
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
HYPOTHESIS AND THEORY article
Front. Microbiol., 07 August 2012
Sec. Evolutionary and Genomic Microbiology
Volume 3 - 2012 | https://doi.org/10.3389/fmicb.2012.00273
This article is part of the Research TopicRecent Advances in the Microbiology of Nitrogen Oxide TransformationsView all 12 articles
Nitric oxide (NO) and nitrous oxide (N2O) are among nature’s most powerful electron acceptors. In recent years it became clear that microorganisms can take advantage of the oxidizing power of these compounds to degrade aliphatic and aromatic hydrocarbons. For two unrelated bacterial species, the “NC10” phylum bacterium “Candidatus Methylomirabilis oxyfera” and the γ-proteobacterial strain HdN1 it has been suggested that under anoxic conditions with nitrate and/or nitrite, monooxygenases are used for methane and hexadecane oxidation, respectively. No degradation was observed with nitrous oxide only. Similarly, “aerobic” pathways for hydrocarbon degradation are employed by (per)chlorate-reducing bacteria, which are known to produce oxygen from chlorite . In the anaerobic methanotroph M. oxyfera, which lacks identifiable enzymes for nitrogen formation, substrate activation in the presence of nitrite was directly associated with both oxygen and nitrogen formation. These findings strongly argue for the role of NO, or an oxygen species derived from it, in the activation reaction of methane. Although oxygen generation elegantly explains the utilization of “aerobic” pathways under anoxic conditions, the underlying mechanism is still elusive. In this perspective, we review the current knowledge about intra-aerobic pathways, their potential presence in other organisms, and identify candidate enzymes related to quinol-dependent NO reductases (qNORs) that might be involved in the formation of oxygen.
In dim anoxic waters of stratified lakes where oxygen-respiring organisms normally cannot survive, a tiny aerobic eukaryote nevertheless makes a living. This heterotrophic ciliate, Histiobalantium natans, can survive without external oxygen because it sequesters chloroplasts from ingested euglinoid flagellates (Phacus suecicus). The chloroplasts, kept active in the ciliate and surrounded by the mitochondria, photosynthesize and produce oxygen that allows the host to thrive in deep waters of stratified lakes, where it avoids metazoan predation and competition with other aerobic ciliates (Esteban et al., 2009). This is just one example of nature’s many twists that allow organisms to take a specific niche: If an essential compound is not available, make it yourself by inventing a variation on a general theme.
For a long time, photosynthesis was the only biological process known to produce oxygen. Cyanobacteria, green plants, and algae use light energy to split water via photosystem II. The electrons obtained serve NADPH and ATP generation for carbon dioxide fixation; oxygen is a mere by-product of this metabolism. This pathway evolved at least 2.7 billion years ago (Canfield, 2005), and, after the vast pools of reduced compounds on early earth were exhausted, oxygen started to accumulate in the atmosphere around 2.45 billion years ago (Holland, 2006). As a consequence, organisms evolved numerous mechanisms to cope with and/or exploit its strong oxidative properties. To prevent oxidative damage by reactive oxygen species (ROS) like superoxide , hydrogen peroxide (H2O2), or the most damaging of all, the hydroxyl radical (OH·), detoxification systems, which often result in the regeneration of oxygen (e.g., by catalase or superoxide dismutase) evolved. These reactions have been studied and reviewed in detail elsewhere (Apel and Hirt, 2004; Murphy, 2009) and are beyond the scope of this article.
On the other hand, a large number of extant organisms are completely dependent on oxygen as the terminal electron acceptor in aerobic respiration. In addition, oxygen is the substrate in an enormous variety of monooxygenase and dioxygenase reactions where one or two oxygen atoms, respectively, are incorporated into the substrate, either for degradation of compounds like aromatic and aliphatic hydrocarbons or biosynthesis of secondary metabolites.
De novo oxygen production can be driven by either light or chemical energy. The second, “dark” way takes advantage of oxidants with a more positive redox potential than the O2/H2O couple. Only a few redox couples are biologically relevant in this respect: hypochlorite(ClO-)/Cl-; , chlorite nitrous oxide nitric oxide and Most of these compounds are intermediates in the respiration of (per)chlorate and nitrate/nitrite, respectively. In this perspective, we review what is known and still to be learned about oxygenic pathways from chloro-oxo species and nitrogen oxides, with a focus on a hypothetical enzymatic mechanism for the hitherto elusive nitrite-driven oxygen production.
The first group of oxygenic chemotrophs identified were perchlorate and chlorate respiring bacteria (Rikken et al., 1996; van Ginkel et al., 1996). These organisms reduce perchlorate and/or chlorate to chlorite . Rather than being further reduced to hypochlorite (ClO-), chlorite is converted into chloride (Cl-) and O2. Perchlorate occurs naturally, but rarely in the environment, with significant concentrations only found in the Chilean salpeter deposits (Beckurts, 1886; Ericksen, 1983). In past decades, anthropogenic contamination from either the use of Chile salpeters as fertilizers, or from chemical waste (e.g., solid rocket fuel spills and explosives) has been a concern and incentive for research on microbial (per)chlorate reduction (Motzer, 2001; Xu et al., 2003). An initial surprise was the wide-spread occurrence of (per)chlorate reduction among microorganisms and in different ecosystems, much broader than could be expected from the known natural sources and the short timeframe of anthropogenic contamination (Coates et al., 1999). It now has become clear that perchlorate is continuously generated in trace amounts in the atmosphere. Accumulation to measurable amounts, however, only occurs where deposition is high, but leaching and microbial reduction is low: in an extremely arid climate (Rajagopalan et al., 2006; Kounaves et al., 2010).
(Per)chlorate respiration in principal only requires two enzymes. At first, (per)chlorate reductase, a molybdopterin-containing respiratory reductase resembling nitrate reductase, catalyzes the reduction of perchlorate to chlorate and of chlorate to chlorite. Then, chlorite is converted in a single exergonic reaction into chloride and oxygen (Eq. 1; van Ginkel et al., 1996). This is a net disproportionation or dismutase reaction in which the chlorine atom becomes reduced and oxygen oxidized.
The reaction is catalyzed at a high rate and with extraordinary specificity by chlorite dismutases (Cld, EC 1.13.11.49), members of the recently defined CDE superfamily of heme enzymes (Goblirsch et al., 2011). This homohexameric or homopentameric heme b enzyme was first purified in the nineties from the β-proteobacterium Azospira oryzae (then called strain GR-1, van Ginkel et al., 1996). The enzyme is now well characterized by the resolution of the atomic structures from several species (de Geus et al., 2009; Mehboob et al., 2009b; Goblirsch et al., 2010; Kostan et al., 2010; Mlynek et al., 2011). Detailed kinetic analysis established that oxygen is not derived from water, but from chlorite itself (Lee et al., 2008; Streit and DuBois, 2008). Catalysis proceeds via an oxoferryl species and a ClO- anion, indicating that, after initial binding to the catalytic heme b, chlorite is first cleaved, after which both oxygen atoms recombine, yielding chloride and oxygen. Most chlorate-reducing organisms found thus far are facultative aerobes that, in absence of extracellular oxygen use the chlorite-derived oxygen for aerobic respiration, analogous to the use of chloroplasts by H. natans in the introduction.
What else to do with oxygen produced by chorite dismutation? Surprisingly, functional chlorite dismutases have also been found in Bacteria and Archaea that cannot grow with chlorate as electron acceptor, e.g., in the nitrite-oxidizing genera Nitrospira (Maixner et al., 2008) and Nitrobacter (Mlynek et al., 2011). In the latter species, Cld is significantly smaller and present as a homodimer. The role of Cld in these organisms, however, is unclear. They may possibly confer insensitivity to chlorite, coupled with the advantage of producing oxygen for nitrite oxidation in microoxic niches. In the archaeon Haloferax volcanii, chlorite dismutase is hypothesized to produce oxygen for a monooxygenase encoded in the same operon, that is involved in the biosynthesis of an antibiotic (Bab-Dinitz et al., 2006). In Pseudomonas chloritidismutans, oxygen likely does not only act as an electron acceptor in respiration, but is also used for alkane activation by a monooxygenase-mediated reaction (Mehboob et al., 2009a). P. chloritidismutans is capable of respiring carbon substrates like fatty acids or alcohols with oxygen, chlorate, and nitrate, but growth on alkanes is not observed with nitrate as electron acceptor. This suggests that oxygen, provided externally or from chlorate reduction, is required for the initial activation of the alkane, a hydroxylation to the corresponding alcohol (Heider, 2007).
The β-proteobacterium Dechloromonas aromatica strain RCB can degrade benzene aerobically and anaerobically with chlorate and nitrate (Coates et al., 2001). However, signature genes of anaerobic hydrocarbon activation (Heider, 2007), like the glycyl-radical enzyme benzyl-succinate synthase cluster, are missing. In contrast, the genome of strain RCB only encodes genes for the aerobic activation of aromatic compounds, including several mono- and dioxygenases (Salinero et al., 2009). Physiological experiments under nitrate-reducing conditions strongly suggest the involvement of a hydroxyl radical-mediated activation leading to phenol as primary intermediate (Chakraborty and Coates, 2005). It is quite unlikely that the very substrate-specific Cld can catalyze O2 production from nitrogen oxide intermediates. This possibility has been negatively tested for NO with the recombinant Cld of Nitrospira defluvii (Maixner et al., 2008), which was also found to be inhibited by NO (179 μM; F. Maixner and K. Ettwig, unpublished results). The open question is: Can the oxidative power for the attack on benzene come from oxygen, also under denitrifying conditions (Weelink et al., 2010)?
The idea that oxygen may be an intermediate of denitrifying, anaerobic bacteria emerged when the genome of the anaerobic methane-oxidizing bacterium “Candidatus Methylomirabilis oxyfera” was assembled from enrichment culture metagenomes.
These freshwater enrichment cultures (Raghoebarsing et al., 2006; Ettwig et al., 2009) couple complete methane oxidation with CO2 as the end product to the reduction of nitrite to dinitrogen (N2) according to Eq. 2.
Methane has the second highest activation energy (after benzene) of all organic compounds. One of the prime questions was how it could be enzymatically activated under anaerobic conditions. Generally, two enzymatic activation mechanisms were already known: Aerobic methane-oxidizing bacteria (MOB) employ a monooxygenase reaction yielding methanol as the first intermediate (Hakemian and Rosenzweig, 2007; Trotsenko and Murrell, 2008). Anaerobic methanotrophic archaea (ANME), that couple methane oxidation to sulfate reduction (most likely performed in association with sulfate-reducing bacteria) reverse the last step of methanogenesis catalyzed by methyl-coenzyme M reductase (Knittel and Boetius, 2009; Scheller et al., 2010). Though energetically costly, a third possibility has also been considered – an activation mechanism involving addition of the methyl group to fumarate, catalyzed by a glycyl radical enzyme homologous to benzyl- or alkyl-succinate synthase (Thauer and Shima, 2008).
Whereas no homologues of the two last mentioned signature genes for anaerobic methane and hydrocarbon degradation could be identified in the genome, surprisingly the entire pathway of aerobic methane oxidation, starting with particulate methane monooxygenase (pMMO), was present, and prominently transcribed and expressed (Ettwig et al., 2010; Luesken et al., 2012). This was consistent with several experimental findings: The M. oxyfera enrichment culture was not sensitive to aerobic handling (Ettwig et al., 2009), but highly sensitive to acetylene (total inhibition at 10 μM; Ettwig et al., 2010), a known inhibitor of pMMO (Prior and Dalton, 1985). Besides methane, the M. oxyfera enrichment culture also oxidized other short-chain alkanes (ethane, propane, butane), a well-known activity of pMMO (Leadbetter and Foster, 1960; Hazeu and de Bruyn, 1980). Finally, using the oxidation of propylene as a proxy for pMMO activity (Prior and Dalton, 1985), comparable rates were obtained for oxygen and nitrite as electron acceptors (Ettwig et al., 2010). Also the analysis of the denitrification pathway caused surprise. In all microbial species studied so far, denitrification proceeds in a step-wise fashion, comprising the subsequent reduction of nitrate to nitrite , nitric oxide (NO), nitrous oxide (N2O), and eventually dinitrogen gas (N2) by dedicated reductases (Figure 1A; Zumft, 1997; Einsle and Kroneck, 2004; Tavares et al., 2006). The last step, nitrous oxide reduction, is not always present, leaving the potent greenhouse gas N2O as the end product (Stein, 2011). Thus, a second startling finding was the apparent lack of an identifiable nitrous oxide reductase in the genome of M. oxyfera, even though it had been shown that dinitrogen gas was the end product of nitrite reduction. Despite the presence of three qNOR paralogs (see below), of which two were highly transcribed and expressed, nitrous oxide was not produced in significant amounts. Now, one of two possibilities might explain the paradoxical results: (1) activation of methane to methanol by NO, yielding N2 as the second product of the pMMO-catalyzed reaction, (2) the disproportionation of NO into N2 and O2 (Eq. 3), analogously to the chlorite dismutase reaction and to be catalyzed by a novel enzyme, NO dismutase (NOD). Again, such disproportionation is exergonic.
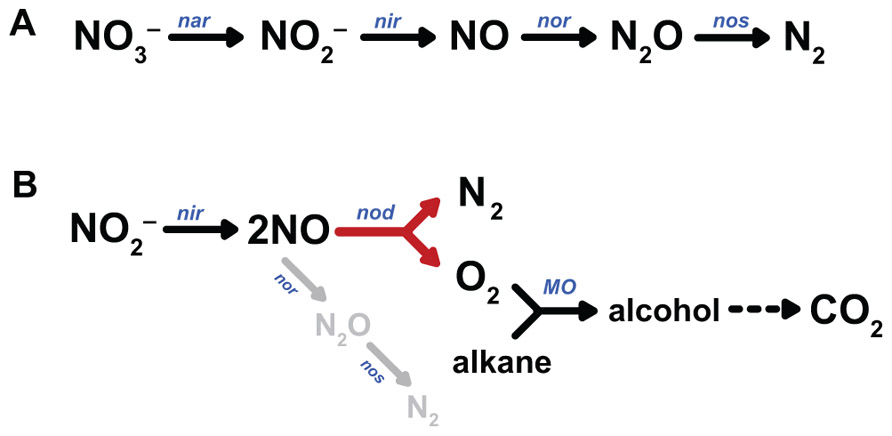
FIGURE 1. Pathways of canonical denitrification (A) and proposed N2 and O2 production by NO dismutation (B). nar, nitrate reductase; nir, nitrite reductase; nor, nitric oxide reductase; nos, nitrous oxide reductase; nod, nitric oxide dismutase, MO, monooxygenase.
Although the first possibility cannot be discarded, it is very unlikely. The pMMO of M. oxyfera shows a high sequence identity to the well-studied enzymes from other organisms with known crystal structures, including amino acids implicated in catalysis (Balasubramanian and Rosenzweig, 2007; Smith et al., 2011). With NO as the direct oxidizing agent, at least some modifications would be expected. Moreover, such role of NO is not in accordance with the experimental reaction stoichiometry (Eq. 2). Next, the bypass of N2O as an intermediate, and the formation of labeled oxygen from 18O-labeled nitrite could be experimentally shown (Ettwig et al., 2010). The hypothetical pathway that is consistent with all observations is shown in Figure 1B. From the overall reaction stoichiometry (Eq. 2) it is inferred that the disproportionation of eight NO molecules would give four oxygen molecules only three of which are consumed in the activation of methane. Residual O2 appears to be respired by one of the terminal oxidases found in the M. oxyfera genome (Wu et al., 2011). Obviously, the most interesting question now is the identity of the enzyme that catalyzes oxygen and nitrogen formation from NO.
The intermediary role for oxygen in the activation of recalcitrant compounds during denitrification may not be limited to M. oxyfera. The facultatively denitrifying γ-proteobacterium strain HdN1 grows on a wide variety of substrates, including C6- to C20-alkanes (Ehrenreich et al., 2000; Zedelius et al., 2010). Growth on hexadecane was observed with oxygen, nitrate, or nitrite as electron acceptors, but not with N2O. In contrast, N2O did serve as a substrate for growth on the corresponding easier-to-degrade C16-alcohol and fatty acid, which do not require oxidative activation (Zedelius et al., 2010). Like M. oxyfera, the HdN1 genome did not contain recognizable genes for the glycyl-radical-catalyzed activation of alkanes, such as alkylsuccinate synthase. Instead, two or possibly three monooxygenases were encoded in the genome. These findings suggest that the activation of the alkane substrate in M. oxyfera and HdN1 take place by a similar mechanism involving oxygen, formed from nitrate or nitrite (Figure 1B).
Like oxygen, NO is a strongly oxidizing compound and most microorganisms that have to deal with it as an intermediate or in their environment have developed a repertory of enzymes that convert it into the harmless N2O as fast as possible (Richardson, 2000; de Vries and Schröder,2002; Watmough et al., 2009). Collectively, the bacterial nitric oxide reductases (NORs) belong to the superfamily of heme-copper oxidases (HCOs; Figure 2). Members of the family share the presence of a heme b (or a) for electron transfer, and a second heme (b3, a3, or o3), that together with an iron (FeB in NOR) or a copper ion (CuB in oxidases) constitute the catalytic center. Both FeB and CuB are ligated to three conserved histidines. The electron-transferring heme is coordinated by two histidines as well, while one more histidine serves as the proximal ligand to the catalytic heme. This histidine sextet is a signature for HCOs.
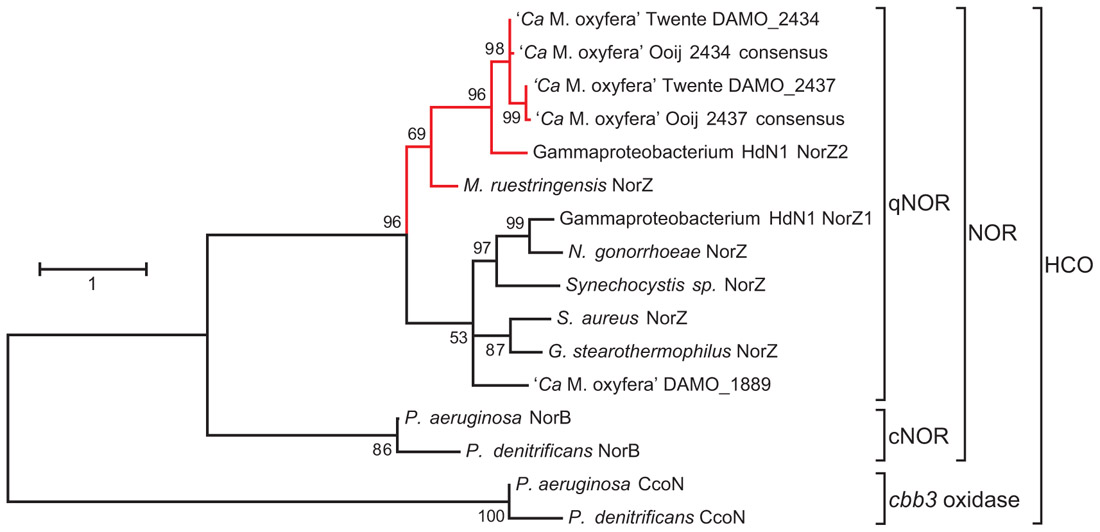
FIGURE 2. Maximum likelihood tree (500 bootstrap replicates) of selected protein sequences from the heme-copper oxidases (HCO) superfamily, with a focus on quinol-dependent nitric oxide reductases (qNORs) including the potential NO dismutases (in red). The tree was calculated with MEGA5 (Tamura et al., 2011) and is based on an alignment created with ClustalW using the default settings. The alignment was manually checked for correct alignment of conserved residues. The sequences Ooij 2434 and Ooij 2437 are consensus sequences based on the contigs obtained by iterative read mapping of the Ooij metagenome (SRR022748.2) to the genome of M. oxyfera (Dutilh et al., 2009). Accession numbers: M. oxyfera DAMO_2434 CBE69496, DAMO_2437 CBE69502, and DAMO_1889 CBE68939; γ-proteobacterium HdN1 NorZ1 CBL45628 and NorZ2 YP_003809511; M. ruestringensis G2PJH6; N. gonorrhoeae ZP_04723508; Synechocystis sp. BAA18795; S. aureus EGL94648; G. stearothermophilus 3AYF_A; P. aeruginosa NorB NP_249215, P. denitrificans NorB BAA32546, P. aeruginosa CcoN NP_252822; P. denitrificans CcoN AAC44516.
Nitric oxide reductases catalyze the two-electron reduction of two molecules of NO into N2O (Eq. 4).
The different NOR types are distinguished on the basis of the electron carrier that supplies nitric oxide reduction with reductant. Best characterized are cNORs which contain an additional cytochrome c subunit for this purpose, and qNORs which use reduced quinone (quinol) as the electron donor. Of both enzymes, atomic structures have been resolved recently (Hino et al., 2010; Matsumoto et al., 2012).
As mentioned above, the M. oxyfera genome contained three qNOR paralogs (EC 1.7.5.2, DAMO_1889, DAMO_2434, and DAMO_2437), in stark contrast to the lack of appreciable N2O production during nitrite-dependent methane oxidation (Raghoebarsing et al., 2006; Ettwig et al., 2008, 2009, 2010). DAMO_1889 was expressed in only low amounts, but the two highly similar DAMO_2434 and DAMO_2437 (84% aa identity) were among the most abundant gene products, both at the transcriptional and protein level (Ettwig et al., 2010). Detailed sequence analysis revealed that DAMO_1889 shared all important features with known qNORs, while DAMO_2434 and DAMO_2437 displayed important differences, which will be discussed in detail below. Strikingly, the unusual characteristics were consistently found in two other protein sequences available in GenBank, putative qNORs from the hexadecane-oxidizing γ-proteobacterial strain HdN1 (Zedelius et al., 2010) and from Muricauda ruestringensis, a Flavobacterium that had been isolated with peptone as a carbon source from a hexadecane-oxidizing, denitrifying enrichment culture (Bruns et al., 2001). A species of the same genus, M. aquimarina, was recently shown to degrade hexadecane and polycyclic aromatic hydrocarbons aerobically (Jiménez et al., 2011). Although the three organisms are only distantly related, their unusual qNOR-like genes form one separate cluster within the qNORs (Figure 2). A similar qNOR, however, is absent from the genome of the benzene-oxidizing D. aromatica strain RCB.
The overall atomic structure of qNOR strongly resembles the one of cNORs and other HCOs (Hino et al., 2010; Matsumoto et al., 2012). The enzyme is composed of a membrane-spanning region with 13 trans-membrane helices (TMHs) that enclose the heme b, heme b3, and FeB moieties, which are coordinated by the conserved histidine sextet. In the qNOR structure, the latter position is occupied by a (redox-insensitive) zinc atom, which most likely is a crystallization artifact (Figure 3). A particular property of qNOR is the presence of an additional (14th) N-terminal TMH that is followed by a long hydrophilic stretch of amino acids. This sequence folds at the periplasmic site as a cyt c domain like in cNOR, although a heme c itself is absent. Instead, the heme c position is filled by a number of voluminous aromatic amino acids. Two hydrophobic channels are observed in the structure that run parallel to the membrane and connect the hydrophobic membrane interior with the active site. These channels might function in substrate (NO) import and product (N2O) export. Two more features distinguish qNOR from cNOR: (1) the presence of a quinol-binding site (Figure 4A) and of a water-filled channel that likely plays a role in the supply of protons for NO reduction (Eq. 4; Matsumoto et al., 2012; Shiro et al., 2012). The channel leads from the bottom of the enzyme in the cytoplasm up to the catalytic site.
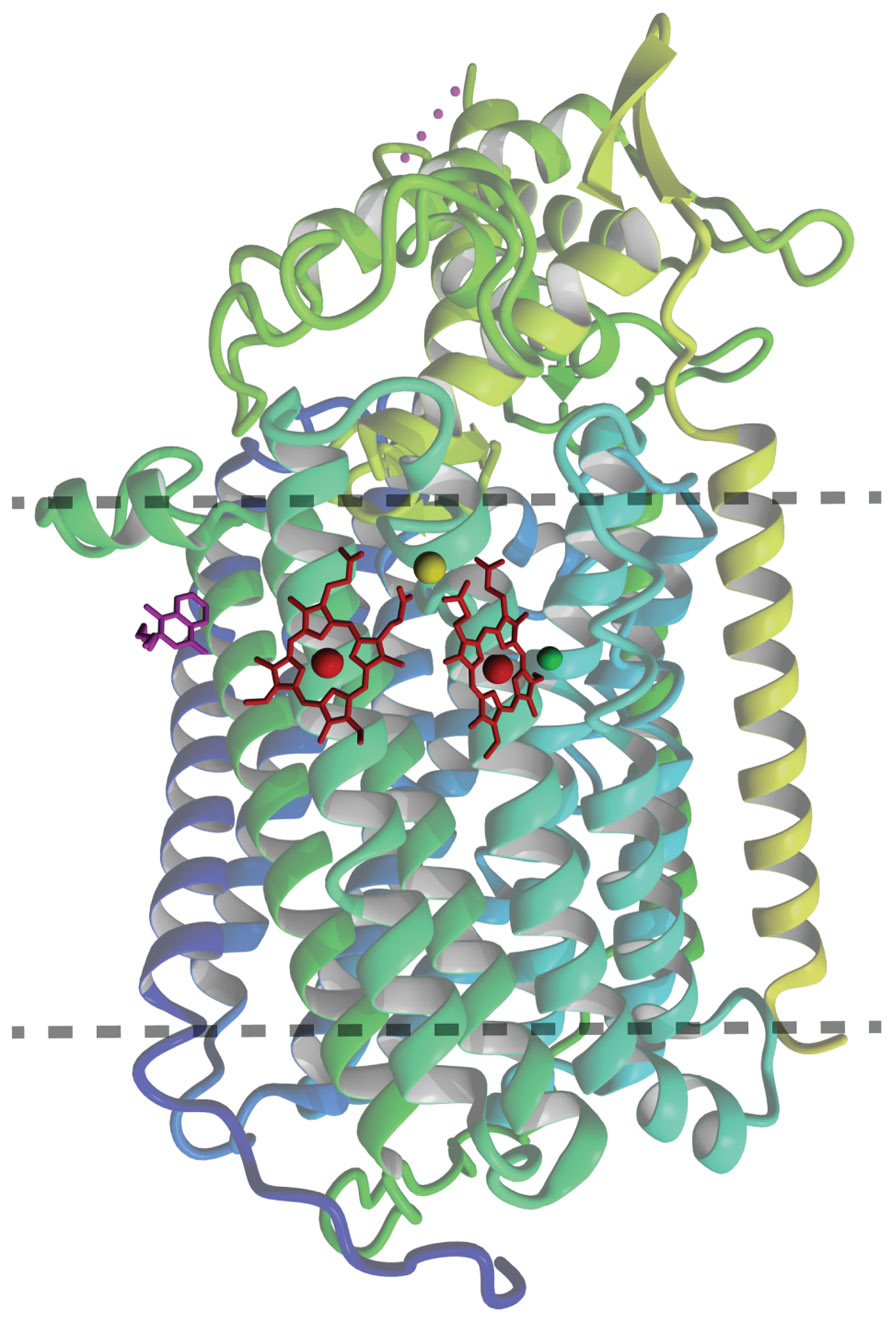
FIGURE 3. Ribbon representation of the overall qNOR structure of Geobacillus stearothermophilus (3AYG, Matsumoto et al., 2012) seen from the plane of the cytoplasmic membrane (between dotted lines). The heme groups are indicated in red, the quinol analog 2-heptyl hydroxyquinoline N-oxide in magenta. The zinc in the catalytic center is shown in green, and the calcium (Ca2+) assisting in coordination of the heme groups in pastel green.
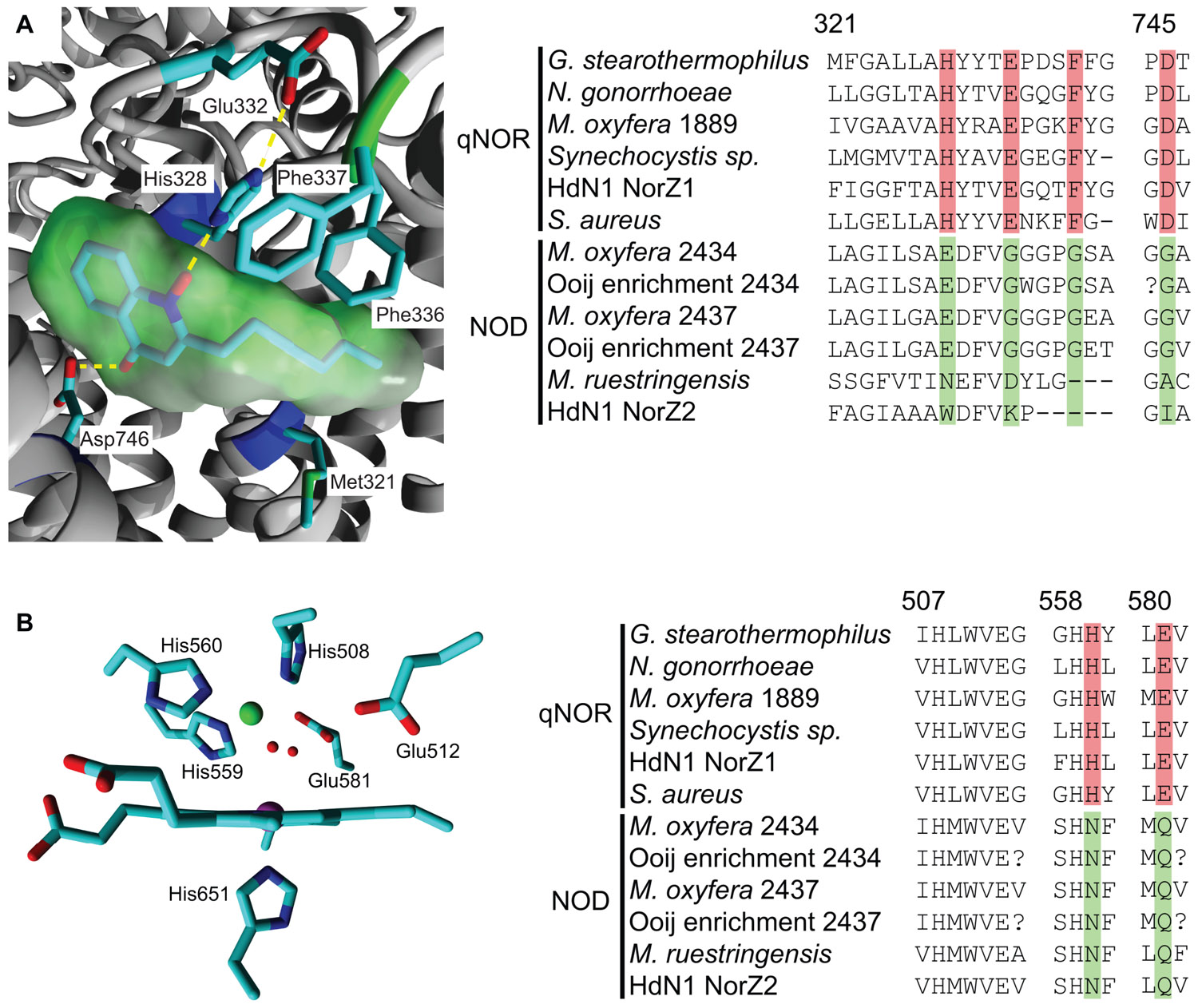
FIGURE 4. Quinol-binding and catalytic sites in the qNOR structure of Geobacillus stearothermophilus (3AYG, Matsumoto et al., 2012;left) and amino acid sequence comparison of these sites in qNORs and putative NODs (right). Sequence accession numbers and alignment are as indicated in Figure 2. Numbering above the alignment refers to the first amino acid and corresponds to the residue numbers of G. stearothermophilus. Specific changes in otherwise strongly conserved residues are highlighted. (A) Quinol binding site with a bound quinol analog, 2-heptyl hydroxyquinoline N-oxide (green molecular surface). His328 and Asp746 form hydrogen bonds with the quinol moiety and the large hydrophobic residues interact with the hydrophobic tail. (B) View of the catalytic site from the plane of the heme b3. The ZnB is indicated in green and two water molecules in the coordination sphere of the ZnB are indicated as small red spheres.
The sequence comparison of the M. oxyfera and the other unusual qNORs establish both resemblances and significant differences with respect to canonical qNORs. In DAMO_1889, all characteristics are conserved, suggesting the protein to be a genuine qNOR. Also in DAMO_2434, DAMO_2437, and their relatives the overall folding is apparently maintained with respect to the one of qNORs, as is inferred from sequence comparison and structural modeling using qNOR of Geobacillus stearothermophilus (PDB 3AYF and 3AYG) as the template (not shown). The arrangement of the 14 TMHs, the hydrophilic domain devoid of heme c, all histidines except one, both putative substrate channels and a portion of the amino acids related with the H+ channel are conserved. This suggests that DAMO_2434 and its relatives, hereafter referred to as putative NOD, bind the electron-transferring heme b, the catalytic heme b3, and non-heme iron (or another catalytic metal). However, in the NODs one of the coordinating histidines is consistently replaced by an asparagine (Figure 4B). Similarly, a glutamate in close vicinity to the catalytic center, which has been implied with catalysis (Thorndycroft et al., 2007; Flock et al., 2009; Hino et al., 2012; Shiro et al., 2012) is substituted by a glutamine residue. Also the amino acids lining the proposed H+ channel in qNOR have undergone several substitutions in the putative NODs. Most importantly, the unusual “qNORs” lack a proper quinol-binding site. Conserved residues that are assumed to constitute the quinol-binding site in qNORs are substituted for amino acids that are unlikely to provide a suitable site for quinol binding in the putative NODs (Figure 4A). In summary, the latter apparently are unable of accepting external electrons, they have a different catalytic site and might be impeded in H+ uptake from outside the protein. Obviously, these properties compromise a role as nitric oxide reductases. The question then is what they do, presuming that they do bear an important biological function – a reasonable assumption given their high expression levels in M. oxyfera. It is tempting to speculate that the modified proteins can bind two NO molecules, rearrange N-O bonds with the aid of the hemes and non-heme metal (iron or otherwise), and recombine both N and O atoms such that N2 and O2 are made. In other words, the enzymes would act as an NO dismutase. At this stage, this is speculation. The proof can only come from the purification and rigorous characterization of these intriguing enzymes.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Apel, K., and Hirt, H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399.
Bab-Dinitz, E., Shmuely, H., Maupin-Furlow, J., Eichler, J., and Shaanan, B. (2006). Haloferax volcanii PitA: an example of functional interaction between the Pfam chlorite dismutase and antibiotic biosynthesis monooxygenase families? Bioinformatics 22, 671–675.
Balasubramanian, R., and Rosenzweig, A. C. (2007). Structural and mechanistic insights into methane oxidation by particulate methane monooxygenase. Acc. Chem. Res. 40, 573–580.
Beckurts, H. (1886). Ueber den Gehalt des Salpeters an chlorsaurem Salz. Arch. Pharm. (Weinheim) 224, 333–337.
Bruns, A., Rohde, M., and Berthe-Corti, L. (2001). Muricauda ruestringensis gen. nov., sp nov., a facultatively anaerobic, appendaged bacterium from German North Sea intertidal sediment. Int. J. Syst. Evol. Microbiol. 51, 1997–2006.
Canfield, D. E. (2005). The early history of atmospheric oxygen. Ann. Rev. Earth Planet. Sci. 33, 1–36.
Chakraborty, R., and Coates, J. D. (2005). Hydroxylation and carboxylation – two crucial steps of anaerobic benzene degradation by Dechloromonas strain RCB. Appl. Environ. Microbiol. 71, 5427–5432.
Coates, J. D., Chakraborty, R., Lack, J. G., O’Connor, S. M., Cole, K. A., Bender, K. S., and Achenbach, L. A. (2001). Anaerobic benzene oxidation coupled to nitrate reduction in pure culture by two strains of Dechloromonas. Nature 411, 1039–1043.
Coates, J. D., Michaelidou, U., Bruce, R. A., O’Connor, S. M., Crespi, J. N., and Achenbach, L. A. (1999). Ubiquity and diversity of dissimilatory (per)chlorate-reducing bacteria. Appl. Environ. Microbiol. 65, 5234–5241.
de Geus, D. C., Thomassen, E. A. J., Hagedoorn, P.-L., Pannu, N. S., Van Duijn, E., and Abrahams, J. P. (2009). Crystal structure of chlorite dismutase, a detoxifying enzyme producing molecular oxygen. J. Mol. Biol. 387, 192–206.
de Vries, S., and Schröder, I. (2002). Comparison between the nitric oxide reductase family and its aerobic relatives, the cytochrome oxidases. Biochem. Soc. Trans. 30, 662–667.
Dutilh, B. E., Huynen, M. A., and Strous, M. (2009). Increasing the coverage of a metapopulation consensus genome by iterative read mapping and assembly. Bioinformatics 25, 2878–2881.
Ehrenreich, P., Behrends, A., Harder, J., and Widdel, F. (2000). Anaerobic oxidation of alkanes by newly isolated denitrifying bacteria. Arch. Microbiol. 173, 58–64.
Einsle, O., and Kroneck, P. M. H. (2004). Structural basis of denitrification. Biol. Chem. 385, 875–883.
Esteban, G. F., Finlay, B. J., and Clarke, K. J. (2009). Sequestered organelles sustain aerobic microbial life in anoxic environments. Environ. Microbiol. 11, 544–550.
Ettwig, K. F., Butler, M. K., Le Paslier, D., Pelletier, E., Mangenot, S., Kuypers, M. M. M., Schreiber, F., Dutilh, B. E., Zedelius, J., De Beer, D., Gloerich, J., Wessels, H. J. C. T., Van Alen, T., Luesken, F., Wu, M. L., Van De Pas-Schoonen, K. T., Op Den Camp, H. J. M., Janssen-Megens, E. M., Francoijs, K. J., Stunnenberg, H., Weissenbach, J., Jetten, M. S. M., and Strous, M. (2010). Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464, 543–548.
Ettwig, K. F., Shima, S., Van De Pas-Schoonen, K. T., Kahnt, J., Medema, M. H., Op Den Camp, H. J. M., Jetten, M. S. M., and Strous, M. (2008). Denitrifying bacteria anaerobically oxidize methane in the absence of Archaea. Environ. Microbiol. 10, 3164–3173.
Ettwig, K. F., Van Alen, T., Van De Pas-Schoonen, K. T., Jetten, M. S. M., and Strous, M. (2009). Enrichment and molecular detection of denitrifying methanotrophic bacteria of the NC10 phylum. Appl. Environ. Microbiol. 75, 3656–3662.
Flock, U., Lachmann, P., Reimann, J., Watmough, N. J., and Adelroth, P. (2009). Exploring the terminal region of the proton pathway in the bacterial nitric oxide reductase. J. Inorg. Biochem. 103, 845–850.
Goblirsch, B., Kurker, R. C., Streit, B. R., Wilmot, C. M., and DuBois, J. L. (2011). Chlorite dismutases, DyPs, and EfeB: 3 microbial heme enzyme families comprise the CDE structural superfamily. J. Mol. Biol. 408, 379–398.
Goblirsch, B. R., Streit, B. R., DuBois, J. L., and Wilmot, C. M. (2010). Structural features promoting dioxygen production by Dechloromonas aromatica chlorite dismutase. J. Biol. Inorg. Chem. 15, 879–888.
Hakemian, A. S., and Rosenzweig, A. C. (2007). The biochemistry of methane oxidation. Annu. Rev. Biochem. 76, 223–241.
Hazeu, W., and de Bruyn, J. C. (1980). Ethane oxidation by methane-oxidizing bacteria. Antonie Van Leeuwenhoek 46, 443–455.
Heider, J. (2007). Adding handles to unhandy substrates: anaerobic hydrocarbon activation mechanisms. Curr. Opin. Chem. Biol. 11, 188–194.
Hino, T., Matsumoto, Y., Nagano, S., Sugimoto, H., Fukumori, Y., Murata, T., Iwata, S., and Shiro, Y. (2010). Structural basis of biological N2O generation by bacterial nitric oxide reductase. Science 330, 1666–1670.
Hino, T., Nagano, S., Sugimoto, H., Tosha, T., and Shiro, Y. (2012). Molecular structure and function of bacterial nitric oxide reductase. Biochim. Biophys. Acta 1817, 680–687.
Holland, H. D. (2006). The oxygenation of the atmosphere and oceans. Philos. Trans. R. Soc. B Biol. Sci. 361, 903–915.
Jiménez, N., Viñas, M., Guiu-Aragonés, C., Bayona, J. M., Albaigés, J., and Solanas, A. M. (2011). Polyphasic approach for assessing changes in an autochthonous marine bacterial community in the presence of Prestige fuel oil and its biodegradation potential. Appl. Microbiol. Biotechnol. 91, 823–834.
Knittel, K., and Boetius, A. (2009). Anaerobic oxidation of methane: progress with an unknown process. Annu. Rev. Microbiol. 63, 311–334.
Kostan, J., Sjöblom, B., Maixner, F., Mlynek, G., Furtmüller, P. G., Obinger, C., Wagner, M., Daims, H., and Djinović-Carugo, K. (2010). Structural and functional characterisation of the chlorite dismutase from the nitrite-oxidizing bacterium “Candidatus Nitrospira defluvii”: identification of a catalytically important amino acid residue. J. Struct. Biol. 172, 331–342.
Kounaves, S. P., Stroble, S. T., Anderson, R. M., Moore, Q., Catling, D. C., Douglas, S., Mckay, C. P., Ming, D. W., Smith, P. H., Tamppari, L. K., and Zent, A. P. (2010). Discovery of natural perchlorate in the Antarctic dry valleys and its global implications. Environ. Sci. Technol. 44, 2360–2364.
Leadbetter, E. R., and Foster, J. W. (1960). Bacterial oxidation of gaseous alkanes. Arch. Mikrobiol. 35, 92–104.
Lee, A. Q., Streit, B. R., Zdilla, M. J., Abu-Omar, M. M., and DuBois, J. L. (2008). Mechanism of and exquisite selectivity for O-O bond formation by the heme-dependent chlorite dismutase. Proc. Natl. Acad. Sci. U.S.A. 105, 15654–15659.
Luesken, F. A., Wu, M. L., Op Den Camp, H. J. M., Keltjens, J. T. M., Stunnenberg, H., Francoijs, K. J., Strous, M., and Jetten, M. S. M. (2012). Effect of oxygen on the anaerobic methanotroph “Candidatus Methylomirabilis oxyfera”: kinetic and transcriptional analysis. Environ. Microbiol. 14, 1024–1034.
Maixner, F., Wagner, M., Lücker, S., Pelletier, E., Schmitz-Esser, S., Hace, K., Spieck, E., Konrat, R., Le Paslier, D., and Daims, H. (2008). Environmental genomics reveals a functional chlorite dismutase in the nitrite-oxidizing bacterium “Candidatus Nitrospira defluvii”. Environ. Microbiol. 10, 3043–3056.
Matsumoto, Y., Tosha, T., Pisliakov, A. V., Hino, T., Sugimoto, H., Nagano, S., Sugita, Y., and Shiro, Y. (2012). Crystal structure of quinol-dependent nitric oxide reductase from Geobacillus stearothermophilus. Nat. Struct. Mol. Biol. 19, 238–245.
Mehboob, F., Junca, H., Schraa, G., and Stams, A. J .M. (2009a). Growth of Pseudomonas chloritidismutans AW-1T on n-alkanes with chlorate as electron acceptor. Appl. Microbiol. Biotechnol. 83, 739–747.
Mehboob, F., Wolterink, A. F. M., Vermeulen, A. J., Jiang, B., Hagedoorn, P.-L., Stams, A. J. M., and Kengen, S. W. M. (2009b). Purification and characterization of a chlorite dismutase from Pseudomonas chloritidismutans. FEMS Microbiol. Lett. 293, 115–121.
Mlynek, G., Sjöblom, B., Kostan, J., Füreder, S., Maixner, F., Gysel, K., Furtmüller, P. G., Obinger, C., Wagner, M., Daims, H., and Djinović-Carugo, K. (2011). Unexpected diversity of chlorite dismutases: a catalytically efficient dimeric enzyme from Nitrobacter winogradsky. J. Bacteriol. 193, 2408–2417.
Prior, S. D., and Dalton, H. (1985). Acetylene as a suicide substrate and active-site probe for methane monooxygenase from Methylococcus capsulatus (Bath). FEMS Microbiol. Lett. 29, 105–109.
Raghoebarsing, A. A., Pol, A., Van De Pas-Schoonen, K. T., Smolders, A. J. P., Ettwig, K. F., Rijpstra, W. I. C., Schouten, S., Damsté, J. S. S., Op Den Camp, H. J. M., Jetten, M. S. M., and Strous, M. (2006). A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 440, 918–921.
Rajagopalan, S., Anderson, T. A., Fahlquist, L., Rainwater, K. A., Ridley, M., and Jackson, W. A. (2006). Widespread presence of naturally occurring perchlorate in high plains of Texas and New Mexico. Environ. Sci. Technol. 40, 3156–3162.
Richardson, D. J. (2000). Bacterial respiration: a flexible process for a changing environment. Microbiology 146, 551–571.
Rikken, G. B., Kroon, A. G. M., and van Ginkel, C. G. (1996). Transformation of (per)chlorate into chloride by a newly isolated bacterium: reduction and dismutation. Appl. Microbiol. Biotechnol. 45, 420–426.
Salinero, K. K., Keller, K., Feil, W., Feil, H., Trong, S., Di Bartolo, G., and Lapidus, A. (2009). Metabolic analysis of the soil microbe Dechloromonas aromatica str. RCB: indications of a surprisingly complex life-style and cryptic anaerobic pathways for aromatic degradation. BMC Genomics 10, 351. doi: 10.1186/1471-2164-10-351
Scheller, S., Goenrich, M., Boecher, R., Thauer, R. K., and Jaun, B. (2010). The key nickel enzyme of methanogenesis catalyses the anaerobic oxidation of methane. Nature 465, 606–608.
Shiro, Y., Sugimoto, H., Tosha, T., Nagano, S., and Hino, T. (2012). Structural basis for nitrous oxide generation by bacterial nitric oxide reductases. Philos. Trans. R. Soc. B Biol. Sci. 367, 1195–1203.
Smith, S. M., Rawat, S., Telser, J., Hoffman, B. M., Stemmler, T. L., and Rosenzweig, A. C. (2011). Crystal structure and characterization of particulate methane monooxygenase from Methylocystis species strain M. Biochemistry (Mosc.) 50, 10231–10240.
Stein, L. Y. (2011). “Surveying N2O-producing pathways in bacteria,” in Methods in Enzymology: Research on Nitrification and Related Processes, Vol. 486, Part A 486, 1st Edn., ed. M. G. Klotz (London: Academic Press), 131–152.
Streit, B. R., and DuBois, J. L. (2008). Chemical and steady-state kinetic analyses of a heterologously expressed heme dependent chlorite dismutase. Biochemistry (Mosc.) 47, 5271–5280.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739.
Tavares, P., Pereira, A. S., Moura, J. J. G., and Moura, I. (2006). Metalloenzymes of the denitrification pathway. J. Inorg. Biochem. 100, 2087–2100.
Thauer, R. K., and Shima, S. (2008). Methane as fuel for anaerobic microorganisms. Ann. N. Y. Acad. Sci. 1125, 158–170.
Thorndycroft, F. H., Butland, G., Richardson, D. J., and Watmough, N. J. (2007). A new assay for nitric oxide reductase reveals two conserved glutamate residues form the entrance to a proton-conducting channel in the bacterial enzyme. Biochem. J. 401, 111–119.
Trotsenko, Y. A., and Murrell, J. C. (2008). Metabolic aspects of aerobic obligate methanotrophy. Adv. Appl. Microbiol. 63, 183–229.
van Ginkel, C. G., Rikken, G. B., Kroon, A. G. M., and Kengen, S. W. M. (1996). Purification and characterization of chlorite dismutase: A novel oxygen-generating enzyme. Arch. Microbiol. 166, 321–326.
Watmough, N. J., Field, S. J., Hughes, R. J. L., and Richardson, D. J. (2009). The bacterial respiratory nitric oxide reductase. Biochem. Soc. Trans. 37, 392–399.
Weelink, S., Van Eekert, M., and Stams, A. J. (2010). Degradation of BTEX by anaerobic bacteria: physiology and application. Rev. Environ. Sci. Biotechnol. 9, 359–385.
Wu, M. L., de Vries, S., Van Alen, T. A., Butler, M. K., Den Camp, H., Keltjens, J. T., Jetten, M. S. M., and Strous, M. (2011). Physiological role of the respiratory quinol oxidase in the anaerobic nitrite-reducing methanotroph “Candidatus Methylomirabilis oxyfera.” Microbiology 157, 890–898.
Xu, J., Song, Y., Min, B., Steinberg, L., and Logan, B. E. (2003). Microbial degradation of perchlorate: principles and applications. Anglais 20, 405–422.
Zedelius, J., Rabus, R., Grundmann, O., Werner, I., Brodkorb, D., Schreiber, F., Ehrenreich, P., Behrends, A., Wilkes, H., Kube, M., Reinhardt, R., and Widdel, F. (2010). Alkane degradation under anoxic conditions by a nitrate-reducing bacterium with possible involvement of the electron acceptor in substrate activation. Environ. Microbiol. Rep. 3, 125–135.
Keywords: oxygen production, nitric oxide, nitric oxide reductase, chlorate reduction, chlorite dismutase, Cld, “Candidatus Methylomirabilis oxyfera”, strain HdN1
Citation: Ettwig KF, Speth DR, Reimann J, Wu ML, Jetten MSM and Keltjens JT (2012) Bacterial oxygen production in the dark. Front. Microbio. 3:273. doi:10.3389/fmicb.2012.00273
Received: 08 June 2012; Paper pending published: 27 June 2012;
Accepted: 10 July 2012; Published online: 07 August 2012.
Edited by:
Boran Kartal, Radboud University, NetherlandsReviewed by:
Natalia Ivanova, Lawrence Berkeley National Laboratory, USACopyright: © 2012 Ettwig, Speth, Reimann, Wu, Jetten and Keltjens. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Katharina F. Ettwig, Department of Microbiology, Institute for Water and Wetland Research, Radboud University Nijmegen, Heyendaalseweg 135, 6525 AJ Nijmegen, Netherlands. e-mail:ay5ldHR3aWdAc2NpZW5jZS5ydS5ubA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.