al., 1996; Berkner and Lipps, 2008; Wagner et al., 2009; Leigh et al., 2011). Transformation is now mainly carried out by electroporation. In S. solfataricus, selection is possible by using a strain with a deletion in lacS, which encodes a β-galactosidase. By using an intact lacS as a marker gene, transformants can be selected by their ability to grow in a minimal medium containing lactose (Worthington et al., 2003). LacS+ colonies can be further identified by blue/white detection using X-Gal (Schelert et al., 2004). Selection based on resistance toward hygromycin B has also been reported using a gene encoding a thermostabilized hygromycin phosphotransferase from E. coli (Cannio et al., 1998). Another system is based on resistance toward butanol or benzyl alcohol using an alcohol dehydrogenase gene from S. solfataricus (Aravalli and Garrett, 1997). In S. acidocaldarius and Sulfolobus islandicus, host strains with defects in pyrE, pyrF or both are utilized, with intact pyrE and pyrF genes as selection markers (Deng et al., 2009; She et al., 2009; Wagner et al., 2009). This strategy has also been utilized in S. solfataricus. Based on these selection strategies, a wide range of Sulfolobus–E. coli shuttle vectors have been developed and are described in detail in the literature (Aravalli and Garrett, 1997; Stedman et al., 1999; Jonuscheit et al., 2003; Albers et al., 2006; Aucelli et al., 2006; Berkner et al., 2007; Berkner and Lipps, 2008).
Application of the Genetic Systems in Sulfolobus
Sulfolobus solfataricus
Gene disruption based on lacS selection has been utilized to examine a wide range of functions in S. solfataricus. Conditions for gene disruption have been carefully examined and optimized (Albers and Driessen, 2007). Genetic and biochemical examination has been performed on genes involved in mercury resistance, demonstrating that the merR gene product represses transcription of an operon that includes the mercuric reductase gene merA (Schelert et al., 2004, 2006). Another study demonstrated that the copR gene product is a transcriptional activator of genes encoding copper-transporting ATPase and copper-binding protein and is necessary for copper tolerance of S. solfataricus (Villafane et al., 2011). A Lrp-like regulator, Ss-LrpB, has been shown to act as an activator of genes including the pyruvate:ferredoxin oxidoreductase gene (Peeters et al., 2009). A genetic study has also been performed on a heat-shock-inducible ribonucleolytic toxin, VapC6, and its antitoxin VapB6. Analysis of disruption strains of these genes has identified possible targets of the ribonucleolytic activity (Maezato et al., 2011). Genetic manipulation is now also possible for the virus Sulfolobus turreted icosahedral virus (STIV) (Snyder et al., 2011; Wirth et al., 2011). An infectious clone of STIV was constructed, and gene disruptions of individual open reading frames and their effects on viral replication have been demonstrated.
Sulfolobus acidocaldarius
In Sulfolobus acidocaldarius, a series of small multicopy, non-integrative shuttle vectors have been developed and their use in overexpression of genes has been demonstrated (Berkner et al., 2007). Promoters for both constitutive and inducible gene expression have been examined. As for constitutive gene expression, the sac7d promoter led to the highest levels of β-galactosidase activity when various promoters were fused upstream of lacS. The mal promoter was the most suitable for induction, displaying a 17-fold increase upon addition of maltose or dextrin (Berkner et al., 2010). In terms of gene disruption, pyrE-deficient host cells have been used with an intact, heterologous pyrE from S. solfataricus to disrupt putative genes involved in UV photoproduct repair (Sakofsky et al., 2011). It should be noted that in this study, the lengths of the homologous regions flanking the selection marker were approximately 50 bp, introduced by PCR in the primer sequences, which may allow high-throughput gene disruption in a genome-wide scale. Another study clarified two in vivo activities of Y-family DNA polymerase in S. acidocaldarius (Sakofsky et al., 2012). One activity promotes slipped strand events within simple repetitive sequences and the other promotes insertion of C opposite a potentially miscoding form of G, which may contribute in preventing G:C to T:A transversions. Genetics have contributed in the identification of sulfolobicins, antimicrobial proteins produced by Sulfolobus species (Ellen et al., 2011). Antimicrobial tests, protein separation, followed by MS led to the identification of candidate genes, and their disruption confirmed that two genes encoding secretion proteins corresponded to the sulfolobicin. Several studies report a genetic examination of genes involved in cell surface structure of S. acidocaldarius. Deletion of individual flagellin genes indicated that all genes were essential for flagellin assembly and that assembly proceeds through hierarchical protein interaction (Lassak et al., 2012). Another study has led to the identification of sulfoquinovose synthase, which is necessary for the synthesis of sulfoquinovose, a component of the N-linked glycans on the surface-layer glycoprotein of S. acidocaldarius (Meyer et al., 2011). Gene disruption confirmed this activity and also demonstrated the importance of N-glycosylation under conditions of increased salt concentrations. A complete genetic analysis of the three type IV pili-like structures in S. acidocaldarius, the flagellum, the UV-induced pili, and the adhesive pili, has also been reported (Henche et al., 2012). The effects of single, double, and triple deletion of the three structures on cell surface structure, surface attachment capability, motility, and biofilm formation were examined. It should be noted that this study utilizes cells expressing a codon adjusted, heat stable green fluorescent protein eCGP123. eCGP123 was used to distinguish strains within a biofilm generated from a mixture of strains with different gene deletions. Another study genetically demonstrates that the UV-inducible type IV pili are involved in intercellular, UV-inducible DNA exchange, a valuable mechanism to maintain chromosome integrity (Ajon et al., 2011).
Sulfolobus islandicus
In Sulfolobus islandicus, several genetic studies have been reported focusing on genes involved in DNA replication and maintenance of DNA topology. In one study, the topoisomerase III gene of S. islandicus was disrupted (Li et al., 2011a). Cells were viable but displayed various defects in chromosome distribution, cell size, and gene transcription. The results suggested that this enzyme plays an important role in chromosome segregation and maintenance of DNA topology for gene transcription. Another study addressed whether any of the three proliferating cell nuclear antigen (PCNA) on the S. islandicus genome are dispensable or not (Zhang et al., 2010). Disruption strains could not be isolated for any of the genes, and an improved knockout system has been described in order to carefully examine the essentiality of each gene. A recent study reported the development of a new gene disruption system in S. islandicus that is based on antibiotic resistance toward simvastatin (Zhang and Whitaker, 2012). The selectable marker gene was a construct promoting overexpression of the HMG-CoA reductase gene.
Thermococcales
Genetic systems have mainly been developed in Thermococcus kodakarensis and Pyrococcus furiosus. Gene disruption has also been demonstrated in Thermococcus onnurineus (Kim et al., 2010). Shuttle vectors are available for Pyrococcus abyssi (Lucas et al., 2002). In T. kodakarensis, gene disruption was accomplished by using host strains deleted of the pyrF and/or trpE genes, and selection with the corresponding intact marker gene (Sato et al., 2003, 2005). A system has also been developed based on the simvastatin/HMG-CoA reductase overexpression system (Matsumi et al., 2007). An improved system utilizing host cells that exhibit agmatine auxotrophy due to deletion of the arginine decarboxylase gene (Fukuda et al., 2008) has also been developed (Santangelo et al., 2010). This system allows selection in complex media and not only accelerates the gene disruption procedure, but should also contribute to the isolation of mutant cells that require nutrient-rich conditions for cell growth. Counterselection is performed with 5-FOA in the pyrF system (Sato et al., 2005), and counterselection in nutrient-rich medium is possible using a hypoxanthine–guanine phosphoribosyltransferase gene which, when present, results in 6-methylpurine sensitivity (Santangelo et al., 2010). In Pyrococcus furiosus, two transformation systems based on shuttle vectors that replicate in P. furiosus and E. coli have been developed. One is based on the shuttle vector system pYS2 from P. abyssi. The selectable marker is an HMG-CoA reductase overexpression cassette, and selection is based on resistance toward simvastatin (Waege et al., 2010). The other system is based on the P. furiosus chromosomal origin and utilizes the pyrF gene as a selectable marker in combination with the P. furiosus COM strain (ΔpyrF). The plasmids existed in a single copy in P. furiosus, and were stable without selective pressure for more than 100 generations (Farkas et al., 2011). The pyrF deletion strain P. furiosus COM1 can also be used as an efficient host for gene disruption (Lipscomb et al., 2011). This strain displays natural competence and a remarkable efficiency in DNA uptake, allowing marker replacement using linear as well as circular DNA. Using pyrF as the selectable marker, construction of markerless deletion mutants via counterselection by 5-FOA resistance has been demonstrated (Lipscomb et al., 2011). In a recent report, the limits of recombination efficiency of P. furiosus COM1 have been examined. It was found that marker replacement was possible with as few as 40 nucleotides of flanking homology to the target region (Farkas et al., 2012), which will surely facilitate genetic studies in this organism. Markerless deletion was utilized to disrupt the trpAB genes, encoding the two subunits of tryptophan synthase. The disruptant displayed tight tryptophan auxotrophy, and the wild-type trpAB genes could be used as a selectable marker in this strain (Farkas et al., 2012).
Application of the Genetic Systems in Thermococcales
Thermococcus kodakarensis and Thermococcus onnurineus
In T. kodakarensis, a wide range of genes has been disrupted in order to understand their physiological functions such as those involved in transcription and its regulation, DNA replication, and metabolism. The functions of individual transcription factors such as TFB1/2 (Santangelo et al., 2007), RNA polymerase subunits E and F (Hirata et al., 2008), the switch 3 loop of subunit B (Santangelo and Reeve, 2010) have been examined, along with sequences that can promote transcription termination (Santangelo et al., 2009). Deletion of transcription regulator genes, followed by transcriptome analysis, has led to the identification of regulons and the function of these regulators (Kanai et al., 2007, 2010). In terms of DNA replication, a large number of proteins expected to be involved in DNA replication were His-tagged. Protein complexes were isolated and their components identified, revealing the various protein networks involved in DNA replication (Li et al., 2010). This led to the discovery of a novel GINS-associated nuclease, GAN (Li et al., 2011b). In other studies, genetic analyses of three mcm genes on the genome have revealed the essentiality/dispensability of the individual genes (Ishino et al., 2011; Pan et al., 2011). Disruption of the reverse gyrase gene indicated that the enzyme provides an advantage to cells when grown at temperatures of 80°C or higher (Atomi et al., 2004). In terms of metabolism, genes examined include those involved in glycolysis (Imanaka et al., 2006; Matsubara et al., 2011), gluconeogenesis (Sato et al., 2004), pentose metabolism (Orita et al., 2006), as well as coenzyme A (Yokooji et al., 2009), polyamine (Morimoto et al., 2010), and compatible solute biosynthesis (Borges et al., 2010). Gene disruption of three putative hydrogenase gene clusters and phenotypic analyses indicated that the cytosolic hydrogenase Hyh and the membrane-bound oxidoreductase complexes Mbh and Mbx are involved in H2 consumption, H2 generation, and H2S generation, respectively (Kanai et al., 2011). Disruption of various pathways related to hydrogen production/consumption has clarified the reductant flux in T. kodakarensis and has demonstrated strategies to elevate hydrogen production in this organism (Santangelo et al., 2011). T. kodakarensis harbors two pairs of genes encoding chaperonins and prefoldins, and gene disruption has been performed to distinguish the functions of the individual proteins at different temperatures (Danno et al., 2008; Fujiwara et al., 2008; Gao et al., 2012). One of multiple homologs of NAD(P)H oxidase has been disrupted to examine its relationship with the oxygen sensitivity of this anaerobic archaeon (Kobori et al., 2012). Systems for gene overexpression, tagging, and protein secretion have been established (Santangelo et al., 2008; Mueller et al., 2009; Yokooji et al., 2009; Takemasa et al., 2011), in many cases relying on the selection/counterselection strategy utilizing the pyrF marker gene (Figure 3). In T. onnurineus, gene disruption has been demonstrated using the simvastatin/HMG-CoA reductase overexpression system. Gene disruption led to the identification of the gene cluster encoding formate hydrogen lyase, cation/proton antiporter and formate transporter, which were responsible for the growth of this organism on formate (Kim et al., 2010).
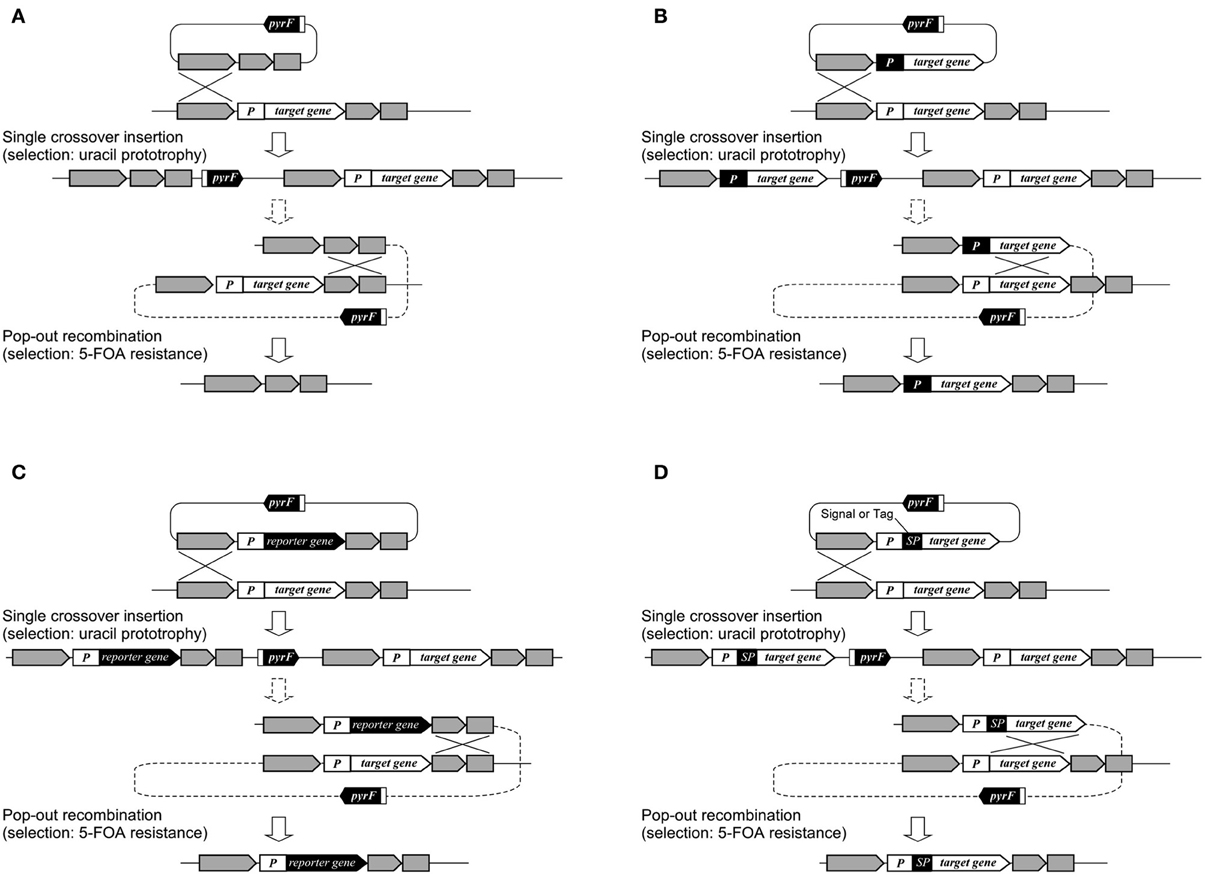
Figure 3. Various applications using selection/counterselection marker genes. (A) Use in markerless gene disruption; (B) use in promoter exchange; (C) use in reporter gene integration; and (D) use in signal peptide (SP) or tag integration. The use of pyrF is shown, but other selection/counterselection marker genes can be applied in a similar manner in a wide range of Archaea.
Pyrococcus furiosus
Using the shuttle vector pYS3, the RNA polymerase subunit D gene with a HisTag sequence was expressed with an inducible promoter deriving from the fructose-1,6-bisphosphatase gene from P. furiosus. This allowed a simple two-step purification of the thermostable RNA polymerase from this organism (Waege et al., 2010). In the process of developing the shuttle vector system based on the P. furiosus chromosomal origin, the minimum replication origin sequence required for autonomous plasmid replication in this organism has been identified (Farkas et al., 2011). Interestingly, the cdc6/orc1 gene adjacent to oriC was not required in cis for replication of the shuttle vector in P. furiosus. The gene disruption system developed with P. furiosus COM1 has been successfully applied in disrupting individual or double gene disruptions of two cytoplasmic hydrogenase genes (Lipscomb et al., 2011). The system was further applied for detailed genetic studies on proteins related to elemental sulfur metabolism, membrane-bound oxidoreductase complex (Mbx), cytoplasmic coenzyme A-dependent NADPH:sulfur oxidoreductase (Nsr), and sulfur-induced protein A (SipA) (Bridger et al., 2011). The mbx disruptant displayed growth defects in the presence of sulfur, and little, if any, sulfide generation was observed, demonstrating that Mbx plays a critical role in elemental sulfur reduction and energy conservation in P. furiosus. Gene manipulation has also been used to overexpress the cytoplasmic [NiFe]-hydrogenase SHI. The promoter of the PF1399 gene, which encodes the S-layer protein, was fused upstream of the four-gene operon (PF0891–PF0894) encoding SHI. In the overexpression strain, a 20-fold higher SHI transcript level was observed, and moreover, a 100-fold higher amount of hydrogenase was obtained when compared with the highest homologous [NiFe]-hydrogenase system previously reported (Chandrayan et al., 2012). In another study, the lactate dehydrogenase gene from the moderately thermophilic Caldicellulosiruptor bescii was introduced into P. furiosus under the control of the cold-induced protein A (cipA, PF0190) promoter. Transcript levels of cipA in P. furiosus are 26-fold higher in cells grown at 72°C compared to those grown at 98°C. This recombinant strain, when grown at 98°C, ferments sugar to produce acetate and hydrogen as end products, as is the case of wild-type P. furiosus. When grown at 72°C, however, the strain generates lactate at concentrations up to 3 mM, demonstrating a temperature-dependent regulation of metabolism (Basen et al., 2012).
Future Perspectives
The genetic systems developed in the halophiles, methanogens, Sulfolobus, and Thermococcales provide the tools to carry out sophisticated genetic analyses in these organisms (Leigh et al., 2011). With the abundance of genome sequence information, functional genomics in these organisms is a realistic approach. On the other hand, the Archaea comprise a diverse group of organisms, and there are still many interesting organisms that cannot be examined genetically. Genetics are limited to Sulfolobus in the Crenarchaeota, and considering the wealth of genome sequence information, the development of genetic systems in Pyrobaculum and Thermoproteus would greatly promote research in these genera. Genetic tools for Ignicoccus can be considered crucial to understand its unique relationship with Nanoarchaeum equitans. In the Euryarchaeota, the thermophilic/acidophilic Thermoplasmatales and the sulfate-reducing Archaeoglobales are major orders in which genetic tools are still not available. Another major group is the (hyper)thermophilic methanogens. Although much needs to be done, the variety of tools that have been developed will surely provide a basis to explore the possibilities of developing genetic systems in other archaea.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Aagaard, C., Leviev, I., Aravalli, R. N., Forterre, P., Prieur, D., and Garrett, R. A. (1996). General vectors for archaeal hyperthermophiles: strategies based on a mobile intron and a plasmid. FEMS Microbiol. Rev. 18, 93–104.
Ajon, M., Fröls, S., Van Wolferen, M., Stoecker, K., Teichmann, D., Driessen, A. J., Grogan, D. W., Albers, S. V., and Schleper, C. (2011). UV-inducible DNA exchange in hyperthermophilic archaea mediated by type IV pili. Mol. Microbiol. 82, 807–817.
Albers, S., and Driessen, A. J. (2007). Conditions for gene disruption by homologous recombination of exogenous DNA into the Sulfolobus solfataricus genome. Archaea 2, 145–149.
Albers, S. V., Jonuscheit, M., Dinkelaker, S., Urich, T., Kletzin, A., Tampe, R., Driessen, A. J., and Schleper, C. (2006). Production of recombinant and tagged proteins in the hyperthermophilic archaeon Sulfolobus solfataricus. Appl. Environ. Microbiol. 72, 102–111.
Allers, T., Barak, S., Liddell, S., Wardell, K., and Mevarech, M. (2010). Improved strains and plasmid vectors for conditional overexpression of His-tagged proteins in Haloferax volcanii. Appl. Environ. Microbiol. 76, 1759–1769.
Allers, T., Ngo, H. P., Mevarech, M., and Lloyd, R. G. (2004). Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Appl. Environ. Microbiol. 70, 943–953.
Aravalli, R. N., and Garrett, R. A. (1997). Shuttle vectors for hyperthermophilic archaea. Extremophiles 1, 183–191.
Argyle, J. L., Tumbula, D. L., and Leigh, J. A. (1996). Neomycin resistance as a selectable marker in Methanococcus maripaludis. Appl. Environ. Microbiol. 62, 4233–4237.
Atomi, H., Matsumi, R., and Imanaka, T. (2004). Reverse gyrase is not a prerequisite for hyperthermophilic life. J. Bacteriol. 186, 4829–4833.
Aucelli, T., Contursi, P., Girfoglio, M., Rossi, M., and Cannio, R. (2006). A spreadable, non-integrative and high copy number shuttle vector for Sulfolobus solfataricus based on the genetic element pSSVx from Sulfolobus islandicus. Nucleic Acids Res. 34, e114.
Baba, T., Ara, T., Hasegawa, M., Takai, Y., Okumura, Y., Baba, M., Datsenko, K. A., Tomita, M., Wanner, B. L., and Mori, H. (2006). Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006.0008.
Basen, M., Sun, J., and Adams, M. W. (2012). Engineering a hyperthermophilic archaeon for temperature-dependent product formation. MBio 3, e00053-00012.
Berghöfer, Y., and Klein, A. (1995). Insertional mutations in the hydrogenase vhc and frc operons encoding selenium-free hydrogenases in Methanococcus voltae. Appl. Environ. Microbiol. 61, 1770–1775.
Berkner, S., Grogan, D., Albers, S. V., and Lipps, G. (2007). Small multicopy, non-integrative shuttle vectors based on the plasmid pRN1 for Sulfolobus acidocaldarius and Sulfolobus solfataricus, model organisms of the (cren-)archaea. Nucleic Acids Res. 35, e88.
Berkner, S., and Lipps, G. (2008). Genetic tools for Sulfolobus spp.: vectors and first applications. Arch. Microbiol. 190, 217–230.
Berkner, S., Wlodkowski, A., Albers, S. V., and Lipps, G. (2010). Inducible and constitutive promoters for genetic systems in Sulfolobus acidocaldarius. Extremophiles 14, 249–259.
Bertani, G., and Baresi, L. (1987). Genetic transformation in the methanogen Methanococcus voltae PS. J. Bacteriol. 169, 2730–2738.
Bitan-Banin, G., Ortenberg, R., and Mevarech, M. (2003). Development of a gene knockout system for the halophilic archaeon Haloferax volcanii by use of the pyrE gene. J. Bacteriol. 185, 772–778.
Blank, C. E., Kessler, P. S., and Leigh, J. A. (1995). Genetics in methanogens: transposon insertion mutagenesis of a Methanococcus maripaludis nifH gene. J. Bacteriol. 177, 5773–5777.
Blaseio, U., and Pfeifer, F. (1990). Transformation of Halobacterium halobium: development of vectors and investigation of gas vesicle synthesis. Proc. Natl. Acad. Sci. U.S.A. 87, 6772–6776.
Boeke, J. D., Lacroute, F., and Fink, G. R. (1984). A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197, 345–346.
Boeke, J. D., Trueheart, J., Natsoulis, G., and Fink, G. R. (1987). 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154, 164–175.
Borges, N., Matsumi, R., Imanaka, T., Atomi, H., and Santos, H. (2010). Thermococcus kodakarensis mutants deficient in di-myo-inositol phosphate use aspartate to cope with heat stress. J. Bacteriol. 192, 191–197.
Bowen, T. L., Lin, W. C., and Whitman, W. B. (1996). Characterization of guanine and hypoxanthine phosphoribosyltransferases in Methanococcus voltae. J. Bacteriol. 178, 2521–2526.
Bowen, T. L., and Whitman, W. B. (1987). Incorporation of exogenous purines and pyrimidines by Methanococcus voltae and isolation of analog-resistant mutants. Appl. Environ. Microbiol. 53, 1822–1826.
Bridger, S. L., Clarkson, S. M., Stirrett, K., Debarry, M. B., Lipscomb, G. L., Schut, G. J., Westpheling, J., Scott, R. A., and Adams, M. W. (2011). Deletion strains reveal metabolic roles for key elemental sulfur-responsive proteins in Pyrococcus furiosus. J. Bacteriol. 193, 6498–6504.
Buan, N., Kulkarni, G., and Metcalf, W. (2011). Genetic methods for Methanosarcina species. Methods Enzymol. 494, 23–42.
Cai, S., Cai, L., Liu, H., Liu, X., Han, J., Zhou, J., and Xiang, H. (2012). Identification of the haloarchaeal phasin (PhaP) that functions in polyhydroxyalkanoate accumulation and granule formation in Haloferax mediterranei. Appl. Environ. Microbiol. 78, 1946–1952.
Cannio, R., Contursi, P., Rossi, M., and Bartolucci, S. (1998). An autonomously replicating transforming vector for Sulfolobus solfataricus. J. Bacteriol. 180, 3237–3240.
Chaban, B., Logan, S. M., Kelly, J. F., and Jarrell, K. F. (2009). AglC and AglK are involved in biosynthesis and attachment of diacetylated glucuronic acid to the N-glycan in Methanococcus voltae. J. Bacteriol. 191, 187–195.
Chandrayan, S. K., McTernan, P. M., Hopkins, R. C., Sun, J., Jenney, F. E. Jr., and Adams, M. W. (2012). Engineering hyperthermophilic archaeon Pyrococcus furiosus to overproduce its cytoplasmic [NiFe]-hydrogenase. J. Biol. Chem. 287, 3257–3264.
Cline, S. W., and Doolittle, W. F. (1992). Transformation of members of the genus Haloarcula with shuttle vectors based on Halobacterium halobium and Haloferax volcanii plasmid replicons. J. Bacteriol. 174, 1076–1080.
Cohen-Kupiec, R., Blank, C., and Leigh, J. A. (1997). Transcriptional regulation in Archaea: in vivo demonstration of a repressor binding site in a methanogen. Proc. Natl. Acad. Sci. U.S.A. 94, 1316–1320.
Coker, J. A., and Dassarma, S. (2007). Genetic and transcriptomic analysis of transcription factor genes in the model halophilic Archaeon: coordinate action of TbpD and TfbA. BMC Genet. 8, 61.
Danno, A., Fukuda, W., Yoshida, M., Aki, R., Tanaka, T., Kanai, T., Imanaka, T., and Fujiwara, S. (2008). Expression profiles and physiological roles of two types of prefoldins from the hyperthermophilic archaeon Thermococcus kodakaraensis. J. Mol. Biol. 382, 298–311.
Deng, L., Zhu, H., Chen, Z., Liang, Y. X., and She, Q. (2009). Unmarked gene deletion and host-vector system for the hyperthermophilic crenarchaeon Sulfolobus islandicus. Extremophiles 13, 735–746.
Dodsworth, J. A., and Leigh, J. A. (2006). Regulation of nitrogenase by 2-oxoglutarate-reversible, direct binding of a PII-like nitrogen sensor protein to dinitrogenase. Proc. Natl. Acad. Sci. U.S.A. 103, 9779–9784.
Dummer, A. M., Bonsall, J. C., Cihla, J. B., Lawry, S. M., Johnson, G. C., and Peck, R. F. (2011). Bacterioopsin-mediated regulation of bacterioruberin biosynthesis in Halobacterium salinarum. J. Bacteriol. 193, 5658–5667.
Ehlers, C., Weidenbach, K., Veit, K., Deppenmeier, U., Metcalf, W. W., and Schmitz, R. A. (2005). Development of genetic methods and construction of a chromosomal glnK1 mutant in Methanosarcina mazei strain Gö1. Mol. Genet. Genomics 273, 290–298.
Elferink, M. G., Schleper, C., and Zillig, W. (1996). Transformation of the extremely thermoacidophilic archaeon Sulfolobus solfataricus via a self-spreading vector. FEMS Microbiol. Lett. 137, 31–35.
Ellen, A. F., Rohulya, O. V., Fusetti, F., Wagner, M., Albers, S. V., and Driessen, A. J. (2011). The sulfolobicin genes of Sulfolobus acidocaldarius encode novel antimicrobial proteins. J. Bacteriol. 193, 4380–4387.
Farkas, J., Chung, D., Debarry, M., Adams, M. W., and Westpheling, J. (2011). Defining components of the chromosomal origin of replication of the hyperthermophilic archaeon Pyrococcus furiosus needed for construction of a stable replicating shuttle vector. Appl. Environ. Microbiol. 77, 6343–6349.
Farkas, J., Stirrett, K., Lipscomb, G. L., Nixon, W., Scott, R. A., Adams, M. W., and Westpheling, J. (2012). Recombinogenic properties of the Pyrococcus furiosus COM1 strain enable rapid selection of targeted mutants. Appl. Environ. Microbiol. 78, 4669–4676.
Fischer, S., John Von Freyend, S., Sabag-Daigle, A., Daniels, C. J., Allers, T., and Marchfelder, A. (2012). Assigning a function to a conserved archaeal metallo-β-lactamase from Haloferax volcanii. Extremophiles 16, 333–343.
Fujiwara, S., Aki, R., Yoshida, M., Higashibata, H., Imanaka, T., and Fukuda, W. (2008). Expression profiles and physiological roles of two types of molecular chaperonins from the hyperthermophilic archaeon Thermococcus kodakarensis. Appl. Environ. Microbiol. 74, 7306–7312.
Fukuda, W., Morimoto, N., Imanaka, T., and Fujiwara, S. (2008). Agmatine is essential for the cell growth of Thermococcus kodakaraensis. FEMS Microbiol. Lett. 287, 113–120.
Gao, L., Danno, A., Fujii, S., Fukuda, W., Imanaka, T., and Fujiwara, S. (2012). Indole-3-glycerol-phosphate synthase is recognized by a cold-inducible group II chaperonin in Thermococcus kodakarensis. Appl. Environ. Microbiol. 78, 3806–3815.
Gardner, W. L., and Whitman, W. B. (1999). Expression vectors for Methanococcus maripaludis: overexpression of acetohydroxyacid synthase and β-galactosidase. Genetics 152, 1439–1447.
Gernhardt, P., Possot, O., Foglino, M., Sibold, L., and Klein, A. (1990). Construction of an integration vector for use in the archaebacterium Methanococcus voltae and expression of a eubacterial resistance gene. Mol. Gen. Genet. 221, 273–279.
Giaever, G., Chu, A. M., Ni, L., Connelly, C., Riles, L., Veronneau, S., Dow, S., Lucau-Danila, A., Anderson, K., Andre, B., Arkin, A. P., Astromoff, A., El-Bakkoury, M., Bangham, R., Benito, R., Brachat, S., Campanaro, S., Curtiss, M., Davis, K., Deutschbauer, A., Entian, K. D., Flaherty, P., Foury, F., Garfinkel, D. J., Gerstein, M., Gotte, D., Guldener, U., Hegemann, J. H., Hempel, S., Herman, Z., Jaramillo, D. F., Kelly, D. E., Kelly, S. L., Kotter, P., Labonte, D., Lamb, D. C., Lan, N., Liang, H., Liao, H., Liu, L., Luo, C., Lussier, M., Mao, R., Menard, P., Ooi, S. L., Revuelta, J. L., Roberts, C. J., Rose, M., Ross-Macdonald, P., Scherens, B., Schimmack, G., Shafer, B., Shoemaker, D. D., Sookhai-Mahadeo, S., Storms, R. K., Strathern, J. N., Valle, G., Voet, M., Volckaert, G., Wang, C. Y., Ward, T. R., Wilhelmy, J., Winzeler, E. A., Yang, Y., Yen, G., Youngman, E., Yu, K., Bussey, H., Boeke, J. D., Snyder, M., Philippsen, P., Davis, R. W., and Johnston, M. (2002). Functional profiling of the Saccharomyces cerevisiae genome. Nature 418, 387–391.
Grochowski, L. L., Xu, H., and White, R. H. (2006). Methanocaldococcus jannaschii uses a modified mevalonate pathway for biosynthesis of isopentenyl diphosphate. J. Bacteriol. 188, 3192–3198.
Heinicke, I., Müller, J., Pittelkow, M., and Klein, A. (2004). Mutational analysis of genes encoding chromatin proteins in the archaeon Methanococcus voltae indicates their involvement in the regulation of gene expression. Mol. Genet. Genomics 272, 76–87.
Henche, A. L., Koerdt, A., Ghosh, A., and Albers, S. V. (2012). Influence of cell surface structures on crenarchaeal biofilm formation using a thermostable green fluorescent protein. Environ. Microbiol. 14, 779–793.
Hirata, A., Kanai, T., Santangelo, T. J., Tajiri, M., Manabe, K., Reeve, J. N., Imanaka, T., and Murakami, K. S. (2008). Archaeal RNA polymerase subunits E and F are not required for transcription in vitro, but a Thermococcus kodakarensis mutant lacking subunit F is temperature-sensitive. Mol. Microbiol. 70, 623–633.
Hohn, M. J., Palioura, S., Su, D., Yuan, J., and Söll, D. (2011). Genetic analysis of selenocysteine biosynthesis in the archaeon Methanococcus maripaludis. Mol. Microbiol. 81, 249–258.
Holmes, M., Pfeifer, F., and Dyall-Smith, M. (1994). Improved shuttle vectors for Haloferax volcanii including a dual-resistance plasmid. Gene 146, 117–121.
Holmes, M. L., and Dyall-Smith, M. L. (1990). A plasmid vector with a selectable marker for halophilic archaebacteria. J. Bacteriol. 172, 756–761.
Holmes, M. L., Nuttall, S. D., and Dyall-Smith, M. L. (1991). Construction and use of halobacterial shuttle vectors and further studies on Haloferax DNA gyrase. J. Bacteriol. 173, 3807–3813.
Imanaka, H., Yamatsu, A., Fukui, T., Atomi, H., and Imanaka, T. (2006). Phosphoenolpyruvate synthase plays an essential role for glycolysis in the modified Embden-Meyerhof pathway in Thermococcus kodakarensis. Mol. Microbiol. 61, 898–909.
Ishino, S., Fujino, S., Tomita, H., Ogino, H., Takao, K., Daiyasu, H., Kanai, T., Atomi, H., and Ishino, Y. (2011). Biochemical and genetical analyses of the three mcm genes from the hyperthermophilic archaeon, Thermococcus kodakarensis. Genes Cells 16, 1176–1189.
Jonuscheit, M., Martusewitsch, E., Stedman, K. M., and Schleper, C. (2003). A reporter gene system for the hyperthermophilic archaeon Sulfolobus solfataricus based on a selectable and integrative shuttle vector. Mol. Microbiol. 48, 1241–1252.
Kanai, T., Akerboom, J., Takedomi, S., Van De Werken, H. J., Blombach, F., Van Der Oost, J., Murakami, T., Atomi, H., and Imanaka, T. (2007). A global transcriptional regulator in Thermococcus kodakaraensis controls the expression levels of both glycolytic and gluconeogenic enzyme-encoding genes. J. Biol. Chem. 282, 33659–33670.
Kanai, T., Matsuoka, R., Beppu, H., Nakajima, A., Okada, Y., Atomi, H., and Imanaka, T. (2011). Distinct physiological roles of the three [NiFe]-hydrogenase orthologs in the hyperthermophilic archaeon Thermococcus kodakarensis. J. Bacteriol. 193, 3109–3116.
Kanai, T., Takedomi, S., Fujiwara, S., Atomi, H., and Imanaka, T. (2010). Identification of the Phr-dependent heat shock regulon in the hyperthermophilic archaeon, Thermococcus kodakaraensis. J. Biochem. 147, 361–370.
Kessler, P. S., Daniel, C., and Leigh, J. A. (2001). Ammonia switch-off of nitrogen fixation in the methanogenic archaeon Methanococcus maripaludis: mechanistic features and requirement for the novel GlnB homologues, NifI(1) and NifI(2). J. Bacteriol. 183, 882–889.
Kessler, P. S., and Leigh, J. A. (1999). Genetics of nitrogen regulation in Methanococcus maripaludis. Genetics 152, 1343–1351.
Kim, W., and Whitman, W. B. (1999). Isolation of acetate auxotrophs of the methane-producing archaeon Methanococcus maripaludis by random insertional mutagenesis. Genetics 152, 1429–1437.
Kim, Y. J., Lee, H. S., Kim, E. S., Bae, S. S., Lim, J. K., Matsumi, R., Lebedinsky, A. V., Sokolova, T. G., Kozhevnikova, D. A., Cha, S. S., Kim, S. J., Kwon, K. K., Imanaka, T., Atomi, H., Bonch-Osmolovskaya, E. A., Lee, J. H., and Kang, S. G. (2010). Formate-driven growth coupled with H2 production. Nature 467, 352–355.
Kixmüller, D., and Greie, J. C. (2012). Construction and characterization of a gradually inducible expression vector for Halobacterium salinarum, based on the kdp promoter. Appl. Environ. Microbiol. 78, 2100–2105.
Kobayashi, K., Ehrlich, S. D., Albertini, A., Amati, G., Andersen, K. K., Arnaud, M., Asai, K., Ashikaga, S., Aymerich, S., Bessieres, P., Boland, F., Brignell, S. C., Bron, S., Bunai, K., Chapuis, J., Christiansen, L. C., Danchin, A., Debarbouille, M., Dervyn, E., Deuerling, E., Devine, K., Devine, S. K., Dreesen, O., Errington, J., Fillinger, S., Foster, S. J., Fujita, Y., Galizzi, A., Gardan, R., Eschevins, C., Fukushima, T., Haga, K., Harwood, C. R., Hecker, M., Hosoya, D., Hullo, M. F., Kakeshita, H., Karamata, D., Kasahara, Y., Kawamura, F., Koga, K., Koski, P., Kuwana, R., Imamura, D., Ishimaru, M., Ishikawa, S., Ishio, I., Le Coq, D., Masson, A., Mauel, C., Meima, R., Mellado, R. P., Moir, A., Moriya, S., Nagakawa, E., Nanamiya, H., Nakai, S., Nygaard, P., Ogura, M., Ohanan, T., O'Reilly, M., O'Rourke, M., Pragai, Z., Pooley, H. M., Rapoport, G., Rawlins, J. P., Rivas, L. A., Rivolta, C., Sadaie, A., Sadaie, Y., Sarvas, M., Sato, T., Saxild, H. H., Scanlan, E., Schumann, W., Seegers, J. F., Sekiguchi, J., Sekowska, A., Seror, S. J., Simon, M., Stragier, P., Studer, R., Takamatsu, H., Tanaka, T., Takeuchi, M., Thomaides, H. B., Vagner, V., Van Dijl, J. M., Watabe, K., Wipat, A., Yamamoto, H., Yamamoto, M., Yamamoto, Y., Yamane, K., Yata, K., Yoshida, K., Yoshikawa, H., Zuber, U., and Ogasawara, N. (2003). Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. U.S.A. 100, 4678–4683.
Kobori, H., Ogino, M., Orita, I., Nakamura, S., Imanaka, T., and Fukui, T. (2012). Characterization of NADH oxidase/NADPH polysulfide oxidoreductase and its unexpected participation in oxygen sensitivity in an anaerobic hyperthermophilic archaeon. J. Bacteriol. 192, 5192–5202.
Ladapo, J., and Whitman, W. B. (1990). Method for isolation of auxotrophs in the methanogenic archaebacteria: role of the acetyl-CoA pathway of autotrophic CO2 fixation in Methanococcus maripaludis. Proc. Natl. Acad. Sci. U.S.A. 87, 5598–5602.
Lam, W. L., and Doolittle, W. F. (1989). Shuttle vectors for the archaebacterium Halobacterium volcanii. Proc. Natl. Acad. Sci. U.S.A. 86, 5478–5482.
Lam, W. L., and Doolittle, W. F. (1992). Mevinolin-resistant mutations identify a promoter and the gene for a eukaryote-like 3-hydroxy-3-methylglutaryl-coenzyme A reductase in the archaebacterium Haloferax volcanii. J. Biol. Chem. 267, 5829–5834.
Lassak, K., Neiner, T., Ghosh, A., Klingl, A., Wirth, R., and Albers, S. V. (2012). Molecular analysis of the crenarchaeal flagellum. Mol. Microbiol. 83, 110–124.
Leigh, J. A., Albers, S. V., Atomi, H., and Allers, T. (2011). Model organisms for genetics in the domain Archaea: methanogens, halophiles, Thermococcales and Sulfolobales. FEMS Microbiol. Rev. 35, 577–608.
Li, X., Guo, L., Deng, L., Feng, D., Ren, Y., Chu, Y., She, Q., and Huang, L. (2011a). Deletion of the topoisomerase III gene in the hyperthermophilic archaeon Sulfolobus islandicus results in slow growth and defects in cell cycle control. J. Genet. Genomics 38, 253–259.
Li, Z., Pan, M., Santangelo, T. J., Chemnitz, W., Yuan, W., Edwards, J. L., Hurwitz, J., Reeve, J. N., and Kelman, Z. (2011b). A novel DNA nuclease is stimulated by association with the GINS complex. Nucleic Acids Res. 39, 6114–6123.
Li, Z., Santangelo, T. J., Čuboňová, L., Reeve, J. N., and Kelman, Z. (2010). Affinity purification of an archaeal DNA replication protein network. MBio 1, e00221-10.
Lie, T. J., Hendrickson, E. L., Niess, U. M., Moore, B. C., Haydock, A. K., and Leigh, J. A. (2010). Overlapping repressor binding sites regulate expression of the Methanococcus maripaludis glnK1 operon. Mol. Microbiol. 75, 755–762.
Lie, T. J., and Leigh, J. A. (2003). A novel repressor of nif and glnA expression in the methanogenic archaeon Methanococcus maripaludis. Mol. Microbiol. 47, 235–246.
Lie, T. J., Wood, G. E., and Leigh, J. A. (2005). Regulation of nif expression in Methanococcus maripaludis: roles of the euryarchaeal repressor NrpR, 2-oxoglutarate, and two operators. J. Biol. Chem. 280, 5236–5241.
Lipscomb, G. L., Stirrett, K., Schut, G. J., Yang, F., Jenney, F. E. Jr. Scott, R. A., Adams, M. W., and Westpheling, J. (2011). Natural competence in the hyperthermophilic archaeon Pyrococcus furiosus facilitates genetic manipulation: construction of markerless deletions of genes encoding the two cytoplasmic hydrogenases. Appl. Environ. Microbiol. 77, 2232–2238.
Liu, H., Han, J., Liu, X., Zhou, J., and Xiang, H. (2011). Development of pyrF-based gene knockout systems for genome-wide manipulation of the archaea Haloferax mediterranei and Haloarcula hispanica. J. Genet. Genomics 38, 261–269.
Lucas, S., Toffin, L., Zivanovic, Y., Charlier, D., Moussard, H., Forterre, P., Prieur, D., and Erauso, G. (2002). Construction of a shuttle vector for, and spheroplast transformation of, the hyperthermophilic archaeon Pyrococcus abyssi. Appl. Environ. Microbiol. 68, 5528–5536.
Maezato, Y., Daugherty, A., Dana, K., Soo, E., Cooper, C., Tachdjian, S., Kelly, R. M., and Blum, P. (2011). VapC6, a ribonucleolytic toxin regulates thermophilicity in the crenarchaeote Sulfolobus solfataricus. RNA 17, 1381–1392.
Mahapatra, A., Patel, A., Soares, J. A., Larue, R. C., Zhang, J. K., Metcalf, W. W., and Krzycki, J. A. (2006). Characterization of a Methanosarcina acetivorans mutant unable to translate UAG as pyrrolysine. Mol. Microbiol. 59, 56–66.
Major, T. A., Liu, Y., and Whitman, W. B. (2010). Characterization of energy-conserving hydrogenase B in Methanococcus maripaludis. J. Bacteriol. 192, 4022–4030.
Matsubara, K., Yokooji, Y., Atomi, H., and Imanaka, T. (2011). Biochemical and genetic characterization of the three metabolic routes in Thermococcus kodakarensis linking glyceraldehyde 3-phosphate and 3-phosphoglycerate. Mol. Microbiol. 81, 1300–1312.
Matsumi, R., Atomi, H., Driessen, A. J., and Van Der Oost, J. (2011). Isoprenoid biosynthesis in Archaea–biochemical and evolutionary implications. Res. Microbiol. 162, 39–52.
Matsumi, R., Manabe, K., Fukui, T., Atomi, H., and Imanaka, T. (2007). Disruption of a sugar transporter gene cluster in a hyperthermophilic archaeon using a host-marker system based on antibiotic resistance. J. Bacteriol. 189, 2683–2691.
Metcalf, W. W., Zhang, J. K., Apolinario, E., Sowers, K. R., and Wolfe, R. S. (1997). A genetic system for Archaea of the genus Methanosarcina: liposome-mediated transformation and construction of shuttle vectors. Proc. Natl. Acad. Sci. U.S.A. 94, 2626–2631.
Meyer, B. H., Zolghadr, B., Peyfoon, E., Pabst, M., Panico, M., Morris, H. R., Haslam, S. M., Messner, P., Schäffer, C., Dell, A., and Albers, S. V. (2011). Sulfoquinovose synthase-an important enzyme in the N-glycosylation pathway of Sulfolobus acidocaldarius. Mol. Microbiol. 82, 1150–1163.
Moore, B. C., and Leigh, J. A. (2005). Markerless mutagenesis in Methanococcus maripaludis demonstrates roles for alanine dehydrogenase, alanine racemase, and alanine permease. J. Bacteriol. 187, 972–979.
Morimoto, N., Fukuda, W., Nakajima, N., Masuda, T., Terui, Y., Kanai, T., Oshima, T., Imanaka, T., and Fujiwara, S. (2010). Dual biosynthesis pathway for longer-chain polyamines in the hyperthermophilic archaeon Thermococcus kodakarensis. J. Bacteriol. 192, 4991–5001.
Mueller, M., Takemasa, R., Schwarz, A., Atomi, H., and Nidetzky, B. (2009). “Short-chain” α-1, 4-glucan phosphorylase having a truncated N-terminal domain: functional expression and characterization of the enzyme from Sulfolobus solfataricus. Biochim. Biophys. Acta 1794, 1709–1714.
Naparstek, S., Guan, Z., and Eichler, J. (2012). A predicted geranylgeranyl reductase reduces the ω-position isoprene of dolichol phosphate in the halophilic archaeon, Haloferax volcanii. Biochim. Biophys. Acta 1821, 923–933.
Ng, S. Y., Wu, J., Nair, D. B., Logan, S. M., Robotham, A., Tessier, L., Kelly, J. F., Uchida, K., Aizawa, S., and Jarrell, K. F. (2011). Genetic and mass spectrometry analyses of the unusual type IV-like pili of the archaeon Methanococcus maripaludis. J. Bacteriol. 193, 804–814.
Orita, I., Sato, T., Yurimoto, H., Kato, N., Atomi, H., Imanaka, T., and Sakai, Y. (2006). The ribulose monophosphate pathway substitutes for the missing pentose phosphate pathway in the archaeon Thermococcus kodakaraensis. J. Bacteriol. 188, 4698–4704.
Ozawa, K., Harashina, T., Yatsunami, R., and Nakamura, S. (2005). Gene cloning, expression and partial characterization of cell division protein FtsZ1 from extremely halophilic archaeon Haloarcula japonica strain TR-1. Extremophiles 9, 281–288.
Pan, M., Santangelo, T. J., Li, Z., Reeve, J. N., and Kelman, Z. (2011). Thermococcus kodakarensis encodes three MCM homologs but only one is essential. Nucleic Acids Res. 39, 9671–9680.
Patel, G. B., Nash, J. H., Agnew, B. J., and Sprott, G. D. (1994). Natural and electroporation-mediated transformation of Methanococcus voltae protoplasts. Appl. Environ. Microbiol. 60, 903–907.
Peck, R. F., Dassarma, S., and Krebs, M. P. (2000). Homologous gene knockout in the archaeon Halobacterium salinarum with ura3 as a counterselectable marker. Mol. Microbiol. 35, 667–676.
Peeters, E., Albers, S. V., Vassart, A., Driessen, A. J., and Charlier, D. (2009). Ss-LrpB, a transcriptional regulator from Sulfolobus solfataricus, regulates a gene cluster with a pyruvate ferredoxin oxidoreductase-encoding operon and permease genes. Mol. Microbiol. 71, 972–988.
Pfeifer, M., Bestgen, H., Bürger, A., and Klein, A. (1998). The vhuU gene encoding a small subunit of a selenium-containing [NiFe]-hydrogenase in Methanococcus voltae appears to be essential for the cell. Arch. Microbiol. 170, 418–426.
Pickl, A., Johnsen, U., and Schönheit, P. (2012). Fructose degradation in the haloarchaeon Haloferax volcanii involves bacterial type phosphoenolpyruvate-dependent phosphotransferase system, fructose-1-phosphate kinase and Class II fructose-1, 6-bisphosphate aldolase. J. Bacteriol. 194, 3088–3097.
Porat, I., Kim, W., Hendrickson, E. L., Xia, Q., Zhang, Y., Wang, T., Taub, F., Moore, B. C., Anderson, I. J., Hackett, M., Leigh, J. A., and Whitman, W. B. (2006). Disruption of the operon encoding Ehb hydrogenase limits anabolic CO2 assimilation in the archaeon Methanococcus maripaludis. J. Bacteriol. 188, 1373–1380.
Prangishvili, D., Albers, S. V., Holz, I., Arnold, H. P., Stedman, K., Klein, T., Singh, H., Hiort, J., Schweier, A., Kristjansson, J. K., and Zillig, W. (1998). Conjugation in archaea: frequent occurrence of conjugative plasmids in Sulfolobus. Plasmid 40, 190–202.
Pritchett, M. A., Zhang, J. K., and Metcalf, W. W. (2004). Development of a markerless genetic exchange method for Methanosarcina acetivorans C2A and its use in construction of new genetic tools for methanogenic archaea. Appl. Environ. Microbiol. 70, 1425–1433.
Rother, M., and Metcalf, W. W. (2005). Genetic technologies for Archaea. Curr. Opin. Microbiol. 8, 745–751.
Sakofsky, C. J., Foster, P. L., and Grogan, D. W. (2012). Roles of the Y-family DNA polymerase Dbh in accurate replication of the Sulfolobus genome at high temperature. DNA Repair (Amst.) 11, 391–400.
Sakofsky, C. J., Runck, L. A., and Grogan, D. W. (2011). Sulfolobus mutants, generated via PCR products, which lack putative enzymes of UV photoproduct repair. Archaea 2011, ID: 864015.
Sandbeck, K. A., and Leigh, J. A. (1991). Recovery of an integration shuttle vector from tandem repeats in Methanococcus maripaludis. Appl. Environ. Microbiol. 57, 2762–2763.
Santangelo, T. J., Čuboňová, L., James, C. L., and Reeve, J. N. (2007). TFB1 or TFB2 is sufficient for Thermococcus kodakaraensis viability and for basal transcription in vitro. J. Mol. Biol. 367, 344–357.
Santangelo, T. J., Čuboňová, L., and Reeve, J. N. (2008). Shuttle vector expression in Thermococcus kodakaraensis: contributions of cis elements to protein synthesis in a hyperthermophilic archaeon. Appl. Environ. Microbiol. 74, 3099–3104.
Santangelo, T. J., Čuboňová, L., and Reeve, J. N. (2010). Thermococcus kodakarensis genetics: TK1827-encoded β-glycosidase, new positive-selection protocol, and targeted and repetitive deletion technology. Appl. Environ. Microbiol. 76, 1044–1052.
Santangelo, T. J., Čuboňová, L., and Reeve, J. N. (2011). Deletion of alternative pathways for reductant recycling in Thermococcus kodakarensis increases hydrogen production. Mol. Microbiol. 81, 897–911.
Santangelo, T. J., Čuboňová, L., Skinner, K. M., and Reeve, J. N. (2009). Archaeal intrinsic transcription termination in vivo. J. Bacteriol. 191, 7102–7108.
Santangelo, T. J., and Reeve, J. N. (2010). Deletion of switch 3 results in an archaeal RNA polymerase that is defective in transcript elongation. J. Biol. Chem. 285, 23908–23915.
Sato, T., Fukui, T., Atomi, H., and Imanaka, T. (2003). Targeted gene disruption by homologous recombination in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Bacteriol. 185, 210–220.
Sato, T., Fukui, T., Atomi, H., and Imanaka, T. (2005). Improved and versatile transformation system allowing multiple genetic manipulations of the hyperthermophilic archaeon Thermococcus kodakaraensis. Appl. Environ. Microbiol. 71, 3889–3899.
Sato, T., Imanaka, H., Rashid, N., Fukui, T., Atomi, H., and Imanaka, T. (2004). Genetic evidence identifying the true gluconeogenic fructose-1, 6-bisphosphatase in Thermococcus kodakaraensis and other hyperthermophiles. J. Bacteriol. 186, 5799–5807.
Saum, R., Schlegel, K., Meyer, B., and Muller, V. (2009). The F1F0 ATP synthase genes in Methanosarcina acetivorans are dispensable for growth and ATP synthesis. FEMS Microbiol. Lett. 300, 230–236.
Schelert, J., Dixit, V., Hoang, V., Simbahan, J., Drozda, M., and Blum, P. (2004). Occurrence and characterization of mercury resistance in the hyperthermophilic archaeon Sulfolobus solfataricus by use of gene disruption. J. Bacteriol. 186, 427–437.
Schelert, J., Drozda, M., Dixit, V., Dillman, A., and Blum, P. (2006). Regulation of mercury resistance in the crenarchaeote Sulfolobus solfataricus. J. Bacteriol. 188, 7141–7150.
She, Q., Zhang, C., Deng, L., Peng, N., Chen, Z., and Liang, Y. X. (2009). Genetic analyses in the hyperthermophilic archaeon Sulfolobus islandicus. Biochem. Soc. Trans. 37, 92–96.
Skowyra, A., and Macneill, S. A. (2012). Identification of essential and non-essential single-stranded DNA-binding proteins in a model archaeal organism. Nucleic Acids Res. 40, 1077–1090.
Snyder, J. C., Brumfield, S. K., Peng, N., She, Q., and Young, M. J. (2011). Sulfolobus turreted icosahedral virus c92 protein responsible for the formation of pyramid-like cellular lysis structures. J. Virol. 85, 6287–6292.
Soppa, J. (2011). Functional genomic and advanced genetic studies reveal novel insights into the metabolism, regulation, and biology of Haloferax volcanii. Archaea 2011, ID: 602408.
Stedman, K. M., Schleper, C., Rumpf, E., and Zillig, W. (1999). Genetic requirements for the function of the archaeal virus SSV1 in Sulfolobus solfataricus: construction and testing of viral shuttle vectors. Genetics 152, 1397–1405.
Stedman, K. M., She, Q., Phan, H., Holz, I., Singh, H., Prangishvili, D., Garrett, R., and Zillig, W. (2000). pING family of conjugative plasmids from the extremely thermophilic archaeon Sulfolobus islandicus: insights into recombination and conjugation in Crenarchaeota. J. Bacteriol. 182, 7014–7020.
Stock, T., Selzer, M., Connery, S., Seyhan, D., Resch, A., and Rother, M. (2011). Disruption and complementation of the selenocysteine biosynthesis pathway reveals a hierarchy of selenoprotein gene expression in the archaeon Methanococcus maripaludis. Mol. Microbiol. 82, 734–747.
Stock, T., Selzer, M., and Rother, M. (2010). In vivo requirement of selenophosphate for selenoprotein synthesis in archaea. Mol. Microbiol. 75, 149–160.
Takemasa, R., Yokooji, Y., Yamatsu, A., Atomi, H., and Imanaka, T. (2011). Thermococcus kodakarensis as a host for gene expression and protein secretion. Appl. Environ. Microbiol. 77, 2392–2398.
Tu, D., Blaha, G., Moore, P. B., and Steitz, T. A. (2005). Gene replacement in Haloarcula marismortui: construction of a strain with two of its three chromosomal rRNA operons deleted. Extremophiles 9, 427–435.
Tumbula, D. L., Bowen, T. L., and Whitman, W. B. (1997). Characterization of pURB500 from the archaeon Methanococcus maripaludis and construction of a shuttle vector. J. Bacteriol. 179, 2976–2986.
Tumbula, D. L., Makula, R. A., and Whitman, W. B. (1994). Transformation of Methanococcus maripaludis and identification of a Pst I-like restriction system. FEMS Microbiol. Lett. 121, 309–314.
Tumbula, D. L., and Whitman, W. B. (1999). Genetics of Methanococcus: possibilities for functional genomics in Archaea. Mol. Microbiol. 33, 1–7.
Vandyke, D. J., Wu, J., Ng, S. Y., Kanbe, M., Chaban, B., Aizawa, S., and Jarrell, K. F. (2008). Identification of a putative acetyltransferase gene, MMP0350, which affects proper assembly of both flagella and pili in the archaeon Methanococcus maripaludis. J. Bacteriol. 190, 5300–5307.
Villafane, A., Voskoboynik, Y., Ruhl, I., Sannino, D., Maezato, Y., Blum, P., and Bini, E. (2011). CopR of Sulfolobus solfataricus represents a novel class of archaeal-specific copper-responsive activators of transcription. Microbiology 157, 2808–2817.
Waege, I., Schmid, G., Thumann, S., Thomm, M., and Hausner, W. (2010). Shuttle vector-based transformation system for Pyrococcus furiosus. Appl. Environ. Microbiol. 76, 3308–3313.
Wagner, M., Berkner, S., Ajon, M., Driessen, A. J., Lipps, G., and Albers, S. V. (2009). Expanding and understanding the genetic toolbox of the hyperthermophilic genus Sulfolobus. Biochem. Soc. Trans. 37, 97–101.
Wang, G., Kennedy, S. P., Fasiludeen, S., Rensing, C., and Dassarma, S. (2004). Arsenic resistance in Halobacterium sp. strain NRC-1 examined by using an improved gene knockout system. J. Bacteriol. 186, 3187–3194.
Weidenbach, K., Gloer, J., Ehlers, C., Sandman, K., Reeve, J. N., and Schmitz, R. A. (2008). Deletion of the archaeal histone in Methanosarcina mazei Gö1 results in reduced growth and genomic transcription. Mol. Microbiol. 67, 662–671.
Welander, P. V., and Metcalf, W. W. (2008). Mutagenesis of the C1 oxidation pathway in Methanosarcina barkeri: new insights into the Mtr/Mer bypass pathway. J. Bacteriol. 190, 1928–1936.
Whitman, W. B., Tumbula, D. L., Yu, J. P., and Kim, W. (1997). Development of genetic approaches for the methane-producing archaebacterium Methanococcus maripaludis. Biofactors 6, 37–46.
Wilbanks, E. G., Larsen, D. J., Neches, R. Y., Yao, A. I., Wu, C. Y., Kjolby, R. A., and Facciotti, M. T. (2012). A workflow for genome-wide mapping of archaeal transcription factors with ChIP-seq. Nucleic Acids Res. 40, e74.
Wirth, J. F., Snyder, J. C., Hochstein, R. A., Ortmann, A. C., Willits, D. A., Douglas, T., and Young, M. J. (2011). Development of a genetic system for the archaeal virus Sulfolobus turreted icosahedral virus (STIV). Virology 415, 6–11.
Worthington, P., Hoang, V., Perez-Pomares, F., and Blum, P. (2003). Targeted disruption of the α-amylase gene in the hyperthermophilic archaeon Sulfolobus solfataricus. J. Bacteriol. 185, 482–488.
Yokooji, Y., Tomita, H., Atomi, H., and Imanaka, T. (2009). Pantoate kinase and phosphopantothenate synthetase, two novel enzymes necessary for CoA biosynthesis in the Archaea. J. Biol. Chem. 284, 28137–28145.
Zhang, C., Guo, L., Deng, L., Wu, Y., Liang, Y., Huang, L., and She, Q. (2010). Revealing the essentiality of multiple archaeal pcna genes using a mutant propagation assay based on an improved knockout method. Microbiology 156, 3386–3397.
Zhang, C., and Whitaker, R. J. (2012). A broadly applicable gene knockout system for the thermoacidophilic Archaea Sulfolobus islandicus based on simvastatin selection. Microbiology 158, 1513–1522.
Zhang, J. K., Pritchett, M. A., Lampe, D. J., Robertson, H. M., and Metcalf, W. W. (2000). In vivo transposon mutagenesis of the methanogenic archaeon Methanosarcina acetivorans C2A using a modified version of the insect mariner-family transposable element Himar1. Proc. Natl. Acad. Sci. U.S.A. 97, 9665–9670.
Zhou, M., Xiang, H., Sun, C., and Tan, H. (2004). Construction of a novel shuttle vector based on an RCR-plasmid from a haloalkaliphilic archaeon and transformation into other haloarchaea. Biotechnol. Lett. 26, 1107–1113.
Zillig, W., Arnold, H. P., Holz, I., Prangishvili, D., Schweier, A., Stedman, K., She, Q., Phan, H., Garrett, R., and Kristjansson, J. K. (1998). Genetic elements in the extremely thermophilic archaeon Sulfolobus. Extremophiles 2, 131–140.
Keywords: Archaea, gene disruption, shuttle vectors, genetics, halophiles, methanogens, Sulfolobus, Thermococcales
Citation: Atomi H, Imanaka T and Fukui T (2012) Overview of the genetic tools in the Archaea. Front. Microbio. 3:337. doi: 10.3389/fmicb.2012.00337
Received: 13 May 2012; Paper pending published: 05 June 2012;
Accepted: 01 September 2012; Published online: 02 October 2012.
Edited by:
Frank T. Robb, Institute of Marine and Environmental Technology, USAReviewed by:
Thijs Ettema, Uppsala University, SwedenImke Schroeder, University of California, Los Angeles, USA
Copyright © 2012 Atomi, Imanaka and Fukui. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Haruyuki Atomi, Department of Synthetic Chemistry and Biological Chemistry, Graduate School of Engineering, Kyoto University, Katsura, Nishikyo-ku, Kyoto 615-8510, Japan. e-mail:YXRvbWlAc2JjaGVtLmt5b3RvLXUuYWMuanA=

